Abstract
We investigated the bacterial diversity of microbial communities in water-filled, human-made and natural container habitats of the mosquitoes Aedes aegypti and Aedes albopictus in suburban landscapes of New Orleans, Louisiana in 2003. We collected water samples from three classes of containers, including tires (n=12), cemetery urns (n=23), and miscellaneous containers that included two tree holes (n=19). Total genomic DNA was extracted from water samples, and 16S ribosomal DNA fragments (operational taxonomic units, OTUs) were amplified by PCR and separated by denaturing gradient gel electrophoresis (DGGE). The bacterial communities in containers represented diverse DGGE-DNA banding patterns that were not related to the class of container or to the local spatial distribution of containers. Mean richness and evenness of OTUs were highest in water samples from tires. Bacterial phylotypes were identified by comparative sequence analysis of 90 16S rDNA DGGE band amplicons. The majority of sequences were placed in five major taxa: Alpha-, Beta- and Gammaproteobacteria, Actinobacteria, Bacteroidetes, Cyanobacteria, Firmicutes, and an unclassified group; Proteobacteria and Bacteroidetes were the predominant heterotrophic bacteria in containers. The bacterial communities in human-made containers consisted mainly of undescribed species, and a phylogenetic analysis based on 16S rRNA sequences suggested that species composition was independent of both container type and the spatial distribution of containers. Comparative PCR-based, cultivation-independent rRNA surveys of microbial communities associated with mosquito habitats can provide significant insight into community organization and dynamics of bacterial species.
Introduction
The mosquitoes Aedes aegypti and Aedes albopictus develop in water-filled, human-made containers that are distributed in urban and suburban landscapes [16]. Container habitats of mosquitoes are ecosystem mesocosms, supporting food webs that are dependent on detritus [20] and microbial communities that metabolize and mineralize organic carbon from detritus [27, 29]. Catabolism of detritus by microbes also produces metabolites that attract gravid mosquitoes and stimulate egg laying [4]. These semiochemicals are thought to cue female mosquitoes to the quality of habitats because bacterial enrichment generally increases the number of gravid females that are attracted to a container as well as the number of eggs that each female lays in a container [5]. Thus, bacterial communities in container habitats potentially exert significant effects on the population dynamics of mosquitoes as well as their spatial distribution in the landscape.
Production of mosquitoes in human-made containers has been extensively investigated (see [12, 47] for reviews), but comparatively little is known about the diversity of bacterial species in container habitats of mosquitoes, and most investigations have lacked substantial phylogenetic and taxonomic resolution of microbial communities. The majority of microbes in environmental samples cannot be cultured in laboratory media [31], and for this reason, cultivation-independent 16S rRNA surveys represent a powerful approach for characterizing natural microbial assemblages [2]. Denaturing gradient gel electrophoresis (DGGE), a genetic fingerprinting technique, has been used in molecular ecology investigations to assess the diversity of various microbial communities [28], and sequence data have been used to characterize the phylogenetic relationships of community members [38].
We conducted a DGGE-based survey of the species composition of bacterial communities in water-filled containers in suburban landscapes in New Orleans, LA, USA. Our hypothesis was that assemblages of bacterial species would be associated more with specific container types that supported populations of A. aegypti and A. albopictus and less with the spatial distribution of the containers. Moreover, although A. albopictus and A. aegypti are sympatric in some regions, they occupy slightly different ecologic niches [16], suggesting that specific assemblages of bacterial species would be found in association with each mosquito species. Accordingly, the objectives of our survey were as follows: (1) to ascertain whether the species structure of bacterial communities could be associated with specific types of containers that were utilized as habitats by immatures of both mosquito species; (2) to determine if the assemblages of bacterial species were spatially dependent; and (3) to characterize the phylogenetic relationships of bacterial species in water-filled containers based on sequences of the most abundant DGGE bands.
Materials and Methods
Collection and Processing of Samples
Our survey of field diversity of bacterial communities in human-made containers (n=52) and natural tree holes (n= 2) was conducted over a 3-day period from September 16–18, 2003. Container-inhabiting Aedes mosquitoes are most abundant from July to October. By early fall, mosquito populations would have been well established in container habitats. Water and mosquito samples were collected from containers in suburban landscapes at 15 separate locations in New Orleans, LA (Supplementary Table 1). Sampling sites were distributed over an area of ~40 km2. When a container was sampled, the water was vigorously homogenized for ca. 5 s with a sterile pipette and a 20 ml sample, collected for bacterial community analysis, was transferred to a labeled sterile plastic centrifuge tube (50 ml, Corning No. 430828, Fisher Scientific). Although this procedure did not specifically sample biofilms, vigorous mixing of the water probably ensured that biofilms were included in the 20 ml samples. The tube was immediately placed on wet ice in the field and then shipped on cold packs to laboratory facilities at North Carolina State University within 2 d of collection. On average, each 20 ml water sample represented about 5–10% of the total volume of water held in each container. Immediately after a water sample was collected, the contents of the container were transferred to an enamel pan, and all mosquito larvae and pupae were transferred to a labeled WhirlPac® bag (Fisher Scientific, Pittsburg, PA, USA), which was placed on wet ice. Within 3 hours of their collection, mosquitoes were killed in hot water and transferred to labeled vials containing 80% ethanol. Subsequently, mosquitoes in each sample were identified to species [41] and counted. In the laboratory, the 20-ml water sample was filtered through a polycarbonate membrane filter (0.22 µm pore size, 47 mm dia., Millipore) and the filter for each sample was cut into quarters and placed into a single cryovial (3.0 ml, Fisher Scientific) after which 1 ml of SET buffer [42] was added. After vortexing, the tubes were stored at −80 °C.
Extraction of Genomic DNA and PCR Amplification of Bacterial Small Subunit rRNA Genes
Samples were thawed at room temperature, and total nucleic acids were extracted and purified using methods described previously by Rivera et al. [36]. Crude DNA was purified with the WIZARD DNA Clean Up System (Promega). Purified DNA was subsequently used as a template to amplify the variable V3 region of 16S rRNA with universal bacterial primers F357-GC (5′-GC-clamp+CCTACGGGAGGCAGCAG-3′) and 518R (5′-ATTACCGCGGCTGCTGG-3′). A 40-bp GC clamp was incorporated into the forward primer to prevent dissociation of the DNA double strand during DGGE analysis [28]. DNA extracted from water samples was amplified with a PCR mixture containing 200 µmol of deoxynucleoside triphosphates, 0.2 µmol of each primer, 5 µl of 10× PCR buffer, 37.5 mM magnesium chloride, 1 U of Taq DNA polymerase, 1 µl (about 5–15 ng) of template DNA, with sterile deionized water added to achieve a final volume of 50 µl. Amplification was made using a touchdown protocol [37]. PCR was performed as follows: the annealing temperature was set at 65 °C and was decreased by 1 °C every cycle until reaching a “touchdown” at 55 °C. The amplification program consisted of 3 min at 94 °C, and 10 touchdown cycles of denaturation at 94 °C for 1 min, annealing at 65 °C (with the temperature decreasing 1 °C each cycle) for 1 min, and extension at 72 °C for 3 min, followed by 40 cycles of 94 °C for 1 min, 55 °C for 1 min, and 72 °C for 3 min. During the last cycle, the length of the extension step was increased to 10 min. Forty amplification cycles was the minimum number needed to yield sufficient PCR product from the water samples for DGGE analyses. With this number of amplification cycles, some biases could have been introduced during amplification due to differences in rRNA gene copy number between bacterial species [10, 43]. PCR products were electrophoresed in a 1.5% agarose gel followed by ethidium bromide staining. Amplicon size and yield were determined by comparison to molecular weight standards (Low DNA Mass Ladder; Gibco BRL).
