Abstract
Cholangiocarcinomas (CCs) are highly lethal malignant tumours arising from the biliary tract epithelium. The disease is notoriously difficult to diagnose and is usually fatal because of its typically late clinical presentation and the lack of effective non-surgical therapeutic modalities. The overall survival rate, including resected patients is poor, with less than 5% of patients surviving 5 years, a rate which has not changed significantly over the past 30 years. Although CC is a relatively uncommon tumor, interest in this disease is rising as incidence and mortality rates for intrahepatic cholangiocarcinoma are increasing markedly worldwide. A variety of risk factors, including primary sclerosing cholangitis, liver fluke infestation, and hepatolithiasis have been described. However, for most CCs the cause is unknown, and affected individuals have no history of exposure to, or association with, known risk factors. Recent advances in molecular pathogenesis have highlighted the importance of epigenetic alterations in the form of promoter region hypermethylation and histone deacetylation in addition to genetic changes in the process of cholangiocarcinogenesis. This review provides a comprehensive overview of the genes reported to be methylated in CC to date and their putative roles in cholangiocarcinogenesis. Future directions in the study of methylated genes and their potential roles as diagnostic and prognostic markers are also discussed.
Keywords: cholangiocarcinoma, epigenetics, histone deacetylation, promoter region hypermethylation
Cholangiocarcinomas (CCs) are rare malignant tumours arising from the biliary tract that were first reported by Durand-Fardel in 1840 (1). The disease is notoriously difficult to diagnose and is usually fatal because of its typically late clinical presentation and the lack of effective non-surgical therapeutic modalities (2). Most patients have unresectable disease at presentation and die within 12 months from the effects of cancer cachexia and a subsequent rapid decline in performance status. Liver failure and recurrent sepsis secondary to biliary obstruction also contribute to the high mortality (3). The overall survival rate, including in resected patients, is poor, with < 5% of patients surviving 5 years, a rate that has not changed significantly over the past 30 years (4).
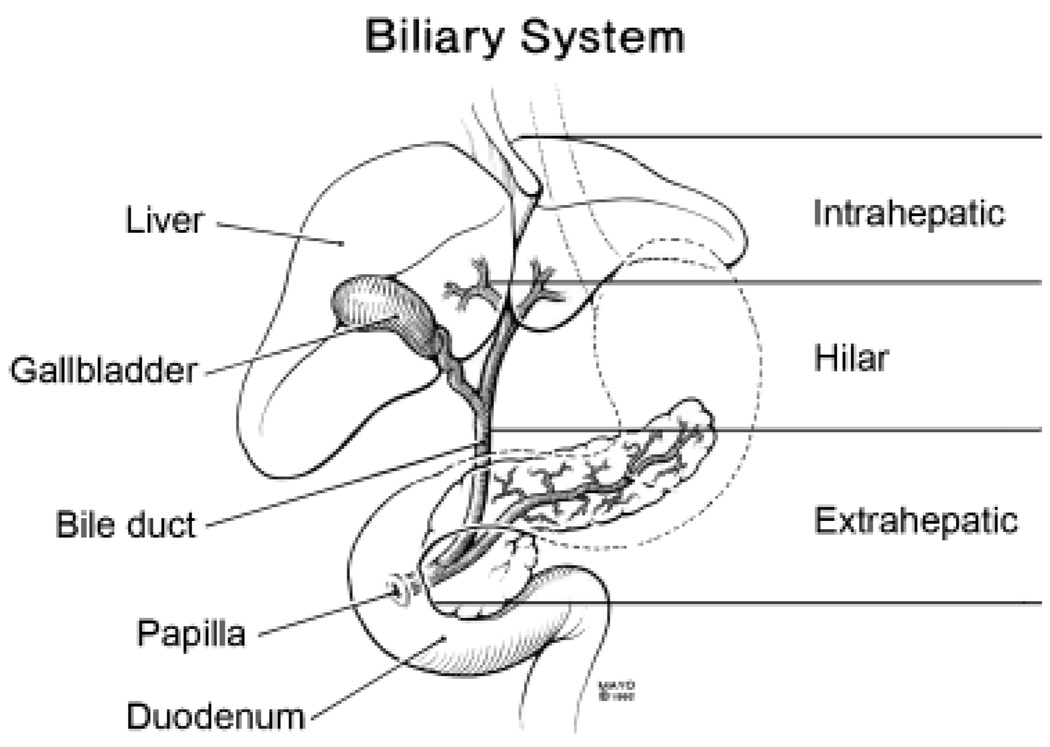
More than 90% of CCs are adenocarcinomas (2). CCs are categorized according to their anatomic location as either intrahepatic (ICC) or extrahepatic (ECC). This classification is rational, as there are clinical, pathological, epidemiological and molecular differences between these two groups. Over 50% of all CCs, however, are ECCs that arise within the perihilar region – some of these tumours extend into the liver and are classified as intrahepatic. These tumours are, therefore, best described as hilar lesions, with the other types being described more specifically as intrahepatic and extrahepatic lesions (5) (Fig. 1).
Fig. 1.
Anatomic classification of cholangiocarcinoma (CC). CCs are classified as intrahepatic, hilar or extrahepatic based on their anatomical location.
Worldwide, CC is relatively uncommon, accounting for 3% of all gastrointestinal cancers (6). Unfortunately, many previous epidemiological studies have grouped ICC together with other primary hepatic tumours, ECC with gallbladder cancers and hilar lesions with intrahepatic extension with ICC. Because of the lack of a consistent classification, the true incidence of these lesions is therefore unknown (7). Nevertheless, hilar cancers (Klatskin tumours) are the most frequent, comprising 55–60% of CCs; 20–30% of CCs are non-hilar ECC, and 10% are ICC (5). Although CC is a relatively rare tumour, interest in this disease is rising as incidence and mortality rates for ICC are increasing markedly worldwide (4, 8–12). On the other hand, the incidence and mortality rates of ECC have been decreasing. The cause for the increasing incidence of ICC remains unclear. The data for ECC are perhaps more difficult to obtain because gallbladder cancers are typically combined with ECC for International Classification of Diseases coding purposes, and the incidence of gallbladder cancers is known to be declining, probably as a result of increasing cholecystectomy rates over the past few decades (4, 8, 11).
Risk factors
Several risk factors have been associated with the development of CC (Fig. 2). However, for most CC cases the cause is unknown, and affected individuals have no history of exposure to or association with known risk factors (13).
Fig. 2.
Proposed aetiopathogenetic mechanism of cholangiocarcinogenesis showing interactions between aetiological agents and cholangiocyte response to injury.
Primary sclerosing cholangitis
Primary sclerosing cholangitis (PSC) is the most common known predisposing condition for CC with a reported lifetime risk between 9 and 23% (14, 15). Among patients with PSC who develop CC, approximately one-third will be diagnosed with CC within 2 years of diagnosis of PSC (16, 17).
Liver fluke infestation
There is a clear established role of infestation with the liver fluke Opisthorchis viverrini (and less definitively Clonorchis sinensis) as an aetiological factor in the development of CC (18). However, the relatively low incidence of CC in countries that have a high prevalence of Opisthorchis and Clonorchis infestations indicates that additional cofactors are important for biliary carcinogenesis. Liver flukes are therefore, for the most part, promoters rather than initiators of CC (19).
Congenital abnormalities
Caroli’s syndrome, congenital hepatic fibrosis, choledochal cysts and anomalous pancreaticobiliary-junction malformations carry a 10–15% lifetime risk of developing CC (20). The lifetime incidence of CC in patients with untreated choledochal cysts is as high as 28% (21, 22). Bile duct adenomas and biliary papillomatosis are also associated with the development of CC (23).
Hepatolithiasis
Although rare in the west, hepatolithiasis (stones in the intra- and/or extrahepatic biliary tract) is relatively common in parts of Asia and has been found to be particularly associated with peripheral ICC (4).
Additional risk factors
Chronic hepatitis C virus infection, hepatitis B virus (HBV) infection, HIV infection, liver cirrhosis and diabetes have all been associated with ICC (24, 25). Carcinogenic toxins, especially thorotrast, have been strongly linked with the development of CC many years after exposure (4, 26). Associations have also been made with exposure to dioxins, vinyl chloride, nitrosamines, alcohol and smoking (27–31). However, in all cases of CC, additional contributing factors may be changes in the patient’s genetic and epigenetic constitution that may predispose to the development of disease.
Prognostic and therapeutic considerations in cholangiocarcinoma
Tumour size, tumour number, lymph node metastasis, tumour stage and tumour histological grade have been identified as factors with a statistically significant prognostic impact (32). The poor outcome of patients with CC is believed to be owing to the occurrence of early lymph node metastasis and perineural invasion (33). The reported positive rate of perihepatic lymph node involvement ranges from 30 to 50% in patients with hilar CC, and from 47 to 58% in those with ICC (34, 35). Recent advances in molecular techniques have revealed that conventional histological examination does not reliably detect micrometastatic foci of biliary tract carcinoma in lymph nodes (36). Perineural invasion is found in as many as 81% of reported cases of CC (37). Although nodal metastases have proven to be a definitive prognostic factor, there is some controversy as to whether perineural infiltration constitutes an adverse prognostic factor. A study from Nagoya, Japan, found a strong correlation between perineural involvement and poor survival; however, this was not confirmed in a study from Germany (37, 38).
Surgical liver resection has long been considered the standard of care for resectable CC; however, recent excellent results from liver transplantation in highly selected patients with hilar CC are challenging this paradigm. Liver transplantation offers several benefits over surgical resection, particularly in that it is not limited by conventional criteria of unresectability such as encasement of major vessels or bilobar tumour extension. There is also a higher chance of achieving tumour-free margins. The major limitation of liver transplantation is the shortage of cadaveric donor organs; this can be somewhat mitigated by the use of living donor transplantation. Using fairly stringent patient selection criteria, the Mayo Clinic protocol of neoadjuvant chemoradiation, followed by liver transplantation has achieved patient survival as high as 92, 82 and 82% at 1, 3 and 5 years respectively (39).
Genetic alterations in cholangiocarcinoma
Tumorigenesis is a multistep process in which defects in various cancer genes accumulate (40, 41). Virtually every tumour type has an enormous complexity of altered gene functions, including activation of growth-promoting genes as well as silencing of genes with tumour growth-suppressing functions, all contributing to uncontrolled proliferation. Hanahan and Weinberg (42) proposed that cancer gene functions can be classified into six essential alterations in cell physiology: self-sufficiency in growth signals, insensitivity to growth-inhibitory signals, evasion of apoptosis, limitless replicative potential, sustained angiogenesis and tissue invasion and metastasis.
