Abstract
Self-renewing embryonic stem (ES) cells have an exceptional need for timely biomass production, yet the transcriptional control mechanisms responsible for meeting this requirement are largely unknown. We report here that Ronin (Thap11), which is essential for the self-renewal of ES cells, binds with its transcriptional coregulator, Hcf-1, to a highly conserved enhancer element that previously lacked a recognized binding factor. The subset of genes bound by Ronin/Hcf-1 function primarily in transcription initiation, mRNA splicing, and cell metabolism; genes involved in cell signaling and cell development are conspicuously underrepresented in this target gene repertoire. Although Ronin/Hcf-1 represses the expression of some target genes, its activity at promoter sites more often leads to the up-regulation of genes essential to protein biosynthesis and energy production. We propose that Ronin/Hcf-1 controls a genetic program that contributes to the unimpeded growth of ES cells.
Keywords: Stem cells, pluripotency, transcriptional control, cell growth
In contrast to all other mammalian cells, embryonic stem (ES) cells are characterized by a truncated cell cycle, relative autonomy from extracellular signal-regulated kinase (Erk) signaling, and an unusually rapid growth rate, analogous to that of cancer cells and primitive unicellular organisms, including bacteria (Orford and Scadden 2008; Ying et al. 2008; Wang et al. 2009). Thus, the task of replicating the cellular biomass, including nucleotides and amino acids, to support the prolific growth of ES cells imposes stringent metabolic demands. Emerging evidence indicates that such requirements are met not by a self-correcting, homeostatic system of housekeeping enzymes, but by a precisely regulated genetic network (Vander Heiden et al. 2009). Although much has been learned about the factors governing the pluripotency of ES cells (Boyer et al. 2005; Bernstein et al. 2006; Loh et al. 2006; Chen et al. 2008; Cole et al. 2008; Kim et al. 2008), relatively little is known about the transcriptional control of ES cell growth. A better understanding of the links between ES cell growth and pluripotency may be key to realizing the full potential of these cells in genetic engineering and regenerative medicine.
We recently discovered a novel zinc finger transcriptional regulator, Ronin (also Thap11), that is essential for the self-renewal of ES cells (Dejosez et al. 2008). Conditional knockout of the Ronin gene induces ES cell death, while its forced expression enables the cells to proliferate transiently without differentiation under conditions that normally do not promote self-renewal. While studying Ronin-overexpressing mouse ES cells, we noticed that they possess a strikingly enlarged nucleolus (Fig. 1A; Supplemental Fig. S1A), the prime site of ribosome synthesis and assembly, suggesting that Ronin overexpression may alter the production of a key growth-related body in ES cells. This observation, coupled with our previous finding that Ronin can interact with the cell growth factor Hcf-1 (Julien and Herr 2003; Wysocka and Herr 2003; Dejosez et al. 2008), which lacks its own DNA-binding domain, led us to predict that Ronin/Hcf-1 may play a pivotal role in meeting the biosynthetic needs of mouse ES cells. Our results support this hypothesis, and provide a new paradigm for understanding the regulation of ES cell growth and its relationship to pluripotency.
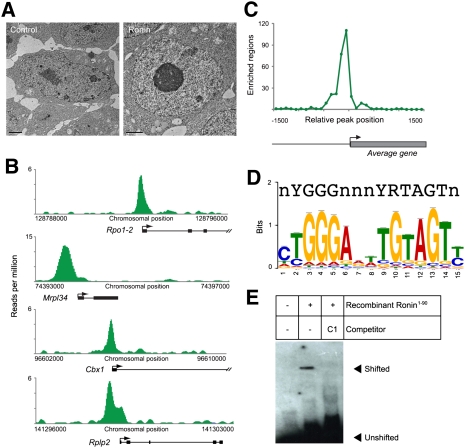
Figure 1.
Ronin binds to a hyperconserved enhancer element in mouse ES cells. (A) Ronin-overexpressing clones are characterized by a rounder cell shape and a more prominent nucleolus than found in control clones (electronmicroscopy; bar, 2 μm). (B) Binding of Ronin at the promoter regions of four representative genes. ChIP-seq results, shown on the Y-axis, are reads per million reads. (C) Histogram showing the distance of the midpoint of each Ronin-binding event from the nearest TSS (arrow). (D) Identification of the consensus RBM depicted as a bit matrix. (Top) The recently discovered M4 sequence in human promoters (Xie et al. 2005) is included for comparison. (E) EMSA analysis of the newly identified RBM using recombinant Ronin1–90 (Thap domain). The specific shift of the RBM is abolished in the presence of a specific competitor (C1).