DGGE Analysis
DGGE, based on the methods of Muyzer et al. [28], was performed using a Dcode™ universal mutation detection system (Bio-Rad) with 8% (wt/vol) acrylamide (acrylamide: bis-acrylamide, 37.5:1.0, wt./wt.) gels, containing a linear chemical gradient that ranged from 40 to 65% (with 100% denaturant consisting of 7 M urea plus 40% [vol/vol] formamide). However, other parameters, such as gel running time and chemical concentration gradient, were modified to give good separation of DGGE OTUs. PCR products were electrophoresed in 1X TAE buffer (40 mM Tris, 20 mM acetate, 1 mM Na2EDTA, pH 7.4) at a constant temperature of 60 °C for 18 h at 50 V. Gels were stained in 0.5X TAE buffer containing SYBR green I and digitally photographed. Each band was considered to be an operational taxonomic unit (OTU).
Construction of a Reference Ladder of Bacterial Species
So that we could assess the quality of DGGE analyses completed on different dates, we developed a reference ladder of 18 bacterial species isolated from an organic infusion made by fermenting senescent white oak leaves (Quercus alba) in well water [45]. Bacteria were cultured on R2A agar plates [35] and isolates that formed colonies with visually distinct morphologies were restreaked several times on R2A agar plates. Pure bacterial colonies, picked from each isolate, were boiled in 50 µl distilled water for 10 min and immediately cooled on ice for 5 min. After centrifugation (30 s at 5,000×g), 4 µl of supernatant was used as DNA template in PCR reactions with V3 primers as described above. Each amplicon was sequenced, and the bacterial isolate was identified using the GenBank database, its migration pattern in DGGE was determined, and all the PCR amplicons of known bacterial species were combined to make a reference ladder consisting of the 18 organisms (see caption for Fig. 1).
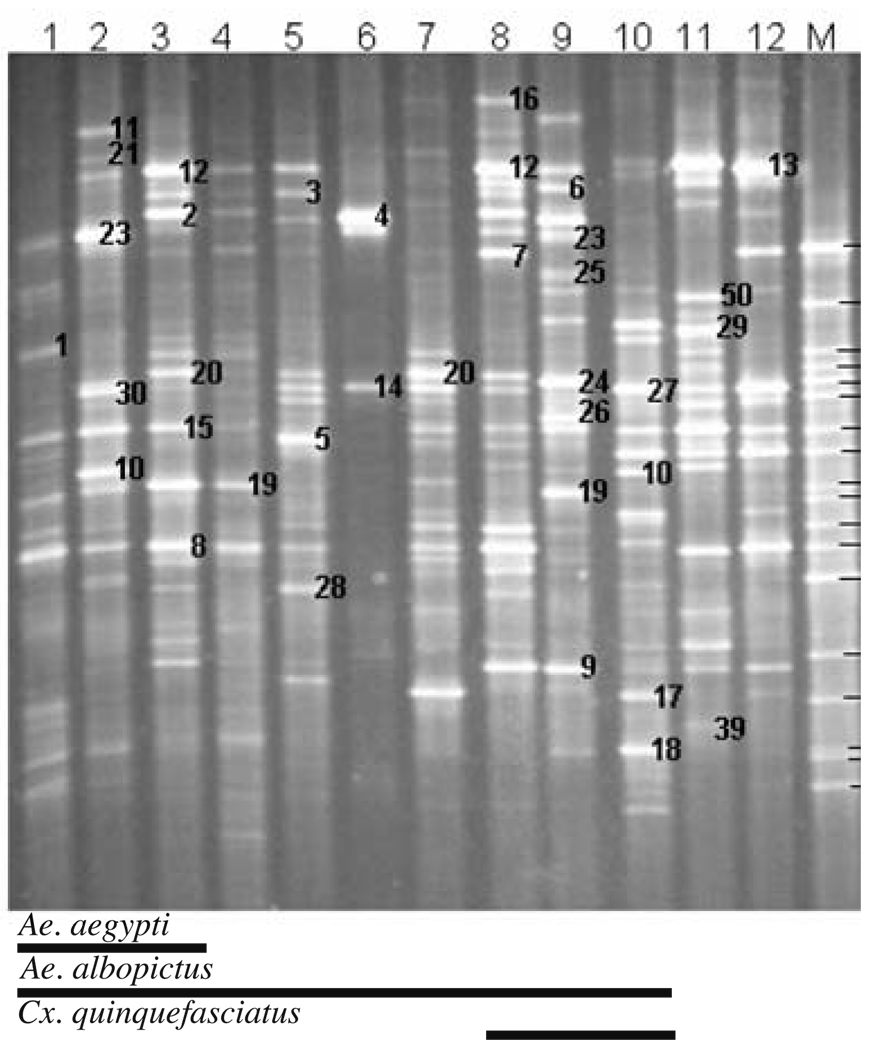
Figure 1.
16S rDNA–DGGE profiles for bacterial communities in tire habitats of mosquitoes that were sampled from New Orleans, LA, USA in 2003. Samples from individual tires are shown in lanes. Lane M contained a DGGE reference ladder of 16S rDNA fragments from bacterial species isolated from white oak leaf infusion. The bands in the marker lanes are as follows (from top to bottom): 1, Gammaproteobacterium; 2, Variovorax sp.; 3, Betaproteobacterium; 4, Agrobacterium tumefaciens; 5, Acidovorax avenae; 6, Bacillus thuringiensis; 7, Betaproteobacterium; 8, Pseudomonas lanceolata; 9, Porphyrobacter sp.; 10, Brevundimonas vesicularis; 11, Caulobacter sp.; 12, Sphingomonas sp.; 13, Betaproteobacterium; 14, Alphaproteobacterium; 15, Rhizobium galegae; 16, Gammaproteobacterium, 17, Azorhizobium caulinodans; 18, Gammaproteobacterium
Analysis of DGGE Banding Patterns
To determine if the type of container affected the diversity of bacterial species, we classified the containers that were sampled into 3 types: tires (n=12), plastic and glass flower urns (n=23), and miscellaneous containers (n=19; including two tree holes). Four different metrics were used to compare the occurrence and abundance of bacterial species within and between the types of containers sampled.
Abundance of bacterial species: To assess the abundance of bacterial species, digital images of banding patterns in each gel were analyzed with 1D Analysis Software (UVP, Upland, CA, USA). Band positions in each sample lane were converted to Rf values which ranged between 0 and 1 using standard positions on all gels that encompassed the uppermost and lowermost band in each sample. Evaluation of band positions for bacterial species in the reference ladder ensured that DGGE analyses were consistent between replicate gels. An intensity profile of the bands in each lane was created using 1D analysis software. Band analysis was performed by setting background band intensity at 10 using the rolling disk method. First, band detection was automatically performed by the software, and then additional bands were assessed by eye. The software carries out a density profile analysis for each lane, detects the bands, and calculates the relative contribution of each band to the total band intensity in the lane. Bands with intensity values <0.05 were excluded from the analysis. The relative contribution of each detected band (=OTU) to the summed intensity of all bands in each lane was calculated separately. In this way, we could estimate the relative abundance and dominance of each bacterial species in each sample [14].
