Abstract
This study expands the capabilities for ultratrace proteomic analysis of our previous work by incorporating on-line sample desalting using a triphasic (reversed phase (RP)/SCX/micro-SPE) trapping column connected to a 3.2 m × 10 μm i.d. poly(styrene-divinylbenzene) (PS-DVB) porous layer open tubular (PLOT) column. To minimize extra sample handling steps, C18 RP packing was incorporated in the capillary tubing upstream of the SCX column for the on-line desalting. For the micro-SPE column, a 50 μm i.d. PS-DVB monolithic column was positioned downstream of the SCX column. High performance separation was achieved on the PLOT column at a mobile phase flow rate of 20 nL/min. The sensitivity and high resolution capability of the new multidimensional platform was evaluated using an in-gel tryptic digested sample of a cervical cancer (SiHa) cell line. For the injected amount of 1200 cells (~500 ng), over 2700 peptides covering greater than 850 unique proteins were identified from the triphasic SCX/PLOT/MS analysis of a single SDS gel section (>40 kDa). The 2D LC/MS platform demonstrated good separation performance, such that more than 85% of the identified peptides were detected from only one salt fraction. In a triplicate analysis of the above >40 kDa gel section, 4497 peptides and 1209 unique proteins were identified when applying stringent filtering criteria, with a false positive rate of 2.4%. When all three SDS-PAGE gel sections of the lysed SiHa cells were analyzed, 5047 peptides (false positive rate 1.8%) and 1857 unique proteins, including cancer related proteins such as MAP kinases, were identified.
Keywords: PLOT column, monolithic column, proteomics, multiple dimensional separations, ultranarrow bore LC column
1. Introduction
Proteomic studies typically involve a broad range of protein identification from samples of high complexity and wide concentration range [1–3]. Shotgun proteomics, a major tool of proteomic analysis, employs a strategy where protein mixtures are proteolytically digested, and the resultant peptides are then separated by RPLC and analyzed by tandem mass spectrometry [4–10]. The samples are of such complexity that a single separation step cannot provide the resolving power required for comprehensive analysis. As a result, multidimensional separations are typically employed. Strong cation exchange (SCX) chromatography is often selected as the initial dimension of peptide fractionation due to its orthogonality to RPLC, which is generally coupled on-line to MS. Other strategies, such as capillary isoelectric focusing (CIEF)/RPLC, have also been used for multidimensional proteomic studies [11,12].
Current LC-ESI-MS approaches typically utilize columns with inner diameters greater than 50 μm, leading to flow rates in the range of hundreds of nL/min to a few μL/min. However, the sensitivity achieved using these columns requires relatively large amounts of sample (microgram level of total protein). New approaches with improved sensitivity that allow analysis of limited sample amounts, e.g. low number of cells, are highly desirable [13]. Since the sensitivity of LC-ESI-MS improves with decreasing flow rate, for columns with same length and packing material [14–16], reduction in column i.d. is an effective means to achieve high sensitivity without sacrificing separation efficiency [17–20]. However, reproducible preparation of higher-efficiency ultranarrow bore LC columns is a significant challenge.
Recently, our laboratory developed a single step approach suitable for preparation of 10 μm i.d. long, high efficiency poly(styrene-divinylbenzene) (PS-DVB) PLOT columns for ultratrace proteomic analysis [21]. The PLOT columns were shown to provide good column to column reproducibility and relatively high loading capacity, operate at 20 nL/min flow rate and allow detection at the low attomole level. A peak capacity of 400 was achieved when the PLOT column was off-line coupled to a 50 μm i.d. PS-DVB monolithic micro-SPE column [21]. Subsequent to this work, our laboratory developed on-line one dimensional (1D) and two dimensional (2D) PLOT/MS platforms to provide robust, high performance, and ultratrace proteomic analysis [22]. The on-line micro-SPE-PLOT column assembly maintained separation performance equivalent to that obtained with direct loading onto the PLOT column, using a newly designed PicoClear tee for connection of a 50 μm i.d. PS-DVB monolithic micro-SPE column to the PLOT column. In comparison with a typical 15 cm × 75 μm i.d. packed capillary column, the number of identified proteins was twice that obtained from on-line micro-SPE-PLOT/MS analysis while using only 5% of the injected sample amount. The resolving power of the micro-SPE-PLOT assembly was greatly increased by incorporating an SCX column for the initial dimension of separation. More than 1000 peptides associated with 536 unique proteins were identified from a single 2D SCX-PLOT/MS analysis of an in-gel digested sample of a gel section (15 to 40 kDa) of the equivalent to 600 cervical cancer (SiHa) cells injected, using 5 ion exchange salt fractions. However, loading on the SCX column was not optimized due to the presence of salt in the sample. A large fraction of the peptides were eluted from the SCX in the loading step.
