Abstract
Hox proteins frequently select and regulate their specific target genes with the help of cofactors like Extradenticle (Exd) and Homothorax (Hth). For the Drosophila Hox protein Sex combs reduced (Scr), Exd has been shown to position a normally unstructured portion of Scr so that two basic amino acid side chains can insert into the minor groove of an Scr-specific DNA-binding site. Here we provide evidence that another Drosophila Hox protein, Deformed (Dfd), uses a very similar mechanism to achieve specificity in vivo, thus generalizing this mechanism. Furthermore, we show that subtle differences in the way Dfd and Scr recognize their specific binding sites, in conjunction with non-DNA-binding domains, influence whether the target gene is transcriptionally activated or repressed. These results suggest that the interaction between these DNA-binding proteins and the DNA-binding site determines the architecture of the Hox–cofactor–DNA ternary complex, which in turn determines whether the complex recruits coactivators or corepressors.
Keywords: DNA binding, Hox genes, activity regulation, homeodomain proteins, transcriptional regulation
A long-standing problem in the field of transcriptional regulation is to understand how different transcription factors select and correctly regulate their target genes. This problem can be conceptually divided into two questions: How do transcription factors specifically recognize their relevant DNA-binding sites (Garvie and Wolberger 2001; Rohs et al. 2010), and how do transcription factor–DNA complexes activate or repress their target genes? There is compelling evidence that these two events may not be independent, and that the DNA-binding site plays a role in both steps (Lefstin and Yamamoto 1998; Leung et al. 2004; Meijsing et al. 2009). Studies with NF-κB (Leung et al. 2004) and glucocorticoid receptor (Meijsing et al. 2009) have reinforced this idea. For example, the quality of the DNA-binding site for both the glucocorticoid receptor and NF-κB affects the regulatory output of the bound complex, suggesting that small structural differences imposed on the protein by DNA binding affects transcriptional output. Nevertheless, although DNA-binding sites may influence the regulatory activity of bound factors, the first step is the recognition of these binding sites—a process that is still not fully understood for many transcription factors (Garvie and Wolberger 2001; Rohs et al. 2010).
The problem of specificity becomes more complex for transcription factors that have very similar DNA-binding domains. For example, the homeotic selector (Hox) genes encode a highly conserved and developmentally important family of transcription factors with very similar DNA-binding homeodomains (Mann et al. 2009). Hox genes play an important role in anterior–posterior (AP) axis determination across animal phyla from insects to vertebrates. Because of their similar homeodomains, Hox proteins bind similar, AT-rich DNA-binding sites in vitro (Gehring et al. 1994; Berger et al. 2008; Noyes et al. 2008). However, in vivo, these factors execute highly specific functions by selecting and regulating different sets of downstream target genes (Gehring et al. 1994; Pearson et al. 2005). Studying these factors provides the opportunity to understand the molecular basis of specificity executed by similar members of the same transcription factor family during animal development.
Hox target genes can be categorized into two general categories: paralog-specific and shared target genes (Mann et al. 2009). Paralog-specific target genes are regulated by only one or a small subset of Hox proteins. Regulation of forkhead (fkh) (Ryoo and Mann 1999) and decapentaplegic (dpp) (Capovilla et al. 1994; Chan et al. 1994; Manak et al. 1994) and autoactivation loops by the Hox genes labial (lab) (Grieder et al. 1997; Ryoo et al. 1999; Marty et al. 2001) and Deformed (Dfd) (Kuziora and McGinnis 1988; Bergson and McGinnis 1990) are examples of paralog-specific interactions. Repression of Distalless (Dll) by abdominal Hox genes (Vachon et al. 1992; Gebelein et al. 2002), but not more anterior Hox genes, represents a semi-paralog-specific target gene (Mann et al. 2009). In contrast, the head-specifying gene optix is repressed by most Hox genes, and provides an example of a shared Hox function (Coiffier et al. 2008). In principle, because a high degree of specificity is required for only paralog-specific target gene regulation, the types of binding sites used by Hox proteins to regulate these distinct types of target genes may be different (Mann et al. 2009).
For Hox proteins, highly specific DNA binding often requires an interaction with two other homeodomain-containing transcription factors: Extradenticle (Exd in Drosophila; Pbx in vertebrates) and Homothorax (Hth in Drosophila and Meis in vertebrates) (Mann and Chan 1996; Mann and Affolter 1998; Moens and Selleri 2006). Biochemical studies have established that Exd binds to DNA cooperatively with Hox proteins to composite Exd–Hox-binding sites that have the general structure TGATN2-3ATN2, where the 5′ TGAT is the Exd half-site (Chan and Mann 1996; Mann et al. 2009). Hox–Exd–DNA complex assembly is enhanced by Hth, which is also required for Exd's nuclear localization and remains associated with Exd in the nucleus (Rieckhof et al. 1997; Abu-Shaar et al. 1999). There are at least two isoforms of Hth: a full-length version containing a homeodomain, and a shorter isoform without a homeodomain (Noro et al. 2006). When the Hox–cofactor complex includes full-length Hth, the target DNA may include a Hth-binding site, present at a variable distance from the Hox–Exd composite site, thus providing additional DNA-binding affinity and specificity (Ryoo et al. 1999; Gebelein et al. 2004; Mann et al. 2009).
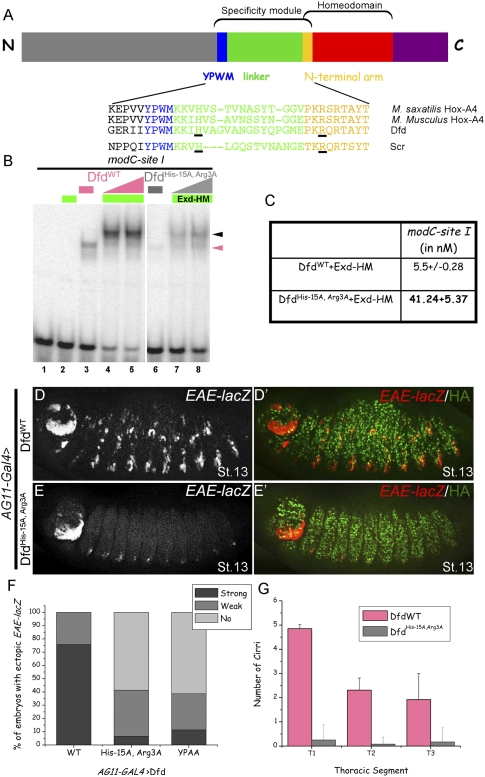
Previous work elucidated the molecular basis by which the Drosophila Hox protein Sex combs reduced (Scr) binds to its paralog-specific target gene, fkh (Joshi et al. 2007). In this case, Scr recognizes its specific binding site in fkh, called fkh250, in part by binding a region of the DNA where the minor groove is unusually narrow. The recognition of fkh250's narrow minor groove, which is an intrinsic feature of this DNA sequence, depends on a protein–protein interaction between Scr and Exd that positions a normally unstructured region of Scr in the minor groove. Two charged residues, a histidine and arginine, in this unstructured region help Scr to read the minor groove topography of fkh250. Like Scr, nearly all Hox proteins have the ability to interact with Exd through a conserved YPWM motif (Mann and Morata 2000). Interestingly, the residues surrounding Hox YPWM motifs, which include the minor groove-interacting His residue of Scr, are conserved in a paralog-specific manner, suggesting that they play a role in specificity (Joshi et al. 2007). Similarly, the N-terminal arms of Hox homeodomains, which in Scr includes two minor groove-inserting Arg residues, are also highly conserved in a paralog-specific manner (Joshi et al. 2007). In another Hox protein, these sequences have also been shown to influence monomeric binding (Liu et al. 2008, 2009). In addition, a recent study showed that sequences N-terminal to the YPWM motif of Scr are largely dispensable for Scr to carry out its specific functions in vivo (Papadopoulos et al. 2010). Based on these observations, the region of a Hox protein from its YPWM motif through the N-terminal arm of its homeodomain, together with the intervening linker region, appears to play a critical role in specificity; we refer to this region as the specificity module (Fig. 3A, below). The paralog-specific conservation of Hox specificity modules, together with functional studies, suggests a general model in which paralog-specific DNA-binding sites are recognized in part by specificity module residues in an Exd-dependent manner.
Figure 3.