- Diversity of bacterial species: The intensity data were used to calculate Shannon–Weaver diversity indices (H′) [40], using the following equation:
where Pi, the proportion of the total diversity represented by the ith species, was calculated as
where ni is the band intensity for the ith band and N is the summed intensities of all bands in a lane. We estimated the evenness (E) of the numbers of bacterial species in each sample using Pielou’s index [48], which was calculated with the following equation:
where H′max was the maximum value of H′ for each class of container (tire, urn, or miscellaneous). Similarity of bacterial communities: Jaccard’s similarity coefficient (Cj) [48], which measures the likeness between paired samples based on the presence and absence of bacterial species, was used to assess the similarity of bacterial communities. A Jaccard’s coefficient was calculated for each sample pair based on the occurrence of bacterial species in the banding profiles of the water samples within and between each class of container. Jaccard’s coefficient was converted into a percentage similarity value by multiplying Cj by 100.
Cluster analysis of bacterial communities: DGGE fingerprint data for OTUs were subjected to cluster analysis to estimate the relative similarity of the bacterial communities in different container samples. OTUs were converted into presence/absence (1/0) values for each sample, creating a matrix of 54 samples×98 OTUs. Phylotypic distances (d) of samples were calculated from Nei-Li’s similarity coefficient (s) [30], where d=(1−s). A dendrogram was constructed using UPGMA (unweighted pair group with mathematical averages) and the distance matrix method described by Nei and Li [30] (PAUP 4.0b10 software, Sinauer Associates, Publishers, Sunderland, MA, USA). The resulting UPGMA dendrograms were used to assess the similarity of bacterial communities in the water samples for the three types of containers.
Identification of Bacterial Species
DGGE gel bands were excised and the DNA extracted and reamplified for nucleotide sequence determination. Based on their occurrence among the samples, best representative bands were chosen. Also, some unique bands in single samples were excised. Only the middle portion of each band was excised with a sterile razor blade to avoid cross-contamination from an adjacent band. Each gel slice was placed in a sterile centrifuge tube (1.5 ml) containing sterile DNA grade water (50 µl) and held overnight at 4 °C to allow passive diffusion of DNA into the water. An aliquot (1 µl) of eluent DNA was reamplified by using the original primer set. A sample (2.5 µl) of each PCR product was subjected to agarose gel electrophoresis to confirm amplification of the appropriate sized DNA fragment and to estimate its concentration. Re-amplified PCR products were subjected again to DGGE analysis to ensure their purity and correct migration within the gels. Seventeen (10.9%) of 156 amplicons produced more than one DGGE band and were discarded. Re-amplified products showing single bands were purified with the QIAquick PCR purification kit (QIAGEN, Valencia, CA, USA) to remove primers and short oligonucleotides. Nucleotide sequencing was carried out in an ABI PRISM 377 automated DNA sequencer (North Carolina State University), with the ABI PRISM BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems) with either the 357F primer (without GC clamp) or the 518R.
Phylogenetic Analyses
V3 fragment sequences were first checked for chimeras using the CHECK-CHIMERA program of the Ribosomal Database Project. The sequences were compared to those in the GenBank database and the Ribosomal Database Project II [24], using the Basic Local Alignment Search Tool (BLAST) [1] and sequence match analysis, respectively. Sequences were aligned with multiple-alignment CLUSTAL X software package [44]. The method of Jukes and Cantor [17] was used to calculate evolutionary distances between the OTUs and phylogenetic trees were constructed by the neighbor-joining method [38]. Tree topologies were evaluated by performing bootstrap analyses [11], consisting of 1,000 iterations with the MEGA3 software package [23].
Statistical Analyses
Scores for each diversity metric [H′, the number of bands (OTUs) per sample, E; and; Cj for paired samples within each container class] were ranked for all containers. A separate nonparametric Kruskal–Wallis test (PROC NPAR1WAY) for each metric was carried out to determine if the distributions of the ranked scores for the classes of containers were significantly different at P≤0.05. If a significant difference was detected, pair-wise comparisons between the types of containers were performed using one-sided Wilcoxon rank sum tests (PROC NPAR1WAY) and the Bonferroni method to control for experimentwise error (α=0.05/3=0.0167). Statistical analyses were performed using SAS statistical analysis software (version 9.1, SAS Institute, Cary, NC, USA).
Nucleotide Sequence Accession Numbers
The partial sequences obtained in this study have been deposited in the GenBank database under accession numbers DQ444526 to DQ444615 (see Supplementary Table 2). The partial sequences of the bacteria used in the reference ladder have been deposited in the GenBank database under accession numbers EF685163 to EF685180.
