Abstract
Myc proteins (c-myc, Mycn and Mycl) target proliferative and apoptotic pathways vital for progression in cancer. Amplification of the MYCN gene has emerged as one of the clearest indicators of aggressive and chemotherapy-refractory disease in children with neuroblastoma, the most common extracranial solid tumor of childhood. Phosphorylation and ubiquitin-mediated modulation of Myc protein influence stability and represent potential targets for therapeutic intervention. Phosphorylation of Myc proteins is controlled in-part by the receptor tyrosine kinase/phosphatidylinositol 3-kinase/Akt/mTOR signaling, with additional contributions from Aurora A kinase. Myc proteins regulate apoptosis in part through interactions with the p53/Mdm2/Arf signaling pathway. Mutation in p53 is commonly observed in patients with relapsed neuroblastoma, contributing to both biology and therapeutic resistance. This review examines Myc function and regulation in neuroblastoma, and discusses emerging therapies that target Mycn.
Keywords: myc, mycn, neuroblastoma, N-myc, mTor, PI3K
Introduction
Neuroblastoma, a neoplasm of peripheral neural crest origin, is the most common malignant extracranial solid tumor of childhood and accounts for 15% of cancer deaths in children (Park et al., 2008). Approximately 650 new cases are diagnosed in the United States annually with peak incidence in early childhood (ages 0–4 years). The most common site of origin is in the adrenal medulla; however, tumors can occur anywhere along the sympathetic chain.
Patients with new diagnoses are typically stratified into risk groups based on age, stage, histopathology, DNA index and genetic/genomic factors. Amplification of the proto-oncogene MYCN occurs in ~25% of tumors and is the best characterized genetic-risk factor for high-risk chemotherapy-refractory disease (Brodeur et al., 1984; Seeger et al., 1985; Riley et al., 2004). Deletion or suppression of caspase 8, loss of chromosomes 1p and 11q, and gain of 17q also correlate with aggressive disease (Bown et al., 1999; Guo et al., 1999; Riley et al., 2004; Attiyeh et al., 2005; Stupack et al., 2006; Maris et al., 2008b).
In contrast with most other treatment-refractory cancers, neuroblastomas, irrespective of risk group, generally respond to initial therapy, which typically includes high doses of chemotherapy (Park et al., 2008). Low- and intermediate-risk patients are subsequently cured and rarely progress. Patients with high-risk disease typically relapse with treatment-refractory tumors. It is conceivable that low- and high-risk neuroblastoma are entirely separate disease entities. Overall, prognosis in the high-risk group is quite poor, with long-term survival of 30–40%.
Familial neuroblastoma
Neuroblastoma may be associated with Hirschsprung disease and neurofibromatosis type 1 (Clausen et al., 1989; Rohrer et al., 2002). Mutations in PHOX2B, a homeodomain-containing transcription factor important for the development in the autonomic nervous system, underlie the congenital central hypoventilation syndrome and confer a heritable predisposition to neuroblastoma; however, these mutations are not found in spontaneous tumors (Mosse et al., 2004; Trochet et al., 2005; Raabe et al., 2008). In fact, familial neuroblastoma is quite rare, and is most often because of dominant gain-of-function mutations in the orphan receptor tyrosine kinase (RTK) anaplastic lymphoma kinase (ALK). ALK, which is linked to MYCN on chromosome 2p23, also shows sporadic gain-of-function mutation in 8% of spontaneous tumors (Chen et al., 2008; George et al., 2008; Janoueix-Lerosey et al., 2008; Mossé et al., 2008). Although direct connections between MYCN and ALK have yet to be elucidated, activation of Alk and other RTKs may contribute to stabilization of Mycn protein (detailed below).
Myc family of proto-oncogenes
Broadly implicated in oncogenesis, the human MYC family of proto-oncogenes is among the most studied genes in cancer (Meyer and Penn, 2008). Early insertional mutagenesis studies in mouse identified c-MYC (homologous to the v-myc gene that drives avian myelocytosis) as capable of transformation by retroviral promoter insertion (Payne et al., 1982). MYCN was subsequently identified as an MYC homolog amplified in neuroblastoma tumors (reviewed in Meyer and Penn, 2008). Amplification of MYCN has emerged as among the clearest genetic indicators of high-risk, aggressive disease (Brodeur et al., 1984; Seeger et al., 1985). MYC family members (c-MYC, MYC and MYCL) show differential expression in normal tissues. Expression of murine N-myc in particular is elevated in normal mammalian developing retina, forebrain, hindbrain, intestine, kidney and has functions in neuronal progenitor cells, developing lung tissues, hematopoetic stem cells and programmed cell death in the developing limb (Zimmerman et al., 1986; Mugrauer et al., 1988; Downs et al., 1989; Hirvonen et al., 1990; Hirning et al., 1991; Knoepfler et al., 2002; Bettess et al., 2005; Okubo et al., 2005; Ota et al., 2007; Martins et al., 2008; Xu et al., 2009).
Myc proteins are basic helix-loop-helix leucine zipper transcription factors. Mycn and c-Myc proteins share several regions of homology and share similar cellular functions. Myc proteins localize to the nucleus and form heterodimers with the basic helix-loop-helix molecule, Max (Blackwood and Eisenman, 1991; Prendergast et al., 1991; Berberich and Cole, 1992; Blackwood et al., 1992; Kato et al., 1992). Myc/Max heterodimers bind to DNA at specific CAC(G/A)TG ‘E-box’ sequences to drive transcription of targets important for proliferation, apoptosis and differentiation (Blackwell et al., 1990, 1993; Amati et al., 1992; Kretzner et al., 1992). Max also heterodimerizes with Mxd or Mnt proteins to influence the transcription of other downstream genes and often to antagonize the proliferative effects of Myc proteins (Ayer et al., 1993; Zervos et al., 1993; Hurlin et al., 1995, 1997; Walkley et al., 2005). In many systems, Mxd/Max or Mnt/Max heterodimers oppose the actions of Myc/Max to repress transcription; however, very little is known about these interactions in neuroblastoma (Grandori et al., 2000; Patel et al., 2004). The complex competition among Myc, Mxd and Mnt proteins for binding to Max further modulates the effects of both c-Myc, and probably Mycn, on gene expression.
Downstream of Mycn
E-boxes are common (~25% of known promoters) with >10 000 sites per cell (Fernandez et al., 2003; Li et al., 2003; Zeller et al., 2006). That there are more E-box sequences than Myc molecules in cells represents a conundrum apparently common to many transcription factors (Farnham, 2009). Adding further complexity to this system, Myc proteins also regulate downstream targets through cap-dependent methylation, altering both global translation as well as the translation of specific proteins (Barna et al., 2008; Cole and Cowling, 2008).
Chromatin immunoprecipitation experiments show that c-Myc and Mycn proteins bind to promoters with variable specificity determined by DNA ultrastructure and cellular context (Fernandez et al., 2003; Li et al., 2003; Mao et al., 2003; Chen et al., 2004; Guccione et al., 2006; Zeller et al., 2006; Kim et al., 2008; Martinato et al., 2008; Westermann et al., 2008). Myc/Max heterodimers bind to E-boxes and interact with a variety of histone modifiers, increasing histone acetylation (Bouchard et al., 2001). These alterations modify chromatin at promoters, affecting gene expression (McMahon et al., 2000; Frank et al., 2001; Vervoorts et al., 2003; Guccione et al., 2006; Martinato et al., 2008; Liu et al., 2009). In fact, histone modification by Mycn effectors is being exploited pharmacologically using histone deacetylase inhibitors. These drugs may alter acetylation in neuroblastomas and other MYC dependent processes thereby modulating transcription (reviewed in Witt et al., 2009).
Several studies have attempted to elucidate specific transcriptional targets for Mycn in neuroblastoma (Alaminos et al., 2003). A large number of targets important for cell cycle control and differentiation have been characterized, including: downregulation of SKP2 and TP53INP1 with resultant decrease in p21WAF1 (Bell et al., 2007), downregulation of DKK1 upstream of the wnt/β-catenin pathway (Koppen et al., 2007), upregulation of NLRR1 both important in neural cell proliferation (Hossain et al., 2008), downregulation of Fyn kinase important in differentiation (Berwanger et al., 2002), regulation of multiple genes responsible for pluripotency (Cotterman and Knoepfler, 2009) and modulation of apoptosis by upregulation of p53 and Mdm2 (Slack et al., 2005b). The multidrug resistance gene MRP1 is regulated by Mycn, driving chemotherapy resistance (Manohar et al., 2004). Importantly, Mycn upregulates oncogenic microRNAs, which have wide ranging effects on cancer (reviewed in Schulte et al., 2009). Mycn also controls several proteins important in ribosome biogenesis (Boon et al., 2001) affecting protein synthesis (reviewed in Ruggero, 2009).
Different groups seeking to stratify neuroblastoma risk using gene expression micoarrays have generated gene lists that are largely non-overlapping (Berwanger et al., 2002; Ohira et al., 2005; Schramm et al., 2005, 2009; Oberthuer et al., 2006). Similar strategies using real-time PCR transcript analysis, are also being performed (Vermeulen et al., 2009). Direct comparison of targets from chromatin immunoprecipitation shows that Mycn and c-Myc have many overlapping targets (Westermann et al., 2008). Further refinement of microarray and chromatin immunoprecipitation techniques and identification of critical transcriptional targets specific to Mycn may provide insights into both biology and therapy for neuroblastoma.
Mycn in cell cycle control
In normal cells, levels of Myc proteins are tightly regulated, with increased levels driven in part through activation of phosphatidylinositol 3-kinase (PI3K), which stabilizes Mycn and c-Myc proteins (Figure 1), enabling entry into the cell cycle (Marqués et al., 2008). In addition to the clearly defined function of Myc family members in control of the cell cycle, c-Myc contributes non-transcriptionally to the initation of DNA replication (Dominguez-Sola et al., 2007). Myc/Max dimers also bind and inhibit Miz-1, a helix-loop-helix transcription factor (reviewed in Herold et al., 2009). Free Miz-1 promotes transcription of p15INK4b and p21Cip1 proteins associated with cell cycle arrest. Inhibition of Miz-1 in response to Myc/Max binding contributes to immortalization, transformation and oncogenesis (Seoane et al., 2001; Staller et al., 2001; Herold et al., 2008). Interactions between Mycn and Miz-1 are incompletely characterized. Expression of Miz-1 has been associated with favorable outcome in neuroblastoma, however, consistent with an interaction among Miz-1, Mycn and Max (Ikegaki et al., 2007).
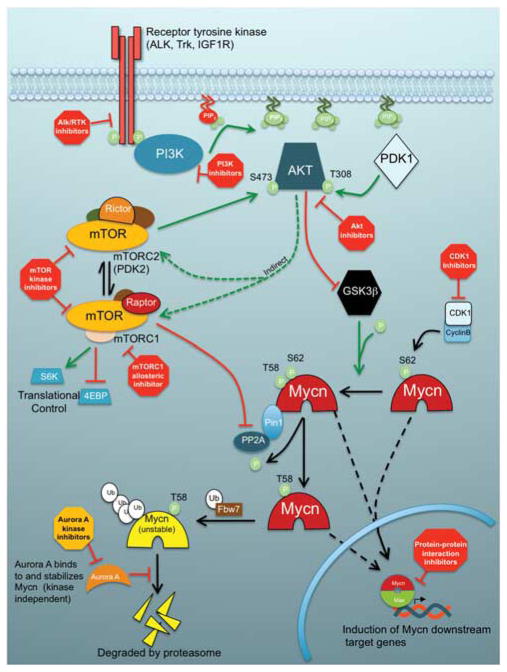
Figure 1.