There is evidence that the neoplastic transformation of biliary epithelial cells and malignant progression of CC are accompanied by a number of genetic and epigenetic alterations (43, 44). Cancer-related genes that have been evaluated thus far include K-ras, p53, p14ARF, p16INK4a and β-catenin. Genetic alterations such as point mutations of K-ras and p53 have been found in a subset of CC (45–48). The reported rates of K-ras mutations in biliary tract cancers vary widely from 0 to 56% for ICC and from 0 to 100% for ECC. These variations are presumably owing to differences in the subsites of cancers, racial and geographical variations in the study populations and the use of different assay techniques in these studies. Most of the K-ras gene mutations occur in codon 12. Mutation or deletion of the cell cycle regulators p14ARF and p16INK4a are not frequent in CC (49, 50). Although overexpression of β-catenin is frequently encountered in CC, mutations of the β-catenin gene have not been identified to date in ICC (51).
Epigenetics and cancer
Epigenetics is the study of heritable changes in gene expression that occur without changes in DNA sequence. DNA methylation and histone modifications constitute the common epigenetic changes in human cancers.
DNA methylation
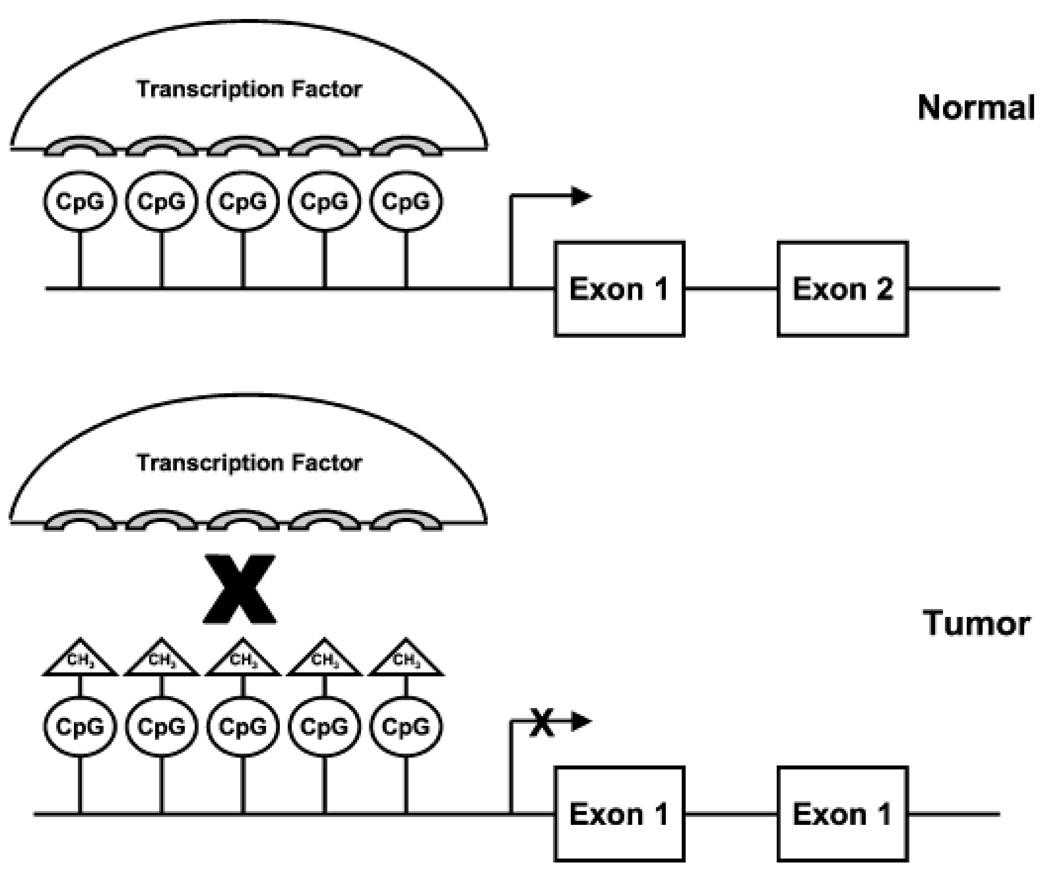
DNA hypermethylation is a naturally occurring reversible process that regulates the expression of cellular genes and has been shown to be essential for embryonic development (52), imprinting (53), X chromosome inactivation (54) and suppression of parasitic DNA sequences (55). DNA methylation occurs via the covalent addition of a methyl group to the 5- position of the cytosine ring within the context of a cytosine nucleotide followed by a guanine nucleotide (CpG dinucleotide or CpG site) (56). CpG dinucleotides appear at about a five-fold lower than expected frequency in the overall genome (57). Small regions of genomic DNA with an increased density of CpG dinucleotides, called CpG islands, represent exceptions to this rule. It has been estimated that almost half of the human gene promoter regions contain these CpG-rich regions, which extend from 0.5 kb to several kb in length and are protected from methylation in normal cells (56, 58). Promoter region hypermethylation typically results in downregulation or silencing of gene transcription; consequently, the expression of a tumour suppressor gene can be silenced through aberrant hypermethylation in cancer (Fig. 3) (59–61). This aberrant hypermethylation of the promoter CpG islands of human tumour suppressor genes is an alternative mechanism of gene inactivation that contributes to the biology of several human neoplasms (62, 63). As genomic methylation has been shown to increase with age, methylation may also be one of the molecular lesions that confer the increased predisposition to cancer with increasing age.
Fig. 3.
Promoter region hypermethylation inhibits transcription factor–DNA interactions. In a normal cell, transcription factors interact with the promoter region of a gene and initiate, enhance or repress gene transcription. Hypermethylation of CpG islands in the promoter region inhibits the interaction between transcription factors and promoter DNA, typically leading to inhibition of gene transcription.
For a variety of genes, a complete paradigm for loss of function in cancer owing to promoter hypermethylation has been established. In each case, several important features are inherent to the epigenetic inactivation (59). First, promoter region methylation almost always occurs in tumours lacking coding region mutations for the involved genes, thereby indicating that the epigenetic change constitutes a true alternative to classical genetic mechanisms for loss of gene function. Second, the expression of each gene can be, at least partially, reactivated by the treatment of cultured tumour cells with the demethylating agent 5-azacytidine. This results in some degree of loss of methylation from the promoter CpG island (64). Third, the selective advantage of losing tumour suppressor gene function for a given gene appears to be identical for coding region mutations vs. promoter region hypermethylation. The methylation change is associated with a fully heritable event to silence wild-type alleles and can even serve as an inactivating event on one allele when the opposite allele is mutated and expressed in the same cells (65). In addition to proven tumour suppressor genes and cancer-causing genes, other genes potentially critical to tumorigenesis are also hypermethylated in human tumours. Prominent among them are p15INK4a, GST-Pi, O6-MGMT, TIMP3, DAP-kinase and p73 (63). Recent studies have suggested that promoter region hypermethylation of a number of key genes occurs early in neoplastic development (66–68).However, the loss of gene function associated with this process may be temporally different from the loss caused by classic promoter region mutations (69, 70).
Despite confirmatory evidence of CpG methylation of CpG-rich promoters having a negative effect on gene expression, the exact mechanism for silencing gene transcription is still under investigation. It is known that the physical status of chromatin differs for promoter region CpG islands that are methylated vs. those that are not. Unmethylated islands in most normal genes are associated with open, transcriptionally favourable chromatin conformations containing highly acetylated histones, irregular nucleosomal spacing and lessening of nucleosomal compaction. All these factors favour the access of transcriptional activators to the promoter (58, 71, 72). Methylated islands, such as those for many transcriptionally repressed genes on the inactive X chromosome of the female, interact with chromatin characterized by tightly packed and uniformly spaced nucleosomes and the presence of de-acetylated histones (58, 73). For the inactivated genes on the inactive X chromosome, it is interesting to note that methylation may follow gene inactivation initiated by the RNA species, X inactive specific transcript, during very early embryogenesis. This methylation may then serve as a final locking mechanism for transcriptional inactivation (74). A number of studies suggest that the modulation of histone acetylation and the presence of proteins that preferentially bind to methylated cytosines may be critical for tumorigenesis (75, 76). Thus, dense CpG island promoter methylation and histone deacetylase (HDAC) activity work together to aberrantly silence genes in cancer (77). In a study of four hypermethylated genes in cultured tumour cells, MLH1, p15, p16 and TIMP3, specific inhibition of cellular HDAC activity with the drug trichostatin (TSA) failed to reactivate any of the genes. In the same cells, the expression of nonmethylated genes was significantly upregulated by TSA. However, if the cells were first pretreated with a low dose of the demethylating agent 5-deoxyazacytidine (DAC) for a short period, producing very modest demethylation of the CpG islands, then subsequent TSA treatment resulted in substantial re-expression of each hypermethylated gene. Promoter methylation and HDAC activity therefore appear to participate as dual layers of control to produce aberrant gene inactivation in cancer cells (77).
Some degree of CpG methylation has been found in almost every type of neoplastic tissue in the gastrointestinal tract. The molecular approaches used to identify the DNA sequences targeted by methylation vary from the use of genome-wide discovery methods for identifying target genes or sequences to a candidate gene approach using studies of methylation in the promoter region of target genes identified by their tumour suppressor effects or by an association between their expression levels and cancer.
Histone modifications
Histones are basic proteins that complex with genomic DNA to form nucleosomes, the basic units of the compacted structure of chromatin. Histones are modified post-translationally by acetylation, methylation and phosphorylation. Histone acetylation appears to be the major means of regulation of histone function. Histones are acetylated on lysine residues at their amino termini by histone acetyltransferases (HATs), and acetylated histones are deacetylated by HDACs. The opposing effects of HATs and HDACs regulate gene expression through chromatin modification. The HDAC-mediated removal of acetyl groups from lysine residues in the amino termini of histones leads to chromatin condensation and transcriptional inactivation of the involved DNA (78, 79). This transcriptional inactivation can contribute to suppression of tumour suppressor gene expression and enhanced tumorigenesis. Similar to the action of DNA methylation inhibitors in removing the epigenetic inactivation of tumour suppressor gene expression, HDAC inhibitors enhance the acetylation state of histones, leading to chromatin decondensation and increased gene expression. HDAC inhibitors can therefore reverse the aberrant epigenetic state associated with cancer and have been shown to act in synergy with DNA methylation inhibitors to inhibit tumour growth (80, 81). There is, as yet, little known about the specific roles of histone modifications in cholangiocarcinogenesis, and this is an area that warrants further investigation.
