Abstract
DNA double-strand breaks (DSBs) are highly cytotoxic lesions that are generated by ionizing radiation (IR) and various DNA-damaging chemicals. Following DSB formation, cells activate the DNA-damage response (DDR) protein kinases ATM, ATR and DNA-PK. These then trigger histone H2AX phosphorylation and the accumulation of proteins such as MDC1, 53BP1, BRCA1, CtIP, RNF8 and RNF168/RIDDLIN into IR-induced foci (IRIF) that amplify DSB signalling and promote DSB repair1,2. Attachment of Small Ubiquitin-related modifier (SUMO) to target proteins controls diverse cellular functions3-6. Here, we show that SUMO1, SUMO2 and SUMO3 accumulate at DSB sites in mammalian cells, with SUMO1 and SUMO2/3 accrual requiring the E3 ligase enzymes PIAS4 and PIAS1. We also establish that PIAS1 and PIAS4 are recruited to damage sites via mechanisms requiring their SAP domains, and are needed for the productive association of 53BP1, BRCA1 and RNF168 with such regions. Furthermore, we show that PIAS1 and PIAS4 promote DSB repair and confer IR resistance. Finally, we establish that PIAS1 and PIAS4 are required for effective Ubiquitin-adduct formation mediated by RNF8, RNF168 and BRCA1 at sites of DNA damage7-11. These findings thus identify PIAS1 and PIAS4 as components of the DDR and reveal how protein recruitment to DSB sites is controlled by coordinated sumoylation and ubiquitylation.
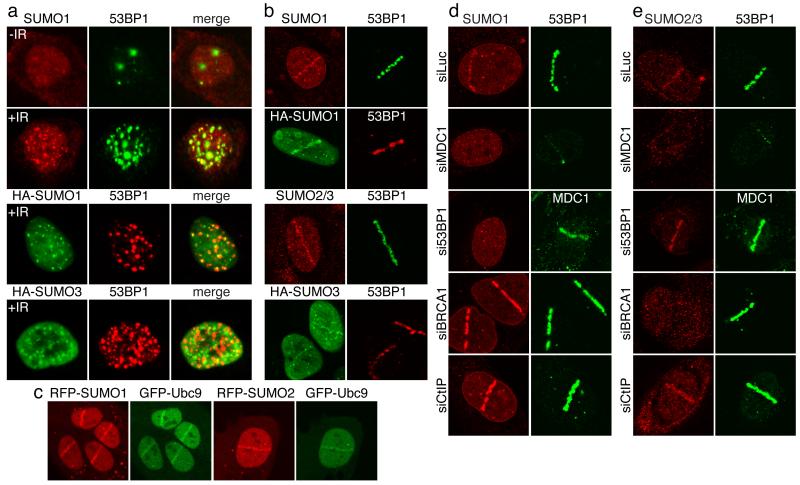
Mammalian cells express SUMO1 and the highly-related proteins SUMO2 and SUMO3 (SUMO2/3). These somewhat functionally-redundant proteins12 are structurally related to Ubiquitin and are covalently attached to target proteins by a SUMO-conjugation system consisting of an E1 activating enzyme (SAE1/SAE2), an E2 ligase (Ubc9) and various E3 ligases with differing target-protein specificities3,4. Involvement of the SUMO pathway in aspects of the DDR was previously reported (for review, see5). Notably, we found that, while SUMO1 exhibited pan-nuclear staining in untreated human cells, four hours after IR treatment, it formed nuclear foci that largely co-localized with 53BP1, suggesting them to be IRIF (Fig. 1a). Similarly, transfected HA-epitope-tagged SUMO1 and SUMO3 formed IRIF (Fig. 1a; SUMO2/3 foci that do not co-localize with 53BP1 presumably reflect SUMO conjugates in other structures, including PML bodies). Next, we employed laser micro-irradiation to induce DNA-damage tracts (laser-lines) in living cells13,14. This revealed that endogenous SUMO1 and SUMO2/3 (the antibody does not discriminate between these) together with HA-tagged SUMO1 and HA-SUMO3 accumulated in laser-lines (Fig. 1b). Moreover, live imaging of cells containing green-fluorescent-protein (GFP)-tagged 53BP1 or red-fluorescent-protein (RFP)-tagged SUMO1, SUMO2 or SUMO3 revealed that all exhibited similar recruitment kinetics: accrual being detectable five minutes after micro-irradiation, peaking in intensity at two to four hours and then gradually diminishing (Supplementary Figs 1a-c, 2a and 2b). Furthermore, we observed SUMO1 and SUMO2/3 accumulation with varying intensities in both G1 and S/G2 cells (Supplementary Fig. 2c). Consistent with sumoylation actively occurring at damage sites, Ubc9 (the only known SUMO E2) accumulated at damaged regions with similar kinetics to SUMO (Fig. 1c and Supplementary Figs 1b, 1d and 2d). Furthermore, we observed faint recruitment of the SUMO E1 component, SAE1 to laser-lines (data not shown), in accord with SAE1 recently being identified as a potential ATM/ATR target15.
Figure 1. SUMOs and Ubc9 accumulate at DNA-damage sites by mechanisms requiring MDC1, 53BP1 and BRCA1.
a, U2OS cells or U2OS cells transfected with HA-SUMO1 or HA-SUMO3 were irradiated (5Gy; +IR) or mock-irradiated (−IR) and probed. b, As in (a) but with laser micro-irradiation. c, U2OS cells co-transfected with GFP-Ubc9 and RFP-SUMO1 or RFP-SUMO2 were micro-irradiated and live cells imaged after 20 minutes. d, U2OS cells transfected with siRNAs were laser micro-irradiated then probed. For siRNA depletions, see Fig. 3e and Supplementary Fig. 10a.