Results and Discussion
Ronin binds to a hyperconserved enhancer element in mouse ES cells that is shared with Hcf-1
We first sought to identify the DNA-binding sites of Ronin using chromatin immunoprecipitation (ChIP) and DNA sequencing (ChIP-seq). Our mapping results revealed 866 Ronin-bound regions (Supplemental Table S1), most of which were located at or immediately upstream of transcription start sites (TSSs) (Fig. 1B,C), suggesting that Ronin participates in transcription initiation. Alignment analysis identified a Ronin-binding motif (CTGGGARWTGTAGTY, designated here as RBM) in 844 of the target gene promoters (Fig. 1D). This functional element was highly enriched compared with random sequences (P ≪ 10−100), but correlated poorly with the Ronin-binding sequence (“3×”) we determined previously by the SELEX method (E = 6.395 × 10−1, derived with a column comparison metric [Pearson correlation] and the Smith-Waterman alignment method) (Dejosez et al. 2008), suggesting that the RBM is more likely to represent the physiologically relevant binding element. Gel shift experiments with either recombinant Ronin protein (Fig. 1E) or nuclear ES cell extracts under low-stringency and high-stringency (physiological) conditions (Supplemental Fig. S1B,C), together with ChIP using a Ronin-specific antibody (in wild-type ES cells) or an anti-Flag-tag antibody (in Ronin-Flag-expressing ES cells), followed by PCR analysis of selected target gene regions (Supplemental Fig. S2), confirmed the identity of each site found to be specifically targeted by Ronin. Finally, a recent report (Sabogal et al. 2010) dealing with the protein structure of the THAP domain predicts a DNA-binding sequence for Ronin that essentially matches our empirically derived sequence.
Intriguingly, the Ronin target sequence closely resembles a promoter sequence (ACTAYRNNNCCCR, the so-called M4 motif) (Fig. 1D) whose conservation rate in humans (61%) is the fourth highest among the top 50 conserved motifs described by Xie et al. (2005). This sequence is notable for another reason: In contrast to most highly conserved regulatory elements in the human genome, its recognition by known binding factors was not reported until recently, when the transcription factors Srebp1 and Ets/Runx were linked to the M4 motif (Hollenhorst et al. 2007; Seo et al. 2009). To assess the noteworthiness of other transcription factors reported to bind to this conserved motif, we performed an FIMO analysis to identify this promoter element in the ES cell genome. With a 0.7× threshold, typical of motif analysis, we identified 4328 candidate motifs (0.6× threshold, 9412 motifs; 0.38× threshold, 66,192). When we aligned the 4328 elements with TSSs, 1094 were found to be associated with high confidence. Since 688 of these TSSs were linked to Ronin at high confidence, the P-random association of Ronin with the RBM was ≤10−100, supporting a strong relationship between Ronin and this motif.
We then compared our data set with the Srebp1 data (Seo et al. 2009) and with the Nanog data set, derived from Boyer et al. (2005). We found that, in comparison with the 688 high-confidence TSSs for Ronin, only 153 were Srebp1 sites and only 60 were Nanog sites. Thus, from these data, Srebp1 appears to be associated with the RBM motif, but only weakly. Similarly, for Nanog, there was evidence of enrichment, although an analysis of co-occupancy with Ronin revealed that only 5% of all Nanog sites harboring the motif were Ronin-bound, no different from the result one would expect by chance. For Ets/Runx, we were able to obtain only human genome binding data, and thus could not perform a direct comparison with the data for mouse Ronin. Even so, a comparison of coordinates from hc18 (University of California at Santa Cruz Genome Browser) with the human motif data (Hollenhorst et al. 2007) showed some enrichment, but again the fraction of sites containing the full motif is much lower than for Ronin in mouse ES cells. The evidence therefore indicates only a weak affinity of Srebp1 and Ets/Runx for the RBM. Thus, neither of these transcription factors appears to compete significantly with Ronin for DNA-binding sites in mouse ES cells.
