Abstract
Many immediate early genes (IEGs) have activity-dependent induction in a subset of brain subdivisions or neuron types. However, none have been reported yet with regulation specific to thalamic-recipient sensory neurons of the telencephalon or in the thalamic sensory input neurons themselves. Here, we report the first such gene, dual specificity phosphatase 1 (dusp1). Dusp1 is an inactivator of mitogen-activated protein kinase (MAPK), and MAPK activates expression of egr1, one of the most commonly studied IEGs, as determined in cultured cells. We found that in the brain of naturally behaving songbirds and other avian species, hearing song, seeing visual stimuli, or performing motor behavior caused high dusp1 upregulation, respectively, in auditory, visual, and somatosensory input cell populations of the thalamus and thalamic-recipient sensory neurons of the telencephalic pallium, whereas high egr1 upregulation occurred only in subsequently connected secondary and tertiary sensory neuronal populations of these same pathways. Motor behavior did not induce high levels of dusp1 expression in the motor-associated areas adjacent to song nuclei, where egr1 is upregulated in response to movement. Our analysis of dusp1 expression in mouse brain suggests similar regulation in the sensory input neurons of the thalamus and thalamic-recipient layer IV and VI neurons of the cortex. These findings suggest that dusp1 has specialized regulation to sensory input neurons of the thalamus and telencephalon; they further suggest that this regulation may serve to attenuate stimulus-induced expression of egr1 and other IEGs, leading to unique molecular properties of forebrain sensory input neurons.
INDEXING TERMS: mkp1, mkp-1, hvh1, ptpn10, cl100, vision, somatosensory, auditory, motor pathways, brain organization, neural activity, motor behavior, brain evolution, parrot, hummingbird, songbird, ring dove, bird, primary sensory, ZENK
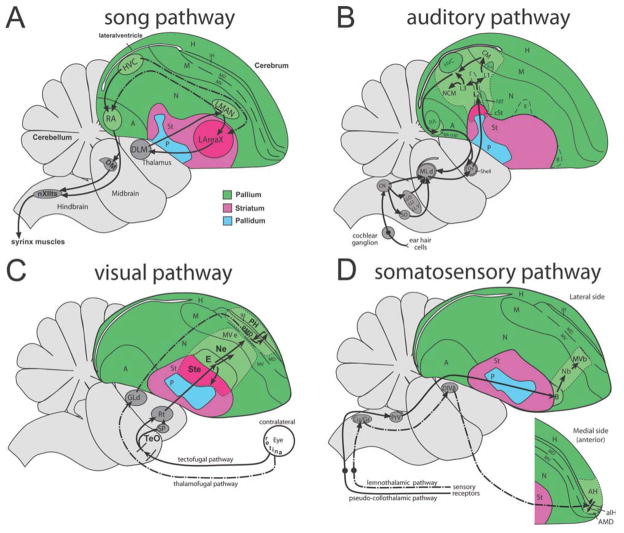
In the brain, immediate early genes (IEGs) are genes whose mRNA expression is dependent on neural activity in the absence of new protein synthesis (Greenberg et al., 1986; Flavell and Greenberg, 2008). As such, these genes are used as markers of neural activity to determine relationships between gene regulation and neural firing, and to map functional domains of the brain (Tischmeyer and Grimm, 1999; Guzowski et al., 2005; Mello and Jarvis, 2008). We have termed this use of IEGs “behavioral molecular brain mapping” (Jarvis, 2004a; Mello and Jarvis, 2008). This approach has been successively used to identify and characterize neural systems involved in perceiving and producing behaviors. For example, in songbirds, hearing- and singing-driven IEG expression helped to discover and/or characterize most nuclei of the vocal learning and auditory pathways, respectively (Fig. 1A,B; Mello et al., 1992; Jarvis and Nottebohm, 1997; Clayton, 2004; Velho et al., 2005; Wada et al., 2006; Pinaud et al., 2008). Likewise, behavioral molecular mapping has recently been used to map visual, somatosensory, and motor pathways in birds (Fig. 1C,D; nonvocal motor pathways not shown; Feenders et al., 2008; Hara et al., 2009).
Figure 1.
Schematic diagrams of songbird brain areas involved in singing, hearing, vision, and somatic sensation. A: Song system. Black solid arrows, vocal motor pathway (from HVC to RA to brainstem motor nuclei) and vocal pallial-basal ganglia-thalamic loop (Area X-DLM-LMAN); dashed arrows, connections between the two vocal pathways. B: Auditory pathways. L2 is the thalamic-recipient auditory zone, followed by secondary (L1 and L3) and tertiary (NCM and CM) connected neurons. In this study, we also named L1, L3, and NCM as N-L2, and CM as MV-L2. C: Two main visual pathways. E is the thalamic-recipient visual zone, followed by secondary (Ne and Ste) and tertiary (MVe) connected neurons in the tectofugal pathway (solid lines). IH is the thalamic-recipient visual zone, followed by secondary and tertiary (PH and PMD) connected neurons in the thalamofugal pathway. D: Somatosensory pathways. aIH and B are the thalamic-recipient somatosensory zones, followed by secondary and tertiary (Nb and MVb or AH and AMD) connected neurons. aIH, AH, and AMD are located medial to B. A and B are modified from Jarvis (2004a), C is from Hara et al. (2009), and D is based on Wild (1987, 1989), Wild and Williams (1999, 2000), and Freund et al. (2008). C and D are more lateral to A and B. See abbreviation list for anatomical terms, and the anatomy section of Materials and Methods for further information on each pathway.
However, of the genes studied thus far, none have been shown to be regulated in the sensory input neurons of the sensory pathways of the avian telencephalon. We use the terminology of sensory input, secondary sensory, and tertiary sensory neurons to describe the order of connections within a brain subdivision (i.e., within the midbrain, thalamus, or telencephalon), which is different from the terminology of first-order (primary), second-order, and third-order neurons that is commonly used to described ascending order of connections starting with sensory neuron receptors in the periphery. Sensory input neurons of the telencephalon are those that receive direct synaptic input from sensory neurons of the thalamus, and in turn sequentially project to higher (secondary, tertiary, etc.) sensory neurons within the same pathway (Fig. 1B–D). For example, for two of the most commonly studied IEGs, the egr1 (a.k.a. zif268, NGF-1A, Krox-24, and ZENK) and c-fos transcription factors, there is little to no sensory-driven induction in avian telencephalic sensory input neurons of auditory (L2), visual (E), or somatosensory (B) pathways, but there is high induction in secondary (surrounding nidopallium) and tertiary (mesopallium) sensory neurons of these pathways when stimuli for each specific sensory modality are processed (Fig. 1B–D; Mello and Clayton, 1994, 1995; Jarvis and Nottebohm, 1997; Velho et al., 2005; Feenders et al., 2008; Hara et al., 2009). A similar lack or paucity of induction of egr1 has been seen in avian and mammalian thalamic sensory input neurons (Mello and Clayton, 1994, 1995; Jarvis and Mello, 2000; Bisler et al., 2002; Soares et al., 2005). This lack of IEG induction occurs even though the sensory input neurons have increased neural firing when processing sensory stimuli (Bigalke-Kunz et al., 1987; Zeigler and Bischof, 1993; Chew et al., 1995; Wild and Williams, 2000). The lack of useful activity-dependent markers for sensory input cell populations hampers the identification and study of neural systems involved in processing sensory stimuli.
In a search for genes with sensory- and motor-driven regulation in the brain during natural stimuli and behavior (Wada et al., 2006), we discovered here that the dusp1 gene shows preferential stimulus-driven regulation in sensory input neurons of the avian thalamus and telencephalon. The sensory-induced dusp1 expression patterns were complementary to the induced egr1 expression patterns in secondary and tertiary sensory neurons of auditory, visual, and somatosensory populations. Dusp1, also known as mitogen-activated protein kinase (MAPK) phosphatase 1 (mkp1), is a negative regulator for MAPK, and MAPK in turn has been shown to upregulate egr1 in cultured cells (Knapska and Kaczmarek, 2004; Machado et al., 2008). Dusp1 has been mainly studied in cultured cells for its role in immunity or cancer (Liu et al., 2007; Boutros et al., 2008). It also has been studied in vivo in mammalian brains, but with strong pharmacological manipulations, in which the patterns of regulation were not linked to behavior (Qian et al., 1994; Takaki et al., 2001; Kodama et al., 2005) or the anatomical and cellular specificity was not well determined (Hu et al., 2009, but see Doi et al., 2007; Pizzio and Golombek, 2008). Our own analyses of the data of these studies in mammalian brain as well as GEN-SAT dusp1 promoter-eGFP knockin mice in the current report indicate that dusp1 is also induced at its highest levels in the thalamic recipient sensory input layers IV and VI of the mammalian cortex (also see Takaki et al., 2001) and in sensory input neurons of the thalamus; layer IV consists of sensory input neurons that receive direct input from sensory nuclei in the thalamus, and layer VI forms direct reciprocal cortical feedback pathways with the thalamus (Karten, 1991; Shipp, 2007). These findings suggest that dusp1 is largely a sensory-driven IEG in the primary sensory areas of the brain, which we suggest could be linked to attenuation of stimulus-induced expression of egr1 and other IEGs in these neurons.
MATERIALS AND METHODS
Animals
We used 33 male zebra finches, 12 budgerigars, and 6 ring doves from our breeding colonies at the Duke University Medical Center. Some of these animals were from prior studies, in which we used brain sections for visual experiments in zebra finches (Hara et al., 2009) and movement experiments in zebra finches, budgerigars, and ring doves (Feenders et al., 2008). All animals were adults. Animal procedures were approved by the Institutional Animal Care and Use Committee of Duke University. We also used images of brain sections from adult mice of the GENSAT project (Gong et al., 2003).
Auditory stimuli experiments
For zebra finches, males were placed individually in sound attenuation boxes overnight. On the following morning, while the lights remained off, two groups of birds were taken: a silent control group that remained in the dark but awake (n = 3 males) and a hearing song group that was presented with digitally recorded zebra finch songs through a speaker for 30 minutes (n = 3). The playbacks consisted of three different songs, totaling 12 seconds in length, presented once every minute, similar to a described protocol (Mello et al., 1992). The songs were from another colony of birds and thus were novel to the hearing group; novel song is known to cause high levels of hearing-induced IEG expression in the auditory pathway (Mello and Clayton, 1994, 1995). The lights were kept off to prevent IEG induction in visual brain areas, in movement-associated brain areas due to the bird’s motivation to hop and make other movements, and in auditory areas by hearing self-singing when the lights are on (Jarvis et al., 1998; Feenders et al., 2008; Hara et al., 2009). For budgerigars, after a 2–3-hour quiet period in a room alone, males were presented with a playback of the recorded warbles for 30 minutes (three repetitions of a 10-minute segment of spontaneous warbles), as previously described (Jarvis and Mello, 2000). Animals that did not sing were sacrificed immediately at the end of the playback period and taken as the hearing song groups.
Visual stimuli experiments
To identify brain areas activated by vision, we used brain sections from a previous study (Hara et al., 2009) of male zebra finches that were unilaterally stimulated with visual stimuli. Briefly, one eye of each bird was covered with several layers of black vinyl electrical tape; the innermost layer was placed so that the smooth surface covered the eye to prevent irritation. The tape was sealed at the edges with super glue to the surrounding skin and feathers to prevent light leakage. We alternated the covering of the right and left eyes in different birds to prevent potential biases in the results. Birds were then individually housed overnight in the dark in sound attenuation boxes. They were divided into three groups: silent alone and kept in the dark for 45 minutes in the morning during waking hours (n = 3 total; right eye covered n = 1, left eye covered n = 2); silent alone with the light turned on for 45 minutes (n = 4; right eye covered n = 2, left eye covered n = 2); and seeing a natural stimulus, a female with the light turned on while singing to her for ~45 minutes (n = 5; right eye covered n = 3 and left eye covered n = 2).
The rationale for the female stimulus group of the previous study was to determine whether there was visually associated IEG regulation in the vocal pathway during singing to females, which was found not to be the case (Hara et al., 2009). A female was placed in the cage with the male, but separated by a cage wall barrier, on the night before the recording session. The cage wall barrier was made from the same metal bar material as the rest of the cage. Thus, the male and female could interact visually and acoustically, but not physically. Another group of male zebra finches with one eye covered were presented with females, and those that did not sing were taken as a “seeing female only” group (n = 5). Behavior was videotaped and audio recorded by using Avisoft Recorder (Avisoft Bioacoustics, Berlin, Germany), to verify that singing or no singing occurred and that the males looked at the females (Hara et al., 2009).
Hopping experiments
To identify activated brain areas involved in nonvocal movements, we used brain sections from a previous study (Feenders et al., 2008) of birds that were induced to repeatedly hop. Briefly, hearing intact or deafened birds were placed in a cylindrical, transparent plexiglass, rotating wheel (zebra finches and budgerigars) or on a treadmill (ring doves). The wheel was inside a sound attenuation chamber and rotated by an attached metal rod that was controlled by a relatively quiet motor, outside of the box, with variable speed control (Feenders et al., 2008). Birds were deafened to prevent hearing-induced expression due to hearing feet hops (or feet steps for the doves) and the mechanical sounds of the rotating wheel (or moving treadmill). Behavior was observed and recorded via an infrared camera, connected to an external video recorder. Before an experiment was started, the wheel was rotated (~20 rpm) or the treadmill run first with lights on for 5 minutes and then in the dark for an additional 10 minutes to get the bird accustomed to the wheel (or treadmill) and to reduce stress in the new environment. The wheel (or treadmill) was then turned off, and the bird was allowed to sit for 2–3 hours in darkness; most birds did not go to sleep, as determined by eyes open and head not resting on the back. The lights were kept off to prevent light- and optic flow-induced gene expression in visual pathways. Thereafter, for zebra finches and budgerigars, three control and experimental groups were taken: 1) hearing intact males that sat still in the dark for an additional 30 minutes (n = 3 each species); 2) hearing intact males that hopped in the rotating wheel in the dark for 30 minutes (n = 3 each species); and 3) deafened males that hopped in the rotating wheel in the dark for 30 minutes (n = 3 each species). For ring doves, two groups were taken: 1) hearing intact males that sat still in the dark for 30 minutes (n = 3); and 2) walking while deaf in the dark for 30 minutes (n = 3).
In addition, in order to verify dusp1 induction in all primary sensory areas in the same animals, and for the double-labeling experiment (see below), freely behaving zebra finches (hearing song in the light for 30–45 minutes) were also taken after being placed individually in the sound-attenuating boxes overnight in the dark (n = 3). Their behavior was monitored to confirm that they did not either stay still or sing.
Cloning of zebra finch dusp1
We cloned a cDNA fragment of dusp1 from whole zebra finch male brain mRNA with degenerate primers and reverse transcriptase-polymerase chain reaction (RT-PCR). First, brain mRNA was reverse transcribed to cDNA by using Superscript Reverse Transcriptase (Invitrogen, Carlsbad, CA) with oligo dT primers. Then dusp1 was amplified by using degenerate primers to conserved regions of the coding sequence from human, mouse, rat, and chicken in the NCBI database (accession nos. X68277, X61940, X84004, and AF026522, respectively). The forward and reverse oligo DNA primers were 5′-CCCWCTSTACGAYCARGGNGG-3′ and 5′-ACRCCGATG-GARACDGGRAARTT-3′, respectively. PCR conditions were 94°C for 1 minute, 58°C for 1 minute, and 72°C 30 seconds, for 25 cycles in 1X PCR buffer (Takara, Otsu, Japan). PCR products were examined on 1% agarose gels, extracted from the gels, ligated into the pGEM-T Easy plasmid (Promega, Madison, WI), and transformed into XL-1 blue E. coli cells. Plasmid DNA was isolated, and the inserted cDNA was sequenced from the 5′- and 3′ ends, by using plasmid sequencing primers. To confirm that dusp1 was cloned, the sequences were BLAST searched against the NCBI nucleotide database, and homologies to other species were found. One of the zebra finch clones (Genbank accession no. AB476742) was identified as a 543-bp fragment (in the forward orientation of the pGEM-T Easy plasmid) that showed 90% and 84% DNA sequence identity, respectively, to the homologous coding region of the chicken and human dusp1 cDNAs.
After the completion of our study, the draft zebra finch genome sequence was made available (UCSC browser; Warren et al., 2010), and a full-length dusp1 sequence was predicted (NCBI accession no. XM_002193132). Our partial cDNA clone is 100% identical to the predicted sequence. It spans exon3 and the beginning of exon4 relative to the human dusp1 gene; however, the zebra finch dusp1 genomic sequence is not yet complete, so it is not possible at this time to determine the total number of exons in songbirds. Our clone shows 84% identity to the zebra finch dusp4 gene, which from our experience is on the borderline of cross-hybridization (85% identity), but not sufficient to show a strong signal at the high-stringency conditions we used. To confirm our prediction, we used the genomic sequence and PCR to clone a zebra finch dusp4 cDNA (Genbank accession no. AB546648), hybridized it to brain sections of silent control and auditory stimulated animals, and found very low dusp4 expression throughout the brain regardless of condition, with a pattern that did not match dusp1 (data not shown), as predicted. The forward and reverse oligo DNA primers were 5′-CCTTTCCATGACCAGGGTG-3′ and 5′-ACACTGG-GAAGCTGAAGACA-3′. Our dusp1 clone showed no other regions of high cross-identity in the draft zebra finch genome.
