Abstract
Brain aging is associated with a progressive decline in cognitive function though the molecular mechanisms remain unknown. Functional changes in brain neurons could be due to age-related alterations in levels of specific proteins critical for information processing. Specialized membrane microdomains known as ‘lipid rafts’ contain protein complexes involved in many signal transduction processes. This study was undertaken to determine if two-dimensional fluorescence difference gel electrophoresis (2D DIGE) analysis of proteins in synaptic membrane lipid rafts revealed age-dependent alterations in levels of raft proteins. Five pairs of young and aged rat synaptic membrane rafts were subjected to DIGE separation, followed by image analysis and identification of significantly altered proteins. Of 1046 matched spots on DIGE gels, 94 showed statistically significant differences in levels between old and young rafts, and 87 of these were decreased in aged rafts. The 41 most significantly altered (p < 0.03) proteins included several synaptic proteins involved in energy metabolism, redox homeostasis, and cytoskeletal structure. This may indicate a disruption in bioenergetic balance and redox homeostasis in synaptic rafts with brain aging. Differential levels of representative identified proteins were confirmed by immunoblot analysis. Our findings provide novel pathways in investigations of mechanisms that may contribute to altered neuronal function in aging brain.
Keywords: 2D DIGE, Brain aging, Energy metabolism, Lipid rafts, Synaptic dysfunction
1. Introduction
Prominent features of brain aging are the progressive decline in cognitive function and impairment in learning and memory formation (Albers and Beal, 2000). However, the mechanisms underlying age-associated changes in neuronal function remain undefined. Of theories proposed to explain the phenotypes observed in aging, the most widely accepted ones include enhanced oxidative stress (Floyd and Hensley, 2002), mitochondrial dysfunction (Albers and Beal, 2000), Ca2+ dyshomeostasis (Khachaturian, 1994), synaptic atrophy (Lee et al., 2000), cytoskeletal abnormalities (Lee et al., 2000), and glial activation (Nichols, 1999). Oxidative stress accompanied by compromised energy metabolism and excessive production of reactive oxygen species (ROS) appears to play a central role in aging. This is supported by the observed decrease in respiratory function and oxidative damage to cellular DNA, proteins and lipids (Albers and Beal, 2000).
It is now clear that there is a loss of ~10–15% of synaptic junctions in aged brain as measured by microdensitometry (Masliah et al., 2006). Synaptic plasma membrane (SPM) preparations isolated from brain contain the synaptic junctions of neurons and offer excellent material for the studies of synaptic protein organization in specific domains, including specialized microdomains enriched in cholesterol, sphingolipids, and signaling proteins. These microdomains, termed ‘lipid rafts’, appear to serve as local organizing sites for the coordination of numerous signaling events by bringing receptors, effectors, and downstream signaling proteins into close proximity and promoting interactions (Simons and Toomre, 2000). It is important to note that the synaptic localization of proteins that affect neuronal plasticity and cognition, the glutamate receptors, is determined by the positioning of lipid raft domains at synapses (Hering et al., 2003; Hou et al., 2008). Given that lipid rafts provide platforms for a diverse array of neuronal processes, identification of age-dependent changes in the protein composition of these microdomains will likely provide insights into the molecular basis for impaired neuronal function with increasing age. The goal of this study was to determine if a proteomic analysis revealed any consistent differences in expression of proteins in lipid rafts isolated from SPMs of young vs. aged rat brains. We used two-dimensional fluorescence difference gel electrophoresis (2D DIGE) to assess age-related changes in the synaptic lipid raft compositions. Many proteomic strategies have been applied to studies of the brain (Rohlff, 2000), particularly in neurodegenerative disorders, such as Alzheimer’s and Parkinson’s disease, as well as schizophrenia (Rohlff, 2000; Jiang et al., 2003). Difference gel electrophoresis is designed to eliminate intergel variability apparent in traditional two-dimensional gel electrophoresis (2DE) and improve reproducibility by allowing co-electrophoresis of up to three different samples in a single gel (Alban et al., 2003). This strategy improves the quantification of differential expression in comparative proteomics.
Results of our analyses comparing five pairs of young vs. aged SPM rafts revealed significant changes in synaptic lipid raft proteins with increasing age. Forty-one raft proteins showing the most significant differences between raft domains from young and old were identified. A substantial number of the identified proteins are associated with energy metabolism. Many of those proteins may be components of the plasma membrane redox system (PMRS) (Ly and Lawen, 2003) and play important roles in energy regulation and maintenance of redox homeostasis. Our observations suggest that disruption in both bioenergetic and redox balance in lipid domains may contribute to altered neuronal function observed in aged brain.
2. Experimental procedures
2.1. Materials
Sources for the various primary antibodies were as follows: anti-Flotillin-1 (FLT-1) and anti-Thy-1, BD Biosciences; anti-Na+/K+-ATPase, anti-ATP synthase beta subunit (ATPB), and anti-voltage-dependent anion-selective channel protein 1 (VDAC1), Affinity Bioreagents; anti-glial fibrillary acidic protein (GFAP), Chemicon International; anti-MnSOD, Stressgen; anti-mitofilin (IMMT), Proteintech Group; anti-NADH dehydrogenase (ubiquinone) Fe-S protein 3 (NDUFS3), MitoSciences; anti-cytochrome c, Santa Cruz Biotechnology; and anti-glutamate dehydrogenase (GDH), Novus Biologicals. Immobilized pH gradient (IPG) strips, Pharmalyte, urea and CyDye DIGE fluor minimal labeling kit were purchased from GE Healthcare. Alkaline phosphatase-conjugated secondary antibodies, ficoll, CHAPS, thiourea, iodoacetamide, trifluoroacetic acid (TFA), dimethylformamide (DMF), α-cyano-4-hydroxycinnamic acid, and horseradish peroxidase-coupled cholera toxin subunit B (CTXB) were from Sigma. Acetonitrile and dithioerythritol (DTT) were obtained from Fisher Scientific International. Amplex Red cholesterol assay kit and Sypro Ruby were from Invitrogen, Bicinchoninic Acid protein assay reagents from Pierce Biotechnology, PVDF membranes and ZipTips from Millipore, trypsin from Promega, and protease inhibitor cocktail from Calbiochem.
2.2. Animals
Eight pairs of Fisher 344/Brown Norway hybrid (F344/BNF1) rats at 5 months (young) and 34 months of age (old) (five pairs for Cy dye labeling and three for immunoblot analysis) were obtained from an NIA colony maintained by Harlan Industries (Indianapolis, IN, USA). All protocols were implemented in accordance with NIH guidelines and approval by the University of Kansas Institutional Animal Care and Use Committee (IACUC).
2.3. Isolation of SPMs, mitochondria and SPM rafts from young and old rat brains
Fisher 344/BNF1 rats were anesthetized by CO2 inhalation according to the IACUC guidelines. Rats were decapitated using a guillotine and brains removed quickly. The brains from one young and one old rat were processed in parallel. Each whole brain was homogenized and processed immediately for the isolation of synaptosomes and mitochondria by discontinuous ficoll density gradient centrifugation as previously described (Michaelis et al., 1983). Synaptosomes were lysed with a hypotonic buffer (3 mM Tris–HCl, pH 8.0, with a cocktail of protease inhibitors) and centrifuged at 64,200 × g for 15 min in a Ti 60 rotor (Beckman Coulter) to isolate the SPMs. The SPM and mitochondrial pellets were both homogenized in buffer containing 10 mM Tris–HCl, 50 µM MgCl2 and 0.32 M sucrose, pH 7.4. Eight pairs of lipid raft preparations were isolated from SPMs using discontinuous sucrose density gradient centrifugation (Jiang et al., 2007). In brief, the SPMs from each animal were solubilized in an equal volume of solubilization buffer (50 mM Tris–HCl, 150 mM NaCl, 5 mM EDTA, pH 7.5, containing 2% Brij 98), to achieve 1% Brij 98 as the final concentration, and incubated on ice for 30 min. The suspension was mixed 1:1 with a 90% sucrose solution and the resultant mixture overlaid with 35% and 5% sucrose solutions. Ultracentrifugation was performed for 18 h at 98,300 × g in a Beckman Optima Max centrifuge in an MLS-50 rotor. A total of eight fractions (0.5 ml each) were collected from the top to the bottom of each tube. Protein concentrations were determined by the Bicinchoninic Acid method according to the manufacturer’s instructions.
2.4. Cholesterol and GM1 analysis
Cholesterol was measured using the Amplex Red cholesterol assay kit and GM1 ganglioside levels determined by dot blotting using CTXB (300 ng/ml) (Jiang et al., 2007). Briefly, 3.5 µl of each fraction was applied onto a PVDF membrane, blocked with 1% BSA for 30 min and exposed to CTXB for 2 h. Color was developed by incubation in a mixture of 1.4 mM 3,3′-diaminobenzidine tetrahydrochloride, 200 mM nickel chloride and 6.2 mM H2O2.
2.5. Immunoblotting
Proteins were separated by SDS-PAGE and transferred to PVDF membranes as we have described previously (Jiang et al., 2007). Non-specific interactions were blocked with 5% milk for 1 h at 25°C and the membranes incubated overnight with the indicated concentrations of primary antibodies. Alkaline phosphatase-conjugated secondary antibodies (1:1000) were added for 2 h at 25°C. Immunoblots were developed using the substrate 5-bromo-4-chloro-3-indoyl phosphate and nitroblue tetrazolium. Blots were scanned and densitometric analysis carried out using Adobe Photoshop 7.0.