Techniques for analysis of DNA methylation
The bisulphite reaction was first described in the early 1970s and was used by Frommer et al. to distinguish between cytosine and 5-methylcytosine (5-mC) in DNA. In this reaction, DNA is first treated with sodium bisulphite to convert cytosine residues to uracil in single-stranded DNA, under conditions in which 5-mC remains essentially non-reactive. The DNA sequence under investigation is then amplified by polymerase chain reaction (PCR) with primers specific for bisulphite-modified DNA. Many methods based on the same principle have been developed, including bisulphite treatment with PCR-single-strand conformation polymorphism analysis [bisulphite-PCR-SSCP (BiPS)], methylation-specific PCR (MSP), combined bisulphite restriction analysis (COBRA) and methylation-sensitive single-nucleotide primer extension, with the first two being the most commonly used. All methods share the same procedure of modifying DNA with sodium bisulphite as the first step, with subsequent PCR amplification using primers specific for bisulphite-modified DNA (82–84).
Methylation-specific PCR is a simple, rapid and inexpensive method to determine the methylation status of CpG islands. This approach allows the determination of methylation patterns from very small amounts of DNA, including those obtained from paraffin-embedded samples, and can be used in the study of abnormally methylated CpG islands in neoplasia, in studies of imprinted genes and in studies of human tumours for clonality by studying genes inactivated on the X chromosome. It is a bisulphite conversion-based PCR technique for the study of DNA CpG methylation. For MSP experiments, two pairs of primers are needed with one pair specific for methylated DNA (M) and the other for unmethylated DNA (U). To achieve discrimination between methylated and unmethylated DNA, one or more CpG sites are included in each primer sequence (or at least one of the pair). Separate PCR reactions are performed using the M and U primer pairs and bisulphite-modified DNA. Successful amplification from the M pair or the U pair indicates methylation or unmethylation respectively (85).
Methylation-sensitive single-nucleotide primer extension is a technique for analysing several CpG dinucleotides in a single reaction. Paired primer extensions with 32P-linked 2′-deoxycytidine 5′-triphosphate (dCTP) or thymidine triphosphate (TTP) reveal whether a cytosine is methylated or not. Primers are made that anneal to the PCR template and subsequently terminate 5′ of the cytosine to be assayed. This quantitatively assesses the ratio of methylated and unmethylated cytosines in the DNA sample at that specific CpG dinucleotide (86).
Combined bisulphite restriction analysis is a relatively easy-to-use quantitative method to determine DNA methylation levels at specific gene loci in small amounts of genomic DNA. In addition, this technique has been reliably applied to DNA obtained from microdissected paraffin-embedded tissue samples (87).
Real-time PCR, also known as kinetic PCR, qPCR, qRT-PCR and RT-qPCR, is a quantitative PCR method for the determination of the copy number of PCR templates such as DNA or cDNA in a PCR reaction. There are two flavours of real-time PCR: probe based and intercalator based. Both methods require a special thermocycler equipped with a sensitive camera that monitors the fluorescence in each well of the 96-well plate at frequent intervals during the PCR reaction. Probe-based real-time PCR, also known as TaqMan PCR, requires a pair of PCR primers as regular PCR does, as well as an additional fluorogenic probe, which is an oligonucleotide with both a reporter fluorescent dye and a quencher dye attached. The intercalator-based method uses a double-stranded DNA dye such as SYBR green in the PCR reaction that fluoresces on binding to newly synthesized double-stranded DNA. The TaqMan method is more specific than the SYBR green method, but is also more expensive. The use of real-time PCR with bisulphite-modified DNA and appropriately designed primers allows quantification of the amount of methylated DNA in a sample.
Genes hypermethylated in cholangiocarcinoma
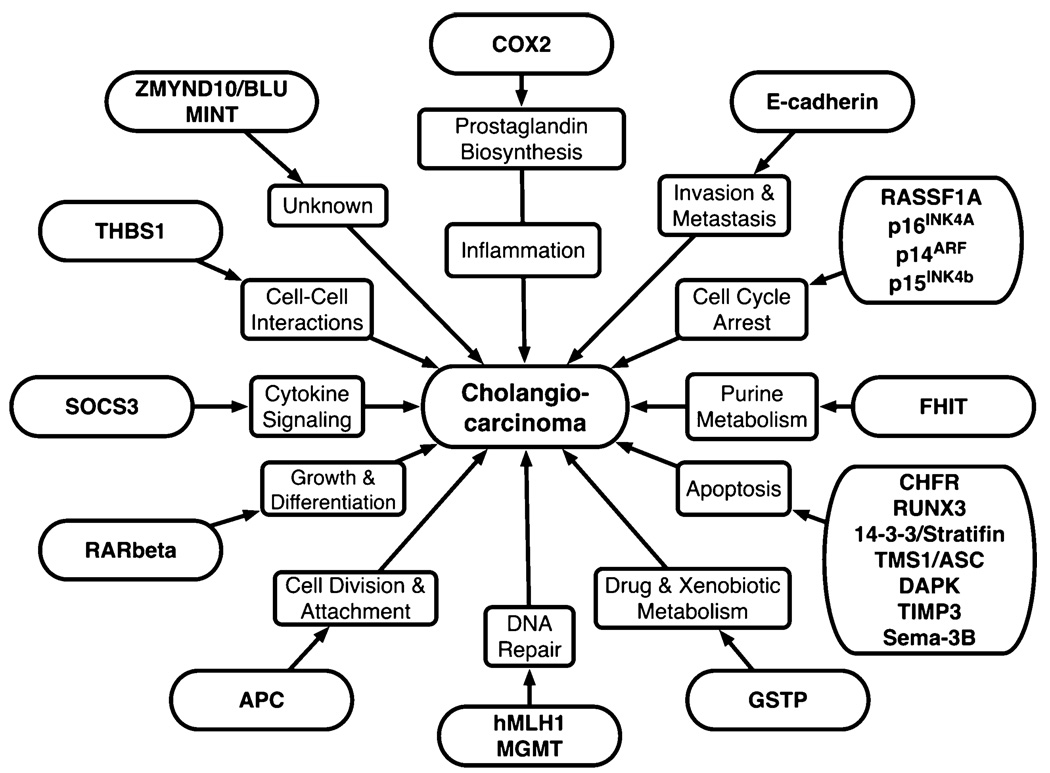
Epigenetic alterations in a number of candidate tumour-related genes have been evaluated in CC. The functions of genes found to be methylated in CC are shown in Figure 4 and their location with reported CpG island methylation frequencies in Table 1.We will briefly summarize the key findings to date on each gene.
Fig. 4.
Genes reported to be methylated in cholangiocarcinoma with their respective potential functions in tumour initiation and progression.
Table 1.
Methylation frequency of candidate genes in different studies of cholangiocarcinoma
| Paper |
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Location | Function | Yang et al. (89) |
Foja et al. (96) |
Wong et al. (95) |
Limpaiboon et al. (105) |
Abraham et al. (104) |
Lee et al. (44) |
Tannapfel et al. (91) |
Liu et al. (114) |
Ahrendt et al. (50) |
Koga et al. (115) |
Tozawa et al. (93) |
Hong et al. (94) |
Tischoff et al. (97) |
Klump et al. (92) |
Isomoto et al. (120) |
|
| 1 | FHIT | 3p14.2 | Purine metabolism | 42% | ||||||||||||||
| 2 | RASSF1A | 3p21.3 | Cell cycle arrest | 65% | 68% | 69% | 27% | |||||||||||
| 3 | hMLH1 | 3p21.3 | DNA mismatch repair |
25% | 44.6% | 0% | 46% | 8.1% | ||||||||||
| 4 | 14-3-3 | 1p36.11 | Apoptosis | 59.5% | ||||||||||||||
| 5 | MINT1 | Unknown | 40.5% | |||||||||||||||
| 6 | MINT2 | Unknown | 0% | |||||||||||||||
| 7 | MINT12 | Unknown | 50.6% | |||||||||||||||
| 8 | MINT25 | 22q11 | Unknown | 15.4% | ||||||||||||||
| 9 | MINT31 | Unknown | 1.3% | |||||||||||||||
| 10 | MINT32 | Unknown | 35.4% | |||||||||||||||
| 11 | APC | 5q21 | Cell division and attachment |
46% | 26.6% | |||||||||||||
| 12 | E-cadherin | 16q22.1 | Proliferation, Invasion and metastasis |
43% | 21.5% | 41% | 27% | |||||||||||
| 13 | INK4a p16 | 9p21 | Cell proliferation and oncogenesis |
50% | 17.7% | 83% | 25% | 70% | 24.3% | 77% | 53.5% | |||||||
| 14 | INK4b p15 | 9p21 | Cell cycle arrest | 50% | ||||||||||||||
| 15 | TIMP3 | 22q12.1 | Apoptosis | 9% | 8.9% | |||||||||||||
| 16 | p14ARF | 9p21 | Cell cycle arrest | 38% | 25% | |||||||||||||
| 17 | p73 | 1p36.3 | Cell cycle regulation | 36% | ||||||||||||||
| 18 | MGMT | 10q26 | DNA mismatch repair |
33% | 49% | |||||||||||||
| 19 | GSTP | 1q43 | Drug/xenobiotic metabolism |
14% | ||||||||||||||
| 20 | RAR-b | 3p24 | Cell growth and differentiation |
14% | ||||||||||||||
| 21 | DAPK | 9q34.1 | Apoptosis | 3% | 32% | 21.4% | ||||||||||||
| 22 | TMS1/ASC | 16p11.2 | Apoptosis | 36.1% | ||||||||||||||
| 23 | Cox-2 | 1q25.2 | Prostaglandin metabolism |
5.1% | ||||||||||||||
| 24 | RUNX3 | 1p36 | Apoptosis | 56.8% | ||||||||||||||
| 25 | CHFR | 12q24.33 | Apoptosis | 16.2% | ||||||||||||||
| 26 | BLU | 3p21.3 | Unknown | 20% | ||||||||||||||
| 27 | Sema-3b | 3p21.3 | Apoptosis | 100% | ||||||||||||||
| 28 | SOCS-3 | 17q25.3 | Cytokine signaling suppression |
88% | ||||||||||||||
| 29 | THBS1 | 15q15 | Cell to cell interactions |
11.4% | 11% | |||||||||||||
p16INK4A
p16INK4A, also called cyclin-dependent kinase inhibitor 2A (CDKN2A), is a tumour suppressor gene located at human chromosome 9p21. The protein product of p16INK4A binds to cyclin-dependent kinases 4 and 6 and inhibits their interaction with cyclin D1 (88). p16 therefore regulates cell proliferation and oncogenesis. Promoter region hypermethylation of the p16INK4A gene in CC has been reported by a number of investigators. The different studies have found a variable methylation frequency of this gene, ranging from as low as 17.7% to as high as 83% (44, 89–93). p16 gene promoter region hypermethylation has been shown to be associated with a poor clinical outcome (44). While the previously referenced studies contained mixed populations of ICC and ECC, Hong et al. (94) found a 77% methylation frequency of the p16INK4A gene in 90 cases of ECC. In this group, promoter hypermethylation of the p16INK4A gene was more commonly observed in tumours with vascular invasion. Ahrendt et al. (50) demonstrated that inactivation of the p16 tumour suppressor gene secondary to promoter hypermethylation is a common event in PSC-associated CC and may play a role in the predisposition to CC in patients with PSC.