In line with SUMO accumulation in IRIF and laser-lines representing responses to DSBs, such accumulation was reduced when cells were pre-incubated with KU-55933, a specific ATM inhibitor16 (Supplementary Fig. 3a), while accumulation of SUMO1 and to a lesser extent SUMO2/3 was enhanced by depletion of CtIP or MMS21, which promote DNA repair17,18 (Figs 1d and 1e, and Supplementary Figs 4a and 4b; see Fig. 3e for CtIP depletion and Supplementary Fig. 10 for other depletions). Furthermore, we observed markedly reduced SUMO1 and SUMO2/3 accumulation at damaged sites in cells that were defective in RNF168 or had been treated with short-interfering RNAs (siRNAs) to deplete MDC1 or RNF8 (Figs 1d and 1e, and Supplementary Figs 3b and 3c). Because MDC1, RNF8 and RNF168 control the retention of 53BP1 and BRCA1 at DNA-damage sites7-9,11,19-23, we tested whether depleting these factors affected SUMO accrual. Indeed, 53BP1 depletion impaired SUMO1 but not SUMO2/3 accumulation in laser-lines (Figs 1d and 1e). Conversely, BRCA1 depletion abolished SUMO2/3 but not SUMO1 accrual (Figs 1d and 1e). Collectively, these data suggested that DNA damage is channelled into 53BP1-SUMO1 or BRCA1-SUMO2/3 pathways.
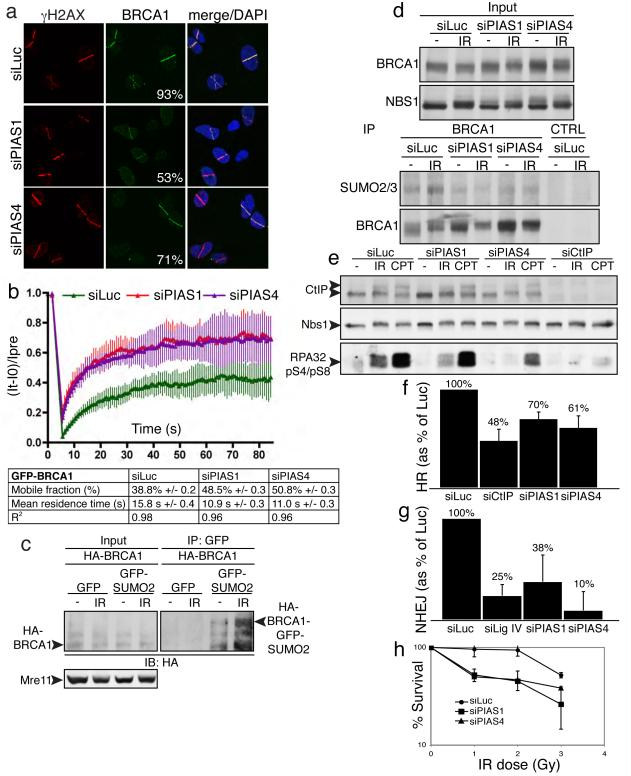
Figure 3. PIAS1 and PIAS4 promote BRCA1 accumulation and sumoylation, RPA phosphorylation, and DSB repair.
a, U2OS cells treated, micro-irradiated and probed as indicated; representative images with % of γH2AX positive cells also positive for BRCA1, each image represents >200 γH2AX-positive cells in two independent experiments. b, U2OS cells stably expressing GFP-BRCA1 and Flag-BARD1 were subjected to FRAP; data from Luciferase (n=7 independent measurements), PIAS1 (n=8) and PIAS4 (n=11); error bars = s.e.d. c, Essentially as Fig. 2h, except cells were co-transfected with HA-BRCA1 and Flag- BARD1. d,e, Extracts were prepared and analyzed 2 h following mock (−) or 10 Gy IR treatment. f-h, Effects of PIAS1/4 depletion on HR-mediated gene-conversion (f), NHEJ (g) and IR sensitivity (h); error bars = s.e.d.; data accumulated over four independent experiments (in each of f-h).
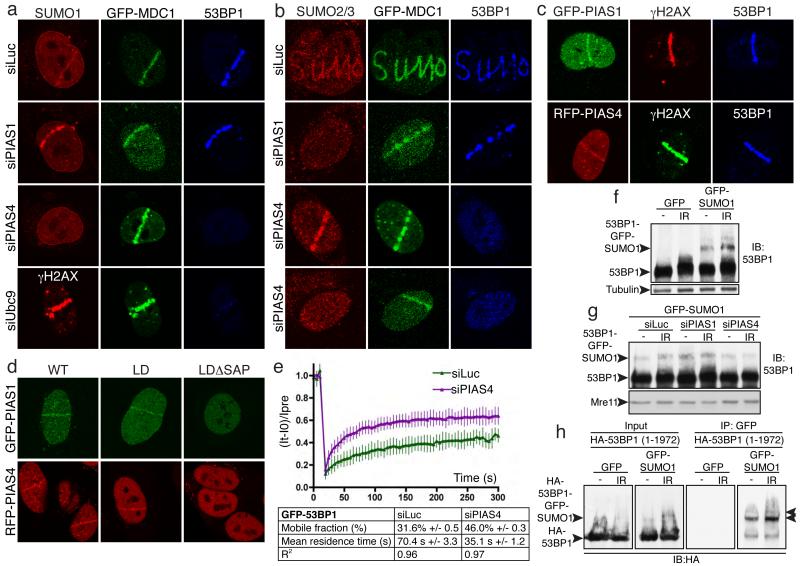
The different accumulation requirements for SUMO1 and SUMO2/3 suggested that their conjugation might require different E3 ligases. By siRNA depletion of various SUMO E3 ligases (Supplementary Fig. 10), we found that most were not required for SUMO1 or SUMO2/3 accrual at DNA-damage sites (Supplementary Figs 4a and 4b). Strikingly, however, depletion of the PIAS4 E3 ligase markedly reduced SUMO1 accrual on laser-lines (Figs 2a and 2b; note that MDC1 recruitment still occurred). Nevertheless, in certain cells, PIAS4 depletion also impaired SUMO2/3 (and 53BP1) accumulation (Fig. 2b, bottom panels), indicating that PIAS4 controls both SUMO1 and SUMO2/3 accrual. Accordingly, PIAS4 depletion impaired the accumulation of GFP-SUMO3 at laser-lines (data not shown). In parallel, we found that PIAS1 depletion markedly reduced SUMO2/3 accumulation at sites of DNA damage in all cells but did not affect SUMO1 accrual (Figs 2a, 2b and Supplementary Figs 4a and 4b). Supporting a model in which PIAS4 and PIAS1 mediate SUMO conjugation at DSB sites, RFP-tagged PIAS4 and GFP-tagged PIAS1 were recruited to laser-lines with similar kinetics to SUMO and Ubc9 (Fig. 2c and 2d). Furthermore, for both PIAS4 and PIAS1, recruitment required their N-terminal SAP domain – originally defined as a DNA/RNA binding motif24 – but was not impaired by mutations predicted to abolish their SUMO E3-ligase functions (Fig. 2d). When expressed alone, however, the SAP domains of PIAS1 and PIAS4 were not detectably recruited to laser-lines, revealing that additional parts of these proteins are required for effective recruitment (data not shown).
