Abstract
The evolutionarily conserved PUF proteins stimulate CCR4 mRNA deadenylation through binding to 3′ UTR sequences of specific mRNA. We have investigated the mechanisms by which PUF3 in Saccharomyces cerevisiae accelerates deadenylation of the COX17 mRNA. PUF3 was shown to affect PAN2 deadenylation of the COX17 mRNA independent of the presence of CCR4, suggesting that PUF3 acts though a general mechanism to affect deadenylation. Similarly, eIF4E, the cap-binding translation initiation factor, known to control CCR4 deadenylation, was shown to affect PAN2 activity in vivo. PUF3 was found to be required for eIF4E effects on COX17 deadenylation. Both eIF4E and PUF3 effects on deadenylation were shown, in turn, to necessitate a functional poly(A) binding protein (PAB1) in which removal of the RRM1 domain of PAB1 blocked both their effects on deadenylation. While removal of the proline-rich region (P domain) of PAB1 substantially reduces CCR4 deadenylation at non-PUF3 controlled mRNA and correspondingly blocked eIF4E effects on deadenylation, PUF3 essentially bypassed this P domain requirement. These results indicate that the PAB1-mRNP structure is critical for PUF3 action. We also found that multiple components of the CCR4-NOT deadenylase complex but not PAN2 interacted with PUF3. PUF3 appears, therefore, both to act independently of CCR4 activity, possibly through effects on PAB1-mRNP structure, and to be capable of retaining the CCR4-NOT complex.
INTRODUCTION
The principal pathway for mRNA degradation in the yeast Saccharomyces cerevisiae proceeds through several steps. There is an initial PAN2/PAN3-dependent trimming of about 15–20 nucleotides (nt) of the poly(A) tail to a default length of about 60–80 nt that is specific for each mRNA (1,2). This trimming requires PAB1 to which PAN3 binds and the translation termination factors eRF1/eRF3 (3,4). The major process of deadenylation requires CCR4 of the CCR4-NOT complex (2,5,6) as it shortens the poly(A) tail down to an end-point size of about 8–12 nt (7). A second deadenylase in the CCR4-NOT complex, CAF1, has been shown to have a much more limited role on deadenylation in yeast (8). Deletion of PAN2/3 has also been shown not to affect the major part of deadenylation (2,6). Poly(A) tail shortening down to the oligo(A) species leads, in turn, to the reduced ability of poly(A) binding protein (PAB1) to bind the poly(A) tail that presumably alters the translation initiation complex association with the mRNA cap structure (9). The LSM complex associates with the oligo(A) tail at this time (10,11), perhaps leading to the cessation of deadenylation of CCR4 (7,8). Changes in the mRNP structure around the cap lead to decapping by DCP1/DCP2 (12–14) and 5′-3′ degradation of the RNA by the exonuclease XRN1 (15). The change from deadenylation to decapping and sequestering of the oligoadenylated mRNA in P-bodies (13,16) may occur at this stage.
Three general models suggest how CCR4 deadenylation may be controlled. In the first model, RNA binding proteins that contact specific mRNA 3′ UTRs make contacts to the CCR4-NOT complex (17–22). Retention of CCR4 at certain mRNA would therefore enhance the rate of deadenylation, as has been additionally verified in yeast by tethering CAF1 to the 3′ UTR (23). A second model for control of deadenylation indicates that the PAB1-mRNP structure must be disrupted and PAB1 removal must be expedited prior to CCR4 action (6,24,25). Factors that control PAB1-mRNP structure would therefore be expected to control CCR4 access to the mRNA. In this case, in vitro studies have shown that the CCR4-NOT complex alone cannot overcome repressive PAB1-mRNP structures (24), suggesting that CCR4-NOT binding to the mRNA may be insufficient by itself to accelerate deadenylation. A third model describing the control of deadenylation would be that the sequestering of mRNA away from the deadenylase in stress granules (13,25–27) restricts deadenylation under certain conditions. However, these models are not mutually exclusive, and factors may act by multiple means to regulate deadenylase function.
The mRNP complex consists of a mature mRNA, PAB1, and translation initiation factors (TIFs), such as the 5′ cap binding complex, eIF4E, the bridging protein eIF4G, and ribosomes. PAB1, being a core component of the mRNP complex, plays a critical role in mRNA deadenylation (6,12,24,25,28). It is composed of four RNA recognition motifs (RRM), an unstructured proline and methionine rich region (P domain), and a globular C terminal domain (C domain). The RRM1 and P domains have been shown to be most important to CCR4 deadenylation (24,25), suggesting that the PAB1-mRNP could play a regulatory role in deadenylation.
How RNA binding proteins specifically control mRNA deadenylation has principally focused on identifying contacts to the CCR4-NOT complex. In yeast, for example, the PUF5 protein has been shown to affect deadenylation by binding of the CCR4-NOT complex (18,19). Whether this is generally valid for other PUF or RNA binding proteins remains to be established. The PUF protein family members all share a conserved RNA binding domain capable of recognizing a consensus repeat consisting of four core ribonucleotides, UGUR (29). PUF proteins are widely conserved and have very diverse functions ranging from roles in mitochondrial biogenesis to affecting embryonic development and germline stem cells (29–32). Despite the wide variety of functions that PUF proteins participate in, they all work by a similar biochemical means, increased degradation and translational repression of the target transcript. In yeast, PUF3 has been shown to control COX17 mRNA deadenylation (33,34), and PUF5 to control HO mRNA deadenylation (18,35).
We have investigated the mechanisms by which PUF3 accelerates deadenylation in yeast. We provide evidence in support of two different models of PUF3-mediated acceleration of mRNA deadenylation. PUF3 affected both CCR4 and PAN2 deadenylation processes, suggesting that it was acting by a general mechanism to accelerate deadenylation. Defects in the PAB1-mRNP structure also restrict deadenylation by either CCR4 or PAN2 (24,25; unpubl. observ.). PUF3 acceleration of deadenylation was shown, in turn, to require a functional PAB1-mRNP structure. In addition, we show that PUF3 is able to co-immunoprecipitate CCR4 and various components of the CCR4-NOT complex. These results provide evidence that PUF3 may act both generally through the PAB1-mRNP structure and specifically through CCR4-NOT complex retention to affect the deadenylation process.
RESULTS
PUF3 acts to affect both CCR4 and PAN2 deadenylases in vivo
Several factors act to increase the rate of deadenylation of mRNA in yeast, resulting in increased mRNA turnover (34,38). Most notable among these effects are defects in TIFs, such as in eIF4E, and the binding of proteins such as PUF3 to specific mRNA 3′ UTR sequences. In both cases, these effects have been shown to be on CCR4 deadenylation (6,19,38), and, in the case of PUF5, retention of the CCR4-NOT complex has been suggested to account for its augmentation of deadenylation rates (18). Because defects in translation initiation factors most likely affect deadenylation generally through effects on the mRNP structure, we sought to investigate whether PUF3 acts also by a general mechanism to affect deadenylation. If PUF3 were acting through a general mechanism to affect poly(A) removal, then it would be expected to be also required for PAN2 deadenylase activity, the second of the two known functional deadenylases in yeast (2,6). No other specific PUF3-induced deadenylase appears to be present in yeast (6). To address this question, we utilized two pairs of strains containing the following genotypes: wild-type and puf3Δ; ccr4Δ and ccr4Δ puf3Δ. By comparing the effect of the puf3Δ on COX17 mRNA deadenylation in the ccr4Δ background when PAN2 is the only deadenylase (2,6) to that of ccr4Δ alone we would be able to assess whether PUF3 was required for PAN2 activity.
