Abstract
Objectives
Decreased susceptibility to fluoroquinolones has become a major problem for the successful therapy of human infections caused by Salmonella enterica, especially the life-threatening typhoid and paratyphoid fevers.
Methods
By using Luminex xTAG beads, we developed a rapid, reliable and cost-effective multiplexed genotyping assay for simultaneously detecting 11 mutations in gyrA, gyrB and parE of S. enterica serovars Typhi and Paratyphi A that result in nalidixic acid resistance (NalR) and/or decreased susceptibility to fluoroquinolones.
Results
This assay yielded unambiguous single nucleotide polymorphism calls on extracted DNA from 292 isolates of Salmonella Typhi (NalR = 223 and NalS = 69) and 106 isolates of Salmonella Paratyphi A (NalR = 24 and NalS = 82). All of the 247 NalR Salmonella Typhi and Salmonella Paratyphi A isolates were found to harbour at least one of the target mutations, with GyrA Phe-83 as the most common one (143/223 for Salmonella Typhi and 18/24 for Salmonella Paratyphi A). We also identified three GyrB mutations in eight NalS Salmonella Typhi isolates (six for GyrB Phe-464, one for GyrB Leu-465 and one for GyrB Asp-466), and mutations GyrB Phe-464 and GyrB Asp-466 seem to be related to the decreased ciprofloxacin susceptibility phenotype in Salmonella Typhi. This assay can also be used directly on boiled single colonies.
Conclusions
The assay presented here would be useful for clinical and reference laboratories to rapidly screen quinolone-resistant isolates of Salmonella Typhi and Salmonella Paratyphi A, and decipher the underlying genetic changes for epidemiological purposes.
Keywords: genotyping, DNA gyrase, mechanisms of resistance, Salmonella
Introduction
Salmonella enterica serovars Typhi and Paratyphi A are invasive, life-threatening human pathogens, causing the systemic diseases typhoid and paratyphoid fever, respectively, which still pose significant threats to public health in many developing countries. In 2000, a population-based study estimated that the global disease burdens associated with typhoid and paratyphoid fever were 21.6 million cases (with 216 000 deaths) and 5.4 million cases, respectively.1 Resistance and decreased susceptibility to fluoroquinolones in Salmonella, often accompanied by resistance to nalidixic acid (NalR), will lead to treatment failures for typhoid and paratyphoid fever.2,3 The most common cause of resistance to nalidixic acid and decreased susceptibility to fluoroquinolones in serovar Typhi is amino acid substitutions in the quinolone resistance-determining region (QRDR) of the DNA gyrase subunit GyrA.4,5 Mutations in the QRDR of the other subunit of DNA gyrase (GyrB) and both subunits of DNA topoisomerases IV (ParC and ParE) will also result in increased resistance to quinolones.6,7
The rapid detection of NalR Salmonella Typhi and Salmonella Paratyphi A is vital in clinical practice to guide antibiotic therapy, and the traditional disc-diffusion protocol is still the most commonly used method for identifying resistant isolates. Microbiologists have attempted to develop molecular assays for rapidly screening NalR isolates and for determining underlying genetic changes in resistant isolates as well. These assays include denaturing HPLC (dHPLC),8 real-time PCR,9,10 restriction fragment length polymorphism,11 pyrosequencing,12 high-resolution melting curve assay13 and mismatch amplification mutation assay.14 However, none of these assays is capable of detecting multiple single nucleotide polymorphisms (SNPs) in different genes simultaneously with medium to high throughput.
We have previously described six different mutations in the QRDR of GyrA within Salmonella Typhi,5 and have also identified additional mutations in GyrB and ParE in unpublished work, some of which are associated with decreased susceptibility to fluoroquinolones without concomitant resistance to nalidixic acid (P. Roumagnac and M. Achtman). In order to efficiently test for the presence of these mutations individually and in combination, we designed a rapid and cost-effective multiplex genotyping assay based on Luminex xTAG beads, which can simultaneously screen for 11 mutations in gyrA, gyrB and parE of Salmonella Typhi and Salmonella Paratyphi A.
Materials and methods
Bacterial isolates
Four sets of isolates including a total of 292 Salmonella Typhi and 106 Salmonella Paratyphi A were tested in this study. Strains of Salmonella Typhi had been isolated between 1991 and 2006 in Asia, Europe and Africa, while Salmonella Paratyphi A strains were isolated between 1917 and 2006 across Asia, Europe, Africa and America. Twelve Salmonella Typhi isolates with known mutations in gyrA, gyrB and parE were used to develop the assay. These mutations are based on genomic sequences for four isolates [CT18, 150(98)S, E02-2759 and AG3]15,16 and on mutations discovered by dHPLC plus sequencing in eight others.5 The assay was validated with a second group of 193 Salmonella Typhi isolates, for which the corresponding regions in gyrA, gyrB and parE had been tested previously by dHPLC (unpublished for gyrB and parE, P. Roumagnac and M. Achtman).5 A third group of 87 isolates of Salmonella Typhi that had not been previously tested was also subjected to the Luminex assay, including 80 NalR and 7 NalS isolates. Lastly, 106 isolates of Salmonella Paratyphi A (24 NalR and 82 NalS) were also included in this study. The MICs for isolates of nalidixic acid (resistant breakpoint 32 mg/L) and ciprofloxacin (resistant breakpoint 4 mg/L) were determined according to the guidelines of the CLSI,17 and genomic DNAs were extracted using standard protocols.
Mutations and oligonucleotides
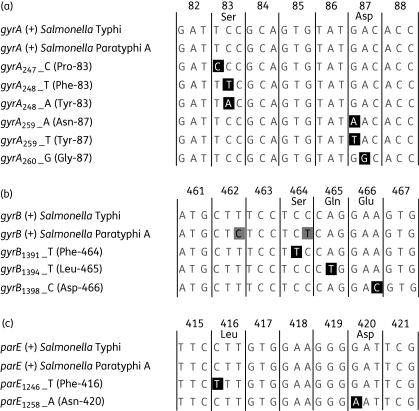
We tested 11 mutations in nine polymorphic sites, including two biallelic polymorphisms (Bips) in gyrA (gyrA247 and gyrA260), three Bips in gyrB (gyrB1391, gyrB1394 and gyrB1398) and two Bips in parE (parE1246 and parE1258), as well as two triallelic polymorphisms (Trips) in gyrA (gyrA248 and gyrA259) (Figure 1).5 Primers were designed with PrimerPlex V1.0 (PREMIER Biosoft International, USA) and are shown in Table 1, including 6 primers for amplifying the three gene fragments in one multiplex PCR and 20 allele-specific primer extension (ASPE) primers for detecting all 11 mutations. The 20 ASPE primers target each of the wild-type and mutant alleles by allele-specific nucleotides at the 3′ end, and contain unique TAGs (specific 24-mer oligonucleotide sequences as ‘tags’ for different ASPE primers) at the 5′ end that are reverse complements to the anti-TAGs on xTAG beads (Luminex Corp., USA). They also contain degenerate nucleotides at internal positions, which will ensure primer extension despite known polymorphisms neighbouring the target SNPs in all isolates from Salmonella Typhi as well as Salmonella Paratyphi A. All oligonucleotides were synthesized by Invitrogen Corp.
