Abstract
A major goal in cell biology is to understand the molecular mechanisms of the biological process under study, which requires functional information about the roles of individual proteins in the cell. For many non-genetic model organisms researchers have relied on the use of inhibitory reagents, such as antibodies that can be microinjected into cells. More recently, the advent of RNA-mediated interference (RNAi) has allowed scientists to knockdown individual proteins and to examine the consequences of the knockdown. In this chapter we present a comparison between microinjection of inhibitory reagents and RNAi for the analysis of protein function in mammalian tissue culture cells, providing both a description of the techniques as well as a discussion of the benefits and drawbacks of each approach. In addition, we present a strategy to employ RNAi for organisms without a sequenced genome. While the focus of our research is on the organization of the mitotic spindle during cell division and thus the examples utilized are from that system, the approaches described here should be readily applicable to multiple experimental models.
Keywords: Microinjection, RNAi, PtK cells, mitosis, siRNA, antibody, inhibition, protein function
1. Introduction
Understanding the molecular mechanism of any biological process requires the complete description of the role of each protein involved in that process. To achieve this goal, it is necessary to have an experimental means to perturb protein function as well as an assay to determine the functional consequences of that perturbation. The choice of approach will be determined by the reagents available to the protein of interest, the equipment available, whether or not the gene sequence is available, as well as the time-course of the experimental process being analyzed.
Microinjection of inhibitory antibodies or dominant-negative reagents has long been a powerful means to inhibit protein function in many cell types (1). However, it has often been criticized because it is questionable whether true loss of protein function is achieved. It is possible that the antibody is exhibiting cross-recognition of other proteins within the cell or that the antibody is simply binding in situ to the protein of interest and causing non-specific blocking of other protein interactions. Often these drawbacks can be overcome by examining the effects of multiple antibodies to the same protein or by complementing antibody injection studies with other inhibition methods.
RNAi has become an extremely useful tool for looking at protein function in many cell types. RNAi has revolutionized how most scientists view protein function studies, and the importance of this discovery is best highlighted by the awarding of a 2006 Nobel Prize to Andrew Z. Fire and Craig C. Mello, the scientists who first described this process (2). To carry out RNAi in vertebrate cells, short dsRNAs are introduced into the cell by transfection (3). This dsRNA then pairs with the endogenous mRNA and induces its degradation by a series of enzymatic activities. Because RNAi knocks out the mRNA, new protein synthesis is inhibited, and the protein levels decrease over the timecourse of the normal turnover of the protein of interest.
In contrast to microinjection, RNAi does not require a purified antibody or dominant-negative reagents, but it does require some information about the individual gene sequence. For organisms in which the genome is sequenced, finding siRNAs to knockout any gene of interest is as easy as searching the website of companies such as Dharmacon or Ambion for their collection of pre-designed RNAs. If a favorite gene is not included in the pre-designed collection, then designing a siRNA only requires entering the accession number of a protein into programs such as Block-IT siRNA Designer (http://rnaidesigner.invitrogen.-com/rnaiexpress/) or Dharmacon siDesign Center (http://www.dharmacon.com/sidesign/default.aspx). In the case of organisms without sequenced genomes, it is still possible to use these siRNA design programs by entering a short amount of sequence obtained by RT-PCR or from a cDNA clone.
Perturbation of protein function by either microinjection of inhibitory antibodies or RNAi should be considered complementary methods of inhibition. Both methodologies have their own strengths and weaknesses that influence their suitability to answer a particular scientific question. For example, microinjection of inhibitory antibodies is quick and will typically display immediate changes in cell behavior and morphology. This allows the experimenter to time the injection relative to the process being analyzed. In contrast, RNAi requires a period of incubation to allow time for the targeted protein to be degraded. With antibody injection the experimenter can inject higher concentrations of the antibody to achieve complete inhibition, whereas with RNAi, sufficient residual protein may remain to carry out all or part of its cellular function. In microinjection, only a small number of cells are often examined, but the exact cell that was injected is known and therefore can be examined phenotypically. In contrast, RNAi is useful to examine a large number of depleted cells. However, since knockdown can vary across a population of cells, it is often difficult to determine if a particular cell shows a phenotypic effect due to depletion unless appropriate antibodies are available. Because of the unique characteristics of each methodology, we use both techniques as complementary approaches to more fully understand the cellular processes we are studying.
2. Materials
2.1. Preparation of Poly-l-Lysine Coated Coverslips
Coverslips: We use 12-mm round No. 1 coverslips (Fisher; 12-545-80) for fixed analysis by immunofluorescence and 22 × 22 mm square No. 1½ coverslips (VWR; 48366-227) for live imaging and microinjection. For live imaging, where the sample will be injected on one microscope and imaged on another, it is useful to use photo-etched coverslips (Electron Microscopy Sciences; 72264-23), which have a marked grid for ease of relocating the injected/treated cell.
Poly-l-lysine: Add 50-mg of poly-l-lysine (Sigma; P1524) to a final volume of 50-ml using sterile ddH2O. Store at −20°C; can be used up to 4 times.
Hybridization oven with hybridization bottles, or alternatively a hot plate.
Platform rocker.
200-ml 1-M HCl (see Note 1.)
3MM Chromatography paper (Whatman; 3030917), 46 × 57 cm.
2.2. Cell Culture
Maintenance media: PtK2 cells are maintained in complete Dulbecco’s Modified Eagle’s Medium (D-MEM) (Invitrogen; 11965-092) supplemented with 10% fetal bovine serum (Invitrogen; 16140-089), 1% penicillin/streptomycin (Invitrogen; 15140-122), and 1% GlutaMAX (Invitrogen; 35050-061); PtK-T cells are maintained in complete F-12 HAM (Invitrogen; 11765-054) medium prepared with the same supplements as listed above for D-MEM. Store at 4°C in the dark.
0.25% Trypsin (Invitrogen; 15050-065). Store at −20°C.
70% ethanol
Sterile Pasteur pipettes, pipettes, pipette tips, and 1.5-ml centrifuge tubes
Hemocytometer
Trypan blue, 0.4% w/v in PBS (12-mM phosphate, 137-mM NaCl, and 3-mM KCl, pH 7.4).
