Abstract
The serotonin1A receptor (5-HT1A R) knock-out mouse (KO) is a widely used animal model for anxiety and cognitive function and regulation of signaling cascades by this receptor has been reported. We aimed to determine individual representatives of signaling cascades in order to screen 5-HT1A R-dependent signaling proteins (SPs).
Hippocampal proteins from wild type and 5-HT1A R KO mice were extracted, run on two-dimensional gel electrophoresis, proteins were identified by MALDI and nano-ESI-LC-MS/MS and SPs were quantified by specific software.
Nucleoside diphosphate kinase A (NDK A, synonym: nm23), Dual specificity mitogen-activated protein kinase kinase 1 (MAPKK1, synonym: MEK), Serine/threonine-protein phosphatase PP1-gamma catalytic subunit (PP-1G), Septin-5, were reduced in the KO mice. Novel phosphorylation sites at T386 on MAPKK1 and at S225 and Y265 on Septin-5 were observed.
MAPKK1 and PP-1G are known 5-HT1A R-dependent signaling compounds and are in agreement with receptor knock-out and septin-5 is involved in serotonin transport, although regulation by 5-HT1A R has not been reported. 5-HT1A R – dependent levels for NDK A have not been demonstrated so far and we herewith propose a role for NDK A in 5-HT1A R signaling.
Reduced SP levels along with findings of two novel phosphorylation sites may be relevant for interpretation of previous and the design of future studies on this receptor system.
Keywords: Serotonin, Serotonin receptor, Signaling, Hippocampus, Knock-out mouse
1. Introduction
The serotonin1A receptor (5-HT1A R) plays an important role in cognitive and integrative functions as well as in emotional states including anxiety – related behavior (Hensler, 2005; Parks et al., 1998; Bailey and Toth, 2004; Sarnyai et al., 2000). 5-HT1A receptors are divided into two functionally different receptor types, the presynaptic inhibitory autoreceptors and the postsynaptic 5-HT1A receptors (Riad et al., 2000). Previous research asserts that mice lacking this receptor primarily show increased anxiety behavior and excitation as well as reduced hippocampal – related cognitive functions (Sarnyai et al., 2000).
The 5-HT1A R is the most and best investigated receptor in the large family of 5-HT receptors. It belongs to a large family of receptors that transduce signals through guanine nucleotide binding and regulatory proteins (G-proteins; Albert and Tiberi, 2001; Raymond et al., 1993). It is an integral membrane protein with seven hydrophobic transmembrane domains connected by three intracellular and three extracellular loops (the amino terminus is oriented extracellularly and the carboxyl terminus towards the cytoplasm). The intracellular domains possess sites for interacting with G-proteins, other regulatory proteins, and sites for phosphorylation by serine–threonine kinases (Mansuy and Shenolikar, 2006).
5-HT1A R-G coupling comprises several and manifold effects impacting associated signaling pathways and mediates “stimulatory” pathways including Gβγ-mediated stimulation of phospholipase Cβ (PLCβ) and mitogen-activated protein kinase (MAPK), leading to cell proliferation and transformation (Albert and Tiberi, 2001).
Activation of ERK and MAPK by 5-HT1A R constitute a sequence of several consecutive phosphorylation and activation steps. Binding of 5-HT to the 5-HT1A R results in the release of βγ-subunits from pertussis toxin-sensitive G-protein and thereby in activation of Src (a non-receptor tyrosine kinase) and phosphorylation of Shc. This in turn leads to recruitment of PI-3 kinase, Grb2 and Sps which is a Ras activator protein. Activated Ras causes the activation of Raf and thereby mitogen- and extracellular signal-regulated kinase (MEK, also known as MAPKK1) in turn activating ERK (Raymond et al., 2001).
A second pathway leading to cell proliferation and transformation is regulation of phospholipases such as PLC. Activated 5-HT1A R activate phospholipases, which cause generation of inositol triphosphate (IP3) – regulating intracellular Ca2+ release – and diacylglycerol (DAG) binding and activating protein kinase C (PKC). Hence mediated by PLC activation, 5-HT1A Rs regulate PKCs including serine–threonine, tyrosine and lipid kinases (Raymond et al., 1999).
Besides its modulating effects on second-messenger pathways the 5-HT1A R also regulates several channels such as G-protein-gated inwardly rectified K+ channels (GIRK), high conductance anion channels, cystic fibrosis transmembrane regulator (CFTR) Cl− channels and Ca2+ channels. GIRK channels play a role in hyperpolarizing postsynaptical potentials in the nervous system during activation of Gi/oα-coupled receptors such as 5-HT1A receptors (Raymond et al., 1999).
Moreover, activation of 5-HT1A R reduces AMPA-evoked currents in prefrontal cortex pyramidal neurons: activated 5-HT1A R decrease autophosphorylated CaKMII and CaMKII phosphorylation of GluR1 – an AMPA receptor subunit – in a protein phosphatase (PP1-)-dependent manner (Schiapparelli et al., 2005). This 5-HT1A R – induced modulation depends on the inhibition of PKA and requires activation of PP1 (Mukhin et al., 2000).
It has been shown that in prefrontal cortex activation of 5-HT1A receptors inhibits N-methyl-D-aspartate (NMDA) receptor-mediated ionic and synaptic currents. The NMDA receptor (NMDAR) is an ionotropic receptor for glutamate and its activation results in the opening of an ion channel which is nonselective to cations. NMDA currents comprise flows of Na+ and K+ ions, and small flows of Ca2+. The NMDA subunit NR2B, which is transported along microtubule to neuronal dendrites turned out to be the primary target of 5-HT1A receptors (Yuen et al., 2005). Serotonergic inhibition of NR2B is regulated by CaMKII and ERK signaling pathways. Accordingly, inhibition of CaMkII and ERK abolishes 5-HT1A regulation of NMDA currents.
Although 5-HT1A R mainly couples with Gi/o-proteins to the inhibition of adenylyl cyclase (AC) it is associated with a multitude of other signaling pathways and effectors.
A large series of these effectors have been reported but so far no systematic approach to search for such effectors was carried out at the protein level forming the rationale for the study. It was the aim of this work to use a proteomic approach for the systematic search for 5-HT1A-dependent effector proteins by comparing hippocampal protein levels between wild type and mice with genetically inactivated 5-HT1A R.
2. Methods
2.1. Animals
Ten wild type mice on a Swiss Webster background and ten 5-HT1A R −/− mice (three months old, Bailey and Toth, 2004) were obtained from M. Toth, Department of Pharmacology, Weill Medical College of Cornell University, New York, USA. Total hippocampi were isolated, taken into liquid nitrogen, transported to Vienna on dry ice and the freezing chain was never interrupted.
2.2. Sample preparation
Hippocampal tissue was powderised in liquid nitrogen and suspended in 2 mL sample buffer (20 mM Tris, 7 M urea, 2 M thiourea, 4% CHAPS, 10 mM 1,4-dithioerythritol, 1 mM EDTA, 1 mM PMSF and 1 tablet Complete™ from Roche Diagnostics). The suspension was sonicated for approximately 30 s and centrifuged at 15,000 × g for 60 min at 12 °C. Desalting was carried out with an Ultrafree-4 centrifugal filter unit (Millipore, Bedford, MA, USA) by centrifugation at 3000 × g at 12 °C until the eluted volume was about 4 mL (Shin et al., 2006). The protein content of the supernatant was determined by the Bradford assay (1976).
2.3. Two-dimensional gel electrophoresis (2-DE)
2-DE was performed essentially as reported (Chen et al., 2006). Samples of 750 μg protein were applied on immobilized pH 3–10 nonlinear gradient strips. Focusing started at 200 V and the voltage was gradually increased to 8000 V at 4 V/min and kept constant for a further 3 h (approximately 150,000 Vh totally). The second-dimensional separation was performed on 10–16% gradient SDS-PAGE. After protein fixation for 12 h in 50% methanol and 10% acetic acid, the gels were stained with colloidal Coomassie blue (Novex, San Diego, CA, USA) for 8 h and excess of dye was washed out from the gels with distilled water. Molecular masses were determined by running standard protein markers (Bio-Rad Laboratories, Hercules, CA, USA), covering the range of 10–250 kDa. Isoelectric point values were used as given by the supplier of the immobilized pH gradient strips.
