Abstract
Objectives
Commercially produced sterile green bottle fly Lucilia sericata maggots are successfully employed by practitioners worldwide to clean a multitude of chronic necrotic wounds and reduce wound bacterial burdens during maggot debridement therapy (MDT). Secretions from the maggots exhibit antimicrobial activity along with other activities beneficial for wound healing. With the rise of multidrug-resistant bacteria, new approaches to identifying the active compounds responsible for the antimicrobial activity within this treatment are imperative. Therefore, the aim of this study was to use a novel approach to investigate the output of secreted proteins from the maggots under conditions mimicking clinical treatments.
Methods
cDNA libraries constructed from microdissected salivary glands and whole maggots, respectively, were treated with transposon-assisted signal trapping (TAST), a technique selecting for the identification of secreted proteins. Several putative secreted components of insect immunity were identified, including a defensin named lucifensin, which was produced recombinantly as a Trx-fusion protein in Escherichia coli, purified using immobilized metal affinity chromatography and reverse-phase HPLC, and tested in vitro against Gram-positive and Gram-negative bacterial strains.
Results
Lucifensin was active against Staphylococcus carnosus, Streptococcus pyogenes and Streptococcus pneumoniae (MIC 2 mg/L), as well as Staphylococcus aureus (MIC 16 mg/L). The peptide did not show antimicrobial activity towards Gram-negative bacteria. The MIC of lucifensin for the methicillin-resistant S. aureus and glycopeptide-intermediate S. aureus isolates tested ranged from 8 to >128 mg/L.
Conclusions
The TAST results did not reveal any highly secreted compounds with putative antimicrobial activity, implying an alternative antimicrobial activity of MDT. Lucifensin showed antimicrobial activities comparable to other defensins and could have potential as a future drug candidate scaffold, for redesign for other applications besides the topical treatment of infected wounds.
Keywords: defensins, Lucilia sericata, MRSA, antimicrobial peptides, wounds
Introduction
Maggots of the blowfly Lucilia sericata have been successfully used as a debridement agent for chronic and infected wounds through history. First used by tribal people, maggot debridement therapy (MDT) was introduced into Western medicine following World War I by Baer,1 and extensively used to treat osteomyelitis and gas gangrenous wounds. Following the rediscovery of Alexander Fleming's penicillin from 1929 by Howard Florey and colleagues in 1939, the use of surgical maggots was abandoned.2 However, due to the appearance of antibiotic resistance and increasing problems with chronic wounds worldwide, the treatment has seen a renaissance in modern medicine through pioneering work by Church, Sherman3 and other biotherapy advocates. The FDA has approved MDT as a medical device, and maggots are produced aseptically and delivered by commercial companies to wound care centres and hospitals worldwide.
MDT has at least two confirmed beneficial effects when applied to wounds. These are debridement (removal of necrotic tissue)4 and the removal of pathogenic bacteria.5–7 The maggots feed through extracorporeal digestion, by excreting a complex cocktail of enzymes in their excretions/secretions (ES), mainly trypsin- and chymotrypsin-like proteases,8 and thereafter ingesting the resulting liquefied material. It was shown that ES possess antimicrobial activity in vitro,9–13 and also break down bacterial biofilms of Staphylococcus epidermidis,14 Staphylococcus aureus and, to a lesser extent, Pseudomonas aeruginosa.15 However, in vitro studies have questioned the antibacterial effect of ES on various bacterial species.16 Recently, we showed that the mere presence of P. aeruginosa cells is toxic to L. sericata maggots in an in vivo plate assay via quorum-sensing-regulated virulence factors.17
Many studies have aimed at determining the elusive antimicrobial compounds from maggot ES.9,10 Low molecular weight compounds and metabolites have been identified and tested in vitro.18 In a recent expressed sequence tag (EST) project, septic-inducible genes in L. sericata were investigated, and several putative peptides and proteins potentially involved in L. sericata immunity were identified.19 Among the septic injury-induced proteins identified were a sapecin-B homologue, three novel proline-rich antimicrobial peptides (AMPs), serine proteases and insect lysozymes. Insect immunity is not only activated as a result of physical insult, but also through oral ingestion of pathogens.20,21 Therefore, compounds involved in insect immunity may be active during MDT as well as the secreted digestive proteins within ES.
In the present study, we use an RNA-based discovery platform, transposon-assisted signal trapping (TAST),22 specifically developed to identify secreted proteins and peptides. We applied this to L. sericata maggots induced with external stimuli mimicking those encountered by the maggots during MDT, without excessive external insults and from actual MDT treatments, with emphasis in this study on proteins with putative antimicrobial activity.
Materials and methods
Lucilia sericata maggot inductions
L. sericata maggots were purchased from a commercial supplier (Zoobiotic, UK or Biomonde GmBh) and reared aseptically in three groups for 1 day (n = 700), 2 days (n = 700) or 3 days (n = 700) at 30°C to first, second and third instar levels, respectively, on blood agar medium [Statens Serum Institut (SSI) art. nr. 677]. In the fourth group, L. sericata maggots (n = 700) were collected at the Copenhagen Wound Healing Center at Bispebjerg Hospital following 3 days of successful treatment of a diabetic patient suffering from large ulcers on both legs. The maggots were collected through two treatments in succession. The patient's ulcers were sampled, and S. aureus and Staphylococcus warnerii were isolated, but no clinical signs of infection throughout the collection period were observed.
Seven hundred viable maggots from each of the 1-, 2- and 3-day-old groups and the MDT-induced group were divided into: (i) a full maggot (FM) pool (n = 4 × 100), where whole maggots were flash frozen in liquid nitrogen and stored at −80°C until further processing; and (ii) a salivary gland/crop (SC) pool (n = 4 × 600). The maggots in the SC pool had the cephalopharyngeal skeleton, salivary glands, crop and proximal part of the midgut microdissected out in RNA later™ (Qiagen, Hilden, Germany) under a stereomicroscope. SC material was stored in RNA later™ solution at −20°C until further processing.
RNA extraction
Fenozol reagent (Active Motif, Rixensart, Belgium) was used under RNase-free conditions to extract total RNA as described by the manufacturer from the FM pool and the SC pool, separately ground under liquid nitrogen with a pestle and mortar. The RNA concentration was measured spectrophotometrically (Ultrospec 3300, Amersham Biosciences) and a purity of 2.1, as estimated by the OD260/OD280 ratio, was accepted. The following steps were performed separately for both FM and SC material, unless otherwise stated.
cDNA library construction
Poly(A) mRNA was extracted from 500 µg of total RNA using the m-trap total kit (Active Motif, Rixensart, Belgium), according to the manufacturer's instructions. Next, double-stranded cDNA was synthesized from 0.4 µg of poly(A) mRNA using a SMART™ PCR cDNA synthesis kit (Clontech, Mountain View, Canada). First-strand synthesis was performed using the primer CDS-III/3: 5′-ATTCTAGAGGCCGAGGCGGCCGAC-ATG-d(T)30 N_1N-3′ in combination with Superscript RT II (Invitrogen, Carlsbad, USA) reverse transcriptase. Second-strand synthesis was performed using the primer SMART IV: 5′-AAGCAGTGGTATCAACGCAGAGTGGCCATTACGGC-CGGG-3′ together with Advantage 2™ polymerase (Clontech) using the LD protocol and 20 amplification cycles, as described by the manufacturer.
