Abstract
Mitochondria serve a critical role in physiology and disease. The genetic basis of mitochondrial regulation in mammalian cells has not yet been detailed. We performed a large-scale RNAi screen to systematically identify genes that affect mitochondrial abundance and function. This screen revealed previously unrecognized roles for >150 proteins in mitochondrial regulation. We report that increased Wnt signals are a potent activator of mitochondrial biogenesis and reactive oxygen species (ROS) generation, leading to DNA damage and acceleration of cellular senescence in primary cells. The signaling protein insulin receptor substrate-1 (IRS-1), shown here to be a transcriptional target of Wnt, is induced in this setting. The increased level of IRS-1 drives activation of mitochondrial biogenesis; furthermore, in insulin-responsive cell types, it enhances insulin signaling, raising the possibility that Wnt proteins may be used to modulate glucose homeostasis. Our results identify a key component of the mitochondrial regulatory apparatus with a potentially important link to metabolic and degenerative disorders.
Keywords: RNAi screen, mitochondria, Wnt signaling, IRS-1
Mitochondrial dysfunction has been implicated as causal or contributory factors in several important human disorders (DiMauro and Schon 2003; Loeb et al. 2005; Lowell and Shulman 2005; Wallace 2005; Kroemer 2006; Lin and Beal 2006). Congenital defects in mitochondrial energy metabolism underlie a multitude of clinical syndromes in the pediatric population (DiMauro and Schon 2003). Several common metabolic and degenerative disorders of adulthood also have strong links to mitochondrial dysfunction (Loeb et al. 2005; Lowell and Shulman 2005; Lin and Beal 2006). An elaborate system of regulatory control over mitochondrial abundance and function is essential, given the wide-ranging impact of mitochondria on many vital cellular processes. The currently known regulators of mitochondria have been identified primarily by gain-of-function approaches and are few in number. As such, there is a compelling need for a better understanding of the genes involved in normal mitochondrial regulation in mammalian cells. To this end, we carried out an RNAi-based genetic screen to identify novel regulators of mitochondrial density and membrane potential. Our results provide a framework for further investigation of mitochondrial biology in mammalian cells.
Results
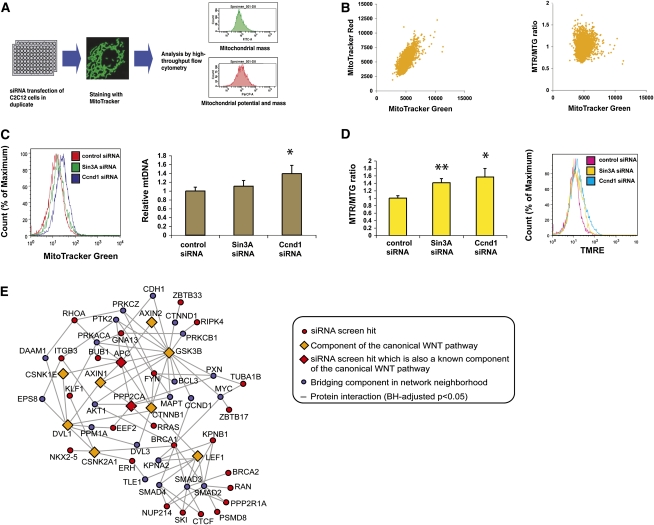
We developed a flow cytometry-based siRNA screen targeting 6363 genes (Fig. 1A). A C2C12 mouse muscle cell line was employed because it is a well-characterized, nontransformed cell line that may have potential relevance with respect to understanding muscle mitochondrial abnormalities seen in type 2 diabetes. Many congenital mitochondrial disorders are associated with prominent neuromuscular phenotypes as well (DiMauro and Schon 2003). Cells were plated in 96-well plates and transfected with siRNAs in duplicate. Five days later, the cells were stained with two different mitochondria-specific dyes, MitoTracker Green and MitoTracker Red CMXRos, and analyzed by high-throughput flow cytometry. This strategy has been employed previously for simultaneous assessment of mitochondrial abundance and membrane potential in live cells (Poot and Pierce 2001; Pendergrass et al. 2004). Both dyes vary with mitochondrial abundance. MitoTracker Red is also sensitive to mitochondrial membrane potential, while MitoTracker Green is minimally affected; thus, the ratio of Red-to-Green can be used to detect changes in the mitochondrial membrane potential while adjusting for any concomitant changes in mitochondrial mass. A positive correlation was noted between MitoTracker Green and MitoTracker Red, as expected, but not between MitoTracker Green and the Red-to-Green ratio (Fig. 1B). As a proof of principle, we confirmed that siRNAs targeting Sin3A and Ccnd1—two genes reported previously to negatively regulate mitochondria (Pile et al. 2003; Wang et al. 2006)—produced an increase in the Red-to-Green ratio or both the MitoTracker Green signal and the Red-to-Green ratio, respectively (Fig. 1C,D; Supplemental Figs. S1, S2A). These changes were further validated by directly measuring mitochondrial DNA by quantitative PCR and by measuring mitochondrial membrane potential with the potentiometric dye tetramethyl rhodamine ethyl ester (TMRE) (Fig. 1D; Supplemental Fig. S2B).
Figure 1.
siRNA screen for mitochondrial regulators. (A) Schematic diagram of the screen. (B) Distribution of MitoTracker Green versus MitoTracker Red CMXRos signals (left plot) and MitoTracker Green versus the Red-to-Green ratio (right plot) in the primary screen. (C) Assessment of mitochondrial content by MitoTracker Green signal and mitochondrial DNA quantitation after siRNA depletion of Sin3A and Ccnd1. (*) P < 0.05 by unpaired t-test. (D) Assessment of mitochondrial membrane potential by the Red-to-Green ratio and TMRE signal after siRNA depletion of Sin3A and Ccnd1. (*) P < 0.05. (E) Network extension of the WNT pathway. The network was constructed by anchoring on canonical WNT pathway components (diamonds) using protein interaction data, incorporating interactions with human orthologs of screen hits (red) via up to one bridging component (blue). The interactions shown fulfilled a Benjamini and Hochberg (BH)-adjusted P-value of <0.05 for the given network size.
The initial screen used pools of four siRNAs per gene. Pools were classified as possible hits if they altered the MitoTracker Green signal (reflecting a change in mitochondrial mass), the MitoTracker Red signal (reflecting a change in either mitochondrial mass, membrane potential, or both), or the Red-to-Green ratio (reflecting a change in mitochondrial membrane potential) by an increase >40% or a decrease >20% compared with the plate mean. Candidate genes were then validated by repeating the transfection experiments with each of the four individual siRNAs. In the validation round, the comparison was made to wells receiving a control siRNA. Confirmation required at least two out of the four oligonucleotides reproducing the phenotype, which decreases the likelihood of off-target effects. About 2.8% of the genes screened were found to significantly alter one of the three parameters in this fashion (Supplemental Table S1).
The validated hits include genes participating in a diverse array of biological processes, and are statistically enriched for genes involved in nucleic acid metabolism, protein metabolism and modification, intracellular protein trafficking, and regulation of the cell cycle (Supplemental Figs. S3, S4; Supplemental Tables S2–S5). In terms of molecular function, there was an enrichment for kinases (Supplemental Fig. S4; Supplemental Table S6). Many of these genes have been implicated previously in various human disorders linked to degenerative, oncologic, or metabolic processes (Supplemental Fig. S3; Supplemental Table S7), while others have been cited in the literature in key functional contexts such as senescence, DNA damage, apoptosis, and diabetes (Supplemental Fig. S5; Supplemental Table S8). Fifteen proteins have previous associations with mitochondria (Supplemental Table S9). An siRNA directed against pyruvate dehydrogenase kinase (PDK1), a negative regulator of pyruvate entry into the TCA cycle, produced an increase in the Red-to-Green ratio, suggesting that it normally reduces mitochondrial activity. Depletion of uncoupling protein 2 (UCP2) likewise increased the Red-to-Green ratio, consistent with the notion that it normally dissipates the mitochondrial membrane potential without significantly affecting mitochondrial mass. The screen also identified Wolfram syndrome 1 (WFS1), a putative transmembrane protein of unknown function discovered recently to be a type 2 diabetes susceptibility gene by genome-wide association studies (Sandhu et al. 2007), suggesting another link between diabetes and mitochondrial regulation.