To overcome the presence of salt in the sample, an on-line desalting step has been added by coupling a triphasic (RP/SCX/micro-SPE) trapping column to a 3.2 m × 10 μm i.d. PLOT column. Following the strategy of others [23], an additional section of C18 particles was packed upstream of the SCX column for on-line desalting. The new platform was used for comprehensive characterization of the proteome of a SiHa cell line with the injection of a limited number of cells. The protein extract of SiHa cells was first separated by 1D SDS-PAGE at the protein level. The gel was then cut into three sections, digested with trypsin, and analyzed by 2D SCX/PLOT/MS. Close to 2000 proteins were identified from the equivalent of 1200 cells (~500 ng), including a number of proteins relevant to cancer, e.g. MAP kinases. The results demonstrate that the newly developed platform is a powerful proteomic approach for multidimensional separation of very limited amount of samples.
2 Materials and methods
2.1 Materials
Styrene, divinylbenzene, ethanol, formic acid (HPLC grade), 3-(trimethoxysilyl)propyl methacrylate, 2,2-diphenyl-1-picrylhydrazyl (DPPH), N,N-dimethylformamide anhydrous (DMF), tetrahydrofuran (THF), azobisoisobutyronitrile (AIBN), ammonium bicarbonate, dithiothreitol (DTT), and iodoacetamide (IAA) were purchased from Sigma-Aldrich (St. Louis, MO). Fused silica capillary tubing was obtained from Polymicro Technologies (Phoenix, AZ). Ammonium acetate, acetonitrile (HPLC grade), and deionized water (HPLC grade) were from Thermo Fisher (Waltham, MA). Trypsin (sequencing grade) was from Promega (Madison, WI). The SiHa cervical cancer cell line was obtained from American Type Culture Collection (ATCC) (Manassas, VA, USA). PEEK microtees were purchased from Upchurch Scientific (Oak Harbor, WA). The PicoClear tee and union were obtained from New Objective (Woburn, MA).
2.2 On-line 2D SCX/PLOT/MS using a 10 μm i.d. PS-DVB PLOT column
Ten micron i.d. PS-DVB PLOT columns were prepared using protocols previously described [21]. Briefly, a 10 μm i.d. capillary, pretreated with 3-(trimethoxysilyl)propyl methacrylate, was filled with a degassed solution containing 5 mg of AIBN, 200 μL styrene, 200 μL divinylbenzene, and 600 μL ethanol. The capillary was sealed with septa at both ends and heated at 74 °C for ~16 h in a water bath. After washing with acetonitrile, the column was ready for use. The ends of the capillary were sealed in water for storage. PS-DVB monolithic micro-SPE columns (50 μm i.d.) were prepared using a published procedure [24] that was slightly modified. A low density PS-DVB monolithic column was synthesized from a polymerization solution containing 5 mg of AIBN, 200 μL styrene, 200 μL divinylbenzene, 40 μL THF, and 550 μL of decanol. The column (4 cm) resulted in a flow rate of 1 μL/min at a back pressure of 2900 psi, allowing rapid sample loading.
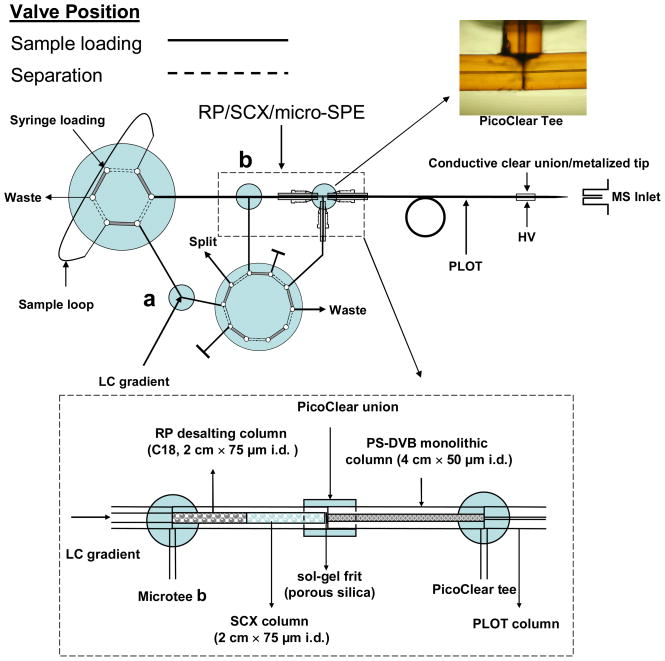
A diagram of the on-line 2D SCX/PLOT/MS system is shown in Figure 1. The triphasic trapping column (RP/SCX/micro-SPE) enabled fast sample loading as well as on-line desalting and washing prior to the two dimensional separation of a complex sample. A frit was first constructed within fused-silica tubing (75 μm i.d./360 μm o.d., 15 cm long) by a modified published procedure [25]. Briefly, the capillary was dipped into a 15% solution of formamide in potassium silicate (Kasil #1, PQ Corp., Valley Forge, PA) and placed in an oven at 100 °C for 3 min. The frit end of the capillary was cut such that 0.5 mm of the frit remained, and the capillary was then rinsed with acetonitrile. The biphasic RP/SCX column was slurry packed with 2 cm of 5 μm 300 Å Polysulfoethyl A strong cation-exchange resin (Nest Group Inc., Southboro, MA), followed by 2 cm of 5 μm Magic C18 packing material (200 Å pore size, Michrom BioResources, Auburn, CA). A 4 cm × 50 μm i.d. PS-DVB monolithic precolumn was carefully butt-to-butt connected to the biphasic RP/SCX column with a PicoClear union (New Objective) to assemble the triphasic RP/SCX/micro-SPE trapping column. A PicoClear tee (New Objective) was used to minimize the dead volume between the triphasic trapping column and the PLOT column (see Figure 1). PEEK microtees were employed to split the mobile phase and thus lower the flow rate. The downstream end of the PLOT column was butt-to-butt connected to a metallized spray tip (360 μm o.d., 20 μm i.d. fused silica with a 5 μm i.d. tip, 2–3 cm length, New Objective), to which the electrospray voltage (1.3 kV) was applied.