Dfd specificity module residues His-15 and Arg3 are required in vitro and in vivo for Dfd-specific functions. (A) Schematic showing Hox specificity module, homeodomain, YPWM motif, and linker. Below are alignments of specificity modules from Dfd orthologs, showing the conserved histidine and an arginine (underlined) at positions analogous to those in Scr's specificity module (Joshi et al. 2007). (B) DfdHis-15A, Arg3A binding to site I is weaker both as a monomer (lane 6) and as cooperative trimer (lanes 7,8) with Exd–HM in comparison with DfdWT (lanes 3–5). Hox monomer and Hox–Exd–HM trimer complexes are indicated by pink and black arrowheads, respectively. (C) Kd measurements for DfdWT–Exd–HM and DfdHis-15A, Arg3A–Exd–HM to site I show an eightfold difference in binding affinity. (D,E) Ubiquitous expression of DfdWT results in ectopic activation of EAE-lacZ (D), while DfdHis-15A, Arg3A (E) is severely compromised in its ability to induce this reporter gene (see F for quantification). (F) Graph showing percentage of total embryos that activate EAE-lacZ ectopically when DfdWT, DfdHis-15A, Arg3A, or DfdYPAA is expressed ubiquitously using AG11-Gal4 (n ≥ 56 for each genotype, n = 2). (G) Graph showing the quantification of ectopic cirri made in thoracic segments of embryonic cuticles when DfdWT or DfdHis-15A, Arg3A is expressed ubiquitously using AG11-Gal4. Error bars represent standard deviations.
In the current study, we test the generality of this model by studying the Hox gene Dfd. During embryogenesis, Dfd is expressed in the maxillary and mandibular segments, which lie adjacent and anterior to the Scr-expressing labial segment. The positive autoregulation of Dfd in the epidermis is the best-characterized paralog-specific target known for Dfd. A 2.7-kb DNA element from Dfd called EAE (epidermal autoregulatory element) mediates this autoregulation (Bergson and McGinnis 1990), and a 570-base-pair (bp) subfragment of EAE called module C (modC) recapitulates expression of the full-length 2.7-kb EAE (Zeng et al. 1994; Pinsonneault et al. 1997). In vivo, this autoregulatory loop has been shown to require both Dfd and exd, but the molecular basis of the Exd input has not been examined. Here we show that, analogous to the regulation of fkh by Scr, the specificity module of Dfd plays a critical role in Dfd autoregulation in an Exd-dependent manner. Interestingly, we also found that Dfd can repress the Scr target, fkh250-lacZ, and that this repression does not require an interaction with Exd or key residues in the specificity module. The difference between Dfd's ability to activate EAE-lacZ and repress fkh250-lacZ depends both on the quality of its specificity module (and thus its ability to recruit Exd) and on sequence motifs close to the N terminus of Dfd. Dfd takes on the properties of Scr (such as activating fkh250-lacZ) only when it has Scr's specificity module and these N-terminal motifs are deleted. Together, in addition to generalizing the role of the Hox specificity module, these data suggest that subtle differences in transcription factor–DNA interactions can have profound consequences on gene regulation in vivo.
Results
Analysis of modC from Dfd
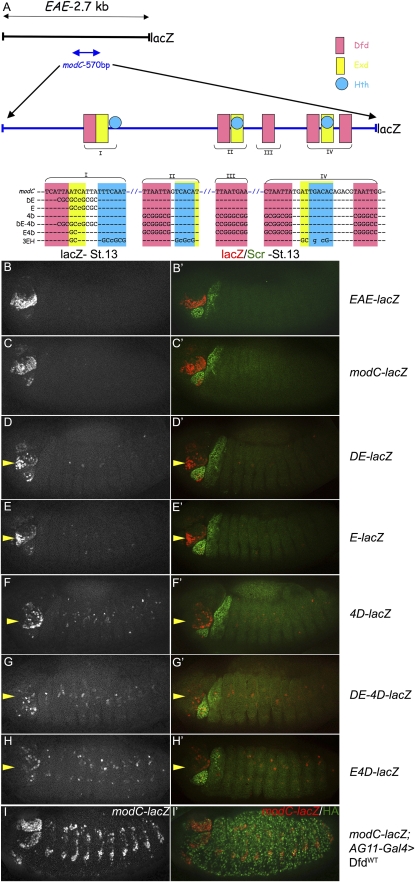
The epidermal expression of Dfd in maxillary and mandibular segments is maintained via an autoregulatory loop mediated by a 2.7-kb EAE enhancer element (Fig. 1A,B; Kuziora and McGinnis 1988; Bergson and McGinnis 1990). A 570-bp subfragment of this enhancer, referred to as modC, recapitulates the expression pattern of the full 2.7-kb element (modC-lacZ) (Fig. 1C; Zeng et al. 1994; Pinsonneault et al. 1997). modC-lacZ, like EAE-lacZ, can be activated in response to ectopic Dfd expression, confirming that it behaves as an autoregulatory element (Fig. 1I).
Figure 1.
Analysis of modC from Dfd. (A) Schematic of 2.7-kb EAE-lacZ and 570-bp modC-lacZ. Sequence of Dfd-, Exd-, and Hth-binding sites in modC are color-coded. Mutant versions of modC are shown below the wild-type sequence. The Dfd–Exd composite binding site (site I) and nearby Hth-binding site is labeled “I.” Other combinations of Dfd-, Exd-, and Hth-binding sites are indicated as II, III, and IV. Note that Exd-binding sites often overlap with Hth-binding sites. (B,C) Epidermal expression of EAE-lacZ (B) and modC-lacZ (C) in maxillary and mandibular segments of the embryo. In these and all subsequent images, costaining for Scr in the right panels is used to identify the adjacent Dfd-expressing segments. Anterior is always to the left, and all embryos are at stage 13. (D,E) Mutagenesis of Dfd–Exd (D) or Exd half-site (E) of site I in modC results in a modest decrease in reporter gene activity (DE-lacZ and E-lacZ, respectively). In these and subsequent images, the yellow arrowheads point to the maxillary segments. (F) Mutagenesis of four Dfd-binding sites in the modC enhancer significantly decreases enhancer activity (4D-lacZ). (G) Mutagenesis of the Dfd–Exd site from site I and four Dfd-binding sites abolishes enhancer activity (DE-4D-lacZ). (H) Mutagenesis of the Exd half-site from site I and four Dfd-binding sites abolishes enhancer activity (E4D-lacZ). (I) modC-lacZ is induced in response to ectopic Dfd by AG11-Gal4.
To identify potential Dfd-binding sites in modC, we used yeast one-hybrid data (Noyes et al. 2008) to generate a position weight matrix of monomeric Dfd-binding sites, and searched modC using TargetExplorer (Sosinsky et al. 2003). We found five Dfd-binding sites in modC. All of these sites bound to Dfd protein, as confirmed using electorphoretic mobility shift assays (EMSAs) (data not shown), and are conserved in 12 Drosophila species (Supplemental Fig. 1A). One of these binding sites has an adjacent Exd-binding site, creating a composite Exd–Hox site (5′-TGATTAATGA-3′) (Fig. 1A, site I).
To test the contribution of these binding sites to the activity of modC, we mutated them and tested the activities of the mutant fragments in an in vivo reporter gene assay. Mutating both the Dfd and Exd half-sites of site I (DE-lacZ) resulted in a decrease in, but not the complete absence of, reporter gene activity (Fig. 1D). A similar level of activity was observed when only the Exd-binding site of site I was mutated (E-lacZ) (Fig. 1E). Mutagenesis of the other four Dfd-binding sites in modC resulted in a greater reduction, but still not the elimination, of reporter gene activity (4D-lacZ) (Fig. 1F). Activity was eliminated only when site I, in addition to the remaining four Dfd-binding sites, was mutated (DE-4D-lacZ; 82% of the embryos showed only background expression) (Fig. 1G). Significantly, mutagenesis of just the Exd-binding site in site I, along with the four remaining Dfd-binding sites, abolished the activity of the enhancer in the maxillary segment (E-4D-lacZ) (Fig. 1H). Together, these results suggest that site I is necessary but not sufficient for full modC activity, and that both the Dfd and Exd inputs are required.
modC-lacZ requires direct Exd–Hth input for activity
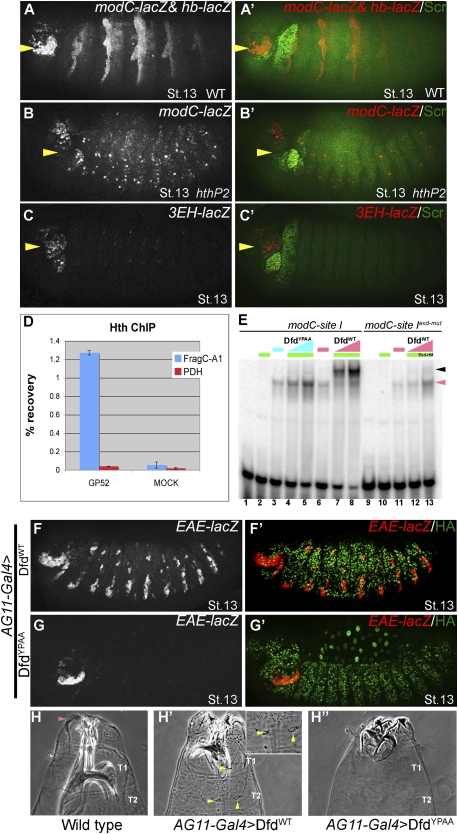
To further test the Exd input into modC, we took three approaches. First, we examined the expression of modC-lacZ in hth mutant embryos (hthP2), where Exd fails to enter the nucleus (Rieckhof et al. 1997). modC-lacZ expression was completely abolished in hthP2 embryos (Fig. 2B). Second, we mutagenized three Exd–Hth-binding sites in modC (identified by TargetExplorer; see the Materials and Methods) that were close to a Dfd-binding site, while leaving the Dfd-binding sites intact (3EH-lacZ) (described in Fig. 1A). The ability of these sites to bind to Exd–Hth (full length) in vitro was confirmed by EMSAs; aside from site I, only one of these bound cooperatively with Dfd, albeit weakly (site II) (Supplemental Fig. 1B). 3EH-lacZ was expressed very weakly in the maxillary segment, suggesting that the Exd–Hth input into modC is direct and essential (Fig. 2C). Third, because Exd remains associated with Hth in nuclei, we used chromatin immunoprecipitation (ChIP) to test if Hth was bound to modC in wild-type embryos. Using anti-Hth antibody, we were able to immunoprecipitate modC DNA compared with a negative control locus, pyruvate dehydrogenase (PDH) (Fig. 2D). Together, these results establish that modC is bound by and depends on Exd–Hth for activity in vivo. We note, however, that these data do not address which isoform of Hth (full length or homeodomainless) is being used to regulate EAE-lacZ in vivo. To address this question, we examined EAE-lacZ activity in embryos that cannot make full-length Hth (hth100-1/hthP2) (Noro et al. 2006). These embryos still express modC-lacZ, suggesting that the homeodomainless isoform of Hth is sufficient for activation (Supplemental Fig. 2).
