Short abstract
We developed a real-time detection (RTD) polymerase chain reaction (PCR) with rapid thermal cycling to detect and quantify Pseudomonas aeruginosa in wound biopsy samples. This method produced a linear quantitative detection range of 7 logs, with a lower detection limit of 103 colony-forming units (CFU)/g tissue or a few copies per reaction. The time from sample collection to result was less than 1h. RTD-PCR has potential for rapid quantitative detection of pathogens in critical care patients, enabling early and individualized treatment.
Keywords: burn wound, polymerase chain reaction, Pseudomonas aeruginosa, quantitation, sepsis
Abstract
Introduction:
Early diagnosis of wound colonisation or prediction of wound sepsis provides an opportunity for therapeutic intervention. There is need for qualitative and quantitative tests that are more rapid than bacterial culture. Pseudomonas aeruginosa results in high morbidity and mortality rates, is inherently resistant to common antibiotics, and is increasingly being isolated as a nosocomial pathogen. We developed three PCR-based methods to detect and quantify P aeruginosa in wound biopsy samples: conventional PCR, enzyme-linked immunosorbent assay (ELISA)-PCR, and RTD-PCR with rapid thermal cycling (LightCycler™ technology), all based on the amplification of the outer membrane lipoprotein gene oprL. We compared the efficacy of these methods to bacterial culture by quantitatively measuring levels of P aeruginosa in serial dilutions, in reconstituted skin samples and 21 burn wound biopsy samples.
Materials and methods:
Serial 10-fold dilutions were made from an overnight P aeruginosa culture and plated out onto Luria-Bertani and cetrimide agar plates. The agar plates were incubated overnight at 37°C, and the colonies were counted in order to estimate the number of CFU per dilution tube. A sample was taken from each dilution tube as a template for the three PCR-based methods.
Serial P aeruginosa dilutions (see above) were added to uninfected cadaveric skin. The reconstituted biopsy samples were homogenized using a tissue tearer and DNA was extracted using XTRAX DNA buffer. The DNA was resuspended in distilled water. A sample was taken as a template for the PCR-based methods.
Twenty-one burn wound biopsy samples were taken from nine patients with suspected P aeruginosa burn wound infection. The biopsy samples were longitudinally divided into two pieces. From one piece, DNA was extracted (using XTRAX DNA buffer) and used as a template for PCR-based techniques (see above). The other piece was homogenized, in physiological water, using a tissue tearer. Serial 10-fold dilutions of the suspension were spread on Luria-Bertani and cetrimide agar plates. Colony counts were performed after overnight incubation at 37°C.
The PCR mixture contained sterile distilled water, PCR buffer, deoxynucleotide mixture or digoxigenin labelling mix, MgCl2, diluted template, primers PAL1 and PAL2, and AmpliTaQ DNA polymerase. The amplification was performed in a GeneAmp® PCR System 2400. An aliquot of the reaction mixture was put on an agarose gel for electrophoresis and visualisation of the PCR product. An image of the gel was made using a digital camera. Image analysis software was used to calculate the band mass of the experimental bands.
An aliquot of the digoxigenin labelling reaction was denatured and then hybridized with the biotinylated capture probe PrL. Some of the resultant solution was transferred to a well of a streptavidin-coated microtitre plate (MTP) and incubated for 3 h at 45°C. The solution was discarded. Peroxidase conjugated antidigoxigenin was added and the MTP was incubated for 30 min at 37°C. The solution was discarded and ABTS substrate was added. The MTP was incubated for 30 min at 37°C. Absorbance was read at 405 nm.
The RTD-PCR mixture contained PCR grade sterile water, diluted template DNA, primers PAL1 and PAL2, 3' fluorescein (FL)-labelled probe oprL-FL, 5' LC Red 640-labelled and 3' phosphorylated probe oprL-LC, MgCl2, and LC DNA Master Hybridisation Probes, containing Taq DNA polymerase, reaction buffer, dNTP mix with dUTP instead of dTTP, and MgCl2. The amplification was performed in a LightCycler™. The fluorescence signal of LC Red 640 was measured during the annealing phase. The measured fluorescence data was processed with analysis software.
Results and discussion:
The three methods showed a good concordance with the culture results. Conventional PCR was at least 100 times less sensitive than bacterial culture and had a low dynamic range (2 logs). With a lower detection limit of 103 CFU/g tissue, ELISA-PCR was ten times more sensitive than conventional PCR. The dynamic range, however, did not increase. ELISA-PCR is very time consuming (8 h). The RTD-PCR produced a linear quantitative detection range of 7 logs with a lower detection limit of 103 CFU/g tissue. More important, however, was that the time from sample collection to result was less than 1 h. Two biopsy specimens scored significantly higher in ELISA-PCR and RTD-PCR than in bacterial culture. This could indicate that DNA from dead bacteria was amplified. One out of ten culture positive biopsy samples was found negative by all PCR-based methods. Topical antimicrobial agents possibly inhibited PCR. These results show that RTD-PCR has potential for the rapid quantitative detection of pathogens in critical care patients, enabling early and individualized treatment. Further study is required to assess the reliability of this new technology, and its impact on patient outcome and hospital costs.
