Abstract
Dysregulated protein phosphorylation is a hallmark of malignant transformation. Transformation can generate major histocompatibility complex (MHC)-bound phosphopeptides that are differentially displayed on tumor cells for specific recognition by T cells. To understand how phosphorylation alters the antigenic identity of self peptides and how MHC class II molecules present phosphopeptides for CD4+ T cell recognition, we determined the crystal structure of a phosphopeptide derived from melanoma antigen recognized by T cells-1 (pMART-1), selectively expressed by human melanomas, in complex with HLA-DR1. The structure revealed that the phosphate moiety attached to the serine residue at position P5 of pMART-1 is available for direct interactions with T cell receptor (TCR), and that the peptide N-terminus adopts an unusual conformation orienting it toward TCR. This structure, combined with measurements of peptide affinity for HLA-DR1 and of peptide–MHC recognition by pMART-1-specific T cells, suggests that TCR recognition is focused on the N-terminal portion of pMART-1. This recognition mode appears to be distinct from that of foreign antigen complexes but is remarkably reminiscent of the way autoreactive TCRs engage self or altered self peptides, consistent with the tolerogenic nature of tumor–host immune interactions.
Keywords: crystal structure, phosphopeptide, MHC class II, T cell receptor, melanoma, tumor antigen
Introduction
A variety of post-translational modifications of naturally processed peptides displayed by major histocompatibility complex (MHC) class I or II molecules have now been described, including glycosylation, deamidation, cysteinylation and phosphorylation.1 Furthermore, peptides bearing these post-translational modifications can be discriminated from their unmodified homologs by T cells. Among the post-translationally modified peptides identified to date, phosphopeptides2-4 are of particular interest since dysregulated phosphorylation is one of the hallmarks of malignant transformation and contributes directly to oncogenic signaling cascades involved in cell growth, differentiation and survival.5,6 Indeed, phosphopeptides directly isolated from the human MHC class I molecule HLA-A2 include those derived from proteins involved in oncogenic signaling and cell cycle regulation.2,3 Because many of these phosphopeptides are differentially displayed on cancer cells,3 they provide a new cohort of targets for cancer immunotherapy.1,7
Until very recently, it was unknown whether MHC class II molecules could present phosphopeptides for specific recognition by CD4+ T cells, whose activation and recruitment are critical for the development of effective and long-lasting anti-tumor immunity.8,9 In one study, mass spectrometric sequencing was used to demonstrate the existence of HLA-DR-associated phosphopeptides on two pairs of autologous human melanoma and Epstein-Barr virus (EBV)-transformed B lymphoblastoid lines.4 The 150 unique phosphopeptides identified derived from 53 different source proteins representing all cellular compartments. As characteristic of nonphosphorylated MHC class II-restricted epitopes, most of the phosphopeptides occurred in nested sets, and their average length was 16 amino acids (range 8–28). Significantly, the majority of source proteins support vital cellular functions, such as metabolism, cell cycle regulation and signal transduction.4 Similar results were reported in a separate study of human MHC class II-restricted phosphopeptides derived from one melanoma and one B lymphoblastoid cell line,10 suggesting the generality of phosphopeptide presentation by MHC class II molecules.
The ability of human CD4+ T cells to specifically recognize MHC class II-restricted phosphopeptides was first demonstrated using as an example an HLA-DR1-restricted phospho-MART-1 (melanoma antigen recognized by T cells-1; also known as Melan-A) peptide (pMART-1100–111; APPAYEKLpSAEQ, where pS is phosphoserine) that was isolated from a cultured melanoma line.4 MART-1 is of special interest because its selective expression by cells of the melanocytic lineage has made it a prime target for immunotherapeutic approaches to melanoma, including vaccines and adoptive T cell transfer.11,12 CD4+ T cells recognizing pMART-1 presented by HLA-DR1 were highly specific for the phosphate moiety of the peptide. Importantly, these T cells recognized intact melanoma cells expressing MART-1 and HLA-DR1, indicating the presence of sufficient quantities of pMART-1 peptide–MHC complexes at the cell surface to trigger T cell signaling.
Establishing the molecular basis for phosphopeptide presentation and recognition will enable the rational design of new cancer immunotherapies targeting this category of tumor-derived epitopes. Recent crystal structures of several phosphopeptide–HLA-A2 complexes showed that the phosphate moiety formed an integral part of these structures, stabilizing interactions with the MHC class I molecule.13,14 Here we report the first structure of a phosphopeptide–MHC class II complex, involving pMART-1100–114 bound to HLA-DR1. This structure, in conjunction with measurements of peptide–MHC affinity and T cell recognition of truncated and substituted pMART-1 peptides, reveals the basis for presentation of tumor-associated MHC class II-restricted phosphopeptides to CD4+ T cells.