2.6. Labeling of synaptic raft proteins with Cy dyes
The buffer in the lipid raft preparation was replaced by a fluor-labeling compatible lysis buffer (7 M urea, 2 M thiourea, 4% CHAPS, pH 8.5) by means of ultrafiltration (Jiang et al., 2004). Differential labeling of each pair (n = 5) of raft proteins isolated from young and aged SPMs was accomplished by using the CyDye DIGE fluor minimal labeling method as per the manufacturer’s instructions. The fluor working solutions (400 pmol/µl) were made in anhydrous DMF. Each SPM raft sample isolated from either a young or an aged rat brain was labeled at a fluor-to-protein ratio of 400 pmol/50 µg protein, with the young preparation labeled with Cy3 and the old with Cy5. The internal standard, comprised of all raft samples of both ages by pooling 25 µg of protein from each of 10 samples, was labeled with Cy2 at the same fluor-to-protein ratio as that used for the individual young and old samples. The labeling reaction was performed on ice for 30 min in the dark and was stopped by the addition of 1 µl of 10 mM lysine and further incubation on ice for 10 min.
2.7. Analysis of synaptic raft proteins showing age-related differences
Analytical gels were prepared for the assessment of potential age-related differences in synaptic raft proteins from five pairs of young vs. old brain SPM rafts, each isolated independently. Aliquots of the labeled protein samples were mixed with an equal volume of 2× sample buffer (7 M urea, 2 M thiourea, 4% CHAPS, 130 mM DTT, 2% Pharmalyte) and incubated on ice for 10 min. The differentially labeled proteins, consisting of the internal standard, one young, and one old raft samples with 50 µg of protein present in each of the three samples, were combined (total protein, 150 µg) into a single microfuge tube. Rehydration buffer (7 M urea, 2 M thiourea, 4% CHAPS, 65 mM DTT, 1% Pharmalyte) was added to give a final volume of 450 µl. This multiplexed sample was then loaded on an immobilized pH 3–10 nonlinear gradient (IPG) strip (24 cm) that was re-swelled overnight using a rehydration loading method in a manifold (GE Healthcare). Five such strips were run, each containing an individual pair of young vs. old rafts, and the same amount of the internal standard. Two-dimensional gel electrophoresis was performed essentially as described (Jiang et al., 2003). The first-dimensional isoelectric focusing in the Ettan IPGphor system (GE Healthcare) was started at 100 V for 8 h and the voltage was gradually increased to 8000 V and kept constant for 12 h. After completion of the first separation, each strip was incubated for 15 min with 65 mM DTT followed by 130 mM iodoacetamide in equilibration buffer (50 mM Tris–HCl, pH 8.8, 6 M urea, 30% glycerol, 2% SDS), and placed on the top of a 10% SDS-polyacrylamide gel (260 mm × 200 mm × 1.0 mm). The second-dimensional separation was performed in an Ettan DALTsix apparatus (GE Healthcare) and run overnight at 1 W/gel. The gels were scanned on a Typhoon Imager (GE Healthcare, Cy2/488 nm, Cy3/532 nm, Cy5/633 nm) using a 100 µm pixel size and the fluorescent images analyzed by Progenesis v2006 2D software (Nonlinear Dynamics). The protein spots on each gel were detected, and gel warping and spot matching across gels was performed. The protein abundance of each spot in the images corresponding to the young and aged samples was determined as a ratio compared with the intensity of the corresponding spot present in the internal standard control image run in the same physical gel. Normalized volumes of the matched spots across all the gels were further used to evaluate the statistical significance of differences between the young and old samples by Student’s two-tailed t-test.
2.8. Mass spectrometry (MS) analysis and protein identification
To identify the proteins in the spots that differed significantly between young and old raft samples in the analytical gels, equal amounts of unlabeled SPM raft proteins isolated from 5-month and 34-month-old rat brains were pooled (0.9 mg total) and applied to a 24-cm IPG pH 3–10 nonlinear strip to make a preparative gel. After the second-dimensional protein separation, gels were fixed in 50% methanol and 7% acetic acid for 2 h, followed by overnight staining with Sypro Ruby. Gels were washed in 10% methanol and 7% acetic acid for 30 min, rinsed twice with ultrapure water, and scanned in a Typhoon imager (457 nm).
The preparative gel was overlaid with the images from the analytical gels to permit the excision of protein spots that differed significantly between young and old samples. Protein identification was performed by matrix-assisted laser desorption/ionization (MALDI)-MS for high-abundance spots as described (Jiang et al., 2003) and by liquid chromatography–MS for low-abundance and/or low-molecular-weight spots as described (Ikehata et al., 2008). The excised spots were destained with 50% acetonitrile in 20 mM ammonium bicarbonate and dehydrated with acetonitrile. Each gel piece was exposed to 10 µl of 10 µg/ml trypsin in 20 mM ammonium bicarbonate. After digestion for 2 h at 37 °C, 30 µl of extraction solution (10% acetonetrile in 20 mM ammonium bicarbonate) was added, and the mixture incubated overnight. The extracted peptides were initially concentrated and desalted with ZipTips, mixed 1:1 with a matrix solution consisting of 1 mg/ml α-cyano-4-hydroxycinnamic acid in 70% acetonitrile and 0.1% TFA, spotted onto a target plate, and analyzed on a 4700 Proteomics Analyzer (MALDI TOF/TOF tandem mass spectrometer), Applied Biosystems. Peak lists were generated using 4700 Explorer (version 2.0) software after spectra were internally calibrated by the known trypsin auto-digest peptide ions (842.50 and 2211.10 m/z). The peptide mass fingerprint search was conducted using Mascot (v2.1, Matrix Science). Significant hits (p < 0.05) defined by the software were considered as protein identities. Parameters set during the search were as follows: the IPI_rat database (v3.31, ~40,000 entries), developed to offer complete non-redundant data sets representing the rat proteomes built from the Swiss-Prot, TrEMBL, Ensembl and RefSeq databases (Kersey et al., 2004), was chosen. Monoisotopic masses were used, the maximum of missed cleavages was one, a mass tolerance of 50 ppm was allowed, and carbamidomethyl on cysteines was considered as fixed modifications and oxidation on methionines as variable modifications. If the peptides from low-abundant proteins could not be identified via MALDI, the digested and extracted peptides were introduced into an LTQ-FT Ultra Hybrid Mass Spectrometer, ThermoFinnigan, via capillary liquid chromatography as described (Ikehata et al., 2008). Survey MS1 mass spectra were acquired in the Fourier transform-ion cyclotron resonance over a mass range of 300–2000 m/z. The three most intense peptide ions in the survey MS1 spectrum were selected and sequentially fragmented for MS2 analysis in the linear ion trap by collision-induced dissociation. Fragmentation of the peptide from the amino terminus produces “b ions” while fragmentation from the C-terminus produces “y ions”, both recorded in the spectrum. MS/MS ion searches were accomplished against IPI_rat (v3.31, ~40,000 entries) using Mascot (v2.1, Matrix Science) to determine the peptide sequence by the mass difference between peaks within the same ion series and to identify the corresponding protein. The search parameters were the same as used for peptide mass fingerprint except for a peptide tolerance of 1.2 Da and MS/MS tolerance of 0.2 Da. As mentioned before, significant hits (p < 0.05) defined by the software were taken as protein identities. Specific isoforms of a protein family were identified and distinguished from other isoforms based on their unique peptide mass fingerprint, experimental molecular weight and pI revealed by their running positions on the gels, and/or amino acid sequences obtained from MS/MS analysis.
3. Results
3.1. Characterization of lipid rafts isolated from young and aged rat brain SPMs
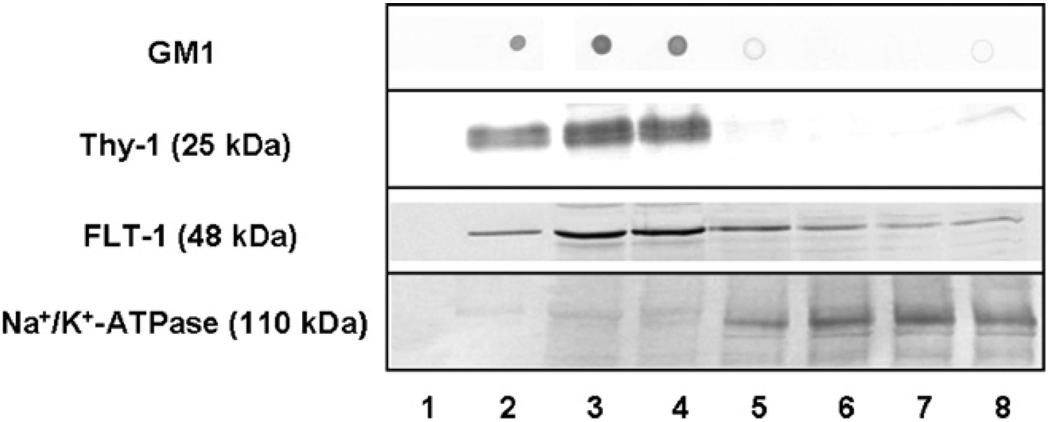
Lipid rafts can be isolated from membranes of different tissues and cell types based on their detergent insolubility at low temperature and low buoyant density upon discontinuous sucrose density gradient centrifugation. We isolated lipid rafts from SPMs prepared from the individual brains of five pairs of 5- and 34-month-old F344/BNF1 rats using 1% Brij 98, a non-ionic detergent known to preserve raft integrity even at physiological temperatures (Lucero and Robbins, 2004). After 18 h of centrifugation at 98,300 × g, a band of white material appeared at the interface of 5% and 35% sucrose. Eight fractions were collected from the top to the bottom of the tube. A small peak representing 13.84% ± 0.87% of the total protein (n = 10, 5 young and 5 old preparations) was present in the low-density fractions, fractions 2–4. A larger protein peak corresponded to the high-density fractions, fractions 5–8. A similar separation pattern was reported previously in the isolation of synaptic membrane raft domains (Hering et al., 2003). Assay of the raft lipid marker cholesterol showed a peak corresponding to 337.56 ± 25.64 µg cholesterol/mg protein in fraction 3 and 70.81 ± 4.81 µg/mg protein in fraction 4. Cholesterol levels in all other fractions were less than 40 µg/mg protein. The other raft lipid marker GM1 ganglioside and the raft protein markers Thy-1 and FLT-1 localized exclusively in fractions 2–4 (Fig. 1). Thus, on the basis of the high cholesterol, GM1 ganglioside, Thy-1 and FLT-1 markers, fractions 2–4 were considered the lipid raft-enriched fractions. Na+/K+-ATPase, a protein that primarily distributes in non-rafts (Hansen et al., 2000), was found exclusively in the high-density fractions, confirming the separation of the raft from the non-raft membrane microdomains.
