Abstract
Inactivation of the yeast nonsense-mediated mRNA decay (NMD) pathway stabilizes nonsense mRNAs and promotes readthrough of premature translation termination codons. Although the latter phenotype is thought to reflect a direct role of NMD factors in translation termination, its mechanism is unknown. Here we show that the reduced termination efficiency of NMD-deficient cells is attributable to increased expression of the magnesium transporter Alr1p and the resulting effects of elevated Mg2+ levels on termination fidelity. Alr1p levels increase because an upstream ORF in ALR1 mRNA targets the transcript for NMD. Our results demonstrate that NMD, at least in yeast, controls Mg2+ homeostasis and, consequently, translational fidelity.
Keywords: NMD, translation termination, magnesium uptake, uORF
Eukaryotic quality control mechanisms detect and degrade aberrant RNAs that arise from genomic mutations or mistakes in transcription and RNA processing. One such mechanism, nonsense-mediated mRNA decay (NMD), targets mRNAs containing premature translation termination codons for rapid degradation, thus ensuring that protein-encoding transcripts encompass proper ORFs (Jacobson and Izaurralde 2007). The destabilization of nonsense-containing transcripts requires their translation and a distinct set of trans-acting factors, including the conserved Upf1, Nmd2 (Upf2), and Upf3 proteins (Jacobson and Izaurralde 2007). Yeast strains with mutations in the UPF1, NMD2, or UPF3 genes show not only elevated nonsense mRNA levels, but also increased translational readthrough of premature stop codons (Weng et al. 1996; Maderazo et al. 2000; Wang et al. 2001; Keeling et al. 2004). This effect is sufficient to promote nonsense suppression, and has been thought to reflect a direct function of the Upf/Nmd proteins in translation termination, possibly as a consequence of their interactions with the release factors eRF1 and eRF3 (Czaplinski et al. 1998; Maderazo et al. 2000; Wang et al. 2001). However, the mechanism by which the Upf/Nmd proteins enhance the efficiency of translation termination is largely unknown. To elucidate the role of NMD factors in translation termination, we screened for mutations that counteract the nonsense suppression phenotype of NMD-deficient cells, and characterized in detail the principal complementation group of the resulting anti-suppressors.
Results and Discussion
Mutations in the ALR1 gene counteract the nonsense suppression phenotype of NMD-deficient cells
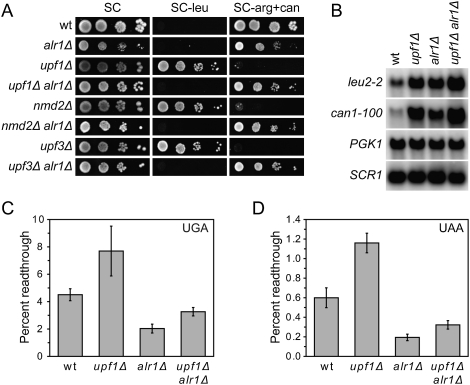
The screen for anti-suppressors was initiated by using Saccharomyces cerevisiae upf1 mutants harboring can1-100 (UAA) and leu2-2 (UGA) nonsense alleles. The lack of NMD in these cells leads to stabilization of the can1-100 and leu2-2 mRNAs and readthrough of their premature stop codons (Weng et al. 1996; Maderazo et al. 2000; Wang et al. 2001), generating functional, full-length Can1p and Leu2p. The production of Can1p and Leu2p can be scored as sensitivity to the toxic arginine analog canavanine and growth on medium lacking leucine, respectively. By selecting for upf1 cells resistant to canavanine (CanR) and subsequently screening for the inability to grow on medium lacking leucine (Leu−), strains harboring mutations that reduced upf1-induced nonsense suppression were identified. One complementation group defined strains with mutations in the ALR1 gene, which codes for a plasma membrane protein required for efficient magnesium uptake (MacDiarmid and Gardner 1998; Graschopf et al. 2001). To demonstrate unambiguously that a mutation in the ALR1 gene generates anti-suppression, we combined an alr1Δ allele with a upf1Δ mutation. The resulting upf1Δ alr1Δ double mutant showed a CanR Leu− phenotype (Fig. 1A), confirming that the lack of Alr1p counteracts the nonsense suppression phenotype of upf1 cells. Furthermore, the alr1Δ allele also counteracted nonsense suppression in cells deleted for NMD2 or UPF3 (Fig. 1A), indicating that the underlying mechanism is relevant for NMD-deficient cells in general.