Results and Discussion
Structure of phosphorylated MART-1 peptide bound to HLA-DR1
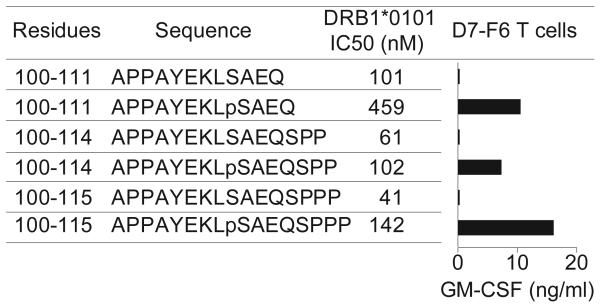
To understand how phosphorylation influences antigenic identity, we determined the crystal structure of pMART-1100–114 (APPAYEKLpSAEQSPP) in complex with HLA-DR1 to 2.1 Å resolution (Table 1; Fig. 1a). This peptide belongs to a nested set of four phosphopeptides derived from the C-terminus of MART-1 that range in length from 12 to 17 residues, all starting at Ala100. The nested peptides were eluted from HLA-DR-peptide complexes isolated from a human melanoma line expressing a single DR molecule (HLA-DRβ1*0101), thereby assuring unambiguous HLA allele restriction, as well as from a second melanoma line expressing HLADRβ1*0101, β1*0404 and β4*0103.4 Phospho-MART-1 peptides were not found on autologous EBV-transformed B cells, consistent with the selective expression of MART-1 in melanocytic lineage cells. The choice of pMART-1100–114 for structural studies was based on its higher affinity for HLA-DR1 (IC50 = 102 nM) compared to three other naturally-processed pMART-1 peptides: pMART-1100–111 (459 nM), pMART-1100–115 (142 nM), and pMART-1100–116 (327 nM). This facilitated assembly of stable pMART-1–HLA-DR1 complexes for crystallization. The affinities of pMART-1 peptides for HLA-DR1 are considered to be in the intermediate range, as defined by the IC50 values of non-phosphorylated peptides for HLA-DR molecules.15
Table 1.
Data collection and refinement statistics
| pMART-1100-114–HLA-DR1 | |
|---|---|
| Data collection statistics | |
| Space group | P21212 |
| Unit cell (Å, °) | a = 91.2, b = 135.5, c = 40.9 |
| Resolution (Å) | 30–2.1 |
| Observations | 43,0714 |
| Unique reflections | 30,332 |
| Completeness (%)a | 100 (100) |
| Mean I/σ(I)a | 51.3 (5.4) |
| Rsym (%)a,b | 7.4 (46.0) |
| Refinement statistics | |
| Resolution range (Å) | 30–2.1 |
| Rwork (%)c | 21.1 |
| Rfree (%)c | 24.8 |
| Protein atoms | 3,134 |
| Water molecules | 198 |
| Average B values (Å2) | |
| Protein main chain | 34.3 |
| Protein side chain | 35.3 |
| Water molecules | 37.6 |
| R.m.s. deviations from ideality | |
| Bond lengths (Å) | 0.010 |
| Bond angles (°) | 1.58 |
| Ramachandran plot statistics | |
| Most favored (%) | 92.5 |
| Additionally allowed (%) | 7.2 |
| Generously allowed (%) | 0.0 |
| Disallowed | 0.3 |
Values in parentheses are statistics for the highest resolution shells.
Rsym = Σ|Ij – <I>|/ΣIj, where Ij is the intensity of an individual reflection and <I> is the average intensity of that reflection.
Rwork = Σ∥Fo| – |Fc∥/Σ|Fo|, where Fc is the calculated structure factor. Rfree is as for Rwork but calculated for a randomly selected 10.0% of reflections not included in the refinement.
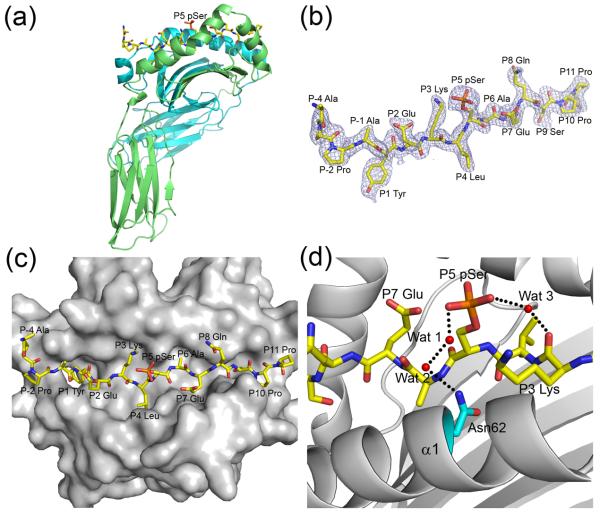
Figure 1.
