Abstract
The salivary transcriptome of the seed-feeding hemipteran, Oncopeltus fasciatus (milkweed bug), is described following assembly of 1,025 ESTs into 305 clusters of related sequences. Inspection of these sequences reveals abundance of low complexity, putative secreted products rich in the amino acids (aa) glycine, serine or threonine, which might function as silk or mucins and assist food canal lubrication and sealing of the feeding site around the mouthparts. Several protease inhibitors were found, including abundant expression of cystatin transcripts that may inhibit cysteine proteases common in seeds that might injure the insect or induce plant apoptosis. Serine proteases and lipases are described that might assist digestion and liquefaction of seed proteins and oils. Finally, several novel putative proteins are described with no known function that might affect plant physiology or act as antimicrobials. Supplemental files mentioned in the text can be obtained from http://exon.niaid.nih.gov/transcriptome.html#non_blood_feeding
Keywords: Feeding, Phytophagy, Salivary glands, Sialotranscriptome
1. Introduction
Arthropod saliva can assist feeding in both the pre- and post-ingestion phases of the meal. Saliva plays an important role in the pre-ingestion phase in predatory, blood-feeding and plant-feeding modes when the arthropod sucks, rather than chews, the meal. In this case, saliva is injected into the source of the meal and contains compounds that paralyze, digest and liquefy the prey tissues, or prevent the vertebrate host hemostasis and inflammation, or digests the plant and interferes with plant defense mechanisms (Ribeiro, 1995). In the case of blood-sucking insects and ticks, where the salivary composition has been better studied, nearly 70 proteins have been identified in mosquitoes (Arca et al., 2005; Ribeiro et al., 2007), and several hundred in ticks (Ribeiro et al., 2006).
A large proportion of the salivary proteins of blood-sucking arthropods were acquired by lineage-specific gene duplication events. Accordingly, in the blood-sucking Nematocera, an expansion of the D7 family of proteins occurred (Valenzuela et al., 2002), among other protein expansions (Arca et al., 2005). Most remarkably, the lipocalin family was greatly expanded in both triatomine bugs and ticks, where they serve multiple functions in the binding of biogenic amines and adenosine nucleotides (Andersen et al., 2003; Francischetti et al., 2002a; Paesen et al., 2000; Ribeiro and Walker, 1994), transporters of nitric oxide (Champagne et al., 1995), anti-clotting (Noeskejungblut et al., 1995; Ribeiro et al., 1995), anti-platelet (Noeske-Jungblut et al., 1994) and anti-complement (Nunn et al., 2005) functions. Indeed there are at least 24 different salivary lipocalins expressed in Rhodnius prolixus salivary glands (Ribeiro et al., 2004a), and a larger number in ticks (Ribeiro et al., 2006). Why did triatomine and ticks acquire such large lipocalin families in their salivary glands? Perhaps these arthropod orders were already rich in such families, increasing the probability of further gene duplication events that were co-opted for a salivary function during the arthropod's adaptation to its mode of feeding or to a particular host.
While the salivary transcriptome of hematophagous insects has been studied to some extent, those of plant feeding insects is not known, although there is evidence that saliva of such insects affects plant physiology in analogous mode to their hematophagous counterparts. For example: saliva or salivary extracts from bugs of the Lygus genus are known to posses plant-growth hormone activity (Hori, 1974) and enzymes (Zeng et al., 2002); aphids secrete a salivary peroxidase that counteracts their hosts noxious catechols (Miles and Peng, 1989; Peng and Miles, 1988), and salivary extracts of caterpillars contain glucose oxidase that detoxify plant defense compounds, and also contain antimicrobial substances (Musser et al., 2005a; Musser et al., 2005b). In this paper, we explored the sialotranscriptome of Oncopeltus fasciatus, a seed-feeding hemipteran of the suborder Heteroptera, infraorder Pentatomorpha, which is the closest related infraorder to the Cimicomorpha to which triatomine bugs and bed bugs belong. We found abundant transcripts coding for protease inhibitors of the cystatin family, serine proteases and lipases, and several novel protein families with unknown function that might interfere with plant defense mechanisms.
2. Materials and methods
2.1. Insects
In this study, we used adult Oncopeltus fasciatus kindly provided by Dr. Alexandre Romeiro (Instituto de Biofísica Carlos Chagas Filho, Universidade Federal do Rio de Janeiro, Rio de Janeiro, Brazil), from which a colony has been established in the laboratory of AHL ( Brazil). The insects were kept in plastic pitchers at 26° C and fed with toasted, peeled sunflower seeds and fresh water. Adult insects were carefully dissected, their intact salivary glands (50 pairs) were extracted and placed into a solution of 75% RNA-Later (Ambion Inc.), 25% 1x PBS (RNase free) and stored in 100% RNA-Later at –20° C for isolating polyA+ RNA at the Laboratory of Malaria and Vector Research (NIAID/NIH).