Schematic model of regulatory pathways involved in Mycn stabilization. In proliferating cells, Mycn is initially phosphorylated at serine 62 by cyclin-dependent kinase 1/CyclinB. The S62-phosphorylated protein is stabilized and competent to enter the nucleus. Nuclear Mycn binds to its partner, the helix-loop-helix transcription factor Max and stimulates the transcription of a target genes important in cell cycle, proliferation, differentiation and apoptosis. This priming phosphorylation at S62 allows the binding of Gsk3β, as well as Pin1 and PP2A in a complex also containing Axin. Active Gsk3β phosphorylates Mycn at threonine 58, producing doubly phosphorylated, stabilized and transcriptionally active Mycn. In a Pin1-mediated process, PP2A dephosphorylates the S62 phosphate, enabling binding of ubiquitin ligase Fbw7. Fbw7 subsequently drives poly-ubiquitination and proteasomal degradation of Mycn. Aurora A kinase can bind to and stabilize polyubiquitinated/phosphorylated Mycn, potentially leaving it competent to bind to Max and activate transcription. In this model, upstream signaling starts at the membrane with receptor tyrosine kinases (RTKs), which are either activated by ligand or by constitutively activating mutations (in Alk, for example). RTKs activate phosphatidylinositol 3-kinase (PI3K), which catalyzes the conversion of phosphatidylinositol-3,4-bisphosphate (PIP2) to phosphatidylinositol-3,4,5-triphosphate (PIP3). PIP3 then binds to Akt, localizing it to the membrane and allowing the phosphorylation and activation of Akt by membrane bound Pdk1 at threonine 308. Active Akt then phosphorylates Glycogen synthase kinase 3β (Gsk3β) and inactivates it, blocking Gsk3β-mediated phosphorylation of Mycn T58 and thereby stabilizing Mycn. Akt also activates mammalian target of rapamycin (mTOR) through several indirect signaling mechanisms including through Tsc2/Tsc1 and Rheb. mTOR exists in two distinct complexes. mTORC1 complex, the rapamycin sensitive form, consists of mTOR bound to multiple effector proteins, including Raptor and Lst8, and is important for the promotion of global translation through S6K and 4EBP. mTORC1 also directly phosphorylates and inhibits PP2A, enabling the accumulation of doubly phosphorylated, active Mycn and contributing to the general proliferation-promoting downstream effects of the PI3K/Akt/Mycn pathway (Cheng et al., 2007). The mTORC2 complex, also known as Pdk2, contains mTOR, rictor, Lst8 and mSin1. mTORC2 phosphorylates Akt at serine 473 further activating this kinase. Inhibitors of this pathway could potentially destabilize Mycn. These inhibitors are currently in clinical trials or in clinical development and are denoted by red octagons. The Aurora A Kinase inhibitor is denoted by an orange octagon, which represents targeted kinase inhibition, but no anticipated activity against Mycn itself. Relevant references are contained within the corresponding text.
Transcription of MYCN is downregulated by the neuroblastoma differentiating agent retinoic acid and upregulated by several known transcription factors including E2F and Sp1/Sp3 (Thiele et al., 1985; Inge et al., 2002; Kramps et al., 2004; Kanemaru et al., 2008). Sonic hedgehog indirectly regulates transcription of MYCN in developing neurons (Kenney et al., 2003, 2004; Oliver et al., 2003; Mill et al., 2005). Levels of Myc mRNA are also by alternate internal ribosomal entry sites (Barna et al., 2008; Cobbold et al., 2008). Although natural antisense transcripts are frequently co-amplified with MYCN in neuroblastoma, the importance of these co-amplified sequences remains unclear (Krystal et al., 1990; Armstrong and Krystal, 1992; Jacobs et al., 2009).
Mycn and apoptosis
In addition to promoting proliferation, the expression of c-Myc and Mycn actually drives apoptosis (Askew et al., 1991; Evan et al., 1992; Shi et al., 1992; Fulda et al., 1999; Paffhausen et al., 2007; Ushmorov et al., 2008). Transformation by Myc proteins, therefore, requires concomitant inhibition of apoptosis. Tissue-specific expression of a switchable allele of c-Myc also induced apoptosis in vivo. In contrast, proliferation and tumorigenesis required lower level, continuous and deregulated expression of c-Myc (Murphy et al., 2008).
Although the association between MYCN amplification and poor outcome has been reproduced in numerous studies over decades, a similar association between expression of Mycn and outcome remains controversial (Chan et al., 1997; Cohn et al., 2000; Tang et al., 2006). Data from preclinical models of switchable myc above suggest that low levels of myc proteins may drive proliferation, with higher levels required to induce apoptosis. If it is true in the context of Mycn and neuroblastoma, then MYCN amplification may serve primarily to dysregulate Mycn during the cell cycle, rather than simply driving high-level expression. This hypothesis is consistent with the claims that amplification and not overexpression is predictive of aggressive disease, although inconsistent with observations that MYCN is often amplified and overexpressed to extreme levels in neuroblastoma.
Mechanisms through which apoptosis is inhibited as a contributer to Mycn-driven transformation are complex and not yet fully elucidated. Silencing of the apoptotic initiator Casp8 is observed frequently in neuroblastoma (Stupack et al., 2006). Crosstalk with the p53 pathway has been implicated in apoptosis mediated by both Mycn and c-Myc (reviewed in Hoffman and Liebermann, 2008; Van Maerken et al., 2009b). Mutations in p53 are rare in primary neuroblastoma (<2%) irrespective of MYCN amplification (Vogan et al., 1993). However, inactivating mutations in p53 and in p53 pathway members are common at relapse (Keshelava et al., 1997, 2000; Tweddle et al., 2001; Carr et al., 2006). Myc proteins indirectly regulate the p53 pathway and p53-dependent apoptosis through the p14ARF-MDM2-p53 axis (reviewed in Van Maerken et al., 2009b).
At baseline, p53 is tightly regulated by Mdm2, which binds to and inhibits the transactivation domain of p53 (Oliner et al., 1993; Thut et al., 1997). Mdm2 also ubiquitinates and targets p53 protein for degradation (reviewed in Coutts et al., 2009). Further regulation is conferred by the tumor suppressor protein p14ARF, which binds to and inhibits Mdm2, allowing activation and stabilization of p53 (Kamijo et al., 1998; Zindy et al., 1998; Weber et al., 1999; Midgley et al., 2000; Lin and Lowe, 2001). The balance between apoptosis and survival remains in equilibrium through multiple feedback and feed-forward loops affected by other signaling pathways including external apoptotic stimuli.
In neuroblastoma cells, Mycn directly stimulates transcription of MDM2 (Slack et al., 2005a; Barbieri et al., 2006; Chen et al., 2009). The resulting inhibition of p53 may in part allow cells to escape Mycn-primed apoptosis. Myc and Mycn also indirectly inhibit p14ARF, through directly driving transcription factors including TWIST1, resulting in activation of MDM2 and escape from apoptosis (Maestro et al., 1999; Valsesia-Wittmann et al., 2004). The p14ARF protein can in-turn feed back and bind to Myc and Mycn proteins, abrogating their ability to activate downstream targets (Qi et al., 2004; Amente et al., 2007). The complex interactions among Myc/Mycn, Mdm2 and p14ARF provide mechanisms through which Mycn both indirectly activates or inactivates p53, altering the sensitivity of cells to apoptotic stimuli (reviewed in Li and Hann, 2009; Van Maerken et al., 2009b).
Neuroblastoma tumors at diagnosis are generally wild type for p53 and respond to chemotherapy. Children with high-risk tumors generally relapse, whereas those with low and intermediate disease are typically cured. As stated above, Mycn-induced downregulation of the p53 axis could potentially underlie the minimal residual disease that drives subsequent relapse in newly diagnosed MYCN-amplified neuroblastomas. This hypothesis is supported by the identification of Mdm2 as an Mycn target, and in studies showing that Mdm2 haploinsufficiency inhibits tumorigenesis in MYCN-driven models for neuroblastoma (Slack et al., 2005a; Chen et al., 2009). In fact, small molecule inhibitors of the p53/Mdm2 interaction (‘Nutlins’) are currently in development and have shown some promise for the preclinical treatment of cancers including neuroblastoma (Barbieri et al., 2006; Chen et al., 2009; Van Maerken et al., 2009a).
Modeling MYCN-amplified neuroblastoma
Mice carrying an MYCN transgene under control of the rat tyrosine hydroxylase promoter develop neuroblastoma tumors several months after birth (Norris et al., 2000; Weiss et al., 2000). Tumors from mice transgenic for TH-MYCN develop in adrenal and mesenteric ganglia and in paraspinous locations. Histolology and genetics show similarities with high-risk neuroblastoma (Weiss et al., 2000; Hackett et al., 2003; Moore et al., 2008). In relatively resistant strains or subspecies (for example C57BL/6, BALBc or Mus musculus castaneus), a range of differentiation was observed in murine tumors. However, when crossed into highly penetrant strains, such as 129 SvJ, the histology was uniformly undifferentiated small round blue cells.
Cell lines derived from TH-MYCN tumors retain the ability to form tumors in nude mice (Cheng et al., 2007). Unlike xenograft models of neuroblastoma, TH-MYCN tumors show native host interactions with the tumor microenvironment including vascular cells (Chesler et al., 2007). When treated with chemotherapy, tumors in this model, similar to their human p53 wild-type counterparts, undergo apoptosis in a p53-dependent manner. Furthermore, tumors from TH-MYCN;p53−/+ mice were refractory to cytoxic chemotherapy (Chesler et al., 2008). These data and corresponding data from human tumors (Keshelava et al., 1998, 2000; Xue et al., 2007) collectively suggest a model in which newly diagnosed neuroblastoma tumors impair the p14ARF-MDM2-p53 axis in the absence of p53 mutation, enabling tumors to arise. Subsequent chemotherapy subsequently provides strong selective pressure for inactivating mutations in p53 or in components of the p53 pathway, resulting in chemotherapy-refractory tumors seen clinically in relapsed patients. Thus, although mutation at p53 rarely contributes to the biology of primary neuroblastoma, therapy-selected mutations in p53 drive a genetically distinct tumor at relapse. This therapy-associated alteration in the biology of neuroblastoma presents a challenge in identifying effective treatments for relapsed tumors.
Mycn as a target for therapy
In light of both the frequency and importance of MYCN amplification in pathogenesis of high-risk neuroblastoma, blockade of Mycn signaling represents an important approach for the developmental therapeutics. The resistance of high-risk relapsed neuroblastoma to conventional chemotherapy and the high morbidity and mortality in these patients present a formidable challenge for clinicians. The prominence of MYCN amplification in the pathogenesis of this disease points to Mycn as a potential therapeutic target.