Tissue culture cells: We use PtK2 cells, which are adherent male marsupial kidney epithelial cells that have a flat morphology and a small number (2n = 12) of large chromosomes. In our studies of mitosis, PtK2 cells provide an ideal system for detailed insight on phenotypes of the spindle components. For live imaging of the microtubule structure using fluorescence microscopy following injection, we use a PtK2 cell line stably expressing GFP-tagged alpha tubulin (PtK-T) (4).
Tissue culture plates: For routine passaging, we grow cells in Falcon 100-mm plates (353003). For plating over 22 × 22 mm coverslips for live imaging or microinjection, we use Corning 60-mm plates (25010). To test antibodies or for phenotypic analysis by immunofluorescence, we use either Corning 35-mm plates (351008) or Corning six-well cell culture plates (3506) (see Note 2).
Poly-l-lysine-coated coverslips: For preparation see Section 3.1.
2.3. Basic Immunofluorescence
Fixative: The type of fixative depends on the antibody being used for immunofluorescence (see Note 3).
Quench: A trace amount of sodium borohydride in 10-ml of TBS (0.15-M NaCl, 0.01-M Tris-HCl pH 7.4). TBS is made as a 10× stock and autoclaved to sterilize. Quench is only necessary if glutaraldehyde is present in the fixative.
TBS-Tx (TBS + 0.1% Triton X-100).
Staining dish: We use a 150-mm gridded Petri plate with parafilm covering the bottom chamber and moist paper towels placed around the inside edge to prevent evaporation of antibody dilutions. The lid should be covered in either black electrician tape or aluminum foil to prevent photo-damage to the fluorescently labeled secondary antibodies.
Abdil-Tx (antibody dilution solution): 2% BSA, 0.1% sodium azide made in TBS-Tx. Filter-sterilize and store at 4°C.
Primary antibody diluted in Abdil-Tx (see Note 4).
Secondary antibody conjugated to a fluorescent tag diluted in Abdil-Tx.
2 µg/ml Hoechst in TBS-Tx. Store at 4°C in the dark.
Mounting media: 0.5% p-phenylenediamine; 20-mM Tris-HCl; 90% glycerol. This is made up by adding 0.1-g p-phenylenediamine (Sigma; P6001) to 266 µl of 1.5-M Tris-HCl, pH 8.8 and 18-ml glycerol in a 50-ml tube. The p-phenylenediamine should be brown and flaky. Invert vigorously to make homogenous. Insert two hypodermic needles through a rubber stopper. Place the stopper securely in the 50-ml tube and wrap parafilm around the junction to seal. To one hypodermic needle, attach a hose hooked to a nitrogen tank and flow gas extremely gently over the surface of the liquid (liquid should only dimple slightly). The other needle should be venting excess gas from the tube. The reaction should take several hours and can go overnight. Finished mounting media should be light amber in color with no flecks. Aliquot and keep frozen at −80°C. A small working aliquot can be kept at −20°C, but it should not be used if it turns brown in color.
Fingernail polish.
2.4. Microinjection
Antibody storage buffer: (10-mM HEPES; pH 7.2; 100-mM KCl). Make up in water and store at 4°C.
Glass injection needles: Pre-pulled needles (World Precision Instruments; TW100F-6) provide a reproducible source of needles without the need to invest in a needle puller if one is not readily available. Make sure to use needles with a filament, which allows efficient delivery of the injection sample to the tip of the needle by capillary action.
Pipette and pipette tips for loading sample into injection needles: We use an Eppendorf Series 2000 Reference Adjustable-Volume pipette and microloader pipette tips, 0.5–20 µl range (Eppendorf North America; 930001007).
Inhibitory reagents for injection: Affinity-purified antibodies at a concentration of at least 1 mg/ml (and sometimes as high as 10–15 mg/ml). A control non-immune immunoglobulin (IgG) raised in the same animal as the experimental antibodies should also be used at an equal or higher concentration than the experimental antibodies.
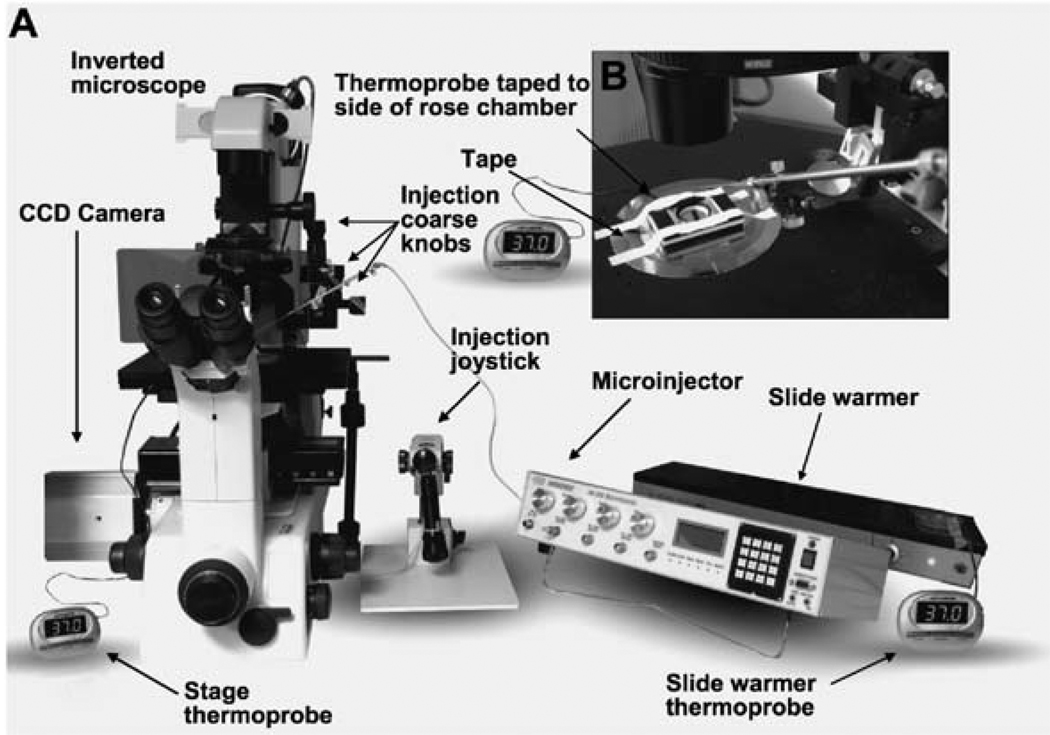
Nikon IM300 microinjector with Nikon/Narishige microinjector controls and a Nikon TE-300 inverted microscope (Fig. 7.1A).
