Abstract
The objective of the workshop was to gain a better understanding of the link between circadian rhythms and human health and disease. The impacts of circadian rhythms on metabolic gene regulation, as well as the effect of nutrient uptake and balance on the molecular components of the clock, were discussed. Topics included the neural circuitry underlying the central clock; the effect of the environment and diet on the central clock as well as peripheral, tissue-specific clocks; and the transcriptional, post-transcriptional, and post-translational (e.g., epigenomic) mechanisms through which these signals are transduced. Evidence presented during the meeting demonstrated that circadian rhythms and metabolism are intricately linked, and that disruption in these rhythms have profound consequences—many times leading to metabolic disease. The mechanisms by which circadian rhythms are maintained and the cross-talk with metabolic signaling are just beginning to be elucidated. However, the interactions between these fields and the knowledge learned will clearly have a profound impact on our understanding of metabolic disease and lead to novel therapeutic approaches in the future
Keywords: Circadian, rhythms, metabolism, core clock, peripheral clocks
In the past decade, it has become clear that the signaling cascades contributing to metabolic regulation respond to both central and cellular timing signals. Disruptions in the normal circadian rhythms of an animal result in changes in sleep, activity, and eating patterns. Change in patterns can lead to the dysfunction of metabolic pathways, and may ultimately lead to a number of diseases, including obesity, metabolic syndrome, type 2 diabetes, cardiovascular disease, and cancer. The environmental cue of light and dark is well described to entrain the central clock. However, it is becoming better appreciated that cues such as nutrient uptake (feeding) and temperature also can impinge on the central clock. Many of the genes responsible for the regulation of the “core” clock have been identified, and their role in peripheral (tissue-specific) clocks is becoming apparent. Evidence from studies on these peripheral clocks, especially as they affect metabolism, is beginning to elucidate the integral role of these clocks in normal physiology as well as disease. Studies demonstrate that the mechanisms by which this dysregulation occurs include signaling at the transcriptional, post-transcriptional, post-translational, and epigenomic levels. One specific point of intersection lies at the level of nuclear receptors (NRs)—hormones, vitamins, and xenobiotic and nutrient-dependent transcription factors involved in reproduction, feeding, and homeostasis. It has now been shown that a number of these NRs are also key components of the clock, providing a direct link between the workings of the clock, gene regulation, and metabolism. Epigenetic mechanisms—specifically the role of clock genes in the modification of chromatin to influence gene regulation—also represent an exciting new overlay to regulation of the clock. A workshop on Circadian Rhythms and Metabolic Disease sponsored by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) in Bethesda, MD, on April 12 and 13, 2010, brought together scientists in the fields of circadian rhythms and metabolism in order to gain a better understanding of the complex interrelationship between clock genes and genes involved in metabolism, and their potential roles in metabolic disease. The workshop brought together investigators from different disciplines to (1) enhance the exchange of information and (2) increase the collaboration between and among investigators from different disciplines with the goal to develop a better understanding of the role(s) of circadian rhythmicity in the regulation and/or modulation of signaling pathways responsible for metabolic regulation.
Mammalian circadian clocks
Dr. Joe Takahashi (University of Texas Southwestern Medical Center, Dallas, TX) began the workshop with a plenary session that provided an overview of the central and cellular clocks in mammals at the genetic and molecular level. As a perspective, he began by emphasizing that circadian rhythms are evolutionarily conserved, and represent an adaptation of the organism to its environment. Traditionally, it is well known that “environment” is reflected by the light/dark cycle, resulting from the rotation of the earth, but, as we learned during the workshop, it also includes wake/feeding and sleep/fasting as additional inputs. Integration of the clock is a complex process that involves regulation of genes on one level, and results in very real behavioral outputs at the organism level. An additional layer of complexity is the presence of peripheral clocks in the cells of almost all tissues throughout the organism. Genetic analysis has allowed identification of a number of core clock genes that generate circadian oscillations (e.g., Clock, Bmal1, Per1, Per2, Cry1, Cry2, and casein kinase ɛ). A number of other genes integral to the clock and circadian behavior (e.g., ror and rev-erbα) and the transcriptional and post-transcriptional regulation were also discussed. Interactions between the proteins encoded by these genes—such as classical feedback mechanisms and post-translational modifications resulting in degradation or increased stability—provide another level of regulation (see Fig. 1; Yoo et al. 2004).
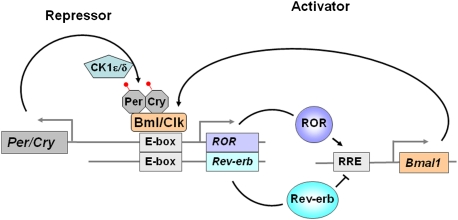
Figure 1.
Interlocked transcriptional feedback loops of the circadian clock. This model figure shows that a key positive loop of the clock is controlled by ROR and the Bmal/Clk heterodimer. This positive loop is opposed by the repressor loop, which involves Per/Cry. (Image courtesy of Ron Evans.) See the text for details.
The overriding core or master clock resides in the brain in the suprachiasmatic nucleus (SCN) of the hypothalamus. Coupling of neurons within the SCN transforms the output from this central clock, and is influenced by outside stimuli such as light/dark and feeding behavior. But Dr. Takahashi went on to describe another level of circadian output that was a common theme throughout the workshop, and that is regulation at the cellular level in every peripheral tissue studied. (Liu et al. 2007) These clocks are influenced by signals from the central clock, but are also regulated in a cell-autonomous level. This regulation can be demonstrated dramatically by monitoring cells in culture as they continue to respond rhythmically, maintaining synchrony. So, not only is there a clock in every cell, but these clocks also have tissue-specific (and species-specific, in some cases) inputs, outputs, and rhythms. Many of these outputs are related to and influenced by metabolism, and a number of the same transcription factors (particularly NRs) are involved in clock regulation as well as metabolism (see Fig. 2). Speakers in the following sessions presented their work on specific aspects of these clocks, and how they influence, and are influenced by, metabolic outputs.