Figure 1.
Multiple alignments of partial gyrA, gyrB and parE fragments in Salmonella Typhi and Salmonella Paratyphi A. Wild-type alleles for each gene are indicated by a plus symbol (+). The mutations tested in this paper are shown in black squares and the wild-type amino acid is indicated on top of the alignments for codons containing target mutations. The amino acids for mutations are shown in brackets after the allele names. (a) Alignment of gyrA showing polymorphisms in codons 83 and 87, of which two are triallelic polymorphisms (Trips). (b) Alignment of gyrB with three mutations. Two silent mutations in Salmonella Paratyphi A are shown in grey squares. (c) Alignment of parE with two mutations.
Table 1.
Detailed information on mutations in gyrA, gyrB and parE, and oligonucleotides used in this study
| Gene | PCR primer pairs | Amplicon | ASPE primer sequencea | Target alleleb | Amino acid | xTAG beadsc |
|---|---|---|---|---|---|---|
| gyrA | GCCCTTCAATGCTGATGTCTTC | 805 bp | CAATAAACTATACTTCTTCACTAAATGGTGTCATACACTGCGDA | gyrA247_T(+) | GyrA Ser-83 | 13 |
| TCTCCTCTGTGTCGCCTCTG | ATACTTCATTCATTCATCAATTCAATGGTGTCATACACTGCGDG | gyrA247_C | GyrA Pro-83 | 15 | ||
| CAATTCATTTACCAATTTACCAATGATGGTGTCATACACTGCGG | gyrA248_C(+) | GyrA Ser-83 | 7 | |||
| TAATCTTCTATATCAACATCTTACGATGGTGTCATACACTGCGA | gyrA248_T | GyrA Phe-83 | 9 | |||
| TACAAATCATCAATCACTTTAATCGATGGTGTCATACACTGCGT | gyrA248_A | GyrA Tyr-83 | 11 | |||
| TACACTTTATCAAATCTTACAATCGCCATACGAACGATGGTGYC | gyrA259_G(+) | GyrA Asp-87 | 3 | |||
| TACATTACCAATAATCTTCAAATCGCCATACGAACGATGGTGYT | gyrA259_A | GyrA Asn-87 | 4 | |||
| CAATTCAAATCACAATAATCAATCGCCATACGAACGATGGTGYA | gyrA259_T | GyrA Tyr-87 | 5 | |||
| CTTTAATCTCAATCAATACAAATCCGCCATACGAACGATGGTGT | gyrA260_A(+) | GyrA Asp-87 | 1 | |||
| CTTTATCAATACATACTACAATCACGCCATACGAACGATGGTGC | gyrA260_G | GyrA Gly-87 | 2 | |||
| gyrB | ACTGGCGGATTGTCAGGAAC | 640 bp | AATCCTTTCTTTAATCTCAAATCAGCTTCGACAAGATGCTTTCCTC | gyrB1391_C(+) | GyrB Ser-464 | 21 |
| ATCGGCTTCGGTCAGAGTTG | CTTTAATCTACACTTTCTAACAATGCTTCGACAAGATGCTTTCCTT | gyrB1391_T | GyrB Phe-464 | 81 | ||
| TCAATTACCTTTTCAATACAATACCGACAAGATGCTTTCCTYYCA | gyrB1394_A(+) | GyrB Gln-465 | 24 | |||
| CTTTTCAATTACTTCAAATCTTCACGACAAGATGCTTTCCTYYCT | gyrB1394_T | GyrB Leu-465 | 25 | |||
| TTACTCAAAATCTACACTTTTTCAAAGATGCTTTCCTYYCWGGAA | gyrB1398_A(+) | GyrB Glu-466 | 26 | |||
| CTTTTCAAATCAATACTCAACTTTAAGATGCTTTCCTYYCWGGAC | gyrB1398_C | GyrB Asp-466 | 27 | |||
| parE | ATACGGTATAGCGGCGGTAG | 493 bp | TCAATCAATTACTTACTCAAATACGCCGAATCCCCTTCCACAAG | parE1246_C(+) | ParE Leu-416 | 19 |
| CGGAACAACTGGCAGAGATG | CTATCTATCTAACTATCTATATCAGCCGAATCCCCTTCCACAAA | parE1246_T | ParE Phe-416 | 78 | ||
| AATCAATCTTCATTCAAATCATCAGGAACCGCCTGCCGAATC | parE1258_G(+) | ParE Asp-420 | 16 | |||
| CTTTAATCCTTTATCACTTTATCAGGAACCGCCTGCCGAATT | parE1258_A | ParE Asn-420 | 17 |
aThe italic nucleotides for each ASPE primer are the TAG sequences provided by Luminex. The nucleotides in bold are degenerate sites (D, A or G or T; Y, C or T; and W, A or T).
bAlleles detected by individual ASPE primers. The subscript numbers indicate the nucleotide positions in gyrA, gyrB and parE, followed by nucleotides at these positions. ‘(+)’ indicates wild-type alleles. Because the primer design was carried out on the CT18 genome and the reading frame of gyrA is on the minus strand of the genome, nucleotides in gyrA alleles are complementary to the 3′-end nucleotides in corresponding ASPE primers. In total, 11 mutations at nine polymorphic sites are shown, including two triallelic polymorphisms (Trips) in gyrA (gyrA248 and gyrA259), two biallelic polymorphisms (Bips) in gyrA, three Bips in gyrB and two Bips in parE.
cDifferent xTAG beads used to call the alleles in the Luminex assay. The numbers are indexed to the coupled anti-TAG, which are reverse complimentary to the italic TAG sequences appended to the ASPE primers.
Multiplex PCR
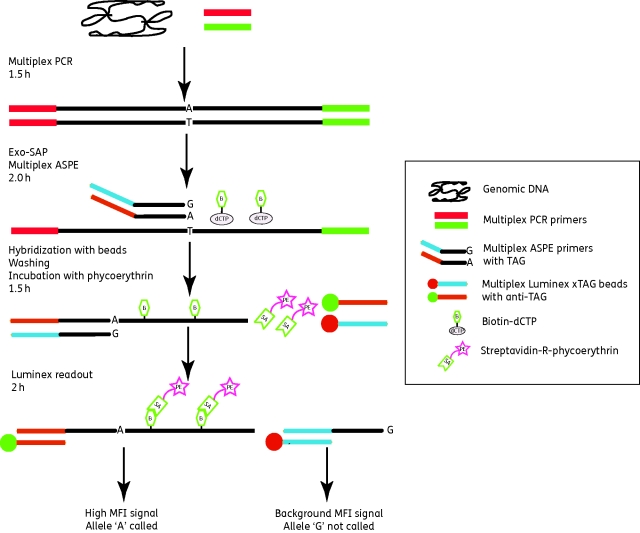
The overall scheme of the assay is shown in Figure 2. The following steps were carried out in 96-well microplates. Each 10 µL multiplex PCR consisted of 5 µL of 2× Multiplex PCR Master Mix (Qiagen, Germany), 1 µL of a mixture of 0.5 µM PCR primers for amplifying gyrA, gyrB and parE, and 4 µL of template DNA (5 ng/µL). One well per microplate contained 4 µL of distilled water (dH2O) instead of template DNA and served as a blank control. PCR cycling was performed in a four-block DNA Engine Tetrad 2 Thermocycler (Bio-Rad Inc., USA), with an initial denaturation at 95°C for 10 min, followed by 30 cycles at 94°C for 30 s, 58°C for 30 s and 72°C for 30 s, and a final extension step at 72°C for 3 min.
