Abstract
Next-generation sequencing technology provides an attractive means to obtain large-scale sequence data necessary for comparative genomic analysis. To analyse the patterns of mutation rate variation and selection intensity across the avian genome, we performed brain transcriptome sequencing using Roche 454 technology of 10 different non-model avian species. Contigs from de novo assemblies were aligned to the two available avian reference genomes, chicken and zebra finch. In total, we identified 6499 different genes across all 10 species, with ~1000 genes found in each full run per species. We found evidence for a higher mutation rate of the Z chromosome than of autosomes (male-biased mutation) and a negative correlation between the neutral substitution rate (dS) and chromosome size. Analyses of the mean dN/dS ratio (ω) of genes across chromosomes supported the Hill–Robertson effect (the effect of selection at linked loci) and point at stochastic problems with ω as an independent measure of selection. Overall, this study demonstrates the usefulness of next-generation sequencing for obtaining genomic resources for comparative genomic analysis of non-model organisms.
Keywords: Next generation sequencing, 454, Avian genomics, Male-mutation bias, Selection, Hill-Robertson effect
Introduction
A major goal in evolutionary biology is to identify and subsequently study those loci that contribute to key phenotypes involved in adaptation, reproduction and survival (Benfey & Mitchell-Olds 2008; Ellegren & Sheldon 2008; Stinchcombe & Hoekstra 2008). Not long ago, this seemed to be a far-fetched goal for biologists working with natural populations of non-model organisms. Large-scale genomic analysis was mainly restricted to model organisms such as Drosophila, Arabidopsis, Saccharomyces, and domestic animals and plants. However, the last few years have seen a change, in that genomic tools have become integral parts of molecular ecology. To mention but a few examples, gene expression (transcriptome) profiling has helped to reveal how variation in gene regulation can be tied to phenotypic variation (Rockman & Kruglyak 2006), the use of genome-wide sets of genetic markers have greatly facilitated mapping of trait loci in pedigrees (linkage mapping) (Beraldi et al. 2007) or populations (association mapping) (Wood et al. 2008), and the focus on candidate genes has offered a direct link to phenotypes in natural populations (Abzhanov et al. 2004).
Ultimately, if we are to fully be able to dissect the relationship between genotypes and phenotypes, genome sequence information will be needed from ecologically relevant organisms. The recent introduction of ‘next-generation’ or massive parallel sequencing technologies (Rothberg & Leamon 2008) in molecular ecology (Vera et al. 2008) holds promise that this will eventually come about (Bonneaud et al. 2008; Ellegren 2008b; Hudson 2008). Importantly, the generation of large amounts of DNA sequence data from related species will allow comparative genomic approaches for the identification of trait loci, and this is particularly so with transcriptome sequencing (‘RNA-seq’; Wang et al. 2009). Transcriptome sequencing reflects the subset of genes from the genome that are functionally active in a selected tissue and species of interest.
Together with expression profiling, genetic mapping and candidate gene approaches, comparative genomics represents one of the main routes for dissecting the genetic basis of phenotypic variation (Ellegren & Sheldon 2008). One common application in comparative genomics is to analyse sequences of orthologous genes from two or more species for rates of divergence and to test whether this divergence deviates from rates expected under a neutral scenario (Ellegren 2008a). Accelerated divergence beyond neutral expectations would indicate evidence of adaptive evolution where positive selection has increased the fixation rate of beneficial alleles (Nielsen 2005; Wright & Andolfatto 2008). In contrast, sequence conservation beyond neutral expectations would indicate evidence of purifying selection due to functional constraints (Ponting 2008). Assessing these rates typically involves analysing the rates of accumulation of non-synonymous substitution (dN) and synonymous substitution (dS) in protein-coding sequence.
So far, there are only a few studies that have utilized genome or transcriptome sequence data from multiple species to make general interferences about natural selection across species. This includes analyses of whole-genome sequences from drosophilids (Begun et al. 2007; Clark et al. 2007) and mammals (Kosiol et al. 2008). The purpose of the present study was to use a next-generation sequencing approach for comparative genomic analyses of mutation and selection processes in avian genomes. Specifically, we performed brain transcriptome sequencing of 10 bird species using Roche 454 technology. This generated a total of more than 1 Gb of raw sequence data, yielding an assembly of 7.27 Mb that could be unambiguously identified as protein-coding sequences.
Materials and methods
Bird species, cDNA library preparation, normalization and transcriptome sequencing
Ten bird species were included in the study (Table 1). They were chosen based on a combination of traits, representing ecologically well-studied species (blue tit, pied flycatcher and crow) and including a few divergent bird lineages (suboscine, hummingbirds, doves, ratites). For nine of the species (not including European crow, see below), RNA was extracted from adult brain tissue preserved in RNA later (QIAGEN). Whole brains were homogenized with a tissue ruptor (Promega or QIAGEN) and total RNA isolation was performed on aliquots of the homogenate following the manufacturer’s instructions for the RNeasy Kit (QIAGEN) or the SV Total RNA Isolation System (Promega). To determine RNA quality, RNA samples were run on 1% agarose gels or a RNA 6000 Nano LabChip using the 2100 Bioanalyzer (Agilent). RNA was prepared from between 1 and 10 individuals of each species, as a means for the identification of single nucleotide polymorphisms (data not shown). When multiple individuals were used, equal amounts of RNA from each individual, as measured by quantification with spectrophotometry (Nanodrop), were pooled prior to sequencing. Individual samples were not tagged.
Table 1.
Summary statistics of brain transcriptome sequencing in 10 bird species
| Species | Common name | Ind* | Plates† | Bases (Mb) | Reads | Contigs | Coverage‡ | Accession number |
|---|---|---|---|---|---|---|---|---|
| Archilochus colubris | Ruby-throated hummingbird | 1 | 1 | 106.1 | 496 627 | 15 204 | 11.43 | SRR029421 |
| Calypte anna | Anna’s hummingbird | 1 | 1 | 96.0 | 464 910 | 16 355 | 10.65 | SRR029422 |
| Corvus brachyrhynchos | American crow | 1 | 1 | 73.4 | 352 032 | 14 814 | 8.83 | SRR029463–64 |
| Corvus corone | European crow | 12 | 2 | 149.5 | 856 675 | 18 133 | 8.11 | SRR019143–44 |
| Dromaius novaehollandiae | Emu | 1 | 2 | 86.0 | 398 710 | 20 603 | 7.60 | SRR029466–67 |
| Ficedula hypoleuca | Pied flycatcher | 10 | 2.5 | 230.0 | 1 056 316 | 45 229 | 8.15 | SRR029159–61 |
| Manacus vitellinus | Golden collared manakin | 1 | 1 | 66.8 | 312 014 | 15 782 | 7.70 | SRR029477–78 |
| Melopsittacus undulatus | Budgerigar | 2 | 2 | 89.2 | 467 567 | 19 198 | 8.28 | SRR029329–30 |
| Parus caeruleus | Blue tit | 7 | 1 | 99.3 | 404 049 | 19 205 | 7.89 | SRR029162 |
| Streptopelia risoria | Ring-necked dove | 1 | 1 | 94.8 | 448 621 | 21 789 | 9.22 | SRR029331 |
Number of individuals that were pooled prior to sequencing.
One plate represent a full Roche 454 run.
Mean number of reads per site.