2.2. Library construction
O. fasciatus salivary gland mRNA was isolated from fifty salivary-gland pairs from adult insects using the Micro-FastTrack mRNA isolation kit (Invitrogen). The polymerase chain reaction (PCR)-based cDNA library was made following the instructions for the SMART cDNA library construction kit (Clontech). Salivary gland polyA+ RNA was used for reverse transcription to cDNA using PowerScript reverse transcriptase (Clontech), the SMART IV oligonucleotide, and the CDS III/3′ primer (Clontech). The reaction was carried out at 42° C for 1 h. Second-strand synthesis was performed by a long-distance (LD), PCR-based protocol using the 5′ PCR primer and the CDS III/3′ primer as sense and anti-sense primers, respectively. These two primers also create Sfi1 A and B restriction enzyme sites at the end of nascent cDNA. Advantage™ Taq polymerase mix (Clontech) was used to carry out the LD PCR reaction on a GeneAmp® PCR System 9700 (Perkin Elmer Corp.). The PCR conditions were: 95° C for 20 s; 24 cycles of 95° C for 5 s; 68° C for 6 min. A small portion of the cDNA was analyzed on a 1.1% agarose/EtBr (0.1 μg/ml) gel to check for the quality and the range of the cDNA synthesized. Double-stranded cDNA was immediately treated with proteinase K (0.8 μg/ml) at 45° C for 20 min. Proteinase K was removed using a Microcon YM-100 mini-column (100,000 MWCO; Millipore) following the manufacturer's recommendations.
The clean, double-stranded cDNA was then digested with SfiI restriction enzyme at 50° C for 2 h, followed by size fractionation on a ChromaSpin–400 drip column (Clontech). The profiles of the fractions were checked on a 1.1% agarose/EtBr (0.1 μg/ml), and fractions containing cDNA of more than 400 bp were pooled and concentrated by mini-column as described above. The cDNAs were then ligated into a λ TriplEx2 vector (Clontech), and the resulting ligation mixture was packaged using GigaPack® III Plus packaging extract (Stratagene) according to the manufacturer's instructions. The packaged library was plated by infecting log-phase XL1-Blue E. coli cells (Clontech). The percentage of re-clones was determined by performing a blue-white selection screening on LB/MgSO4 plates containing X-gal/IPTG. Recombinants were also determined by PCR, using vector primers (5′ λ TriplEx2 and 3′ λ TriplEx2 sequencing primers) flanking the inserted cDNA and visualizing the products on a 1.1% agarose/EtBr gel.
2.3. Sequencing of the Oncopeltus fasciatus cDNA library
The Oncopeltus fasciatus (Of) salivary gland cDNA library was plated on LB/MgSO4 plates containing X-gal/IPTG to an average of 250 plaques per 150-mm Petri dish. Recombinant (white) plaques were randomly selected and transferred to 96-well Microtest ™ U-bottom plates (BD BioSciences) containing 100 μl of SM buffer (0.1 M NaCl, 0.01 M MgSO4.7 H2O, 0.035 M Tris-HCl [pH 7.5], 0.01% gelatin) per well. The plates were covered and placed on a gyrating shaker for 30 min at room temperature. The phage suspension was either immediately used for PCR or stored at 4° C for future use.
To amplify the cDNA using PCR, 4 μl of the phage sample was used as a template. The primers were sequences from the λ TriplEx2 vector and named pTEx2 5seq (5′-TCCGAG ATC TGG ACG AGC-3′) and pTEx2 3LD (5′-ATA CGA CTC ACT ATA GGG CGA ATT GGC-3′), positioned at the 5′ and 3′ end of the cDNA insert, respectively. The reaction was carried out in 96-well flexible PCR plates (Fisher Scientific) using Platinum SuperMix (Invitrogen) on a GeneAmp® PCR system 9700. The PCR conditions were: 1 hold at 95° C for 3 min; 25 cycles of 95° C for 1 min, 61° C for 30 sec; 72° C for 2 min. Amplified products were analyzed on a 1.5% agarose/EtBr gel. cDNA library clones (1100 clones) were PCR amplified; those showing a single band were selected for sequencing. Approximately 200–250 ng of each PCR product was used for DNA sequencing. cDNA sequencing was carried out using a BigDye® Terminator v3.1 cycle sequencing kit (Applied Biosystems), and reaction products were analyzed on an ABI 3730xl DNA analyzer (Applied Biosystems). A total of 1,017 cDNA library clones were sequenced, of which 916 were used in this work.