Figure 2.
Overview of the Luminex xTAG assay developed in this paper. More details of all steps can be found in the Materials and methods section. The estimated time for each step is shown on the left, based on testing two 96-well plates of samples. Only one fragment with one biallelic polymorphism site (Bip) is shown for illustrative purposes.
Exo-SAP treatment
To remove the unincorporated PCR primers and dNTPs, 1 µL of Exo-SAP containing 5 U of exonuclease I (Exo) and 0.5 U of shrimp alkaline phosphatase (SAP) (USB Corp., Germany) was added to each multiplex PCR product and mixed. The mixture was then incubated at 37°C for 30 min, followed by 10 min at 80°C to inactivate the enzymes.
Multiplex ASPE
The ASPE used here follows the protocol recommended by Luminex (http://www.luminexcorp.com/support/protocols/xtag_protocols.html). Briefly, each 10 µL multiplex ASPE mixture reaction contained 5 µL of Exo-SAP-treated multiplex PCR product, 0.3 U of Tsp DNA polymerase (Invitrogen), 25 nM ASPE primer mixture for all 11 SNPs (Table 1), 5 µM dATP/dTTP/dGTP and biotin-dCTP (Invitrogen), 20 mM Tris-HCl (pH 8.4), 50 mM KCl and 1.25 mM MgCl2. Thermocycling was performed at 95°C for 5 min, followed by 25 cycles of 94°C for 30 s, 55°C for 30 s and 72°C for 1 min, and a final extension at 72°C for 3 min.
Hybridization
A mixture of 20 distinct Luminex xTAG beads (shown in the last column of Table 1) containing 250 beads/µL for each bead type was prepared in 2× Tm hybridization buffer (0.4 M NaCl/0.2 M Tris/0.16% Triton X-100, pH 8.0). Then, 2 µL of the bead mixture was added per well containing 10 µL of ASPE products, followed by 23 µL of 2× Tm hybridization buffer and 15 µL of dH2O for a total volume of 50 µL. A bead control consisted of 2 µL of bead mixture, 23 µL of 2× Tm hybridization buffer and 25 µL of dH2O. The hybridization mixture was then incubated at 95°C for 1 min and 37°C for 30 min.
Washing and detecting
The hybridization mixture was centrifuged at 3220 g for 3 min and the supernatant was discarded. The xTAG bead pellets were then washed twice by adding 75 µL of 1× Tm hybridization buffer (0.2 M NaCl/0.1 M Tris/0.08% Triton X-100, pH 8.0), centrifuged at 3220 g for 3 min and the supernatant discarded. Then, 75 µL of 1× Tm hybridization buffer containing 2 mg/L streptavidin-R-phycoerythrin (Invitrogen, USA) was added to each well and the mixture was incubated at 37°C for 15 min. For detection, 50 µL of the final solution was injected into a Luminex 200 station (Luminex Corp., USA), at a sample plate temperature of 37°C. Fluorescence signals were measured for 100 beads for each of the 20 types of beads.
SNP calling
The Luminex station provides median fluorescence intensity (MFI) values for 100 beads of each bead type for all tested samples. The MFI values for samples were corrected by subtracting the values of the bead control, and the blank control was used to ensure that contamination and unspecific signals were not introduced during the multiplex PCR and ASPE process. SNPs were called with the software MasterPlex (MiraiBio, USA) only when the background-corrected MFI was >300 and the ratio of MFIcalled allele/(MFIwild allele + MFImutant allele) was > 0.9. For the two Trips in gyrA (Figure 1 and Table 2), the denominator consisted of the MFI for the wild-type plus both of the mutant alleles.
Table 2.
Raw data of background-corrected MFI values for the 12 Salmonella Typhi isolates used for setting up the assaya
|
Salmonella Typhi isolates |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Allele | Amino acid | CT18b | MP0150 | 150(98)Sb | MP0247 | MP0248 | MP0327 | 8(04)Nb | E02-2759b | MP0068 | ct 42 | MP0562 | AG3b | H2O |
| gyrA247_C | GyrA Pro-83 | 27.5 | 2331.5 | 17.0 | 26.0 | 22.0 | 0.5 | 16.0 | 19.0 | 22.0 | 25.5 | 17.0 | 11.0 | 15.0 |
| gyrA247_T(+) | GyrA Ser-83 | 2321.0 | 28.0 | 1639.0 | 1594.0 | 1497.0 | 1244.0 | 1850.0 | 1373.0 | 2349.0 | 2055.0 | 1864.5 | 1942.0 | 16.0 |
| gyrA248_T | GyrA Phe-83 | 25.0 | 10.0 | 2302.0 | 24.0 | 13.0 | 0.5 | 5.0 | 16.5 | 15.5 | 13.0 | 2784.0 | 2823.0 | 12.0 |
| gyrA248_A | GyrA Tyr-83 | 12.0 | 0.5 | 12.0 | 1610.0 | 12.5 | 0.5 | 0.5 | 1.0 | 13.5 | 22.5 | 17.0 | 19.0 | 15.0 |
| gyrA248_C(+) | GyrA Ser-83 | 1453.5 | 1129.5 | 7.5 | 5.0 | 1252.0 | 1100.5 | 1675.0 | 1972.0 | 1547.0 | 1845.0 | 31.5 | 27.0 | 16.5 |
| gyrA259_A | GyrA Asn-87 | 11.0 | 13.0 | 19.0 | 18.0 | 1969.0 | 1.0 | 12.0 | 13.0 | 19.0 | 18.5 | 14.0 | 15.0 | 18.0 |
| gyrA259_T | GyrA Tyr-87 | 16.0 | 25.0 | 23.0 | 22.5 | 17.0 | 1251.5 | 21.5 | 21.5 | 13.5 | 12.0 | 18.0 | 17.0 | 29.0 |
| gyrA259_G(+) | GyrA Asp-87 | 1621.5 | 1225.5 | 1226.5 | 1250.5 | 21.5 | 0.5 | 1657.5 | 1060.0 | 1673.5 | 2024.5 | 1554.5 | 1618.5 | 23.0 |
| gyrA260_G | GyrA Gly-87 | 25.0 | 24.0 | 19.0 | 22.0 | 19.0 | 9.0 | 3812.0 | 22.5 | 22.5 | 28.5 | 25.0 | 28.0 | 15.0 |
| gyrA260_A(+) | GyrA Asp-87 | 4591.5 | 3483.5 | 3562.5 | 3515.0 | 5238.0 | 1679.5 | 24.5 | 2769.5 | 4425.0 | 3078.5 | 4326.5 | 4733.5 | 14.5 |
| gyrB1391_T | GyrB Phe-464 | 15.0 | 18.0 | 19.0 | 26.0 | 26.0 | 9.0 | 12.0 | 1140.0 | 27.0 | 19.0 | 15.0 | 18.0 | 20.0 |