Structure of the pMART-1100–114–HLA-DR1 complex. (a) Side view of the pMART-1100–114–HLA-DR1 complex. MHC α chain is cyan and β chain is green. The peptide is drawn in stick representation with carbon atoms in yellow, oxygen atoms in red, nitrogen atoms in blue, and the phosphate atom in orange. (b) Electron density for the bound pMART-1100–114 peptide. The 2Fo– Fc map at 2.1 Å resolution is contoured at σ. (c) Top view of the pMART-1100–114–HLA-DR1 complex. (d) Interactions of P5 pSer with P3 Lys and the HLA-DR1 α chain. Bound water molecules (Wat 1–3) are drawn as red spheres. Hydrogen bonds are indicated by broken black lines.
The pMART-1–HLA-DR1 complex was assembled in two steps in order to maximize the yield of recombinant protein. First, HLA-DR1 bearing CLIP87–101 peptide (PVSKMRMATPLIMQA) was prepared by in vitro folding from bacterial inclusion bodies. Second, pMART-1100–114 phosphopeptide (APPAYEKLpSAEQSPP) was loaded into CLIP–HLA-DR1 using the peptide-exchange catalyst HLA-DM. Briefly, the extracellular portions of the HLA-DR1 α and β chains (residues 1–181 and 1–192, respectively) were expressed separately as inclusion bodies in Escherichia coli BL21(DE3) cells (Novagen). Inclusion bodies were dissolved in 8 M urea, 50 mM Tris-HCl (pH 8.0), and 10 mM DTT, followed by purification on a Poros HQ20 anion exchange column (Perspective Biosystems) in 50 mM Tris-HCl, 8 M urea, and 1 mM DTT at pH 8.0 (DRα) or pH 8.5 (DRβ), using a linear NaCl gradient.26 For in vitro folding, the purified subunits were diluted to a final concentration of 40 μg/ml each in a folding solution containing 50 mM Tris-HCl, 30% (w/v) glycerol, 0.5 mM EDTA, 3 mM reduced glutathione, and 0.9 mM oxidized glutathione (pH 8.0). CLIP peptide (GenScript) was added to a final concentration of 5 μM, and the folding mixture was kept for two weeks at 4 °C. The final folding solution was concentrated and dialyzed against 50 mM Mes (pH 6.0). Purification was carried out with sequential Superdex S-200 Mono Q FPLC columns (GE Healthcare). The CLIP–HLA-DR1 complex was concentrated to 0.8 mg/ml and loaded with pMART-1100–114 (Pi Proteomics) by overnight incubation at 37 °C in 100 mM sodium citrate-HCl (pH 5.8) containing 200 μM phosphopeptide and 0.2 mg/ml soluble HLA-DM. The HLA-DR1–pMART-1 complex was crystallized at room temperature in hanging drops by mixing equal volumes of the protein solution at 5 mg/ml and a reservoir solution of 20% (w/v) PEG 8000 and 0.1 M sodium cacodylate (pH 6.5). For data collection, crystals were transferred to a cryoprotectant solution (mother liquor containing 30% (w/v) PEG 8000), prior to flash-cooling. X-ray diffraction data to 2.1 Å resolution were recorded at beamline X29 of the Brookhaven National Synchrotron Light Source with an ADSC Quantum-315 CCD detector. All data were indexed, integrated, and scaled with the program HKL2000.27 Data collection statistics are shown in Table 1. The structure was solved by molecular replacement with the program Phaser28 using MIG1–HLA-DR1 (PDB accession code 1T5W),29 with all the peptide residues truncated to alanine, as a search model. The DR1 molecule was located by automatic rotation and translation searches with Z-score of 40.0; initial rigid body refinement gave Rfree of 37.2% and Rwork of 37.7%. Refinement was carried out using CNS1.2,30 including iterative cycles of simulated annealing, positional refinement and B factor refinement, interspersed with model rebuilding into σA-weighted Fo– Fc and 2Fo– Fc electron density maps using XtalView.31 Stereochemical parameters were evaluated with PROCHECK.32 Refinement statistics are summarized in Table 1.
Continuous and unambiguous electron density was observed for the entire 15-residue pMART-1100–114 peptide, including the N- and C-termini and the phosphate moiety attached to Ser108 (Fig. 1b). The phosphopeptide adopts an extended conformation in the HLA-DR1 binding groove resembling that of non-phosphorylated peptides bound to MHC class II molecules (Fig. 1c). The primary anchor residues are Tyr104 (P1) and Ala109 (P6), in agreement with the requirement for an aromatic or large hydrophobic residue at P1 and a small uncharged residue at P6 for efficient binding to HLA-DR1.15,16 The secondary anchors are Leu107 (P4), Glu110 (P7), and Ser112 (P9). This binding register is likely to be maintained in nonphosphorylated counterparts of pMART-1100–114, since the side chain of the phosphorylated Ser108 residue at P5 projects into solvent, away from the peptide-binding groove of HLA-DR1 (Fig. 1a,c).