Figure 1.
A mutation in the ALR1 gene counteracts the termination defect of NMD-deficient cells. (A) Nonsense suppression in NMD- and Alr1p-deficient cells. The indicated strains (MJY142, MJY67, MJY130, MJY447, MJY169, MJY193, MJY150, and MJY151) were grown overnight in liquid synthetic complete (SC) medium at 30°C. Cells were serially diluted; spotted onto SC, SC-leu, and SC-arg + can (200 μg/mL can) plates; and incubated for 3 d at 30°C. (B) Northern analysis of total RNA isolated from wild-type (MJY142), upf1Δ (MJY67), alr1Δ (MJY130), and upf1Δ alr1Δ (MJY447) cells grown in SC medium at 30°C. The blot was probed for leu2-2, can1-100, PGK1, and SCR1 transcripts using randomly labeled DNA fragments. The noncoding SCR1 transcript serves as a loading control. (C,D) Readthrough levels of UGA (C) and UAA (D) stop codons in the wild-type (MJY142), upf1Δ (MJY67), alr1Δ (MJY130), and upf1Δ alr1Δ (MJY447) strains. The values represent the average of five independent experiments. The standard deviation is indicated.
The fidelity of translation termination is enhanced by the lack of Alr1p
The lack of nonsense suppression in upf1 alr1 double mutants could be caused by either a reduction in leu2-2 and can1-100 mRNA levels or improved termination efficiency. The former possibility was excluded by the finding that the levels of leu2-2 and can1-100 mRNAs were in fact higher in upf1Δ alr1Δ cells than in upf1Δ cells (Fig. 1B; Supplemental Table S1). To assess directly whether a lack of Alr1p improves termination efficiency, we used a dual-luciferase readthrough reporter system (Keeling et al. 2004) that controls for differences in mRNA abundance and efficiency of translation initiation. The level of readthrough was quantified by determining the ratio of firefly and Renilla luciferase activities in a strain harboring a construct in which the respective luciferase genes are separated by a stop codon, and comparing the ratio to that obtained with a construct in which a sense codon replaced the stop codon (Keeling et al. 2004). Analyses of UGA and UAA readthrough demonstrated that the alr1Δ allele counteracted the termination defect of upf1Δ cells (Fig. 1C,D). The alr1Δ allele also generated a reduction in readthrough in NMD-proficient cells, implying that the lack of Alr1p leads to a general enhancement of termination fidelity. Importantly, growth on medium containing guanidine hydrochloride (Tuite et al. 1981) did not eliminate the effect of the alr1Δ allele, excluding the possibility that lack of Alr1p reduces readthrough by modulating the prion form of eRF3 [PSI+] or some other element cured by guanidine hydrochloride treatment (Supplemental Material; Supplemental Fig. S1).
NMD-deficient cells show elevated Alr1p and Alr2p levels
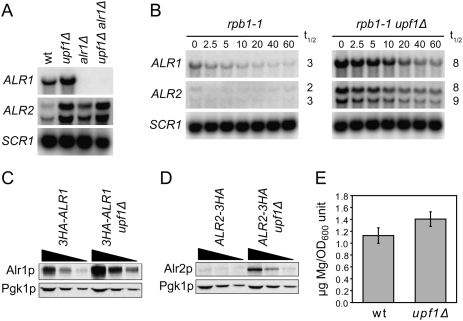
It is well established that the fidelity of in vitro translation systems is influenced by Mg2+ concentration; i.e., the levels of misreading at sense codons and readthrough at nonsense codons increases with increasing Mg2+ concentrations (Szer and Ochoa 1964; Capecchi 1967; Schlanger and Friedman 1973). Since a strain deleted for ALR1 shows reduced intracellular magnesium (Graschopf et al. 2001), and there is no evidence to suggest a direct role for Alr1p in translation termination, it seemed possible that the alr1Δ anti-suppressor effect could simply reflect reduced Mg2+ without explaining the mechanism by which NMD factors prevent translational readthrough. However, our earlier genome-wide approaches (He et al. 2003; Johansson et al. 2007) have shown that Upf1p associates