Double-stranded cDNA was purified using Illustra GFX™ DNA purification spin columns (GFX) (GE Healthcare, Buckinghamshire, UK) and subsequently cut using the restriction enzyme SfiI (New England Biolabs, Frankfurt, Germany) for 2 h at 50°C in the supplied NE buffer. The digested cDNA was size separated on a 0.8% agarose gel (Sea Plaque LMP) to enrich for transcripts of >400 bp. The reconcentrated cDNA fraction >400 bp was excised from the gel and GFX purified. This was subsequently ligated (16°C overnight) into SfiI cut and GFX purified signal trapping vector pMHas7i (kanamycinr) and as described in international patent application WO 01/7731522 using TN4 ligase (New England Biolabs, Frankfurt, Germany) in the supplied NE buffer. The ligation mixture was electroporated into Escherichia coli DH10B (Invitrogen) and then diluted in 1 mL of the supplied SOC medium at 37°C (Invitrogen). Subsequently, bacteria were incubated at 37°C for 1 h while shaking at 225 rpm and selected on Luria–Bertani (LB) agar plates supplemented with 50 mg/mL of kanamycin (37°C overnight). Lastly, isolation of plasmid pool DNA was done for the E. coli colonies (100 000 in total) using the JETSTAR 2.0 midi kit (GENOMED GmbH, Löhne, Germany), according to the manufacturer's instructions.
TAST
The plasmid used in cloning the cDNA library, pMHas7i, is described in international patent application WO 01/77315. Notable features of this plasmid are the SfiIA–SfiIB restriction sites proximal to the Shine–Dalgarno region of the lac promoter. This allows SfiI-adapted cDNAs to be cloned into the vector, and the resulting constructs to be actively transcribed and translated in the E. coli host. The signal-trapping transposon TnSig is also described in international patent application WO 01/77315. Briefly, the transposon is a derivative of the MuA mini transposon Entranceposon (Finnzymes, Oy). Directly adjacent to the left transposon border, an open reading frame (ORF) of the E. coli β-lactamase gene product, lacking a signal peptide, is inserted. Insertion of the transposon in a secreted gene can result in an in-frame fusion, secretion of the β-lactamase fusion peptide and, consequently, ampicillin-resistant E. coli colonies. The transposon also contains a chloramphenicol resistance gene, so the resulting DH10B E. coli cells containing a plasmid with a cDNA sequence with a transposon inserted in-frame with the signal peptide will be kanamycinr/ampicillinr/chloramphenicolr; designated ‘trappants’.
The transposon tagging reaction was carried out by mixing 80 ng of purified TnSig transposon,22 1 µg of plasmid cDNA library and 1 µL of Hyper MuA transposase (Epicentre Madison, USA) in 1× Hyper MuA buffer in a reaction volume of 20 µL. The reaction was incubated at 30°C for 3 h, and subsequently inactivated by the addition of 1 µL of Hyper MuA stop reagent and incubation at 75°C for 10 min. The reaction mixture was ethanol/sodium acetate precipitated, washed twice in 70% ethanol to remove excess salt and resuspended in 10 µL of TE buffer, pH 8. An aliquot of 2 µL of the resulting suspension was transformed into DH10B electrocompetent E. coli cells. The reaction mixture was diluted in 1 mL of SOC medium pre-heated to 37°C and incubated at 37°C at 225 rpm for 1 h. Three aliquots of 20 µL were plated onto LB kanamycin, LB kanamycin/chloramphenicol and LB kanamycin/chloramphenicol/ampicillin for controls, while the rest of the 1 mL was plated on triple antibiotic LB plates with the following antibiotic concentrations: 50 mg/L kanamycin; 10 mg/L chloramphenicol; and 15 mg/L ampicillin. Plates were incubated for 2 days at 30°C and then 1200 colonies were reisolated on new triple antibiotic LB plates (kanamycin/chloramphenicol/ampicillin) to ensure the isolation of surviving, actively growing ‘trappants’. After additional incubation (24–48 h, 30°C), 1000 ‘trappants’ from both the FM and SC library were inoculated into 96-well growth blocks with 1 mL of terrific broth (TB)23 containing kanamycin (50 mg/L) and chloramphenicol (10 mg/L), and incubated under humid conditions (37°C, 2 days). ‘Trappant’ culture broth was supplemented to a total concentration of 15% glycerol and stored at −20°C until shipment.
Sequencing
Glycerol stock of 1000 confirmed ‘trappants’ from each library was shipped to GATC biotech (Konstanz, Germany), and the plasmid inserts were sequenced with the transposon-anchored TnSig-specific primers seqA (5′-AGCGTTTGCGGCCGCGATCC-3′) and seqB (5′-TTATTCGGTCGAAAAGGATCC-3′).
Bioinformatics analysis
The removal of the vector sequence, trimming of the low quality sequence and assembly were done with phredPhrap package (http://www.phrap.org), as described by Becker et al.22 The resulting contigs were annotated using the PEDANT genome database,24,25 manually inspected using the VnTi software (Invitrogen), and searched against public and commercial databases. Sequences with putative antimicrobial activity are listed in Table 1.
Table 1.
TAST hits with putative antimicrobial activity or involvement in insect immunity
| Accession - BankIt1355267 | BlastP UniProt | Bit score | Amino acid identity | Best hit putative function | Sequence | Clones recovered |
|---|---|---|---|---|---|---|
| ZY200177_HM243535 | 1.9E-40 | 168 | 77/94 (82%) | defensin | full length | 1 FM |
| ZY200113_HM243536 | 6.9E-97 | 358 | 173/258 (68%) | cuticle protein | full length | 1 FM |
| ZY200117_HM243534 | 2,80E-73 | 279 | 149/300 (50%) | attacin | full length | 1 FM |
| ZY200244_HM243537 | 4,50E-19 | 97 | 46/84 (54%) | chitin binding protein | partial | 1 SC |
| ZY200249_HM243538 | 2,30E-74 | 280 | 129/141 (91%) | lysozyme 1 | full length | 1 SC |
| ZY200358_HM243539 | 3,40E-51 | 204 | 93/142 (65%) | lysozyme 1 | full length | 3 SC |
| ZY200466_HM243544 | 2,90E-32 | 140 | 62/70 (89%) | lysine-rich lysozyme 2 | partial | 1 FM |
| ZY200108_HM243545 | 2,30E-13 | 78 | 48/171 (28%) | lectin (alpha subunit) | partial | 1 FM |
| ZY200235_HM243540 | 5,20E-81 | 304 | 135/272 (50%) | lectin alpha subunit | full length | 1 SC |
| ZY200304_HM243546 | 2,70E-51 | 205 | 97/123 (79%) | ferritin heavy chain-like | partial | 1 SC 2 FM |
| ZY200341_HM243541 | 3,30E-92 | 342 | 166/205 (81%) | ferritin heavy chain-like | full length | 1 SC 3 FM |
| ZY200373_HM243542 | 7,90E-96 | 354 | 177/225 (79%) | ferritin light chain like | full length | 2 SC 2 FM |
| ZY200234_HM243543 | 2,10E-81 | 308 | 146/185 (78%) | peptidoglycan recognition protein | full length | 1 SC |
Cloning and recombinant expression of L. sericata AMP ZY200177 (lucifensin)
The ZY200177 coding sequence was amplified from cDNA using the Extensor high-fidelity PCR system (AB Gene, Surrey, UK) and ZY200177-specific oligonucleotide primers ZY200177-1: 5′-CCCCCCGGTACCGACGACGACGACAAGGCTACTTGCGATTTATTGAGTGGTAC-3′ and ZY200177-2: 5′-CCCCCCGAATTCTTATTAATTACGACACACGCAAATAGC-3′. The 165 bp PCR product was digested first with KpnI in NE buffer I with BSA added, GFX purified overnight and then EcoRI digested (New England Biolabs) in the supplied NE buffer, which cut in the overhangs introduced by the PCR primers.