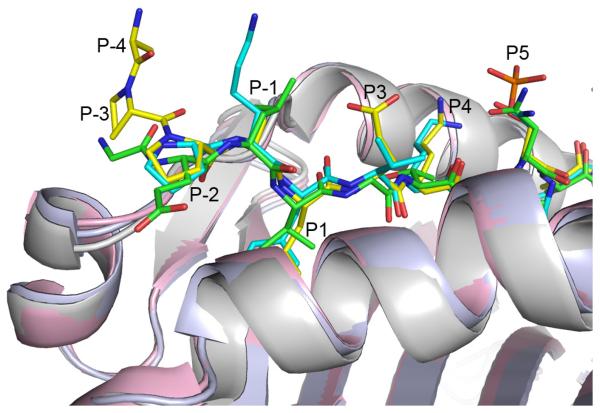
The conformation of pMART-1100–114 was directly compared with those of peptides from other peptide–MHC class II complexes16-20 by superposition of the α1/β1 domains of the class II molecules (Fig. 2). An unusual feature of the phospho-MART-1 peptide is that N-terminal residues P-3 and P-2, which lie outside the binding groove, are well ordered in the structure (Fig. 1b), despite the absence of crystal contacts to the N-terminus. In other peptide–MHC class II complexes, by contrast, these residues (when present) are generally not clearly defined in the electron density, indicating flexibility. Another notable feature of pMART-1 is that its main chain makes a nearly 90° turn at P-2 Pro102, causing P-4 Ala100 and P-3 Pro101 to point upward from the α1/β1 platform of HLA-DR1 (Fig. 2). This orientation suggests the possibility of TCR contacts to the ordered N-terminus of the peptide (Fig. 1b), as discussed later.
Figure 2.
Superposition of peptides bound to human MHC class II molecules. Phospho-MART-1 peptide bound to HLA-DR1 is yellow (carbon atoms). Hemagglutinin peptide bound to HLA-DR1 is cyan (PDB accession code 1DLH).16 Triose phosphate isomerase peptide bound to HLA-DR1 is green (1KLU).20 The peptide backbones superpose well from P-1 to P5, but diverge at P-2.
Phosphate-mediated interactions between phosphopeptide and HLA-DR1
No direct contacts were observed between the phosphate moiety of P5 pSer108 and the phospho-MART-1 peptide or HLA-DR1. However, P5 pSer108 is stabilized by ordered water molecules that form bridging hydrogen bonding interactions (Fig. 1d). In particular, the P5 pSer108 O2P atom forms a water-mediated hydrogen bond with the main chain oxygen atom of P3 Lys106. Similarly, the P5 pSer108 O1P atom interacts with the Nδ2 atom of HLA-DR1 Asn62α via two bridging water molecules. By contrast, the reported phosphopeptide–HLA-A2 complexes include multiple direct contacts between the phosphate group and the α1 and α2 helices of the MHC class I molecule, in addition to water-mediated interactions.13,14
Phosphorylated MART-1 peptides bind HLA-DR1 with 2–4-fold lower affinity than their non-phosphorylated counterparts (Fig. 3). This modest reduction may be attributed to electrostatic repulsion between the negatively charged phosphate group of P5 pSer108 and the nearby carboxylate group of P7 Glu110 (Fig. 1d). In phosphopeptide–HLA-A2 complexes, by comparison, phosphorylation was sometimes (but not always) found to increase affinity for MHC class I,13,14 suggesting that certain MHC class II-restricted phosphopeptides might also display phosphorylation-dependent binding to MHC.
Figure 3.
Specific recognition of phosphorylated versus non-phosphorylated MART-1 peptides by CD4+ T cells. The phospho-MART-1-specific T cell clone D7-F6 was co-incubated overnight with HLA-DR1+ 2048-EBV cells pre-pulsed with peptides (25 μM). GM-CSF secretion was measured by ELISA. Background GM-CSF secretion from T cells + APC + HA307-319 control peptide was <15 pg/ml. Results are representative of four separate experiments with D7-F6 and the parent T cell line D7 showing specific T cell recognition of phosphopeptides but not non-phosphorylated peptides, despite the comparatively lower MHC affinities (higher IC50 values) of the phosphorylated MART-1 peptides.