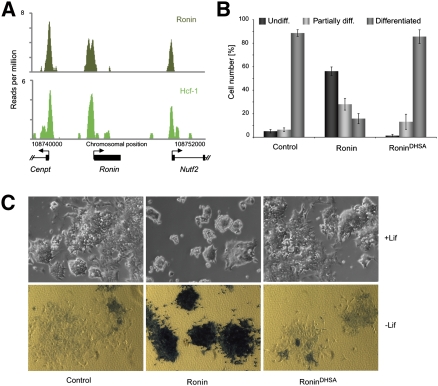
Because Ronin lacks a transactivation domain and can interact directly with a well-defined transcriptional coregulator of cell growth, the Hcf-1 protein (Dejosez et al. 2008), we considered that both factors might be needed at the RBM to initiate gene transcription. We therefore performed ChIP-seq with an Hcf-1 antibody (Wilson et al. 1993), identifying 743 genomic loci occupied by Hcf-1 at a high confidence level (see Supplemental Table S2). These regions overlapped with 56% of the target promoters that were also bound by Ronin (Fig. 2A; Supplemental Table S3). Even when the Hcf-1 signal did not attain significance by our conservative criteria, we were still able to detect a distinct binding peak, indicating that Hcf-1 generally co-occupies Ronin-bound target sites. To assess the functional significance of the interaction between Ronin and Hcf-1 at a common regulatory motif, we took advantage of our previous finding that Ronin contains a DHSY copy (Dejosez et al. 2008) of the previously defined HCF-1-binding motif, D/EHxY (Freiman and Herr 1997; Lu et al. 1998), and mutated a conserved residue essential for binding: 246Y → 246A, resulting in RoninDHSA (Supplemental Fig. S3A,B). By yeast two-hybrid assay, this change completely abolished the ability of Ronin to interact with Hcf-1 (Supplemental Fig. S3B). To interrogate how the inability of Ronin to recruit Hcf-1 might affect its transcriptional activity in ES cells, we generated stably transfected ES cell lines overexpressing either wild-type Ronin (EF1α-Ronin) or the mutant form, which is not capable of binding Hcf-1 (EF1α-RoninDHSA) (see Supplemental Fig. S3C). In such cells, the association of Hcf-1 with the selected target sites shown in Figure 5C (below) was reduced, indicating that Ronin is indeed primarily responsible for binding to those promoters. Because expression of Ronin effectively supports the self-renewal of ES cells in leukemia inhibitory factor (Lif)-free medium (Dejosez et al. 2008), we tested the effect of Hcf-1 on this property by plating control, EF1α-Ronin, and EF1α-RoninDHSA ES cells at clonal densities in medium without Lif. As reported previously (Dejosez et al. 2008), Ronin overexpression robustly made Lif nonessential for ES cells, while EF1α-RoninDHSA ES cells still differentiated (Fig. 2B,C), suggesting that Ronin must interact with Hcf-1 to produce its anti-differentiation effect. Rescue experiments using Ronin knockout ES cells revealed that transient overexpression of wild-type Ronin has a positive effect on self-renewal, while expression of the Ronin mutant incapable of binding to Hcf-1 did not show this effect (Supplemental Fig. S4A). Finally, careful analysis of RoninloxP/loxP MEFs for changes in morphology, proliferative capacity, and cell cycle phase distribution did not reveal any obvious phenotype after Cre-mediated excision of Ronin that could be linked to loss of Ronin expression (Supplemental Fig. S4B,C), suggesting that Ronin function is restricted to certain cell types.
Figure 2.
Ronin and Hcf-1 bind together to specific genes. (A) ChIP-seq results obtained in mouse ES cells after immunoprecipitation with Ronin or Hcf-1 antibodies. A representative region containing three Ronin-bound genes shows substantial overlap between Ronin- and Hcf-1-binding peaks in the promoter regions of all three genes. (B) Quantification of the experiment shown in the bottom panel of C. Values are means ± SD of triplicate experiments. (C, top panel) Morphology of control, Ronin-overexpressing, and RoninDHSA-overexpressing mouse ES cells after 3 d of culture in the presence of Lif (10× magnification). (Bottom panel) Cells were stained for alkaline phosphatase activity after 4 d of culture in the absence of Lif (20× magnification).