Figure 1.
Diagram of the advanced on-line 2D SCX/PLOT/MS system using a 3.2 m × 10 μm i.d. PLOT column and an on-line triphasic trapping column. For the latter column, a sol-gel frit was first made in 75 μm i.d. fused-silica tubing, followed by 2 cm of SCX resin (Polysulfoethyl A™) and then 2 cm of C18 RP packing. A 4 cm × 50 μm i.d. PS-DVB monolithic column was connected to the biphasic RP/SCX column to assemble the triphasic RP/SCX/RP trapping column. A PicoClear tee was used to minimize the dead volume between the triphasic trapping column and the PLOT column, while allowing fast sample loading. Microtees a and b were employed to split the mobile phase during SCX fractionation and PLOT separation, respectively. The sample was first injected onto the C18 column for desalting, then gradient eluted and trapped on the SCX packing, and finally stepwise eluted and trapped onto the monolithic micro-SPE column. The trapped sample was switched on-line to the PLOT column for separation. See text for the details.
Reversed-phase gradient elution was performed using an Ultimate 3000 Pumping System (Dionex, Sunnyvale, CA) with mobile phase A as 0.1% (v/v) formic acid in water and mobile phase B as 0.1% (v/v) formic acid, 90% (v/v) acetonitrile in water. The buffers for the salt gradient steps (5–250 mM) were prepared from a 1 M ammonium acetate stock solution in solvent A containing 5% acetonitrile. A sharp RP gradient, mobile phase B increased from 0% to 45% in 20 min, was used to elute the peptides from C18 desalting column onto the SCX column, while a longer RP gradient, mobile phase B increasing from 0% to 45% in 60 min, was used for the PLOT/MS analyses.
2.3 Mass spectrometry and data analysis
NanoESI-MS was performed on a linear ion trap mass spectrometer (LTQ, Thermo Fisher, San Jose, CA). Each spectral scan consisted of an m/z range of 400–2000, followed by MS/MS scans of the seven most abundant ions from the MS scan. A collision energy setting of 35% was applied for ion fragmentation, and dynamic exclusion was applied to discriminate against previously analyzed ions (data-dependent analysis). Data generated from the LC/MS experiments were analyzed using the Sequest Cluster search engine (ver. 3.0) and stored in the CPAS system [26]. The database search was conducted against a human protein database (Swiss-Prot release 52 with 15,498 protein sequences downloaded in March 2007) with combined normal and reversed sequences to facilitate the estimation of the peptide false positive rate, which was calculated using 2 times the number of significant unique peptide assignments in the reversed database divided by the total number of unique peptide identifications. [27]. Trypsin was specified as the digestion enzyme with one missed cleavage, and carboxyamidomethylation of cysteines was designated as a fixed modification. In order to minimize the level of false positive identifications, stringent criteria were used for selection of peptides identifications. Peptides were assigned combined scores based on a PeptideProphet [28] probability ≥0.90, and ΔCn ≥0.10, Xcorr ≥1.9, 2.2 and 3.75 for singly, doubly, and triply charged ions, respectively. By using the above stringent criteria, the peptides including the single-hit peptides were confidently identified. ProteinProphet was employed to assemble the minimal list of protein groups based on identified peptides. Protein annotation was obtained using DAVID 2007 suite [29] and Ingenuity Pathway Analysis ver. 5.0 (Ingenuity Systems, Redwood City, CA).