Results
Diversity, Abundance, and Similarity of Bacterial Species in Various Container Types
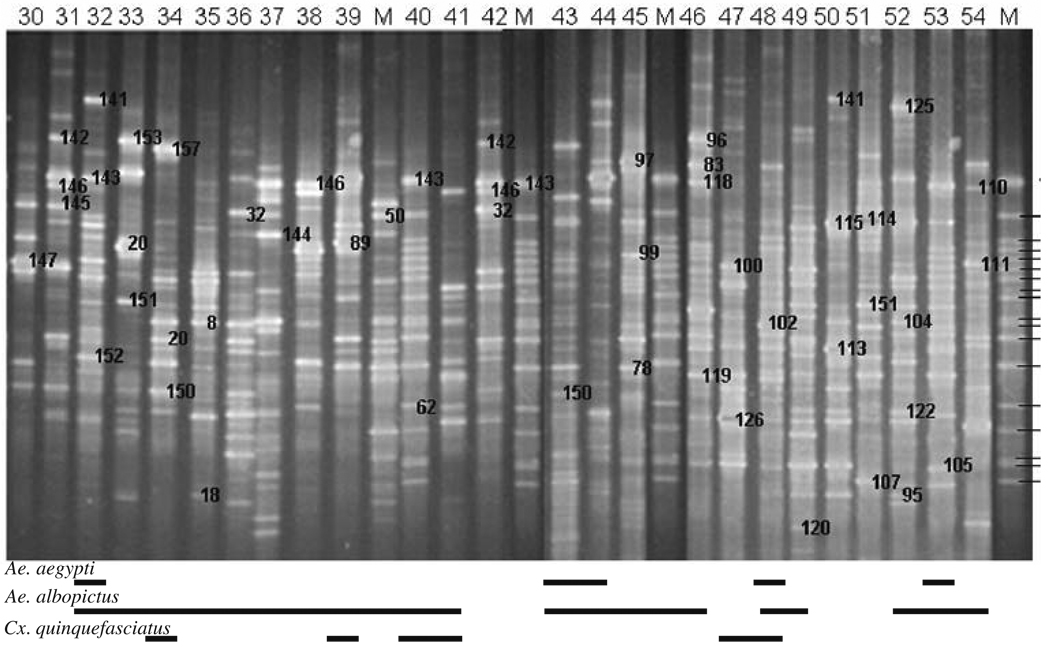
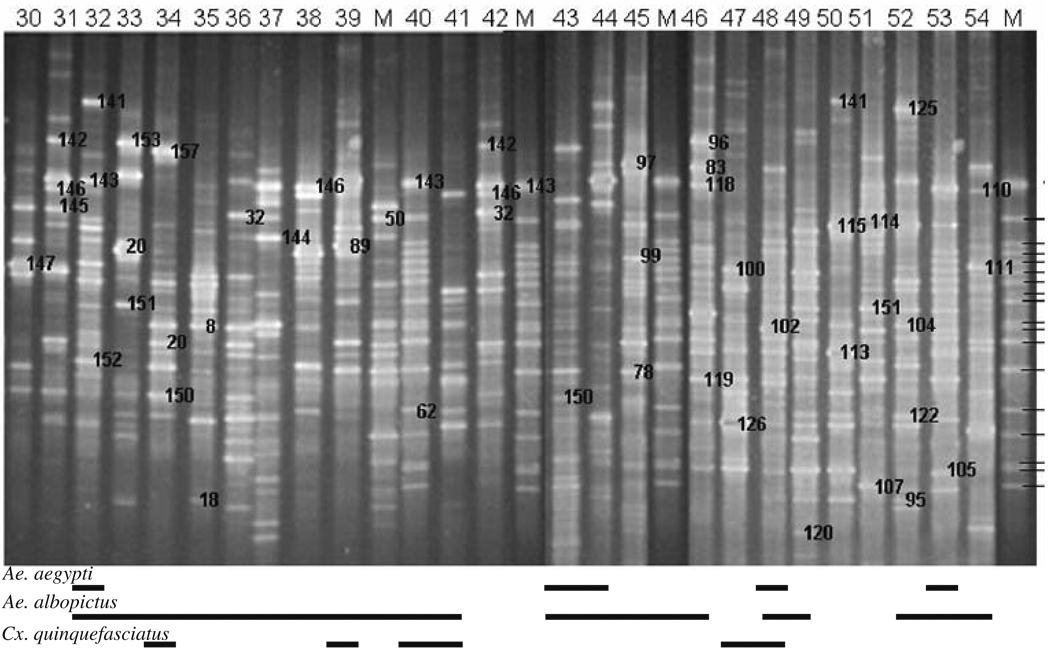
DGGE analysis of 16S rRNA gene PCR products for 54 water samples revealed that DNA banding patterns were reproducible in replicated analyses. Each band (OTU) was assumed to represent at least one unique phylotype, so the richness of bacterial species in a sample was reflected in the number of DGGE-DNA bands. Similarly, the intensity of a band was presumed to reflect the relative abundance of that species in a sample. Diverse DNA banding patterns were found in water samples collected from tires (9–36 bacterial species, n=12; Fig. 1), plastic and glass flower urns (16–33 bacterial species, n=23; Fig. 2), and miscellaneous containers (including tree holes; 15–52 bacterial species, n=19; Fig. 3). A significant difference in the mean number of bacterial species, in mean species diversity (H′), in mean species evenness (E), and in mean similarity (Cj) per sample was found among the three types (Kruskal–Wallis test; df=2; χ2=10.06–17.33; P=0.0066–0.0001). Generally, mean diversity of bacterial species found in water samples from tires was always significantly larger than in urns (one-sided Wilcoxon ranked sum test; P=0.0017–0.0001) but tires did not differ significantly from miscellaneous containers (P=0.0309–0.001) in all diversity metrics (Table 1). Likewise, differences in bacterial diversity in water samples collected from urns and miscellaneous containers were not different for every diversity metric (Table 1).
Figure 2.
16S rDNA–DGGE profiles for bacterial communities in water collected from glass and plastic urn habitats of mosquitoes in New Orleans, LA, USA in 2003. Sites where containers were located are listed in Supplementary Table 1. The bands in the marker lanes are listed in Fig. 1
Figure 3.
16S rDNA–DGGE profiles for bacterial communities in water samples collected from miscellaneous container habitats of mosquitoes in New Orleans, LA, USA in 2003. Sites where containers were located are listed in Supplementary Table 1. The bands in the marker lanes are listed in Fig. 1
Table 1.
Metrics (mean±SD) used to compare bacterial species diversity in water samples collected from container habitats of mosquitoes in New Orleans, LA during 2003
| Metric | Containera | ||
|---|---|---|---|
| Tire (n=12) | Urn (n=23) | Miscellaneous (n=19) | |
| No. DGGE-DNA bands | 28.4a (6.70) | 23.1b (4.20) | 25.9b (7.80) |
| H′ | 3.10a (0.36) | 2.92b (0.22) | 3.07ab (0.27) |
| E | 0.921a (0.107) | 0.865b (0.066) | 0.811c (0.071) |
| Cj | 0.157a (0.080) | 0.113b (0.060) | 0.133a (0.062) |
Paired comparisons between container types for each metric were made using one-sided Wilcoxon ranked sum tests and the Bonferroni method to control for experimentwise error (α=0.05/3=0.0167). Means within each row followed by the same letters are not significantly different
In general, the abundance of each bacterial species in the water samples did not exceed 10% with the exception of Pseudomonas putida (= band 4) in tire water sample NO-6, which accounted for 47.6% of the band intensity profile (Fig. 1). Notably, this species was not detected in any other water sample taken from tires. The most abundant bacterial species varied among containers, even for containers found at the same location. For example, an undescribed bacterium (band 10, 5.2% of total intensity), Pseudomonas sp. (band 23, 7.3%), and Pseudomonas fulva (band 30, 8.6%) were predominant in tire sample NO-2, but in tire sample NO-3 Pseudomonas fluorescens (band 2, 6.7%), and Pseudomonas putida (band 12, 7.3%) were the most abundant bacterial species. These tires were located at sampling sites that were approximately 10 km apart.
Cluster Analyses
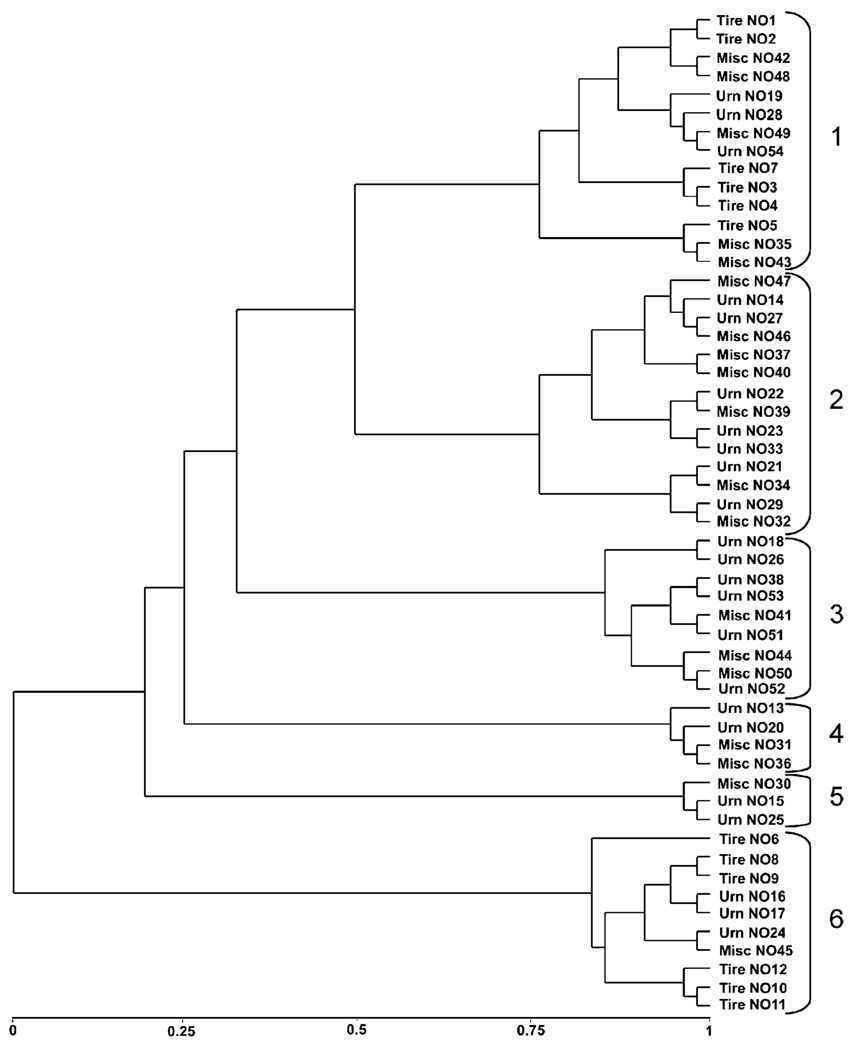
We generated dendrograms through UPGMA analyses of the matrix of distance coefficients so that we could determine if the species similarity of bacterial communities in containers resulted in clustering patterns that could be related to the class of container, sampling location, or presence–absence of mosquitoes. The dendrogram separated into 6 major clusters at phylotypic distances ranging from 1.0–0.5 (Fig. 4). Each cluster was composed of 3–14 containers that were grouped into 1–4 subclusters that separated at phylotypic distances from 0.25–0.15. Containers clustered based on the commonality of OTUs independently of the spatial distribution of the containers. Among subclusters within each major cluster, percentage similarity of bacterial communities between pairs of containers ranged from 7.1–47.6%. Only two (4%) of 54 water samples were from natural habitats, and the bacterial communities in these adjacent pecan tree holes (NO-31 and NO-32) clustered separately, exhibiting only 8% similarity. Notably, bacterial communities from tires were clustered at opposite ends of the dendrogram in clusters 1 and 6. Clusters 2–5 were grouped into subclusters composed of urns and miscellaneous containers (Fig. 4).
