Abstract
In their native environments, proteins perform their biological roles in highly concentrated viscous solutions and in complex networks with numerous partners. Yet for many years, the normal practice has been to purify a protein of interest in order to characterize its structural and functional properties. In this Commentary, we discuss how protein scientists are now tackling the theoretical and methodological challenges of studying proteins in their physiological context.
We have arrived at the post-reductionist era of biochemistry. For protein science, this means the collective consciousness has been elevated and there is wide acceptance that we must consider the physiological environment of a protein when investigating its function. Moreover, we accept that understanding a given protein demands that we explore its complex networks of interactions in the cell. The tables have turned so radically over the last decade that it is ironically now necessary to defend detailed studies of individual, well-defined molecules or complexes. Nonetheless, most would agree that we need detailed studies of individual biomolecules for in-depth mechanistic insights; but we must also admit the complexity of their in vivo worlds and consider the impact of their native environments on their functions.
Most importantly, the era of blind, fervent reductionism, wherein biochemists and biophysicists purified and purified to enable studies of isolated biomolecules, is over. It is abundantly evident that proteins do not work as separate entities and that their most basic properties—such as affinities for ligands, catalytic activities and stabilities—are influenced by their interactions and solution environment. Minimally, one must examine a bioactive component in complex with its usual partners, and ideally, we need to develop ways to include the full complexity of the cellular environment when we explore the chemical origins of biological function. This is extremely challenging: the old Mother Goose nursery rhyme about Humpty Dumpty pointed out that after Humpty “had a great fall; all the king’s horses, and all the king’s men, couldn’t put Humpty together again.” It is virtually impossible to reassemble an in vivo environment—that is, to put Humpty back together again. Instead, one can reassemble complexes and pathways and simply accept that this is an approximation of the cellular complexity. Optimally, we can take on the challenge and develop more and more powerful approaches to examine biochemical events in situ without disruption of the cellular complexity—that is, we can study Humpty before he falls.

As much as the protein research community acknowledges that post-reductionism is integral to current-day protein science, it is not clear that we have prepared ourselves for the intellectual and technical challenges implicit in carrying out post-reductionist research. This brief Commentary is intended to point out issues that challenge protein science in the post-reductionist era and to describe technical obstacles along with promising advances that should help us to successfully address these issues. While not providing a full coverage review, we describe selected examples of recent studies that illustrate successful forays into protein chemistry in the cell. We selfishly hope to encourage our colleagues to grapple with the hard questions and to develop powerful new methods that are applicable to the study of proteins in situ.
How does the intracellular environment impact protein science?
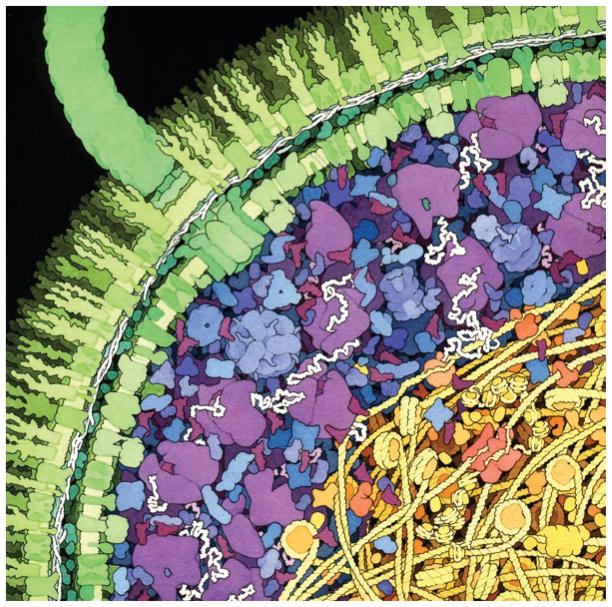
The in-cell environment is crowded and inhomogeneous. When one takes even a cursory look at the physical chemical properties of the intracellular environment (and to a similar extent, the extracellular environment—though here we will focus on the in-cell world), it is clear that it is far from a dilute, ideal solution, and therefore most of the physical chemistry we learned in college does not apply to the intracellular environment (Fig. 1). The interior of cells is replete with macromolecules—from 200 to 400 grams per liter. One impact of this high concentration is macromolecular crowding: little space is available for soluble species to roam, and the available space is irregular and inhomogeneous, such that a given molecule may sample only a fraction of the free space depending on its size, shape and flexibility. Many theoretical treatments have predicted the impact of macromolecular crowding on the properties and interactions of proteins, and there are a growing number of ‘bottom-up’ studies exploring the effects of crowding1. Though detailed results may vary from one study to another, there is general agreement that crowding favors both specific and nonspecific intermolecular associations, biases conformational distributions toward compact states and stabilizes proteins. However, most of the research done so far is still in environments that are very far from the complexity inside the cell.