Figure 2.
Dfd-specific functions depend on Exd–Hth. (A,B) modC-lacZ expression is normal in the maxillary segment (yellow arrowheads) of hthP2 heterozygotes (A; visualized by hb-lacZ marked balancer), but is abolished in hthP2 homozygotes (B). The ectopic expression driven by modC in hthP2 embryos suggests that Hth may have a repressive role in thoracic and abdominal segments. (C) modC-lacZ requires direct Exd–Hth input. Mutagenesis of three Exd–Hth-binding sites in modC (3EH-lacZ) (Fig. 1A) results in a large decrease in enhancer activity. Note that Dfd monomer binding to this element is weakly affected by this mutation. (D) Hth binds to modC in vivo. Anti-Hth antibody was used to precipitate embryonic chromatin. Immunoprecipitated chromatin was assayed for the presence of modC (spanning site I) and a negative control locus, PDH. Data are presented as a percentage of the signal obtained relative to input chromatin and represent the averages and standard deviation of two independent ChIPs. (E, lanes 7,8) DfdWT binds cooperatively to site I with Exd–HM. (Lanes 4,5) DfdYPAA fails to bind cooperatively with Exd–HM to site I. (Lanes 12,13) DfdWT fails to bind cooperatively with Exd–HM to site I that has a mutant Exd half-site (site Iexd-mut). Hox monomer and Hox–Exd–HM trimer complexes are indicated by the pink and black arrowheads, respectively. (F,G) Ectopic expression of DfdWT (F) results in ectopic activation of EAE-lacZ, while ectopic expression of DfdYPAA (G) does not (see Fig. 3F for quantification). (H) Head and thorax of a wild-type embryo showing the normal position of cirri in the head segment (indicated by pink arrowhead). Ubiquitous expression of DfdWT (H′) results in ectopic cirri (yellow arrowheads, see inset in panel) and mouth hooks in thoracic segments (see Fig. 3G for quantification), but expression of DfdYPAA failed to induce any Dfd-specific structures (H″).
Dfd must interact with Exd to carry out its specific functions in vivo
When Exd–Hox heterodimers bind to composite DNA-binding sites, the Hox YPWM motif binds to a hydrophobic pocket in Exd's homeodomain (Passner et al. 1999; Piper et al. 1999; LaRonde-LeBlanc and Wolberger 2003; Joshi et al. 2007; Mann et al. 2009). To test if this interaction is important for Dfd to carry out its in vivo functions, the tryptophan (W) and methionione (M) residues of Dfd's YPWM motif were mutagenized to alanines. DfdYPAA and DfdWT were first tested for their ability to bind cooperatively with Exd to site I of modC. To assess Hox–Exd cooperativity, we used full-length Exd in complex with a homeodomainless isoform of Hth called HM, which enhances Exd's ability to bind DNA (Noro et al. 2006). On its own, Exd–HM did not bind to site I (Fig. 2E, lane 2) and DfdWT bound as a monomer (Fig. 2E, lane 6), but together they bound very cooperatively (Fig. 2E, lanes 7,8). Formation of this Dfd–Exd–HM complex required the Exd half-site of site I (Fig. 2E, lanes 12,13; Supplemental Fig. 3). In contrast to DfdWT, DfdYPAA failed to bind cooperatively in the presence of Exd–HM (Fig. 2E, lanes 4,5), even though both proteins bound similarly as monomers to site I (Fig. 2E, lanes 3,6). Thus, Dfd's YPWM motif and the Exd half-site are both required for Dfd and Exd–HM to bind cooperatively to site I.
To test the role of Dfd's YPWM motif in vivo, we generated transgenes capable of inducible expression of DfdWT and DfdYPAA (using the phiC31 and UAS-GAL4 systems) (Brand and Perrimon 1993; Bischof et al. 2007). Ubiquitous expression of DfdWT using AG11-Gal4 resulted in the ectopic expression of EAE-lacZ in the epidermis of stage 13 embryos (Fig. 2F). In contrast, ubiquitous expression of DfdYPAA did not induce this reporter gene in the majority of the embryos (Figs. 2G, 3F [for quantification]).
Dfd is normally expressed in the embryonic maxillary and mandibular segments, where it dictates the identity of larval head structures such as cirri (Fig. 2H) and mouth hooks. Ubiquitous expression of DfdWT resulted in the appearance of additional cirri in thoracic segments (Figs. 2H′ [and inset], 3G [for quantification]). In contrast, DfdYPAA was incapable of inducing ectopic cirri (Fig. 2H″). These results establish that a Dfd protein that cannot interact with Exd in vitro is incapable of carrying out Dfd-specific functions in vivo.
Dfd specificity module residues His-15 and Arg3 are required
Our previous studies demonstrated that two basic residues in Scr's specificity module are required for Scr to execute its specific functions in vivo (Joshi et al. 2007). One of these residues is an arginine at position 3 of the homeodomain (Arg3 in the N-terminal arm), and the second is a histidine located four residues away from the M of the YPWM motif (His-12, numbered relative to the start of the homeodomain) (Fig. 3A). Interestingly, although the context is different, Dfd has a histidine (His-15) and an arginine (Arg3) at the same positions (Fig. 3A, underlined residues). These residues are highly conserved among Dfd orthologs (Fig. 3A). To test if they play a role in specificity, we mutated these residues to alanines. In vitro, DfdHis-15A, Arg3A bound significantly weaker as a monomer to site I compared with DfdWT (Fig. 3B, cf. lanes 3,6). Cooperative binding of DfdHis-15A, Arg3A with Exd–HM was also weaker by about an order of magnitude compared with DfdWT (Fig. 3B; cf. lanes 4,5 and 7,8); Kd measurements show that Exd–HM–DfdHis-15A, Arg3A had an approximately eightfold lower affinity for site I compared with Exd–HM–DfdWT (Fig. 3C).
To test this mutant in vivo, we generated a transgenic line capable of inducible expression of DfdHis-15A, Arg3A. Compared with DfdWT (Fig. 3D,F), DfdHis-15A, Arg3A was impaired in its ability to activate EAE-lacZ (Fig. 3E,F) and in its ability to induce Dfd-specific transformations of the embryonic cuticle (Fig. 3G). However, as shown below, DfdHis-15A, Arg3A functions as a transcriptional repressor as well as DfdWT, demonstrating that it retains some activity in vivo. These results show that, for Dfd to execute its paralog-specific functions, it requires not only its YPWM motif, but also two basic residues in its specificity module. Together, these findings suggest that, like Scr, Dfd requires these basic residues to recognize its specific binding sites in an Exd-dependent manner.