2.4. In-gel digestion
Spots were excised with a spot picker (PROTEINEER sp™, Bruker Daltonics, Leipzig, Germany), placed into 96-well microtiter plates (Bruker Daltonics, Leipzig, Germany) and in-gel digestion and sample preparation for MALDI analysis were performed by an automated procedure (PROTEINEER dp™, Bruker Daltonics; John et al., 2007). Briefly, spots were excised and washed with 10 mM ammonium bicarbonate and 50% acetonitrile in 10 mM ammonium bicarbonate. After washing, gel plugs were shrunk by addition of acetonitrile and dried by blowing out the liquid through the pierced well bottom. The dried gel pieces were reswollen with 40 ng/μL trypsin (Promega, Madison, WI, USA) in digestion buffer (consisting of 5 mM Octyl b-D-glucopyranoside (OGP) and 10 mM ammonium bicarbonate) and incubated for 4 h at 30 °C. Extraction was performed with 15 μL of 1% TFA in 5 mM OGP at 20 °C for 30 min.
2.5. MALDI-TOF–TOF mass spectrometry and data processing
A target (AnchorChip™, Bruker Daltonics, Bremen, Germany) was wiped using paper tissues in sequence with acetone and N-heptane, followed by ultrasonication in isopropanol and subsequently in HPLC grade water and dried in air. Four micro-liters of extracted peptides were directly applied onto the target that was loaded with α-cyano-4-hydroxy-cinnamic acid (Bruker Daltonics, Bremen, Germany) matrix thinlayer. The mass spectrometer used in this work was an Ultraflex™ TOF/TOF (Bruker Daltonics, Bremen, Germany) operated in the reflector mode for MALDI-TOF peptide mass fingerprint (PMF) or LIFT mode for MALDI-TOF–TOF MS/MS with a fully automated mode using the FlexControl™ software. Samples were analyzed by one PMF from MALDI-TOF, followed by additional LIFT-TOF/TOF MS/MS analysis of three peptides. Data were accumulated from 200 consecutive laser shots to produce PMF and MS/MS spectra. Accelerating voltage was 25 kV for PMF. For MS/MS, accelerating voltage was 8 kV in TOF1 stage to promote metastable fragmentation. After selection of jointly migrating parent and fragment ions in a timed ion gate, ions were lifted by 19 kV to high potential energy in the LIFT cell under nitrogen gas. Peptide standards were used for external calibration. The m/z range was 700–4000 for PMF and 40–2560 for MS/MS. PMF and MS/MS spectra were interpreted primarily with the FlexAnalysis software (Bruker Daltonics, Bremen, Germany). The signal to noise ratio threshold was set as three. Autoproteolysis products of trypsin were used for internal calibration. Database searches, through the MASCOT 2.1 (Matrix Science, London, UK) against MSDB 20051115 database, using combined one PMF and three MS/MS datasets were performed via BioTools 2.3 software (Bruker Daltonics, Bremen, Germany). Species searches were limited to rodents. Trypsin was used and one maximum missing cleavage site was tolerated. A mass tolerance of 25 ppm for PMF, MS/MS tolerance of 0.5 Da, modification of carbamidomethyl (C), and variable modification of oxidation of methionine and phosphorylation (Tyr, Thr, and Ser) were considered. A randomized MSDB database (http://www.matrixscience.com/help/decoy_help.html) was searched simultaneously and false positive results were omitted. Protein identification rejected from MASCOT were manually examined and filtered to create a confirmed protein identification list. Positive protein identifications were based on a significant MOWSE score.
2.6. Analysis of peptides by nano-LC-ESI-MS/MS and data processing
Spots that were not identified by MALDI-TOF–TOF were analyzed by a second mass spectrometrical approach (Weitzdorfer et al., 2006). Solutions remaining from MALDI analyses were transferred to new 0.5 mL protein lobind tubes (Eppendorf, Hamburg, Germany). Protein spots were extracted twice using 15 μL 0.1% formic acid, 2% acetonitrile. Extracted solutions were pooled and concentrated in a Speed-Vac (Eppendorf, Hamburg, Germany) until the volume reached 8 μL, and 6.4 μL were used for nano-LC-ESI-MS/MS investigation. The HPLC used was an Ultimate 3000 system (Dionex Corporation, Sunnyvale, CA, USA) equipped with a PepMap C-18 analytic column (75 μm × 150 mm). The flow rate was 300 nL/min. The gradient was (A = 0.1% formic acid in water, B = 80% ACN/0.08% formic acid in water) 4–60% B from 0 min to 30 min, 90% B from 30 min to 35 min, 4%B from 35 min to 60 min. A QSTAR XL (Applied Biosystems, Foster City, CA, USA) mass spectrometer was used to record peptide spectra over the mass range of m/z 350–1600, and MS/MS spectra in information dependent data acquisition over the mass range of m/z 50–1600. Repeatedly, MS spectra were recorded followed by two MS/MS spectra generation of highest intensity precursor ions; the accumulation time was 1 s for MS spectra and 2 s for MS/MS spectra. The voltage between ion spray tip and orifice was set to 3200 V. The collision energy was set automatically according to the mass and charge state of the peptides chosen for fragmentation. Doubly or triply charged peptides were chosen for MS/MS experiments due to their good fragmentation characteristics. MS/MS spectra were interpreted and peak lists were generated by a MASCOT module (mascot.dll 1.6b21; Matrix Science, London, UK) in Analyst QS 1.1 (Applied Biosystems, Foster City, CA, USA). Searches were done by using the MASCOT 2.1 (Matrix Science, London, UK) against MSDB 20051115 database for protein identification. Searching parameters were set as follows: enzyme selected as trypsin with one maximum missing cleavage site, species limited to rodents, a mass tolerance of 500 ppm for peptide tolerance, 0.15 Da for MS/MS tolerance, fixed modification of carbamidomethyl(C) and variable modification of methionine oxidation and phosphorylation (Tyr, Thr, and Ser). A randomized MSDB database (http://www.matrixscience.com/help/decoy_help.html) was searched simultaneously and false positive results were omitted. Protein identification returned from MASCOT were manually examined and filtered to create a confirmed protein identification list. Positive protein identifications were based on a significant MOWSE score.
2.7. Quantification
Protein spots representing signaling proteins from all gels (10 per group; total n = 20) were outlined and matched, first automatically and then manually, and quantified using the Proteomweaver software (Definiens, Munich, Germany). The level of each protein was evaluated by its intensity (spot volume) in the 2-DE gel (Frischer et al., 2006).
2.8. Western blots
Sample preparation for immunoblotting was carried out as given in detail previously (Cheon et al., 2008). Electrophoresis and transfer to membranes was performed as given recently (Sunyer et al., 2009). The amount of protein applied to gels was 10 μg. First antibodies used were as follows: rabbit monoclonal antibody against MEK1 (Abcam, ab32091, in a dilution of 1:2000; Cambridge, UK); rabbit polyclonal antibody against nm23 (Abcam, ab55417, in a dilution of 1:2000; Cambridge, UK); chicken polyclonal antibody against PP-1G (Abcam, ab26175, in a dilution of 1:2000; Cambridge, UK). Anti-actin (Abcam ab3280-500, in a dilution of 1:5000) was used as a loading control.
Secondary antibodies used were anti-rabbit IgG, HRP linked (Cell Signaling, #7074; Danvers, MA) and rabbit anti-chicken IgY, HRP conjugate (Upstate, JBC1381990; Billerica, MA). All antibodies were used in a dilution of 1:2000. Controls were run without primary antibodies. The secondary antibody against mouse IgG, HRP-linked Antibody #7076 was from Cell Signaling and was used in a dilution of 1:5000.
2.9. Statistics
2.9.1. Proteomics
GraphPad Instat 3.05 was used to perform statistical analysis. Each protein spot volume and summarized levels of multiple spot proteins were input as raw data (arbitrary units of optical density). Mean and standard deviation were calculated. Subsequently, one-way analysis of variance (ANOVA) was performed to find significantly regulated proteins. Only those proteins which had ANOVA P values less than 0.05 were analyzed with Tukey multiple comparison test to compare all pairs of groups and to correct for multiple testing.
2.9.2. Western blotting
Statistical analysis to compare the two groups was performed by unpaired Student’s t-test. If data violated a principal assumption of a parametric distribution non-parametric Mann–Whitney-U-test was carried out. All calculations were performed using SPSS 14.0 (SPSS Inc., Chicago, IL).