Library construction and normalization was performed using a variation of the Clontech SMART system as described previously (Warren et al. 2008). The features of this library are that the synthesis starts from the 3′ end of the mRNA using an oligo dT primer, and the cDNA are expected to be full-length. The normalized cDNA was sequenced according to the standard 454-FLX library protocol (Roche). Sequences were processed to remove adaptors and short reads prior to assembly. Raw read data have been deposited on the NCBI short read archive. Accession numbers can be found in Table 1.
European crow (Corvus corone) sequencing was carried out at a different 454 FLX platform, and was based on non-normalized cDNA library (Wolf et al. 2010).
Sequence assembly, contig annotation and estimation of substitution rates
Reads from the different species were assembled de novo separately using the Newbler assembler (Roche). Reads that could not be assembled and thus remained as singletons were disregarded from further analyses, which thus were based on sequence contigs only. By excluding singletons we sought to reduce the impact of sequencing errors (Huse et al. 2007).
All contigs were first mapped onto the zebra finch genome using BLAT (Kent 2002) (minimum score = 40, step Size = 5) and the best hit for each contig was considered for annotating the genomic region that was covered. To specifically detect protein-coding genes in the transcriptome data, we downloaded zebra finch coding sequences from the BioMart database (ENSEMBL 54) using BLASTX and FASTY for mapping. We followed a reciprocal blast approach to minimize false positive orthologous sequences. All contigs with a blast hit >10−20 were discarded, a stringent criterion that leads to a higher identification of orthologous as opposed to paralogous sequences. Quality scores for the contigs were not taken into account. Sequences were assumed to be orthologous if the zebra finch protein sequence had the best hit to a contig and that same contig had the best hit to that particular zebra finch sequence (reciprocal blast criterion). Additionally, sequences that contained frame shifts were discarded. Pair-wise and multiple alignments were generated for all species based on protein sequences using MAFFT Version 6.704b (Katoh & Toh 2008) and back-translated to DNA sequences for subsequent analysis. Alignments are available upon request. Substitution rates were estimated separately for synonymous (dS) and non-synonymous substitutions (dN) using a maximum likelihood method implemented in the CO-DEML program of the PAML package Version 4.1 (Yang 2007). Pair-wise maximum likelihood analysis were performed in runmode-2. We excluded all alignments that were shorter than 150 bp or that had dS larger than 2 to minimize statistical artefacts from short sequences and saturation effects in dS. To calculate the mean ratio between dN and dS (denoted mean ω) per chromosome or for two species, we divided the mean non-synonymous rate by the mean synonymous rate.
Data on chromosomal location of zebra finch genes was taken from ENSEMBL. For the purpose of this study, we assumed the same location of genes in the other species. Although we know this assumption can not be correct in every single case, we believe it is justified by the high overall degree of chromosome conservation across birds (Griffin et al. 2007). To estimate the male mutation bias (αm), we used the following equation (see Miyata et al. 1987):
where Z represents mean dS of the Z chromosome and A the mean dS of autosomes.
Statistical analyses
We used weighted multiple regression analysis based on linear mixed effect modelling to decipher the relationship between mean ω per chromosome, mean dS per chromosome and chromosome length. We defined mean ω as the response variable, mean dS and chromosome length as fixed explanatory variables. Visual inspection of the relationship between mean ω and chromosome length suggested adding a quadratic term to the model equation. All variables were normalized by logarithmic transformation prior to analysis. Data were further centred (z-transformed), as parameter estimates of the fixed effects then represent standardized semi-partial regression coefficients that allow direct comparisons of effect sizes across variables. To appropriately account for the study design and pseudo-replication, species identity and differences in slopes of all variables were included as random factors. Model selection was based on a backward selection and Akaike’s information criteria that qualitatively yielded the same results. Chromosomes that harbour only a few genes were given less weight by weighting data points by the reciprocal standard error thus taking into account both variance of data points and samples sizes. R 2.9.1 (R DCT 2006) was used to perform all statistical analyses.
Results
Sequencing and assembly
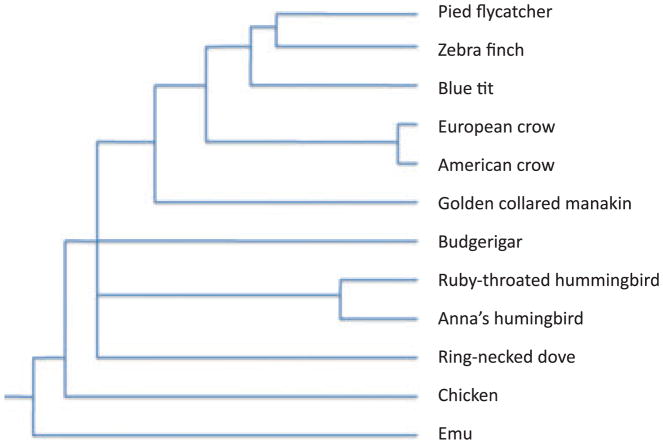
We prepared cDNA from adult brain tissue of 10 different bird species (Table 1, Fig. 1). Prior to sequencing on a Roche 454 GS-FLX platform, the cDNAs were normalized to in theory increase the number of different transcripts expected to be detected by random sequencing of templates (cf. Hale et al. 2009). About 1–2.5 full GS-FLX plate runs were performed for each species and the total amount of raw sequence data obtained varied from 53 Mb for Manacus vitellinus (1 plate) to 188 Mb for Ficedula hypoleuca (2.5 plate runs were performed for the latter species). The mean read length varied between 211 and 247 bp.
Fig. 1.
A schematic phylogenetic tree indicating the approximate relationships between species included in the study, following Hackett et al. (2008). As the precise topology is not critical for the purpose of this study, controversial nodes are not shown resolved. Branch lengths are not drawn according to scale.
Before further data analyses, the reads were assembled into larger contigs. Due to the fact that a reference genome is not available for any of the species, it was necessary to perform de novo assembly. The average contig length across species was 453 bp (range 78–5911 bp). The number of contigs obtained from the assembly step varied between 15 204 in Archilochus colubris to 45 229 in F. hypoleuca. Mean coverage per base varied between 7.60 in Dromaius novaehollandiae and 11.43 in A. colubris (Table 2). Somewhat surprisingly, we did not find a significant correlation between coverage per base and the total amount of raw sequence data ( , P = 0.80). However, the number of contigs correlated well with the total amount of raw data ( , P = 1.7 × 10−5.
Table 2.