2.4. Bioinformatic tools and procedures utilized
EST were trimmed of primer and vector sequences, clusterized, and compared with other databases as previously described (Valenzuela et al., 2003). For functional annotation of the transcripts, we used the program BlastX (Altschul et al., 1997) to compare nucleotide sequences to the nonredundant (NR) protein database of the National Center for Biotechnology Information (NCBI) and to the gene ontology database (Ashburner et al., 2000). The tool rpsBlast (Schaffer et al., 2001) was used to search for conserved protein domains in the Pfam (Bateman et al., 2000), SMART (Letunic et al., 2002), Kog (Tatusov et al., 2003) and conserved domain databases (Marchler-Bauer et al., 2002). We also compared the transcripts with other subsets of mitochondrial and rRNA nucleotide sequences downloaded from NCBI. All BLAST comparisons were done with the complexity filter off, but segments of polymonucleotides of 20 bases or larger were masked. All six-frame translations were used in the case of BlastX or rpsBlast. To identify possible transcripts coding for secreted proteins, segments of the three-frame translations of all ESTs (because the libraries are unidirectional, we did not use six-frame translations) starting with a methionine found in the first 100 predicted aa, or the predicted protein translation in the case of complete coding sequences, were submitted to the SignalP server (Nielsen et al., 1997) to help identify translation products that could be secreted. O-glycosylation sites on the proteins were predicted with the program NetOGlyc (http://www.cbs.dtu.dk/services/ NetOGlyc/) (Hansen et al., 1998). Functional annotation of the transcripts was based on all the comparisons above. Following inspection of all these results, transcripts were classified as either Secretory (S), Housekeeping (H), or of Unknown (U) function, with further subdivisions based on function and/or protein families. Sequence alignments were performed with the ClustalX (Thompson et al., 1997) software package.
3. Results and discussion
3.1. cDNA library characteristics
A total of 1,025 clones were used to assemble a clusterized database (Supplemental Table S1), yielding 305 clusters of related sequences, 169 of which contained only one EST. Accordingly, additional library sequencing would lead to less than 20% of the new sequences to be novel. The consensus sequence of each cluster is named either a contig (deriving from two or more sequences) or a singleton (deriving from a single sequence); in this paper, for simplicity sake, we will use the denomination cluster to address sequences deriving both from consensus sequences and from singletons. The 305 clusters were compared by BlastX, BlastN, or rpsBlast (Altschul et al., 1997) to several databases and to the SignalP server. The EST assembly, BLAST, and signal peptide results were transferred into an Excel spreadsheet for manual annotation.
Four categories of expressed genes derived from the manual annotation of the contigs (Table 1). The S category (transcripts coding for possible secreted polypeptides) contained 9.8% of the clusters and 41.2% of the sequences, with an average number of 14.1 sequences per cluster. The H category (transcripts coding for possible housekeeping polypeptides) had 23.9% and 20.4% of the clusters and sequences, respectively, and an average of 2.9 sequences per cluster. Sixty-six percent of the clusters, containing 38.2% of all sequences, were classified as U (unknown) because no assignment for their function could be made; they had an average of 1.95 sequence per cluster. Five of the clusters in the U class contained 10 or more EST sequences, with a total of 97 ESTs, and possibly represent 3′ truncated mRNA sequences coding for low complexity polypeptides, possibly of the S class. The remaining U-class clusters could have derived from truncated 3′ or 5′ untranslated regions of genes of the above two categories, as was recently indicated for a sialotranscriptome of An. gambiae (Arca et al., 2005). The large ratio of sequences per cluster in the secreted class is typical for sialotranscriptomes completed previously. Finally, one singleton coded for a probable transposable element sequence. Transposable element transcripts have been a regular finding in most sialotranscriptomes to date.
Table 1.
Library classification according to transcript class
| Class | Clusters | Seqs | Seqs / cluster | % Clusters | % Seqs |
|---|---|---|---|---|---|
| Secreted | 30 | 423 | 14.1 | 9.8 | 41.3 |
| Housekeeping | 73 | 209 | 2.9 | 23.9 | 20.4 |
| TE | 1 | 1 | 1.0 | 0.3 | 0.1 |
| Unknown | 201 | 392 | 2.0 | 65.9 | 38.2 |
| Total | 305 | 1025 |
3.2. H genes
The 73 clusters (comprising 209 EST) attributed to H genes expressed in the salivary glands of O. fasciatus were further characterized into 14 subgroups according to function (Table 2). As observed in previous sialotranscriptomes (Francischetti et al., 2002b; Ribeiro et al., 2004a; Ribeiro et al., 2004b), the larger set was associated with protein synthesis machinery (126 EST in 32 clusters). The second larger group (14 clusters and 34 sequences) codes for conserved proteins of unknown function, presumably associated with cellular metabolism. ESTs coding for proteins associated with energy metabolism, signal transduction, protein modification, and protein export machineries were also found. Additional inspection of each cluster for further information can be done online with Supplemental Table S1.
Table 2.