with the ALR1 mRNA, and that this transcript accumulates more than twofold in upf1Δ, nmd2Δ, and upf3Δ cells. Those studies also showed that the transcript for the closely related Alr2 protein—which, if overexpressed, improves growth of alr1Δ cells (MacDiarmid and Gardner 1998)—accumulates in upf1Δ, nmd2Δ, and upf3Δ cells, and is rapidly degraded upon NMD reactivation. Decay rate measurements revealed that the increased abundance of ALR1 and ALR2 mRNAs in upf1Δ cells correlates to an increase in their half-lives (Fig. 2A,B), demonstrating that they are bona fide substrates of the NMD pathway. These findings implied that the reduced termination fidelity of NMD-deficient cells may be caused by increased Alr1p and Alr2p expression, and the consequent effects of elevated Mg2+ levels. Figure 2, C–E, demonstrates that Alr1p and Alr2p do accumulate in upf1Δ cells, correlating with an increase in free cellular Mg2+. The finding that NMD down-regulates not only expression of Alr1p, but also that of Alr2p, explained: (1) the partial suppression of the growth defect of an alr1Δ mutant by upf1Δ, nmd2Δ, or upf3Δ alleles; (2) the slightly higher readthrough levels in upf1Δ alr1Δ cells versus alr1Δ cells; and (3) the elimination of the suppressing effect of the upf1Δ allele on growth of alr1Δ cells by introduction of an alr2Δ allele (Fig. 1; Supplemental Fig. S3; data not shown). Consistent with the notion that Alr2p plays a minor role in Mg2+ uptake (MacDiarmid and Gardner 1998; Wachek et al. 2006), an alr2Δ allele does not counteract the nonsense suppression phenotype of upf1Δ cells or prevent accumulation of Mg2+ in the presence of elevated Alr1p levels (data not shown; see below). Collectively, these results suggest a model in which the readthrough phenotype of NMD-deficient cells is caused by elevated Mg2+ levels, primarily through increased Alr1p expression, and their effects on translational fidelity.
Figure 2.
NMD-deficient cells show increased expression of magnesium transporters. (A) Northern analysis of total RNA isolated from wild-type (MJY142), upf1Δ (MJY67), alr1Δ (MJY130), and upf1Δ alr1Δ (MJY447) cells grown in SC medium at 30°C. The blot was probed for ALR1, ALR2, and SCR1 transcripts using randomly labeled DNA fragments. (B) mRNA decay rates in rpb1-1 (MJY249) and rpb1-1 upf1Δ (MJY251) cells. The strains were grown at 24°C in SC medium followed by inhibition of RNA polymerase II transcription by a shift to 37°C. Time points (minutes) after the shift are indicated above the lanes. The signal in each lane was normalized to the corresponding SCR1 signal, and the half-life (t1/2, in minutes) was determined from the initial slope of the curve. (C,D) Western blot analyses of twofold serial dilutions of cell extracts from strains in which the DNA sequence for three tandem influenza virus hemagglutinin epitopes (3HA) was fused to the endogenous ALR1 (C, MJY290 and MJY309) or ALR2 (D, MJY167 and MJY183) ORF. Monoclonal antibodies against HA or Pgk1p were used to detect the indicated proteins. Control experiments showed that the sequence for the 3HA tag did not prevent accumulation of the transcripts in upf1Δ cells (Supplemental Fig. S2). (E) Mg2+ levels in supernatants of permeabilized wild-type (MJY142) and upf1Δ (MJY67) cells. The values, determined by using eriochrome blue SE, represent the average from four independent cultures of each strain. The standard deviation is indicated. A two-tailed Student's t-test revealed that the value for the upf1Δ strain is significantly different from that of the wild type (P = 0.02).