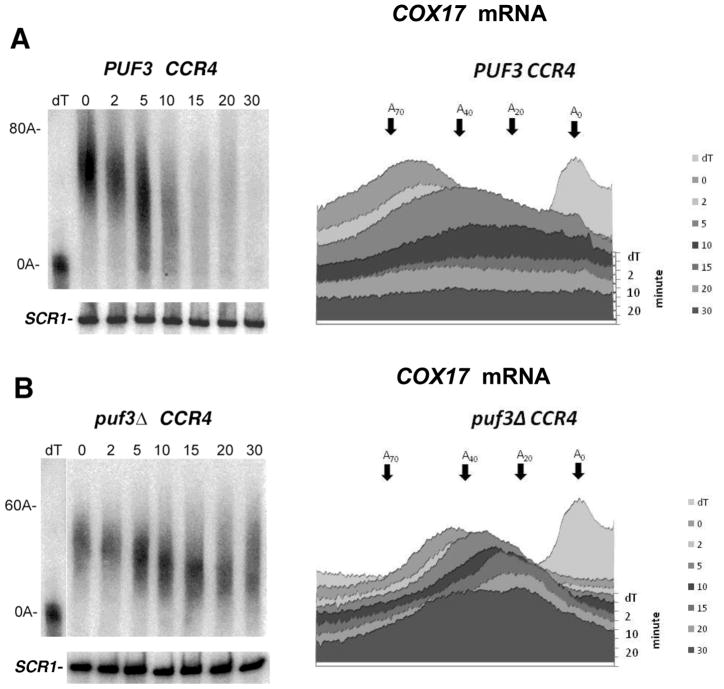
A pulse-chase experiment was utilized to examine COX17 mRNA poly(A) tails in each of these strain backgrounds. A GAL10-COX17 plasmid was expressed in each strain in which the COX17 mRNA expression was under the control of the GAL10 promoter (34). Following the induction of COX17 mRNA expression for a short time (the pulse), mRNA synthesis was shut off with the addition of glucose (the chase) and the decay of the poly(A) tail was followed by Northern analysis. As shown in Figure 1A, in the wild-type strain at the zero time COX17 mRNA contains a poly(A) tail of around 40–75 A’s, in agreement with previous observations (6,33,34). Within 5 min following the cessation of COX17 mRNA synthesis, some completely deadenylated COX17 mRNA were detected (see also densitometric analyses, right panels), indicative of an extremely rapid deadenylation by CCR4 (6). It should be noted that at 5 min both completely deadenylated COX17 mRNA poly(A) tails coexist with nearly full-length poly(A) tails, characteristic of extremely rapid deadenylation processes that could be occurring through a processive means (7,25,34). Correspondingly, it was observed that COX17 mRNA disappeared rapidly following 5 min (an SCR1 loading control indicated equivalent levels of RNA had been loaded in all lanes, Figure 1), consistent with rapid deadenylation causing enhanced decapping and XRN1 5′ exonuclease activity (7,34). In a puf3Δ background, CCR4 deadenylation was slowed considerably in which it took up to 10 to 15 min before the oligo(A) form was detected in appreciable amounts (Figure 1B), as previously observed (34). Decapping and subsequent disappearance of the mRNA was also slowed in the puf3Δ background (34).
Figure 1. PUF3 affects both PAN2 and CCR4 deadenylation.
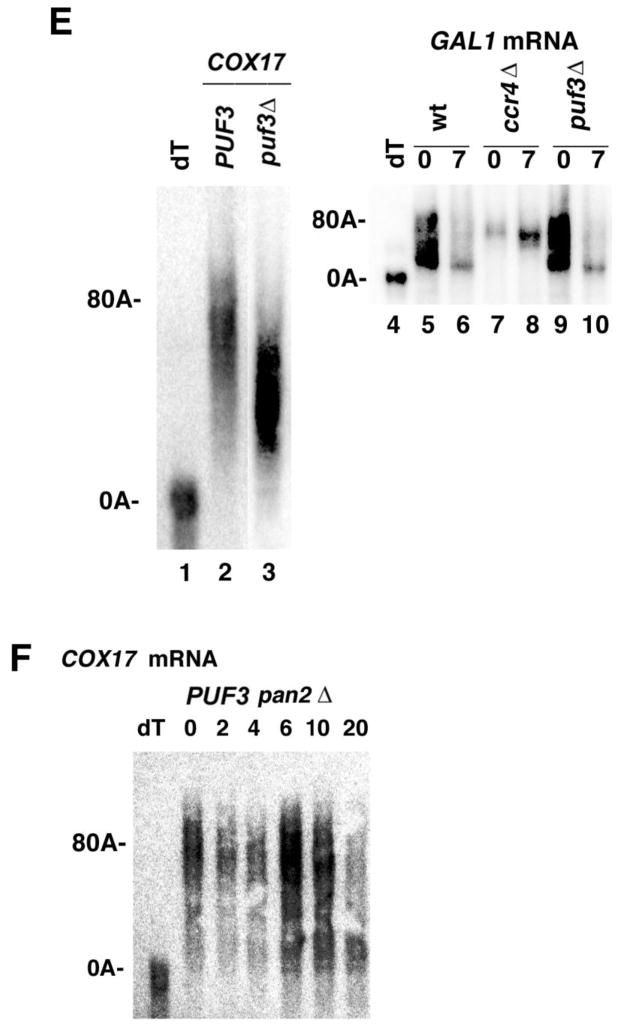
Transcriptional pulse-chase analyses on COX17 mRNA for panels A–D and F were conducted as previously described (34). Following induction of the GAL10-COX17 mRNA (expressed from the pRP1007 plasmid) with galactose, transcription was shut off with glucose and, at the times (in min) indicated above the Figure, RNA was extracted and Northern analyses were conducted as described (25). dT refers to the RNA sample probed with oligo (dT) followed by RNase H digestion to remove the poly (A) tail. Roughly equivalent amounts of RNA were loaded into each lane across a panel, as detected by probing for SCR1 RNA and by A260 spectrophotometric analysis. Panels on the right represent densitometric scans of each lane represented in the corresponding left panel. The demarcations on the right of the plot skip every other time point for clarity. The oligo(A) lengths were determined using the following standards: the size of SCR1, the length of the completely deadenylated length of COX17 poly(A) tail (dT sample), and the length of the completely undeadenylated poly(A) tail at time zero based on other experiments using different GAL1 poly(A) tail lengths as standards. For panel E, poly(A) tail lengths were determined for COX17 mRNA (lanes 1–3) or for GAL1 mRNA (lanes 4–10) following twenty min induction of mRNA synthesis (time zero for lanes 2, 3, 5, 7, and 9) or after shifting to glucose containing medium for 7 min after galactose induction (lanes 6, 8, and 10). GAL1 mRNA analysis was conducted as previously described (25). A. Strain YAS319/YC504 (wild-type); B. Strain 1738-3/YC504 (puf3Δ); C. Strain RP840-1a (ccr4Δ); D. Strain RP1241-1a (puf3Δ ccr4Δ); and F. Strain RP1619 (pan2Δ). All strains carried the pRP1007 plasmid. E. Lane 1: strain RP840/pRP1007; lane 2: strain RP1241/pRP1007; lanes 5–6; strain RP840 (wild-type); lanes 7–8: RP840-1a; and lanes 9–10: RP1241 (puf3Δ).
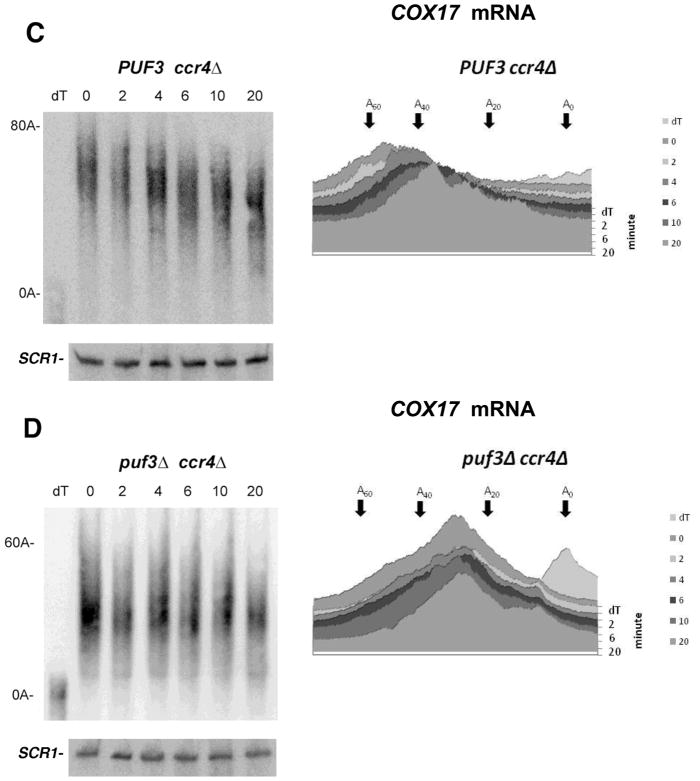
In a ccr4Δ background when PAN2 was the only deadenylase, COX17 mRNA deadenylation was slowed considerably due to the fact that CCR4 is responsible for the vast majority of mRNA deadenylation in yeast (2,6,25) (Figure 1C). This result is the same as previously observed (6), and the residual deadenylation that is observed has been shown previously to be the result of PAN2 activity (6). Relatedly, in a pan2Δ background, deadenylation of the COX17 mRNA to the oligo(A) form took between 4 and 6 min, indicating that a pan2 deletion does not significantly affect COX17 mRNA deadenylation (Figure 1F). In contrast, in a ccr4Δ puf3Δ background little or no appreciable deadenylation of the COX17 poly(A) tail by PAN2 was observed (Figure 1D). This last result indicates that PUF3 is also required for PAN2 deadenylation at the COX17 mRNA. Since no specific PUF3-induced deadenylase appears to be present in yeast (6), PUF3 may therefore be acting by a general mechanism to control both CCR4 and PAN2 activities.