In situ hybridizations
After each of the above behavioral sessions, birds were decapitated, and their brains were removed, embedded in OCT compound (Sakura Fine Technical, Tokyo, Japan) inside tissue block molds, frozen in a dry ice ethanol bath, and stored at −80°C. In situ hybridizations were performed as previously described (Wada et al., 2006). In brief, 12-μm frozen sections were cut in the sagittal plane to maximize the amount of brain tissue per section; for the monocular visual experiments, coronal sections were used to compare differences of gene regulation in corresponding regions between hemispheres. Sections of all birds of a given experiment were simultaneously fixed in 3% paraformaldehyde, washed in phosphate-buffered saline (PBS; pH 7.4), acetylated, dehydrated in an ascending ethanol series (70%, 95%, and 100% for 2 minutes each), air dried, and processed for in situ hybridization with antisense and sense 35S-UTP-labeled riboprobes of zebra finch dusp1 or egr1. The egr1 clone is described in Wada et al. (2006). To generate the riboprobes, the dusp1 (543-bp) and egr1 (1,100-bp) inserts in the pGEM-T Easy vector were PCR amplified with plasmid primers, and the amplified products were purified. With the amplified DNA, SP6 RNA polymerase was used to synthesize the antisense 35S-riboprobes, and T7 RNA polymerase was used to synthesize the sense 35S-riboprobes. Then 1 × 106 cpm of the 35S-probe was added to the hybridization solution. Hybridization and washes were at 65°C. Slides were dehydrated in an ascending ethanol series, exposed to X-ray film (Biomax MR, Kodak, Rochester, NY) for 1–4 days (dusp1) or 1–2 days (egr1), then dipped into autoradiographic emulsion (NTB2, Kodak), incubated for 1–2 weeks at 4°C, processed with D-19 developer (Kodak) and fixer (Kodak), Nissl-stained with cresyl-violet acetate solution (Sigma, St. Louis, MO), placed in xylene, and coverslipped with Permount mounting medium (Sigma). We observed no specific signals with the sense probes (not shown).
For double labeling in situ detection of dusp1 and egr1, a 35S-UTP-labeled riboprobe of dusp1 or egr1 was used in combination with a digoxigenin (DIG)-UTP-labeled ribo-probe of the other gene. The two probes were added simultaneously to the hybridization solution. After hybridization, the double-labeled slides were not dehydrated in EtOHs, but were washed in buffer 1 (100 mM Tris, pH 7.5, 150 mM NaCl, 0.1% Tween 20) at room temperature (RT) twice for 30 minutes and incubated in blocking solution (5% lamb serum and 1% bovine serum albumin [BSA] in buffer 1), and then with an anti-DIG-alkaline phosphatase (AP) antibody (1:2,000 in buffer 1) at 4°C overnight. The sections were then washed in buffer 1 at RT three times for 30 minutes each and in buffer 2 (100 mM Tris, pH 9.5, 100 mM NaCl, 50 mM MgCl2) twice for 5 minutes each. Thereafter, the slides were reacted with either NBT/BCIP solution (NBT/BCIP Ready-to-Use Tablets, Roche, Indianapolis, IN) or BM purple (Roche) for 5–6 hours in the dark and washed once in stop buffer (2 mM Tris, pH 8.0, 1 mM EDTA, pH 8.0), and then twice in PBS for 3 minutes each and in water for 10 seconds. The slides were dried overnight and dipped into Ilford autoradiography emulsion (Ilford K5, Polysciences, Warrington, PA). We did not use Kodak NTB emulsion (either NTB2 or 3) because it removes the AP chromogenic product from the DIG probe (Young, 1989; Kerner et al., 1998). The slides were incubated for 1–2 weeks at 4°C, processed with D-19 developer and fixer (Kodak), and coverslipped with mounting medium (VECTASHIELD with DAPI, Vector, Burlingame, CA).
Quantification and statistics
Brain images on X-ray films were digitally scanned from a dissecting microscope connected to a SPOT-III CCD camera by using SPOT imaging software (Diagnostic Instruments, Sterling Heights, MI). For quantifications, care was taken to use the same light settings across all images of the same gene. We used Adobe Photoshop CS3 (Adobe Systems, San Jose, CA) to measure the mean pixel intensities on a 256 gray scale in the brain areas of interest from at least two adjacent sections. We then quantified fold gene induction by measuring expression levels of each gene in the region of interest in stimulated animals divided by the average expression levels in control animals for a given experiment. For these comparisons, statistical differences were determined by unpaired t-test (asterisks inside bar graphs). A value of ~1 represents no induction relative to controls; statistically significant values above or below 1 represent induced or reduced expression, respectively. We also made comparisons between genes (dusp1 and egr1) within the same brain region from adjacent sections of the same animals by using paired t-test (asterisks above bar graphs). For the vision experiments, we additionally performed ratio comparisons of stimulated gene expression between hemispheres (contralateral to eye covered side/contralateral to eye open side), by using paired t-test, as a stringent test for differences within the same animals.
For the double-label dusp1 and egr1 experiments, we used a compound microscope at 60 × magnification and Slide book software (Olympus, Tokyo, Japan) to acquire images of the regions of interest. The total numbers of cells (range 51–61, n = 3 birds) within a given field from at least two adjacent sections were counted. Of this total, the subsets of single- and double-labeled dusp1 and egr1 cells were estimated and corrected with the Abercrombie equation [N = n(T/(T + D)], where N is the corrected number of the labeled cells, n the estimated number of the labeled cells, T the thickness of the section (12 μm), and D the mean diameter of the nuclei; Guillery and Herrup, 1997). We only considered a cell labeled if we could find a clear nucleus stained by DAPI or counterstained by the chromogenic background signal (purple reaction product) associated with the DIG reaction product. From the total number of cells, the mean percentage of dusp1+, egr1+, and dusp1+/egr1+ relative to labeled cells were determined and statistically compared within and across adjacent brain regions by ANOVA among regions, e.g., L2 vs. L1, followed by Fisher’s PLSD post hoc test.
Nomenclature
We used the new avian brain nomenclature (Reiner et al., 2004; Jarvis et al., 2005) with modifications that have been discussed in several previous reports (Mouritsen et al., 2005; Feenders et al., 2008; Hara et al., 2009; Kubikova et al., 2010). In particular, based on gene markers and other evidence, the formally named dorsal hyperstriatum (HD) was originally revised to hyperpallium densocellulare (HD) and the ventral hyperstriatum (HV) originally revised as simply mesopallium (Reiner et al., 2004; Jarvis et al., 2005). Our subsequent reports using mesopallium-specific markers (GluR1, FoxP1, D1B, and D3) in multiple avian species (Wada et al., 2004; Mouritsen et al., 2005; Feenders et al., 2008; Hara et al., 2009; Kubikova et al., 2010) led us to modify this change; we argue that the formally named dorsal hyperstriatum (HD) is the dorsal mesopallium (MD) and the formally named ventral hyperstriatum (HV) is the ventral mesopallium (MV). This nomenclature is a minority view alternative to what others consider HD as a distinct brain subdivision, not part of or related to the mesopallium. Additional studies are necessary to resolve this issue. Secondary and tertiary sensory areas of the telencephalon were given names associated with the name of sensory input cell populations in which the projection is from (Feenders et al., 2008). Thus, for the auditory areas adjacent to or near Field L2 we called them N-L2 (for L1 and L3) and MV-L2 (for caudal mesopallium [CM]). For visual areas adjacent to or near the entopallium (E) that have been called lateral nidopallium (LN) and lateral ventral mesopallium (LMV), we called them nidopallium adjacent to the entopallium (Ne) and ventral mesopallium near the entopallium (MVe). For somatosensory areas adjacent to or near basorostralis (B), we called them Nb and MVb as well. This naming scheme allowed us to universally compare functionally activated, homologous brain areas across species (Feenders et al., 2008).
Figure preparation
The photomicrographs were adjusted in Adobe Photoshop CS3. The Levels function was used to expand the range of image pixels within the full 250 range. The intensity of the background outside the tissue was reduced equally for all brain sections, in order to see the brain section with or without gene expression. Color images were further adjusted by the color adjustment function so that the signals in white color had enough contrast within the visible spectrum. All images of the same gene in control and experimental groups were adjusted in the same way to avoid unintentional modification in gene expression images across groups.
RESULTS
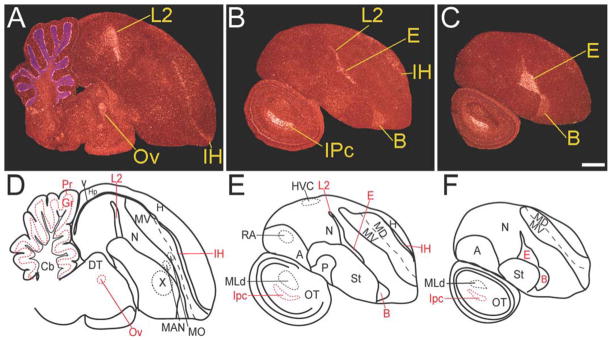
In situ hybridizations of brain sections from freely behaving zebra finches revealed that relative to the rest of the brain, there was higher dusp1 expression in thalamic-recipient sensory input cell populations of the telencephalon. These populations included Field L2 (auditory), entopallium (E, visual), basorostralis (B, somatosensory), and the intercalated layer of the hyperpallium (IH; visual and somatosensory; Fig. 2A–C). Moreover, L2, E, and B formed one continuum of labeled cells between the nidopallium and striatum, whereas IH formed one continuum between the hyperpallium (H) and dorsal mesopallium (MD). In these sensory input and higher sensory neuronal populations, we found specific and complementary regulation of dusp1 and egr1, by using stimulus and behavioral manipulations.
Figure 2.
Dusp1 mRNA expression in a zebra finch brain from a freely behaving animal. A–C: Darkfield images of in situ hybridizations from medial to lateral. White silver grains, dusp1 mRNA expression. Red, cresyl violet cellular stain. Besides the high dusp1 expression in the thalamic-recipient sensory zones of the telencephalon (L2, E, B, and IH), there is higher expression also in the thalamic auditory nucleus Ov, midbrain visual nucleus IPc, and the purkinje (Pr) and granular (Gr) neuron layers of the cerebellum. Sections are sagittal; anterior is right, and dorsal is up. D–E: Anatomical profiles of brain areas in A–C. For abbreviations, see list. Scale bar = 1 mm in C (applies to A–C).
Hearing-induced regulation in auditory input neural populations
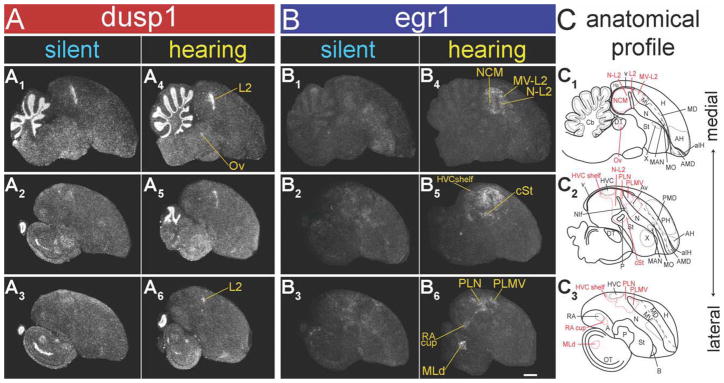
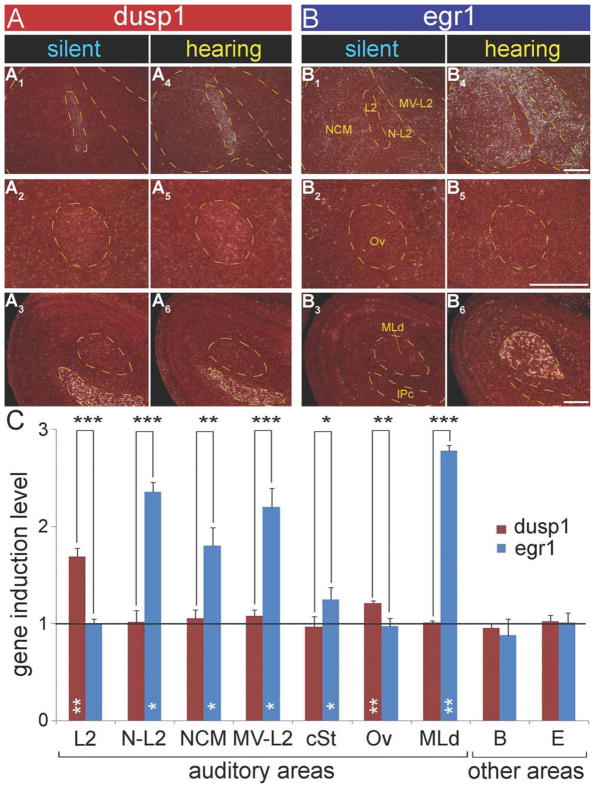
Relative to silent control zebra finches sitting still in the dark, animals that heard 30 minutes of song playbacks and also sat still in the dark had increased dusp1 expression throughout Field L2 (Figs. 3A1–6, 4A1,4,C, red bars, * inside bar). In the secondary and tertiary auditory neuron populations that are known to express high levels of egr1 in response to hearing song (Mello and Clayton, 1994; Jarvis and Nottebohm, 1997), there was no detectable activation of dusp1. These populations included the nidopallium adjacent to L2 (N-L2, consisting of L1, L3, PLN, and the HVC shelf; secondary sensory neurons), the caudal medial nidopallium (NCM), the caudal ventral mesopallium near L2 (MV-L2, consisting of CM and PLMV; tertiary sensory neurons), the caudal striatum (cSt) adjacent to L2, and the RA cup (also tertiary sensory neurons) in the arcopallium adjacent to RA (Figs. 3A1–6, 4A1,4,C, red bars). To be certain that these higher sensory neurons expressed egr1 in our birds, we hybridized adjacent sections to egr1 and found robust hearing song-induced expression (Figs. 3B1–6, 4B1,4,C, blue bars, * inside bars). The anatomical contrast in activation between the two genes was prominent, such that the dusp1 and egr1 expression domains formed complementary images of each other in primary vs. higher (secondary, tertiary, etc.) telencephalic auditory areas (Figs. 3A vs. B and 4A vs. B). This differential regulation between the two genes in the telencephalic auditory areas was significant (Fig. 4C, * above bars). Thus, the lack of dusp1 induction in the higher (secondary, tertiary, etc.) auditory neurons was not due to a lack of activity in these neurons.
Figure 3.
Dusp1 and egr1 mRNA expression patterns of zebra finch after auditory stimulation with song. Shown are negative-image film autoradiographs of in situ hybridizations with dusp1 (A) and egr1 (B), from a silent control male bird (no auditory stimulus) in darkness in a sound attenuation chamber (A1–3: dusp1; B1–3: egr1), and a male bird that heard 30 minutes of conspecific songs while sitting still in the dark in the chamber (A4–6: dusp1; B4–6: egr1). Adjacent sagittal sections were used for each gene. White, gene expression. Lines and names in yellow, auditory areas where each mRNA was upregulated. The right-most column shows anatomical profiles of brain areas in which auditory areas are highlighted in red and others in black (C). Sections are sagittal. For abbreviations, see list. Scale bar = 1 mm in B6 (applies to A1–B6).
Figure 4.
Magnified images and quantification of dusp1 and egr1 expression in auditory areas of zebra finch brain after song playbacks. A: Dusp1 expression in auditory regions from silent control (A1–3) and hearing song (A4–6) animals. B: Egr1 expression in auditory regions from adjacent sections of the silent control (B1–3) and hearing song (B4–6) animals. Yellow dashed lines show the Nissl-stained boundary of areas, as labeled in B1–3. Sections are sagittal; anterior is right, and dorsal is up. C: Quantification of dusp1 (red bars) and egr1 (blue bars) expression in seven auditory areas, and visual (E) and somatosensory (B) areas as control regions. Each bar shows an average value ± SD. Values are normalized by the average level of expression in the same brain areas of silent control birds. A value of ~1 indicates no change in expression levels relative to silent controls. Values significantly above 1 indicate induced expression in animals that heard song (n = 3) relative to silent controls (n = 3, white stars inside bars; unpaired t-test). Black stars above bars indicate significant differences between amount of dusp1 and egr1 induction (paired t-test between the same brain regions of the same animals). *, P < 0.05; **, P < 0.01; and ***, P < 0.001. For abbreviations, see list. Scale bar = 500 μm in B4 (applies to A1,A4,B1,B4), B5 (applies to A2,A5,B2,B5), and B6 (applies to A3,A6,B3,B6).