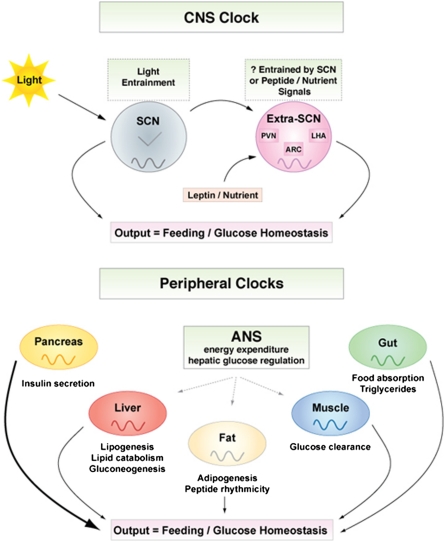
Figure 2.
Clock controlled output. The central and peripheral clocks together regulate metabolism, inflammation, and energy homeostasis. (Image courtesy of Billie Marcheva and Joseph Bass.) See the text for examples.
Central and neural circuitry
The first session of the workshop addressed the central clock, the neuronal connections in this clock, hypothalamic circuitry, and the link between feeding and circadian rhythms. Dr. Rae Silver (Barnard College and Columbia University, New York, NY) began by describing a food-entrained oscillator (FEO) that is distinct from the light-entrained oscillator (LEO) found in the SCN. Both of these regulate behavior, but one is entrained by food (eating) and the other is entrained by light (light/dark cycle). When the FEO and LEO cues are in synchrony, then the food- and light-entrained behaviors are indistinguishable. However, if the cues are out of synchrony, then the behaviors are distinguishable and the organism becomes entrained by food anticipatory activity (Antle and Silver 2009). Indeed, experimental mouse models demonstrate that, when the LEO is weakened (e.g., by SCN lesion, clock gene mutation, or knockouts), then the FEO is enhanced. When FEO is disrupted, the SCN component is enhanced, indicating that there is a distinct cue responsible for signaling feeding behavior. Dr. Silver then presented data that supported the role of ghrelin as this cue for food, and ghrelin-producing cells as FEOs. Ghrelin is the only gastrointestinal hormone that is increased in anticipation of a meal. It is secreted by oxyntic cells in the fundus of the stomach, and these same cells also express the clock proteins PER1 and PER2. Many peripheral tissues as well as brain regions contain ghrelin receptors. Ghrelin is secreted right before a meal and is shifted by the timing of the meal, and, importantly, ghrelin rhythms are not shifted by light cues. Furthermore, ghrelin receptor knockout mice show diminished food anticipatory activity (LeSauter et al. 2009).
Dr. Erik Herzog (Washington University, St. Louis, MO) discussed his work on identifying factors responsible for synchronizing the output from neuronal cells in the SCN, the master circadian pacemaker. Using electrical activity measurements from single neurons, Dr. Herzog demonstrated the precise daily synchrony when SCN cells communicate with each other. In fact, there is evidence that the cells of the SCN can maintain their rhythm for >1 yr in culture. In contrast, SCN cells that are prevented from communicating with each other display individualized periods in culture. These results led Dr. Herzog to ask: “How do cells communicate in order to keep this synchronous rhythm?” His results demonstrate that VIP (vasoactive intestinal polypeptide) was required for SCN synchrony (Aton et al. 2005). VIP is secreted on a daily basis and in response to light, and new data indicate that it entrains SCN neurons to each other in a predictable phase relationship. Since the receptors for VIP (VPAC2R) are widely expressed, this same peptide could also play a role in the rhythmicity of clocks in the brain and body. By cross-correlating neuronal spikes at millisecond resolution, Dr. Herzog's group is also mapping the connectivity of neurons in the SCN. Their measurements have revealed both fast inhibitory and excitatory connections in roughly equal numbers, which are relatively rare, compared with the possible number of connections, and varying in strength and number over the circadian day. Finally, these connections may regulate not only the timing of melatonin release from the pineal gland, but also glucose production from the liver, providing evidence for the connection between the master and peripheral clocks and effects at the metabolic level (SJ Aton, GM Freeman, S An, C Tsai, RM Krock, and ED Herzog, unpubl.).
Dr. Clifford Saper (Harvard Medical School, Boston, MA) continued the discussion of neuronal links and circuitry in and from the SCN. He provided evidence showing that neuronal projections from the SCN passed through the subparaventricular zone to the dorsomedial nucleus of the hypothalamus (DMH). Targeted disruption of the DMH led to disruptions in sleep pattern, as well as in the circadian rhythm of locomotor activity and feeding, and the secretion of corticosteroids; however, body temperature and melatonin secretion were not affected (Chou et al. 2004). In another set of experiments, he showed that food restriction not only led to a shift in the rhythm of locomotor activity and body temperature, but also reset gene expression in the DMH (Gooley et al. 2006). Lesions of the DMH prevented entrainment in animals on a restricted feeding paradigm. To identify the clock that drives food entrainment, Mieda et al. (2006) and Fuller et al. (2008) looked at clock gene expression in the brain during restricted feeding and induction of a robust rhythm of clock gene expression in the compact part of the DMH, but only under conditions of food entrainment. To test whether the clock gene expression in the DMH was capable of driving entrainment of locomotor activity and body temperature rhythms, Fuller et al. (2008) then studied mice that were deficient in the core clock protein BMAL1. Mice lacking BMAL1 had no baseline circadian rhythms, but restoration of BMAL1 to the SCN resulted in normal locomotor and body temperature rhythms. However, these animals did not entrain to food. Restoring BMAL1 to the DMH did not affect baseline circadian rhythms, but did allow entrainment of body temperature and locomotor activity during restricted feeding. Together, these results indicate that the DMH is required for food entrainment, and that the DMH clock is capable of driving food-entrained rhythms. However, other signals—such as from the ghrelin-containing cells of the stomach, described by Dr. Silver—could still contribute to food entrainment in the intact animal. The overall effect of these studies has been to show how intimately interconnected signaling in brain regions is to enable responses to environmental cues.