Figure 1.
A cross-section of a bacterial cell. Image was painted by David Goodsell (downloaded from http://mgl.scripps.edu/people/goodsell/illustration/public; copyright 1999). The composition of macromolecules is depicted to scale, with an effort to show the impact of native concentrations of macromolecules on the environment: for example, ribosomes are in purple, white strands are mRNA and enzymes are in blue. Macromolecular crowding and confinement and their likely impact on diffusion can be appreciated from this image.
In addition to its expected effects on energetics, the restriction in available space inside the cell is predicted to have a profound impact on macromolecular diffusion. Hence, movement of molecules inside cells is complex. Some macromolecules effectively do not move, leading to ‘confinement’—that is, restriction of solutes to smaller volumes because of effective boundaries to diffusion. Macromolecular crowding and confinement together cause restricted translational diffusion inside cells. There is a large body of literature reporting on efforts to measure translational diffusion of macromolecules in cells2, much of it coming from fluorescence recovery after photobleaching (FRAP) or from fluorescence correlation spectroscopy (FCS), and at this juncture the resulting picture is not completely consistent. In dilute solutions, protein translational diffusion can be predicted by the Stokes-Einstein equation, where the mean square displacement in a random walk scales linearly with time. In cells, however, many researchers find that translational diffusion is anomalous—that is, it does not vary linearly with time. Rotational diffusion is also hindered, which offers one explanation for why the otherwise tantalizing approach of in-cell NMR has been confounded by invisibility of resonances for proteins above 60 to 100 residues in size (Q. Wang and L.M.G., unpublished data)3,4. We need more observations to obtain reliable data and deeper theoretical analyses to understand this complex intracellular world. Moreover, the impact of the in vivo environment on diffusion differs in different cellular compartments and also depends on the nature of the diffusing molecule.
About 20 years ago, a few prescient scientists like Paul Srere attempted to convince their peers that biochemical pathways are organized and that proteins inside cells are involved in highly nonrandom interactions biased by weak associations5. At the same time, McConkey proposed that these privileged weak interactions, which he termed ‘quinary’, are a special attribute of living systems, and he pointed out that they are very readily perturbed by even the gentlest of cell disruption protocols6. Though there was not widespread acceptance of these ideas at the time, we now see how right these scientists were. For example, Durek and Walther in a recent study compared two types of interaction networks: the metabolic pathway map and the protein-protein interaction network (PIN)7. The coincidence of the networks provided a compelling argument that protein-protein interactions have evolved to favor efficient fluxes of substrates through metabolic networks, exactly as Paul Srere had argued. As Bruce Alberts so eloquently put it: “…as it turns out, we can walk and we can talk because the chemistry that makes life possible is much more elaborate and sophisticated than anything we students had ever considered. Proteins make up most of the dry mass of a cell. But instead of a cell dominated by randomly colliding individual protein molecules, we now know that nearly every major process in a cell is carried out by assemblies of 10 or more protein molecules. And, as it carries out its biological functions, each of these protein assemblies interacts with several other large complexes of proteins. Indeed, the entire cell can be viewed as a factory that contains an elaborate network of interlocking assembly lines, each of which is composed of a set of large protein machines”8.
The surfaces of proteins are not inert; they are sticky, with exposed side chains and backbone groups that may interact with a variety of other surfaces. The groups on the surface of a protein comprise the face that a protein presents to its neighbors and offer electrostatic, van der Waals, hydrogen bonding and hydrophobic interactions. The resulting interactions favor association between some proteins and disfavor association with others. These biased weak interactions are subject to evolutionary selection: they must be ‘tuned’ to increase the probability of productive encounters and facilitate the self-organization of cellular machines and networks. As noted above, components of signal transduction pathways, metabolic networks, gene expression regulators, protein folding facilitators and so on are associated, often by weak transient interactions, into large multiprotein complexes. But even beyond these functional complexes and pathways, there is a remarkable ability of cellular components to self-organize, and reciprocally, the lion’s share of constituents of cells and subcellular compartments are arrayed in nonrandom fashion, such that free diffusion and random mixing are inappropriate concepts when applied to the interior of cells.