Figure 4.
Cluster analysis of DGGE-DNA bands for bacterial communities in water samples taken from human-made containers using UPGMA and Nei-Li’s distance coefficient. Containers clustered into six major groups
Relationship Among Mosquito Species, Bacteria, and Container Types
Overall, 44 (81.5%) of the 54 samples contained mosquito larvae or pupae. A. albopictus immatures were found most frequently (39 containers, 72.2%), followed by Culex quinquefasciatus (11 containers, 20.4%) and A. aegypti (8 containers, 14.8%). The three species were never collected from the same container. Likewise, A. aegypti immatures were never found with C. quinquefasciatus in the containers that we sampled, but A. albopictus co-occurred with A. aegypti and C. quinquefasciatus in 8 (14.8%) and 7 containers (13.0%), respectively.
All three mosquito species were collected from each of the three types of containers (Supplementary Table 1). However, there was no relationship between the presence–absence of mosquitoes and either the number of bacterial species or specific assemblages of bacterial species (Figs. 1–3). Likewise, differences in H′ based on the presence–absence of mosquito larvae in the different classes of containers were not apparent. Mean H′ in tire samples with mosquitoes (3.06±0.12) and without mosquitoes (3.30±0.05) was not significantly different (P=0.0857).
Sequence Analyses
We eluted DNA from 156 excised DGGE gel bands for reamplification by PCR. V3 amplicons could not be detected for 7 (4.4%), 17 bands (10.9%) represented more than one phylotype and were discarded, 15 (9.6%) of the remaining 132 amplified sequences had too many ambiguous nucleotide positions, which prevented these bacteria from being identified, and 6 sequences were found to be chimeras and were discarded. Twenty-one (15.9%) of 132 bands from different samples, but with the same Rf values, were found to contain 100% nucleotide sequence homology. These amplicons were identified to the same bacterial taxa and have been assigned the same numbers in Figs. 1, 2, and 3. Partial sequence analysis of the remaining 90 bands, and their tentative phylogenetic affiliations or species identifications, is given in Table 2 of the supplementary material. For the sequenced bands, 54 sequences were more than 98% identical to sequences in the GenBank database; 20 of the 54 sequences were undescribed species. Thirty-six (40%) of 90 sequences exhibited 92–97% identity with sequences in the GenBank database, but these sequences represented unclassified bacterial species. A total of 14 genera were identified from the band sequences (see Supplementary Table 2). Pseudomonas spp. were the most abundant bacteria, accounting for 16.6% of the number of cultivable species identified.
Phylogenetic Affiliation of Predominant Bacteria
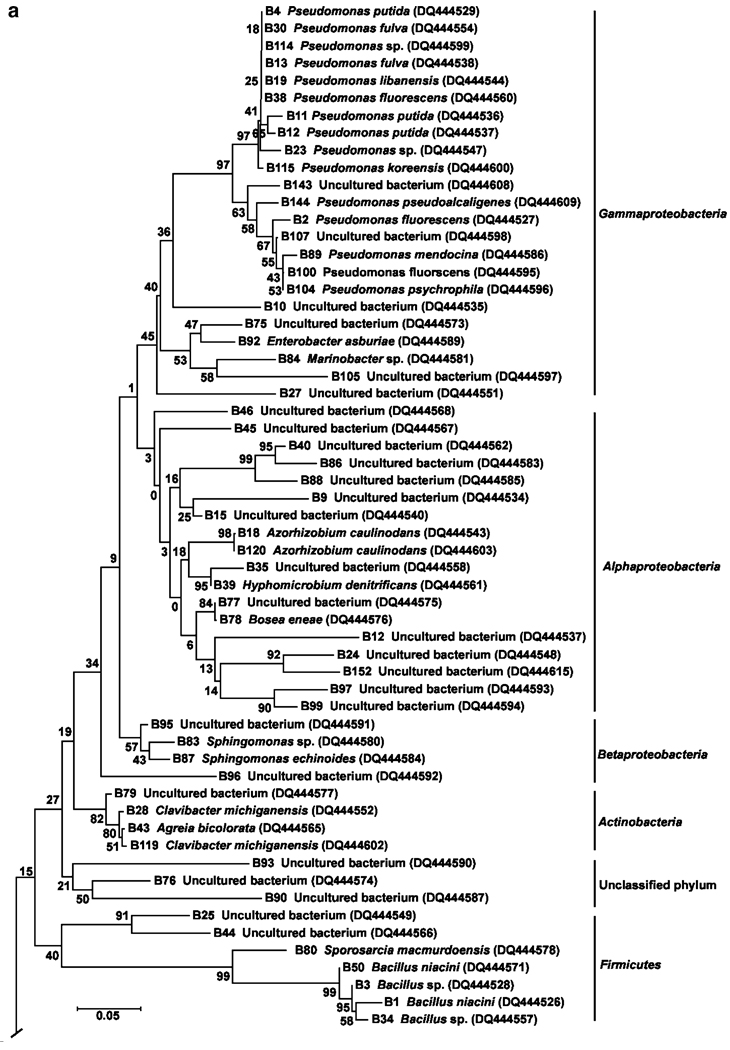
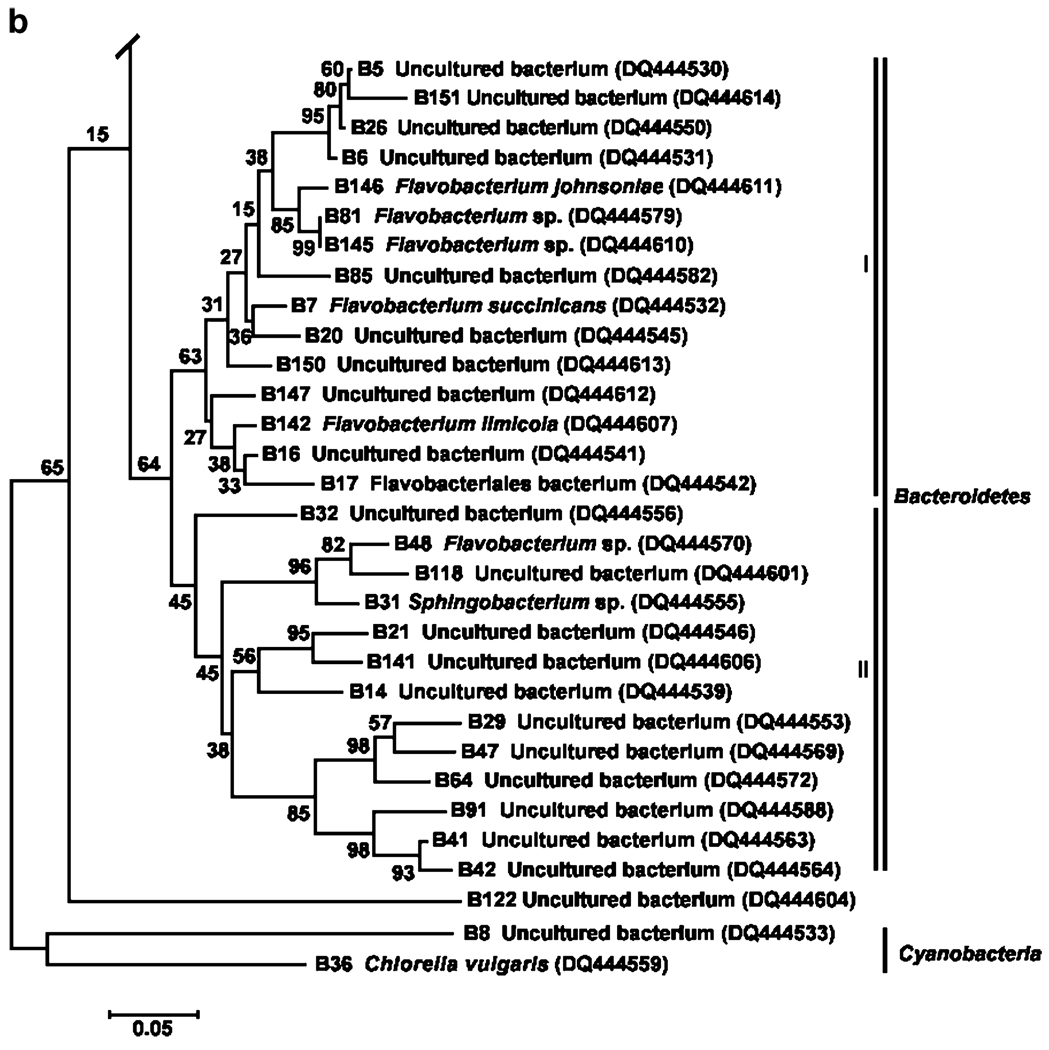
The phylogenetic relationships between the bacterial taxa identified from 16S rDNA sequences are shown in Fig. 5a and b. Bacteria were placed in five major taxa: Alpha-, Beta-, and Gammaproteobacteria, Actinobacteria, Bacteroidetes, Cyanobacteria, Firmicutes, and an unclassified group. However, Proteobacteria were the most prevalent group of bacteria based on sequence analyses (50% of the total number of sequences), and comprised of 23 bacterial sequences within the Gammaproteobacteria (6 undescribed bacteria), 18 sequences in the phylum Alphaproteobacteria (14 undescribed bacteria), and 4 sequences in the Betaproteobacteria (2 undescribed bacteria). Other phyla were represented by few bacterial sequences: Actinobacteria (4.4%), Firmicutes (7.7%), and Cyanobacteria (2.2%). The second most dominant group represented by the phylum Bacteroidetes formed two subclusters (Fig. 5b) with 21 of the 29 (72.4%) sequences belonging to uncultivable bacteria. A small number of band sequences (3.3%) could not be assigned to any phylum (bootstrap value=21%).
Figure 5.
a Neighbor-joining tree showing phylogenetic positions of 16S rRNA gene sequences (phyla Proteobacteria, Firmicutes, and Actinobacteri) from water collected from human-made container habitats of mosquitoes. Bootstrap values for a total of 1,000 iterations are shown at the nodes of the tree. Bar=0.05 nucleotide substitution per sequence position. b Neighbor-joining tree showing phylogenetic positions of 16S rRNA gene sequences (phyla-Bacteroidetes and Cynobacteria) from mosquito container habitats. Bootstrap values for a total of 1,000 iterations are shown at the nodes of the tree. Bar=0.05 substitution per sequence position