The digested DNA fragment was ligated into the E. coli expression plasmid pET32a(+) (Novagen), in an expression approach similar to that used by Xu and colleagues for recombinant Musca domestica cecropin.26,27 The resulting plasmid (pANAS03) encoded a translational fusion peptide containing an N-terminal thioredoxin part followed by a his-tag, an enterokinase (EK) cleavage site and, lastly, the mature peptide sequence of the ZY200177, and was transformed into DH10B E. coli and the plasmid, with correctly inserted sequence verified by sequencing. GFX purified (pANAS03) from a verified clone was transformed into the E. coli strain BL21 (DE3) Singles competent cells [F–ompT hsdSB(rB− mB−) gal dcm lacY1] (Novagen), as prescribed by the manufacturer. Ten transformants were reisolated under selective and non-inducing conditions on LB agar with ampicillin (100 mg/L).
To test the expression of the 21.18 kDa Trx–lucifensin fusion protein, 10 transformants were grown in 100 mL of TB supplemented with 1% glucose and ampicillin (100 mg/L), at 37°C and 200 rpm in 300 mL shakeflasks to an OD600 of ∼5–10. Shakeflasks were subsequently transferred to 15°C for 1 h at 270 rpm. Then, 0.5 mL of each culture was sampled as an uninduced control, and the cultures were subsequently induced by adding IPTG to 0.5 mM and incubated at 15°C, 270 rpm overnight. Next, 0.5 mL of each culture was drawn as induced samples. Cells were pelleted by centrifugation at 10 000 g for 5 min and the supernatant removed. The cells were resuspended in 150 µL of Bugbuster (Novagen) and lysed for 25 min while shaking at 600 rpm at room temperature. The samples were centrifuged at 10 000 g for 5 min and the volume of the supernatant adjusted for differences in OD600 between sampling points and checked on an SDS gel. Samples were denatured at 95°C for 5 min in 1 × Novex Tris–glycine SDS sample buffer with NuPAGE sample reducing agent added. Pre-cast NuPAGE Novex Bis-Tris mini gels were run on the XCell Sure Lock Mini Cell System in 1 × NuPAGE MES SDS running buffer, with 500 µL of NuPAGE antioxidant added to the inner buffer chamber, at 200 V for 30 min. All NuPAGE articles are from Invitrogen (Carlsbad, USA). Gels were stained for 1 h in InstantBlue (Expedeon, Cambridge, UK) and destained in sterile milli-Q water overnight. A highly expressing clone was selected, and a single colony plated and grown in 4 L of TB as described for initial inductions; culture broth was pelleted by centrifugation and stored at −20°C until further processing.
Immobilized metal affinity chromatography (IMAC)
Pelleted cells were lysed by suspension in one-quarter of the cell culture volume in a buffer containing 50 mM Tris–HCl, pH 7.5, 0.04% lysozyme (Sigma), 0.01% benzonase (Sigma), 1% 3-(N,N-dimethylmyristylammonio)propanesulphonate (SB3-14) (Sigma) and 0.1% 3-(4-heptyl)phenyl-3-hydroxypropyl)dimethylammoniopropanesulphonate (C7BzO) (Sigma) for 45 min at room temperature. The resulting lysate was filtered through a 0.22 µm filter (Nalgene super Mach).
The Trx–lucifensin fusion protein was captured from the soluble fraction using IMAC on an Äkta Explorer fitted with a 25 mL TALON column (GE Healthcare) calibrated with 50 mM Tris–HCl, pH 7.5/1 M urea. The lysate was added to the column and subsequently washed with 50 mM Tris–HCl, pH 7.5/1 M urea and eluted in a buffer containing 50 mM Tris–HCl, pH 7.5, 500 mM imidazole and 1 M urea. Fractions containing the 21.2 kDa fusion protein were pooled in a Spectra/Por® dialysis tube (Spectrum Labs) and dialysed against 50 mM Tris–HCl, pH 7.0/5 mM CaCl2 at 4°C overnight.
EK digestion and reverse-phase chromatography (RP-HPLC)
To release ZY200177 (hereafter named lucifensin) from the Trx-fusion protein, EK (Novozymes) (1 : 75, v/v) was added and the suspension was incubated at 30°C overnight at 200 rpm. The digest was adjusted with formic acid and loaded onto a Gemini 10 µ C18 100 Å 250 × 10 mm column (GE Healthcare) calibrated with 1% formic acid. The bound protein was subsequently eluted with a linear gradient of ethanol (0%–80%, v/v). The fractions containing the 4.1 kDa lucifensin were identified by SDS gel and antimicrobial activity was verified in radial diffusion assays (RDAs) with Staphylococcus carnosus. The fractions containing lucifensin were pooled and lyophilized in a Genevac EZ-2 solvent evaporation system (Genevac) and resuspended in 0.1% acetic acid. The purity of lucifensin was estimated to >95% by the appearance of a single band on an SDS gel. The concentration of the suspended purified lucifensin was determined using RP-HPLC, where the peptide is quantified based on known peptide standards corrected for the extension coefficient of lucifensin (calculated molar absorbance lucifensin E280 0.398 mg/mL). The lucifensin peptide was purified to homogeneity using two consecutive rounds of RP-HPLC.
The monoisotopic molecular mass was determined by UPLC-MS using a Q-TOF Premier (Waters) connected to an Acquity UPLC (Waters). The mass spectrometer was operated in ESI+ mode with leucine enkephalin for lock mass. The separation was done on an Acquity BEH300 C4 column (2.1 × 150 mm; 1.7 µm; 300 Å) using a water-acetonitrile gradient (0.1% TFA).