Stunning examples of the impact of biased weak interactions in living systems include the re-organization of the native interior structure of Euglena gracilis after centrifugation-induced stratification of cellular components9, the establishment and remodeling of the three-dimensional arrangement of nuclear constituents10 and the maintenance of the vesicular Golgi network even after chemical disruption11. As was recently pointed out, the requirement to avoid nonfunctional interactions (and conversely retain functional interactions) places a substantial constraint on both proteome diversity and protein expression level in cells12. These and other observations comprise compelling evidence that the information content in evolutionarily selected macromolecules includes three-dimensional patterns of preferred and nonpreferred interactions, enabling self-assembly at levels of organization well beyond the already impressive process of an individual protein folding to its native structure.
Cells are temporally as well as spatially complex. To extend Alberts’ analogy, the cellular factory is not static. Molecular cargos move about the factory floor, and the cell’s assembly lines produce and degrade proteins, oligonucleotides and small molecules. Even biomolecular interactions once thought to be long-lived, such as those between transcription factors and DNA, are dynamic and may have lifetimes of seconds or less10. These transient interactions, which are likely key to the cell’s ability to respond to its environment, may be facilitated by spatial confinement. This interplay between temporal and spatial complexity can only be studied in situ.
Promising technologies to tackle in-cell protein science
Although implementing post-reductionist protein science is indeed daunting, spectacular progress in methods suited to the complexities of physiological environments offers hope for future breakthroughs (Fig. 2). There have been tremendous advances in optical imaging in recent years, and the dream of seeing inside cells with molecular resolution is no longer out of reach13. Subdiffraction resolution is achievable by a variety of clever optical methods, a wide array of labeling options is now available and new and powerful visualization methods are emerging. Cryo-electron tomography holds promise of complete three-dimensional reconstructions of cells, with exquisite detail in the subcellular architecture14. Methods of combining the microscopic view with fits to X-ray structures of proteins and machines enable the visualization of cellular interiors and macromolecular assemblies with atomic resolution15. We anticipate soon being able to localize proteins of interest in living cells with extraordinary spatial resolution; the next goal will be temporal resolution.
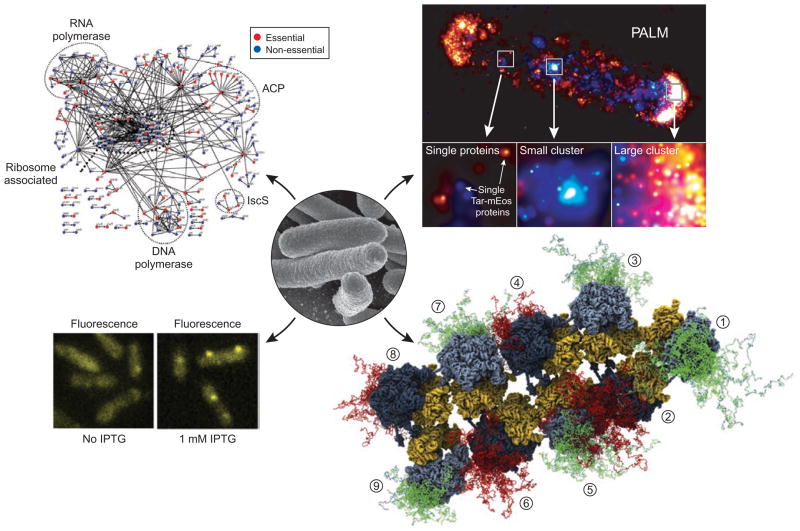
Figure 2.
Examples of powerful new methods to study proteins in their native environments, networks and complexes. A scanning electron micrograph of E. coli cells is shown in the center (from the US National Institute of Allergy and Infectious Diseases; http://www3.niaid.nih.gov/topics/biodefenserelated/biodefense/publicmedia/image_library.html). Clockwise around the cells are shown, beginning with the upper left: an E. coli protein interaction network35; an image of the chemotaxis receptors of E. coli obtained using subdiffraction fluorescence microscopy (photoactivation light microscopy, PALM)33; a reconstructed schematic of E. coli polysomes obtained by fitting ribosomal structures to cryo-electron tomography images and modeling the nascent chains (shown in green or red) emerging from individual ribosomes (numbered from 1 to 8)15; and localization of T7 RNA polymerase molecules (labeled with a fluorescent protein marker) to promoters on DNA upon IPTG induction of transcription31.
The combination of increased computational horsepower, improved computational algorithms and growing boldness about taking on complex systems is opening up the possibility of simulating and dissecting molecular behaviors in cellular environments. Recent promising examples include full treatments of protein emergence from the ribosome and exploration of conformational space cotranslationally16, integration of information from different time and length scales17, and correlation of network behaviors with molecular mechanisms18.