| gyrB1391_C(+) | GyrB Ser-464 | 1971.0 | 1718.0 | 1374.5 | 1478.0 | 1522.0 | 1592.0 | 1338.5 | 16.0 | 1974.0 | 1755.0 | 1427.0 | 1750.0 | 23.0 |
| gyrB1394_T | GyrB Leu-465 | 17.0 | 21.5 | 14.5 | 21.5 | 10.0 | 11.5 | 18.5 | 17.5 | 3005.5 | 24.5 | 18.5 | 11.5 | 16.0 |
| gyrB1394_A(+) | GyrB Gln-465 | 5961.0 | 5578.0 | 4700.0 | 5516.5 | 5372.0 | 2709.0 | 5198.0 | 1721.0 | 14.5 | 4967.0 | 5478.0 | 5935.0 | 15.0 |
| gyrB1398_C | GyrB Asp-466 | 27.5 | 18.5 | 22.0 | 2.5 | 21.5 | 10.0 | 10.5 | 0.5 | 16.5 | 3984.5 | 21.0 | 22.5 | 15.0 |
| gyrB1398_A(+) | GyrB Glu-466 | 4463.0 | 4221.0 | 3633.0 | 3550.0 | 3923.0 | 1634.0 | 3694.5 | 1486.5 | 1140.0 | 18.5 | 4045.0 | 4349.0 | 20.0 |
| parE1246_T | ParE Phe-416 | 20.0 | 22.5 | 16.0 | 20.0 | 22.0 | 14.5 | 17.0 | 16.0 | 20.0 | 21.0 | 2722.0 | 16.0 | 17.0 |
| parE1246_C(+) | ParE Leu-416 | 4119.5 | 3655.0 | 3842.0 | 3692.0 | 3833.0 | 2455.0 | 3866.0 | 3801.5 | 5039.0 | 4362.0 | 23.0 | 2382.0 | 25.0 |
| parE1258_A | ParE Asn-420 | 22.5 | 11.5 | 2.0 | 21.0 | 18.0 | 6.5 | 8.5 | 8.0 | 11.5 | 17.5 | 21.0 | 1918.5 | 24.0 |
| parE1258_G(+) | ParE Asp-420 | 1498.5 | 1388.0 | 1189.0 | 1178.5 | 1462.5 | 1762.0 | 1417.5 | 1158.0 | 1760.0 | 1636.5 | 1532.0 | 15.0 | 29.0 |
aThe MFI values were corrected by subtracting the value of the bead control; values of <0 were arbitrarily set to 0.5. For easier visualization, the values of called SNPs are shown in bold. The isolates MP0562 and AG3 contain dual mutations in gyrA and parE.
Reproducibility
DNA from isolate CT18 was used to evaluate the reproducibility of the assay in five independent repeats, following the steps described above.
Direct detection from bacterial colonies
Twelve random isolates of Salmonella Paratyphi A (4 NalS and 8 NalR) were streaked to single colonies on trypticase soy agar plates. For each isolate, a single colony was resuspended in 100 µL of H2O and incubated at 95°C for 15 min in order to kill the bacteria and release genomic DNA. After centrifugation, the supernatants were transferred to new tubes and used as templates for multiplex PCR.
Results
Development of the Luminex xTAG assay
The Luminex xTAG genotyping assay consists of four steps (Figure 2), as follows.
(i) A multiplex PCR to amplify three gene fragments of gyrA, gyrB and parE containing 11 target SNPs (Table 1), after which unincorporated dNTPs and PCR primers are removed by Exo-SAP treatment.
(ii) A multiplex primer extension step using 20 ASPE primers, during which biotin-dCTP is incorporated if there is a perfect match at the 3′ end. This step includes oligonucleotide primers specific for each of the wild-type and mutated alleles at each of the Bips and Trips (Figure 1), and which contain allele-specific unique TAG sequences. Tsp polymerase is so specific that only one of the alternative primers is extended for each nucleotide position and the other primers for that position serve as internal controls.
(iii) The ASPE products are hybridized to a mixture of 20 types of Luminex xTAG beads, each containing one anti-TAG sequence that is complementary to one of the TAG sequences appended to an ASPE primer. After washing the beads, the fluorescent dye streptavidin-R-phycoerythrin is added to bind to biotin-dCTP in the ASPE products.
(iv) The ASPE products/xTAG beads/streptavidin-R-phycoerythrin complex is quantified with a Luminex station, which calculates the MFI for 100 beads of each type. The entire procedure was conducted in two 96-well plates per test, with one plate for step (i) and the second one for the other three steps.
MFI values were corrected by subtracting the bead control and arbitrarily set to 0.5 if this correction resulted in negative values. Examples of such corrected data for the 12 reference isolates used to develop this assay are shown in Table 2. SNPs were called when corrected MFI was > 300 and the ratio of MFIcalled allele/(MFIwild allele + MFImutant allele) was > 0.9. These calls agreed completely with prior assignments based on dHPLC and sequencing.5 In Table 2, the MFI values of called alleles ranged from >1000 to 5961. In contrast, the MFI of uncalled alleles ranged from 0.5 to 31.5, only slightly greater than the values for the H2O control (12.0–29.0). These results demonstrate that the Luminex assay provides a tool to unambiguously call all the targeted SNPs.
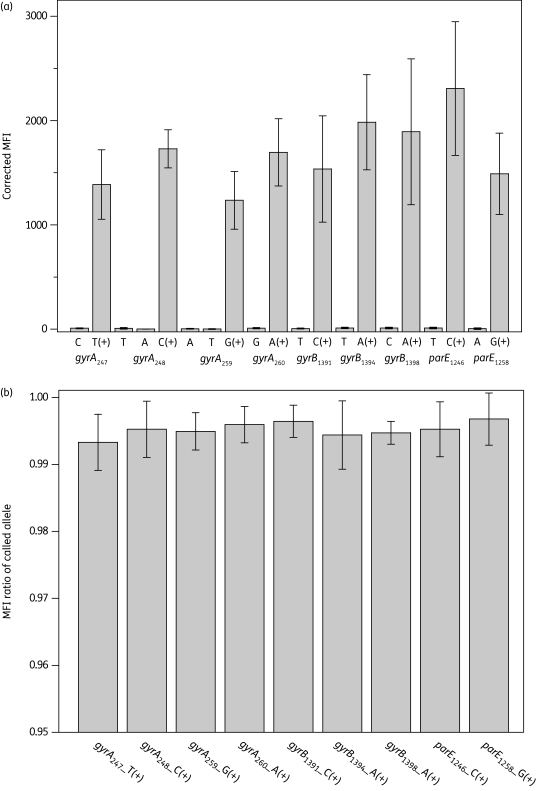
Reproducibility of the assay was tested by five independent trials with isolate CT18 (Figure 3). CT18 is wild-type for all the nine polymorphic positions in gyrA, gyrB and parE. MFI values for mutant alleles were almost negligible whereas corrected MFI values for wild-type alleles were strong. Positive MFI results showed considerable variation between individual tests, possibly reflecting variability between different runs in PCR and ASPE efficiencies. However, the ratios of MFIcalled allele/(MFIwild allele + MFImutant allele) were very uniform, with an average ratio of called MFI of >0.99 and a coefficient of variation of <0.5%. The MFI of called alleles was always >50 times higher than that of the uncalled ones. The use of MFI ratios instead of net MFI values minimizes the variation among different tests and yields unambiguous SNP calling results.