Ras association domain family 1A (RASSF1A)
The Ras effector homologue gene (RASSF1A) located at 3p21.3 encodes a protein similar to the RAS effector proteins. Loss or altered expression of this gene has been implicated in the pathogenesis of a variety of cancers, which suggests a tumour suppressor function for this gene. Inactivation of this gene was found to be correlated with the hypermethylation of its CpG-island promoter region. RASSF1A interacts with the DNA repair protein XPA. RASSF1A also inhibits the accumulation of cyclin D1, and thus induces cell cycle arrest. Seven alternatively spliced transcript variants of this gene encoding distinct isoforms have been reported. In studies of a mixture of ICCs and ECCs, RASSF1A promoter methylation has been reported to occur at a frequency of 27–69% (89, 93, 95–97). Significantly, when ICCs and ECCs were considered separately, methylation of the RASSF1A promoter was more common in ECC than ICC (83 vs. 47%, P = 0.003) (89).
Human mutL homologue 1
Human mutL homologue 1 (hMLH1) is a DNA mismatch repair gene located at 3p21.3. Mismatch repair is an important mechanism by which cells correct errors in DNA replication during proliferation to maintain the fidelity of the genome (98, 99). Genetic and epigenetic alterations of hMLH1 have been reported in various cancers (64, 100–103). Mutations in the MLH1 gene result in hereditary nonpolyposis colorectal cancer. In sporadic CC, reported methylation frequencies of the hMLH1 gene promoter are 25 and 8.1% (89, 93). However, Abraham et al. (104) determined hMLH1 methylation status in biliary papillary neoplasms and found no instances of aberrant methylation of the hMLH1 promoter (0 of 10 cases). In a study of 65 cases of liver fluke-related CC, Limpaiboon et al. (105) reported hypermethylation of the hMLH1 promoter in 29 (44.6%) of the cases. In addition, they found an association between promoter hypermethylation of hMLH1 and the poorly differentiated subtype of CC with vascular invasion.
Fragile histidine triad
The fragile histidine triad (FHIT) gene covers more than 1.5 Mb of genomic DNA at human chromosome 3p14.2, but encodes a transcript only 1.1 kb in size. The FHIT gene locus coincides with the extremely aphidicolin-sensitive common chromosomal fragile site FRA3B. The familial renal cell carcinoma t(3;8)(p14.2;q24.1) translocation site is located in intron 3 of the FHIT gene (106). The FHIT protein is a diadenosine 5′,5‴-P1,P3-triphosphate hydrolase that is involved in purine metabolism. FHIT has been shown to have a tumour suppressor function in multiple cancers and aberrant transcripts from this gene have been found in about half of all oesophageal, stomach and colon carcinomas. Foja et al. (96) utilized MSP and COBRA in their study of 19 ICC cases and detected epigenetic silencing of the FHIT promoter region in eight of 19 (42%) ICC specimens. These results represented the first demonstration of FHIT hypermethylation in ICC.
14-3-3
The 14-3-3 proteins constitute a family of closely related regulatory genes that are highly conserved in eukaryotes (107, 108). 14-3-3 sigma (stratifin) was first identified as an epithelial marker (human epithelial marker 1) that was inactivated during neoplastic transformation (109). However, it is now clear that sigma influences many biological processes, including the cell cycle and cell death (apoptosis) (110, 111) and functions as a tumour suppressor gene. Inactivation of this gene by CpG island hypermethylation has been found to occur during tumorigenesis. Lee et al. (44) reported methylation of the 14-3-3 sigma gene in 59.5% of 79 ICC samples.
Target of Methylation-mediated Silencing/Apoptosis Speck like protein containing a CARD (TMS1/ASC)
The TMS1/ASC gene is composed of three exons and encompasses a 1.5 kb region at chromosome 16p11.2. It is a member of the caspase (CASP) recruitment domain family of proapoptotic mediators and also acts in concert with CASP9 to recruit other activators downstream in this cascade. The TMS1/ASC gene was originally identified as a target of methylation-induced silencing in cell lines that overexpress DNA methyltransferase 1 (DNMT1) (112, 113). Liu et al. (114) reported aberrant methylation of the TMS1/ASC gene in 13 (36.1%) of 36 CCs and concluded that epigenetic inactivation of the TMS1/ASC gene could be associated with cholangiocarcinogenesis.
Adenomatous polyposis coli
The adenomatous polyposis coli (APC) gene is located on the long arm of chromosome 5 at 5q21–q22. APC is a tumour suppressor gene that controls cell division, cell–cell interactions and cell migration and invasion. APC also regulates conservation of chromosomal number during cell division. APC protein functions through interactions with other proteins, particularly proteins that are involved in cell attachment and signalling. APC gene hypermethylation ranged from 26.6 to 46% in different studies of CC (44, 89). In addition, APC gene hypermethylation is associated with a worse clinical outcome in CC patients (44).
Death-associated protein kinase
The death-associated protein kinase (DAPK) gene is located at chromosome 9q34.1 and is a positive mediator of interferon-γ (IFN-γ)-induced programmed cell death. DAPK encodes a structurally unique 160 kD calmodulin-dependent serine–threonine kinase that carries eight ankyrin repeats and two putative P-loop consensus sites. DAPK is a tumour suppressor candidate and is hypermethylated, with methylation frequency ranging from 3 to 21.4% in CCs. Additionally, DAPK gene hypermethylation is associated with poorly differentiated CCs and with a poor prognosis (89, 93).
Epithelial cadherin
The epithelial (E) cadherin gene is located at chromosome 16q22.1. This gene is a classical cadherin from the cadherin superfamily. The encoded protein is a calcium-dependent cell–cell adhesion glycoprotein comprised of five extracellular cadherin repeats, a transmembrane region and a highly conserved cytoplasmic tail. Genetic and epigenetic alterations (hypermethylation of the promoter region) in this gene lead to loss of function, which in turn is thought to contribute to progression of cancer by increasing proliferation, invasion and metastasis. The methylation frequency of this gene in CC ranges from 21.5 to 43% (44, 89, 93, 115).
Retinoic acid receptor-β
The retinoic acid receptor-β (RAR-β) gene is located at chromosome 3p24. The gene is also known as HAP, RRB2 and NR1B2 and encodes a member of the thyroid–steroid hormone receptor superfamily of nuclear transcriptional regulators. This receptor localizes to the cytoplasm and to subnuclear compartments. RAR-β binds to retinoic acid, the biologically active form of vitamin A, which mediates cellular signalling in embryonic morphogenesis, cell growth and differentiation. It is thought that this protein limits the growth of many cell types by regulating gene expression. The gene was first identified in a hepatocellular carcinoma in which it flanked a HBV integration site. Gene silencing by promoter region hypermethylation of this gene is associated with the development of cancer. Yang et al. (89) found a methylation frequency of 14% of CCs.
p73
The p73 gene is located at chromosome 1p36.3. The structure of p73 is similar to p53. The important core-binding region of the protein enables the p53 protein to bind DNA, to stick to other p53 proteins and to activate particular genes. p73 also contains the 10 amino acids that are most frequently mutated in p53, predisposing the carrier to cancer. However, there are also important differences. Unlike p53, p73 does not respond when the cell’s DNA sustains damage from ultraviolet light. Both overexpression and underexpression of this gene have been implicated in the process of oncogenesis. Yang et al. (89) found that 36% of CCs had p73 promoter methylation.
O6-methylguanine-DNA methyltransferase
The O6-methylguanine-DNA methyltransferase (MGMT) gene is located at chromosome 10q26. This gene is responsible for repairing alkylation DNA damage and also has a role in inhibiting oestrogen receptor-mediated cell proliferation. Methylation of discrete regions of the MGMT CpG island is associated with heterochromatinization of the MGMT transcription start site and silencing of the gene. Koga et al. (115) found a high methylation frequency of 49% in MGMT gene whereas Yang et al. (89) reported a 33% methylation frequency of MGMT gene in CC. MGMT gene methylation has also been associated with an increased frequency of GC to AT transitions in oncogenes and tumour suppressor genes and a poor prognosis (115).
Glutathione S-transferase pi
The glutathione S-transferase pi (GSTP) gene is located at chromosome 1q43. It is important in drug and xenobiotic metabolism. Promoter region hypermethylation of the GSTP gene has been reported in CC, hepatocellular carcinomas and prostatic adenocarcinomas (89, 116, 117). In a study of 79 ICCs, the GSTP gene methylation frequency was reported to be < 15% (44). Interestingly, GSTP gene hypermethylation was more frequently seen in ICC than in ECC (89).
Checkpoint with forkhead and ring finger domains
The checkpoint with forkhead and ring finger domains (CHFR) gene is located at 12q24.33. CHFR is a putative tumour suppressor gene that has been shown to be silenced by promoter hypermethylation but its specific role in suppressing tumorigenesis remains to be fully elucidated. CHFR is a mitotic stress checkpoint gene, whose product mediates a delay of entry into metaphase after treatment with microtubule inhibitors. CHFR may induce chromosomal instability owing to delayed chromosome condensation (118) and resultant DNA aneuploidy. The frequency of CHFR methylation was 16.2% in biliary tract carcinomas in a study by Tozawa et al. (93).