Discussion
Our study is the first investigation of the species structure of bacterial communities in human-made container habitats of mosquitoes. Container habitats of mosquitoes are ecosystem mesocosms that are analogous to aquatic ecosystems that naturally occur in tree holes and other plant cavities [20]. These aquatic habitats support food webs that are dependent on detritus [8] and microbial communities that metabolize and mineralize organic carbon from detritus [29]. Based on sequence analysis of 16S rDNA amplicons, our investigation revealed that human-made water-filled containers and tree holes hold bacterial communities of mainly undescribed species. Additionally, DGGE banding patterns that were highly variable showed that species composition was independent of both container type and their spatial distribution. Although the three types of containers that we evaluated were inhabited by significantly different numbers of bacterial species, we also found no relationship between the presence of mosquitoes and either the number of bacterial species or specific assemblages of bacterial species.
Bacteria are an integral part of the diet of larval mosquitoes [26]. Shifts in the species composition of bacterial communities have been observed in experimental microcosms when mosquito larvae were added [19]. Change in community composition was not due to nutrient enrichment from excretory products of mosquito larvae that stimulated differential growth of bacterial species [9] but resulted from an increase in the abundance of bacteria that were recalcitrant to digestion [19]. Nutrient inputs from exogenous sources have been observed to influence concentrations of inorganic ions and organic compounds in the microcosms but had less-pronounced effects on microbial community structure than the presence of mosquito larvae [19]. Similarly, the feeding activity of other bacterial predators has been shown to enhance bacterial species richness and alter the rates of decomposition of organic matter in experimental aquatic ecosystems [22]. In view of these studies, it is likely that the mosquito larvae in the container habitats that we sampled influenced the species structure of bacterial communities. However, understanding the impacts of mosquito larvae on bacterial community structure in human-made containers requires experimental research under more controlled conditions.
The relationship between mosquito larvae and bacteria extends beyond trophic interactions. Bacteria are also a primary functional group of decomposers, catabolizing and recycling organic matter, and metabolites derived from the breakdown of detritus act as semiochemicals, mediating the oviposition behavior of mosquitoes [4] by attracting or repelling gravid mosquitoes [5, 21, 34, 46], and arresting and stimulating them to lay eggs [5]. Thus, we hypothesized that the spatial distribution of mosquitoes among available habitats would be influenced by microbial communities in water-filled containers because the attraction and egg-laying responses of gravid mosquitoes are mediated in part by the resource status of their oviposition sites. Because A. albopictus and A. aegypti are thought to occupy slightly different ecological niches as reflected by the spatial separation of these mosquitoes in some geographic areas where the species are sympatric [16], we also hypothesized that specific assemblages of bacterial species would be found in association with each mosquito species inhabiting human-made containers in suburban landscapes of New Orleans. However, both of these ecologically appealing hypotheses were not supported by results of our analysis of bacterial and mosquito species. While bacterial assemblages varied markedly among containers, mosquitoes were collected from most (~81%) containers examined, suggesting that bacterial communities in these containers are likely to be composed of species that are functionally similar in terms of their catabolic activity [13, 32]. Moreover, we collected A. albopictus immatures from all of the containers in which A. aegypti was found, and all three mosquito species were found in some of the containers comprising each of the six major groups that were separated by cluster analysis of bacterial OTUs. These findings suggest that the species structure of bacterial communities in the human-made containers that we sampled was not a critical factor determining the occurrence of mosquitoes.