Wnt signaling activates mitochondrial biogenesis and oxidative phosphorylation (OXPHOS) gene expression
Among the validated hits, we noted the presence of two genes with known connections to the Wnt signaling pathway. The APC gene scored during the validation round in the MitoTracker Red category, suggesting that Wnt signaling may enhance mitochondrial biogenesis and/or activity. The transcriptional regulator ZBTB33 (also known as kaiso), reported previously to negatively regulate Wnt signaling in Xenopus, scored in the MitoTracker Green and Red categories (Ruzov et al. 2004; Park et al. 2005). We further tested siRNAs directed against β-catenin and Axin2, two well-established components of this pathway, and found results consistent with a role for Wnt signaling as a positive activator of mitochondrial proliferation (data not shown). Based on these observations, we constructed an extended Wnt network using protein interaction data (Keshava et al. 2009), which revealed connections to a number of other hits from the screen (Fig. 1E). Wnt signaling has been a subject of intensive investigation in recent years, especially in the contexts of development, tissue renewal, degenerative diseases, and cancer (Moon et al. 2004; Clevers 2006)—all processes in which mitochondria are thought to play prominent roles. We hypothesized that some of the biological phenotypes of Wnt signaling may be mediated through its effects on mitochondria, and, given the importance of the processes controlled by Wnt signaling, further reasoned that a careful analysis of this pathway in the context of mitochondrial regulation may yield valuable insights.
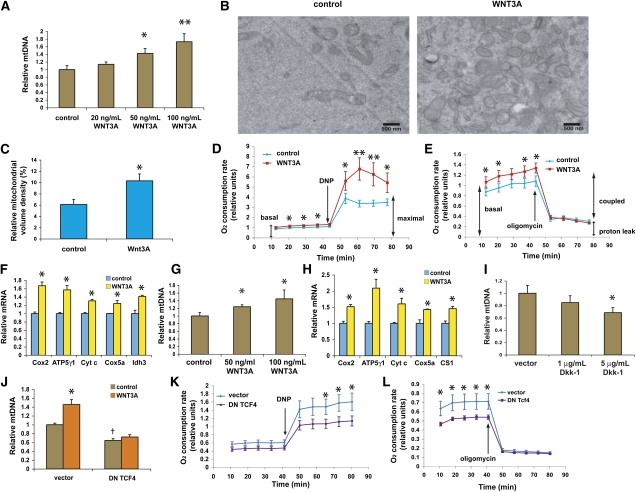
In order to test if a direct modulation of the Wnt signaling pathway could affect mitochondria, we added recombinant Wnt3A protein to the C2C12 cell culture media. We found that mitochondrial biogenesis was markedly induced within 3 d in a dose-dependent fashion, as assessed by MitoTracker Green staining and direct quantitation of mitochondrial DNA (Fig. 2A). To further evaluate structural changes in mitochondria, we examined these cells using transmission electron microscopy. We observed a range of mitochondrial morphologies that largely overlapped between the control and Wnt3A-treated cells. Quantitative analysis revealed that Wnt3A-treated cells showed a nearly twofold increase in mitochondrial volume density compared with controls (Fig. 2B,C; Supplemental Fig. S6). Individual mitochondria of unusually large size or length were seen occasionally in Wnt3A-treated cells but not in controls (Supplemental Fig. S7).
Figure 2.
Increased Wnt signals activate mitochondrial biogenesis and OXPHOS gene expression. Cells received either 0.1% BSA in PBS (control) or the recombinant Wnt3A protein at the doses indicated. (*) P < 0.05 by unpaired t-test; (**) P < 0.01. (A) Mitochondrial proliferation in C2C12 cells following Wnt3A treatment as assessed by mitochondrial DNA quantitation. (B) Electron micrographs of C2C12 cells treated with control or 50 ng/mL Wnt3A protein for 3 d. (C) Wnt3A-treated cells show a significant increase in mitochondrial volume density (P < 0.05), as quantified by using a grid superimposed on electron micrographs (N = 15 each). The number of points falling within mitochondria was expressed as a percentage of the number of points falling within cytoplasm. A small increase in cristae surface density was also noted (data not shown). Error bars represent SEM. (D,E) Increased cellular oxygen consumption results from Wnt3A treatment. Drugs were added at the indicated times. (F) Activation of mitochondrial OXPHOS gene expression from Wnt3A treatment. (G,H) Mitochondrial proliferation and increased OXPHOS gene expression in MEFs following 3 d of Wnt3A exposure. (I) Treatment with the Wnt3A antagonist Dkk-1 (5 μg/mL) for 1 wk reduces mitochondrial DNA in MEFs. (J) Stable expression of dominant-negative TCF4 (DN TCF4) cDNA reduces mitochondrial DNA and abrogates the Wnt3A effect on mitochondrial proliferation. C2C12 cells were transduced with retroviruses carrying an empty vector or the dominant-negative TCF4 cDNA, and were selected for puromycin resistance. (*) P < 0.05 for control versus Wnt3A; (†) P < 0.05 for vector versus dominant-negative TCF4 (DN TCF4). (K,L) Dominant-negative TCF4 cDNA expression reduces oxygen consumption. DNP and oligomycin were injected at the indicated times to measure the maximal respiratory capacity and uncoupled respiration, respectively.
To determine if the increased mitochondria were functional, we examined cellular oxygen consumption in Wnt3A-treated C2C12 cells. Significant enhancement of basal cellular respiration was seen in Wnt3A-treated cells (Fig. 2D,E); addition of the uncoupler 2,4-dinitrophenol (DNP) elicited a much greater increase in Wnt3A-treated cells compared with untreated cells, consistent with a bona fide increase in functional mitochondrial abundance and the maximal respiratory capacity (Fig. 2D). There was no significant difference in the inner mitochondrial membrane proton leak, measured by adding the ATP synthase inhibitor oligomycin (Fig. 2E), indicating that nearly all of the changes in mitochondrial bioenergetics produced by Wnt3A treatment involved respiration coupled to OXPHOS. Consistent with this idea, we also found an induction in the expression of key mitochondrial OXPHOS genes (Fig. 2F), such as ATP synthase γ subunit (ATP5γ1), cytochrome c (Cyc1), and mitochondrially encoded cytochrome oxidase subunit 2 (Cox2). These increases were detectable at 48 h after initiation of treatment, and were more pronounced at 72 h. We observed essentially the same results in primary mouse embryonic fibroblasts (MEFs) (Fig. 2G,H), indicating that this pathway is likely to be functionally important across different cell types. Analysis of the OXPHOS complexes at the protein level (Supplemental Fig. S8) and assays of citrate synthase activity (Supplemental Fig. S9) provided further confirmation of the increases in respiratory capacity. Intact mitochondria purified from Wnt3A-treated cells also showed enhanced state III oxidation with adenosine disphosphate (ADP) (Supplemental Fig. S10).
We also investigated the effect of adding recombinant Dkk-1 protein, a soluble antagonist of Wnt signaling (Moon et al. 2004; Clevers 2006), and found a reduction in MitoTracker Green fluorescence and mitochondrial DNA content (Fig. 2I). This shows that attenuation of endogenous Wnt signaling produces significant effects on mitochondria, and is consistent with the notion that Wnt signals may represent a control point for mitochondrial regulation.
