Abstract
Pre-rRNA transcription by RNA Polymerase I (Pol I) is very robust on active rDNA repeats. Loss of yeast Topoisomerase I (Top1) generated truncated pre-rRNA fragments, which were stabilized in strains lacking TRAMP (Trf4/Trf5–Air1/Air2–Mtr4 polyadenylation complexes) or exosome degradation activities. Loss of both Top1 and Top2 blocked pre-rRNA synthesis, with pre-rRNAs truncated predominately in the 18S 5′ region. Positive supercoils in front of Pol I are predicted to slow elongation, while rDNA opening in its wake might cause R-loop formation. Chromatin immunoprecipitation analysis showed substantial levels of RNA/DNA hybrids in the wild type, particularly over the 18S 5′ region. The absence of RNase H1 and H2 in cells depleted of Top1 increased the accumulation of RNA/DNA hybrids and reduced pre-rRNA truncation and pre-rRNA synthesis. Hybrid accumulation over the rDNA was greatly exacerbated when Top1, Top2, and RNase H were all absent. Electron microscopy (EM) analysis revealed Pol I pileups in the wild type, particularly over the 18S. Pileups were longer and more frequent in the absence of Top1, and their frequency was exacerbated when RNase H activity was also lacking. We conclude that the loss of Top1 enhances inherent R-loop formation, particularly over the 5′ region of the rDNA, imposing persistent transcription blocks when RNase H is limiting.
Keywords: rRNA synthesis, RNA polymerase I, transcription elongation, topoisomerase, RNase H, R-loops
Yeast rDNA is comprised of ∼150–200 tandem repeats, about half of which are actively transcribed. The primary transcript of 35S pre-rRNA is processed into the mature 18S rRNA component of the 40S ribosome subunit and 5.8S and 25S rRNA components of the 60S subunit (see Fig. 1A; Supplemental Fig. S1). Active rDNA repeats are heavily transcribed by RNA Polymerase I (Pol I), as visualized by “Miller” chromatin spreads (French et al. 2003), and this was proposed to involve “transcription factories” where the DNA template is reeled through a polymerase array with constrained mobility (for review, see Cook 1999; Sutherland and Bickmore 2009).
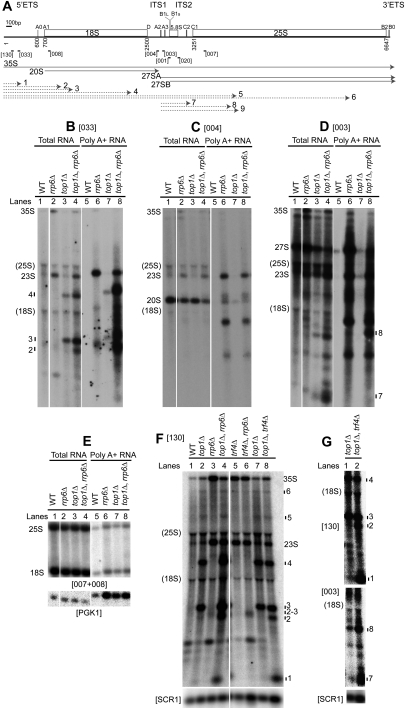
Figure 1.
Aberrant pre-rRNAs generated in the absence of Top1 are targeted for degradation by TRAMP and exosome complexes. (A) Schematic representation of a 35S rRNA gene in S. cerevisiae. Pre-rRNA processing sites A0, A1, D, A2, A3, B1, C2, C1, B2, and B0 are labeled. Probes used in Northern analyses are indicated by bars. The 35S, 20S, 27SA, and 27SB pre-rRNAs are depicted by solid arrows. Truncated pre-rRNA fragments generated in the absence of Top1 are depicted by dotted arrows and are labeled 1–9. Numbers of nucleotides along the rDNA unit relative to the start of transcription (0) are indicated. The pre-rRNA processing pathway is depicted in Supplemental Figure S1. (B–E) Polyadenylated pre-rRNA fragments accumulate in double top1Δ rrp6Δ mutants. Total RNA was extracted from wild-type (WT), rrp6Δ, top1Δ, and top1Δ rrp6Δ strains grown at 25°C. Polyadenylated RNAs were purified and resolved alongside total RNAs on a 1.2% glyoxal-agarose gel. Pre-rRNA fragments were detected by Northern analysis: probe 033, hybridizing at +278 in 5′-ETS (B); probe 004, hybridizing upstream of site A2 (C); and probe 003, hybridizing between A2 and A3 (D). (E) As controls for poly(A)+ RNA specificity, 25S and 18S rRNA and PGK1 mRNA were analyzed. (F,G) TRAMP participates in pre-rRNA surveillance in top1Δ mutants. Total RNA was extracted from wild-type and single and double top1Δ, trf4Δ, and rrp6Δ strains grown at 25°C and analyzed using probe 130 (hybridizes in the 5′-ETS upstream of probe 033) and 003. F and G are from two independent experiments. (Bottom panels) SCR1 was used as a loading control. Intact pre-rRNAs and truncated pre-rRNA fragments are labeled. “(18S)” and “(25S)” indicate the positions of migration of 18S and 25S rRNAs. Probe names are bracketed. The 23S RNA is an aberrant pre-rRNA processing intermediate extending from +1 to site A3, shown previously to be polyadenylated and targeted for degradation by TRAMP and Rrp6 (Dez et al. 2006).
Electron microscopy (EM) analyses (Osheim et al. 2004) and metabolic labeling (Kos and Tollervey 2010) demonstrate that ∼50%–70% of yeast pre-rRNA transcripts are cleaved cotranscriptionally. This indicates that yeast preribosome assembly is at least partly cotranscriptional. Consistent with this, ribosome assembly is impaired in mutants with defects in Pol I elongation, indicating a coupling between these processes (Schneider et al. 2007; Zhang et al. 2009). Surveillance systems monitor yeast ribosome synthesis, and the pre-rRNA components of aberrant preribosomes are polyadenylated by the Trf4/Trf5–Air1/Air2–Mtr4 polyadenylation complexes (TRAMP) and targeted for degradation by the exosome nuclease complex (Dez et al. 2006; Schneider et al. 2007; Wery et al. 2009).
During transcription, either the DNA or the polymerase must rotate once for every ∼10 nucleotides (nt) transcribed. DNA rotation generates positive supercoils (more tightly wound DNA) ahead of the transcription bubble, whereas negative supercoils (more loosely wound DNA) are left behind the polymerase (for review, see Wang and Lynch 1993; Cook 1999). Eukaryotic Topoisomerase I (Top1) Top2 are able to relax both positive and negative DNA supercoils (for review, see Wang 2002), and play essential roles during rDNA transcription in Saccharomyces cerevisiae (Brill et al. 1987; Schultz et al. 1992).
The accumulation of positive supercoiling ahead of the transcription bubble resists opening of the DNA, which can slow or impede transcription elongation by Pol I (Zhang et al. 1988), as seen following treatment with the Top1 inhibitor camptothecin (Koster et al. 2007). In contrast, negative supercoiling behind the transcription bubble can lead to opening of the DNA. When this happens, the nascent RNA may hybridize to the transcribed strand, creating RNA–DNA hybrids known as R-loops (Drolet 2006). R-loops can also impede transcription elongation, as well as exposing the nontemplate strand to cleavage and recombination (for review, see Aguilera and Gomez-Gonzalez 2008). Yeast top1Δ mutants show hyperrecombination of the rDNA array (Houseley et al. 2007), while top1Δ top2-ts double mutants undergo major excisions of the rDNA repeats due to a failure to relax negative supercoils (Trigueros and Roca 2002).
R-loops can be removed by cleavage by RNase H1 and H2, which have overlapping functions and similar catalytic mechanisms (Arudchandran et al. 2000). Yeast RNase H1 is a single protein, whereas RNase H2 is a complex of three proteins: Rnh2A (Rnh201), Ydr279 (Rnh202), and Ylr154 (Rnh203) (Jeong et al. 2004). Seventy percent of RNase H activity is lost upon deletion of RNH201 (Jeong et al. 2004). However, rnh1Δ rnh201Δ double-mutant strains are viable, indicating that they retain some RNase H activity. Moreover, a residual activity was still able to degrade poly(rA):poly(dT) substrates in double-mutant cell extract (Arudchandran et al. 2000), confirming that an additional RNase H activity exists. Yeast RNase H is reported to alleviate transcriptional blocks in THO complex mutants (Huertas and Aguilera 2003), and to destroy RNA–DNA hybrids formed by telomeric TERRA transcripts that accumulate in the absence of Rat1 5′–3′ exonuclease activity (Luke et al. 2008). In higher eukaryotes, overexpression of RNase H can suppress the genomic instability otherwise induced by R-loop formation following depletion of the pre-mRNA splicing factor ASF/SF2 (Li and Manley 2005) or Top1 (Tuduri et al. 2009).
TOP1 deletion was reported to be synthetic-lethal with deletion of the genes encoding the RNA Pol I subunits Rpa34/Rpa49 (Beckouet et al. 2008), a heterodimer that functions during Pol I transcription initiation and elongation (Kuhn et al. 2007; Beckouet et al. 2008). The Trf4 component of the TRAMP complex also interacts genetically with Top1, suggesting further links between pre-rRNA metabolism and rDNA structure (Houseley et al. 2007).