Session on model organisms
In order to study circadian rhythms, particularly by the controlled disruption/modulation of these rhythms, a number of nonmammalian model organisms have proven quite useful in providing mechanistic insights into the workings of circadian clocks. In this session, we heard from three investigators who use either the fruit fly (Drosophilia melanogaster) or the eastern North American monarch butterfly (Danaus plexippus) as model organisms. Dr. Steven Reppert (Univeristy of Massachusetts Medical School, Worcester, MA) described an elegant model to study the molecular clock of the monarch butterfly. Using a flight simulator and molecular approaches, his laboratory has been able to dissect the components of the light-entrained circadian clock and the time-compensated sun compass of the butterfly. The clock and sun compass integrate to prompt and direct the southern migration of the butterflies during the fall season. The circadian clock located in the butterfly brain expresses period and timeless, which are under transcriptional control by the CLOCK protein. The butterfly clock also expresses two cryptochrome proteins: Cry1 (Drosophila-like), which acts as a photoreceptor, and Cry2 (vertebrate-like), which is a clock component and transcriptional repressor; Cry2 is expressed in all nondrosophilid insects so far examined. The expression of Cry1 and Cry2 in the butterfly likely represents an ancestral clock mechanism common among lepidopteran species (Zhu et al. 2008). During evolution, Drosophila lost cry2, and express only cry1, while other insects and vertebrates lost cry1 and express cry2 (Yuan et al. 2007). Specific antibodies against monarch Cry1 and Cry2 label clock circuits in the brain that may, in turn, regulate migratory behaviors (Sauman et al. 2005; Zhu et al. 2008). Using the monarch butterfly model, the Reppert laboratory has determined that the major timing mechanism for sun compass orientation resides in the butterfly antennae, not the brain (Merlin et al. 2009). These studies provide a unique model in which a peripheral clock regulates brain-generated behavior. To continue studies on circadian clocks in the monarch butterfly, Dr. Reppert and his colleagues are developing a genomic toolbox that consists of expressed sequence tags, the entire genomic sequence of the butterfly, and a zinc finger nuclease strategy to knock out specific genes to help elucidate the underlying molecular mechanisms of clock function.
Using the Drosophila model, Dr. Michael Rosbash (Brandeis University, Waltham, MA) has undertaken a detailed investigation of the genes that are regulated by the CLOCK protein (CLK/CYC). The current molecular model for regulation of rhythms in flies is that the CLK/CYC protein regulates the expression of the period and timeless genes, and, in a classical feedback loop, the PERIOD and TIMELESS proteins then inhibit the expression of the clock gene (Menet et al. 2010). While it generally has been thought that there are few direct target genes of CLK besides period and timeless, Dr. Rosbash provided convincing evidence that there are many more genes regulated by CLK in Drosophila. Using chromatin immunoprecipitation (ChIP) with antibodies against not only CLK, but also PER and RNA Polymerase II, he identified >2200 peaks reflecting CLK binding. Further analysis using tiling arrays demonstrated that there were at least 700 direct CLK target genes within the genome, with not all of the binding motifs reflected by the canonical E-box, and that, in most cases, PolII was cycling on these same genes. He then went on to describe a series of experiments that provided evidence for the heretofore unappreciated number of direct CLK target genes assessed previously by analysis of cycling mRNAs. Studies revealed that almost 50% of CLK target genes have alternative start sites. Analysis of neuron-specific genes resulted in the identification of a number of cycling clock genes above and beyond those identified in Drosophila (Nagoshi et al. 2010). He also showed that the number of cycling mRNAs identified could be increased by performing RNA sequencing (RNA-seq) analysis at six separate time points (Kadener et al. 2009). In summary, while the overall mechanism of transcriptional regulation by CLK genes is correct, more sophisticated analysis, as described by Dr. Rosbash, has shown that this regulation is much more complicated than originally thought.
Dr. Amita Sehgal (University of Pennsylvania, Philadelphia, PA) continued discussion of circadian clocks in Drosophila, particularly with respect to the circadian control of feeding and metabolic activity. In Drosophila, in addition to the neuronal clock, the fat body (analogous to liver and adipose tissue) also has a clock, and disruption of this clock affects feeding. Disruption of the fat body clock decreases energy stores, increases sensitivity to starvation, and increases feeding. In contrast, disruption of the neuronal clock increases the storage of nutrients and decreases the sensitivity to starvation. Interestingly, disruption of the two clocks together has minimal effects on metabolic activity. Inhibition of the central clock results in storage of triglycerides in the fat body. Microarray analysis from wild-type fat body versus CLOCK-impaired fat body demonstrates that a number of metabolic genes are regulated by CLOCK in this tissue. In addition to showing effects of the central and peripheral clocks on metabolic pathways, Dr. Sehgal ended by showing data that show that disruption of signaling pathways involved in metabolism (e.g., mTOR) can affect circadian rhythms in the fly, thus providing further evidence of the reciprocal and integral relationship in this model system between circadian rhythms and metabolism (Xu et al. 2008). Clearly, even the most primitive vertebrates must respond to the environment in which they live, with dependent pathways that are broadly conserved throughout evolution.