Number and proportion of contigs from transcriptome data that align to chicken and zebra finch respectively
| Species | Chicken |
Zebra finch |
||
|---|---|---|---|---|
| Contigs | Proportion | Contigs | Proportion | |
| Archilochus colubris | 8819 | 0.58 | 10 330 | 0.68 |
| Calypte anna | 9907 | 0.61 | 11 423 | 0.70 |
| Corvus brachyrhynchos | 9252 | 0.62 | 13 434 | 0.91 |
| Corvus corone | 12 647 | 0.70 | 17 168 | 0.95 |
| Dromaius novaehollandiae | 11 964 | 0.58 | 11 838 | 0.57 |
| Ficedula hypoleuca | 24 071 | 0.53 | 43 049 | 0.95 |
| Manacus vitellinus | 10 426 | 0.66 | 14 188 | 0.90 |
| Melopsittacus undulatus | 12 360 | 0.64 | 14 585 | 0.76 |
| Parus caeruleus | 10 927 | 0.57 | 18 516 | 0.96 |
| Streptopelia risoria | 8605 | 0.39 | 10 122 | 0.46 |
Contig annotation
With the recent completion of draft zebra finch genome sequencing (Warren et al., unpublished), there are now two bird genomes available, also including chicken (Hillier et al. 2004). We aligned all contigs against the assembled chicken and zebra finch genomes as to annotate the transcriptome sequences from non-model avian species. As expected, the proportion of contigs that aligned to the reference genomes was inversely correlated with genetic distance (Fig. 1, Table 2). For example, when zebra finch (which belongs to Passeriformes) was used as a reference, 91–96% of the contigs from the other oscine passerines, 90% of the suboscine passerines (most basal clade within Passeriformes), 68–76% of the parrot and 46–57% of the dove and emu contigs aligned. For emu, which is equally distant to chicken and zebra finch, a very similar proportion of contigs align to the two reference genomes (Table 2). In the following analyses, we focused on the zebra finch as reference given that most species sequenced are more closely related to it than to chicken (Fig. 1).
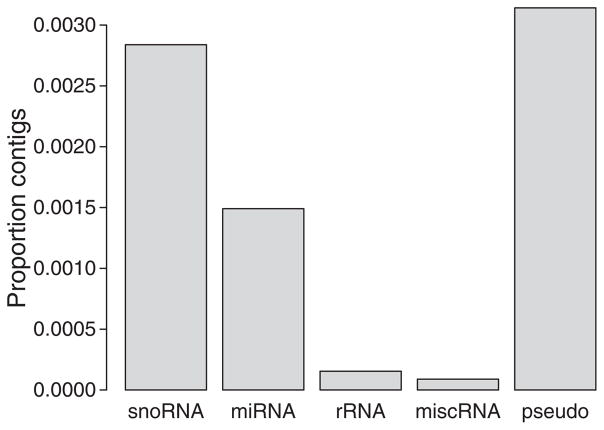
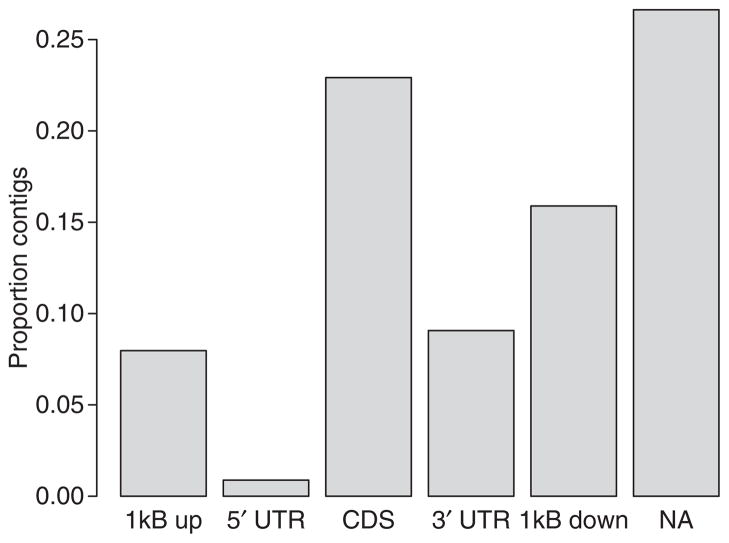
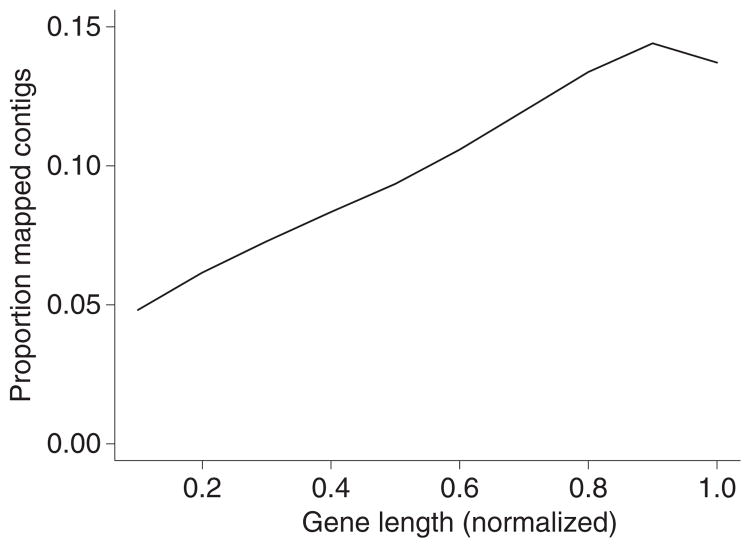
Around 65% of contig sequences aligning to the zebra finch genome were from protein-coding genes, including coding regions (CDS), untranslated regions (5′ UTR and 3′ UTR) and regions immediately upstream of ENSEMBL-annotated 5′ UTRs and downstream of 3′ UTRs (Fig. 2). About 26.9% of the contigs were assigned at least partly to putative intergenic regions in the zebra finch genome (NA in Fig. 2). Only a minor proportion of the aligned contigs (on average <1%) represented other ENSEMBL-annotated RNA-types and pseudogenes (Fig. 3). We found that there is a propensity for reads to come from the 3′ end of the coding regions (CDS) of genes (Fig. 4). This is also evident as an excess of 3′ UTR reads over 5′ UTR reads (two-sample t-test, P = 3.5 × 10−5) and an excess of the 1 kb downstream 3′ UTR region over the 1 kb upstream 5′ UTR region (two-sample t-test, P = 0.0015).
Fig. 2.
Proportion of contigs that aligned to different parts of protein-coding genes, including coding region (CDS), 5′ and 3′ untranslated (UTR) regions, and regions 1 kb upstream and downstream, and not annotated regions respectively of zebra finch-defined UTR regions. snoRNA, small nuclear RNA; miR-NA, micro-RNA; rRNA, ribosomal RNA; miscRNA, miscellaneous RNA; pseude, pseudogenes.
Fig. 3.
Proportion of contigs that aligned to different parts of non-coding RNA-genes and pseudogenes, as defined from alignment to the ENSEMBL-modelled zebra finch genome predictions.
Fig. 4.
The distribution of contig coverage in different parts of the CDS region of protein-coding genes. The length of all genes was normalized and the coverage calculated in 10% bins.
We focused our subsequent analysis on contigs that represent CDS regions. Between 635 (A. colubris) and 3662 (F. hypoleuca) ENSEMBL modelled protein coding genes matched those in the zebra finch (Table 3). The number of genes obtained is highly correlated with the amount of raw sequence data ( , P = 0.0016). On average, ~27% of the CDS of zebra finch protein-coding genes are covered by one or more contigs per species. This value ranges from 20.0% in Melopsittacus undulatus to 37.1% in Corvus corone.
Table 3.
