Abstract
Holliday junction (HJ) resolution is essential for chromosome segregation at meiosis and the repair of stalled/collapsed replication forks in mitotic cells. All organisms possess nucleases that promote HJ resolution by the introduction of symmetrically related nicks in two strands at, or close to, the junction point. GEN1, a member of the Rad2/XPG nuclease family, was isolated recently from human cells and shown to promote HJ resolution in vitro and in vivo. Here, we provide the first biochemical/structural characterization of GEN1, showing that, like the Escherichia coli HJ resolvase RuvC, it binds specifically to HJs and resolves them by a dual incision mechanism in which nicks are introduced in the pair of continuous (noncrossing) strands within the lifetime of the GEN1–HJ complex. In contrast to RuvC, but like other Rad2/XPG family members such as FEN1, GEN1 is a monomeric 5′-flap endonuclease. However, the unique feature of GEN1 that distinguishes it from other Rad2/XPG nucleases is its ability to dimerize on HJs. This functional adaptation provides the two symmetrically aligned active sites required for HJ resolution.
Keywords: Structure-specific nuclease, recombination, DNA repair, DNA replication, FEN1, EXO1, electron microscopy
During meiosis, genetic recombination leads to the formation of four-way intermediate structures (Holliday junctions [HJs]) that link recombining DNA helices at points of strand exchange (Holliday 1964). The resolution of these intermediates can lead to crossover formation, in which flanking markers are exchanged, or noncrossover formation and gene conversion. As crossover formation is critical for the bipolar segregation of homologous chromosomes in meiosis I, HJ resolution represents a critical step in cell biology (Roeder 1997). HJs can also form in mitotic cells during the processes of double-strand break repair (DSBR) or replication fork (RF) restart (Paques and Haber 1999; Cox et al. 2000).
Prokaryotes encode a specialized nuclease called RuvC that promotes the resolution of HJs that arise during recombinational DNA repair (West 1997). RuvC is a dimeric protein that promotes HJ resolution by introducing a pair of symmetrically related nicks in two strands that lie diametrically opposed across the junction point. HJs are known to adopt an anti-parallel stacked-X structure, such that one pair of strands is continuous while the other pair crosses over from one helical axis to the other (Lilley 2000). RuvC-mediated incisions occur in the continuous strands and take place within the lifetime of the RuvC–HJ complex. This mechanism of cleavage is also common to that mediated by the bacteriophage resolvase T7 endonuclease I and yeast mitochondrial resolvase Cce1. Another phage resolvase, T4 endonuclease VII, promotes similar resolution reactions, although in this case the incisions occur in the pair of crossing strands (Declais and Lilley 2008). The products of resolution are nicked duplex molecules that can be readily repaired by DNA ligation.
Despite their functional and mechanistic similarities, these HJ resolvases show little amino acid sequence homology and have evolutionary roots in at least four different structural folds (Aravind et al. 2000; Garcia et al. 2000; Kvaratskhelia et al. 2000; Lilley and White 2000; Nishino et al. 2001). It appears that convergent evolution has endowed them with strikingly similar features that allow them to pursue a common strategy for HJ resolution. All form homodimers (sometimes underpinned by domain swapping) to coordinate two active sites for resolution, and use large, twofold symmetrical, basic surfaces to bind all four arms of the HJ (Lilley and White 2001; Declais and Lilley 2008).
Eukaryotes possess alternative, and more mechanistically varied, ways to process HJs, possibly reflecting the critical importance of this step for cell viability and mutation avoidance. For example, HJs that arise in somatic human cells may be “dissolved” by the actions of the BLM complex (which includes BLM helicase, topoisomerase IIIα, RMI1, and RMI2). This topoisomerase-mediated dissolution reaction always results in noncrossover products, and provides a mechanism that is essential for the avoidance of sister chromatid exchanges (Wu and Hickson 2003). Inactivation of HJ dissolution through genetic mutation leads to the cancer predisposition syndrome known as Bloom's syndrome (Wu and Hickson 2006). Eukaryotic cells also contain a variety of enzymes that can process recombination intermediates, including HJs, by nucleolytic cleavage. One such enzyme is MUS81–EME1, a member of the XPF family of heterodimeric nucleases (Ciccia et al. 2008). The yeast homologs of MUS81–EME1, Saccharomyces cerevisiae Mus81–Mms4 and Schizosaccharomyces pombe Mus81–Eme1, play important roles in the maturation of recombination intermediates leading to crossover formation in meiosis (Interthal and Heyer 2000; Boddy et al. 2001; Smith et al. 2003). They are also required for efficient DNA repair in mitotic cells, in particular after treatment with agents that cause RF stress (Doe et al. 2002). In contrast to the RuvC-like resolvases, MUS81–EME1 preferentially cleave 3′-flap substrates and nicked HJs (nHJs), rather than fully intact four-way intermediates, and HJ cleavage occurs by an asymmetric nick and counternick mechanism that leaves gaps and flaps in the product molecules (Ciccia et al. 2003; Gaillard et al. 2003; Ehmsen and Heyer 2008). Symmetric HJ cleavage has also been observed at high protein concentrations, and may be dependent on the formation of a MUS81–EME1 heterotetramer (Gaskell et al. 2007; Taylor and McGowan 2008). Moreover, MUS81–EME1 also forms part of a larger nuclease complex containing SLX1–SLX4 and the nucleotide excision repair nuclease XPF-ERCC1, raising the possibility that these nucleases cooperate to process HJs (Andersen et al. 2009; Fekairi et al. 2009; Munoz et al. 2009; Svendsen et al. 2009).
Recently, yeast and human HJ resolvases (Yen1 and GEN1, respectively) were identified that promote HJ resolution by RuvC-like symmetrical cleavage (Ip et al. 2008; West 2009). Yen1/GEN1 is a member of the Rad2/XPG family of monomeric, structure-specific nucleases (Harrington and Lieber 1994a; Johnson et al. 1998). This protein family is characterized by an N-terminal (N) and an internal (I) XPG nuclease motif, and a helix–hairpin–helix domain (Lieber 1997; Hosfield et al. 1998). Prominent members of the family include the nucleotide excision repair protein XPG; the Okazaki fragment processing/DNA repair protein FEN1; and EXO1, an enzyme required for DNA replication, DNA repair, and meiotic recombination (Friedberg et al. 2006). The signature activity of the family is the ability to cleave 5′-flaps. However, Yen1/GEN1 has a unique place in the Rad2/XPG family, in that it is also an HJ resolvase.
Deletion of YEN1 in S. cerevisiae severely enhances the sensitivity of mus81Δ cells to RF damage, and this defect is dependent on RAD52 (Blanco et al. 2010; Tay and Wu 2010). This indicates that Mus81 and Yen1 function in overlapping pathways to process replication-associated recombination intermediates in budding yeast. Also, expression of human GEN1 alleviates the severe chromosome segregation defect/meiotic lethality exhibited by mus81 mutants in fission yeast (an organism that lacks a GEN1 homolog) (Lorenz et al. 2010). These data indicate that GEN1 is able to resolve meiotic HJ intermediates to produce crossovers in vivo.
In the work described here, we provide the first mechanistic analysis of HJ resolution by the human GEN1 protein, showing that the reaction shares many of the hallmark features of HJ resolution catalyzed by RuvC. However, there are significant differences that arise from the evolutionary relationships of GEN1 to the Rad2/XPG family of nucleases. The functional adaptation of GEN1 from a monomeric 5′-flap endonuclease into an HJ resolvase has required a novel gain of function. The critical step in this adaptation is the ability of GEN1 to bind and dimerize on the HJ in order to provide the twin active sites necessary to catalyze the symmetrical and simultaneous dual incision reaction required for efficient HJ resolution.
Results
Substrate specificity of GEN1
GEN1 protein (908 amino acids) contains the XPG-N, XPG-I, and helix–hairpin–helix domains essential for nuclease activity, linked to a C-terminal tail region that appears to be naturally disordered (Supplemental Fig. 1A). Using a variety of expression systems, we were unable to generate soluble full-length GEN1 (possibly due to the presence of this disordered tail), leading us to express and purify an active truncated form of the protein, GEN11–527, from Escherichia coli for further analysis. GEN11–527 is similar in length to an active form of GEN1 (∼60 kDa) that was first identified during a search for human HJ-resolving enzymes from HeLa cell-free extracts (Ip et al. 2008).