Neuroblastoma presents both common and unique challenges for therapy. The initial response to chemotherapy even in high-risk disease is somewhat unusual. However, the acquisition of p53 mutant, therapy-refractory disease is common to many cancers (McDermott et al., 2008). Deregulated expression of Mycn may contribute to genomic instability and, in combination with the strong selective pressures of chemotherapy and radiation, select for mutation at p53 through interactions with Mdm2 and p14Arf detailed above, and by driving the cell cycle and activating p53-dependent check points. Alleviation of check-point activation by blocking Mycn itself could conceivably impair the acquisition of p53 mutant, refractory disease.
Transcription factors have long been considered as targets for cancer therapy; however, clinical approaches to block this class of molecules remain elusive. Clearly, the most direct means of silencing these molecules would be by disrupting Myc synthesis directly through RNAi methods and there are numerous examples of siRNA used successfully on cultured cells. However, although these approaches are extremely useful tools in the laboratory, they have yet to achieve regular use in the clinic largely because of difficulty with delivery (Whitehead et al., 2009).
It is difficult to develop drugs with activities sufficient to block protein–protein interactions or binding of such factors to target sequences. Molecules developed against c-Myc have been primarily directed against the Myc/Max interaction domain and so far have fairly low potency. Progress is being made in this area, however, and the development of inhibitors, which dissociate Myc/Max heterodimers is important (Yin et al., 2003; Wang et al., 2006; Berg, 2008; Brooks and Hurley, 2009). Chemists are good at developing kinase inhibitors, however, and the stability of Myc molecules is regulated at a number of levels by kinases and critical phospho-residues.
Post-transcriptional modification and stabilization of Myc proteins
In neuroblastoma and in neural progenitor cells, Mycn protein stability is regulated by a complex signaling network involving both feedback and feed-forward loops (Figure 1) (Kenney et al., 2004; Sjostrom et al., 2005; Chesler et al., 2006; Kang et al., 2008). Although both c-Myc and Mycn are widely phosphorylated, sites critical to stabilization of Myc proteins are located in the N-terminal Myc Box I, which is commonly mutated in c-Myc in Burkitt lymphoma (Bhatia et al., 1993; Smith-Sørensen et al., 1996; Bahram et al., 2000). Comparable mutations in c-Myc and Mycn are rare in solid tumors including neuroblastoma. The important residues in the Myc Box I region of c-Myc and Mycn are threonine 58 and serine 62 (Henriksson et al., 1993; Pulverer et al., 1994; Lutterbach and Hann, 1999; Sjostrom et al., 2005).
Phosphorylation at S62 stabilizes Mycn protein and primes it for phosphorylation at T58. Candidate kinases proposed to phosphorylate c-Myc at S62 include extracellular signal-regulated kinase, the canonical downstream effector of the Ras/Raf/MAPK; c-Jun N-terminal kinase and cyclin-dependent kinase 1 pathway (Henriksson et al., 1993; Lutterbach and Hann, 1999; Sears et al., 2000; Benassi et al., 2006). As shown in cerebellar neural progenitor cells and synchronized mitotic SJ8 neuroblastoma cells, cyclin-dependent kinase 1 is thus far the only candidate priming kinase for Mycn at S62 (Sjostrom et al., 2005).
Myc and Mycn proteins monophosphorylated at S62 are substrates for a second phosphorylation at T58, controlled by glycogen synthase kinase 3β (Gsk3β). Gsk3β signals in the Wnt pathway (reviewed in MacDonald et al., 2009), and also downstream of the PI3K/Akt/mTOR pathway (reviewed in Engelman, 2009; Ma and Blenis, 2009; Memmott and Dennis, 2009). The S62 priming phosphorylation site in Myc/Mycn promotes interaction with a complex containing Gsk3β, Pin1, PP2A and Axin. This complex induces the phosphorylation at T58 by Gsk3β in both Myc and Mycn. For c-Myc, Pin1 has been shown to regulate dephosphorylation of S62 by PP2A, a phosphatase and tumor suppressor that itself is regulated by mTOR (Peterson et al., 1999; Gingras et al., 2001; Hartley and Cooper, 2002; Yeh et al., 2004; Arnold and Sears, 2006; Arnold et al., 2009). Regulation of Mycn dephosphorylation by Pin1 and PP2A has not been fully characterized.
Myc proteins monophosphorylated at T58, then bind to the E3 ligase Fbxw7 and are targeted for ubiquitination and degradation (Saksela et al., 1992; Lutterbach and Hann, 1994; Sears et al., 1999; Gregory and Hann, 2000; Oliver et al., 2003; Herbst et al., 2004; Kenney et al., 2004; Welcker et al., 2004; Yada et al., 2004; Otto et al., 2009). Aurora kinase A, which has critical functions in cell cycle regulation and spindle assembly, contributes at this step to the stabilization of phosphorylated and ubiquitinated Mycn, but not of equivalently modified c-myc (Otto et al., 2009). Consistent with these findings, expression of Aurora A kinase is a negative prognostic factor in neuroblastoma (Shang et al., 2009). Although the emerging function of inhibitors of Aurora kinase A in cancer therapy suggested that inhibitors of Aurora kinase A might have unique and multifaceted functions in the therapy of neuroblastoma, stabilization of Mycn apparently requires a scaffold function of Aurora kinase and was independent of Aurora A kinase activity (Maris, 2009; Otto et al., 2009).
Myc proteins as downstream targets of PI3K
The phosphorylation of c-Myc and Mycn is thus regulated directly by Gsk3β, and indirectly by upstream signaling through RTKs, PI3K, Akt and mTOR. RTKs are important to PI3K signaling in neuroblastoma. As mentioned above, activating mutations in the orphan RTK ALK are found in ~9% of neuroblastoma. Other RTKs that could potentially regulate stabilization of Mycn include the insulin-like growth factor receptor and the Trk family of neurotropin receptors (TrkA, TrkB and TrkC). Although mutation in Trk genes is not typically observed in neuroblastoma, expression of TrkA correlates with favorable prognosis, whereas expression of TrkB correlates with amplification of MYCN and poor outcome (reviewed in Brodeur et al., 2009). Although neither Trk nor Alk proteins have been directly linked to stabilization of Mycn, these kinases are all upstream activators of PI3K, implying a possible connection (Bai et al., 2000; Norris et al., 2000; Ho et al., 2002; Marzec et al., 2007).
RTKs activate PI3K, which catalyzes the conversion of phosphatidylinositol-3,4-bisphosphate (PIP2) to phosphatidylinositol-3,4,5-triphosphate (PIP3) (reviewed in Ma and Blenis, 2009; Memmott and Dennis, 2009). PIP3 binds to Akt and localizes it to the membrane, enabling phosphorylation at T308 by the kinase PDK1. Activation of Akt leads to phosphorylation and inactivation of Gsk3β, stabilizing Mycn by blocking phosphorylation at T58. Activated Akt also phosphorylates and inhibits the tuberous sclerosis 2 (Tsc2) tumor suppressor protein. Tsc2 binds to Tsc1, enabling the Ras-related GTPase Rheb to stimulate mTOR bound in the mTORC1 complex of proteins. As mentioned above, mTORC1 downregulates PP2A, which normally dephosphorylates Myc/Mycn at S62, targeting it for ubiquitination, and promoting stabilization of Myc and Mycn. Thus, activation of RTKs and PI3K converge on Akt to phosphorylate and inhibit Gsk3β, blocking phosphorylation at T58 and promoting stabilization. In addition, Akt activates mTORC1, inhibiting dephosphorylation of T58/S62-phosphorylated Myc/Mycn at S62, leading to further stabilization of Myc/Mycn.
The critical importance of Mycn phosphorylation and stability as a downstream target of PI3K/Akt/mTOR in neuroblastoma cells is apparent when neuroblastoma cells are treated with a broad spectrum PI3K inhibitor. Activation of Akt predicts poor outcome in neuroblastoma patients (Opel et al., 2007). Treated cells show a decreased proliferation, which is largely rescued when they are engineered to express T58/S62 phosphorylation site-deficient Mycn mutants (Sjostrom et al., 2005; Chesler et al., 2006). These data show that degradation of Mycn is a critical downstream factor in the efficacy of PI3K/mTOR pathway and suggest that clinical inhibitors of PI3K should show activity in Mycn-driven neuroblastoma (detailed review in Fulda, 2009).
Mycn stability as a therapeutic target in neuroblastoma
The post-translational modification and stabilization of Myc and Mycn proteins represents an area with promise for currently available and emerging-targeted therapies. Inhibitors of RTKs (Alk and Trk), PI3K and Akt should activate Gsk3β (which is negatively regulated by phosphorylation), whereas inhibitors of mTOR kinase activity should inhibit the de-activation of PP2A by mTORC1, enabling PP2A to dephosphorylate S62, collectively driving degradation of Mycn in neuroblastoma (Figure 1). Specific Alk and Trk inhibitors are currently in development and have shown promise preclinically and in phase I trials (reviewed in Chiarle et al., 2008; Li and Morris, 2008; Brodeur et al., 2009; Mossé et al., 2009).
Downstream of RTKs, inhibitors of PI3K, mTOR, dual inhibitors of PI3K/mTOR and inhibitors of Akt are all in clinical trials (reviewed in Garcia-Echeverria and Sellers, 2008; Engelman, 2009). Allosteric inhibitors of mTORC1 (so-called rapalogs) block mTOR independently of ATP binding, are currently being tested in neuroblastoma and have shown mixed results preclincally (Houghton et al., 2008; Johnsen et al., 2008; Maris et al., 2008a; Wagner and Danks, 2009). These agents only affect some outputs of mTORC1. In comparison, ATP-competitive inhibitors of mTOR more completely block mTORC1 and also block mTORC2 (Feldman et al., 2009; Thoreen et al., 2009; Yu et al., 2009; Zask et al., 2009). The availability of inhibitors targeting RTKs, PI3K, Akt and mTOR, and the use of these to destabilize Mycn protein, represent important areas of investigation. Further, because of the complex interrelation of pathway members, inhibition at one point often induces feedback activation in other signaling pathways, justifying the need to test these agents in combination using preclinical models.
The regulation of Mycn phosphorylation also involves a priming phosphorylation at S62. As efficient destabilization of Mycn would require activators of kinases responsible for these phosphorylation steps, the ability to finesse phosphorylation at S62 presents a therapeutic challenge. Aurora kinase A represents an additional therapeutic target, as Aurora kinase A stabilizes Mycn at later steps. Inhibitors of Aurora A kinase are currently in clinical trials in neuroblastoma (Shang et al., 2009). However, asMycn is stabilized by a kinase-independent activity of Aurora A, these inhibitors are unlikely to affect Mycn protein (Gautschi et al., 2008; Maris, 2009). An allosteric and ATP-competitive small molecule inhibitor of Aurora A–Mycn interactions, if it could be developed, should retain the ability to block kinase-dependent functions, whereas also twisting Aurora A kinase, thereby disrupting a scaffolding function and degrading Mycn protein. The interplay among RTKs, PI3K, Akt, mTORC1/2, Aurora A and Mycn is quite complex. However, the broad functions for these kinases in cancer biology, and the specific function in regulating the stabilization of Mycn proteins suggests functions in both Mycn-driven and Mycn-independent cancers including neuroblastoma. The current availability of clinical Alk, Trk, PI3K, mTOR and Aurora inhibitors presents an important translational opportunity to test these agents in children with high-risk neuroblastoma.
