Abstract
Objectives
Measurement of transglutaminase autoantibodies (TGAA) is considered to be the most efficient single serologic test for celiac disease (CD) by the American Gastroenterological Association Institute. We hypothesized that a large international collaborative effort toward improving and standardizing TGAA measurement is both feasible and necessary. The primary aim of this workshop is to compare TGAA assays among various research and clinical laboratories and examine assay concordance and improve (and eventually standardize) the TGAA assay.
Methods
A total of 20 laboratories (5 commercial laboratories, 15 research and clinical laboratories) participated that included enzyme-linked immunosorbent assay (ELISA) and radiobinding assays. A total of 150 serum samples were distributed to each laboratory, with each laboratory receiving an equal aliquot that was coded and blinded, composed of 100 healthy control sera and 50 CD sera.
Results
Laboratory sensitivity ranged from 69% to 93% and specificity ranged from 96% to 100%. By receiver operator characteristic analysis, the area under the curve (C index) ranged from 0.9488 to 0.9904. When analyzing for linear correlation, r-squared was as high as 0.8882 but as low as 0.4244 for the celiac samples between different laboratories performing ELISA.
Conclusions
This transglutaminase autoantibody workshop allows for larger-scale international participation for the purposes of improving and eventually standardizing the TGAA assay with subsequent workshops.
Introduction
Since the initial 1997 report of the role of tissue transglutaminase in celiac disease (CD), nearly 600 articles have been published on transglutaminase autoantibodies (TGAA) in CD (1). Measurement of TGAA currently serves to (i) aid in the diagnosis of CD, (ii) identify individuals with the presence of celiac autoimmunity who are considered to be at risk of developing CD, and (iii) guide clinicians in managing patients with CD on a gluten-free diet. Furthermore, TGAA can serve as a means of selecting individuals for clinical trials aimed at disease prevention or natural history observational studies. In view of the increasing use and the increasing number of laboratories worldwide that offer TGAA testing, there is a clear need for standardization (2,3). Ideally, reference sera for developing and standardizing new assays using international units should be available to all laboratories.
A prior TG autoantibody workshop in 2000 led to the availability of standardized reference sera created by the European working group and helped standardize measurement of TGAA by enzyme-linked immunosorbent assay (ELISA) at that time (4). This current workshop format is different in that it seeks not to standardize certain aspects of the assay procedure (although it may be necessary in the future), but to allow each participating laboratory to perform and report the assay in their usual fashion, so that comparison of different assay formats is possible. The data are shared with all participating laboratories to allow each laboratory to independently improve their assay based on our shared findings.
It has previously been suggested that TGAA measurement by radiobinding assay (RBA) is more sensitive and specific than ELISA (5). Autoantibody workshops in type 1 diabetes have shown that RBA is superior to ELISA in measuring the major diabetes autoantibodies of insulin and IA-2 (6). On the other hand, ELISA assays measuring glutamic acid decarboxylase autoantibodies currently perform better in these workshops, but few studies have used these ELISAs in longitudinal studies to correlate with disease development. RBAs have also been used in systemic lupus erythematosis to measure anti-dsDNA (7) and anti-RNP autoantibodies (8). Furthermore, RBA has also been used as a molecular-based method of detecting autoantibodies against cytochrome CYP2D6 (also known as anti-liver-kidney microsomal antibodies) in autoimmune hepatitis (9) reportedly with better sensitivity. The aim of the current workshop is to provide feedback to a participating laboratory in terms of assay performance, and to allow for individual laboratory comparison with other laboratory peers. Secondary aims include examining assay concordance (to eventually facilitate standardization efforts) and comparing the fluid-phase RBA to the solid-phase ELISA assays, as part of an effort to identify the best-performing assay formats. Commercial laboratories do not generally perform RBA due to the necessity of handling radioactive waste products and because prior assay formats were prohibitive for large-scale use. However, newer assay formats allow high-throughput analysis on a 96-well assay plate, and have virtually become the standard for measuring certain autoantibodies. We hypothesized that a large international collaborative effort is feasible and could lead to improvement and eventual standardization of TGAA assays.
Methods
Participants
A total of 20 laboratories (5 commercial laboratories, 15 research and clinical laboratories) participated (see APPENDIX for list of participating laboratories). The 15 laboratories performed measurements using 16 ELISAs and 5 RBAs. Nine of the assays used commercial ELISA kits performed in their own institution, one laboratory utilized a modified version of a commercial kit and one laboratory used an in-house ELISA. The five commercial producers performed their own ELISA assay. All results from participating laboratories were de-identified and coded as Labs A through T to ensure confidentiality. All laboratories performed their assay and reported the results in their usual fashion, with their standard interpretation as POSITIVE or NEGATIVE. Some laboratories also reported some results as “indeterminate” or “indifferent,” and those results were considered NEGATIVE in the final analysis. Other information regarding general assay methods (i.e., laboratory cutoff for positivity, type of TG substrate, detection method, fluid-phase or solid-phase, etc.) was also provided by each laboratory.