The proportion of contigs aligning to the zebra finch genome and the number of genes, and coverage of those genes, identified through alignment to zebra finch
| Species | Aligning contigs (%) | Genes | Coverage* |
|---|---|---|---|
| Archilochus colubris | 68 | 635 | 23.57 |
| Calypte anna | 70 | 898 | 23.72 |
| Corvus brachyrhynchos | 91 | 869 | 25.41 |
| Corvus corone | 95 | 2208 | 37.11 |
| Dromaius novaehollandiae | 57 | 1395 | 24.43 |
| Ficedula hypoleuca | 95 | 3661 | 31.10 |
| Manacus vitellinus | 90 | 1206 | 29.37 |
| Melopsittacus undulatus | 76 | 1125 | 20.00 |
| Parus caeruleus | 96 | 2291 | 27.94 |
| Streptopelia risoria | 46 | 941 | 25.36 |
Mean sequence coverage of each gene.
The ability to perform many molecular evolutionary analyses is dependent on that sequence information is available from the same gene in different species. In total, we were able to retrieve sequence data for 6499 protein-coding genes combined from all species. However, due to the random sampling process of 454 sequencing, there was limited overlap of genes across species. Of the 6499 genes, only 55 were sequenced across all 10 species plus zebra finch. Moreover, even if there is sequence data from the same gene in two or more species, it does not necessarily mean that the same region of the gene has been sequenced in all species. When we exclude all genes that do not have overlapping sequence data in all species, of the 55 genes, we obtain only 15 genes (ATP5A1, B5KFQ7, C1orf151, DYNLRB1, GPM6A, ITGB1BP3, KRAS, LSM3, PVALB-2, RANP1, RPL5, RPL7, RPL23A, SOD2, UQCRB; i.e. three rRNA genes) that are sequenced in all 10 species.
Substitution rate variation
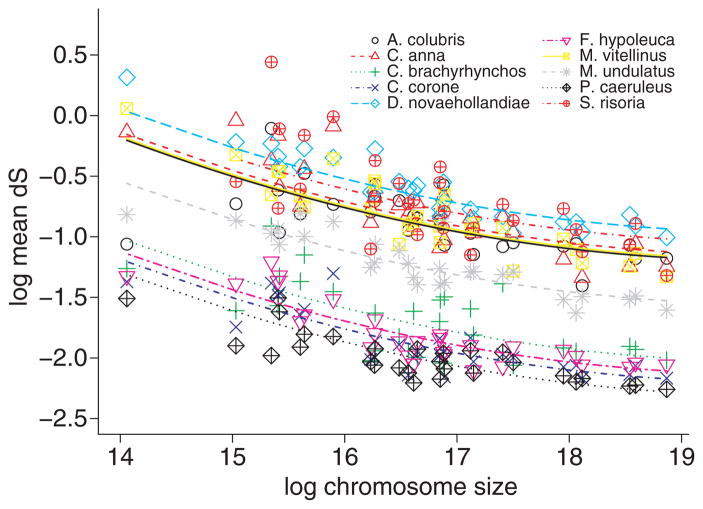
Based on the alignment of orthologous gene sequences, we made pair-wise estimates of the synonymous substitution rate (dS) in comparisons between zebra finch and each of the 10 species (Table 4). Mean dS increased with genetic distance between species, which is expected under the assumption of a molecular clock. When mean dS is estimated for each chromosome in the zebra finch karyotype, we find a clear negative correlation between chromosome size and dS in all species comparisons (Fig. 5). Although we do not have information on the chromosomal location of genes or karyotype organization in other species than zebra finch and chicken, using data on chromosome size from zebra finch can be justified on basis of the overall high degree of chromosomal conservation in avian genomes (Griffin et al. 2007). Thus, small chromosomes appear to have elevated mutation rates.
Table 4.
Estimates of the mean rates of synonymous (dS) and non-synonymous substitution rate (dN) and their ratio (ω) in comparisons with zebra finch
| Species | No. genes | Mean length | dN | dS | ω |
|---|---|---|---|---|---|
| Archilochus colubris | 617 | 390 | 0.0286 | 0.3793 | 0.0753 |
| Calypte anna | 867 | 400 | 0.0283 | 0.3795 | 0.0746 |
| Corvus brachyrhynchos | 837 | 377 | 0.0173 | 0.1665 | 0.1037 |
| Corvus corone | 2168 | 565 | 0.0160 | 0.1400 | 0.1142 |
| Dromaius novaehollandiae | 1315 | 400 | 0.0378 | 0.4658 | 0.0811 |
| Ficedula hypoleuca | 3575 | 523 | 0.0191 | 0.1520 | 0.1254 |
| Manacus vitellinus | 1180 | 410 | 0.0217 | 0.2616 | 0.0828 |
| Melopsittacus undulatus | 1094 | 336 | 0.0337 | 0.3740 | 0.0900 |
| Parus caeruleus | 2249 | 495 | 0.0146 | 0.1238 | 0.1179 |
| Streptopelia risoria | 764 | 397 | 0.0356 | 0.4233 | 0.0841 |
Alignments shorter than 150 bp or with dS > 2 were excluded from the estimates.
Fig. 5.
The relationship between substitution rate (log mean dS) and chromosome size (log2) in pairwise comparisons including in each case zebra finch.
We also find that the Z chromosome tends to have higher dS than autosomes. The male mutation bias (αm) can be estimated by contrasting the rate of mutation in autosomes and sex chromosomes. As there were no W-linked genes in the sets of genes identified by transcriptome sequencing in different species, we estimated αm by comparisons of autosomal and Z-linked rates (see Materials and methods). For eight of the 10 species comparisons αm was higher than one (Table 5). Altogether, mean αm was 2.4, suggesting that the male mutation rate is at least twice as high the female mutation rate across broad avian comparisons.
Table 5.
Estimates of the male mutation bias (αm) based on comparisons of synonymous substitution rates (dS) in autosomes and the Z chromosome according to the zebra finch genome mapping
| Species | dS Z | dS auto | αm |
|---|---|---|---|
| Archilochus colubris | 0.449 | 0.368 | 4.888 |
| Calypte anna | 0.354 | 0.380 | 0.0283 |
| Corvus brachyrhynchos | 0.198 | 0.162 | 5.000 |
| Corvus corone | 0.158 | 0.137 | 2.703 |
| Dromaius novaehollandiae | 0.476 | 0.464 | 1.168 |
| Ficedula hypoleuca | 0.163 | 0.147 | 1.970 |
| Manacus vitellinus | 0.290 | 0.259 | 2.210 |
| Melopsittacus undulatus | 0.415 | 0.368 | 2.242 |
| Parus caeruleus | 0.145 | 0.122 | 3.604 |
| Streptopelia risoria | 0.376 | 0.423 | 0.500 |
Selection
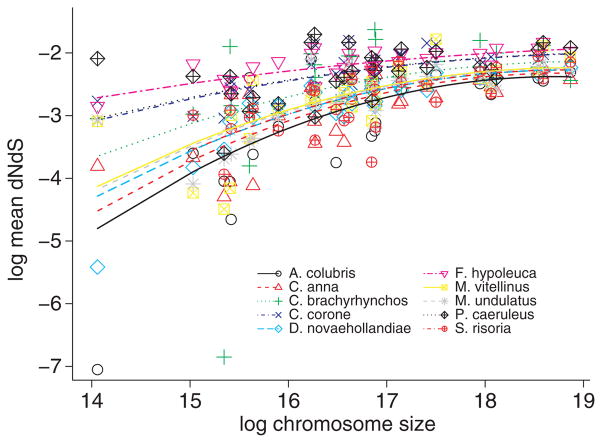
We sought to study the overall patterns of natural selection in avian genomes by estimating the dN/dS ratio (ω) of orthologous genes in each comparison with zebra finch and one of the species subject to transcriptome sequencing. In birds, there is evidence for striking differences in recombination rate among chromosomes, with a strong negative correlation between recombination rate and chromosomes size (Hillier et al. 2004; Groenen et al. 2009). Theory predicts that differences in recombination rates should translate into differences in effective population size (Hill & Robertson 1966). We therefore expect that selection is more efficient in small than in large chromosomes. Under the assumption that most mutations that affect fitness are slightly deleterious and that selection is more efficient in chromosomes with high recombination rates, dN/dS ratios should be smaller in small chromosomes. Indeed, we found that within each of the 10 species comparisons with zebra finch, mean ω per chromosome was positively correlated with chromosome size, supporting the idea that genes in small chromosomes evolve under stronger constraint (Fig. 6). An alternative explanation would be that adaptive evolution is more important for genes in larger chromosomes.