The specificity of recombinant GEN11–527 was determined using a series of branched and linear DNA substrates produced by annealing partially complementary oligos. All were related by a common 5′-32P end-labeled DNA strand (Fig. 1A; orange). Substrates included linear duplex, splayed arm, 3′-flap, 5′-flap, RF, and HJ DNAs. GEN11–527 cleaved the HJ (substrate VI) most efficiently, followed by the 5′-flap and the RF (substrates IV and V), as indicated by the appearance of fast-migrating products during neutral PAGE (Fig. 1B, lanes 7–12). We did not observe the cleavage of linear duplex, splayed arm, or 3′-flap (substrates I–III) DNAs (Fig. 1B, lanes 1–6).
Figure 1.
Substrate specificity of GEN1. (A) Schematic representation of substrates I–VI showing DNA branch lengths (nucleotides). Sites of incision, determined in C, are indicated. (B) Substrates I–VI, 5′-32P end-labeled in oligo 1 (orange in A), were incubated in the presence or absence of GEN11–527 (15 nM) for 2 min. Reaction products were resolved by neutral PAGE. (C) Substrates I–VI, 5′-32P end-labeled in oligonucleotide 1 (lanes 2–7), or in oligo 5 (substrate I), oligo 4 (substrates II–V), and oligo 3 (substrate VI) (lanes 9–14, respectively; blue in A), were incubated with GEN11–527 as in B, except products were analyzed by denaturing PAGE. (Lanes 1,8) 60-nt marker.
To allow detection/mapping of the sites of incision, the products of cleavage were also analyzed by denaturing PAGE (Fig. 1C). In this case, each substrate was 5′-32P end-labeled in either of two component strands (Fig. 1A, indicated in orange or blue). In accord with data presented in Figure 1B, we did not observe any nicking of linear duplex, splayed arm, or 3′-flap DNA. In contrast, the HJ was cut with perfect symmetry at sites located across the junction and positioned 1 nucleotide (nt) to the 3′ side of the junction point (Fig. 1C, lanes 7,14). With the 5′-flap substrate, we observed removal of the flap strand by incision at one of three sites to release 5′-32P-labeled ssDNA products that were 26, 31, or 32 nt in length (Fig. 1C, lane 12). No incisions were observed in the strand opposing the flap (Fig. 1C, lane 5). The RF substrate (V) was processed by GEN11–527 in a similar fashion (Fig. 1C, lanes 6,13) at a site located 2 nt to the 3′ side of the branch point. The major incision sites on all substrates are summarized schematically in Figure 1A.
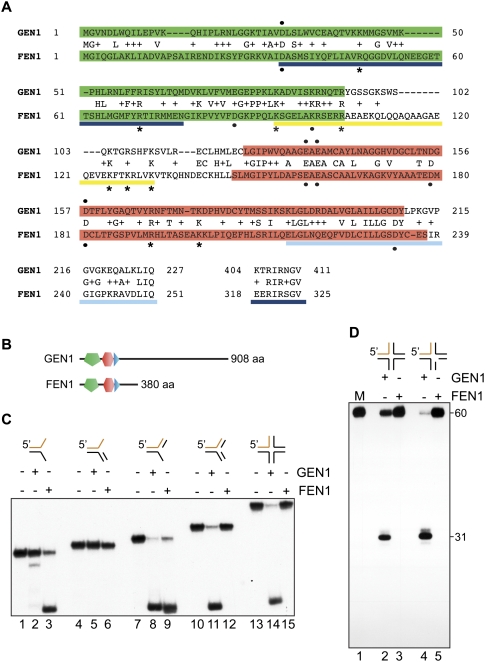
Comparison of GEN1 and FEN1
The ability of GEN11–527 to cut dsDNA substrates containing secondary structures, such as HJs and RFs, appears to distinguish it from other nucleases of the Rad2/XPG family. To define the unique position that GEN1 has within this nuclease family, we compared the activities of GEN11–527 with FEN1, since these enzymes show significant (50%) sequence similarity in their N termini, which contain the bipartite nuclease domain and the helix–hairpin–helix DNA-binding domain (Fig. 2A,B). Because FEN1 is an Mn2+-stimulated nuclease (Harrington and Lieber 1994b), the enzymes were compared at protein concentrations that displayed similar levels of 5′-flap endonuclease activity in an Mn2+-containing buffer. We found that FEN1 cleaved the splayed arm and 5′-flap substrates, whereas no activity was detected with 3′-flap, RF, or HJ substrates (Fig. 2C; Supplemental Fig. 2). As expected, GEN11–527 again showed specificity for the HJ, RF, and 5′-flap substrates. However, in the presence of Mn2+, substrate selectivity was relaxed so that the preference for HJs over the RF and 5′-flap substrates was reduced, and a minor cleavage product (5 nt in length) (see Supplemental Fig. 2) was observed with the splayed arm substrate. When the activities of GEN11–527 and FEN1 were compared with static (X0), mobile (X26), and nicked HJ DNAs, we observed that only GEN1 exhibited cleavage activity with these substrates, with a pre-existing nick accelerating the rate of junction cleavage approximately fivefold (Fig. 2D; data not shown). A similar rate increase with nHJs has also been observed with the RuvC protein, and may relate to increased junction flexibility (Fogg and Lilley 2000).
Figure 2.
Comparative analysis of GEN1 and FEN1. (A) Sequence alignment of the N-terminal regions of GEN1 and FEN1 showing regions of homology. The XPG-N (green) and XPG-I (red) domains are indicated. Regions important for FEN1 DNA binding are indicated: helix–hairpin–helix domain (light blue), helical clamp (yellow), and additional interacting regions (dark blue). Residues important for DNA binding (asterisks) and nuclease activity (black circles) are indicated below for FEN1. Some residues required for GEN1 activity (black circles) are indicated above. Identical and conserved (+) residues are indicated. (B) Position of the XPG-N (green), XPG-I (red), and helix–hairpin–helix (blue) domains in GEN1 and FEN1. (C) Nuclease activities of GEN11–527 and FEN1. The indicated substrates, 5′-32P end-labeled on common oligo 1 (orange), were incubated with GEN11–527 (150 nM) or FEN1 (0.01 U) for 1 min. Reaction products were analyzed by neutral PAGE. (D) HJ X0 (lanes 2,3) and nicked junction X0 (lanes 4,5), 5′-32P-labeled at oligo 1 (orange), were incubated with GEN11–527 (150 nM) or FEN1 (0.01 U) for 1 min. Products were analyzed by denaturing PAGE. (Lane 1) 60-nt marker.
In addition to their structure-specific endonuclease activities, some members of the Rad2/XPG family, such as FEN1, also possess 5′-to-3′ exonuclease activity (Harrington and Lieber 1994b). However, little or no exonuclease activity could be attributed to GEN11–527 when analyzed using 3′-tailed or linear duplex DNA (Supplemental Fig. 3).
To summarize, the nuclease activities of GEN1 and FEN1 are quite distinct despite their evolutionary relationships within the Rad2/XPG family. Although GEN1 retains the characteristic Rad2/XPG ability to cleave 5′-flaps that occur at single-strand/double-strand junctions, it also possesses a property that is unique among this nuclease family, in that it has the ability to cleave DNA substrates with double-stranded branches, and, in particular, to promote HJ resolution.