Here we show that Pol I transcription in the absence of Top1 is facilitated by RNase H activity via cleavage of RNA–DNA hybrids engaged in R-loops. We also report that the 5′ end of the 18S rDNA poses problems during pre-rRNA transcription even in the wild type that are exacerbated when both Top1 and Top2 are absent, and represents the major site for R-loop formation.
Results
Aberrant pre-rRNA fragments are generated in top1Δ mutants, and are targeted for degradation by the exosome and TRAMP complexes
To investigate the effects of loss of Top1 on rRNA synthesis, total RNA extracted from wild-type and top1Δ strains was analyzed by Northern. Several truncated species were identified (labeled 1–9 in Fig. 1), which were mapped by comparison of hybridization patterns using multiple probes across the 35S pre-rRNA (see Fig. 1A).
Two major truncated pre-rRNAs, species 3 and 4, were detected with probes 130 and 033, which hybridize to different regions of the 5′-ETS (Fig. 1B [lane 3], F [lanes 2,7]), but not with probe 004, which hybridizes between sites D and A2 (Fig. 1C, lane 3). The RNAs were separated on agarose gels, which have limited resolution, but species 3 and 4 have estimated sizes of ∼1.1 kb and ∼2.1 kb, respectively. This indicates that species 3 extends from the 5′ end of the pre-rRNA to a position ∼0.4 kb into the 18S rRNA region, while species 4 extends ∼1.4 kb into 18S. Two less abundant RNAs, species 5 (∼4 kb) and 6 (∼6 kb), are predicted to extend from the 5′ end of the pre-rRNA to sites within the 25S rRNA (Fig. 1F, lanes 2,7). Species 7 and 8 have 5′ ends at the cotranscriptional cleavage site A2, since they were detected with probe 003 (which lies 3′ to site A2) but not with 004 (5′ to A2) (Fig. 1D [lane 3], G [lane 1]; Supplemental Fig. S3B, lane 2). Species 7 was ∼0.4–0.5 kb in size, corresponding to 3′ ends within ITS2, whereas species 8 was ∼1.2 kb, corresponding to a 3′ end ∼0.4 kb into the 25S rRNA region.
The presence of these truncated pre-rRNA species strongly suggested that the absence of Top1 impedes Pol I transcription, leading to cleavage or release of nascent pre-rRNAs.
In other mutants, aberrant preribosomes are targeted cotranscriptionally by the TRAMP nuclear surveillance complex and degraded by the exosome (Dez et al. 2006; Schneider et al. 2007; Wery et al. 2009). We therefore deleted the gene encoding the nonessential, nuclear exosome component Rrp6 in top1Δ strains. The top1Δ rrp6Δ double-mutant strain showed strongly increased accumulation of RNA species 3, 4 (Fig. 1B [lane 4], F [lane 4]), 7, and 8 (Fig. 1D, lane 4; Supplemental Fig. S3B, lane 4) relative to the top1Δ single mutant. Additional pre-rRNA species were also detected; probe 130, which lies close to the 5′ end of the pre-rRNA primary transcript, detected a small RNA of ∼240 nt, designated species 1 (Fig. 1F, lane 4; Supplemental Fig. S3A, lane 4), while 5′-ETS probes 130 and 033 detected an RNA with a size <1 kb, designated species 2 (Fig. 1B [lane 4], F [lane 4]).
Surveillance of aberrant pre-rRNAs by the TRAMP complex is associated with RNA polyadenylation. Purification of poly(A)+ RNAs from the top1Δ rrp6Δ strain showed that species 2, 3, 4 (Fig. 1B, lane 8), and 8 (Fig. 1D, lane 8) are indeed polyadenylated. In contrast, species 7 was not detectably polyadenylated (Fig. 1D, lane 8), although it remains possible that short oligo(A) tails were present and escaped our purification method. Deletion of TRF4 in a top1Δ background led to stabilization of species 1, 2 (probe 130) (Fig. 1F [lane 8], G [top panel, lane 2]), 7, and 8 (probe 003) (Fig. 1G, lane 2).
Pre-rRNA fragments extending from the 5′ end of the rDNA into the 5′ region of the 18S were detected with probes 130 and 033 in both single rrp6Δ and double trf4Δ rrp6Δ mutants (TOP1+) (species labeled 2 and 3) (Fig. 1F, lanes 3,6). Thus, Pol I transcription through the 5′ region of the 18S rDNA might be a limiting step even in otherwise wild-type cells, as proposed previously (Schneider et al. 2007).
In conclusion, the absence of Top1 leads to the accumulation of two major groups of RNAs: species 1–4 extend from the transcription start site to sites within the 5′-ETS and 18S rDNA, whereas species 7 and 8 extend from the A2 site of cotranscriptional cleavage to positions in ITS2 and the 5′ region of the 25S rDNA (see Fig. 1A). These aberrant pre-rRNAs are polyadenylated by TRAMP and degraded by the exosome.
Pre-rRNA fragments are increased in top1Δ mutants at low growth temperatures
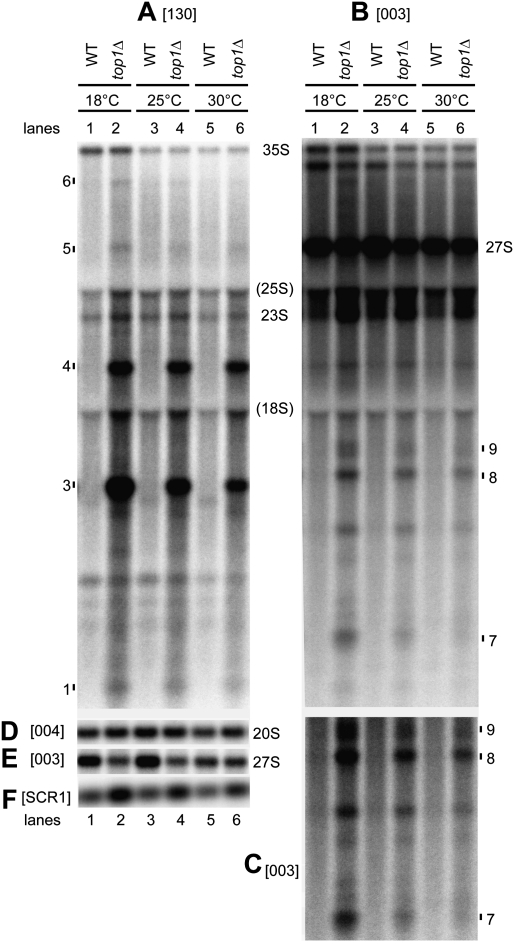
If positive supercoiling before, and/or negative supercoiling behind, transcribing Pol I restrict elongation in top1Δ strains, this might be exacerbated by a more rigid DNA structure at lower growth temperatures (Drolet 2006). To assess this, top1Δ strains were grown at 18°C, 25°C, or 30°C. Northern analyses showed that accumulation of truncated RNA species 3–6 (probe 130) (Fig. 2A) and 7–9 (probe 003) (Fig. 2B,C) increased progressively from 30°C to 25°C to 18°C. The ratio between species 3 and 4 increased as growth temperature decreased (Fig. 2A, cf. lanes 6,4,2). Comparison of the levels of truncated pre-rRNA species with those of 35S pre-rRNA (Fig. 2A–C) showed that species 3 and 4 are more abundant than species 7, 8, and 9, arguing against the possibility that all of these fragments might be generated by cleavage of a single truncated pre-rRNA species.
Figure 2.
Pre-rRNA synthesis defects increase in top1Δ mutant at low growth temperatures. Total RNA from wild-type and single top1Δ strains grown at 18°C, 25°C, or 30°C was resolved on an agarose gel and analyzed by Northern hybridization, using probes 130 (A), 003 (B,C,E), and 004 (D). (F) Cytoplasmic SCR1 RNA was used as a loading control. C shows a stronger exposure of the lower region (below 18S) of B. Intact pre-rRNAs and truncated pre-rRNA fragments are labeled. “(18S)” and “(25S)” indicate the position of migration of 18S and 25S rRNAs. Probe names are bracketed (see Fig. 1A for location of probes).
At 18°C, synthesis of 27S pre-rRNA was reduced, whereas levels of 20S pre-rRNA were affected little (Fig. 2D,E, probes 004 and 003; see Supplemental Fig. S1 for the rRNA maturation pathway). Fast tritium-labeling experiments performed according to Kos and Tollervey (2010) at 25°C showed that the kinetics of cotranscriptional and post-transcriptional processing of pre-rRNA species 35S, 20S, and 27S are delayed in the top1Δ mutant relative to the wild type (data not shown). At lower temperatures, increased transcriptional blockages, in particular in the 5′ region of 18S, would affect the synthesis of all pre-rRNA species. However, blockages downstream from site A2 (Supplemental Fig. S1) would affect only 27S and 35S synthesis, potentially leading to the observed increase in 20S to 27S ratios.