Two thermoprobes. These are digital thermometers with a remote probe (Acu-Rite; Model # 00890A1), which are used to monitor the temperature of both the microscope stage and the plate warmer.
Rose chamber: This is a cell imaging chamber, designed for high resolution and long-term live imaging (Fig. 7.2). The chamber consists of three main layers: two metal planchets and one silicon spacer. The rose chamber is designed so that imaging can be carried out through the thin glass of the coverslip while providing enough space for holding up to 1 ml of media on top of the cells and a thin layer of mineral oil that covers the media surface preventing evaporation of media. The rose chamber can be replaced with glass bottom culture dishes (MatTek Corporation; P35GC-1.5-14-C). For a detailed discussion of different viewing chambers refer to (5).
Heating tray used as a slide or culture dish warmer.
Air stream incubator (ASI) (Nevtek, Burnsville, VA): This is a device that maintains a flow of air at constant temperature, which is necessary to keep mammalian cultured cells at 37°C. The ASI can be replaced with any available temperature-controlled stage device.
Observation/imaging media: For phase contrast imaging of PtK2 cells, we use D-MEM with no phenol red, (Invitrogen; 11039-021), supplemented with 20-mM HEPES (titrated to pH 7.2 with potassium hydroxide) before use to maintain the appropriate pH of the media in the absence of CO2. When imaging fluorescent protein-expressing cells, it is also necessary to add an oxygen scavenging mix such as oxyrase to prevent photo-damage. Oxyrase (Oxyrase Inc.; EC-0050) is used at a final concentration of 0.3 units/ml.
Fig. 7.1. The microinjection setup.
(A) The microinjection setup includes a Nikon IM300 microinjector with Nikon/Narishige micromanipulators mounted on a Nikon TE-300 inverted microscope with an attached CCD camera. A slide warmer on the right side is used for short-term incubation (<1 h). (B) A close-up view of the assembled rose chamber mounted on top of the microscope stage secured with tape. A thermoprobe is placed directly under the tape on top of the rose chamber to monitor its temperature during live imaging.
Fig. 7.2. Assembly of rose chamber for long-term live imaging.
(A) A disassembled chamber is shown to illustrate the order of placement of each component of the assembled chamber. (B) An assembled rose chamber.
2.5. Oligofectamine Transfection (RNAi)
Incomplete D-MEM: D-MEM with no supplements.
RNAi media: D-MEM supplemented only with 10% fetal bovine serum and 1% GlutaMAX but without antibiotics.
Oligofectamine (Invitrogen; 12252-011) (see Note 5).
siRNAs at 20 pmol/µl: siRNAs designed to the genes of interest as well as negative and positive control siRNAs. For a negative control, Dharmacon’s non-targeting siRNA #2 works well in PtK2 cells with no noticeable non-specific effects. For a positive transfection control for mitotic cells, Eg5 siRNA (5′ CAAGGAUGAAGUCUAUCAAdTdT) (6) is a good choice because the loss-of-function phenotype (monopolar spindles) is easily identifiable, the phenotype manifests itself after a short incubation time, and because the kinesin-5 Eg5 is found across a wide phylogeny of organisms (7,8) (see Note 6).
3. Methods
3.1. Preparation of Coverslips
Tissue culture cells generally adhere to glass somewhat poorly. Treating glass coverslips with poly-l-lysine helps cells remain attached to the surface keeping them flatter, which improves the ability to follow the intracellular events more clearly. The coverslips are first extensively washed with acid, which etches the glass, and then coated with poly-l-lysine.
3.1.1. Acid Wash
Place 1–2 boxes of coverslips in 1-M HCl and heat to 60°C for 4–16 h. A hybridization oven with an internal rotisserie works well for this step, since it can easily be set to the desired temperature. The coverslips can be divided equally between two hybridization bottles containing 50–100 ml of acid. If a hybridization oven is not available, a hot plate and glass dish can be used (see Note 1).
Let cool to room temperature and carefully decant the acid into a separate container and dispose of appropriately.
Rinse coverslips 3× 10 min with dH2O then with 3× 10 min with ddH2O. If the acid wash is performed in a glass container on a hot plate, transfer the glass container to a rocker for the rinses and rock gently to allow the H2O to evenly reach the coverslips. Do not shake or rock too vigorously, or the coverslips will break.
3.1.2. Poly-l-lysine Coating
To coat coverslips replace the ddH2O with 50-ml of 1 mg/ml poly-l-lysine while rocking or rotating gently for at least 30-min.
Decant poly-l-lysine and store at −20°C. This solution may be reused up to four times.
Rinse coverslips 5× 10 min with dH2O, then 5× 10 min with ddH2O to remove any free poly-l-lysine, which is toxic to the cells.
Dry coverslips by laying them out individually on Whatman paper. They must be separated or they will dry stuck together.
To sterilize coverslips before plating cells, cover with 100% ethanol, aspirate off, and let dry for a few minutes in the hood.
3.2. Maintenance of Cultured Cells
All of the subsequent protocols should be performed in a sterile tissue culture hood using sterile techniques and solutions to avoid contamination and its potential spread to other cultures (see Note 7.). For more in-depth information on media, cells, and maintenance techniques, please refer to (9,10).
Prewarm trypsin and maintenance media in a 37°C water bath.
Remove semiconfluent PtK2 plate(s) in log phase from the 37°C, 5% CO2 incubator. Aspirate off media from cells and rinse each plate 2× with 1.5-ml of trypsin. Incubate in 1 ml of trypsin for 5-min in the incubator until the cells detach from the plate. Add 9-ml of maintenance media to each trypsinized plate and combine multiple plates. Pipette cells up and down and across the plate to resuspend the cells.