Acknowledgments
We thank Chris Hackett and Theo Nicolaides for critical review. We acknowledge support from NIH grants CA133091, NS055750, CA102321, CA097257, CA128583; Burroughs Wellcome Fund, American Brain Tumor Association, The Brain Tumor Society, Accelerate Brain Cancer Cure; Alex’s Lemonade Stand, Children’s National Brain Tumor, Wallace H. Coulter, Katie Dougherty, Pediatric Brain Tumor, Samuel G Waxman and V Foundations.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- Alaminos M, Mora J, Cheung N-KV, Smith A, Qin J, Chen L, et al. Genome-wide analysis of gene expression associated with MYCN in human neuroblastoma. Cancer Res. 2003;63:4538–4546. [PubMed] [Google Scholar]
- Amati B, Dalton S, Brooks MW, Littlewood TD, Evan GI, Land H. Transcriptional activation by the human c-Myc oncoprotein in yeast requires interaction with Max. Nature. 1992;359:423–426. doi: 10.1038/359423a0. [DOI] [PubMed] [Google Scholar]
- Amente S, Gargano B, Diolaiti D, Della Valle G, Lania L, Majello B. p14ARF interacts with N-Myc and inhibits its transcriptional activity. FEBS Lett. 2007;581:821–825. doi: 10.1016/j.febslet.2007.01.062. [DOI] [PubMed] [Google Scholar]
- Armstrong BC, Krystal GW. Isolation and characterization of complementary DNA for N-cym, a gene encoded by the DNA strand opposite to N-myc. Cell Growth Differ. 1992;3:385–390. [PubMed] [Google Scholar]
- Arnold HK, Sears RC. Protein phosphatase 2A regulatory subunit B56alpha associates with c-myc and negatively regulates c-myc accumulation. Mol Cell Biol. 2006;26:2832–2844. doi: 10.1128/MCB.26.7.2832-2844.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold HK, Zhang X, Daniel CJ, Tibbitts D, Escamilla-Powers J, Farrell A, et al. The Axin1 scaffold protein promotes formation of a degradation complex for c-Myc. EMBO J. 2009;28:500–512. doi: 10.1038/emboj.2008.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askew DS, Ashmun RA, Simmons BC, Cleveland JL. Constitutive c-myc expression in an IL-3-dependent myeloid cell line suppresses cell cycle arrest and accelerates apoptosis. Oncogene. 1991;6:1915–1922. [PubMed] [Google Scholar]
- Attiyeh EF, London WB, Mossé YP, Wang Q, Winter C, Khazi D, et al. Chromosome 1p and 11q deletions and outcome in neuroblastoma. N Engl J Med. 2005;353:2243–2253. doi: 10.1056/NEJMoa052399. [DOI] [PubMed] [Google Scholar]
- Ayer DE, Kretzner L, Eisenman RN. Mad: a heterodimeric partner for Max that antagonizes Myc transcriptional activity. Cell. 1993;72:211–222. doi: 10.1016/0092-8674(93)90661-9. [DOI] [PubMed] [Google Scholar]
- Bahram F, von der Lehr N, Cetinkaya C, Larsson LG. c-Myc hot spot mutations in lymphomas result in inefficient ubiquitination and decreased proteasome-mediated turnover. Blood. 2000;95:2104–2110. [PubMed] [Google Scholar]
- Bai RY, Ouyang T, Miething C, Morris SW, Peschel C, Duyster J. Nucleophosmin-anaplastic lymphoma kinase associated with anaplastic large-cell lymphoma activates the phosphatidylinositol 3-kinase/Akt antiapoptotic signaling pathway. Blood. 2000;96:4319–4327. [PubMed] [Google Scholar]
- Barbieri E, Mehta P, Chen Z, Zhang L, Slack A, Berg S, et al. MDM2 inhibition sensitizes neuroblastoma to chemotherapy-induced apoptotic cell death. Mol Cancer Ther. 2006;5:2358–2365. doi: 10.1158/1535-7163.MCT-06-0305. [DOI] [PubMed] [Google Scholar]
- Barna M, Pusic A, Zollo O, Costa M, Kondrashov N, Rego E, et al. Suppression of Myc oncogenic activity by ribosomal protein haploinsufficiency. Nature. 2008;456:971–975. doi: 10.1038/nature07449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell E, Lunec J, Tweddle DA. Cell cycle regulation targets of MYCN identified by gene expression microarrays. Cell Cycle. 2007;6:1249–1256. doi: 10.4161/cc.6.10.4222. [DOI] [PubMed] [Google Scholar]
- Benassi B, Fanciulli M, Fiorentino F, Porrello A, Chiorino G, Loda M, et al. c-Myc phosphorylation is required for cellular response to oxidative stress. Mol Cell. 2006;21:509–519. doi: 10.1016/j.molcel.2006.01.009. [DOI] [PubMed] [Google Scholar]
- Berberich SJ, Cole MD. Casein kinase II inhibits the DNA-binding activity of Max homodimers but not Myc/Max heterodimers. Genes Dev. 1992;6:166–176. doi: 10.1101/gad.6.2.166. [DOI] [PubMed] [Google Scholar]
- Berg T. Inhibition of transcription factors with small organic molecules. Curr Opin Chem Biol. 2008;12:464–471. doi: 10.1016/j.cbpa.2008.07.023. [DOI] [PubMed] [Google Scholar]
- Berwanger B, Hartmann O, Bergmann E, Bernard S, Nielsen D, Krause M, et al. Loss of a FYN-regulated differentiation and growth arrest pathway in advanced stage neuroblastoma. Cancer Cell. 2002;2:377–386. doi: 10.1016/s1535-6108(02)00179-4. [DOI] [PubMed] [Google Scholar]
- Bettess MD, Dubois N, Murphy MJ, Dubey C, Roger C, Robine S, et al. c-Myc is required for the formation of intestinal crypts but dispensable for homeostasis of the adult intestinal epithelium. Mol Cell Biol. 2005;25:7868–7878. doi: 10.1128/MCB.25.17.7868-7878.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia K, Huppi K, Spangler G, Siwarski D, Iyer R, Magrath I. Point mutations in the c-Myc transactivation domain are common in Burkitt’s lymphoma and mouse plasmacytomas. Nat Genet. 1993;5:56–61. doi: 10.1038/ng0993-56. [DOI] [PubMed] [Google Scholar]
- Blackwell TK, Kretzner L, Blackwood EM, Eisenman RN, Weintraub H. Sequence-specific DNA binding by the c-Myc protein. Science. 1990;250:1149–1151. doi: 10.1126/science.2251503. [DOI] [PubMed] [Google Scholar]
- Blackwell TK, Huang J, Ma A, Kretzner L, Alt FW, Eisenman RN, et al. Binding of myc proteins to canonical and noncanonical DNA sequences. Mol Cell Biol. 1993;13:5216–5224. doi: 10.1128/mcb.13.9.5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwood EM, Eisenman RN. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991;251:1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- Blackwood EM, Lüscher B, Eisenman RN. Myc and Max associate in vivo. Genes Dev. 1992;6:71–80. doi: 10.1101/gad.6.1.71. [DOI] [PubMed] [Google Scholar]
- Boon K, Caron HN, van Asperen R, Valentijn L, Hermus MC, van Sluis P, et al. N-myc enhances the expression of a large set of genes functioning in ribosome biogenesis and protein synthesis. EMBO J. 2001;20:1383–1393. doi: 10.1093/emboj/20.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard C, Dittrich O, Kiermaier A, Dohmann K, Menkel A, Eilers M, et al. Regulation of cyclin D2 gene expression by the Myc/Max/Mad network: Myc-dependent TRRAP recruitment and histone acetylation at the cyclin D2 promoter. Genes Dev. 2001;15:2042–2047. doi: 10.1101/gad.907901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bown N, Cotterill S, Lastowska M, O’Neill S, Pearson AD, Plantaz D, et al. Gain of chromosome arm 17q and adverse outcome in patients with neuroblastoma. N Engl J Med. 1999;340:1954–1961. doi: 10.1056/NEJM199906243402504. [DOI] [PubMed] [Google Scholar]
- Brodeur GM, Seeger RC, Schwab M, Varmus HE, Bishop JM. Amplification of N-myc in untreated human neuroblastomas correlates with advanced disease stage. Science. 1984;224:1121–1124. doi: 10.1126/science.6719137. [DOI] [PubMed] [Google Scholar]
- Brodeur GM, Minturn JE, Ho R, Simpson AM, Iyer R, Varela CR, et al. Trk receptor expression and inhibition in neuroblastomas. Clin Cancer Res. 2009;15:3244–3250. doi: 10.1158/1078-0432.CCR-08-1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks T, Hurley L. The role of supercoiling in transcriptional control of MYC and its importance in molecular therapeutics. Nat Rev Cancer. 2009;9:849–861. doi: 10.1038/nrc2733. [DOI] [PubMed] [Google Scholar]
- Carr J, Bell E, Pearson ADJ, Kees UR, Beris H, Lunec J, et al. Increased frequency of aberrations in the p53/MDM2/p14(ARF) pathway in neuroblastoma cell lines established at relapse. Cancer Res. 2006;66:2138–2145. doi: 10.1158/0008-5472.CAN-05-2623. [DOI] [PubMed] [Google Scholar]
- Chan HS, Gallie BL, DeBoer G, Haddad G, Ikegaki N, Dimitroulakos J, et al. MYCN protein expression as a predictor of neuroblastoma prognosis. Clin Cancer Res. 1997;3:1699–1706. [PubMed] [Google Scholar]
- Chen Q-R, Bilke S, Wei JS, Whiteford CC, Cenacchi N, Krasnoselsky AL, et al. cDNA array-CGH profiling identifies genomic alterations specific to stage and MYCN-amplification in neuroblastoma. BMC Genomics. 2004;5:70. doi: 10.1186/1471-2164-5-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Takita J, Choi YL, Kato M, Ohira M, Sanada M, et al. Oncogenic mutations of ALK kinase in neuroblastoma. Nature. 2008;455:971–974. doi: 10.1038/nature07399. [DOI] [PubMed] [Google Scholar]
- Chen Z, Lin Y, Barbieri E, Burlingame S, Hicks J, Ludwig A, et al. Mdm2 deficiency suppresses MYCN-driven neuroblastoma tumorigenesis in vivo. Neoplasia. 2009;11:753–762. doi: 10.1593/neo.09466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng AJ, Cheng NC, Ford J, Smith J, Murray JE, Flemming C, et al. Cell lines from MYCN transgenic murine tumours reflect the molecular and biological characteristics of human neuroblastoma. Eur J Cancer. 2007;43:1467–1475. doi: 10.1016/j.ejca.2007.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesler L, Schlieve C, Goldenberg DD, Kenney A, Kim G, McMillan A, et al. Inhibition of phosphatidylinositol 3-kinase destabilizes Mycn protein and blocks malignant progression in neuroblastoma. Cancer Res. 2006;66:8139–8146. doi: 10.1158/0008-5472.CAN-05-2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesler L, Goldenberg DD, Seales IT, Satchi-Fainaro R, Grimmer M, Collins R, et al. Malignant progression and blockade of angiogenesis in a murine transgenic model of neuroblastoma. Cancer Res. 2007;67:9435–9442. doi: 10.1158/0008-5472.CAN-07-1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesler L, Goldenberg DD, Collins R, Grimmer M, Kim GE, Tihan T, et al. Chemotherapy-induced apoptosis in a transgenic model of neuroblastoma proceeds through p53 induction. Neoplasia. 2008;10:1268–1274. doi: 10.1593/neo.08778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiarle R, Voena C, Ambrogio C, Piva R, Inghirami G. The anaplastic lymphoma kinase in the pathogenesis of cancer. Nat Rev Cancer. 2008;8:11–23. doi: 10.1038/nrc2291. [DOI] [PubMed] [Google Scholar]