These data indicate that rRNA synthesis in top1Δ strains is more affected at low growth temperatures, where excess supercoils may be more frequent and/or stable (Baaklini et al. 2008). The predominance of species 3 at lower temperatures suggests increased transcription blockage in the 5′ region of 18S rDNA, with reduced synthesis of downstream RNA species. Increased transcription elongation defects may also produce the elevated 20S to 27S pre-rRNA ratio seen at low temperatures.
rRNA transcription stalls in the 5′ region of the 18S rDNA in the absence of both Top1 and Top2
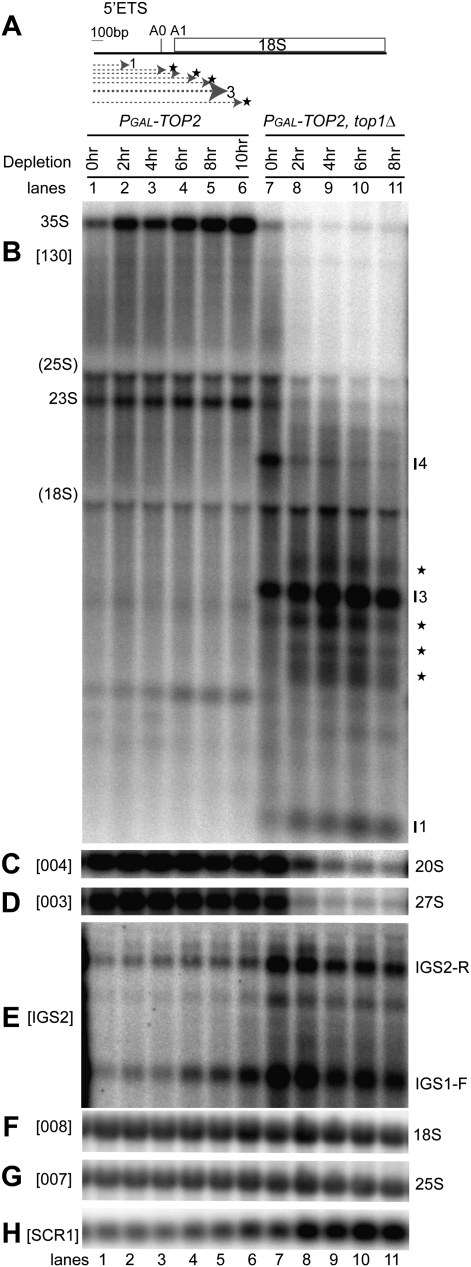
Although pre-rRNAs were truncated and degraded in the absence of Top1, synthesis of full-length pre-rRNA was only partially inhibited. Top1 is partially redundant with Top2, and rRNA synthesis is fully inhibited in double top1Δ top2-ts strains at nonpermissive temperatures (Brill et al. 1987; Schultz et al. 1992). To analyze rRNA synthesis and pre-rRNA fragment accumulation in the absence of Top2 or both Top1 and Top2, we constructed single PGAL-TOP2 and double PGAL-TOP2 top1Δ strains. As expected, both mutant strains showed growth inhibition in glucose medium, since Top2 is essential for viability in yeast. Northern analyses detected no major rRNA synthesis defects in the single PGAL-TOP2 strain even after several hours of depletion (Fig. 3B–H, lanes 1–6; Supplemental Fig. S3A–F, lanes 9–14). In double PGAL-TOP2 top1Δ strains, depletion of Top2 reduced formation of truncated species 4 relative to the top1Δ single mutant (Fig. 3B, cf. lanes 7 and 8–11). Formation of the normal 35S, 20S, 27S (Fig. 3B–D, lanes 7–11), and 7S (Supplemental Fig. S3C, lanes 15–19) pre-rRNAs was also inhibited. However, this was not the case for truncated species 1 and 3 (Fig. 3B, lanes 7–11; Supplemental Fig. S3A, lanes 15–19). In addition, multiple heterogeneous species were detected in the double mutant, with predicted 3′ ends at sites encompassing the region from site A0 (+600 nt in 5′-ETS) to ∼500 nt into the 18S rDNA region (Fig. 3A,B; Supplemental Fig. S3A, species labeled by stars).
Figure 3.
Pre-rRNAs are truncated predominately within the 5′ region of 18S rRNA when both Top1 and Top2 are absent. (A) Diagram representing the 5′-ETS and 18S rDNA sequences. Heterogeneous, truncated pre-rRNA fragments that accumulate in the absence of both Top1 and Top2 are depicted by dotted arrows followed by a star. (B–H) Strains PGAL-TOP2 and PGAL-TOP2 top1Δ were shifted from galactose-containing medium (0 h) to glucose-containing medium at 30°C (2–10 h). Total RNA was analyzed by Northern hybridization. The membrane was hybridized successively with probes 130 (B), 004 (C), 003 (D), 008 (F), and 007 (G). (E) Random primed probe IGS2 was used to detect ncRNAs transcribed by RNA Pol II from the intergenic rDNA spacers IGS1 and IGS2, located between the 35S rDNA units. (H) Cytoplasmic SCR1 RNA was used as a loading control. Intact pre-rRNAs and truncated pre-rRNA fragments are labeled. Heterogeneous truncated pre-rRNA fragments are labeled by stars. “(18S)” and “(25S)” indicate the position of migration of 18S and 25S rRNAs. Probe names are bracketed (see Fig. 1A for location of probes).
The accumulation of these truncated fragments, together with the loss of fragment 4 (see Fig. 3A) and all normal pre-rRNAs, strongly indicates that Pol I elongation is arrested predominately within the 5′ region of the 18S rDNA in strains lacking both Top1 and Top2.
Northern analyses on RNA from a top1Δ top2-ts strain gave similar results, showing the preferential loss of truncated species 4 relative to 1 and 3 at the restrictive temperature of 37°C, as well as significant reductions in the 20S and 27S pre-rRNAs (Supplemental Fig. S4A–C, lanes 1,2).
Together, the data indicate that, in the absence of Top1 alone, Pol I transcription elongation is affected mainly over the 18S sequences. In the absence of Top1 at lower growth temperatures, or when Top1 is lacking and Top2 is limiting, transcriptional blocks become predominant in the 5′ region of 18S, leading to the inhibition of rRNA synthesis.
The absence of both Top1 and RNase H inhibits rRNA synthesis and increases non-protein-coding RNA transcript (ncRNA) expression from intergenic spacers (IGSs)
If the absence of Top1, or both Top1 and Top2, results in rDNA opening behind Pol I, the associated, nascent pre-rRNA transcripts may form R-loops with the DNA template. R-loops are predicted targets for RNase H activity, which might therefore generate the observed pre-rRNA fragments.
A recent analysis reported that overexpression of the Rnh201 subunit of RNase H2 increased degradation of RNA–DNA hybrids over telomeric regions in the 5′–3′ exonuclease rat1-1 mutant (Luke et al. 2008). However, overexpression of Rnh201 in a top1Δ mutant did not alter the abundance or distribution of pre-rRNA fragments (Supplemental Fig. S2). This was also the case for overexpression of Rnh1 or Rnh201 in top1Δ top2-ts mutants at 25°C and 37°C (Supplemental Fig. S4).
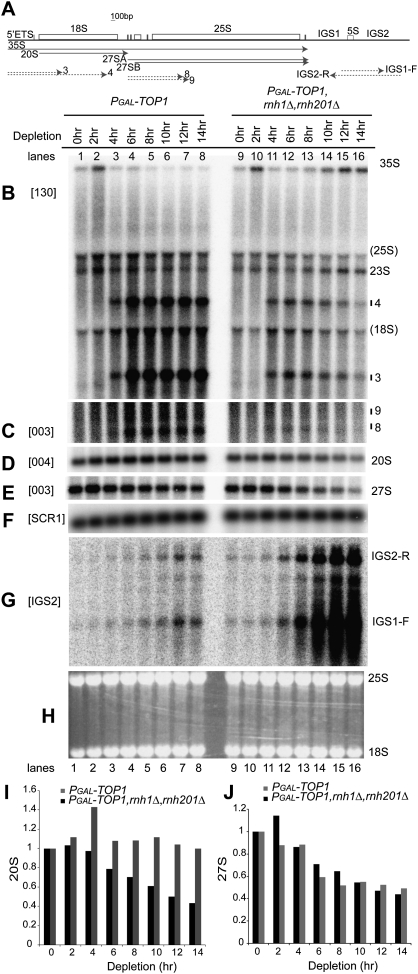
To test whether pre-rRNA fragments accumulated in top1Δ mutants are generated by RNase H cleavage, we initially attempted to delete both RNH1 and RNH201 in top1Δ strains. Deletion of both RNH1 and RNH201 in TOP1 strains had no detectable effect on growth (Arudchandran et al. 2000; data not shown) or rRNA synthesis (Fig. 4B–H, cf. lanes 1 and 9). However, we could not delete both RNH1 and RNH201 in top1Δ strains, suggesting that top1Δ, rnh1Δ, and rnh201Δ are lethal in combination. We therefore placed TOP1 expression under the control of a conditional PGAL promoter and deleted both RNH1 and RNH201 in this strain. Northern analyses were performed using total RNA extracted from single PGAL-TOP1 and triple PGAL-TOP1 rnh1Δ rnh201Δ mutants, which were shifted from permissive galactose-containing meduim (0 h) to nonpermissive glucose-containing medium for 2–14 h at 30°C.