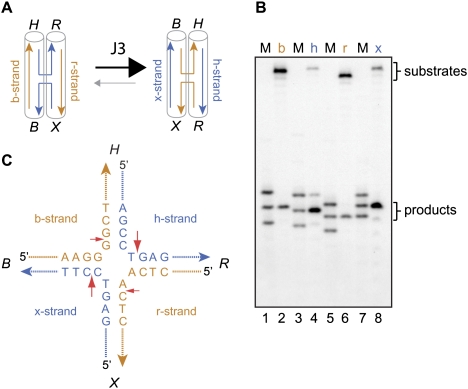
Structural basis of HJ resolution
Previously, it was shown that some junction-specific nucleases (e.g., RuvC, Cce1, and T7 endonuclease I) promote HJ resolution by the introduction of pairwise incisions in the continuous (noncrossing) strands, whereas others (e.g., T4 endonuclease VII) incise the pair of strands that cross from one helical axis to the other (West 1997; Declais and Lilley 2008). To determine the mode of cleavage by GEN1, similar analyses were performed using a well-characterized junction known as J3. In the presence of divalent metal ions, this junction adopts an anti-parallel stacked-X structure with a fourfold conformer bias, such that there is coaxial stacking of helix B on X and H on R (Duckett et al. 1988; Lilley 2000). Thus, in populations of J3, the h and x strands are fourfold more likely to be continuous than exchanging, while the b and r strands are fourfold more likely to be the exchanging pair of strands (Fig. 3A; McKinney et al. 2003). Using four J3 junctions, each 5′-32P-labeled in a different strand, we found that incisions were introduced with a 4:1 ratio into strands h and x (∼80% cleavage) versus strands b and r (∼20% cleavage), closely reflecting the conformer bias of J3, and indicating that GEN11–527 cuts the continuous pair of strands, as observed with RuvC and Cce1 (Fig. 3B). In all cases, cleavage occurred at symmetrically related sites located 1 nt to the 3′ side of the branch point (summarized in Fig. 3C). Similar experiments were also carried out with a different junction (J1) containing sequence changes at the crossover point (Duckett et al. 1988). This junction adopts a different conformer bias compared with J3, and again we observed GEN1-mediated cleavage in the pair of continuous strands (data not shown).
Figure 3.
Incision of the continuous strands of the HJ. (A) Stacking conformers of junction J3. In the preferred form, shown at right, strands x and h (blue) are continuous, while b and r (orange) exchange between helices. (B) Junction J3, 5′-32P-labeled in strands b, h, r, or x, was incubated with GEN11–527 (150 nM) for 2 min. DNA products were analyzed by denaturing PAGE and autoradiography. Lanes 1, 3, 5, and 7 contained a mix of marker oligonucleotides (M) of 15, 16, and 17 nt, representing the 5′ sequence of strands b, h, r, and x, respectively. (C) Central sequence of junction J3. Individual strands are named b, h, r, and x, and corresponding helical segments are named B, H, R, and X. Red arrows indicate GEN11–527 cleavage sites deduced in B.
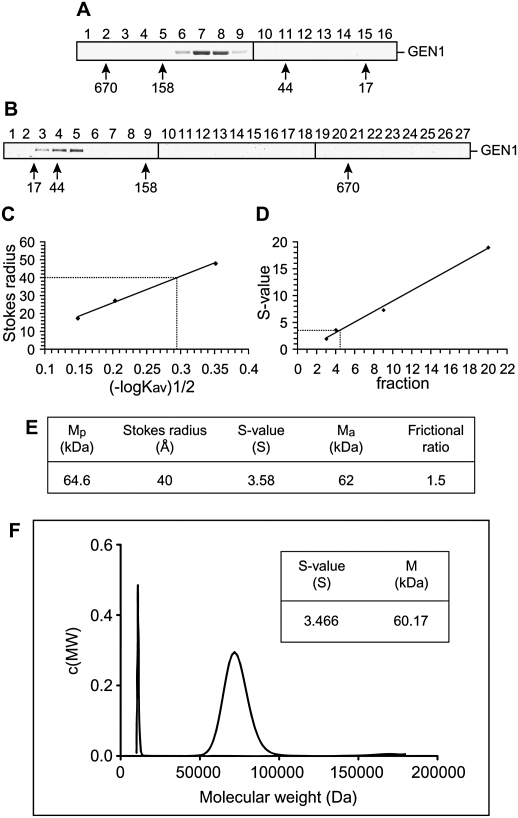
Solution state of GEN11–527
HJ resolution requires a dual incision reaction mediated by two symmetrically related active sites. It is therefore not surprising that all HJ resolvases are dimeric proteins. In contrast, however, Rad2/XPG family members are generally monomeric, raising the question of whether GEN1 retains the characteristic monomeric form, or has developed the ability to dimerize in order to promote HJ resolution. To determine the solution state of GEN1, we carried out a variety of hydrodynamic analyses. When analyzed by size exclusion chromatography and sedimentation velocity ultracentrifugation, GEN11–527 (predicted mass, 64.6 kDa) exhibited a molecular weight of ∼103 kDa or ∼50 kDa, respectively (Fig. 4A,B). These data indicate a Stokes radius of 40 Å and a Svedberg coefficient (S-value) of 3.58 (Fig. 4C,D). We calculated a native molecular mass of 62 kDa for GEN11–527 (Siegel and Monty 1966), in good agreement with the predicted mass of the monomeric protein. That GEN11–527 appears larger than expected (fast elution) in size exclusion chromatography and smaller than expected (slow sedimentation) in density gradient ultracentrifugation is reflected in the frictional ratio of 1.5 (Fig. 4E), suggestive of a deviation from a perfectly globular shape. This may relate to the unstructured C-terminal extension of the protein, given that amino acid residues 422–527 fall into the region predicted to be naturally disordered (Supplemental Fig. 1A).
Figure 4.
Hydrodynamic analysis of GEN11–527. (A,B) Size exclusion chromatography (A) and density gradient ultracentrifugation (B) of GEN11–527. Fractions were analyzed by SDS-PAGE. Mass standards are indicated. (C) Stokes radius of GEN11–527 determined from A. (D) S-value of GEN11–527 from B. (E) Summary of GEN11–527 parameters. Mp and Ma are the predicted and apparent native molecular mass. (F) Molecular weight distribution c(MW) of GEN11–527 determined by analytical ultracentrifugation. The inset shows the S-value and molecular mass of GEN11–527, as calculated using DCDT+.
The native molecular mass of GEN11–527 was confirmed by analytical ultracentrifugation, a more direct method of mass analysis, showing that the protein exists as a single species corresponding to a molecular weight of 60.17 kDa with an S-value of 3.446 (Fig. 4F). We therefore conclude that, like other Rad2/XPG nucleases, GEN11–527 is a monomer in solution.
To remove any concerns that our in vitro observations with GEN11–527 were not truly representative of the actions of GEN1 in vivo, full-length C-terminally FLAP-tagged GEN1 (containing GFP and a Flag epitope) was expressed in HeLa cells and immunoprecipitated using anti-Flag and anti-GFP antibodies. We did not observe the copurification of endogenous GEN1, indicating that the proteins do not self-associate to form dimers in vivo (Supplemental Fig. 4). This observation contrasts with MUS81-FLAP, which copurified with endogenous EME1 under the same conditions.
GEN1–HJ complexes
To gain insight into the way that GEN1 interacts with DNA, we used electron microscopy (EM) to visualize complexes formed between GEN11–527 and HJ575, a HJ that has four arms 575 bp in length. Using reaction conditions that resulted in 52% of HJ575 being bound by GEN11–527, we observed a strong preference for junction-specific binding (Fig. 5A). Scoring 127 individual protein–DNA complexes revealed that 77% of the HJs were bound at the intersection of the four duplex arms, 11% were bound along the duplex portion of one arm (internal binding), and 12% were bound at the end of an arm (Fig. 5B).
Figure 5.
Electron microscopic visualization of GEN11–527. (A) Binding of GEN11–527 to HJ DNA with 575-bp arms. Representative images of the protein (arrows) binding to the junction point, to one of the arms, or at a DNA end are shown. (B) Quantitative analysis of the data represented in A (n = 127). (C) Free GEN11–527 mounted side by side with streptavidin as size reference. (D) HJ575-bound GEN11–527 (arrows) with streptavidin in the background. (E) Size analysis of GEN11–527. Distribution of projected areas of free GEN11–527 (n = 134) and streptavidin (SA, n = 82), and of DNA-bound GEN11–527 (n = 49) and free SA (n = 100), measured from EM images as shown in C (26,000× magnification) and D (42,000× magnification), respectively.