Differential dusp1 and egr1 activation also occurred at earlier stations of the auditory pathway. The thalamic auditory nucleus ovoidalis (Ov), which does not show hearing-induced egr1 expression (Figs. 3B1,4, 4B2,5,C; Mello and Clayton, 1994; Jarvis and Nottebohm, 1997), showed hearing-induced dusp1 expression (Figs. 3A1,4, 4A2,5,C). The upregulation of dusp1 in Ov, though, was less robust than it was in L2 (Fig. 4C). Conversely, the midbrain auditory nucleus MLd, which showed high levels of hearing-induced egr1 expression (Figs. 3B3,6, 4B3,6,C), did not show detectable hearing-induced dusp1 expression within the same medial part of MLd (Figs. 3A3,6, 4A3,6,C). The lateral part of MLd showed dusp1 expression in some birds regardless of hearing or silent condition. Likewise, the Ov shell, which receives descending auditory feedback from RA cup in the telencephalon (Fig. 1B; Mello et al., 1998) and shows some hearing-induced egr1 expression (Fig. 4B5; Mello and Clayton, 1994), did not appear to show cells with induced dusp1 expression in response to hearing song (Fig. 4A5).
The hearing-song-induced regulation of dusp1 was specific to the auditory pathway, as we did not detect significant induction above silent control levels in telencephalic sensory input neurons of the visual (E) and somatosensory (B) pathways (Fig. 4C). We also did not detect any increase in egr1 expression in the higher sensory neurons in visual and somatosensory nidopallium (secondary sensory) and ventral mesopallium (tertiary sensory) adjacent to E (Ne and MVe) and B (Nb and MVb), respectively (data not shown). In summary, the results suggest that hearing song specifically causes induction of dusp1 gene expression in auditory input cell populations in which egr1 is not or is minimally regulated, and vice versa for higher auditory populations in which egr1 gene expression is induced. The two genes combined functionally map the entire auditory pathway from the midbrain to the forebrain.
Visually induced regulation in visual input neural populations
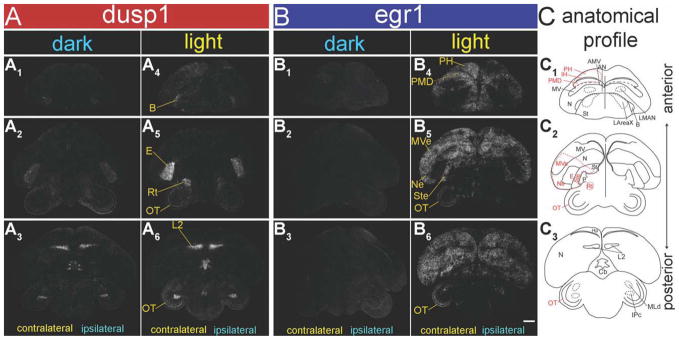
To determine whether dusp1 can be regulated in sensory input neurons other than auditory, we used brain sections from a monocular occlusion experiment that we recently showed reduced egr1 induction in the visual pathways (Fig. 1C; Hara et al., 2009). This reduction occurs because in birds with laterally placed eyes, such as the zebra finch, the visual pathways are almost completely crossed at the optic chiasm (Weidner et al., 1985); thus blocking visual input from one eye significantly reduces the activation in visual pathway regions of the contralateral hemisphere (Hara et al., 2009). We therefore examined dusp1 expression in zebra finches with one eye covered. First, we found that relative to animals that sat still in the dark, those that were then exposed to light for 45 minutes had higher dusp1 expression throughout most of the sensory input neuron populations of the telencephalon of both hemispheres (L2, E, B, and IH; Figs. 5A1–6, 6C, red bars). This increase is not surprising considering that when the lights are turned on, the birds perform movements that can activate somatosensory pathways (i.e., B) and make sounds that can activate auditory pathways (i.e., L2). However, when expression was compared between hemispheres, dusp1 expression was higher contralateral to the open eye only within E, the visual input neurons of the tectofugal visual pathway and for some animals in posterior IH, the visual input neurons of the thalamofugal visual pathway (Figs. 5A1–6, 6A1,2,4,5,C, * on x-axis between bars). Although barely detectable in expression, there was a weak quantitative hemispheric difference in dusp1 induction in some higher sensory neurons in the visual regions adjacent to E (MVe and Ste; Figs. 5A2,5, 6A2,5,C, * on x-axis between red bars). There was no significant difference in dusp1 levels in all other higher sensory neurons in the visual regions adjacent to E or IH (Ne, the posterior hyperpallium [PH], and the posterior dorsal mesopallium [PMD]; Fig. 6C, red * on x-axis between red bars).
Figure 5.
Dusp1 and egr1 mRNA expression patterns in zebra finch brain after visual stimulation with light. Shown are negative-image film autoradiographs of in situ hybridizations with dusp1 (A) and egr1 (B), from a sitting still control male bird in the dark with the left eye covered (A1–3: dusp1; B1–3: egr1), and a male bird stimulated with lights on for 45 minutes also with the left eye covered (A4–6: dusp1; B4–6: egr1). Adjacent sagittal sections were used for each gene. Contralateral hemisphere is opposite of the open eye; ipsilateral hemisphere is the same side as the open eye. White, gene expression. Lines and names in yellow, visual areas where each mRNA was induced. The right-most column shows anatomical profiles of brain areas, in which visual areas are highlighted in red and others in black (C). Sections are coronal. Dorsal is up, and right hemisphere is on the right. For abbreviations, see list. Scale bar = 1 mm in B6 (applies to A1–B6).
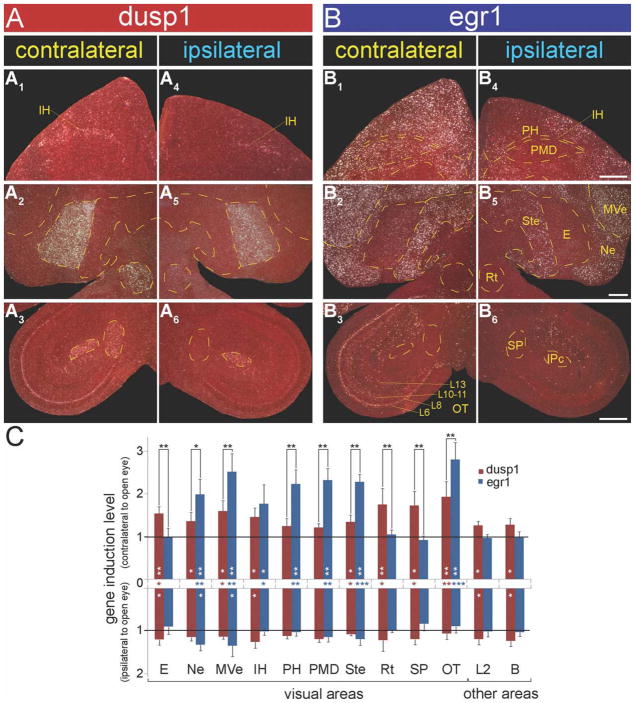
Figure 6.
Magnified images and quantification of dusp1 and egr1 expression in visual areas of zebra finch brain after light stimulation. A: Dusp1 expression in visual regions of both brain hemispheres of an animal with one eye covered. Contralateral hemisphere is opposite the open eye (A1–3); ipsilateral hemisphere is the same side as the open eye (A4–6). B: Egr1 expression in visual regions (B1–3: contralateral hemisphere; B4–6: ipsilateral hemisphere) from adjacent sections of the animal in A. Yellow dashed lines show the boundary of areas, as labeled in B4–6. C: Quantification of dusp1 (red bars) and egr1 (blue bars) gene expression in 10 visual areas, and auditory (L2) and somatosensory (B) areas as control regions. Each bar shows an average value ± SD. Values are normalized by the average level of expression in the same brain areas of dark control birds; contralateral side to the open eye is above the x-axis and ipsilateral is below the x-axis. A value ~1 indicates no change in expression levels relative to silent controls. Values significantly above 1 indicate induced expression in a hemisphere region of the light-stimulated animals (n = 4) relative to dark-housed animals (n = 3, white stars inside bars; unpaired t-test). Significant differences in brain regions between hemispheres within the same bird are indicated by red (dusp1) or blue (egr1) stars on the x-axis between bars (paired t-test within animal). Significant differences between amount of dusp1 and egr1 induction are indicated by black stars above (contralateral) bars (paired t-test between the same brain regions of the same animals). The ipsilateral side did not show significant differences between two genes. *, P < 0.05; **, P < 0.01; and ***, P < 0.001. For abbreviations, see list. Scale bar = 500 μm in B4 (applies to A1,A4,B1,B4), B5 (applies to A2,A5,B2,B5), and B6 (applies to A3,A6,B3,B6).
In contrast, there was robust induced expression of egr1 in these higher visual areas (Ne, MVe, Ste, PH, and PMD) contralateral to the open eye (Figs. 5B1–6, 6B1,2,4,5,C, white * in blue bars; and blue * on x-axis). PMD is a dumbbell-shaped visual region in frontal sections formally called HD (hyperstriatum dorsale or hyperpallium densocellulare) in past studies (Shimizu and Bowers, 1999; Medina and Reiner, 2000; Kruztfeld and Wild, 2004) that we now designate as dorsal mesopallium (MD). PH is the overlying visual Wulst part of the posterior hyperpallium. These regional differences between light-induced dusp1 in sensory input and egr1 in higher sensory neurons of the visual pathways were significant (Fig. 6C, black * above bars). In animals housed in the dark with one eye covered, there was no hemispheric difference in dusp1 or egr1 expression in any of the brain regions measured (Figs. 5A1–3,B1–3; P = 0.72–0.92 paired t-test), demonstrating that light stimulation was necessary for the observed hemispheric differences in the light-stimulated group.
Within the thalamus, there was robust dusp1 induction in multiple visual nuclei contralateral to the open eye. These included: 1) nucleus rotundus (Rt), which is a thalamic sensory input nucleus that sends input to E; and 2) the subpretectal nucleus (SP), which is an inhibitory thalamic nucleus and projects to Rt (Figs. 1C, 5A2,5, 6A2,3,5,6,C, red bars; Benowitz and Karten, 1976; Deng and Rogers, 1998b; Theiss et al., 2003). SP is not thought to be a sensory input nucleus of the thalamus; however, its path of connectivity is similar to that of Rt (Fig. 1C), and thus technically it could be considered sensory input. These thalamic nuclei did not show egr1 induction in response to light stimulation (Figs. 5B2,5, 6B2,3,5,6,C blue bars). Within the midbrain, light stimulation caused an intense band of dusp1 induction in layer 8 of the optic tectum (OT) contralateral to the open eye, but no detectable induction in other layers (Figs. 5A2,3,5,6, 6A3,6,C, red bars). However, unlike the non-overlap of dusp1 and egr1 induction in the midbrain auditory nucleus MLd, egr1 induction was also found in the OT layer 8 (as well as layers 6, 10–11, and 13) contralateral to the open eye (Figs. 5B2,3,5,6, 6B3,6,C, blue bars).
Another midbrain nucleus, the isthmi pars parvocellularis (IPc), which encodes both visual and auditory responses and is reciprocally connected with the OT (Maczko et al., 2006), showed high basal dusp1 expression bilaterally (Figs. 5A,3,6, 6A3,6; P = 0.4801; paired t-test between ipsilateral and contralateral hemispheres of light stimulated animals). Dusp1 expression in IPc was high even in animals in the dark and no different from animals stimulated with light (P = 0.8077; unpaired t-test between animals in the dark and with light stimulated). There was no detectable egr1 expression in IPc of any of the groups (Figs. 5B3,6, 6B3,6), consistent with differential regulation of the two genes.
We wondered whether differential IEG induction occurred in visual areas in different social contexts, such as looking at a female vs. alone. In a previous report (Hara et al., 2009), we found higher levels of induced egr1 expression in PH and lower induction in Ne contralateral to the open eye when males viewed females relative to light alone. Interestingly, in three of five males with one eye covered that sang to and viewed females with the open eye, there was higher dusp1 induction in IH of the hemisphere “ipsilateral” to the open eye (Fig. 7A,C). This finding is in stark contrast to the higher egr1 induction in the adjacent PH and PMD “contralateral” to the open eye of the same animals (Fig. 7B,C), as well as in the light-stimulated-only condition. This differential expression pattern (higher dusp1 induction on the ipsilateral side) was specific to IH of the thalamofugal visual pathway, as dusp1 induction in Rt of the tectifugal visual pathway was higher contralaterally to the open eye in these same males (Fig. 7C), similar to the light-stimulation-only condition (Fig. 6C). In males that viewed females with both eyes open, and also did not sing (n = 3), dusp1 was higher bilaterally in IH (data not shown).
Figure 7.
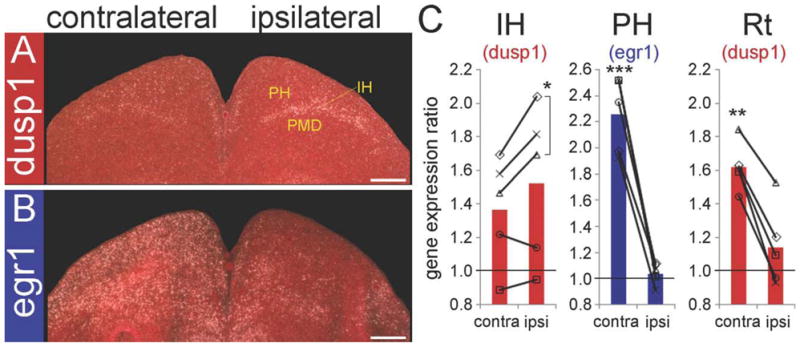
Social context-dependent dusp1 and egr1 expression in visual area IH of zebra finch brain. A: Brain section with dusp1 gene expression in a male that looked at and sang to females for 30 minutes with one eye covered. Contralateral is opposite and ipsilateral is the same side as the open eye. Note the higher expression of dusp1 in the side ipsilateral to the open eye. B: Egr1 expression in adjacent sections showing induced expression in the PH and PMD contralateral to the open eye. C: Quantifications of dusp1 and egr1 in IH and other visual areas when birds looked at females. Each bar shows an average value of dusp1 (red bars) or egr1 (blue bars) gene expression in all animals (n = 5). Each symbol indicates one bird. Values are normalized by the average level of expression in the same brain areas of dark control birds (n = 3). A value of ~1 indicates no change in expression levels relative to silent controls. Significant differences in brain regions between hemispheres within the same bird are indicated by black stars (paired t-test within animals). *, P < 0.05; **, P < 0.01; and ***, P < 0.001. For abbreviations, see list. Scale bar = 500 μm in A,B.
In summary, the results suggest that light stimulation specifically causes dusp1 upregulation in visual input cell populations in which egr1 is not or is minimally regulated and vice versa for higher visual cell populations in which egr1 is highly upregulated. Further, seeing a female for some animals appears to cause a more robust upregulation of dusp1 in visual input cells of the thalamofugal visual pathway (IH) ipsilateral to the open eye, whereas the egr1 induction in the adjacent higher visual regions is blocked by this condition, suggesting an inverse excitatory-inhibitory relationship between IH and the surrounding visual regions when viewing females. The two genes together effectively define and map most if not all known regions of the visual pathways from the midbrain to the forebrain.