Figure 4.
RNase H facilitates pre-rRNA transcription in the absence of Top1. (A) Schematic showing the locations of pre-rRNAs (solid arrows), truncated pre-rRNA fragments (dashed arrows, left), and RNA Pol II transcribed ncRNAs synthesized from the IGS regions (dashed arrows, right). (B–J) Strains PGAL-TOP1 and PGAL-TOP1 rnh1Δ rnh201Δ were shifted from galactose-containing medium (0 h) to glucose-containing medium at 30°C (2–14 h). Total RNA was analyzed by Northern hybridization. The membrane was hybridized successively with probes 130 (B), 003 (C,E), and 004 (D). (F) SCR1 was used as a loading control. (G) Random primed probe IGS2 was used to detect ncRNAs transcribed from the intergenic rDNA spacers IGS1 and IGS2. (H) Ethidium staining of 25S and 18S rRNAs. Intact pre-rRNAs and truncated pre-rRNA fragments are labeled. “(18S)” and “(25S)” indicate the position of migration of 18S and 25S rRNAs. Probe names are bracketed (see Fig. 1A for locations of probes). (I,J) Quantification of 20S and 27S pre-rRNAs detected by probes 004 (D) and 003 (E). (F) Signals were normalized to the loading control SCR1, and were expressed relative to 0-h samples, which were set to 1.
Growth of the single PGAL-TOP1 mutant strain was only slightly inhibited after the shift to glucose, consistent with the viability of top1Δ mutants (data not shown). Growth of the PGAL-TOP1 rnh1Δ rnh201Δ triple-mutant strain slowed after 10 h of Top1 depletion in glucose-containing medium, and completely ceased after ∼14–16 h (data not shown).
Northern hybridization showed a marked reduction in accumulation of truncated pre-rRNA fragments 3, 4, 8, and 9 in the triple mutant relative to the single mutant during Top1 depletion (Fig. 4B,C). Accumulation of 27S but not 20S was decreased in the single mutant during Top1 depletion (Fig. 4D,E [lanes 1–8], I,J), as was observed in top1Δ strains (see Fig. 2D,E). In contrast, both 20S and 27S accumulation decreased in the triple mutant (Fig. 4D,E [lanes 9–16], I,J).
At early times of Top1 depletion (6 h), the triple mutant showed reductions in both pre-rRNA truncation and synthesis of 20S and 27S pre-rRNA (Fig. 4B–D, lane 12). In the absence of both Top1 and RNase H1 and H201, stable RNA–DNA hybrids may impose transcriptional blocks along the rDNA, further reducing pre-rRNA transcription, as shown in vitro (Tous and Aguilera 2007). This is supported by the almost complete disappearance of pre-rRNA fragments in the triple mutant after 14 h of Top1 depletion (Fig. 4B,C, lane 16). The reduced level of pre-rRNA fragments still seen in the triple PGAL-TOP1 rnh1Δ rnh201Δ mutant after 6 h of Top1 depletion could arise from the residual RNase H activity observed previously in extracts from strains lacking both RNase H1 and H201 (Arudchandran et al. 2000), or from other RNA cleavage activities (see the Discussion).
RNA Pol II generates ncRNA from the rDNA IGSs (IGS1 and IGS2). Northern hybridization showed that ncRNAs IGS1-F and IGS2-R accumulated in mutants lacking Top1 (Fig. 3E, lane 7; Fig. 4G, lanes 7,8), as reported previously (Houseley et al. 2007). Accumulation of these transcripts increased strongly in the triple mutant (Fig. 4G, lanes 9–16), but this was not the case in the PGAL-TOP2 top1Δ strain depleted of Top2 (Fig. 3E, lanes 7–11). Transcription of IGS1-F and IGS2-R is normally limited by a repressive chromatin structure that requires the histone deacetylase Sir2 (Li et al. 2006). IGS transcription in mutants lacking Top1 is possibly due to disruption of the chromatin structure caused by accumulation of supercoils in the IGS1 and IGS2 regions. The increase in ncRNA levels seen in the triple-mutant strain indicates a specific role for RNase H in the synthesis or degradation of IGS transcripts.
RNA–DNA hybrids accumulate over the Pol I promoter, the 5′-ETS, and the IGS regions of the rDNA when both Top1 and RNase H are absent
The accumulation of pre-rRNA fragments with 3′ ends within the 5′ region of the 18S rRNA, in the absence of Top1 alone or both Top1 and Top2, strongly suggested that Pol I might have to overcome a particular problem that requires topoisomerase activity during transcription of that region.
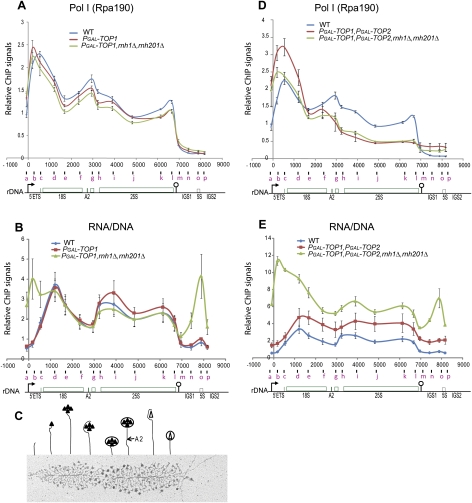
To gain more insight into the roles of topoisomerase and RNase H activities during Pol I transcription elongation, we analyzed by chromatin immunoprecipitation (ChIP) the distribution of both Pol I (Rpa190) and RNA/DNA hybrids along the rDNA in mutants depleted of Top1, or Top1 and Top2, in the presence or absence of RNase H1 and H201 (Fig. 5). Wild-type, PGAL-TOP1, and PGAL-TOP1 rnh1Δ rnh201Δ strains were harvested after 6 h of Top1 depletion in glucose medium at 30°C, at which time growth was affected little in the triple mutant (data not shown). In the wild type (Fig. 5A, blue), the Pol I (Rpa190) signal was maximal over the 5′-ETS (Fig. 5A, probe c), dropped across the 18S, and declined to background levels over the terminator region (Fig. 5A, probe m). ChIP with antibodies directed against the smallest subunit of Pol I (Rpa34-13MYC) gave similar profiles (data not shown). In the single and triple mutants (Fig. 5A, red and green), Pol I signal over the 5′-ETS was shifted toward the promoter region (Fig. 5A, probes a,b), and the overall Pol I signal was decreased in the single and decreased more in the triple mutant.
Figure 5.
ChIP analyses of Pol I and RNA/DNA hybrids over the rDNA unit. ChIP analyses of the occupancy of Pol I (A,D) and RNA/DNA hybrids (B,E) over the rDNA were performed at 6 h after shift from galactose-containing medium to glucose-containing medium at 30°C, using a combination of either wild-type, PGAL-TOP1, and PGAL-TOP1 rnh1Δ rnh201Δ strains (A,B); or wild-type, PGAL-TOP1 PGAL-TOP2, and PGAL-TOP1 PGAL-TOP2 rnh1Δ rnh201Δ strains (D,E). Sonicated cross-linked chromatin from each strain served for the immunoprecipitation of Pol I (Rpa190) (A,D) and RNA/DNA hybrids (B,E). Numbers of base pairs along the rDNA unit relative to the start of 35S rRNA transcription (0) are plotted on the X-axis. The middle position of each PCR fragment quantified is plotted below the X-axis and is indicated by a letter (a–p) (for coordinates of quantitative PCR primers, see Supplemental Table S3). An rDNA cartoon is shown below each graph, with the promoter and terminator depicted by an arrow and a lollipop, respectively. The means of three to five independent experiments are shown with standard errors. Relative ChIP signals are plotted on the Y-axis (for normalizations, see the Materials and Methods). (C) Schematic representation of the ribosome assembly pathway as visualized by EM Miller spreads of active rDNA genes (adapted from Figure 8 in Wery et al. 2009, with permission from the RNA Society, © 2009); see also Osheim et al. 2004). Filled and empty triangles represent preribosomal proteins and synthesis factors assembling cotranscriptionally into nascent preribosomes. Cleavage at site A2 is indicated above the 25S gene. The EM image shows an rRNA gene from a top1Δ strain.
Together, these data suggest that Pol I occupancy is highest over the 5′ region of the rDNA. We propose that R-loops and accumulation of positive supercoils in the 5′ region of the 18S rDNA slow transcription elongation. Stalling or pausing of any polymerase results in other polymerases becoming backed up over the upstream regions, and this is particularly frequent in the absence of Top1.
To assess the occurrence of R-loops along the rDNA, we performed ChIP using the S9.6 antibody, which specifically recognizes RNA–DNA hybrids (see Supplemental Fig. S5 for specificity controls; Boguslawski et al. 1986; Hu et al. 2006). In the wild type (Fig. 5B, blue), RNA–DNA hybrids were detected all along the rDNA, but peaked in the 5′ region of 18S (Fig. 5B, probe d, ∼0.3 kb into the 18S rRNA coding region), dropped over the rest of 18S and ITS (Fig. 5B, probes e–g), increased over the 25S, and declined over the terminator region (Fig. 5B, probe m). In rnh1Δ rnh201Δ (TOP1+) cells, the RNA–DNA hybrid profile was similar to the wild type (data not shown), suggesting that RNA/DNA hybrids detected in the wild type are transient.