Figure 2. PIAS1 and PIAS4 are recruited to DNA-damage sites and mediate 53BP1 recruitment and sumoylation.
a,b, U2OS cells stably expressing GFP-MDC1 were treated, micro-irradiated and probed (Supplementary Figs. 4c and 10a-c for quantifications and depletions, respectively). c, Cells expressing GFP-PIAS1 or RFP-PIAS4 were micro-irradiated and probed. d, Cells expressing GFP-PIAS1 or RFP-PIAS4 wild-type (WT), ligase dead (LD), delta SAP (data not shown) or ligase-dead delta SAP (LDΔSAP) were micro-irradiated and imaged. e, Cells stably expressing GFP-53BP1 subjected to FRAP (n=11 independent measurements; error bars = s.e.d.). f-h, U2OS cells stably expressing GFP-SUMO1 or GFP (f, g) or HEK293 cells co-transfected with full-length (1-1972) HA-53BP1 and GFP-SUMO1 or GFP (h) were treated with or without IR (10 Gy).
Strikingly, PIAS4 depletion by either of two independent siRNA oligonucleotides, but not depletion of any other E3 enzyme tested, severely impaired 53BP1 accumulation in laser-lines and in IRIF (Figs 2a, 2b and Supplementary Figs 4a-d; demonstration of siRNA specificity is provided by use of a point-mutated siRNA-resistant PIAS4 construct in Supplemental Figs 5a and 5b). In accord with this, Ubc9 depletion impaired 53BP1 accumulation, while histone H2AX phosphorylation (γH2AX) and MDC1 recruitment still ensued (Fig. 2a). Furthermore, fluorescence-recovery after photo-bleaching (FRAP) assays established that PIAS4 depletion significantly reduced the residence time of 53BP1 in laser-lines and increased the mobile fraction of 53BP1 molecules in these locations (Fig. 2e; representative images are shown in Supplementary Fig. 5d). By contrast, RFP-PIAS4 accrual in laser lines was not impaired by 53BP1 depletion (Supplementary Fig. 5c), implying that PIAS4 acts upstream of 53BP1.
During the above studies, we noted that the IR-induced shift in 53BP1 electrophoretic mobility on SDS-polyacrylamide gels was reduced by PIAS4 depletion (data not shown). Consistent with this mobility-shift at least in part reflecting 53BP1 sumoylation, the migration of endogenously-expressed 53BP1 on SDS-gels was shifted yet further in cells expressing GFP-tagged SUMO1 (Fig. 2f); and furthermore, this shift was diminished by PIAS4 depletion but not by PIAS1 depletion (Fig. 2g). To directly test for 53BP1 sumoylation, we transiently co-expressed HA-tagged 53BP1 with GFP-SUMO1 or GFP in cells, then performed GFP-immunoprecipitations. Western immunoblotting of resulting samples with an anti-HA antibody established that 53BP1 was indeed sumoylated in an IR-inducible manner (Fig. 2h), a conclusion supported by reciprocal immunoprecipitation-western experiments (Supplementary Fig. 6a) and by experiments with endogenous 53BP1 and SUMO1 (Supplementary Fig. 6b; this also showed that 53BP1 sumoylation was reduced by depleting PIAS4 but not PIAS1). Studies with cells expressing 53BP1 truncations revealed that both the N-terminal (residues 1-1052) and C-terminal (1052-1972) regions of 53BP1 can be sumoylated and suggested that C-terminal sumoylation occurs between residues 1052 and 1710 (Supplementary Figs 6c-e). While these data indicated that DNA-damage induced 53BP1 sumoylation occurs, we note that this cannot account for all the PIAS-dependent sumoylation signals observed in IRIF or laser-lines. Consequently, there must be additional DDR factors (some of which might have been identified in previous studies5) that are targeted for DNA-damage induced, PIAS-mediated sumoylation.