It should also be noted that a puf3 deletion resulted in significantly shorter initial COX17 poly(A) tail lengths following the short burst of mRNA synthesis than that which was observed in the wild-type background (compare 0 min time points for Figure 1A with 1B and also see Figure 1E, lanes 2 and 3, in which samples were analyzed on the same gel). These observations of effects on the default poly(A) tail length of COX17 mRNA are consistent with analyses by previous researchers, although it has not previously been remarked upon (6, 34). The puf3 defect does not generally affect the initial poly(A) tail length of mRNA to which it does not bind, for example, GAL1 mRNA (Figure 1E, lane 5 compared to lane 9). Since the initial default poly(A) tail length is determined by PAN2 and by poly(A) polymerase, PUF3 could be affecting either of these enzymatic processes for mRNA to which it binds, independently of its effects on the cytoplasmic deadenylation of the poly(A) tail once the default poly(A) tail length is obtained (6, 24).
A defect in eIF4E also affects both CCR4 and PAN2 deadenylation activity
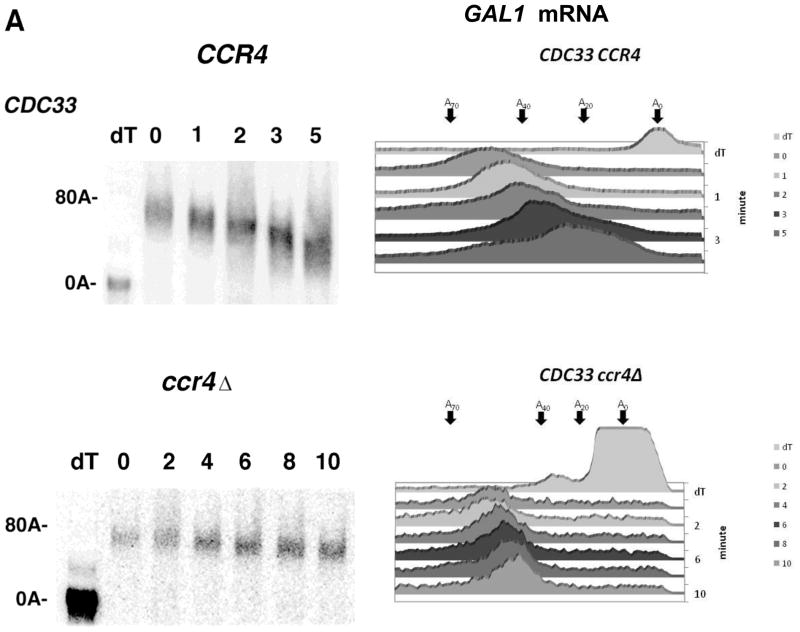
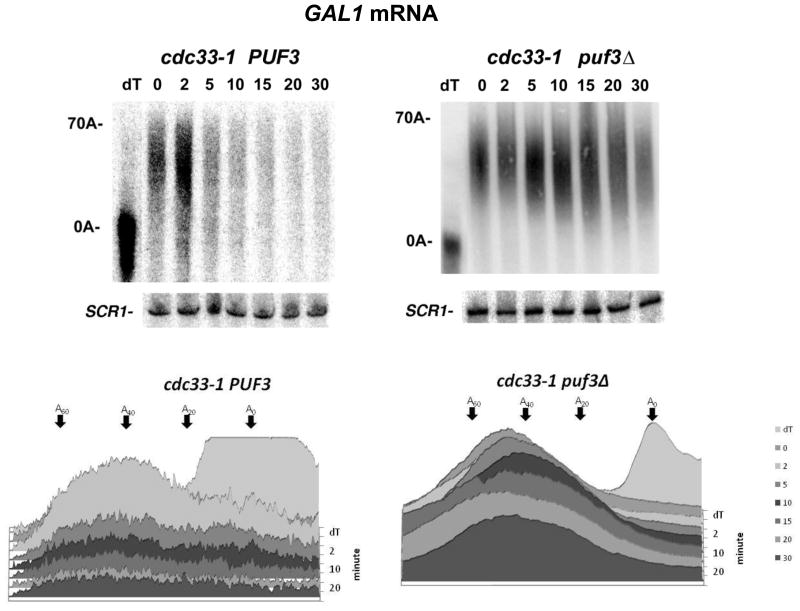
If TIFs affect deadenylation by a general mechanism, it would be expected that defects in TIFs, in addition to affecting deadenylation by CCR4 (38), should be able to accelerate deadenylation of mRNA by PAN2. To examine this model we used the cdc33-1 allele (a temperature sensitive mutation in eIF4E blocking its ability to bind the mRNA cap, 39) that accelerates deadenylation at the restrictive temperature of 37°C (38). At the restrictive temperature the cdc33-1 allele reduces protein synthesis by about 70% (36,38). We first tested the effect of the cdc33-1 allele on general deadenylation using the GAL1 mRNA that is not controlled by PUF3 (Figure 1E, compare lanes 5 and 6 (PUF3) to that of lanes 9 and 10 (puf3Δ); data not shown). Following a pulse-chase analysis of GAL1 conducted at the restrictive temperature, in the wild-type strain the poly(A) tail just began to be shortened to the oligo(A) form in about 3–5 min (Figure 2A, top panel) in which at 5 min a significant portion of the mRNA species still contained about 25 A’s (see densitometric scans). In contrast, in the isogenic cdc33-1-containing strain, GAL1 poly(A) tails appeared in the oligo(A) species within 2 min after cessation of transcription, with a sizeable amount of the mRNA existing in the oligo(A) form by 3 min and almost all of it in the oligo(A) form by 5 min (Figure 2B, cdc33-1, left top panel, see also densitometric scans). These results are very similar to what was previously observed for cdc33-1 effects on the deadenylation of the non-PUF3 controlled MFA2pG and PGK1pG mRNAs (38). In the ccr4Δ cdc33-1 background when PAN2 is the only deadenylase, GAL1 deadenylation was also very fast (Figure 2B, right panel as compared to ccr4Δ in the CDC33 wild-type control strain, Figure 2A, bottom panel), indicating that a defect in the translation initiation complex can alter the rate of deadenylation even when CCR4 is not present. These results indicate that both PUF3 and defects in TIFs can affect deadenylation independently of CCR4 presence.
Figure 2. A defect in eIF4E affects PAN2 deadenylation.
Transcriptional pulse chase analysis was conducted on the GAL1 gene as described in Figure 1, except that all strains (lacking pRP1007) were grown at 37°C for 1 hr prior to the galactose induction (25). At the times (in min) indicated above the Figure, RNA was isolated. All lanes had roughly equivalent levels of total RNA as assessed by A260 spectrophotometric readings, which agree well with total RNA levels determined by either ethidium bromide staining of rRNA levels (25,36,48) or by detection of SCR1 mRNA levels by Northern analyses (see Figures 1, 3, and 4). A. Top panel- Strain YAS319/YC504 [PAB1-CEN-TRP1]; Bottom panel- Strain YAS319-1a/YC504 (ccr4Δ). Densitometric scans of these Northerns are presented in the right panel. B. Top left panel- Strain YAS1881/YC504 (cdc33-1). Top right panel- Strain YAS1881-1a/YC504 (cdc33-1 ccr4Δ). Densitometric scans of each Northern are presented in the bottom panel. C. Left panel- Strain YAS1881/YC505 [PAB1-ΔRRM1-CEN-TRP1]; right panel- Strain YAS1881/YC510 [PAB1-ΔP-CEN-TRP1]. Densitometric scans are presented in the bottom panel.
PUF3 acts downstream of eIF4E in affecting deadenylation
Since both PUF3 and defects in TIFs accelerate deadenylation, we examined the epistasis of these factors on poly(A) removal by assessing whether PUF3 was required for the acceleration of deadenylation caused by the cdc33-1 allele at the COX17 mRNA. In the cdc33-1 background at the restrictive temperature, COX17 mRNA deadenylation is faster than in the wild-type background, as the oligo(A) form of COX17 mRNA appeared in the first 2 min following synthesis of the mRNA (Figure 3, left panel) as compared to about 5 min in the wild-type strain (see Figure 1A). However, in a puf3Δ background, cdc33-1 was incapable of accelerating deadenylation (Figure 3, right panel, cdc33-1 puf3Δ, as compared to Figure 3, left panel, cdc33-1) in which the oligo(A) form did not appear until at least 10–15 min or later after the turning off of mRNA synthesis. PUF3 is therefore required for the cdc33-1 defect to enhance the deadenylation process at the COX17 mRNA. It should also be noted that deletion of PUF3 also blocked the rapid decapping and degradation of the COX17 mRNA that occurred as a result of a defect in eIF4E, suggesting that the enhancement of both deadenylation and decapping by the eIF4E defect (38) are mechanistically linked processes.