Sera
Six laboratories contributed sera (2 ml) from individuals with CD as defined by accepted clinical criteria including small bowel biopsy, for the workshop: Liping Yu (University of Colorado), Claudio Tiberti (University of Rome), Daniel Agardh (Lund University), Markku Maki (University of Tampere), Olli Simell (University of Turku), and Ingrid van Hoogstraten (Vrije Universiteit Medical Center). One laboratory (Yu) contributed 20 samples, whereas the remaining 5 laboratories contributed a total of 21 samples. The healthy control sera were provided by Dr Patricia Mueller from the CDC, from individuals not known to have CD or type 1 diabetes. These control sera were originally used as negative controls for the Diabetes Antibody Standardization Program workshops, held in collaboration between the Immunology of Diabetes Society and the CDC (10). A total of 150 serum samples were distributed to each laboratory, with each laboratory receiving an equal aliquot (70–100 μl of serum) that was coded and blinded regarding clinical information, composed of 100 healthy control sera and 50 CD sera. The 50 CD sera included 30 samples from untreated patients, 11 samples from patients on a gluten-free diet (duration 2 months–2 years), 4 samples as replicates of one of the samples from the untreated patients, and 5 samples that were serial dilutions of one sample, at dilutions of 1:16, 1:32, 1:64, 1:128, and 1:256. Sera were diluted with sera from a healthy negative control sample. All sera were sent by express courier to each laboratory, with the sera randomized and coded 0 through 150. There was the potential for any sera in question to be sent back to a “reference” laboratory for testing, to control for any variables with shipping, but this was not needed during the study. Of the celiac sera, 28 samples were from children age 18 or less (the youngest was 3 years 9 months old), and 18 samples were from individuals also known to have type 1 diabetes.
Ethical consideration
All laboratories contributing sera were required to have IRB or appropriate review board approval for human research. This study was performed in compliance with all HIPAA regulations.
Analysis
All laboratories were aware of only their laboratory identifying code (A through T), and the results of all the assays were shared by all participating laboratories. There were three independent analyzers (EL, EB, MS) who then shared their analysis before presenting the final analysis to the entire group. Information regarding sensitivity, specificity, receiver operator characteristic curves, laboratory concordance, coefficient of variation, and limits of detection were included in the analysis and performed using GraphPad Prism version 4.0. Correlation is measured by Pearson's r-test.
Results
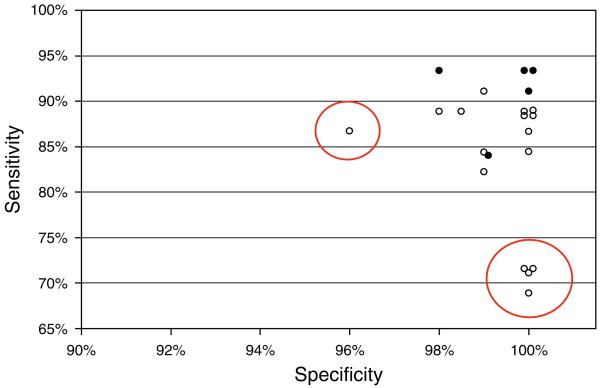
There was 100% participation and follow-up. All laboratories reported TGAA results in their own units and provided interpretation for each serum. The specificity ranged from 96% to 100%. The highest sensitivity (93%) with 100% specificity was achieved by two RBA. The sensitivity and specificity for each laboratory is shown in Figure 1 (for all sera excluding those that were serially diluted). We identified some laboratories with substantially lower sensitivity or specificity and were able to provide them with that information.
Figure 1.
Specificity and sensitivity for all 20 laboratories (21 assays) enrolled in the TGAA workshop. Empty symbols indicate ELISA assay, Solid symbols indicate RBA assay. Circled laboratories indicate assays that performed with lower specificity or sensitivity compared to the rest of the group.
The overall analysis is shown in Table 1, providing the sensitivity/specificity for each laboratory, as well as area under the curve by receiver operator characteristic analysis. Coefficients of variation (CV) from the five samples in replicate are provided, but the sera saturated the signals in some laboratory assays (reported values as “greater than X”), so that coefficients of variation could not be assessed for all laboratories.