For routine passaging, dilute trypsinized cell solution into prewarmed maintenance media. Return the newly passed cells to the 37°C incubator (see Note 8).
For plating for analysis of fixed cells by immunofluorescence, plate diluted cell solution over sterile poly-l-lysine coated coverslips (For preparation of coverslips, see Section 3.1.). The size of the plate will depend on the number of coverslips that need to be analyzed. (For plating volumes and the number of coverslips that will fit into different size dishes, see Note 9.)
To plate cells for microinjection, plate as if for immunofluorescence, except use 60-mm dishes over sterile 22 × 22 mm coverslips.
For siRNA transfections, the number of wells to be plated for RNAi will depend on how the cells will be analyzed (see Note 10). The plating density of the cells also depends on a variety of factors, one of which is incubation time prior to cell analysis. Since this will vary for each protein being knocked down, this will have to be optimized for each oligonucleotide. For our experiments, PtK2 cells typically are plated at 15,000–20,000 cells/ml per 35-mm dish. A 100-mm plate of cells at log phase is sufficient to plate at least 24–35-mm wells. We also typically plate four wells per oligonucleotide: one for immunofluorescence and three for immunoblotting (see Note 11 for further considerations.).
To perform cell counts for plating for RNAi, place 90-µl of the trypsinized cell suspension in a 1.5-ml microcentrifuge tube. Add 10-µl of trypan blue, which stains dead cells blue and let sit for a few minutes (no longer than 10-min). Pipette up and down to break up any cell clumps and to resuspend any settled cells and add 10-µl of cells to each side of a hemocytometer. Count the number of non-blue cells and calculate the cell density.
3.3. Basic Immunofluorescence
In the study of cellular processes, the analysis of fixed cells by fluorescence microscopy is usually the first step to assessing the initial phenotypic effects on the cell by either microinjection or RNAi. Specific steps for these procedures are outlined below for the study of PtK2 cells. For more indepth information, please refer to (11).
Pull plates with cells growing on 12-mm coverslips from the 37°C, 5% CO2 incubator.
Aspirate off media and rinse cells quickly with PBS.
Add fixative and incubate for an appropriate amount of time based on the fixative used. Alcohol-based fixatives, such as cold methanol, are incubated at RT for 5-min. Aldehyde fixatives are incubated at RT for 20-min.
If cells were fixed with glutaraldehyde replace fixative with 1-ml of Quench and incubate for 5-min.
Rinse cells 2× with TBS-Tx.
Move the coverslips to a parafilm-lined staining dish, keeping each coverslip wet with TBS-Tx. Wash 1× with a stream of TBS-Tx. Always finish by leaving a drop of buffer on the coverslip to keep the cells hydrated.
Aspirate off all TBS-Tx, and overlay each coverslip with 75-µl Abdil-Tx for minimum of 30-min at RT.
Aspirate off Abdi-Tx and replace with 35-µl of primary antibody diluted in Abdil-Tx, and incubate for 30-min. Wash 3× with TBS-Tx.
Aspirate off last wash and incubate coverslip for 30-min in 35-µl of secondary antibody conjugated to a fluorescent tag. Wash 3× with TBS-Tx.
The cells can be co-stained with additional antibodies by repeating steps 8 and 9.
Stain the DNA by incubating coverslips for 5–15 min in 50-µl of 2 µg/ml Hoechst made in TBS-Tx. Wash coverslips 3× with TBS-Tx.
To mount, blot each coverslip briefly by touching its edge to a kimwipe, then place cell-side down on a 2-µl drop of mounting media on a slide. Aspirate the top of the coverslip, seal the circumference with fingernail polish, and let dry.
Coverslips must be washed gently with either water or lens cleaner before viewing on the fluorescence microscope. This is particularly important when using oil immersion objectives where any remaining salts on the coverslip will damage the lens.
Slides can be stored at −20°C in the dark for several weeks before the fluorescence fades.
3.4. Microinjection
Microinjection of either antibodies or dominant-negative reagents into vertebrate tissue culture cells is a powerful way to analyze protein function. The cells can be injected during interphase or during mitosis so it is possible to achieve temporal resolution of the experiment. The first part of this section describes the preparation of the injectate (Section 3.4.1.) and the microscope and rose chamber (Section 3.4.2.) followed by an outline of the microinjection steps (Section 3.4.3.). We then present the application of the microinjection for either a fixed time-point experiment (Section 3.4.4.) or for live imaging (Section 3.4.5.).
3.4.1. Preparation of Injectate
Antibodies are purified by affinity chromatography and eluted using Glycine-HCl, pH 2.0, followed immediately by neutralization in 2-M Tris-HCl, pH 8.5 (12).
The antibodies are then dialyzed into antibody storage buffer, quantified by A280, flash frozen in 10 to 30-µl aliquots, and stored at −80°C.
For injection, the antibodies and control IgG are thawed on ice and diluted to the desired concentration in antibody storage buffer (see Note 12).
Prepare antibody/injection sample by spinning at 14K rpm for 15-min and transfer the supernatant to a clean tube for injection. Place on ice for the duration of the experiment.
3.4.2. Preparation of Microinjection Microscope and Assembly of Rose Chamber
Prewarm slide warmer and imaging media (and rose chamber parts, if needed) to 37°C.
Place a thermoprobe on the stage beside where either the rose chamber or culture will be mounted (Fig. 7.1B).
Turn on ASI and adjust the air flow at the stage. Allow the temperature to stabilize to the desired temperature (see Note 13), readjusting the ASI as necessary.
For imaging of live cells, cells grown on a 22 × 22 mm coverslip are transferred to a 35-mm dish containing 2-ml of prewarmed imaging media and placed on the 37°C warming tray.
Assemble the rose chamber (Fig. 7.2). This needs to be done quickly to prevent the cells from cooling too much. Blot to remove excess media, and quickly place the coverslip onto the bottom metal planchet of the chamber, positioning it in the center with the cells facing up. Quickly place the silicon spacer over the coverslip and then the top planchet on the silicon spacer. Keep both in place by pressing down firmly on the top planchet. Using a transfer pipette, add approximately 1-ml of prewarmed media into the chamber on top of the cells. While still pressing firmly, screw down the corners of the chamber. Wipe the bottom of the coverslip with a kimwipe, wet with dH2O, then wipe with a kimwipe wet with 70% EtOH. Using water first will prevent the ethanol from precipitating proteins and salts from the media. Cover the observation media with a layer of mineral oil to prevent evaporation, and place the assembled chamber onto the stage. Tape it down as shown in Fig. 7.1B to prevent it from moving during the subsequent steps.