Dfd represses fkh250-lacZ in an Exd-independent manner
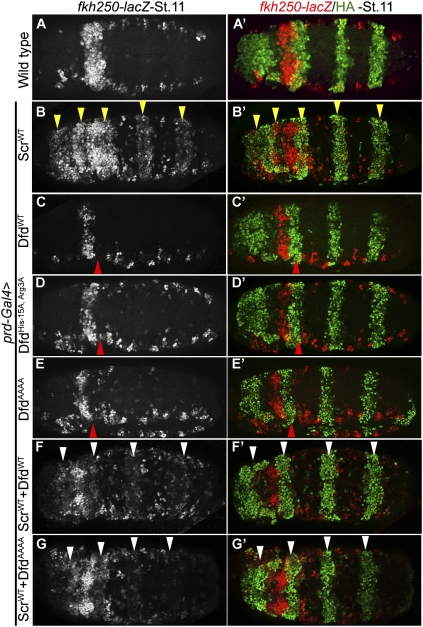
As noted above, both Dfd and Scr have conserved histidine and arginine residues at the same positions in their specificity modules, and these residues are important for both of these Hox proteins to activate their paralog-specific target genes (EAE-lacZ and fkh250-lacZ, respectively) in vivo (Fig. 7A, below). Given these similarities, we next tested if Dfd could also activate the Scr target, fkh250-lacZ. This was unlikely because neither fkh250-lacZ nor fkh is expressed in the maxillary or mandibular segments of wild-type embryos, where Dfd is expressed. To test if Dfd could regulate fkh250-lacZ, we expressed DfdWT or ScrWT using prd-Gal4. In wild-type embryos, fkh250-lacZ is expressed in parasegment 2 (PS2), where endogenous Scr is expressed (Fig. 4A; Ryoo et al. 1999). Ectopic expression of Scr via prd-Gal4 activated fkh250-lacZ, as described previously (Fig. 4B; Joshi et al. 2007). In contrast, Dfd repressed fkh250-lacZ (Fig. 4C). Next, we tested the role of the YPWM motif, Arg, and His residues in this repression by ectopic expression of DfdYPAA or DfdHis-15A, Arg3A. Surprisingly, both of these mutant proteins had the capacity to repress fkh250-lacZ (Fig. 4D; data not shown). To rule out the possibility that mutation of YPWM to YPAA in DfdYPAA may not have completely abolished the interaction between Dfd and Exd, we tested an additional mutant in which YPWM was changed to AAAA. DfdAAAA also repressed fkh250-lacZ, suggesting that this repression was indeed independent of this motif (Fig. 4E). Finally, because expression of Dfd by prd-Gal4 results in repression of Scr (data not shown), we considered the possibility that repression of fkh250-lacZ may be an indirect consequence of repressing Scr, which is required for fkh250-lacZ expression. To address this possibility, we coexpressed Dfd and Scr using prd-Gal4. Although the Scr transgenic line used in this experiment expressed Scr at higher levels than the Dfd transgene (see the Materials and Methods), the coexpression of both proteins resulted in repression of fkh250-lacZ (Fig. 4F). In posterior segments, we observed very weak activation when both proteins were coexpressed (Fig. 4F), in contrast to strong activation when only Scr was expressed (Fig. 4B). Moreover, although DfdAAAA is crippled in its ability to interact with Exd, it was also able to repress fkh250-lacZ when coexpressed with Scr (Fig. 4G). These results suggest that Dfd directly represses fkh250-lacZ, and that this repression does not require Dfd to interact with Exd, nor does it require key residues in its specificity module.
Figure 7.
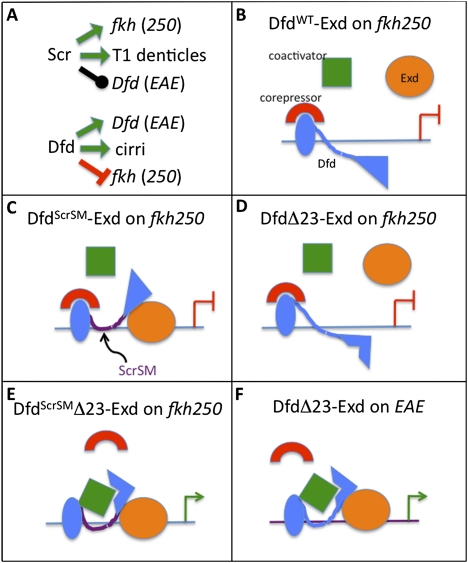
Interplay between DNA-binding specificity and transcriptional output. (A) Specific in vivo functions of Scr and Dfd. Green arrows indicate specific functions, the repression bar is indicated in red, and the black line indicates no effect. (B–F) Schematized is a subset of the various regulatory scenarios examined in this study. Activation is indicated by a green arrow, and the repression bar is indicated in red. A comparison between DfdScrSM–Exd bound to fkh250 (C), which represses transcription, and DfdScrSMΔ23–Exd bound to fkh250 (E), which activates transcription, suggests that motifs 2 and 3 are required to either recruit a corepressor or block the recruitment of a coactivator. A comparison between DfdΔ23–Exd bound to fkh250 (D), which represses transcription, and DfdScrSMΔ23–Exd bound to fkh250 (E), which activates transcription, suggests that differences in the specificity module, and thus how these proteins bind to fkh250, determines the sign of the regulation. Finally, a comparison between DfdΔ23–Exd bound to fkh250 (D), which represses transcription, and DfdΔ23–Exd bound to EAE (F), which activates transcription, suggests that differences between binding sites also contribute to differences in regulatory output.
Figure 4.
Dfd represses fkh250-lacZ in an Exd-independent manner. (A) Wild-type expression patterns of fkh250-lacZ and prd-GAL4 (driving GFP) to show overlap between these two expression domains. The strongest expression of fkh250-lacZ is in PS2. (B) Ectopic expression of ScrWT activates fkh250-lacZ (yellow arrowheads). (C) Ectopic expression of DfdWT represses fkh250-lacZ (red arrowhead). (D) Ectopic expression of DfdHis-15A, Arg3A represses fkh250-lacZ (red arrowhead), showing that repression of fkh250-lacZ by Dfd is independent of these residues. (E) Ectopic expression of DfdAAAA represses fkh250-lacZ (red arrowhead), suggesting that repression of fkh250-lacZ by Dfd does not require recruitment of Exd by its YPWM motif. (F) Coexpression of both DfdWT and ScrWT represses fkh250-lacZ in PS2 and largely blocks its activation in posterior segments (white arrowheads), showing that fkh250-lacZ repression by DfdWT is direct. (G) Coexpression of both DfdAAAA and ScrWT represses fkh250-lacZ in PS2 and largely blocks its activation in posterior segments (white arrowheads), showing that repression of fkh250-lacZ by Dfd is direct and unlikely to require Exd recruitment.
Enhancing Exd recruitment by Dfd on fkh250 is not sufficient for activation
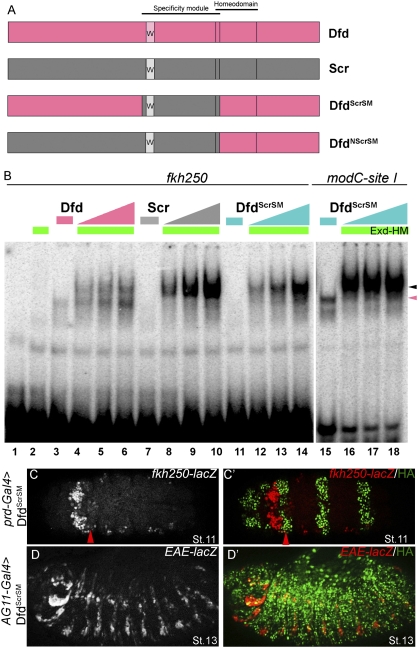
Exd has been suggested to function as an activating cofactor when recruited cooperatively by Dfd (Pinsonneault et al. 1997). Consistent with this proposal, in the experiments described above, we found that Dfd represses fkh250-lacZ in an Exd-independent manner and activates EAE-lacZ in an Exd-dependent manner. To examine Dfd's ability to bind fkh250 in vitro, we carried out EMSAs. Unlike ScrWT, DfdWT bound to fkh250 as monomer (Fig. 5B, lane 3). In the presence of Exd–HM, DfdWT did not bind to fkh250 as cooperatively as ScrWT (Figure 5B, cf. lanes 4–6 and 8–10). Together, these observations raise the possibility that Dfd's weaker ability to bind cooperatively with Exd to fkh250 may explain why it does not activate fkh250-lacZ in vivo. To test this idea, we replaced the specificity module of Dfd with that of Scr to generate a chimeric protein, DfdScrSM (schematized in Fig. 5A). DfdScrSM bound with Exd–HM to fkh250 in a highly cooperative manner (Kd = 32 ± 7 nM), comparable with ScrWT (Fig. 5B; Joshi et al. 2007). This observation supports the idea that Hox specificity modules contribute to the DNA-binding properties of these factors. DfdScrSM also bound in a highly cooperative manner with Exd–HM to site I of modC (Kd = 5.0 ± 0.3 nM) (Fig. 5B), similar to DfdWT (Kd = 5.5 ± 0.3 nM) (Fig. 3B) and ScrWT (Kd = 11 ± 0.3 nM) (data not shown). Thus, although both Dfd and Scr specificity modules have similarly positioned histidine and arginine residues—which are important for the activation of EAE-lacZ and fkh250-lacZ, respectively—they confer distinct DNA-binding properties to these proteins. We speculate that these differences are due to residues in the vicinity of the histidine and arginine, which differ in Scr and Dfd.
Figure 5.