Functional classification of transcripts coding for putative housekeeping proteins expressed in the salivary glands of Oncopeltus fasciatus
| Class | Clusters | Seqs | Seqs / cluster | % Clusters | % Seqs |
|---|---|---|---|---|---|
| Protein synthesis | 32 | 126 | 3.9 | 43.8 | 60.3 |
| Conserved | 14 | 34 | 2.4 | 19.2 | 16.3 |
| Energy metabolism | 12 | 25 | 2.1 | 16.4 | 12.0 |
| Signal transduction | 3 | 6 | 2.0 | 4.1 | 2.9 |
| Protein export | 2 | 3 | 1.5 | 2.7 | 1.4 |
| Protein modification | 2 | 3 | 1.5 | 2.7 | 1.4 |
| Transcription machinery | 1 | 2 | 2.0 | 1.4 | 1.0 |
| Nuclear regulation | 1 | 2 | 2.0 | 1.4 | 1.0 |
| Nucleotide metabolism | 1 | 2 | 2.0 | 1.4 | 1.0 |
| Amino acid metabolism | 1 | 2 | 2.0 | 1.4 | 1.0 |
| Transporters | 1 | 1 | 1.0 | 1.4 | 0.5 |
| Transcription factor | 1 | 1 | 1.0 | 1.4 | 0.5 |
| Proteasome machinery | 1 | 1 | 1.0 | 1.4 | 0.5 |
| Intermediate metabolism | 1 | 1 | 1.0 | 1.4 | 0.5 |
| Total | 73 | 209 |
3.3. S proteins and peptides
Inspection of the clusters coding for putative secreted proteins in the Supplemental Table S1 reveals a remarkable lack of any protein sequence resembling any of the known lipocalins. Instead, 47% of the S class sequences, representing 40% of the S-class clusters appear associated to products of low complexity, being Ser, or Thr, or Gly or Pro rich (Table 3). The second most abundant group of secreted polypeptides are represented by cystatins and pacifastin, which are protease inhibitors that might inhibit the abundant cysteine proteases found in seeds. Peptides containing GGY motifs and found to function as antimicrobials were also found, representing 18% of the sequences and 26% of the clusters of the S class. Transcripts coding for enzymes were also found, including a truncated clone coding for a serine protease. Salivary trypsin-like activity has been previously described and cloned from the salivary glands of the plant-feeding bugs of the Lygus genus (Knop Wright et al., 2006; Zeng et al., 2002; Zhu et al., 2003), where they may assist in the exodigestion of plant tissues. We additionally found evidence for 5 ESTs coding for a triglyceride lipase that may assist digestion of lipidic seed constituents. A cluster with 2 ESTs codes for a protein with weak similarity to a Tenebrio protein annotated as chitinase, and containing a chitin-binding site predicted by the CDD. These transcripts may code for either an enzyme acting on sugar residues or to a lectin like protein. Additionally, we found seven clusters with 54 sequences that code for various polypeptides having no homology to the databases, except for one that shows an ankyrin repeat.
Table 3.
Classification of transcripts coding for putative secreted products expressed in the salivary glands of Oncopeltus fasciatus
| Class | Clusters | Seqs | Seqs / cluster | % Clusters | % Seqs |
|---|---|---|---|---|---|
| Low complexity polypeptides | 12 | 197 | 16.4 | 40.0 | 46.6 |
| Protease inhibitors | 6 | 87 | 14.5 | 20.0 | 20.6 |
| Antimicrobial | 3 | 77 | 25.7 | 10.0 | 18.2 |
| Other | 6 | 54 | 9.0 | 20.0 | 12.8 |
| Enzymes | 3 | 8 | 2.7 | 10.0 | 1.9 |
| Total | 30 | 423 |
3.4. Analysis of the Oncopeltus fasciatus sialotranscriptome
Several clusters of sequences coding for H and putative S polypeptides, indicated in Supplemental Table S1, are abundant and complete enough to extract consensus sequences of novel sequences. Additionally, we have performed primer extension studies in several clones to obtain full- or near full-length sequences of products of interest. A total of 58 novel sequences, 33 of which code for S proteins, are grouped together in Supplemental Table S2.
A more detailed description of the transcripts found in the salivary glands of Oncopeltus fasciatus follows.
3.4.1. Secreted proteins
Cystatins
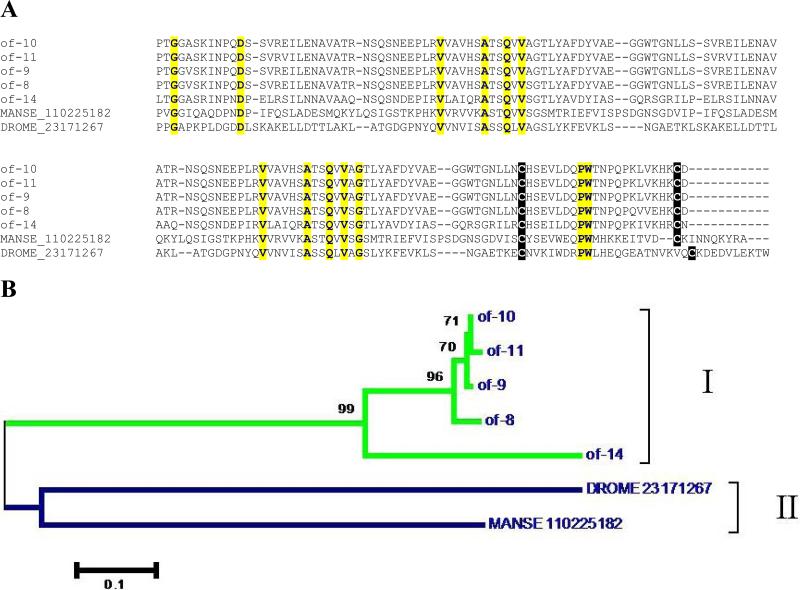
Full-length information was obtained for five similar cystatins in the O. fasciatus sialotranscriptome. These sequences are similar to other insect cystatins, particularly a multicystatin from the lepidopteran Manduca sexta and one protein from Drosophila melanogaster (Fig. 1). All five bug cystatins contain signal peptides indicative of secretion, and the conserved motif QXVXG and the dipeptide PW near the aminoterminus. Additionally, the putative proteins contain two cysteines that might form a disulfide bond in the carboxyterminal regions (Fig. 1A). The dendrogram (Fig. 1B) shows the expected divergence between the bug, the fly, the butterfly sequences, and the proximity of the bug cystatins. The proximity of the bug cystatins may result either from relatively recent gene duplication events, or they may be alleles. The aa distance of OF-14, however, indicates that at least two different genes code for the five messages found.