If injecting cells for fixed-time-point analysis, cells grown on a 12-mm coverslip are placed in a 35-mm tissue culture dish with 2 ml-of prewarmed observation media. Place the dish on the slide warmer to keep at 37°C until ready to inject. For injection, the dish is simply moved to the microscope stage ring designed for a 35-mm dish. Injection is carried out directly in the 35-mm dish.
3.4.3. Microinjection of Cells
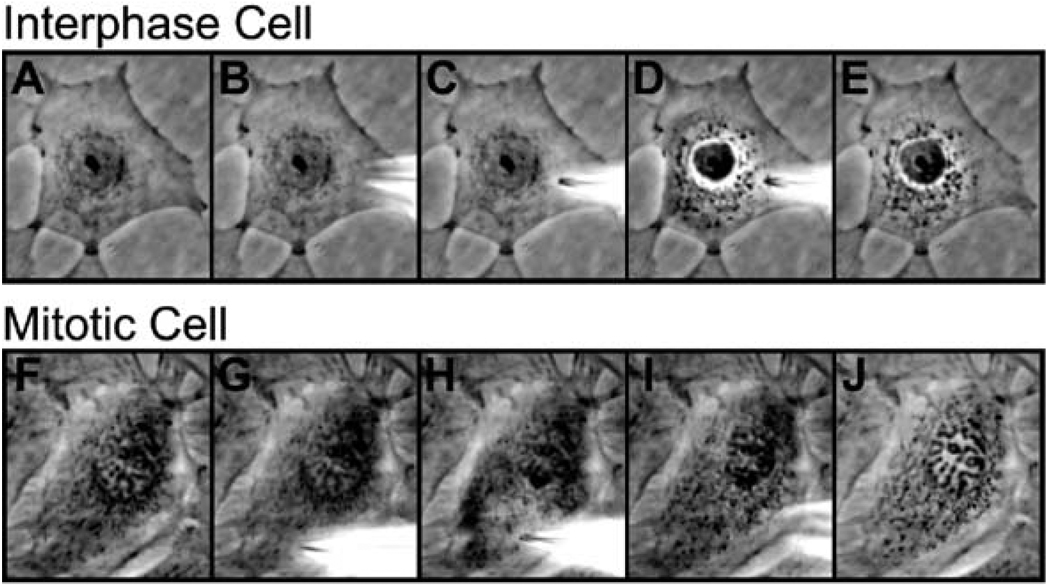
Scan the surface of the coverslip to find the cell of interest. For interphase cells find a group of cells to inject (Fig. 7.3A). For mitotic cells find a cell at prophase, right before nuclear envelope breakdown has taken place (Fig. 7.3F). Center the cell in the cross-bars of the eyepiece.
Adjust the needle holder, which is attached to the stage to an approximately 45° angle and tighten it in place.
For a system with an automated injector, open the N2 tank valve and adjust the output pressure to 60–80 psi.
Using the eppendorf microloader tip and pipette, carefully backload the needle with ~2-µl of the sample. Avoid touching the needle against anything for it is extremely fragile and will easily break. Attach the filled needle to the needle holder on the stage and secure.
Turn off the ASI before injection. Failure to do so will lead to warming the needle to above 37°C and killing the injected cell (see Note 14).
By eye, position the needle with the coarse X and Y adjustment knobs to place the needle tip at the center of the transmitted light on the surface of the media. Using the Z-axis coarse knob, carefully lower the tip of the needle so that it breaks the surface of the media, but do not lower it so far that it hits the bottom of the dish and breaks.
At this point the needle tip should be in the general vicinity of the cells, and its position is now carefully fine-tuned. To first detect the position of the needle, look through the eyepiece and move the needle using coarse knobs in the X- and Y-axes, to move laterally in relatively large increments. A shadow of the needle moving across the field of view should be detectable. Move the needle using the Y-axis knob until the shadow is at the center of the field of view and adjust with the X-axis knob so that the tip of the needle, which should appear as the narrowest part, is at the center of the field of view. While the tip of the needle is in sight, carefully lower the needle in the Z-axis so that the needle is just above the cell surface (Fig. 7.3B and G).
Adjust the balance pressure to approximately 1–3 psi, which will create a constant flow of sample from the tip. The sample should now be detected through the eyepiece as a stream of liquid flowing from the tip.
To inject in the cytoplasmic area of the cell, center the target cell and aim at a region that is relatively thick. It is usually easiest to inject cells near the periphery of the nucleus because the cell is thickest in this area (Fig. 7.3C and H).
Quickly move the tip down in the Z-plane using the fine adjustment of the joystick and then back up (a twisting movement of the joystick back and forth). As the cytoplasm is injected, a fast but gentle movement of sample is seen moving throughout the cell (Fig. 7.3D and I).
Before removing the dish/imaging chamber, move the tip up with the Z-plane coarse knob to avoid breaking the needle tip, which can be used for the next set of cells as long as it is intact and functional.
For trouble shooting injections, see Note 15.
Fig. 7.3. Microinjection of interphase and mitotic PtK2 cells are shown in sequence.
(A) An interphase cell is shown. (B) The same interphase cell is shown with the microinjection tip just out of focus above the cell. (C) The microinjection tip is in focus above the cell prior to injection. (D) As the cell is injected, the injectate can be seen as a wave traveling through the cell. (E) The cell is shown post-injection. (F) A prophase cell is shown with intact nuclear envelope and condensed chromosomes prior to injection. (G) A prophase cell is shown with the microinjection tip just out of focus above the cell. (H) The microinjection tip is in focus above the cell prior to injection at a thick, organelle-rich area, away from the nucleus. (I) As the cell is injected, the injectate can be seen as a wave traveling through the cell. (J) An image of the cell is shown post-injection.