2.4 Sample preparation
An in-gel tryptic digest of a lysate of a SiHa cervical cancer cell line was used to evaluate the performance of the triphasic 2D SCX/PLOT/MS platform. In-gel digestion was performed using procedures previously described [30]. A total of 60,000 SiHa cells were lysed with 2% SDS in 50 mM NH4HCO3 via five bursts of 20-second sonication followed by 20-second ice cooling. Then, 20% of the protein extract, corresponding to 12,000 cells, was loaded on an SDS PAGE gel (4%–12% gradient). After electrophoresis, the gel was cut into 3 sections with molecular weight ranges less than 15 kDa (section 1), between 15 kDa and 40 kDa (section 2), and greater than 40 kDa (section 3), see Figure 2. Each section was further minced into small pieces (approximately 0.5 mm2) and subjected to 2 to 3 cycles of gel dehydration with acetonitrile, followed by rehydration with ammonium bicarbonate buffer (0.1 M, pH 8.0). After reduction with dithiothreitol (DTT) and alkylation with iodoacetamide (IAA), an 8 ng/μL trypsin solution (pH 8.0) was added to each section, and in-gel digestion was conducted overnight at 37°C. The supernatant was removed and saved. Gel pieces were further extracted with 5% formic acid (200 μL) in an extraction buffer (acetonitrile:50 mM NH4HCO3 = 2:1) at 37°C for 15 minutes. The formic acid solution, containing tryptic peptides, was combined with the previous supernatant and dried with a SpeedVac. The sample, prepared from each gel section, was reconstituted in 20 μL of solvent A, and 2 μL of the sample solution (equivalent to 1200 cells) was injected for on-line SCX/PLOT/MS analysis.
Figure 2.
A Coomassie blue-stained SDS-PAGE gel of the protein extract of 12,000 SiHa cells. The gel was cut into 3 sections with molecular weight ranges less than 15 kDa (section 1), between 15 kDa and 40 kDa (section 2), and greater than 40 kDa (section 3).
3 RESULTS AND DISCUSSION
Highly complex peptide mixtures with a wide concentration range that result from proteomic samples are typically resolved and characterized by a combination of high-performance RPLC coupled to an orthogonal separation method, e.g. strong cation exchange (SCX), and analyzed by tandem mass spectrometry. In our previous paper, we developed an on-line SCX/PLOT/MS system with a 10 μm i.d. PLOT column for ultratrace proteomic analysis [22]; however, this platform did not include sample desalting prior to the SCX/RPLC analysis. This step is important for effective binding of peptides to the SCX column, as salts, typically introduced during sample preparation, compete with the peptides for the cation exchange sites. Though desalting is commonly performed off-line with an SPE column, in this study, in order to minimize sample handling steps and thus potential sample loss, a triphasic (RP/SCX/micro-SPE) trapping column was developed to desalt the sample on-line, following the strategy of others[23]. As diagrammed in the inset of Figure 1, RP particles (C18) were packed in the capillary tube upstream of the SCX column. A PicoClear union was used to connect the biphasic 75 μm i.d. RP/SCX column to a 50 μm i.d. monolithic precolumn in order to minimize the dead volume at the joint. The monolithic micro-SPE column was positioned downstream of the RP/SCX section. Further downstream, the monolithic micro-SPE section of the RP/SCX/micro-SPE triphasic column was connected to the PLOT column by a PicoClear tee. Note that the fused silica capillary, connected to the PicoClear tee in the 90° arm, must be carefully positioned in order to permit mobile phase flow during the sample loading step.
To inject the sample onto the triphasic column, the 10-port valve was first set in the load position (solid line in Figure 1). In this position, microtee a and the PicoClear tee served as mobile phase splitters and microtee b as a connector. The entire 2 μL solution from the sample loop was loaded onto the RP/SCX/micro-SPE triphasic column at a flow rate of 0.6 μL/min. Peptides were trapped onto the RP desalting column, while salts and other components were directed to waste through the fused silica capillary attached to the PicoClear tee. After washing with 30 trapping column volumes of solvent A, the 10-port valve was switched to the separation position, and the triphasic trapping column was connected on-line to the PLOT separation column. The functions of the PicoClear tee and the other two microtees were thus reversed with microtee a and the PicoClear tee serving as connectors, while the microtee b acted as a splitter. The mobile phase flow was split immediately before the triphasic/PLOT connection in order to minimize the gradient delay time.