Fig. 1.
Characterization of synaptic lipid rafts isolated from rat brain. The levels of GM1, raft marker proteins Thy-1 and FLT-1, and the non-raft marker Na+/K+-ATPase were determined in the density gradient fractions using dot blots or immunoblots. Dilutions of the antibodies were: anti-Thy-1, 1:250; anti-FLT-1, 1:250; and anti-Na+/K+-ATPase, 1:500. The amounts of protein loaded on the gels were: Thy-1 and FLT-1 (15 µg), and Na+/K+-ATPase (10 µg). Representative blots from five independent experiments with similar results are shown.
Comparison of the distribution of total protein and both raft lipid and protein markers in each fraction showed no statistically significant differences between preparations isolated from young and old rat brains, suggesting that the overall protein content of lipid rafts did not change with increasing age. Fig. 1 shows a representative blot from a young brain synaptic raft preparation, and essentially similar patterns were observed in all preparations. For subsequent DIGE studies and immunoblot validation, fraction 3 was selected as the primary representative of lipid rafts as it was highly enriched in both lipid and protein raft markers.
3.2. Differential levels of proteins in synaptic rafts
Potential differences in the levels of various proteins in young vs. aged SPM rafts were investigated by 2D DIGE, followed by gel image analysis and protein identification. Equal aliquots of the three differentially labeled samples, i.e., Cy2 internal standard, Cy3 young, and Cy5 old, were combined and separated by 2DE in a single gel. Five such gels were run with an equal amount of the internal standard present in each gel. Thus, from the five gels 15 gel images were generated in which the protein profile of each sample could be visualized at its proper Cy dye wavelength.
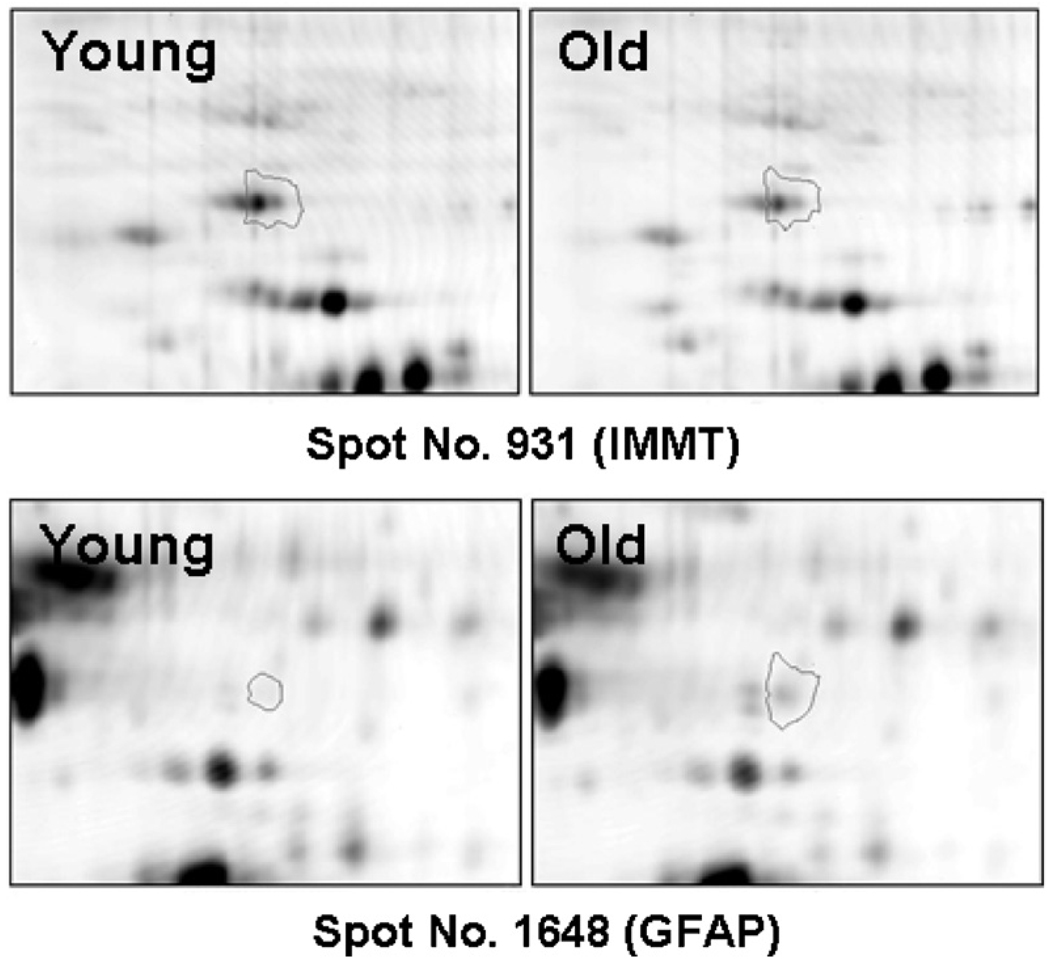
Progenesis image analysis software identified 1046 spots that matched across all five gels. Taking p < 0.05 as an initial threshold for significance, 94 raft protein spots exhibited statistically significant differences in levels with age, 87 of which were decreased and 7 increased in the aged brain samples. Fig. 2 shows high-resolution gel images of a protein that decreased in aged rafts (IMMT) and one that increased with age (GFAP) as representatives of the fluorescence differences in young vs. aged rafts.
Fig. 2.
Enlarged images of representative protein spots illustrating age-dependent differences in protein levels. Progenesis software was used to generate magnified images of the protein spots and images are shown of two spots exhibiting differential expression with age. Spot no. 931 identified as IMMT, was decreased with age. Spot no. 1648 corresponding to GFAP was increased in aged samples. Only one representative example of a protein that decreased and one that increased with age is shown for clarity. However, Fig. 1 in Supplementary Material shows a full array of differentially expressed proteins that were identified and are listed in Table 1.
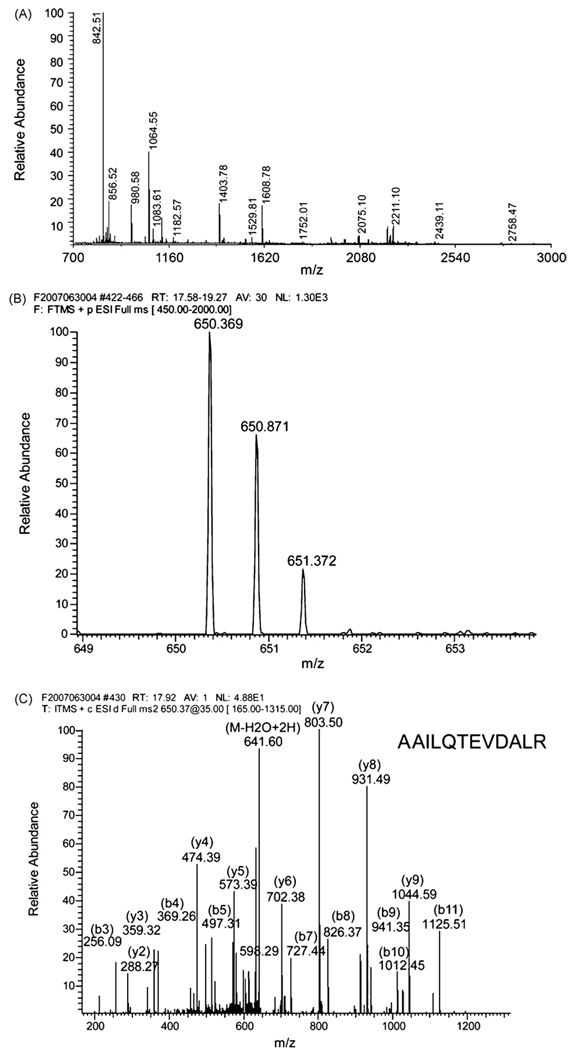
For protein identification by MS analysis, a significance level of (p < 0.03) was used to select spots to be excised from the gel (Table 1). Forty-one proteins were identified via MALDI-TOF and/or LTQ-FT MS analysis (Supplementary Fig. 1 shows a representative gel with the identified spots numbered). Fig. 3 shows the MS spectrum of NDUS1 or NADH dehydrogenase (ubiquinone) Fe-S protein 1 (Fig. 3A) and MS1 and MS2 spectra of RB6I2 or ELKS/Rab6-interacting/CAST (ERC) protein (Fig. 3B and C) as representatives of protein identification via MALDI and LTQ-FT, respectively. Fig. 3C represents an example of the single-peptide-based protein identification. The ERC family predominantly contains three members in brain: ERC1 and isoforms 1 and 2 of ERC2. The fragmentation MS/MS spectrum was matched to the AAILQTEVDALR peptide that occurs in either ERC1 or isoform 1 of ERC2, but not in isoform 2 of ERC2. Because the amino acid sequence of this peptide is present in both ERC1 and isoform 1 of ERC2, it is not possible to determine which of these two protein species was seen in our samples.
Table 1.