3. Results
3.1. Protein identification
As shown in Table 1 a series of signaling proteins, some with several expression forms, from several signaling cascades has been identified. Table 1 reveals identification of signaling proteins from both groups. Statistically significant differential protein levels are shown in bold letters in the table and the level of significance was set as P < 0.05.
Table 1.
Protein identification and differential hippocampal protein levels. UniProtKB accession numbers are provided and proteins in bold are reflecting the level of significance p < 0.05.
| Accession number | Protein name | Spot no. | WT | KO | MS score | Matched peptides | Sequence coverage % | Theor. MW | Theor. pI | Obs. pI | MS/MS-peptide sequences |
|---|---|---|---|---|---|---|---|---|---|---|---|
| O35864 | COP9 signalosome complex subunit 5 | 0.19 ± 0.08 | 0.18 ± 010 | 138 | 17 | 47 | 37,417.59 | 6.11 | 6.0 | K.GYKPPDEGPSEYQTIPLNK.I | |
| O55131 | Similar to Septin-7, O55131, (93%) | 0.22 ± 0.10 | 0.15 ± 0.08 | 67 | 8 | 31 | 50,549.86 | 8.73 | 8.8 | ||
| O88441 | Metaxin-2 (Involved in transport of proteins into the mitochondrion.) | 0.28 ± 0.10 | 0.31 ± 0.06 | 98 | 9 | 55 | 29,758.12 | 5.44 | 5.5 | ||
| O88544 | COP9 signalosome complex subunit 4 | 0.12 ± 0.09 | 0.11 ± 0.06 | 380 | 21 | 63 | 46,284.81 | 5.57 | 5.65 | R.VISFEEQVASIR.Q K.HALHCTILASAGQQR.S R.LYLEDDDPVQAEAYINR.A |
|
| P12658 | Calbindin | 1.10 ± 0.24 | 1.25 ± 0.21 | 64 | 5 | 32 | 29,862.88 | 4.71 | 4.9 | ||
| P15532 | Nucleoside diphosphate kinase A | 0.80 ± 0.11 | 0.70 ± 0.12 | 390 | 13 | 83 | 17,207.80 | 6.84 | 6.8 |
R.TFIAIKPDGVQR.G K.EISLWFQPEELVEYK.S K.YMHSGPVVAMVWEGLNVVK.T K.YMHSGPVVAMVWEGLNVVK.T + Oxidation (M) |
|
| P18872 | Guanine nucleotide- binding protein G(o) subunit alpha 1 | 1.18 ± 0.57 | 1.57 ± 1.02 | 90 | 12 | 28 | 39,953.35 | 5.34 | 5.4 | R.IGAGDYQPTEQDILR.T | |
| P29387 | Guanine nucleotide- binding protein subunit beta 4 | 0.26 ± 0.07 | 0.2 ± 0.03 | 81 | 8 | 39 | 37,247.83 | 5.74 | 5.7 | ||
| P31938 | Dual specificity mitogen-activated protein kinase kinase 1 | (1) | 0.45 ± 0.07 | 0.38 ± 0.08 | 333 | 14 | 44 | 43,342.85 | 6.26 | 6.1 |
K.LPSGVFSLEFQDFVNK.C K.LCDFGVSGQLIDSMANSFVGTR.S K.ELELLFGCHVEGDAAETPPRPR.T |
| (2) | 0.26 ± 0.07 | 0.27 ± 0.07 | 264 | 8 | 29 | 43,342.85 | 4.62 | 6.2 | K.LPSGVFSLEFQDFVNK.C K.LCDFGVSGQLIDSMANSFVGTR.S K.ELELLFGCHVEGDAAETPPRPR.T |
||
| P42208 | Septin-2 | (1) | 0.20 ± 0.07 | 0.20 ± 0.04 | 358 | 15 | 51 | 41,525.52 | 6.10 | 6.1 | K.TIISYIDEQFER.Y R.LTVVDTPGYGDAINCR.D K.STLINSLFLTDLYPER.I |
| (2) | 0.09 ± 0.05 | 0.09 ± 0.06 | 67 | 7 | 32 | 41,525.52 | 6.10 | 6.0 | |||
| P53810 | Phosphatidylinositol transfer protein alpha isoform | 0.45 ± 0.14 | 0.40 ± 0.11 | 252 | 13 | 44 | 31,762.20 | 5.98 | 6.1 | K.AWNAYPYCR.T K.HVEAIYIDIADR.S K.IETWHKPDLGTQENVHK.L R.VILPVSVDEYQVGQLYSVAEASK.N |
|
| P53994 | Ras-related protein Rab-2A | 0.27 ± 0.08 | 0.23 ± 0.08 | 125 | 5 | 33 | 23,547.52 | 6.08 | 6.1 | R.FQPVHDLTIGVEFGAR.M | |
| P54227 | Stathmin | (1) | 0.51 ± 0.08 | 0.56 ± 0.08 | 71 | 1 | 8 | 17,143.27 | 5.76 | 5.7 | R.ASGQAFELILSPR.S |
| (2) | 0.43 ± 0.06 | 0.44 ± 0.08 | 66 | 1 | 8 | 17,143.27 | 5.76 | 5.5 | R.ASGQAFELILSPR.S | ||
| P61982 | 14-3-3 protein gamma | (1) | 0.49 ± 0.28 | 0.61 ± 0.34 | 363 | 12 | 53 | 28,171.40 | 4.80 | 5.2 | K.TAFDEAIAELDTLNEESYK.D |
| (2) | 0.58 ± 0.24 | 0.74 ± 0.21 | 566 | 16 | 62 | 4.9 | K.DSTLIMQLLR.D K.EHMQPTHPIR.L K.NVTELNEPLSNEER.N K.TAFDDAIAELDTLNEDSYK.D |
||||
| (3) | 0.50 ± 0.26 | 0.52 ± 0.20 | 412 | 12 | 53 | 28,171.40 | 4.80 | 5.2 | K.NVTELNEPLSNEER.N K.TAFDDAIAELDTLNEDSYK.D |
||
| (4) | 0.25 ± 0.24 | 0.24 ± 0.10 | 124 | 8 | 36 | 28,171.40 | 4.80 | 5.2 | K.ELEAVCQDVLSLLDNYLIK.N | ||
| P62137 | Serine/threonine- protein phosphatase PP1-alpha catalytic subunit | 0.16 ± 0.04 | 0.16 ± 0.04 | 425 | 17 | 60 | 37,540.09 | 5.94 | 5.9 | R.YPENFFLLR.G R.AHQVVEDGYEFFAK.R K.ICGDIHGQYYDLLR.L R.EIFLSQPILLELEAPLK.I |
|
| P62141 | Serine/threonine- protein phosphatase PP1-beta catalytic subunit | 0.19 ± 0.08 | 0.23 ± 0.08 | 256 | 16 | 59 | 37,055.64 | 5.85 | 5.8 | K.ICGDIHGQYTDLLR.L K.TFTDCFNCLPIAAIVDEK.I |