- Clausen N, Andersson P, Tommerup N. Familial occurrence of neuroblastoma, von Recklinghausen’s neurofibromatosis, Hirschsprung’s agangliosis and jaw-winking syndrome. Acta Paediatr Scand. 1989;78:736–741. doi: 10.1111/j.1651-2227.1989.tb11135.x. [DOI] [PubMed] [Google Scholar]
- Cobbold LC, Spriggs KA, Haines SJ, Dobbyn HC, Hayes C, de Moor CH, et al. Identification of internal ribosome entry segment (IRES)-trans-acting factors for the Myc family of IRESs. Mol Cell Biol. 2008;28:40–49. doi: 10.1128/MCB.01298-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn SL, London WB, Huang D, Katzenstein HM, Salwen HR, Reinhart T, et al. MYCN expression is not prognostic of adverse outcome in advanced-stage neuroblastoma with nonamplified MYCN. J Clin Oncol. 2000;18:3604–3613. doi: 10.1200/JCO.2000.18.21.3604. [DOI] [PubMed] [Google Scholar]
- Cole MD, Cowling VH. Transcription-independent functions of MYC: regulation of translation and DNA replication. Nat Rev Mol Cell Biol. 2008;9:810–815. doi: 10.1038/nrm2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotterman R, Knoepfler PS. N-Myc regulates expression of pluripotency genes in neuroblastoma including lif, klf2, klf4, and lin28b. PLoS One. 2009;4:e5799. doi: 10.1371/journal.pone.0005799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutts AS, Adams CJ, La Thangue NB. p53 ubiquitination by Mdm2: a never ending tail? DNA Repair (Amst) 2009;8:483–490. doi: 10.1016/j.dnarep.2009.01.008. [DOI] [PubMed] [Google Scholar]
- Dominguez-Sola D, Ying CY, Grandori C, Ruggiero L, Chen B, Li M, et al. Non-transcriptional control of DNA replication by c-Myc. Nature. 2007;448:445–451. doi: 10.1038/nature05953. [DOI] [PubMed] [Google Scholar]
- Downs KM, Martin GR, Bishop JM. Contrasting patterns of myc and N-myc expression during gastrulation of the mouse embryo. Genes Dev. 1989;3:860–869. doi: 10.1101/gad.3.6.860. [DOI] [PubMed] [Google Scholar]
- Engelman JA. Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat Rev Cancer. 2009;9:550–562. doi: 10.1038/nrc2664. [DOI] [PubMed] [Google Scholar]
- Evan GI, Wyllie AH, Gilbert CS, Littlewood TD, Land H, Brooks M, et al. Induction of apoptosis in fibroblasts by c-myc protein. Cell. 1992;69:119–128. doi: 10.1016/0092-8674(92)90123-t. [DOI] [PubMed] [Google Scholar]
- Farnham PJ. Insights from genomic profiling of transcription factors. Nat Rev Genet. 2009;10:605–616. doi: 10.1038/nrg2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman ME, Apsel B, Uotila A, Loewith R, Knight ZA, Ruggero D, et al. Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS Biol. 2009;7:e38. doi: 10.1371/journal.pbio.1000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez PC, Frank SR, Wang L, Schroeder M, Liu S, Greene J, et al. Genomic targets of the human c-Myc protein. Genes Dev. 2003;17:1115–1129. doi: 10.1101/gad.1067003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank SR, Schroeder M, Fernandez P, Taubert S, Amati B. Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev. 2001;15:2069–2082. doi: 10.1101/gad.906601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulda S, Lutz W, Schwab M, Debatin KM. MycN sensitizes neuroblastoma cells for drug-induced apoptosis. Oncogene. 1999;18:1479–1486. doi: 10.1038/sj.onc.1202435. [DOI] [PubMed] [Google Scholar]
- Fulda S. The PI3K/Akt/mTOR pathway as therapeutic target in neuroblastoma. Curr Cancer Drug Targets. 2009;9:729–737. doi: 10.2174/156800909789271521. [DOI] [PubMed] [Google Scholar]
- Garcia-Echeverria C, Sellers WR. Drug discovery approaches targeting the PI3K/Akt pathway in cancer. Oncogene. 2008;27:5511–5526. doi: 10.1038/onc.2008.246. [DOI] [PubMed] [Google Scholar]
- Gautschi O, Heighway J, Mack PC, Purnell PR, Lara PN, Gandara DR. Aurora kinases as anticancer drug targets. Clin Cancer Res. 2008;14:1639–1648. doi: 10.1158/1078-0432.CCR-07-2179. [DOI] [PubMed] [Google Scholar]
- George RE, Sanda T, Hanna M, Fröhling S, Luther W, Zhang J, et al. Activating mutations in ALK provide a therapeutic target in neuroblastoma. Nature. 2008;455:975–978. doi: 10.1038/nature07397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras AC, Raught B, Gygi SP, Niedzwiecka A, Miron M, Burley SK, et al. Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genes Dev. 2001;15:2852–2864. doi: 10.1101/gad.912401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandori C, Cowley SM, James LP, Eisenman RN. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu Rev Cell Dev Biol. 2000;16:653–699. doi: 10.1146/annurev.cellbio.16.1.653. [DOI] [PubMed] [Google Scholar]
- Gregory MA, Hann SR. c-Myc proteolysis by the ubiquitin-proteasome pathway: stabilization of c-Myc in Burkitt’s lymphoma cells. Mol Cell Biol. 2000;20:2423–2435. doi: 10.1128/mcb.20.7.2423-2435.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guccione E, Martinato F, Finocchiaro G, Luzi L, Tizzoni L, Dall’ Olio V, et al. Myc-binding-site recognition in the human genome is determined by chromatin context. Nat Cell Biol. 2006;8:764–770. doi: 10.1038/ncb1434. [DOI] [PubMed] [Google Scholar]
- Guo C, White PS, Weiss MJ, Hogarty MD, Thompson PM, Stram DO, et al. Allelic deletion at 11q23 is common in MYCN single copy neuroblastomas. Oncogene. 1999;18:4948–4957. doi: 10.1038/sj.onc.1202887. [DOI] [PubMed] [Google Scholar]
- Hackett CS, Hodgson JG, Law ME, Fridlyand J, Osoegawa K, de Jong PJ, et al. Genome-wide array CGH analysis of murine neuroblastoma reveals distinct genomic aberrations which parallel those in human tumors. Cancer Res. 2003;63:5266–5273. [PubMed] [Google Scholar]
- Hartley D, Cooper GM. Role of mTOR in the degradation of IRS-1: regulation of PP2A activity. J Cell Biochem. 2002;85:304–314. doi: 10.1002/jcb.10135. [DOI] [PubMed] [Google Scholar]
- Henriksson M, Bakardjiev A, Klein G, Lüscher B. Phosphorylation sites mapping in the N-terminal domain of c-myc modulate its transforming potential. Oncogene. 1993;8:3199–3209. [PubMed] [Google Scholar]
- Herbst A, Salghetti SE, Kim SY, Tansey WP. Multiple cell-type-specific elements regulate Myc protein stability. Oncogene. 2004;23:3863–3871. doi: 10.1038/sj.onc.1207492. [DOI] [PubMed] [Google Scholar]
- Herold S, Hock A, Herkert B, Berns K, Mullenders J, Beijersbergen R, et al. Miz1 and HectH9 regulate the stability of the checkpoint protein, TopBP1. EMBO J. 2008;27:2851–2861. doi: 10.1038/emboj.2008.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold S, Herkert B, Eilers M. Facilitating replication under stress: an oncogenic function of MYC? Nat Rev Cancer. 2009;9:441–444. doi: 10.1038/nrc2640. [DOI] [PubMed] [Google Scholar]
- Hirning U, Schmid P, Schulz WA, Rettenberger G, Hameister H. A comparative analysis of N-myc and c-myc expression and cellular proliferation in mouse organogenesis. Mech Dev. 1991;33:119–125. doi: 10.1016/0925-4773(91)90078-k. [DOI] [PubMed] [Google Scholar]
- Hirvonen H, Mäkelä TP, Sandberg M, Kalimo H, Vuorio E, Alitalo K. Expression of the myc proto-oncogenes in developing human fetal brain. Oncogene. 1990;5:1787–1797. [PubMed] [Google Scholar]
- Ho R, Eggert A, Hishiki T, Minturn JE, Ikegaki N, Foster P, et al. Resistance to chemotherapy mediated by TrkB in neuroblastomas. Cancer Res. 2002;62:6462–6466. [PubMed] [Google Scholar]
- Hoffman B, Liebermann DA. Apoptotic signaling by c-MYC. Oncogene. 2008;27:6462–6472. doi: 10.1038/onc.2008.312. [DOI] [PubMed] [Google Scholar]
- Hossain MS, Ozaki T, Wang H, Nakagawa A, Takenobu H, Ohira M, et al. N-MYC promotes cell proliferation through a direct transactivation of neuronal leucine-rich repeat protein-1 (NLRR1) gene in neuroblastoma. Oncogene. 2008;27:6075–6082. doi: 10.1038/onc.2008.200. [DOI] [PubMed] [Google Scholar]
- Houghton PJ, Morton CL, Kolb EA, Gorlick R, Lock R, Carol H, et al. Initial testing (stage 1) of the mTOR inhibitor rapamycin by the pediatric preclinical testing program. Pediatr Blood Cancer. 2008;50:799–805. doi: 10.1002/pbc.21296. [DOI] [PubMed] [Google Scholar]
- Hurlin PJ, Quéva C, Koskinen PJ, Steingrímsson E, Ayer DE, Copeland NG, et al. Mad3 and Mad4: novel Max-interacting transcriptional repressors that suppress c-myc dependent transformation and are expressed during neural and epidermal differentiation. EMBO J. 1995;14:5646–5659. doi: 10.1002/j.1460-2075.1995.tb00252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurlin PJ, Quéva C, Eisenman RN. Mnt, a novel Max-interacting protein is coexpressed with Myc in proliferating cells and mediates repression at Myc binding sites. Genes Dev. 1997;11:44–58. doi: 10.1101/gad.11.1.44. [DOI] [PubMed] [Google Scholar]
- Ikegaki N, Gotoh T, Kung B, Riceberg JS, Kim DY, Zhao H, et al. De novo identification of MIZ-1 (ZBTB17) encoding a MYC-interacting zinc-finger protein as a new favorable neuroblastoma gene. Clin Cancer Res. 2007;13:6001–6009. doi: 10.1158/1078-0432.CCR-07-0071. [DOI] [PubMed] [Google Scholar]