Table 1. Overall laboratory performance, arranged by sensitivity.
| Lab | Assay | True positives | Sensitivity (%) | Specificity (%) | CV (intra-assay) | AUC |
|---|---|---|---|---|---|---|
| K | RBA | 42/45 | 93 | 100 | 10.9% | 0.9642 |
| P | RBA | 42/45 | 93 | 100 | 16.2% | 0.9612 |
| F | RBA | 42/45 | 93 | 98 | 7.3% | 0.982 |
| O2 | RBA | 41/45 | 91 | 100 | 3.1% | 0.975 |
| J | ELISA | 41/45 | 91 | 99 | 11.4% | 0.9798 |
| T | ELISA | 40/45 | 89 | 100 | NA | 0.9904 |
| G | ELISA | 40/45 | 89 | 100 | NA | 0.9884 |
| A | ELISA | 40/45 | 89 | 100 | NA | 0.9831 |
| E | ELISA | 40/45 | 89 | 100 | NA | 0.9751 |
| N | ELISA | 40/45 | 89 | 100 | NA | 0.9572 |
| Q | ELISA | 40/45 | 89 | 98 | 7.0% | 0.9638 |
| B | ELISA | 39/45 | 87 | 100 | 5.1% | 0.9872 |
| O1 | ELISA | 41/45 | 87 | 96 | 4.7% | 0.9607 |
| D | ELISA | 38/45 | 84 | 100 | 19.5% | 0.9801 |
| L | RBA | 38/45 | 84 | 99 | 12.7% | 0.9736 |
| I | ELISA | 38/45 | 84 | 99 | 26.1% | 0.9416 |
| M | ELISA | 37/45 | 82 | 99 | 12.4% | 0.9601 |
| S | ELISA | 32/45 | 71 | 100 | NA | 0.9824 |
| R | ELISA | 32/45 | 71 | 100 | 9.6% | 0.9663 |
| C | ELISA | 32/45 | 71 | 100 | 11.6% | 0.9631 |
| H | ELISA | 31/45 | 69 | 100 | NA | 0.9488 |
AUC, area under the curve; CV, coefficient of variation; ELISA, enzyme-linked immunosorbent assay; Lab, laboratory; NA, not applicable; RBA, radiobinding assay.
There were 11 sera from individuals reportedly on a gluten-free diet. Three sera from CD patients on gluten-free diet for either 2 months, 1 year, and 2 years duration, were undetectable by all laboratories. These three sera were the only CD sera reported negative by the three laboratories (Labs K, P, and F) with the highest sensitivity, all of which utilize the RBA. Therefore, no laboratory in this workshop had a detection rate greater than 42% of 45%, or 93% sensitivity. Lab P had also reported a modified version of their standard RBA (not included in this analysis), which did detect one of these three sera, and yielded a sensitivity with 95%, specificity of 100%, and an area under the curve of 0.987. Of the remaining eight sera from individuals on a gluten-free diet, one sample was reported as negative by 17 of the 21 assays, but all of the remaining samples were positive by all laboratories. RBA more frequently identified the sera from celiac patients as positive than ELISA. If TGAA titers do indeed decrease in individuals on a gluten-free diet, then differences in detecting TGAA in such sera could represent differences in assay sensitivity. On the other hand, it may be that some assays do not detect TGAA in gluten-free treated sera because of possible conformational changes in autoantibody specificity.
Figure 2 illustrates the frequency of which a particular laboratory reported a celiac sample as negative, as compared to all of the other participating laboratories.
Figure 2.
A graphic representation of all of the celiac sera (excluding the diluted samples) in the workshop, organized by laboratories performing ELISA (group on left, n=16) or RBA (group on right, n=5) in columns. All values are normalized to “1” by dividing the TGAA level by the assay's cutoff, such that a number >1 is interpreted as positive (clear box) and a number <1 is interpreted as negative (shaded box) by that assay. Sera are arranged from top to bottom in order of increasing normalized TGAA value according to the reference laboratory.
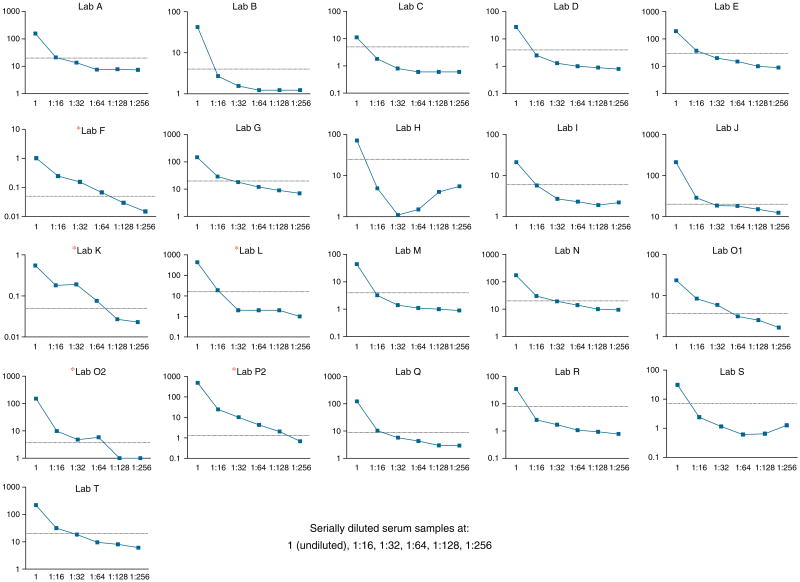
To assess limits of detection, serial dilutions were performed for a single sample. The limit of detection ranged from the 1:128 dilution to the 1:16 dilution. The RBA provided the highest sensitivity in ability to detect diluted samples, but none were able to detect sera diluted at 1:256 (Figure 3).