Alr1p levels modulate the fidelity of translation termination
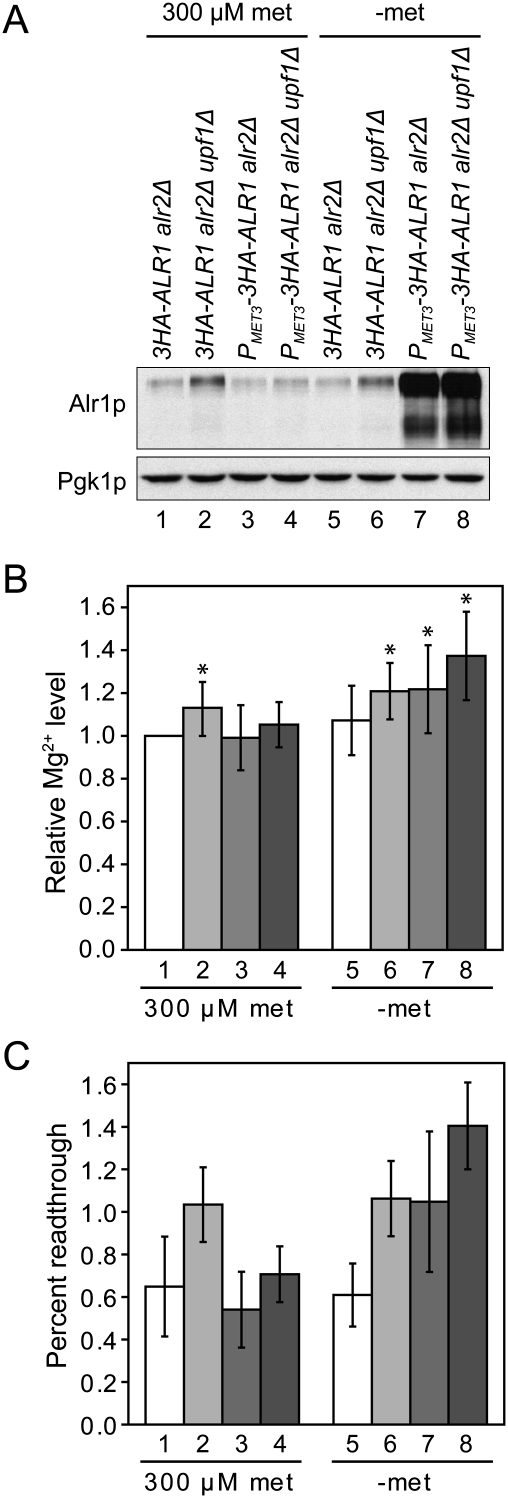
If elevated Mg2+ levels are the basis of increased readthrough in NMD-deficient cells, then the phenotype should be abolished by restoring normal Alr1p expression levels. Furthermore, wild-type cells with increased expression of Alr1p should show elevated levels of readthrough. To test these predictions, we constructed wild-type and upf1Δ cells in which the sequence upstream of the chromosomal 3HA-ALR1 ORF was replaced with the promoter and 5′-untranslated region (5′-UTR) of the regulatable MET3 gene (Mao et al. 2002). To exclude the possibility that differences in Alr2p expression would influence the results, we also introduced an alr2Δ allele into the strains. The 3HA-ALR1 transcript harboring the MET3 5′-UTR is largely resistant to NMD, although the level of the transcript was consistently somewhat higher in upf1Δ cells (Supplemental Fig. S4; Supplemental Table S2). By titrating the methionine concentration in the medium, we found that the presence of 300 μM methionine generated Alr1p and Mg2+ levels comparable with those observed in the control 3HA-ALR1 alr2Δ strain (Fig. 3A,B). The methionine concentration required to mimic the Alr1p levels in the 3HA-ALR1 alr2Δ upf1Δ strain was too low for reproducible analyses of the readthrough levels; i.e., slight differences in the number of cell divisions yielded large variations in the Alr1p levels. We therefore analyzed the strains with full derepression of PMET3 with the caveat that this generates Alr1p levels that are considerably higher than those observed in the 3HA-ALR1 alr2Δ upf1Δ strain (Fig. 3A). Consistent with the somewhat higher abundance of the MET3 5′-UTR-containing 3HA-ALR1 transcript in upf1Δ cells, the derepression of PMET3-3HA-ALR1 yielded slightly higher Alr1p and Mg2+ levels in the upf1Δ derivative (Fig. 3A,B; data not shown). As predicted, the efficiency of translation termination correlated inversely with Mg2+ levels; i.e., the readthrough phenotype of upf1Δ cells was abolished by restoring normal Alr1p expression levels, and elevated Alr1p expression induced readthrough even in NMD-proficient cells (Fig. 3C). The relative abundance of the can1-100 and leu2-2 nonsense transcripts was unaffected by Alr1p levels irrespective of the NMD status of the cells (Supplemental Fig. S4A; Supplemental Table S2), indicating that the accumulation of nonsense transcripts in upf1Δ cells is unrelated to the Alr1p expression level, and that NMD occurs independently of its effects on Mg2+ uptake. Collectively, these results show that Mg2+ levels modulate the efficiency of termination in vivo, and they strongly support the notion that the reduced termination fidelity of NMD-deficient yeast cells is an indirect consequence of increased Alr1p expression. While our findings do not contradict the notion that NMD may be triggered by the intrinsic inefficiency of premature termination (Amrani et al. 2004), they suggest that the detection of altered termination fidelity in NMD-deficient cells is a reflection of the set of endogenous transcripts subject to NMD regulation. Since the sets of endogenous NMD substrates differ in yeast and metazoans (Rehwinkel et al. 2006), this also explains why down-regulation of NMD in mammalian cells can lead to unchanged or even reduced levels of translational readthrough (Mendell et al. 2002; Ivanov et al. 2008).