Transcription and peripheral clocks
The common theme of transcription and peripheral clocks was discussed next. Topics addressed included the role of NRs in circadian rhythms, post-transcriptional modifications as a mechanism of clock regulation, and the overlay of epigenetic modifications as a regulatory mechanism. Dr. Ron Evans (Salk Institute, La Jolla, CA) began by putting the situation in perspective when he said that both physiology and circadian rhythms share many of the same features. Both are “self-regulating, adaptive, and recurrent processes that must reset themselves every day.” He presented the phylosphere of the 48 nuclear hormone receptors, dividing these receptors into six groups by their known or deduced functions (Bookout et al. 2006). One group of NRs encompasses those involved in both circadian and basal metabolic functions (e.g., RORα/β and REV-ERBα/β) (Yang et al. 2006). The metabolic and physiologic functions of NRs are well described in gluconeogenesis, adipogenesis, inflammation, reproduction, etc. The theme of Dr. Evans' talk was to demonstrate the integral link between NRs and circadian rhythms, as well as the reciprocal nature of this interaction. He showed that the cellular energy state-sensing AMP kinase acts as a master metabolic switch, as it is influenced by energy inputs such as exercise, feeding, and nutrients, and is able to transmit this signal to the tissue-specific peripheral clocks. At a mechanistic level, Dr. Evans showed data supporting the role of AMP kinase in phosphorylating CRY1 at serine residue 71 (Ser71). Ser71 is conserved in all CRY1 proteins that act as transcriptional repressors (e.g., vertebrates), but not in evolutionarily distinct CRY1 that acts as a photoreceptor (such as in monarch butterflies). He showed that AMP kinase-regulated phosphorylation of Ser71 results in the degradation of the CRY1 protein and an alteration of the circadian clock in hepatocytes and fibroblasts. The role of AMP kinase in peripheral clocks is demonstrated further by the fact that this kinase can alter the set time of the hepatocyte clock, and its activation alters circadian rhythms in fibroblasts (Lamia et al. 2009). To demonstrate further the link between clock and metabolism, he hypothesized that CRY knockout mice would show different sets of glucocorticoid-regulated genes compared with those with intact CRY1 with potential disruptions in glucose homeostasis (RM Evans, unpubl.). Since 12 out of 49 NRs are known to interact with the CRY1 protein, there are certainly many other NR- and tissue-specific mechanisms that integrally link the circadian clock and metabolic pathways.
Dr. Mitch Lazar (University of Pennsylvania, Philadelphia, PA) continued the topic of NRs and the circadian clock by discussing the epigenetic regulation of gene expression in the liver. NRs respond to cues from the environment that are transduced through ligand binding. Ligand-bound NR interacts directly with DNA and recruits a series of coactivators or corepressors that exhibit or recruit enzymatic activity. As an example, the corepressor NCoR/SMRT recruits the histone deacetylase HDAC3, leading to epigenetic changes and regulation of gene expression (Alenghat et al. 2008). This regulation impacts both circadian as well as metabolic genes. In the liver, one of the NRs that functions in this way is Rev-erbα (Yin et al. 2010). Ongoing studies using cistromic approaches reveal that the recruitment of Rev-erbα together with HDAC3 at thousands of sites in the mouse liver genome modulates the expression of genes regulating hepatic lipid metabolism and circadian rhythm. The Rev-erbα interaction with the NCoR–HDAC3 complex is gated by heme, which functions as a ligand for Rev-erbα (Yin et al. 2007) and may serve as the physiological link between the circadian and metabolic pathways.
In two related talks, the role of Rev-erbα and related receptors were discussed further. Dr. Lei Yin (University of Michigan, Ann Arbor, MI) described her work on the regulation of the Rev-erbα protein through degradation that is stimluated by lithium, which itself is known to regulate circadian rhythm (Yin et al. 2006) . She identified two E3 ligases that cause ubiquitination and subsequent degradation of the Rev-erbα protein. Depletion of either of these ligases impacts the regulation of clock genes by Rev-erbα and resultant circadian function in the mouse liver (L Yin, unpubl.). Dr. Thomas Burris (Scripps Research Institute, Jupiter, FL) described a number of agonists of the Rev-erbα receptor that have been developed through a chemical biology approach. He provided data to show that these agonists induce adipogenesis, suppress glucose output from hepatocytes, and inhibit PEPCK gene transcription (Kumar et al. 2010). Both better agonists as well as antagonists are under development for Rev-erbα in addition to ROR (α/β/γ) (Wang et al. 2010). Given the integral role of NRs in physiology and circadian rhythms, these synthetic ligands could be very useful for treatment of both metabolic disorders and defects in circadian rhythms.
Dr. Carla Green (University of Texas Southwestern Medical Center, Dallas, TX) described her ongoing research on the discovery of nocturnin, a gene that increases expression at night and is the only one of five identified deadenylases that is rhythmically expressed (Baggs amd Green 2003). While nocturnin is not part of the clock mechanism, it is an output of the clock, and has a high-amplitude rhythm in the liver. While nocturnin knockout animals have no obvious metabolic defects, when put on a high-fat diet, these animals are resistant to weight gain and are protected from hepatic steatosis (Green et al. 2007). There is less fat storage in their livers, and while there is a change in the expression of genes involved in lipid metabolism, there is no change in the oxidation of lipids. Further investigation indicates that the mice accumulate large droplets of lipid in the cells lining their intestines. Future work will investigate the mRNAs that are regulated by nocturnin, and may lead to important avenues for therapeutic intervention in fatty liver disease and other components of the metabolic syndrome.
Dr. John Hogenesch (University of Pennsylvania, Philadelphia, PA) gave a comprehensive view of the transcriptional output of the mouse liver clock. Using a combination of exon-based DNA arrays and RNA-seq, his group sampled mouse livers every 2 h and analyzed RNA expression. The exon-based arrays were able to identify hundreds of genes whose transcription were not overtly cyclic at the whole-transcript level, but had cycling of one or more exons. The RNA-seq data were taken at a 6-h frequency, and identified many alternative splicing events as well as noncoding RNA expression that appears to be clock regulated. He showed that genome-wide RNA profiling from mouse livers at 2-h intervals for 2 d resulted in the detection of circadian as well as ultradian rhythms. Some genes, such as Bmal1, showed a 24-h frequency, while others cycled at a 12-h frequency (e.g., hspa1b) and some cycled at an 8-h frequency (Hughes et al. 2009). Therefore, this analysis gave a more comprehensive view of circadian gene regulation. Given the large expanse of data to be analyzed from this type of analysis, Dr. Hogenesch brought on board mathematician Karl Kornacker (Ohio State University), who developed an algorithm (“JTK cycle”) that is many times faster than previous algorithms, is resistant to outliers, and accurately reports cycling, period, amplitude, and phase. In collaboration, they identified >600 genes that were not identified by previous analysis, demonstrating again the importance of the integration of disciplines in addressing this type of research. Dr. Hogeneschs' work aptly summed this session, pointing to changes in expression of individual genes and networks of genes as the end point of circadian patterns of response. These, in turn, are reflected as changes in behavior of the whole organism.