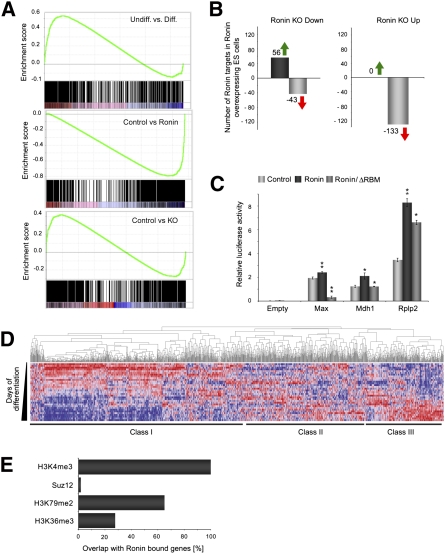
Figure 5.
Ronin/Hcf-1 can either activate or repress its transcriptional targets in mouse ES cells. (A) GSEA analyses of Ronin/Hcf-1 target genes, showing enrichment of bound genes in undifferentiated versus differentiated mouse ES cells (top panel), in Ronin-overexpressing versus control ES cells (middle panel), and in control compared with Ronin knockout ES cells (bottom panel). (B) Overlap between the numbers of Ronin-bound genes that are up-regulated or down-regulated in Ronin-overexpressing ES cells in relation to their status in Ronin knockout ES cells. (C) Luciferase reporter assays with pGL3-based promoter constructs, showing effects on the expression of selected target genes. (*) P < 0.05; (**) P < 0.01. (ΔRBM) Mutant reporter construct with deleted RBM. (D) Relative expression levels of Ronin target genes during the differentiation of mouse ES cells. (Red) Up-regulated genes; (blue) down-regulated genes. (E) Prevalence of H3K4me3-, Suz12-, H3K79me2-, or H3K36me3-enriched regions at Ronin-binding sites.
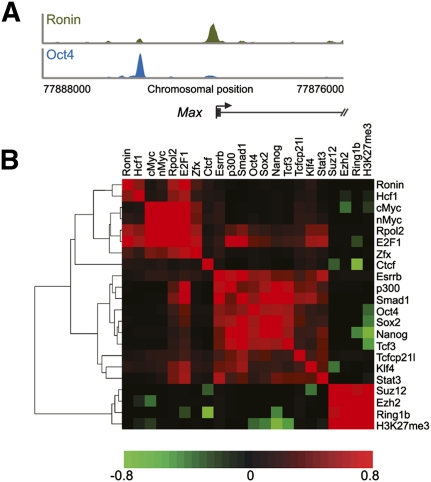
Approximately 40% of the 866 promoters bound by Ronin were also occupied by one or more of the transcription factors Oct4, Sox2, and Nanog, which have central roles in pluripotency control (Boyer et al. 2005; Bernstein et al. 2006; Loh et al. 2006; Chen et al. 2008; Cole et al. 2008; Kim et al. 2008; Marson et al. 2008). We find it interesting that Ronin consistently occupied sites within promoter regions that were only 50–100 base pairs (bp) upstream of TSSs, in contrast to the more distant sites occupied by Oct4 (Fig. 3A) and other core transcription factors (data not shown). To assess the genome-wide binding preferences of Ronin versus those of other regulatory factors in ES cells, we calculated target similarity scores for genomic regions identified as highly enriched in ChIP-seq experiments (Chen et al. 2008; Ku et al. 2008; Marson et al. 2008; Seila et al. 2008), and subjected the matrix of scores to hierarchical clustering analysis. Ronin and Hcf-1 clustered together rather than with canonical pluripotency factors (Fig. 3B).
Figure 3.
Comparison of binding characteristics of Ronin and canonical pluripotency factors. (A) ChIP-seq results obtained by precipitation with antibodies against Ronin or Oct4. (B) Hierarchical clustering analysis of 22 prominent transcriptional regulators in mouse ES cells, based on target similarity scores calculated with a Pearson correlation similarity metric.