Figure 3.
Increased Alr1p expression accounts for the reduced termination fidelity of NMD-deficient cells. (A) Western blot analysis of the 3HA-ALR1 alr2Δ (MJY455), 3HA-ALR1 alr2Δ upf1Δ (MJY456), PMET3-3HA-ALR1 alr2Δ (MJY430), and PMET3-3HA-ALR1 alr2Δ upf1Δ (MJY432) strains. The strains were grown in SC-cys medium containing 300 μM met, harvested and washed with water, diluted to OD600 ∼0.1 in fresh SC-cys (300 μM met) and SC-met-cys medium, and allowed to grow for two generations. Monoclonal antibodies against HA or Pgk1p were used to detect the indicated proteins. (B) Mg2+ levels in selected strains. Cells were grown as described above and assessed for Mg2+ levels by using eriochrome blue SE, and the resulting values were expressed relative to that obtained for the 3HA-ALR1 alr2Δ strain (300 μM met), which was set to 1. Strains are numbered as in A. The values represent the average of 10 independent experiments. The standard deviation is indicated. An asterisk indicates that the value is significantly different from 1 using the Wilcoxon signed rank test (P < 0.05). (C) Readthrough of the UAA codon in strains shifted to medium with or without 300 μM met. Cells were manipulated as in A, except that the medium also lacked uracil to allow selection for the reporter plasmids. Strains are numbered as in A. The values represent the average of five independent experiments. The standard deviation is indicated.
NMD regulation of Alr1p levels requires an upstream ORF (uORF) in the ALR1 5′-UTR
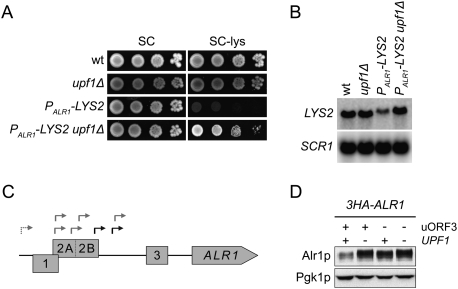
The observation that the MET3 5′-UTR-containing ALR1 transcript is largely resistant to NMD (Supplemental Fig. S4B) suggested that the ALR1 5′-UTR may be responsible for triggering decay. To assess this possibility, we replaced the sequences upstream of the endogenous LYS2 ORF with the promoter and 5′-UTR of the ALR1 gene. Introduction of a upf1Δ allele into cells harboring this PALR1-LYS2 allele improved growth on medium lacking lysine, which correlated with increased levels of ALR15′-UTR-LYS2 transcripts (Fig. 4A,B). Determination of ALR1 5′-UTR lengths and subsequent mutational analyses of upstream AUG codons revealed that the NMD reactivity of ALR1 transcripts is caused by the inclusion of one to three uORFs (Fig. 4C; Supplemental Material; Supplemental Fig. S5). Although these analyses showed that alternative extended transcripts encompassing uORF1 and uORF2A/2B contributed to the pool of NMD-regulated ALR1 transcripts, they also suggested that the shorter uORF3-containing transcripts accounted for the NMD-dependent regulation of Alr1p levels (Fig. 4D). This notion was supported further by the finding that uORF3-dependent NMD regulation of ALR1 mRNA and Alr1p levels was largely intact in cells in which the 5′-UTR sequence upstream of uORF3 had been replaced with the 5′-UTR sequence of the MET3 transcript (Supplemental Fig. S6).
Figure 4.