Introduction
Although effective topical antimicrobial chemotherapy and early excision of burn wounds have significantly reduced the occurrence of invasive burn wound infections, sepsis is still a major problem [1,2,3]. The risk of septicaemia increases in proportion to the degree of cutaneous infection [4,5,6]. Many investigators have reported [6,7,8,9,10,11] that quantitative biopsy culture was the best method for early detection of sepsis. Heininger et al [12] stressed that only 4-12% of blood cultures is found positive. Conversely, McManus et al [13] reported in 1987 that high tissue counts did not necessarily indicate invasion, and that the principal value of quantitative biopsy culture was the demonstration of the predominant burn wound flora. Even so, when sepsis ensues, while awaiting the results of blood cultures, a knowledge of the organisms that colonize a burn wound can facilitate prompt and appropriate antibiotic treatment that is based on the expected sensitivity of the identified germs, rather than initiating a purely empirical therapy. There is need for qualitative and quantitative tests that are more rapid than bacterial culture.
We decided to develop such a test for the rapid detection and quantitation of Pseudomonas aeruginosa in burn wound biopsy samples. This bacterium is ubiquitous, is inherently resistant to common antibiotics, and therefore is one of the most problematic pathogens in modern hospitals [14,15,16,17]. Burn wound patients, mechanically ventilated patients and cystic fibrosis patients are particularly susceptible to P aeruginosa infection [1,18,19]. Biopsy samples are, with respect to homogenization and DNA extraction, very tough clinical samples and thus would be an excellent test case for the applicability of the method on direct clinical samples.
In 1997 our group developed a PCR test for the direct detection and identification of P aeruginosa in clinical samples that is based on the amplification of the outer membrane lipoprotein gene oprL [20,21,22]. Since then we developed several quantitative variants of this test, exploiting the technology available at the time.
In the first instance, we amplified the oprL gene by means of conventional PCR and visualized the PCR product by ethidium bromide (EtBr) staining of agarose gels. The intensity of the fluorescence produced by EtBr was quantified.
Second, we developed an ELISA-mediated PCR in order to quantify the amplified oprL gene. PCR products were labelled with digoxigenin during the amplification process and quantitatively detected by absorbance reading in microtitre plates.
Finally, we exploited the recently developed `real-time' quantitative PCR technology [23,24,25,26,27]. We opted for the LightCycler™ [28] system (Roche Diagnostics, Brussels, Belgium) because it features rapid capillary tube resistive thermal cycling, reducing the amplification time dramatically. Two adjacent hybridization probes, labelled with different fluorescent dyes, are used to monitor the appearance of PCR product. The emitted light signal is proportional to the amount of specific DNA product available for hybridization, and thus increases every cycle [29]. The probes were designed to be complementary to a conserved region of the oprL gene, as determined by sequence analysis of the oprL gene of 85 nonrelated clinical P aeruginosa isolates.
In the present report we compare the assay performance of the above-mentioned methods, in terms of practicability, to bacterial culture. For this purpose three types of samples were assayed: serial P aeruginosa dilutions, uninfected skin spiked with P aeruginosa and 21 burn wound biopsy samples. All methods were useful, but only Light-Cycler™ RTD-PCR allowed rapid quantitative detection of P aeruginosa in skin biopsies with an adequate detection limit and a wide log-linear range.
Materials and methods
Sample preparation
Serial Pseudomonas aeruginosa dilutions
Serial 10-fold dilutions were made from an overnight P aeruginosa culture (strain PAO-1, ATCC 15692) in order to look for the lower detection limit and the dynamic range of the PCR-based methods. Dilutions were made in physiological water. Aliquots (100 μl) were plated out, in triplicate, onto Luria-Bertani (Gibco-BRL Life Technologies, Paisley, Scotland) and cetrimide (Sanofi Pasteur, Brussels, Belgium) agar plates. The agar plates were incubated overnight at 37°C, and the colonies were counted in order to estimate the number of CFU per dilution tube. A sample (5 μl) was taken from each dilution tube as a template for the three PCR-based methods. Standards ranging from 1 to 106 CFU/5 μl were assayed.