After washing the triphasic column, peptides adsorbed on the C18 desalting column were eluted onto the SCX column with a sharp reversed phase gradient. The 10-port valve was switched back to the loading position (dashed line in Figure 1), and a step series of 7 solutions of increasing ammonium acetate salt concentration were sequentially loaded into the loop of the 6-port injection valve, and pumped through the SCX column. After each step, the eluted peptides from the SCX column were trapped onto the PS-DVB monolithic micro-SPE column, with the salt and buffer being diverted to waste through the fused silica capillary attached to the PicoClear tee. The triphasic trapping column was then washed with 30 column volumes of solvent A, and subsequently the gradient LC separation on the PLOT column was performed. The design presented in Figure 1 shows a powerful platform for ultratrace proteomic analysis of highly complex peptide mixtures. The incorporation of the C18 packing upstream of the SCX column provided effective on-line sample desalting while minimizing sample loss.
An in-gel tryptic digest sample of gel section 3 (> 40 kDa) of the SiHa cervical cancer cell line was used to evaluate the triphasic on-line 2D SCX/PLOT/MS platform. Figure 3A presents the base peak chromatogram of the unretained SCX fraction, following sample elution from the C18 desalting column onto the SCX column. Only 39 peptides were identified in the flow through fraction, even though the peak intensity of the eluted analytes was high. Removal of these contaminants in the elution of peptides from desalting column improved the PLOT/MS analysis of the subsequent SCX fractions (due to decreased sample contamination and ion suppression).
Figure 3.
On-line 2D RP/SCX/PLOT/MS/MS analyses of an in-gel tryptic digest sample of a gel section (>40 kDa) of 1200 SiHa cells: (A) PLOT/MS analysis of the peptides that were unretained on the SCX column; (B–F) PLOT/MS analyses of eluted sample after each salt step for ammonium acetate concentrations of 5, 10, 15, 20, and 250 mM, respectively. 2 μL of ammonium acetate solution was delivered to the SCX column for each salt step elution. RP gradient: 0% mobile phase B (0.1% (v/v) formic acid, 10% (v/v) water in acetonitrile), 0 min; 45% B, 60 min; 80% B, 65 min; 100% A (0.1% (v/v) formic acid in water), 70 min. Data collection was initiated at the start of the gradient. Flow rate: ~20 nL/min with a column inlet pressure of ~2300 psi. The sample was first loaded onto the C18 desalting column, then gradient eluted onto the SCX column, and finally stepwise eluted and trapped onto the monolithic micro-SPE column. The trapped sample was switched on-line to the PLOT column for separation.
In our previous work [22], we used an on-line 2D SCX/PLOT/MS system without the upstream RP trap column for the analysis of a similar cell line sample. Although the sample was diluted 100-fold before injection, the salt present in the sample still led to a poorer SCX separation. Indeed, a large fraction of peptides (40%) were found in the unretained fraction. In comparison, using the triphasic RP/SCX/micro-SPE column with on-line desalting, less than 2% of the peptides were identified in the flow through fraction.
Figures 3B - F present base-peak PLOT column chromatograms for 5 ammonium acetate concentration steps in the analysis of the SiHa cell line. Importantly, since the same chemistry was used in the micro-SPE column and PLOT columns (PS-DVB), there was minimal change in peptide elution order on the two columns. It is worth noting that C18 packing was also tested for the micro-SPE prior to PLOT column, but significantly broader peaks were observed in the base-peak chromatograms when identical sample and experimental procedures were employed (data not shown). Peptides were eluted from the C18 micro-SPE column at a higher percentage of organic solvent and, therefore, were not well retained on the less hydrophobic PS-DVB PLOT column.
The resolving power of the 2D platform in Figure 1 can be seen from the fact that more than 85% of all peptides were identified in only one salt fraction. Using the criteria described in the Materials and Methods Section, 2724 peptides covering 862 unique proteins were identified from a single 2D SCX/PLOT/MS analysis of the in-gel tryptic digest of the >40 kDa gel section, from the injection of the equivalent to 1200 SiHa cells. The mean value of the number of peptides identified per protein was three, with 253 proteins (29% of the total) being identified from a single peptide.
In a subsequent triplicate SCX/PLOT/MS analysis of the gel section 3, the number of the confidently identified proteins increased. A total of 4497 peptides and 1209 unique proteins were identified in the combined analysis of the three replicates (false positive rate of 2.4%). A total of 1096 peptides (42%) and 510 proteins (63%) were found in all three analyses.