Genes targeted by Ronin/Hcf-1 in mouse ES cells function in protein biosynthesis and energy production
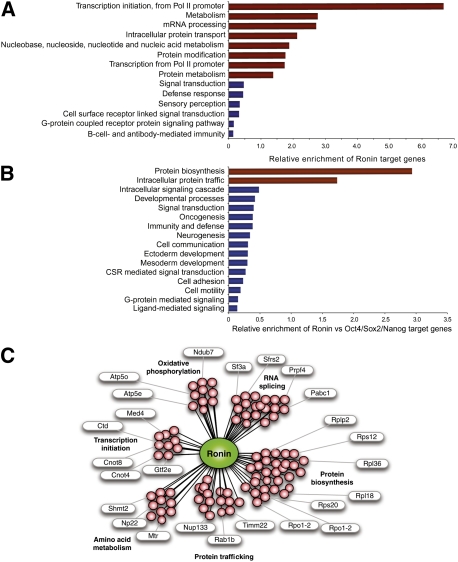
The prominent nucleolus of Ronin-overexpressing cells (Fig. 1A; Supplemental Fig. S1A) and the co-occupancy of a hyperconserved DNA-binding motif by Ronin and Hcf-1 led us to consider that these factors may be involved in the regulation of biomass production supporting ES cell growth. To test this hypothesis, we focused on the subset of genes whose promoters were bound solely by Ronin/Hcf-1. Using the PANTHER tool, we determined the functional categories of all genes that met this stringent requirement. Transcription initiation, mRNA splicing, and metabolism were among the most overrepresented categories, while cell signaling and cell development were underrepresented (Fig. 4A,B; also Supplemental Tables S4,S5). Close inspection of the individual genes within these categories yielded a more informative functional portrait (Fig. 4C). Ronin/Hcf-1 recognized as many as 30% of genes encoding ribosomal proteins and two key subunits of RNA polymerase I, Rpo1-2 and Rpo1-4 (protein biosynthesis); Ctd, Cnot4/8, and Med4 (transcription initiation); Rab1b, Nup133, and Timm22 (protein trafficking); and Frap1 (mTor), Eif4a1, Eif4ebp1, Eef2, Tsc2, Rps6kb2, and Rps6 (mTor signaling pathway). These results are important because alterations in ribosomal biosynthesis, transcription initiation, protein transport, and overall control of growth and metabolism can have profound effects on the metabolome (Warner 1999; Moss and Stefanovsky 2002; Tsai and McKay 2002). Moreover, identification of key constituents of the mTor signaling pathway in this analysis, including the mTor protein itself, provides a mechanism by which Ronin/Hcf-1 could exert a profound effect on cell growth and metabolism through regulation of a relatively limited number of target genes. Ronin/Hcf-1 also bound specifically to genes encoding mitochondrial ribosomal proteins (Mrpl19, Mrpl32, Mrpl50, and Mrpl54), mitochondrial translation factors (Tufm), and rate-limiting members of the oxidative phosphorylation cascade (Atp5e, NADH dehydrogenase, and Atp5e), suggesting involvement in the control of energy production in ES cells. Finally, some of the target genes (e.g., Mtr) encode threonine catabolic enzymes, whose increased expression in ES cells facilitates a high-flux metabolic state characterized by enhanced threonine catabolism (Wang et al. 2009). Thus, Ronin/Hcf-1 appears to transcriptionally regulate a subset of genes with specific functions in protein biosynthesis and energy production, but not cell development (Fig. 4).
Figure 4.
Cellular functions of genes targeted by Ronin/Hcf-1 in mouse ES cells. (A) PANTHER analysis of target genes. Categories with enrichment values >1 are significantly overrepresented (red), while those with lower values are significantly underrepresented (blue). (B) PANTHER analysis of Ronin versus Oct4/Sox2/Nanog targets, as in A. (C) Summary of Ronin/Hcf-1 target genes by major functional categories.
Ronin can either activate or repress its transcriptional targets in mouse ES cells
To explore the different dimensions of target gene regulation by Ronin/Hcf-1, we conducted gene set enrichment analysis (GSEA) of RNA from Ronin targeted genes in wild-type ES cells compared with those in differentiated, Ronin-overexpressing or Ronin knockout cells (Fig. 5A). The results show that genes occupied by these factors are generally highly transcribed, and that the transcripts are significantly overrepresented in ES cells. Thus, Ronin/Hcf-1 up-regulates the expression of many (although not all) of its target genes, consistent with the ability of Hcf-1 to either positively or negatively affect transcription, depending on the cellular context (Wysocka and Herr 2003). To test the reverse prediction, we transfected RoninloxP/− ES cells with the gene encoding Cre recombinase, sorted Cre-positive cells at 18 h post-transfection, extracted the RNA, and performed microarray analysis of gene expression. Interestingly, the entire subset of 133 genes found to be up-regulated in Ronin knockout cells (Fig. 5B, right) were down-regulated in our Ronin-overexpressing clones, whereas, in the converse situation, only 43 of 99 genes found to be down-regulated after knockout (Fig. 5B, left) were down-regulated in the Ronin-overexpressing clones. Additional evidence for direct transcriptional control of Ronin targets was obtained in experiments in which we cloned a set of the Ronin targeted promoters and performed luciferase reporter assays. As shown in Figure 5C, Ronin gain of function had a positive effect on gene expression, while loss of the RBM diminished or abolished the results of Ronin overexpression. To track the expression of Ronin/Hcf-1-controlled genes more closely, we analyzed the results of DNA microarrays over 14 d of ES cell differentiation (Fig. 5D). As expected, the largest class of Ronin target genes (Class I) showed rapid down-regulation after induction of differentiation, while the two remaining classes either were up-regulated (Class III) or demonstrated complex regulation (Class II).