Figure 1.
Comparisons of 5 salivary cystatins from Oncopeltus fasciatus with 2 other insect cystatins from the fly Drosophila melanogaster (gi|23171267) and the moth Manduca sexta (gi|110225182). A) Clustal alignment: Conserved amino acids are marked in yellow and cysteines in reverse black background. The signal peptide of the sequences are omitted. OF-10 and OF-11 differ by only one amino acid in the signal peptide region (not shown). B) Dendogram of the alignment. Bootstrap values higher than 50% are shown in the tree branches. The bar indicates 10% amino acid substitution. Clade I refers to O. fasciatus branch, II to other insects.
The seeds of leguminous plants are rich in cysteine proteases needed in protein digestion during plant germination (Muntz and Shutov, 2002), and also in inducing apoptosis following tissue damage (Solomon et al., 1999). It is possible that the active cysteine proteases may be deleterious to the bug's survival, possibly by producing undesired proteolysis of gut tissues, or that it may trigger plant defense mechanisms that might disrupt feeding, accounting for the adaptive role of the salivary cystatins. It is also interesting that bugs are hosts to gut inhabiting trypanosomatids, such as members of the Trypanosoma, Herpetomonas and Phytomonas genera, and that these protozoa have a cysteine protease on their surface that is quite important for their survival (Caffrey et al., 2000; Cazzulo et al., 1997; Santos et al., 2006). To the extent that the ingested salivary cystatins can inhibit the protozoa proteases, these polypeptides may also be important for parasite colonization of the insect (Friend and Smith, 1982).
Pacifastin
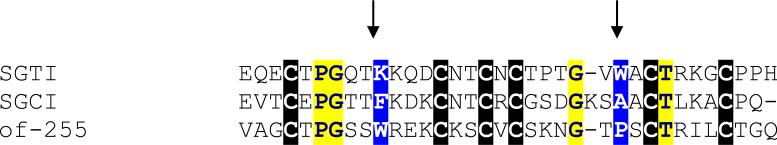
Pacifastins are cysteine-rich peptides found in arthropods, usually containing several repeats of ~ 35 aa (Simonet et al., 2003). The smaller 35 aa unit has a typical 3 Beta sheet structure (Kellenberger and Roussel, 2005) and are known to be serine protease inhibitors. Alignment of a truncated member of this family found in this work with the trypsin (SGTI) and chymotrypsin (SGCI) inhibitors of Schistocerca gregaria shows conservation of the domain in the protein named Of-255 (Fig. 2 and Supplemental Table S2). Interestingly, the P1 position for inhibition of serine proteases in SGCI is a phenylalanine, and for SGTI is a lysine, reflecting the preferred substrates of chymotrypsin and trypsin, respectively. Of-255 has a tryptophan in this position, indicating it may act as a chymotrypsin inhibitor (Kellenberger and Roussel, 2005). Alternatively, the pacifastin fold may have been recruited to perform some other function, possibly as an antimicrobial, as occur with cysteine-rich protease inhibitors of ticks (Fogaca et al., 2005).
Fig. 2.
Clustal alignment of the salivary pacifastin of Oncopeltus fasciatus (Of-255) with the crayfish trypsin (SGTI) and chymotrypsin (SGCI) inhibitors. Conserved amino acids are shown in yellow, cysteines in black background. Residues with blue background (shown by arrows) were shown to be important for enzyme specificity of SGCI and SGTI (Kellenberger and Roussel, 2005).
Possible mucins
Four putative proteins having 10 or more O-galactosylation sites are described in Supplemental Table S2. One of them is a serine-rich protein with 54 predicted galactosylation sites. Two others, Of-23 and Of-24 containing PSX repeats, may represent alternatively spliced variants of the same gene. They differ by a 3 aa insertion in the middle of the sequence. Mucins may be important in feeding, by lubricating the mouthparts and by helping to seal the mouthparts in the feeding lesion.
Other low complexity proteins
Four additional proteins contain over 20% of the aa serine or threonine, although they do not appear to be heavily galactosylated as the mucins described above. One of them, Of-30, which is a truncated peptide and presumed secreted, has 54% of its residues composed by glycine. The function of these proteins is unknown, although one may postulate that they may play a role in the formation of the feeding sheath that forms during penetration of the mouthparts of seed feeding insects; this may help to seal the junction between the feeding stylets and the plant lesion (Forbes, 1977; Miles, 1972).