Enhancing Exd recruitment by Dfd to fkh250 is not sufficient for activation. (A) Schematics of DfdScrSM and DfdNScrSM chimeras. Extent of specificity modules and homeodomains are indicated (for details, see the text). (B) DfdWT binds less cooperatively with Exd–HM to fkh250 (lanes 4–6) compared with ScrWT (lanes 8–10). (Lanes 12–14) DfdScrSM binds very cooperatively with Exd–HM to fkh250. (Lanes 16–18) DfdScrSM binds very cooperatively with Exd–HM on site I. Hox monomer and Hox–Exd–HM trimer complexes are indicated by pink and black arrowheads, respectively. (C,D) Ectopic expression of DfdScrSM represses fkh250-lacZ (C; red arrowhead) and activates EAE-lacZ (D), suggesting that an enhancement of Exd recruitment is not sufficient for fkh250-lacZ activation.
Next, we asked if DfdScrSM, which binds very cooperatively with Exd–HM to fkh250, can activate fkh250-lacZ in vivo. We found that, like DfdWT, DfdScrSM repressed fkh250-lacZ (Fig. 5C) and activated EAE-lacZ (cf. Figs. 5D and 2F). These results argue that enhanced recruitment of Exd is not sufficient for target gene activation, and that other aspects of the protein–DNA complex contribute to gene activation versus repression. Consistently, repression of fkh250-lacZ by DfdScrSM was independent of Exd, because it did not require an intact YPWM motif (i.e., DfdScrSM,AAAA repressed fkh250-lacZ) (data not shown).
Changing the functional specificity of Dfd
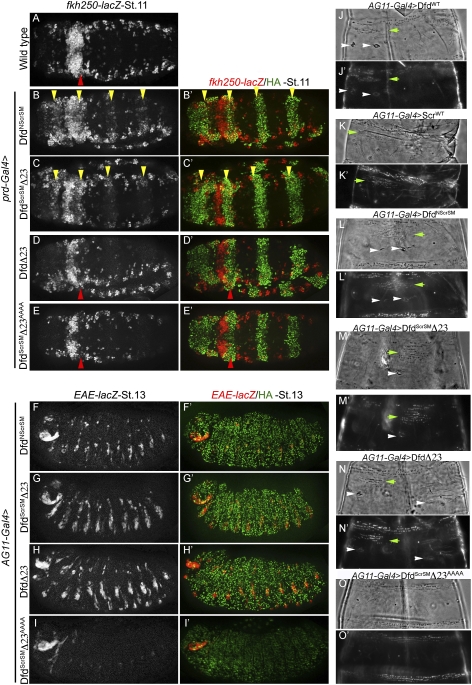
The repression of fkh250-lacZ by DfdScrSM suggested the presence of a repression domain in Dfd that functioned whether or not Exd was recruited to the fkh250-binding site. To identify this repression domain, we made another chimera in which all sequences N-terminal to the Dfd homeodomain, including its specificity module, were replaced by the equivalent region from Scr (DfdNScrSM) (schematized in Fig. 5A). In contrast to DfdScrSM, DfdNScrSM activated fkh250-lacZ (Fig. 6B), although this activation was weaker than that induced by ScrWT (Fig. 4B). DfdNScrSM also activated EAE-lacZ (Fig. 6F). Another chimera, DfdNScr—which has Dfd's specificity module, homeodomain, and C terminus, but Scr's N terminus—also activated EAE-lacZ, but had no affect on fkh250-lacZ expression (Supplemental Fig. 4). These results suggest that Dfd's ability to repress fkh250-lacZ depends on sequences N-terminal to its YPWM motif. In the N-terminal region of Dfd, there are three well-conserved sequence motifs in addition to YPWM (Supplemental Fig. 5). Motif 1, which is also called the MxSYF motif, is found at the beginning of nearly all Hox proteins and has been suggested to play a role in transcriptional activation (Zhao et al. 1996; Tour et al. 2005), while motifs 2 and 3 are specific to Dfd orthologs. Based on these observations, we focused on motifs 2 and 3. Deletion of motif 2 from DfdScrSM did not affect this protein's ability to repress fkh250-lacZ (data not shown). However, deletion of both motifs 2 and 3 (DfdScrSMΔ23) resulted in the activation of fkh250-lacZ (Fig. 6C). Like DfdWT and DfdScrSM, DfdScrSMΔ23 was also able to activate EAE-lacZ (Fig. 6G). Importantly, the activation of EAE-lacZ and fkh250-lacZ by DfdScrSMΔ23 required its YPWM motif, because mutating this motif in DfdScrSMΔ23 resulted in a protein (DfdScrSMΔ23AAAA) that no longer activated these targets (Fig. 6E,I). In fact, DfdScrSMΔ23AAAA repressed fkh250-lacZ, suggesting that, when the interaction with Exd is eliminated, the default activity is to repress this target gene (see the Discussion; Fig. 7). Note that neither motif 2 nor 3 contributed to cooperative or monomeric DNA binding to either fkh250 or modC (Supplemental Fig. 6), suggesting that they function to recruit additional factors in a context-dependent manner (see the Discussion).
Figure 6.
Changing the functional specificity of Dfd. (A) Wild-type expression pattern of fkh250-lacZ. (B,C) Ectopic expression of DfdNScrSM (B) or DfdScrSMΔ23 (C) activates fkh250-lacZ (yellow arrowheads), showing a change in specificity. (D,E) Ectopic expression of DfdΔ23 (D) or DfdScrSMΔ23AAAA (E) represses fkh250-lacZ in PS2 (red arrowhead), emphasizing the importance of Scr's SM- and Exd-mediated cooperative binding in activation of fkh250-lacZ. (F–H) Ectopic expression of DfdNScrSM (F), DfdScrSMΔ23 (G), or DfdΔ23 (H) activates EAE-lacZ. These proteins retain the ability to activate Dfd, like DfdWT. (I) Ectopic expression of DfdScrSMΔ23AAAA does not activate EAE-lacZ, emphasizing the importance of Exd recruitment in EAE-lacZ activation. (J–O) Cuticle phenotypes resulting from ubiquitous expression of DfdWT (J), ScrWT (K), DfdNScrSM (L), DfdScrSMΔ23 (M), DfdΔ23 (N), and DfdScrSMΔ23AAAA (O). Scr-specific structures (ectopic T1 beard) are indicated with green arrowheads, and Dfd-specific structures (cirri) are indicated by white arrowheads. Dark-field and phase-contrast images for the T2 segment are shown for all cuticles.
Consistent with the above results, when we analyzed the embryonic cuticle phenotype resulting from the expression of DfdNScrSM and DfdScrSMΔ23 (Fig. 6L,M), we found that they produced a mixture of both Scr- and Dfd-specific identities (Fig. 6J,K). Denticle types usually limited to the first thoracic segment (T1) were seen in T2 and T3 (an Scr-like transformation), but ectopic cirri were also observed in these segments (a Dfd-like transformation). However, DfdWT was also able to induce the T1-specific denticles, arguing that this phenotype is not a strict Scr-specific readout (Fig. 6J, green arrowhead). DfdScrSMΔ23AAAA, like DfdYPAA, did not produce either of these transformations (Fig. 6O), demonstrating that interaction with Exd is critical for this chimera to produce these transformations. Thus, DfdScrSMΔ23 has many of the properties normally associated with Scr (such as activation of fkh250-lacZ), but also retains functions normally associated with Dfd (such as activation of EAE-lacZ and induction of cirri).
To determine the role of motifs 2 and 3 in an otherwise wild-type Dfd protein, we deleted these motifs from Dfd to generate DfdΔ23. Surprisingly, in all of our assays, DfdΔ23 behaved like DfdWT: It repressed fkh250-lacZ (Fig. 6D), activated EAE-lacZ (Fig. 6H), and generated ectopic cirri in thoracic segments (Fig. 6N). Thus, a change of functional specificity from Dfd- to Scr-specific readouts was only observed when both the Scr specificity module was present and Dfd motifs 2 and 3 were deleted. These observations emphasize the importance of the specificity module in both DNA binding and activity regulation, but they also reveal the interdependence of these two processes.
Discussion
The role of Hox genes in the specification of cellular and regional identities during animal development is well established, but many questions remain concerning the mechanism by which they select and regulate their target genes. Part of the reason is that very few directly targeted enhancers have been characterized at high resolution. In this work, we analyze the regulation of a regulatory element that is activated directly by Dfd, and compare this mechanism to what has been described previously for Scr. Dfd and Scr have distinct properties in vivo (Fig. 7A). The comparison between these two Hox proteins not only revealed common themes, but also gave new insights into how the regulatory properties of Hox proteins are determined both by the quality of the binding site and by subtle differences in the DNA recognition properties of these proteins (summarized in Fig. 7).
Generality of the Hox specificity model
Previous work on Scr's ability to specifically regulate its target gene, fkh, revealed that the N-terminal arm of its homeodomain and preceding linker region are positioned in such a manner as to allow the insertion of two basic side chains into the minor groove of the target DNA, fkh250 (Joshi et al. 2007). Importantly, the correct positioning of these residues depends on an interaction between Scr's YPWM motif and the cofactor Exd. Here, we show that an analogous mechanism is required for Dfd to bind productively to a Hox–Exd-binding site in the EAE element and to activate EAE-lacZ in vivo. Specifically, we found that Dfd's YPWM motif is required for cooperative binding to EAE's site I in vitro, and for executing Dfd-specific functions in vivo. Like Scr, Dfd has the same two basic residues—a histidine (likely to be protonated when bound to DNA) (see Joshi et al. 2007) and an arginine—at the equivalent positions relative to its YPWM motif and homeodomain. Moreover, these residues are also required for Dfd to execute its specific functions in vivo. Thus, the activation of fkh by Scr and the activation of Dfd by Dfd appear to use analogous mechanisms, whereby linker and N-terminal arm residues are used to bind paralog-specific binding sites in an Exd-dependent manner (Fig. 7).