- Inge TH, Casson LK, Priebe W, Trent JO, Georgeson KE, Miller DM, et al. Importance of Sp1 consensus motifs in the MYCN promoter. Surgery. 2002;132:232–238. doi: 10.1067/msy.2002.125387. [DOI] [PubMed] [Google Scholar]
- Jacobs JFM, van Bokhoven H, van Leeuwen FN, Hulsbergen-van de Kaa CA, de Vries IJM, Adema GJ, et al. Regulation of MYCN expression in human neuroblastoma cells. BMC Cancer. 2009;9:239. doi: 10.1186/1471-2407-9-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janoueix-Lerosey I, Lequin D, Brugières L, Ribeiro A, de Pontual L, Combaret V, et al. Somatic and germline activating mutations of the ALK kinase receptor in neuroblastoma. Nature. 2008;455:967–970. doi: 10.1038/nature07398. [DOI] [PubMed] [Google Scholar]
- Johnsen JI, Segerström L, Orrego A, Elfman L, Henriksson M, Kågedal B, et al. Inhibitors of mammalian target of rapamycin downregulate MYCN protein expression and inhibit neuroblastoma growth in vitro and in vivo. Oncogene. 2008;27:2910–2922. doi: 10.1038/sj.onc.1210938. [DOI] [PubMed] [Google Scholar]
- Kamijo T, Weber JD, Zambetti G, Zindy F, Roussel MF, Sherr CJ. Functional and physical interactions of the ARF tumor suppressor with p53 and Mdm2. Proc Natl Acad Sci USA. 1998;95:8292–8297. doi: 10.1073/pnas.95.14.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanemaru KK, Tuthill MC, Takeuchi KK, Sidell N, Wada RK. Retinoic acid induced downregulation of MYCN is not mediated through changes in Sp1/Sp3. Pediatr Blood Cancer. 2008;50:806–811. doi: 10.1002/pbc.21273. [DOI] [PubMed] [Google Scholar]
- Kang J, Rychahou PG, Ishola TA, Mourot JM, Evers BM, Chung DH. N-myc is a novel regulator of PI3K-mediated VEGF expression in neuroblastoma. Oncogene. 2008;27:3999–4007. doi: 10.1038/onc.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato GJ, Lee WM, Chen LL, Dang CV. Max: functional domains and interaction with c-Myc. Genes Dev. 1992;6:81–92. doi: 10.1101/gad.6.1.81. [DOI] [PubMed] [Google Scholar]
- Kenney AM, Cole MD, Rowitch DH. Nmyc upregulation by sonic hedgehog signaling promotes proliferation in developing cerebellar granule neuron precursors. Development. 2003;130:15–28. doi: 10.1242/dev.00182. [DOI] [PubMed] [Google Scholar]
- Kenney AM, Widlund HR, Rowitch DH. Hedgehog and PI-3 kinase signaling converge on Nmyc1 to promote cell cycle progression in cerebellar neuronal precursors. Development. 2004;131:217–228. doi: 10.1242/dev.00891. [DOI] [PubMed] [Google Scholar]
- Keshelava N, Seeger RC, Reynolds CP. Drug resistance in human neuroblastoma cell lines correlates with clinical therapy. Eur J Cancer. 1997;33:2002–2006. doi: 10.1016/s0959-8049(97)00213-x. [DOI] [PubMed] [Google Scholar]
- Keshelava N, Seeger RC, Groshen S, Reynolds CP. Drug resistance patterns of human neuroblastoma cell lines derived from patients at different phases of therapy. Cancer Res. 1998;58:5396–5405. [PubMed] [Google Scholar]
- Keshelava N, Zuo JJ, Waidyaratne NS, Triche TJ, Reynolds CP. p53 mutations and loss of p53 function confer multidrug resistance in neuroblastoma. Med Pediatr Oncol. 2000;35:563–568. doi: 10.1002/1096-911x(20001201)35:6<563::aid-mpo15>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- Kim J, Lee J-h, Iyer VR. Global identification of Myc target genes reveals its direct role in mitochondrial biogenesis and its E-box usage in vivo. PLoS One. 2008;3:e1798. doi: 10.1371/journal.pone.0001798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoepfler PS, Cheng PF, Eisenman RN. N-myc is essential during neurogenesis for the rapid expansion of progenitor cell populations and the inhibition of neuronal differentiation. Genes Dev. 2002;16:2699–2712. doi: 10.1101/gad.1021202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppen A, Ait-Aissa R, Hopman S, Koster J, Haneveld F, Versteeg R, et al. Dickkopf-1 is down-regulated by MYCN and inhibits neuroblastoma cell proliferation. Cancer Lett. 2007;256:218–228. doi: 10.1016/j.canlet.2007.06.011. [DOI] [PubMed] [Google Scholar]
- Kramps C, Strieder V, Sapetschnig A, Suske G, Lutz W. E2F and Sp1/Sp3 Synergize but are not sufficient to activate the MYCN gene in neuroblastomas. J Biol Chem. 2004;279:5110–5117. doi: 10.1074/jbc.M304758200. [DOI] [PubMed] [Google Scholar]
- Kretzner L, Blackwood EM, Eisenman RN. Myc and Max proteins possess distinct transcriptional activities. Nature. 1992;359:426–429. doi: 10.1038/359426a0. [DOI] [PubMed] [Google Scholar]
- Krystal GW, Armstrong BC, Battey JF. N-myc mRNA forms an RNA-RNA duplex with endogenous antisense transcripts. Mol Cell Biol. 1990;10:4180–4191. doi: 10.1128/mcb.10.8.4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Morris SW. Development of anaplastic lymphoma kinase (ALK) small-molecule inhibitors for cancer therapy. Med Res Rev. 2008;28:372–412. doi: 10.1002/med.20109. [DOI] [PubMed] [Google Scholar]
- Li Z, Van Calcar S, Qu C, Cavenee WK, Zhang MQ, Ren B. A global transcriptional regulatory role for c-Myc in Burkitt’s lymphoma cells. Proc Natl Acad Sci USA. 2003;100:8164–8169. doi: 10.1073/pnas.1332764100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Hann S. The Myc-nucleophosmin-ARF network: a complex web unveiled. Cell Cycle. 2009;8:2703–2707. doi: 10.4161/cc.8.17.9418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin AW, Lowe SW. Oncogenic ras activates the ARF-p53 pathway to suppress epithelial cell transformation. Proc Natl Acad Sci USA. 2001;98:5025–5030. doi: 10.1073/pnas.091100298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Liu PY, Tee AEL, Haber M, Norris MD, Gleave ME, et al. Over-expression of clusterin is a resistance factor to the anticancer effect of histone deacetylase inhibitors. Eur J Cancer. 2009;45:1846–1854. doi: 10.1016/j.ejca.2009.03.002. [DOI] [PubMed] [Google Scholar]
- Lutterbach B, Hann SR. Hierarchical phosphorylation at N-terminal transformation-sensitive sites in c-Myc protein is regulated by mitogens and in mitosis. Mol Cell Biol. 1994;14:5510–5522. doi: 10.1128/mcb.14.8.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutterbach B, Hann SR. c-Myc transactivation domain-associated kinases: questionable role for map kinases in c-Myc phosphorylation. J Cell Biochem. 1999;72:483–491. doi: 10.1002/(sici)1097-4644(19990315)72:4<483::aid-jcb4>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009;10:307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maestro R, Dei Tos AP, Hamamori Y, Krasnokutsky S, Sartorelli V, Kedes L, et al. Twist is a potential oncogene that inhibits apoptosis. Genes Dev. 1999;13:2207–2217. doi: 10.1101/gad.13.17.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manohar CF, Bray JA, Salwen HR, Madafiglio J, Cheng A, Flemming C, et al. MYCN-mediated regulation of the MRP1 promoter in human neuroblastoma. Oncogene. 2004;23:753–762. doi: 10.1038/sj.onc.1207151. [DOI] [PubMed] [Google Scholar]
- Mao DYL, Watson JD, Yan PS, Barsyte-Lovejoy D, Khosravi F, Wong WW-L, et al. Analysis of Myc bound loci identified by CpG island arrays shows that Max is essential for Myc-dependent repression. Curr Biol. 2003;13:882–886. doi: 10.1016/s0960-9822(03)00297-5. [DOI] [PubMed] [Google Scholar]
- Maris JM, Courtright J, Houghton PJ, Morton CL, Kolb EA, Lock R, et al. Initial testing (stage 1) of sunitinib by the pediatric preclinical testing program. Pediatr Blood Cancer. 2008a;51:42–48. doi: 10.1002/pbc.21535. [DOI] [PubMed] [Google Scholar]
- Maris JM, Mosse YP, Bradfield JP, Hou C, Monni S, Scott RH, et al. Chromosome 6p22 locus associated with clinically aggressive neuroblastoma. N Engl J Med. 2008b;358:2585–2593. doi: 10.1056/NEJMoa0708698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maris JM. Unholy matrimony: Aurora A and N-Myc as malignant partners in neuroblastoma. Cancer Cell. 2009;15:5–6. doi: 10.1016/j.ccr.2008.12.008. [DOI] [PubMed] [Google Scholar]
- Marqués M, Kumar A, Cortés I, Gonzalez-García A, Hernández C, Moreno-Ortiz MC, et al. Phosphoinositide 3-kinases p110alpha and p110beta regulate cell cycle entry, exhibiting distinct activation kinetics in G1 phase. Mol Cell Biol. 2008;28:2803–2814. doi: 10.1128/MCB.01786-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinato F, Cesaroni M, Amati B, Guccione E. Analysis of Myc-induced histone modifications on target chromatin. PLoS One. 2008;3:e3650. doi: 10.1371/journal.pone.0003650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins RAP, Zindy F, Donovan S, Zhang J, Pounds S, Wey A, et al. N-myc coordinates retinal growth with eye size during mouse development. Genes Dev. 2008;22:179–193. doi: 10.1101/gad.1608008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzec M, Kasprzycka M, Liu X, El-Salem M, Halasa K, Raghunath PN, et al. Oncogenic tyrosine kinase NPM/ALK induces activation of the rapamycin-sensitive mTOR signaling pathway. Oncogene. 2007;26:5606–5614. doi: 10.1038/sj.onc.1210346. [DOI] [PubMed] [Google Scholar]
- McDermott U, Iafrate AJ, Gray NS, Shioda T, Classon M, Maheswaran S, et al. Genomic alterations of anaplastic lymphoma kinase may sensitize tumors to anaplastic lymphoma kinase inhibitors. Cancer Res. 2008;68:3389–3395. doi: 10.1158/0008-5472.CAN-07-6186. [DOI] [PubMed] [Google Scholar]
- McMahon SB, Wood MA, Cole MD. The essential cofactor TRRAP recruits the histone acetyltransferase hGCN5 to c-Myc. Mol Cell Biol. 2000;20:556–562. doi: 10.1128/mcb.20.2.556-562.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memmott RM, Dennis PA. Akt-dependent and -independent mechanisms of mTOR regulation in cancer. Cell Signal. 2009;21:656–664. doi: 10.1016/j.cellsig.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer. 2008;8:976–990. doi: 10.1038/nrc2231. [DOI] [PubMed] [Google Scholar]