The oligoclonal pMART-1-specific CD4+ T cell line designated “D7” was raised by repetitive in vitro stimulation of peripheral blood mononuclear cells (PBMC) from a melanoma patient expressing HLA-DRβ1*0101. Briefly, T cells were grown under microculture conditions and stimulated every 10–14 days with irradiated autologous PBMC or HLA-DR1+ allogeneic EBV-B cells pulsed with pMART-1100-111 (APPAYEKLpSAEQ). Long-term CD4+ T cell cultures were maintained in RPMI 1640 + 10% heat-inactivated human AB serum, IL-2 120 IU/ml, and IL-7 and IL-15 at 25 ng/ml each. The T cell clone designated “D7-F6” was subcultured from D7 T cells under limiting dilution conditions in microtiter plates, and stimulated repetitively with pMART-1100-115 (APPAYEKLpSAEQSPPP). To assess T cell recognition of peptides, 1.2–5 × 104 T cells/well were co-cultured overnight in flat-bottom 96-well plates with 1 × 105 HLA-DR1+ EBV-B cells which had been pre-pulsed with peptides. Medium consisted of RPMI 1640 + 10% human AB serum, with IL-2 120 IU/ml. Culture supernatants were harvested, and GM-CSF and IFN-γ secretion by activated T cells was measured using commercially available ELISA kits (R&D Systems). Competition assays to quantitatively measure peptide binding to HLA-DRβ1*0101 were based on inhibition of binding of a high affinity radiolabeled peptide to purified MHC molecules.33 HLA class II molecules were purified from the EBV-transformed homozygous B lymphoblastoid cell line LG2. Peptide binding assays were performed by incubating purified human class II molecules (5–500 nM) with various concentrations of unlabeled peptide inhibitors and 0.1–1 nM 125I-radiolabeled probe peptide for 48 h in PBS containing 0.05–0.15% Nonidet P-40 and a protease inhibitor cocktail.15,33 MHC binding of the radiolabeled peptide (HA307–319; PKYVKQNTLKLAT) was determined by capturing peptide–MHC complexes on L243 (anti-HLA-DRA) antibody coated Lumitrac 600 plates (Greiner Bio-one) and measuring bound cpm. The concentration of peptide yielding 50% inhibition of the binding of the radiolabeled probe peptide (IC50) was then calculated. Peptides were typically tested at six different concentrations covering a 100,000-fold dose range, and in three or more independent assays. Under the conditions utilized, where [label] < [MHC] and IC50 ≥ [MHC], the measured IC50 values are reasonable approximations of the KD values.15
Recognition of phospho-MART-1 peptides by CD4+ T cells
The phosphate group of P5 pSer is fully exposed to solvent and therefore potentially available for direct interactions with TCR. Indeed, the phosphate is a critical determinant for TCR recognition, as demonstrated using a CD4+ T cell clone (D7-F6) specific for pMART-1 (Fig. 3). These cells secreted the cytokine granulocyte-macrophage colony-stimulating factor (GM-CSF) in response to phosphorylated MART-1 peptides pulsed onto DR1-expressing antigen-presenting cells (APCs), but not to their non-phosphorylated homologs.
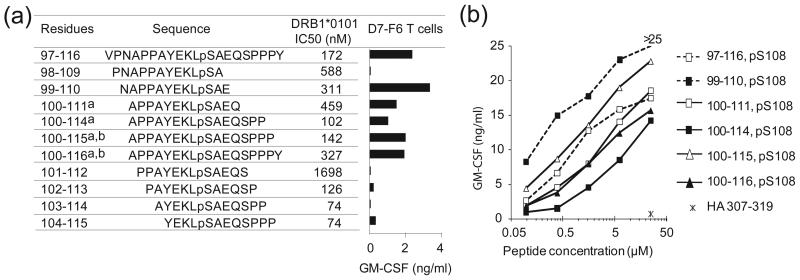
Having established that phosphorylation is essential for TCR recognition, we next examined recognition of a series of synthetic overlapping phospho-MART-1 peptides, in order to identify other determinants of T cell reactivity (Fig. 4a,b). Similar to pMART-1100–116, a series of naturally occurring C-terminally truncated phosphopeptides (pMART-1100–115, pMART-1100–114, pMART-1100–111) and the synthetic peptide pMART-199–110 were also capable of stimulating T cell reactivity, indicating that residues Gln111 (P8) through Tyr116 (P13) are not involved in critical interactions with TCR. In the pMART-1100–114–HLA-DR1 structure, P8 Gln111 and P11 Pro114 (like P5 pSer108) are ~70% accessible to solvent and therefore potentially available for contacts to TCR, whereas P9 Ser112 and P10 Pro113 are ~80% buried (P9 Ser112 is a secondary anchor residue) and unlikely to contact TCR directly. Indeed, removal of P9 Ser112 and P10 Pro113 (compare pMART-1100–114 with pMART-1100–111) resulted in a 4-fold loss of affinity for HLA-DR1 but a paradoxical increase in T cell activation (Fig. 4b). Residues P12 Pro115 and P13 Tyr116, although absent from the pMART-1100–114–HLA-DR1 structure, are expected to extend beyond the peptide-binding groove.
Figure 4.