Interactions between central and peripheral clocks
Dr. Satchidananda Panda (Salk Institute, La Jolla, CA) discussed the contributions of both the central core clock components as well as input from time of feeding on hepatic metabolic genes. Feeding time determines the targets and phases of rhythmic transcripts. He demonstrated that, in Cry-deficient mice, consolidated feeding could restore rhythmicity in many liver gene transcripts (∼600 out of 3000 genes transcripts) (Vollmers et al. 2009). Conversely, in fasting mice, only 368 out of 2629 genes remain rhythmic. Thus, there is a synergistic interaction between clock and temporally restricted feeding patterns in many important metabolic genes, including those involved in mitochondrial function. Moving forward, it will be important to determine the role of nutrient quality, quantity, and feeding pattern in maintaining full rhythmicity of liver gene expression and normal physiological states.
Dr. Vijay Yechoor (Baylor College of Medicine, Houston, TX) discussed the role of the clock gene Bmal1 in hypoinsulinemia. Bmal1 knockout mice (global, but not liver-specific) have impaired glucose tolerance early in life that develops into frank diabetes (Lamia et al. 2008). Glucose-stimulated insulin secretion (GSIS) is defective (the first phase of insulin secretion is virtually absent). He noticed that these mice have an increased β-cell area in the pancreas, but that the stimulation of insulin secretion and the expression of the glucose transporter Glut2 were normal. However, he did find that the islets from these mice have increased expression of uncoupling protein 2 (UCP2), and that impaired GSIS can be partially rescued by a UCP2 inhibitor (J Lee, M Kim, R Li, V Liu, X Xia, K Ma, L Fu, DD Moore, and V Yehoor, unpubl.). Thus, he concludes that the core clock component Bmal1 is necessary for normal mitochondrial function in the β cells of the islet, providing another integral link between the clock and metabolism, specifically in the β cell. Further research will investigate whether this is due to central or peripheral clock genes by studying a β-cell-specific Bmal1 knockout. Results of these studies may have important clinical implications regarding the treatment of diabetes.
Dr. Joseph Bass (Northwestern University, Evanston, IL) also discussed the synchronization of feeding, metabolism, and sleep, and the overlap of circadian and metabolic transcription networks (Marcheva et al. 2009). This connection is well demonstrated in clock knockout mice that show a behavioral and metabolic phenotype with loss of eating rhythms, increased body weight, and metabolic syndrome reflected by hyperglycemia and hyperlipidemia (Turek et al. 2005). He described the circadian clock as a “hub” that regulates metabolism, inflammation, and energy homeostasis (Maury et al. 2010). He then discussed the contribution of the central clock versus the peripheral clock, and underscored the intensive interest in delineating tissue-specific clock function in metabolic homeostasis. He described genetic strategies that his group and others are now exploiting to dissect the impact of clock expression in brain or periphery. He also highlighted how further insight into clock function within local tissue physiology may open opportunities to target the clock as part of a therapeutic strategy in metabolic disorders (Marcheva et al. 2010). Thus, building on the work in transcription, the maintenance of homeostasis is paramount, and disruptions of normal physiology at any level leads to metabolic dysfunction.
The second day of the meeting started with a plenary talk by Dr. Ueli Schibler (University of Geneva, Switzerland) entitled “Signaling within the Mammalian Circadian Timing System.” Dr. Schibler continued the discussion of central and peripheral clocks (Dibner et al. 2010). He provided evidence that body temperature increases the strength of circadian oscillations through the cold-inducible binding protein (CIRP), and that these oscillations are synchronized through a heat-shock transcription factor (HSF-1). In fact, a low-amplitude change in body temperature results in a large change in gene expression rhythms. HSF-1 is a circadian transcription factor, and is post-transcriptionally activated by elevated temperature (Reinke et al. 2008). He showed that, in the liver, HSF-1 mRNA and protein accumulate throughout the day, although the nuclear abundance (and DNA-binding activity) is strongly circadian. Furthermore, the expression of the clock gene (Bmal1) in a cultured fibroblast cell line can be synchronized by simulating body temperature oscillations in an HSF-1-dependent manner. CIRP expression is regulated by temperature body rhythms in the liver, and simulated body temperature rhythms in cultured fibroblasts in a manner that is independent of the core clock. The free-running circadian clock is strongly dampened in cells from CIRP knockout mice. Since Cirp mRNA and protein accumulate to threefold to fivefold higher levels when the body temperature reaches nadir values, Schibler proposed that body temperature rhythms enhance the strength of peripheral circadian oscillators in a CIRP-dependent fashion. In conclusion, a decrease in body temperature results in increased CIRP, which enhances the amplitudes and magnitudes of circadian oscillators, whereas the periodic increase of body temperature rhythms leads to HSF-1 activity cycles that in turn participate in the synchronization of peripheral clocks (C Saini, J Morf, and U Schibler, unpubl.).
Dr. Paolo Sassone-Corsi (University of California at Irvine, Irvine, CA) discussed the connection between epigenetics and regulation of metabolism by the circadian clock. Post-translational modification can involve methylation, acetylation, phosphorylation, ubiquitination, and even glycosylation, all of which are “sensors” of metabolism (via specific metabolites). He provided two examples of the intricate interplay between transcription and enzymatic activity of these protein modifiers. The CLOCK protein is itself a histone acetylase (HAT), capable of acetylating not only histone H3, but also the clock protein BMAL1. In addition, the HDAC activity of SIRT1 is regulated in a circadian manner. Since these enzymatic activities are dependent on metabolites (such as NAD+ in the case of SIRT-1), there exists a precise feedback mechanism that intricately links energy production and metabolism with the clock (Grimaldi et al. 2009; Yasukazu et al. 2009). For example, SIRT-1 regulates nampt, which codes for nicotinamide phosphoribosyltransferase, the rate-limiting enzyme in the NAD+ salvage pathway. Thus, Sirt-1 functions as a rheostat in the control of circadian function, and ablation of sirt-1 activity results in disturbances in the circadian cycle (Nakahata et al. 2008, 2009). Another example is the trimethylation of K4 in histone H3, which is an activating modification (unlike most methylations on H3, which are silencing). The methylase responsible is being characterized regarding its regulation and interaction with many coregulators, including those that are recruited in a circadian manner (P Sassone Corsi, unpubl.). The exciting convergence of molecular mechanisms of circadian rhythmicity and the epigenome opens up many new avenues for understanding how the genome responds to environmental cues.