Further analysis indicated some variation in the size of the protein bound to the DNA junctions, but not of protein bound to internal sites or DNA ends (Fig. 5A; data not shown), suggesting that GEN11–527 may interact with HJs in different monomeric/oligomeric states. To address this question, we used EM to analyze the mass of GEN11–-527 free in solution (Fig. 5C) and bound to the HJ substrate (Fig. 5D), using streptavidin, a 52-kDa protein, as a size marker. The projected area of free and DNA-bound GEN11–527 was derived by digital image analysis and compared with the streptavidin control (Fig. 5E). We found that the projected areas of free GEN11–527 and streptavidin were very similar, a result consistent with hydrodynamic analyses indicating that GEN11–527 is monomeric. In contrast, the projected area of GEN11–527 bound to the HJ varied such that the average projected area of DNA-bound GEN11–527 was 9137 pixels compared with 7382 pixels for streptavidin. This translates to a calculated molecular weight of ∼96 kDa for GEN11–527. Although these estimates include a small amount of mass associated with the DNA bound within the GEN11–527 particles, our EM analysis supports the notion that GEN11–527 binds to the intersection of the four duplex arms of HJs in both monomeric and dimeric forms. In contrast, we found that RF substrate RF5 was bound almost exclusively by monomer-size GEN1 particles, and that binding occurred at the branch point of the fork and at internal/DNA end positions with approximately the same frequency (Supplemental Fig. 5 A,B).
The association of GEN1 with HJ DNA was also analyzed by band-shift assays using 32P-labeled synthetic HJ and linear duplex substrates. We found that GEN11–527 readily formed complexes with linear dsDNA (Fig. 6A), but that these complexes could be competed away by the addition of excess unlabeled poly[dI–dC] (Fig. 6C). In contrast, addition of poly[dI–dC] to complexes formed between GEN11–527 and HJ DNA revealed the presence of stable junction-specific complexes (Fig. 6B,D). In the presence of poly[dI–dC], three distinct protein–DNA complexes were observed (Fig. 6D, I–III). These complexes were formed in a concentration-dependent manner, indicative of the recruitment of multiple protein units.
Figure 6.
EMSA analysis of GEN11–527–DNA complexes. GEN11–527 (12.5 nM, 25 nM, 50 nM, 100 nM, 200 nM, 400 nM, 800 nM, or 1.6 μM) was incubated with 5′-32P-labeled linear duplex DNA (A) and HJ X26 (B), and complexes were analyzed by neutral PAGE. (C,D) As A and B, but with poly[dI–dC]. Three specific GEN11–527–HJ complexes are indicated (I–III). (E) Purified GEN11–527 and GEN11–527–MBP (0.5 pmol) were analyzed by 4%–12% SDS-PAGE and stained with Instant Blue. Molecular mass markers (kilodaltons) are indicated. (F) GEN11–527, GEN11–527–MBP, or a mix of both proteins (400 nM total in each case) was incubated with HJ X26 in the presence of poly[dI–dC]. Complexes were analyzed as in A. GEN11–527–DNA complexes I–III and GEN11–527–MBP–DNA complexes I*–III* are indicated. The arrow denotes an intermediate complex present only in the protein mixture.
To determine whether GEN1 forms dimeric complexes with HJ DNA, we adopted a strategy used previously in studies of T4 endonuclease VII and RuvC (Pöhler et al. 1996; Shah et al. 1997). For this, we prepared a construct that expressed a maltose-binding protein (MBP) fused to GEN11–527. The fusion of MBP adds an additional mass of 40 kDa to the protein (Fig. 6E). GEN11–527–MBP was purified and found to form distinct complexes with HJ DNA that exhibited greater retardation than GEN11–527 (Fig. 6F, cf. lanes 2 and 4). Importantly, when GEN11–527–MBP and GEN11–527 were mixed and allowed to bind to the HJ, we observed the formation of an intermediate complex that migrated between GEN11–527 complex II and the corresponding GEN11–527–MBP complex II* (Fig. 6F, lane 3, arrow). These results indicate that GEN11–527 and GEN11–527–MBP associate to form a heterodimeric complex on the HJ DNA, and support the concept of DNA-driven subunit associations, as indicated by the EM studies. Recruitment of more than one GEN11–527 subunit to RFs requires significantly higher protein concentrations (Supplemental Fig. 5C), indicating that the four-way junction itself facilitates the binding reaction.
Mechanism of HJ resolution by GEN1
Bona fide HJ resolvases promote resolution by the introduction of two symmetrically related incisions within the lifetime of the HJ–resolvase complex (Giraud-Panis and Lilley 1997; Fogg et al. 2000). With RuvC, near-simultaneous cleavage is ensured by an increased rate of cleavage of the second scissile bond compared with the first (Fogg and Lilley 2000). This mechanism requires the presence of two active sites, and is distinct from one that involves the introduction of a single nick, followed by protein dissociation, reassociation, and counternicking.
To determine whether GEN11–527 promotes resolution within the lifetime of the enzyme–junction complex, we used a mutant derivative of the inverted repeat-containing plasmid pIRbke8 (Lilley 1985), which we designate pIRbkemut. When supercoiled, this plasmid extrudes the inverted repeat to form a cruciform structure (Fig. 7A). Coordinated cleavage results in the formation of a linear duplex product, whereas uncoordinated cleavage leads to the formation of a nicked duplex plasmid. The nicked plasmid cannot serve as a substrate for resolvases, as the loss of superhelical stress results in cruciform reabsorbtion (Fig. 7A). We found that GEN11–527 resolved pIRbke8mut to form linear products (Fig. 7B). Similar results were obtained over a range of protein concentrations (data not shown). These results indicate that GEN11–527 catalyzes dual incision of the DNA junction within the lifetime of the enzyme–DNA complex, most likely by the coordination of two active centers. Moreover, the formation of very few nicked plasmid products indicates that the monomeric GEN11–527–junction complexes seen by EM are unlikely to be functionally active.
Figure 7.
Mechanism of HJ cleavage by GEN11–527. (A) Schematic of the assay to determine whether resolution occurs within the lifetime of the protein–HJ complex. Supercoiled plasmid pIRbke8mut extrudes its inverted repeat into a cruciform structure. When extruded, the EcoRI site is inaccessible. Incision of one strand at the junction releases superhelical tension, and produces a nicked duplex product in which the cruciform is reabsorbed. Cleavage of both strands at the junction gives rise to a linear duplex product. (B) Analysis of the cleavage mechanism of plasmid pIRbke8mut by GEN11–527 (0, 66 nM, 112 nM, 325 nM, 750 nM, 1.5 μM, or 3 μM). Reaction products were analyzed by agarose gel electrophoresis and quantified. Percent product was corrected for the background of linear DNA observed in the mock reaction and normalized to reflect 75% cruciform extrusion (the substrate fraction refractory to EcoRI cleavage) as 100%. (C) Time course of cleavage of HJ X0 (○) and 5′-flap DNA (◆) by GEN11–527 (10 nM) at the indicated substrate concentrations. Reaction products were resolved by neutral PAGE, quantified by phosphorimaging, and expressed as a percentage of total DNA (±SD, n = 3).
The observed substrate-dependent dimerization of GEN1 appears to be fundamentally different from that seen with other HJ resolvases, which are constitutive dimers. To confirm that this was indeed the case, we explored the possibility that the resolution reaction may be susceptible to substrate inhibition when increasing amounts of substrate were added to sequester protein monomers and thereby prevent dimerization. We found that increases to the substrate to protein ratio decreased the rate of HJ cleavage by GEN11–-527 compared with that of the 5′-flap substrate (Fig. 7C). Substrate inhibition was also observed with the mobile HJ substrate X26 (Supplemental Fig. 6).
These results indicate that monomers of GEN1 cleave 5′-flap structures, consistent with the mode of action of other members of the Rad2/XPG family, and that efficient HJ resolution requires dimerization in order to provide the two active sites required for near simultaneous dual incision (Supplemental Fig. 7). This monomer/dimer substrate-driven switch distinguishes GEN1 from other HJ resolvases, and also from the other members of the Rad2/XPG family of nucleases.