|
| P62259 | 14-3-3 protein epsilon | 0.34 ± 0.19 | 0.53 ± 0.45 | 404 | 14 | 50 | 29,173.90 | 4.63 | 4.85 | R.YLAEFATGNDR.K K.AASDIAMTELPPTHPIR.L K.AASDIAMTELPPTHPIR.L + Oxidation (M) K.AAFDDAIAELDTLSEESYK.D |
|
| P62880 | Guanine nucleotide- binding protein G(I)/G(S)/G(T) subunit beta 2 | 3.10 ± 3.33 | 1.02 ± 0.24 | 475 | 16 | 45 | 37,199.84 | 5.60 | 5.5 | K.IYAMHWGTDSR.L R.ELPGHTGYLSCCR.F K.ACGDSTLTQITAGLDPVGR.I R.KACGDSTLTQITAGLDPVGR.I |
|
| P62881 | Guanine nucleotide- binding protein subunit beta 5 | 0.10 ± 0.05 | 0.10 ± 0.08 | 169 | 6 | 23 | 43,565.27 | 6.01 | 5.7 | K.LHDVELHQVAER.V K.ESIIFGASSVDFSLSGR.L |
|
| P63028 | Translationally-controlled tumor protein | 0.21 ± 0.06 | 0.23 ± 0.11 | 116 | 5 | 40 | 19,462.17 | 4.76 | 5.2 | R.DLISHDELFSDIYK.I | |
| P63085 | Mitogen-activated protein kinase 1 | 0.85 ± 0.16 | 0.80 ± 0.18 | 306 | 13 | 52 | 41,144.41 | 6.53 | 7.0 | K.ISPFEHQTYCQR.T R.FRHENIIGINDIIR.A R.VADPDHDHTGFLTEYVATR.W |
|
| P63087 | Serine/threonine- protein phosphatase PP1-gamma catalytic subunit | 0.15 ± 0.05 | 0.12 ± 0.03 | 212 | 12 | 45 | 36,983.79 | 6.12 | 6.1 | R.YPENFFLLR.G K.ICGDIHGQYYDLLR.L |
|
| P63101 | 14-3-3 protein zeta/delta | 0.66 ± 0.26 | 1.12 ± 0.56 | 300 | 15 | 58 | 27,771.14 | 4.73 | 4.9 | K.DSTLIMQLLR.D K.DSTLIMQLLR.D + Oxidation (M) K.GIVDQSQQAYQEAFEISK.K K.TAFDEAIAELDTLSEESYK.D |
|
| P70296 | Phosphatidylethanolamine-binding protein 1 | ||||||||||
| (1) | 1.17 ± 0.61 | 1.22 ± 0.58 | 274 | 13 | 72 | 20,699.25 | 5.19 | 5.2 | K.LYTLVLTDPDAPSR.K K.GNDISSGTVLSDYVGSGPPSGTGLHR.Y |
||
| (2) | 1.73 ± 0.32 | 1.80 ± 0.23 | 80 | 7 | 63 | 20,699.25 | 5.19 | 5.4 | |||
| (3) | 0.80 ± 0.37 | 0.66 ± 0.27 | 298 | 9 | 65 | 20,699.25 | 5.19 | 4.9 | K.LYTLVLTDPDAPSR.K K.GNDISSGTVLSDYVGSGPPSGTGLHR.Y |
||
| Q01768 | Nucleoside diphosphate kinase B | 0.82 ± 0.28 | 0.91 ± 0.16 | 369 | 10 | 69 | 17,363.07 | 6.97 | 7.2 | DRPFFPGLVK.Y R.TFIAIKPDGVQR.G R.VMLGETNPADSKPGTIR.G K.EIHLWFKPEELIDYK.S |
|
| Q06138 | Similar to Calcium- binding protein 39, Q06138 (100%) | 0.10 ± 0.08 | 0.10 ± 0.03 | 138 | 13 | 39 | 39,841.97 | 6.69 | 7.0 | K.DVAQIFNNILR.R R.NIQFEAFHVFK.V K.LLSAEFLEQHYDR.F |
|
| Q08331 | Calretinin | 0.50 ± 0.17 | 0.44 ± 0.18 | 172 | 10 | 42 | 31,372.64 | 4.94 | 5.2 | R.LLPVQENFLLK.F K.ELENFFQELEK.A |
|
| Q3TBA0 | Similar to SEPTIN6 type II homolog | 0.40 ± 0.10 | 0.40 ± 0.09 | 110 | 10 | 36 | 48,951.79 | 6.36 | 6.4 | K.FESDPATHNEPGVR.L | |
| Q3TBI9 | Guanosine diphosphate (GDP) dissociation inhibitor 1, full insert sequence | 0.42 ± 0.31 | 0.42 ± 0.12 | 554 | 31 | 66 | 50,539.61 | 4.95 | 5.2 | K.FLVFVANFDENDPK.T K.SPYLYPLYGLGELPQGFAR.L K.FDLGQDVIDFTGHALALYR.T K.NTNDANSCQIIIPQNQVNR.K |
|
| Q3THY7 | Guanine nucleotide- binding protein, beta 2, related sequence 1, full insert sequence | 0.41 ± 0.14 | 0.39 ± 0.16 | 158 | 12 | 64 | 35,048.67 | 7.06 | 7.5 | K.HLYTLDGGDIINALCFSPNR.Y | |
| Q3U1B1 | Guanine nucleotide- binding protein, beta 1, full insert sequence | 1.82 ± 1.03 | 1.95 ± 0.97 | 271 | 12 | 54 | 37,404.04 | 5.75 | 5.5 | R.ELAGHTGYLSCCR.F K.ACADATLSQITNNIDPVGR.I |
|
| Q3U5X4 | Annexin A5, full insert sequence | 0.23 ± 0.11 | 0.19 ± 0.09 | 137 | 11 | 37 | 35,752.44 | 4.83 | 5.2 | K.GLGTDEDSILNLLTSR.S | |
| Q3UKW2 | Calmodulin 1, full insert sequence | 3.71 ± 0.60 | 3.95 ± 1.01 | 133 | 5 | 27 | 21,559.82 | 4.21 | 4.1 | R.VFDKDGNGYISAAELR.H | |
| Q3UL78 | Cell division cycle 42 homolog (Saccharomyces cerevisiae), full insert sequence | (1) | 0.33 ± 0.07 | 0.35 ± 0.09 | 165 | 5 | 61 | 16,503.07 | 6.14 | 5.8 | K.TPFLLVGTQIDLR.D |
| (2) | 0.21 ± 0.04 | 0.18 ± 0.05 | 163 | 5 | 61 | 16,503.07 | 6.14 | 6.4 | K.TPFLLVGTQIDLR.D | ||
| Q3UM45 | Protein phosphatase 1 regulatory subunit 7 | 0.13 ± 0.05 | 0.16 ± 0.09 | 154 | 14 | 49 | 41,291.95 | 4.85 | 5.15 | K.LQNLDALTNLTVLSVQSNR.L | |
| Q3UUX9 | Guanosine diphosphate (GDP) dissociation inhibitor 3, full insert sequence | (1) | 0.14 ± 0.06 | 0.17 ± 0.05 | 81 | 10 | 33 | 50,537.13 | 5.93 | 5.95 | |
| (2) | 0.24 ± 0.11 | 0.21 ± 0.10 | 315 | 25 | 62 | 50,537.13 | 5.93 | 5.9 | K.SPYLYPLYGLGELPQGFAR.L K.FDLGQDVIDFTGHSLALYR.T |