In parallel work, we found that both PIAS1 or PIAS4 depletion reduced the proportion of damaged (γH2AX positive) cells displaying BRCA1 accumulation and decreased BRCA1-staining intensity in those cells still exhibiting BRCA1 accrual (Figs 3a and Supplementary Fig. 7b; cells with weak BRCA1 staining in Fig. 3a were counted positive). By employing cDNA-complementation studies, we established that BRCA1 accrual required the SAP domain and E3-ligase activity of PIAS1 (Supplemental Figs 7 c-e). Furthermore FRAP analyses revealed that PIAS1 or PIAS4 depletion reduced the residence time of GFP-BRCA1 at damaged sites and increased the mobile fraction of BRCA1 molecules (Fig. 3b; see representative images in Supplementary Fig. 7b). Through using epitope-tagged SUMO2 and BRCA1 in immunoprecipitation-western studies, we also established that BRCA1 is sumoylated and that this is enhanced upon IR treatment (Fig. 3c). Accordingly, probing western blots of BRCA1 immunoprecipitates for SUMO2/3 revealed that IR enhanced BRCA1 sumoylation in a manner promoted by both PIAS1 and PIAS4 (Fig. 3d).
DSBs can be processed into single-stranded DNA that is bound by replication protein A (RPA) to promote ATR signalling and DSB repair by homologous recombination (HR)17. Notably, RPA accumulation in laser-lines (whether normalized or not to cell cycle profiles in Supplementary Fig. 10e) was impaired by PIAS1 or PIAS4 depletion (Supplementary Figs 8a-c). Furthermore, phosphorylation of the 34 kDa subunit of RPA on Ser-4 and Ser-8 (pS4/pS8) in response to IR or camptothecin treatment was diminished by PIAS4 depletion, while PIAS1 depletion impaired IR-induced but not camptothecin-induced RPA phosphorylation (Fig. 3e; CtIP depletion also impaired RPA phosphorylation, as previously reported17). Consistent with these findings and the involvement of BRCA1 and RPA in DNA repair by HR17,25,26, PIAS1 or PIAS4 depletion reduced HR in a cell-based gene conversion assay27 (Fig. 3f). PIAS1 and PIAS4 depletion also impaired DSB repair by non-homologous end-joining (NHEJ) as assessed by a cell-based plasmid-integration assay28 (Fig. 3g) and resulted in IR hypersensitivity (Fig. 3h).
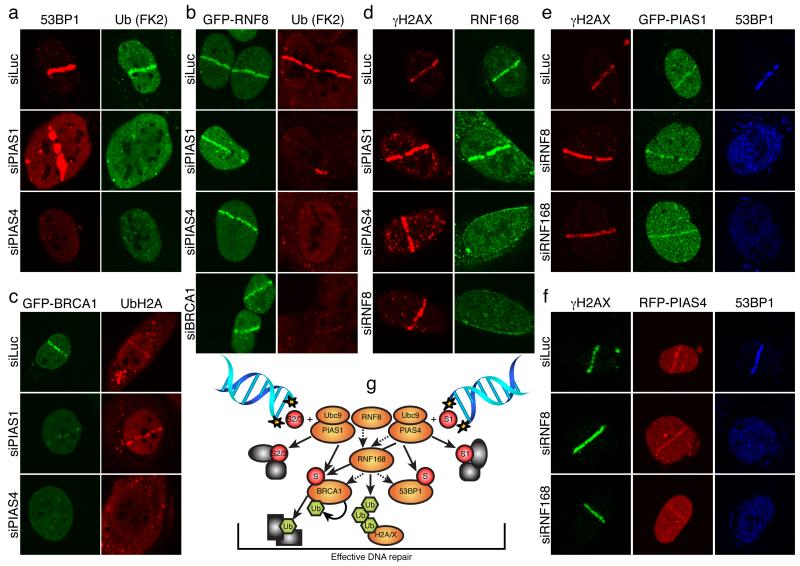
Accumulation of 53BP1, BRCA1 and Ubiquitin conjugates at DSB sites requires the Ubiquitin E3 ligases, RNF8 and RNF168, which ubiquitylate histones H2A and H2AX7-9,11,22. Furthermore, it has been reported that in both Caenorhabditis elegans and mammalian cells, Ubiquitin-conjugate formation at DNA-damage sites requires BRCA1 E3-Ubiquitin ligase activity10,29, although other groups have reported the effect of BRCA1 depletion on Ubiquitin accrual to be only minor7,9,11. In our assays, we found that, as for BRCA1 depletion, PIAS1 or PIAS4 depletion dramatically impaired Ubiquitin-conjugate accumulation (as detected by the FK2 antibody) in laser-lines, while GFP-RNF8 accumulation appeared normal (Figs 4a and 4b and Supplementary Fig. 9a). Furthermore, PIAS4 depletion but not PIAS1 depletion markedly impaired histone H2A ubiquitylation at damaged sites (Fig. 4c and Supplementary Fig. 9b). Consistent with PIAS4 being required for DNA-damage-induced accrual of 53BP1, BRCA1, FK2-Ubiquitin conjugates and Ubiquitin-H2A, the recruitment of endogenous RNF168 to damaged regions was impaired in PIAS4 depleted cells (Fig. 4d and Supplementary Figs 9c and 9d). By contrast, RNF168 still assembled at damage sites in PIAS1-depleted cells (Fig. 4d; as shown previously7,11, RNF168 accrual was RNF8 dependent). Because 53BP1 still accumulated under conditions where the FK2-Ubiquitin signal was severely impaired (upon BRCA1 or PIAS1 depletion; Figs 1d, 1e, 2a, 2b and 4a), these data implied that 53BP1 recruitment does not require Ubiquitin conjugates recognized by the FK2 antibody but, instead, relies on other ubiquitylated proteins (most likely H2A and H2AX). Significantly, depletion of RNF8 or RNF168, while abolishing 53BP1 accrual at sites of DNA damage, did not affect accumulation of GFP-PIAS1 or RFP-PIAS4 (Figs 4e and 4f). We therefore conclude that PIAS1 and PIAS4 function in parallel with RNF8 to orchestrate RNF8-, RNF168- and BRCA1-dependent accumulation of Ubiquitin conjugates at DNA-damage sites. Only PIAS4, however, is needed for RNF8- and RNF168-mediated H2A and possibly H2AX ubiquitylation.