Figure 3.
Serial dilutions of a single celiac disease serum sample. Results were plotted for each individual laboratory, with the sample undiluted, and then diluted at 1:16, 1:32, 1:64, 1:128, and 1:256. RBA in this figure are indicated by the asterisk next to the laboratory code.
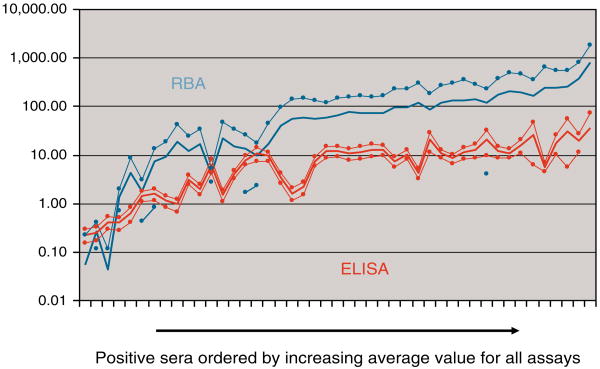
When the data are analyzed according to signal-to-noise ratio (dividing the signal for a positive test by the cutoff for the assay), the RBA in general provided the highest ratio of a positive signal relative to background. This is illustrated by showing the mean data and standard deviations for all positive sera expressed as a ratio of the signal divided by the assay cutoff, for RBA and ELISA assays (Figure 4). The ELISA assays were unable to provide further increased signal for high-titered sera, typically saturating at signals that were only 10 times the upper limit of normal. On the other hand, the RBA is able to continue to provide a steady increase in signal over 100 times the upper limit of normal for high-tittered sera. It is important to note, however, that the ELISA assays together provided less variation in signal-to-noise, suggesting that there is more “uniformity” among ELISAs as compared to RBA.
Figure 4.
The average of the ratios of signal-to-assay cutoff for ELISA and RBA were plotted in increasing order along the x axis for the 45 undiluted celiac disease sera. Sera were ordered by increasing average ratio for all assays along the x axis. RBA showed a wide confidence interval as (indicated by thin line), whereas ELISA showed a narrow interval.
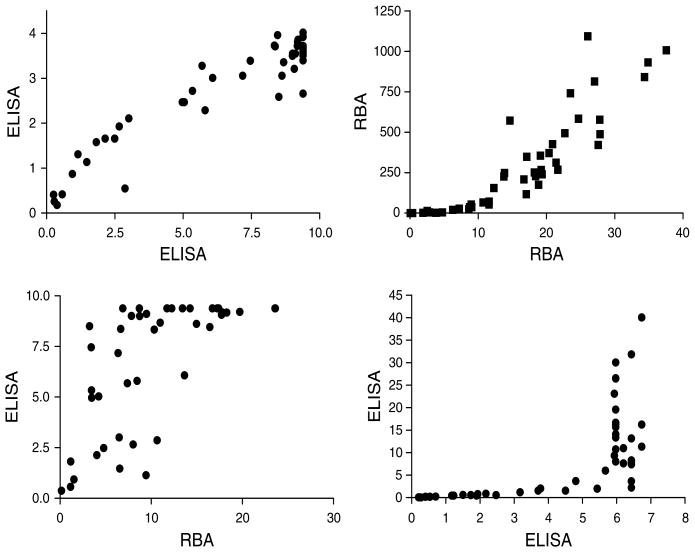
In assessing the correlation between various assays, some assays did indeed provide excellent correlation, whereas others demonstrated a more nonlinear relationship upon direct comparison (Figure 5). This indicates that assay units vary and are not strictly comparable, and that increments in units do not always follow a linear relationship between assays. Therefore, changes in titer will have different meanings in the various assays. The five commercial producers showed correlations with each other ranging from 0.8466 to as high as 0.9514 (r-squared). It is important to note also that for participating laboratories utilizing kits from the same manufacturer, there was excellent correlation ranging from 0.7577 for one kit to 0.9682 for another kit (r-squared). Other laboratory assays did not fare so well when compared with other kits, with r-squared values as low as 0.4244 on head to head comparison.