Profiles of Pol I and RNA/DNA hybrids over the rDNA in the wild type do not completely overlap (Fig. 5A,B, blue). We propose that the profile of RNA/DNA hybrids largely reflects the cotranscriptional ribosome assembly pathway (Fig. 5B [blue], C). Short nascent pre-rRNA transcripts located toward the 5′ end of the gene may be less amenable to R-looping than longer transcripts located further downstream, in particular over the 5′ region of 18S. The drop in R-loop occupancy over the second half of 18S and ITS spacers is consistent with the packaging of transcripts to form large “terminal balls” (Osheim et al. 2009; Wery et al. 2009). RNA–DNA hybrids were increased substantially over the Pol I promoter and 5′-ETS sequences in the triple mutant after 6 h of Top1 depletion (Fig. 5B, green, probes a–c). This was not the case in the single mutant, which showed a profile similar to the wild type (Fig. 5B, red and blue), suggesting that RNase H1 and H2 are responsible for the cleavage of these RNA/DNA hybrids in the absence of Top1.
RNA–DNA hybrids also increased over the IGS region in the triple mutant (Fig. 5B, green, probes n–p). This is consistent with the accumulation of Pol II ncRNAs from the rDNA IGSs in the absence of Top1 and RNase H (Fig. 4G, lanes 9–16). The simplest hypothesis would be that cleavage of these RNA–DNA hybrids by RNase H1 and H2 normally limits accumulation of the ncRNAs.
To analyze the distributions of Pol I and RNA/DNA hybrids in the absence of both topoisomerases, Top1 and Top2, we placed TOP2 expression under the PGAL promoter in PGAL-TOP1 and PGAL-TOP1 rnh1Δ rnh201Δ strains. Wild-type, PGAL-TOP1 PGAL-TOP2 (double mutant), and PGAL-TOP1 PGAL-TOP2 rnh1Δ rnh201Δ (quadruple mutant) strains were shifted from galactose to glucose medium for 6 h at 30°C and processed for ChIP analyses. Growth rates decreased in both mutants beginning 4–5 h post-shift due to depletion of Top1 and Top2 (data not shown). Pol I ChIP signals in the double and quadruple mutants dropped sharply from the 18S center (Fig. 5D, red and green, probe e). This sharp decrease in Pol I signal over the 18S and downstream regions is consistent with the conclusion from rRNA analyses (Fig. 3). Transcription elongation in the absence of both Top1 and Top2 is largely blocked in the 5′ region of 18S, possibly due to high levels of positive supercoils in this region.
Elevated levels of RNA/DNA hybrids were observed in the top1 top2 double mutant (Fig. 5E, red), but this increase was much more dramatic in the quadruple mutant also lacking RNase H1 and H2 activity, particularly over the 5′ region (Fig. 5E, green). We postulate that the accumulation of negative supercoils in the absence of both Top1 and Top2 provokes DNA strand separation and the formation of RNA/DNA hybrids, which cannot be cleared readily when RNase H is also deficient. Persistent R-loops are predicted to slow Pol I transcription elongation (Tous and Aguilera 2007), and will exacerbate transcriptional blocks imposed by positive supercoiling in the absence of Top1, Top2, and RNase H.
EM reveals polymerase pileups over the 5′-ETS and 5′ region of 18S rDNA in the absence of Top1, RNase H1, and H201
To directly test the model that stable R-loops impose transcriptional blocks along the rDNA, we analyzed Pol I transcription by EM analysis of chromatin spreads. These were prepared from wild-type, PGAL-TOP1 (single mutant), and PGAL-TOP1 rnh1Δ rnh201Δ (triple mutant) strains 6 h after transfer to glucose medium at 30°C.
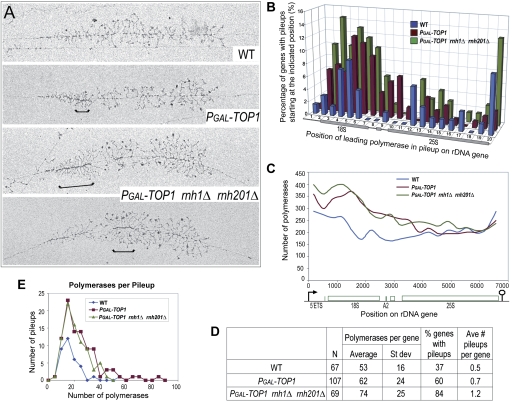
Inspection of Pol I distribution revealed the presence of “pileups,” which were defined as arrays of five or more polymerases with no visible intervening DNA (examples are bracketed in Fig. 6A). We attribute these to transcriptional pausing or stalling of the leading polymerase (situated at the right end of a bracket in Fig. 6A; see Supplemental Fig. S6A for schematic), hindering the progression of subsequent polymerases. In wild-type cells, analysis of all visualized, active rDNA genes (data not shown) revealed pileups on 37% of genes (the number of genes analyzed, N = 105), showing that transient transcriptional pausing is surprisingly frequent. The fraction of genes showing pileups increased to 55% in the PGAL-TOP1 single mutant (N = 180), and to 81% in the triple mutant (N = 209).
Figure 6.
Polymerase pileups over the 5′-ETS and 18S increase when Top1 and RNase H1 and H201 are absent. (A) Representative rRNA genes from Miller spreads of wild-type, PGAL-TOP1, and PGAL-TOP1 rnh1Δ rnh201Δ strains after 6 h of Top1 depletion in glucose at 30°C. Brackets indicate sites of polymerase pileups. These were defined as at least five tightly packed polymerases. The leading polymerase corresponds to the polymerase situated at the right end of a bracket (Supplemental Fig. S6A). (B) Sites of Pol I pausing across the 35S rDNA gene. The gene was divided into 20 equal segments (∼337 bp each), and the position of the leading polymerase in each pileup was plotted onto the segment in which it occurred. The Y-axis shows the percentage of all rDNA genes for each strain with a pileup starting at the indicated position along the gene (X-axis). All rDNA genes that could be visualized from the 5′ to 3′ ends were included in the analysis, and their lengths were normalized. (C) EM analysis of Pol I occupancy over the rDNA unit. For each of the three strains, polymerase positions were measured along 77 rDNA genes, yielding the position of 15,115 polymerases. Each gene was divided into 20 equal segments, and the number of polymerases in each segment was determined. Data were plotted using the midpoints of the 20 gene segments for positioning on the X-axis using smoothed lines. (D) Total frequencies of pileup occurrence for wild-type and mutant strains. (N) Number of rDNA genes analyzed. The same sample of genes was used in B. (E) Plot of pileup lengths (number of polymerases per pileup) for wild-type and mutant strains. The same sample of genes was used in B.
To determine whether polymerases were preferentially paused at particular sites, we divided the 35S gene into 20 equal segments and analyzed all genes that could be unambiguously traced from the 5′ to 3′ ends. We mapped the position of the leading polymerase along a schematic 35S gene, in all pileups (five or more polymerases) (Fig. 6B) or in only longer pileups, which presumably reflect more persistent pauses (20 or more polymerases) (Supplemental Fig. S6B). Notably, the distribution of inferred pause sites along the rDNA was similar in all strains (Fig. 6B; Supplemental Fig. S6B), indicating that pausing is a normal feature of Pol I transcription, which is exacerbated in the mutant strains. In wild type, the major pause sites fell within the 18S rDNA, whereas lower levels of pausing were observed at sites in the 25S rDNA (Fig. 6B, blue). As Top1 was depleted, pausing strongly increased along the 18S rDNA (Fig. 6B, red), and this was exacerbated when RNase H was also limiting (Fig. 6B, green). The increase in pausing was more prominent in the 5′ region of the rDNA when both Top1 and RNase H were deficient, with stalled polymerases at the 5′ region of 18S backed up into the 5′-ETS. The proportion of 35S genes with pileups increased from wild-type to single- to triple-mutant strains (Fig. 6D). Both mutants showed similar numbers of polymerases per pileup, but this was greater than the wild type (Fig. 6E).
Figure 6B shows the locations of the leading polymerase in each pileup on active rDNA genes, but we also quantified the distribution of all polymerases (Fig. 6C). Pol I occupancy in the wild type was predominant over the 5′-ETS and the 5′ region of 18S (Fig. 6C, blue), consistent with Pol I ChIP data (cf. blue graphs in Figs. 5A and 6C). In both mutants, Pol I occupancy increased from the promoter to approximately +4000 nt downstream (Fig. 6C, red and green), with more polymerases across the 5′-ETS and 5′ region of 18S in the triple mutant. The increase in pileup frequency and Pol I occupancy (Fig. 6B,C), together with the increased accumulation of RNA/DNA hybrids (Fig. 5B), across the 5′-ETS and 5′ region of the 18S in the triple mutant suggested that stable R-loops exacerbate Pol I pausing in these regions. Persistent polymerase pileups are predicted to impose transcriptional blocks that would lead to reduced rRNA synthesis (Tous and Aguilera 2007; Klumpp and Hwa 2008), consistent with RNA analyses in the triple mutant (Fig. 4).