RDAs and MIC determination
Initially, the antimicrobial activity of the fractions containing lucifensin was confirmed using an RDA described previously by Lehrer et al.,28 with several modifications. Briefly, 30 mL of melted 1/10 Mueller–Hinton broth (MHB)23/1% agarose was cooled to 42°C, supplemented to 5.0 × 105 cfu/mL with S. carnosus ATCC 51365 and was poured into a single-well omnitray (Nunc). The omnitray was overlayed with a TSP plate (Nunc) and left to solidify. After ∼1 h, the TSP plate was removed, leaving 96 1-mm wells in which 10 µL of the fractions of interest could be tested. Depending on the solvent involved, fractions were supplemented with 1 M Tris–HCl, pH 7.5 (1 : 1, v/v) prior to analysis in order to neutralize the antimicrobial activity of the solvent. For the Candida utilis RDA, a similar set-up was used with the modification that 1/3 potato dextrose (PD) agarose was used. S. carnosus and C. utilis were chosen as reporter strains due to their high susceptibility towards AMPs in the RDA assays.
The strain collection listed in Table 2 was tested for susceptibility to lucifensin by determining the strain MICs using a microbroth dilution assay.29 Colonies of the respective microorganism from a 5% blood agar plate (SSI) incubated overnight were suspended in MHB, pH 7.4 and diluted in MHB to a concentration of 5.0 × 105 cfu/mL. An aliquot of 90 µL of the bacterial suspension was incubated with 10 µL of lucifensin or vancomycin, gentamicin, ampicillin, ceftriaxone and linezolid as reference antibiotics in differing concentrations in polypropylene 96-well plates (Nunc) for 18–24 h at 37°C. The lucifensin peptide and reference antibiotics solutions were made fresh on the day of the assay and diluted 2-fold. The concentrations assayed ranged from 0.125 to 128 mg/L and the MIC was recorded as the lowest peptide concentration at which visual growth was inhibited.
Table 2.
MIC values of lucifensin
| Species | ATCC no. | Lucifensin MIC (mg/L) |
|---|---|---|
| Streptococcus pneumoniae | 49619 | 2 |
| Streptococcus pyogenes | 12344 | 2 |
| Staphylococcus carnosus | 51365 | 2 |
| Staphylococcus aureus | 29737 | 8 |
| Staphylococcus epidermidis | 12228 | 32 |
| Enterococcus faecalis | 29212 | 32 |
| Escherichia coli | 25922 | >128 |
| Pseudomonas aeruginosa | 9027 | >128 |
Results
Identification of putative antimicrobial proteins using TAST
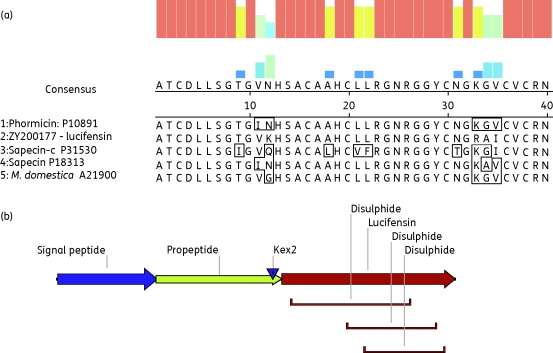
The TAST technique employs insertionally mutagenizing a cDNA library of interest with a transposon containing a β-lactamase gene without a signal peptide, which therefore encodes an inactive protein since it is not secreted. Subsequent clones containing a cDNA fragment with the transposon inserted in-frame to a signal peptide from a secreted protein will confer ampicillin resistance through secretion of the β-lactamase and allow clones containing cDNA from a secreted protein to be selected and the inserted cDNA sequenced.22 The bioinformatic analysis of the TAST results from the SC and FM L. sericata libraries revealed many novel L. sericata secreted proteins, mainly trypsin- and chymotrypsin-like proteases, lipases and odorant-binding proteins (results not shown). More interestingly, several sequences of putative proteins with predicted antimicrobial activity or with putative functions in insect immunity and, therefore, with a possible role in the antimicrobial effect of MDT were identified (Table 1). One partial lysozyme 2 and two full-length lysozyme 1 sequences were identified. Both lysozyme 1 homologues ZY200358 and ZY200249 were trapped exclusively from the SC library, indicating that they may be secreted during MDT. A partial (ZY200118) and a full-length (ZY200235) sequence of the L. sericata lectin α-subunit were identified. Lectins have been shown to be induced from the fat body in response to injury in Sarcophaga peregrina.30 One full-length sequence (ZY200117) represented a sarcotoxin IIA homologue belonging to the attacin family. These large proteins are similarly expressed upon injury31 and were only recovered from one FM clone. Among the TAST hits we also identified the full-length 276 bp ORF ZY200177 with high homology to known insect defensins with six conserved cysteine residues (Figure 1a). This comprised a novel L. sericata defensin, which we initially named sericasin. Based on recent successes with the development of defensins for medical applications32,33 and the overall potential of this molecule group, the encoded peptide was recombinantly expressed and purified for activity testing. During the revision of this manuscript a study by Cerovsky et al.34 was published in which they had successfully identified the same peptide from L. sericata ES, gut, salivary glands, fat body and haemolymph, which they named lucifensin; we adopted this name for the peptide.
Figure 1.
(a) ClustalW alignment of mature processed insect defensins from: 1, Phormia teranova P10891 (PDB:1ica); 2, TAST-identified Lucilia sericata ZY200177-lucifensin; 3, Sarcophaga peregrina P31530; 4, S. peregrina P18313 (PDB:1L4V); and 5, Musca domestica A21900. Boxes in the sequences indicate amino acid differences compared with lucifensin and the coloured bars indicate the level of overall homology. (b) Schematic representation of the full-length translation of the ZY200177 ORF containing the putative signal peptide (predicted by SignalP 3.0 Server), the deduced propeptide with a Kex2 cleavage site and the mature lucifensin defensin with disulphide bridges inferred by homology.
The putative 92 amino acid protein sequence comprised a signal peptide (amino acids 1–23) predicted by SignalP v. 3.0 (http://www.cbs.dtu.dk/services/SignalP/), a propeptide (amino acids 24–52) with a Kex2 cleavage site (amino acids 51–52) and a mature peptide named lucifensin (amino acids 53–92) (Figure 1b). Lucifensin was identified from the FM library and recovered from 1 clone out of 2000 sequenced containing the entire ORF. If the peptide was very active during MDT, we would have expected it to be recovered in high numbers from both the FM and SC libraries.
Expression and purification of Trx–lucifensin fusion
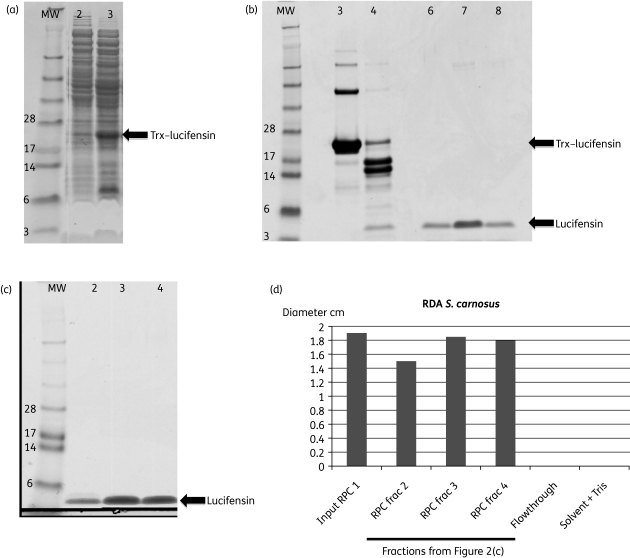
The plasmid pANAS03 containing the Trx–lucifensin fusion was transformed into E. coli BL21 cells. Upon IPTG induction, lucifensin was expressed as a fusion protein to thioredoxin (Figure 2). The soluble fraction of lysed cells harbouring the recombinant plasmid was analysed by SDS–PAGE (Figure 2a) for the expression of the fusion protein. A protein with an apparent molecular weight of 22 kDa was expressed upon induction with IPTG, which is close to the calculated molecular weight of 21.8 kDa for the fusion protein. Using IMAC, the fusion protein was captured (Figure 2b).