Reconstituted biopsy samples
Reconstituted biopsy samples were prepared in sterile 5-ml tubes (Nunc; Roskilde, Denmark) by adding 10 μl serial P aeruginosa dilutions (see above) to 50 mg uninfected cadaveric skin, at final concentrations of 10-109 CFU/g tissue. XTRAX DNA extraction buffer (1 ml; Gull Laboratories, Salt Lake City, UT, USA) was added, and the reconstituted biopsy samples were homogenized for 1 min, at 30 000 rpm, using a tissue tearer (BioSpec Products, Bartlesville, Oklahoma, USA). The suspensions were transferred to a 1.8-ml cryotube (Nunc) with a loosened cap and were microwaved for 13 s at 600 W. The tubes were gently shaken to mix the contents and were microwaved for a further 6 s. The tubes were cooled for 3 min at room temperature and centrifuged for 1min at 10 000 g in order to pellet the proteins.
Supernatant (500 μl) was transferred to a microcentrifuge tube (Eppendorf, Hamburg, Germany). Molecular grade isopropanol (500 μl; Sigma Aldrich, Deisenhofen, Germany) was added, and the contents of the tube were mixed thoroughly by inverting the tube 10 times. The tube was centrifuged for 1 min at 10 000 g in order to pellet the DNA. The supernatant was discarded by decanting, and the tube was allowed to stand upside down for 2 min to allow the remaining fluid to drain. The DNA was resuspended in 30 μl distilled water. A sample (5 μl) was taken as a template for conventional PCR and ELISA-PCR, and 3 μl was taken for RTD-PCR. Standards ranging from 103 to 109 CFU/g skin tissue were assayed.
Clinical burn wound biopsy samples
Twenty-one burn wound biopsy samples, weighing 20-170 mg (mean 54 mg), were assayed. The biopsy specimens were aseptically taken from nine patients with suspected P aeruginosa burn wound infection, using a 4-mm punch biopsy needle (Labo Stiefel, Leuven, Belgium; n =15) or a lancet (n =6).
The samples were longitudinally divided into two pieces using a sterile scalpel and weighed. From one piece DNA was extracted using XTRAX DNA buffer and used as template for the PCR-based techniques (see above). The other piece was collected in a sterile 5-ml tube containing 10 μl physiological water/mg tissue, and was homogenized, on ice, for 1 min at 30 000 rpm using a tissue tearer. Serial 10-fold dilutions of the homogenized wound biopsy samples were spread, in triplicate, on Luria-Bertani and cetrimide agar plates. Colony counts were performed after overnight incubation at 37°C.
Conventional polymerase chain reaction and digoxigenin labelling
The PCR was completed in 200-μl microcentrifuge tubes. The PCR mixture (50 μl final volume) contained the following: 26.5 μl sterile distilled water; 5 μl 10×PCR buffer (500 mmol/l KCl and 100 mmol/l Tris-HCl: pH 8.3); 5 μl of a deoxynucleotide mixture (dGTP, dTTP, dATP and dCTP; 2 mmol/l each) or 5 μl digoxigenin-labelling mix from Boehringer-Mannheim (Brussels, Belgium; 2 mmol/l dGTP, dATP and dCTP, and 1.9 mmol/l digoxigenin-dUTP); 6 μl MgCl2 (2.5 mmol/l); 5 μl diluted template DNA; 1 μl primer PAL1 (25 μmol/l); 1 μl primer PAL2 (25 μmol/l); and 0.5 μl AmpliTaQ DNA polymerase (5 U/μl). The primers had the following sequences: PAL1, 5'-ATGGAAATGCTGAAATTCGGC-3'; and PAL2, 5'-CTTCTTCAGCTCGACGCGACG-3'. All PCR reagents were purchased from PE Applied Biosystems (Nieuwerkerk a/d Ijssel, The Netherlands).
A reaction mixture containing all of the ingredients except the template was made. The amplification was performed in a GeneAmp® PCR System 2400 (PE Applied Biosystems). The amplification program was set at 35 cycles of denaturation at 94°C for 30 s or 45 s (digoxigenin labelling), annealing at 57°C for 30 s or 1 min (digoxigenin labelling), and elongation at 72°C for 30 s or 2 min (digoxigenin labelling).
Some of the reaction mixture (10 μl) was put on an agarose gel of 2% (weight/volume) for electrophoresis and visualization of the PCR product after staining with EtBr on a ultraviolet transilluminator. An image of the gel was made using a DC40 digital camera (Eastman Kodak Company, Rochester, New York, USA). Using the intensity measured for the bands originating from the standards, P aeruginosa serial dilutions or reconstituted samples, Kodak Digital Science 1D image analysis software was used to calculate the band mass of the experimental bands using linear regression.