Figure 3.
Reproducibility of the Luminex assay in five independent tests with Salmonella Typhi CT18. (a) Raw data of average corrected MFI values (MFI minus the MFI of bead control). Error bars indicate the standard deviation. (b) Ratios of MFIcalled allele/(MFIwild allele + MFImutant allele). The average ratio for the called allele is shown, with error bars indicating the standard deviation.
The assays just described were performed with purified DNA as template. We also tested whether boiled single colonies from agar plates could be used as a template with eight NalR and four NalS isolates of Salmonella Paratyphi A. The results were congruent with results obtained with purified DNA from these isolates, namely all four NalS isolates contained only wild-type alleles, seven NalR isolates were called as GyrA Phe-83 and one NalR isolate was called as GyrA Tyr-83. This result indicates that this assay could be used by clinical microbiologists immediately after cultivation of Salmonella isolates. Starting with template DNA, testing 188 samples, two H2O controls and two bead controls in two 96-well microplates took a total of ∼7 h (Figure 2), with hands-on time of <1 h. Hence, the assay can be completed within 1 working day, producing 2068 datapoints (188 samples × 11 SNPs).
Validation with a larger panel of isolates
A panel of 205 isolates of Salmonella Typhi (including the 12 isolates used to develop the assay) was tested to validate the Luminex assay. The mutation profiles for gyrA, gyrB and parE of these isolates had previously been determined, either by whole genome sequencing or dHPLC (Table 3). 5,15,16
Table 3.
Overview of mutation profiles for gyrA, gyrB and parE of all isolates included in this study
| Validation panela |
Salmonella Typhi |
Salmonella Paratyphi A |
|||||
|---|---|---|---|---|---|---|---|
| Mutation profile | Amino acid(s) | NalR (n = 143) | NalS (n = 62) | NalR (n = 80) | NalS (n = 7) | NalR (n = 24) | NalS (n = 82) |
| Wild-type | wild-type | 54 | 7 | 82 | |||
| gyrA247_C | GyrA Pro-83 | 1 | |||||
| gyrA248_T | GyrA Phe-83 | 77 | 66 | 18 | |||
| gyrA248_A | GyrA Tyr-83 | 18 | 9 | 3 | |||
| gyrA259_A | GyrA Asn-87 | 4 | 2 | 2 | |||
| gyrA259_T | GyrA Tyr-87 | 1 | 1 | ||||
| gyrA260_G | GyrA Gly-87 | 10 | |||||
| gyrB1391_T | GyrB Phe-464 | 6 | |||||
| gyrB1394_T | GyrB Leu-465 | 1 | |||||
| gyrB1398_C | GyrB Asp-466 | 1 | |||||
| gyrA248_T, parE1246_T | GyrA Phe-83, ParE Phe-416 | 1 | |||||
| gyrA248_T, parE1258_A | GyrA Phe-83, ParE Asn-420 | 31 | 2 | ||||
| gyrA248_T, gyrA259_A | GyrA Phe-83, GyrA Asn-87 | 1 | |||||
The results with the 62 NalS isolates were fully consistent with previous dHPLC data: 54 were wild-type for all the targeted SNPs; six contained GyrB Phe-464; one was GyrB Leu-465; and one was GyrB Asp-466. All of the 143 NalR isolates harboured at least one of the target mutations. The mutation profiles of these isolates were: GyrA Phe-83 (77 isolates); GyrA Tyr-83 (18 isolates); GyrA Pro-83 (1 isolate); GyrA Asn-87 (4 isolates); GyrA Tyr-87 (1 isolate); GyrA Gly-87 (10 isolates); GyrA Phe-83 + ParE Asn-420 (31 isolates); and GyrA Phe-83 + ParE Phe-416 (1 isolate). These results were only 88.8% (127/143) concordant with the prior dHPLC results, because 11 isolates were GyrA Phe-83 according to dHPLC whereas they were GyrA Phe-83 + ParE Asn-420 based on the Luminex assay, and five isolates were GyrA Tyr-83 in dHPLC and GyrA Phe-83 in the Luminex assay. The 16 discrepancies were resolved by direct sequencing using the primers listed in Table 1. The direct sequencing results confirmed the Luminex SNP calls for all 16 isolates, showing that the Luminex assay made the correct calls, including multiple SNPs, for all Salmonella Typhi isolates (both NalS and NalR). We were surprised at the large number of prior false calls with dHPLC, which was thought to be highly reliable until now. Possibly, these false dHPLC calls reflect the fact that dHPLC was performed with multiplexed DNA templates of 5 or 10 isolates in order to increase throughput and it was not sufficiently sensitive to signals from mixtures of alleles.5
Mutation profiles for GyrA, GyrB and ParE of Salmonella Typhi and Salmonella Paratyphi A
We also tested additional isolates of serovars Typhi (87 isolates) and Paratyphi A (106 isolates) for which levels of susceptibility to nalidixic acid were known (Table 3), but which had not been previously evaluated in regard to their DNA gyrase and topoisomerase SNP profiles. All NalS isolates of serovars Typhi (7 isolates) and Paratyphi A (82 isolates) were wild-type for all alleles that were tested. In contrast, mutations in GyrA or GyrA + ParE were found in all NalR isolates of serovars Typhi (80 isolates) and Paratyphi A (24 isolates). One of the Typhi isolates even contained two mutations in GyrA (Table 3).
Overall, we have tested a total of 292 isolates of Salmonella Typhi (NalR = 223 and NalS = 69) and 106 isolates of Salmonella Paratyphi A (NalR = 24 and NalS = 82) with the Luminex assay. All 247 NalR Salmonella Typhi and Salmonella Paratyphi A isolates were found to harbour at least one of the target mutations, which demonstrated 100% sensitivity for identifying NalR mutations in this strain collection. The most common mutation in the NalR Salmonella Typhi isolates was GyrA Phe-83 (143/223 = 64.1%). Three other less common mutation profiles were GyrA Phe-83 + ParE Asn-420 (33/223 = 14.8%), GyrA Tyr-83 (27/223 = 12.1%) and GyrA Gly-87 (10/223 = 4.5%). These four mutation profiles account for 95.5% (213/223) of all NalR Salmonella Typhi isolates that were tested. The predominating mutation profile of Salmonella Paratyphi A NalR isolates was also GyrA Phe-83 (18/24 = 75.0%).
GyrB mutations
The Luminex assay also recognized mutations in GyrB within eight NalS Salmonella Typhi isolates (Table 4). Six of these isolates are mutated at GyrB Phe-464, and one each at GyrB Leu-465 and GyrB Asp-466. These isolates were designated NalS, because their MICs of nalidixic acid are below the breakpoint of 32 mg/L associated with resistance to nalidixic acid.17 However, as shown in Table 4, all of them, except isolate E98-4364, showed decreased susceptibility to ciprofloxacin, which is of clear significance in clinical practice. Different groups have already reported such decreased ciprofloxacin susceptibility (DCS) isolates in Salmonella Typhi, for which the ciprofloxacin MIC is 0.12–1 mg/L.18–20 Patients infected with DCS Salmonella Typhi isolates normally experience longer times to fever clearance and more frequent treatment failure than those without DCS.18 According to Table 4, mutations GyrB Phe-464 and GyrB Asp-466 seem to be responsible for the DCS phenotype in some isolates, but this needs to be further confirmed by functional assays. It had already been reported that isolates of Salmonella Typhimurium with GyrB Phe-464 + GyrA Phe-83 + GyrA Asn-87 mutations were fully resistant to ciprofloxacin (MIC 16–32 mg/L).21 We therefore considered it worthwhile to include these SNPs in the Luminex assay. Thus, this assay recognizes two major sources of decreased susceptibility to fluoroquinolones, only one of which is recognized by tests for resistance to nalidixic acid.