Fig. 6.
The relationship between log mean ω (dN/dS) and (log2) chromosome size in pairwise comparisons including in each case zebra finch. Lines represent predicted values of the full model including different slopes (model 1, Table 6, that despite of being less parsimonious allows a more detailed inspection of the data.
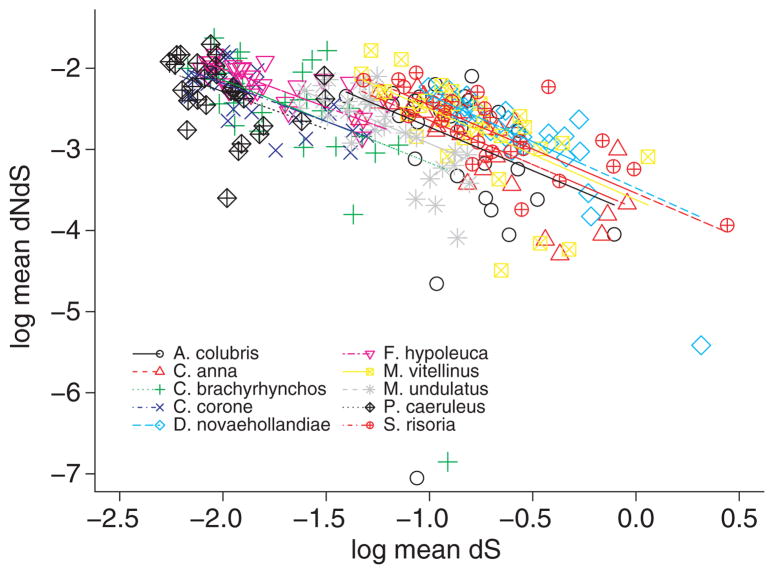
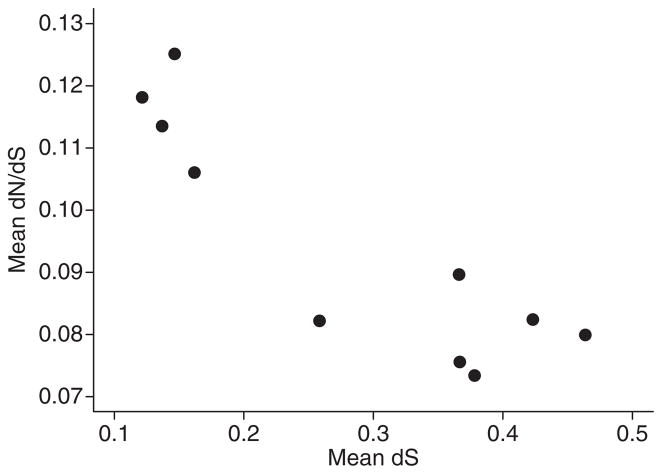
However, as stated before mean dS per chromosome was also found to be negatively correlated with chromosome size. Recently, there have been theoretical and empirical claims that mean dN/dS may not be independent of mean dS (Rocha et al. 2006; Kryazhimskiy & Plotkin 2008; Wolf et al. 2009). This notion is reflected in our observation that mean ω of each species comparison (0.075–0.125) is negatively correlated with genetic distance between zebra finch and the compared species, at least when dS is taken as a proxy for this distance (Fig. 7). The relationship between mean dN/dS and chromosome size (and hence recombination rate) may thus not reflect differences in selection efficiency but be simply explained by differences in mutation rate (measured by mean dS) among chromosomes. To account for this possibility, we included both mean dS and chromosome size as explanatory variables of mean dN/dS in the statistical models. Closer inspection of the models suggests that mean ω is indeed negatively correlated with mean dS (Fig. 8). The two best statistical models include both chromosome length (linear and quadratic terms) and mean dS as explanatory variables (Table 6). Inspection of standardized semi-partial regression coefficients suggests that the effects of mean dS and chromosome size are similar (Table 6). Despite considerable co-variation in mean dS and chromosome length (r = −0.35), we thus conclude that both dS and chromosome size are correlated with mean ω.
Fig. 7.
The relationship between mean ω (dN/dS) and dS in pairwise comparisons including in each case zebra finch.
Fig. 8.
The relationship between log mean ω (dN/dS) and (log2) chromosome size in pairwise comparisons including in each case zebra finch. Lines represent predicted values of the full model including different slopes (model 1, Table 6) that despite of being less parsimonious allows a more detailed inspection of the data.
Table 6.
Comparison of linear mixed effect models fitting the relationship of mean ω per chromosome vs. chromosome length (CL and CL2) and mean dS per chromosome (dS) as explanatory variables
| Model (d.f.) | Model selection criteria |
Parameter estimates of fixed effects |
Standard deviation of random effects |
Residual standard deviation | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| AIC | BIC | LRT | CL | (CL)2 | dS | Intercept Species | Slopes |
||||
| CL | (CL)2 | dS | |||||||||
| 1 (15) | 2409 | 2432 | 0.3579*** | −0.1015*** | −0.4074*** | 0.0062 | — | — | — | 0.4953 | |
| 2 (6) | 2412 | 2468 | n.s. | 0.3873*** | −0.1173*** | −0.3892*** | 0.0054 | 0.0079 | 0.0046 | 0.0013 | 0.4737 |
| 3 (5) | 2442 | 2461 | *** | 0.5059*** | −0.1248*** | — | 0.0173 | — | — | — | 0.5125 |
| 4 (4) | 2454 | 2469 | *** | — | — | −0.9778*** | 0.0274 | — | — | — | 0.5225 |
Listed are model names and their degrees of freedom (d.f.), information criteria used for model selection (AIC, Akaike’s information criterion; BIC, Bayesian information criterion; LRT, likelihood ratio test), parameter estimates of fixed effects and variance components of random effects. As data has been z-transformed prior to analysis, parameter estimates represent standardized semi-partial regression coefficients that can be directly compared between variables and give an impression on relative effect sizes
α < 0.001,
α < 0.01,
α < 0.05.