Considering that Ronin's transcriptional activity depends on its interaction with Hcf-1, we performed Western blot analysis to detect Hcf-1 in wild-type control and Ronin-overexpressing ES cells. Hcf-1 protein was clearly up-regulated in the Ronin-overexpressing cells. The Ronin Hcf-1-binding mutant, on the other hand, appeared to disrupt the Ronin–Hcf-1 complex, leading to Hcf-1 overflow that was consequently counteracted by down-regulation of Hcf-1 expression in the Ronin mutant cell line (Supplemental Fig. S3E). Although the Hcf-1 gene was not directly targeted by Ronin, its only known regulator, Hpip, was (Supplemental Table S1), supporting the notion that Ronin and Hcf-1 form a single functional unit under the control of an autoregulatory loop.
Results of the present analysis confirm the dependence of Ronin on interaction with Hcf-1, and broaden our understanding of how this transcriptional modulation influences Ronin action. Most critical, perhaps, is the demonstration that Ronin must bind to Hcf-1 in order to be functionally active. Although Ronin retained some activity in the absence of Hcf-1, its interaction with this coregulator clearly amplified its induced effects. Finally, we would stress that recruitment of an Hcf-1-containing complex enables Ronin to either up-regulate or repress target genes, thus increasing the versatility of its regulatory action. However, Hcf-1 has many more binding targets than Ronin does, suggesting that it mediates other activities in ES cells, possibly through interaction with site-specific DNA-binding factors such as E2F and Luman (Lu et al. 1997, 1998; Tyagi et al. 2007). Further analysis of the Ronin-occupied promoter regions (Fig. 5E) showed that Ronin is closely associated with promoters containing histone H3K4me3-modified nucleosomes, a mark of genes that undergo transcription initiation (100% overlap), and with H3K36me3 (28%) and H3K79me2 (80%), both marks of genes that are fully transcribed. In contrast, there was essentially no overlap with Suz12 (0.02%), a component of Polycomb-Repressive Complex 2, which catalyzes the H3K27me3 mark associated with transcriptionally repressed bivalent domains (Bernstein et al. 2006), indicative of a very strong negative correlation (binomial P < 10−9). These results agree with the well-documented ability of Hcf-1 to recruit Ash2/Set1 to target gene promoters (Wysocka and Herr 2003).
Concluding remarks
Our results indicate that Ronin/Hcf-1 contributes to ES cell pluripotency by binding to a hyperconserved enhancer element and regulating the transcription of genes involved in key metabolic processes that sustain the growth of self-renewing ES cells until they exit the undifferentiated state. The highly conserved nature of this DNA sequence and its tissue specificity (Xie et al. 2005) suggest that genes controlled through Ronin binding are apt to perform essential functions in ES cells. Moreover, as the only transcriptional regulator known to bind to this conserved motif in ES cells, Ronin separates itself from the canonical pluripotency factors. We acknowledge that Oct4, Sox2, Nanog, and Tcf3 co-occupy the promoters of certain genes with Ronin, but the sites they recognize are entirely different from the RBM (Fig. 3A). Although these canonical factors can interact among themselves to regulate target gene expression (Kim et al. 2008), there is no evidence to suggest that they cooperate with Ronin in transcriptional regulation. This dissociation is underscored by (1) the apparent lack of Ronin binding to Suz12-enriched genomic regions (Fig. 5E), which are often targeted by Oct4 and other canonical factors (Boyer et al. 2006); (2) the difference in global genomic binding pattern between Ronin and Oct4, Sox2, Nanog, and Tcf3 (Fig. 3A); (3) the absence of canonical factors at approximately half of the gene promoters bound by Ronin; and (4) the marginal overlap between Ronin targets identified in this study and those down-regulated upon Oct4 knockdown in a previous report (Ivanova et al. 2006). The observation that Ronin/Hcf-1 binding to target sites more commonly leads to gene activation than repression revises our earlier suggestion (Dejosez et al. 2008) that Ronin is primarily a global repressor, based on the assumption that acute up-regulation of Ronin under otherwise steady-state conditions exerts a dominant-negative effect on Ronin function, similar to observations for other proteins that harbor the Thap domain (Cayrol et al. 2007). We interpret the reported increase in H3K9 methylation (Dejosez et al. 2008) as a secondary or indirect effect of Ronin. Gene activation by Ronin/Hcf-1 is most likely mediated through an epigenetic mechanism involving Hcf-1/Ash2/Set1.