Figure 3. PUF3 is required for eIF4E effects on deadenylation at the COX17 mRNA.
Transcriptional pulse chase analysis was conducted as described in Figure 1, except that all strains were grown at 37°C for 1 hr prior to the galactose induction. Similar levels of RNA were loaded into each lane, as detected by probing for SCR1 RNA. Top left panel- Strain YAS1881/YC504 (cdc33-1); top right panel- Strain 1777-6/YC504 (cdc33-1 puf3Δ). Densitometric scans of each Northern are present in the bottom panels.
The PAB1 RRM1 domain is required for PUF3 acceleration of deadenylation
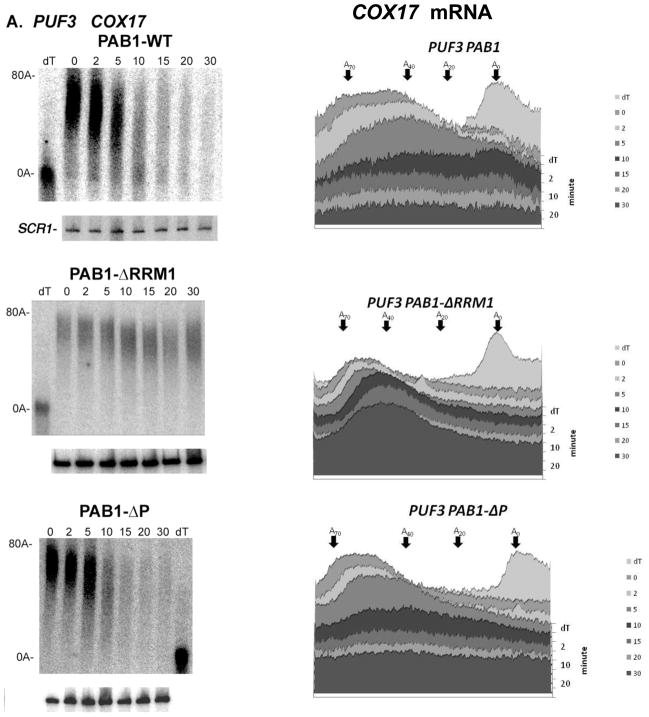
We have previously shown that specific domains of PAB1 are required for deadenylation (25), indicating that the PAB1-mRNP structure is critical to the process of poly(A) removal (24). The RRM1 and P domains of PAB1, when deleted, severely block deadenylation and appear to do so by affecting the PAB1-mRNP structure (24,25). Strains with RRM1 or P domain deletions are viable, grow nearly like wild-type (25), do not affect mRNA transport (24,40), and display normal polysomal RNA profiles, although deleting RRM1 causes a 28% decrease in the rate of protein synthesis (in contrast, the cdc33-1 allele causes a 70% drop in protein synthesis) (25,36). We consequently first examined whether the ability of a defect in eIF4E to allow rapid deadenylation was dependent on PAB1. For this analysis we studied the deadenylation rate of GAL1 mRNA in a cdc33-1 background in strains carrying PAB1-ΔRRM1 or PAB1-ΔP. As shown in Figure 2C, in a cdc33-1 background when either the RRM1 or P domain of PAB1 was deleted, GAL1 mRNA deadenylation was slowed considerably and the oligo(A) form did not appear until around 10 min. In contrast, in a PAB1 wild-type background cdc33-1 allowed rapid deadenylation of GAL1 mRNA in that the oligo(A) form of the mRNA appeared within 2 min after cessation of transcription Figure 2B, top left panel. These results indicate that alterations in PAB1 mRNP structure are critical to deadenylation even in the presence of a defect in eIF4E. It should be noted, however, that removal of either the RRM1 domain or the P domain of PAB1 normally causes a very significant blockage in GAL1 mRNA deadenylation in which the oligo(A) form does not appear until at least 20 min (25). Therefore, removal of either the RRM1 or P domain of PAB1 significantly reduces the accelerated deadenylation caused by the cdc33-1 allele, but this blockage is not complete, suggesting that some of the effects of the cdc33-1 allele on deadenylation may be independent of PAB1.
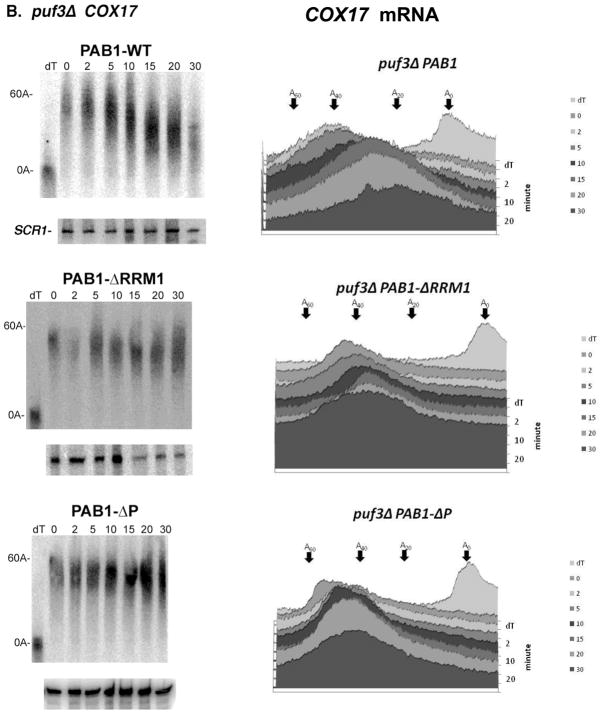
We subsequently assessed using pulse-chase analysis whether PAB1 acts downstream of PUF3 in the control of COX17 mRNA deadenylation by determining if PAB1 was required for PUF3-dependent deadenylation of COX17. In a wild-type PAB1 background, COX17 mRNA was rapidly deadenylated (Figure 4A, top panel) and within 5 minutes of transcriptional shut off some completely deadenylated species were observed, as described above, and by 10 min the completely deadenylated species are the predominate product (see densitometric scans). Deletion of PUF3 slowed this rapid deadenylation (Figure 4B, top panel) as previously mentioned (34; Figure 1B). In contrast, when COX17 mRNA deadenylation was analyzed in a PAB1-ΔRRM1 background, COX17 displayed extremely slow deadenylation (Figure 4A, middle panel). In the wild-type PUF3 background, oligo(A) mRNA did not appear even after 30 min. These observations indicate that the RRM1 domain of PAB1 is necessary for deadenylation of the COX17 poly(A) tail and that the resultant PAB1-ΔRRM1 mRNP structure substantially blocks the ability of PUF3 to accelerate deadenylation. Enhanced decapping by PUF3 was also blocked by removal of the RRM1 domain. Deletion of other domains of PAB1, such as RRM3, RRM4, or the C domain had no major effect on PUF3 acceleration of COX17 mRNA deadenylation (data not shown), although deleting the C domain appeared to result in a slight slowing of COX17 mRNA deadenylation, consistent with its effects on GAL1 and MFA2pG deadenylation (25).
Figure 4. The RRM1 region of PAB1 is required for PUF3 acceleration of deadenylation.
Transcriptional pulse chase analysis was conducted as described in Figure 1. Roughly equivalent amounts of RNA were loaded into each of the different panels, as detected by probing for SCR1 RNA. Therefore, in section A for the PAB1-WT and PAB1-ΔP panels, the lack of RNA at times after 10 to 15 minutes indicates that complete deadenylation has occurred followed by decapping and degradation. All strains carried the pRP1007 plasmid. Strains carried the PAB1 variant as indicated: PAB1-WT (YC504); PAB1-ΔRRM1 (YC505); PAB1-ΔP (YC510), A. Strain YAS319/YC504 (wild-type); B. Strain 1738-3/YC504 (puf3Δ.). Densitometric scans of each Northern are in the right panels.