Discussion
The work presented here shows that GEN1 possesses properties distinct from other HJ resolvases, and also from the nuclease family from which it is derived. Members of the Rad2/XPG family are monomeric nucleases that play diverse roles in replication, recombination, and repair, using their 5′-flap endonuclease activities to incise and trim a variety of DNA intermediates. GEN1 has characteristic features of this family, consistent with its evolutionary heritage, but has been adapted from a simple 5′-flap endonuclease into an HJ resolvase. No other member of the Rad2/XPG family exhibits HJ resolution activity. The critical step in this adaptation is GEN1's ability to bind its substrate to form a dimeric complex that contains the two active sites required for the dual incision reaction that is both symmetric and near simultaneous. We showed that dual incision occurs within the lifetime of the HJ–GEN1 complex, and that resolution occurs by cleavage of the continuous, rather than the exchanging, pair of strands. In these respects, GEN1 is a bona fide HJ resolvase that is functionally similar to RuvC.
In our analysis of GEN1, we used a truncated version of the protein spanning amino acids 1–527. This truncation mimics the ∼60-kDa N-terminal fragment of GEN1 originally purified from HeLa cell extracts and identified by mass spectrometry (Ip et al. 2008). It has been shown that the C-terminal tail of GEN1 is dispensable for HJ resolution activity in vitro (Ip et al. 2008), and that expression of GEN11–527 in fission yeast rescues the severe meiotic lethality of mus81Δ cells, indicating that the truncated protein resolves HJs in vivo (Lorenz et al. 2010). At the present time, we have not been able to purify full-length GEN1, in part due to protein insolubility, and also because it is susceptible to protein degradation—findings that may relate to natural disorder within the C terminus. These observations are reminiscent of those found with EXO1, another member of the Rad2/XPG family with a large C-terminal tail, which may also be unstructured (Supplemental Fig. 1). Full-length EXO1 is unstable, and its activities have been characterized using a stable N-terminal fragment (Wilson et al. 1998; Qiu et al. 1999). Interestingly, the C terminus of human EXO1 contains multiple phosphorylation sites that, in response to RF blockage, serve to regulate the stability of EXO1 in vivo (El-Shemerly et al. 2005, 2008). Presently, the function of the C terminus of GEN1 is unknown, but, by analogy to EXO1, it may be an acceptor for post-translational modification. In this regard, it is noteworthy that budding yeast Yen1, the homolog of GEN1, is phosphorylated in a cell cycle-specific manner (Ubersax et al. 2003), and that a phosphorylation site at the C terminus is involved in nuclear import/export of the protein (Kosugi et al. 2009).
In vitro analysis shows that GEN11–527 is a structure-specific nuclease, acting preferentially on HJs, 5′-flaps, and RF substrates. The nucleolytic cleavage of fully double-stranded RFs indicates a departure from the activities typically associated with the Rad2/XPG nucleases. XPG, FEN1, and EXO1 possess 5′-flap endonuclease activity, and all cleave splayed arm substrates but not RFs (Harrington and Lieber 1994b; Lee and Wilson 1999; Hohl et al. 2003). In contrast, simple intersections of ssDNA and dsDNA, such as those in splayed arm substrates, are not sufficient to induce cleavage by GEN11–-527 under normal reaction conditions. Sequence comparisons of GEN1 and FEN1 show a strong degree of homology across the nuclease and helix–hairpin–helix domains. However, two apparent deletions in GEN1 correspond to regions that are implicated in precise and efficient cleavage of 5′-flap substrates by FEN1. One comprises a conserved threonine residue (T61 in FEN1) that binds an unpaired nucleotide on the 3′ side of the flapped DNA strand interruption. The other is part of a helical arch (indicated in yellow in Fig. 2A) located above the active site, which appears to encircle or clamp down on the 5′-flap, creating a groove suitable for ssDNA interactions (Shen et al. 2005). These deletions would be consistent with the concept that the relevant targets of GEN1 are double-stranded, as a 3′-end binding pocket would be obsolete, and the helical clamp may have to be altered to create a space large enough for duplex DNA. Side-by-side analysis of GEN11–527 and FEN1 illustrates that the 5′-flap endonuclease activity of GEN11–527 is less precise than that of FEN1, and is not restricted to the site of the ssDNA–dsDNA junction, as cleavage also occurs within the flap itself. It is possible that the 5′-flap is not a cognate GEN1 substrate, and that amino acid residues that would normally position an accurately fitting dsDNA substrate, when unoccupied, may lead to slippage of the 5′-flap and result in spurious cleavage. Under our assay conditions, FEN1 was completely inactive on fixed, mobile, and nicked HJs. This rules out the possibility that HJ cleavage by GEN1 relates to a simple extension of its 5′-flap endonuclease activity induced by breathing (exposure of single-stranded regions) at the junction core.
The adaptation of GEN1 to accept duplex strands would appear a natural prerequisite for HJ resolution. Moreover, this ability to promote HJ resolution appears to be an acquired and specific activity of GEN1 that sets the enzyme apart from all other Rad2/XPG family members. In contrast to all single-scission endonuclease activities associated with GEN1 and other Rad2/XPG nucleases, HJ resolution requires the accurate coordination of two incisions. GEN11–527 specifically places these incisions in the continuous strand of the HJ, as also observed with RuvC, Cce1, and T7 endonuclease I, but in contrast with T4 endonuclease VII. This difference is thought to reflect the distance between the two active sites within the dimer: The active sites in T7 endonuclease I are separated by a distance of ∼30 Å to match the spacing of scissile bonds in continuous strands proximal to the junction point (Hadden et al. 2007), whereas the active sites in T4 endonuclease VII are positioned to accommodate the significantly shorter distance of ∼15 Å between scissile bonds in the exchanged strands (Biertumpfel et al. 2007). To a first approximation, the specificity of GEN11–527 for continuous strand cleavage close to the junction point indicates that the active sites are likely to be positioned ∼30 Å apart.
Typical of other Rad2/XPG family members, we found that GEN1 is monomeric in solution, and yet has the capacity to dimerize on the HJ so that two active sites can coordinate a dual incision reaction. Our studies suggest two modes of action for GEN1, according to which single-cleavage substrates (such as 5′-flaps) are processed by monomeric GEN1, while HJs are cut only after the assembly of GEN1 dimers (Supplemental Fig. 7). Given that we did not find any substantial nicking of HJs by monomers of GEN1, it is possible that dimer assembly results in a conformational change that provides a trigger for dual incision. The requirement for dimer assembly could serve as a licensing step that prevents futile HJ nicking by a GEN1 monomer.
In summary, two functional adaptations appear to enable GEN1 to function as a resolvase: (1) the ability to accommodate fully dsDNA substrates, and (2) the ability to assemble a functional protein dimer via substrate-directed binding. The latter deviation from normal Rad2/XPG function allows GEN1 to act on HJs like a classical RuvC-like resolvase, albeit by a distinct mechanism. The end products of the cleavage reaction, however, are indistinguishable from those produced by the prokaryotic HJ resolvases, suggesting that the adaptation of GEN1 represents a new solution to the problem of HJ resolution.
Materials and methods
Sequence analysis
The sequences for human GEN1 (NP_872431), FEN1 (NP_004102), and EXO1 (NP_006018) were retrieved from the NCBI database (http://www.ncbi.nlm.nih.gov), and were analyzed using BLAST. Protein disorder prediction was performed with iPDA (Su et al. 2007).
Proteins
Human GEN11–527 (Ip et al. 2008) and GEN11–527–MBP (carrying MBP at its C terminus) were expressed in E. coli BL21(RIL)DE3 from a pET-DEST42 derivative with a C-terminal V5/His tag and purified by HisTrap, heparin, ssDNA-cellulose, and monoS chromatography. The MBP sequence (malE from E. coli) was taken from pMAL-c4X (New England Biolabs). Human FEN1 was purchased from Trevigen.
DNA substrates
Synthetic DNA substrates were prepared by annealing the oligonucleotides shown in Supplemental Table 1. HJ X0 (Benson and West 1994) was assembled from oligos 1–4 and contains four heterologous arms. Other structures derived from the X0 sequence contained oligo 1 in different combinations: linear duplex (oligos 1 and 5), splayed arm (oligos 1 and 4), 3′-flap (oligos 1, 4, and 6), 5′-flap (oligos 1, 4, and 7), RF (oligos 1, 4, 6, and 7), nHJ (oligos 1, 2, 4, 8, and 9), and duplex with a recessed 5′ end (oligos 1 and 10). HJ J3 (Duckett et al. 1988) contained oligos 11–14, and HJ X26 (Constantinou et al. 2001), with a 26-bp homologous core, was composed of oligos 15–18. Substrates were prepared as described (Rass and West 2006). One oligonucleotide was 5′-end-labeled using T4 polynucleotide kinase (New England Biolabs) and γ-32P ATP (PerkinElmer).