Figure 2.
(a) SDS–PAGE analysis of the soluble fraction of the Trx–lucifensin fusion protein in BL21 cells. Lane 2, uninduced; and lane 3, IPTG induced. (b) SDS–PAGE analysis. Lane 3, IMAC-captured Trx–lucifensin fusion protein; lane 4, enterokinase digest showing ∼4 kDa released recombinant lucifensin; and lanes 6–8, first round of RP-HPLC fractions containing mature lucifensin. (c) SDS–PAGE analysis of second RP-HPLC purification. Lanes 2–4, fractions containing mature lucifensin (>95% pure). (d) RDAs on S. carnosus of pooled fractions containing lucifensin from the first RP-HPLC (input RPC) and corresponding to (c), lanes 2–4. All samples were adjusted with Tris, pH 7.5, to abrogate the antimicrobial effect of the solvent formic acid/ethanol.
In order to generate mature lucifensin, the purified fusion protein was treated with EK at 30°C overnight at 200 rpm (Figure 2b). Subsequently, the lucifensin peptide was purified to homogeneity using two consecutive rounds of RP-HPLC in a formic acid/ethanol gradient. The second RP-HPLC run removed additional impurities (Figure 2c) and the antimicrobial activity of the fractions containing lucifensin assayed by S. carnosus RDA showed consistency with SDS–PAGE gel band intensities of the ∼4 kDa peptide (Figure 2d).
The concentration of recombinant lucifensin was determined via RP-HPLC and the monoisotopic molecular mass was confirmed by electrospray Q-TOF MS/MS to be 4113.8783 Da, in agreement with the calculated mass of 4113.89 Da (results not shown). Approximately 4 mg of purified recombinant lucifensin (>95% purity as judged by SDS gel, Figure 2c) was produced from IPTG-induced cells pertaining from 1 L of TB culture medium.
Antimicrobial activity of lucifensin
The antimicrobial activity of recombinant lucifensin was tested against both Gram-negative and Gram-positive bacteria (Table 2) using a CLSI (formerly NCCLS) MIC assay in which the reference antibiotics included were within the limits of acceptance, thereby complying with CLSI guidelines.35–37 Lucifensin was active against S. carnosus, Streptococcus pyogenes and Streptococcus pneumoniae with MIC values of 2 mg/L, and against Enterococcus faecalis and S. aureus with MIC values of 32 and 16 mg/L, respectively, but did not show any antimicrobial activity towards the Gram-negative bacteria tested at concentrations <128 mg/L. Subsequently, to evaluate the activity of the peptide against S. aureus, MIC assays were performed for a selection of 15 methicillin-resistant S. aureus (MRSA) and glycopeptide-intermediate S. aureus (GISA) isolates, including the predominant European and American clones. The MIC values for lucifensin against the MRSA and GISA isolates tested ranged from 8 to >128 mg/L (results not shown). The peptide did not show antifungal activity, as tested in a RDA with C. utilis (results not shown); however, activity against other fungi cannot be ruled out.
Discussion
The emergence of antibiotic resistance in a number of bacterial species and the alarmingly few new antibiotics being developed through to the clinic38 has turned research towards alternative modes of antimicrobial therapy and the development of novel antimicrobials.32 Maggots of L. sericata have been used to treat infected wounds for centuries and are used currently when other treatments fail. Therefore, it is apparent to try to identify the antibacterial mechanisms in maggots and purify components that could be used in modern medicine.
Previous studies into the antimicrobial activity involved in MDT have mainly been performed using chromatography-based approaches, applied to ES of uninjured maggots where substances with low molecular weight (<500 Da) were identified.9 A similar approach was performed using HPLC to investigate whole maggot extracts of immune-challenged maggots, and identified three known low molecular weight components: p-hydroxybenzoic acid; p-hydroxyphenylacetic acid; and octahydro-dipyrrolo[1,2-a;1′,2′-d] pyrazine-5,10-dione, also known as the cyclic dimer of proline or proline diketopiperazine or cyclo[Pro,Pro].18 A recent EST project focused selectively on septically inducible genes in L. sericata using the subtracted cDNA library approach identified several putative peptides and proteins potentially involved in L. sericata immunity.19 Among the septic injury induced proteins, a sapecin-B homologue, a diptericin homologue, novel proline-rich AMPs, serine proteases, transferrin, ferritin and insect lysozymes were identified, but none of the identified partial-length sequences were translated into recombinantly expressed antimicrobial compounds. However, in parallel to our study, the amino acid sequence of the mature lucifensin peptide was identified. The peptide was detected in trace amounts from L. sericata, ES, gut, salivary glands, fat body and haemolymph, and the sequence of the mature peptide was deduced by Edman degradation and shown to have antimicrobial activity against Micrococcus luteus.34 These results thus confirmed the sequence identity of the lucifensin peptide identified by the TAST technology.
In this study, we utilized a proprietary mRNA-based discovery platform (TAST) designed to selectively identify secreted proteins containing a signal peptide.22 The method has previously been used to successfully identify potent AMPs.32 The method has the clear advantage that the amount of maggots needed is limited compared with the amounts used when applying a strict HPLC approach to purify compounds of interest either from ES or whole maggot extracts. Compared with other EST approaches, TAST has the distinct benefit that it selects for secreted peptides and proteins. As the beneficial activities of the maggots in MDT are derived from compounds secreted into the wound environment either in secretions or excretions, this is ideal. We constructed cDNA libraries from whole maggots and maggot SCs induced under conditions mimicking the conditions and challenges that the maggots are faced with during MDT. Furthermore, different instar levels were represented, as they are present also during the time course of MDT. Sterility of the maggots in relation to the RNA isolation was not essential, as the ratio of L. sericata to bacterial RNA was such that any bacterial sequences trapped could easily be removed in the bioinformatics and annotation analysis.
One thousand clones containing a signal-trapped gene from each library were sequenced, and as only 12 clones from the FM library and 10 clones from the SC library contained sequences with homology to insect immunity-associated antimicrobial proteins, these sequences can with some degree of certainty be ruled out as antimicrobial effector molecules in MDT, as they are most likely not highly expressed during MDT. The number of times that a sequence was ‘trapped’ in individual clones is an estimate of the actual expression and should be confirmed in future studies of clinical samples. The sequences reported here can be used as scaffolds for real-time PCR studies of the actual expression in different L. sericata tissues. The list of putative secreted proteins identified during this study comprises a number of full-length sequences homologous to proteins involved in immunity in Diptera39 and other insect orders.