Digoxigenin detection
Some of the digoxigenin labelling reaction (35 μl) was transferred to a sterile microcentrifuge tube (Eppendorf). Denaturation solution (40 μl) was added and the tube was incubated for 10 min at room temperature. The tube was filled up to 500 μl with hybridization solution, which contained 50 pmol/ml biotinylated capture probe PrL with the following sequence: 5'-AAGCCGGAAGCCATGCGCG-CT-3'. Some of the resultant solution (200 μl) was transferred to a well of a streptavidin-coated microtitre plate (MTP). The MTP was incubated for 3 h at 45°C on a MTP shaker. The solution was discarded and the well was washed five times with 250 μl washing solution. A quantity (200 μl) of a peroxidase conjugated antidigoxigenin solution (10 mU/ml) was added, and the MTP was incubated for 30 min at 37°C on a MTP shaker. The solution was discarded and the well was washed five times with washing solution. ABTS substrate solution (200 μl) was added and the MTP was incubated for 30 min at 37°C on a MTP shaker. Absorbance was read at 405 nm. All digoxigenin detection reagents were purchased from Boehringer-Mannheim.
Real-time detection polymerase chain reaction
The RTD-PCR mixture (20 μl final volume) contained the following: 3.8 μl PCR grade sterile water; 3 μl diluted template DNA; 2 μl primer PAL1 (5 μmol/l); 2 μl primer PAL2(5 μmol/l); 2 μl of the 3' fluorescein-labelled probe oprL-FL (2 μmol/l); 4 μl of 5' LC Red 640-labelled and 3' phosphorylated probe oprL-LC (2 μmol/l); 1.2 μl MgCl2 (25 mmol/l); 2 μl LC DNA Master Hybridisation Probes containing Taq DNA polymerase; reaction buffer; dNTP mix with dUTP instead of dTTP; and 10 mmol/l MgCl2. All reagents were purchased from Roche Molecular Biochemicals (Brussels, Belgium). The probes were manufactured by the TIB MOLBIOL synthesis laboratory in Berlin, Germany, and had the following sequences: oprL-FL, 5'-TGCGATCACCACCTTCTACTTCGAGT-FL-3'; and oprL-LC, 5'-LC Red 640-CGACAGCTCCGACCTGAAG-p-3'.
Samples were spun into glass capillary tubes, capped and placed in the LightCycler™. After an initial denaturation at 95°C for 30 s, amplification was performed for 45 cycles of denaturation at 95°C for 2 s, annealing at 59°C for 10 s and elongation at 72°C for 10 s. The fluorescence signal of LC Red 640 was measured during the annealing phase. The measured fluorescence data was processed with analysis software.
Results and discussion
Conventional polymerase chain reaction
With a lower detection level between 104 and 105 CFU/g, wound biopsy tissue conventional PCR was at least 100 times less sensitive than bacterial culture (Table 1). This problem was related to the limited amount of sample, as compared with bacterial culture, that can be incorporated into one PCR reaction. Concentration of DNA or division of the sample over multiple PCR reactions could resolve this problem, but would result in a more elaborate method. Conventional PCR had a low dynamic range; bands that were very close to the background level gave poor results, and bands that were saturated also skewed the results (Fig. 1).
Table 1.
Summary of the characteristics of the tested quantitation methods
| Lower detection limit* | |||||||
| Log-linear range | Cost per test | Set up | |||||
| Method | (CFU/g) | (CFU/reaction) | (CFU/g) | Time to result (h) | Ease of use | (US$)† | cost ($) |
| Culture | 102 | 1 | 102-109 | 24 | Easy | 6 | 3500 |
| PCR | 104-105 | 10-102 | 106-108 | 4 | Moderate | 12‡ | 20 000 |
| ELISA-PCR | 103-104 | 1-10 | 105-107 | 8 | Elaborate | 25‡ | 20 000 |
| RTD-PCR (LC) | 103-104 | 1-10 | 103-109 | 1 | Moderate | 9‡ | 73 000 |
*The lower detection limit is variable due to variations in inter-run amplification efficiency. †Not including labour costs, only reagents and disposables. ‡Cost per clinical sample, based on the analysis of batches of 10 clinical samples and eight standards. LC, LightCycler™.
Figure 1.