Circadian rhythms and human disease
A number of talks addressed the role of circadian misalignment as it influences human disease. Dr. Charles Czeisler (Harvard Medical School, Boston, MA) discussed the metabolic disorders prevalent when circadian and behavioral patterns (of sleep and eating) are misaligned. Misalignment of the endogenous circadian phase can occur with respect to retinal light exposure, meal times, and the times of wake/activity, work/school, exercise, and sleep. Examples that Dr. Czeisler cited are those misalignments that occur as a result of jet lag (a transient misalignment), as well as an example of a more chronic misalignment that occurs during shift work. Citing some of his earlier work, he showed a linear correlation between bed time and melatonin release (Shanahan et al. 1999). More recent studies of Navy personnel on submarines that have an 18-h day (and a 6-h sleep time) show nonentrained circadian rhythms of melatonin release (Gronfier et al. 2007). Interestingly, the onset of melatonin secretion can be used to diagnose a human condition known as delayed sleep phase syndrome (Duffy and Czeisler 2009). Dr. Czeisler demonstrated that a bright light pulse (but not one of dim light) can result in the entrainment of circadian rhythm. He reviewed for us a large body of literature demonstrating that shift work increases the risk for a number of diseases, including coronary heart disease, gastrointestinal disorders, diabetes, and cancer. His talk provided the foundation and background for many of the presentations that followed.
Dr. R. Daniel Rudic (Medical College of Georgia, Augusta, GA) described the role of core clock genes Bmal1, Clock, Period, and Cry in the vasculature, where these components also oscillate. He showed that a clock mutant lead to an exacerbation of injury to an artery induced by ligation (Westgate et al. 2008). Again, there is evidence for influence from the central circadian clock, as well as peripheral clocks expressed in vessel walls (Anea et al. 2009).
Dr. Steven A. Shea (Harvard Medical School, Boston, MA) addressed the concept of episodic diseases, those that frequently occur at certain times of the day, such as asthma exacerbations, heart attacks, and epileptic seizures. Using a forced desynchrony protocol, he demonstrated that there is a circadian rhythm in the response to stressors. Dr. Shea's research is aimed at determining the mechanisms by which misalignment between circadian rhythms and behavior (sleeping, eating, and exercise) can affect physiological measures (cardiopulmonary and metabolic measures) and, ultimately, susceptibility to disease. For example, with colleagues, he found that circadian misalignment resulted in decreased leptin levels (i.e., signaling negative energy balance that could promote appetite) and decreased glucose tolerance (increased plasma glucose without a decrease in insulin), as well as a 20% decrease in sleep efficiency. It appears that circadian misalignment rather than the sleep disruption has the greater effect on these outcomes (Scheer et al. 2009). Mechanistic studies also indicate that the changes in leptin are not due to either a decrease in insulin, glucose, or cortisol, or by increasing catecholamines. Furthermore, these changes were induced by acute interventions during the experimental protocol, and were readily reversible. More research is needed to determine the longer-term effects (years of shift work) and the genetic and/or other factors that make some people susceptible to adverse effects of less or altered sleep while others are not.
Finally, there were a number of talks that focused around the role of sleep in circadian rhythms and metabolism, and, consequently, disease. Dr. Fred Turek (Northwestern University, Evanston, IL) began by telling us that sleep, circadian rhythms, and fuel metabolism cannot be separated, such that sleep, diet, and exercise are all intertwined with the clock (Turek 2008). There are many examples in genetic mutant mice as well as evidence from humans where shift workers have increased body mass index (BMI) and also increased incidence of diseases such as type 2 diabetes. Chronic sleep loss leads to obesity, diabetes, and cardiovascular disease, and circadian dysregulation leads to metabolic dysfunction and disease (Arble et al. 2009). The disruption of core clock genes is known to have important implications for human health. So, what are the sleep genes? Analysis indicates that there is overlap in sleep and circadian genes, and their identification is under way (Laposky et al. 2008). As Dr. Turek concluded, “sleep is good for clocks, and clocks are good for sleep.”
Drs. Louis Ptacek and Ying-Hui Fu discussed their investigation of human circadian and sleep variants. They found that mutations in a number of genes affect sleep quantity. However, the mechanisms that control how much sleep people need are unknown. Study of these genes/mutations is an opportunity to begin to increase our understanding. Drs. Ptacek and Fu and their collaborator Dr. Chris Jones identified families with natural short sleep (FNSS) and families with natural long sleep (FNLS). They identified a mutation in Dec2 (a gene that codes for a transcription factor) that causes FNSS and, when put into mice, also causes less sleep and an as yet not fully characterized metabolic syndrome. Advanced sleep phase syndrome (ASPS) is an alteration in biological sleep and wake preferences in humans. This group identified an autosomal-dominant form of familial ASPS (FASPS), and identified a number of causative mutations, including one at a hPer2 at a serine site that is phosphorylated by a metabolic regulatory enzyme, casein kinase 1δ. Another FASPS family carries a casein kinase 1δ mutation, and mutation carriers also have a propensity to migraines with aura (Toh et al. 2001; Xu et al. 2005). They identified 70 families with FASPS and 20 with FDSPS (familial delayed sleep phase syndrome). Mouse models of some of the mutations have already been made, and more are being developed in order to study the molecular consequences of these mutations in vivo. Thus, the integration of neural, cellular, and molecular responses to both central and peripheral clocks becomes a central concern in maintaining proper physiological responses and health.