Insects use an innate immune system to fend off infection, and innate immunity relies on the detection and recognition of common microbial components, such as lipopolysaccharides or peptidoglycans, in order to mount a suitable systemic response.40 Lectins and peptidoglycan-recognition proteins (PGRPs) are important mediators in insect immunity. Their functions include the detection and neutralization of pathogenic and non-self material, and have been identified in many insect species.41–43 The C-type lectin CLEM-36 has been shown to be specifically expressed in the tip of the mouthparts in adult Sarcophaga peregrina.44 Flies are exposed to invasion and colonization of their mouthparts and crop during feeding, from potentially pathogenic bacteria. The presence of PGRPs is a perfect defence mechanism in order to avoid colonization in adult flies and maggots, especially for L. sericata maggots taking into account their natural habitat in decomposing cadavers or when used for MDT. We identified a novel putative full-length PGRP (ZY200234) and a putative full-length lectin α-subunit homologue (ZY200235). Both proteins were identified from the SC library indicating functions as reported in S. peregrina; additionally, a partial lectin sequence (ZY200108), not identical to ZY200235, was also identified in the FM library.
Furthermore, a full-length and a partial lysozyme 1 homologue were retrieved exclusively from the SC library, indicating that lysozyme could be present in the secretions of the maggots and play a part in the antimicrobial activity during MDT. A partial lysine-rich lysozyme 2 sequence was identified from the FMs, similar to an enzyme identified from whole immune-induced maggots.19 However, the sequences were not identical. The sequence variation of lysozyme secreted by L. sericata should be further investigated in relation to AMP activity in MDT.
A large AMP precursor belonging to the attacin family with homology to sarcotoxin IIA was identified in the FM library. Sarcotoxin IIA has been reported to inhibit cell wall synthesis and septum formation in E. coli,45 and attacins in general are active against Gram-negative bacteria. The rather large size of the attacin polypeptides has rendered activity studies difficult and, for this reason, limited information is available.31
The sequence ZY200177 showed high homology to known insect defensins, including sapecin.46 Furthermore, using fold recognition and homology modeling, the peptide was predicted to adapt to the conserved cysteine-stabilized α-helix and β-sheet (CSαβ) structural fold characteristic of peptides belonging to members of the defensin family. We, thus, decided to recombinantly produce the encoded lucifensin peptide. The activity of lucifensin was seen to be specific against Gram-positive microorganisms and, most notably, against S. carnosus, S. pneumoniae and S. pyogenes. MDT has been reported to be effective in eradicating MRSA,47 and as MRSAs are an expanding problem within the healthcare sector worldwide and specifically in the treatment of chronic wounds, the peptide was tested against several MRSA and GISA isolates. Notably, lucifensin did show antimicrobial effect against both MRSA and GISA isolates, comparable to that reported for other defensins.33
An interesting question is whether the lucifensin peptide holds the potential to be developed into a pharmaceutical drug candidate itself or whether its benefit will remain restricted to the direct application of maggots in wounds only. In general, the development of AMPs into therapeutics has been hampered by their potential for toxic side effects, suboptimal efficacy, salt sensitivity, liability to serum proteases, and, most notably, the lack of cost-effective and commercially viable production systems.48 However, recent attempts in the development of defensins for medical applications successfully addressed these issues.32,33 In combination with the potent activity of lucifensin against clinically relevant Streptococcus species at physiological salt levels, it appears that the peptide is a promising anti-infective candidate for further drug development.
Our TAST results did not contain any obvious protein candidate sequences with antimicrobial homology that were differentially recovered from the SC library, so the present study cannot be used to pinpoint lucifensin as the elusive antimicrobial molecule of MDT. This is in contrast to the parallel work by Cerovsky et al.,34 in which they were able to isolate lucifensin from salivary glands, fat body, ES and haemolymph, indicating that the peptide could be constitutively expressed as part of the systemic immune mechanism of L. sericata. In their study, all material was derived from mid-third instar 4-day-old maggots reared under non-sterile conditions, so the bacterial challenge leading to the output of lucifensin was most likely through oral challenge. These conditions are comparable to our inductions, though only 50% of the material used in our study was from 3-day-old third instar maggots. If the expression of lucifensin is late onset, perhaps in preparation for pupation, this may explain the discrepancy between the studies. The actual amount of the lucifensin that Cerovsky et al.34 were able to recover was not reported, only that they could identify its presence via antimicrobial activity against the reporter strain M. luteus. Extrapolating from these results into what the local concentration of lucifensin in the wound fluid and tissue could be during MDT is therefore difficult. In the EST project performed by Altincicek et al.,19 in which they looked at septic injury-inducible genes in L. sericata second instar maggots, a number of AMPs were identified; however, lucifensin was not among these. This would support the theory that a time-dependent expression of the peptide may occur during the maggot life cycle and, therefore, also during MDT. The Altincicek et al.19 study was, like ours, based on a transcriptomics approach in which the cDNA of the peptides was harboured on plasmids in E. coli. Although lucifensin does not show antimicrobial activity against E. coli, counterselection against AMP sequence-containing clones in E. coli during propagation of the cDNA library cannot be ruled out. In order to definitively evaluate whether lucifensin is indeed the elusive antimicrobial activity within ES, future studies should validate the expression of lucifensin via Q-PCR at different timepoints in the maggot life cycle. From the present study we cannot establish the nature of the elusive antimicrobial effect of MDT, but can, however, report a number of novel full-length components of L. sericata immunity among which we recombinantly expressed, purified and tested lucifensin. Recombinant lucifensin could have potential as a future drug candidate scaffold, for redesign for other applications besides the topical treatment of infected wounds, as has been established for other defensins.32
Funding
This work was supported by a clinical PhD scholarship from the University of Copenhagen, Faculty of Health Sciences (to A. S. A.) and by a grant from the Danish Research Council for Technology and Production (grant number 274-05-0435 to K. A. K).
Transparency declarations
D. S., K. M. S., T. K. and S. N. are employed by Novozymes A/S Denmark and have employee stocks in the company. A. S. A. was affiliated with Novozymes A/S during the study as an unpaid consultant. All other authors: none to declare.
Acknowledgements
We would like to acknowledge the competent nursing staff at the Copenhagen Wound Healing Center, Bispebjerg Hospital for logistical aid in obtaining maggots from actual MDT treatments. We thank Dorotea Raventos (Novozymes A/S) for invaluable assistance with an initial yeast expression strategy and RDAs, Steen Buskov (Novozymes A/S) for expert assistance with quantification and MS work, and Hans-Henrik Kristensen (Novozymes A/S) for insightful comments on the final manuscript. We thank Margaretha Hasselbalch and Mari-Ann Allerslev (Novozymes) for technical assistance on cDNA library construction and TAST. We also thank Ida Ellingsgaard, Jeanette Theil, Marianne Ravn Markvardsen, Müzdan Hamidovski and Jeanette Dam Larsen (all Novozymes A/S) for technical assistance with regard to the expression, purification and activity testing of lucifensin.