Hopping-induced regulation in somatosensory input neural populations
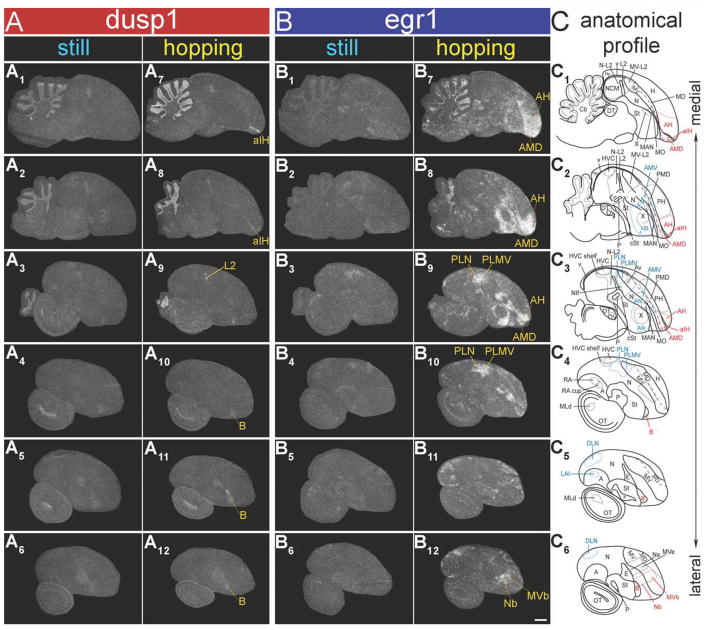
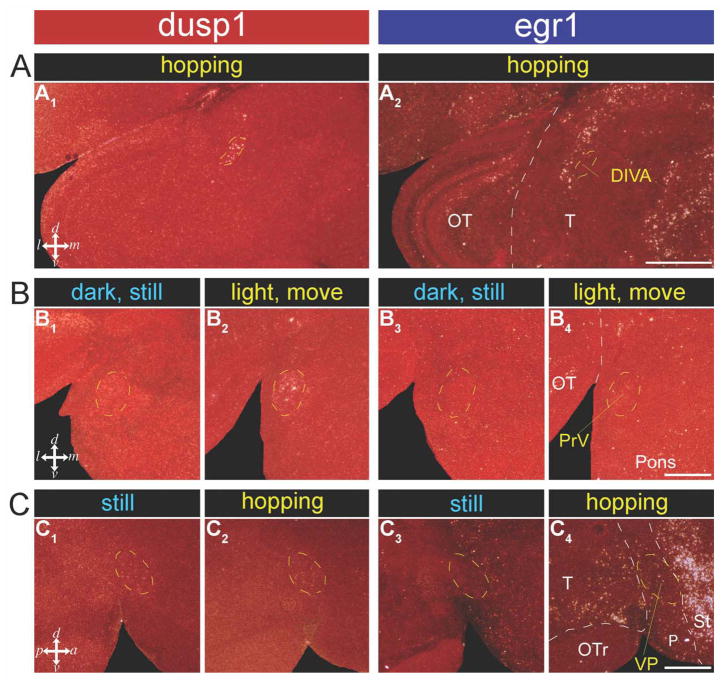
To determine whether high dusp1 induction is restricted to sensory pathways or can be induced in motor systems, we examined dusp1 expression in animals that hopped in a rotating wheel in the dark. Hopping in song birds results in movement-associated egr1 upregulation in both in putative motor pathway areas adjacent to the telencephalic song nuclei and somatosensory pathways (Fig. 1A,D; Feenders et al., 2008). These experiments have to be conducted with animals moving in the dark and while deaf to prevent IEG induction in visual and auditory areas, respectively, from optic flow and hearing the hopping sounds during movement (Feenders et al., 2008). Confirming this requirement, we found that analogous to the egr1 findings in higher sensory neurons for hearing intact animals, hopping in the dark resulted in dusp1 induction in auditory input (Ov and L2) populations as well as somatosensory input populations—the anterior portion of IH (aIH) of the lemnothalamic somatosensory pathway and B of the pseudo-collothalamic somatosensory pathway (aIH shown in Fig. 8A1; Wild and Farabaugh, 1996; Wild and Williams, 1999). The second pathway is called pseudo-collothalamic, because it skips both the midbrain (collo) and thalamus, and projects directly from the trigeminal principle sensory nucleus V (PrV) in the pons to B in the telencephalon (Fig. 1D; Jarvis, 2009).
Figure 8.
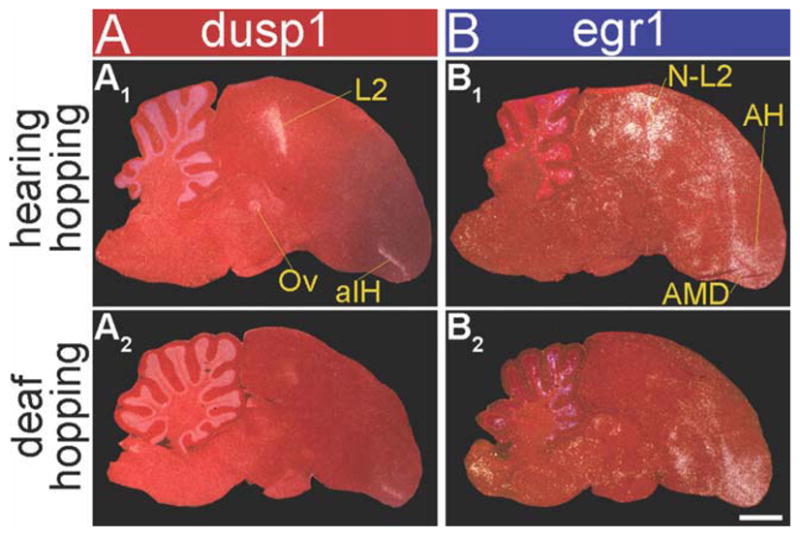
Dusp1 and egr1 mRNA expression patterns in zebra finch brain after hopping in a rotating wheel with hearing intact and while deaf, both in the dark. Shown are darkfield images of medial brain sections with either dusp1 (A) or egr1 (B) mRNA expression. White silver grains, mRNA expression. Red, cresyl violet cellular stain. A1: Dusp1 mRNA expression in a bird that hopped for 30 minutes with hearing intact. B1: Egr1 mRNA expression in an adjacent section from the same bird in A1. A2: Dusp1 mRNA expression in a deafened bird that hopped for 30 minutes. B2: Egr1 mRNA expression in an adjacent section from the same bird in A2. Compare these levels with the basal dusp1 and egr1 expression of animals that sat still and silent in the dark, in Figures 3A1,B1 and 9A1,2,B1,2. Note that deafening eliminated the hopping-induced dusp1 and egr1 expression in auditory areas (L2 for dusp1, P < 0.01; N-L2 for egr1, P < 0.01), but not in somatosensory areas (aIH for dusp1, P = 0.634; AH and AMD for egr1, P = 0.527) (n = 3/group; unpaired t-test). Difference in the red color in the anterior side of the section in A1 was due to unintended variation in cresyl violet staining, and did not affect the radioactive signal intensity. For abbreviations, see list. Scale bar = 1 mm in B2 (applies to A1–B2).
Deafening eliminated the dusp1 induction in Ov and most of L2 of the hopping animals, but not the induction in aIH and B (Figs. 8A1,2, 9A1–12; higher power in Fig. 10A1–4 and quantification in Fig. 10C, red bars). As observed in the sensory input neurons in the auditory and visual systems, dusp1 induction in the somatosensory input populations of the hopping animals was complementary to the patterns of egr1 induction in higher somatosensory populations. These higher populations included the anterior hyperpallium (AH) and anterior dorsal mesopallium (AMD) of the lemnothalamic somatosensory pathway surrounding aIH, the nidopallium adjacent to B (Nb), and ventral mesopallium near B (MVb) of the pseudo-collothalamic somatosensory pathway (Figs. 8A2,B2, 9A7–12,B7–12, 10A3,4,B3,4). There was some low-level, detectable dusp1 induction in the higher somatosensory populations (AH, AMD, Nb, and MVb) and likewise some egr1 induction in aIH (Figs. 9A,B, 10A–C). Despite this overlap of induction, the differences between dusp1 (higher in somatosensory input populations) and egr1 (higher in secondary and tertiary somatosensory populations) expression were large and significant (Fig. 10C, * above bars). There was no detectable dusp1 induction in E of the visual pathway in the deaf animals that hopped in the dark (Figs. 10A2,4,C), consistent with the lack of visual input.
Figure 9.
dusp1 and egr1 mRNA expression patterns in zebra finch brain after hopping in a rotating wheel when deaf. Shown are negative-image film autoradiographs of in situ hybridizations with dusp1 (A) and egr1 (B), from a silent control hearing intact bird (sitting in the rotating wheel in the dark; A1–6: dusp1; B1–6: egr1), and a hopping deaf bird (hopping for 30 minutes in the wheel in the dark; A7–12: dusp1, B7–12: egr1). Adjacent sagittal sections were used for each gene. White, gene expression. Lines and names in yellow, somatosensory areas where each mRNA was upregulated. The right-most column shows anatomical profiles of brain areas, in which somatosensory areas are highlighted in red, putative motor areas in light blue, and others in black (C). For abbreviations, see list. Scale bar = 1 mm in B12 (applies to A1–B12).
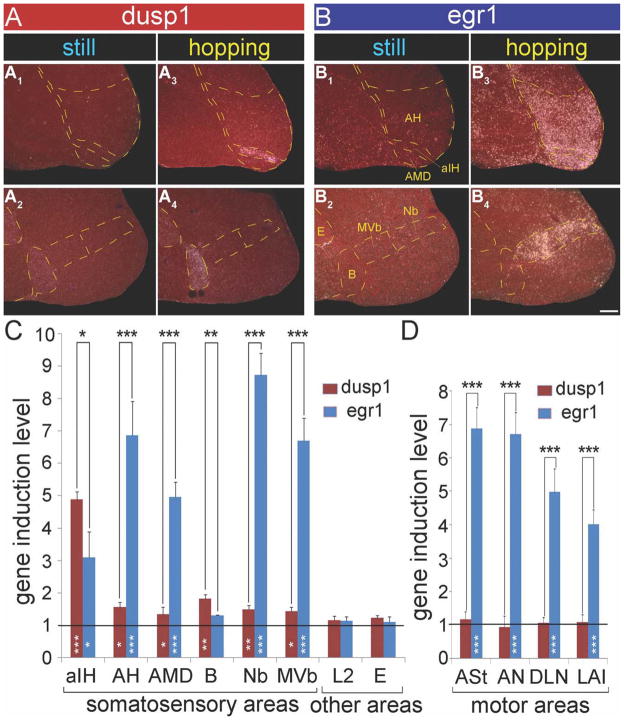
Figure 10.
Magnified images and quantification of dusp1 and egr1 expression in somatosensory areas and several putative motor areas of zebra finch brain after hopping. A: Dusp1 expression in somatosensory regions from a sitting still control male bird in the dark (A1,2), and a hopping deaf animal in the dark (A3,4). B: Egr1 expression in adjacent sections of the sitting still control (B1,2) and the hopping deaf (B3,4) animal. Yellow dashed lines show the boundary of areas, as labeled in B1,2. Sections are sagittal; anterior is right, and dorsal is up. C: Quantification of dusp1 (red bars) and egr1 (blue bars) expression in six somatosensory areas, and auditory (L2) and visual (E) areas as controls. D: Quantification of dusp1 (red bars) and egr1 (blue bars) expression in four motor-associated areas as examples of regulation in the motor system. For C and D, values are average expression levels in hopping animals normalized by the average level in the same brain areas of sitting still control birds, ± SD. A value of ~1 indicates no change in expression levels relative to controls. Values significantly above 1 indicate induced expression in animals that hopped (n = 3) relative to still controls (n = 3, white stars inside bars; unpaired t-test). Black stars above bars indicate significant differences between amount of dusp1 and egr1 induction (paired t-test between the same brain regions of the same animals). *, P < 0.05; **, P < 0.01; and ***, P < 0.001. For abbreviations, see list. Scale bar = 500 μm in B4 (applies to A1–B4)
Interestingly, a lateral portion of L2 showed dusp1 expression induced by both hearing (Fig. 3A6) and hopping (Fig. 9A9). This lateral portion of L2 was directly adjacent to the posterior lateral nidopallium (PLN) and posterior lateral ventral mesopallium (PLMV), which we previously found (Feenders et al., 2008) showed both hearing- and hopping-induced egr1 expression (Figs. 3B6, 9B9). This part of L2 also abuts the NIf song nucleus, and NIf shows both robust auditory and singing-associated motor activity and is necessary for auditory input into the song motor system (Jarvis and Nottebohm, 1997; Cardin and Schmidt, 2004; Cardin et al., 2005; Bauer et al., 2008). These findings further support the idea that this lateral portion of the auditory pathway (lateral L2, PLN, and PLMV) adjacent to the NIf and Avalanche (Av adjacent to PLMV) song nuclei could be a source of auditory input into the putative avian motor pathway (Feenders et al., 2008).
For the somatosensory nuclei of the brainstem, the dorsal intermediate ventral anterior (DIVA) nucleus of the thalamus and PrV in the pons (Fig. 1D), we did not have a sufficient number of animals with these regions in our sagittal brain dissections to assess dusp1 and egr1 regulation quantitatively. However, we had frontal sections of one dark hopping animal with DIVA, as well as three visually stimulated animals moving in the light, and three sitting still control animals in the dark with PrV. We found that in these two nuclei there was bilateral induced dusp1 expression (P < 0.001; unpaired t-test for PrV) and no noticeable egr1 expression in the moving animals relative to the sitting still animals (Fig. 11A,B).
Figure 11.
Dusp1 and egr1 expression in somatosensory brainstem areas and the ventral pallidum (VP) of zebra finch brain. A: DIVA of a hopping animal, frontal section (A1: dusp1, A2: egr1). B: PrV of an animal sitting in the dark with one eye covered (B1: dusp1, B3: egr1), and a light stimulated animal moving around in the cage, also with one eye covered (B2: dusp1, B4: egr1). There is higher dusp1 expression bilaterally in the stimulated animal (other hemisphere not shown), indicating that the increased expression is presumably not due to light stimulation but to another factor- presumably movement. C: VP of still (C1: dusp1, C3: egr1) and hopping animals (C2: dusp1, C4: egr1), sagittal sections. The amount of dusp1 expression in labeled cells does not appear to differ between the sitting still and hopping animals. Abbreviations not in the main text: OTr, optic tract; T, thalamus. For abbreviations, see list. Scale bar = 1 mm in A2 (applies to A1,A2), and 500 μm in B4 (applies to B1–B4), and C4 (applies to C1–C4).
In contrast to the known somatosensory areas, we did not find detectable dusp1 induction in the motor-associated areas adjacent to the song nuclei. These areas include the anterior striatum (ASt) adjacent to Area X, the anterior nidopallium (AN) adjacent to MAN, PLN and DLN adjacent to HVC, the anterior ventral mesopallium (AMV) adjacent to MO, the PLMV adjacent to Av, and the lateral intermediate arcopallium (LAI) adjacent to RA (Fig. 9A2–6,8–12, 10D, red bars). An assessment of specialized dusp1 expression in song nuclei of vocal learners will be reported separately (Horita et al., in preparation). Intriguingly, egr1 is not induced by activity in the pallidum (P, Fig. 9B4–6,10–12; Feenders et al., 2008), and we found higher dusp1 expression in isolated cells of the ventral pallidum (VP) at a location (Fig. 11C1,2) that was recently shown to receive a direct projection from the striatum adjacent to Area X (Person et al., 2008). However, we did not note apparent differences in VP between the sitting still and hopping animals, unlike the robust upregulation of egr1 in the striatum (St) between VP and Area X (Fig. 11C3,4). In another structure involved in motor behavior, the cerebellum, there was increased dusp1 expression throughout the granular layer in the hopping animals, whereas egr1 was increased in the granular layer of specific anterior (I–VII) and posterior (X) lobes (Fig. 9A1,2,7,8,B1,2,7,8; Feenders et al., 2008). We do not know whether the overlap of expression in the granular layer results from the same or different cells expressing dusp1 and egr1.
In summary, the results suggest that in the zebra finch brain, high levels of induced dusp1 expression occurs in sensory input neurons of the thalamus and the telencephalon where egr1 expression is minimal or does not occur. Conversely, low to no dusp1 induction occurs in higher sensory neurons of the thalamus and telencephalon, and telencephalic motor pathway neurons, where egr1 induction is robust. The activated brain regions are specific to a given stimulus category and behavior. Exceptions are layer 8 of the OT and the granule cell layer of the cerebellum, where both dusp1 and egr1 were induced to high levels.
Segregation of dusp1- and egr1-expressing cells
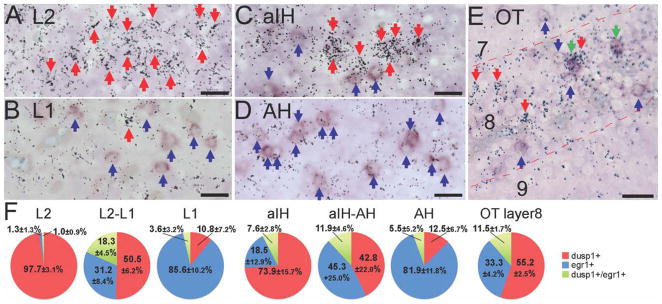
To assess whether there is a distinct separation or co-expression of some cells with induced dusp1 and egr1 expression in the adjacent or the same brain regions, we performed double-labeling experiments. We used brain sections from zebra finch males that had heard song and freely moved within ~45 minutes after lights were on in the morning, in order to maximize dusp1 and egr1 induction in multiple brain areas of the same animal. We found that within the central portions of the sensory input neuronal populations (L2, E, B, and aIH), of the labeled cells, almost all (~98% in L2) or most (~74% in aIH) were dusp1 positive only (Fig. 12A,C,F; Entopallium and basorostralis not shown). At the boundaries of these regions with the surrounding nidopallium (e.g., L2-L1 and aIH-AH), there was an intermingling of dusp1 only (~43–51%) and egr1 only (~31–45%) expressing cells (Fig. 12F). On the other side of the boundaries (e.g., in higher sensory L1 and AH), of the labeled cells, the majority (>82%) were egr1 positive only (Fig. 12B,D,F). A recognizable minority (12–18%) at the borders (L2-L1, aIH-AH) co-expressed both dusp1 and egr1, whereas a small minority did so within the sensory input populations (L2, and aIH; Fig. 12A–D,F).
Figure 12.