Identification of raft proteins altered in aging brain. Threshold was initially set as p < 0.03 and 41 proteins were identified (actin at p < 0.05 was included making the total 42). Considering the post-translational modifications, three appended spots (*) were identified. The fold difference was displayed as a ratio of young-to-old. The scores indicating significance (p < 0.05) were given by Mascot (Mowse score). For single-peptide-based protein identification, the precursor m/z, the observed charge and the identified sequence were reported in the lower row.
| Spot no. | Protein ID | Accession# in IPI | Theoretical mass (kDa)/pI | Number of matched peptides (sequence coverage) | MS method | Mowse score | Fold difference (Y/O) | P value |
|---|---|---|---|---|---|---|---|---|
| Down-regulated proteins | ||||||||
| Synaptic proteins | ||||||||
| 707 | RB6I2 [ELKS/RAB6-interacting/CAST family member 1 (ERC protein 1 or CAST2)] or isoform 1 of ERC2 [ERC protein 2 (CAST1)]. | IPI00201791 or IPI00204418 |
111.81/6.1 or 110.72/6.6 |
1 (1%) | LTQ | 60 | 2.03 | 0.00014 |
| 650.37/+2/R.AAILQTEVDALR.L | ||||||||
| 1906 | SEPT5 [Septin-5, isoform 1 or 2 or 3]. | IPI00655290 or IPI00201737 or IPI00559449 |
43.28/6.3 or 44.35/6.3 or 41.42/6.2 |
3 (8%) | LTQ | 136 | 2.33 | 0.013 |
| 1185 | SYN2 [Synapsin-2, isoform IIa or IIb]. | IPI00210036 or IPI00231408 |
63.70/8.7 or 52.82/8.1 |
4 (14%) | LTQ | 134 | 2.39 | 0.0022 |
| 1289 | STXB1 [Syntaxin-binding protein 1 (Munc18-1), isoform 1 or 2]. | IPI00205372 or IPI00230938 |
67.93/6.5 or 69.09/6.3 |
14 (21%) | LTQ | 497 | 2.17 | 0.011 |
| Signaling proteins | ||||||||
| 2300 | 1433E [14-3-3 protein epsilon]. | IPI00325135 | 29.33/4.6 | 15 (49%) | MALDI | 88 | 2.08 | 0.025 |
| 2333 | 1433G [14-3-3 protein gamma]. | IPI00230835 | 28.46/4.8 | 9 (34%) | MALDI | 80 | 1.99 | 0.007 |
| Bioenergetic and redox homeostasis proteins | ||||||||
| 1096 | NDUS1 [NADH dehydrogenase (ubiquinone) Fe-S protein 1]. | IPI00358033 | 80.36/5.6 | 18 (30%) | MALDI | 89 | 1.58 | 0.013 |
| 1104 | 24 (40%) | MALDI | 147 | 1.45 | 0.0081 | |||
| 1109 | 8 (15%) | LTQ | 362 | 1.53 | 0.0060 | |||
| 1664 | NDUFV1 [NADH dehydrogenase (ubiquinone) flavoprotein 1]. | IPI00191913 | 51.40/9.0 | 14 (31%) | MALDI | 66 | 1.48 | 0.021 |
| 1666 | 24 (45%) | MALDI | 153 | 1.48 | 0.0026 | |||
| 1780 | NDUS2 [NADH dehydrogenase (ubiquinone) Fe-S protein 2]. | IPI00471647 | 52.50/6.5 | 22 (40%) | MALDI | 97 | 1.58 | 0.014 |
| 2375 | NDUFS3 [NADH dehydrogenase (ubiquinone) Fe-S protein 3]. | IPI00202616 | 30.38/7.1 | 15 (41%) | MALDI | 121 | 1.78 | 0.016 |
| 2504 | NDUFS8 [Similar to NADH dehydrogenase (ubiquinone) Fe-S protein 8]. | IPI00188330 | 24.41/5.9 | 10 (33%) | LTQ | 201 | 1.70 | 0.0071 |
| 2387 | LOC681867 [Similar to NADH-ubiquinone oxidoreductase PDSW subunit]. | IPI00202238 | 21.13/7.6 | 5 (19%) | LTQ | 143 | 1.55 | 0.0044 |
| 2019 | NDUAA [NADH dehydrogenase (ubiquinone) 1 alpha subcomplex subunit 10]. | IPI00561513 | 40.75/7.6 | 10 (21%) | LTQ | 452 | 1.75 | 0.015 |
| 1280 | DHSA [Succinate dehydrogenase (ubiquinone) flavoprotein subunit]. | IPI00200659 | 71.54/6.8 | 17 (36%) | MALDI | 89 | 1.69 | 0.014 |
| 2379 | DHSB [Succinate dehydrogenase Ip subunit]. | IPI00208288 | 32.61/9.0 | 5 (12%) | LTQ | 130 | 1.51 | 0.0063 |
| 2369 | UCRI [Cytochrome B-C1 complex subunit Rieske]. | IPI00362949 | 29.71/9.0 | 6 (27%) | LTQ | 177 | 1.71 | 0.015 |
| 1747 | UQCR1 [Cytochrome B-C1 complex subunit 1]. | IPI00471577 | 53.50/5.6 | 1 (4%) | LTQ | 66 | 1.61 | 0.013 |
| 786.10/+3/K.VVELLADIVQNISLEDSQIEK.E | ||||||||
| 1756 | 5 (12%) | LTQ | 221 | 1.48 | 0.026 | |||
| 1757 | 1 (4%) | LTQ | 69 | 1.68 | 0.026 | |||
| 785.76/+3/K.VVELLADIVQNISLEDSQIEK.E | ||||||||
| 1875 | UQCR2 [Cytochrome B-C1 complex subunit 2]. | IPI00188924 | 48.42/9.2 | 17 (26%) | LTQ | 500 | 1.43 | 0.000049 |
| 1585 | ATPA [ATP synthase subunit alpha]. | IPI00396910 | 59.83/9.2 | 3 (8%) | LTQ | 160 | 2.55 | 0.0088 |
| 1586 | 18 (31%) | MALDI | 66 | 1.40 | 0.0060 | |||
| 1588 | 19 (41%) | MALDI | 95 | 1.79 | 0.0070 | |||
| 1589 | 19 (31%) | MALDI | 74 | 1.34 | 0.0047 | |||
| 1621 | ATPB [ATP synthase subunit beta]. | IPI00551812 | 56.32/5.2 | 29 (55%) | MALDI | 164 | 1.58 | 0.0066 |
| 1626 | 22 (54%) | MALDI | 154 | 1.42 | * 0.041 | |||
| 2501 | ATP5H [ATP synthase subunit D]. | IPI00230838 | 18.81/6.2 | 15 (44%) | LTQ | 420 | 1.57 | 0.00034 |
| 2502 | 3 (25%) | LTQ | 62 | 1.80 | 0.0001 | |||
| 762 | ODO1 [2-Oxoglutarate dehydrogenase E1 component (Alpha-ketoglutarate dehydrogenase)]. | IPI00215093 | 117.42/6.3 | 16 (15%) | MALDI | 71 | 1.71 | 0.0037 |
| 765 | 18 (18%) | MALDI | 62 | 1.48 | 0.025 | |||
| 1844 | KCRB [Creatine kinase B-type]. | IPI00470288 | 42.98/5.4 | 11 (28%) | LTQ | 406 | 1.87 | 0.024 |
| 1269 | ODP2 [Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex]. | IPI00231714 | 67.64/8.8 | 3 (5%) | LTQ | 75 | 1.84 | 0.015 |
| 2356 | PGAM1 [Phosphoglycerate mutase 1]. | IPI00421428 | 28.93/6.7 | 7 (16%) | LTQ | 124 | 1.99 | 0.012 |
| 2358 | 6 (15%) | LTQ | 132 | 1.99 | 0.013 | |||
| 628 | PYC [Pyruvate carboxylase]. | IPI00210435 | 130.44/6.3 | 1 (0%) | LTQ | 76 | 1.91 | 0.027 |
| 682.84/+2/K.IAEEFEVELER.G | ||||||||
| 2162 | VDAC1 [Voltage-dependent anion-selective channel protein 1]. | IPI00421874 | 30.85/8.6 | 11 (52%) | MALDI | 79 | 1.34 | * 0.032 |
| 2163 | 12 (55%) | MALDI | 88 | 1.47 | * 0.048 | |||
| 2171 | 7 (25%) | LTQ | 273 | 1.24 | 0.017 | |||
| Cytoskeletal proteins | ||||||||
| 1881 | ACTB [Actin, cytoplasmic 1 (Beta-actin)] or ACTG [Actin, cytoplasmic 2 (Gamma-actin)]. | IPI00189819 or IPI00767505 |
42.05/5.3 or 42.11/5.3 |
16 (39%) | MALDI | 81 | 1.38 | ** 0.047 |
| 2209 | CAPZB [F-actin-capping protein subunit beta]. | IPI00365283 | 30.95/5.7 | 5 (13%) | LTQ | 230 | 1.47 | 0.019 |
| 1487 | TBA1A [Tubulin alpha-1A chain] or TBA1B [Tubulin alpha-1B chain]. | IPI00189795 or IPI00339167 |
50.12/4.9 or 50.10/4.9 |
17 (46%) | MALDI | 109 | 1.33 | 0.0039 |
| 1530 | TBB2A [Tubulin beta-2A chain] or TBB2B [Tubulin beta-2B chain]. | IPI00475639 or IPI00655259 |
49.91/4.9 or 49.95/4.9 |
26 (45%) | MALDI | 141 | 1.38 | 0.0019 |
| 2022 | STML2 [Stomatin-like protein 2]. | IPI00203528 | 38.50/8.7 | 4 (16%) | LTQ | 224 | 1.82 | 0.0095 |
| Transporters | ||||||||
| 1249 | SLC25A13 [Similar to calcium-binding mitochondrial carrier protein Aralar2]. | IPI00358163 | 74.75/8.9 | 3 (4%) | LTQ | 155 | 1.34 | 0.022 |
| 2248 | VDAC2 [Voltage-dependent anion-selective channel protein 2] or Isoform 1 or 2 of VDAC3 [Voltage-dependent anion-selective channel protein 3]. | IPI00198327 or IPI00556929 or IPI00231067 |
31.73/7.4 or 31.18/8.9 or 31.31/8.9 |
1 (3%) | LTQ | 66 | 2.61 | 0.00085 |
| 508.80/+2/K.LTLSALVDGK.N | ||||||||
| Heat shock proteins | ||||||||
| 1229 | HSP7C [Heat shock cognate 71 kDa protein]. | IPI00208205 | 70.83/5.4 | 12 (17%) | LTQ | 638 | 1.49 | 0.026 |
| 1191 | GRP75 [Stress-70 protein (GRP 75)]. | IPI00363265 | 73.81/6.0 | 17 (24%) | LTQ | 802 | 1.40 | 0.026 |
| Others | ||||||||
| 1545 | ANXA11 [Annexin A11]. | IPI00364621 | 54.47/7.5 | 1 (1%) | LTQ | 51 | 1.70 | 0.0032 |
| 551.25/+2/R.LLISLSQGNR.D | ||||||||
| 926 | IMMT [Inner membrane protein (mitofilin)]. | IPI00566985 | 78.59/5.4 | 16 (28%) | MALDI | 74 | 1.54 | 0.017 |
| 931 | 18 (23%) | MALDI | 106 | 1.62 | 0.0035 | |||
| 2073 | PP1A [Serine/threonine-protein phosphatase PP1-alpha catalytic subunit]. | IPI00208265 | 38.23/5.9 | 2 (6%) | LTQ | 98 | 1.65 | 0.003 |
| 1239 | TKT [Transketolase]. | IPI00231139 | 71.91/7.5 | 2 (6%) | LTQ | 69 | 1.34 | 0.0086 |
| 2405 | UCHL1 [Ubiquitin carboxyl-terminal hydrolase isozyme L1]. | IPI00204375 | 25.17/5.1 | 3 (21%) | LTQ | 92 | 1.77 | 0.0011 |
| Up-regulated proteins | ||||||||
| 1648 | GFAP [Glial fibrillary acidic protein, isoform 1 or 2]. | IPI00190943 or IPI00210211 |
49.98/5.4 or 49.52/5.7 |
30 (60%) | MALDI | 163 | 0.44 | 0.029 |
Fig. 3.