Plasmid pIRbke8mut (3683 bp) was generated from pIRbke8 (Lilley and Markham 1983) by mutating one of its two EcoRI sites to leave a single site at the center of the inverted repeat. Plasmid DNA was propagated in E. coli DH5R and isolated in supercoiled form. Cruciform extrusion was stimulated by incubation for 90 min at 37°C in 50 mM Tris-HCl (pH 7.5), 50 mM NaCl, and 0.1 mM EDTA. HJ575 and RF5 DNA were prepared as described (Lee et al. 1997; Subramanian and Griffith 2005).
Nuclease assays
Unless stated otherwise, reactions (10 μL) contained 32P-labeled synthetic substrate DNA (1 nM) in phosphate buffer: 60 mM sodium phosphate (pH 7.4), 1 mM DTT, 0.1 mg/mL BSA, and 5 mM Mg(OAc)2. After incubation at 37°C, DNA products were deproteinized for 15 min at 37°C using 2 mg/mL proteinase K and 0.4% SDS. The comparison of FEN1 and GEN11–527 was carried out in buffer REC12 (Trevigen) containing 50 mM Tris-HCl (pH 8), 10 mM MnCl2, 1 mM DTT, 0.1 mg/mL BSA, and 5% glycerol. Products were analyzed by 10% neutral PAGE or 12% denaturing PAGE (containing 7 M urea), followed by autoradiography. Quantifications were carried out using a Typhoon scanner for phosphorimaging and ImageQuant image analysis software (GE Healthcare).
Cleavage of plasmid pIRbke8mut (20 nM) was carried out in phosphate buffer. Reactions were prewarmed to 37°C and initiated by enzyme addition, and, after 5 min, reactions were terminated/deproteinized. DNA products were resolved by 1% agarose gel electrophoresis, stained with SYBR Green (Roche), and analyzed using a Typhoon scanner and ImageQuant software. Complete digestion with EcoRI was used to determine the fraction of plasmid DNA that did not extrude the cruciform.
Hydrodynamic analysis
Size exclusion chromatography was carried out at 4°C on a Superdex 200 PC 3.2/30 column (GE Healthcare) equilibrated in 50 mM potassium phosphate (pH 7.2), 150 mM KCl, 10% glycerol, 1 mM EDTA, and 1 mM DTT. GEN11–527 (50 μL, 7.5 μg, 2.32 μM) was applied to the column, and 50-μL fractions were collected, analyzed by 10% SDS-PAGE, and stained using Instant Blue (Expedeon). The elution volume for GEN11–-527 was determined and converted into parameter Kav, and the Stokes radius was estimated using a linear calibration plot of Stokes radii versus (−log Kav)1/2 values obtained for a gel filtration standard (Bio-Rad) containing thyroglobulin (670 kDa), γ-globulin (158 kDa), ovalbumin (44 kDa), and myoblobin (17 kDa). The frictional ratio was calculated as described (Tsai 2006).
Sedimentation velocity ultracentrifugation was performed using a 15%–35% (v/v) 4.8-mL glycerol density gradient made in size exclusion buffer and layered in 13 × 51-mm open-top centrifugation tubes (Beckman Coulter). GEN11–527 (150 μL, 7.5 μg, 0.77 μM) was loaded and centrifugation was carried out in a SW55Ti swinging bucket rotor at 42,000 rpm for 16 h at 4°C (Optima LE-80K Ultracentrifuge, Beckman Coulter). Bio-Rad gel filtration standards were used for calibration. Fractions (180 μL) were collected and aliquots (12 μL) were analyzed by 10% SDS-PAGE.
Analytical ultracentrifugation was carried out in 50 mM potassium phosphate (pH 7.2) containing 100 mM KCl using a Beckman XL-I Ultracentrifuge with absorbance optics at 280 nm. The velocity sedimentation of GEN11–527 (400 μL, 160 μg, 6.19 μM) was measured at 16°C, and data were analyzed using the improved dc/dt method as implemented in DCDT+ (Philo 2006) and as a continuous sedimentation distribution in SEDFIT (Schuck 2000). Good fits to experimental data were obtained with an even distribution of residuals by both methods.
EM
GEN11–527 (50 nM) and HJ575 (1 nM) or RF5 (2 nM) DNA were incubated for 30 min at 37°C in 50 mM Tris-HCl (pH 8.0), 1 mM DTT, and 5 mM EDTA. Protein–DNA complexes were fixed with 0.6% glutaraldehyde, and either diluted in 10 mM Tris-HCl (pH 7.6) and 0.1 mM EDTA, or passed over a column containing 2% agarose beads (Agarose Bead Technology) equilibrated with 10 mM Tris-HCl (pH 7.6) and 0.1 mM EDTA to remove unbound GEN11–527. Protein–DNA complexes were mixed with a buffer containing 2.5 mM spermidine, and were incubated on glow charged carbon grids for 3 min (Griffith and Christiansen 1978). Samples were washed with a series of water–ethanol washes, air-dried, and rotary shadowcast with tungsten at 1 × 10−6 Torr. Samples were analyzed using an FEI Tecnai 12 transmission electron microscope (FEI, Inc.) at 40 kV, and images were captured on a Gatan Ultrascan 4000 slow-scan CCD camera with supporting software (Gatan, Inc.).
For mass analysis, free GEN11–527 or streptavidin were diluted to 10 ng/μL in 20 mM HEPES (pH 7.5) and 0.1 mM EDTA, mixed with an equal volume of 1.2% glutaraldehyde, and incubated on ice for 10 min prior to preparation for EM. For GEN11–527–DNA complexes, streptavidin was added at a concentration of 1 ng/μL. Fields of free or DNA-bound GEN11–527 were captured on digital micrographs. Projected surface areas were measured using ImageJ software (Abramoff et al. 2004), and the molecular weight of DNA-bound GEN11–527, the mean, and SEM (standard error of the mean) were calculated. A scatter plot showing the distribution of measured particles was constructed using GraphPad Prism version 5. To determine the molecular weight from the projected area, with streptavidin as the standard, we used the equation Mwsample/Mwstandard = (Projected areasample/Projected areastandard)3/2 (Griffith et al. 1995).
DNA-binding assays
Reactions (10 μL) contained 32P-labeled DNA substrates (1 nM) in 60 mM sodium phosphate (pH 7.4), 1 mM DTT, 0.1 mg/mL BSA, 5 mM EDTA, and 5% glycerol. Poly[dI–dC] (60 ng) was added as indicated. Incubation was for 10 min at room temperature. Reactions were then put on ice and analyzed immediately by 4% neutral PAGE at 4°C, followed by autoradiography.
BAC-mediated protein expression
BACs harboring GEN1 (clone 2190M2) and MUS81 (clone 2084A20) were obtained from Invitrogen. A localization and affinity purification (LAP) cassette encoding GFP, or a FLAP cassette additionally encoding the Flag epitope, was inserted as a C-terminal fusion using recombineering (Gene Bridges) (Poser et al. 2008). HeLa cells expressing LAP- or FLAP-tagged versions of GEN1 or MUS81 were harvested and, after lysis in LAP buffer (Poser et al. 2008), cleared from insoluble material by ultracentrifugation. Lysates were normalized for total protein content and analyzed by Western blotting. Tagged proteins were immunoprecipitated using anti-Flag M2 affinity gel (Sigma) for 2 h at 4°C, eluted using Flag peptide, and incubated with GFP-Trap matrix (Chromotek) for 1 h at 4°C. Immunoprecipitated material was eluted by boiling in NuPAGE LDS sample buffer, and analyzed by Western blotting using the following antibodies: mouse anti-GFP (Roche), mouse anti-MUS81 (MTA30 210/3, Abcam), mouse anti-EME1 (MTA31 7H2/1, Santa Cruz Biotechnology), and rabbit polyclonal anti-GEN1 raised against C-terminal peptide 890–908.