Runt-related transcription factor 3
Runt-related transcription factor 3 (RUNX3) is located at chromosome 1p36. This gene encodes a member of the runt domain-containing family of transcription factors. A heterodimer of this protein and a β subunit form a complex that binds to the core DNA sequence 5′-PYGPYGGT-3′ found in a number of enhancers and promoters, and can either activate or suppress transcription. It also interacts with other transcription factors. RUNX3 functions as a tumour suppressor, and the gene is frequently deleted or transcriptionally silenced in cancer. Multiple transcript variants encoding different isoforms have been found for this gene. It is a major growth regulator of gastric epithelial cells, owing to activation of transforming growth factor-β (TGF-β)-induced apoptosis (118). RUNX3 was found to be methylated in 56.8% of biliary tract cancers. In addition, RUNX3 methylation was associated with poorer survival (93). In the RUNX3 knockout mouse, the gastric mucosa exhibits hyperplasia caused by stimulated proliferation and suppressed apoptosis of epithelial cells, and the cells are resistant to the growth-inhibitory and apoptosis-inducing action of TGF-β (119). RUNX3 may also regulate proliferation of the biliary tract epithelium, and its silencing by epigenetic alteration may play an important role in biliary tract carcinogenesis (93).
Tissue inhibitor of metalloproteinase 3
The tissue inhibitor of metalloproteinase 3 (TIMP3) gene belongs to the TIMP family and is located at chromosome 22q12.3. The proteins encoded by this gene family are inhibitors of the matrix metalloproteinases, a group of peptidases involved in degradation of the extracellular matrix (ECM). Expression of this gene is induced in response to mitogenic stimulation and this netrin domain-containing protein is localized to the ECM. This gene could play a role in the induction of apoptosis. In CCs, two groups have reported methylation frequencies of 8.9 and 9%, respectively, for this gene (44, 89). Additionally, patients with CpG island methylation of TIMP3 gene had worse survival (44).
Semaphorin 3B
The semaphorin 3B (SEMA3B) candidate tumour suppressor gene is located on chromosome 3 at 3p21.3. The semaphorin/collapsin family of molecules plays a critical role in the guidance of growth cones during neuronal development. The secreted protein encoded by this gene family member is important in axonal guidance and has been shown to act as a tumour suppressor by inducing apoptosis. Tischoff et al. (97) reported a 100% methylation frequency of this gene in 15 cases of CC.
Blu protein/Zinc finger MYND domain containing protein 10 (BLU/ZMYND10)
The Blunt (BLU) gene is located at chromosome 3p21.3. The exact function of this gene is not known but it is postulated to be a tumour suppressor gene. The methylation frequency of this gene was found to be 20% in a study of 15 CCs (97).
Thrombospondin 1
The thrombospondin 1 (THBS1) gene is located at chromosome 15q15. The protein encoded by this gene is a subunit of a disulphide-linked homotrimeric protein. This protein is an adhesive glycoprotein that mediates cell-to-cell and cell-to-matrix interactions. THBS1 can bind to fibrinogen, fibronectin, laminin, type V collagen and α-v-β-1 integrins. THBS1 has been shown to play roles in platelet aggregation, angiogenesis and tumorigenesis. The THBS1 gene promoter has been found to be methylated in 11% of 15 CCs (44, 97).
Suppressor of cytokine signalling 3
The suppressor of the cytokine signalling 3 (SOCS 3) gene is at chromosome 17q25.3. This gene encodes a member of the STAT-induced STAT inhibitor (SSI), also known as SOCS family. SSI family members are cytokine-inducible negative regulators of cytokine signalling. The expression of this gene is induced by various cytokines, including interleukin-6 (IL-6), IL-10 and IFN-γ. The protein encoded by this gene can bind to and inhibit the activity of janus kinase 2 (JAK2). A recent study has shown an 88% methylation frequency of this gene in eight CCs (120).
p14ARF
The p14 alternate reading frame (p14ARF) gene is located on chromosome region 9p21 and is encoded by the β transcript of CDKN2A (p16/CDKN2A). The p14 gene has a 5′ exon that is spliced into common exons 2 and 3 and has the ability to elicit a p53 response, manifest in the increased expression of both CDKN1A, also called coilin-interacting protein (CIP1), and mouse double minute 2 (MDM2), and resulting in a distinctive cell cycle arrest in both the G1 and the G2/M phases. The two unrelated proteins encoded by the p16INK4A–p14ARF locus function as tumour suppressors. p14ARF binds to MDM2 and promotes the rapid degradation of MDM2. This interaction is mediated by the E1-β-encoded N-terminal domain of ARF and the C-terminal region of MDM2. ARF-promoted MDM2 degradation is associated with MDM2 modification and concurrent p53 stabilization and accumulation. The functional consequence of ARF-regulated p53 levels via MDM2 proteolysis is evidenced by the ability of ectopically expressed ARF to restore a p53-imposed G1 cell cycle arrest that is otherwise abrogated by MDM2. Thus, deletion of the p16INK4A–p14ARF locus simultaneously impairs the p16INK4A-cyclin D/CDK4-RB and the p14ARF-MDM2-p53 pathways (121). Yang et al. (89) and Tannapfel et al. (91) reported methylation frequencies of 38 and 25%, respectively, in CC (89, 91).
p15INK4b
Hannon and Beach isolated a member of the p16INK4 family, which they referred to as p15INK4b or p15. The expression of CDKN2B protein was induced approximately 30-fold in human keratinocytes by treatment with TGF-β, suggesting that p15 may act as an effector of TGF-β-mediated cell cycle arrest. This gene is located adjacent to p16INK4a on 9p21 and is codeleted in a high proportion of established human cancer cell lines (122). Promoter hypermethylation of this gene has been studied in CC and a methylation frequency of 50% has been reported (89).
Cyclooxygenase 2
The cyclooxygenase 2 (COX-2) or prostaglandin-endoperoxide synthase 2 (PTGS2) gene is located at 1q25.2–q25.3. COX is the key enzyme in prostaglandin biosynthesis, and acts both as a dioxygenase and as a peroxidase. There are two isozymes of COX (PTGS): a constitutive COX-1 (PTGS1) and an inducible COX-2 (PTGS2), which differ in their regulation of expression and tissue distribution. This gene encodes COX-2, which shows 86–89% amino acid sequence identity with mouse, rat, sheep, bovine, horse and rabbit COX-2 proteins. Human COX-2 is expressed in a limited number of cell types and is regulated by specific stimulatory events, suggesting that it is responsible for the prostanoid biosynthesis involved in inflammation and mitogenesis. The expression of this gene is deregulated in epithelial tumours. A methylation frequency of 5.1% of 79 resected cases of CC was found in the COX-2 gene (44).
Methylated in tumour genes
MINT refers to CpG islands that are found to be methylated in tumours (MINT). A variety of MINT CpG islands are associated with carcinogenesis of the biliary tract epithelium and other epithelial cancers such as gastric cancer. MINT1 and MINT2 correspond to CpG islands that are in the 5′ region of cDNAs with open-reading frames that have no known protein homology. The CpG island of MINT25 maps to chromosome 22q11–13, but the associated gene is unknown at this point. MINT31 is 2 kb upstream of the CACNA1G, a T-type calcium channel gene (123). In a study to determine CpG island methylator phenotype (CIMP) in CCs, six MINT loci were selected for analysis. Four MINT loci were methylated at a frequency more than 15%, with MINT2 not being methylated at all, and MINT31 methylated only once. Tumours with methylation at three or more of the five MINT loci, except for MINT2, were defined as CIMP+. CIMP− tumours were defined as those with methylation at less than three MINT loci. Eighteen (22.8%) of the 79 cases were classed as CIMP+ tumours. With respect to clinicopathological factors, no significant association with CIMP was found, except for the histological type. All the CCs of the papillary type (n = 10) were CIMP− tumours, and showed a lower than average methylation index, 0.11; the average methylation index of the other histological types was 0.19 (P = 0.064). These cases were identical to the CCs of intraductal growth type on macroscopic classification (44).
Unanswered questions and future directions in the study of methylated genes in cholangiocarcinoma
Cholangiocarcinoma continues to be a highly lethal disease with an extremely poor response to conventional anticancer therapies and a poor survival rate. The identification of genes that are differentially methylated in CC may provide valuable information on potential markers for detection of early-stage curable disease, markers prognostic of response to specific treatments and overall prognosis and new targets for the design of rational therapies.
Molecular markers such as CA 19-9, carcinoembryonic antigen, CA-125, platelet-derived growth factor and basic fibroblast growth factor have been studied the most as markers for the diagnosis of CC, but none of these is a sensitive and specific marker for early-stage disease. Thus, there is a significant need to develop reliable markers for early detection of CC. CpG hypermethylation is a promising technology for this purpose. Genes such as p16 and RASSF1A, which are frequently methylated in CC, could serve as potential molecular markers in the early diagnosis of CC in at-risk individuals. These genes are also hypermethylated in multiple other epithelial cancers, rendering them less specific for identifying CCs, especially in a context in which DNA methylation markers are measured in analytes such as stool, urine or serum, which contain integrated DNA samples derived from the entire body. There is therefore a critical need for experiments focused on identifying CC-specific methylation markers using newer technologies for genome-wide evaluation of hypermethylated promoter regions.
Regarding the use of CpG hypermethylation as a prognostic predictor in CC, studies performed thus far have shown that p16, hMLH1, DAPK, MGMT, APC, TIMP3 and RUNX3 gene methylation are associated with vascular invasion and/or poor survival in CC. Additional refinements can be expected in our ability to predict phenotypic and treatment response using methylation markers.
Unlike gene mutation events, epigenetic events can be reversed to restore the function of key pathways in malignant transformation. Combinations of demethylating agents such DAC and histone deacetylatase inhibitors may be effective in reversing the epigenetic changes in susceptible gene promoters and thereby reversing or inhibiting cholangiocarcinogenesis, tumour progression or metastasis.
DNA methylation is used as a clinical biomarker in acute leukaemias, and inhibitors of DNA methylation are currently in clinical use for treatment of myelodysplastic syndrome and are in clinical trials for other haematopoietic malignancies and bone tumours such as osteosarcoma; however, the potential of DNA methylation as a diagnostic and therapeutic target is only now beginning to be explored in CC. With advancements in our understanding of the molecular pathogenesis and epigenetics of CC, clinical results with methylation markers as diagnostic markers and demethylating agents as therapeutic targets are likely to be reported in the near future.