Gaps (DNA free of Pol I) were generally observed in front of pileups, but there was no clear correlation between the length of the pileup and gap length (Supplemental Fig. S7). This may simply reflect the stochastic nature of transcription initiation, but might also indicate that paused polymerases impact on the elongation of neighboring polymerases. However, a complication is that the observed pileups presumably represent a mix of those in the process of formation, in which new polymerases are joining, and those in the process of resolution, in which previously stalled polymerases are moving away from the pause site.
In EM analyses, the Pol I distribution observed in spreads of wild-type cells was in good agreement with the occupancy inferred from the ChIP data. The match was less close for PGAL-TOP1 and PGAL-TOP1rnh1Δ rnh201Δ strains, but it should be noted that only polymerases on active genes are counted in EM analyses, whereas ChIP efficiencies reflect the averaged distribution across all rDNA genes. Moreover, the efficiency of immunoprecipitation of cross-linked DNA fragments in ChIP analyses may not be fully proportional to the number of associated polymerases, since each fragment can be precipitated only once even if many polymerases are bound (for further discussion of this point, see Kim et al. 2009).
R-loops associated with stalled polymerases were not visible in the chromatin spreads, suggesting that they may be limited in size by the subsequent polymerases. It is, however, unclear how R-loops would appear in chromatin spreads, or whether regions of rRNA genes harboring R-loops would be amenable to dispersal, since they may be subject to strong topological constraints.
We conclude that RNA Pol I frequently pauses or stalls during rDNA transcription even in wild-type strains, in particular over the 18S region. This could be caused by the accumulation of positive supercoils in front of the polymerase and/or R-loop formation in its wake. Pausing was greatly increased by loss of Top1, and was augmented further when combined with reduced RNase H activity.
Discussion
We report that pre-rRNA transcription is affected by the absence of Top1, particularly over the 5′ region of 18S rDNA. Pre-rRNA fragments that accumulated in the absence of Top1 were consistent with cleavage of nascent transcripts. The truncated pre-rRNA fragments had predicted 5′ ends at the transcription initiation site or at cotranscriptional cleavage site A2 within ITS1 (Fig. 1A). The most abundant fragments extended from the initiation site into the 5′ region of the 18S rRNA. Topoisomerase activity is essential for transcription by Pol I, since rRNA synthesis was abolished in top2-ts top1Δ mutants at 37°C (Brill et al. 1987; Schultz et al. 1992). We showed that, in strains lacking both Top1 and Top2, transcription blockage occurs predominantly within the 18S 5′ region (Fig. 3; Supplemental Fig. S4), again indicating the stringent requirement for topoisomerase activity in this region.
Negative torsion behind transcribing Pol I can lead to unpairing of the DNA helix followed by hybridization of the nascent pre-rRNA to the template strand, forming an R-loop (Drolet 2006; Aguilera and Gomez-Gonzalez 2008). R-loops are targets for RNase H, suggesting that this activity might generate pre-rRNA fragments observed in the absence of Top1 (see model in Fig. 7). Consistent with this, decreased RNase H activity reduced pre-rRNA truncation in strains lacking Top1 (Fig. 4). rRNA synthesis was also reduced in these strains, supporting the conclusion from in vitro data (Tous and Aguilera 2007) that stable R-loops block transcription elongation. Pre-rRNA fragment accumulation was not abolished completely in the absence of Top1 and RNase H1 and H201. This might reflect the residual RNase H activity in these strains (Arudchandran et al. 2000), intrinsic RNase H cleavage activity of A34/A49 heterodimer of RNA Pol I (Kuhn et al. 2007), endonuclease activity of the exosome subunit Rrp44 (Lebreton et al. 2008; Schaeffer et al. 2009; Schneider et al. 2009), or dissociation of the duplex by RNA–DNA helicases like Sen1, which plays a role in Pol I transcription termination (Kawauchi et al. 2008).
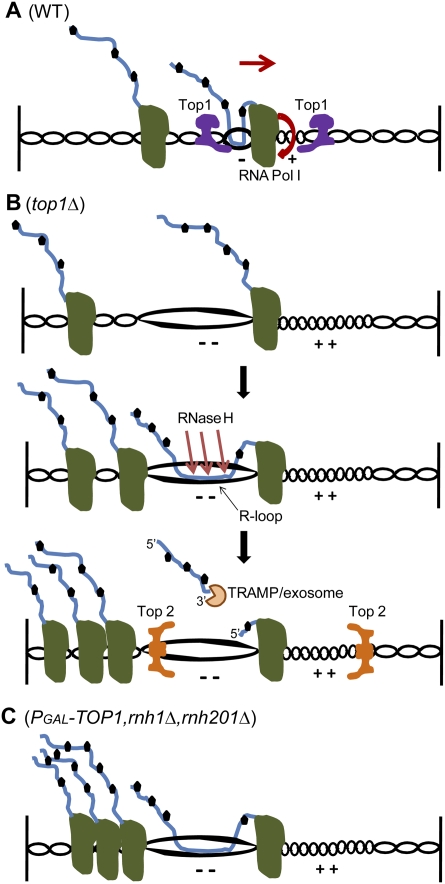
Figure 7.
Model for the role of R-loops in blocking pre-rRNA transcription. (A) Polymerase movement during transcription forces the rDNA to rotate, building up positive torsion (+) in front and negative torsion (−) in its rear (Cook 1999). Torsion ahead of Pol I causes positive supercoiling, which resists strand opening. This slows or pauses transcription elongation, generating polymerase pileups, in particular over the 5′ region of the 18S rDNA. Torsion behind the polymerase leads to negative supercoiling, which facilitates DNA strand opening and stimulates formation of R-loops with the nascent pre-rRNA (Roy et al. 2010). These structures also slow transcription elongation (Tous and Aguilera 2007) and trigger pileup formation. In wild-type strains, pileups are normally transient, with Top1 resolving both negative and positive supercoiling and facilitating transcription. rDNA rotation and direction of transcription are depicted by a bent arrow and a straight arrow, respectively. (B) In strains lacking Top1, which provides the major topoisomerase activity during rDNA transcription, more torsion is accumulated (− − and ++) and R-loops occur more frequently, leading to an increase in pileup formation. RNase H1 and H2 cleave the RNA–DNA hybrids, releasing truncated pre-rRNA fragments that are targeted and degraded by the TRAMP and exosome complexes. Top2 resolves positive and negative supercoiling. Both activities should lead to the release of transcriptional blocks, but Top2 is not predicted to resolve strand separation induced by negative torsion (Lavelle 2008; SL French, ML Sikes, RD Hontz, YN Osheim, TE Lambert, A El Hage, MM Smith, D Tollervey, JS Smith, and AL Beyer, in prep.). (C) In the absence of both Top1 and RNase H1 and H201, persistent R-loops block rotation of the rDNA and cause severe polymerase arrests and pileups.
Truncated pre-rRNA fragments were stabilized in top1Δ strains defective in TRAMP (trf4Δ) or the exosome (rrp6Δ), indicating that these complexes degrade the 5′ pre-rRNA fragments released by RNase H cleavage. The fate of the 3′ fragments resulting from RNase H cleavage is unclear, but rapid degradation by the nuclear 5′-to-3′ exonuclease Rat1 would be predicted. Rat1 functions in transcription termination at the 3′ end of the rDNA (El Hage et al. 2008; Kawauchi et al. 2008), and might also play a role in the release of stalled polymerases associated with transcripts that are engaged in R-loops. Consistent with this model, Rat1 has been shown to function in the termination of RNA Pol II molecules engaged in the production of uncapped mRNA (Jimeno-Gonzalez et al. 2010).
Strains carrying rpa49Δ and rpa34Δ are defective in Pol I elongation (Kuhn et al. 2007), and also accumulated pre-rRNA fragments that terminated in the 5′ region of the 18S rDNA when combined with the exosome mutation rrp6Δ (data not shown), as seen previously in rpa135(D784G) rrp6Δ double mutants (Schneider et al. 2007). Thus, transcription through the 5′ region of the 18S rDNA appears to pose a particular challenge to elongation-compromised Pol I. Moreover, pre-rRNA fragments with 3′ ends that extended to the same region were detected in TRAMP and exosome mutants with intact topoisomerase and RNA polymerase activity, indicating that some level of pre-rRNA truncation normally occurs here in wild-type strains.
In EM analyses, pileups of five or more polymerases were seen on 37% of active wild-type rDNA transcription units, principally over the 18S rDNA. We interpret these as resulting from the collision of multiple transcribing polymerases with a single stalled or paused Pol I. In strains lacking Top1, the proportion of rDNA units with stalled Pol I was increased to 53%, and this rose to 84% in strains lacking both Top1 and RNase H. Notably, the distribution of the leading stalled polymerase was not clearly altered in the different strains, suggesting that the loss of these activities exacerbates an underlying pause that is present in the wild type. In the absence of Top1, both the frequency of pileups and the numbers of contiguous polymerases increased, presumably reflecting increased duration of stalling of the leading polymerase. In strains lacking both Top1 and RNase H activity, pileup frequency increased further. The presence of persistent R-loops might slow down local rotation of the rDNA, reducing the speed of elongation of Pol I in the same pileup, which would increase the residency times of pileups and impede their resolution (see model in Fig. 7). The density of R-loops was increased dramatically in strains lacking both Top1 and RNase H activity, particularly over the promoter and 5′-ETS region (Fig. 5). This may reflect both the increased time available for R-loop formation due to the stalled polymerases, and R-loop stabilization due to reduced RNase H activity.