Binding of phospho-MART-1 peptides to HLA-DR1 and recognition by CD4+ T cells. (a) Affinities of overlapping phosphorylated MART-1 peptides for HLA-DR1, as measured by 50% inhibitory concentrations (IC50), are shown. Synthetic phosphopeptides include four naturally expressed sequences isolated from cultured melanoma cells: a)1363-mel and b) 2048-mel. Bar graph demonstrates specific recognition of phospho-MART-1 peptides (15 μM) by the CD4+ T cell clone D7-F6. GM-CSF secretion from T cells was measured by ELISA; background secretion from T cells + APC + HA307-319 control peptide was <0.02 ng/ml. Similar results were obtained with IFNγ secretion, and using the parent T cell line D7. Results are representative of four separate experiments. (b) Recognition of titrated overlapping phospho-MART-1 peptides (0.1–25 μM) by the specific CD4+ T cell line D7. Background GM-CSF secretion from T cells + APC + HA307-319 control peptide (25 μM) was 0.7 ng/ml. Results are representative of three separate experiments. Similar results were observed with IFNγ secretion.
In sharp contrast to C-terminal residues P8 to P13, N-terminal residues P-4 to P7 contain important determinants for T cell recognition, besides the critical P5 pSer108 residue. Thus, removal of P7 Glu110 (compare pMART-199–110 with pMART-198–109) abrogated T cell stimulation, even though this truncation reduced affinity for HLA-DR1 by less than 2-fold (Fig. 4a). In the pMART-1100–114–HLA-DR1 structure (Fig. 1c), the side chain of P7 Glu110 projects from the surface of the MHC molecule and could interact with TCR. Removal of N-terminal residues P-4 Ala100, P-3 Pro101, P-2 Pro102, and P-1 Ala103 (hereafter referred to as the APPA motif) effectively abolished TCR recognition, as evident from comparing T cell stimulation by pMART-1100–114 versus pMART-1103–114, and pMART-1100–115 versus pMART-1104–115 (Fig. 4a). Since truncation of residues P-4 to P-1 did not reduce binding to HLA-DR1 (indeed, a significant increase in MHC binding affinity was noted with removal of P-4 Ala100), these four N-terminal residues must include structural determinants required for TCR recognition.
To assess the contribution of individual residues of the APPA motif to TCR recognition, P-4 Ala100 and P-1 Ala103 of pMART-1100–114 were each replaced by isoleucine, while P-3 Pro101 and P-2 Pro102 were replaced by alanine. Except for the replacement at P-4, these substitutions significantly reduced T cell stimulatory capacity compared to wild type pMART-1100–114, suggesting interactions with TCR (Supplementary Fig. 1). Most pronounced was the effect of substituting P-3 Pro101, which contributes directly to the unusual conformation of the APPA motif, whereby the peptide main chain makes a 90° turn at P-2 Pro102 directing N-terminal residues P-4 Ala100 and P-3 Pro101 towards the TCR (Fig. 2). In addition, although replacing P-4 Ala100 by the chemically similar isoleucine residue did not significantly affect T cell recognition, removal of P-4 Ala100 caused a total loss of T cell reactivity (Fig. 4a).
The marked focus on the N-terminal portion of pMART-1100–114 by TCR D7-F6 closely resembles peptide recognition by autoreactive TCRs specific for self antigens, including tumor antigens, presented by MHC class II molecules.21 In particular, X-ray crystallographic studies of autoimmune TCRs specific for myelin basic protein associated with multiple sclerosis,22,23 and of a tumor-specific TCR that recognizes a somatically mutated human melanoma antigen,24 have revealed substantial alterations in the topology of TCR binding to peptide–MHC compared to anti-foreign TCRs.21 In the self-reactive TCR–peptide–MHC complexes, the TCR is skewed toward the peptide N-terminus and the MHC class II β chain helix relative to its central position in anti-foreign TCR–peptide–MHC complexes, resulting in suboptimal TCR binding that may have enabled escape from negative thymic selection. The concentration on the N-terminal half of pMART-1 is therefore highly reminiscent of the way autoreactive TCRs engage self or altered self peptides.21
MART-1 protein plays an essential role in regulating mammalian pigmentation25 but was not known to be phosphorylated prior to the discovery of phospho-MART-1 peptides complexed with HLA-DR1.4 Tissue expression patterns of phospho-MART-1 protein versus its non-phosphorylated counterpart have yet to be determined. If MART-1 is expressed in the thymus and phosphorylation occurs after thymic development, then this post-translational modification will create a neo-epitope, most likely by increasing TCR affinity for MART-1–HLA-DR1 ligands beyond the threshold required for efficient T cell activation in the periphery. Alternatively, if pMART-1 is expressed in the thymus during T cell development, then surviving reactive TCRs are predicted to have suboptimal peptide–MHC binding characteristics. Future efforts to determine the structure of TCR D7-F6 bound to pMART-1100–114–HLA-DR1 and to measure the biophysical properties of this complex should provide relevant insights.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grants AI073654 (to S.L.T. and R.A.M.), AI036900 (to R.A.M.), CA134060 (to V.H.E.) and AI33993 (to D.F.H.), and by a grant from the Melanoma Research Alliance Foundation (to V.H.E., D.F.H. and S.L.T.). We thank H. Robinson (Brookhaven National Synchrotron Light Source) for X-ray data collection. Support for beamline X29 comes from the Offices of Biological and Environmental Research and of Basic Energy Sciences of the U.S. Department of Energy, and from the National Center for Research Resources of the National Institutes of Health.