- Midgley CA, Desterro JM, Saville MK, Howard S, Sparks A, Hay RT, et al. An N-terminal p14ARF peptide blocks Mdm2-dependent ubiquitination in vitro and can activate p53 in vivo. Oncogene. 2000;19:2312–2323. doi: 10.1038/sj.onc.1203593. [DOI] [PubMed] [Google Scholar]
- Mill P, Mo R, Hu MC, Dagnino L, Rosenblum ND, Hui C-C. Shh controls epithelial proliferation via independent pathways that converge on N-Myc. Dev Cell. 2005;9:293–303. doi: 10.1016/j.devcel.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Moore HC, Wood KM, Jackson MS, Lastowska MA, Hall D, Imrie H, et al. Histological profile of tumours from MYCN transgenic mice. J Clin Pathol. 2008;61:1098–1103. doi: 10.1136/jcp.2007.054627. [DOI] [PubMed] [Google Scholar]
- Mossé Y, Wood A, Maris J. Inhibition of ALK signaling for cancer therapy. Clin Cancer Res. 2009;15:5609–5614. doi: 10.1158/1078-0432.CCR-08-2762. [DOI] [PubMed] [Google Scholar]
- Mosse YP, Laudenslager M, Khazi D, Carlisle AJ, Winter CL, Rappaport E, et al. Germline PHOX2B mutation in hereditary neuroblastoma. Am J Hum Genet. 2004;75:727–730. doi: 10.1086/424530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossé YP, Laudenslager M, Longo L, Cole KA, Wood A, Attiyeh EF, et al. Identification of ALK as a major familial neuroblastoma predisposition gene. Nature. 2008;455:930–935. doi: 10.1038/nature07261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugrauer G, Alt FW, Ekblom P. N-myc proto-oncogene expression during organogenesis in the developing mouse as revealed by in situ hybridization. J Cell Biol. 1988;107:1325–1335. doi: 10.1083/jcb.107.4.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy DJ, Junttila MR, Pouyet L, Karnezis A, Shchors K, Bui DA, et al. Distinct thresholds govern Myc’s biological output in vivo. Cancer Cell. 2008;14:447–457. doi: 10.1016/j.ccr.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris MD, Burkhart CA, Marshall GM, Weiss WA, Haber M. Expression of N-myc and MRP genes and their relationship to N-myc gene dosage and tumor formation in a murine neuroblastoma model. Med Pediatr Oncol. 2000;35:585–589. doi: 10.1002/1096-911x(20001201)35:6<585::aid-mpo20>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- Oberthuer A, Berthold F, Warnat P, Hero B, Kahlert Y, Spitz R, et al. Customized oligonucleotide microarray gene expression-based classification of neuroblastoma patients outperforms current clinical risk stratification. J Clin Oncol. 2006;24:5070–5078. doi: 10.1200/JCO.2006.06.1879. [DOI] [PubMed] [Google Scholar]
- Ohira M, Oba S, Nakamura Y, Isogai E, Kaneko S, Nakagawa A, et al. Expression profiling using a tumor-specific cDNA microarray predicts the prognosis of intermediate risk neuroblastomas. Cancer Cell. 2005;7:337–350. doi: 10.1016/j.ccr.2005.03.019. [DOI] [PubMed] [Google Scholar]
- Okubo T, Knoepfler PS, Eisenman RN, Hogan BLM. Nmyc plays an essential role during lung development as a dosage-sensitive regulator of progenitor cell proliferation and differentiation. Development. 2005;132:1363–1374. doi: 10.1242/dev.01678. [DOI] [PubMed] [Google Scholar]
- Oliner JD, Pietenpol JA, Thiagalingam S, Gyuris J, Kinzler KW, Vogelstein B. Oncoprotein MDM2 conceals the activation domain of tumour suppressor p53. Nature. 1993;362:857–860. doi: 10.1038/362857a0. [DOI] [PubMed] [Google Scholar]
- Oliver TG, Grasfeder LL, Carroll AL, Kaiser C, Gillingham CL, Lin SM, et al. Transcriptional profiling of the Sonic hedgehog response: a critical role for N-myc in proliferation of neuronal precursors. Proc Natl Acad Sci USA. 2003;100:7331–7336. doi: 10.1073/pnas.0832317100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opel D, Poremba C, Simon T, Debatin K-M, Fulda S. Activation of Akt predicts poor outcome in neuroblastoma. Cancer Res. 2007;67:735–745. doi: 10.1158/0008-5472.CAN-06-2201. [DOI] [PubMed] [Google Scholar]
- Ota S, Zhou Z-Q, Keene DR, Knoepfler P, Hurlin PJ. Activities of N-Myc in the developing limb link control of skeletal size with digit separation. Development. 2007;134:1583–1592. doi: 10.1242/dev.000703. [DOI] [PubMed] [Google Scholar]
- Otto T, Horn S, Brockmann M, Eilers U, Schüttrumpf L, Popov N, et al. Stabilization of N-Myc is a critical function of Aurora A in human neuroblastoma. Cancer Cell. 2009;15:67–78. doi: 10.1016/j.ccr.2008.12.005. [DOI] [PubMed] [Google Scholar]
- Paffhausen T, Schwab M, Westermann F. Targeted MYCN expression affects cytotoxic potential of chemotherapeutic drugs in neuroblastoma cells. Cancer Lett. 2007;250:17–24. doi: 10.1016/j.canlet.2006.09.010. [DOI] [PubMed] [Google Scholar]
- Park JR, Eggert A, Caron H. Neuroblastoma: biology, prognosis, and treatment. Pediatr Clin North Am. 2008;55:97–120. doi: 10.1016/j.pcl.2007.10.014. [DOI] [PubMed] [Google Scholar]
- Patel JH, Loboda AP, Showe MK, Showe LC, McMahon SB. Analysis of genomic targets reveals complex functions of MYC. Nat Rev Cancer. 2004;4:562–568. doi: 10.1038/nrc1393. [DOI] [PubMed] [Google Scholar]
- Payne GS, Bishop JM, Varmus HE. Multiple arrangements of viral DNA and an activated host oncogene in bursal lymphomas. Nature. 1982;295:209–214. doi: 10.1038/295209a0. [DOI] [PubMed] [Google Scholar]
- Peterson RT, Desai BN, Hardwick JS, Schreiber SL. Protein phosphatase 2A interacts with the 70-kDa S6 kinase and is activated by inhibition of FKBP12-rapamycinassociated protein. Proc Natl Acad Sci USA. 1999;96:4438–4442. doi: 10.1073/pnas.96.8.4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prendergast GC, Lawe D, Ziff EB. Association of Myn, the murine homolog of max, with c-Myc stimulates methylation-sensitive DNA binding and ras cotransformation. Cell. 1991;65:395–407. doi: 10.1016/0092-8674(91)90457-a. [DOI] [PubMed] [Google Scholar]
- Pulverer BJ, Fisher C, Vousden K, Littlewood T, Evan G, Woodgett JR. Site-specific modulation of c-Myc cotransformation by residues phosphorylated in vivo. Oncogene. 1994;9:59–70. [PubMed] [Google Scholar]
- Qi Y, Gregory MA, Li Z, Brousal JP, West K, Hann SR. p19ARF directly and differentially controls the functions of c-Myc independently of p53. Nature. 2004;431:712–717. doi: 10.1038/nature02958. [DOI] [PubMed] [Google Scholar]
- Raabe EH, Laudenslager M, Winter C, Wasserman N, Cole K, Laquaglia M, et al. Prevalence and functional consequence of PHOX2B mutations in neuroblastoma. Oncogene. 2008;27:469–476. doi: 10.1038/sj.onc.1210659. [DOI] [PubMed] [Google Scholar]
- Riley RD, Heney D, Jones DR, Sutton AJ, Lambert PC, Abrams KR, et al. A systematic review of molecular and biological tumor markers in neuroblastoma. Clin Cancer Res. 2004;10:4–12. doi: 10.1158/1078-0432.ccr-1051-2. [DOI] [PubMed] [Google Scholar]
- Rohrer T, Trachsel D, Engelcke G, Hammer J. Congenital central hypoventilation syndrome associated with Hirschsprung’s disease and neuroblastoma: case of multiple neurocristopathies. Pediatr Pulmonol. 2002;33:71–76. doi: 10.1002/ppul.10031. [DOI] [PubMed] [Google Scholar]
- Ruggero D. The Role of Myc-Induced Protein Synthesis in Cancer. Cancer Res. 2009;69:8839–8843. doi: 10.1158/0008-5472.CAN-09-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saksela K, Mäkelä TP, Hughes K, Woodgett JR, Alitalo K. Activation of protein kinase C increases phosphorylation of the L-myc trans-activator domain at a GSK-3 target site. Oncogene. 1992;7:347–353. [PubMed] [Google Scholar]
- Schramm A, Schulte JH, Klein-Hitpass L, Havers W, Sieverts H, Berwanger B, et al. Prediction of clinical outcome and biological characterization of neuroblastoma by expression profiling. Oncogene. 2005;24:7902–7912. doi: 10.1038/sj.onc.1208936. [DOI] [PubMed] [Google Scholar]
- Schramm A, Mierswa I, Kaderali L, Morik K, Eggert A, Schulte JH. Reanalysis of neuroblastoma expression profiling data using improved methodology and extended follow-up increases validity of outcome prediction. Cancer Lett. 2009;282:55–62. doi: 10.1016/j.canlet.2009.02.052. [DOI] [PubMed] [Google Scholar]
- Schulte JH, Horn S, Schlierf S, Schramm A, Heukamp LC, Christiansen H, et al. MicroRNAs in the pathogenesis of neuroblastoma. Cancer Lett. 2009;274:10–15. doi: 10.1016/j.canlet.2008.06.010. [DOI] [PubMed] [Google Scholar]
- Sears R, Leone G, DeGregori J, Nevins JR. Ras enhances Myc protein stability. Mol Cell. 1999;3:169–179. doi: 10.1016/s1097-2765(00)80308-1. [DOI] [PubMed] [Google Scholar]
- Sears R, Nuckolls F, Haura E, Taya Y, Tamai K, Nevins JR. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev. 2000;14:2501–2514. doi: 10.1101/gad.836800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger RC, Brodeur GM, Sather H, Dalton A, Siegel SE, Wong KY, et al. Association of multiple copies of the N-myc oncogene with rapid progression of neuroblastomas. N Engl J Med. 1985;313:1111–1116. doi: 10.1056/NEJM198510313131802. [DOI] [PubMed] [Google Scholar]
- Seoane J, Pouponnot C, Staller P, Schader M, Eilers M, Massagué J. TGFbeta influences Myc, Miz-1 and Smad to control the CDK inhibitor p15INK4b. Nat Cell Biol. 2001;3:400–408. doi: 10.1038/35070086. [DOI] [PubMed] [Google Scholar]
- Shang X, Burlingame S, Okcu M, Ge N, Russell H, Egler R, et al. Aurora A is a negative prognostic factor and a new therapeutic target in human neuroblastoma. Mol Cancer Ther. 2009;8:2461–2469. doi: 10.1158/1535-7163.MCT-08-0857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Glynn JM, Guilbert LJ, Cotter TG, Bissonnette RP, Green DR. Role for c-myc in activation-induced apoptotic cell death in T cell hybridomas. Science. 1992;257:212–214. doi: 10.1126/science.1378649. [DOI] [PubMed] [Google Scholar]