Table 4.
Characteristics of Salmonella Typhi isolates with GyrB mutations
| Isolate | Year | Country | NAL MIC (mg/L)a | CIP MIC (mg/L)b | GyrB mutation |
|---|---|---|---|---|---|
| AG-22 | 2004 | Vietnam | 6.0 | 0.250 | Phe-464 |
| AS-22541 | 2003 | Bangladesh | 8.0 | 0.125 | Phe-464 |
| DT-33 | 2002 | Vietnam | 8.0 | 0.500 | Phe-464 |
| DT-94 | 2002 | Vietnam | 16.0 | 0.125 | Phe-464 |
| dty1-121 | 1997 | Vietnam | 8.0 | 0.190 | Phe-464 |
| E02-2759 | 2002 | India | 4.0 | 0.125 | Phe-464 |
| ct 42 | 1994 | Vietnam | 16.0 | 0.160 | Asp-466 |
| E98-4364 | 1998 | Mexico | 2.0 | 0.030 | Leu-465 |
aMIC of nalidixic acid (NAL), which ranges from 0.75 to 4.0 mg/L for 25 isolates without mutations.
bMIC of ciprofloxacin (CIP), which ranges from 0.012 to 0.032 mg/L for 25 isolates without mutations.
Discussion
Luminex bead-based technology is a recently developed platform for the detection of multiplexed signals. This system incorporates 5.6 µm polystyrene microspheres that are internally labelled with two spectrally distinct fluorochromes. Mixing these two fluorochromes in various proportions yields 100 types of distinguishable, commercially available beads, on the basis of characteristic fluorescence signals. In the past, Luminex assays, also known as bead-based suspension arrays, have been used predominantly for quantitative measurements of multiplexed protein–ligand interactions or high-throughput nucleic acid detection through DNA hybridization.22,23 For example, a Luminex hybridization assay has been introduced to detect the six most common serogroups of S. enterica in the USA.24 Luminex assays are now being increasingly used for multiplexed genotyping on the basis of individual SNPs.25 For example, Ducey et al.26 and Ward et al.27 have presented Luminex-based assays for genotyping Listeria monocytogenes, which can simultaneously detect 51 or 64 SNPs on the basis of calibrated MFI values.
We have developed a somewhat different strategy for the SNP typing of bacterial DNA in this assay. Instead of attempting to calibrate a range of acceptable MFI values that indicate a positive reaction, we designed ASPE primers that are specific for both the mutated and the wild-type alleles at each targeted polymorphic position. For Bips this involves one mutant-specific and one wild-type-specific ASPE primer, while for Trips, such as in gyrA, it involves two mutant-specific and one wild-type-specific ASPE primer. The assay described here uses 20 ASPE primers and 20 types of beads to screen three gene fragments for 11 mutations, seven Bips and two Trips, and results in calls for nine alternative alleles in each assay (Figure 3). This approach has the disadvantage that two or three ASPE primers are needed for each polymorphic site, but this disadvantage is accompanied by very high reliability when measured as MFI ratios (Figure 3). This characteristic removes the necessity for including positive controls for analysing real samples, which improves the overall throughput. In other assays that have been described, such as dHPLC8,28 and high-resolution melting curve assays,13 SNPs are called by examining the curves (including shape and melt temperature/retention time), which can be subject to human errors. Positive controls for every genotype should also be included in every assay batch when using these two technologies, to ensure reliable comparisons and SNP callings.
Another underlying advantage of using ASPE specific for both the mutation and wild-type alleles is that this approach provides a limited ability for mutation discovery. Failure to call any SNP for all ASPE primers within one of the three multiplexed gene fragments implies polymorphisms within the PCR primer-binding region, which we have not yet observed. On the other hand, if only certain mutations are not called in one fragment, this indicates the existence of novel sequence diversity within the ASPE primer binding sites. In its original form, when the assay had been developed only for Salmonella Typhi, it failed to call SNPs in codons 465 and 466 in gyrB for Salmonella Paratyphi A isolates. This proved to reflect two silent mutations in the gyrB of Salmonella Paratyphi A isolates (Figure 1b), which significantly reduced the ASPE efficiency for neighbouring target mutations. After introducing appropriate degenerate nucleotides into the ASPE primers, the assay now also works well for Salmonella Paratyphi A. Similarly, the Luminex assay failed to call SNPs in codon 464 of gyrB in two Salmonella Typhi isolates, which subsequently turned out to harbour a novel mutation in this codon (F.-X. Weill, unpublished data).
Glass slide-based microarray technology can potentially offer similar or even much higher multiplexing capacity in terms of numbers of targeted mutations. For instance, a microarray has been described that simultaneously scans all potential mutations in codons 83 and 87 of gyrA in extraintestinal pathogenic Escherichia coli, as well as mutations inside the virulence-related gene fimH.29 However, the glass slide-based microarray can only test one sample per slide, with a maximum throughput of dozens of samples per day. The Luminex assay presented here offers higher throughput (11 SNPs for 188 isolates per day). Although the current Luminex protocol is not simple enough yet for routine use in hospitals with limited numbers of samples, it will be ideal for reference laboratories and central clinical laboratories with large sample collections. The total cost of this assay per sample (including oligos, reagents and xTAG beads) was ∼€2.40 (US$ 3.50), which means ∼€0.20 per SNP. For testing large numbers of samples, it might be possible to reduce costs dramatically by using an oligonucleotide ligation assay with very low numbers of beads.30
The Luminex assay identified one or two mutations in each NalR isolate that was tested (100% sensitivity), resulting in nine distinct mutation profiles (genotypes) of NalR Salmonella Typhi and four profiles of NalR Salmonella Paratyphi A (Table 3). All the single mutations of gyrA were located in codons 83 and 87, which are known to represent ‘hotspots’ of NalR mutations in Salmonella.4,6 The three dual-mutation profiles of Salmonella Typhi might reflect the accumulation of point mutations in GyrA and ParE under the selection of fluoroquinolone usage for therapy.31
Several papers reported mutations of ParC in ciprofloxacin-resistant Salmonella Typhi and Salmonella Paratyphi A isolates.32–34 We failed to identify any resistant strain with a ParC mutation in our current Salmonella Typhi and Salmonella Paratyphi A collections; therefore, we excluded ParC from the assay due to the lack of control isolates for assay validation. We also did not include probes for plasmid-mediated quinolone resistance [qnrA, qnrB, qnrC, qnrS, qepA and aac(6′)-Ib-cr]35,36 or quinolone resistance related to decreased antibiotic uptake (mar and acr genes),6 which were only reported in some serovars of S. enterica other than Typhi and Paratyphi A. However, the assay presented in this paper can potentially be extended to detect additional quinolone-resistance-related mutations in Salmonella. Luminex is currently bringing a hardware upgrade to market that can distinguish 500 analytes (capable of testing 250 Bips) and it would also be possible to combine multiple SNP typing with direct-hybridization tests in the same assays. Probably, the most limiting feature of the Luminex assay is the number of multiplexed PCR products that can simultaneously be amplified. One human genetic study reported successful 15-plex and 40-plex SNP genotyping,30 and we have succeeded in developing an 18-plex assay for genotyping Salmonella Typhi (Y. Song and M. Achtman, unpublished data). For extending this assay to detect SNPs in QRDRs of some other common serovars of Salmonella (Typhimurium, Enteritidis, Dublin etc.), multiplex PCR is not likely to be a problem, as it will only need to amplify four genes (gyrA, gyrB, parC and parE). Since there are more mutations in QRDRs of other Salmonella serovars than what we have tested here,6 careful alignments of the targeted regions must be performed before designing ASPE primers (Figure 1). Degenerate nucleotides will be necessary to cover the variations inside the upstream regions (∼8 bp) of certain targeted mutations; this has been proven as an effective strategy for gyrA and gyrB in this study (Table 1).