Abbreviations used
- MHC
major histocompatibility complex
- MART-1
melanoma antigen recognized by T cells-1
- pMART-1
MART-1 phosphopeptide
- TCR
T cell receptor
- HA
hemagglutinin peptide
- mutTPI
mutant triose phosphate isomerase peptide, PBMC, peripheral blood mononuclear cells
- EBV
Epstein-Barr virus
- GM-CSF
granulocyte-macrophage colony-stimulating factor
- APC
antigen-presenting cell
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession number
Atomic coordinates and structure factors for the pMART-1100–114–HLA-DR1 complex have been deposited in the Protein Data Bank under accession code 3L6F.
References
- 1.Engelhard VH, Altrich-Vanlith M, Ostankovitch M, Zarling AL. Post-translational modifications of naturally processed MHC-binding epitopes. Curr. Opin. Immunol. 200618:92–97. doi: 10.1016/j.coi.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 2.Zarling AL, Ficarro SB, White FM, Shabanowitz J, Hunt DH, Engelhard VH. Phosphorylated peptides are naturally processed and presented by major histocompatibility complex class I molecules in vivo. J. Exp. Med. 2000192:1755–1762. doi: 10.1084/jem.192.12.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zarling AL, Polefrone JM, Evans AM, Mikesh LM, Shabanowitz J, Lewis ST, et al. Identification of class I MHC-associated phosphopeptides as targets for cancer immunotherapy. Proc. Natl. Acad. Sci. USA. 2006103:14889–14894. doi: 10.1073/pnas.0604045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Depontieu F, Qian J, Zarling AL, McMiller TL, Salay TM, Norris A, et al. Identification of tumor-associated MHC class II-restricted phosphopeptides as targets for immunotherapy. Proc. Natl. Acad. Sci. USA. 2009106:12073–12078. doi: 10.1073/pnas.0903852106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blume-Jensen P, Hunter T. Oncogenic kinase signaling. Nature. 2001411:355–365. doi: 10.1038/35077225. [DOI] [PubMed] [Google Scholar]
- 6.Ibrahim N, Haluska FG. Molecular pathogenesis of cutaneous melanocytic neoplasms. Annu. Rev. Pathol. 2009;4:551–579. doi: 10.1146/annurev.pathol.3.121806.151541. [DOI] [PubMed] [Google Scholar]
- 7.Williamson NA, Rossjohn J, Purcell AE. Tumors reveal their secrets to cytotoxic T cells. Proc. Natl. Acad. Sci. USA. 2006103:14649–14650. doi: 10.1073/pnas.0606951103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pardoll DM, Topalian SL. The role of CD4+ T cell responses in antitumor immunity. Curr. Opin. Immunol. 199810:588–594. doi: 10.1016/s0952-7915(98)80228-8. [DOI] [PubMed] [Google Scholar]
- 9.Gerloni M, Zanetti M. CD4 T cells in tumor immunity. Springer Semin. Immunopathol. 200527:37–48. doi: 10.1007/s00281-004-0193-z. [DOI] [PubMed] [Google Scholar]
- 10.Meyer VS, Drews O, Gunder M, Hennenlotter J, Rammensee H-G, Stevanovic S. Identification of natural MHC class II presented phosphopeptides and tumor-derived MHC class I phospholigands. J. Proteome Res. 20098:3666–3674. doi: 10.1021/pr800937k. [DOI] [PubMed] [Google Scholar]
- 11.Kawakami Y, Eliyahu S, Sakaguchi K, Robbins PF, Rivoltini L, Yannelli JR, et al. Identification of the MART-1 human melanoma antigen recognized by the majority of HLA-A2-restricted tumor infiltrating lymphocytes. J. Exp. Med. 1994180:347–352. doi: 10.1084/jem.180.1.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosenberg SA, Yang JC, Restifo NP. Cancer immunotherapy: moving beyond current vaccines. Nat. Med. 200410:909–915. doi: 10.1038/nm1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mohammed F, Cobbold M, Zarling AL, Salim M, Barrett-Wilt GA, Shabanowitz J, et al. Phosphorylation-dependent interaction between antigenic peptides and MHC class I: a molecular basis for the presentation of transformed self. Nat. Immunol. 20089:1236–1243. doi: 10.1038/ni.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petersen J, Wurzbacher SJ, Williamson NA, Ramarathinam SH, Reid HH, Nair AKN, et al. Phosphorylated self-peptides alter human leukocyte antigen class I-restricted antigen presentation and generate tumor-specific epitopes. Proc. Natl. Acad. Sci. USA. 2009106:2776–2781. doi: 10.1073/pnas.0812901106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Southwood S, Sidney J, Kondo A, del Guernico M-F, Appella E, Hoffman S, et al. Several common HLA-DR types share largely overlapping peptide binding repertoires. J. Immunol. 1998160:3363–3373. [PubMed] [Google Scholar]