NMD-regulation of Alr1p levels is attributable to a uORF in the ALR1 5′-UTR. (A) Growth in the absence of lysine. The indicated strains (MJY142, MJY67, MJY363, and MJY365) were grown overnight in SC medium at 30°C, serially diluted, spotted onto SC and SC-lys plates, and incubated for 3 d at 30°C. (B) Northern analysis of total RNA isolated from indicated strains (MJY142, MJY67, MJY363, and MJY365) grown in SC medium at 30°C. The blot was probed for LYS2 and SCR1 transcripts using randomly labeled DNA fragments. (C) The chromosomal region upstream of the ALR1 ORF. uORF1, uORF2A/2B, and uORF3 are shown as boxes. Transcriptional start sites, as determined by 5′ RLM-RACE, are indicated with gray arrows (identified in upf1Δ [MJY67] cells) and black arrows (identified in both wild-type [MJY142] and upf1Δ cells). The dashed arrow indicates the presence of an extended subspecies encompassing uORF1 and uORF2A (see the Supplemental Material). (D) Western analysis of the indicated strains (MJY290, MJY309, MJY422, and MJY424) grown in SC medium at 30°C. uORF3 was eliminated by changing the sequence for the AUG codon to AAG. Monoclonal antibodies against HA or Pgk1p were used to detect the indicated proteins.
Concluding remarks
The importance of Mg2+ as a cofactor/counterion and its role in establishing and maintaining structures of nucleic acids, proteins, and membranes implies that cells must strictly control intracellular Mg2+ concentrations. It was shown previously that expression of S. cerevisiae Alr1p is regulated at the levels of protein stability and mRNA abundance (Graschopf et al. 2001), and our results demonstrate that ALR1 (and ALR2) mRNA levels are at least partially controlled by the NMD pathway. Since uORF3 is conserved in other Saccharomyces species (Cvijovic et al. 2007), NMD regulation of Mg2+ uptake and translational fidelity may be a widespread phenomenon. However, it remains to be seen whether altered Mg2+ levels contribute to other phenotypes reported for NMD-deficient yeast cells.
Elevated Mg2+ levels have been shown previously to decrease translational fidelity in cell-free systems (Szer and Ochoa 1964; Capecchi 1967; Schlanger and Friedman 1973), but, to the best of our knowledge, comparable effects have not been demonstrated in vivo. Given the importance of Mg2+ ions to ribosome structure (Selmer et al. 2006), one possible mechanism for Mg2+-mediated mistranslation would involve a conformational distortion of the A site that, much like aminoglycoside-induced nonsense suppression, would falsely trigger flipping of bases that normally monitor mRNA:tRNA base-pairing (Vicens and Westhof 2003).
Materials and methods
Strains, plasmids, media, and genetic procedures
Yeast strains used in this study are listed in Supplemental Table S3. Yeast transformations, media, and genetic procedures have been described (Amberg et al. 2005). Unless stated otherwise, all experiments were performed using exponentially growing cells at an optical density at 600 nm (OD600) of ∼0.5. Standard methods (Longtine et al. 1998; Tong et al. 2001; Amberg et al. 2005) were used to delete genes and to integrate sequences for markers, promoters, tags, and mutant alleles. Deletions of genes and integration of markers were confirmed by PCR using primers that annealed outside of sequences present in the transformed DNA fragment(s). Integrations of promoters, tags, and mutant alleles were confirmed by DNA sequencing of the relevant PCR products. The genetic screen and details of strain and plasmid constructions are described in the Supplemental Material.
RNA methods
mRNA levels and mRNA half-lives were determined essentially as described previously (Johansson et al. 2007; He et al. 2008). Transcriptional start sites were mapped by using the FirstChoice RLM-RACE kit (Ambion) combined with the TOPO TA Cloning Kit for Sequencing (Invitrogen), and subsequent DNA sequencing of seven to 10 clones for each strain.
Cell extracts and Western blotting procedures
Cells representing four OD600 units were harvested, washed once with water, and resuspended in 200 μL of a slightly modified version of a previously described sample buffer (8.75 mM Tris-HCl at pH 6.8, 8 M urea, 0.875% sodium dodecyl sulfate [SDS], 1% β-mercaptoethanol, 1× protease inhibitor cocktail [Roche Applied Science], bromphenol blue) (Schandel and Jenness 1994). Samples were transferred to 2-mL tubes containing ∼0.15 g of glass beads (425–600 μm, Sigma-Aldrich), vortexed six times for 30 sec with 1 min on ice between steps, and incubated for 10 min at 37°C. Following centrifugation, a fraction of the samples, typically 10 μL, was resolved by 8% SDS-PAGE, transferred to Immobilon-P (Millipore) membranes, and incubated with monoclonal anti-HA antibodies (12CA5, Roche Applied Science) and then with horseradish peroxidase-conjugated goat anti-mouse IgG (Zymed laboratories). Blots were stripped and reprobed with monoclonal anti-Pgk1p antibodies (22C5-D8, Molecular Probes). Proteins were detected using ECL Western blotting detection reagents (GE Healthcare) and Kodak BioMax light film.