The YPWM-to-YPAA mutation severely impaired Dfd's ability to carry out its specific in vivo functions, such as activation of EAE-lacZ and production of cirri. Thus, the YPWM motif of Dfd is critical for Dfd function in vivo. This situation contrasts with other apparently more complex scenarios. For example, mutation of the YPWM motif of the Hox protein Ultrabithorax (Ubx) did not significantly impair some of its in vivo functions (Galant et al. 2002; Merabet et al. 2007). In this case, it appears that other sequence motifs, in particular a domain C-terminal to the Ubx homeodomain, are important for Ubx to carry out its specific functions in vivo (Chan and Mann 1993; Merabet et al. 2007). These Ubx sequences also appear to help recruit Exd to DNA, and therefore may be used for binding site selection in conjunction with YPWM at a subset of Ubx target-binding sites. Interestingly, a sequence motif immediately C-terminal to Dfd's homeodomain also plays a role in in vivo specificity, although its impact on DNA binding has not been examined (Lin and McGinnis 1992). As these sequences are still present in DfdScrSMΔ23, it may explain why this chimera retains some Dfd-specific functions, such as the formation of cirri and ability to activate EAE-lacZ. The picture that emerges from all of these data is that Hox proteins may use different motifs to interact with cofactors such as Exd, depending on the specific in vivo function and target gene being regulated.
Role of Hox specificity modules
In general, the sequences surrounding Hox YPWM motifs and the N-terminal arms of their homeodomains are highly conserved, from invertebrates to vertebrates, in a paralog-specific manner (Joshi et al. 2007). Thus, based on the results presented here, we hypothesize that these sequences, which we refer to here as Hox specificity modules, may in general be used for the recognition of specific DNA-binding sites in a cofactor-dependent manner. In the case of Scr binding to fkh250, an X-ray crystal structure revealed that the histidine and arginine side chains recognize an unusually narrow minor groove that is an intrinsic feature of the fkh250-binding site. Without the benefit of a Dfd–Exd–site I crystal structure, we cannot know with certainty if Dfd's His-15 and Arg3 also read the shape of a narrow minor groove. However, the fact that the same two basic residues are required for both Scr and Dfd suggests the possibility that this is the case for Dfd binding to EAE site I as well.
DfdScrSMΔ23, which has the specificity module of Scr in place of Dfd's, exhibited clear Scr-like functions in vivo, as assayed by fkh250-lacZ activation and larval cuticle transformation. Other attempts to swap Hox specificities by generating chimeric Hox proteins have had variable success. For example, when the linker and N-terminal arm of Scr is used to replace the equivalent region of Antennapedia (Antp), the chimera behaved like Scr (Gibson et al. 1990; Furukubo-Tokunaga et al. 1993). This finding supports the importance of specificity modules in conferring Hox specificity. When the homeodomain and C-terminal region of Ubx were replaced by the equivalent domains from Antp, the chimera behaved like Antp (Chan and Mann 1993), suggesting that the identity of the linker region may not be critical in all cases. Other Hox chimeras have generated less clear changes of specificity. For example, chimeras between Ubx and Dfd generated a cuticle phenotype that was dissimilar to that produced by either parent protein (Kuziora and McGinnis 1989; Lin and McGinnis 1992). Similarly, a chimera between Ubx and Abd-B had novel properties that were unlike those produced by either parent protein (Zhu and Kuziora 1996). It is noteworthy that the cleanest changes in specificity occurred when the chimera was generated between Hox genes that are adjacent to each other in the Hox complex. This correlation may be due to the fact that adjacent Hox genes are more similar to each other, both in sequence and in function, than nonadjacent Hox genes (Gehring et al. 2009). This higher degree of similarity is likely a consequence of how these genes are thought to have duplicated during evolution (Gehring et al. 2009).
Architecture of Hox cofactor targeted enhancers
In our previous work on the regulation of fkh by Scr, the reporter gene used to study the activity of the Exd–Scr-binding site had a multimerized version of the minimal 37-bp fkh250 element (Ryoo and Mann 1999). In contrast, in the work described here, we characterize an intact regulatory element from the Dfd gene, revealing significantly more complexity. In particular, the 570-bp modC element contains a single “classical” Exd–Hox composite site, but also four additional Dfd sites and several additional Exd–Hth-binding sites. Our mutagenesis studies suggest that all of these inputs are important for the full activity of this enhancer. Also noteworthy is that there are additional Dfd–Exd-binding sites in the larger 2.7-kb EAE element that, in principle, could also be used in vivo (data not shown). Thus, the picture that emerges from this analysis is that native enhancer elements may use a combination of classical Exd–Hox-binding sites together with additional arrangements that may not always conform to the classical spacing of the Exd and Hox half-sites (Fig. 1A). This picture raises the question of how the linker and N-terminal arm residues are positioned correctly in these nonclassical arrangements. The answer may lie in the fact that, in vivo, the assembly of the complete multiprotein complex—which is likely to include factors in addition to Dfd, Exd, and Hth—promotes the recognition of Dfd-binding sites in ways that are not fully revealed by experiments that examine binding to individual or small groups of binding sites in isolation.
Hox functional specificity depends on both DNA-binding specificity and activity regulation
Depending on the context, most transcription factors have the capacity to activate and repress transcription. In most cases, it is not understood how this choice is made. One established scenario is that other proteins that get recruited to an enhancer element determine the sign of the regulation (Grienenberger et al. 2003; Gebelein et al. 2004; Hersh and Carroll 2005; Taghli-Lamallem et al. 2007; Li-Kroeger et al. 2008). However, this type of model is not sufficient to explain the results presented here. Our results suggest that the DNA-binding properties of the Exd–Hox complex influence the regulatory output of the bound protein–DNA complex (Fig. 7). Deletion of two motifs (Δ23) from the N-terminal region of DfdScrSM converted this protein from a repressor of fkh250-lacZ to an activator of fkh250-lacZ, while deletion of the same motifs from DfdWT did not change the regulatory output: The protein retained its ability to repress fkh250-lacZ. The only difference between DfdΔ23 (represses) and DfdScrSMΔ23 (activates) is the specificity module, and the only difference between DfdScrSMΔ23 (activates) and DfdScrSM (represses) is the presence or absence of motifs 2 and 3. These results imply that the relevance of motifs 2 and 3, which are far from the DNA-binding domain, depends on the identity of the specificity module. These findings lead us to suggest that the DNA-binding site, together with how it is read by the specificity module, plays an important role in determining the overall conformation of the Hox–Exd complex (Fig. 7), which eventually determines whether there will be recruitment of a coactivator or corepressor. This idea fits well with a DNA allostery model that was supported recently by cell culture experiments with the glucocorticoid receptor (Lefstin and Yamamoto 1998; Meijsing et al. 2009). In these experiments, it was discovered that small differences in the DNA-binding site lead to differences in conformation and the degree of transcriptional activation. Here, we extend this idea by showing that Hox proteins with different specificity modules, and therefore with slightly different DNA recognition properties, result in unique regulatory outputs in an in vivo context (Fig. 7B,E). Furthermore, in our experiments, we observed a complete change in the sign of the regulation from repression to activation, instead of a more subtle change of activation amplitude. Thus, the transcriptional output of a Hox–cofactor complex depends both on the ability of these complexes to bind to their binding sites with high specificity, in part by reading structural features of the DNA, and on the three-dimensional architecture of the bound complex, which is a consequence of both protein–DNA and protein–protein interactions. An important goal for the future will be to use structural biology methods to see how different Hox specificity modules result in distinct conformations of Exd–Hox complexes.