In contrast to the effects of the RRM1 deletion, in the PAB1-ΔP background, COX17 deadenylation was rapid. At ten minutes some nearly completely deadenylated poly(A) tails were observed along with much longer poly(A) tails (Figure 4A, bottom panel). Deadenylation was slightly slower with PAB1-ΔP than in wild-type PAB1 in which at 5 minutes the nearly completely deadenylated species were first observed. Significantly decreased total levels of mRNA were observed by fifteen minutes (Figure 4A, bottom panel), indicative of rapid decapping and degradation that follows upon deadenylation. The SCR1 control indicated that nearly identical levels of total RNA had been loaded in each lane. These data confirm that PAB1-ΔP does not block PUF3-enhanced deadenylation, whereas PAB1-ΔRRM1 does. Therefore, the mRNP structure created by deleting the PAB1 RRM1 domain, unlike that of deleting the P domain, is resistant to PUF3 action.
In the puf3Δ background, both PAB1-ΔRRM1 and PAB1-ΔP showed a block in deadenylation more severe than that observed for wild-type PAB1 (Figure 4B). For example, after 30 minutes little deadenylation has occurred in the PAB1-ΔP puf3Δ strain. These observations suggest that in the absence of PUF3 the deadenylation of COX17 is blocked by removing either the PAB1 RRM1 or the P domain, as previously observed for the non-PUF3 controlled MFA2pG and GAL1 mRNA (25). However, in the presence of PUF3, the P domain of PAB1 is not essential for deadenylation. This PAB1 allele-specific effect on PUF3 deadenylation of COX17 mRNA but not on the deadenylation of other mRNA, including that of COX17 when PUF3 is not functional, suggests that PUF3 acts redundantly with the P domain of PAB1, either in conjunction with it or at the site of the PAB1-mRNP structure, to affect deadenylation.
One model consistent with these results is that PUF3 interacts directly with PAB1 to rearrange the PAB1-mRNP structure. However, experiments to identify possible PUF3 physical interactions with PAB1, either when PAB1 was bound to poly(A) or when it was by itself, did not demonstrate any specific interactions between the two proteins (data not shown).
PUF3 co-immunoprecipitates the CCR4-NOT complex
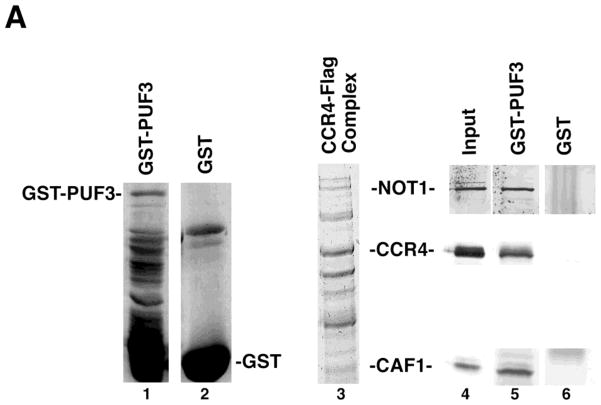
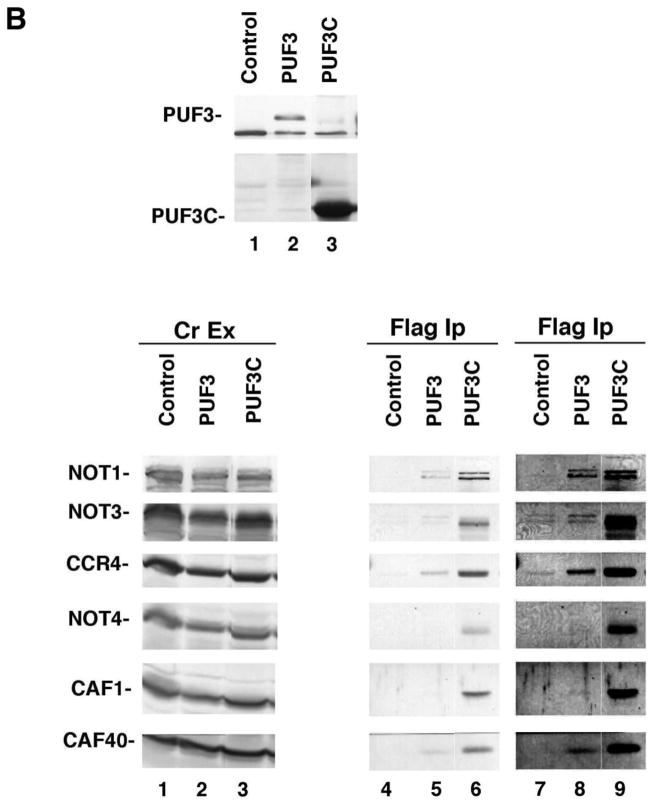
Because it has been previously shown that PUF5 can associate with the CCR4-NOT complex, we also examined whether PUF3 could contact components of the CCR4-NOT complex in vitro. Using a GST-PUF3 fusion bound to glutathione agarose beads, we tested whether components of the purified the CCR4-NOT complex isolated from yeast could be retained by GST-PUF3. As shown in Figure 5A, NOT1, CCR4, and CAF1 could be retained on the glutathione agarose beads containing GST-PUF3 (lane 5), but were not able to bind to GST alone (lane 6). We subsequently investigated if PUF3 could co-immunoprecipitate the CCR4-NOT complex from yeast crude extracts. A full-length PUF3 tagged with the FLAG epitope was expressed in yeast (Figure 5B, top panel, lane 2) and immunoprecipitated with FLAG beads. Multiple components of the CCR4-NOT complex were found to co-immunoprecipitate with FLAG-PUF3 as identified by Western analysis, including NOT1, NOT3, CCR4, and CAF40 (Figure 5B, lower panel, lane 5), but not in the control lacking FLAG-PUF3 (lane 4). NOT4 and CAF1 were also faintly visualized in the original Western for the FLAG-PUF3 immunoprecipitation (see darker exposure, Figure 5B, lower panel, lanes 7–9) and in other experiments, suggesting that the whole CCR4-NOT complex was capable of interacting with PUF3 from yeast crude extracts. We also analyzed the ability of just the C-terminal region of PUF3 in interacting with the CCR4-NOT complex. PUF3C, containing the C-terminal region of PUF3, has been shown previously to be sufficient for recapitulating the ability of PUF3 to control the deadenylation of COX17 mRNA (33). Our results indicated PUF3C was able to immunoprecipitate all CCR4-NOT components tested (Figure 5B, lower panel, lane 6). Using FLAG-PUF3C we were also able to show that the PUF3 contacts to CCR4 and CAF1 were essentially RNA independent, as the treatment with RNase A, which destroys the PAB1-mRNP structure (unpubl. observ.), did not interfere with their interactions (data not shown). We also assessed whether PUF3C could interact with PAN2. In this case, however, PUF3C was unable to specifically immunoprecipitate PAN2 (data not shown). It should be noted, however, that extensive immunoprecipitation analysis using various components of the CCR4-NOT components as the primary baits have not demonstrated any specific interaction with PUF3 or other PUFs (42–45).
Figure 5. PUF3 can immunoprecipitate the CCR4-NOT complex.
A. GST-PUF3 can interact with the CCR4-NOT complex in vitro. Purified GST-PUF3 was bound to glutathione agarose beads and the purified CCR4-NOT complex was assayed for retention to the beads. Equivalent amounts of GST and GST-PUF3 were used. Lanes 1 and 2, respectively- Coomassie stained GST-PUF3 and GST; lane 3- Coomassie stained CCR4-NOT complex; lanes 4–6- Western analysis using NOT1, CCR4, and CAF1 antibodies, as indicated. Five μL of the CCR4-NOT complex was used as input (lane 4) and 50 μL was used to bind the glutathione beads (lanes 5 and 6).
B. PUF3 interacts with CCR4-NOT proteins in crude extracts. Top panel- Flag-PUF3 and Flag-PUF3C were purified by Flag-bead chromatography. PUF3 proteins were detected with Flag antibody. Lane 1- yeast strain EGY191 in which a 6His-Flag-PUF3 variant was not expressed; lane 2- strain EGY191 with 6His-Flag-PUF3; lane 3- strain EGY191 with 6His-Flag-PUF3C, containing residues 466–589 of PUF3.
Lower panel- Following Flag purification of PUF3 proteins, Western analysis was conducted with antibodies directed against each of the CCR4-NOT proteins as indicated (41). Lanes 1 to 3- CCR4-NOT proteins in crude extract (Cr Ex); Lanes 4 to 6- CCR4-NOT proteins detected in immunoprecipitations with FLAG antibody; Lanes 7 to 9 are duplicative of lanes 4 to 6 except the exposure is increased. Control, PUF3, and PUF3C are as indicated in section A, above.