Discussion
Retrieving gene sequence data from non-model organisms
There has recently been a burst of studies, in a range of species, reporting on transcriptome sequencing efforts using next-generation sequencing technology (Cheung et al. 2006, 2008; Novaes et al. 2008; Vera et al. 2008; Barakat et al. 2009; Hahn et al. 2009; Hale et al. 2009; Meyer et al. 2009). Although these studies have generally focused on a single species, our approach was to perform transcriptome sequencing across several divergent bird lineages, ultimately with the purpose of being able to study the role of natural selection in avian gene sequence evolution. There was no reference genome available for any of the species sequenced so we made use of the genome of the closest relative, the zebra finch, for compiling and annotating genes.
We identified roughly 1000 genes per full GS-FLX run and there was no obvious saturation in gene discovery in those cases where 1.0–1.5 additional runs were performed. This finding supports the common belief that brain tissue represents a rich source of different transcripts and that even larger-scale sequencing should identify more genes; more sequencing should also yield increased sequence depth and gene coverage.
About three-quarters of the contigs that aligned to the zebra finch genome are from protein-coding genes. This includes a significant proportion (almost one-quarter) of regions 1 kb upstream or downstream respectively of ENSEMBL-defined zebra finch UTRs. This may represent genes where the annotation of UTRs in the zebra finch is incomplete in the ENSEMBL modules or where the UTRs of the other species differ from zebra finch. The overabundance of sequence data from 3′ end of genes is inherent to cDNA sequencing, even though the protocol for cDNA synthesis is optimized for full-length construction.
Another one-quarter of the contigs that align to the zebra finch genome are from intergenic regions that are not annotated. There are a number of explanations to this observation. For instance, such regions can represent unknown genes, alternatively spliced exons or various types of non-coding RNA transcripts. The fact that we only found <1% of the contigs to correspond to known ENSEMBL predicted non-coding RNA-genes indicates either such transcripts are rarely expressed in avian brains or that that the ENSEMBL models have grossly underestimated the occurrence of such transcripts, given the recent realization of the high abundance of transcribed non-coding RNAs in vertebrate genomes (Bertone et al. 2004; Fantom et al. 2005; Sultan et al. 2008). In turn, this has bearing to the fraction of contigs that do not align to the zebra finch genome. Although only 5–10% of contigs from the species most closely related to zebra finch failed to align, this proportion increased to ~50% in the more distantly related species. This result is not unexpected, as increasing genetic distance will lead to decreasing levels of homologous alignments and thus difficult to detect orthologous sequence relationship of regions evolving under limited constraints (Ponting 2008). We also note that the decreasing proportion of contigs aligning to the reference genome with increasing genetic distance argues against that non-aligning sequences would consist of a significant amount of xenobiotic contamination or represent technical artefacts.
Nucleotide substitution and chromosome size
Two general conclusions about substitution rate variation in birds can be drawn from our study. These are evidence for male-biased mutation and a negative relationship between chromosome size and mutation rate. As there were so few genes for which we could retrieve sequence data in multiple species, all substitution rate estimates come from pair-wise comparisons of zebra finch and one of the species from the transcriptome sequencing effort. As zebra finch is included in every such comparison, this means that the observed substitution rates are not independent. Moreover, as several of the species subject to transcriptome sequencing belong to the same family or order of birds, they share substitutions from internal branches. This should be kept in mind when the results are examined.
If synonymous substitutions are considered effectively neutral and thus accepted to reflect the underlying mutation rate, it is clear for all species comparisons that the incidence of mutation is higher in small chromosomes (Fig. 5). This mirrors what was observed in chicken-human (Hillier et al. 2004) and chicken-turkey comparisons (Axelsson et al. 2005), and therefore seems to represent a general trend in avian genomes. It is often observed, across taxonomic groups, that several genomic parameters like base composition, recombination rate and substitution rate covary, although it may be difficult to reveal the causality of such relationships (e.g. (Hardison et al. 2003). GC content and recombination rate correlate negatively with chromosome size in birds (Hillier et al. 2004) and both offer plausible explanations to the elevated rate of synonymous substitutions in small chromosomes. The incidence of the highly mutable CpG dinucleotide is positively correlated with GC content and there is significant correlation between GC and substitution rate in birds (Axelsson et al. 2005; Webster et al. 2006). We surmise that recombination might indirectly be responsible for the correlation between dS and chromosome size as high recombination tends to give high GC content, likely due to biased gene conversion (Pozzoli et al. 2008). Alternatively, there might be a more direct link due to recombination being mutagenic, i.e. that recombination and mutation rates correlate positively (Hellmann et al. 2003). There are several important implications to the within-genome heterogeneity seen in avian mutation rates. For example, attempts towards molecular dating of divergence times need to make sure that rates are calibrated using appropriately selected sequence data.
Male-biased mutation
The number of mitotic cell divisions in germ line is typically much higher in males than in females. If germ line mutations are somehow replication-associated, due for example to replication-errors, the majority of mutations can be expected to have a paternal origin (Ellegren 2007). Previous work in birds, in most cases based on data from a limited number of species, have revealed a moderate male mutation bias, most often in the range of two to three times higher mutation rate in males than in females (e.g. Axelsson et al. 2004). Our study confirms this level of male excess in avian germ line mutation, at least broadly speaking (mean αm = 2.4). However, the estimates of αm in individual species comparisons vary considerably (0.03–5.00). This can most likely be attributed to sampling error as estimates of αm from autosome–Z chromosome comparisons are sensitive to parameter estimation of substitution rates in individual chromosome categories (more so than in autosome–W or Z–W chromosome comparisons; Ellegren 2007). Alternatively, as it has been demonstrated that the magnitude of the male mutation bias can be affected by life history (Bartosch-Harlid et al. 2003), the variation might at least in part be biologically meaningful. Specifically, increased sperm production from sexual selection would, in theory, increase the male mutation bias. However, we find no obvious reasons to believe that sexual selection would be more intense in those lineages represented by particularly high αm estimates (American crow, Ruby-throated hummingbird, Blue tit) compared to those with very low estimates (Anna’s hummingbird, Ring-necked dove).
Selection
Theory predicts that slightly deleterious mutations should be more effectively purged in regions of high recombination, the so-called Hill-Robertson effect (Hill & Robertson 1966). As slightly deleterious mutations are likely to contribute to the accumulation of non-synonymous substitutions in populations with low-moderate effective population size (Ne) (Hahn 2008; Ellegren 2009), ω is expected to be lower in regions of high recombination and higher in regions of low recombination (see Bullaughey et al. 2008). The rate of recombination varies considerably among avian chromosomes and is strongly negatively correlated with chromosome size (Hillier et al. 2004; Groenen et al. 2009). Previously, ω was found to be smaller in microchromosomes than in macrochromosomes in the chicken-turkey comparison, and it was hypothesized that this was the result of Hill–Robertson inference (Axelsson et al. 2005). Given that chromosome size is a good predictor for recombination rates, our observation, seen across all species comparisons, of decreasing ω with decreasing chromosome size would be in accordance with this interpretation.