Conclusions and perspectives
In summary, this highly interactive and informative workshop used wide-ranging presentations, from flies to humans, to demonstrate the importance of circadian rhythmicity to homeostasis of the entire organism. Building on observations ranging from molecular studies to integrated whole-body physiology, a better understanding of the central and peripheral clocks is emerging. Progress to date has allowed for the development of a level of maturity that facilitates formulation of informed hypotheses to test the role of multiple zeitgebers and other environmental factors as integrators of clock functions. Translating this emerging system of biological clocks to human behavior, and linking its dysfunction to metabolic disease, is both a great challenge and the next logical step.
Acknowledgments
Wethank the co-organizers for the workshop, Drs. Mitch Lazar and Charles Czeisler, and Drs. Ron Evans and Joe Bass for contributing model figures.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1948310.
References
- Alenghat T, Meyers K, Leitner K, Adeniji-Adele A, Mullican SE, Avila J, Bucan M, Ahima RS, Kaestner KH, Lazar MA 2008. Nuclear receptor corepressor-histone deacetylase 3 governs circadian metabolic physiology. Nature 456: 997–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anea CB, Zhang M, Stepp DW, Simkins GB, Reed G, Fulton DJ, Rudic RD 2009. Vascular disease in mice with a dysfunctional circadian clock. Circulation 119: 1510–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antle MC, Silver R 2009. Neural basis of timing and anticipatory behaviors. Eur J Neurosci 30: 1643–1649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arble DM, Bass J, Laposky AD, Vitaterna MH, Turek FW 2009. Circadian timing of food intake contributes to weight gain. Obesity (Silver Spring) 17: 2100–2102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aton SJ, Colwell CS, Harmar AJ, Waschek J, Herzog ED 2005. Vasoactive intestinal polypeptide mediates circadian rhythmicity and synchrony in mammalian clock neurons. Nat Neurosci 8: 476–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baggs J, Green CB 2003. Nocturnin, a deadenylase in Xenopus laevis retina: A mechanism for posttranscriptional control of circadian-related mRNA. Curr Biol 13: 189–198 [DOI] [PubMed] [Google Scholar]
- Bookout AL, Jeong Y, Downes M, Yu RT, Evans RM, Mangelsdorf DJ 2006. Anatomical profiling of nuclear receptor expression reveals a hierarchical transcriptional network. Cell 126: 789–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou TC, Rotman SR, Saper CB 2004. Lateral hypothalamic acetylcholinesterase-immunoreactive neurons co-express either orexin or melanin concentrating hormone. Neurosci Lett 370: 123–126 [DOI] [PubMed] [Google Scholar]
- Dibner C, Schibler U, Albrecht U 2010. The mammalian circadian timing system: Organization and coordination of central and peripheral clocks. Annu Rev Physiol 72: 517–549 [DOI] [PubMed] [Google Scholar]
- Duffy JF, Czeisler CA 2009. Effect of light on human circadian physiology. Sleep Med Clin 4: 165–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller PM, Lu J, Saper CB 2008. Differential rescue of light- and food-entrainable circadian rhythms. Science 320: 1074–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gooley JJ, Schomer A, Saper CB 2006. The dorsomedial hypothalamic nucleus is critical for the expression of food-entrainable circadian rhythms. Nat Neurosci 9: 398–407 [DOI] [PubMed] [Google Scholar]
- Green CB, Douris N, Kojima S, Strayer CA, Fogerty J, Lourim D, Keller S, Besharse JC 2007. Loss of nocturnin, a circadian deadenylase, confers resistance to hepatic steatosis and diet-induced obesity. Proc Natl Acad Sci 104: 9888–9893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaldi B, Nakahata Y, Kaluzova M, Masubuchi S, Sassone-Corsi P 2009. Chromatin remodeling, metabolism and circadian clocks: The interplay of CLOCK and SIRT1. Int J Biochem Cell Biol 41: 81–86 [DOI] [PubMed] [Google Scholar]
- Gronfier C, Wright KP Jr, Kronauer RE, Czeisler CA 2007. Entrainment of the human circadian pacemaker to longer-than-24-h days. Proc Natl Acad Sci 104: 9081–9086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes ME, DiTacchio L, Hayes KR, Vollmers C, Pulivarthy S, Baggs JE, Panda S, Hogenesch JB 2009. Harmonics of circadian gene transcription in mammals. PLoS Genet 5: e1000442 doi: 10.1371/journal.pgen.1000442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadener S, Menet JS, Sugino K, Horwich MD, Weissbein U, Nawathean P, Vagin VV, Zamore PD, Nelson SB, Rosbash M 2009. A role for microRNAs in the Drosophila circadian clock. Genes Dev 23: 2179–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar N, Solt LA, Wang Y, Rogers PM, Bhattacharyya G, Kamenecka TM, Stayrook KR, Crumbley C, Floyd ZE, Gimble JM, et al. 2010. Regulation of adipogenesis by natural and synthetic REV-ERB ligands. Endocrinology doi: 10.1210.en.2009-0800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamia KA, Storch KF, Weitz CJ 2008. Physiological significance of a peripheral tissue circadian clock. Proc Natl Acad Sci 105: 15172–15177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamia KA, Sachdeva UM, DiTacchio L, Williams EC, Alvarez JG, Egan DF, Vasquez DS, Juguilon H, Panda S, Shaw RJ, et al. 2009. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science 326: 437–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laposky AD, Bass J, Kohsaka A, Turek FW 2008. Sleep and circadian rhythms: Key components in the regulation of energy metabolism. FEBS Lett 582: 142–151 [DOI] [PubMed] [Google Scholar]