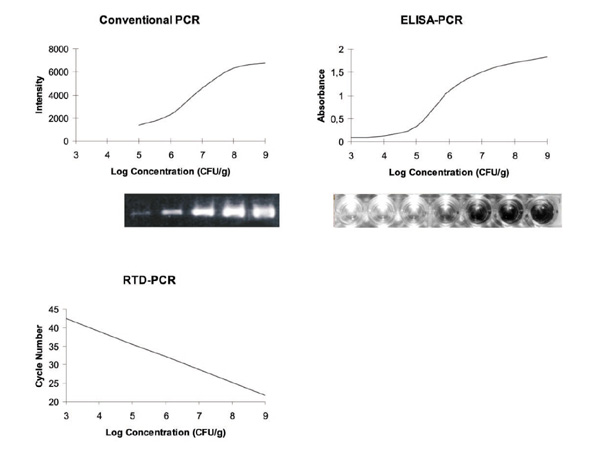
Standard curves for reconstituted biopsy samples.
Ten out of twenty-one burn wound biopsy samples turned out to be culture positive; five of them, with a density lower than 104 CFU/g, were not detected by conventional PCR. All culture-negative samples were also negative in PCR.
Enzyme-linked immunosorbent assay polymerase chain reaction
With a detection limit of 103 CFU/g tissue, ELISA-PCR was at least 10 times more sensitive than conventional PCR (Table 1). However, the linear response range did not increase, but shifted to lower concentrations along with the detection limit (Fig. 1). This technique is very time-consuming and involves handling that increase the risk of contamination of PCR product (Fig. 2). Only one culture-positive wound biopsy, out of 10, containing 3000 CFU/g tissue, was found to be negative by ELISA-PCR. Topical antimicrobial agents possibly inhibited PCR.
Figure 2.
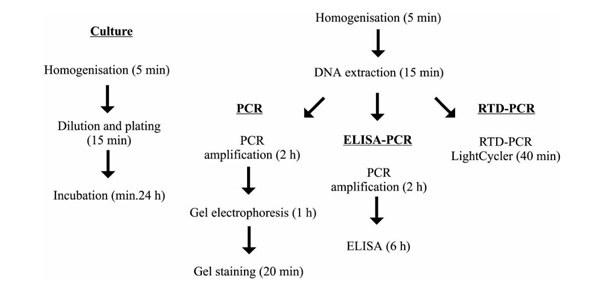
Comparison of the procedural steps involved in the quantitation methods.
Two biopsy specimens with approximately 200 and 400 CFU/g tissue, as determined by bacterial culture, scored significantly higher in ELISA-PCR: 1200 and 1450 CFU/g tissue, respectively. This could indicate that DNA from nonproliferating or dead bacteria was amplified. The question regarding whether quantitative estimates of bacteria, determined using PCR-based methods, correlate with the number of viable bacteria has been raised before [30]. For this reason, PCR-based tests are probably not appropriate for the monitoring of treatment efficacy. They should be considered as complementary indicative tests in critical care settings. On the other hand, PCR-based methods could detect bacteria that were inhibited in their growth by residual concentrations of antimicrobials in the sample [31], overgrown by other bacteria present in the sample, autolyzed during incubation [32], or difficult to cultivate in vitro [33], and are thus under-diagnosed by bacterial culture. PCR-ELISA has merit with regard to its sensitivity, but is too time-consuming and expensive to implement in a clinical laboratory (Fig. 2).
Real-time detection polymerase chain reaction
RTD-PCR produced a linear quantitative detection range of 7 logs with a lower detection limit of 103 CFU/g tissue, or a few copies per reaction (Table 1). RTD-PCR uses a kinetic approach rather than an end-point approach. Kinetic quantification in the log phase of the reaction, the phase of constant efficiency, clearly generates a larger linear response than end-point detection in the plateau phase of the reaction (Fig. 1). Also, there is no need for any post-PCR sample manipulation, eliminating PCR contamination concerns. The recent introduction of a second dye for the LightCycler™ system offers the prospect of an internal control, addressing the problem of tube-to-tube variations and PCR inhibition, and thus improving the reliability of the results. However, the greatest merit of Light-Cycler™ RTD-PCR is its rapidity. Rapid thermal cycling reduced the time of amplification, detection and analysis of DNA from several hours to 30 min (Fig. 2). The RTD-PCR results for the burn wound biopsy samples were concordant with those obtained using ELISA-PCR; the false-negative result and the two over-diagnosed biopsies were also observed using RTD-PCR. The use of capillaries instead of tubes permits smaller reaction volumes, thus lowering the reagent costs. The set-up costs, however, may be beyond the capabilities of some laboratories (Table 1).
Conclusion
Up until now, RTD-PCR has been applied for the detection of food-borne pathogens [34], cancer [35,36,37], genetic diseases [38] and infectious diseases [39,40,41,42].