We next asked whether this effect of Wnt3A is mediated through the canonical signaling cascade involving β-catenin and TCF, or through noncanonical pathways. C2C12 cells stably expressing the dominant-negative TCF4 cDNA (Supplemental Fig. S11), which binds the TCF target sites but is not regulated by β-catenin, showed reduced levels of mitochondrial DNA, and were resistant to the stimulatory effects of Wnt3A (Fig. 2J). These cells also showed a significant reduction in basal and stimulated respiration, while the proton leak was unaffected (Fig. 2K,L). These results suggest that the Wnt3A effect on mitochondria is primarily through the canonical pathway involving TCF4. Interestingly, we also found that the addition of recombinant Wnt5A protein, which has been reported to signal through noncanonical pathways in a variety of assays (Ishitani et al. 2003; Topol et al. 2003), antagonizes the Wnt3A effect on mitochondria in a dose-dependent fashion (Supplemental Fig. S12). This raises the possibility of complex functional interactions between the Wnt3A-mediated mitochondrial proliferation pathway and other signaling cascades activated by extracellular modulators.
We also examined the transcriptional regulator ZBTB33 by generating cell lines stably expressing shRNA against ZBTB33. Two independent shRNAs that produced a significant knockdown of the ZBTB33 transcript (>60%) substantially increased the mitochondrial DNA, mitochondrial OXPHOS gene expression, and cellular oxygen consumption (Supplemental Fig. S13A–C). Overexpression of the cDNA significantly reduced cellular oxygen consumption (Supplemental Fig. S13D). Taken together, these results reinforce the notion that Wnt signaling is a key regulator of mitochondrial function.
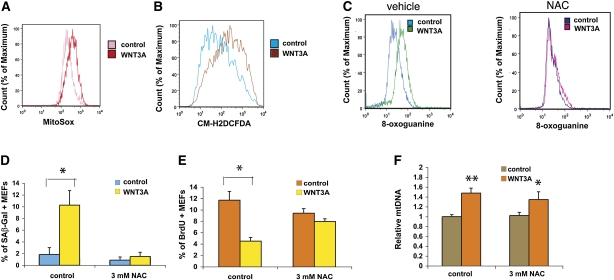
Wnt-mediated cellular senescence is dependent on mitochondrial reactive oxygen species (ROS) production
Increased mitochondrial density implies an increased number of sites capable of generating ROS. We therefore measured two major oxygen radical species, mitochondrial superoxide and hydrogen peroxide, using the dyes MitoSox Red and CM-H2CFDA in conjunction with flow cytometry. Wnt3A-treated C2C12 cells showed markedly increased superoxide and hydrogen peroxide levels (Fig. 3A,B; Supplemental Fig. S14), and similar results were also seen in MEFs. Direct biochemical measurements of cellular peroxide levels confirmed these results (Supplemental Fig. S15). These increases in ROS were dependent on mitochondrial proliferation, as blunted responses were seen in cells stably expressing an shRNA against DNA polymerase γ (POLG), which is required for mitochondrial DNA replication (Supplemental Fig. S16A,B). Stable expression of the dominant-negative TCF4 cDNA, which effectively blocks Wnt3A-induced mitochondrial proliferation (Fig. 2J), also prevented increases in ROS (Supplemental Fig. S16C). Chronic elevation of ROS species may result in accumulation of ROS-induced cellular damage, which may in turn predispose to cellular senescence, cell death, or transformation.
Figure 3.
Wnt-driven effects on mitochondria lead to increased oxidative stress. (*) P < 0.05 by unpaired t-test; (**) P < 0.01. (A,B) Wnt3A treatment increases mitochondrial superoxide and hydrogen peroxide levels. This is observed after 3 d of Wnt3A treatment (shown), as well as following long-term exposure. (C) Continuous exposure of MEFs to WNT3A results in increased oxidative DNA damage, as measured by 8-oxoguanine levels, which is prevented by concominant NAC treatment. NAC also blocks the Wnt3A-induced increase in the SAβ-gal staining (D) and decrease in BrdU incorporation of cells given a 1-h pulse of BrdU (E). In C–E, MEFs were examined after 2 wk of Wnt3A treatment in 3% oxygen. (F) Wnt3A treatment induces mitochondrial proliferation after 3 d of treatment with or without NAC present.
In light of the recent reports that continuous Wnt exposure can trigger cellular senescence in culture and in animals (Brack et al. 2007; Liu et al. 2007), we sought to determine whether this is dependent on the increased oxidative stress. Extended treatment of MEFs with Wnt3A protein over a 2-wk period resulted in markedly increased oxidative DNA damage, as measured by 8-oxoguanine content (Fig. 3C; Supplemental Fig. S17). In order to minimize oxidative stress from atmospheric oxygen, we propagated MEFs in physiological (3%) oxygen rather than in standard tissue culture oxygen (20%) (Parrinello et al. 2003). Growth in 3% oxygen allows MEFs to be maintained for extended periods of time without undergoing significant replicative senescence, while senescence is routine in 20% oxygen (Parrinello et al. 2003). Under the low-oxygen conditions, endogenous ROS generation, which largely takes place within mitochondria, is the major determinant of oxidative stress in cells. We saw a clear increase in senescence-associated β-galactosidase (SAβ-gal) staining, and a decrease in proliferation measured by BrdU incorporation in Wnt3A-treated cells (Fig. 3D,E; Supplemental Fig. S18). Importantly, all of these parameters were effectively normalized by the concomitant addition of the peroxide scavenger N-acetylcysteine (NAC) (Fig. 3C–E; Supplemental Fig. S18). Stable expression of the mitochondrial superoxide dismutase (SOD2) cDNA had a similar but less pronounced effect (Supplemental Fig. S19A–C). Mitochondrial biogenesis was still stimulated by Wnt under these circumstances (Fig. 3F), consistent with the idea that it is directly activated by Wnt rather than as a secondary response to an increase in ROS levels. The induction of Wnt-mediated senescence, in contrast, required elevated ROS levels. These data strongly suggest that Wnt induces replicative senescence in MEFs via increased mitochondrial biogenesis and the consequent increase in ROS-mediated damage.
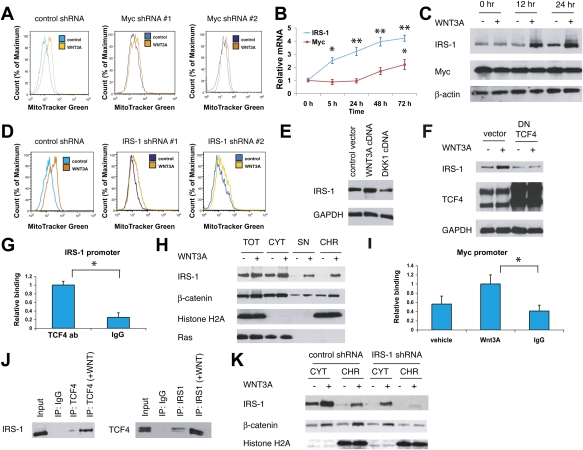
Wnt-mediated mitochondrial proliferation involves IRS-1 and Myc
In light of the above data suggesting that Wnt3A acts on mitochondria through the canonical pathway (Fig. 2J–L), we hypothesized that transcriptional targets of TCF4 are likely to mediate the mitochondrial phenotype. We examined known targets of TCF4 and focused on the transcription factor Myc, which has been shown to bind and activate genes involved in mitochondrial biogenesis and function, and to increase ROS (He et al. 1998; Vafa et al. 2002; Li et al. 2005; Kim et al. 2008). These effects of Myc on mitochondrial proliferation are consistent with its role as a pleiotropic regulator of cellular functions, including growth, differentiation, and apoptosis. Indeed, we found that siRNA or shRNA-based depletion of Myc reduced Wnt-mediated mitochondrial proliferation at day 3 by more than half (Fig. 4A; Supplemental Figs. S20, S21). Interestingly, while Myc is known to be a direct target of TCF4 in colon cancer cells (He et al. 1998; Hatzis et al. 2008), we found the Myc transcript levels did not rise until ∼36 h after treatment with Wnt3A protein (Fig. 4B), and the Myc protein levels initially remained the same as well (Fig. 4C; Supplemental Fig. S22).