The similarities in the distribution of stalled Pol I, R-loops, and pre-rRNA cleavage sites suggest that underlying features of transcription in the 5′ region of the 18S rDNA are distinct from other segments of the 6.7-kb pre-rRNA transcript. This region lies ∼1 kb, or ∼100 turns of the rDNA, from the start of transcription, and it is feasible that, simply due to physical features of transcription, Top1 activity first becomes crucial for Pol I transcription over this region. However, the existence of a checkpoint for ribosome assembly, which would slow Pol I elongation until the pre-rRNA is folded and/or bound by r-proteins and pre-40S assembly factors, was suggested previously (Moss et al. 2007; Schneider et al. 2007). In yeast, the U3 snoRNA binds to the 5′-ETS and to the extreme 5′ end of the 18S rRNA sequence, where it may play a major role in the mechanism or timing of formation of the central pseudoknot in 18S rRNA (for review, see Henras et al. 2008). It could be envisaged that binding and/or release of U3 and other early-acting ribosome synthesis factors is synchronized with the transcription elongation rate to promote productive pre-rRNA folding and reduce the tendency of exposed, unfolded RNA sequences to generate R-loops. Such surveillance of preribosome assembly might occur preferentially at a location where Pol I is slowed by physical processes.
In the absence of Top1, levels of Pol II ncRNA transcripts derived from the rDNA IGS regions and rDNA recombination are both increased relative to wild-type strains (Bryk et al. 1997; Houseley et al. 2007). We speculate that underwound DNA in the IGS regions disrupts transcriptional silencing and forms R-loops with ncRNAs, which might be poorly packaged. This leaves the complementary strand single-stranded and prone to cleavage and recombination (Aguilera and Gomez-Gonzalez 2008). In strains lacking Top1 and RNase H, RNA–DNA hybrids accumulated over IGS1 and IGS2, and ncRNA levels were increased relative to top1Δ single mutants, suggesting that RNase H-mediated cleavage is a major pathway of ncRNA degradation in the top1Δ strains.
In yeast, cleavage of the nascent transcripts generated by RNA Pol II is essential to avoid transcriptional roadblocks and for cell viability (Sigurdsson et al. 2010). Similarly, cleavage of the pre-rRNA in top1Δ and Pol I elongation mutants might be essential to avoid transcriptional blocks, in particular over the 5′ region of 18S. Persistent accumulation of RNA polymerase pileups along the rDNA was also predicted to reduce rRNA transcription in modeling data from bacteria (Klumpp and Hwa 2008). R-loop formation also occurs in Top1 mutants in Escherichia coli, and leads to growth defects, impaired transcription elongation on the rDNA, and extensive RNA degradation by RNase H (Hraiky et al. 2000; Drolet 2006; Baaklini et al. 2008). This supports the conclusion (Li and Manley 2006) that the deleterious effects that R-loops can have on transcription have been conserved over a long evolutionary distance.
Materials and methods
Strains, plasmids, and growth conditions
All mutants and tagged yeast strains were constructed using strain BY4741 (derived from S288C). The strains and plasmids used in this study are listed in Supplemental Table S1. Regulated PGAL expression and deletions were constructed by one-step PCR using template plasmids pFA6a-kanMX6-PGAL1-3HA and pFA6a-kanMX6/NatMX6/HphMX6, respectively. Growth and handling of S. cerevisiae were by standard techniques. Cells were grown in YPD medium at 18°C, 25°C, or 30°C. For RNA and ChIP analyses, strains where TOP1 or TOP2 was placed under PGAL1 promoter were grown at 30°C to OD600 ∼0.3–0.4 in complete synthetic medium containing 2% galactose and 2% sucrose, then transferred to the same prewarmed medium containing 2% glucose. For EM analysis, wild-type, PGAL-TOP1, and PGAL-TOP1 rnh1Δ rnh201Δ strains were grown at 30°C in YP-Gal, then shifted to YPD for 6 h.
RNA analyses
RNA extraction, Northern hybridization, and quantifications were as described (Supplemental Material; El Hage et al. 2008). Poly(A) RNA extraction was as described (Supplemental Material; Dez et al. 2006). Primers for RNA analysis are listed in Supplemental Table S2.
ChIP
ChIP experiments were performed essentially as described (Supplemental Material; El Hage et al. 2008). RNA–DNA hybrids were detected using Ab S9.6, kindly provided by Dr. S. Leppla, or purified by Eurogentec from supernatants of a mouse hybridoma cell line purchased from American Type Culture Collection. Pol I subunit Rpa190 was detected with rabbit polyclonal antibody (kind gift from David Schneider). Pol I subunit Rpa34-13MYC was detected using mouse monoclonal antibody c-Myc 9E11 (Santa Cruz Biotechnology). Primers used for quantitative PCRs are listed in Supplemental Table S3.
For detection of both RNA–DNA hybrids and Rpa190 in Figure 5, sonicated DNA from the same strain was split into three equal parts that served for no antibody control, S9.6, and anti-Rpa190. Values for ChIPs were determined using the formula ΔCt = 2−(Ct IP − Ct Background). “Ct IP” is the cycle number for immunoprecipitate, and “Ct Background” is the cycle number for control without antibody. To normalize Pol I ChIP values for Figure 5, A and D, each set of Rpa190-ΔCt values was normalized to 25S (= average of ΔCt values of primers j and k) in the same data set. The means of three to five independent experiments (replicates) were calculated with standard errors. This shows the relative distribution of Pol I along the length of the rDNA in each strain. However, because the values are normalized internally, differences in overall Pol I ChIP signals between different strains will be lost. To assess this, we divided the average of the Rpa190-ΔCt values from each mutant to the average of the Rpa190-ΔCt values from the wild type. The normalized ChIP values obtained for each data set for the mutants were then multiplied by this correction factor.
A different correction was applied in Figure 5, B and E, to account for differences in ChIP efficiencies and Pol I loading. The ΔCt values for RNA/DNA hybrids were each normalized to the average of Rpa190-ΔCts within the same strain and replicate. The means of three to five independent experiments were calculated with standard errors.
EM
Miller chromatin spreads and EM analyses were performed as described (Supplemental Material; Osheim et al. 2009).
Acknowledgments
We thank Steven Leppla, David Schneider, Andrés Aguilera, and Brian Luke for providing reagents. We thank Jean Beggs, Steve Innocente, Laura Milligan, Olivier Cordin, and Hannah Mischo for fruitful discussions; Lea Harrington and Laura Guardano for helping with the production of S9.6 antibodies; and Alexandra Moreira and Steve West for critical reading of the manuscript. This work was supported by the Wellcome Trust and by U.S. Public Health Service grant GM-63952 to A.L.B.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.573310.
Supplemental material is available at http://www.genesdev.org.