Although a limited number of clinical specimens were tested, the present results indicate that RTD-PCR, and more specifically LightCycler™ technology, has potential for quantitative applications in the clinical laboratory. In particular, it has applications for the critical care population, at the point of care, and it is important that the test is subjected to further optimization and evaluation. Early infection diagnosis remains a difficult problem for patients with burn wounds or cystic fibrosis, and for critical care patients in general. Prognosis and survival are often dependent on an early, individualized treatment. Automated extraction of DNA from a variety of clinical samples (blood, expectorations, urine, etc.) and subsequent rapid, online, quantitative detection of pathogens (P aeruginosa, Staphylococcus aureus, Haemophilus influenzae, among others) by RTD-PCR is now possible, allowing early therapeutic decisions to be made. Multiple colour detection will open the door to multiplex RTD-PCR. Further studies are necessary to assess the impact of rapid RTD-PCR on patient outcome and hospital costs [43,44].
Acknowledgments
Acknowledgements
This work was supported by grants of ESSO Benelux and VZW `De Vrienden van de Huidbank'.
References
- Pruitt BA, Jr, McManus AT, Kim SH, Goodwin CW. Burn wound infections: current status. World J Surg. 1998;22:135–145. doi: 10.1007/s002689900361. [DOI] [PubMed] [Google Scholar]
- Lee JJ, Marvin JA, Heimbach DM, Grube BJ, Engrave LH. Infection control in a burn center. J Burn Care Rehabil. 1992;11:575–580. doi: 10.1097/00004630-199011000-00018. [DOI] [PubMed] [Google Scholar]
- McManus AT, Mason AD, Jr, McManus WF, Pruitt BA., Jr Twenty-five year review of Pseudomonas aeruginosa bacteremia in a burn center. Eur J Clin Microbiol. 1985;4:219–223. doi: 10.1007/BF02013601. [DOI] [PubMed] [Google Scholar]
- Teplitz C, Davis D, Mason AD, Jr, Moncrief JA. Pseudomonas burn wound sepsis. I. Pathogenesis of experimental Pseudomonas burn wound sepsis. J Surg Res. 1964;4:200–222. doi: 10.1016/s0022-4804(64)80026-3. [DOI] [PubMed] [Google Scholar]
- Moncrief JA, Lindberg RB, Switzer WE, Pruitt BA., Jr Use of topical antibacterial therapy in the treatment of the burn wound. . Arch Surg. 1966;92:558–565. doi: 10.1001/archsurg.1966.01320220114019. [DOI] [PubMed] [Google Scholar]
- Perez-Cappelano R, Manelli JC, Palayret D, Carlin G, Echinard C, Jouglard JP. Evaluation of the septicaemic risk by a quantitative study of the cutaneous flora in patients with burns. Burns. 1976;3:42–45. [Google Scholar]
- Lawrence JC, Lilly HA. A quantitative method for investigating the bacteriology of the skin: its applicability to burns. Br J Exp Pathol. 1972;53:550–558. [PMC free article] [PubMed] [Google Scholar]
- Loebl EC, Marvin JA, Heck EL, Curreri PW, Baxter CR. The method of quantitative burn-wound biopsy cultures and its routine use in the care of the burned patient. Am J Clin Pathol. 1974;61:20–24. doi: 10.1093/ajcp/61.1.20. [DOI] [PubMed] [Google Scholar]
- McManus WF, Goodwin CW, Mason AD, Jr, Pruitt BA., Jr Burn wound infection. J Trauma. 1981;21:753–756. doi: 10.1097/00005373-198109000-00001. [DOI] [PubMed] [Google Scholar]
- Taddonio TE, Thomson PD, Tait MJ, Prasad JK, Feller I. Rapid quantification of bacterial and fungal growth in burn wounds: biopsy homogenate Gram stain versus microbial culture results. Burns. 1988;14:180–184. doi: 10.1016/0305-4179(88)90035-6. [DOI] [PubMed] [Google Scholar]
- Bharadwaj R, Joshi BN, Phadke SA. Assessment of burn wound sepsis by swab, full thickness biopsy culture and blood culture: a comparative study. Burns. 1983;10:124–126. doi: 10.1016/0305-4179(83)90010-4. [DOI] [PubMed] [Google Scholar]
- Heininger A, Binder M, Schmidt S, Unertl K, Botzenhart K, Döring G. PCR and blood culture for detection of Escherichia coli bacteremia in rats. J Clin Microbiol. 1999;37:2479–2482. doi: 10.1128/jcm.37.8.2479-2482.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus AT, Kim SH, McManus WF, Mason AD, Jr, Pruitt BA., Jr Comparison of quantitative microbiology and histopathology in divided burn-wound biopsy specimens. Arch Surg. 1987;122:74–76. doi: 10.1001/archsurg.1987.01400130080012. [DOI] [PubMed] [Google Scholar]