Figure 4.
Wnt-driven mitochondrial biogenesis involves Myc and IRS-1. (A) Depletion of Myc by shRNA reduces Wnt-driven mitochondrial proliferation. Cells transduced with viruses expressing Myc shRNAs or controls were treated with Wnt3A for 3 d and assayed for MitoTracker Green signal. (B) IRS-1 mRNA is rapidly induced in MEFs by Wnt3A treatment, while the Myc mRNA induction shows delayed kinetics. Similar induction was seen in C2C12 cells (not shown). (C) The IRS-1 protein is induced within 12 h following Wnt3A treatment. (D) Depletion of IRS-1 by shRNA reduces Wnt-driven mitochondrial proliferation. Cells expressing IRS-1 shRNAs or controls were treated with Wnt3A for 3 d and were analyzed as in A. (E) Stable expression of Wnt3A induces IRS-1 in C2C12 cells, while expressing Dkk-1 has the opposite effect. Cells were transduced with viruses expressing the Wnt3A cDNA or the Dkk-1 cDNA, selected, and examined. (F) Stable expression of dominant-negative TCF4 cDNA reduces the IRS-1 protein levels and abrogates Wnt3A-mediated induction of IRS-1. Cells were treated with recombinant Wnt3A protein for 3 d. (G) TCF4 binds the IRS-1 promoter, as determined by ChIP and quantitative PCR. C2C12 cells were treated with Wnt3A protein for 1 d and subjected to chemical cross-linking, and the DNA was sheared and immunoprecipitated with rabbit IgG or the TCF4 antibody. (H) The IRS-1 protein accumulates in the chromatin fraction following Wnt3A treatment in C2C12 cells. (TOT) Total lysate; (CYT) cytoplasmic proteins; (SN) soluble nuclear proteins; (CHR) chromatin fraction. Histone H2A and Ras are chromatin and cytosolic markers, respectively. (I) IRS-1 occupies the Myc promoter in a Wnt-dependent fashion. C2C12 cells were treated for 1 d with vehicle or Wnt3A protein, cross-linked, and processed for ChIP with rabbit IgG or the IRS-1 antibody. (J) Reciprocal coimmunoprecipitation studies of TCF4 and IRS-1 in Wnt3A-treated C2C12 cells. (K) Depletion of IRS-1 impairs Wnt3A-induced chromatin accumulation of β-catenin.
The delay in Myc induction kinetics suggested the possible existence of another Wnt target that is activated earlier and facilitates the subsequent induction of Myc. We therefore examined gene expression patterns in C2C12 cells following Wnt3A treatment using DNA microarrays. We looked for genes that were elevated at least twofold at both 5 h and 24 h after Wnt3A treatment, and compared this group to a recently published list of ∼7000 high-confidence TCF4 chromatin occupancy sites (Hatzis et al. 2008) to identify potential direct TCF4 targets. The overlapping genes' mRNAs were then individually depleted in C2C12 cells with siRNA or shRNA, and treated with Wnt3A to look for a diminution of response. The docking protein insulin receptor substrate-1 (IRS-1) emerged as an attractive candidate, producing a substantial reduction in Wnt-mediated mitochondrial proliferation when depleted by RNAi (Fig. 4D; Supplemental Fig. S23). The IRS-1 mRNA and protein were induced rapidly in both C2C12 cells and MEFs following Wnt3A treatment, and remained elevated throughout (Fig. 4B,C), suggesting that it is likely a direct transcriptional target. The IRS-1 protein was also induced by stable expression of the Wnt3A cDNA—about twofold—and was suppressed by ∼40% when the Dkk-1 cDNA was expressed (Fig. 4E), consistent with the notion that endogenous Wnt signaling regulates the basal expression of IRS-1. In addition, stable expression of the dominant-negative TCF4 cDNA reduced the basal IRS-1 levels and abolished its induction by Wnt3A (Fig. 4F); TCF4 shRNAs likewise attenuated Wnt-dependent IRS-1 induction (Supplemental Fig. S24). Chromatin immunoprecipitation (ChIP) analysis further confirmed that TCF4 is bound to the promoter of IRS-1 (Fig. 4G). Stable overexpression of IRS-1 led to increased mitochondrial proliferation, elevated superoxide levels, and increased Myc transcript in the absence of exogenous Wnt, while depleting IRS-1 reduced the Myc transcript levels and largely prevented its induction with Wnt3A (Supplemental Figs. S25, S26).
We next attempted to understand how IRS-1 increases the Myc transcript. While the cytosolic/membrane localization of IRS-1 in association with the insulin and insulin-like growth factor 1 (IGF-1) receptors is widely known, nuclear accumulation of IRS-1 following stimulation with IGF-1 has also been reported in the literature (Tu et al. 2002). Once inside the nucleus, IRS-1 is capable of associating with and activating specific promoters, although IRS-1 does not contain a DNA-binding motif (Wu et al. 2008). We reasoned that Wnt3A treatment may lead to an analogous set of events if the overall induction of IRS-1 increases the nuclear IRS-1 concentration as a secondary consequence. We in fact saw a pronounced elevation in nuclear IRS-1 following Wnt treatment in both C2C12 cells and MEFs (Fig. 4H; data not shown). Much of the nuclear IRS-1 was chromatin-bound, consistent with a role in transcriptional regulation. We were able to confirm by ChIP analysis the binding of IRS-1 to the Myc promoter in a Wnt-dependent fashion (Fig. 4I). TCF4 also bound the same region of the Myc promoter, which led us to speculate that IRS-1 may associate with the TCF4 transcriptional complex. Reciprocal coimmunoprecipitation experiments using antibodies to IRS-1 and TCF4 revealed a physical interaction between these two proteins (Fig. 4J). Depletion of IRS-1 significantly impaired the chromatin association of the coactivator β-catenin following Wnt3A treatment (Fig. 4K), suggesting that IRS-1 may enhance transcription in part by modulating recruitment of other coactivators.
These data are consistent with the model that Wnt3A rapidly induces IRS-1, which then accumulates in the nucleus and increases transcription from the Myc promoter (Fig. 5D), in part by enhancing the recruitment of β-catenin to the TCF complex occupying this promoter. Myc, in turn, increases mitochondrial proliferation and ROS production. In addition, the cytosolic targets of IRS-1 are also likely to play a significant contributory role.
Figure 5.
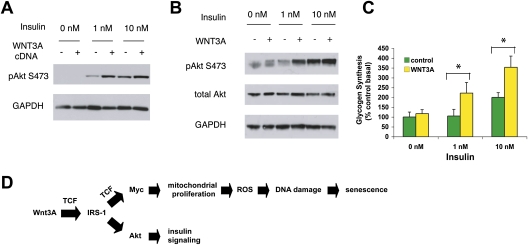
Wnt regulates insulin signaling in muscle cells in culture. (A) Stable expression of Wnt3A cDNA enhances insulin-stimulated Akt activation. C2C12 cells virally transduced to express the Wnt3A cDNA were incubated in serum-free medium for 3 h, treated with different doses of insulin, and examined for the phosphorylation of Akt at Ser473. (B) Treatment with recombinant Wnt3A protein augments insulin-stimulated Akt activation. C2C12 cells were treated with control or 50 ng/mL Wnt3A protein for 1 d, incubated in serum-free medium, and stimulated with insulin. (C) Treatment with Wnt3A protein enhances insulin-stimulated glycogen synthesis in C2C12 myocytes. Cells were treated with control or Wnt3A protein for 1 d, stimulated with varying doses of insulin, incubated in medium containing 14C-glucose, and lysed, and the glycogen was extracted for measurement of incorporated 14C radioactivity. (*) P < 0.05 by unpaired t-test. (D) A model of Wnt3A-mediated regulation of mitochondria and insulin signaling.