Acknowledgments
We thank Anthony Hyman for providing the BAC modification cassettes. This work was supported by grants to S.C.W. (Cancer Research UK, the Breast Cancer Campaign, the Louis-Jeantet Foundation, the Swiss Bridge Foundation, and the European Research Council), and to J.D.G. and S.A.C. (National Institutes of Health grants GM31819 and ES31773). M.G.B. was supported by the Angeles Alvariño program of the Xunta de Galicia (Spain).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.585310.
Supplemental material is available at http://www.genesdev.org.
References
- Abramoff MD, Magelhaes PJ, Ram SJ 2004. Image processing with ImageJ. Biophotonics Int 11: 36–42 [Google Scholar]
- Andersen SL, Bergstralh DT, Kohl KP, LaRocque JR, Moore CB, Sekelsky J 2009. Drosophila MUS312 and the vertebrate ortholog BTBD12 interact with DNA structure-specific endonucleases in DNA repair and recombination. Mol Cell 35: 128–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Makarova KS, Koonin EV 2000. Holliday junction resolvases and related nucleases: Identification of new families, phyletic distribution and evolutionary trajectories. Nucleic Acids Res 28: 3417–3432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson FE, West SC 1994. Substrate specificity of the Escherichia coli RuvC protein: Resolution of 3- and 4-stranded recombination intermediates. J Biol Chem 269: 5195–5201 [PubMed] [Google Scholar]
- Biertumpfel C, Yang W, Suck D 2007. Crystal structure of T4 endonuclease VII resolving a Holliday junction. Nature 449: 616–620 [DOI] [PubMed] [Google Scholar]
- Blanco MG, Matos J, Rass U, Ip SCY, West SC 2010. Functional overlap between the structure-specific nucleases Yen1 and Mus81–Mms4 for DNA damage repair in S. cerevisiae. DNA Repair (Amst) 9: 394–402 [DOI] [PubMed] [Google Scholar]
- Boddy MN, Gaillard PHL, McDonald WH, Shanahan P, Yates JR, Russell P 2001. Mus81–Eme1 are essential components of a Holliday junction resolvase. Cell 107: 537–548 [DOI] [PubMed] [Google Scholar]
- Ciccia A, Constantinou A, West SC 2003. Identification and characterization of the human MUS81/EME1 endonuclease. J Biol Chem 278: 25172–25178 [DOI] [PubMed] [Google Scholar]
- Ciccia A, McDonald N, West SC 2008. Structural and functional relationships of the XPF/MUS81 family of proteins. Annu Rev Biochem 77: 259–287 [DOI] [PubMed] [Google Scholar]
- Constantinou A, Davies AA, West SC 2001. Branch migration and Holliday junction resolution catalyzed by activities from mammalian cells. Cell 104: 259–268 [DOI] [PubMed] [Google Scholar]
- Cox MM, Goodman MF, Kreuzer KN, Sherratt DJ, Sandler SJ, Marians KJ 2000. The importance of repairing stalled replication forks. Nature 404: 37–41 [DOI] [PubMed] [Google Scholar]
- Declais AC, Lilley DMJ 2008. New insight into the recognition of branched DNA structure by junction-resolving enzymes. Curr Opin Struct Biol 18: 86–95 [DOI] [PubMed] [Google Scholar]
- Doe CL, Ahn JS, Dixon J, Whitby MC 2002. Mus81–Eme1 and Rqh1 involvement in processing stalled and collapsed replication forks. J Biol Chem 277: 32753–32759 [DOI] [PubMed] [Google Scholar]
- Duckett DR, Murchie AIH, Diekmann S, Von Kitzing E, Kemper B, Lilley DMJ 1988. The structure of the Holliday junction and its resolution. Cell 55: 79–89 [DOI] [PubMed] [Google Scholar]
- Ehmsen KT, Heyer WD 2008. Saccharomyces cerevisiae Mus81–Mms4 is a catalytic, DNA structure-selective endonuclease. Nucleic Acids Res 36: 2182–2195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Shemerly M, Janscak P, Hess D, Jiricny J, Ferrari SR 2005. Degradation of human exonuclease 1β upon DNA synthesis inhibition. Cancer Res 65: 3604–3609 [DOI] [PubMed] [Google Scholar]
- El-Shemerly M, Hess D, Pyakurel AK, Moselhy S, Ferrari SR 2008. ATR-dependent pathways control hEXO1 stability in response to stalled forks. Nucleic Acids Res 36: 511–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekairi S, Scaglione S, Chahwan C, Taylor ER, Tissier A, Coulon S, Dong MQ, Ruse C, Yates JR, Russell P, et al. 2009. Human SLX4 is a Holliday junction resolvase subunit that binds multiple DNA repair/recombination endonucleases. Cell 138: 78–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogg JM, Lilley DMJ 2000. Ensuring productive resolution by the junction-resolving enzyme RuvC: Large enhancement of the second-strand cleavage rate. Biochemistry 39: 16125–16134 [DOI] [PubMed] [Google Scholar]
- Fogg JM, Schofield MJ, Declais AC, Lilley DMJ 2000. Yeast resolving enzyme CCE1 makes sequential cleavages in DNA junctions within the lifetime of the complex. Biochemistry 39: 4082–4089 [DOI] [PubMed] [Google Scholar]
- Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T 2006. DNA repair and mutagenesis ASM Press, Washington, DC [Google Scholar]
- Gaillard P-HL, Noguchi E, Shanahan P, Russell P 2003. The endogenous Mus81–Eme1 complex resolves Holliday junctions by a nick and couternick mechanism. Mol Cell 12: 747–759 [DOI] [PubMed] [Google Scholar]
- Garcia AD, Aravind L, Koonin EV, Moss B 2000. Bacterial-type DNA Holliday junction resolvases in eukaryotic viruses. Proc Natl Acad Sci 97: 8926–8931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaskell LJ, Osman F, Gilbert RJ, Whitby MC 2007. Mus81 cleavage of Holliday junctions: A failsafe for processing meiotic recombination intermediates? EMBO J 26: 1891–1901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud-Panis MJE, Lilley DMJ 1997. Near simultaneous DNA cleavage by the subunits of the junction-resolving enzyme T4 endonuclease VII. EMBO J 16: 2528–2534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith JD, Christiansen G 1978. Electron microscopic visalization of chromatin and other DNA–protein complexes. Annu Rev Biophys Bioeng 7: 19–35 [DOI] [PubMed] [Google Scholar]
- Griffith JD, Makhov A, Zawel L, Reinberg D 1995. Visualization of TBP oligomers binding and bending the HIV-1 and adeno promoters. J Mol Biol 246: 576–584 [DOI] [PubMed] [Google Scholar]
- Hadden JM, Declais AC, Carr SB, Lilley DMJ, Phillips SEV 2007. The structural basis of Holliday junction resolution by T7 endonuclease. Nature 449: 621–624 [DOI] [PubMed] [Google Scholar]
- Harrington JJ, Lieber MR 1994a. Functional domains within FEN-1 and RAD2 define a family of structure-specific endonucleases: Implications for nucleotide excision repair. Genes Dev 8: 1344–1355 [DOI] [PubMed] [Google Scholar]
- Harrington JJ, Lieber MR 1994b. The characterization of a mammalian DNA structure specific endonuclease. EMBO J 13: 1235–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohl M, Thorel F, Clarkson SG, Scharer OD 2003. Structural determinants for substrate binding and catalysis by the structure-specific endonuclease XPG. J Biol Chem 278: 19500–19508 [DOI] [PubMed] [Google Scholar]
- Holliday R 1964. A mechanism for gene conversion in fungi. Genet Res Camb 5: 282–304 [DOI] [PubMed] [Google Scholar]
- Hosfield DJ, Mol CD, Shen BH, Tainer JA 1998. Structure of the DNA repair and replication endonuclease and exonuclease FEN-1: Coupling DNA and PCNA binding to FEN-1 activity. Cell 95: 135–146 [DOI] [PubMed] [Google Scholar]
- Interthal H, Heyer WD 2000. MUS81 encodes a novel helix–hairpin–helix protein involved in the response to UV- and methylation-induced DNA damage in Saccharomyces cerevisiae. Mol Gen Genet 263: 812–827 [DOI] [PubMed] [Google Scholar]
- Ip SCY, Rass U, Blanco MG, Flynn HR, Skehel JM, West SC 2008. Identification of Holliday junction resolvases from humans and yeast. Nature 456: 357–361 [DOI] [PubMed] [Google Scholar]