- Masuda N, Sakagawa E, Ohya S. Outer membrane proteins responsible for multiple drug resistance in Pseudomonas aeruginosa. . Antimicrob Agents Chemother. 1995;39:645–649. doi: 10.1128/AAC.39.3.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H. Prevention of drug access to bacterial targets: permeability barriers and active efflux. Science. 1994;264:382–388. doi: 10.1126/science.8153625. [DOI] [PubMed] [Google Scholar]
- Burdon DW, Whitby JL. Contamination of hospital disinfectants with Pseudomonas species. . Br Med J. 1967;2:153–155. doi: 10.1136/bmj.2.5545.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrott PL, Terry PM, Whitworth EN, Frawley LW, Coble RS, Wachsmuth IK, McGowan JE., Jr Pseudomonas aeruginosa peritonitis associated with contaminated poloxamer-iodine solution. Lancet. 1982;ii:683–685. doi: 10.1016/s0140-6736(82)90712-7. [DOI] [PubMed] [Google Scholar]
- Craven DE, Steger KA. Nosocomial pneumonia in mechanically ventilated adult patients: epidemiology and prevention in 1996. Semin Respir Infect. 1996;11:32–53. [PubMed] [Google Scholar]
- Koch C, Høiby N. Pathogenesis of cystic fibrosis. . Lancet. 1993;341:1065–1069. doi: 10.1016/0140-6736(93)92422-p. [DOI] [PubMed] [Google Scholar]
- Cornelis P, Bouia A, Belarbi A, Guyonvarch A, Kammerer B, Hannaert V, Hubert JC. Cloning and analysis of the gene for the major outer membrane lipoprotein from Pseudomonas aeruginosa. . Mol Microbiol. 1989;3:421–428. doi: 10.1111/j.1365-2958.1989.tb00187.x. [DOI] [PubMed] [Google Scholar]
- Lim A, Jr, De Vos D, Brauns M, Mossialos D, Gaballa A, Qing D, Cornelis P. Molecular and immunological characterization of OprL, the 18-kDa outer membrane peptidoglycan-associated (PAL) lipoprotein of Pseudomonas aeruginosa. . Microbiology . 1997;143:1709–1716. doi: 10.1099/00221287-143-5-1709. [DOI] [PubMed] [Google Scholar]
- De Vos D, Lim A, Jr, Pirnay JP, Struelens M, Vandenvelde C, Duinslaeger L, Vanderkelen A, Cornelis P. Direct detection of Pseudomonas aeruginosa in clinical samples such as skin biopsy specimens and expectorations by multiplex PCR based on two outer membrane lipoprotein genes, oprI and oprL. J Clin Microbiol. 1997;35:1295–1299. doi: 10.1128/jcm.35.6.1295-1299.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland PM, Abramson RD, Watson R, Gelfand DH. Detection of specific polymerase chain reaction product by utilizing the 5' -3' exonuclease activity of Thermus aquaticus DNA polymerase. Proc Natl Acad Sci USA. 1991;88:7276–7280. doi: 10.1073/pnas.88.16.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyagi S, Kramer FR. Molecular beacons: probes that fluoresce upon hybridization. Nature Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- Wittwer CT, Herrmann MG, Moss AA, Rasmussen RP. Continuous fluorescence monitoring of rapid cycle DNA amplifications. BioTechniques. 1997;22:130–138. doi: 10.2144/97221bi01. [DOI] [PubMed] [Google Scholar]
- Nazarenko IA, Bhatnagar SK, Hohman RJ. A closed tube format for amplification and detection of DNA based on energy transfer. Nucleic Acids Res. 1997;25:2516–2521. doi: 10.1093/nar/25.12.2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitcombe D, Theaker J, Guy SP, Brown T, Little S. Detection of PCR products using self-probing amplicons and fluorescence. Nature Biotechnol. 1999;17:804–807. doi: 10.1038/11751. [DOI] [PubMed] [Google Scholar]
- Wittwer C, Ririe K, Andrew R, David D, Gundry R, Balis U. The Light-Cycler™: a microvolume multisample fluorimeter with rapid temperature control. BioTechniques. 1997;22:176–181. doi: 10.2144/97221pf02. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Flood SJA, Marmaro J, Giusti W, Deetz K. Oligonucleotides with fluorescent dyes at opposite ends provide a quenched probe system useful for detecting PCR product and nucleic acid hybridization. PCR Methods Appl. 1995;4:357–362. doi: 10.1101/gr.4.6.357. [DOI] [PubMed] [Google Scholar]