References
- 1.Baer WS. The treatment of chronic osteomyelitis with the maggot (larva of the blow fly) J Bone Joint Surg. 1931;13:438. doi: 10.1007/s11999-010-1416-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pechter EA, Sherman RA. Maggot therapy: the surgical metamorphosis. Plast Reconstr Surg. 1983;72:567–70. doi: 10.1097/00006534-198310000-00032. [DOI] [PubMed] [Google Scholar]
- 3.Sherman RA, Pechter EA. Maggot therapy: a review of the therapeutic applications of fly larvae in human medicine, especially for treating osteomyelitis. Med Vet Entomol. 1988;2:225–30. doi: 10.1111/j.1365-2915.1988.tb00188.x. doi:10.1111/j.1365-2915.1988.tb00188.x. [DOI] [PubMed] [Google Scholar]
- 4.Dumville JC, Worthy G, Bland JM, et al. Larval therapy for leg ulcers (VenUS II): randomised controlled trial. BMJ. 2009;338:b773. doi: 10.1136/bmj.b773. doi:10.1136/bmj.b773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jaklic D, Lapanje A, Zupancic K, et al. Selective antimicrobial activity of maggots against pathogenic bacteria. J Med Microbiol. 2008;57:617–25. doi: 10.1099/jmm.0.47515-0. doi:10.1099/jmm.0.47515-0. [DOI] [PubMed] [Google Scholar]
- 6.Steenvoorde P, Jukema GN. The antimicrobial activity of maggots: in-vivo results. J Tissue Viability. 2004;14:97–101. doi: 10.1016/s0965-206x(04)43005-8. [DOI] [PubMed] [Google Scholar]
- 7.Mumcuoglu KY, Miller J, Mumcuoglu M, et al. Destruction of bacteria in the digestive tract of the maggot of Lucilia sericata (Diptera: Calliphoridae) J Med Entomol. 2001;38:161–6. doi: 10.1603/0022-2585-38.2.161. [DOI] [PubMed] [Google Scholar]
- 8.Chambers L, Woodrow S, Brown AP, et al. Degradation of extracellular matrix components by defined proteinases from the greenbottle larva Lucilia sericata used for the clinical debridement of non-healing wounds. Br J Dermatol. 2003;148:14–23. doi: 10.1046/j.1365-2133.2003.04935.x. doi:10.1046/j.1365-2133.2003.04935.x. [DOI] [PubMed] [Google Scholar]
- 9.Bexfield A, Bond AE, Roberts EC, et al. The antibacterial activity against MRSA strains and other bacteria of a <500Da fraction from maggot excretions/secretions of Lucilia sericata (Diptera: Calliphoridae) Microbes Infect. 2008;10:325–33. doi: 10.1016/j.micinf.2007.12.011. doi:10.1016/j.micinf.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 10.Bexfield A, Nigam Y, Thomas S, et al. Detection and partial characterisation of two antibacterial factors from the excretions/secretions of the medicinal maggot Lucilia sericata and their activity against methicillin-resistant Staphylococcus aureus (MRSA) Microbes Infect. 2004;6:1297–304. doi: 10.1016/j.micinf.2004.08.011. doi:10.1016/j.micinf.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 11.Daeschlein G, Mumcuoglu KY, Assadian O, et al. In vitro antibacterial activity of Lucilia sericata maggot secretions. Skin Pharmacol Physiol. 2007;20:112–5. doi: 10.1159/000097983. doi:10.1159/000097983. [DOI] [PubMed] [Google Scholar]
- 12.Kerridge A, Lappin-Scott H, Stevens JR. Antibacterial properties of larval secretions of the blowfly Lucilia sericata. Med Vet Entomol. 2005;19:333–7. doi: 10.1111/j.1365-2915.2005.00577.x. doi:10.1111/j.1365-2915.2005.00577.x. [DOI] [PubMed] [Google Scholar]
- 13.Thomas S, Andrews AM, Hay NP, et al. The anti-microbial activity of maggot secretions: results of a preliminary study. J Tissue Viability. 1999;9:127–32. doi: 10.1016/s0965-206x(99)80032-1. [DOI] [PubMed] [Google Scholar]
- 14.Harris LG, Bexfield A, Nigam Y, et al. Disruption of Staphylococcus epidermidis biofilms by medicinal maggot Lucilia sericata excretions/secretions. Int J Artif Organs. 2009;32:555–64. doi: 10.1177/039139880903200904. [DOI] [PubMed] [Google Scholar]
- 15.van der Plas MJ, Jukema GN, Wai SW, et al. Maggot excretions/secretions are differentially effective against biofilms of Staphylococcus aureus and Pseudomonas aeruginosa. J Antimicrob Chemother. 2008;61:117–22. doi: 10.1093/jac/dkm407. doi:10.1093/jac/dkm407. [DOI] [PubMed] [Google Scholar]
- 16.Cazander G, van Veen KE, Bernards AT, et al. Do maggots have an influence on bacterial growth? A study on the susceptibility of strains of six different bacterial species to maggots of Lucilia sericata and their excretions/secretions. J Tissue Viability. 2009;18:80–7. doi: 10.1016/j.jtv.2009.02.005. doi:10.1016/j.jtv.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 17.Andersen AS, Joergensen B, Bjarnsholt T, et al. Quorum-sensing-regulated virulence factors in Pseudomonas aeruginosa are toxic to Lucilia sericata maggots. Microbiology. 2010;156:400–7. doi: 10.1099/mic.0.032730-0. doi:10.1099/mic.0.032730-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huberman L, Gollop N, Mumcuoglu KY, et al. Antibacterial substances of low molecular weight isolated from the blowfly, Lucilia sericata. Med Vet Entomol. 2007;21:127–31. doi: 10.1111/j.1365-2915.2007.00668.x. doi:10.1111/j.1365-2915.2007.00668.x. [DOI] [PubMed] [Google Scholar]
- 19.Altincicek B, Vilcinskas A. Septic injury-inducible genes in medicinal maggots of the green blow fly Lucilia sericata. Insect Mol Biol. 2009;18:119–25. doi: 10.1111/j.1365-2583.2008.00856.x. doi:10.1111/j.1365-2583.2008.00856.x. [DOI] [PubMed] [Google Scholar]
- 20.Vodovar N, Vinals M, Liehl P, et al. Drosophila host defense after oral infection by an entomopathogenic Pseudomonas species. Proc Natl Acad Sci USA. 2005;102:11414–9. doi: 10.1073/pnas.0502240102. doi:10.1073/pnas.0502240102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Freitak D, Wheat CW, Heckel DG, et al. Immune system responses and fitness costs associated with consumption of bacteria in larvae of Trichoplusia ni. BMC Biol. 2007;5:56. doi: 10.1186/1741-7007-5-56. doi:10.1186/1741-7007-5-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Becker F, Schnorr K, Wilting R, et al. Development of in vitro transposon assisted signal sequence trapping and its use in screening Bacillus halodurans C125 and Sulfolobus solfataricus P2 gene libraries. J Microbiol Methods. 2004;57:123–33. doi: 10.1016/j.mimet.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 24.Walter MC, Rattei T, Arnold R, et al. PEDANT covers all complete RefSeq genomes. Nucleic Acids Res. 2009;37:D408–11. doi: 10.1093/nar/gkn749. doi:10.1093/nar/gkn749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frishman D, Albermann K, Hani J, et al. Functional and structural genomics using PEDANT. Bioinformatics. 2001;17:44–57. doi: 10.1093/bioinformatics/17.1.44. doi:10.1093/bioinformatics/17.1.44. [DOI] [PubMed] [Google Scholar]