||
| Q3UWP8 | Calreticulin, full insert sequence. [Fragment] | 0.45 ± 0.15 | 0.54 ± 0.19 | 84 | 10 | 41 | 42,195.92 | 4.57 | 4.7 | ||
| Q3V067 | Protein phosphatase 3, regulatory subunit B, alpha isoform (calcineurin B, type I), full insert sequence | (1) | 0.63 ± 0.11 | 0.75 ± 0.24 | 484 | 10 | 65 | 18,207.72 | 4.73 | 4.9 | K.DGYISNGELFQVLK.M R.VIDIFDTDGNGEVDFK.E R.ISFEEFCAVVGGLDIHK.K |
| (2) | 0.70 ± 0.13 | 0.69 ± 0.14 | 431 | 9 | 65 | 18,207.72 | 4.73 | 4.9 | K.DGYISNGELFQVLK.M R.VIDIFDTDGNGEVDFK.E R.ISFEEFCAVVGGLDIHK.K |
||
| Q543N6 | Protein phosphatase 2a, regulatory subunit B′ (PP2A, subunit B′, PR53 isoform) | 0.10 ± 0.04 | 0.10 ± 0.04 | 74 | 7 | 32 | 36,710.15 | 5.95 | 6.0 | ||
| Q545R3 | N-myc downstream regulated 1, full insert sequence | 0.11 ± 0.02 | 0.10 ± 0.02 | 67 | 10 | 29 | 43,008.65 | 5.96 | 5.78 | ||
| Q60631 | Growth factor receptor-bound protein 2 | (1) | 0.46 ± 0.01 | 0.05 ± 0.01 | 165 | 5 | 61 | 16,503.07 | 6.14 | 5.8 | K.TPFLLVGTQIDLR.D |
| (2) | 0.18 ± 0.03 | 0.17 ± 0.03 | 163 | 5 | 61 | 16,503.07 | 6.14 | 5.8 | K.TPFLLVGTQIDLR.D | ||
| Q60829 | Dopamine- and cAMP- regulated neuronal phosphoprotein | 0.41 ± 0.30 | 0.38 ± 0.13 | 71 | 5 | 45 | 21,780.53 | 4.60 | 5.0 | ||
| Q60932 | Voltage-dependent anion-selective channel protein 1 | (1) | 1.03 ± 0.33 | 1.16 ± 0.41 | 743 | 5 | 72 | 32,351.49 | 8.55 | 8.4 | K.KLETAVNLAWTAGNSNTR.F K.VNNSSLIGLGYTQTLKPGIK.L R.EHINLGCDVDFDIAGPSIR.G K.WNTDNTLGTEITVEDQLAR.G K.TDEFQLHTNVNDGTEFGGSIYQK.V |
| (2) | 1.00 ± 0.44 | 0.94 ± 0.46 | 532 | 13 | 59 | 32,351.49 | 8.55 | 8.5 | R.WTEYGLTFTEK.W R.EHINLGCDVDFDIAGPSIR.G K.WNTDNTLGTEITVEDQLAR.G K.TDEFQLHTNVNDGTEFGGSIYQK.V |
||
| (3) | 1.12 ± 0.45 | 1.10 ± 0.34 | 442 | 13 | 60 | 32,351.49 | 8.55 | 7.5 | K.YQVDPDACFSAK.V K.WNTDNTLGTEITVEDQLAR.G K.TDEFQLHTNVNDGTEFGGSIYQK.V |
||
| (4) | 0.46 ± 0.12 | 0.43 ± 0.14 | 100 | 8 | 49 | 32,351.49 | 8.55 | 6.8 | |||
| (5) | 0.95 ± 0.29 | 0.97 ± 0.20 | 360 | 12 | 55 | 32,351.49 | 8.55 | 6.9 | R.EHINLGCDVDFDIAGPSIR.G K.WNTDNTLGTEITVEDQLAR.G |
||
| (6) | 0.32 ± 0.07 | 0.28 ± 0.08 | 321 | 10 | 48 | 32,351.49 | 8.55 | 6.5 | R.WTEYGLTFTEK.W R.EHINLGCDVDFDIAGPSIR.G K.WNTDNTLGTEITVEDQLAR.G K.TDEFQLHTNVNDGTEFGGSIYQK.V |
||
| Q62048 | Astrocytic phosphoprotein PEA-15 | 0.30 ± 0.11 | 0.28 ± 0.11 | 109 | 5 | 36 | 15,054.13 | 4.94 | 5.2 | K.DNLSYIEHIFEISR.R | |
| Q63844 | Mitogen-activated protein kinase 3 | 0.06 ± 0.04 | 0.08 ± 0.05 | 75 | 8 | 34 | 42,935.30 | 6.16 | 6.2 | ||
| Q6P1F6 | Similar to Serine/threonine-protein phosphatase 2A 55 kDa regulatory subunit B, alpha isoform, Q6P1F6 (95%) | 0.25 ± 0.08 | 0.24 ± 0.07 | 147 | 16 | 41 | 51,692.08 | 5.82 | 6.0 | R.INLWHLEITDR.S | |
| Q6PI18 | Similar to Valosin containing protein, Q6PI18 (91%) | 0.22 ± 0.12 | 0.24 ± 0.22 | 138 | 17 | 30 | 89,321.80 | 5.14 | 5.65 | ||
| Q76MZ3 | Serine/threonine- protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform | 0.47 ± 0.23 | 0.42 ± 0.16 | 150 | 20 | 38 | 65,191.42 | 5.00 | 5.2 | ||
| Q78MH6 | Voltage-dependent anion channel 2 | (1) | 0.87 ± 0.27 | 0.72 ± 0.16 | 101 | 6 | 37 | 31,732.84 | 7.44 | 7.2 | K.VNNSSLIGVGYTQTLRPGVK.L |
| (2) | 0.43 ± 0.09 | 0.40 ± 0.09 | 417 | 10 | 53 | 6.1 | K.WCEYGLTFTEK.W K.VNNSSLIGVGYTQTLRPGVK.L K.VCEDFDTSVNLAWTSGTNCTR.F R.TGDFQLHTNVNNGTEFGGSIYQK.V |
||||
| Q8BG73 | SH3 domain-binding glutamic acid-rich-like protein 2 | 0.17 ± 0.10 | 0.15 ± 0.09 | 233 | 4 | 53 | 12,254.96 | 5.49 | 5.7 | R.YCGDYDSFFESK.E K.KPAQGNPLPPQIFNGDR.Y |
|
| Q8BN07 | Protein phosphatase 2a, catalytic subunit, beta isoform, full insert sequence. [Fragment] | 0.13 ± 0.10 | 0.26 ± 0.25 | 126 | 12 | 50 | 32643.82 | 5.37 | 5.4 | K.YSFLQFDPAPR.R | |
| Q8BNF8 | Valosin containing protein, full insert sequence | 0.09 ± 0.08 | 0.13 ± 0.18 | 135 | 15 | 34 | 80,149.37 | 5.12 | 5.4 | ||
| Q8BPI0 | RHOGDI-1 homolog [Fragment] | (1) | 0.29 ± 0.07 | 0.60 ± 0.01 | 245 | 12 | 56 | 22,947.89 | 5.20 | 5.4 | R.VAVSADPNVPNVIVTR.L R.AEEYEFLTPMEEAPK.G R.AEEYEFLTPMEEAPK.G + Oxidation (M) R.LTLVCSTAPGPLELDLTGDLESFK.K |