Wnt-mediated induction of IRS-1 enhances insulin signaling
Given the role of IRS-1 as a core signaling element in insulin action, we examined whether Wnt-mediated induction of IRS-1 alters insulin action in insulin-responsive cells. Both treatment with recombinant Wnt3A protein and stable expression of Wnt3A cDNA enhanced insulin-stimulated Akt activation in C2C12 cells, as indicated by increased phosphorylation of Akt at Ser473 (Fig. 5A,B). These differences were most pronounced at physiological (<1 nM) concentrations of insulin, although they were still readily appreciable at concentrations as high as 10 nM. In muscle and liver, activated Akt is known to phosphorylate glycogen synthesis kinase 3, and thus stimulate glycogen synthesis, one of the hallmarks of insulin action on cellular glucose metabolism (Taniguchi et al. 2006). Consistent with this, Wnt3A treatment produced significant increases in insulin-stimulated glycogen synthesis in C2C12 myocytes, as assessed by incorporation of radiolabeled glucose into the total cellular glycogen pool (Fig. 5C). These results demonstrate that Wnt-mediated induction of IRS-1 can bring about both enhanced insulin sensitivity and increased oxidative stress in muscle cells, which is notable because these two entities are often viewed as being in opposition (Houstis et al. 2006).
Discussion
In this study, we identified >150 candidate proteins as potential regulators of mitochondrial biogenesis and function. These proteins fall into diverse functional classes, including modifiers of proteins and nucleic acids, signaling molecules, and transcriptional regulators, illustrating the complexity of mitochondrial biology. The disease associations of some of these genes, such as the type 2 diabetes gene WFS1, may help generate novel testable hypotheses about their functions, and serve to further highlight the utility of genetic approaches in studying complex biological processes.
We focused in particular on the functional role of the Wnt signaling pathway in mitochondrial regulation. Wnt signaling has been implicated in numerous critical aspects of physiology and disease, including embryonic development, stem cell self-renewal and aging, and oncogenesis (Clevers 2006). More recently, connections have also been made to the pathogenesis of type 2 diabetes in humans (Grant et al. 2006; Christodoulides et al. 2008), with a common variant of the TCF7L2 gene (also known as TCF4) conferring on individuals an increased risk of developing the disease. We show here for the first time that Wnt signaling strongly activates mitochondrial biogenesis, which in turn produces elevated levels of ROS and oxidative damage. It is tempting to speculate that this effect on mitochondria and ROS generation may be responsible for some of the major biological consequences of aberrantly activated Wnt signaling, such as depletion of stem cells or predisposition to certain tumors. At least in the case of Wnt-induced cellular senescence, ROS appears to be a critical factor, as antioxidant therapy or ectopic SOD2 expression can produce amelioration of the phenotype. It is conceivable that, under other circumstances, chronic exposure to elevated ROS levels predisposes the cell to additional growth-promoting DNA mutations that ultimately lead to cancer.
Our data placing IRS-1 immediately downstream from Wnt activation are especially intriguing given its role as a core signaling molecule in insulin and IGF-1 action. Insulin signaling is known to enhance mitochondrial respiration and mitochondrial protein synthesis (Boirie 2003), and there is a compelling body of evidence implicating the insulin/IGF-1 pathway in the regulation of life span and aging across different species (Russell and Kahn 2007). These ideas together point to previously unappreciated parallels between Wnt-mediated and insulin/IGF-1-mediated cellular aging. Furthermore, we showed that Wnt-mediated induction of IRS-1 can directly modulate insulin signaling, as illustrated by enhancement of insulin-stimulated glycogen synthesis in C2C12 myocytes. Disruption of the TCF4 activity with the dominant-negative allele impairs Wnt-mediated induction of IRS-1 and reduces basal IRS-1 levels, suggesting a potential mechanism by which a loss of TCF4 function may predispose to the development of insulin resistance in target tissues. Of interest, it has been reported recently that a genetic variant near the IRS1 gene is associated with reduced basal levels of the IRS-1 protein and peripheral insulin resistance (Rung et al. 2009). As a caveat, these concepts may not necessarily be applicable in human disease associated with TCF4 polymorphisms, as there could be significant differences between humans and mice regarding the TCF4 protein expression and contribution to physiology at key sites—one example being the β cell (Yi et al. 2005). The enhancement of insulin signaling by Wnt3A in muscle cells is also significant because it raises the possibility of harnessing this pathway to modulate cellular metabolism. The generation of mouse models should enable detailed characterization of potential consequences on systemic glucose and lipid homeostasis. The observed improvement in insulin sensitivity is noteworthy, especially in the context of accompanying increases in ROS generation. It has been suggested that elevated ROS levels cause insulin resistance (Houstis et al. 2006), and thus the present example of Wnt3A signaling in muscle may represent an interesting exception.
As a prototypical adaptor protein, IRS-1 is thought to carry out its function largely by activating the Erk and PI3-kinase pathways. An emerging notion, reinforced by our findings here, is that of IRS-1 as a nuclear transcriptional regulator that responds to extracellular signals. IRS-1 has been reported previously to interact with the estrogen receptor α on estrogen-responsive promoters, and to activate IGF-1-responsive promoters (Morelli et al. 2004; Wu et al. 2008). IRS-1 is known to signal downstream from many classical hormones, including growth hormone, prolactin, and gonadal steroid hormones, as well as various cytokines and growth factors, and it is conceivable that this paradigm of nuclear coactivation by IRS-1 is conserved in other contexts. The mechanism whereby Wnt enhances nuclear localization of IRS-1 remains to be elucidated. We have thus far been unable to find any evidence that IRS-2 undergoes similar regulation in response to Wnt, suggesting a possible basis for functional differences between IRS-1 and IRS-2.
The sequential induction of IRS-1 and Myc following Wnt stimulation provides an example of how a cascade of gene expression can result from activation of an adaptor protein. After the initial induction by Wnt3A and TCF4, IRS-1 accumulates and helps further activate additional TCF4 targets such as Myc, which then triggers expression of a third tier of genes. The induction of a coactivator in this case enables one transcription factor to drive multiple temporal stages of a response.
The functional antagonism of the Wnt3A effect on cultured muscle cells by noncanonical Wnt5A signaling suggests that cross-talk with other regulatory pathways exist, which may be further defined and exploited. A deeper understanding of the interplay between these pathways could lead to new insights into mechanisms of aging, cancer, diabetes mellitus, and other degenerative disorders characterized by mitochondrial dysfunction. Likewise, other genes in our collection of candidate mitochondrial regulators may contribute significantly to aspects of human biology, potentially displaying functional allelic variations in the human population, as is the case with WFS1. Further work on the mitochondrial phenotypes of these genes may uncover important links to physiological and pathological processes associated with mitochondrial abnormalities.
Materials and methods
Constructs and reagents
Human ZBTB33 and dominant-negative TCF4 cDNAs were kindly provided by A. Prokhortchouk and E. Fearon, respectively. The mouse Wnt3A cDNA was from American Type Culture Collection, the SOD2 and Dkk-1 cDNAs were from Open Biosystems, and the IRS-1 and Myc cDNAs were from Addgene. The cDNAs were subcloned into pMSCV or pHAGE expression vectors. Antibodies were purchased from commercial vendors, including TCF4 (Cell Signaling), IRS-1 (Bethyl Laboratories), Myc (Cell Signaling), Wnt3A (Cell Signaling), Histone H2A (Upstate Biotechnologies), Ras (Upstate Biotechnologies), β-catenin (BD Biosciences), ATP synthase subunit α (MitoSciences), Complex I subunit NDUF8 (MitoSciences), GADPH (Santa Cruz Biotechnology), and β-actin (Santa Cruz Biotechnology). Purified Wnt3A, Wnt5A, and Dkk-1 proteins were purchased from R&D Systems. NAC was from Sigma.