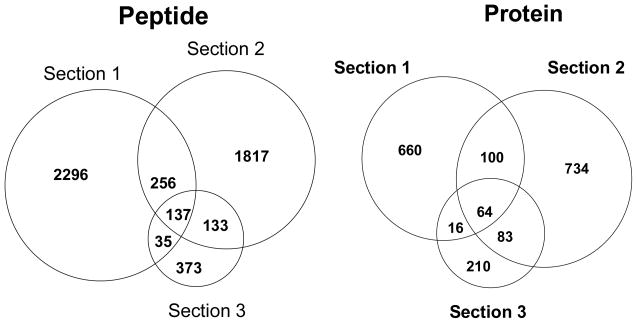
We next used the triphasic 2D SCX/PLOT/MS platform for comprehensive characterization of the SiHa cell proteome (all three gel sections in Figure 2), injecting 1200 cells. Figure 4 presents the distribution of peptides and proteins identified from each salt fraction by the SCX/PLOT/MS analysis of the three sections. In total, 5047 peptides covering 1857 unique proteins were identified by 2D SCX/PLOT/MS analysis from the injection of the equivalent to 1200 SiHa cells (false positive rate 1.8%). Recently, CIEF-nanoLC-ESI-MS/MS analysis of microdissected tumor tissue of glioblastoma has been reported and roughly 1800 unique proteins were identified from three CIEF-nanoLC-ESI-MS/MS analyses injection a total of 30 μg protein (10 μg protein/run), an amount corresponding to 60,000 selectively isolated cells [12]. Given the small size sample used in this study (~500 ng total protein for an amount equivalent to 1200 cells), these results are highly promising in demonstrating the high sensitivity and high resolving power of the platform. Finally, Figure 5 shows the distribution of the peptides and proteins identified from the 2D SCX/PLOT/MS analysis of the three sections of the SDS-PAGE gel. The low number of overlapped proteins between the sections indicated that the SDS-PAGE separation of proteins was an effective approach to fractionate the complex SiHa cell line sample on the protein level.
Figure 4.
Distribution, as a function of SCX salt fraction, of peptides and proteins identified from the 2D SCX/PLOT/MS analysis of the in-gel tryptic digest of the three SDS gel sections from 1200 SiHa cells injected. See Figures 2 and 3 for conditions.
Figure 5.
Venn diagram of the number of peptides and proteins identified by the 2D RP/SCX/PLOT/MS analysis of the in-gel tryptic digest of three SDS-gel sections from 1200 SiHa cells injected. See Figures 2 and 3 for conditions.
Annotation of the identified proteins was performed using DAVID 2007 Bioinformatics resources and IPA 5.0. The analysis showed that out of the 1857 identified proteins, 1634 were annotated as 958 cytosolic, 488 nuclear, 125 plasma membrane and 36 extracellular proteins. As expected, the identified proteins were significantly enriched for cytosolic proteins. Gene Ontology (GO) terms corresponding to these proteins were grouped and a subset of the results for terms corresponding to the cellular components is presented in Table 1. The table lists the number of proteins in a particular cellular component, e.g. mitochondrion (shown in italics), followed by subcategories, e.g. proteins found in mitochondrial envelope, matrix or associated with mitochondrial electron transport. The results in Table 1 demonstrate that the 2D SCX/PLOT/MS platform allows identification of all major cellular organelles including various membrane related proteins. Interestingly, a large number of proteins associated with the nucleus (247) were identified, encompassing several categories typically found in rapidly dividing cells, as expected for a cancer cell line. It should be further noted that since some proteins are associated with multiple annotation terms, the same protein could belong to several categories shown in Table 1.
Table 1.
Selected annotations of cellular component of identified proteins from a SiHa cervical cancer cell line analyzed by the 2D SCX/PLOT-MS platform.
| Protein group | Number of proteins |
|---|---|
| Mitochondrion | 174 |
| Envelope | 41 |
| Matrix | 27 |
| Electron transport | 11 |
| Cytoskeleton | 116 |
| Actin cytoskeleton | 54 |
| Microtubule | 26 |
| Filament | 6 |
| Nucleus | 247 |
| Nuclear pore | 23 |
| Nucleolus | 33 |
| Nuclear lumen | 72 |
| Chromosome | 46 |
| Nuclear envelope | 22 |
| Centrosome | 4 |
| Kinetochore | 5 |
| ER | 95 |
| Membrane | 19 |
| Network | 8 |
| Lumen | 8 |
| Golgi apparatus | 59 |
| Golgi vesicles | 20 |
| Ribosome | 75 |
| Large subunit | 24 |
| Small subunit | 10 |
| Proteosome | 28 |
| Core | 14 |
Note: The table summarizes the total number of proteins associated with a particular GeneOntology term. Numbers and terms in italic represents major cellular compartments followed by subcategories, see text for details.