GGY peptide
Of-3 represents a secreted polypeptide with mature molecular weight of 7.9 kDa and containing two GGY domains. Peptides of about this size and containing GGY repeats have been described in worms as possessing antimicrobial activity (Couillault et al., 2004).
Other secreted peptides
Supplemental Table S2 presents six additional putative secreted polypeptides, and one truncated clone coding for a low complexity protein that we presume to be also secreted. They do not have significant similarities to other proteins in the non-redundant database and their functions are unknown. They may act by preventing plant defense mechanisms or by having antimicrobial activity.
3.4.2 Housekeeping proteins
Supplemental Table S2 additionally presents 15 putative salivary proteins with a presumed housekeeping function. These include 4 ribosomal proteins, 4 enzymes (chitinase, carboxypeptidase B, lysosomal acid lipase and cytochrome c oxidase subunit 3, all missing their carboxy terminal end), 6 proteins that are similar to other known proteins of unknown function, including one with an ankyrin repeat, and a Glu-Val-rich protein of unknown function.
4. Concluding remarks
The sialotranscriptome of Oncopeltus fasciatus reveals the salivary strategy of this seed-feeding bug, by first showing the abundance of transcripts associated with protease inhibitors of the cystatin family, which target enzymes, known to be rich in seeds, and the presence of serine proteases and lipases that might assist external digestion of seed proteins and oils. On the other hand, the role of the abundant transcripts coding for putative secreted polypeptides of low complexity, rich in Gly and/or Ser and Thr, is not clear. They may be related to silk or mucin proteins that may be important to form the feeding sheath that seals the food lesion (Miles, 1972).
Supplementary Material
Acknowledgments
This work was supported by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health. We thank NIAID intramural editor Nancy Shulman for assistance, and Chuong Huynh from NCBI for help with posting the data.
Abbreviations
- aa
amino acid
- cDNA
DNA complementary to RNA
- EST
expressed sequence tag
- Of
Oncopeltus fasciatus
- S
Putative secreted transcripts
- H
Putative housekeeping transcripts
- U
Unknown function transcripts
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen JF, Francischetti IM, Valenzuela JG, Schuck P, Ribeiro JM. Inhibition of hemostasis by a high affinity biogenic amine-binding protein from the saliva of a blood-feeding insect. J. Biol. Chem. 2003;278:4611–7. doi: 10.1074/jbc.M211438200. [DOI] [PubMed] [Google Scholar]
- Arca B, Lombardo F, Valenzuela JG, Francischetti IM, Marinotti O, Coluzzi M, Ribeiro JM. An updated catalogue of salivary gland transcripts in the adult female mosquito, Anopheles gambiae. J. Exp. biol. 2005;208:3971–86. doi: 10.1242/jeb.01849. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A, Birney E, Durbin R, Eddy SR, Howe KL, Sonnhammer EL. The Pfam protein families database. Nucleic Acids Res. 2000;28:263–6. doi: 10.1093/nar/28.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffrey CR, Scory S, Steverding D. Cysteine proteinases of trypanosome parasites: novel targets for chemotherapy. Curr Drug Targets. 2000;1:155–62. doi: 10.2174/1389450003349290. [DOI] [PubMed] [Google Scholar]
- Cazzulo JJ, Stoka V, Turk V. Cruzipain, the major cysteine proteinase from the protozoan parasite Trypanosoma cruzi. Biol Chem. 1997;378:1–10. doi: 10.1515/bchm.1997.378.1.1. [DOI] [PubMed] [Google Scholar]
- Champagne D, Nussenzveig RH, Ribeiro JMC. Purification, characterization, and cloning of nitric oxide-carrying heme proteins nitrophorins from salivary glands of the blood sucking insect Rhodnius prolixus. J. Biol. Chem. 1995;270:8691–8695. doi: 10.1074/jbc.270.15.8691. [DOI] [PubMed] [Google Scholar]
- Couillault C, Pujol N, Reboul J, Sabatier L, Guichou JF, Kohara Y, Ewbank JJ. TLR-independent control of innate immunity in Caenorhabditis elegans by the TIR domain adaptor protein TIR-1, an ortholog of human SARM. Nat. Immunol. 2004;5:488–94. doi: 10.1038/ni1060. [DOI] [PubMed] [Google Scholar]
- Fogaca AC, Almeida IC, Eberlin MN, Tanaka AS, Bulet P, Daffre S. Ixodidin, a novel antimicrobial peptide from the hemocytes of the cattle tick Boophilus microplus with inhibitory activity against serine proteinases. Peptides. 2005;27:667–74. doi: 10.1016/j.peptides.2005.07.013. [DOI] [PubMed] [Google Scholar]