References
- Aguilera A, Gomez-Gonzalez B 2008. Genome instability: A mechanistic view of its causes and consequences. Nat Rev Genet 9: 204–217 [DOI] [PubMed] [Google Scholar]
- Arud chandran A, Cerritelli S, Narimatsu S, Itaya M, Shin DY, Shimada Y, Crouch RJ 2000. The absence of ribonuclease H1 or H2 alters the sensitivity of Saccharomyces cerevisiae to hydroxyurea, caffeine and ethyl methanesulphonate: Implications for roles of RNases H in DNA replication and repair. Genes Cells 5: 789–802 [DOI] [PubMed] [Google Scholar]
- Baaklini I, Usongo V, Nolent F, Sanscartier P, Hraiky C, Drlica K, Drolet M 2008. Hypernegative supercoiling inhibits growth by causing RNA degradation. J Bacteriol 190: 7346–7356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckouet F, Labarre-Mariotte S, Albert B, Imazawa Y, Werner M, Gadal O, Nogi Y, Thuriaux P 2008. Two RNA polymerase I subunits control the binding and release of Rrn3 during transcription. Mol Cell Biol 28: 1596–1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boguslawski SJ, Smith DE, Michalak MA, Mickelson KE, Yehle CO, Patterson WL, Carrico RJ 1986. Characterization of monoclonal antibody to DNA.RNA and its application to immunodetection of hybrids. J Immunol Methods 89: 123–130 [DOI] [PubMed] [Google Scholar]
- Brill SJ, DiNardo S, Voelkel-Meiman K, Sternglanz R 1987. Need for DNA topoisomerase activity as a swivel for DNA replication for transcription of ribosomal RNA. Nature 326: 414–416 [DOI] [PubMed] [Google Scholar]
- Bryk M, Banerjee M, Murphy M, Knudsen KE, Garfinkel DJ, Curcio MJ 1997. Transcriptional silencing of Ty1 elements in the RDN1 locus of yeast. Genes Dev 11: 255–269 [DOI] [PubMed] [Google Scholar]
- Cook PR 1999. The organization of replication and transcription. Science 284: 1790–1795 [DOI] [PubMed] [Google Scholar]
- Dez C, Houseley J, Tollervey D 2006. Surveillance of nuclear-restricted pre-ribosomes within a subnucleolar region of Saccharomyces cerevisiae. EMBO J 25: 1534–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drolet M 2006. Growth inhibition mediated by excess negative supercoiling: The interplay between transcription elongation, R-loop formation and DNA topology. Mol Microbiol 59: 723–730 [DOI] [PubMed] [Google Scholar]
- El Hage A, Koper M, Kufel J, Tollervey D 2008. Efficient termination of transcription by RNA polymerase I requires the 5′ exonuclease Rat1 in yeast. Genes Dev 22: 1069–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- French SL, Osheim YN, Cioci F, Nomura M, Beyer AL 2003. In exponentially growing Saccharomyces cerevisiae cells, rRNA synthesis is determined by the summed RNA polymerase I loading rate rather than by the number of active genes. Mol Cell Biol 23: 1558–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henras AK, Soudet J, Gerus M, Lebaron S, Caizergues-Ferrer M, Mougin A, Henry Y 2008. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell Mol Life Sci 65: 2334–2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J, Kotovic K, El Hage A, Tollervey D 2007. Trf4 targets ncRNAs from telomeric and rDNA spacer regions and functions in rDNA copy number control. EMBO J 26: 4996–5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hraiky C, Raymond MA, Drolet M 2000. RNase H overproduction corrects a defect at the level of transcription elongation during rRNA synthesis in the absence of DNA topoisomerase I in Escherichia coli. J Biol Chem 275: 11257–11263 [DOI] [PubMed] [Google Scholar]
- Hu Z, Zhang A, Storz G, Gottesman S, Leppla SH 2006. An antibody-based microarray assay for small RNA detection. Nucleic Acids Res 34: e52 doi: 10.1093/nar/gkl142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas P, Aguilera A 2003. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol Cell 12: 711–721 [DOI] [PubMed] [Google Scholar]
- Jeong HS, Backlund PS, Chen HC, Karavanov AA, Crouch RJ 2004. RNase H2 of Saccharomyces cerevisiae is a complex of three proteins. Nucleic Acids Res 32: 407–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimeno-Gonzalez S, Haaning LL, Malagon F, Jensen TH 2010. The yeast 5′–3′ exonuclease Rat1p functions during transcription elongation by RNA polymerase II. Mol Cell 37: 580–587 [DOI] [PubMed] [Google Scholar]
- Kawauchi J, Mischo H, Braglia P, Rondon A, Proudfoot NJ 2008. Budding yeast RNA polymerases I and II employ parallel mechanisms of transcriptional termination. Genes Dev 22: 1082–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Suh H, Cho EJ, Buratowski S 2009. Phosphorylation of the yeast Rpb1 C-terminal domain at serines 2, 5, and 7. J Biol Chem 284: 26421–26426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klumpp S, Hwa T 2008. Stochasticity and traffic jams in the transcription of ribosomal RNA: Intriguing role of termination and antitermination. Proc Natl Acad Sci 105: 18159–18164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kos M, Tollervey D 2010. Yeast Pre-rRNA processing and modification occur cotranscriptionally. Mol Cell 37: 809–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster DA, Palle K, Bot ES, Bjornsti MA, Dekker NH 2007. Antitumour drugs impede DNA uncoiling by topoisomerase I. Nature 448: 213–217 [DOI] [PubMed] [Google Scholar]
- Kuhn CD, Geiger SR, Baumli S, Gartmann M, Gerber J, Jennebach S, Mielke T, Tschochner H, Beckmann R, Cramer P 2007. Functional architecture of RNA polymerase I. Cell 131: 1260–1272 [DOI] [PubMed] [Google Scholar]
- Lavelle C 2008. DNA torsional stress propagates through chromatin fiber and participates in transcriptional regulation. Nat Struct Mol Biol 15: 123–125 [DOI] [PubMed] [Google Scholar]
- Lebreton A, Tomecki R, Dziembowski A, Seraphin B 2008. Endonucleolytic RNA cleavage by a eukaryotic exosome. Nature 456: 993–996 [DOI] [PubMed] [Google Scholar]
- Li X, Manley JL 2005. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell 122: 365–378 [DOI] [PubMed] [Google Scholar]
- Li X, Manley JL 2006. Cotranscriptional processes and their influence on genome stability. Genes Dev 20: 1838–1847 [DOI] [PubMed] [Google Scholar]
- Li C, Mueller JE, Bryk M 2006. Sir2 represses endogenous polymerase II transcription units in the ribosomal DNA nontranscribed spacer. Mol Biol Cell 17: 3848–3859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke B, Panza A, Redon S, Iglesias N, Li Z, Lingner J 2008. The Rat1p 5′ to 3′ exonuclease degrades telomeric repeat-containing RNA and promotes telomere elongation in Saccharomyces cerevisiae. Mol Cell 32: 465–477 [DOI] [PubMed] [Google Scholar]
- Moss T, Langlois F, Gagnon-Kugler T, Stefanovsky V 2007. A housekeeper with power of attorney: The rRNA genes in ribosome biogenesis. Cell Mol Life Sci 64: 29–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osheim YN, French SL, Keck KM, Champion EA, Spasov K, Dragon F, Baserga SJ, Beyer AL 2004. Pre-18S ribosomal RNA is structurally compacted into the SSU processome prior to being cleaved from nascent transcripts in Saccharomyces cerevisiae. Mol Cell 16: 943–954 [DOI] [PubMed] [Google Scholar]
- Osheim YN, French SL, Sikes ML, Beyer AL 2009. Electron microscope visualization of RNA transcription and processing in Saccharomyces cerevisiae by Miller chromatin spreading. Methods Mol Biol 464: 55–69 [DOI] [PubMed] [Google Scholar]
- Roy D, Zhang Z, Lu Z, Hsieh CL, Lieber MR 2010. Competition between the RNA transcript and the nontemplate DNA strand during R-loop formation in vitro: A nick can serve as a strong R-loop initiation site. Mol Cell Biol 30: 146–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer D, Tsanova B, Barbas A, Reis FP, Dastidar EG, Sanchez-Rotunno M, Arraiano CM, van Hoof A 2009. The exosome contains domains with specific endoribonuclease, exoribonuclease and cytoplasmic mRNA decay activities. Nat Struct Mol Biol 16: 56–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider DA, Michel A, Sikes ML, Vu L, Dodd JA, Salgia S, Osheim YN, Beyer AL, Nomura M 2007. Transcription elongation by RNA polymerase I is linked to efficient rRNA processing and ribosome assembly. Mol Cell 26: 217–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C, Leung E, Brown J, Tollervey D 2009. The N-terminal PIN domain of the exosome subunit Rrp44 harbors endonuclease activity and tethers Rrp44 to the yeast core exosome. Nucleic Acids Res 37: 1127–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz MC, Brill SJ, Ju Q, Sternglanz R, Reeder RH 1992. Topoisomerases and yeast rRNA transcription: Negative supercoiling stimulates initiation and topoisomerase activity is required for elongation. Genes Dev 6: 1332–1341 [DOI] [PubMed] [Google Scholar]
- Sigurdsson S, Dirac-svejstrup AB, Svejstrup JQ 2010. Evidence that transcript cleavage is essential for RNA polymerase II transcription and cell viability. Mol Cell 38: 202–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland H, Bickmore WA 2009. Transcription factories: Gene expression in unions? Nat Rev Genet 10: 457–466 [DOI] [PubMed] [Google Scholar]
- Tous C, Aguilera A 2007. Impairment of transcription elongation by R-loops in vitro. Biochem Biophys Res Commun 360: 428–432 [DOI] [PubMed] [Google Scholar]
- Trigueros S, Roca J 2002. Failure to relax negative supercoiling of DNA is a primary cause of mitotic hyper-recombination in topoisomerase-deficient yeast cells. J Biol Chem 277: 37207–37211 [DOI] [PubMed] [Google Scholar]
- Tuduri S, Crabbe L, Conti C, Tourriere H, Holtgreve-Grez H, Jauch A, Pantesco V, De Vos J, Thomas A, Theillet C, et al. 2009. Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nat Cell Biol 11: 1315–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JC 2002. Cellular roles of DNA topoisomerases: A molecular perspective. Nat Rev Mol Cell Biol 3: 430–440 [DOI] [PubMed] [Google Scholar]
- Wang JC, Lynch AS 1993. Transcription and DNA supercoiling. Curr Opin Genet Dev 3: 764–768 [DOI] [PubMed] [Google Scholar]
- Wery M, Ruidant S, Schillewaert S, Lepore N, Lafontaine DL 2009. The nuclear poly(A) polymerase and exosome cofactor Trf5 is recruited cotranscriptionally to nucleolar surveillance. RNA 15: 406–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Wang JC, Liu LF 1988. Involvement of DNA topoisomerase I in transcription of human ribosomal RNA genes. Proc Natl Acad Sci 85: 1060–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Sikes ML, Beyer AL, Schneider DA 2009. The Paf1 complex is required for efficient transcription elongation by RNA polymerase I. Proc Natl Acad Sci 106: 2153–2158 [DOI] [PMC free article] [PubMed] [Google Scholar]