In conclusion, the contribution of epigenetic silencing through promoter region hypermethylation of various genes to the inactivation of key pathways involved in the development of CC is increasingly being appreciated. We have provided a comprehensive review of the genes currently known to be regulated by hypermethylation. Unfortunately, the currently available information is fragmented, as most groups have studied methylation of one or, at most, only a limited number of genes in CC. As multigene and genome-wide approaches to evaluating gene methylation come on-stream, it is to be expected that we will gain a more complete view of the role of gene methylation in CC.
Correlation of the molecular information on methylation of genes or networks of genes with clinical and prognostic variables will allow classification of tumours with particular alterations in gene methylation into clinically and biologically relevant hierarchies. In particular, because of the stable and amplifiable nature of DNA, DNA-based methylation markers can be relatively easily transferred from the research laboratory setting into clinical diagnostic use. These genes are also potentially important targets for therapeutic intervention. Hence, methylation markers in CC hold promise not only for diagnostic but for therapeutic advances as well.
Acknowledgements
This work was supported by NIH grant CA100882, a generous gift from The Richard M. Schulze Family Foundation and by the Miles and Shirley Fiterman Center for Digestive Diseases at the Mayo Clinic, Rochester, MN, USA (to L. R. R.). The authors thank Stacy Roberson and Karlyn Smith for secretarial assistance.
References
- 1.Olnes MJ, Erlich R. A review and update on cholangiocarcinoma. Oncology. 2004;66:167–179. doi: 10.1159/000077991. [DOI] [PubMed] [Google Scholar]
- 2.Ishak KG, Anthony PP, Sobin LH. Histological typing of tumours of the liver. Berlin: Springer Verlag; WHO International Histological Classification of Tumours. 1994
- 3.Carriaga MT, Henson DE. Liver, gallbladder, extrahepatic bile ducts, and pancreas. Cancer. 1995;75 Suppl.:171–190. doi: 10.1002/1097-0142(19950101)75:1+<171::aid-cncr2820751306>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 4.Shaib Y, El-Serag HB. The epidemiology of cholangiocarcinoma. Semin Liver Dis. 2004;24:115–125. doi: 10.1055/s-2004-828889. [DOI] [PubMed] [Google Scholar]
- 5.Khan SA, Davidson BR, Goldin R, et al. Guidelines for the diagnosis and treatment of cholangiocarcinoma: consensus document. Gut. 2002;51 Suppl. 6:VI1–VI9. doi: 10.1136/gut.51.suppl_6.vi1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vauthey JN, Blumgart LH. Recent advances in the management of cholangiocarcinomas. Semin Liver Dis. 1994;14:109–114. doi: 10.1055/s-2007-1007302. [DOI] [PubMed] [Google Scholar]
- 7.Patel T. Cholangiocarcinoma. Nat Clin Pract Gastroenterol Hepatol. 2006;3:33–42. doi: 10.1038/ncpgasthep0389. [DOI] [PubMed] [Google Scholar]
- 8.Khan SA, Taylor-Robinson SD, Toledano MB, et al. Changing international trends in mortality rates for liver, biliary and pancreatic tumours. J Hepatol. 2002;37:806–813. doi: 10.1016/s0168-8278(02)00297-0. [DOI] [PubMed] [Google Scholar]
- 9.Patel T. Increasing incidence and mortality of primary intrahepatic cholangiocarcinoma in the United States. Hepatology. 2001;33:1353–1357. doi: 10.1053/jhep.2001.25087. [DOI] [PubMed] [Google Scholar]
- 10.Patel T. Worldwide trends in mortality from biliary tract malignancies. BMC Cancer. 2002;2:10. doi: 10.1186/1471-2407-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taylor-Robinson SD, Toledano MB, Arora S, et al. Increase in mortality rates from intrahepatic cholangiocarcinoma in England and Wales 1968–1998. Gut. 2001;48:816–820. doi: 10.1136/gut.48.6.816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shaib YH, Davila JA, McGlynn K, et al. Rising incidence of intrahepatic cholangiocarcinoma in the United States: a true increase? J Hepatol. 2004;40:472–477. doi: 10.1016/j.jhep.2003.11.030. [DOI] [PubMed] [Google Scholar]
- 13.Lazaridis KN, Gores GJ. Cholangiocarcinoma. Gastroenterology. 2005;128:1655–1667. doi: 10.1053/j.gastro.2005.03.040. [DOI] [PubMed] [Google Scholar]
- 14.Farrant JM, Hayllar KM, Wilkinson ML, et al. Natural history and prognostic variables in primary sclerosing cholangitis. Gastroenterology. 1991;100:1710–1717. doi: 10.1016/0016-5085(91)90673-9. [DOI] [PubMed] [Google Scholar]
- 15.Broome U, Olsson R, Loof L, et al. Natural history and prognostic factors in 305 Swedish patients with primary sclerosing cholangitis. Gut. 1996;38:610–615. doi: 10.1136/gut.38.4.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bergquist A, Broome U. Hepatobiliary and extra-hepatic malignancies in primary sclerosing cholangitis. Best Pract Res Clin Gastroenterol. 2001;15:643–656. doi: 10.1053/bega.2001.0210. [DOI] [PubMed] [Google Scholar]
- 17.Bergquist A, Ekbom A, Olsson R, et al. Hepatic and extrahepatic malignancies in primary sclerosing cholangitis. J Hepatol. 2002;36:321–327. doi: 10.1016/s0168-8278(01)00288-4. [DOI] [PubMed] [Google Scholar]
- 18.Watanapa P, Watanapa WB. Liver fluke-associated cholangiocarcinoma. Br J Surg. 2002;89:962–970. doi: 10.1046/j.1365-2168.2002.02143.x. [DOI] [PubMed] [Google Scholar]
- 19.Khurana S, Dubey ML, Malla N. Association of parasitic infections and cancers. Indian J Med Microbiol. 2005;23:74–79. doi: 10.4103/0255-0857.16044. [DOI] [PubMed] [Google Scholar]
- 20.Simeone DM. Gallbladder and biliary tree: anatomy and structural anomalies. In: Yamada T, editor. Textbook of Gastroenterology. Philadelphia: Lippincott Williams and Wilkins; 1999. pp. 2244–2257. [Google Scholar]
- 21.Scott J, Shousha S, Thomas HC, et al. Bile duct carcinoma: a late complication of congenital hepatic fibrosis. Case report and review of literature. Am J Gastroenterol. 1980;73:113–119. [PubMed] [Google Scholar]
- 22.Lipsett PA, Pitt HA, Colombani PM, et al. Choledochal cyst disease. A changing pattern of presentation. Ann Surg. 1994;220:644–652. doi: 10.1097/00000658-199411000-00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khan SA, Thomas HC, Davidson BR, et al. Cholangiocarcinoma. Lancet. 2005;366:1303–1314. doi: 10.1016/S0140-6736(05)67530-7. [DOI] [PubMed] [Google Scholar]
- 24.Shaib YH, El-Serag HB, Davila JA, et al. Risk factors of intrahepatic cholangiocarcinoma in the United States: a case-control study. Gastroenterology. 2005;128:620–626. doi: 10.1053/j.gastro.2004.12.048. [DOI] [PubMed] [Google Scholar]
- 25.Kobayashi M, Ikeda K, Saitoh S, et al. Incidence of primary cholangiocellular carcinoma of the liver in Japanese patients with hepatitis C virus-related cirrhosis. Cancer. 2000;88:2471–2477. doi: 10.1002/1097-0142(20000601)88:11<2471::aid-cncr7>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 26.Sahani D, Prasad SR, Tannabe KK, et al. Thorotrast-induced cholangiocarcinoma: case report. Abdom Imaging. 2003;28:72–74. doi: 10.1007/s00261-001-0148-y. [DOI] [PubMed] [Google Scholar]
- 27.Hardell L, Bengtsson NO, Jonsson U, et al. Aetiological aspects on primary liver cancer with special regard to alcohol, organic solvents and acute intermittent porphyria – an epidemiological investigation. Br J Cancer. 1984;50:389–397. doi: 10.1038/bjc.1984.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walker NJ, Crockett PW, Nyska A, et al. Dose-additive carcinogenicity of a defined mixture of ‘dioxin-like compounds’. Environ Health Perspect. 2005;113:43–48. doi: 10.1289/ehp.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bond GG, McLaren EA, Sabel FL, et al. Liver and biliary tract cancer among chemical workers. Am J Ind Med. 1990;18:19–24. doi: 10.1002/ajim.4700180103. [DOI] [PubMed] [Google Scholar]
- 30.Bergquist A, Glaumann H, Persson B, et al. Risk factors and clinical presentation of hepatobiliary carcinoma in patients with primary sclerosing cholangitis: a case–control study. Hepatology. 1998;27:311–316. doi: 10.1002/hep.510270201. [DOI] [PubMed] [Google Scholar]
- 31.Chalasani N, Baluyut A, Ismail A, et al. Cholangiocarcinoma in patients with primary sclerosing cholangitis: a multicenter case–control study. Hepatology. 2000;31:7–11. doi: 10.1002/hep.510310103. [DOI] [PubMed] [Google Scholar]
- 32.Paik KY, Jung JC, Heo JS, et al. What prognostic factors are important for resected intrahepatic cholangiocarcinoma? J Gastroenterol Hepatol. 2007 doi: 10.1111/j.1440-1746.2007.05040.x. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 33.Rosai J. Ackerman’s Surgical Pathology. Vol. 960. Mosby: St. Louis; 1996. p. 914.p. 915. [Google Scholar]
- 34.Kitagawa Y, Nagino M, Kamiya J, et al. Lymph node metastasis from hilar cholangiocarcinoma: audit of 110 patients who underwent regional and paraaortic node dissection. Ann Surg. 2001;233:385–392. doi: 10.1097/00000658-200103000-00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamamoto M, Takasaki K, Yoshikawa T. Lymph node metastasis in intrahepatic cholangiocarcinoma. Jpn J Clin Oncol. 1999;29:147–150. doi: 10.1093/jjco/29.3.147. [DOI] [PubMed] [Google Scholar]