- Desjardin LE, Chen Y, Perkins MD, Teixeira L, Cave MD, Eisenach KD. Comparison of the ABI 7700 system (TaqMan) and competitive PCR for quantification of IS6110 DNA in sputum during treatment of tuberculosis. J Clin Microbiol. 1998;36:1964–1968. doi: 10.1128/jcm.36.7.1964-1968.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto T, Tanaka M, Ogata N, Mizunoe Y, Takahashi K, Kumazawa J. Significance of urinary endotoxin concentration in patients with urinary tract infection. Urol Res. 1991;19:293–295. doi: 10.1007/BF00299061. [DOI] [PubMed] [Google Scholar]
- Fischer GW, Smith LP, Hemming VG, Longfield R, Valdes-Dapena AA, Lopreiato JO. Avoidance of false-negative blood culture results by rapid detection of pneumococcal antigen. JAMA. 1984;252:1742–1743. [PubMed] [Google Scholar]
- Razin S. DNA probes and PCR in diagnosis of mycoplasma infections. Mol Cell Probes. 1994;8:497–511. doi: 10.1006/mcpr.1994.1071. [DOI] [PubMed] [Google Scholar]
- Oberst RD, Hays MP, Bohra LK, Phebus RK, Yamashiro CT, PaszkoKolva C, Flood SJ, Sargeant JM, Gillespie JR. PCR-based DNA amplification and presumptive detection of Escherichia coli O157:H7 with an internal fluorogenic probe and the 5' nuclease (TaqMan) assay. Appl Environ Microbiol. 1998;64:3389–3396. doi: 10.1128/aem.64.9.3389-3396.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieche I, Olivi M, Champeme MH, Vidaud D, Lidereau R, Vidaud M. Novel approach to quantitative polymerase chain reaction using real-time detection: application to the detection of gene amplification in breast cancer. Int J Cancer. 1998;78:661–666. doi: 10.1002/(sici)1097-0215(19981123)78:5<661::aid-ijc22>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- Laurendeau I, Bahuau M, Vodovar N, Larramendy C, Olivi M, Bieche I, Vidaud M, Vidaud D. TaqMan PCR-based gene dosage assay for predictive testing in individuals from a cancer family with INK4 locus haploinsufficiency. Clin Chem. 1999;45:982–986. [PubMed] [Google Scholar]
- Mensink E, van de Locht A, Schattenberg A, Linders E, Schaap N, Geurts van Kessel A, De Witte T. Quantitation of minimal residual disease in Philadelphia chromosome positive chronic myeloid leukemia using real-time quantitative RT-PCR. Br J Haematol. 1998;102:768–774. doi: 10.1046/j.1365-2141.1998.00823.x. [DOI] [PubMed] [Google Scholar]
- Von Ahsen N, Schutz E, Armstrong VW, Oellerich M. Rapid detection of prothrombotic mutations of prothrombin (G20210A), factor V (G1691A) and methylenetetrahydrofolate reductase (C677T) by real-time fluorescence PCR with the LightCycler. Clin Chem. 1999;45:694–696. [PubMed] [Google Scholar]
- Lewin SR, Vesanen M, Kostrikis L, Hurley A, Duran M, Zhang L, Ho DD, Markowitz M. Use of real-time PCR and molecular beacons to detect virus replication in human immunodeficiency virus type 1-infected individuals on prolonged effective antiretroviral therapy. J Virol. 1999;73:6099–6103. doi: 10.1128/jvi.73.7.6099-6103.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pahl A, Kuhlbrandt U, Brune K, Rollinghoff M, Gessner A. Quantitative detection of Borrelia burgdorferi by real-time PCR. J Clin Microbiol. 1999;37:1958–1963. doi: 10.1128/jcm.37.6.1958-1963.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martell M, Gomez J, Esteban JI, Sauleda S, Quer J, Cabot B, Esteban R, Guardia J. High-throughput real-time reverse transcription-PCR quantitation of hepatitis C virus RNA. J Clin Microbiol. 1999;37:327–332. doi: 10.1128/jcm.37.2.327-332.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura H, Morita M, Yabuta Y, Kuzushima K, Kato K, Kojima S, Mat-suyama T, Morishima T. Quantitative analysis of Epstein-Barr virus load by using a real-time PCR assay. J Clin Microbiol. 1999;37:132–136. doi: 10.1128/jcm.37.1.132-136.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doern G, Vautour R, Gaudet M, Levy B. Clinical impact of rapid in vitro susceptibility testing and bacterial identification. J Clin Microbiol. 1994;32:1757–1762. doi: 10.1128/jcm.32.7.1757-1762.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barenfanger J, Drake C, Kacich G. Clinical and financial benefits of rapid bacterial identification and antimicrobial susceptibility testing. J Clin Microbiol. 1999;37:1415–1418. doi: 10.1128/jcm.37.5.1415-1418.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]