Assessment of single- and double-labeled dusp1 and egr1 cells in zebra finch brain. A–E: Examples of single- and double-labeled cells in L2 (A) and L1 (B; auditory), aIH (C) and AH (D; somatosensory), and OT layer 8 (E; visual). Boundaries (L2-L1, aIH-AH, not shown) were determined by cellular morphology. The sensory input neurons have a small granular morphology relative to the neurons of the surrounding nidopallium (for L2 and E), hyperpallium and mesopallium (for aIH). Red arrows, single-labeled dusp1 cells. Blue arrows, single-labeled egr1 cells. Green arrows, double-labeled cells. Dusp1 was measured by radioactive in situ hybridizations (silver grains), and egr1 was measured by DIG chromogenic in situ hybridizations (purple). F: Proportion of cells that express only dusp1, only egr1, or both dusp1 and egr1. Numbers in the pie charts indicate mean percentage ± SD of labeled cells; n = 169 cells in L2, 77 in L2-L1 boundary, 82 in L1, 66 in aIH, 113 in aIH-AH boundary, 103 in AH, and 146 in OT layer 8 (n = 5 birds). There are significant difference in the relative distribution of dusp1 single-labeled and egr1 single-labeled cells among areas of a given pathway (P < 0.01, ANOVA followed by Fisher’s PLSD post hoc test, e.g., L2 vs L1) except for the aIH-AH boundary, which had large SD due to large variation of gene expression in this region across birds. For abbreviations, see list. Scale bar = 20 μm in A–E.
Within the OT, a similar result was found. Although layer 8 had induction of both dusp1 and egr1, the majority of the cells (~89%) were intermingled single-labeled dusp1 (~33.5%) or egr1 (~55.2%) cells (Fig. 12E,F). The minority (~11%) was double labeled (Fig. 12E,F). We attempted to measure expression in the cerebellum, which had high levels of dusp1 in the animals we collected; however, the egr1 expression was too low to detect reliably. In summary, we find that it is possible for cells to express both genes, but the majority of cells that express high levels of one gene or the other.
Other avian species
We wondered whether the pattern of differential dusp1 induction in sensory input neurons was specific to the zebra finch, a songbird, or whether it was present in other avian groups. Thus, we examined dusp1 relative to egr1 induction in two other avian species: budgerigar, a parrot that like songbirds belongs to a vocal learning order, and ring dove, which belongs to a vocal nonlearning order (Nottebohm, 1972; Jarvis, 2004b). We assessed dusp1 expression in adjacent sections that had been hybridized to egr1 from experiments that mapped hearing and/or movement-associated brain areas in these species (Jarvis and Mello, 2000; Feenders et al., 2008).
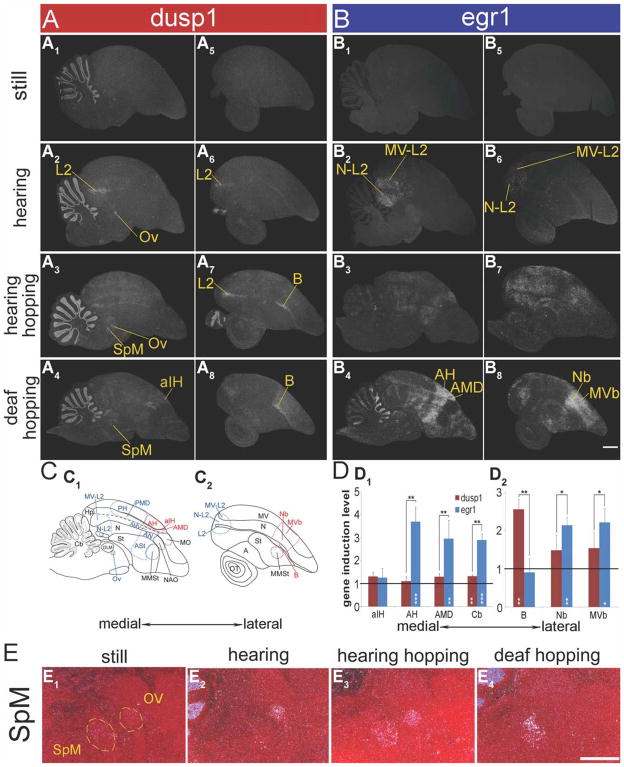
For quiet control budgerigars that sat relatively still in normal room light or in the dark alone for more than 3 hours, dusp1 expression was low throughout the telencephalic sensory input neural populations (Fig. 13A1,5). This is unlike the zebra finch, in which basal levels were often higher in the sensory input neural populations. However, similar to the zebra finch, the budgerigar cerebellum had high basal expression of dusp1 (Fig. 13A1–4). Budgerigars that sat relatively still while hearing 30 minutes of warble song playbacks showed distinct and robust upregulation of dusp1 throughout L2 in the telencephalon and Ov in the thalamus (Fig. 13A1,2,5,6). Those that hopped in the rotating wheel in the dark, but with hearing intact, also showed increased dusp1 expression in the lateral portion of L2 and in Ov (Fig. 13A1,3,5,7), as well as in nucleus B, the sensory input neurons of the pseudo-collothalamic somatosensory pathway (Fig. 13A5,7). Deafening eliminated the hopping-associated dusp1 induction in L2 and Ov, but not in B (Fig. 13A1,4,5,8,D). Interestingly, unlike zebra finches, hopping budgerigars did not show robust dusp1 induction in aIH, the sensory input neurons of the lemnothalamic somatosensory pathway (Fig. 13A1,4,D). We also found high movement-associated dusp1 induction in a budgerigar thalamic nucleus, what appears to be the medial spiriform nucleus (SpM; Fig. 13E; Roberts et al., 2001). We did not note dusp1 induction in such a nucleus in zebra finches.
Figure 13.
Dusp1 and egr1 mRNA expression in a parrot brain. Shown are medial to lateral serial sagittal sections hybridized to dusp1(A) and egr1 (B) respectively, from four groups of budgerigars: 1) a silent control male bird sitting relatively still in dim light (A1,5, B1,5); 2) a male bird that heard a 30-minute playback of natural conspecific warble song (A2,6, B2,6); 3) a hearing intact male bird that hopped for 30 minutes in a rotating wheel while in the dark (A3,7, B3,7); and 4) a deafened male bird that hopped for 30 minutes in the rotating wheel while in the dark (A4,8, B4,8). Note that deafening eliminated most of the dusp1 and egr1 induction in caudal areas of the brain in the hopping animals. Yellow lines and names indicate areas where each mRNA was upregulated. C: Anatomical profiles of brain areas of medial and lateral sections. D: Quantifications of dusp1 (red bars) and egr1 (blue bars) gene expression in six somatosensory areas and the anterior cerebellum (Cb) of deaf hopping (n = 3) and sitting still (n = 3) birds. For the quantifications, each bar shows an average value ± SD. Values are normalized by the average level of expression in the same brain areas of sitting still control birds. A value of ~1 indicates no change in expression levels relative to sitting still controls. Values significantly above 1 indicate induced expression in animals that hopped relative to still control (white stars inside bars; unpaired t-test). Black stars above bars indicate significant differences between amount of dusp1 and egr1 induction (paired t-test between the same regions of the same animals). *, P < 0.05; **, P < 0.01; and ***, P < 0.001. For abbreviations, see list. Scale bar = 1 mm in B8 (applies to A1–B8) and E4 (applies to E1–E4).
As observed in zebra finch brains, the patterns of sensory-driven dusp1 expression in the budgerigar brain were complementary to the patterns of egr1 expression. Hearing song resulted in induced egr1 expression in the higher sensory neurons adjacent to or near L2 (Fig. 13B1,2,5,6) and movement-induced egr1 expression in higher somatosensory populations in the nidopallium (Nb) and ventral mesopallium (MVb) adjacent to B (Fig. 13B5,8,D). Further, although aIH did not have high dusp1 induction in budgerigars (Fig. 13A4,D), the adjacent somatosensory regions of AH and AMD had high egr1 induction (Fig. 13B4,D; Feenders et al 2008). Similar to the zebra finch, the deaf hopping budgerigars had some dusp1 induction throughout the granule layer of the cerebellum and high egr1 induction in the granular layer of anterior cerebellum lobules II–VI (Fig. 13A4,B4,D). Unlike the zebra finch, in budgerigars, low levels of dusp1 were found in the motor-associated areas adjacent to budgerigar song nuclei. However, the expression relative to sitting still animals was not significant (P = 0.101, 0.359, 0.491 for ASt, AN, and AMV adjacent to song nuclei MMSt, NAO, and MO; unpaired t-test; n = 3/group). These dusp1 levels in budgerigar motor-associated regions were far lower than the robust egr1 induction (Fig. 13A4,B4). Consistent with the differential telencephalic dusp1 and egr1 expression, we did not find high levels of egr1 induction in thalamic nuclei of Ov or SpM of the hearing song or hopping animals (Fig. 13B3).
Next, we wanted to determine whether the lack or paucity of dusp1 induction in motor areas was a feature specialized to vocal learners, so we examined these areas in the brains of a vocal nonlearner, ring doves. We compared dusp1 and egr1 expression in the brain of deaf ring doves that walked on a treadmill for 30 minutes in the dark with expression in those that sat relatively still in darkness (Feenders et al., 2008). Similar to the zebra finch and budgerigar, we did not find high dusp1 induction in the putative motor areas of the ring dove telencephalon (AN, AMV, ASt, PLN, PLMV, DLN, and AI; Fig. 14A1,3,4,6). These brain areas show movement-associated egr1 induction (Fig. 14B1,3,4,6; Feenders et al., 2008). In the cerebellum (Cb), in contrast to the findings in budgerigars and the zebra finch, there was robust induction of both dusp1 and egr1 in the ring dove anterior lobules (Fig. 14A1,4,B1,4). As for somatosensory pathways, similar to budgerigars, there was no robust induction of dusp1 in ring dove aIH, although egr1 was induced in surrounding AH and AMD (Fig. 14A1,4,B1,4,E). There was no detectable induction in B of the second somatosensory pathway, which was not unexpected as egr1 was not induced in the adjacent Nb and MVb of the walking ring doves (Fig. 14A2,5,B2,5,E; Feenders et al., 2008).
Figure 14.
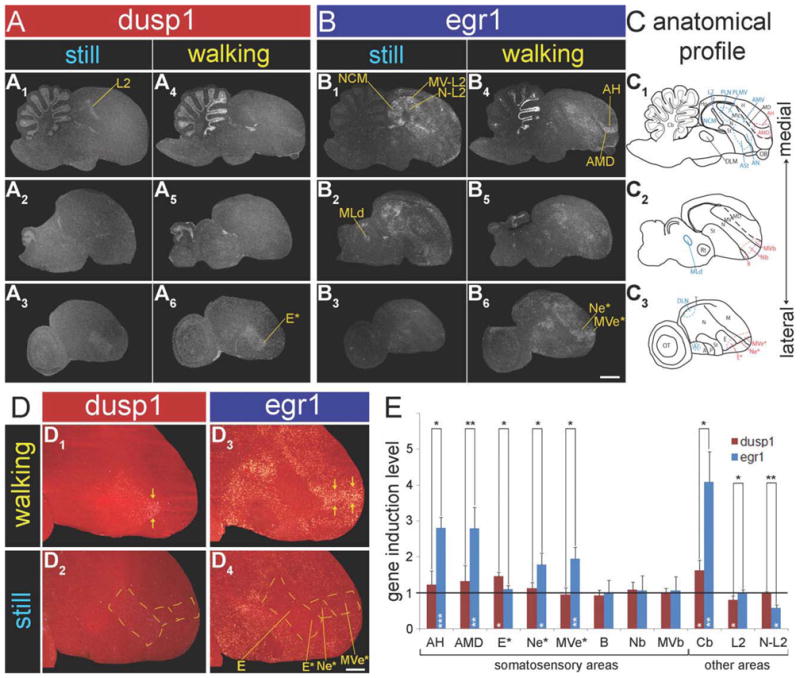
Dusp1 and egr1 mRNA expression in ring doves. Shown are medial to lateral serial sagittal brain sections hybridized to dusp1 (A) and egr1 (B) from two groups of doves: still—silent control male birds sitting relatively still in the dark (A1–3, B1–3) and walking—deafened male bird that walked for 30 minutes in the rotating wheel while in the dark. Note that deafening eliminated most of the dusp1 and egr1 induction in auditory areas such as N-L2. Areas where each mRNA was induced are indicated by yellow lines and names. C: Anatomical profiles of brain areas. Blue, motor-associated areas; red, somatosensory areas. D: Magnified images of dusp1 and egr1 expression in a portion of E, Ne, and MVe laterally adjacent to B (highlighted by *) of a deafened ring dove, after walking in the dark. E: Quantification of dusp1 (red bars) and egr1 (blue bars) expression in eight somatosensory areas (three areas [E*, Ne*, and MVe*] were identified as putative somatosensory), the anterior cerebellum, and two auditory areas of walking deaf birds. Each bar shows an average value ± SD. Values are normalized by the average level of expression in the same brain areas of sitting still control birds. A value ~1 indicates no change in expression levels relative to sitting still controls. Values significantly above 1 indicate induced expression in animals that walked (n = 3) relative to still controls (n = 3, white stars inside bars; unpaired t-test). Black stars above bars indicate significant differences between amount of dusp1 and egr1 induction (paired t-test between the same regions of the same animals). *, P < 0.05; **, P < 0.01; and ***, P <0.001. For abbreviations, see list. Scale bar = 2 mm in B6 (applies to A1–B6); 1 mm in D4 (applies to D1–D4).
Instead, a region of the brain with the highest increased dusp1 expression was an anterior-ventral portion of E laterally adjacent to B that we name here E* (Fig. 14A3,6,D1,2,E); likewise, there was strip of cells with egr1 induction within the nidopallium and mesopallium adjacent to E* (Ne* and MVe* in Fig. 14B3,6,D3,4,E). There was no noticeable dusp1 induction in the thalamic visual nucleus Rt that projects to E, and thus the increase in the anterior-ventral part of E* could be due to somatosensory or some other sensory processing. There was no dusp1 induction in L2 and in fact deafening reduced the basal expression in L2 (Fig. 14A1,4,E), consistent with the reduced egr1 expression in the adjacent N-L2 and MV-L2 (Fig. 14B1,4,E; Feenders et al., 2008).
In summary, with some exceptions, the pattern of dusp1 gene induction is similar in distantly related avian groups. The exceptions relative to the zebra finch are that in the budgerigar the induction in the sensory input neural populations is more prominent due to the lower basal levels in control animals, but induction in aIH does not occur with hopping; there is some expression in budgerigar motor-associated areas, but the levels are still much lower than that seen for egr1; in the dove, the induction in aIH is also less, and the movement-induced expression of dusp1 and egr1 in the cerebellum is anatomically coincident.
Dusp1 expression in a mammalian brain
We wondered whether differential dusp1 expression in sensory input neural populations was specific to birds, or whether it could be found in other vertebrate groups, such as mammals. Most prior experiments on dusp1 regulation in mammalian neurons have been conducted with cells in culture, and some have been conducted in the brain of rodents, but mainly in animals that received strong insults such as seizures, brain lesions, and pharmacological manipulations, in which the patterns of regulation cannot be clearly linked to a behavior (Qian et al., 1994; Takaki et al., 2001; Kodama et al., 2005). In a study that used physiological levels of methamphetamine (a serotonin and dopamine receptor agonist) in rats, the authors noted that high levels of dusp1 induction was restricted to layers IV and VI of the cortex, followed by the thalamus (individual nuclei not specified), and moderate induction occurred in the striatum (Fig. 3 of Takaki et al., 2001). Although that study did not point out any relationships with sensory input neurons, we note here that layer IV neurons in mammalian cortex are sensory input neurons that receive direct input from sensory nuclei of the thalamus; layer VI neurons are reciprocally connected in a feedback pathway with the thalamus (Shipp, 2007).