A huge amount of effort has been invested in experimental determination of protein-protein interaction maps, thereby leading to massive amounts of data19. There has been extensive analysis on the significance of protein interactomes derived from genetic or physical mapping. It is important to keep in mind that the interactions one identifies using a particular method satisfy the criteria imposed by that method. For example, physical interaction maps require that a given protein-protein interaction is stable enough to be isolated (for example, by affinity tagging20 or by immuno-precipitation), to reconstitute an active protein21 or to form a complex that modulates gene expression (for example, yeast two-hybrid methods22). Nonetheless, the richness of information about possible protein-protein interactions that we now have at our fingertips is vast and changes forever how we ask questions about protein function.
The recent implementation of large-scale quantitative epistasis mapping23 has yielded abundant information about functional modules24, thereby enabling networks of interacting proteins to be defined based on genetic linkages as opposed to physical interaction. Together, these data and physical interaction maps will shed light on both the spatial and functional organization of macromolecules inside an organism.
As noted in Bruce Alberts’ comments above, proteins work in teams. Identifying the team members and the temporal and environmental changes in the line-up will be crucial to understanding biochemistry in the cell. New isotope labeling methods developed to follow the in vitro assembly of large biomolecular complexes such as the ribosome25 should be applicable in vivo. Hydroxyl radical footprinting of RNA to study ribosome-RNA interactions has already been applied to frozen cells26. Other new and powerful mass spectrometric methods27 promise to unveil details of macromolecular complexes as they form and disassemble in vivo.
Protein function, localization and association are all affected by the heterogeneous, dynamic chemical environment in the cell, which makes quantitation and localization of large and small molecules and ions essential. Dynamic changes in pH and Ca2+ are routinely imaged using fluorescent probes, but obtaining more comprehensive information is difficult. Promising recent advances in mass spectrometry have enabled the determination of metabolite concentrations for cell populations28, and methods for spatially resolving metabolite concentrations in single cells are under development29.
A powerful way to dissect the function of a protein without disrupting the integrity of the cellular context in which it carries out that function is to create specific chemical probes that enable the switching of the protein’s action—either turning it on or turning it off. These approaches have flourished with the great excitement about chemical biology and the synthetic prowess that has been brought to bear on these strategies30.
The holy grail is to study a protein in situ. Though progress here is slow and pitfalls numerous, there have nonetheless been successes. For example, Xie et al. have developed methods to observe biochemical events such as cytoskeletal rearrangements and transcriptional regulation at the single-molecule level inside cells31. Werner et al. have recently correlated in-cell localization with genetically defined interactions for a major fraction of the proteome of Caulobacter crescentus32. Greenfield et al. have observed the clustering of chemotactic receptors in Escherichia coli in real time at subdiffraction resolution33. And we have developed fluorescent labeling approaches that report on the folding status of a protein inside a cell34.
Perspective.
The challenges of doing post-reductionist protein science are real, but the importance is indisputable. We urge scientists who can develop and deploy the physics of complex systems and who can devise methods and improved computational strategies to observe, measure and model phenomena inside cells to help all of us face the complexity inherent in post-reductionist protein science. The resulting insights will elevate our science: the knowledge gained from post-reductionist protein science will reveal new activities, modes of regulation and functional networks, while the ability to manipulate the cellular environment will allow us to probe pressing biological questions with enhanced physiological relevance. The post-reductionist perspective will irrevocably redefine how we think about and study the protein machinery of nature.
Contributor Information
Lila M Gierasch, Email: gierasch@biochem.umass.edu, Department of Biochemistry and Molecular Biology and the Department of Chemistry, University of Massachusetts, Amherst, Amherst, Massachusetts, USA.
Anne Gershenson, Email: gershenson@biochem.umass.edu, Department of Biochemistry and Molecular Biology, University of Massachusetts, Amherst, Amherst, Massachusetts, USA.