- Forbes A. The mouthparts and feeding mechanism of aphids. In: Maramorosch K. F. H. a. K., editor. Aphids as Virus Vectors. Academic Press, Inc.; New York: 1977. pp. 83–103. [Google Scholar]
- Francischetti IM, Andersen JF, Ribeiro JM. Biochemical and functional characterization of recombinant Rhodnius prolixus platelet aggregation inhibitor 1 as a novel lipocalin with high affinity for adenosine diphosphate and other adenine nucleotides. Biochemistry. 2002a;41:3810–8. doi: 10.1021/bi011015s. [DOI] [PubMed] [Google Scholar]
- Francischetti IM, Valenzuela JG, Pham VM, Garfield MK, Ribeiro JM. Toward a catalog for the transcripts and proteins sialome from the salivary gland of the malaria vector Anopheles gambiae. J. Exp. biol. 2002b;205:2429–51. doi: 10.1242/jeb.205.16.2429. [DOI] [PubMed] [Google Scholar]
- Friend WG, Smith JJB. ATP analogs and other phosphate compounds as gorging stimulants for Rhodnius prolixus. J. Insect Physiol. 1982;28:371–376. [Google Scholar]
- Hansen JE, Lund O, Tolstrup N, Gooley AA, Williams KL, Brunak S. NetOglyc: prediction of mucin type O-glycosylation sites based on sequence context and surface accessibility. Glycoconj J. 1998;15:115–30. doi: 10.1023/a:1006960004440. [DOI] [PubMed] [Google Scholar]
- Hori K. Plant growth-promoting factor in the salivary gland of the bug, Lygus disponsi. J Insect Physiol. 1974;20:1623–7. doi: 10.1016/0022-1910(74)90091-2. [DOI] [PubMed] [Google Scholar]
- Kellenberger C, Roussel A. Structure-activity relationship within the serine protease inhibitors of the pacifastin family. Protein Pept Lett. 2005;12:409–14. doi: 10.2174/0929866054395239. [DOI] [PubMed] [Google Scholar]
- Knop Wright M, Brandt SL, Coudron TA, Wagner RM, Habibi J, Backus EA, Huesing JE. Characterization of digestive proteolytic activity in Lygus hesperus Knight Hemiptera: Miridae. J Insect Physiol. 2006;52:717–28. doi: 10.1016/j.jinsphys.2006.03.012. [DOI] [PubMed] [Google Scholar]
- Letunic I, Goodstadt L, Dickens NJ, Doerks T, Schultz J, Mott R, Ciccarelli F, Copley RR, Ponting CP, Bork P. Recent improvements to the SMART domain-based sequence annotation resource. Nucleic Acids Res. 2002;30:242–4. doi: 10.1093/nar/30.1.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Panchenko AR, Shoemaker BA, Thiessen PA, Geer LY, Bryant SH. CDD: a database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res. 2002;30:281–3. doi: 10.1093/nar/30.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles PW. The saliva of Hemiptera. Adv. Insect Physiol. 1972;9:183–255. [Google Scholar]
- Miles PW, Peng Z. Studies on the salivary physiology of plant bugs: Detoxification of phytochemicals by the salivary peroxidase of aphids. J. Insect Physiol. 1989;35:865–872. [Google Scholar]
- Muntz K, Shutov AD. Legumains and their functions in plants. Trends Plant Sci. 2002;7:340–4. doi: 10.1016/s1360-1385(02)02298-7. [DOI] [PubMed] [Google Scholar]
- Musser RO, Cipollini DF, Hum-Musser SM, Williams SA, Brown JK, Felton GW. Evidence that the caterpillar salivary enzyme glucose oxidase provides herbivore offense in solanaceous plants. Arch Insect Biochem Physiol. 2005a;58:128–37. doi: 10.1002/arch.20039. [DOI] [PubMed] [Google Scholar]
- Musser RO, Kwon HS, Williams SA, White CJ, Romano MA, Holt SM, Bradbury S, Brown JK, Felton GW. Evidence that caterpillar labial saliva suppresses infectivity of potential bacterial pathogens. Arch Insect Biochem Physiol. 2005b;58:138–44. doi: 10.1002/arch.20031. [DOI] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- Noeske-Jungblut C, Kratzschmar J, Haendler B, Alagon A, Possani L, Verhallen P, Donner P, Schleuning WD. An inhibitor of collagen-induced platelet aggregation from the saliva of Triatoma pallidipennis. J. Biol. Chem. 1994;269:5050–3. [PubMed] [Google Scholar]
- Noeskejungblut C, Haendler BDP, Alagon A, Possani L, Scheleuning WD. Triabin, a highly potent exosite inhibitor of thrombin. J. Biol. Chem. 1995;270:28629–28634. doi: 10.1074/jbc.270.48.28629. [DOI] [PubMed] [Google Scholar]
- Nunn MA, Sharma A, Paesen GC, Adamson S, Lissina O, Willis AC, Nuttall PA. Complement inhibitor of C5 activation from the soft tick Ornithodoros moubata. J Immunol. 2005;174:2084–91. doi: 10.4049/jimmunol.174.4.2084. [DOI] [PubMed] [Google Scholar]
- Paesen GC, Adams PL, Nuttall PA, Stuart DL. Tick histamine-binding proteins: lipocalins with a second binding cavity. Biochim. Biophys. Acta. 2000;1482:92–101. doi: 10.1016/s0167-4838(00)00168-0. [DOI] [PubMed] [Google Scholar]