- LeSauter J, Hoque N, Weintraub M, Pfaff DW, Silver R 2009. Stomach ghrelin-secreting cells as food-entrainable circadian clocks. Proc Natl Acad Sci 106: 13582–13587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu AC, Welsh DK, Ko CH, Tran HG, Zhang EE, Priest AA, Buhr ED, Singer O, Meeker K, Verma IM, et al. 2007. Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell 129: 605–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcheva B, Ramsey KM, Affinati AH, Bass J 2009. Clock genes and metabolic disease. J Appl Physiol 107: 1638–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcheva B, Ramsey KM, Buhr ED, Kobayashi Y, Su H, Ko CH, Ivanova G, Omura C, Mo S, Vitaterna MH, et al. 2010. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature doi:10.1038/nature09253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maury E, Ramsey KM, Bass J 2010. Molecular clocks and cardiometabolic syndrome. Circ Res 106: 447–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menet JS, Abruzzi KC, Desrochers J, Rodriguez J, Rosbash M 2010. Dynamic PER repression mechanisms in the Drosophila circadian clock: From on-DNA to off-DNA. Genes Dev 24: 358–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlin C, Gegear RJ, Reppert SM 2009. Antennal circadian clocks coordinate sun compass orientation in migratory monarch butterflies. Science 325: 1700–1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mieda M, Williams SC, Richardson JA, Tanaka K, Yanagisawa M 2006. The dorsomedial hypothalamic nucleus as a putative food-entrainable circadian pacemaker. Proc Natl Acad Sci 103: 12150–12155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagoshi E, Sugino K, Kula E, Okazaki E, Tachibana T, Nelson S, Rosbash M 2010. Dissecting differential gene expression within the circadian neuronal circuit of Drosophila. Nat Neurosci 13: 60–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente LP, Sassone-Corsi P 2008. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell 134: 329–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P 2009. Circadian control of the NAD+-salvage pathway by CLOCK-SIRT1. Science 324: 654–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke H, Saini C, Fleury-Olela F, Dibner C, Benjamin IJ, Schibler U 2008. Differential display of DNA-binding proteins reveals heat-shock factor 1 as a circadian transcription factor. Genes Dev 22: 331–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauman I, Briscoe AD, Zhu H, Shi D, Froy O, Stalleicken J, Yuan Q, Casselman A, Reppert SM 2005. Connecting the navigational clock to sun compass input in monarch butterfly brain. Neuron 46: 457–467 [DOI] [PubMed] [Google Scholar]
- Scheer FA, Hilton MF, Mantzoros CM, Shea SA 2009. Adverse metabolic and cardiovascular consequences of circadian misalignment. Proc Natl Acad Sci 106: 4453–4458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanahan TL, Kronauer RE, Duffy JF, Williams GH, Czeisler CA 1999. Melatonin rhythm observed throughout a three-cycle bright-light stimulus designed to reset the human circadian pacemaker. J Biol Rhythms 14: 237–253 [DOI] [PubMed] [Google Scholar]
- Toh KL, Jones CR, He Y, Eide EJ, Hinz WA, Virshup DM, Ptáček LJ, Fu YH 2001. An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science 291: 1040–1043 [DOI] [PubMed] [Google Scholar]
- Turek FW 2008. Circadian clocks: Tips from the tip of the iceberg. Nature 456: 881–883 [DOI] [PubMed] [Google Scholar]
- Turek FW, Joshu C, Kosaka A, Lin E, McDearmon E, Laposky A, Olson S, Easton A, Jensen D, Eckel R, et al. 2005. Obesity and metabolic syndrome in Clock mutant mice. Science 308: 1043–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmers C, Gill S, DiTacchio L, Pulivarthy SR, Le HD, Panda S 2009. Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proc Natl Acad Sci 106: 21453–21458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Solt LA, Burris TP 2010. Regulation of FGF21 expression and secretion by the retinoic acid receptor-related orphan receptorα. J Biol Chem 285: 15668–15673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westgate EJ, Cheng Y, Reilly DF, Price TS, Walisser JA, Bradfield CA, FitzGerald GA 2008. Genetic components of the circadian clock regulate thrombogenesis in vivo. Circulation 117: 2087–2095 [DOI] [PubMed] [Google Scholar]
- Xu Y, Padiath QS, Shapiro RE, Jones CR, Wu SC, Saigoh N, Saigoh K, Ptácek LJ, Fu YH 2005. Functional consequences of a CKIδ mutation causing familial advanced sleep phase syndrome. Nature 434: 640–644 [DOI] [PubMed] [Google Scholar]
- Xu K, Zheng X, Sehgal A 2008. Regulation of feeding and metabolism by neuronal and peripheral clocks in Drosophila. Cell Metab 8: 289–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Downes M, Yu RT, Bookout AL, He W, Straume M, Mangelsdorf DJ, Evans RM 2006. Nuclear receptor expression links the circadian clock to metabolism. Cell 126: 801–810 [DOI] [PubMed] [Google Scholar]
- Yasukazu N, Saurabh S, Giuseppe A, Milota K, Sassone-Corsi P 2009. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science 324: 654–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L, Wang J, Klein PS, Lazar MA 2006. Nuclear receptor Rev-erbα is a critical lithium-sensitive component of the circadian clock. Science 311: 102–105 [DOI] [PubMed] [Google Scholar]
- Yin L, Wu N, Curtin JC, Qatanani M, Szwergold NR, Reid RA, Waitt GM, Pearce KH, Wisely GB, Lazar MA 2007. Rev-erbα is a heme sensor that coordinates metabolic and circadian pathways. Science 318: 1789. [DOI] [PubMed] [Google Scholar]
- Yin L, Wu N, Lazar MA 2010. Nuclear receptor Rev-erbα: A heme receptor that coordinates circadian rhythm and metabolism. Nucl Recept Signal 8: e001 doi: 10.1621/nrs.08001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, Siepka SM, Hong HK, Oh WJ, Yoo OJ, et al. 2004. PERIOD2:LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci 101: 5339–5346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Q, Metterville D, Briscoe AD, Reppert SM 2007. Insect cryptochromes: Gene duplication and loss define diverse ways to construct insect circadian clocks. Mol Biol Evol 24: 948–955 [DOI] [PubMed] [Google Scholar]
- Zhu H, Sauman I, Yuan Q, Casselman A, Emery-Le M, Emery P, Reppert SM 2008. Cryptochromes define a novel circadian clock mechanism in monarch butterflies that may underlie sun compass navigation. PLoS Biol 6: e4 doi: 10.1371/journal.pbio.0060004 [DOI] [PMC free article] [PubMed] [Google Scholar]