- Sjostrom SK, Finn G, Hahn WC, Rowitch DH, Kenney AM. The Cdk1 complex plays a prime role in regulating N-myc phosphorylation and turnover in neural precursors. Dev Cell. 2005;9:327–338. doi: 10.1016/j.devcel.2005.07.014. [DOI] [PubMed] [Google Scholar]
- Slack A, Chen Z, Tonelli R, Pule M, Hunt L, Pession A, et al. The p53 regulatory gene MDM2 is a direct transcriptional target of MYCN in neuroblastoma. Proc Natl Acad Sci USA. 2005a;102:731–736. doi: 10.1073/pnas.0405495102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack A, Lozano G, Shohet JM. MDM2 as MYCN transcriptional target: implications for neuroblastoma pathogenesis. Cancer Lett. 2005b;228:21–27. doi: 10.1016/j.canlet.2005.01.050. [DOI] [PubMed] [Google Scholar]
- Smith-Sørensen B, Hijmans EM, Beijersbergen RL, Bernards R. Functional analysis of Burkitt’s lymphoma mutant c-Myc proteins. J Biol Chem. 1996;271:5513–5518. doi: 10.1074/jbc.271.10.5513. [DOI] [PubMed] [Google Scholar]
- Staller P, Peukert K, Kiermaier A, Seoane J, Lukas J, Karsunky H, et al. Repression of p15INK4b expression by Myc through association with Miz-1. Nat Cell Biol. 2001;3:392–399. doi: 10.1038/35070076. [DOI] [PubMed] [Google Scholar]
- Stupack DG, Teitz T, Potter MD, Mikolon D, Houghton PJ, Kidd VJ, et al. Potentiation of neuroblastoma metastasis by loss of caspase-8. Nature. 2006;439:95–99. doi: 10.1038/nature04323. [DOI] [PubMed] [Google Scholar]
- Tang XX, Zhao H, Kung B, Kim DY, Hicks SL, Cohn SL, et al. The MYCN enigma: significance of MYCN expression in neuroblastoma. Cancer Res. 2006;66:2826–2833. doi: 10.1158/0008-5472.CAN-05-0854. [DOI] [PubMed] [Google Scholar]
- Thiele CJ, Reynolds CP, Israel MA. Decreased expression of N-myc precedes retinoic acid-induced morphological differentiation of human neuroblastoma. Nature. 1985;313:404–406. doi: 10.1038/313404a0. [DOI] [PubMed] [Google Scholar]
- Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, et al. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem. 2009;284:8023–8032. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thut CJ, Goodrich JA, Tjian R. Repression of p53-mediated transcription by MDM2: a dual mechanism. Genes Dev. 1997;11:1974–1986. doi: 10.1101/gad.11.15.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trochet D, O’Brien LM, Gozal D, Trang H, Nordenskjöld A, Laudier B, et al. PHOX2B genotype allows for prediction of tumor risk in congenital central hypoventilation syndrome. Am J Hum Genet. 2005;76:421–426. doi: 10.1086/428366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tweddle DA, Malcolm AJ, Bown N, Pearson AD, Lunec J. Evidence for the development of p53 mutations after cytotoxic therapy in a neuroblastoma cell line. Cancer Res. 2001;61:8–13. [PubMed] [Google Scholar]
- Ushmorov A, Hogarty MD, Liu X, Knauss H, Debatin KM, Beltinger C. N-myc augments death and attenuates protective effects of Bcl-2 in trophically stressed neuroblastoma cells. Oncogene. 2008;27:3424–3434. doi: 10.1038/sj.onc.1211017. [DOI] [PubMed] [Google Scholar]
- Valsesia-Wittmann S, Magdeleine M, Dupasquier S, Garin E, Jallas A-C, Combaret V, et al. Oncogenic cooperation between H-Twist and N-Myc overrides failsafe programs in cancer cells. Cancer Cell. 2004;6:625–630. doi: 10.1016/j.ccr.2004.09.033. [DOI] [PubMed] [Google Scholar]
- Van Maerken T, Ferdinande L, Taildeman J, Lambertz I, Yigit N, Vercruysse L, et al. Antitumor activity of the selective MDM2 antagonist Nutlin-3 against chemoresistant neuroblastoma with wild-type p53. J Natl Cancer Inst. 2009a;101:1562–1574. doi: 10.1093/jnci/djp355. [DOI] [PubMed] [Google Scholar]
- Van Maerken T, Vandesompele J, Rihani A, De Paepe A, Speleman F. Escape from p53-mediated tumor surveillance in neuroblastoma: switching off the p14(ARF)-MDM2-p53 axis. Cell Death Differ. 2009b;16:1563–1572. doi: 10.1038/cdd.2009.138. [DOI] [PubMed] [Google Scholar]
- Vermeulen J, De Preter K, Naranjo A, Vercruysse L, Van Roy N, Hellemans J, et al. Predicting outcomes for children with neuroblastoma using a multigene-expression signature: a retrospective SIOPEN/COG/GPOH study. Lancet Oncol. 2009;10:663–671. doi: 10.1016/S1470-2045(09)70154-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vervoorts J, Lüscher-Firzlaff JM, Rottmann S, Lilischkis R, Walsemann G, Dohmann K, et al. Stimulation of c-MYC transcriptional activity and acetylation by recruitment of the cofactor CBP. EMBO Rep. 2003;4:484–490. doi: 10.1038/sj.embor.embor821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogan K, Bernstein M, Leclerc JM, Brisson L, Brossard J, Brodeur GM, et al. Absence of p53 gene mutations in primary neuroblastomas. Cancer Res. 1993;53:5269–5273. [PubMed] [Google Scholar]
- Wagner LM, Danks MK. New therapeutic targets for the treatment of high-risk neuroblastoma. J Cell Biochem. 2009;107:46–57. doi: 10.1002/jcb.22094. [DOI] [PubMed] [Google Scholar]
- Walkley CR, Fero ML, Chien W-M, Purton LE, McArthur GA. Negative cell-cycle regulators cooperatively control self-renewal and differentiation of haematopoietic stem cells. Nat Cell Biol. 2005;7:172–178. doi: 10.1038/ncb1214. [DOI] [PubMed] [Google Scholar]
- Wang W, Kim S-H, El-Deiry WS. Small-molecule modulators of p53 family signaling and antitumor effects in p53-deficient human colon tumor xenografts. Proc Natl Acad Sci USA. 2006;103:11003–11008. doi: 10.1073/pnas.0604507103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber JD, Taylor LJ, Roussel MF, Sherr CJ, Bar-Sagi D. Nucleolar Arf sequesters Mdm2 and activates p53. Nat Cell Biol. 1999;1:20–26. doi: 10.1038/8991. [DOI] [PubMed] [Google Scholar]
- Weiss WA, Godfrey T, Francisco C, Bishop JM. Genome-wide screen for allelic imbalance in a mouse model for neuroblastoma. Cancer Res. 2000;60:2483–2487. [PubMed] [Google Scholar]
- Welcker M, Orian A, Jin J, Grim JE, Grim JA, Harper JW, et al. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc Natl Acad Sci USA. 2004;101:9085–9090. doi: 10.1073/pnas.0402770101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann F, Muth D, Benner A, Bauer T, Henrich K, Oberthuer A, et al. Distinct transcriptional MYCN/c-MYC activities are associated with spontaneous regression or malignant progression in neuroblastomas. Genome Biol. 2008;9:R150. doi: 10.1186/gb-2008-9-10-r150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead KA, Langer R, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witt O, Deubzer HE, Lodrini M, Milde T, Oehme I. Targeting histone deacetylases in neuroblastoma. Curr Pharm Des. 2009;15:436–447. doi: 10.2174/138161209787315774. [DOI] [PubMed] [Google Scholar]
- Xu XL, Fang Y, Lee TC, Forrest D, Gregory-Evans C, Almeida D, et al. Retinoblastoma has properties of a cone precursor tumor and depends upon cone-specific MDM2 signaling. Cell. 2009;137:1018–1031. doi: 10.1016/j.cell.2009.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue C, Haber M, Flemming C, Marshall GM, Lock RB, MacKenzie KL, et al. p53 determines multidrug sensitivity of childhood neuroblastoma. Cancer Res. 2007;67:10351–10360. doi: 10.1158/0008-5472.CAN-06-4345. [DOI] [PubMed] [Google Scholar]
- Yada M, Hatakeyama S, Kamura T, Nishiyama M, Tsunematsu R, Imaki H, et al. Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. EMBO J. 2004;23:2116–2125. doi: 10.1038/sj.emboj.7600217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E, Cunningham M, Arnold H, Chasse D, Monteith T, Ivaldi G, et al. A signalling pathway controlling c-Myc degradation that impacts oncogenic transformation of human cells. Nat Cell Biol. 2004;6:308–318. doi: 10.1038/ncb1110. [DOI] [PubMed] [Google Scholar]
- Yin X, Giap C, Lazo JS, Prochownik EV. Low molecular weight inhibitors of Myc-Max interaction and function. Oncogene. 2003;22:6151–6159. doi: 10.1038/sj.onc.1206641. [DOI] [PubMed] [Google Scholar]
- Yu K, Toral-Barza L, Shi C, Zhang W-G, Lucas J, Shor B, et al. Biochemical, cellular, and in vivo activity of novel ATP-competitive and selective inhibitors of the mammalian target of rapamycin. Cancer Res. 2009;69:6232–6240. doi: 10.1158/0008-5472.CAN-09-0299. [DOI] [PubMed] [Google Scholar]
- Zask A, Verheijen J, Curran K, Kaplan J, Richard D, Nowak P, et al. ATP-competitive inhibitors of the mammalian target of rapamycin: design and synthesis of highly potent and selective pyrazolopyrimidines. J Med Chem. 2009;52:5013–5016. doi: 10.1021/jm900851f. [DOI] [PubMed] [Google Scholar]
- Zeller KI, Zhao X, Lee CWH, Chiu KP, Yao F, Yustein JT, et al. Global mapping of c-Myc binding sites and target gene networks in human B cells. Proc Natl Acad Sci USA. 2006;103:17834–17839. doi: 10.1073/pnas.0604129103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zervos AS, Gyuris J, Brent R. Mxi1, a protein that specifically interacts with Max to bind Myc-Max recognition sites. Cell. 1993;72:223–232. doi: 10.1016/0092-8674(93)90662-a. [DOI] [PubMed] [Google Scholar]
- Zimmerman KA, Yancopoulos GD, Collum RG, Smith RK, Kohl NE, Denis KA, et al. Differential expression of myc family genes during murine development. Nature. 1986;319:780–783. doi: 10.1038/319780a0. [DOI] [PubMed] [Google Scholar]
- Zindy F, Eischen CM, Randle DH, Kamijo T, Cleveland JL, Sherr CJ, et al. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 1998;12:2424–2433. doi: 10.1101/gad.12.15.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]