Representative mass spectra of MALDI-TOF and LTQ-FT used for protein identification by peptide mass fingerprint and MS/MS ion search. (A) The MALDI-TOF spectrum of NADH dehydrogenase (ubiquinone) Fe-S protein 1 identified in the peptide mass fingerprint search. The MS spectrum of spot #1104 was internally calibrated using the two ions of trypsin autolytic peptides at 842.51 and 2211.10 m/z. The monoisotopic mass list was submitted for a peptide mass fingerprint search in Mascot, resulting in one significant hit with a top score of 147 for NADH dehydrogenase (ubiquinone) Fe-S protein 1 as reported (Table 1). Matched peptide masses are assigned to the peaks in the range of 800–2500 m/z. (B and C) The LTQ-FT spectra of the ERC protein in the MS/MS ion search. The protein identification was based on a single peptide with a resultant Mascot score of 60 (Table 1). The double-charged precursor ion at 650.369 m/z was detected in the MS1 with a deviation of 3 ppm (B) and fragmented in the MS2 (C). The MS/MS data with a series of b and y ions were searched and scored in Mascot. As a result, the fragmentation spectrum was matched to the peptide AAILQTEVDALR of either ERC1 or isoform 1 of ERC2. In (C) the matched b and y ions as well as their detected masses are marked. The unique sequence of this peptide was matched only to isoform 1 of ERC2 protein.
Table 1 summarizes the MS data with the name of each identified protein, the fold change in protein levels in the samples from the aged rats, the corresponding P value, and the corresponding spot number shown in supplementary Fig. 1. The proteins in Table 1 are grouped according to categories of molecular or cellular function. It appeared that some spots derived from the same protein migrated slightly differently, possibly due to post-translational modifications. These proteins showed similar fold changes and, taken altogether, are reflected in the assessment of the altered level of the protein. For example, spot no. 1621, identified as ATPB or ATP synthase subunit beta, was found to be in relatively lower abundance in old vs. young. Considering phosphorylation as the post-translational modification previously observed on ATPBs (Hojlund et al., 2003), the single spot identified may not fully reflect the total level of the given protein in the samples. Therefore, some adjacent spots were additionally analyzed and, as expected, spot no. 1626 was also identified as ATPB. Interestingly, that spot also exhibited a statistically significant decrease in aging rafts (p < 0.05).
Almost half of identified SPM raft proteins that were reduced with increasing age are typically associated with bioenergetics and metabolic homeostasis (Table 1, 3rd section). Of these, 15 belong to oxidative phosphorylation complexes associated with mitochondria, among which 7 are components of complex I (NADH dehydrogenase), 2 of complex II (succinate dehydrogenase), 3 of complex III (cytochrome c oxidoreductase), and 3 of complex V (ATP synthase). The change in expression averaged 1.62 ± 0.05-fold for all these components with small standard errors, indicating a possible functional coordination between them.
Differential levels of four presynaptic proteins were also observed: RB6I2 (ERC1 or ERC2) designated as a novel cytomatrix-at-the-active-zone (CAZ) protein, SEPT5 (septin 5) identified as a neuron-specific GTPase compartmentalized in synaptic vesicles (Beites et al., 1999), SYN2 (synapsin 2) and STXB1 (Munc18-1). Interestingly, the abundance of these proteins in lipid rafts was relatively low, even in young brain; however, the magnitude of the decrease with increasing age was >2-fold, even greater than changes in the energy-related proteins.
Several proteins believed to be associated with the cytoskeletal network also showed markedly lower levels in aged rafts (Table 1, 4th section). The levels of both alpha and beta chains of tubulin, the proteins that form microtubules, and actin proteins differed by ~1.3–1.5-fold. Statistical analysis of one of the relatively abundant protein spots which we identified as an actin, i.e., spot no. 1881, indicated a 1.4-fold difference (p < 0.05) (Table 1). Despite slightly less significance, actin was included in the spots analyzed because the results are consistent with lowered mRNA levels of beta-actin previously observed in the aged rat brain (Nishibayashi et al., 1994). Of the 41 proteins identified, only one, GFAP, was found to be increased with advancing age. GFAP is a class-III intermediate filament protein highly expressed in glial cells.
3.3. Assessment of mitochondrial contamination in the SPM rafts
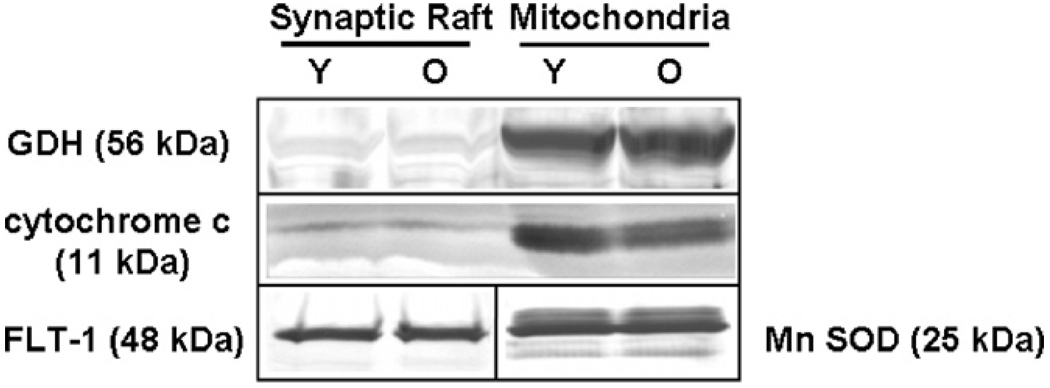
The observation that several proteins believed to be localized to mitochondria were found to be highly represented in lipid rafts is consistent with earlier findings from different types of cells (Bae et al., 2004; Kim et al., 2006). Such proteins may actually be components of specialized domains in the plasma membrane in addition to the mitochondria. To evaluate further the extent of mitochondrial membrane fragment contaminants in our raft preparations, mitochondrial fractions and three new pairs of synaptic rafts were isolated from young and old rat brains. Proteins were separated by SDS-PAGE, transferred to PVDF membranes, and probed for the levels of two mitochondrial markers not associated with the respiratory complex, cytochrome c (Bae et al., 2004; Kim et al., 2006) and GDH (Lai et al., 1986). The latter protein is localized not only in the mitochondrial matrix but also on the inner membrane where the respiratory chain apparatus is located. The immunoblots as illustrated in Fig. 4 clearly show that these two proteins were highly enriched in mitochondrial preparations, with nearly undetectable presence in lipid rafts. Densitometric analysis of the immunoblots of separate preparations from three pairs of young and old rats showed minimal levels of these mitochondrial proteins in rafts and a high mitochondrion-to-raft ratio in protein levels (cytochrome c: 10.74; GDH: 18.3) (data not shown). There was no significant difference between young and aged animals in levels of the loading controls used in these experiments, i.e., flotillin for lipid rafts and MnSOD for mitochondria (e.g., Fig. 4). The very low content of the mitochondrial markers found in lipid rafts indicated that our raft preparations had only negligible contamination from mitochondria. The relatively abundant levels of energy-regulating proteins observed in our SPM rafts were not likely due to mitochondrial contamination and appear to support the extra-mitochondrial localization of such proteins in the plasma membrane as noted by others, e.g., (Bae et al., 2004).
Fig. 4.
Evaluation of mitochondrial contamination by immunoblotting. Representative immunoblots show the levels of two mitochondrial markers in synaptic rafts vs. in mitochondria, both isolated from young (Y) and old (O) rat brains. Fifty micrograms of protein per lane were separated on an SDS-PAGE gel, transferred to PVDF membranes, and probed with anti-cytochrome c (1:200) and anti-GDH (1:800) antibodies. Flotillin (1:250) and MnSOD (1:2500) were used as loading controls for SPM raft and mitochondrial preparations, respectively.