Dual-luciferase assays
Readthrough assays were performed using the dual-luciferase reporter assay system (Promega) and a TD-20/20 luminometer (Turner Biosystems). An appropriate number of cells in 5 μL of the relevant medium was transferred to 100 μL of passive lysis buffer followed by 12 sec of vortexing and immediate use of 20 μL of lysate for the assay. When performing readthrough assays in [psi−] strains, the firefly and Renilla activities were determined using different numbers of cells, ensuring that the values were within the linear range of the respective assay. Each individual culture was assayed three times, and the values were derived from five independent experiments, performed on different days, using independent transformants for each experiment.
Magnesium measurements
Cells representing ∼15 OD600 units were harvested and washed once with water and twice with buffer E (30 mM MOPS at pH 7.1, 100 mM KCl), and aliquots from the last wash step were used for a final OD600 reading. Following centrifugation of a defined volume of the remaining sample, the cell pellet was resuspended in buffer ET (bufffer E containing 0.05% Triton X-100), frozen on dry ice, and stored at −70°C. The frozen cell suspension was transferred for 30 min to 30°C, vortexed briefly, and centrifuged for 5 min. The supernatants were transferred to fresh 1.5-mL tubes, and the Mg2+ concentration was determined using eriochrome blue SE essentially as described previously (Scarpa 1974). Briefly, the Mg2+ levels were determined in buffer E containing 30 μM eriochrome blue SE (Sigma-Aldrich) by measuring the difference in absorbance at 592 and 554 nm. A standard curve was obtained using dilutions of an MgCl2 stock solution. Each sample was assayed three times, and the magnesium concentration was calculated per OD600 unit. Mock samples consisting of the same volume of medium as the cultures were subjected to all steps of the protocol to ensure that the background was negligible.
Acknowledgments
We thank A. Byström and D. Bedwell for strains and plasmids. P. Rydén and members of A.J.'s laboratory are acknowledged for helpful comments on the manuscript. This work was supported by a grant to A.J. from the National Institutes of Health. During his stay in A.J.'s laboratory, M.J.O.J. was supported in part by a post-doctoral fellowship from the Swedish Research Council.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1930710.
Supplemental material is available at http://www.genesdev.org.
References
- Amberg DC, Burke DJ, Strathern JN 2005. Methods in yeast genetics Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Amrani N, Ganesan R, Kervestin S, Mangus DA, Ghosh S, Jacobson A 2004. A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature 432: 112–118 [DOI] [PubMed] [Google Scholar]
- Capecchi MR 1967. Polarity in vitro. J Mol Biol 30: 213–217 [DOI] [PubMed] [Google Scholar]
- Cvijovic M, Dalevi D, Bilsland E, Kemp GJ, Sunnerhagen P 2007. Identification of putative regulatory upstream ORFs in the yeast genome using heuristics and evolutionary conservation. BMC Bioinformatics 8: 295 doi: 10.1186/1471-2105-8-295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaplinski K, Ruiz-Echevarria MJ, Paushkin SV, Han X, Weng Y, Perlick HA, Dietz HC, Ter-Avanesyan MD, Peltz SW 1998. The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes Dev 12: 1665–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graschopf A, Stadler JA, Hoellerer MK, Eder S, Sieghardt M, Kohlwein SD, Schweyen RJ 2001. The yeast plasma membrane protein Alr1 controls Mg2+ homeostasis and is subject to Mg2+-dependent control of its synthesis and degradation. J Biol Chem 276: 16216–16222 [DOI] [PubMed] [Google Scholar]
- He F, Li X, Spatrick P, Casillo R, Dong S, Jacobson A 2003. Genome-wide analysis of mRNAs regulated by the nonsense-mediated and 5′ to 3′ mRNA decay pathways in yeast. Mol Cell 12: 1439–1452 [DOI] [PubMed] [Google Scholar]
- He F, Amrani N, Johansson MJO, Jacobson A 2008. Qualitative and quantitative assessment of the activity of the yeast nonsense-mediated mRNA decay pathway. Methods Enzymol 449: 127–147 [DOI] [PubMed] [Google Scholar]