The containers that we sampled supported bacterial communities representing a broad phylogenetic diversity, which provides insight into the functional properties of species that cannot be gained by enumerating sample OTUs [25]. The Proteobacteria were the predominant group in our investigation, accounting for 45 (50%) of 90 sequences, consistent with other investigations reporting high prevalence (20–50%) of Proteobacteria in bacterial communities in aquatic habitats [2, 3, 18] such as seawater [15, 33], decaying salt marsh grass [7], and wastewater [7]. In potable water distribution systems, many bacterial species, including members of the Proteobacteria, Actinobacteria, low-G+C-content gram-positive bacteria, and Cytophaga– Flavobacterium–Bacterioides group, readily adhere to surfaces to form multi-species biofilms [39]. It is worth noting that although we vigorously mixed the water in the containers prior to sampling, we did not explicitly collect biofilms. Bacteria in the phylum Bacteriodes accounted for 29 (32.2%) of 90 sequences in container habitats that we sampled, and Flavobacteria (7.7% of band sequences) was the most commonly detected genus. The genus Bacillus comprised a small number (4.4%) of OTUs, which is surprising since spore-forming bacilli are common on the surface of green and senescent oak leaves [6], which were common in the study area.
A noteworthy feature of the bacterial communities in these habitats was the presence of potentially aerobic phylotypes. In experimental microcosms, the presence of mosquito larvae contributes to enriched and anoxic conditions favorable to the growth of facultative anaerobes in the Enterobacteriaceae [19], and Enterobacter cloacae was isolated from larval holding water that was highly attractive to gravid A. aegypti [5]. Surprisingly, Enterobacteriaceae were rare in the containers that we sampled in the field, underscoring previous observations that a variety of bacterial species produce chemicals that attract gravid mosquitoes and may stimulate them to lay eggs in human-made containers.
Results of cluster analysis suggested a stronger basis for similar bacterial community structure in tires relative to other types of containers. Bacterial communities from tires clustered into two groups at opposite ends of the dendrogram. The species structure of bacterial communities in tires reflected in cluster analysis raises intriguing questions about the ecological processes that result in certain bacterial assemblages in particular containers and the successional paths that these communities undergo. This preliminary observation is the foundation of our current research to measure the time-course of container occupancy by mosquitoes to define physical, chemical, and biological parameters of tire habitats. Measurements of such experimental mesocosms under natural conditions, coupled with evaluations of their suitability for mosquito development will in turn present unique opportunities to formulate new hypotheses on the interaction of mosquitoes and their microbial environment.
Supplementary Material
Acknowledgement
Partial support for our research was provided by NIH, NIAID through cooperative agreement U01-AI-58303-01.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00248-008-9379-6) contains supplementary material, which is available to authorized users.
Contributor Information
Loganathan Ponnusamy, Department of Entomology, North Carolina State University, Campus Box 7647, Raleigh, NC 27695-7647, USA.
Ning Xu, Department of Entomology, North Carolina State University, Campus Box 7647, Raleigh, NC 27695-7647, USA.
Gil Stav, Department of Tropical Medicine, Tulane Health Sciences Center, Tulane University, New Orleans, LA 70112, USA.
Dawn M. Wesson, Department of Tropical Medicine, Tulane Health Sciences Center, Tulane University, New Orleans, LA 70112, USA
Coby Schal, Department of Entomology, North Carolina State University, Campus Box 7647, Raleigh, NC 27695-7647, USA.
Charles S. Apperson, Email: charles_apperson@ncsu.edu, Department of Entomology, North Carolina State University, Campus Box 7647, Raleigh, NC 27695-7647, USA.
References
- 1.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann RI, Ludwig W, Schleifer KH. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Araya RK, Tani Takagi T, Yamaguchi N, Nasu M. Bacterial activity and community composition in stream water and biofilm from an urban river determined by fluorescent in situ hybridization. FEMS Microbiol Ecol. 2003;43:111–119. doi: 10.1111/j.1574-6941.2003.tb01050.x. [DOI] [PubMed] [Google Scholar]
- 4.Bentley MD, Day JF. Chemical ecology and behavioral aspects of mosquito oviposition. Ann Rev Entomol. 1989;34:401–421. doi: 10.1146/annurev.en.34.010189.002153. [DOI] [PubMed] [Google Scholar]
- 5.Benzon GL, Apperson CS. Reexamination of chemically mediated oviposition behavior in Aedes aegypti (L.) (Diptera: Culicidae) J Med Entomol. 1988;25:158–164. doi: 10.1093/jmedent/25.3.158. [DOI] [PubMed] [Google Scholar]
- 6.Brunel B, Perissol C, Fernandez M, Boeufgras JM, LePetit J. Occurrence of Bacillus species on evergreen oak leaves. FEMS Micro Ecol. 1994;14:331–342. [Google Scholar]
- 7.Carrero-Colón M, Nakatsu CH, Konopka A. Effect of nutrient periodicity on microbial community dynamics. Appl Environ Microbiol. 2006;72:3175–3183. doi: 10.1128/AEM.72.5.3175-3183.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cochran-Stafira DL, von Ende CN. Integrating bacteria into food webs: studies with Sarracenia purpurea inquilines. Ecology. 1998;79:880–898. [Google Scholar]
- 9.DeAngelis DL. Dynamics of nutrient cycling and food webs. New York: Chapman and Hall; 1992. p. 270. [Google Scholar]
- 10.Farrelly V, Rainey FA, Stackebrandt E. Effect of genome size and rrn gene copy number on PCR amplification of 16S rRNA genes from a mixture of bacterial species. Appl Environ Microbiol. 1995;61:2798–2801. doi: 10.1128/aem.61.7.2798-2801.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 12.Focks DA, Alexander N. Multicountry study of Aedes aegypti pupal productivity survey methodology: findings and recommendations. 2006 WHO/TDR/IRM/DEN/06.1. [Google Scholar]
- 13.Franklin RB, Mills AL. Structural and functional responses of a sewage microbial community to dilution-induced reductions in diversity. Microb Ecol. 2006;52:280–288. doi: 10.1007/s00248-006-9033-0. [DOI] [PubMed] [Google Scholar]
- 14.Fromin N, Hamelin J, Tarnawski S, Roesti D, Jourdain-Miserez K, Forestier N, Teyssier-Cuvelle S, Gillet F, Aragno M, Rossi P. Statistical analysis of denaturing gel electrophoresis (DGE) fingerprinting patterns. Environ Micobiol. 2002;4:634–643. doi: 10.1046/j.1462-2920.2002.00358.x. [DOI] [PubMed] [Google Scholar]