Materials and methods
Drosophila strains and genetics
Wild-type flies were yw. Standard methods were used to recombine and balance chromosomes bearing mutations, markers, or transgenes. The hthP2 mutation used (Rieckhof et al. 1997) was balanced over hb-lacZ-marked TM3-Sb balancer to identify homozygous embryos. The transgenic lines generated for all of the experiments were made using the phiC31-based integration system (Bischof et al. 2007). All of the UAS lines were inserted into attP site 51D, except for UAS-ScrWT used for the competition experiment shown in Figure 4, which was generated using standard P-element transgenesis (Joshi et al. 2007). All UAS lines were HA-tagged, and anti-HA staining was used to detect protein expression in vivo. The UAS-ScrWT transgenic line expressed Scr at least three times more than the UAS-DfdWT transgenic insertion at site attP 51D, as assayed by comparative anti-HA antibody staining (data not shown). AG11-GAL4 on the second chromosome and prd-GAL4 on the third chromosome were used for ectopic expression in all of the experiments. All of the lacZ reporter genes were inserted at attP site 86Fa; single transgenic adults were picked and used to set up multiple lines for each insertion. modC-lacZ and its mutant variants were generated by amplifying this 570 bp from the EAE-lacZ construct (provided by W. Mcginnis) (Bergson and McGinnis 1990; Zeng et al. 1994). EAE-lacZ line on the X chromosome (Bergson and McGinnis 1990; Zeng et al. 1994) was used as a reporter for all of the ectopic expression experiments, except for Figure 1I, for which modC-lacZ at 86Fa was used. fkh250-lacZ on the second chromosome was used (Ryoo and Mann 1999). All of the experiments were done with 12-h egg collections at 25°C, except for those done for ChIP experiments (see below). Embryonic cuticle preparations were generated as described (Rieckhof et al. 1997). Cirri quantification was done by counting and adding the number of branches in each ectopic cirri for each of the thoracic segments. These numbers were then compared between wild-type Dfd and its mutant versions. At least 30 embryonic cuticles were scored for each genotype.
Antibodies and immunohistochemistry
Guinea pig anti-Scr (GP111 dilution; 1:1000), rabbit anti-βgal (Cappel; 1:5000), rat anti-HA (3F10 from Roche; 1:500), and guinea pig anti-Dfd (1:50; kind gift from W. Mcginnis, University of California at San Diego) were used for staining embryos as described previously (Noro et al. 2006). Secondary antibodies used were AlexaFluor488 (1:500) and AlexaFluor555 (1:1000) conjugates from Molecular Probes, and embryos were mounted with Vectashield. Optical section single images or Z-series were collected on Zeiss AxioScope/ApoTome; Z-series were analyzed by ImageJ (http://rsbweb.nih.gov/ij). Other image analyses were done with Photoshop CS3. EAE-lacZ expression was referred to as weak in the graph shown in Figure 3F if it could be visualized by eye, but did not show any significant levels on acquisition using the exposure time of 20 msec in red channel 555 nm.
ChIP
Five-hour to 6 h embryo collections were done at 25°C, and embryos were aged for another 6 h at 25°C. The embryos were then dechorionated in bleach and fixed for 25 min in 2% formaldehyde, 100 mM NaCl, 1 mM EDTA (pH 8), 0.5 mM EGTA (pH 8), 100 mM HEPES (pH 7.6), and an equal volume of heptane at room temperature with vigorous shaking. Fixation was quenched using 0.125 M glycine in 1× PBS with 0.02% Triton, and washing in 1× PBS and 0.02% Triton. Standard chromatin preparation and immunoprecipitation protocol was used as described previously (Agelopoulos and Thanos 2006). Twenty micrograms of chromatin was used for ChIP in all the experiments. Guinea pig anti-Hth antibody (GP-52) was used in dilution of 1:100 for each immunoprecipitation. Primers AC-2L and AC-3R, which amplify a 360-bp region of the modC enhancer, were used to detect EAE sequences, and primers from the 5′ region of PDH were used as a negative control. A “no antibody” control or mock immunoprecipitation (with unprogrammed IgG) was done for each experiment, and failed to show any enrichment of either the modC or PDH amplicons. The data shown represent an average of two experiments done with two different chromatin preparations. Real-time PCR was used to quantify these immunoprecipitations. Sequences of the primers are available on request.
Protein–DNA-binding assays
Each putative binding site was examined for binding by EMSA. The modC site I probe was GGCGGCGCGTTCATTAATCATTATAGCTGTGGGACGAGG, and the fkh250 probe was GATCTCAATGTCAAGATTAATCGCCAGCTGTGGGACGAGG; the sequences of the other probes are available on request. EMSAs were carried out as described (Ryoo and Mann 1999). All DNA-binding experiments were carried out with nearly full-length 6XHis-tagged forms of Scr (residues 2–406) and full-length Exd, which was copurified with the HM domain of Hth from Escherichia coli and used at 150 ng per reaction. Dfd protein, Dfd mutants, and the DfdScrSM chimera extended from residues 130–586 of Dfd protein. The specificity modules of Scr and Dfd are shown in Figure 3A. modC-site I mutants are shown in Figure 1A. Hox concentrations used in different experiments are as follows: 5 and 20 ng per reaction in Figure 2E for both DfdWT and DfdYPAA; 10 and 20 ng per reaction in Figure 3B for both DfdWT and DfdHIs-14A,Arg3A; and 10, 20, and 40 ng per reaction in Figure 5B for all three: DfdWT, DfdScrSM, and ScrWT. All binding reactions were done in a 20-μL volume. For Kd measurements, increasing amounts of the Hox protein (from 5 to 500 ng) were added to a reaction mix with 150 ng of HM–Exd, and the data were analyzed as described (LaRonde-LeBlanc and Wolberger 2003).
Acknowledgments
We thank W. McGinnis for flies and reagents; D. Baronnier for help with molecular biology and generating anti-Scr antibody; M. Agelopoulos and M. Slattery for help with ChIP experiments; O. Hobert, R. Rohs, and members of the Mann laboratory for comments on the manuscript; and L.S. Shashidhara for discussions. This work was supported by NIH grant GM058575 to R.S.M.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1936910.
Supplemental material is available at http://www.genesdev.org.
References
- Abu-Shaar M, Ryoo HD, Mann RS 1999. Control of the nuclear localization of Extradenticle by competing nuclear import and export signals. Genes Dev 13: 935–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agelopoulos M, Thanos D 2006. Epigenetic determination of a cell-specific gene expression program by ATF-2 and the histone variant macroH2A. EMBO J 25: 4843–4853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger MF, Badis G, Gehrke AR, Talukder S, Philippakis AA, Pena-Castillo L, Alleyne TM, Mnaimneh S, Botvinnik OB, Chan ET, et al. 2008. Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cell 133: 1266–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergson C, McGinnis W 1990. An autoregulatory enhancer element of the Drosophila homeotic gene Deformed. EMBO J 9: 4287–4297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof J, Maeda RK, Hediger M, Karch F, Basler K 2007. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc Natl Acad Sci 104: 3312–3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, Perrimon N 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415 [DOI] [PubMed] [Google Scholar]
- Capovilla M, Brandt M, Botas J 1994. Direct regulation of decapentaplegic by Ultrabithorax and its role in Drosophila midgut morphogenesis. Cell 76: 461–475 [DOI] [PubMed] [Google Scholar]
- Chan SK, Mann RS 1993. The segment identity functions of Ultrabithorax are contained within its homeo domain and carboxy-terminal sequences. Genes Dev 7: 796–811 [DOI] [PubMed] [Google Scholar]
- Chan SK, Mann RS 1996. A structural model for a homeotic protein-extradenticle-DNA complex accounts for the choice of HOX protein in the heterodimer. Proc Natl Acad Sci 93: 5223–5228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SK, Jaffe L, Capovilla M, Botas J, Mann RS 1994. The DNA binding specificity of Ultrabithorax is modulated by cooperative interactions with extradenticle, another homeoprotein. Cell 78: 603–615 [DOI] [PubMed] [Google Scholar]
- Coiffier D, Charroux B, Kerridge S 2008. Common functions of central and posterior Hox genes for the repression of head in the trunk of Drosophila. Development 135: 291–300 [DOI] [PubMed] [Google Scholar]
- Furukubo-Tokunaga K, Flister S, Gehring WJ 1993. Functional specificity of the Antennapedia homeodomain. Proc Natl Acad Sci 90: 6360–6364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galant R, Walsh CM, Carroll SB 2002. Hox repression of a target gene: Extradenticle-independent, additive action through multiple monomer binding sites. Development 129: 3115–3126 [DOI] [PubMed] [Google Scholar]
- Garvie CW, Wolberger C 2001. Recognition of specific DNA sequences. Mol Cell 8: 937–946 [DOI] [PubMed] [Google Scholar]
- Gebelein B, Culi J, Ryoo HD, Zhang W, Mann RS 2002. Specificity of Distalless repression and limb primordia development by abdominal Hox proteins. Dev Cell 3: 487–498 [DOI] [PubMed] [Google Scholar]