Figure 4. Linkage between PIAS1/4 and RNF8/168.
a, U2OS cells were treated and probed as indicated. b,c, As (a) except cells stably expressed GFP-RNF8 or GFP-BRCA1/Flag-BARD1; see Supplementary Figs. 9a and 9b for quantifications. d, U2OS cells were treated and probed as indicated; see Supplementary Figs. 9d 10c for quantifications and siRNA efficiencies, respectively. e,f, U2OS cells stably expressing GFP-PIAS1 (e) or RFP-PIAS4 (f) were treated and probed as indicated. g, Model; dashed arrows indicate protein requirements for accumulation, solid arrows indicate target-protein modifications.
Our findings invoke a model in which PIAS1 and PIAS4 act in parallel but overlapping SUMO-conjugation pathways to control the DDR (Fig. 4g). In this regard, we note that mouse knock-out studies have revealed that PIAS1 or PIAS4 loss is tolerated, while deletion of both leads to embryonic lethality and an inability to derive viable cells30. Significantly, while both PIAS1 and PIAS4 promote Ubiquitin-FK2-adduct accumulation, only PIAS4 is needed for accrual of RNF168 and ubiquitylated H2A at DNA-damage sites. An attractive explanation for these and other data is that, after being recruited by RNF8-, PIAS1- and PIAS4-dependent mechanisms, BRCA1 (together with BARD1) is itself the major Ubiquitin E3 ligase for generating FK2-reactive Ubiquitin conjugates. Significantly, after PIAS1 or PIAS4 depletion, we still detect weak association of BRCA1 at damage sites but not the accumulation of BRCA1-dependent FK2-Ubiquitin conjugates. Consequently, we speculate that PIAS1- and PIAS4-dependent sumoylation of BRCA1 – and in all likelihood various other DDR proteins – not only mediates the stable association of BRCA1 with DNA-damage sites but also promote BRCA1 Ubiquitin-ligase activity. Furthermore, we found that GFP-RNF8 recruitment still occurred upon PIAS1 or PIAS4 depletion, revealing that RNF8 recruitment is insufficient to effectively recruit RNF168 and mediate effective Ubiquitin-conjugate production at DSB sites. Thus, we speculate that RNF8, RNF168 and/or BRCA1/BARD1 might require pre-sumoylation of their targets and/or that sumoylation regulates their Ubiquitin-ligase activities. Future studies will be required to define the precise mechanisms by which the Ubiquitin- and SUMO-conjugation systems cooperate at DSB sites, and determine how PIAS1 and PIAS4 impinge on chromatin structure, promote DSB signalling and repair, and potentially regulate yet other aspects of the DDR.
Methods summary
U2OS-based lines were maintained under standard conditions. cDNA cloning was by standard procedures. siRNA transfections were with Lipofectamine RNAiMAX (Invitrogen). IR was administered with a Faxitron X-ray machine (Faxitron X-ray Corporation). ATM inhibition was by KU-55933 (KuDOS Pharmaceuticals). Laser micro-irradiation was with a FluoView 1000 confocal microscope (Olympus) with 37°C heating stage (Ibidi) and 405 nm diode (6 mW). FRAP was performed when laser-track accumulation of GFP-tagged protein reached maximal steady-state level. For immunofluorescence, cells were pre-extracted or not, fixed with 2% paraformaldehyde, permeabilized and stained. For whole cell extracts, cells were lysed on plates with 2% SDS, 50 mM Tris-HCl pH 7.5, 20 mM N-ethylmaleimide (Sigma-Aldrich) and protease inhibitor cocktail (Roche). To immunoprecipitate 53BP1, BRCA1 and sumoylated proteins, different lysis and binding buffers were used (Supplementary Information). HR and NHEJ assays were as previously described17,28. For IR survival, cells were transfected with siRNA and exposed to IR. After 10-14 days, colonies were stained with 0.5% crystal violet/20% ethanol, counted and normalized to plating efficiencies. For Florescence-Activated Cell Sorting (FACS) of propidium iodide-stained cells, data were analyzed by FlowJo software. All error-bars represent STDEV. Detailed descriptions of methods are provided in Supplementary Information.
Supplementary Material
Acknowledgements
We thank SPJ lab members for support and encouragement, in particular Jeanine Harrigan, Pablo Huertas, Serge Gravel, Kate Dry and Ross Chapman. We also thank Claudia Lukas for U2OS cells expressing GFP-BRCA1/FLAG-BARD1, Daniel Durocher and Grant Stewart for hTERT RIDDLE syndrome fibroblasts complemented with vector or HA-RNF168 and RNF168 antibody, Thanos Halazonetis for RNF8 antibody, Richard Baer for the Flag-BARD1 construct, Paul Harkin for the HA-BRCA1 construct, Kuniyoshi Iwabuchi for HA-53BP1; full length, N, C, CΔBRCT and BRCT constructs, and Rachael Walker for help with FACS. Research in the SPJ lab is supported by grants from Cancer Research UK and the European Union (Integrated Project DNA repair, LSHG-CT-2005-512113, and Genomic Instability in Cancer and Precancer, HEALTH-F2-2007-201630).