Cell culture
C2C12 cells (American Type Culture Collection) were maintained in DMEM containing 10% fetal calf serum (Invitrogen). For differentiation, confluent cells were kept in DMEM with 2% horse serum for 3 d. Primary MEFs were obtained at passage 1 (Stem Cell Technologies) and maintained in DMEM with 10% fetal calf serum in a low-oxygen (3%) incubator.
siRNA screen
An arrayed siRNA library targeting 6363 genes in the mouse genome was obtained from Dharmacon, and was reformatted into 96-well plates. The siRNA pools, consisting of four distinct siRNAs per gene, were transiently transfected into C2C12 cells with Lipofectamine 2000 (Invitrogen) at a 100 nM final concentration. After 5 d, cells were stained with MitoTracker Green and MitoTracker Red CMXRos (Invitrogen) for 1 h, washed with PBS, and analyzed using a BD LSRII flow cytometer equipped with a high-throughput sampler designed for 96-well plates. The MitoTracker Green signal was collected using the FITC channel at 530 nm, while the MitoTracker Red signal was detected using the PerCP channel at 660 nm. Cell counts, forward scatter, and side scatter were also collected. Firefly luciferase siRNA, which lacks significant homology with known mouse genes, and a nontargeting control siRNA were present on each plate. A fluorescently labeled siGLO oligonucleotide and the cytotoxic Plk1 siRNA served as visual and functional transfection controls. Ccnd1 siRNA and Sin3A siRNA pools were used as positive controls. The screen was performed in duplicate. The replicate MitroTracker Green and MitroTracker Red values and the Red-to-Green ratios were normalized to the plate mean, and an average increase >40% or a decrease >20% in any of the three parameters was chosen as a cutoff for the primary hit list. The intention was to include as many candidates as possible at the initial stage, with both positive and negative regulators represented. This hit list was then filtered for correlated toxicity, defined as a reduction >80% in cell count relative to control during a set sampling period. This resulted in 345 candidates in the primary hit list that were then subjected to the validation round. The setup of the validation round was essentially identical, except that the four individual oligonucleotides from each pool used in the primary screen were placed into separate wells. The mean MitoTracker signals from duplicate wells were compared with the mean signals from the wells receiving a control siRNA, arbitrarily set at 5000. An individual siRNA was considered to score if it sufficiently altered the MitoTracker Green signal (<4000 or >7000), MitoTracker Red signal (<4000 or >9000), or the Red-to-Green ratio (<0.9 or >1.4). Validation of a gene required at least two out of four independent siRNAs scoring in this fashion, with a phenotype consistent with that from the primary screen. This resulted in 176 validated hits, for a 51% confirmation rate.
Classification of screen hits
Hits were classified into molecular function and biological process categories using the Panther classification system (Thomas et al. 2003). Statistical enrichment was assessed using the hypergeometric distribution, and a Benjamini and Hochberg-adjusted P-value of <0.05 was considered significant.
Extension of the WNT network
The network was constructed using human protein interaction data (Keshava et al. 2009), and anchoring on components of the canonical WNT signaling pathway (KEGG ID: hsa04310). Screen hits were mapped to human orthologs using mapping data from NCBI. The network uses graph theoretic representations—which abstract components (gene products) as nodes, and interactions (Bejamini and Hochberg-adjusted P-value of <0.05) between components as edges—implemented in the Perl programming language.
Viral packaging and infection
Viral supernatants were prepared by transfecting 293T cells with a retroviral or lentiviral vector along with packaging plasmids. pMSCV-PM and pLKO shRNA vectors (Open Biosystems) were used for gene knockdown, while pMSCV and pHAGE vectors were used for overexpression. The control shRNAs 1 and 2 included a nontargeting scrambled sequence (CCTTCGATTCCCTCAAAGACA) or no hairpin insert. ZBTB33 shRNAs 1 and 2 included the hairpin sequences CGTGTTAATTTGATCTACCAT and GCCACCAAACAAGCGTATGAA, respectively. IRS-1 shRNAs 1 and 2 included the sequences CGGAACAATTAGTGTGCATAA and CGGTCCTCTCTTACTACTCAT. Myc shRNAs 1 and 2 included the hairpin sequences GCTTCGAAACTCTGGTGCATA and CATCCTATGTTGCGGTCGCTA. POLG shRNA included the hairpin sequence CGATACTATGAGCATGCACAT. Viruses were collected 48 h after transfection, filtered with a 0.45-μm filter, and used to infect C2C12 cells or MEFs in the presence of 5 μg/mL polybrene. The transduced cell populations were selected with 2 μg/mL puromycin.
Isolation of intact mitochondria
Cells were washed with cold PBS, collected, resuspended in ice-cold isolation buffer (10 mM Tris-MOPS, 1 mM EGTA, 200 mM sucrose), and homogenized with a glass-Teflon motorized homogenizer. Mitochondria were obtained using a standard density centrifugation protocol and were used immediately.
Oxygen consumption measurement
Real-time measurements of oxygen tension and pH in cultured cells were obtained in 24-well microplates using the XF24 flux analyzer (Seahorse Bioscience). The oxygen consumption rate and the proton extrusion rates were calculated from these measurements and normalized to protein content from cell lysates. When appropriate,DNP (final concentration 40 μM) or oligomycin (final concentration 2 μg/mL) was added to assess the maximum respiratory capacity and the inner mitochondrial membrane proton leak, respectively. For measurement of mitochondrial respiration, isolated mitochondria were diluted in mitochondrial assay solution (115 mM KCl, 10 mM KH2PO4, 2 mM MgCl2, 3 mM HEPES, 1 mM EGTA, 0.2% fatty acid-free BSA) and attached to the bottom of the 24-well microplates by centrifugation at 3000g for 20 min, and the oxygen tension was recorded using the XF24 flux analyzer. State III respiration was measured by adding 0.25 mM ADP in the presence of 5 mM succinate.
ROS and oxidative DNA damage measurements
For quantitation of superoxide or peroxide levels, cells were stained with MitoSox Red or CM-H2CFDA (Invitrogen) according to instructions and analyzed with the LSRII flow cytometer (BD Biosciences). As an alternative method based on xylenol orange–iron complex formation, cells were washed, lysed, and assayed with Peroxide Assay Kit (Biomedical Research Service). For quantitation of oxidative DNA damage, cells were dissociated, fixed, permeabilized overnight, stained with an 8-oxoguanine antibody in the OxyDNA Kit (BD Biosciences), and analyzed by flow cytometry.
Cellular senescence assays
BrdU assays were performed with a BrdU Labeling Kit (BD Biosciences) according to the manufacturer's instructions. Briefly, cells were pulsed with 0.1 mg/mL BrdU for 1 h, fixed, permeabilized, stained with anti-BrdU-FITC antibody, and analyzed by flow cytometry to measure the number of cells in S phase. SAβ-gal staining was performed with a β-galactosidase Staining Kit (Sigma). Percentage positive staining was determined by dividing the number of β-gal-positive cells into the total cell number within 10 random fields from duplicate dishes.
Mitochondrial DNA measurement
Total cellular DNA was isolated from cells or tissues with DNAeasy Blood and Tissue Kit (Qiagen). Mitochondrial DNA content was determined by quantitative real-time PCR by comparing the mitochondrially encoded Cox2 gene to an intron of the nuclear-encoded β-globin gene. An Applied Biosystems Prism 7500 Sequence Detection System and a SYBR Green PCR Kit were used.