The situation is complicated by there being substantial uncertainties associated with the basic properties of the apparent dN/dS ratio, including time dependency (Rocha et al. 2006), effects of within-population variation (Kryazhimskiy & Plotkin 2008), gene conversion (Berglund et al. 2009), estimation procedure, sequencing and annotation errors (Schneider et al. 2009). Using simulations and empirical genomic data, we have recently demonstrated that ω taken as phase value can show a strong negative relationship with dS (Wolf et al. 2009). Most of this variation is explained by the correlated sampling variance in dS, which affects the denominator of ω and dS likewise. This leads to a negative correlation between ω and dS that will disappear when taking the correct mean of ω (sum of dN/sum of dS) for a large number of genes. The influence of correlated sampling variances will persist, however, if the mean is based on only a few genes, as is the case for most small chromosomes in our data set. This suggests that at least part of the correlation between mean ω and mean dS reflects a stochastic artefact. Due to co-linearity between dS and chromosome size, this artefact will also be visible in the relationship between mean ω and chromosome size and can lead to incorrect inferences regarding the role of selection. We here treated this problem with a semi-partial regression, finding that the effects of mean dS and chromosome size on ω are similar in size, and that chromosome size therefore is an important factor in explaining the role of natural selection on avian genes. The most obvious explanation to this would be through an effect of recombination, i.e. Hill–Robertson inference. To formally test this hypothesis, recombination rate data from the species under investigation would be needed and better ways of controlling for the inter-dependency of mean ω and mean dS must be found (see, e.g. Piganeau & Eyre-Walker 2009).
Acknowledgments
We thank Diethard Tautz for support and Garth Spellman for help with samples. H.E. acknowledges funding from the Swedish Research Council and the Knut and Alice Wallenberg Foundation, and B.A.S. acknowledges NSF IBN-0646459 Grant. Postdoctoral support for D.E.J. was provided by National Science Foundation Grant MCB-0817687 to N. Valenzuela and SVE and NIH No. 5F32GM072494.
Footnotes
Axel Künstner’s PhD thesis focuses on molecular evolution of avian genomes. Research in the Ellegren laboratory integrates genomics and evolutionary biology.
Conflicts of interest
The authors have no conflict of interest to declare and note that the sponsors of the issue had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- Abzhanov A, Protas M, Grant BR, Grant PR, Tabin CJ. Bmp4 and morphological variation of beaks in Darwin’s finches. Science. 2004;305:1462–1465. doi: 10.1126/science.1098095. [DOI] [PubMed] [Google Scholar]
- Axelsson E, Smith NGC, Sundstrom H, Berlin S, Ellegren H. Male-biased mutation rate and divergence in autosomal, Z-linked and W-linked introns of chicken and turkey. Molecular Biology and Evolution. 2004;21:1538–1547. doi: 10.1093/molbev/msh157. [DOI] [PubMed] [Google Scholar]
- Axelsson E, Webster MT, Smith NGC, Burt DW, Ellegren H. Comparison of the chicken and turkey genomes reveals a higher rate of nucleotide divergence on microchromosomes than macrochromosomes. Genome Research. 2005;15:120–125. doi: 10.1101/gr.3021305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barakat A, DiLoreto DS, Zhang Y, et al. Comparison of the transcriptomes of American chestnut (Castanea dentata) and Chinese chestnut (Castanea mollissima) in response to the chestnut blight infection. BMC Plant Biology. 2009;9:11. doi: 10.1186/1471-2229-9-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartosch-Harlid A, Berlin S, Smith NGC, Moller AP, Ellegren H. Life history and the male mutation bias. Evolution. 2003;57:2398–2406. doi: 10.1554/03-036. [DOI] [PubMed] [Google Scholar]
- Begun DJ, Holloway AK, Stevens K, et al. Population genomics: whole-genome analysis of polymorphism and divergence in Drosophila simulans. PLoS Biology. 2007;5:2534–2559. doi: 10.1371/journal.pbio.0050310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benfey PN, Mitchell-Olds T. Perspective - From genotype to phenotype: systems biology meets natural variation. Science. 2008;320:495–497. doi: 10.1126/science.1153716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beraldi D, McRae AF, Gratten J, et al. Mapping quantitative trait loci underlying fitness-related traits in a free-living sheep population. Evolution. 2007;61:1403–1416. doi: 10.1111/j.1558-5646.2007.00106.x. [DOI] [PubMed] [Google Scholar]
- Berglund J, Pollard KS, Webster MT. Hotspots of biased nucleotide substitutions in human genes. PLoS Biology. 2009;7:45–62. doi: 10.1371/journal.pbio.1000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertone P, Stolc V, Royce TE, et al. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242–2246. doi: 10.1126/science.1103388. [DOI] [PubMed] [Google Scholar]
- Bonneaud C, Burnside J, Edwards SV. High-speed developments in avian genomics. BioScience. 2008;58:587–595. [Google Scholar]
- Bullaughey K, Przeworski M, Coop G. No effect of recombination on the efficacy of natural selection in primates. Genome Research. 2008;18:544–554. doi: 10.1101/gr.071548.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung F, Haas BJ, Goldberg SMD, et al. Sequencing Medicago truncatula expressed sequenced tags using 454 Life Sciences technology. BMC Genomics. 2006;7:10. doi: 10.1186/1471-2164-7-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung F, Win J, Lang JM, et al. Analysis of the Pythium ultimum transcriptome using Sanger and pyrosequencing approaches. BMC Genomics. 2008;9:10. doi: 10.1186/1471-2164-9-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AG, Eisen MB, Smith DR, et al. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- Ellegren H. Characteristics, causes and evolutionary consequences of male-biased mutation. Proceedings of the Royal Society of London B: Biological Sciences. 2007;274:1–10. doi: 10.1098/rspb.2006.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren H. Comparative genomics and the study of evolution by natural selection. Molecular Ecology. 2008a;17:4586–4596. doi: 10.1111/j.1365-294X.2008.03954.x. [DOI] [PubMed] [Google Scholar]
- Ellegren H. Sequencing goes 454 and takes large-scale genomics into the wild. Molecular Ecology. 2008b;17:1629–1631. doi: 10.1111/j.1365-294X.2008.03699.x. [DOI] [PubMed] [Google Scholar]
- Ellegren H. A selection model of molecular evolution incorporating the effective population size. Evolution. 2009;63:301–305. doi: 10.1111/j.1558-5646.2008.00560.x. [DOI] [PubMed] [Google Scholar]
- Ellegren H, Sheldon BC. Genetic basis of fitness differences in natural populations. Nature. 2008;452:169–175. doi: 10.1038/nature06737. [DOI] [PubMed] [Google Scholar]
- Fantom CT, Carninci P, Kasukawa T, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- Griffin DK, Robertson LBW, Tempest HG, Skinner BM. The evolution of the avian genome as revealed by comparative molecular cytogenetics. Cytogenetic and Genome Research. 2007;117:64–77. doi: 10.1159/000103166. [DOI] [PubMed] [Google Scholar]
- Groenen MAM, Wahlberg P, Foglio M, et al. A high-density SNP-based linkage map of the chicken genome reveals sequence features correlated with recombination rate. Genome Research. 2009;19:510–519. doi: 10.1101/gr.086538.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett SJ, Kimball RT, Reddy S, et al. A phylogenomic study of birds reveals their evolutionary history. Science. 2008;320:1763–1768. doi: 10.1126/science.1157704. [DOI] [PubMed] [Google Scholar]