Figure 5.
Examples of laboratory correlation for the celiac disease samples. Top left: Two ELISAs demonstrate excellent correlation (r-squared = 0.8882). Top right, Two RBAs with good correlation (r-squared = 0.7751). Bottom two graphs: examples of poor and unusual correlation curves between different assays (bottom left r-squared = 0.5037, bottom right r-squared = 0.4244).
Finally, as receiver operator characteristic analysis was performed for each laboratory, the optimum cutoff for each assay was determined for this workshop (Table 2). On the basis of this analysis, one can see that it is possible to adjust the cutoff of most ELISA assays to provide sensitivities and specificities as good as that of the RBA, suggesting that most ELISA assays have sufficient differences between a positive and negative signal that allow for the potential of optimization. However, some assays still performed suboptimally (adjusted sensitivity less than 90%) despite adjustments in the cutoff to provide 99–100% specificity.
Table 2. Adjusted specificity and sensitivity based on ROC analysis optimizing each laboratory's cutoff for positivity.
| Lab | Assay | Adjusted specificity (%) | Adjusted sensitivity (%) |
|---|---|---|---|
| G | ELISA | 100 | 96 |
| T | ELISA | 100 | 93 |
| A | ELISA | 100 | 93 |
| F | RBA | 100 | 93 |
| D | ELISA | 99 | 93 |
| J | ELISA | 99 | 93 |
| E | ELISA | 100 | 93 |
| L | RBA | 99 | 93 |
| K | RBA | 100 | 93 |
| P | RBA | 100 | 93 |
| B | ELISA | 100 | 91 |
| O2 | RBA | 100 | 91 |
| N | ELISA | 100 | 91 |
| S | ELISA | 99 | 89 |
| Q | ELISA | 99 | 89 |
| I | ELISA | 99 | 89 |
| R | ELISA | 99 | 84 |
| M | ELISA | 99 | 84 |
| O1 | ELISA | 100 | 82 |
| H | ELISA | 99 | 82 |
| C | ELISA | 100 | 76 |
ELISA, enzyme-linked immunosorbent assay; Lab, laboratory; RBA, radiobinding assay.
Discussion
Measurement of immunoglobulin A antibody to human recombinant tissue transglutaminase is recommended for initial testing for CD and is considered the most efficient single serologic test for the detection of CD by the American Gastroenterological Association Institute (11). In addition, the North American Society for Pediatric Gastroenterology, Hepatology and Nutrition's Celiac Disease Guideline Committee recommends the measurement of transglutaminase autoantibodies after the institution of a GFD “to demonstrate a decrease in antibody titer as an indirect indicator of dietary adherence and recovery (12). Therefore, proficient and quantitative assays for measurement of TGAA are necessary for effective screening and monitoring in CD. Given the countless laboratories that perform the TGAA assay and the importance of this single test, the need for standardization of the TGAA assay is clear, as indicated by the NIH Consensus Development Panel on Celiac Disease in 2004 (13). This is a report of the first of a series of TGAA workshops that are planned over the next several years. The purpose of these workshops is to foster a collaborative environment as a forum to improve assay performance for the measurement of TGAA, and to set the groundwork for international standardization. To sponsor an objective and nonthreatening autoantibody workshop, all of the sera were coded and blinded to the participating laboratories, and the laboratory identities were hidden from their reported results, even though all laboratories had access to the entire dataset. This format has allowed direct comparison between different TGAA assays as performed by each individual laboratory or commercial producer, in particular, it allows for an objective comparison between ELISA and RBA formats. One participating laboratory has already reported an improvement in assay performance by changing their source of human recombinant TG. Another laboratory discovered problems with their reagents and equipment software.
The results of this workshop highlight several major points:
There is a widespread interest and willingness to collaborate toward the comparison, improvement and eventually standardization of the TGAA assay.
Not all TGAA assays perform equally—in fact, some performed poorly with unacceptably low sensitivity.
RBA is more sensitive than ELISA in detecting low-titer sera (including serial dilutions).
RBA in general provides much higher signal-to-noise ratios over ELISA, with significantly higher positive signals and higher titer saturation levels, but inter-laboratory variability of signals was less for ELISA than RBA.
Although RBA had higher sensitivity, some ELISA assays showed the potential for antibody proficiency comparable to RBA after adjusting the cutoff levels by receiver operator characteristic analysis.