Appendix
Methods
Cell culture
U2OS cells were grown in DMEM (Sigma-Aldrich) supplemented with 10% foetal bovine serum (FBS; BioSera), 100 units/ml of penicillin, and 100 mg/ml of streptomycin (Sigma-Aldrich). U2OS cells stably expressing GFP-MDC128, GFP-53BP1, GFP-CtIP17, GFP-RNF8, GFP-SUMO1, GFP-SUMO2, GFP-SUMO3, GFP-Ubc9, GFP-PIAS1 (WT, LD, ΔSAP and LDΔSAP) and RFP-PIAS4 (WT, WT siRNA resistant, LD, ΔSAP and LDΔSAP) were grown in standard U2OS medium supplemented with 1mg/ml of G418 (Gibco, Invitrogen). U2OS cells stably expressing GFP-BRCA1 and Flag-BARD19 were provided by Claudia Lukas and were grown with 0.4 mg/ml of G418. hTERT RIDDLE syndrome fibroblasts complemented with vector or HA-RIDDLIN/RNF16811 were provided by Daniel Durocher and Grant Stewart, and were grown in standard medium supplemented with 0.5mg/ml of G418.
siRNA transfection and sequences
siRNA duplexes were obtained from MWG biotech or QIAGEN (Supplementary Table 1). Two consecutive rounds of siRNA transfections were carried out with Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer’s protocol unless otherwise specified. siRNA-transfected cells were assayed 48 h after transfection. For co-transfection with siRNA and expression constructs, cells were first transfected with siRNA followed by plasmid transfection 24 h later by using Fugene6 (Roche) according to manufacturer’s protocol. Cells were assayed 48 h post plasmid transfections. siRNA-mediated down-regulation of over-expressed protein was achieved by a first round of siRNA transfection as described above with an additional siRNA transfection 24 h after plasmid transfection. All PIAS4 siRNA mediated down regulation experiments were carried out using PIAS4-1 siRNA unless otherwise specified.
Laser micro-irradiation and imaging of live and fixed cells
For generation of localized damage in cellular DNA by exposure to a UV-A laser beam13,14, cells were plated on glass-bottomed dishes (Willco-Wells) and pre-sensitized with 10 μM 5-bromo-2′-deoxyuridine (BrdU, Sigma-Aldrich) in phenol red-free medium (Invitrogen) for 24 h at 37°C. Laser micro-irradiation was performed by using a FluoView 1000 confocal microscope (Olympus) equipped with a 37°C heating stage (Ibidi) and a 405 nm laser diode (6 mW) focused through a 60x UPlanSApo/1.35 oil objective to yield a spot size of 0.5-1 mm. The time of cell exposure to the laser beam was around 250 msec (fast scanning mode). Laser settings (0.40 mW output, 50 scans) were chosen that generate a detectable damage response restricted to the laser path in a pre-sensitization-dependent manner without noticeable cytotoxicity. Imaging of live and fixed cells was done on the same microscope by using objective and software described above.
Fluorescent recovery after photo-bleaching (FRAP)
FRAP analyses were performed on the microscope used for laser micro-irradiation when the accumulation of the GFP-tagged protein on the laser track reached its maximal steady-state level. After a series of three pre-bleach images, a rectangular region placed over the laser-damaged line was subject to a bleach pulse (five scans with 488 nm argon laser focused through a 60x UPlanSApo/1.35 oil objective, main scanner, 100% AOTF, slow scanning mode), followed by image acquisition in 5 sec intervals for GFP-53BP1 and at fastest speed for GFP-BRCA1. Average fluorescent intensities in the bleached region were normalized against intensities in an undamaged nucleus in the same field after background subtraction to correct for overall bleaching of the GFP signal due to repetitive imaging. For mathematical modelling of GFP-tagged protein mobility, (It-I0)/Ipre values were plotted as a function of time, where I0 is the fluorescence intensity immediately after bleaching and Ipre is the average of the 3 pre-bleach measurements. Estimation of mobile protein fraction (A) and residence time (τ) were performed using Prism 4 software assuming the existence of one protein population using the following equation: y(t)= A(1−exp(−t/τ)) .
Immunofluorescence
Cells were washed three times with PBS 0.1% Tween-20 followed by pre-extraction for 10 min (pre-extraction buffer: 25 mM Hepes 7.4, 50 mM NaCl, 1 mM EDTA, 3 mM MgCl2, 300 mM sucrose and 0.5% TritonX-100). Cells were fixed with 2% formaldehyde (w/v) in PBS for 20 min. Following three washes with PBS 0.1% Tween-20, cells were blocked for 1 h with 5% BSA in PBS 0.1% Tween-20 co-stained with the appropriate antibodies (Supplementary Table 3) in blocking solution over night and then co-immunostained with the appropriate secondary antibodies (Supplementary Table 3) in blocking solution. For imaging RNF168/RIDDLIN pre-extraction buffer: 20 mM HEPES, 20 mM NaCl, 5 mM MgCl2, 1 mM DTT, 0.5% NP-40, serine/threonine phosphatases inhibitor cocktail (Sigma-Aldrich), protease inhibitor cocktail (Roche). Pre-extraction step was omitted and permeabilization (0.5% Triton X-100 in PBS) was performed after fixation for imaging of Ubc9, PIAS1, PIAS4, CtIP and RNF8.
Treatment with small-molecule inhibitors and DNA-damaging agents
Camptothecin was obtained from Sigma-Aldrich, ATM KU-55933 inhibitor was provided by KuDOS Pharmaceuticals Ltd. Ionizing radiation (IR) treatment was performed by using a Faxitron X-ray machine (Faxitron X-ray Corporation, Illinois, USA). Where appropriate, ATM inhibitor (20 μM) was applied to the culture medium 1 h prior to laser micro-irradiation.