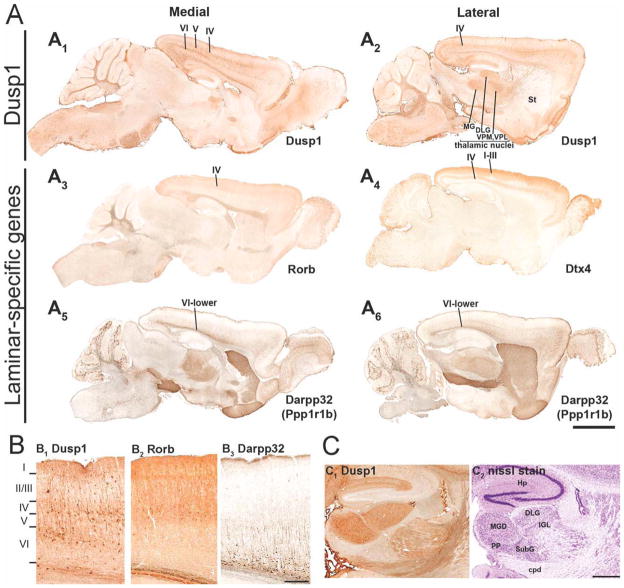
Thus, to determine the pattern of dusp1 in rodents that freely behave, we examined dusp1 expression in mice from the Gene Expression Nervous System Atlas (GEN-SAT) database (Gong et al., 2003). These transgenic mice have been constructed with a BAC transgene containing the dusp1 promoter driving enhanced green fluorescent protein (eGFP) expression. First, we noted that dusp1-eGFP expression was present in the mouse cortex and differentially so in two layers (Fig. 15A1,2,B1). We compared this layered pattern with other layer-specific markers (Rorb for layer IV, Dtx for layers I–III and part of IV, and Darpp32 for lower layer VI; Fig. 15A3–6,B2,3; Molyneaux et al., 2007). The analyses revealed that the two layers of dusp1 expression in the GENSAT mice were layer IV and upper layer VI, with minimal expression in layer V between them (Fig. 15B). Second, not all brain sections or all brain regions had equal dusp1 expression levels in layers IV and VI (Fig. 15A1,2), indicative of immediate early gene activation. Third, similar to birds, dusp1-eGFP expression was low throughout most of the remaining mouse telencephalon, including the striatum. Fourth, within the thalamus, dusp1-eGFP soma expression was mostly absent, except in the sensory input nuclei, including auditory (medial geniculate, MGD), visual (lateral geniculate, DLG), and somatosensory (ventral posterior lateral and medial, VPL and VPM) nuclei (Fig. 15A2,C).
Figure 15.
Dusp1-eGFP expression in GENSAT mouse brains. A: Immunocytochemistry detection of enhanced GFP (eGFP) driven by the dusp1 promoter (A1,2) and promoters of cortical laminar-specific genes in the mouse brain (A3: Rorb for layer IV; A4: Dtx4 for layers I–III and IV; A5,6: Darpp32, a.k.a. Ppp1r1b for layer VI). Darpp32 is also expressed in the striatum at a high level and parts of the thalamus at an intermediate level. B: Magnified images of dusp1 (B1), Rorb (B2), and Darpp32 (B3) in the visual cortex (from A, caudal cortical region). C: Magnified images of dusp1 expression (C1) and adjacent Nissl-stained section (C2) in sensory input thalamic nuclei (MGD and DLG). Note the absence of expression in the higher order nuclei of the thalamus, including the peripeduncular nucleus (PP), the subgeniculate nucleus (SubG), and the intergeniculate leaflet (IGL). Gene abbreviations: Rorb, RAR-related orphan receptor β; Dtx4, deltex 4 homolog (Drosophila); Darpp32, dopamine- and adenosine 3′,5′-monophosphate-regulated phosphoprotein or Ppp1r1b, protein phosphatase 1, regulatory (inhibitor) subunit 1B. Images are from the GENSAT database (GENSAT Project, NINDS Contract #N01NS02331 to The Rockefeller University, New York, NY). For abbreviations, see list. Scale bar = 2 mm in A6 (applies to A1–A6); 200 μm in B3 (applies to B1–B3); 500 μm in C2 (applies to C1,C2).
In summary, this analysis suggests that as in birds, dusp1 expression in mammals, under normal, behavioral, physiological conditions, is expressed at its highest levels in sensory input neurons of the thalamus and telencephalon. Future work will be necessary to determine whether induced expression occurs in specific brain areas by specific behaviors and to confirm the cortical layer cell types with double-labeling experiments of layer-specific markers or tracers and dusp1/egr1 expression.
DISCUSSION
In this study, we examined dusp1 regulation in brains from awake behaving animals. We found that dusp1 is regulated in distinct neuronal populations in which egr1 and a number of other IEGs are not or are minimally regulated. These areas are the sensory input populations of the thalamus and telencephalon (Fig. 15A). Below we discuss the implications of our findings.
Functional molecular mapping of brain pathways
Our results show that the combination of dusp1 and egr1 can be used as a molecular tool kit to map neurons of nearly entire brain systems anatomically and functionally. This is because the two genes, at least in birds, were induced mainly in complementary populations of neurons. In doing so, we were able to identify and characterize nearly all avian brain regions from the midbrain to the telencephalon of five pathways of three major sensory systems: one auditory, two visual, and two somatosensory (Fig. 16A). The only sensory nuclei in which we did not find evidence of dusp1 or egr1 induction were within the thalamic GLd complex of the thalamofugal visual pathway (gray in Fig. 16A). However, the avian GLd complex consists of ~6 noncontiguous small nuclei (Deng and Rogers, 1998a; Heyers et al., 2008) that are difficult to find in all of our sections, and thus the status of dusp1 and egr1 regulation in them is uncertain. Additional experiments are necessary, such as placing tracers into visual IH and assessing dusp1 mRNA expression in the specific nuclei of the GLd complex that project to IH. We would not be surprised to find that a specific GLd nucleus would show expression, because the mouse homolog, the MG, shows distinct dusp1-eGFP expression. The somatosensory pathway to the basorostralis of the telencephalon in birds does not have a thalamic component (Wild and Farabaugh, 1996), which are mammalian VPM and VPL. Instead, the basorostralis in birds receives a direct projection from the cranial sensory nucleus PrV, bypassing the midbrain and thalamus, and PrV in turn receives somatosensory input from the face and neck. We find that PrV, like thalamic sensory input populations and mouse VPM and VPL, shows dusp1 and not egr1 expression (Fig. 16A). In this manner, avian PrV behaves like a thalamic sensory input cell group in its connectivity and its dusp1/egr1 expression, in support of the pseudo-collothalamic hypothesis (Jarvis, 2009).
Figure 16.
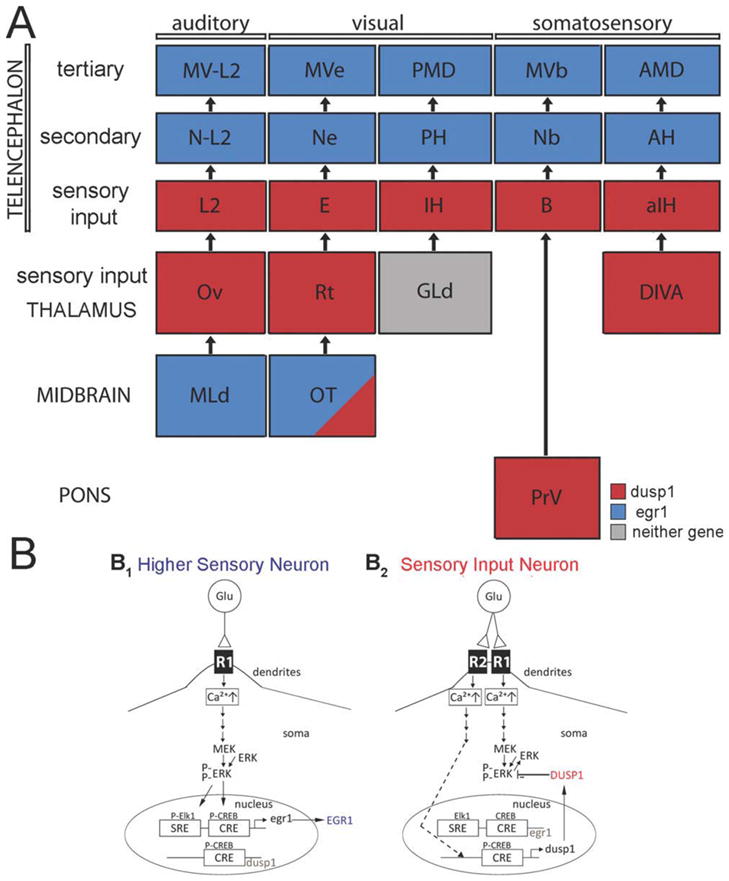
Summary of results of this study and proposed putative mechanisms of differential dusp1 and egr1 regulation. A: Summary of dusp1 and egr1 molecular profiles in the cellular stations of five sensory pathways of the avian brain. Red, areas that show activity-dependent dusp1 induction. Blue, areas that show activity-dependent egr1 induction. Gray, areas where we could not identify regulation of either gene or find apparent expression. OT shows induction of both genes in layer 8 and only egr1 induction in some other layers, and is thus filled in both blue and red, as most of the neurons do not express high levels of both genes. B: A proposed putative signaling mechanisms of how dusp1 and egr1 could be differentially regulated in different neuron types: higher sensory (B1) vs. sensory input (B2). Only a proposed dependent mechanism is shown. In the higher sensory neurons, upregulation of egr1 is occurs via one type of receptor (R1; B1). In the sensory input neurons, this pathway is also initiated, but it is suppressed by overexpression of the DUSP1 protein induced via another receptor (R2; B2). Specific receptors are not shown, as this needs to be determined in neuron types of intact brains as opposed to cells in culture. Dashed line indicates undetermined intermediate signaling steps. Multiple lines with arrows indicate multiple molecular steps. Abbreviations: CRE, cAMP response element (a promoter); CREB, cAMP response element binding protein; dusp1, dual specificity phosphatase 1 (protein capitalized); egr1, early growth response gene 1 (protein capitalized); ERK, extracellular signal-regulated kinase; Elk1, Ets-domain transcription factor; MEK, MAP kinase kinase; P, phosphate; SRE, serum response element (a promoter). For other abbreviations, see list.
Within motor systems, we found low dusp1 expression in the ventral pallidum, where egr1 is not upregulated. However the dusp1 expression in the pallidum generally did not appear to be regulated by movement activity in birds. The pallidum in mammals (and presumed in birds) modulates movements through parallel cortical-basal-ganglia-thalamic-cortical loops (Csillag and Montagnese, 2005; Doupe et al., 2005; Nambu, 2008). Neurons throughout the pallidum show high spontaneous firing rates (Bengtson and Osborne, 2000). It is possible that the high firing rates could lead to a constitutive dusp1 expression in the absence of movement.
The avian visual systems showed interesting patterns of dusp1 vs. egr1 regulation. First, unilateral eye occlusion did not completely block light-induced dusp1 expression in E and IH of the hemisphere contralateral to the open eye. This finding is consistent with the presence of some bilateral projections from the OT to Rt (then Rt unilaterally to E) and from the GLd complex to IH (Miceli and Reperant, 1982; Zeigler and Bischof, 1993; Gunturkun et al., 1998). Second, differential hemispheric expression occurred in IH of the thalamofugal pathway when males viewed and sang to females, but the upregulation was higher ipsilateral to the open eye, whereas the egr1 regulation was higher in secondary and tertiary sensory neurons contralateral to the open eye. These findings suggest that there could be some bilateral feedback in the thalamofugal visual system, which would allow one population of neurons to be more active when the other eye sees a natural stimulus of interest. Deciphering such a mechanism requires more experimentation with manipulation of stimuli, neuronal tracing studies, and electrophysiological studies.
Differences among species
We found species differences in dusp1 expression. In the zebra finch, the basal levels were higher in sensory input populations of the telencephalon. In parrot, and apparently in mice, the basal levels can be low in the telencephalic sensory input neurons. In the parrot and ring dove, aIH did not show robust dusp1 expression in response to hopping or walking, although the surrounding AH and AMD showed robust egr1 induction. The aIH is expected to be active in hopping and walking in birds, as it receives somatosensory input from the feet and shows neuronal firing with feet somatic stimulation in owls (Manger et al., 2002). It is possible that aIH was active during hopping and walking in the budgerigars and doves, but dusp1 levels remained low, or that other input to the surrounding AH and AMD activated egr1 in these regions. Also in ring doves, dusp1 induction in response to walking occurred in the anterior-lateral part of the entopallium, a supposed visual system nucleus, even though the animals were in the dark. As far as we know, there is no report of somatosensory input into the lateral part of the entopallium, but this idea is testable. Finally, in zebra finches dusp1 levels were high in the midbrain’s IPc, which receives auditory and visual input (Maczko et al., 2006), but we did not note high basal levels in the other species. In zebra finches, it is possible that the high levels are due to activity by both auditory and visual input. However, this is unlikely to be the main explanation, as IPc still had high dusp1 expression in the animals that sat still in the dark (no auditory and no visual stimuli). Although IPc does not appear to be a sensory input nucleus, its high dusp1 and undetectable egr1 expression levels are consistent with the general relationship of these two genes throughout the brain. This result emphasizes that whether differences between species are due to true species differences or small differences in the stimuli presented or behaviors performed, a general principle is that when dusp1 is high, egr1 is low.
A possible general property of the vertebrate brain
Our preliminary analyses of dusp1 in the GENSAT mice and that of Takaki et al (2001) in rats stimulated with methamphetamine suggest that in the mammalian brain, dusp1 is expressed in similar types of neural populations as in the avian brain. In the two studies that examined dusp1 in brain sections containing the cortex in response to seizures (Qian et al., 1994; Kodama et al., 2005), we noted apparent increases of dusp1 in most, if not all, cortical layers, but the layers with the highest increases appear to us to be IV and VI. It is possible that such strong stimulation leads to more spreading of dusp1 induction to other cortical layers.
A corollary of these findings is that little to no egr1 induction occurs in sensory input thalamic nuclei, such as in the rat VPM after somatosensory stimulation of whiskers (Bisler et al., 2002) and in the cebus monkey’s DLG after light stimulation (Soares et al., 2005). However, within the cortex, contradictory results have been reported for egr1 expression. In the visual cortex, for example, light stimulation has been reported to cause much less (Soares et al., 2005) or much higher (Pinaud et al., 2003) egr1 expression in layers IV and VI relative to layers II and III. The differences between studies could be due to differences in subdivisions of layer IV (which have different connectivity), developmental age of the animals, or possibly species differences (reviewed in Kaczmarek and Chaudhuri, 1997). In support of the connectivity hypothesis, a recent study in primates (Takahata et al., 2009) has shown that layer IVCα, the source of visual input from magnocellular neurons of the lateral geniculate in the thalamus (for form vision), has little if any light-stimulated egr1 expression, whereas layer IVCβ, the source of visual input from parvocellular neurons of the lateral geniculate in the thalamus (for color vision), has high levels of light-stimulated egr1 expression. Perhaps the avian telencephalic sensory input neurons that lack egr1 and express high levels of dusp1 are analogous to mammalian layer IVCα neurons. To be certain that the inverse regulation exists in mammals, double-label dusp1 and egr1 experiments in mammalian brains are necessary. Nevertheless, our analyses of the overall findings suggest that in mammals, thalamic sensory input and thalamo-recipient sensory cell populations of the telencephalon express the highest levels of dusp1, and a subset of these populations expresses very little egr1.
Given these partial parallels between birds and mammals, preferential activity-dependent regulation of dusp1 in sensory input neurons may be a general principle of vertebrate brains. This parallel is consistent with the nuclear to layered hypothesis of vertebrate brain evolution, whereby different layers of mammalian cortex are proposed to be homologous to different subdivisions of the avian pallium, including homology of avian telencephalic sensory input neurons to mammalian layer IV neurons (Karten, 1991; Jarvis et al., 2005).
Potential mechanisms of differential regulation
To explain the differential regulation of dusp1 and egr1, we propose two types of mechanisms: 1) one in which the two genes are regulated dependently (Fig. 16B); and 2) another in which they are independent of each other. For a dependent mechanism, the lack of significant overlap of dusp1 and egr1 expression in the brain of naturally behaving animals is consistent with recent findings in cultured mammalian neuroblastoma cells, which showed that dusp1 is a potent inhibitor of egr1 gene expression (Rossler et al., 2008); overexpression of dusp1 completely blocks stimulus-induced egr1 expression (Rossler et al., 2008). This block occurs through a MAP kinase signaling pathway. Dusp1, also known as MAP kinase phosphatase 1 (mkp1), is a negative regulator for specific MAP kinases (i.e., ERK1) that in turn activate the ETS-domain transcription factor (Elk1) and CREB, which in turn bind to the egr1 promoter to upregulate egr1 mRNA expression (Fig. 16B; Knapska and Kaczmarek, 2004; Machado et al., 2008). Dusp1 inactivates ERK1 and other MAP kinases by dephosphorylating them at two amino acid resides, a tyrosine and a threonine (Farooq and Zhou, 2004; Liu et al., 2007), the reason why it is called a dual specificity phosphatase.
MAP kinases comprise three major subtypes: the extracellular signal-regulated kinase (ERK) that induces cell growth and proliferation, the c-jun aminoterminal kinase (JNK), and the p38 kinase that induces apoptosis and cell stress reactions. ERK1 activates the Elk1 and CREB transcription factors via phosphorylation. ERK1/2 can also activate dusp1 expression via CREB, and thus dusp1 theoretically can inhibit its own expression via a feedback inhibitory loop. For ERK, there are five gene variants in mammals and each is expressed throughout most cortical layers (Di Benedetto et al., 2007), but the highest activity-dependent activation (phosphorylation) of ERK1/2 appears to occur, in our interpretation, in layer IV (Sgambato et al., 1998). This dependent mechanism is consistent with other findings in songbirds, in which ERK1 activation is necessary for the hearing-induced egr1 expression in the songbird higher auditory neurons (i.e., in NCM; Cheng and Clayton, 2004).