In summary, we describe a flexible, rapid, medium throughput and cost-effective multiplexed Luminex xTAG assay, which can simultaneously detect 11 mutations in gyrA, gyrB and parE of Salmonella Typhi and Salmonella Paratyphi A. This assay yields unambiguous SNP calls and possesses a limited mutation discovery potential. It can be used on extracted DNA or on single colonies of Salmonella Typhi and Salmonella Paratyphi A, which makes it an ideal platform for clinical and reference laboratories to rapidly screen quinolone-resistant Salmonella Typhi and Salmonella Paratyphi A isolates for guiding therapy, and detect the underlying genetic changes for molecular epidemiological analysis. The genotyping assay presented here can also be readily adapted to decipher evolutionary events and detailed population genetics in a variety of other genetically monomorphic bacterial pathogens, such as Salmonella Typhi, Yersinia pestis, Bacillus anthracis and Mycobacterium tuberculosis, in which SNPs can serve as reliable genetic markers.37
Funding
This work was supported by Science Foundation Ireland (05/FE1/B882 to M. A.) and The Wellcome Trust (to K. E. H. via her doctoral supervisor, Dr Gordon Dougan).
Transparency declarations
No conflicts of interest to declare.
Acknowledgements
We are grateful to Dr Keith Turner of The Wellcome Trust Sanger Institute, UK and Dr Sherry Dunbar of Luminex Corp., USA for their kind help in the early stages of developing the assay, and to Dr Gordon Dougan for his intellectual support. We would like to thank The Hospital for Tropical Diseases, Ho Chi Minh City, Dong Thap Provincial Hospital, Cao Lanh, Dong Thap province and An Giang Provincial Hospital, Long Xuyen, An Giang, Vietnam for their contribution to clinical typhoid fever research. We would also like to thank the International Vaccine Institute, based in Seoul, Korea and its DOMI programme for supplying some Salmonella Typhi strains.
References
- 1.Crump JA, Luby SP, Mintz ED. The global burden of typhoid fever. Bull World Hlth Org. 2004;82:346–53. [PMC free article] [PubMed] [Google Scholar]
- 2.Parry CM. The treatment of multidrug-resistant and nalidixic acid-resistant typhoid fever in Viet Nam. Trans R Soc Trop Med Hyg. 2004;98:413–22. doi: 10.1016/j.trstmh.2003.10.014. doi:10.1016/j.trstmh.2003.10.014. [DOI] [PubMed] [Google Scholar]
- 3.Bhan MK, Bahl R, Bhatnagar S. Typhoid and paratyphoid fever. Lancet. 2005;366:749–62. doi: 10.1016/S0140-6736(05)67181-4. doi:10.1016/S0140-6736(05)67181-4. [DOI] [PubMed] [Google Scholar]
- 4.Chau TT, Campbell JI, Galindo CM, et al. Antimicrobial drug resistance of Salmonella enterica serovar Typhi in Asia and molecular mechanism of reduced susceptibility to the fluoroquinolones. Antimicrob Agents Chemother. 2007;51:4315–23. doi: 10.1128/AAC.00294-07. doi:10.1128/AAC.00294-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roumagnac P, Weill F-X, Dolecek C, et al. Evolutionary history of Salmonella Typhi. Science. 2006;314:1301–4. doi: 10.1126/science.1134933. doi:10.1126/science.1134933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hopkins KL, Davies RH, Threlfall EJ. Mechanisms of quinolone resistance in Escherichia coli and Salmonella: recent developments. Int J Antimicrob Agents. 2005;25:358–73. doi: 10.1016/j.ijantimicag.2005.02.006. doi:10.1016/j.ijantimicag.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 7.Jacoby GA. Mechanisms of resistance to quinolones. Clin Infect Dis. 2005;41(Suppl 2):S120–6. doi: 10.1086/428052. [DOI] [PubMed] [Google Scholar]
- 8.Eaves DJ, Liebana E, Woodward MJ, et al. Detection of gyrA mutations in quinolone-resistant Salmonella enterica by denaturing high-performance liquid chromatography. J Clin Microbiol. 2002;40:4121–5. doi: 10.1128/JCM.40.11.4121-4125.2002. doi:10.1128/JCM.40.11.4121-4125.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Esaki H, Noda K, Otsuki N, et al. Rapid detection of quinolone-resistant Salmonella by real time SNP genotyping. J Microbiol Methods. 2004;58:131–4. doi: 10.1016/j.mimet.2004.03.010. doi:10.1016/j.mimet.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 10.Walker RA, Saunders N, Lawson AJ, et al. Use of a LightCycler gyrA mutation assay for rapid identification of mutations conferring decreased susceptibility to ciprofloxacin in multiresistant Salmonella enterica serotype Typhimurium DT104 isolates. J Clin Microbiol. 2001;39:1443–8. doi: 10.1128/JCM.39.4.1443-1448.2001. doi:10.1128/JCM.39.4.1443-1448.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang TM, Chang YF, Chang CF. Detection of mutations in the gyrA gene and class I integron from quinolone-resistant Salmonella enterica serovar Choleraesuis isolates in Taiwan. Vet Microbiol. 2004;100:247–54. doi: 10.1016/j.vetmic.2004.03.003. doi:10.1016/j.vetmic.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 12.Hopkins KL, Arnold C, Threlfall EJ. Rapid detection of gyrA and parC mutations in quinolone-resistant Salmonella enterica using pyrosequencing technology. J Microbiol Methods. 2007;68:163–71. doi: 10.1016/j.mimet.2006.07.006. doi:10.1016/j.mimet.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 13.Slinger R, Bellfoy D, Desjardins M, et al. High-resolution melting assay for the detection of gyrA mutations causing quinolone resistance in Salmonella enterica serovars Typhi and Paratyphi. Diagn Microbiol Infect Dis. 2007;57:455–8. doi: 10.1016/j.diagmicrobio.2006.09.011. doi:10.1016/j.diagmicrobio.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 14.Lin CC, Chen TH, Wang YC, et al. Analysis of ciprofloxacin-resistant Salmonella strains from swine, chicken, and their carcasses in Taiwan and detection of parC resistance mutations by a mismatch amplification mutation assay PCR. J Food Prot. 2009;72:14–20. doi: 10.4315/0362-028x-72.1.14. [DOI] [PubMed] [Google Scholar]
- 15.Holt KE, Parkhill J, Mazzoni CJ, et al. High-throughput sequencing provides insights into genome variation and evolution in Salmonella Typhi. Nature Genet. 2008;40:987–93. doi: 10.1038/ng.195. doi:10.1038/ng.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parkhill J, Dougan G, James KD, et al. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature. 2001;413:848–52. doi: 10.1038/35101607. doi:10.1038/35101607. [DOI] [PubMed] [Google Scholar]