- 15.Gonzalez J, Moran MA. Numerical dominance of a group of bacteria in the alpha-subclass of the class Proteobacteria in coastal seawater. Appl Environ Microbiol. 1997;63:4237–4242. doi: 10.1128/aem.63.11.4237-4242.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hawley WA. The biology of Aedes albopictus. J Am Mosq Control Assoc (Suppl.) 1988;4:2–39. [PubMed] [Google Scholar]
- 17.Jukes TH, Cantor CR. Evolution of protein molecules. In: Munro HN, editor. Mammalian protein metabolism. New York: Academic; 1969. pp. 21–132. [Google Scholar]
- 18.Kampfer P, Erhart R, Beimfohr C, Bohringer J, Wagner M, Amann R. Characterization of bacterial communities from activated sludge: culture-dependent numerical identification versus in situ identification using group- and genus-specific rRNA-targeted oligonucleotide probes. Microb Ecol. 1996;32:101–121. doi: 10.1007/BF00185883. [DOI] [PubMed] [Google Scholar]
- 19.Kaufman MG, Walker ED, Smith TW, Merritt RW, Klug MJ. Effects of larval mosquitoes (Aedes triseriatus) and stemflow on microbial community dynamics in container habitats. Appl Environ Microbiol. 1999;65:2661–2673. doi: 10.1128/aem.65.6.2661-2673.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kitching RL. Food webs and container habitats: The natural history and ecology of phytotelmata. Cambridge, UK: Cambridge Univ. Press; 2000. [Google Scholar]
- 21.Knols BGJ, Sumba LA, Guda TO, Deng AL, Hassanali A, Beier JC. Mediation of oviposition site selection in the African malaria mosquito Anopheles gambiae (Diptera: Culicidae) by semiochemicals of microbial origin. Int J Trop Insect Sci. 2004;24:260–265. [Google Scholar]
- 22.Kneitel JM, Miller TE. Resource and toppredator regulation in the pitcher plant (Sarracenia purpurea) inquiline community. Ecol. 2002;83:680–688. [Google Scholar]
- 23.Kumar S, Tamura K, Nei M. MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 24.Maidak BL, Cole JR, Lilburn TG, Parker CT, Jr, Saxman PR, Farris RJ, Garrity GM, Olsen GJ, Schmidt TM, Tiedje JM. The RDP-II (Ribosomal Database Project) Nucleic Acids Res. 2001;29:173–174. doi: 10.1093/nar/29.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin AP. Phylogenetic approaches for describing and comparing the diversity of microbial communities. Appl Environ Microbiol. 2002;68:3673–3682. doi: 10.1128/AEM.68.8.3673-3682.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Merritt RW, Dadd RH, Walker ED. Feeding behavior, natural food, and nutritional relationships of larval mosquitoes. Ann Rev Entomol. 1992;37:349–377. doi: 10.1146/annurev.en.37.010192.002025. [DOI] [PubMed] [Google Scholar]
- 27.Moore JC, Berlow EL, Coleman DC, de Ruiter PD, Dong Q, Hastings A, Johnson NC, McCann KS, Melville K, Morin PJ, Nadelhoffer K, Rosemond AD, Post DM, Sabo JL, Scow KM, Vanni MJ, Wall DH. Detritus, trophic dynamics and biodiversity. Ecol Letters. 2004;7:584–600. [Google Scholar]
- 28.Muyzer GE, de Waal C, Uitterlinden AG. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified gene coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neely CL, Beare MH, Hargrove WL, Coleman DC. Relationships between fungal and bacterial substrate-induced respiration, biomass, and plant residue decomposition. Soil Biol Biochem. 1991;23:947–954. [Google Scholar]
- 30.Nei M, Li W-H. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci. 1979;76:5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pace MR. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 32.Petchey OL, Gaston KJ. Functional diversity (FB), species richness and community composition. Ecol Letters. 2002;5:402–411. [Google Scholar]
- 33.Rappe MS, Kemp PF, Giovannoni SJ. Phylogenetic diversity of marine coastal picoplankton 16S rRNA genes cloned from the continental shelf off Cape Hatteras, North Carolina. Limnol Oceanogr. 1997;42:811–826. [Google Scholar]
- 34.Rejmankova E, Harbin-Ireland A, Lege M. Bacterial abundance in larval habitats of four species of Anopheles (Diptera: Culicidae) in Belize, Central America. J Vector Ecol. 2000;25:229–238. [PubMed] [Google Scholar]
- 35.Reasoner DJ, Geldreicht EE. A new medium for the enumeration and subculture of bacteria from potable water. Appl Environ Microbiol. 1985;49:1–7. doi: 10.1128/aem.49.1.1-7.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rivera ING, Lipp EK, Gil A, Choopun N, Huq A, Colwell RR. Method of DNA extraction and application of multiplex polymerase chain reaction to detect toxigenic Vibrio cholerae O1 and O139 from aquatic ecosystems. Environ Microbiol. 2003;5:599–606. doi: 10.1046/j.1462-2920.2003.00443.x. [DOI] [PubMed] [Google Scholar]
- 37.Roux KH. Optimization and troubleshooting in PCR. In: Dieffenbach CW, Dveksler GS, editors. PCR primer: a laboratory manual. Plainview, NY: Cold Spring Harbor Laboratory; 1995. pp. 53–62. [Google Scholar]
- 38.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 39.Schmeisser C, Stockigt C, Raasch C, Wingender J, Timmis KN, Wenderoth DF, Flemming HC, Liesegang H, Schmitz RA, Jaeger KE, Streit WR. Metagenome survey of biofilms in drinking-water networks. Appl Environ Microbiol. 2003;69:7298–7309. doi: 10.1128/AEM.69.12.7298-7309.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shannon CE, Weaver W. The mathematical theory of communication. Urbana, IL: Univ. of Illinois Press; 1963. [PubMed] [Google Scholar]
- 41.Slaff M, Apperson CS. NC State Univ. Agric. Ext. Ser. Pub. no. AG-412. Raleigh, NC: 1989. A key to the mosquitoes of North Carolina and the Mid-Atlantic states. [Google Scholar]
- 42.Sommerville CC, Knight IT, Straube WL, Colwell RR. Simple, rapid method for direct isolation of nucleic acids from aquatic environments. Appl Environ Microbiol. 1989;55:548–554. doi: 10.1128/aem.55.3.548-554.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suzuki MT, Giovannoni SJ. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol. 1996;62:625–630. doi: 10.1128/aem.62.2.625-630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The ClustalX Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trexler JD, Apperson CS, Schal C. Laboratory and field evaluations of the oviposition response of Aedes albopictus and Aedes triseriatus (Diptera: Culicidae) to oak leaf infusions. J Med Entomol. 1998;35:967–976. doi: 10.1093/jmedent/35.6.967. [DOI] [PubMed] [Google Scholar]
- 46.Trexler JD, Apperson CS, Zurek L, Gemeno C, Schal C, Kaufman M, Walker E, Watson DW, Wallace L. Role of bacteria in mediating the oviposition responses of Aedes albopictus (Diptera: Culicidae) J Med Entomol. 2003;40:841–848. doi: 10.1603/0022-2585-40.6.841. [DOI] [PubMed] [Google Scholar]
- 47.Vezzani D. Review: Artificial container-breeding mosquitoes and cemeteries: a perfect match. Trop Med Int Health. 2007;12:299–313. doi: 10.1111/j.1365-3156.2006.01781.x. [DOI] [PubMed] [Google Scholar]
- 48.Washington HG. Diversity, biotic and similarity indices: a review with special relevance to aquatic ecosystems. Water Res. 1984;18:653–694. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.