DISCUSSION
In this study we addressed the mechanisms by which PUF3 accelerates the deadenylation process. Two general models were examined: 1) retention of the deadenylases at the mRNA through PUF3 binding to the mRNA and 2) effects on the mRNP structure. We provide evidence that PUF3 may act by both mechanisms. We showed that PUF3 is capable of immunoprecipitating multiple components of the CCR4-NOT complex, consistent with its ability to retain CCR4 at the mRNA. Since it has been previously shown that PUF5 is able to physically interact with CAF1 of the CCR4-NOT complex and to accelerate deadenylation by binding of CCR4, it is likely that PUF3 can act by a similar mechanism (18).
However, several observations support the model that PUF3 is also affecting the mRNP structure. We showed that PUF3 was responsible for accelerating the rate of deadenylation when PAN2 was the only deadenylase, as no PUF3-induced deadenylase appears to exist in yeast (6). While it is possible that PUF3 retains both CCR4 and PAN2 at the mRNA, either separately or together in a larger complex, no specific physical interaction was observed between PUF3 and PAN2 and a number of studies have demonstrated no interactions between PAN2 and the CCR4-NOT complex (42–45; unpublished observations). An alternative view is that since a pan2 defect can block mRNA transport out of the nucleus in 50% of the cells observed (46), the combination of a ccr4 deletion and this transport defect is the cause for the lack of COX17 mRNA deadenylation in the ccr4 pan2 strain background. However, this possibility also seems unlikely in that combining a defect that blocks nuclear transport by 30% (as in a PAB1 RRM4 deletion) (40) with that of a ccr4 deletion has no additive effect over that of ccr4 alone on deadenylation (25; unpubl. results). The simpler model is that PUF3 acts to affect a step common to both the CCR4 and PAN2 deadenylation processes. Such a common point of action could be affecting the mRNP structure that is known to influence mRNA deadenylation rates (12,25,38). A defect in eIF4E also allowed increased deadenylation by either CCR4 or PAN2, suggesting that PUF3 in controlling deadenylation is acting either similarly to eIF4E or at a step affected by eIF4E. Because in a puf3 deletion background, COX17 mRNA deadenylation remained slow even in the presence of a defect in eIF4E, PUF3 action at COX17 appears to be downstream of eIF4E. We believe that the site of PUF3 function is in the vicinity of PAB1 in that the RRM1 domain of PAB1 was required for PUF3 action at the COX17 mRNA. Since other domains of PAB1 were not similarly required for PUF3 function, these results are consistent with the model that rearrangements of the PAB1-mRNP structure, as dictated by RRM1 of PAB1, mediate the ability of PUF3 to enhance deadenylation.
The closed loop structure, involving eIF4E, eIF4G and PAB1 contacts linking the 5′ cap to the poly(A) tail, could be one site of action for PUF3. However, we do not believe it is simply the closed loop structure that PUF3 interferes with, thereby accelerating deadenylation. Because removal of the P domain of PAB1 substantially blocked the enhanced deadenylation caused by a defect in eIF4E (cdc33-1 allele) but not the deadenylation enhanced by PUF3, it appears that PUF3 does not act in the same manner as a defect in eIF4E. Therefore, while such a relaxation of closed loop contacts would be expected to accelerate deadenylation, as previously observed (38), PUF3 does not appear to act solely by this method.
In contrast, because of the differential effects of removing the RRM1 and P domains on PUF3 action, it appears that PUF3 acts redundantly with the P domain of PAB1. This redundant action may be at a step involving rearrangement of the PAB1 mRNP structure. We have previously established that the RRM1 and P domains of PAB1 are critical to deadenylation of several mRNA (25), and the rearrangement of the PAB1-mRNP appears to represent a key step in allowing CCR4 deadenylation (24,25). More recent data has indicated that removal of the RRM1 domain of PAB1 alters the PAB1-mRNP structure (47). Most critically, this effect is of such a nature that addition of exogenous PAB1 could not rectify the resultant structure (47), perhaps because PAB1-ΔRRM1 is unable to dissociate from the mRNA (25). These results imply a PAB1-ΔRRM1 mRNP configuration that is resistant to rearrangement and, therefore, understandably, resistant to deadenylation.
While PAB1-ΔP generally blocks non-PUF3-controlled deadenylation as severely as does PAB1-ΔRRM1 (Figure 4B; 25), PUF3 is able to specifically bypass this P domain defect. The PAB1-ΔP-mRNP structure must be of a significantly different nature than that of the PAB1-ΔRRM1 mRNP structure to allow PUF3 to allow deadenylation. These results suggest that PUF3 acts in a manner similar to that or redundant with the PAB1 P domain in terms of effects on mRNP structure and deadenylation.
Another possible model for PUF3 effects on both CCR4 and PAN2 deadenylation would be that PUF3 binds the CCR4-NOT complex and then components of this complex, even in the absence of CCR4 enzyme activity, bind additional factors that rearrange the PAB1-mRNP structure and augment PAN2-induced poly(A) removal. However, two published experiments do not support this model. First, it has been shown that recruitment of CAF1 and the corresponding CCR4-NOT complex to an mRNA 3′UTR in vivo greatly accelerates deadenylation in a manner similar to the PUF3 augmentation of deadenylation at COX17 (23). This increased deadenylation is completely dependent on CCR4 enzyme activity, indicating that simply recruiting the CCR4-NOT complex is insufficient to rearrange the PAB1-mRNP and allow PAN2 deadenylase function to be augmented. That is, CCR4-NOT recruitment either by itself or in conjunction with some other protein bound to the complex cannot simply augment deadenylation without CCR4 enzyme activity being present. Second, it has been shown in vitro that CCR4-NOT deadenylation in the presence of only PAB1 bound to the mRNA requires the P domain of PAB1 (24). The CCR4-NOT complex by itself is unable to rearrange the PAB1-mRNA structure without the P domain of PAB1. In contrast, PUF3 in vivo can accelerate deadenylation when the P domain of PAB1 is removed. Therefore, whatever effect PUF3 is having in vivo, it is not simply that of delivering CCR4-NOT to the mRNA. PUF3 presence is required for some interaction, whether it be the rearrangement of the PAB1-mRNP structure or the binding of some other protein to the RNA that in turn affects the PAB1-mRNP structure.
Our results also indicate that the enhancement of both deadenylation and decapping by PUF3 and the eIF4E defect are mechanistically linked. This conclusion is supported by the observation that in every situation that deadenylation was accelerated or blocked, decapping was correspondingly accelerated or blocked. The PAB1-mRNP structure appears to be the critical factor in controlling both these processes.
We additionally observed that PUF3 affected the initial default length of the COX17 mRNA poly(A) tails: removing PUF3 shortened this default length. We ascribe this phenomenon to PUF3 binding the mRNA, as a similar effect on the default length of GAL1 poly(A) tails was not observed. PUF3, through its mRNA contacts, could be either required for proper PAN2 trimming of the poly(A) tail or for proper polyadenylation by the poly(A) polymerase. Although the poly(A) trimming process has not been extensively studied for individual mRNA, it is quite possible that the wide array of RNA binding proteins in the cell, specific to different subsets of mRNA, are critical in regulating this process and causing the well known differences in this default length. Since poly(A) tail lengths play such an important role in PAB1 binding and ultimately affect the rates of mRNA degradation and protein translation, it can be envisaged that specific RNA binding proteins are involved more globally than previously considered to control these lengths both at the first step of forming the mature mRNA and at the later more commonly analyzed mRNA metabolic steps of translation and deadenylation. Close scrutiny of in vivo deadenylation analysis of HO mRNA poly(A) tail lengths indicates that puf5Δ also results in significantly shorter default poly(A) tail lengths for HO mRNA (18).
MATERIALS AND METHODS
Yeast Strains
Saccharomyces cerevisiae strains are listed in Table 1. Yeast strains carrying pAS77 [PAB1-CEN-URA3]) (25) were used for transforming PAB1 variants expressed under their own promoter: plasmid YC504 (pRS314: PAB1-CEN-TRP1), plasmid YC505 (contains PAB1-ΔRRM1), and plasmid YC510 (contains PAB1-ΔP) (25). Plasmid AS77 was subsequently lost from each strain following selection on plates containing 5-fluororotic acid (25). Plasmid RP1007 [GAL10-COX17-LEU2], following transformation into yeast, was used for quantitating rates of deadenylation for COX17 (34). The 6His-Flag-PUF3 plasmid and the 6His-Flag-PUF3C (amino acids 466-589 of PUF3) were expressed from a PGK1 promoter on plasmids YC430 (TRP1) and YC406 (TRP1), respectively.
Table 1.