- 17.Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: Eighteenth Informational Supplement M100-S18. Wayne, PA, USA: CLSI; 2008. [Google Scholar]
- 18.Crump JA, Kretsinger K, Gay K, et al. Clinical response and outcome of infection with Salmonella enterica serotype Typhi with decreased susceptibility to fluoroquinolones: a United States foodnet multicenter retrospective cohort study. Antimicrob Agents Chemother. 2008;52:1278–84. doi: 10.1128/AAC.01509-07. doi:10.1128/AAC.01509-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cooke FJ, Wain J, Threlfall EJ. Fluoroquinolone resistance in Salmonella Typhi. BMJ. 2006;333:353–4. doi: 10.1136/bmj.333.7563.353-b. doi:10.1136/bmj.333.7563.353-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cooke FJ, Day M, Wain J, et al. Cases of typhoid fever imported into England, Scotland and Wales (2000–2003) Trans R Soc Trop Med Hyg. 2007;101:398–404. doi: 10.1016/j.trstmh.2006.07.005. doi:10.1016/j.trstmh.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 21.Casin I, Breuil J, Darchis JP, et al. Fluoroquinolone resistance linked to GyrA, GyrB, and ParC mutations in Salmonella enterica Typhimurium isolates in humans. Emerg Infect Dis. 2003;9:1455–7. doi: 10.3201/eid0911.030317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eriksson R, Jobs M, Ekstrand C, et al. Multiplex and quantifiable detection of nucleic acid from pathogenic fungi using padlock probes, generic real time PCR and specific suspension array readout. J Microbiol Methods. 2009;78:195–202. doi: 10.1016/j.mimet.2009.05.016. doi:10.1016/j.mimet.2009.05.016. [DOI] [PubMed] [Google Scholar]
- 23.Nazarenko I, Kobayashi L, Giles J, et al. A novel method of HPV genotyping using Hybrid Capture® sample preparation method combined with GP5+/6+ PCR and multiplex detection on Luminex® XMAP®. J Virol Methods. 2008;154:76–81. doi: 10.1016/j.jviromet.2008.09.002. doi:10.1016/j.jviromet.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 24.Fitzgerald C, Collins M, van DS, et al. Multiplex, bead-based suspension array for molecular determination of common Salmonella serogroups. J Clin Microbiol. 2007;45:3323–34. doi: 10.1128/JCM.00025-07. doi:10.1128/JCM.00025-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dunbar SA. Applications of Luminex xMAP technology for rapid, high-throughput multiplexed nucleic acid detection. Clin Chim Acta. 2006;363:71–82. doi: 10.1016/j.cccn.2005.06.023. doi:10.1016/j.cccn.2005.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ducey TF, Page B, Usgaard T, et al. A single-nucleotide-polymorphism-based multilocus genotyping assay for subtyping lineage I isolates of Listeria monocytogenes. Appl Environ Microbiol. 2007;73:133–47. doi: 10.1128/AEM.01453-06. doi:10.1128/AEM.01453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ward TJ, Ducey TF, Usgaard T, et al. Multilocus genotyping assays for single nucleotide polymorphism-based subtyping of Listeria monocytogenes. Appl Environ Microbiol. 2008;74:7629–42. doi: 10.1128/AEM.01127-08. doi:10.1128/AEM.01127-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Randall LP, Coldham NG, Woodward MJ. Detection of mutations in Salmonella enterica gyrA, gyrB, parC and parE genes by denaturing high performance liquid chromatography (DHPLC) using standard HPLC instrumentation. J Antimicrob Chemother. 2005;56:619–23. doi: 10.1093/jac/dki293. doi:10.1093/jac/dki293. [DOI] [PubMed] [Google Scholar]
- 29.Barl T, Dobrindt U, Yu X, et al. Genotyping DNA chip for the simultaneous assessment of antibiotic resistance and pathogenic potential of extraintestinal pathogenic Escherichia coli. Int J Antimicrob Agents. 2008;32:272–7. doi: 10.1016/j.ijantimicag.2008.04.020. doi:10.1016/j.ijantimicag.2008.04.020. [DOI] [PubMed] [Google Scholar]
- 30.Bruse S, Moreau M, Azaro M, et al. Improvements to bead-based oligonucleotide ligation SNP genotyping assays. BioTechniques. 2008;45:559–71. doi: 10.2144/000112960. doi:10.2144/000112960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Turner AK, Nair S, Wain J. The acquisition of full fluoroquinolone resistance in Salmonella Typhi by accumulation of point mutations in the topoisomerase targets. J Antimicrob Chemother. 2006;58:733–40. doi: 10.1093/jac/dkl333. doi:10.1093/jac/dkl333. [DOI] [PubMed] [Google Scholar]
- 32.Adachi T, Sagara H, Hirose K, et al. Fluoroquinolone-resistant Salmonella Paratyphi A. Emerg Infect Dis. 2005;11:172–4. doi: 10.3201/eid1101.040145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gaind R, Paglietti B, Murgia M, et al. Molecular characterization of ciprofloxacin-resistant Salmonella enterica serovar Typhi and Paratyphi A causing enteric fever in India. J Antimicrob Chemother. 2006;58:1139–44. doi: 10.1093/jac/dkl391. doi:10.1093/jac/dkl391. [DOI] [PubMed] [Google Scholar]
- 34.Dimitrov T, Dashti AA, Albaksami O, et al. Ciprofloxacin-resistant Salmonella enterica serovar Typhi from Kuwait with novel mutations in gyrA and parC genes. J Clin Microbiol. 2009;47:208–11. doi: 10.1128/JCM.01161-08. doi:10.1128/JCM.01161-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim HB, Park CH, Kim CJ, et al. Prevalence of plasmid-mediated quinolone resistance determinants over a 9-year period. Antimicrob Agents Chemother. 2009;53:639–45. doi: 10.1128/AAC.01051-08. doi:10.1128/AAC.01051-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robicsek A, Jacoby GA, Hooper DC. The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect Dis. 2006;6:629–40. doi: 10.1016/S1473-3099(06)70599-0. doi:10.1016/S1473-3099(06)70599-0. [DOI] [PubMed] [Google Scholar]
- 37.Achtman M. Evolution, population structure and phylogeography of genetically monomorphic bacterial pathogens. Annu Rev Microbiol. 2008;62:53–70. doi: 10.1146/annurev.micro.62.081307.162832. doi:10.1146/annurev.micro.62.081307.162832. [DOI] [PubMed] [Google Scholar]