3.4.4. Fixed-Time-Point Assay
When first analyzing the function of a protein by microinjection of antibodies, it is easiest to assess the phenotype by injecting the antibody, waiting a set amount of time and then fixing the cells and processing them for immunofluorescence. If it is not known whether the affected process is during interphase or mitosis, it is simpler to start with interphase cells because they are easier to inject and there are more of them on a coverslip. When looking for a mitotic defect, it is easiest to inject only cells at prophase and then allow the cells to incubate at 37°C to progress through mitosis. For interphase cells, we usually start with a 2-h incubation in antibody and have extended that time period up to 24-h post-injection before analysis. For mitotic cells, we usually fix at 30-min post-injection for prometaphase/metaphase defects and 40-min post-injection for anaphase defects. The cellular morphology as well as the cell cycle stage is scored under a fluorescence microscope.
Place 35-mm dish with cells growing on 12-mm coverslips onto the stage ring plate designed for 35-mm dishes.
Gently inject the cells as instructed in Section 3.4.3. Inject each coverslip for a period of 5-min and return to the 37°C warming tray. Keeping the injection time period short is a way to maintain cell cycle synchrony in the experiment, at least for mitotic cells.
Allow the cells to incubate for the desired amount of time. For a short incubation (less than 1-h), it is fine to leave the cells on the warming tray. For longer incubations, return the cells to the CO2 incubator, which will better maintain temperature and humidity.
Directly following incubation, aspirate off the observation media and add 2-ml of fixative. Fix for the appropriate time period and follow by staining and mounting the coverslip on a microscope slide for observation (see Section 3.3 for basic immunological protocols).
3.4.5. Live Imaging
Place assembled rose chamber with live cells on the stage.
Select live imaging acquisition settings on the imaging software available with your scope (see Note 16).
Inject the cell as instructed in Section 3.4.3. As soon as the cell has been injected, quickly remove the needle and turn the ASI back on.
Turn off the microscope light and allow at least one minute for cell to recover, then view live on the screen to center the cell and refocus.
Begin time-lapse imaging, and record the time of injection and the time when imaging commenced.
Throughout imaging, make sure that the ASI is at 36–38°C. The rose chamber and coverslip with no change of media can be used for a maximum of 5-h. For more detailed discussion of live imaging refer to (5).
3.5. Oligofectamine Transfection (RNAi)
Knocking down protein levels by RNAi is a fairly simple way to address protein function. However it relies on the availability of cDNA sequence to design the siRNA. We describe a fairly straight-forward approach to obtain a sufficient amount of sequence for siRNA design to apply this technology to model systems without a sequenced genome. We then describe a basic protocol for transfecting adherent cells grown in culture. All steps should be performed in a sterile tissue culture hood using sterile techniques and solutions.
3.5.1. Designing siRNAs for Knockdown
The rat kangaroo genome has yet to be sequenced, therefore no databases exist to search for gene sequences of interest. Traditional methods of cloning cDNAs are laborious and time consuming; however, comparison of homologous sequences across different mammalian species often shows a high degree of DNA identity through portions of the coding region if not the whole sequence (6) that can be used to design primers for RT-PCR, which in turn are used to generate siRNAs.
Search databases for homologous sequences to the desired gene. We used homologous sequences from mammalian species since these theoretically should have the most identity to the rat kangaroo genome.
Generate sequence alignments using the mammalian homologues on a program such as Sequencher (Gene Codes Corporation), which allows hand manipulation to refine the alignments to gain the highest identity possible between the sequences used.
Design degenerate primers that (1) minimize the degree of degeneracy needed as much as possible, (2) produce a PCR product between 400 and 1000 bp long, and (3) that overlap, where possible, any published siRNAs of any of the homologous sequences.
Use total RNA isolated from PtK2 tissue culture cells as template in an RT-PCR reaction using the designed degenerate primers (see Note 17).
Purify the RT-PCR product, either the whole or a partial gene sequence. Sequence both strands of the PCR product and compare the results to the original alignments to verify that the product does indeed have a high degree of identity to the homologous sequences used in the alignment.
Use the confirmed sequence to design siRNAs for subsequent RNAi transfections using programs such as the Dharmacon siDesign Center program.
3.5.2. Oligofectamine Transfection
This is a modified version of the Invitrogen protocol “Transfecting siRNA into HeLa Cells Using Oligofectamine” (http://www.invitrogen.com/content/sfs/protocols/sirna_oftsf_proc.pdf) (3,13).
Prewarm all media to 37°C.
-
For each 35-mm well to be transfected, prepare the RNAi complexes (see Notes 18–20):
Before use, gently mix Oligofectamine, then dilute 3 µl of transfection reagent into 12-µl of incomplete D-MEM in a 1.5-ml sterile microcentrifuge tube. Pipette gently up and down to mix and let incubate at room temperature for 5-min.
Meanwhile, in a second sterile 1.5-ml microcentrifuge tube, add 200-pmol of siRNA to 175-µl of incomplete media. Pipette up and down gently to mix. Add the diluted siRNA to the Oligofectamine complexes after their 5-min incubation and mix gently.
Let the siRNA:lipid complexes form at room temperature for 20-min.
While the complexes are incubating, rinse cells 2× with 1-ml of incomplete D-MEM media, and then cover with 1-ml of RNAi complete media.
Add 800-µl of D-MEM complete to the siRNA/lipid complexes. Remove media from wells and replace with the 1-ml of siRNA/lipid RNAi media. Incubate at 37°C, 5% CO2 incubator for 24-h.
Add 1-ml of D-MEM complete media back to each 35-mm well. Return to 37°C, 5% CO2 incubator.
Assay for gene activity at 24–72 h after transfection as is appropriate for the target gene (see Notes 21–22).
Acknowledgments
The authors would like to thank Susan Kline for early instruction in microinjection and suggestions on transfections of PtK2 cells. The authors would also like to thank Chantal LeBlanc for editing of the manuscript. Work in the Walczak lab is supported by NIH R01GM059618, an ACS Scholar award RSG CSM-106128, and in part by the Indiana METACyt Initiative of Indiana University, funded in part through a major grant from the Lilly Endowment, Inc. Rania Rizk is supported by a predoctoral fellowship from the American Heart Association.