| (2) | 0.42 ± 0.12 | 0.46 ± 0.20 | 192 | 9 | 46 | 22,947.89 | 5.20 | 5.3 | R.VAVSADPNVPNVIVTR.L K.SIQEIQELDKDDESLR.K |
||
| (3) | 0.11 ± 0.08 | 0.13 ± 0.09 | 164 | 11 | 52 | 22,947.89 | 5.20 | 5.2 | R.VAVSADPNVPNVIVTR.L | ||
| Q8BXS6 | SH3 domain protein 2A, full insert sequence | 0.23 ± 0.06 | 0.29 ± 0.10 | 152 | 8 | 35 | 39,941.27 | 5.20 | 5.5 | K.QAVQILQQVTVR.L R.ALYDFEPENEGELGFK.E |
|
| Q8C1B7 | Septin-11 | (1) | 0.17 ± 0.06 | 0.15 ± 0.04 | 411 | 18 | 47 | 49,563.45 | 6.26 | 6.8 | R.SLFNYHDTR.I K.FESDPATHNEPGVR.L R.NLSLSGHVGFDSLPDQLVNK.S |
| (2) | 0.27 ± 0.05 | 0.25 ± 0.05 | 159 | 12 | 44 | 49,563.45 | 6.26 | 6.4 | K.FESDPATHNEPGVR.L | ||
| Q8C2L2 | 2 days neonate thymus thymic cells cDNA, RIKEN full-length enriched library, clone:E430014P14 product:septin 6, full insert sequence. | 0.134 ± 0.03 | 0.12 ± 0.04 | 94 | 9 | 48 | 39,698.18 | 5.56 | 6.2 | R.TVPLAGHVGFDSLPDQLVNK.S | |
| Q8CEG4 | Similar to valosin containing protein, full insert sequence, Q8CEG4 (97%) | 0.28 ± 0.10 | 0.27 ± 0.09 | 143 | 7 | 19 | 89,363.88 | 5.14 | 5.65 | R.EDEEESLNEVGYDDIGGCR.K | |
| Q8CHH9 | Septin-8 | (1) | 0.16 ± 0.04 | 0.19 ± 0.06 | 91 | 13 | 41 | 49,681.19 | 5.68 | 5.8 | |
| (2) | 0.13 ± 0.05 | 0.12 ± 0.03 | 71 | 9 | 32 | 49,681.19 | 5.86 | 5.8 | |||
| (3) | 0.12 ± 0.04 | 0.13 ± 0.03 | 99 | 10 | 40 | 49,681.19 | 5.68 | 5.7 | |||
| Q8JZR2 | Crk protein | 0.36 ± 0.11 | 0.38 ± 0.12 | 70 | 6 | 51 | 22,889.53 | 5.33 | 5.4 | ||
| Q99K69 | RAS protein activator like 1 | 0.04 ± 0.03 | 0.04 ± 0.03 | 104 | 12 | 25 | 89,394.80 | 5.95 | 6.1 | ||
| Q9CQI6 | Coactosin-like protein | 0.23 ± 0.05 | 0.25 ± 0.05 | 104 | 3 | 39 | 15,812.72 | 5.28 | 5.2 | R.DDGSAVIWVTFR.Y dimethylaminohydrolase 1 | |
| Q9CWS0 | NG,NG- dimethylarginine | 0.43 ± 0.13 | 0.45 ± 0.11 | 399 | 14 | 46 | 31,249.78 | 5.64 | 5.7 | K.VDGLLTCCSVFINK.K K.DENATLDGGDVLFTGR.E K.DYAVSTVPVADSLHLK.S K.LTVPDDMAANCIYLNIPSK.G |
|
| Q9D7X3 | Dual specificity protein phosphatase 3 | 0.15 ± 0.063 | 0.12 ± 0.06 | 114 | 7 | 45 | 20,472.15 | 6.07 | 7.5 | K.ANDTQEFNLSAYFER.A R.EIGPNDGFLAQLCQLNDR.L |
|
| Q9DC07 | LIM and SH3 protein (LASP Homolog), full insert sequence | 0.70 ± 0.19 | 0.62 ± 0.20 | 91 | 10 | 43 | 31,113.09 | 8.54 | 8.1 | R.AMYDYSAQDEDEVSFR.D + Oxidation (M) | |
| Q9R1T4 | Septin-6 | 0.18 ± 0.03 | 0.19 ± 0.04 | 107 | 9 | 42 | 49,488.37 | 6.04 | 6.2 | ||
| Q9Z2Q6 | Septin-5 | (1) | 0.56 ± 0.18 | 0.41± 0.07 | 450 | 17 | 50 | 42,747.94 | 6.21 | 6.2 |
K.DVTCDVHYENYR.A K.QYVGFATLPNQVHR.K K.ESAPFAVIGSNTVVEAK.G R.LYPWGIVEVENQAHCDFVK.L |
| (2) | 0.19 ± 0.05 | 0.18 ± 0.05 | 65 | 5 | 21 | 42,747.94 | 6.21 | 6.1 | |||
| (3) | 0.09 ± 0.04 | 0.10 ± 0.04 | 68 | 7 | 32 | 41,525.52 | 6.10 | 6.0 | |||
| (4) | 0.18 ± 0.06 | 0.19 ± 0.06 | 65 | 7 | 33 | 42,747.94 | 6.21 | 6.2 |
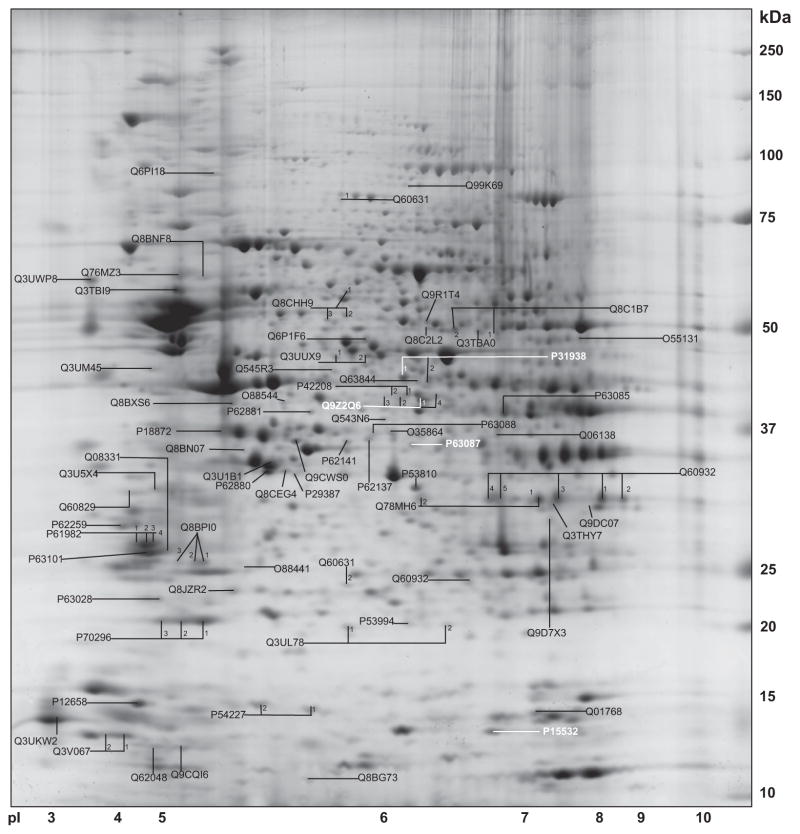
The Fig. 1 shows a combined identification map of signaling proteins.
Fig. 1.
A combined map of proteins from WT and KO mice is given. Spots are identified and UniProtKB accession numbers are provided. Proteins with significantly different levels are shown in “white”.
3.2. Protein quantification
As shown above, four proteins showed statistically significant differential hippocampal levels between groups and Table 1 demonstrates means and standard deviations as well as statistical results.
Nucleoside diphosphate kinase A (EC 2.7.4.6) was represented by a single spot and levels were reduced in KO mice by about 12%.
Dual specificity mitogen-activated protein kinase 1 (syn.: MAP kinase kinase 1, MAPKK1, ERK activator kinase 1, MEK1) with the enzyme classification number EC 2.7.12.2, was reduced in hippocampus of KO mice by about 16% and was represented by two spots.
Serine/threonine-protein phosphatase 1 (EC 3.1.3.16) levels were decreased by about 20% and the enzyme was represented by a single spot.
Septin-5 was represented by four spots and spot volume of one of them was reduced in hippocampus of KO mice by about 27%.
Gel images of the spots with different protein levels between groups are shown in Fig. 2.
Fig. 2.
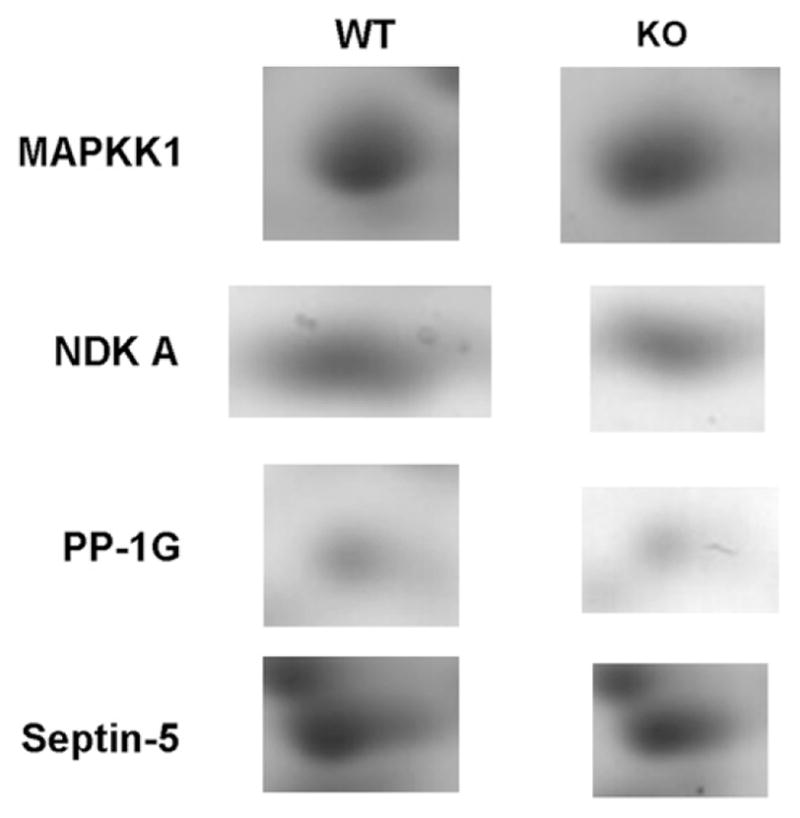
Images of individual spots on 2-DE in WT and KO mice.