Yeast strains.
| RP840 | Mata leu2 his4 trp1 ura3 cup1::LEU2/PGK1pG/MFA2pG |
| RP840-1a | Isogenic to RP840 except ccr4::URA3 |
| RP1241 | Isogenic to RP840 except puf3::Neo |
| RP1241-1a | Isogenic to RP1241 except puf3::Neo ccr4::URA3 |
| YAS319/YC504 | Matα ura3 his3 trp1 leu2 pab1::HIS3 YC504 [PAB1-CEN-TRP1] |
| YAS1881/YC504 | Isogenic to AS319 except cdc33-1 |
| 1738-3/YC504 | Matα ura3 his3 trp1 leu2 pab1::HIS3 puf3::Neo YC504 [PAB1-CEN-TRP1] |
| 1777-6/YC504 | Mata ura3 his3 trp1 leu2 pab1::HIS3 puf3::Neo cdc33-1 YC504 [PAB1-CEN-TRP1] |
| YAS1881-1a/YC504 | Isogenic to AS1881 except ccr4::URA3 |
| YAS319-1a-uN/YC504 | Isogenic to YAS319 except ccr4::ura3::Neo |
| EGY191 | Matα ura3 his3 trp1LexA-leu2 |
| RP1619 | Isogenic to RP840 except his4-539 pan2::URA3 |
Protein analyses
In vivo immunoprecipitations were conducted as previously described (36). RNase A (0.1 mg/mL) treatment of extracts was conducted for 30 min prior to the immunoprecipitation. In vitro binding analyses with GST-PUF3 were conducted essentially as described (37). Briefly, GST and GST-PUF3, both expressed from plasmid pGEX-KG, were bound to glutathione agarose beads. Fifty μL of the Flag-CCR4-NOT complex, isolated as described following purification on Flag beads (5), was incubated with 15 μL of glutathione agarose beads containing the designated GST protein. Following four washes with the CCR4-NOT extraction buffer (50 mM Tris, pH 7.9, 150 mM NaCl, 0.1% NP40, and 1 mM MgCl2) and two washes with the same buffer containing 1 M NaCl, the bound proteins were eluted by boiling.
RNA analyses
Pulse-chase analyses for GAL1 and COX17 mRNA were conducted as previously described (2,8,36). Following growth of cells on non-inducing medium containing 2% raffinose, the GAL-controlled mRNA were induced for 10–20 min with 2% galactose and mRNA expression was shut off with 4% glucose. At the indicated time points RNA was isolated and subjected to Northern analysis. Experiments utilizing the cdc33-1 temperature sensitive allele were conducted following shifting of cells to 37°C for one hour prior to the pulse-chase experiment (done at the same temperature). SCR1 mRNA levels were used as a control for mRNA levels (as determined following initial A260 quantitation) in the analysis of COX17 mRNA levels as described (18). All pulse-chase experiments were conducted at least in triplicate.
Acknowledgments
We would like to thank Roy Parker and Wendy Olivas for providing strains and plasmids used in this study. This research was supported by Hatch grant H291 and by NIH grant GM78078. This is Scientific Contribution Number 2416 from the New Hampshire Agricultural Experiment Station.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Brown CE, Sachs AB. Poly(A) tail length control in Saccharomyces cerevisiae occurs by message-specific deadenylation. Mol Cell Biol. 1998;18:6548–59. doi: 10.1128/mcb.18.11.6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tucker M, Valencia-Sanchez MA, Staples RR, Chen J, Denis CL, Parker R. The transcription factor associated proteins, Ccr4p and Caf1p, are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell. 2001;104:377–386. doi: 10.1016/s0092-8674(01)00225-2. [DOI] [PubMed] [Google Scholar]
- 3.Hoshino S, Imai M, Kobayashi T, Uchida N, Katada T. The eukaryotic polypeptide chain releasing factor (eRF3/GSPT) carrying the translation termination signal to the 3′-poly(A) tail of mRNA. Direct association of erf3/GSPT with polyadenylate-binding protein. J Biol Chem. 1999;274:16677–16680. doi: 10.1074/jbc.274.24.16677. [DOI] [PubMed] [Google Scholar]
- 4.Hosoda N, Kobayashi, Uchida N, Funakoshi Y, Kikuchi Y, Hoshino S, Katada T. Translation termination factor eRF3 mediates mRNA decay through the regulation of deadenylation. J Biol Chem. 2003;278:38287–38291. doi: 10.1074/jbc.C300300200. [DOI] [PubMed] [Google Scholar]
- 5.Chen J, Chiang YC, Denis CL. CCR4, a 3′-5′ poly (A) RNA and ssDNA exonuclease, is the catalytic component of the cytoplasmic deadenylase. EMBO J. 2002;21:1414–1426. doi: 10.1093/emboj/21.6.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tucker M, Staples RR, Valencia-Sanchez MA, Muhlrad D, Parker R. CCR4p is the catalytic sub-unit of Ccr4p/Pop2p/Notp mRNA deadenylase complex in Saccharomyces cerevisiae. EMBO J. 2002;21:1427–1436. doi: 10.1093/emboj/21.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Decker CJ, Parker R. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev. 1993;7:1632–1643. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- 8.Viswanathan P, Ohn T, Chiang YC, Chen J, Denis CL. Mouse CAF1 can function as a processive deadenylase 3′-5′-exonuclease in vitro but in yeast the deadenylase function of CAF1 is not required for mRNA poly(A) removal. J Biol Chem. 2004;279:23988–23995. doi: 10.1074/jbc.M402803200. [DOI] [PubMed] [Google Scholar]
- 9.Tarun SZ, Jr, Sachs AB. Association of the yeast poly(A) tail binding protein with translation initiation factor elF-4G. EMBO J. 1996;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- 10.Chowdhury A, Mukhopadhyay J, Tharun S. The decapping activator Lsm1p-7p-Pat1p complex has the intrinsic ability to distinguish between oligoadenylated and polyadenylated RNAs. RNA. 2007;13:998–1016. doi: 10.1261/rna.502507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tharun S, Parker R. Targeting an mRNA for decapping: displacement of translation factors and association of the Lsm1p-7p complex on deadenylated yeast mRNAs Mol. Cell. 2001;8:1075–1083. doi: 10.1016/s1097-2765(01)00395-1. [DOI] [PubMed] [Google Scholar]
- 12.Caponigro G, Parker R. Multiple functions for the poly(A)-binding protein in mRNA decapping and deadenylation in yeast. Genes Dev. 1995;9:2421–2432. doi: 10.1101/gad.9.19.2421. [DOI] [PubMed] [Google Scholar]
- 13.Coller J, Parker R. Eukaryotic mRNA decapping. Annu Rev Biochem. 2004;73:861–890. doi: 10.1146/annurev.biochem.73.011303.074032. [DOI] [PubMed] [Google Scholar]
- 14.Tucker M, Parker R. Mechanisms and control of mRNA decapping in Saccharomyces cerevisiae. Annu Rev Biochem. 2000;69:571–595. doi: 10.1146/annurev.biochem.69.1.571. [DOI] [PubMed] [Google Scholar]
- 15.Muhlrad D, Decker CJ, Parker R. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′-3′ digestion of the transcript. Genes Dev. 1994;8:855–866. doi: 10.1101/gad.8.7.855. [DOI] [PubMed] [Google Scholar]
- 16.Sheth U, Parker R. P bodies and the control of mRNA translation and degradaton. Mol Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 17.Chicoine J, Benoit P, Gamberi C, Paliouras M, Simonelig M, Lasko Pl. Bicaudal-C recruits CCR4-NOT deadenylase to target mRNAs and regulates oogenesis, cytoskelatal organization, and its own expression. Dev Cell. 2007;13:691–704. doi: 10.1016/j.devcel.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 18.Goldstrohm AC, Hook BA, Seay DJ, Wickens M. PUF proteins bind Pop2p to regulate messenger RNAs. Nat Struct Mol Biol. 2006;13:533–539. doi: 10.1038/nsmb1100. [DOI] [PubMed] [Google Scholar]
- 19.Goldstrohm AC, Seay DJ, Hook BA, Wickens M. PUF protein-mediated deadenylation is catalyzed by Ccr4p. J Biol Chem. 2007;282:109–114. doi: 10.1074/jbc.M609413200. [DOI] [PubMed] [Google Scholar]
- 20.Lykke-Andersen J, Wagner E. Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains tin the proteins TTP and BRF-1. Genes Dev. 2005;19:351–361. doi: 10.1101/gad.1282305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rendl LM, Bieman MA, Smibert CA. S. cerevisiae Vts1p induces deadenylation-dependent transcript degradation and interacts with Ccr4-Pop2p-Not deadenylase complex. RNA. 2008;14:1–9. doi: 10.1261/rna.955508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Semotok JL, Cooperstuck RL, Pinder BD, Vari HK, Lipshitz HD, Smibert CA. Smaug recruits the CCR4/POP2/NOT deadenylase complex to trigger maternal transcript localization in the early Drosophila embryo. Curr Biol. 2005;15:284–294. doi: 10.1016/j.cub.2005.01.048. [DOI] [PubMed] [Google Scholar]