3.4. Confirmation of age-associated changes in protein levels by immunoblot analysis
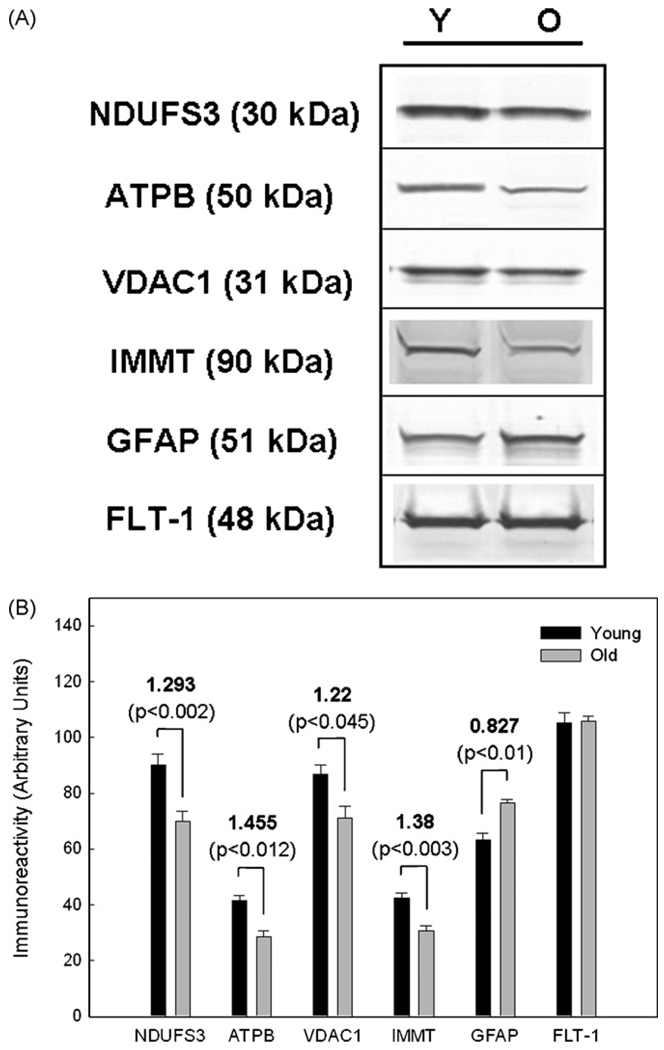
The next set of experiments was undertaken to confirm our proteomics results through immunoblot analyses of a few representative proteins that were statistically different in young vs. aged rafts. Lipid rafts prepared from new pairs of young and old rat SPMs were run on SDS-PAGE, probed with antibodies to several of the identified proteins, and subjected to densitometric analysis to determine if differential protein levels could be directly confirmed in the old vs. young rafts. The proteins NDUFS3, ATPB, VDAC1, IMMT, and GFAP representing a range of fold-changes were selected for immunoblot analysis. The protein level of the lipid raft marker FLT-1 served as a gel loading control as numerous experiments revealed no age-dependent change in levels of this protein. The absence of differences in FLT-1 levels in young vs. aged samples confirmed the stability of this protein, consistent with its role as a scaffold in rafts that organizes multi-protein signaling complexes (Stuermer and Plattner, 2005).
Representative immunoblots of five proteins assessed in young vs. old rafts are presented in Fig. 5A. Densitometric analysis of the band intensity from these experiments using three pairs of young and old raft preparations showed statistically significant changes in the protein levels of NDUFS3, ATPB, VDAC1, IMMT and GFAP with increasing age (Fig. 5B). The fold change calculated from these data was in good agreement with the DIGE results.
Fig. 5.
Immunoblot confirmation of the age-associated differences in levels of selected proteins. (A) Representative immunoblots showing age-related differences in protein levels in young (Y) vs. old (O) rafts. (B) Densitometric analysis of the protein alterations shown in (A). Data represent mean ± S.E.M. immunoreactivities in three separate preparations, with the indicated fold difference (Y/O) and P value above each set of samples. The dilutions used for the various antibodies were: anti-NDUFS3 (1:1,600), anti-ATPB (1:400), anti-VDAC1 (1:1,600), anti-IMMT (1:100), anti-GFAP (1:500), and anti-FLT-1 (1:250). The protein loading amounts were: NDUFS3 (25 µg), ATPB, VDAC1 and GFAP (30 µg), IMMT and FLT-1 (50 µg).
3.5. Functional categories of the altered proteins
We have identified significant changes in several synaptic raft proteins with advanced age, but the roles these proteins may play in raft-mediated synaptic function and possible implications for synaptic alterations in normal aging are undefined. Nevertheless, it seems important to consider the known biological functions of the major classes of raft proteins that changed with age. Thus, the identified proteins were analyzed for association with relevant biological processes by manual PubMed search and by using the online Onto-Express tool. Important caveats in this analysis are the limited number of raft proteins identified thus far and the likelihood that these represent the most abundant ones. Nevertheless, the raft-associated proteins that appear to change with increasing age are dominated by proteins involved in redox homeostasis and energy metabolism, with modest changes in proteins responsible for cell structure and, proteins expected to be at the synapses and involved in neurotransmission (Table 1).
4. Discussion
Only recently have reports appeared describing the raft proteomes from various organs and cell types including neurons (Martosella et al., 2006), hepatocytes (Bae et al., 2004), HeLa cells (Foster et al., 2003), and immune system cells (Li et al., 2004). Proteomic information has also now been obtained regarding the protein composition of several types of subcellular fractions obtained from the brain, including synaptic membranes (Stevens et al., 2003), postsynaptic densities (Peng et al., 2004; Li et al., 2005), synaptosomes and synaptic vesicles (Jia et al., 2006; Burre and Volknandt, 2007). In a quantitative proteomics report on rafts prepared from intact synaptosomes, the authors identified numerous proteins involved in synaptic vesicle cycling that were quite abundant in these preparations (Jia et al., 2006). Although, our starting material was an isolated SPM fraction rather than intact synaptosomes, we identified raft proteins that show considerable overlap with those in rafts from synaptosomes (Jia et al., 2006). These included proteins such as synapsin 2, syntaxin binding protein, actin, tubulin, VDAC1, NADH dehydrogenase, ATP synthase, ERCs and Septins. The substantial overlap between the proteins identified in synaptosomal rafts and those we identified in isolated SPM rafts do begin to provide overall characterization of the protein composition of synaptic rafts.
To our knowledge, the present study is the first report of age-dependent differences in levels of several proteins present in SPM lipid rafts isolated from rat brain. Our most striking finding with regard to aging as reflected in the SPM rafts was that the majority of proteins significantly decreased in aged SPM rafts are involved in bioenergetics and redox homeostasis. Although several such proteins are typically associated with the inner mitochondrial membrane, many of the same proteins are also localized in the transplasma membrane electron transport system or ‘PMRS’, a system that plays both pro-oxidant and anti-oxidant roles with a high degree of cell-specific regulation (del Castillo-Olivares et al., 2000; Ly and Lawen, 2003). Localization of electron transport proteins in plasma membranes has been reported in numerous cell types (Baker et al., 2004; Ushio-Fukai, 2006) including neurons (Bae et al., 2004; Kim et al., 2006), suggesting an extra-mitochondrial localization for several of these proteins. The cell surface localization of components of the respiratory chain and the ATP synthase complex has been further confirmed by immunohistochemistry (Kim et al., 2006), immunofluoresence (Bae et al., 2004; Kim et al., 2006), and cell surface biotinylation (Bae et al., 2004).
The PMRS contains at least two critical enzyme activities, NADH:ferricyanide reductase from VDAC1 (Baker et al., 2004) and the NADH oxidase, both of which play a role in the survival of cells devoid of mitochondrial DNA, i.e., ρ0 cells, or under partial anaerobic conditions (Ly and Lawen, 2003). These protein complexes apparently control cytoplasmic redox states by maintaining NADH/NAD+ ratios and consequently sustain ATP levels in the absence of actual mitochondria. The presence of PMRS proteins in lipid rafts is supported by the studies showing that known PMRS components, NADPH oxidase and VDAC1 (aka ‘Porin’), are expressed and activated in raft-like domains (Bathori et al., 1999; Ushio-Fukai, 2006). Our analysis further supports the presence of PMRS proteins in raft domains and, more importantly, indicates that these PMRS proteins are the ones most frequently altered with brain aging. Based on the known cellular functions of the PMRS, it is possible that, although mitochondria supply 90% of total cellular ATP (Vyatkina et al., 2004), the raft-localized proteins may contribute to the production of ATP through the ATP synthase complex or the cytosolic glycolytic machinery utilizing NAD+ regenerated by the PMRS. This would allow for highly localized responses to rapidly increased energy demands at synapses.
In addition to its energy-regulating activity, the PMRS serves as both a pro-oxidant system that generates ROS at the surface of phagocytic cells and, more importantly for neurons, an anti-oxidant system that protects the plasma membrane against oxidative damage (del Castillo-Olivares et al., 2000; Hyun et al., 2006a). Apparently this protection is due, at least in part, to the activity of the PMRS in transferring electrons from NAD(P)H and ascorbate to external ROS, thereby maintaining levels of reduced Coenzyme Q and α-tocopherol that protect the membrane against lipid peroxidation (Ly and Lawen, 2003; De Cabo et al., 2004; Hyun et al., 2006a). Of particular relevance to our study are the reports of significant age-related decreases in some PMRS enzyme activities observed in both aging liver and brain (De Cabo et al., 2004; Hyun et al., 2006b). Quite remarkably, caloric restriction, the only non-genetic means of extending life-span in mammals, led to dramatically higher levels and activities of several PMRS enzymes, Co-enzyme Q and α-tocopherol, in restricted vs. ad libitum-fed aged rats (De Cabo et al., 2004; Hyun et al., 2006b). Thus, our unbiased proteomic analysis of SPM rafts from young and old rats led to results fully consistent with the hypothesis that biological aging is associated with a diminution in the properties of the PMRS. An age-related reduction in this system could reduce antioxidant capacity, leading to the formation of markers of oxidative stress so well documented in aged tissues, particularly brain. A similar analysis comparing aged calorically restricted with ad libitum-fed rats might be quite instructive in assessing the possible prevention of the protein losses from the raft domains.