- Ivanov PV, Gehring NH, Kunz JB, Hentze MW, Kulozik AE 2008. Interactions between UPF1, eRFs, PABP and the exon junction complex suggest an integrated model for mammalian NMD pathways. EMBO J 27: 736–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson A, Izaurralde E 2007. Nonsense-mediated mRNA decay: From yeast to metazoans. In Translational control in biology and medicine (ed. Mathews MB et al. ), pp. 655–688 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Johansson MJO, He F, Spatrick P, Li C, Jacobson A 2007. Association of yeast Upf1p with direct substrates of the NMD pathway. Proc Natl Acad Sci 104: 20872–20877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling KM, Lanier J, Du M, Salas-Marco J, Gao L, Kaenjak-Angeletti A, Bedwell DM 2004. Leaky termination at premature stop codons antagonizes nonsense-mediated mRNA decay in S. cerevisiae. RNA 10: 691–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961 [DOI] [PubMed] [Google Scholar]
- MacDiarmid CW, Gardner RC 1998. Overexpression of the Saccharomyces cerevisiae magnesium transport system confers resistance to aluminum ion. J Biol Chem 273: 1727–1732 [DOI] [PubMed] [Google Scholar]
- Maderazo AB, He F, Mangus DA, Jacobson A 2000. Upf1p control of nonsense mRNA translation is regulated by Nmd2p and Upf3p. Mol Cell Biol 20: 4591–4603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X, Hu Y, Liang C, Lu C 2002. MET3 promoter: A tightly regulated promoter and its application in construction of conditional lethal strain. Curr Microbiol 45: 37–40 [DOI] [PubMed] [Google Scholar]
- Mendell JT, ap Rhys CM, Dietz HC 2002. Separable roles for rent1/hUpf1 in altered splicing and decay of nonsense transcripts. Science 298: 419–422 [DOI] [PubMed] [Google Scholar]
- Rehwinkel J, Raes J, Izaurralde E 2006. Nonsense-mediated mRNA decay: Target genes and functional diversification of effectors. Trends Biochem Sci 31: 639–646 [DOI] [PubMed] [Google Scholar]
- Scarpa A 1974. Indicators of free magnesium in biological systems. Biochemistry 13: 2789–2794 [DOI] [PubMed] [Google Scholar]
- Schandel KA, Jenness DD 1994. Direct evidence for ligand-induced internalization of the yeast α-factor pheromone receptor. Mol Cell Biol 14: 7245–7255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlanger G, Friedman SM 1973. Ambiguity in a polypeptide-synthesizing extract from Saccharomyces cerevisiae. J Bacteriol 115: 129–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmer M, Dunham CM, Murphy FV, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V 2006. Structure of the 70S ribosome complexed with mRNA and tRNA. Science 313: 1935–1942 [DOI] [PubMed] [Google Scholar]
- Szer W, Ochoa S 1964. Complexing ability and coding properties of synthetic polynucleotides. J Mol Biol 8: 823–834 [DOI] [PubMed] [Google Scholar]
- Tong AH, Evangelista M, Parsons AB, Xu H, Bader GD, Page N, Robinson M, Raghibizadeh S, Hogue CW, Bussey H, et al. 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294: 2364–2368 [DOI] [PubMed] [Google Scholar]
- Tuite MF, Mundy CR, Cox BS 1981. Agents that cause a high frequency of genetic change from [psi+] to [psi−] in Saccharomyces cerevisiae. Genetics 98: 691–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicens Q, Westhof E 2003. RNA as a drug target: The case of aminoglycosides. Chembiochem 4: 1018–1023 [DOI] [PubMed] [Google Scholar]
- Wachek M, Aichinger MC, Stadler JA, Schweyen RJ, Graschopf A 2006. Oligomerization of the Mg2+-transport proteins Alr1p and Alr2p in yeast plasma membrane. FEBS J 273: 4236–4249 [DOI] [PubMed] [Google Scholar]
- Wang W, Czaplinski K, Rao Y, Peltz SW 2001. The role of Upf proteins in modulating the translation read-through of nonsense-containing transcripts. EMBO J 20: 880–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng Y, Czaplinski K, Peltz SW 1996. Genetic and biochemical characterization of mutations in the ATPase and helicase regions of the Upf1 protein. Mol Cell Biol 16: 5477–5490 [DOI] [PMC free article] [PubMed] [Google Scholar]