Mitochondrial gene expression
Total RNA was extracted from cells or tissues with either an RNAeasy Plus RNA Isolation Kit (Qiagen) or Trizol (Invitrogen). cDNA was prepared using SuperScript III RT (Invitrogen), and was subjected to quantitative real-time PCR using a SYBR Green PCR Kit on an Applied Biosystems 7500 Sequence Detection System. Primer sequences used for real-time PCR are provided in Supplemental Table S2.
Citrate synthase assay
Cells were lysed and processed using the reagents provided in the Citrate Synthase Assay Kit (Sigma). A colorimetric reaction was used to measure the rate of reaction between acetyl coenzyme A and oxaloacetic acid.
Electron microscopy
The samples were fixed for 1 h in a mixture of 2.5% glutaraldehyde and 2% formaldehyde in 0.1 M sodium cacodylate buffer (pH 7.4), washed with 0.1 M cacodylate buffer, post-fixed with 1% osmium tetroxide/1.5% potassium ferrocyanide for 1 h, washed with water, stained with 1% uranyl acetate, washed again with water, and dehydrated with 70% ethanol for 15 min, 90% ethanol for 15 min, and then 100% ethanol for 15 min twice. Samples were infiltrated and embedded in Epon, and ultrathin sections were cut on a microtome, placed on copper grids, and viewed on a Tecnai G2 microscope. The quantification of mitochondrial volume density and cristae surface density was performed as described (Weibel 1979).
Microarray analysis
RNA samples from Wnt3A-treated C2C12 cells were reverse-transcribed and used to generate Cy3- and Cy5-labeled cRNA using the Two-Color Linear Amplification Kit (Agilent). Mouse gene expression microarray (Agilent) was hybridized, scanned, and feature-extracted following the manufacturer's instructions.
Western analysis
Whole-cell extracts were prepared by cell lysis, and equal amounts of lysates were resolved by SDS/PAGE, transferred to Immobilon-P membrane (Millipore), and probed with the appropriate antibodies. The proteins were visualized by ECL chemiluminescence (Pierce).
Coimmunoprecipitation
C2C12 cells were treated with 0.1% BSA in PBS (control) or 50 ng/mL Wnt3A for 24 h, washed with PBS, and lysed in 1× lysis buffer (20 mM Tris-HCl at pH 8, 137 mM NaCl, 10% glycerol, 1% NP-40, 2 mM EDTA). Immunoprecipitation with IRS-1 antibody, TCF4 antibody, or rabbit IgG was performed using protein A-conjugated beads as described previously.
ChIP
Chromatin/protein complexes were prepared from C2C12 cells treated with 0.1% BSA in PBS or 50 ng/mL Wnt3A for 24 h. Chemical cross-linking was carried out as described previously (Cunningham et al. 2007), consisting of a 20-min treatment with 1.5 mM DSP at 37°C prior to a 10-min treatment with 1% formaldehyde at room temperature. The DNA was enzymatically sheared with Micrococcal nuclease to generate fragments of 150–900 base pairs (bp). Following immunoprecipitation, purified DNA was analyzed by PCR. Ratios of input DNA to bound DNA were obtained, and data were normalized to the Wnt-treated sample. Primer sequences used for PCR amplification of the Myc and IRS-1 promoters are provided in Supplemental Table S10.
Cell fractionation
Chromatin fractionations were performed as described previously (Zou et al. 2002). Briefly, 3 × 106 cells were washed and resuspended in a HEPES-based buffer containing protease and phosphatase inhibitors and were lysed with 0.1% Triton, and cytoplasmic proteins were separated from nuclei by low-speed centrifugation. Isolated nuclei were subsequently lysed, and soluble nuclear proteins were separated from chromatin by multiple centrifugation steps.
Glycogen synthesis assay
C2C12 cells grown in six-well plates were treated with 50 ng/mL Wnt3A for 24 h and incubated in serum-free DMEM for 4 h. Cells were washed gently with warm Krebs-Ringer-HEPES-BSA (KRHB) buffer, incubated in 1 mL of KRHB buffer for 15 min, and stimulated with an appropriate concentration of insulin for 1 h. A mixture of 14C-glucose (specific activity 250 mCi/mmol) and deoxyglucose was added to cells to the final concentrations of 2.5 uCi/mL and 5 mM, respectively, which were then incubated for another hour at 37°C. Cells were washed three times with ice-cold PBS, lysed by adding 300 μL of 1 M KOH, incubated for 30 min at room temperature, vortexed, boiled for 30 min, and cooled on ice. After removing a small aliquot for protein assay, 50 μL of glycogen carrier and 900 μL of cold ethanol were added, and the samples were kept overnight at −20°C before being spun at 17,000g for 10 min. The glycogen pellet was washed with cold ethanol and resuspended in water, and the incorporation of radioactivity was measured using a scintillation counter. The glycogen synthesis rate was normalized to protein content.
Acknowledgments
We thank J. Cunningham, P. Puigserver, and members of the Elledge laboratory for advice and discussions; M. Rosene and Y. Leng for technical assistance; and the ICCB and the EM Core for assistance with the siRNA library and electron microscopy, respectively. This work was supported by a fellowship from the Damon Runyon Cancer Research Foundation awarded to J.C.Y.; a fellowship from the Crohn's and Colitis Foundation of America to A.N.; NIH career development grants to J.C.Y. and B.H.K.; NIH research grants to A.B., R.J.X., and S.J.E.; and a CMRC grant to S.J.E. S.J.E. is an Investigator with the Howard Hughes Medical Institute.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1924910.
Supplemental material is available at http://www.genesdev.org.