The DIGE analysis of young and aged SPM rafts also revealed significant age-dependent reductions in some proteins involved in synaptic activity (Table 1). In fact, the magnitude of the change in these proteins was the largest of all differences observed, ≥2.03. Multiple binding interactions among several elements in the ‘cytomatrix at the active zone’ (CAZ) complex, and the ERC family of proteins, key components of the CAZ complex, are physically and functionally associated with soluble NSF-attachment protein receptor (SNARE) complex (Takao-Rikitsu et al., 2004). Interestingly for our focus on rafts, the ERC (aka CAST) proteins, appear in the Triton X-100 fraction of transfected cells and recruit other binding partners to the insoluble microdomains, suggesting that at least a portion of the CAZ complexes partition into the lipid rafts (Higa et al., 2007). Aged rafts also showed reduced levels of synapsins, Septin 5, syntaxin binding protein 1 and cytoskeletal proteins, e.g., (Kartmann and Roth, 2001; Birthelmer et al., 2003; Giovedi et al., 2004; Arunachalam et al., 2008) all of which are implicated in neurotransmitter secretion. Whether reductions in CAZ proteins contribute to the attenuated neurotransmitter release that accompanies biological aging is not known (Birthelmer et al., 2003).
Cytoskeletal proteins were also highly represented among those most significantly reduced in aged SPM rafts (Table 1). Plasma membrane lipid microdomains are closely associated with the cytoskeletal network that provides scaffolding for the organization of diverse pre- and post-synaptic signaling complexes (Sudhof et al., 1989; Bhaskar et al., 2004; Frank and Lisanti, 2006; Spiliotis and Nelson, 2006). In the aging brain, decreased levels of crucial synaptic proteins, including key cytoskeletal proteins, could influence the organization of the active zones.
Although GFAP is widely used as an astroglial cell marker, GFAP is found in synapses possibly due to the association of astrocytic processes with the synaptic cleft. An elevation in GFAP is considered to be a biomarker of enhanced astrogliosis associated with brain aging and dementias (Wallin et al., 1996; Nichols, 1999). The presence of GFAP in our raft preparations may be a contaminant coming from regions where synaptic and glial membranes interact very closely. The higher GFAP levels in aged SPM rafts may simply reflect the enhanced gliosis present in the aged brain.
Our overall goal was to apply an unbiased proteomics strategy to probe for age-related differences in levels of proteins localized in brain SPM rafts as organizing microdomains for protein complexes central to neuronal signaling (Simons and Toomre, 2000). Our results were somewhat unexpected in revealing that the majority of proteins significantly reduced in aged SPM rafts are those of the multifunctional PMRS system. The marked reduction in levels of these proteins could lead to both local energy deficiencies and to a decrease in anti-oxidant defenses at the plasma membrane, a combination likely to enhance oxidative stress. Clearly transcriptional, translational, post-translational, or combinations of such processes may be responsible for the age-dependent reductions, and more targeted biochemical studies are needed to identify the mechanisms and the functional implications of the altered protein levels. The use of calorically restricted animals and more detailed studies of PMRS-associated anti-oxidants such as co-enzyme Q and α-tocopherol could provide insights into the process through which aging in the brain leads to progressive, if often subtle, cognitive impairment and enhanced vulnerability to neurodegenerative diseases. Our results indicate that the little-studied PMRS warrants further assessment as a pathway that contributes to altered signaling in aging brain and a potential source of insights for influencing the state of oxidative stress that develops with increasing age.
Supplementary Material
Acknowledgements
This work was supported by NIH grants RR-P20 017708 and AG12993, J.R. and Inez Jay Funds, University of Kansas, and funds from the Kansas City University of Medicine and Biosciences (A.Z.).
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.neurobiolaging.2008.11.005.
Footnotes
Conflict of interest
There is no conflict of interest.
References
- Alban A, David SO, Bjorkesten L, Andersson C, Sloge E, Lewis S, Currie I. A novel experimental design for comparative two-dimensional gel analysis: two-dimensional difference gel electrophoresis incorporating a pooled internal standard. Proteomics. 2003;3:36–44. doi: 10.1002/pmic.200390006. [DOI] [PubMed] [Google Scholar]
- Albers DS, Beal MF. Mitochondrial dysfunction and oxidative stress in aging and neurodegenerative disease. J. Neural. Transm. Suppl. 2000;59:133–154. doi: 10.1007/978-3-7091-6781-6_16. [DOI] [PubMed] [Google Scholar]
- Arunachalam L, Han L, Tassew NG, He Y, Wang L, Xie L, Fujita Y, Kwan E, Davletov B, Monnier PP, Gaisano HY, Sugita S. Munc18-1 Is critical for plasma membrane localization of syntaxin1 but not of SNAP-25 in PC12 cells. Mol. Biol. Cell. 2008;19:722–734. doi: 10.1091/mbc.E07-07-0662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae TJ, Kim MS, Kim JW, Kim BW, Choo HJ, Lee JW, Kim KB, Lee CS, Kim JH, Chang SY, Kang CY, Lee SW, Ko YG. Lipid raft proteome reveals ATP synthase complex in the cell surface. Proteomics. 2004;4:3536–3548. doi: 10.1002/pmic.200400952. [DOI] [PubMed] [Google Scholar]
- Baker MA, Lane DJ, Ly JD, De Pinto V, Lawen A. VDAC1 is a transplasma membrane NADH-ferricyanide reductase. J. Biol. Chem. 2004;279:4811–4819. doi: 10.1074/jbc.M311020200. [DOI] [PubMed] [Google Scholar]
- Bathori G, Parolini I, Tombola F, Szabo I, Messina A, Oliva M, De Pinto V, Lisanti M, Sargiacomo M, Zoratti M. Porin is present in the plasma membrane where it is concentrated in caveolae and caveolae-related domains. J. Biol. Chem. 1999;274:29607–29612. doi: 10.1074/jbc.274.42.29607. [DOI] [PubMed] [Google Scholar]
- Beites CL, Xie H, Bowser R, Trimble WS. The septin CDCrel-1 binds syntaxin and inhibits exocytosis. Nat. Neurosci. 1999;2:434–439. doi: 10.1038/8100. [DOI] [PubMed] [Google Scholar]
- Bhaskar K, Shareef MM, Sharma VM, Shetty AP, Ramamohan Y, Pant HC, Raju TR, Shetty KT. Co-purification and localization of Munc18-1 (p67) and Cdk5 with neuronal cytoskeletal proteins. Neurochem. Int. 2004;44:35–44. doi: 10.1016/s0197-0186(03)00099-8. [DOI] [PubMed] [Google Scholar]
- Birthelmer A, Stemmelin J, Jackisch R, Cassel JC. Presynaptic modulation of acetylcholine, noradrenaline, and serotonin release in the hippocampus of aged rats with various levels of memory impairments. Brain Res. Bull. 2003;60:283–296. doi: 10.1016/s0361-9230(03)00042-x. [DOI] [PubMed] [Google Scholar]
- Burre J, Volknandt W. The synaptic vesicle proteome. J. Neurochem. 2007;101:1448–1462. doi: 10.1111/j.1471-4159.2007.04453.x. [DOI] [PubMed] [Google Scholar]
- De Cabo R, Cabello R, Rios M, Lopez-Lluch G, Ingram DK, Lane MA, Navas P. Calorie restriction attenuates age-related alterations in the plasma membrane antioxidant system in rat liver. Exp. Gerontol. 2004;39:297–304. doi: 10.1016/j.exger.2003.12.003. [DOI] [PubMed] [Google Scholar]
- del Castillo-Olivares A, Nunez de Castro I, Medina MA. Dual role of plasma membrane electron transport systems in defense. Crit. Rev. Biochem. Mol. Biol. 2000;35:197–220. doi: 10.1080/10409230091169203. [DOI] [PubMed] [Google Scholar]
- Floyd RA, Hensley K. Oxidative stress in brain aging. Implications for therapeutics of neurodegenerative diseases. Neurobiol. Aging. 2002;23:795–807. doi: 10.1016/s0197-4580(02)00019-2. [DOI] [PubMed] [Google Scholar]
- Foster LJ, De Hoog CL, Mann M. Unbiased quantitative proteomics of lipid rafts reveals high specificity for signaling factors. Proc. Natl. Acad. Sci. U.S.A. 2003;100:5813–5818. doi: 10.1073/pnas.0631608100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank PG, Lisanti MP. Defining lipid raft structure and function with proximity imaging. Am. J. Physiol. Heart Circ. Physiol. 2006;290:H2165–H2166. doi: 10.1152/ajpheart.00041.2006. [DOI] [PubMed] [Google Scholar]
- Giovedi S, Vaccaro P, Valtorta F, Darchen F, Greengard P, Cesareni G, Benfenati F. Synapsin is a novel Rab3 effector protein on small synaptic vesicles. I. Identification and characterization of the synapsin I-Rab3 interactions in vitro and in intact nerve terminals. J. Biol. Chem. 2004;279:43760–43768. doi: 10.1074/jbc.M403293200. [DOI] [PubMed] [Google Scholar]
- Hansen GH, Niels-Christiansen LL, Thorsen E, Immerdal L, Danielsen EM. Cholesterol depletion of enterocytes. Effect on the Golgi complex and apical membrane trafficking. J. Biol. Chem. 2000;275:5136–5142. doi: 10.1074/jbc.275.7.5136. [DOI] [PubMed] [Google Scholar]