There are important differences between ELISA and RBA that likely affect the performance of these assays. ELISA assays are performed with TG substrate bound to a 96-well plate. A patient's serum is added to the well, which allows TGAA bind onto the plate. Therefore, autoantibody-antigen binding occurs in the solid phase. The autoantibody is then detected using various detection methods such as horseradish peroxidase and alkaline phosphatase. The RBA is typically performed in the following fashion: patient sera are preincubated with 35S-labeled TG to allow for autoantibody-antigen binding in the fluid phase. The mixture is then placed on a 96-well filtration plate loaded with anti-immunoglobulin A beads to bind the autoantibody-antigen complex, and then excess radiolabeled TG is removed by filtration. The plates are read on a β-counter with results expressed as counts per minute. The two major differences between ELISA and RBA are solid vs. fluid-phase binding, and the method of detection. Potential antigen binding sites may be hidden or undergo conformation changes when plate bound but not in solution, decreasing the effectiveness of an assay. One example of this is illustrated by the current difficulties in measuring human insulin autoantibodies by ELISA but not by RBA. Furthermore, the use of radiolabeled substrate over enzymatic reaction for detection of autoantibodies allows for the potential of unlimited signal for a high-titered sera, as it is measured as counts per minute.
One of the limitations with this workshop is that most celiac samples supplied were high-titered sera for TGAA, and less from the low to medium-range of TGAA, which could affect sensitivity, area under the curve and other assay performance determinations. Nearly half of the celiac samples for the workshop were contributed by a single laboratory (identified through both clinical and research-based assays, measured, and identified by either a commercial ELISA or in-house RBA) so some selection bias might be present as well. This could potentially affect the overall comparisons for which that particular laboratory was included, but would not affect results that highlight individual laboratory performances. In addition, some of the CD sera were from individuals that also had type 1 diabetes, and were otherwise asymptomatic. It is unknown whether the coexistence of additional autoimmune diseases would affect TGAA characteristics or their measurement, but the objective in this workshop is to detect TGAA regardless of clinical status provided there was biopsy confirmation of CD, and to assess laboratory consensus for positivity. For example, despite sampling sera from individuals reportedly on a gluten-free diet, most still had TGAA detectable by all laboratories. Even in the three CD sera that were negative by all laboratories, one should still be cautious in declaring that these individuals were absolutely TGAA negative, as their presence or absence could be a function of the assay's limits of detection. Furthermore, the reported sensitivity/specificity in this paper applies only to our findings in this workshop, particularly as 11 individuals on a GFD were included in the analysis. Exluding the 11 subjects in contention, the overall sensitivity and specificity is as shown in Table 3. Four laboratories had a 100% sensitivity when adjusting for untreated patients, with specificities of 98–100%. Three out of these four laboratories were RBA.
Table 3. Sensitivity and specificity excluding the 11 sera from individuals on a gluten-free diet.
| Lab | Assay | Sensitivity (%) | Specificity (%) |
|---|---|---|---|
| K | RBA | 100 | 100 |
| P | RBA | 100 | 100 |
| J | ELISA | 100 | 99 |
| F | RBA | 100 | 98 |
| A | ELISA | 97 | 100 |
| E | ELISA | 97 | 100 |
| G | ELISA | 97 | 100 |
| N | ELISA | 97 | 100 |
| O2 | RBA | 97 | 100 |
| T | ELISA | 97 | 100 |
| Q | ELISA | 97 | 98 |
| B | ELISA | 94 | 100 |
| O1 | ELISA | 94 | 96 |
| D | ELISA | 91 | 100 |
| I | ELISA | 91 | 99 |
| L | RBA | 91 | 99 |
| M | ELISA | 88 | 99 |
| C | ELISA | 74 | 100 |
| R | ELISA | 74 | 100 |
| S | ELISA | 74 | 100 |
| H | ELISA | 71 | 100 |
ELISA, enzyme-linked immunosorbent assay; Lab, laboratory; RBA, radiobinding assay.
Individual assay dynamics have been previously analyzed in terms of quantitative ability. In this workshop, most celiac samples that were negative in ELISA assays showed low levels of TGAA in a RBA. The prognostic value of these low-intermediate TGAA levels remains to be established. In addition, some ELISA assays became saturated at higher levels of TGAA. Although this could be easily adjusted for by performing further dilutions to quantify high-titered sera, it might not be routinely done even though quantitative discrimination at the high values might be important for monitoring autoantibody responses to a gluten-free diet. On the other hand, the adjusted sensitivity and specificity indicate that some ELISAs have the capacity to perform well and possibly even better than the RBAs. In addition, there is less variability in the ELISAs overall. Therefore, there is the potential for TGAA measurement by ELISA to be among the best-performing assays. This remains to be determined through future TGAA workshops.