- Johnson RE, Kovvali GK, Prakash L, Prakash S 1998. Role of yeast Rth1 nuclease and its homologs in mutation avoidance, DNA repair, and DNA replication. Curr Genet 34: 21–29 [DOI] [PubMed] [Google Scholar]
- Kosugi S, Hasebe M, Tomita M, Yanagawa H 2009. Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc Natl Acad Sci 106: 10171–10176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvaratskhelia M, Wardleworth BN, Norman DG, White MF 2000. A conserved nuclease domain in the archaeal Holliday junction resolving enzyme Hjc. J Biol Chem 275: 25540–25546 [DOI] [PubMed] [Google Scholar]
- Lee BI, Wilson DM 1999. The RAD2 domain of human exonuclease 1 exhibits 5′ to 3′ exonuclease and flap structure-specific endonuclease activities. J Biol Chem 274: 37763–37769 [DOI] [PubMed] [Google Scholar]
- Lee SM, Cavallo L, Griffith J 1997. Human p53 binds Holliday junctions strongly and facilitates their cleavage. J Biol Chem 272: 7532–7539 [DOI] [PubMed] [Google Scholar]
- Lieber MR 1997. The FEN-1 family of structure-specific nucleases in eukaryotic DNA replication, recombination and repair. Bioessays 19: 233–240 [DOI] [PubMed] [Google Scholar]
- Lilley DMJ 1985. The kinetic properties of cruciform extrusion are determined by DNA base-sequence. Nucleic Acids Res 13: 1443–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley DMJ 2000. Structures of helical junctions in nucleic acids. Q Rev Biophys 33: 109–159 [DOI] [PubMed] [Google Scholar]
- Lilley DM, Markham AF 1983. Dynamics of cruciform extrusion in supercoiled DNA: Use of a synthetic inverted repeat to study conformational populations. EMBO J 2: 527–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley DMJ, White MF 2000. Resolving the relationships of resolving enzymes. Proc Natl Acad Sci 97: 9351–9353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley DMJ, White MF 2001. The junction-resolving enzymes. Nat Rev Mol Cell Biol 2: 433–443 [DOI] [PubMed] [Google Scholar]
- Lorenz A, West SC, Whitby MC 2010. The human Holliday junction resolvase GEN1 rescues the meiotic phenotype of a Schizosaccharomyces pombe mus81 mutant. Nucleic Acids Res 38: 1866–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney SA, Declais AC, Lilley DMJ, Ha T 2003. Structural dynamics of individual Holliday junctions. Nat Struct Biol 10: 93–97 [DOI] [PubMed] [Google Scholar]
- Munoz IM, Hain K, Declais AC, Gardiner M, Toh GW, Sanchez-Pulido L, Heuckmann JM, Toth R, Macartney T, Eppink B, et al. 2009. Coordination of structure-specific nucleases by human SLX4/BTBD12 is required for DNA repair. Mol Cell 35: 116–127 [DOI] [PubMed] [Google Scholar]
- Nishino T, Komori K, Tsuchiya D, Ishino Y, Morikawa K 2001. Crystal structure of the archaeal Holliday Junction resolvase Hjc and implications for DNA recognition. Structure 9: 197–204 [DOI] [PubMed] [Google Scholar]
- Paques F, Haber JE 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev 63: 349–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philo JS 2006. Improved methods for fitting sedimentation coefficient distributions derived by time-derivative techniques. Anal Biochem 354: 238–246 [DOI] [PubMed] [Google Scholar]
- Pöhler JRG, Giraud-Panis MJE, Lilley DMJ 1996. T4 endonuclease VII selects and alters the structure of the four-way junction; Binding of a resolution-defective mutant enzyme. J Mol Biol 260: 687–696 [DOI] [PubMed] [Google Scholar]
- Poser I, Sarov M, Hutchins JR, Heriche JK, Toyoda Y, Pozniakovsky A, Weigl D, Nitzsche A, Hegemann B, Bird AW, et al. 2008. BAC TransgeneOmics: A high-throughput method for exploration of protein function in mammals. Nat Methods 5: 409–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J, Qian Y, Chen V, Guan MX, Shen B 1999. Human exonuclease I functionally complements its yeast homologues in DNA recombination, RNA primer removal, and mutation avoidance. J Biol Chem 274: 17893–17900 [DOI] [PubMed] [Google Scholar]
- Rass U, West SC 2006. Synthetic junctions as tools to identify and characterise Holliday junction resolvases. Meth Enzymol 408: 485–501 [DOI] [PubMed] [Google Scholar]
- Roeder GS 1997. Meiotic chromosomes: It takes two to tango. Genes Dev 11: 2600–2621 [DOI] [PubMed] [Google Scholar]
- Schuck P 2000. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugaton and lamm equation modeling. Biophys J 78: 1606–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah R, Cosstick R, West SC 1997. The RuvC dimer resolves Holliday junctions by a dual incision mechanism that involves base-specific contacts. EMBO J 16: 1464–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B, Singh P, Liu R, Qui J, Zheng L, Finger LD, Alas S 2005. Multiple but dissectible functions for FEN-1 nucleases in nucleic acid processing, genome stability and diseases. Bioessays 27: 717–729 [DOI] [PubMed] [Google Scholar]
- Siegel LM, Monty KJ 1966. Determination of molecular weights and frictional ratios of proteins in impure systems by use of gel filtration and density gradient centrifugation. Application to crude preparations of sulfite and hydroxylamine reductases. Biochim Biophys Acta 112: 346–362 [DOI] [PubMed] [Google Scholar]
- Smith GR, Boddy MN, Shanahan P, Russell P 2003. Fission yeast Mus81–Eme1 Holliday junction resolvase is required for meiotic crossing over but not for gene conversion. Genetics 165: 2289–2293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su C-T, Chen C-Y, Hsu C-M 2007. iPDA: Integrated protein disorder analyzer. Nucl Acids Res 35: W465–W472 doi: 10.1093/nar/gkm353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian D, Griffith JD 2005. p53 monitors replication fork regression by binding to ‘chickenfoot’ intermediates. J Biol Chem 280: 42568–42572 [DOI] [PubMed] [Google Scholar]
- Svendsen JM, Smogorzewska A, Sowa ME, O'Connell BC, Gygi SP, Elledge SJ, Harper JW 2009. Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell 138: 63–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay YD, Wu L 2010. Overlapping roles for Yen1 and Mus81 in cellular Holliday junction processing. J Biol Chem 285: 11427–11432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor ER, McGowan CH 2008. Cleavage mechanism of human MUS81–EME1 acting on Holliday-junction structures. Proc Natl Acad Sci 105: 3757–3762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CS 2006. Biomacromolecules: Introduction to structure, function and informatics Wiley, New York [Google Scholar]
- Ubersax JA, Woodbury EL, Quang PN, Paraz M, Blethrow JD, Shah K, Shokat KM, Morgan DO 2003. Targets of the cyclin-dependent kinase Cdk1. Nature 425: 859–864 [DOI] [PubMed] [Google Scholar]
- West SC 1997. Processing of recombination intermediates by the RuvABC proteins. Annu Rev Genet 31: 213–244 [DOI] [PubMed] [Google Scholar]
- West SC 2009. The search for a human Holliday junction resolvase. Biochem Soc Trans 37: 519–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DM, Carney JP, Coleman MA, Adamson AW, Christensen M, Lamerdin JE 1998. Hex1: A new human Rad2 nuclease family member with homology to yeast exonuclease 1. Nucleic Acids Res 26: 3762–3768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Hickson ID 2003. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature 426: 870–874 [DOI] [PubMed] [Google Scholar]
- Wu L, Hickson ID 2006. DNA Helicases required for homologous recombination and repair of damaged replication forks. Annu Rev Genet 40: 279–306 [DOI] [PubMed] [Google Scholar]