- Hahn MW. Toward a selection theory of molecular evolution. Evolution. 2008;62:255–265. doi: 10.1111/j.1558-5646.2007.00308.x. [DOI] [PubMed] [Google Scholar]
- Hahn DA, Ragland GJ, Shoemaker DD, Denlinger DL. Gene discovery using massively parallel pyrosequencing to develop ESTs for the flesh fly Sarcophaga crassipalpis. BMC Genomics. 2009;10:9. doi: 10.1186/1471-2164-10-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale MC, McCormick CR, Jackson JR, DeWoody JA. Next-generation pyrosequencing of gonad transcriptomes in the polyploid lake sturgeon (Acipenser fulvescens): the relative merits of normalization and rarefaction in gene discovery. BMC Genomics. 2009;10:11. doi: 10.1186/1471-2164-10-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardison RC, Roskin KM, Yang S, et al. Covariation in frequencies of substitution, deletion, transposition, and recombination during eutherian evolution. Genome Research. 2003;13:13–26. doi: 10.1101/gr.844103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmann I, Ebersberger I, Ptak SE, Paabo S, Przeworski M. A neutral explanation for the correlation of diversity with recombination rates in humans. American Journal of Human Genetics. 2003;72:1527–1535. doi: 10.1086/375657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill WG, Robertson A. The effect of linkage on limits to artificial selection. Genetical Research. 1966;8:269–294. [PubMed] [Google Scholar]
- Hillier LW, Miller W, Birney E, et al. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- Hudson ME. Sequencing breakthroughs for genomic ecology and evolutionary biology. Molecular Ecology Resources. 2008;8:3–17. doi: 10.1111/j.1471-8286.2007.02019.x. [DOI] [PubMed] [Google Scholar]
- Huse SM, Huber JA, Morrison HG, Sogin ML, Welch DM. Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biology. 2007;8:R143. doi: 10.1186/gb-2007-8-7-r143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Toh H. Improved accuracy of multiple ncRNA alignment by incorporating structural information into a MAFFT-based framework. BMC Bioinformatics. 2008;9:13. doi: 10.1186/1471-2105-9-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ. BLAT – the BLAST-like alignment tool. Genome Research. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosiol C, Vinar T, da Fonseca RR, et al. Patterns of positive selection in six mammalian genomes. PLoS Genetics. 2008;4:17. doi: 10.1371/journal.pgen.1000144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryazhimskiy S, Plotkin JB. The Population genetics of dN/dS. PLoS Genetics. 2008;4:10. doi: 10.1371/journal.pgen.1000304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer E, Aglyamova GV, Wang S, et al. Sequencing and de novo analysis of a coral larval transcriptome using 454 GSFlx. BMC Genomics. 2009;10:18. doi: 10.1186/1471-2164-10-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata T, Hayashida H, Kuma K, Mitsuyasu K, Yasunaga T. Male-driven molecular evolution – a model and nucleotide-sequence analysis. Cold Spring Harbor Symposia on Quantitative Biology. 1987;52:863–867. doi: 10.1101/sqb.1987.052.01.094. [DOI] [PubMed] [Google Scholar]
- Nielsen R. Molecular signatures of natural selection. Annual Review of Genetics. 2005;39:197–218. doi: 10.1146/annurev.genet.39.073003.112420. [DOI] [PubMed] [Google Scholar]
- Novaes E, Drost DR, Farmerie WG, et al. High-throughput gene and SNP discovery in Eucalyptus grandis, an uncharacterized genome. BMC Genomics. 2008;9:14. doi: 10.1186/1471-2164-9-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piganeau G, Eyre-Walker A. Evidence for variation in the effective population size of animal mitochondrial DNA. PLoS ONE. 2009;4:e4396. doi: 10.1371/journal.pone.0004396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP. The functional repertoires of metazoan genomes. Nature Reviews Genetics. 2008;9:689–698. doi: 10.1038/nrg2413. [DOI] [PubMed] [Google Scholar]
- Pozzoli U, Menozzi G, Fumagalli M, et al. Both selective and neutral processes drive GC content evolution in the human genome. BMC Evolutionary Biology. 2008;8:12. doi: 10.1186/1471-2148-8-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R DCT. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2006. [Google Scholar]
- Rocha EPC, Smith JM, Hurst LD, et al. Comparisons of dN/dS are time dependent for closely related bacterial genomes. Journal of Theoretical Biology. 2006;239:226–235. doi: 10.1016/j.jtbi.2005.08.037. [DOI] [PubMed] [Google Scholar]
- Rockman MV, Kruglyak L. Genetics of global gene expression. Nature Reviews Genetics. 2006;7:862–872. doi: 10.1038/nrg1964. [DOI] [PubMed] [Google Scholar]
- Rothberg JM, Leamon JH. The development and impact of 454 sequencing. Nature Biotechnology. 2008;26:1117–1124. doi: 10.1038/nbt1485. [DOI] [PubMed] [Google Scholar]
- Schneider A, Souvorov A, Sabath N, et al. Estimates of positive Darwinian selection are inflated by errors in sequencing, annotation, and alignment. Genome Biology and Evolution. 2009;1:114–118. doi: 10.1093/gbe/evp012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinchcombe JR, Hoekstra HE. Combining population genomics and quantitative genetics: finding the genes underlying ecologically important traits. Heredity. 2008;100:158–170. doi: 10.1038/sj.hdy.6800937. [DOI] [PubMed] [Google Scholar]
- Sultan M, Schulz MH, Richard H, et al. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science. 2008;321:956–960. doi: 10.1126/science.1160342. [DOI] [PubMed] [Google Scholar]
- Vera JC, Wheat CW, Fescemyer HW, et al. Rapid transcriptome characterization for a nonmodel organism using 454 pyrosequencing. Molecular Ecology. 2008;17:1636–1647. doi: 10.1111/j.1365-294X.2008.03666.x. [DOI] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nature Reviews Genetics. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren WC, Hillier LW, Graves JAM, et al. Genome analysis of the platypus reveals unique signatures of evolution. Nature. 2008;453:U175–U171. doi: 10.1038/nature06936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster MT, Axelsson E, Ellegren H. Strong regional biases in nucleotide substitution in the chicken genome. Molecular Biology and Evolution. 2006;23:1203–1216. doi: 10.1093/molbev/msk008. [DOI] [PubMed] [Google Scholar]
- Wolf JBW, Künstner A, Nam K, Jakobsson M, Ellegren H. Nonlinear dynamics of nonsynonymous (dN) and synonymous substitution rates affects inference of selection. Genome Biology and Evolution. 2009;1:308–319. doi: 10.1093/gbe/evp030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf JBW, Bayer T, Haubold B. Nucleotide divergence versus gene expression differentiation: comparative transcriptome sequencing in natural isolates from the carrion crow and its hybrid zone with the hooded crow. Molecular Ecology. 2010;19 (Suppl 1):162–175. doi: 10.1111/j.1365-294X.2009.04471.x. [DOI] [PubMed] [Google Scholar]
- Wood HM, Grahame JW, Humphray S, Rogers J, Butlin RK. Sequence differentiation in regions identified by a genome scan for local adaptation. Molecular Ecology. 2008;17:3123–3135. doi: 10.1111/j.1365-294X.2008.03755.x. [DOI] [PubMed] [Google Scholar]
- Wright SI, Andolfatto P. The impact of natural selection on the genome: emerging patterns in Drosophila and Arabidopsis. Annual Review of Ecology Evolution and Systematics. 2008;39:193–213. [Google Scholar]
- Yang ZH. PAML 4: phylogenetic analysis by maximum likelihood. Molecular Biology and Evolution. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]