Plasmids and Cloning
SUMO1, SUMO2, SUMO3, Ubc9, PIAS1, PIAS2, PIAS3 and PIAS4 were PCR amplified from a human foetal brain cDNA library and cloned into pCS2-mRFP (Roger Y. Tsien) and pEGFP-C1 (Clontech). PIAS1ΔSAP and PIAS4ΔSAP were sub-cloned from the original clones while PIAS1C351A and PIAS4C342A/C347A were created using QuikChange Site-Directed Mutagenesis Kit (Stratagene). PIAS4 siRNA resistant clone was obtained by inserting the 7 nucleotide mismatches underlined (GATCCAAAGTCCGGACTGAA) into PIAS4 cDNA using QuikChange Site-Directed Mutagenesis Kit (Stratagene). MMS21 was cloned into pCL-NCX (Imgenex) initially modified to contain a 7His-3Flag tag using an adaptor duplex. RNF8 was cloned into pCDNA3.1 (Invitrogen) initially modified to contain GFP. PIAS1 and PIAS4 were also cloned into pCDNA3.1(−) (Invitrogen) initially modified to contain 3XFlag-S-tag in the same reading frame as pEGFP-C1. Mammalian expression plasmids encoding HA-53BP1 (full length, N, C, CΔBRCT and BRCT) were provided by Kuniyoshi Iwabuchi. Mammalian expression plasmids encoding Flag-BARD1 and HA-BRCA1 were provided by Richard Baer and Paul Harkin respectively. Primers were obtained from Sigma-Aldrich (Supplementary Table 2).
Immunoprecipitation and Immunoblotting
Cell extracts were prepared on plates by using lysis buffer containing 2% SDS, 50 mM Tris-HCl pH 7.5, 20 mM N-ethylmaleimide (Sigma-Aldrich) and protease inhibitor cocktail (Roche). Sonicating or passing the extracts 10 times through a 19G needle mounted syringe reduced viscosity. For 53BP1 immunoprecipitation, cell extracts were prepared as mentioned above and then diluted 1:20 with lysis buffer containing 150mM NaCl and 1% NP40 instead of SDS. For BRCA1 immunoprecipitation, cell extracts were prepared by using lysis buffer containing 20 mM HEPES pH 7.4, 450 mM NaCl, 1.5 mM MgCl2, 1 mM EGTA, 1% Tween20, 10% glycerol, serine/threonine phosphatase inhibitor cocktail (Sigma-Aldrich), protease inhibitor cocktail (Roche) and 10 mM N-ethylmaleimide (Sigma-Aldrich), the extracts were then sonicated and diluted 1:2 with the same buffer lacking NaCl. For GFP and HA-53BP1 immunoprecipitation using GFP-Trap-A (ChromoTek GmbH) and anti HA antibodies, cell extracts were prepared by using lysis buffer containing 20 mM HEPES pH 7.4, 500 mM NaCl, 1.5 mM MgCl2, 1 mM EGTA, 1% Triton X-100, 10% glycerol, serine/threonine phosphatase inhibitor cocktail (Sigma-Aldrich), protease inhibitor cocktail (Roche) and 10 mM N-ethylmaleimide (Sigma-Aldrich), the extracts were then sonicated and diluted 1:2 with the same buffer lacking NaCl. In all cases the extracts were cleared using centrifugation 14,000 rpm for 45 min at 4°C. Antibodies against 53BP1 (Sigma-Aldrich) and BRCA1 (Santa-Cruz 1:1 mix of rabbit polyclonals, same mix was used for immunoblotting) were pre-bound to Dynabeads ProteinG (Invitrogen) and incubated for 2 h at room temperature (53BP1) or overnight at 4°C (BRCA1 and GFP-Trap-A and HA) followed by 5 washes with immunoprecipitation buffer (2 washes with lysis buffer were added for the GFP-Trap-A immunoprecipitation) and 5 min boiling in 1.5X SDS sample buffer. Proteins were resolved by 4-18% gradient SDS-PAGE (unless otherwise specified) and transferred to PVDF membrane (GE Healthcare). Immunoblotting was performed with the appropriate antibodies (Supplementary Table 3).
Random plasmid integration assay
Assays were performed as previously described28 with minor modifications. Briefly, one day after transfection with siRNA, U2OS cells were transfected with BamHI-XhoI linearized pEGFP-C1 (Clontech). The following day, cells were collected, counted and plated on three plates, one of which contained 0.5 mg/ml G418. One day after plating, the cells on a plate lacking G418 were fixed to assess transfection efficiency and the other two plates were incubated for 10-14 days at 37°C to allow colony formation. Colonies were stained with 0.5% crystal violet/20% ethanol and counted. Random-plasmid integration events were normalized to transfection and plating efficiencies.
Homologous recombination assay
A U2OS clone with the integrated HR reporter DR-GFP was generated as described previously17,31. One day after transfection with siRNA, U2OS-DR-GFP cells were co-transfected with an I-SceI expression vector (pCBA-I-SceI) and a vector expressing monomeric red fluorescent protein (pCS2-mRFP). The latter plasmid was added in a 1:5 ratio to mark the I-SceI-positive cells. Cells were harvested one day after I-SceI transfection and subjected to flow cytometric analysis to examine recombination induced by I-SceI digestion. Only RFP positive cells were analyzed for HR efficiency to circumvent possible differences in transfection efficiencies. Fluorescence-activated cell sorting data were analyzed by using Summit V4.3 software to reveal the percentage of GFP-positive cells relative to the number of transfected cells (RFP positive). The data were related to a control siRNA treatment in each individual experiment. The dividing line between GFP (HR) positive and negative cells was set to 0.5 % background level of GFP positive cells in the internal control (RFP positive, not transfected with I-SceI). This gate was then applied to the RFP/I-SceI positive samples to determine HR efficiency. Results are presented as a percentage of control siRNA.
IR survival assays
U2OS cells were transfected with siRNA and exposed to IR. Cells were left for 10-14 days at 37°C to allow colony formation. Colonies were stained with 0.5% crystal violet/20% ethanol and counted. Results were normalized to plating efficiencies.