The fact that a minority of cells showed co-expression of both dusp1 and egr1 suggest that an independent pathway could be possible. For an independent mechanism, neuronal activity could be linked to different signal transduction pathways for each gene (Fig. 16B, sensory input neuron), but it may also require an ERK independent or an alternative ERK mechanism between the two genes, to prevent dusp1 suppression of egr1 induction (Fig. 16B).
The presence of double-labeled cells could also be explained by a dependent mechanism whereby most if not all neurons would have a balance between dusp1 and egr1 expression, but that the balance is heavily tipped in one direction depending on the cell type (sensory input or higher sensory). What would be responsible for tipping the balance? We believe that neurotransmitter receptors are good candidates (Fig. 16B). The regulation of specific IEGs by neuronal activity is controlled by neurotransmitter release from presynaptic terminals onto specific neurotransmitter receptor types (Flavell and Greenberg, 2008). We have previously shown that the glutamate receptors mGluR1, GluR1 (an AMPA receptor subunit), GluR5 (a kainate receptor subunit), and NR2A (an NMDA receptor subunit) are expressed at higher or lower levels in sensory input neurons relative to the adjacent higher sensory neurons of the avian telencephalon (Wada et al., 2004). NMDA receptors are required for dusp1 expression (Qian et al., 1994), and NMDA receptors activate dusp1 in mammalian cortex but not in the striatum (Takaki et al., 2001). NMDA receptors also activate egr1 expression in both the cortex and striatum (Gerfen, 2000). Thus, differential expression of one specific receptor subtype (Fig. 16B) may over- or underactivate dusp1 in sensory input vs. higher sensory neurons, which would then determine the level of suppression of egr1 in those neurons. This hypothesis can be tested by performing triple-labeling experiments with dusp1, egr1, and ion channel receptors, from the brains of sensory stimulated animals that have been manipulated with pharmacological and genetic agents against the specific receptors.
In summary, we suggest a dependent mechanism where high dusp1 expression leads to low egr1 expression. However, this does not mean that the converse is true. Low dusp1 expression will not automatically lead to high egr1 expression, not until an increase occurs in the firing rate of those neurons. Once the firing rate increases, we argue that high dusp1 will inhibit ERK and thus egr1 induction, whereas low dusp1 will allow egr1 induction.
Possible functional consequences
Our results raise a question as to what is unique about sensory input neurons that would make them favor dusp1 over egr1 expression. The answer to this question may be related to the biological role of dusp1. Dusp1 is thought to play an important role in the cellular response to environmental stress and subsequent programmed cell death, by inactivating cellular survival responses induced by ERK and subsequent IEGs (Liu et al., 2007). We also argue it could play a role in dampening neural plasticity of sensory input neurons by downregulating egr1 and other IEGs. Concordant with this first hypothesis, the sensory input neurons of the avian telencephalon and thalamus and layer IVC of the primate visual cortex show some of the highest levels of cytochrome oxidase (CO) in the brain, indicative of their higher metabolic activity relative to the rest of the telencephalon (Braun et al., 1985a,b; Adret and Margoliash, 2002; Takahata et al., 2009). In fact all of the brain regions we noted with high constitutive dusp1 expression in zebra finches (E, L2, B, Ov, Rt, IPc, and SP) have the highest CO activity in zebra finches (Braun et al., 1985a,b). In support of this idea, it was recently discovered that the two genes, dusp1 and CO, in humans have the identical promoter binding site for the stress-induced transcription factor p53 and are simultaneously upregulated by p53 in response to cellular stress (Liu et al., 2008).
Alternatively or concordant with the second hypothesis, the sensory input neurons of the songbird auditory pathway (L2; Chew et al., 1995) and mammalian layer IV neurons of the adult visual pathway (Thompson, 2000) show the least neural plasticity in response to hearing novel songs or to visual pathway manipulations, respectively, relative to higher sensory neurons (i.e., cortical layers). Perhaps the sensory input neurons need to perform basic services that require greater metabolic activity and dusp1 is needed to protect against this stress and reduce plasticity. These ideas can be tested by inactivating dusp1 in sensory input neurons of birds and mammals and determining whether activity-dependent induction of cellular stress and/or plasticity is converted to the type seen for higher sensory neurons.
In conclusion, by using natural behavioral stimuli and behaviors, we have identified and characterized an activity-dependent gene, dusp1, in the brains of awake behaving animals, which shows complementary expression patterns relative to the commonly studied egr1 gene. The activated expression patterns allowed us to generate novel, testable hypotheses on the mechanisms of how dusp1 and egr1 regulation are linked in the intact brain, and the functions of the brain areas. Further, the results have revealed unique properties of gene activation in sensory systems.
Acknowledgments
We thank Gesa Feenders for use of brain sections processed in the study of Feenders et al. (2008), and Masahiko Kobayashi for in situ hybridization with dusp4 and sense probe of dusp1. We thank Dr. Kotaro Oka for critical reading of the manuscript and supervision of H.H., Maurice Anderson for animal care and breeding, and an anonymous reviewer for pointing us to the GENSAT mice images. H.H. performed most of the experiments and the analyses, and wrote the paper; K.W. cloned the dusp1 gene, performed pilot in situ hybridizations, helped supervise the project, and wrote the paper; M.R. performed egr1 in situ hybridizations on non-songbird species; E.H. performed the vision behavior experiments; and E.D.J. performed behavior experiments of non-songbird species, helped supervise the project, and wrote the paper.
Grant sponsor: National Institutes of Health; Grant number: R01DC7218 (to E.D.J.); Grant sponsor: Grant-in-Aid for Scientific Research in Japan; Grant sponsor: Takeda Science Foundation; Grant sponsor: Kanae Foundation for promotion of medical science (fellowship to K.W.); Grant sponsor: Japan Student Services Organization (fellowship to H.H.).
Abbreviations
- A
arcopallium
- AH
anterior hyperpallium
- aIH
anterior part of the intercalated layer of the hyperpallium
- AMD
anterior dorsal mesopallium
- AMV
anterior ventral mesopallium
- AN
anterior nidopallium
- Area X
a vocal nucleus
- ASt
anterior striatum
- Av
nucleus avalanche
- B
basorostralis
- Cb
cerebellum
- CM
caudal mesopallium
- CN
cochlea nucleus
- cpd
cerebral peduncle
- cSt
caudal striatum
- Cu
cuneate nucleus
- DIVA
dorsal intermediate ventral anterior nucleus of the thalamus
- DLG
dorsal lateral geniculate nucleus
- DLN
dorsal lateral nidopallium
- DLM
dorsal lateral medial nucleus of the thalamus
- DM
dorsal medial nucleus of the midbrain
- DT
dorsal thamalus
- E
entopallium
- GLd
dorsolateral geniculate nucleus
- Gr
gracile nucleus
- H
hyperpallium
- Hp
hippocampus
- HVC
a vocal nucleus (no acronym)
- IGL
intergeniculate leaflet of the thalamus
- IH
intercalated layer of the hyperpallium
- IPc
nucleus isthmi pars parvocellularis
- LAI
lateral intermediate arcopallium
- LAM
lateral nucleus of the anterior mesopallium
- LLD
lateral lemniscus, dorsal part
- LLI
lateral lemniscus, intermediate part
- LLV
lateral lemniscus, ventral part
- M
mesopallium
- MAN
magnocellular nucleus of the anterior nidopallium
- MG
medial geniculate body
- MGD
medial geniculate body, dorsal nucleus
- MLd
dorsal part of the lateral mesencephalic nucleus
- MMSt
magnocellular nucleus of the medial striatum
- MO
oval nucleus of the mesopallium
- MD
dorsal mesopallium
- MV
ventral mesopallium
- MVb
ventral mesopallium near B
- MVe
ventral mesopallium near to E
- MV-L2
ventral mesopallium near L2 (same as CM)
- N
nidopallium
- Nb
nidopallium adjacent to B
- Ne
nidopallium adjacent to E
- N-L2
nidopallium adjacent to L2
- NAO
oval nucleus of the anterior nidopallium
- NIf
interfacial nucleus of the nidopallium
- nXIIts
12th nucleus, tracheosyringeal part
- Ov
nucleus ovoidalis
- P
pallidum
- PH
posterior hyperpallium
- PLMV
posterior lateral ventral mesopallium
- PLN
posterior lateral nidopallium
- PP
peripeduncular nucleus
- PrV
principal sensory trigeminal nucleus
- RA
robust nucleus of the arcopallium
- Rt
nucleus rotundus
- SO
superior olivary nucleus
- SP
subpretectal nucleus
- SpM
medial spiriform nucleus
- St
striatum
- Ste
striatum adjacent to E
- SubG
subgeniculate nucleus
- TeO
optic tectum
- Uva
nucleus uvaeformis
- v
ventricle
- VP
ventral palidum
- VPL
ventral posterior lateral nucleus of the thalamus
- VPM
ventral posterior medial nucleus of the thalamus
LITERATURE CITED
- Adret P, Margoliash D. Metabolic and neural activity in the song system nucleus robustus archistriatalis: effect of age and gender. J Comp Neurol. 2002;454:409–423. doi: 10.1002/cne.10459. [DOI] [PubMed] [Google Scholar]
- Bauer EE, Coleman MJ, Roberts TF, Roy A, Prather JF, Mooney R. A synaptic basis for auditory-vocal integration in the songbird. J Neurosci. 2008;28:1509–1522. doi: 10.1523/JNEUROSCI.3838-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtson CP, Osborne PB. Electrophysiological properties of cholinergic and noncholinergic neurons in the ventral pallidal region of the nucleus basalis in rat brain slices. J Neurophysiol. 2000;83:2649–2660. doi: 10.1152/jn.2000.83.5.2649. [DOI] [PubMed] [Google Scholar]
- Benowitz LI, Karten HJ. Organization of the tectofugal visual pathway in the pigeon: a retrograde transport study. J Comp Neurol. 1976;167:503–520. doi: 10.1002/cne.901670407. [DOI] [PubMed] [Google Scholar]
- Bigalke-Kunz B, Rubsamen R, Dorrscheidt GJ. Tonotopic organization and functional characterization of the auditory thalamus in a songbird, the European starling. J Comp Physiol [A] 1987;161:255–265. doi: 10.1007/BF00615245. [DOI] [PubMed] [Google Scholar]
- Bisler S, Schleicher A, Gass P, Stehle JH, Zilles K, Staiger JF. Expression of c-Fos, ICER, Krox-24 and JunB in the whisker-to-barrel pathway of rats: time course of induction upon whisker stimulation by tactile exploration of an enriched environment. J Chem Neuroanat. 2002;23:187–198. doi: 10.1016/s0891-0618(01)00155-7. [DOI] [PubMed] [Google Scholar]
- Boutros T, Chevet E, Metrakos P. Mitogen-activated protein (MAP) kinase/MAP kinase phosphatase regulation: roles in cell growth, death, and cancer. Pharmacol Rev. 2008;60:261–310. doi: 10.1124/pr.107.00106. [DOI] [PubMed] [Google Scholar]
- Braun K, Scheich H, Schachner M, Heizmann CW. Distribution of parvalbumin, cytochrome oxidase activity and 14C-2-deoxyglucose uptake in the brain of the zebra finch. I. Auditory and vocal motor systems. Cell Tissue Res. 1985a;240:101–115. [Google Scholar]
- Braun K, Scheich H, Schachner M, Heizmann CW. Distribution of parvalbumin, cytochrome oxidase activity and 14C-2-deoxyglucose uptake in the brain of the zebra finch. II. Visual system. Cell Tissue Res. 1985b;240:117–127. [Google Scholar]
- Cardin JA, Schmidt MF. Auditory responses in multiple sensorimotor song system nuclei are co-modulated by behavioral state. J Neurophysiol. 2004;91:2148–2163. doi: 10.1152/jn.00918.2003. [DOI] [PubMed] [Google Scholar]
- Cardin JA, Raksin JN, Schmidt MF. Sensorimotor nucleus NIf is necessary for auditory processing but not vocal motor output in the avian song system. J Neurophysiol. 2005;93:2157–2166. doi: 10.1152/jn.01001.2004. [DOI] [PubMed] [Google Scholar]
- Cheng HY, Clayton DF. Activation and habituation of extracellular signal-regulated kinase phosphorylation in zebra finch auditory forebrain during song presentation. J Neurosci. 2004;24:7503–7513. doi: 10.1523/JNEUROSCI.1405-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew SJ, Mello C, Nottebohm F, Jarvis E, Vicario DS. Decrements in auditory responses to a repeated conspecific song are long-lasting and require two periods of protein synthesis in the songbird forebrain. Proc Natl Acad Sci U S A. 1995;92:3406–3410. doi: 10.1073/pnas.92.8.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton DF. Songbird genomics: methods, mechanisms, opportunities, and pitfalls. Ann N Y Acad Sci. 2004;1016:45–60. doi: 10.1196/annals.1298.028. [DOI] [PubMed] [Google Scholar]
- Csillag A, Montagnese CM. Thalamotelencephalic organization in birds. Brain Res Bull. 2005;66:303–310. doi: 10.1016/j.brainresbull.2005.03.020. [DOI] [PubMed] [Google Scholar]
- Deng C, Rogers LJ. Bilaterally projecting neurons in the two visual pathways of chicks. Brain Res. 1998a;794:281–290. doi: 10.1016/s0006-8993(98)00237-6. [DOI] [PubMed] [Google Scholar]
- Deng C, Rogers LJ. Organisation of the tectorotundal and SP/IPS-rotundal projections in the chick. J Comp Neurol. 1998b;394:171–185. [PubMed] [Google Scholar]
- Di Benedetto B, Hitz C, Holter SM, Kuhn R, Vogt Weisenhorn DM, Wurst W. Differential mRNA distribution of components of the ERK/MAPK signalling cascade in the adult mouse brain. J Comp Neurol. 2007;500:542–556. doi: 10.1002/cne.21186. [DOI] [PubMed] [Google Scholar]
- Doi M, Cho S, Yujnovsky I, Hirayama J, Cermakian N, Cato AC, Sassone-Corsi P. Light-inducible and clock-controlled expression of MAP kinase phosphatase 1 in mouse central pacemaker neurons. J Biol Rhythms. 2007;22:127–139. doi: 10.1177/0748730406298332. [DOI] [PubMed] [Google Scholar]
- Doupe AJ, Perkel DJ, Reiner A, Stern EA. Birdbrains could teach basal ganglia research a new song. Trends Neurosci. 2005;28:353–363. doi: 10.1016/j.tins.2005.05.005. [DOI] [PubMed] [Google Scholar]
- Farooq A, Zhou MM. Structure and regulation of MAPK phosphatases. Cell Signal. 2004;16:769–779. doi: 10.1016/j.cellsig.2003.12.008. [DOI] [PubMed] [Google Scholar]
- Feenders G, Liedvogel M, Rivas M, Zapka M, Horita H, Hara E, Wada K, Mouritsen H, Jarvis ED. Molecular mapping of movement-associated areas in the avian brain: a motor theory for vocal learning origin. PLoS ONE. 2008;3:e1768. doi: 10.1371/journal.pone.0001768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell SW, Greenberg ME. Signaling mechanisms linking neuronal activity to gene expression and plasticity of the nervous system. Annu Rev Neurosci. 2008;31:563–590. doi: 10.1146/annurev.neuro.31.060407.125631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freund N, Gunturkun O, Manns M. A morphological study of the nucleus subpretectalis of the pigeon. Brain Res Bull. 2008;75:491–493. doi: 10.1016/j.brainresbull.2007.10.031. [DOI] [PubMed] [Google Scholar]
- Gerfen CR. Molecular effects of dopamine on striatal-projection pathways. Trends Neurosci. 2000;23:S64–70. doi: 10.1016/s1471-1931(00)00019-7. [DOI] [PubMed] [Google Scholar]
- Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, Heintz N. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- Greenberg ME, Hermanowski AL, Ziff EB. Effect of protein synthesis inhibitors on growth factor activation of c-fos, c-myc, and actin gene transcription. Mol Cell Biol. 1986;6:1050–1057. doi: 10.1128/mcb.6.4.1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillery RW, Herrup K. Quantification without pontification: choosing a method for counting objects in sectioned tissues. J Comp Neurol. 1997;386:2–7. doi: 10.1002/(sici)1096-9861(19970915)386:1<2::aid-cne2>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- Gunturkun O, Hellmann B, Melsbach G, Prior H. Asymmetries of representation in the visual system of pigeons. Neuroreport. 1998;9:4127–4130. doi: 10.1097/00001756-199812210-00023. [DOI] [PubMed] [Google Scholar]