3.3. Novel phosphorylation sites
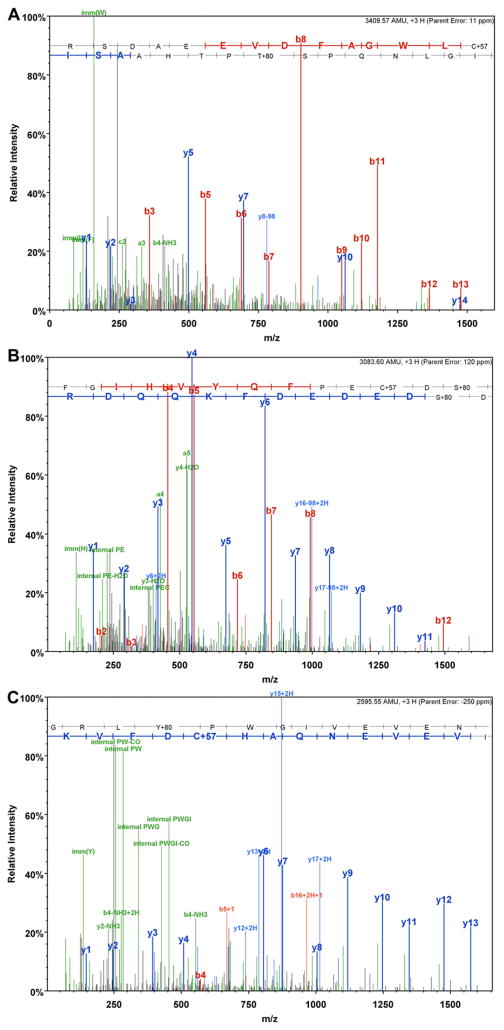
nano-ESI-LC/MS/MS revealed a spectrum of Dual specificity mitogen-activated protein kinase kinase 1(P31938) with a peptide 363RSDAEEVDFAGWLCSTIGLNQPSTPTHAASI393 (m/z = 1137.53, 3+) containing T386 phosphorylation (Fig. 3a).
Fig. 3.
Identification of phosphorylated tryptic peptides by nano-ESI-LC/MS/MS. a) MS/MS spectrum of dual specificity mitogen-activated protein kinase kinase 1 (P31938) peptide 363RSDAEEVDFAGWLCSTIGLNQPSTPTHAASI393 (m/z = 1137.53, 3+) containing T386 phosphorylation.; b) MS/MS spectrum of Septin-5 (Q9Z2Q6) peptide 213FGIHVYQFPECDSDEDEDFKQQDR236 (m/z = 1028.87, 3+) containing S225 phosphorylation; c) MS/MS spectrum of Septin-5 (Q9Z2Q6) peptide 262GRLYPWGIVEVENQAHCDFVK282 (m/z = 866.19, 3+) containing Y265 phosphorylation.
Another two spectra were obtained for Septin-5 peptides, showing the sequence 213FGIHVYQFPECDSDEDEDFKQQDR236 (m/z = 1028.87, 3+,) containing S225 phosphorylation (Fig. 3b) and the peptide 262GRLYPWGIVEVENQAHCDFVK282 (m/z = 866.19, 3+) demonstrating Y265 phosphorylation (Fig. 3c).
3.4. Western blotting
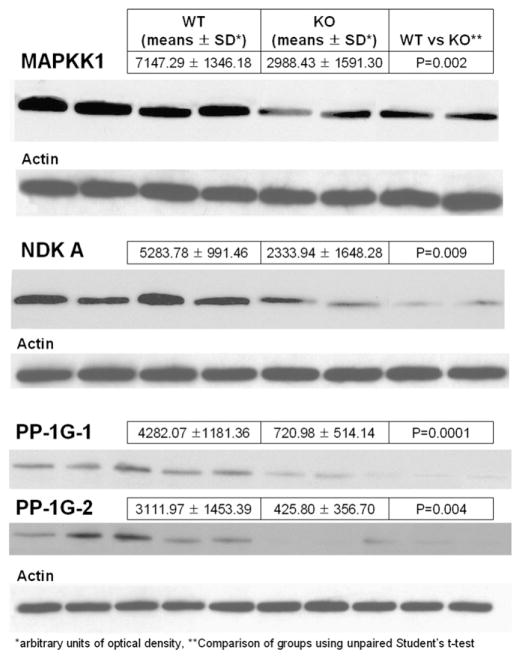
As shown in Fig. 4 results from 2-DE could be verified for MAPKK1, NDKA, and the two expression forms of PP-1G.
Fig. 4.
Western blots showing immunoreactive bands for MAPKK1, NDKA and the two expression forms of PP-1G including results from immunoblotting. A single actin band was comparable between individuals and groups and used as loading control.
Actin used as a loading control was comparable between lanes.
4. Discussion
The major outcome of the study is derangement of protein levels for signaling proteins Nucleoside diphosphate kinase A (NDK A), Dual specificity mitogen-activated protein kinase kinase 1 (MAPKK 1), Serine/threonine-protein phosphatase PP1-gamma catalytic subunit (PP-1G), Septin-5, in (5-HT1A R) KO mice, findings that may well be linked to the reported cognitive deficits. Moreover, the protein chemical finding of novel phosphorylation sites at T386 on MAPKK 1 and at S225 and Y265 on Septin-5 may be of functional relevance.
Protein levels of Nucleoside diphosphate kinase A but not the B form were significantly reduced in the KO animals. This is in agreement with a previous report studying brain areas from patients with Alzheimer’s disease demonstrating decreased nucleoside diphosphate kinase A levels and activity (Kim et al., 2002). Decreased levels in the KO strain may be providing a clue for a possible role of this enzyme in the mechanisms of cognitive function or at least indicate 5-HT1A R-dependent modulation of protein levels.
MAPKK1 is a major signaling compound essential for generation of long-term synaptic plasticity and memory (Kelleher et al., 2004). Moreover, MAPKK 1 is involved in regulation of the NMDA receptor currents, an indeed, the NMDA receptor is the key element for generation of cognitive functions (Gu et al., 2005). Reduced hippocampal levels for one expression form of (spot 1 with a slightly lower pI) MAPKK 1, the phosphorylated form containing the novel phosphorylation site T386, may be well responsible for cognitive decline as reported for this (5-HT1A R) KO mouse. The fact that only one expression form is reduced shows specificity of the finding and there was no phosphorylation site for MAPKK 1 described so far and only predicted based upon similarity.
Serine/threonine-protein phosphatase 1 (PP1) has a key role in the formation of learning and memory (Mansuy and Shenolikar, 2006) in health and neurodegenerative disease and discriminates poor and good learning mouse strains (Pollak et al., 2006). The one expression form observed in our study revealed moderate but significantly reduced PP-1G hippocampal levels. The biological significance of reduced PP-1G levels remains elusive but fits the context of the signaling derangement in this KO mouse.
Septin-5 is a functionally poorly described protein analyzed by mass spectrometry in synaptosomes and brain (Munton et al., 2007; http://www.expasy.org/uniprot/Q9Z2Q6#ref4 and http://www.expasy.org/uniprot/Q9Z2Q6#ref5) with predicted function as a cell division control-related protein based upon domains and we herein report a role in the effector chain of the 5-HT1A R, an association with impaired cognitive function in the mouse or at least a link to the 5-HT1A R knock-out state. Only one out of four (spot 1) expression forms was presented with significantly reduced hippocampal protein levels. On this expression form two novel phosphorylation sites, at S225 and at Y265 were herein observed (so far only phosphorylation of S327 and T336 were reported) and decreased hippocampal levels may be compatible with cognitive decline in the 5-HT1A R – KO mouse as well – In human fetal Down syndrome cortex, a member of the septin family, Septin-7, was significantly and remarkably reduced that may support a role of this protein for involvement in cognitive functions (Engidawork et al., 2003).
We conclude that genetic inactivation of the 5-HT1A R in the mouse is linked to aberrant signaling involving different effectors, protein kinase and – phosphatase systems. These signaling systems were already associated with cognitive deficits in dementing disorders and it is herein linked to the 5-HT1A R in the KO mouse that shows severe cognitive impairment. And indeed, the 5-HT1A R has been shown to be linked to impaired cognitive function in neurodegenerative disease (Borg, 2008). As to the novel findings of phosphorylation sites, we may conclude that these post-translational modifications may change the function of the corresponding proteins significantly and the activity of signaling systems do very much depend on the phosphorylation state. We are currently following up complete signaling cascades the effector proteins are belonging to and carry out studies complementing information on the novel phosphorylation sites including quantification. This study provides the first evidence for a link between the above mentioned signaling proteins and the 5-HT1A receptor.
Acknowledgments
We are highly indebted to the Verein zur Durchführung der wissenschaftlichen Forschung auf dem Gebiet der Neonatologie und Kinderintensivmedizin “Unser Kind”.