Footnotes
HCl is extremely corrosive, inhalation of vapor can cause serious injury, and liquid acids can cause severe damage to skin and eyes. Use appropriate precautions when making and handling the hot acid, and dispose of the acid properly. If a hot plate and glass container are used to acid-wash the coverslips, the temperature and acid level will have to be monitored much more closely to make sure the acid does not overheat, and the glass container should be loosely covered to prevent excessive evaporation. The use of a hot plate requires that this step be carried out under a ventilated hood.
All tissue culture plates are not created equal. We have experienced problems with unhealthy cells when we have attempted to try plates from different manufacturers. When changing any reagents involved with tissue culture, it is always prudent to test the new batch on a subpopulation of cells before completely switching over.
The antibodies used will determine the specific fixative needed. To look at microtubule structure with DM1α anti-tubulin antibody (Sigma), we find that fixation with 4% formaldehyde; 0.1% glutaraldhyde in PHEM buffer (60-mM PIPES, 25-mM HEPES, 10-mM EGTA, and 4-mM MgSO4, pH 7) works well. However, many of our other polyclonal antibodies do not work well with glutaraldehyde fixation, and we typically use 2–4% formaldehyde in PHEM buffer without the glutaraldehyde. Cold (−20°C) 100% methanol can also be used to fix cells in which microtubules are to be visualized, and this works for some of our other antibodies as well. It should be noted that methanol fixation can cause some distortion in the condensed chromatin of mitotic cells. Dispose used fixative in accordance to local and state regulations.
Antibody dilutions can be stored at 4°C; however, the concentrations and the lengths of time these dilutions can be used will vary from antibody to antibody and will have to be empirically determined by each lab.
For siRNA transfection in PtK2 cells, several popular lipid-based reagents were tested, many of which caused vesicularization in cells, which can interfere with subsequent imaging by microscopy. We found that Oligofectamine (Invitrogen) produced the most consistent results with the least degree of cytotoxic effects compared to other transfection reagents tested.
siRNAs purchased from Dharmacon have been more successful for us than siRNAs from other companies both in their reliability and efficiency of knockdown. For a negative control, Dharmacon’s non-targeting siRNA #2 designed to Luciferase works well in PtK2 cells, whereas the GFP siRNA (GCAAGCUGACCCUGAAGUUCAU) (14) produced cytotoxic effects in our PtK2 cells.
It cannot be stressed enough the importance of an ever-present diligence to avoid contamination. Cultures should be routinely examined under the microscope to assess the health of the culture and inadvertent cross-contamination of other cell lines, or contamination by fungi or bacteria. Cells should also routinely be tested for contaminants such as mycoplasma, either by PCR or immunofluorescent assays.
Many cell lines are sensitive to cell density; plating at densities either too low or too high can have an adverse effect on the cells.
A 35-mm plate holds 2-ml of media and four 12-mm coverslips; a 60-mm plate holds 4-ml of media and 12 12-mm coverslips or two 22 × 22 mm coverslips and four 12-mm coverslips; and one 100-mm plate holds 10-ml of media and up to 30 12-mm coverslips.
The number of wells to be plated for RNAi will depend on how cells will be analyzed. We typically plate the cells in 35-mm dishes or six-well plates (each well is equivalent to a 35-mm dish). For live imaging, cells are plated on 22 × 22 mm coverslips in 60-mm dishes. For immunoblots, cells from three wells for each experimental condition are trypsinized, washed, and counted. The number of wells harvested for immunoblots can also be dependent on the antibody used.
The density at which cells will be plated for RNAi experiments depends on many factors such as the growth rate of the cells, the media in which the cells are grown, and the number of days required to achieve knockdown of the protein as determined by either immunofluorescence, immunoblot, or qRTPCR. For Eg5 RNAi in our PtK2 cells, the effect is seen in as little as 24-h after transfection though we fix cells for immunofluorescence by 48-h. For kinesin-13 MCAK RNAi, cells are processed after 72-h for efficient knockdown (6). Other factors include cell density at processing; overly confluent cells at processing will retard the number of mitotic cells and cause difficulty in imaging. However, the transfection efficiency is at times better if the cells are more confluent versus less, so sometimes a balance needs to be maintained. The optimal plating density will need to be empirically determined.
The concentration of antibody used must be empirically determined. We often start with a needle concentration of 1–2 mg/ml and have rarely needed to go above 5 mg/ml. There are, however, reports in the literature in which people have used much higher concentrations. The buffer in which the antibody is stored must be at physiological pH, cannot have too high a salt concentration and must not contain sodium azide or other preservatives, which will kill cells.
While it would be ideal to have a thermoprobe directly monitoring the temperature of the media while imaging, this is not possible due to space restraints and perturbation of the cells. Instead, we attach the thermoprobe to the top-side of the rose chamber. Because the temperature at the top of the chamber will be different than inside the media-filled chamber, it is necessary to predetermine this difference before injections. To do this, place the assembled rose chamber on the stage (Fig. 7.1B). Attach one probe directly on top of the glass coverslip, using tape. Add ~1-ml of media and a thin layer of mineral oil on top of the media. Place a second thermoprobe at the top-side of the rose chamber (Fig. 7.1B). Turn the ASI on and allow the temperature reported by the probe inside the rose chamber to reach the desired temperature and remain constant. This probe reflects the closest estimation to the temperature of the cells during imaging. Now record the temperature of the probe placed at the side of the chamber. This is the temperature that will be used to maintain cells at the actual desired temperature. In our setup, we have determined that the temperature of the coverslip within the rose chamber is typically 1°C lower than that of the probe on the top of the rose chamber.
As the needle is lowered into the imaging media, it is in a direct flow of the ASI and will heat up to temperatures several degrees warmer than the media. This increased temperature of the glass needle will kill the injected cell, making it imperative that the ASI be turned off right before injections.
Troubleshooting injections: For a clogged needle, press the clear button, or gently scrape the tip of the needle along a cell-free area of the coverslip to unclog it. Be careful because this latter method can also damage the needle tip. A clogged needle can also be due to a high concentration of the injectate or aggregates in the injectate. To alleviate these problems, try diluting the sample or centrifuging at a higher speed before loading the needle.