In summary, TGAA is the most widely used antibody assay for the diagnosis, and monitoring of CD. The best TGAA assays in the world should be identified so that current methods can be improved upon, rather than continuing to insist on utilizing inaccurate tests. One would not accept a “clock that tells time poorly (14)”. Likewise, one should not tolerate assays that perform suboptimally regardless of the field of study. Workshops such as this one designed to compare assays and to promote efforts to share methods to improve the assay are necessary to advance the field. Therefore, it may be necessary to develop a common fundamental protocol, thus standardizing many aspects of the assay. In addition, through use of standardized sera, there will be the potential for standardizing the reporting internationally, so that all assays will be comparable from laboratory to laboratory. Referring physicians are often faced with samples that are sent to different laboratories producing different assay results. Thus, it is very difficult to compare TGAA values obtained at different time points for the same patient when the samples are sent to different laboratories. It is also difficult for clinicians to be able to relate to different normal ranges and different assay units and, indeed, tests with difference performance characteristics. It would greatly improve the simplicity for testing if there was a standardization of units for reporting and if assay performance were fairly tightly correlated. The participants of this Transglutaminase Autoantibody Workshop would like to make an appeal to government institutions and centralized agencies to take on this challenge, and to institute a Celiac Disease Antibody Standardization Program, similar to one currently being sponsored by the CDC for type 1 diabetes (10).
Study Highlights.
What is Current Knowledge
Transglutaminase autoantibodies best test for celiac disease.
Transglutaminase autoantibody assays vary greatly qualitatively.
Transglutaminase autoantibody assay reporting is unstandardized.
What is New Here
An international autoantibody workshop for transglutaminase is feasible.
Radiobinding assays formats are more sensitive than ELISA for transglutaminase autoantibodies.
ELISA assays have the potential to perform as well as radiobinding assays.
Acknowledgments
Financial support: the project was funded by Edwin Liu's K08 Career Development Award. We also acknowledge the following support for their contributions to the center, which is necessary for the entire workshop mechanism to exist: National Institutes of Health grant M01RR00069, General Clinical Research Center program in the National Center for Research Resources, NIH grants DK RO1-DK50979, DK32083, DK32493, Autoimmunity Center of Excellence grant U19AI46374, Diabetes Endocrine Research Center P3057516, Autoimmunity Prevention Center grant 5U19AI50864, and Immune Tolerance Network Autoantibody Core Laboratory.
There was no support from any private or commercial group.
Appendix
| Joe Murray | Mayo Clinic, Minnesota, USA |
| Kristin Leiferman/John Zone | University of Utah, USA |
| Dongmei Miao/Liping Yu | Barbara Davis Center for Childhood Diabetes, University of Colorado, USA |
| Martin Stern | University Children's Hospital, Tübingen, Germany |
| Claudio Tiberti/Margherita Bonamico | Department of Clinical Sciences and Paediatrics, University of Rome, Italy |
| Ezio Bonifacio | Diabetes Research Institute, Munich, Germany |
| Kaija Laurila/Markku Maki | University of Tampere, Tampere, Finland |
| Mary von Blomberg/Ingrid van Hoogstraten | Vrije Universiteit Medical Center, Amsterdam, Netherlands |
| Joseph McMann/Ritu Verma | Children's Hospital of Philadelphia, Pennsylvania, USA |
| Ake Lernmark/Daniel Agardh | Lund University, Malmö, Sweden |
| Philip Bufler/Sibylle Koletzko | Dr V. Haunershes Kinderspital, University of Munich, Germany |
| Jo Wilson/Devasenan Devendra | Central Middlesex Hospital & Imperial College, London, UK |
| Alistair Williams | University of Bristol, UK |
| Hannah Kakkula/Olli Simell | University of Turku, Finland |
| James Goeken | University of Iowa, USA |
| Eurospital* | Trieste, Italy |
| INOVA* | San Diego, California, USA |
| Prometheus* | San Diego, California, USA |
| Radim Diagnostics* | Pomezia, Italy |
| The Binding Site* | Birmingham, UK |
Commercial laboratories.
Footnotes
Conflict of Interest: All of the authors have no financial disclosures or conflicts of interest with the publication of this paper.
Potential competing interests: there are none. All laboratories are blinded and deidentified.
Guarantor of the article: Edwin Liu, MD.
Specific author contributions: Marcella Li: performed the bulk of the labor with organizing/aliquoting the sera, shipping, and performing related assays; Liping Yu: responsible for the quality control of our radioassay and also helped with data analysis, trouble shooting, planning details of the workshop. Our laboratory was used as a “reference lab”; Claudio Tiberti: contributed sera to the workshop, helped plan initial stages of workshop and paper preparation; Margherita Bonamico: participated in data analysis and paper preparation; Iman Taki: in charge of collecting the bulk of our sera at the institution and organizing the workshop; Dongmei Miao: technician performed radioassay and also performed the assay to serve as a reference for other laboratories; Joseph A. Murray: organized the workshop and paper preparation; Marian J. Rewers: contributed sera for our laboratory and helped plan the workshop; Edward J. Hoffenberg: mentor for the senior author, provided help in planning the workshop and recruiting participants; Daniel Agardh: contributed sera for the workshop, data analysis, and paper preparation; Patricia Mueller: contributed the control sera for the workshop, data analysis, and paper preparation; Martin Stern and Ezio Bonifacio: planned the details of the workshop, data analysis, and paper preparation; Edwin Liu: organized the workshop, communication with participants, overseeing our laboratory, data analysis, and paper preparation.