References
- Albert PR, Tiberi M. Receptor signaling and structure: insights from serotonin-1 receptors. Trends Endocrinol Metab. 2001;12:453–460. doi: 10.1016/s1043-2760(01)00498-2. [DOI] [PubMed] [Google Scholar]
- Bailey SJ, Toth M. Variability in the benzodiazepine response of serotonin 5-HT1A receptor null mice displaying anxiety-like phenotype: evidence for genetic modifiers in the 5-HT-mediated regulation of GABA(A) receptors. J Neurosci. 2004;24:6343–6351. doi: 10.1523/JNEUROSCI.0563-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borg J. Molecular imaging of the 5-HT(1A) receptor in relation to human cognition. Behav Brain Res. 2008;195:103–111. doi: 10.1016/j.bbr.2008.06.011. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Chen WQ, Kang SU, Lubec G. Protein profiling by the combination of two independent mass spectrometry techniques. Nat Protoc. 2006;1:1446–1452. doi: 10.1038/nprot.2006.246. [DOI] [PubMed] [Google Scholar]
- Cheon MS, Dierssen M, Kim SH, Lubec G. Protein expression of BACE1, BACE2 and APP in Down syndrome brains. Amino Acids. 2008;35:339–343. doi: 10.1007/s00726-007-0618-9. [DOI] [PubMed] [Google Scholar]
- Engidawork E, Gulesserian T, Fountoulakis M, Lubec G. Aberrant protein expression in cerebral cortex of fetus with Down syndrome. Neuroscience. 2003;122:145–154. doi: 10.1016/s0306-4522(03)00605-5. [DOI] [PubMed] [Google Scholar]
- Frischer T, Myung JK, Maurer G, Eichler I, Szepfalusi Z, Lubec G. Possible dysregulation of chaperon and metabolic proteins in cystic fibrosis bronchial tissue. Proteomics. 2006;6:3381–3388. doi: 10.1002/pmic.200500487. [DOI] [PubMed] [Google Scholar]
- Gu Z, Jiang Q, Fu AK, Ip NY, Yan Z. Regulation of NMDA receptors by neuregulin signaling in prefrontal cortex. J Neurosci. 2005;25:4974–4984. doi: 10.1523/JNEUROSCI.1086-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensler JG. Serotonin. In: Siegel G, Albers RW, Brady S, Price D, editors. Basic Neurochemistry: Molecular, Cellular and Medical Aspects. 7. Academic Press; New York: 2005. pp. 227–247. [Google Scholar]
- John JP, Pintsov O, Petter-Puchner A, Redl H, Pollak A, Chen WQ, Lubec G. Nitric oxide and oxygen radical attack on GDP-dissociation inhibitor 2 (GDI-2) in spinal cord injury of the rat. J Proteome Res. 2007;6:1500–1509. doi: 10.1021/pr060620k. [DOI] [PubMed] [Google Scholar]
- Kelleher RJ, 3rd, Govindarajan A, Jung HY, Kang H, Tonegawa S. Translational control by MAPK signaling in long-term synaptic plasticity and memory. Cell. 2004;116:467–479. doi: 10.1016/s0092-8674(04)00115-1. [DOI] [PubMed] [Google Scholar]
- Kim SH, Fountoulakis M, Cairns NJ, Lubec G. Human brain nucleoside diphosphate kinase activity is decreased in Alzheimer’s disease and Down syndrome. Biochem Biophys Res Commun. 2002;296:970–975. doi: 10.1016/s0006-291x(02)02035-1. [DOI] [PubMed] [Google Scholar]
- Mansuy IM, Shenolikar S. Protein serine/threonine phosphatases in neuronal plasticity and disorders of learning and memory. Trends Neurosci. 2006;29:679–686. doi: 10.1016/j.tins.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Mukhin YV, Garnovskaya MN, Collinsworth G, Grewal JS, Pendergrass D, Nagai T, Pinckney S, Greene EL, Raymond JR. 5-Hydroxy-tryptamine1A receptor/Gibetagamma stimulates mitogen-activated protein kinase via NAD(P)H oxidase and reactive oxygen species upstream of src in Chinese hamster ovary fibroblasts. Biochem J. 2000;347:61–67. [PMC free article] [PubMed] [Google Scholar]
- Munton RP, Tweedie-Cullen R, Livingstone-Zatchej M, Weinandy F, Waidelich M, Longo D, Gehrig P, Potthast F, Rutishauser D, Gerrits B, Panse C, Schlapbach R, Mansuy IM. Qualitative and quantitative analyses of protein phosphorylation in naive and stimulated mouse synaptosomal preparations. Mol Cell Proteomics. 2007;6:283–293. doi: 10.1074/mcp.M600046-MCP200. [DOI] [PubMed] [Google Scholar]
- Parks CL, Robinson PS, Sibille E, Shenk T, Toth M. Increased anxiety of mice lacking the serotonin1A receptor. Proc Natl Acad Sci U S A. 1998;95:10734–10739. doi: 10.1073/pnas.95.18.10734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollak DD, John J, Schneider A, Hoeger H, Lubec G. Strain-dependent expression of signaling proteins in the mouse hippocampus. Neuroscience. 2006;138:149–158. doi: 10.1016/j.neuroscience.2005.11.004. [DOI] [PubMed] [Google Scholar]
- Raymond JR, Olsen CL, Gettys TW. Cell-specific physical and functional coupling of human 5-HT1A receptors to inhibitory G protein alpha-subunits and lack of coupling to Gs alpha. Biochemistry. 1993;32:11064–11073. doi: 10.1021/bi00092a016. [DOI] [PubMed] [Google Scholar]
- Raymond JR, Mukhin YV, Gettys TW, Garnovskaya MN. The recombinant 5-HT1A receptor: G protein coupling and signalling pathways. Br J Pharmacol. 1999;127:1751–1764. doi: 10.1038/sj.bjp.0702723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond JR, Mukhin YV, Gelasco A, Turner J, Collinsworth G, Gettys TW, Grewal JS, Garnovskaya MN. Multiplicity of mechanisms of serotonin receptor signal transduction. Pharmacol Ther. 2001;92:179–212. doi: 10.1016/s0163-7258(01)00169-3. [DOI] [PubMed] [Google Scholar]
- Riad M, Garcia S, Watkins KC, Jodoin N, Doucet E, Langlois X, el Mestikawy S, Hamon M, Descarries L. Somatodendritic localization of 5-HT1A and preterminal axonal localization of 5-HT1B serotonin receptors in adult rat brain. J Comp Neurol. 2000;417:181–194. [PubMed] [Google Scholar]
- Sarnyai Z, Sibille EL, Pavlides C, Fenster RJ, McEwen BS, Toth M. Impaired hippocampal-dependent learning and functional abnormalities in the hippocampus in mice lacking serotonin(1A) receptors. Proc Natl Acad Sci U S A. 2000;97:4731–4736. doi: 10.1073/pnas.97.26.14731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiapparelli L, Del Río J, Frechilla D. Serotonin 5-HT receptor blockade enhances Ca(2+)/calmodulin-dependent protein kinase II function and membrane expression of AMPA receptor subunits in the rat hippocampus: implications for memory formation. J Neurochem. 2005;94:884–895. doi: 10.1111/j.1471-4159.2005.03193.x. [DOI] [PubMed] [Google Scholar]
- Shin JH, Krapfenbauer K, Lubec G. Mass-spectrometrical analysis of proteins encoded on chromosome 21 in human fetal brain. Amino Acids. 2006;31:435–447. doi: 10.1007/s00726-005-0257-y. [DOI] [PubMed] [Google Scholar]
- Sunyer B, Shim KS, An G, Hoger H, Lubec G. Hippocampal levels of phosphorylated protein kinase a (phosphor-S96) are linked to spatial memory enhancement by SGS742. Hippocampus. 2009;19:90–98. doi: 10.1002/hipo.20484. [DOI] [PubMed] [Google Scholar]
- Weitzdorfer R, Hoger H, Pollak A, Lubec G. Changes of hippocampal protein levels during postnatal brain development in the rat. J Proteome Res. 2006;5:3205–3212. doi: 10.1021/pr0602545. [DOI] [PubMed] [Google Scholar]
- Yuen EY, Jiang Q, Chen P, Gu Z, Feng J, Yan Z. Serotonin 5-HT1A receptors regulate NMDA receptor channels through a microtubule-dependent mechanism. J Neurosci. 2005;25:5488–5501. doi: 10.1523/JNEUROSCI.1187-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]