Analysis using IPA revealed that multiple proteins involved in major metabolic pathways, such as oxidative phosphorylation (45 proteins out of 162, canonical), citrate cycle (22 out of 57), glycolysis (34 out of 140) and purine metabolism (71 out of 412), were identified. With respect to regulatory pathways, proteins involved in protein ubiquitination (62 out of 200) including 14 out of 16 proteasome PA700/20S subunits each with at least 2 unique peptides, NRF2-mediated oxidative stress (36 out of 142) including the family of MAP kinases (MAPK1, MAP2K2, MAP2K MAP2K3 and MAP2K6), actin cytoskeleton signaling (50 out of 254) including ras homolog gene family, member A (RHOA) with 3 unique peptides and dedicator of cytokinesis 1 (DOCK1), VEGF (19 out of 90) including HuR antigen with 7 unique peptides as well as integrin signaling (45 out 212) including integrin-linked kinase (ILK) (1 unique peptide) and PI3K/AKT signaling 29 out of 144, were identified, demonstrating the depth of proteomic coverage using PLOT columns. Disease annotation provided a strong association with hematological disease and with cancer, in particular, with hepatocellular cancer (p-value = 5 × 10−8). Since the IPA knowledgebase consists of manually curated information found in the literature, the association with hepatocellular rather than cervical cancer may be a result of the available literature. Proteins linked to cell transformation (p-value = 6 × 10−6) and cell invasions (p-value =3 × 10−4) were also detected. In summary, a great deal of information was available from the results of this study using PLOT columns because of the deep proteomic coverage.
4 Conclusions
An advanced 2D SCX/PLOT/MS platform using 10 μm i.d. PLOT columns has been presented for multidimensional separation-MS analysis of very small amounts of proteomic samples. For sample recovery and simplified operation, we implemented a triphasic RP/SCX/micro-SPE trapping column for on-line desalting and fractionation. Sample loading, salt step elution, and desalting were operated at a flow rate of 0.6 μL/min, which is 30 fold greater than the flow rate for the PLOT column. The RP/SCX/micro-SPE/PLOT column assembly demonstrated high separation efficiency at a mobile phase flow rate of 20 nL/min using a zero dead volume connection with a PicoClear union and tee. In addition, the triphasic trapping column served as a filter to minimize clogging of the PLOT column and ESI tip. To improve further the resolving power of the system, 1D SDS-PAGE fractionation of proteins was incorporated as the initial sample pretreatment step leading to identification of close to 2000 proteins using a protein amount equivalent to only 1200 SiHa cells (~500 ng). The platform has thus been demonstrated to provide effective analysis of very limited sample amounts. However, minimization of losses during sample preparation prior to sample injection require careful experimental control. While we have focused in this study on 2D SCX/PLOT/MS analysis at the peptide level, it should be noted that the high sensitivity and high resolving power of PLOT column is clearly also applicable to protein level separation. Work is continuing on the development of PLOT columns with different surface chemistry for other separation modes, e.g. hydrophilic interaction chromatography (HILIC) and application of PLOT columns to very limited amount of tissue sample, such as cells collected using laser capture microdissecttion (LCM).
Acknowledgments
The authors thank NIH GM 15847 for support of this work. We are grateful to Mr. Dongdong Wang for helpful discussions and New Objective for providing a PicoClear tee. Contribution number 908 from the Barnett Institute.
Abbreviations
- SCX
strong cation exchange
- PLOT
porous layer open tubular
- SPE
solid phase extraction
- SDS
sodium dodecyl sulfate
- PAGE
polyacrylamide gel electrophoresis
- RP
reversed phase
- ESI
eletrospray ionization
- PS-DVB
poly(styrene-divinylbenzene)
Footnotes
Dedicated to Professor Stellan Hjerten on his 80th birthday.