References
- 1.Dieterich W, Ehnis T, Bauer M, et al. Identification of tissue transglutaminase as the autoantigen of celiac disease. Nat Med. 1997;3:797–801. doi: 10.1038/nm0797-797. [DOI] [PubMed] [Google Scholar]
- 2.Liu E, Li M, Bao F, et al. Need for quantitative assessment of transglutaminase autoantibodies for celiac disease in screening-identified children. J Pediatr. 2005;146:494–9. doi: 10.1016/j.jpeds.2004.11.028. [DOI] [PubMed] [Google Scholar]
- 3.Van Meensel B, Hiele M, Hoffman I, et al. Diagnostic accuracy of ten second-generation (human) tissue transglutaminase antibody assays in celiac disease. Clin Chem. 2004;50:2125–35. doi: 10.1373/clinchem.2004.035832. [DOI] [PubMed] [Google Scholar]
- 4.Stern M. Comparative evaluation of serologic tests for celiac disease: a European initiative toward standardization. J Pediatr Gastroenterol Nutr. 2000;31:513–9. doi: 10.1097/00005176-200011000-00012. [DOI] [PubMed] [Google Scholar]
- 5.Bonamico M, Tiberti C, Picarelli A, et al. Radioimmunoassay to detect antitransglutaminase autoantibodies is the most sensitive and specific screening method for celiac disease. Am J Gastroenterol. 2001;96:1536–40. doi: 10.1111/j.1572-0241.2001.03754.x. [DOI] [PubMed] [Google Scholar]
- 6.Bonifacio E, Atkinson M, Eisenbarth GS, et al. International workshop on autoimmunity in animal models of autoimmune diabetes identifies insulin, but not GAD or IA-2 as specific antigens of humoral autoimmunity in the non-obese diabetic mouse. Diabetes. 2001;50:2451–58. doi: 10.2337/diabetes.50.11.2451. [DOI] [PubMed] [Google Scholar]
- 7.Yu L, Wang J, O'Dell JR, et al. Anti-dsDNA antibody assay: high specificity and sensitivity with a filtration radioassay in comparison to low specificity with the standard ELISA. J Rheumatol. 2007;34:734–9. [PubMed] [Google Scholar]
- 8.Yamamoto AM, Amoura Z, Johannet C, et al. Quantitative radioligand assays using de novo-synthesized recombinant autoantigens in connective tissue diseases: new tools to approach the pathogenic significance of anti-RNP antibodies in rheumatic diseases. Arthritis Rheum. 2000;43:689–98. doi: 10.1002/1529-0131(200003)43:3<689::AID-ANR27>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 9.Yamamoto AM, Johanet C, Duclos-Vallee JC, et al. A new approach to cytochrome CYP2D6 antibody detection in autoimmune hepatitis type-2 (AIH-2) and chronic hepatitis C virus (HCV) infection: a sensitive and quantitative radioligand assay. Clin Exp Immunol. 1997;108:396–400. doi: 10.1046/j.1365-2249.1997.4071302.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bingley PJ, Bonifacio E, Mueller PW. Diabetes antibody standardization program: first assay proficiency evaluation. Diabetes. 2003;52:1128–36. doi: 10.2337/diabetes.52.5.1128. [DOI] [PubMed] [Google Scholar]
- 11.AGA Institute. AGA Institute Medical Position Statement on the Diagnosis and Management of Celiac Disease. Gastroenterology. 2006;131:1977–80. doi: 10.1053/j.gastro.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hill ID, Dirks MH, Liptak GS, et al. Guideline for the diagnosis and treatment of celiac disease in children: recommendations of the North American Society for Pediatric Gastroenterology, Hepatology and Nutrition. J Pediatr Gastroenterol Nutr. 2005;40:1–19. doi: 10.1097/00005176-200501000-00001. [DOI] [PubMed] [Google Scholar]
- 13.NIH Consensus Development Program. NIH Consensus Development Conference on Celiac Disease. NIH Consens State Sci Statements. 2004;21:1–23. [PubMed] [Google Scholar]
- 14.Liu E, Eisenbarth GS. Accepting clocks that tell time poorly: fluid-phase versus standard ELISA autoantibody assays. Clin Immunol. 2007;125:120–6. doi: 10.1016/j.clim.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]