Recently, the life cycle of ES cells was compared with that of yeast cells and other unicellular metazoans, in the sense that it follows a relatively primitive set of behavioral rules that differ from those of more mature cells (Silva and Smith 2008; Ying et al. 2008; Wang et al. 2009). We suggest that the growth and metabolic capacity of ES cells may represent a previously unrecognized level of pluripotency control. Indeed, the genes bound by Ronin could well account for most of the protein and metabolic budget expended by ES cells engaged in self-renewal, a process that may consume as much as 50% of the total energy produced by a cell (Moss and Stefanovsky 2002). Thus, any shortfall in energy could cause ES cells to lose their full self-renewal capacity, leading to apoptotic death or perhaps a rapid transition to differentiation. Hence, it will be important to determine if the transcriptional activity of Ronin/Hcf-1 is intrinsically self-maintaining, or is modulated by signaling from upstream molecules.
Materials and methods
Cell culture, differentiation, and alkaline phosphatase staining
Mouse ES cells (line R1 and derivatives), were cultured in DMEM + GlutaMax I (Invitrogen) supplemented with 1000 U/mL Lif (Millipore).
Establishment of cell lines with stable integration of control, Ronin, or RoninDHSA vectors
R1 mouse ES cell lines overexpressing control (EF1a-Neo), Ronin (EF1a-Ronin-Flag-Ires-Neo), or RoninDHSA (EF1a-RoninDHSA-Flag-Ires-Neo) constructs were established as described previously (Dejosez et al. 2008).
Electrophoretic mobility shift assays (EMSAs)
EMSAs were performed essentially as described (Dejosez et al. 2008) using the Lightshift Chemiluminescent EMSA kit (Pierce).
ChIP bioinformatic analysis and ChIP-PCR
Ronin and HCF-1 ChIP-derived reads were aligned to the mouse genome (NCBI build 36, University of California at Santa Cruz build mm8) using an iterative version of ELAND to improve the read count, and all mapped reads were analyzed as described previously (Marson et al. 2008).
Directional yeast two-hybrid analysis
Yeast two-hybrid analysis was performed as described previously (Dejosez et al. 2008) using the ProQuest Two-Hybrid system (Invitrogen) and gateway technology, according to the manufacturer's recommendations.
See the Supplemental Material for full materials and methods.
Acknowledgments
We are grateful to M.K. Brenner for critical reading of the manuscript, and to W. Herr for helpful discussions and experimental suggestions. We also thank J. Gilbert for editorial advice and comments. This work was supported by the Diana Helis Henry Medical Research Foundation (T.P.Z.), the Huffington Foundation (T.P.Z.), and National Institutes of Health grants R01 EB005173-01, 1R01 GM077442-01, P20 EB007076, P01 GM81627, and 2R01 HG002668 (R.A.Y.).
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1935210.
Supplemental material is available at http://www.genesdev.org.