- Guzowski JF, Timlin JA, Roysam B, McNaughton BL, Worley PF, Barnes CA. Mapping behaviorally relevant neural circuits with immediate-early gene expression. Curr Opin Neurobiol. 2005;15:599–606. doi: 10.1016/j.conb.2005.08.018. [DOI] [PubMed] [Google Scholar]
- Hara E, Kubikova L, Hessler NA, Jarvis ED. Assessing visual requirements for social context-dependent activation of the songbird song system. Proc Biol Sci. 2009;276:279–289. doi: 10.1098/rspb.2008.1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyers D, Manns M, Luksch H, Gunturkun O, Mouritsen H. Calcium-binding proteins label functional streams of the visual system in a songbird. Brain Res Bull. 2008;75:348–355. doi: 10.1016/j.brainresbull.2007.10.029. [DOI] [PubMed] [Google Scholar]
- Hu S, Ying Z, Gomez-Pinilla F, Frautschy SA. Exercise can increase small heat shock proteins (sHSP) and pre-and post-synaptic proteins in the hippocampus. Brain Res. 2009;1249:191–201. doi: 10.1016/j.brainres.2008.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis ED. Brains and birdsong. In: Marler P, Slabbekoorn H, editors. Nature’s music: the science of bird Song. San Diego: Elsevier-Academic Press; 2004a. pp. 226–271. [Google Scholar]
- Jarvis ED. Learned birdsong and the neurobiology of human language. Ann N Y Acad Sci. 2004b;1016:749–777. doi: 10.1196/annals.1298.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis ED. Evolution of the pallium in birds and reptiles. In: Binder MD, Hirokawa N, Windhorst U, Butler As-e, editors. Encyclopedia of neuroscience. Berlin: Springer-Verlag; 2009. pp. 1390–1400. [Google Scholar]
- Jarvis ED, Mello CV. Molecular mapping of brain areas involved in parrot vocal communication. J Comp Neurol. 2000;419:1–31. doi: 10.1002/(sici)1096-9861(20000327)419:1<1::aid-cne1>3.0.co;2-m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis ED, Nottebohm F. Motor-driven gene expression. Proc Natl Acad Sci U S A. 1997;94:4097–4102. doi: 10.1073/pnas.94.8.4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis ED, Scharff C, Grossman MR, Ramos JA, Nottebohm F. For whom the bird sings: context-dependent gene expression. Neuron. 1998;21:775–788. doi: 10.1016/s0896-6273(00)80594-2. [DOI] [PubMed] [Google Scholar]
- Jarvis ED, Gunturkun O, Bruce L, Csillag A, Karten H, Kuenzel W, Medina L, Paxinos G, Perkel DJ, Shimizu T, Striedter G, Wild JM, Ball GF, Dugas-Ford J, Durand SE, Hough GE, Husband S, Kubikova L, Lee DW, Mello CV, Powers A, Siang C, Smulders TV, Wada K, White SA, Yamamoto K, Yu J, Reiner A, Butler AB. Avian brains and a new understanding of vertebrate brain evolution. Nat Rev Neurosci. 2005;6:151–159. doi: 10.1038/nrn1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczmarek L, Chaudhuri A. Sensory regulation of immediate-early gene expression in mammalian visual cortex: implications for functional mapping and neural plasticity. Brain Res Brain Res Rev. 1997;23:237–256. doi: 10.1016/s0165-0173(97)00005-2. [DOI] [PubMed] [Google Scholar]
- Karten HJ. Homology and evolutionary origins of the ‘neocortex’. Brain Behav Evol. 1991;38:264–272. doi: 10.1159/000114393. [DOI] [PubMed] [Google Scholar]
- Kerner JA, Standaert DG, Penney JB, Jr, Young AB, Landwehrmeyer GB. Simultaneous isotopic and nonisotopic in situ hybridization histochemistry with cRNA probes. Brain Res Brain Res Protoc. 1998;3:22–32. doi: 10.1016/s1385-299x(98)00017-8. [DOI] [PubMed] [Google Scholar]
- Knapska E, Kaczmarek L. A gene for neuronal plasticity in the mammalian brain: Zif268/Egr-1/NGFI-A/Krox-24/TIS8/ZENK? Prog Neurobiol. 2004;74:183–211. doi: 10.1016/j.pneurobio.2004.05.007. [DOI] [PubMed] [Google Scholar]
- Kodama M, Russell DS, Duman RS. Electroconvulsive seizures increase the expression of MAP kinase phosphatases in limbic regions of rat brain. Neuropsychopharmacology. 2005;30:360–371. doi: 10.1038/sj.npp.1300588. [DOI] [PubMed] [Google Scholar]
- Krutzfeldt NO, Wild JM. Definition and connections of the entopallium in the zebra finch (Taeniopygia guttata) J Comp Neurol. 2004;468:452–465. doi: 10.1002/cne.10972. [DOI] [PubMed] [Google Scholar]
- Kubikova L, Wada K, Jarvis ED. Dopamine receptors in a songbird brain. J Comp Neurol. 2010;518:741–769. doi: 10.1002/cne.22255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Shepherd EG, Nelin LD. MAPK phosphatases—regulating the immune response. Nat Rev Immunol. 2007;7:202–212. doi: 10.1038/nri2035. [DOI] [PubMed] [Google Scholar]
- Liu YX, Wang J, Guo J, Wu J, Lieberman HB, Yin Y. DUSP1 is controlled by p53 during the cellular response to oxidative stress. Mol Cancer Res. 2008;6:624–633. doi: 10.1158/1541-7786.MCR-07-2019. [DOI] [PubMed] [Google Scholar]
- Machado HB, Vician LJ, Herschman HR. The MAPK pathway is required for depolarization-induced “promiscuous” immediate-early gene expression but not for depolarization-restricted immediate-early gene expression in neurons. J Neurosci Res. 2008;86:593–602. doi: 10.1002/jnr.21529. [DOI] [PubMed] [Google Scholar]
- Maczko KA, Knudsen PF, Knudsen EI. Auditory and visual space maps in the cholinergic nucleus isthmi pars parvocellularis of the barn owl. J Neurosci. 2006;26:12799–12806. doi: 10.1523/JNEUROSCI.3946-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manger PR, Elston GN, Pettigrew JD. Multiple maps and activity-dependent representational plasticity in the anterior Wulst of the adult barn owl (Tyto alba) Eur J Neurosci. 2002;16:743–750. doi: 10.1046/j.1460-9568.2002.02119.x. [DOI] [PubMed] [Google Scholar]
- Medina L, Reiner A. Do birds possess homologues of mammalian primary visual, somatosensory and motor cortices? Trends Neurosci. 2000;23:1–12. doi: 10.1016/s0166-2236(99)01486-1. [DOI] [PubMed] [Google Scholar]
- Mello CV, Clayton DF. Song-induced ZENK gene expression in auditory pathways of songbird brain and its relation to the song control system. J Neurosci. 1994;14:6652–6666. doi: 10.1523/JNEUROSCI.14-11-06652.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello CV, Clayton DF. Differential induction of the ZENK gene in the avian forebrain and song control circuit after metrazole-induced depolarization. J Neurobiol. 1995;26:145–161. doi: 10.1002/neu.480260112. [DOI] [PubMed] [Google Scholar]
- Mello CV, Jarvis ED. Behavior-dependent expression of inducible genes in vocal learning birds. In: Zeigler HP, Marler P, editors. Neuroscience of birdsong. Cambridge: Cambridge University Press; 2008. pp. 381–397. [Google Scholar]
- Mello CV, Vicario DS, Clayton DF. Song presentation induces gene expression in the songbird forebrain. Proc Natl Acad Sci U S A. 1992;89:6818–6822. doi: 10.1073/pnas.89.15.6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello CV, Vates GE, Okuhata S, Nottebohm F. Descending auditory pathways in the adult male zebra finch (Taeniopygia guttata) J Comp Neurol. 1998;395:137–160. [PubMed] [Google Scholar]
- Miceli D, Reperant J. Thalamo-hyperstriatal projections in the pigeon (Columbia livia) as demonstrated by retrograde double-labeling with fluorescent tracers. Brain Res. 1982;245:365–371. doi: 10.1016/0006-8993(82)90819-8. [DOI] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Menezes JR, Macklis JD. Neuronal subtype specification in the cerebral cortex. Nat Rev Neurosci. 2007;8:427–437. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- Mouritsen H, Feenders G, Liedvogel M, Wada K, Jarvis ED. Night-vision brain area in migratory songbirds. Proc Natl Acad Sci U S A. 2005;102:8339–8344. doi: 10.1073/pnas.0409575102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambu A. Seven problems on the basal ganglia. Curr Opin Neurobiol. 2008;18:595–604. doi: 10.1016/j.conb.2008.11.001. [DOI] [PubMed] [Google Scholar]
- Nottebohm F. The origins of vocal learning. Am Naturalist. 1972;106:116–140. [Google Scholar]
- Person AL, Gale SD, Farries MA, Perkel DJ. Organization of the songbird basal ganglia, including area X. J Comp Neurol. 2008;508:840–866. doi: 10.1002/cne.21699. [DOI] [PubMed] [Google Scholar]
- Pinaud R, Vargas CD, Ribeiro S, Monteiro MV, Tremere LA, Vianney P, Delgado P, Mello CV, Rocha-Miranda CE, Volchan E. Light-induced Egr-1 expression in the striate cortex of the opossum. Brain Res Bull. 2003;61:139–146. doi: 10.1016/s0361-9230(03)00100-x. [DOI] [PubMed] [Google Scholar]
- Pinaud R, Osorio C, Alzate O, Jarvis ED. Profiling of experience-regulated proteins in the songbird auditory fore-brain using quantitative proteomics. Eur J Neurosci. 2008;27:1409–1422. doi: 10.1111/j.1460-9568.2008.06102.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzio GA, Golombek DA. Photic regulation of map kinase phosphatases MKP1/2 and MKP3 in the hamster suprachiasmatic nuclei. J Mol Neurosci. 2008;34:187–192. doi: 10.1007/s12031-007-9021-y. [DOI] [PubMed] [Google Scholar]
- Qian Z, Gilbert M, Kandel ER. Temporal and spatial regulation of the expression of BAD2, a MAP kinase phosphatase, during seizure, kindling, and long-term potentiation. Learn Mem. 1994;1:180–188. [PubMed] [Google Scholar]
- Reiner A, Perkel DJ, Bruce LL, Butler AB, Csillag A, Kuenzel W, Medina L, Paxinos G, Shimizu T, Striedter G, Wild M, Ball GF, Durand S, Guturkun O, Lee DW, Mello CV, Powers A, White SA, Hough G, Kubikova L, Smulders TV, Wada K, Dugas-Ford J, Husband S, Yamamoto K, Yu J, Siang C, Jarvis ED. Revised nomenclature for avian telencephalon and some related brainstem nuclei. J Comp Neurol. 2004;473:377–414. doi: 10.1002/cne.20118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts TF, Brauth SE, Hall WS. Distribution of iron in the parrot brain: conserved (pallidal) and derived (nigral) labeling patterns. Brain Res. 2001;921:138–149. doi: 10.1016/s0006-8993(01)03110-9. [DOI] [PubMed] [Google Scholar]
- Rossler OG, Henss I, Thiel G. Transcriptional response to muscarinic acetylcholine receptor stimulation: regulation of Egr-1 biosynthesis by ERK, Elk-1, MKP-1, and calcineurin in carbachol-stimulated human neuroblastoma cells. Arch Biochem Biophys. 2008;470:93–102. doi: 10.1016/j.abb.2007.11.008. [DOI] [PubMed] [Google Scholar]
- Sgambato V, Pages C, Rogard M, Besson MJ, Caboche J. Extracellular signal-regulated kinase (ERK) controls immediate early gene induction on corticostriatal stimulation. J Neurosci. 1998;18:8814–8825. doi: 10.1523/JNEUROSCI.18-21-08814.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T, Bowers AN. Visual circuits of the avian telencephalon: evolutionary implications. Behav Brain Res. 1999;98:183–191. doi: 10.1016/s0166-4328(98)00083-7. [DOI] [PubMed] [Google Scholar]
- Shipp S. Structure and function of the cerebral cortex. Curr Biol. 2007;17:R443–449. doi: 10.1016/j.cub.2007.03.044. [DOI] [PubMed] [Google Scholar]
- Soares JG, Pereira AC, Botelho EP, Pereira SS, Fiorani M, Gattass R. Differential expression of Zif268 and c-Fos in the primary visual cortex and lateral geniculate nucleus of normal Cebus monkeys and after monocular lesions. J Comp Neurol. 2005;482:166–175. doi: 10.1002/cne.20361. [DOI] [PubMed] [Google Scholar]
- Takahata T, Higo N, Kaas JH, Yamamori T. Expression of immediate-early genes reveals functional compartments within ocular dominance columns after brief monocular inactivation. Proc Natl Acad Sci U S A. 2009;106:12151–12155. doi: 10.1073/pnas.0905092106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaki M, Ujike H, Kodama M, Takehisa Y, Nakata K, Kuroda S. Two kinds of mitogen-activated protein kinase phosphatases, MKP-1 and MKP-3, are differentially activated by acute and chronic methamphetamine treatment in the rat brain. J Neurochem. 2001;79:679–688. doi: 10.1046/j.1471-4159.2001.00615.x. [DOI] [PubMed] [Google Scholar]
- Theiss MP, Hellmann B, Gunturkun O. The architecture of an inhibitory sidepath within the avian tectofugal system. Neuroreport. 2003;14:879–882. doi: 10.1097/00001756-200305060-00021. [DOI] [PubMed] [Google Scholar]
- Thompson I. Cortical development: binocular plasticity turned outside-in. Curr Biol. 2000;10:R348–R350. doi: 10.1016/s0960-9822(00)00464-4. [DOI] [PubMed] [Google Scholar]
- Tischmeyer W, Grimm R. Activation of immediate early genes and memory formation. Cell Mol Life Sci. 1999;55:564–574. doi: 10.1007/s000180050315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velho TA, Pinaud R, Rodrigues PV, Mello CV. Co-induction of activity-dependent genes in songbirds. Eur J Neurosci. 2005;22:1667–1678. doi: 10.1111/j.1460-9568.2005.04369.x. [DOI] [PubMed] [Google Scholar]
- Wada K, Sakaguchi H, Jarvis ED, Hagiwara M. Differential expression of glutamate receptors in avian neural pathways for learned vocalization. J Comp Neurol. 2004;476:44–64. doi: 10.1002/cne.20201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada K, Howard JT, McConnell P, Whitney O, Lints T, Rivas MV, Horita H, Patterson MA, White SA, Scharff C, Haesler S, Zhao S, Sakaguchi H, Hagiwara M, Shiraki T, Hirozane-Kishikawa T, Skene P, Hayashizaki Y, Carninci P, Jarvis ED. A molecular neuroethological approach for identifying and characterizing a cascade of behaviorally regulated genes. Proc Natl Acad Sci U S A. 2006;103:15212–15217. doi: 10.1073/pnas.0607098103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren WC, Clayton DF, Ellegren H, Arnold AP, Hillier LW, Kunstner A, Searle S, White S, Vilella AJ, Fairley S, Heger A, Kong L, Ponting CP, Jarvis ED, et al. The genome of a songbird. Nature. 2010 doi: 10.1038/nature08819. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidner C, Reperant J, Miceli D, Haby M, Rio JP. An anatomical study of ipsilateral retinal projections in the quail using radioautographic, horseradish peroxidase, fluorescence and degeneration techniques. Brain Res. 1985;340:99–108. doi: 10.1016/0006-8993(85)90778-4. [DOI] [PubMed] [Google Scholar]
- Wild JM. The avian somatosensory system: connections of regions of body representation in the forebrain of the pigeon. Brain Res. 1987;412:205–223. doi: 10.1016/0006-8993(87)91127-9. [DOI] [PubMed] [Google Scholar]
- Wild JM. Avian somatosensory system: II. Ascending projections of the dorsal column and external cuneate nuclei in the pigeon. J Comp Neurol. 1989;287:1–18. doi: 10.1002/cne.902870102. [DOI] [PubMed] [Google Scholar]
- Wild JM, Farabaugh SM. Organization of afferent and efferent projections of the nucleus basalis prosencephali in a passerine, Taeniopygia guttata. J Comp Neurol. 1996;365:306–328. doi: 10.1002/(SICI)1096-9861(19960205)365:2<306::AID-CNE8>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Wild JM, Williams MN. Rostral wulst of passerine birds: II. Intratelencephalic projections to nuclei associated with the auditory and song systems. J Comp Neurol. 1999;413:520–534. doi: 10.1002/(sici)1096-9861(19991101)413:4<520::aid-cne3>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- Wild JM, Williams MN. Rostral wulst in passerine birds. I. Origin, course, and terminations of an avian pyramidal tract. J Comp Neurol. 2000;416:429–450. [PubMed] [Google Scholar]
- Young WS., 3rd Simultaneous use of digoxigenin- and radiolabeled oligodeoxyribonucleotide probes for hybridization histochemistry. Neuropeptides. 1989;13:271–275. doi: 10.1016/0143-4179(89)90081-4. [DOI] [PubMed] [Google Scholar]
- Zeigler HP, Bischof H-J. Vision, brain, and behavior in birds. Cambridge, MA: MIT Press; 1993. [Google Scholar]