- 26.Xu X, Jin F, Yu X, et al. Expression and purification of a recombinant antibacterial peptide, cecropin, from Escherichia coli. Protein Expr Purif. 2007;53:293–301. doi: 10.1016/j.pep.2006.12.020. doi:10.1016/j.pep.2006.12.020. [DOI] [PubMed] [Google Scholar]
- 27.Jin F, Dong X, Xu X, et al. cDNA cloning and recombinant expression of the general odorant binding protein II from Spodoptera litura. Sci China C Life Sci. 2009;52:80–7. doi: 10.1007/s11427-009-0001-z. [DOI] [PubMed] [Google Scholar]
- 28.Lehrer RI, Rosenman M, Harwig SS, et al. Ultrasensitive assays for endogenous antimicrobial polypeptides. J Immunol Methods. 1991;137:167–73. doi: 10.1016/0022-1759(91)90021-7. doi:10.1016/0022-1759(91)90021-7. [DOI] [PubMed] [Google Scholar]
- 29.Clinical and Laboratory Standards Institute. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically—Seventh Edition: Approved Standard M7-A7. Wayne, PA, USA: CLSI; 2006. [Google Scholar]
- 30.Tanji T, Shiraishi H, Natori S, et al. Differential activation of the lectin and antimicrobial peptide genes in Sarcophaga peregrina (the flesh fly) Arch Insect Biochem Physiol. 2008;69:189–98. doi: 10.1002/arch.20280. doi:10.1002/arch.20280. [DOI] [PubMed] [Google Scholar]
- 31.Ando K, Natori S. Molecular cloning, sequencing, and characterization of cDNA for sarcotoxin IIA, an inducible antibacterial protein of Sarcophaga peregrina (flesh fly) Biochemistry. 1988;27:1715–21. doi: 10.1021/bi00405a050. doi:10.1021/bi00405a050. [DOI] [PubMed] [Google Scholar]
- 32.Mygind PH, Fischer RL, Schnorr KM, et al. Plectasin is a peptide antibiotic with therapeutic potential from a saprophytic fungus. Nature. 2005;437:975–80. doi: 10.1038/nature04051. doi:10.1038/nature04051. [DOI] [PubMed] [Google Scholar]
- 33.Gottlieb CT, Thomsen LE, Ingmer H, et al. Antimicrobial peptides effectively kill a broad spectrum of Listeria monocytogenes and Staphylococcus aureus strains independently of origin, sub-type, or virulence factor expression. BMC Microbiol. 2008;8:205. doi: 10.1186/1471-2180-8-205. doi:10.1186/1471-2180-8-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cerovsky V, Zdarek J, Fucik V, et al. Lucifensin, the long-sought antimicrobial factor of medicinal maggots of the blowfly Lucilia sericata. Cell Mol Life Sci. 2010;67:455–66. doi: 10.1007/s00018-009-0194-0. doi:10.1007/s00018-009-0194-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.National Committee for Clinical Laboratory Standards. Methods for Determining Bactericidal Activity of Antimicrobial Agents: Approved Standard M26-A. Wayne, PA, USA: NCCLS; 1999. [Google Scholar]
- 36.National Committee for Clinical Laboratory Standards. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically. Wayne, PA, USA: NCCLS; 2000. 5th Edn: Approved Standard M7-A5. [Google Scholar]
- 37.National Committee for Clinical Laboratory Standards. Performance Standards for Antimicrobial Susceptibility Testing: Twelfth Informational Supplement M100-S12. Wayne, PA, USA: NCCLS; 2002. [Google Scholar]
- 38.Gould IM. Antibiotic resistance: the perfect storm. Int J Antimicrob Agents. 2009;34(Suppl 3):S2–5. doi: 10.1016/S0924-8579(09)70549-7. [DOI] [PubMed] [Google Scholar]
- 39.Natori S, Shiraishi H, Hori S, et al. The roles of Sarcophaga defense molecules in immunity and metamorphosis. Dev Comp Immunol. 1999;23:317–28. doi: 10.1016/s0145-305x(99)00014-2. doi:10.1016/S0145-305X(99)00014-2. [DOI] [PubMed] [Google Scholar]
- 40.Imler JL, Bulet P. Antimicrobial peptides in Drosophila: structures, activities and gene regulation. Chem Immunol Allergy. 2005;86:1–21. doi: 10.1159/000086648. [DOI] [PubMed] [Google Scholar]
- 41.Hetru C, Troxler L, Hoffmann JA. Drosophila melanogaster antimicrobial defense. J Infect Dis. 2003;187(Suppl 2):S327–34. doi: 10.1086/374758. [DOI] [PubMed] [Google Scholar]
- 42.Jomori T, Natori S. Molecular cloning of cDNA for lipopolysaccharide-binding protein from the hemolymph of the American cockroach, Periplaneta americana. Similarity of the protein with animal lectins and its acute phase expression. J Biol Chem. 1991;266:13318–23. [PubMed] [Google Scholar]
- 43.Chen C, Billingsley PF. Detection and characterization of a mannan-binding lectin from the mosquito, Anopheles stephensi (Liston) Eur J Biochem. 1999;263:360–6. doi: 10.1046/j.1432-1327.1999.00513.x. doi:10.1046/j.1432-1327.1999.00513.x. [DOI] [PubMed] [Google Scholar]
- 44.Yamamoto-Kihara M, Kotani E. Isolation and characterization of a C-type lectin cDNA specifically expressed in the tip of mouthparts of the flesh fly Sarcophaga peregrina. Insect Mol Biol. 2004;13:133–40. doi: 10.1111/j.0962-1075.2004.00468.x. doi:10.1111/j.0962-1075.2004.00468.x. [DOI] [PubMed] [Google Scholar]
- 45.Ando K, Natori S. Inhibitory effect of sarcotoxin IIA, an antibacterial protein of Sarcophaga peregrina, on growth of Escherichia coli. J Biochem. 1988;103:735–9. doi: 10.1093/oxfordjournals.jbchem.a122337. [DOI] [PubMed] [Google Scholar]
- 46.Matsuyama K, Natori S. Mode of action of sapecin, a novel antibacterial protein of Sarcophaga peregrina (flesh fly) J Biochem. 1990;108:128–32. doi: 10.1093/oxfordjournals.jbchem.a123151. [DOI] [PubMed] [Google Scholar]
- 47.Bowling FL, Salgami EV, Boulton AJ. Larval therapy: a novel treatment in eliminating methicillin-resistant Staphylococcus aureus from diabetic foot ulcers. Diabetes Care. 2007;30:370–1. doi: 10.2337/dc06-2348. doi:10.2337/dc06-2348. [DOI] [PubMed] [Google Scholar]
- 48.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–95. doi: 10.1038/415389a. doi:10.1038/415389a. [DOI] [PubMed] [Google Scholar]