- Peng Z, Miles PW. Studies on the salivary physiology of plant bugs: Function of the catechol oxidase of the rose aphid. J. Insect Physiol. 1988;34:1027–1033. [Google Scholar]
- Ribeiro JM, Alarcon-Chaidez F, Francischetti IM, Mans BJ, Mather TN, Valenzuela JG, Wikel SK. An annotated catalog of salivary gland transcripts from Ixodes scapularis ticks. Insect Biochem. Mol. Biol. 2006;36:111–29. doi: 10.1016/j.ibmb.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Ribeiro JM, Andersen J, Silva-Neto MA, Pham VM, Garfield MK, Valenzuela JG. Exploring the sialome of the blood-sucking bug Rhodnius prolixus. Insect Biochem. Mol. Biol. 2004a;34:61–79. doi: 10.1016/j.ibmb.2003.09.004. [DOI] [PubMed] [Google Scholar]
- Ribeiro JM, Arca B, Lombardo F, Calvo E, Phan VM, Chandra PK, Wikel SK. An annotated catalogue of salivary gland transcripts in the adult female mosquito, Aedes aegypti. BMC Genomics. 2007;8:6. doi: 10.1186/1471-2164-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro JM, Charlab R, Pham VM, Garfield M, Valenzuela JG. An insight into the salivary transcriptome and proteome of the adult female mosquito Culex pipiens quinquefasciatus. Insect Biochem. Mol. Biol. 2004b;34:543–63. doi: 10.1016/j.ibmb.2004.02.008. [DOI] [PubMed] [Google Scholar]
- Ribeiro JMC. Insect saliva: Function, biochemistry and physiology. In: Boer R. F. C. a. G. d., editor. Regulatory mechanisms of insect feeding. Chapman & Hall; London: 1995. pp. 74–97. [Google Scholar]
- Ribeiro JMC, Schneider M, Guimaraes JA. Purification and characterization of Prolixin S Nitrophorin 2, the salivary anticoagulant of the blood sucking bug, Rhodnius prolixus. Biochem. J. 1995;308:243–249. doi: 10.1042/bj3080243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro JMC, Walker FA. High affinity histamine-binding and anti-histaminic activity of the salivary NO-carrying heme protein Nitrophorin of Rhodnius prolixus. J. Exp. Med. 1994;180:2251–2257. doi: 10.1084/jem.180.6.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos AL, d'Avila-Levy CM, Dias FA, Ribeiro RO, Pereira FM, Elias CG, Souto-Padron T, Lopes AH, Alviano CS, Branquinha MH, et al. Phytomonas serpens: cysteine peptidase inhibitors interfere with growth, ultrastructure and host adhesion. Int J Parasitol. 2006;36:47–56. doi: 10.1016/j.ijpara.2005.09.004. [DOI] [PubMed] [Google Scholar]
- Schaffer AA, Aravind L, Madden TL, Shavirin S, Spouge JL, Wolf YI, Koonin EV, Altschul SF. Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res. 2001;29:2994–3005. doi: 10.1093/nar/29.14.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonet G, Claeys I, Franssens V, De Loof A, Broeck JV. Genomics, evolution and biological functions of the pacifastin peptide family: a conserved serine protease inhibitor family in arthropods. Peptides. 2003;24:1633–44. doi: 10.1016/j.peptides.2003.07.014. [DOI] [PubMed] [Google Scholar]
- Solomon M, Belenghi B, Delledonne M, Menachem E, Levine A. The involvement of cysteine proteases and protease inhibitor genes in the regulation of programmed cell death in plants. Plant Cell. 1999;11:431–44. doi: 10.1105/tpc.11.3.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–82. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela JG, Charlab R, Gonzalez EC, Miranda-Santos IKF, Marinotti O, Francischetti IM, Ribeiro JMC. The D7 family of salivary proteins in blood sucking Diptera. Insect Mol. Biol. 2002;11:149–55. doi: 10.1046/j.1365-2583.2002.00319.x. [DOI] [PubMed] [Google Scholar]
- Valenzuela JG, Francischetti IM, Pham VM, Garfield MK, Ribeiro JM. Exploring the salivary gland transcriptome and proteome of the Anopheles stephensi mosquito. Insect Biochem. Mol. Biol. 2003;33:717–32. doi: 10.1016/s0965-1748(03)00067-5. [DOI] [PubMed] [Google Scholar]
- Zeng F, Zhu Y, Cohen A. Partial characterization of trypsin-like protease and molecular cloning of a trypsin-like precursor cDNA in salivary glands of Lygus lineolaris. Comp Biochem Physiol B Biochem Mol Biol. 2002;131:453–63. doi: 10.1016/s1096-4959(01)00514-0. [DOI] [PubMed] [Google Scholar]
- Zhu YC, Zeng F, Oppert B. Molecular cloning of trypsin-like cDNAs and comparison of proteinase activities in the salivary glands and gut of the tarnished plant bug Lygus lineolaris Heteroptera: Miridae. Insect Biochem. Mol. Biol. 2003;33:889–99. doi: 10.1016/s0965-1748(03)00094-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.