- 23.Finoux AL, Seraphin B. In vivo targeting of the yeast Pop2 deadenylase subunit to reporter transcripts induces their rapid degradation and generates new decay intermediates. J Biol Chem. 2005;281:25940–25947. doi: 10.1074/jbc.M600132200. [DOI] [PubMed] [Google Scholar]
- 24.Simon E, Seraphin B. A specific role for the C-terminal region of the poly(A)-binding protein in mRNA decay. Nucl Acids Res. 2007;35:6017–6028. doi: 10.1093/nar/gkm452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yao G, Chiang YC, Zhang C, Lee D, Laue TM, Denis CL. PAB1 self-association precludes its binding to poly (A), thereby accelerating CCR4 deadenylation in vivo. Mol Cell Biol. 2007;27:6243–6253. doi: 10.1128/MCB.00734-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buchan JR, Muhlrad D, Parker R. P bodies promote stress granule assembly in Saccharomyces cerevisiae. J Cell Biol. 2008;183:441–455. doi: 10.1083/jcb.200807043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoyle NP, Castelli LM, Campbell SG, Holmes LEA, Ashe MP. Stress-dependent relocalization of translationally primed mRNPs to cytoplasmic granules that are kinetically and spatially distinct from P-bodies. J Cell Biol. 2007;179:65–74. doi: 10.1083/jcb.200707010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morrissey JP, Deardorff JA, Hebron C, Sachs AB. Decapping of stabilized, polyadenylated mRNA in yeast pab1 mutants. Yeast. 1999;15:687–702. doi: 10.1002/(SICI)1097-0061(19990615)15:8<687::AID-YEA412>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 29.Spassov DS, Jurecic R. The PUF family of RNA-binding proteins: does evolutionarily conserved structure equal conserved function? IUBMB Life. 2003;55:359–366. doi: 10.1080/15216540310001603093. [DOI] [PubMed] [Google Scholar]
- 30.Lin H, Spradling AC. A novel group of pumilio mutations affects the asymetmetric division of germline stgem cells in the Drosophila ovary. Development. 1997;124:2463–2476. doi: 10.1242/dev.124.12.2463. [DOI] [PubMed] [Google Scholar]
- 31.Crittenden SL, Bernstein DS, Bachorik JL, Thompson BE, Gallegos M, Petcherski AG, Moulder G, Barstead R, Wickens M, Kimble J. A conserved RNA-binding protein controls germline stem cells in Caenorhabditis elegans. Nature. 2002;417:660–663. doi: 10.1038/nature754. [DOI] [PubMed] [Google Scholar]
- 32.Nakahata S, Kotani T, Mita K, Kawasaki T, Katsu Y, Nagahama Y, Yamashita M. Involvement of Xenopus Pumilio in the translational regulation that is specific to cyclin B1 mRNA during oocyte maturation. Mech Dev. 2003;120:865–880. doi: 10.1016/s0925-4773(03)00160-6. [DOI] [PubMed] [Google Scholar]
- 33.Jackson JS, jr, Houshmandi SS, leban FL, Olivas WM. Recruitment of the Puf3 protein to its mRNA target for regulation of mRNA decay in yeast. RNA. 2004;10:1625–1636. doi: 10.1261/rna.7270204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olivas W, Parker R. The Puf3 protein is a transcript-specific regulator of mRNA degradation in yeast. EMBO J. 2000;19:6602–661. doi: 10.1093/emboj/19.23.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tadauchi T, Matsumoto K, Herskowitz I, Irie K. Post-transcriptional regulation through the HO 3′-UTR by Mpt5, a yeast homolog of Pumilio and FBF. EMBO. 2001;20:552–561. doi: 10.1093/emboj/20.3.552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ohn T, Chiang YC, Lee DJ, Yao G, Zhang C, Denis CL. CAF1 plays an important role in mRNA deadenylation separate from its contact to CCR4. Nucl Acids Res. 2007;35:3002–3015. doi: 10.1093/nar/gkm196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chiang YC, Komarnitsky P, Chase D, Denis CL. ADR1 activation domains contact the histone acetyltransferase GCN5 and the core transcriptional factor TFIIB. J Biol Chem. 1996;271:32359–32365. doi: 10.1074/jbc.271.50.32359. [DOI] [PubMed] [Google Scholar]
- 38.Schwartz DC, Parker R. Mutations in translation initiation factors lead to increased rates of deadenylation and decapping of mRNAs in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:5247–5256. doi: 10.1128/mcb.19.8.5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Altmann M, Schmitz N, Berset C, Trachsel H. A novel inhibitor of cap-dependent translation initiation in yeast: p20 competes with elF4G for binding to elF4E. EMBO J. 1997;16:1114–1121. doi: 10.1093/emboj/16.5.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brune C, Munchel SE, Fischer N, Podtelejnikov AV, Weis K. Yeast poly(A)-binding protein Pab1 shuttles between nucleus and the cytoplasm and functions in mRNA export. RNA. 2005;11:517–531. doi: 10.1261/rna.7291205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Viswanathan P, Chen J, Chiang YC, Denis CL. Identification of multiple RNA features that influence CCR4 deadenylation activity. J Biol Chem. 2003;278:14949–14955. doi: 10.1074/jbc.M211794200. [DOI] [PubMed] [Google Scholar]
- 42.Chen J, Rappsilber J, Chiang YC, Russell P, Mann M, Denis CL. Purification and characterization of the 1.0 MDa CCR4-NOT complex identifies two novel components of the complex. J Mol Biol. 2001;314:683–694. doi: 10.1006/jmbi.2001.5162. [DOI] [PubMed] [Google Scholar]
- 43.Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, Remor M, Hofert C, Schelder M, Brajenovic M, Ruffner H, Merino A, Klein K, Hudak M, Dickson D, Ho Yuen, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutiller K, Yang L, Wolting C, Donaldson I, Schandorff S, Shwearane J, Vo M, Taggart J, Goudreault M, Muskat B, Alfarano C, Dewar D, Lin A, Michalickov K, Willams AR, Sassi H, Nielsen PA, Rasmussen KJ, Andersen JR, Johansen LE, Hansen LH, Jespersen H, Podtelejnikov A, Nielsen E, Crawford J, Poulsen V, Sorensen BD, Matthiesen J, Hendrickson RC, Gleeson F, Pawson T, Moran MF, Durocher D, Mann M, Hogue CWV, Figeys D, Tyers M. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 44.Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutlier K, Yang L, Wolting C, Donaldson I, Schandorff S, Shewnarane J, Vo M, Taggart J, Goudreault M, Muskat B, Alfarano C, Dewar D, Lin Z, Michalickova K, Williams AR, Sassi H, Nielsen PA, Rasmussen KJ, Andersen JR, Johansen LE, Hansen LH, Jespersen H, Podtelejnikov A, Nielsen E, Crawford J, Poulsen V, Sorensen B, Matthiesen J, Hendrickson RC, Gleeson F, Pawson T, Moran MF, Durocher D, Mann M, Hogue CWV, Figeys D, Tyers M. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 45.Liu HY, Badarinarayana V, Audino DC, Rappsilber J, Mann M, Denis CL. The NOT proteins are part of the CCR4 transcriptional complex and affect gene expression both positively and negatively. EMBO J. 1998;17:1096–1106. doi: 10.1093/emboj/17.4.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dunn EF, Hammell CM, Hodge CA, Cole CN. Yeast poly(A)-binding protein, Pab1, and PAN, a poly(A) nuclease complex recruited by Pab1, connect mRNA biogenesis to export. Genes Dev. 2005;19:90–103. doi: 10.1101/gad.1267005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Amrani N, Ghosh S, Mangus DA, Jacobson A. Translation factors promote the formation of two states of the closed-loop mRNP. Nature. 2008;453:1276–1280. doi: 10.1038/nature06974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Denis CL, Gallo C. Constitutive RNA synthesis for the yeast activator ADR1 and identification of the ADR1-5c mutation: implications in post-translational control of ADR1. Mol Cell Biol. 1986;6:4026–4030. doi: 10.1128/mcb.6.11.4026. [DOI] [PMC free article] [PubMed] [Google Scholar]