References
- 1.Zhou HX, Rivas G, Minton AP. Annu Rev Biophys. 2008;37:375–397. doi: 10.1146/annurev.biophys.37.032807.125817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dix JA, Verkman AS. Annu Rev Biophys. 2008;37:247–263. doi: 10.1146/annurev.biophys.37.032807.125824. [DOI] [PubMed] [Google Scholar]
- 3.Pielak GJ, et al. Biochemistry. 2009;48:226–234. doi: 10.1021/bi8018948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Serber Z, Corsini L, Durst F, Dötsch V. Methods Enzymol. 2005;394:17–41. doi: 10.1016/S0076-6879(05)94002-0. [DOI] [PubMed] [Google Scholar]
- 5.Srere PA. Trends Biochem Sci. 2000;25:150–153. doi: 10.1016/s0968-0004(00)01550-4. [DOI] [PubMed] [Google Scholar]
- 6.McConkey EH. Proc Natl Acad Sci USA. 1982;79:3236– 3240. doi: 10.1073/pnas.79.10.3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Durek P, Walther D. BMC Syst Biol. 2008;2:100. doi: 10.1186/1752-0509-2-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alberts B. Cell. 1998;92:291–294. doi: 10.1016/s0092-8674(00)80922-8. [DOI] [PubMed] [Google Scholar]
- 9.Kempner ES, Miller JH. Cell Motil Cytoskeleton. 2003;56:219–224. doi: 10.1002/cm.10145. [DOI] [PubMed] [Google Scholar]
- 10.Misteli T. Cell. 2007;128:787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 11.Lippincott-Schwartz J, Roberts TH, Hirschberg K. Annu Rev Cell Dev Biol. 2000;16:557–589. doi: 10.1146/annurev.cellbio.16.1.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang J, Maslov S, Shakhnovich EI. Mol Syst Biol. 2008;4:210. doi: 10.1038/msb.2008.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilt BA, et al. Annu Rev Neurosci. 2009;32:435–506. doi: 10.1146/annurev.neuro.051508.135540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoenger A, McIntosh JR. Curr Opin Cell Biol. 2009;21:89–96. doi: 10.1016/j.ceb.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brandt F, et al. Cell. 2009;136:261–271. doi: 10.1016/j.cell.2008.11.016. [DOI] [PubMed] [Google Scholar]
- 16.Elcock AH. PLoS Comput Biol. 2006;2:e98. doi: 10.1371/journal.pcbi.0020098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alber F, Forster F, Korkin D, Topf M, Sali A. Annu Rev Biochem. 2008;77:443–477. doi: 10.1146/annurev.biochem.77.060407.135530. [DOI] [PubMed] [Google Scholar]
- 18.Stein M, Gabdoulline RR, Wade RC. Curr Opin Struct Biol. 2007;17:166–172. doi: 10.1016/j.sbi.2007.03.014. [DOI] [PubMed] [Google Scholar]
- 19.Bader S, Kuhner S, Gavin AC. FEBS Lett. 2008;582:1220–1224. doi: 10.1016/j.febslet.2008.02.015. [DOI] [PubMed] [Google Scholar]
- 20.Puig O, et al. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- 21.Remy I, Michnick SW. Methods Mol Biol. 2004;261:411–426. doi: 10.1385/1-59259-762-9:411. [DOI] [PubMed] [Google Scholar]
- 22.Fields S, Sternglanz R. Trends Genet. 1994;10:286–292. doi: 10.1016/0168-9525(90)90012-u. [DOI] [PubMed] [Google Scholar]
- 23.Collins SR, Schuldiner M, Krogan NJ, Weissman JS. Genome Biol. 2006;7:R63. doi: 10.1186/gb-2006-7-7-r63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roguev A, et al. Science. 2008;322:405–410. doi: 10.1126/science.1162609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sykes MT, Williamson JR. Annu Rev Biophys. 2009;38:197–215. doi: 10.1146/annurev.biophys.050708.133615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adilakshmi T, Lease RA, Woodson SA. Nucleic Acids Res. 2006;34:e64. doi: 10.1093/nar/gkl291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharon M, Robinson CV. Annu Rev Biochem. 2007;76:167–193. doi: 10.1146/annurev.biochem.76.061005.090816. [DOI] [PubMed] [Google Scholar]
- 28.Bennett BD, et al. Nat Chem Biol. 2009;5:593–599. doi: 10.1038/nchembio.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gutstein HB, Morris JS, Annangudi SP, Sweedler JV. Mass Spectrom Rev. 2008;27:316–330. doi: 10.1002/mas.20161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Specht KM, Shokat KM. Curr Opin Cell Biol. 2002;14:155–159. doi: 10.1016/s0955-0674(02)00317-4. [DOI] [PubMed] [Google Scholar]
- 31.Xie XS, Choi PJ, Li GW, Lee NK, Lia G. Annu Rev Biophys. 2008;37:417–444. doi: 10.1146/annurev.biophys.37.092607.174640. [DOI] [PubMed] [Google Scholar]
- 32.Werner JN, et al. Proc Natl Acad Sci USA. 2009;106:7858–7863. doi: 10.1073/pnas.0901781106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Greenfield D, et al. PLoS Biol. 2009;7:e1000137. doi: 10.1371/journal.pbio.1000137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ignatova Z, et al. Biopolymers. 2007;88:157–163. doi: 10.1002/bip.20665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Butland G, et al. Nature. 2005;433:531–537. doi: 10.1038/nature03239. [DOI] [PubMed] [Google Scholar]