For live imaging of mitotic PtK2 cells under phase contrast microscope we use 100-ms exposures at 30-s intervals for 120-min, which allows us to follow the events of mitosis without damaging the cell. This can be optimized depending on the nature of the experiment.
Using degenerate primers is not typically recommended in RT-PCR. However we have had some success if we can limit the degree of degeneracy of the primers. Additionally, in some instances our RT-PCR gave multiple products, which required isolating the different bands and sequencing each band. By comparing the sequences back to the original alignments, we identified the band with the most identity to the homologous gene sequences as the intended product. Additionally, if we could obtain at least a partial sequence of the RT-PCR product, we could further amplify the PCR product by doing nested PCR on the RT-PCR product.
If transfecting cells in multiple wells with the same siRNA, cocktail mixes can be used. In this case, multiply each reagent added by a fraction over the number of wells to be transfected. For example, if transfecting three wells with luciferase siRNA, add 3.2-fold more of each reagent to the appropriate tubes: 9.6-µl of Oligofectamine to 38.4-µl of incomplete media and 32-µl of siRNA to 560-µl of incomplete media. The siRNA/lipid complexes can then be added to 2560-µl of RNAi media in a 5-ml sterile tube instead of the 800-µl of RNAi media being added to the complexes. Aliquots of this cocktail are then used to replace the rinse media.
The ratio of lipid to siRNA may need to be optimized for each siRNA used to achieve the most efficient knockdown. Using excess lipid or siRNA will increase non-specific cytotoxic effects of the RNAi transfection. Some gene targets may require multiple siRNAs for efficient knockdown or may require that the concentration of oligonucleotide be adjusted to increase the transfection efficiency. Dharmacon has a useful complementary guide book, RNA Interference – Technical Reference & Application Guide (www.dharmacon.com) that has many useful suggestions on how to troubleshoot RNAi.
The length of time a cell line has been subcultured affects the efficiency of the knockdown. This seems to be particularly true for PtK2 cells, and only those cells subcultured for 8 weeks or less should be used for RNAi.
The percentage of knockdown that needs to be attained to see a phenotypic effect can vary greatly depending on the gene of interest. For example, we see spindle defects in PtK2 cells when we have reduced MCAK by 70% as shown by western immunoblot. However, CENP-A has to be reduced by >90% to cause mislocalization of other centromere proteins (15).
The ideal situation is that both the analysis of the phenotypic effects of knockdown and the assessment of knockdown efficiency by immunofluorescence would be performed on the same cell. We can do that for Eg5 RNAi, since both the Eg5 antibody and the anti-tubulin antibodies used to determine spindle defects work in methanol fix. However, this is not always possible because the antibodies used to assess both the knockdown and the phenotypic effects may require different fixation methods. In this case, we plate four coverslips per well so that two coverslips are used to determine knockdown and use the other two coverslips for the phenotypic analysis in order to reduce as many experimental differences as possible in the treatment of these cells.
References
- 1.Scholey JM. Functions of motor proteins in echinoderm embryos: an argument in support of antibody inhibition experiments. Cell Motil. Cytoskel. 1998;39:257–260. doi: 10.1002/(SICI)1097-0169(1998)39:4<257::AID-CM1>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 2.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 3.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 4.Khodjakov A, Copenagle L, Gordon MB, Compton DA, Kapoor TM. Minus-end capture of preformed kinetochore fibers contributes to spindle morphogenesis. J. Cell. Biol. 2003;160:671–683. doi: 10.1083/jcb.200208143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khodjakov A, Rieder CL. Imaging the division process in living tissue culture cells. Methods. 2006;38:2–16. doi: 10.1016/j.ymeth.2005.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stout JR, Rizk RS, Kline SL, Walczak CE. Deciphering protein function during mitosis in PtK cells using RNAi. BMC Cell Biol. 2006;7:26. doi: 10.1186/1471-2121-7-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mayer TU, Kapoor TM, Haggarty SJ, King RW, Schreiber SL, Mitchison TJ. Small molecule inhibitor of mitotic spindle bipolarity identified in a phenotype-based screen. Science. 1999;286:971–974. doi: 10.1126/science.286.5441.971. [DOI] [PubMed] [Google Scholar]
- 8.Weil D, Garcon L, Harper M, Dumenil D, Dautry F, Kress M. Targeting the kinesin Eg5 to monitor siRNA transfection in mammalian cells. Biotechniques. 2002;33:1244–1248. doi: 10.2144/02336st01. [DOI] [PubMed] [Google Scholar]
- 9.Freshney RI. In: Culture of Animal Cells: A Manual of Basic Techniques. Freshney RI, editor. New Jersey: Wiley-Liss; 2005. [Google Scholar]
- 10.Phelan MC. Basic techniques for mammalian cell tissue culture. In: Bonifacino JS, Dasso M, Harford JB, Lippincott-Schwartz J, Yamada KM, editors. Current Protocols in Cell Biology. New Jersey: Wiley-Liss; 2003. [Google Scholar]
- 11.Spector DL, Goldman RD, Leinwand LA. Cells: A Laboratory Manual, 3. New York: Cold Spring Harbor Laboratory Press; 1998. Visualization of Organelles, Proteins, and Gene Expression. 3 vols. [Google Scholar]
- 12.Harlow E, Lane D. Antibodies: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 13.Harborth J, Elbashir SM, Bechert K, Tuschl T, Weber K. Identification of essential genes in cultured mammalian cells using small interfering RNAs. J Cell Sci. 2001;114:4557–4565. doi: 10.1242/jcs.114.24.4557. [DOI] [PubMed] [Google Scholar]
- 14.Caplen NJ, Parrish S, Imani F, Fire A, Morgan RA. Specific inhibition of gene expression by small double-stranded RNAs in invertebrate and vertebrate systems. Proc Natl Acad Sci U S A. 2001;98:9742–9747. doi: 10.1073/pnas.171251798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu ST, Rattner JB, Jablonski SA, Yen TJ. Mapping the assembly pathways that specify formation of the trilaminar kinetochore plates in human cells. J Cell Biol. 2006;175:41–53. doi: 10.1083/jcb.200606020. [DOI] [PMC free article] [PubMed] [Google Scholar]