- 16.Stern LJ, Brown JH, Jardetzky TS, Gorga JC, Urban RG, Strominger JL, et al. Crystal structure of the human class II MHC protein HLA-DR1 complexed with an influenza virus peptide. Nature. 1994368:215–221. doi: 10.1038/368215a0. [DOI] [PubMed] [Google Scholar]
- 17.Ghosh P, Amaya M, Mellins E, Wiley DC. The structure of an intermediate in class II MHC maturation: CLIP bound to HLA-DR3. Nature. 1995378:457–462. doi: 10.1038/378457a0. [DOI] [PubMed] [Google Scholar]
- 18.Dessen A, Lawrence CM, Cupo S, Zaller DM, Wiley DC. X-ray crystal structure of HLA-DR4 (DRA*0101, DRB*0401) complexed wih a peptide from human collagen II. Immunity. 19977:473–481. doi: 10.1016/s1074-7613(00)80369-6. [DOI] [PubMed] [Google Scholar]
- 19.Smith KJ, Pyrdol J, Gauthier L, Wiley DC, Wucherpfennig KW. Crystal structure of HLA-DR2 (DRA*0101, DRB1*1501) complexed with a peptide from human myelin basic protein. J. Exp. Med. 1998188:1511–1520. doi: 10.1084/jem.188.8.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sundberg EJ, Sawicki MW, Southwood S, Andersen PS, Sette A, Mariuzza RA. Minor structural changes in a mutated human melanoma antigen correspond to dramatically enhanced stimulation of a CD4+ tumor-infiltrating lymphocyte line. J. Mol. Biol. 2002319:449–461. doi: 10.1016/S0022-2836(02)00370-4. [DOI] [PubMed] [Google Scholar]
- 21.Wucherpfennig KW, Call MJ, Deng L, Mariuzza RA. Structural alterations in peptide–MHC recognition by self-reactive T cell receptors. Curr. Opin. Immunol. 200921:590–595. doi: 10.1016/j.coi.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hahn M, Nicholson MJ, Pyrdol J, Wucherpfennig KW. Unconventional topology of self peptide-major histocompatibility complex binding by a human autoimmune T cell receptor. Nat. Immunol. 20056:490–496. doi: 10.1038/ni1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y, Huang Y, Lue J, Quandt JA, Martin R, Mariuzza RA. Structure of a human autoimmune TCR bound to a myelin basic protein self-peptide and a multiple sclerosis-associated MHC class II molecule. EMBO J. 200524:2968–2979. doi: 10.1038/sj.emboj.7600771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deng L, Langley RJ, Brown PH, Xu G, Teng L, Wang Q, et al. Structural basis for the recognition of mutant self by a tumor-specific, MHC class II-restricted T cell receptor. Nat. Immunol. 20078:398–408. doi: 10.1038/ni1447. [DOI] [PubMed] [Google Scholar]
- 25.Hoashi T, Watabe H, Muller J, Yamaguchi Y, Vieira WD, Hearing VJ. MART-1 is required for the function of the melanosomal matrix protein PMEL17/GP100 and the maturation of melanomasomes. J. Biol. Chem. 2005280:14006–14016. doi: 10.1074/jbc.M413692200. [DOI] [PubMed] [Google Scholar]
- 26.Frayser M, Sato AK, Xu L, Stern LJ. Empty and peptide-loaded class II major histocompatibility complex proteins produced by expression in Escherichia coli and folding in vitro. Protein Expr. Purif. 199915:105–114. doi: 10.1006/prep.1998.0987. [DOI] [PubMed] [Google Scholar]
- 27.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 28.Storoni LC, McCoy AJ, Read RJ. Likelihood-enhanced fast rotation functions. Acta Crystallogr. D. 200460:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- 29.Zavala-Ruiz Z, Sturg I, Anderson MW, Gorski J, Stern LJ. A polymorphic pocket in the P10 position contributes to peptide binding specificity in class II MHC proteins. Chem. Biol. 200411:1395–1402. doi: 10.1016/j.chembiol.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 30.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, et al. Crystallography and NMR systems: a new software suite for macromolecular structure determination. Acta Crystallogr. D. 199854:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 31.McRee DE. XtalView/Xfit–A versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 1999125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 32.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: A program to check the stereo chemical quality of protein structures. J. Appl. Crystallogr. 199326:283–291. [Google Scholar]
- 33.Sidney J, Southwood S, Oseroff C, del Guercio MF, Sette A, Grey HM. The measurement of MHC/peptide interactions by gel infiltration. Curr. Protocols Immunol. 1998:18.3.1–18.3.19. doi: 10.1002/0471142735.im1803s31. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.