- 36.Okami J, Dohno K, Sakon M, et al. Genetic detection for micrometastasis in lymph node of biliary tract carcinoma. Clin Cancer Res. 2000;6:2326–2332. [PubMed] [Google Scholar]
- 37.Bhuiya MR, Nimura Y, Kamiya J, et al. Clinicopathologic studies on perineural invasion of bile duct carcinoma. Ann Surg. 1992;215:344–349. doi: 10.1097/00000658-199204000-00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klempnauer J, Ridder GJ, von Wasielewski R, et al. Resectional surgery of hilar cholangiocarcinoma: a multivariate analysis of prognostic factors. J Clin Oncol. 1997;15:947–954. doi: 10.1200/JCO.1997.15.3.947. [DOI] [PubMed] [Google Scholar]
- 39.Rea DJ, Heimbach JK, Rosen CB, et al. Liver transplantation with neoadjuvant chemoradiation is more effective than resection for hilar cholangiocarcinoma. Ann Surg. 2005;242:451–458. doi: 10.1097/01.sla.0000179678.13285.fa. discussion 458-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 41.Vogelstein B, Kinzler KW. The Genetic Basis of Human Cancer. New York: McGraw-Hill, Health Professions Division; 1998. [Google Scholar]
- 42.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 43.Rashid A. Cellular and molecular biology of biliary tract cancers. Surg Oncol Clin North Am. 2002;11:995–1009. doi: 10.1016/s1055-3207(02)00042-x. [DOI] [PubMed] [Google Scholar]
- 44.Lee S, Kim WH, Jung HY, et al. Aberrant CpG island methylation of multiple genes in intrahepatic cholangiocarcinoma. Am J Pathol. 2002;161:1015–1022. doi: 10.1016/S0002-9440(10)64262-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kang YK, Kim WH, Lee HW, et al. Mutation of p53 and K-ras, and loss of heterozygosity of APC in intrahepatic cholangiocarcinoma. Lab Invest. 1999;79:477–483. [PubMed] [Google Scholar]
- 46.Sturm PD, Bass IO, Clement MJ, et al. Alterations of the p53 tumor-suppressor gene and K-ras oncogene in perihilar cholangiocarcinomas from a high-incidence area. Int J Cancer. 1998;78:695–698. doi: 10.1002/(sici)1097-0215(19981209)78:6<695::aid-ijc5>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 47.Kiba T, Tsuda H, Pairojkul C, et al. Mutations of the p53 tumor suppressor gene and the ras gene family in intrahepatic cholangiocellular carcinomas in Japan and Thailand. Mol Carcinog. 1993;8:312–318. doi: 10.1002/mc.2940080415. [DOI] [PubMed] [Google Scholar]
- 48.Wattanasirichaigoon S, Tasanakhajorn U, Jesadapatarakul S. The incidence of K-ras codon 12 mutations in cholangiocarcinoma detected by polymerase chain reaction technique. J Med Assoc Thai. 1998;81:316–323. [PubMed] [Google Scholar]
- 49.Tannapfel A, Benicke M, Katalinic A, et al. Frequency of p16(INK4A) alterations and K-ras mutations in intrahepatic cholangiocarcinoma of the liver. Gut. 2000;47:721–727. doi: 10.1136/gut.47.5.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ahrendt SA, Eisenberger CF, Yip L, et al. Chromosome 9p21 loss and p16 inactivation in primary sclerosing cholangitis-associated cholangiocarcinoma. J Surg Res. 1999;84:88–93. doi: 10.1006/jsre.1999.5615. [DOI] [PubMed] [Google Scholar]
- 51.Sugimachi K, Taguchi K, Aishima S, et al. Altered expression of beta-catenin without genetic mutation in intrahepatic cholangiocarcinoma. Mod Pathol. 2001;14:900–905. doi: 10.1038/modpathol.3880409. [DOI] [PubMed] [Google Scholar]
- 52.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 53.Li E, Beard C, Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993;366:362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- 54.Panning B, Jaenisch R. RNA and the epigenetic regulation of X chromosome inactivation. Cell. 1998;93:305–308. doi: 10.1016/s0092-8674(00)81155-1. [DOI] [PubMed] [Google Scholar]
- 55.Walsh CP, Chaillet JR, Bestor TH. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat Genet. 1998;20:116–117. doi: 10.1038/2413. [DOI] [PubMed] [Google Scholar]
- 56.Bird A. The essentials of DNA methylation. Cell. 1992;70:5–8. doi: 10.1016/0092-8674(92)90526-i. [DOI] [PubMed] [Google Scholar]
- 57.Cooper DN, Krawczak M. Cytosine methylation and the fate of CpG dinucleotides in vertebrate genomes. Hum Genet. 1989;83:181–188. doi: 10.1007/BF00286715. [DOI] [PubMed] [Google Scholar]
- 58.Antequera F, Bird A. CpG islands. In: Jost JP, Saluz HP, editors. DNA Methylation: Molecular Biology and Biological Significance. Basel: Birkhauser Verlag; 1993. pp. 169–185. [Google Scholar]
- 59.Baylin SB, Herman JG, Graff JR, et al. Alterations in DNA methylation: a fundamental aspect of neoplasia. In: Klein G, Van de Woude GF, editors. Advances in Cancer Research. San Diego: Academic Press; 1998. pp. 141–196. [PubMed] [Google Scholar]
- 60.Jones PA. DNA methylation errors and cancer. Cancer Res. 1996;56:2463–2467. [PubMed] [Google Scholar]
- 61.Jones PA, Laird PW. Cancer epigenetics comes of age. Nat Genet. 1999;21:163–167. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- 62.Herman JG, Baylin SB. Promoter-region hypermethylation and gene silencing in human cancer. Curr Top Microbiol Immunol. 2000;249:35–54. doi: 10.1007/978-3-642-59696-4_3. [DOI] [PubMed] [Google Scholar]
- 63.Baylin SB, Herman JG. DNA hypermethylation in tumor-igenesis: epigenetics joins genetics. Trends Genet. 2000;16:168–174. doi: 10.1016/s0168-9525(99)01971-x. [DOI] [PubMed] [Google Scholar]
- 64.Herman JG, Umar A, Polyak K, et al. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci USA. 1998;95:6870–6875. doi: 10.1073/pnas.95.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Myohanen SK, Baylin SB, Herman JG. Hypermethylation can selectively silence individual p16ink4A alleles in neoplasia. Cancer Res. 1998;58:591–593. [PubMed] [Google Scholar]
- 66.Ahuja N, Li Q, Mohan AL, et al. Aging and DNA methylation in colorectal mucosa and cancer. Cancer Res. 1998;58:5489–5494. [PubMed] [Google Scholar]
- 67.Issa JP, Ottaviano YL, Celano P, et al. Methylation of the oestrogen receptor CpG island links ageing and neoplasia in human colon. Nat Genet. 1994;7:536–540. doi: 10.1038/ng0894-536. [DOI] [PubMed] [Google Scholar]
- 68.Issa JP, Vertino PM, Boehm CD, et al. Switch from mono-allelic to biallelic human IGF2 promoter methylation during aging and carcinogenesis. Proc Natl Acad Sci USA. 1996;93:11757–11762. doi: 10.1073/pnas.93.21.11757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hsieh CL. Dependence of transcriptional repression on CpG methylation density. Mol Cell Biol. 1994;14:5487–5494. doi: 10.1128/mcb.14.8.5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vertino PM, Yen RW, Gao J, et al. De novo methylation of CpG island sequences in human fibroblasts overexpressing DNA (cytosine-5-)-methyltransferase. Mol Cell Biol. 1996;16:4555–4565. doi: 10.1128/mcb.16.8.4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lewis J, Bird A. DNA methylation and chromatin structure. FEBS Lett. 1991;285:155–159. doi: 10.1016/0014-5793(91)80795-5. [DOI] [PubMed] [Google Scholar]
- 72.Tazi J, Bird A. Alternative chromatin structure at CpG islands. Cell. 1990;60:909–920. doi: 10.1016/0092-8674(90)90339-g. [DOI] [PubMed] [Google Scholar]
- 73.Jeppesen P, Turner BM. The inactive X chromosome in female mammals is distinguished by a lack of histone H4 acetylation, a cytogenetic marker for gene expression. Cell. 1993;74:281–289. doi: 10.1016/0092-8674(93)90419-q. [DOI] [PubMed] [Google Scholar]
- 74.Marahrens Y, Loring J, Jaenisch R. Role of the Xist gene in X chromosome choosing. Cell. 1998;92:657–664. doi: 10.1016/s0092-8674(00)81133-2. [DOI] [PubMed] [Google Scholar]
- 75.Jones PL, Veenstra GJ, Wade PA, et al. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 76.Nan X, Ng HH, Johnson CA, et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 77.Cameron EE, Bachman KE, Myohanen S, et al. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat Genet. 1999;21:103–107. doi: 10.1038/5047. [DOI] [PubMed] [Google Scholar]
- 78.Roth SY, Denu JM, Allis CD. Histone acetyltransferases. Annu Rev Biochem. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- 79.Thiagalingam S, Cheng KH, Lee HJ, et al. Histone deacetylases: unique players in shaping the epigenetic histone code. Ann NY Acad Sci. 2003;983:84–100. doi: 10.1111/j.1749-6632.2003.tb05964.x. [DOI] [PubMed] [Google Scholar]
- 80.Baylin SB, Ohm JE. Epigenetic gene silencing in cancer – a mechanism for early oncogenic pathway addiction? Nat Rev Cancer. 2006;6:107–116. doi: 10.1038/nrc1799. [DOI] [PubMed] [Google Scholar]
- 81.Pandolfi PP. Histone deacetylases and transcriptional therapy with their inhibitors. Cancer Chemother Pharmacol. 2001;48 Suppl. 1:S17–S19. doi: 10.1007/s002800100322. [DOI] [PubMed] [Google Scholar]
- 82.Hayatsu H, Wataya Y, Kai K, et al. Reaction of sodium bisulfite with uracil, cytosine, and their derivatives. Biochemistry. 1970;9:2858–2865. doi: 10.1021/bi00816a016. [DOI] [PubMed] [Google Scholar]