References
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326 [DOI] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. 2005. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122: 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. 2006. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441: 349–353 [DOI] [PubMed] [Google Scholar]
- Cayrol C, Lacroix C, Mathe C, Ecochard V, Ceribelli M, Loreau E, Lazar V, Dessen P, Mantovani R, Aguilar L, et al. 2007. The THAP-zinc finger protein THAP1 regulates endothelial cell proliferation through modulation of pRB/E2F cell-cycle target genes. Blood 109: 584–594 [DOI] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. 2008. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133: 1106–1117 [DOI] [PubMed] [Google Scholar]
- Cole MF, Johnstone SE, Newman JJ, Kagey MH, Young RA 2008. Tcf3 is an integral component of the core regulatory circuitry of embryonic stem cells. Genes Dev 22: 746–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejosez M, Krumenacker JS, Zitur LJ, Passeri M, Chu LF, Songyang Z, Thomson JA, Zwaka TP 2008. Ronin is essential for embryogenesis and the pluripotency of mouse embryonic stem cells. Cell 133: 1162–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freiman RN, Herr W 1997. Viral mimicry: Common mode of association with HCF by VP16 and the cellular protein LZIP. Genes Dev 11: 3122–3127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenhorst PC, Shah AA, Hopkins C, Graves BJ 2007. Genome-wide analyses reveal properties of redundant and specific promoter occupancy within the ETS gene family. Genes Dev 21: 1882–1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova N, Dobrin R, Lu R, Kotenko I, Levorse J, DeCoste C, Schafer X, Lun Y, Lemischka IR 2006. Dissecting self-renewal in stem cells with RNA interference. Nature 442: 533–538 [DOI] [PubMed] [Google Scholar]
- Julien E, Herr W 2003. Proteolytic processing is necessary to separate and ensure proper cell growth and cytokinesis functions of HCF-1. EMBO J 22: 2360–2369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Chu J, Shen X, Wang J, Orkin SH 2008. An extended transcriptional network for pluripotency of embryonic stem cells. Cell 132: 1049–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, et al. 2008. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet 4: e1000242 doi: 10.1371/journal.pgen.1000242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, et al. 2006. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet 38: 431–440 [DOI] [PubMed] [Google Scholar]
- Lu R, Yang P, O'Hare P, Misra V 1997. Luman, a new member of the CREB/ATF family, binds to herpes simplex virus VP16-associated host cellular factor. Mol Cell Biol 17: 5117–5126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Yang P, Padmakumar S, Misra V 1998. The herpesvirus transactivator VP16 mimics a human basic domain leucine zipper protein, luman, in its interaction with HCF. J Virol 72: 6291–6297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, et al. 2008. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell 134: 521–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss T, Stefanovsky VY 2002. At the center of eukaryotic life. Cell 109: 545–548 [DOI] [PubMed] [Google Scholar]
- Orford KW, Scadden DT 2008. Deconstructing stem cell self-renewal: Genetic insights into cell-cycle regulation. Nat Rev Genet 9: 115–128 [DOI] [PubMed] [Google Scholar]
- Sabogal A, Lyubimov AY, Corn JE, Berger JM, Rio DC 2010. THAP proteins target specific DNA sites through bipartite recognition of adjacent major and minor grooves. Nat Struct Mol Biol 17: 117–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA 2008. Divergent transcription from active promoters. Science 322: 1849–1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo YK, Chong HK, Infante AM, Im SS, Xie X, Osborne TF 2009. Genome-wide analysis of SREBP-1 binding in mouse liver chromatin reveals a preference for promoter proximal binding to a new motif. Proc Natl Acad Sci 106: 13765–13769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J, Smith A 2008. Capturing pluripotency. Cell 132: 532–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai RY, McKay RD 2002. A nucleolar mechanism controlling cell proliferation in stem cells and cancer cells. Genes Dev 16: 2991–3003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyagi S, Chabes AL, Wysocka J, Herr W 2007. E2F activation of S phase promoters via association with HCF-1 and the MLL family of histone H3K4 methyltransferases. Mol Cell 27: 107–119 [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB 2009. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 324: 1029–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Alexander P, Wu L, Hammer R, Cleaver O, McKnight SL 2009. Dependence of mouse embryonic stem cells on threonine catabolism. Science 325: 435–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JR 1999. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci 24: 437–440 [DOI] [PubMed] [Google Scholar]
- Wilson AC, LaMarco K, Peterson MG, Herr W 1993. The VP16 accessory protein HCF is a family of polypeptides processed from a large precursor protein. Cell 74: 115–125 [DOI] [PubMed] [Google Scholar]
- Wysocka J, Herr W 2003. The herpes simplex virus VP16-induced complex: The makings of a regulatory switch. Trends Biochem Sci 28: 294–304 [DOI] [PubMed] [Google Scholar]
- Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M 2005. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature 434: 338–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A 2008. The ground state of embryonic stem cell self-renewal. Nature 453: 519–523 [DOI] [PMC free article] [PubMed] [Google Scholar]