- Hering H, Lin CC, Sheng M. Lipid rafts in the maintenance of synapses, dendritic spines, and surface AMPA receptor stability. J. Neurosci. 2003;23:3262–3271. doi: 10.1523/JNEUROSCI.23-08-03262.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higa S, Tokoro T, Inoue E, Kitajima I, Ohtsuka T. The active zone protein CAST directly associates with Ligand-of-Numb protein X. Biochem. Biophys. Res. Commun. 2007;354:686–692. doi: 10.1016/j.bbrc.2007.01.036. [DOI] [PubMed] [Google Scholar]
- Hojlund K, Wrzesinski K, Larsen PM, Fey SJ, Roepstorff P, Handberg A, Dela F, Vinten J, McCormack JG, Reynet C, Beck-Nielsen H. Proteome analysis reveals phosphorylation of ATP synthase beta-subunit in human skeletal muscle and proteins with potential roles in type 2 diabetes. J. Biol. Chem. 2003;278:10436–10442. doi: 10.1074/jbc.M212881200. [DOI] [PubMed] [Google Scholar]
- Hou Q, Huang Y, Amato S, Snyder SH, Huganir RL, Man HY. Regulation of AMPA receptor localization in lipid rafts. Mol. Cell Neurosci. 2008;38:213–223. doi: 10.1016/j.mcn.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun DH, Hernandez JO, Mattson MP, de Cabo R. The plasma membrane redox system in aging. Ageing Res. Rev. 2006a;5:209–220. doi: 10.1016/j.arr.2006.03.005. [DOI] [PubMed] [Google Scholar]
- Hyun DH, Emerson SS, Jo DG, Mattson MP, de Cabo R. Calorie restriction up-regulates the plasma membrane redox system in brain cells and suppresses oxidative stress during aging. Proc. Natl. Acad. Sci. U.S.A. 2006b;103:19908–19912. doi: 10.1073/pnas.0608008103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikehata K, Duzhak TG, Galeva NA, Ji T, Koen YM, Hanzlik RP. Protein targets of reactive metabolites of thiobenzamide in rat liver in vivo. Chem. Res. Toxicol. 2008;21:1432–1442. doi: 10.1021/tx800093k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia JY, Lamer S, Schumann M, Schmidt MR, Krause E, Haucke V. Quantitative proteomics analysis of detergent-resistant membranes from chemical synapses: evidence for cholesterol as spatial organizer of synaptic vesicle cycling. Mol. Cell. Proteomics. 2006;5:2060–2071. doi: 10.1074/mcp.M600161-MCP200. [DOI] [PubMed] [Google Scholar]
- Jiang L, He L, Fountoulakis M. Comparison of protein precipitation methods for sample preparation prior to proteomic analysis. J. Chromatogr. A. 2004;1023:317–320. doi: 10.1016/j.chroma.2003.10.029. [DOI] [PubMed] [Google Scholar]
- Jiang L, Fernandes D, Mehta N, Bean JL, Michaelis ML, Zaidi A. Partitioning of the plasma membrane Ca(2+)-ATPase into lipid rafts in primary neurons: effects of cholesterol depletion. J. Neurochem. 2007;102:378–388. doi: 10.1111/j.1471-4159.2007.04480.x. [DOI] [PubMed] [Google Scholar]
- Jiang L, Lindpaintner K, Li HF, Gu NF, Langen H, He L, Fountoulakis M. Proteomic analysis of the cerebrospinal fluid of patients with schizophrenia. Amino Acids. 2003;25:49–57. doi: 10.1007/s00726-003-0356-6. [DOI] [PubMed] [Google Scholar]
- Kartmann B, Roth D. Novel roles for mammalian septins: from vesicle trafficking to oncogenesis. J. Cell Sci. 2001;114:839–844. doi: 10.1242/jcs.114.5.839. [DOI] [PubMed] [Google Scholar]
- Kersey PJ, Duarte J, Williams A, Karavidopoulou Y, Birney E, Apweiler R. The International Protein Index: an integrated database for proteomics experiments. Proteomics. 2004;4:1985–1988. doi: 10.1002/pmic.200300721. [DOI] [PubMed] [Google Scholar]
- Khachaturian ZS. Calcium hypothesis of Alzheimer’s disease and brain aging. Ann. NY. Acad. Sci. 1994;747:1–11. doi: 10.1111/j.1749-6632.1994.tb44398.x. [DOI] [PubMed] [Google Scholar]
- Kim KB, Lee JW, Lee CS, Kim BW, Choo HJ, Jung SY, Chi SG, Yoon YS, Yoon G, Ko YG. Oxidation-reduction respiratory chains and ATP synthase complex are localized in detergent-resistant lipid rafts. Proteomics. 2006;6:2444–2453. doi: 10.1002/pmic.200500574. [DOI] [PubMed] [Google Scholar]
- Lai JC, Sheu KF, Kim YT, Clarke DD, Blass JP. The subcellular localization of glutamate dehydrogenase (GDH): is GDH a marker for mitochondria in brain? Neurochem. Res. 1986;11:733–744. doi: 10.1007/BF00965341. [DOI] [PubMed] [Google Scholar]
- Lee CK, Weindruch R, Prolla TA. Gene-expression profile of the ageing brain in mice. Nat. Genet. 2000;25:294–297. doi: 10.1038/77046. [DOI] [PubMed] [Google Scholar]
- Li K, Hornshaw MP, van Minnen J, Smalla KH, Gundelfinger ED, Smit AB. Organelle proteomics of rat synaptic proteins: correlation-profiling by isotope-coded affinity tagging in conjunction with liquid chromatography-tandem mass spectrometry to reveal postsynaptic density specific proteins. J. Proteome Res. 2005;4:725–733. doi: 10.1021/pr049802+. [DOI] [PubMed] [Google Scholar]
- Li N, Shaw AR, Zhang N, Mak A, Li L. Lipid raft proteomics: analysis of in-solution digest of sodium dodecyl sulfate-solubilized lipid raft proteins by liquid chromatography-matrix-assisted laser desorption/ionization tandem mass spectrometry. Proteomics. 2004;4:3156–3166. doi: 10.1002/pmic.200400832. [DOI] [PubMed] [Google Scholar]
- Lucero HA, Robbins PW. Lipid rafts-protein association and the regulation of protein activity. Arch. Biochem. Biophys. 2004;426:208–224. doi: 10.1016/j.abb.2004.03.020. [DOI] [PubMed] [Google Scholar]
- Ly JD, Lawen A. Transplasma membrane electron transport: enzymes involved and biological function. Redox Rep. 2003;8:3–21. doi: 10.1179/135100003125001198. [DOI] [PubMed] [Google Scholar]
- Martosella J, Zolotarjova N, Liu H, Moyer SC, Perkins PD, Boyes BE. High recovery HPLC separation of lipid rafts for membrane proteome analysis. J. Proteome Res. 2006;5:1301–1312. doi: 10.1021/pr060051g. [DOI] [PubMed] [Google Scholar]
- Masliah E, Crews L, Hansen L. Synaptic remodeling during aging and in Alzheimer’s disease. J. Alzheimers Dis. 2006;9:91–99. doi: 10.3233/jad-2006-9s311. [DOI] [PubMed] [Google Scholar]
- Michaelis EK, Michaelis ML, Chang HH, Kitos TE. High affinity Ca2+-stimulated Mg2+-dependent ATPase in rat brain synaptosomes, synaptic membranes, and microsomes. J. Biol. Chem. 1983;258:6101–6108. [PubMed] [Google Scholar]
- Nichols NR. Glial responses to steroids as markers of brain aging. J. Neurobiol. 1999;40:585–601. [PubMed] [Google Scholar]
- Nishibayashi S, Ogawa N, Asanuma M, Kondo Y, Mori A. Tubulin and actin mRNAs in the young-adult and the aged rat brain: effects of repeated administration with bifemelane hydrochloride. Arch. Gerontol. Geriatr. 1994;19:265–272. doi: 10.1016/0167-4943(94)00572-9. [DOI] [PubMed] [Google Scholar]
- Peng J, Kim MJ, Cheng D, Duong DM, Gygi SP, Sheng M. Semiquantitative proteomic analysis of rat forebrain postsynaptic density fractions by mass spectrometry. J. Biol. Chem. 2004;279:21003–21011. doi: 10.1074/jbc.M400103200. [DOI] [PubMed] [Google Scholar]
- Rohlff C. Proteomics in molecular medicine: applications in central nervous systems disorders. Electrophoresis. 2000;21:1227–1234. doi: 10.1002/(SICI)1522-2683(20000401)21:6<1227::AID-ELPS1227>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Simons K, Toomre D. Lipid rafts and signal transduction. Nat. Rev. Mol. Cell Biol. 2000;1:31–39. doi: 10.1038/35036052. [DOI] [PubMed] [Google Scholar]
- Spiliotis ET, Nelson WJ. Here come the septins: novel polymers that coordinate intracellular functions and organization. J. Cell Sci. 2006;119:4–10. doi: 10.1242/jcs.02746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens SM, Jr, Zharikova AD, Prokai L. Proteomic analysis of the synaptic plasma membrane fraction isolated from rat forebrain. Brain Res. Mol. Brain Res. 2003;117:116–128. doi: 10.1016/s0169-328x(03)00282-1. [DOI] [PubMed] [Google Scholar]
- Stuermer CA, Plattner H. The ‘lipid raft’ microdomain proteins reggie-1 and reggie-2 (flotillins) are scaffolds for protein interaction and signalling. Biochem. Soc. Symp. 2005;72:109–118. doi: 10.1042/bss0720109. [DOI] [PubMed] [Google Scholar]
- Sudhof TC, Czernik AJ, Kao HT, Takei K, Johnston PA, Horiuchi A, Kanazir SD, Wagner MA, Perin MS, De Camilli P, et al. Synapsins: mosaics of shared and individual domains in a family of synaptic vesicle phosphoproteins. Science. 1989;245:1474–1480. doi: 10.1126/science.2506642. [DOI] [PubMed] [Google Scholar]
- Takao-Rikitsu E, Mochida S, Inoue E, Deguchi-Tawarada M, Inoue M, Ohtsuka T, Takai Y. Physical and functional interaction of the active zone proteins, CAST, RIM1, and Bassoon, in neurotransmitter release. J. Cell Biol. 2004;164:301–311. doi: 10.1083/jcb.200307101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushio-Fukai M. Localizing NADPH oxidase-derived ROS. Sci. STKE. 2006;2006:re8. doi: 10.1126/stke.3492006re8. [DOI] [PubMed] [Google Scholar]
- Vyatkina G, Bhatia V, Gerstner A, Papaconstantinou J, Garg N. Impaired mitochondrial respiratory chain and bioenergetics during chagasic cardiomyopathy development. Biochim. Biophys. Acta. 2004;1689:162–173. doi: 10.1016/j.bbadis.2004.03.005. [DOI] [PubMed] [Google Scholar]
- Wallin A, Blennow K, Rosengren LE. Glial fibrillary acidic protein in the cerebrospinal fluid of patients with dementia. Dementia. 1996;7:267–272. doi: 10.1159/000106891. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.