References
- 1.Wickware P, Smaglik P. Nature. 2001;413:869–875. doi: 10.1038/35057438. [DOI] [PubMed] [Google Scholar]
- 2.Pandey A, Mann M. Nature. 2000;405:837–846. doi: 10.1038/35015709. [DOI] [PubMed] [Google Scholar]
- 3.Aebersold R, Goodlett DR. Chemical Reviews. 2001;101:269–95. doi: 10.1021/cr990076h. [DOI] [PubMed] [Google Scholar]
- 4.Liu H, Lin D, Yates JR. Biotechniques. 2002;32:898–911. doi: 10.2144/02324pt01. [DOI] [PubMed] [Google Scholar]
- 5.Issaq HJ, Chan KC, Janini GM, Conrads TP, Veenstra TD. J Chromatogr B. 2005;817:35–47. doi: 10.1016/j.jchromb.2004.07.042. [DOI] [PubMed] [Google Scholar]
- 6.Link AJ, Eng J, Schieltz DM, Carmack E, Mize GJ, Morris DR, Garvik BM, Yates JR. Nature Biotechnol. 2001;19:676–682. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- 7.Stoll DR, Cohen JD, Carr PW. J Chromatogr A. 2006;1122:123–137. doi: 10.1016/j.chroma.2006.04.058. [DOI] [PubMed] [Google Scholar]
- 8.Peng J, Elias JE, Thoreen CC, Licklider LJ, Gygi SP. J Proteome Res. 2003;2:43–50. doi: 10.1021/pr025556v. [DOI] [PubMed] [Google Scholar]
- 9.Wagner K, Miliotis T, Marko-Varga G, Bischoff R, Unger KK. Anal Chem. 2002;74:809–820. doi: 10.1021/ac010627f. [DOI] [PubMed] [Google Scholar]
- 10.Washburn MP, Wolters D, Yates JR. Nature Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 11.Chen J, Balgley BM, DeVoe DL, Lee CS. Anal Chem. 2003;75:3145–3152. doi: 10.1021/ac034014+. [DOI] [PubMed] [Google Scholar]
- 12.Wang Y, Rudnick PA, Evans EL, Li J, DeVoe DL, Lee CS, Balgley BM. Anal Chem. 2005;77:6549–6556. doi: 10.1021/ac050491b. [DOI] [PubMed] [Google Scholar]
- 13.Bonner RF, Emmert-Buck M, Cole K, Pohida T, Chuaqui R, Goldstein S, Liotta LA. Science. 1997;278:1481–1483. doi: 10.1126/science.278.5342.1481. [DOI] [PubMed] [Google Scholar]
- 14.Wilm M, Mann M. Anal Chem. 1996;68:1–8. doi: 10.1021/ac9509519. [DOI] [PubMed] [Google Scholar]
- 15.Cech NB, Enke CG. Mass Spectrom Rev. 2001;20:362–387. doi: 10.1002/mas.10008. [DOI] [PubMed] [Google Scholar]
- 16.Smith RD, Shen Y, Tang K. Accounts Chem Res. 2004;37:269–278. doi: 10.1021/ar0301330. [DOI] [PubMed] [Google Scholar]
- 17.Shen Y, Moore RJ, Zhao R, Blonder J, et al. Anal Chem. 2003;75:3596–3605. doi: 10.1021/ac0300690. [DOI] [PubMed] [Google Scholar]
- 18.Ivanov AR, Zang L, Karger BL. Anal Chem. 2003;75:5306–5316. doi: 10.1021/ac030163g. [DOI] [PubMed] [Google Scholar]
- 19.Luo Q, Shen Y, Hixson KK, Zhao R, Yang F, Moore RJ, Mottaz HM, Smith RD. Anal Chem. 2005;77:5028–5035. doi: 10.1021/ac050454k. [DOI] [PubMed] [Google Scholar]
- 20.Luo Q, Tang K, Yang F, Elias A, Shen Y, Moore RJ, Zhao R, Hixson KK, Rossie SS, Smith RD. J Proteome Res. 2006;5:1091–1097. doi: 10.1021/pr050424y. [DOI] [PubMed] [Google Scholar]
- 21.Yue G, Luo Q, Zhang J, Wu S, Karger BL. Anal Chem. 2007;79:938–946. doi: 10.1021/ac061411m. [DOI] [PubMed] [Google Scholar]
- 22.Luo Q, Yue G, Valaskovic GA, Gu Y, Wu S, Karger BL. Anal Chem. 2007;79:6174–6181. doi: 10.1021/ac070583w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McDonald WH, Ohi R, Miyamoto DT, Mitchison TJ, Yates JR. Int J Mass Spectrom. 2002;219:245–251. [Google Scholar]
- 24.Premstaller A, Oberacher H, Huber CG. Anal Chem. 2000;72:4386–4393. doi: 10.1021/ac000283d. [DOI] [PubMed] [Google Scholar]
- 25.Cortes HJ, Pfeiffer CD, Richter BE, Steven TS. J High Resolut Chromatogr. 1987;10:446–448. [Google Scholar]
- 26.Rauch A, Bellew M, Eng J, Fitzgibbon M, Holzman T, Hussey P, Igra M, Maclean B, Lin CW, Detter A, Fang R, Faca V, Gafken P, Zhang H, Whitaker J, States D, Hanash S, Paulovich A, McIntosh MW. J Proteome Res. 2006;5:112–121. doi: 10.1021/pr0503533. [DOI] [PubMed] [Google Scholar]
- 27.Elias JE, Gygi SP. Nat Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 28.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Anal Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 29.Dennis G, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:R60. [PubMed] [Google Scholar]
- 30.Gu Y, Wu S, Meyer JL, Hancock WS, Burg LJ, Hanlon DW, Karger BL. J Proteome Res. ASAP. [DOI] [PubMed] [Google Scholar]