- Gebelein B, McKay DJ, Mann RS 2004. Direct integration of Hox and segmentation gene inputs during Drosophila development. Nature 431: 653–659 [DOI] [PubMed] [Google Scholar]
- Gehring WJ, Qian YQ, Billeter M, Furukubo-Tokunaga K, Schier AF, Resendez-Perez D, Affolter M, Otting G, Wuthrich K 1994. Homeodomain-DNA recognition. Cell 78: 211–223 [DOI] [PubMed] [Google Scholar]
- Gehring WJ, Kloter U, Suga H 2009. Evolution of the Hox gene complex from an evolutionary ground state. Curr Top Dev Biol 88: 35–61 [DOI] [PubMed] [Google Scholar]
- Gibson G, Schier A, LeMotte P, Gehring WJ 1990. The specificities of Sex combs reduced and Antennapedia are defined by a distinct portion of each protein that includes the homeodomain. Cell 62: 1087–1103 [DOI] [PubMed] [Google Scholar]
- Grieder NC, Marty T, Ryoo HD, Mann RS, Affolter M 1997. Synergistic activation of a Drosophila enhancer by HOM/EXD and DPP signaling. EMBO J 16: 7402–7410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grienenberger A, Merabet S, Manak J, Iltis I, Fabre A, Berenger H, Scott MP, Pradel J, Graba Y 2003. Tgfβ signaling acts on a Hox response element to confer specificity and diversity to Hox protein function. Development 130: 5445–5455 [DOI] [PubMed] [Google Scholar]
- Hersh BM, Carroll SB 2005. Direct regulation of knot gene expression by Ultrabithorax and the evolution of cis-regulatory elements in Drosophila. Development 132: 1567–1577 [DOI] [PubMed] [Google Scholar]
- Joshi R, Passner JM, Rohs R, Jain R, Sosinsky A, Crickmore MA, Jacob V, Aggarwal AK, Honig B, Mann RS 2007. Functional specificity of a Hox protein mediated by the recognition of minor groove structure. Cell 131: 530–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuziora MA, McGinnis W 1988. Autoregulation of a Drosophila homeotic selector gene. Cell 55: 477–485 [DOI] [PubMed] [Google Scholar]
- Kuziora MA, McGinnis W 1989. A homeodomain substitution changes the regulatory specificity of the deformed protein in Drosophila embryos. Cell 59: 563–571 [DOI] [PubMed] [Google Scholar]
- LaRonde-LeBlanc NA, Wolberger C 2003. Structure of HoxA9 and Pbx1 bound to DNA: Hox hexapeptide and DNA recognition anterior to posterior. Genes Dev 17: 2060–2072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefstin JA, Yamamoto KR 1998. Allosteric effects of DNA on transcriptional regulators. Nature 392: 885–888 [DOI] [PubMed] [Google Scholar]
- Leung TH, Hoffmann A, Baltimore D 2004. One nucleotide in a κB site can determine cofactor specificity for NF-κB dimers. Cell 118: 453–464 [DOI] [PubMed] [Google Scholar]
- Li-Kroeger D, Witt LM, Grimes HL, Cook TA, Gebelein B 2008. Hox and senseless antagonism functions as a molecular switch to regulate EGF secretion in the Drosophila PNS. Dev Cell 15: 298–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L, McGinnis W 1992. Mapping functional specificity in the Dfd and Ubx homeo domains. Genes Dev 6: 1071–1081 [DOI] [PubMed] [Google Scholar]
- Liu Y, Matthews KS, Bondos SE 2008. Multiple intrinsically disordered sequences alter DNA binding by the homeodomain of the Drosophila hox protein ultrabithorax. J Biol Chem 283: 20874–20887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Matthews KS, Bondos SE 2009. Internal regulatory interactions determine DNA binding specificity by a Hox transcription factor. J Mol Biol 390: 760–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manak JR, Mathies LD, Scott MP 1994. Regulation of a decapentaplegic midgut enhancer by homeotic proteins. Development 120: 3605–3619 [DOI] [PubMed] [Google Scholar]
- Mann RS, Affolter M 1998. Hox proteins meet more partners. Curr Opin Genet Dev 8: 423–429 [DOI] [PubMed] [Google Scholar]
- Mann RS, Chan SK 1996. Extra specificity from extradenticle: The partnership between HOX and PBX/EXD homeodomain proteins. Trends Genet 12: 258–262 [DOI] [PubMed] [Google Scholar]
- Mann RS, Morata G 2000. The developmental and molecular biology of genes that subdivide the body of Drosophila. Annu Rev Cell Dev Biol 16: 243–271 [DOI] [PubMed] [Google Scholar]
- Mann RS, Lelli KM, Joshi R 2009. Hox specificity unique roles for cofactors and collaborators. Curr Top Dev Biol 88: 63–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marty T, Vigano MA, Ribeiro C, Nussbaumer U, Grieder NC, Affolter M 2001. A HOX complex, a repressor element and a 50 bp sequence confer regional specificity to a DPP-responsive enhancer. Development 128: 2833–2845 [DOI] [PubMed] [Google Scholar]
- Meijsing SH, Pufall MA, So AY, Bates DL, Chen L, Yamamoto KR 2009. DNA binding site sequence directs glucocorticoid receptor structure and activity. Science 324: 407–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merabet S, Saadaoui M, Sambrani N, Hudry B, Pradel J, Affolter M, Graba Y 2007. A unique Extradenticle recruitment mode in the Drosophila Hox protein Ultrabithorax. Proc Natl Acad Sci 104: 16946–16951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens CB, Selleri L 2006. Hox cofactors in vertebrate development. Dev Biol 291: 193–206 [DOI] [PubMed] [Google Scholar]
- Noro B, Culi J, McKay DJ, Zhang W, Mann RS 2006. Distinct functions of homeodomain-containing and homeodomain-less isoforms encoded by homothorax. Genes Dev 20: 1636–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noyes MB, Christensen RG, Wakabayashi A, Stormo GD, Brodsky MH, Wolfe SA 2008. Analysis of homeodomain specificities allows the family-wide prediction of preferred recognition sites. Cell 133: 1277–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos DK, Vukojevic V, Adachi Y, Terenius L, Rigler R, Gehring WJ 2010. Function and specificity of synthetic Hox transcription factors in vivo. Proc Natl Acad Sci 107: 4087–4092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passner JM, Ryoo HD, Shen L, Mann RS, Aggarwal AK 1999. Structure of a DNA-bound Ultrabithorax-Extradenticle homeodomain complex. Nature 397: 714–719 [DOI] [PubMed] [Google Scholar]
- Pearson JC, Lemons D, McGinnis W 2005. Modulating Hox gene functions during animal body patterning. Nat Rev Genet 6: 893–904 [DOI] [PubMed] [Google Scholar]
- Pinsonneault J, Florence B, Vaessin H, McGinnis W 1997. A model for extradenticle function as a switch that changes HOX proteins from repressors to activators. EMBO J 16: 2032–2042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper DE, Batchelor AH, Chang CP, Cleary ML, Wolberger C 1999. Structure of a HoxB1-Pbx1 heterodimer bound to DNA: Role of the hexapeptide and a fourth homeodomain helix in complex formation. Cell 96: 587–597 [DOI] [PubMed] [Google Scholar]
- Rieckhof GE, Casares F, Ryoo HD, Abu-Shaar M, Mann RS 1997. Nuclear translocation of extradenticle requires homothorax, which encodes an extradenticle-related homeodomain protein. Cell 91: 171–183 [DOI] [PubMed] [Google Scholar]
- Rohs R, Jin X, West SM, Joshi R, Honig B, Mann RS 2010. Origins of specificity in protein–DNA recognition. Annu. Rev. Biochem 79: 233–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryoo HD, Mann RS 1999. The control of trunk Hox specificity and activity by Extradenticle. Genes Dev 13: 1704–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryoo HD, Marty T, Casares F, Affolter M, Mann RS 1999. Regulation of Hox target genes by a DNA bound Homothorax/Hox/Extradenticle complex. Development 126: 5137–5148 [DOI] [PubMed] [Google Scholar]
- Sosinsky A, Bonin CP, Mann RS, Honig B 2003. Target Explorer: An automated tool for the identification of new target genes for a specified set of transcription factors. Nucleic Acids Res 31: 3589–3592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taghli-Lamallem O, Gallet A, Leroy F, Malapert P, Vola C, Kerridge S, Fasano L 2007. Direct interaction between Teashirt and Sex combs reduced proteins, via Tsh's acidic domain, is essential for specifying the identity of the prothorax in Drosophila. Dev Biol 307: 142–151 [DOI] [PubMed] [Google Scholar]
- Tour E, Hittinger CT, McGinnis W 2005. Evolutionarily conserved domains required for activation and repression functions of the Drosophila Hox protein Ultrabithorax. Development 132: 5271–5281 [DOI] [PubMed] [Google Scholar]
- Vachon G, Cohen B, Pfeifle C, McGuffin ME, Botas J, Cohen SM 1992. Homeotic genes of the Bithorax complex repress limb development in the abdomen of the Drosophila embryo through the target gene Distal-less. Cell 71: 437–450 [DOI] [PubMed] [Google Scholar]
- Zeng C, Pinsonneault J, Gellon G, McGinnis N, McGinnis W 1994. Deformed protein binding sites and cofactor binding sites are required for the function of a small segment-specific regulatory element in Drosophila embryos. EMBO J 13: 2362–2377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao JJ, Lazzarini RA, Pick L 1996. Functional dissection of the mouse Hox-a5 gene. EMBO J 15: 1313–1322 [PMC free article] [PubMed] [Google Scholar]
- Zhu A, Kuziora MA 1996. Functional domains in the Deformed protein. Development 122: 1577–1587 [DOI] [PubMed] [Google Scholar]