References
- Boirie Y 2003. Insulin regulation of mitochondrial proteins and oxidative phosphorylation in human muscle. Trends Endocrinol Metab 14: 393–394 [DOI] [PubMed] [Google Scholar]
- Brack AS, Conboy MJ, Roy S, Lee M, Kuo CJ, Keller C, Rando TA 2007. Increased Wnt signaling during aging alters muscle stem cell fate and increases fibrosis. Science 317: 803–810 [DOI] [PubMed] [Google Scholar]
- Christodoulides C, Lagathu C, Sethi J, Vidal-Puig A 2008. Adipogenesis and WNT signaling. Trends Endocrinol Metab 20: 16–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H 2006. Wnt/β-catenin signaling in development and disease. Cell 127: 469–480 [DOI] [PubMed] [Google Scholar]
- Cunningham JT, Rodgers JT, Arlow DH, Vazquez F, Mootha VK, Puigserver P 2007. mTOR controls mitochondrial oxidative function through a YY1–PGC1α transcriptional complex. Nature 450: 736–740 [DOI] [PubMed] [Google Scholar]
- DiMauro S, Schon EA 2003. Mitochondrial respiratory-chain diseases. N Engl J Med 348: 2656–2668 [DOI] [PubMed] [Google Scholar]
- Grant SF, Thorleifsson G, Reynisdottir I, Benediktsson R, Manolescu A, Sainz J, Helgason A, Stefansson H, Emilsson V, Helgadottir A, et al. 2006. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet 38: 320–323 [DOI] [PubMed] [Google Scholar]
- Hatzis P, van der Flier LG, van Driel MA, Guryev V, Nielsen F, Denissov S, Nijman IJ, Koster J, Santo EE, Welboren W, et al. 2008. Genome-wide pattern of TCF7L2/TCF4 chromatin occupancy in colorectal cancer cells. Mol Cell Biol 28: 2732–2744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He TC, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT, Morin PJ, Vogelstein B, Kinzler KW 1998. Identification of c-MYC as a target of the APC pathway. Science 281: 1509–1512 [DOI] [PubMed] [Google Scholar]
- Houstis N, Rosen ED, Lander ES 2006. Reactive oxygen species have a causal role in multiple forms of insulin resistance. Nature 440: 944–948 [DOI] [PubMed] [Google Scholar]
- Ishitani T, Kishida S, Hyodo-Miura J, Ueno N, Yasuda J, Waterman M, Shibuya H, Moon RT, Ninomiya-Tsuji J, Matsuomoto K 2003. The TAK1–NLK mitogen activated protein kinase cascade functions in the Wnt-5a/Ca2+ pathway to antagonize Wnt/β-catenin signaling. Mol Cell Biol 22: 131–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshava PTS, Goel R, Kandasamy K, Keerthikumar S, Kumar S, Mathivanan S, Telikicheria D, Raju R, Shafreen B, Venugopal A, et al. 2009. Human protein reference database—2009 update. Nucleic Acids Res 37: D767–D772 doi: 10.1093/nar/gkn892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Lee JH, Iyer VR 2008. Global identification of Myc target genes reveals its direct role in mitochondrial biogenesis and its E-box usage in vivo. PLoS One 3: e1798 doi: 10.1371/journal.pone.0001798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroemer G 2006. Mitochondria in cancer. Oncogene 25: 4630–4632 [DOI] [PubMed] [Google Scholar]
- Li F, Wang Y, Zeller KI, Potter JJ, Wonsey DR, O'Donnell KA, Kim JW, Yustein JT, Lee LA, Dang CV 2005. Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis. Mol Cell Biol 25: 6225–6234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MT, Beal MF 2006. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature 443: 787–795 [DOI] [PubMed] [Google Scholar]
- Liu H, Fergusson MM, Castilho RM, Liu J, Cao L, Chen J, Malide D, Rovira II, Schimel D, Kuo J, et al. 2007. Augmented Wnt signaling in a mammalian model of accelerated aging. Science 317: 803–806 [DOI] [PubMed] [Google Scholar]
- Loeb LA, Wallace DC, Martin GM 2005. The mitochondrial theory of aging and its relationship to reactive oxygen species damage and somatic mtDNA mutations. Proc Natl Acad Sci 102: 18769–18770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowell BB, Shulman GI 2005. Mitochondrial dysfunction and type 2 diabetes. Science 307: 384–387 [DOI] [PubMed] [Google Scholar]
- Moon RT, Kohn AD, De Ferrari GV, Kaykas A 2004. Wnt and β-catenin signaling: Diseases and therapies. Nat Rev Genet 5: 689–699 [DOI] [PubMed] [Google Scholar]
- Morelli C, Garfalo C, Sisci D, del Rincon S, Cascio S, Tu X, Vecchione A, Sauter ER, Miller WH Jr, Surmacz E 2004. Nuclear insulin receptor substrate 1 interacts with estrogen receptor α at ERE promoters. Oncogene 23: 7517–7526 [DOI] [PubMed] [Google Scholar]
- Park JI, Kim SW, Lyons JP, Ji H, Nguyen TT, Cho K, Barton MC, Deroo T, Vleminckx K, Moon RT, et al. 2005. Kaiso/p120-catenin and TCF/β-catenin complexes coordinately regulate canonical Wnt gene targets. Dev Cell 8: 843–854 [DOI] [PubMed] [Google Scholar]
- Parrinello S, Samper E, Krtolica A, Goldstein J, Melov S, Campisi J 2003. Oxygen sensitivity severely limits the replicative lifespan of murine fibroblasts. Nat Cell Biol 5: 741–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendergrass W, Wolf N, Poot M 2004. Efficacy of MitoTracker Green and CMXRosamine to measure changes in mitochondrial membrane potentials in living cells and tissues. Cytometry A 61: 162–169 [DOI] [PubMed] [Google Scholar]
- Pile L, Spellman PT, Katzenberger RJ, Wassarman DA 2003. The SIN3 deacetylase complex represses genes encoding mitochondrial proteins. J Biol Chem 278: 37840–37848 [DOI] [PubMed] [Google Scholar]
- Poot M, Pierce RH 2001. Analysis of mitochondria by flow cytometry. Methods Cell Biol 64: 118–128 [DOI] [PubMed] [Google Scholar]
- Rung J, Cauchi S, Albrechtsen A, Shen L, Rocheleau G, Cavalcanti-Proenca C, Bacot F, Balkau B, Bellisle A, Broch-Johnsen K, et al. 2009. Genetic variant near IRS1 is associated with type 2 diabetes, insulin resistance, and hyperinsulinemia. Nat Genet 41: 1110–1115 [DOI] [PubMed] [Google Scholar]
- Russell SJ, Kahn CR 2007. Endocrine regulation of ageing. Nat Rev Mol Cell Biol 8: 681–691 [DOI] [PubMed] [Google Scholar]
- Ruzov A, Dunican DS, Prokhortchouk A, Pennings S, Stancheva I, Prokhortchouk E, Meehan RR 2004. Kaiso is a genome-wide repressor of transcription that is essential for amphibian development. Development 131: 6185–6194 [DOI] [PubMed] [Google Scholar]
- Sandhu MS, Weedon MN, Fawcett KA, Waasson J, Debenham SL, Daly A, Lango H, Frayling TM, Neumann RJ, Sherva R, et al. 2007. Common variants in WFS1 confer risk of type 2 diabetes. Nat Genet 39: 951–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi CM, Emanuelli B, Kahn CR 2006. Critical nodes in signaling pathways: Insights into insulin action. Nat Rev Mol Cell Biol 7: 85–96 [DOI] [PubMed] [Google Scholar]
- Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, Diemer K, Muruganujan A, Narechania A 2003. Panther: A library of protein families and subfamilies indexed by function. Genome Res 13: 2129–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topol L, Jiang X, Choi H, Garrett-Beal L, Carolan PJ, Yang Y 2003. Wnt-5a inhibits the canonical Wnt pathway by promoting GSK-3-independent β-catenin degradation. J Cell Biol 162: 899–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu X, Batta P, Innocent N, Prisco M, Casaburi I, Belletti B, Baserga R 2002. Nuclear translocation of insulin receptor substrate-1 by oncogenes and Igf-1. J Biol Chem 277: 44357–44365 [DOI] [PubMed] [Google Scholar]
- Vafa O, Wade M, Kern S, Beeche M, Pandita TK, Hampton GM, Wahl GM 2002. c-Myc can induce DNA damage, increase reactive oxygen species, and mitigate p53 function: A mechanism for oncogene-induced genetic instability. Mol Cell 9: 1031–1044 [DOI] [PubMed] [Google Scholar]
- Wallace DC 2005. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: A dawn for evolutionary medicine. Annu Rev Genet 39: 359–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Li Z, Lu Y, Du R, Katiyar S, Yang J, Fu M, Leader JE, Quong A, Novikoff PM, et al. 2006. Cyclin D1 repression of nuclear factor 1 integrates nuclear DNA synthesis and mitochondrial function. Proc Natl Acad Sci 103: 11567–11572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weibel E 1979. Stereological methods: Practical methods for biological morphometry Academic Press, London [Google Scholar]
- Wu A, Chen J, Baserga R 2008. Nuclear insulin receptor substrate-1 activates promoters of cell cycle progression genes. Oncogene 27: 397–403 [DOI] [PubMed] [Google Scholar]
- Yi F, Brubaker PL, Jin T 2005. TCF4 mediates cell type-specific regulation of proglucagon gene expression by β-catenin and glycogen synthase kinase-3β. J Biol Chem 280: 1457–1464 [DOI] [PubMed] [Google Scholar]
- Zou L, Cortez D, Elledge SJ 2002. Regulation of ATR substrate selection by Rad17- dependent loading of Rad9 complexes onto chromatin. Genes Dev 16: 198–208 [DOI] [PMC free article] [PubMed] [Google Scholar]