Florescence-activated cell sorting (FACS)
To determine cell-cycle distribution, cells were fixed with 70% ethanol, incubated for 30 min with RNase A (250 μg/ml) and propidium iodide (10 μg/ml) at 37°C and analyzed by FACS. Data were analyzed by using FlowJo software to reveal the percentage of cells in each phase of the cell cycle.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
References
- 1.Stucki M, Jackson SP. MDC1/NFBD1: a key regulator of the DNA damage response in higher eukaryotes. DNA Repair (Amst) 2004;3:953–957. doi: 10.1016/j.dnarep.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 2.Downs JA, Nussenzweig MC, Nussenzweig A. Chromatin dynamics and the preservation of genetic information. Nature. 2007;447:951–958. doi: 10.1038/nature05980. [DOI] [PubMed] [Google Scholar]
- 3.Hay RT. SUMO: a history of modification. Mol Cell. 2005;18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 4.Meulmeester E, Melchior F. Cell biology: SUMO. Nature. 2008;452:709–711. doi: 10.1038/452709a. [DOI] [PubMed] [Google Scholar]
- 5.Bergink S, Jentsch S. Principles of ubiquitin and SUMO modifications in DNA repair. Nature. 2009;458:461–467. doi: 10.1038/nature07963. [DOI] [PubMed] [Google Scholar]
- 6.Geoffroy MC, Hay RT. An additional role for SUMO in ubiquitin-mediated proteolysis. Nat Rev Mol Cell Biol. 2009;10:564–568. doi: 10.1038/nrm2707. [DOI] [PubMed] [Google Scholar]
- 7.Doil C, et al. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell. 2009;136:435–446. doi: 10.1016/j.cell.2008.12.041. [DOI] [PubMed] [Google Scholar]
- 8.Huen MS, et al. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007;131:901–914. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mailand N, et al. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;131:887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- 10.Morris JR, Solomon E. BRCA1: BARD1 induces the formation of conjugated ubiquitin structures, dependent on K6 of ubiquitin, in cells during DNA replication and repair. Hum Mol Genet. 2004;13:807–817. doi: 10.1093/hmg/ddh095. [DOI] [PubMed] [Google Scholar]
- 11.Stewart GS, et al. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell. 2009;136:420–434. doi: 10.1016/j.cell.2008.12.042. [DOI] [PubMed] [Google Scholar]
- 12.Evdokimov E, Sharma P, Lockett SJ, Lualdi M, Kuehn MR. Loss of SUMO1 in mice affects RanGAP1 localization and formation of PML nuclear bodies, but is not lethal as it can be compensated by SUMO2 or SUMO3. J Cell Sci. 2008;121:4106–4113. doi: 10.1242/jcs.038570. [DOI] [PubMed] [Google Scholar]
- 13.Lukas C, Falck J, Bartkova J, Bartek J, Lukas J. Distinct spatiotemporal dynamics of mammalian checkpoint regulators induced by DNA damage. Nat Cell Biol. 2003;5:255–260. doi: 10.1038/ncb945. [DOI] [PubMed] [Google Scholar]
- 14.Limoli CL, Ward JF. A new method for introducing double-strand breaks into cellular DNA. Radiat Res. 1993;134:160–169. [PubMed] [Google Scholar]
- 15.Matsuoka S, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 16.Hickson I, et al. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 2004;64:9152–9159. doi: 10.1158/0008-5472.CAN-04-2727. [DOI] [PubMed] [Google Scholar]
- 17.Sartori AA, et al. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Potts PR, Yu H. Human MMS21/NSE2 is a SUMO ligase required for DNA repair. Mol Cell Biol. 2005;25:7021–7032. doi: 10.1128/MCB.25.16.7021-7032.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kolas NK, et al. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science. 2007;318:1637–1640. doi: 10.1126/science.1150034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lou Z, et al. MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol Cell. 2006;21:187–200. doi: 10.1016/j.molcel.2005.11.025. [DOI] [PubMed] [Google Scholar]
- 21.Stewart GS, Wang B, Bignell CR, Taylor AM, Elledge SJ. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature. 2003;421:961–966. doi: 10.1038/nature01446. [DOI] [PubMed] [Google Scholar]
- 22.Wang B, Elledge SJ. Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. Proc Natl Acad Sci U S A. 2007;104:20759–20763. doi: 10.1073/pnas.0710061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xie A, et al. Distinct roles of chromatin-associated proteins MDC1 and 53BP1 in mammalian double-strand break repair. Mol Cell. 2007;28:1045–1057. doi: 10.1016/j.molcel.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aravind L, Koonin EV. SAP - a putative DNA-binding motif involved in chromosomal organization. Trends Biochem Sci. 2000;25:112–114. doi: 10.1016/s0968-0004(99)01537-6. [DOI] [PubMed] [Google Scholar]
- 25.Durant ST, Nickoloff JA. Good timing in the cell cycle for precise DNA repair by BRCA1. Cell Cycle. 2005;4:1216–1222. doi: 10.4161/cc.4.9.2027. [DOI] [PubMed] [Google Scholar]
- 26.Gudmundsdottir K, Ashworth A. The roles of BRCA1 and BRCA2 and associated proteins in the maintenance of genomic stability. Oncogene. 2006;25:5864–5874. doi: 10.1038/sj.onc.1209874. [DOI] [PubMed] [Google Scholar]
- 27.Pierce AJ, Hu P, Han M, Ellis N, Jasin M. Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes Dev. 2001;15:3237–3242. doi: 10.1101/gad.946401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stucki M, et al. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005;123:1213–1226. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- 29.Polanowska J, Martin JS, Garcia-Muse T, Petalcorin MI, Boulton SJ. A conserved pathway to activate BRCA1-dependent ubiquitylation at DNA damage sites. EMBO J. 2006;25:2178–2188. doi: 10.1038/sj.emboj.7601102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tahk S, et al. Control of specificity and magnitude of NF-kappa B and STAT1-mediated gene activation through PIASy and PIAS1 cooperation. Proc Natl Acad Sci U S A. 2007;104:11643–11648. doi: 10.1073/pnas.0701877104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.