- 83.Frommer M, McDonald LE, Millar DS, et al. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci USA. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wong IH. Qualitative and quantitative polymerase chain reaction-based methods for DNA methylation analyses. Methods Mol Biol. 2006;336:33–43. doi: 10.1385/1-59745-074-X:33. [DOI] [PubMed] [Google Scholar]
- 85.Sasaki M, Anast J, Bassett W, et al. Bisulfite conversion-specific and methylation-specific PCR: a sensitive technique for accurate evaluation of CpG methylation. Biochem Biophys Res Commun. 2003;309:305–309. doi: 10.1016/j.bbrc.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 86.Gonzalgo ML, Jones PA. Rapid quantitation of methylation differences at specific sites using methylation-sensitive single nucleotide primer extension (Ms-SNuPE) Nucleic Acids Res. 1997;25:2529–2531. doi: 10.1093/nar/25.12.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xiong Z, Laird PW. COBRA: a sensitive and quantitative DNA methylation assay. Nucleic Acids Res. 1997;25:2532–2534. doi: 10.1093/nar/25.12.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Serrano M, Hannon GJ, Beach D. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature. 1993;366:704–707. doi: 10.1038/366704a0. [DOI] [PubMed] [Google Scholar]
- 89.Yang B, House MG, Guo M, et al. Promoter methylation profiles of tumor suppressor genes in intrahepatic and extrahepatic cholangiocarcinoma. Mod Pathol. 2005;18:412–420. doi: 10.1038/modpathol.3800287. [DOI] [PubMed] [Google Scholar]
- 90.Ueki T, Hsing AW, Gao YT, et al. Alterations of p16 and prognosis in biliary tract cancers from a population-based study in China. Clin Cancer Res. 2004;10:1717–1725. doi: 10.1158/1078-0432.ccr-1137-3. [DOI] [PubMed] [Google Scholar]
- 91.Tannapfel A, Sommerer F, Benicke M, et al. Genetic and epigenetic alterations of the INK4a-ARF pathway in cholangiocarcinoma. J Pathol. 2002;197:624–631. doi: 10.1002/path.1139. [DOI] [PubMed] [Google Scholar]
- 92.Klump B, Hsieh CJ, Dette S, et al. Promoter methylation of INK4a/ARF as detected in bile-significance for the differential diagnosis in biliary disease. Clin Cancer Res. 2003;9:1773–1778. [PubMed] [Google Scholar]
- 93.Tozawa T, Tamura G, Honda T, et al. Promoter hypermethylation of DAP-kinase is associated with poor survival in primary biliary tract carcinoma patients. Cancer Sci. 2004;95:736–740. doi: 10.1111/j.1349-7006.2004.tb03254.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hong SM, Choi J, Ryu K, et al. Promoter hypermethylation of the p16 gene and loss of its protein expression is correlated with tumor progression in extrahepatic bile duct carcinomas. Arch Pathol Lab Med. 2006;130:33–38. doi: 10.5858/2006-130-33-PHOTPG. [DOI] [PubMed] [Google Scholar]
- 95.Wong N, Li L, Tsang K, et al. Frequent loss of chromosome 3p and hypermethylation of RASSF1A in cholangiocarcinoma. J Hepatol. 2002;37:633–639. doi: 10.1016/s0168-8278(02)00269-6. [DOI] [PubMed] [Google Scholar]
- 96.Foja S, Goldberg M, Schagdarsurengin U, et al. Promoter methylation and loss of coding exons of the fragile histidine triad (FHIT) gene in intrahepatic cholangiocarcinomas. Liver Int. 2005;25:1202–1208. doi: 10.1111/j.1478-3231.2005.01174.x. [DOI] [PubMed] [Google Scholar]
- 97.Tischoff I, Markwarth A, Witzigmann H, et al. Allele loss and epigenetic inactivation of 3p21.3 in malignant liver tumors. Int J Cancer. 2005;115:684–689. doi: 10.1002/ijc.20944. [DOI] [PubMed] [Google Scholar]
- 98.Fishel R, Kolodner RD. Identification of mismatch repair genes and their role in the development of cancer. Curr Opin Genet Dev. 1995;5:382–395. doi: 10.1016/0959-437x(95)80055-7. [DOI] [PubMed] [Google Scholar]
- 99.Kolodner RD, Marsischky GT. Eukaryotic DNA mismatch repair. Curr Opin Genet Dev. 1999;9:89–96. doi: 10.1016/s0959-437x(99)80013-6. [DOI] [PubMed] [Google Scholar]
- 100.Kane MF, Loda M, Gaida GM, et al. Methylation of the hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair-defective human tumor cell lines. Cancer Res. 1997;57:808–811. [PubMed] [Google Scholar]
- 101.Esteller M, Levine R, Baylin SB, et al. MLH1 promoter hypermethylation is associated with the microsatellite instability phenotype in sporadic endometrial carcinomas. Oncogene. 1998;17:2413–2417. doi: 10.1038/sj.onc.1202178. [DOI] [PubMed] [Google Scholar]
- 102.Fleisher AS, Esteller M, Wang S, et al. Hypermethylation of the hMLH1 gene promoter in human gastric cancers with microsatellite instability. Cancer Res. 1999;59:1090–1095. [PubMed] [Google Scholar]
- 103.Fang DC, Wang RQ, Yang SM, et al. Mutation and methylation of hMLH1 in gastric carcinomas with microsatellite instability. World J Gastroenterol. 2003;9:655–659. doi: 10.3748/wjg.v9.i4.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Abraham SC, Lee JH, Boitnott JK, et al. Microsatellite instability in intraductal papillary neoplasms of the biliary tract. Mod Pathol. 2002;15:1309–1317. doi: 10.1097/01.MP.0000038461.80167.34. [DOI] [PubMed] [Google Scholar]
- 105.Limpaiboon T, Khaenam P, Chinnasri P, et al. Promoter hypermethylation is a major event of hMLH1 gene inactivation in liver fluke related cholangiocarcinoma. Cancer Lett. 2005;217:213–219. doi: 10.1016/j.canlet.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 106.Ohta M, Inoue H, Cotticelli MG, et al. The FHIT gene, spanning the chromosome 3p14.2 fragile site and renal carcinoma-associated t (3;8) breakpoint, is abnormal in digestive tract cancers. Cell. 1996;84:587–597. doi: 10.1016/s0092-8674(00)81034-x. [DOI] [PubMed] [Google Scholar]
- 107.Tzivion G, Avruch J. 14-3-3 proteins: active cofactors in cellular regulation by serine/threonine phosphorylation. J Biol Chem. 2002;277:3061–3064. doi: 10.1074/jbc.R100059200. [DOI] [PubMed] [Google Scholar]
- 108.Subramanian RR, Masters SC, Zhang H, et al. Functional conservation of 14-3-3 isoforms in inhibiting bad-induced apoptosis. Exp Cell Res. 2001;271:142–151. doi: 10.1006/excr.2001.5376. [DOI] [PubMed] [Google Scholar]
- 109.Prasad GL, Valverius EM, McDuffie E, et al. Complementary DNA cloning of a novel epithelial cell marker protein, HME1, that may be down-regulated in neoplastic mammary cells. Cell Growth Differ. 1992;3:507–513. [PubMed] [Google Scholar]
- 110.Fu H, Subramanian RR, Masters SC. 14-3-3 proteins: structure, function, and regulation. Annu Rev Pharmacol Toxicol. 2000;40:617–647. doi: 10.1146/annurev.pharmtox.40.1.617. [DOI] [PubMed] [Google Scholar]
- 111.van Hemert MJ, Steensma HY, van Heusden GP. 14-3-3 proteins: key regulators of cell division, signalling and apoptosis. Bioessays. 2001;23:936–946. doi: 10.1002/bies.1134. [DOI] [PubMed] [Google Scholar]
- 112.Conway KE, McConnell BB, Bowring CE, et al. TMS1, a novel proapoptotic caspase recruitment domain protein, is a target of methylation-induced gene silencing in human breast cancers. Cancer Res. 2000;60:6236–6242. [PubMed] [Google Scholar]
- 113.Teitz T, Wei T, Valentine MB, et al. Caspase 8 is deleted or silenced preferentially in childhood neuroblastomas with amplification of MYCN. Nat Med. 2000;6:529–535. doi: 10.1038/75007. [DOI] [PubMed] [Google Scholar]
- 114.Liu XF, Zhu SG, Zhang H, et al. The methylation status of the TMS1/ASC gene in cholangiocarcinoma and its clinical significance. Hepatobiliary Pancreat Dis Int. 2006;5:449–453. [PubMed] [Google Scholar]
- 115.Koga Y, Kitajima Y, Miyoshi A, et al. Tumor progression through epigenetic gene silencing of O(6)-methylguanine-DNA methyltransferase in human biliary tract cancers. Ann Surg Oncol. 2005;12:354–363. doi: 10.1245/ASO.2005.07.020. [DOI] [PubMed] [Google Scholar]
- 116.Yang B, Guo M, Herman JG, et al. Aberrant promoter methylation profiles of tumor suppressor genes in hepatocellular carcinoma. Am J Pathol. 2003;163:1101–1107. doi: 10.1016/S0002-9440(10)63469-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kollermann J, Kempkensteffen C, Helpap B, et al. Impact of hormonal therapy on the detection of promoter hyper-methylation of the detoxifying glutathione-S-transferase P1 gene (GSTP1) in prostate cancer. BMC Urol. 2006;6:15. doi: 10.1186/1471-2490-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 119.Li QL, Ito K, Sakakura C, et al. Causal relationship between the loss of RUNX3 expression and gastric cancer. Cell. 2002;109:113–124. doi: 10.1016/s0092-8674(02)00690-6. [DOI] [PubMed] [Google Scholar]
- 120.Isomoto H, Mott JL, Kobayashi S, et al. Sustained IL-6/STAT-3 signaling in cholangiocarcinoma cells due to SOCS-3 epigenetic silencing. Gastroenterology. 2007;132:384–396. doi: 10.1053/j.gastro.2006.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhang Y, Xiong Y, Yarbrough WG. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell. 1998;92:725–734. doi: 10.1016/s0092-8674(00)81401-4. [DOI] [PubMed] [Google Scholar]
- 122.Hannon GJ, Beach D. p15INK4B is a potential effector of TGF-beta-induced cell cycle arrest. Nature. 1994;371:257–261. doi: 10.1038/371257a0. [DOI] [PubMed] [Google Scholar]
- 123.Lee JH, Park SJ, Abraham SC, et al. Frequent CpG island methylation in precursor lesions and early gastric adenocarcinomas. Oncogene. 2004;23:4646–4654. doi: 10.1038/sj.onc.1207588. [DOI] [PubMed] [Google Scholar]