Summary
Integrative and conjugative elements (ICEs), a.k.a. conjugative transposons, are mobile genetic elements involved in many biological processes, including pathogenesis, symbiosis, and the spread of antibiotic resistance. Unlike conjugative plasmids that are extra-chromosomal and replicate autonomously, ICEs are integrated in the chromosome and replicate passively during chromosomal replication. It is generally thought that ICEs do not replicate autonomously. We found that when induced, Bacillus subtilis ICEBs1 undergoes autonomous plasmid-like replication. Replication was unidirectional, initiated from the ICEBs1 origin of transfer, oriT, and required the ICEBs1-encoded relaxase NicK. Replication also required several host proteins needed for chromosomal replication, but did not require the replicative helicase DnaC or the helicase loader protein DnaB. Rather, replication of ICEBs1 required the helicase PcrA that is required for rolling circle replication of many plasmids. Transfer of ICEBs1 from the donor required PcrA, but did not require replication, indicating that PcrA, and not DNA replication, facilitates unwinding of ICEBs1 DNA for horizontal transfer. Although not needed for horizontal transfer, replication of ICEBs1 was needed for stability of the element. We propose that autonomous plasmid-like replication is a common property of ICEs and contributes to the stability and maintenance of these mobile genetic elements in bacterial populations.
Keywords: Bacillus subtilis, conjugative transposon, integrative and conjugative element, rolling circle replication, horizontal gene transfer
Introduction
Horizontal gene transfer helps drive evolution and is important in pathogenesis, symbiosis, and the spread of antibiotic resistance (Keeling & Palmer, 2008, Ochman et al., 2000). In bacteria, horizontal gene transfer is often mediated by mobile genetic elements, including phages, plasmids, and transposons (Frost et al., 2005). These elements are typically comprised of various functional modules, apparently assembled by acquisition from or exchange with other mobile and non-mobile genetic elements (Osborn & Boltner, 2002, Toussaint & Merlin, 2002, Burrus et al., 2002).
Conjugative plasmids and conjugative transposons, a.k.a., integrative and conjugative elements (ICEs), transfer directly from one cell to another by mating. Many elements transfer single stranded DNA into recipient cells (reviewed in Waters & Guiney, 1993, Lanka & Wilkins, 1995, Llosa et al., 2002). The transferred strand is unwound from a double-stranded circular element starting at a nick in the origin of transfer (oriT). A DNA relaxase (nickase) makes the single strand cut and is covalently attached to the 5′ end of the strand to be transferred. For plasmids, the transferred strand is replaced by replication in the donor. Although replacement strand synthesis has been proposed as one mechanism to promote plasmid unwinding and single-strand transfer, it has been difficult to address the role of replication in plasmid conjugation (Meyer, 2009), since replication is needed for plasmid maintenance.
Many ICE’s and conjugative plasmids appear to use similar machinery and mechanisms to transfer DNA (Toussaint & Merlin, 2002). Both plasmids and ICEs mate as extra-chromosomal elements. However, whereas conjugative plasmids start out as extra-chromosomal elements, ICEs are typically found integrated in the host genome and excise to form a circular extra-chromosomal element prior to mating.
In contrast to plasmids that replicate autonomously to ensure their inheritance during cell growth and cell division, ICEs are generally thought to be incapable of autonomous replication, instead relying on their integration in the host genome and replication and segregation along with the host chromosome for genetic stability (Burrus & Waldor, 2004). However, some reports indicate that certain ICE and ICE-like elements are capable of autonomous replication (Ramsay et al., 2006, Wang et al., 2001, te Poele et al., 2008). We investigated whether ICEBs1 (Fig. 1A), an ~21 kb conjugative transposon in Bacillus subtilis (Auchtung et al., 2005, Burrus et al., 2002), is capable of autonomous replication.
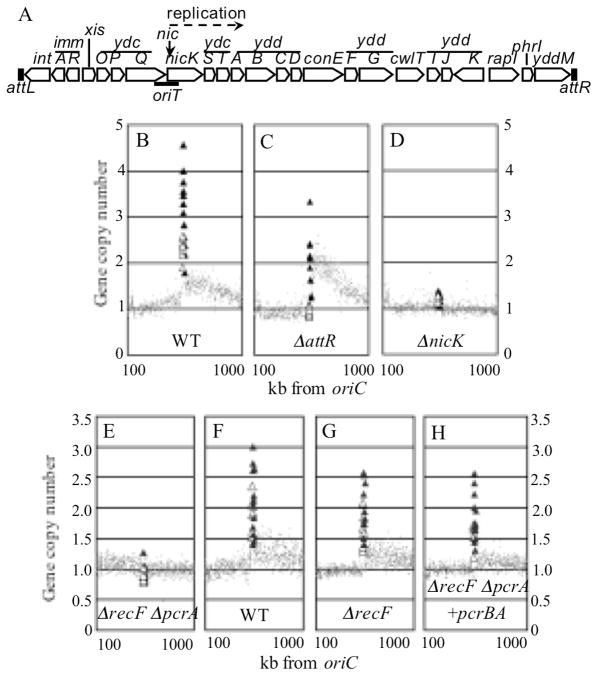
Figure 1. Unidirectional replication of ICEBs1 is activated by RapI and requires nicK and pcrA.
A. Schematic diagram of ICEBs1 integrated in the B. subtilis chromosome. Genes (open arrows), the left and right attachment sites (attL, attR), origin of transfer (oriT), the site in oriT nicked by NicK (nic), and the location and direction of replication initiation (dashed arrow) are indicated.
B–H. The effect of RapI-overexpression on gene copy number was determined using genomic microarrays, comparing induced vs uninduced samples, and plotted versus position in the chromosome. Values for ICEBs1 genes are represented as large triangles, with open triangles representing genes to the left of nicK (int-ydcQ) and closed triangles representing nicK and genes to its right (ydcS-yddM). Values for non-ICEBs1 chromosomal genes are shown as gray dots. For simplicity, we plot ICEBs1 genes in their integrated chromosomal location even though, once excised, ICEBs1 is no longer co-linear with the chromosome. Data for genes from 100 to 1000 kb to the right of the B. subtilis oriC are shown.
B–D. Strains were grown in minimal D-glucose medium at 37°C and expression of RapI (Pspank(hy)-rapI) was induced by addition of IPTG for 1 hr. Uninduced rapI-null cells (IRN342) were grown under the same conditions and used as controls. Cy-labeled samples were hybridized to DNA microarrays of spotted PCR products. Data are the averages of two induced and uninduced samples. B. wild type (JMA168); C. ΔattR (CAL108); D. ΔnicK (CAL306).
E–H. Strains were grown in minimal L-arabinose medium at 30°C and expression of RapI (Pxyl-rapI) was induced by addition of D-xylose for 2 hours. Uninduced rapI-null cells (IRN342) were grown under the same conditions. Cy-labeled samples were hybridized to DNA microarrays of spotted oligonucleotides. Data are from the averages of three induced and two uninduced samples. Replication of ICEBs1 was lower than observed in previous experiments (Fig. 1, Table 1). Activation of ICEBs1 by RapI expression from lacA::{(Pxyl-rapI) tet} is less efficient than from the other RapI-expression constructs (B. Bose, ADG unpublished results). E. ΔrecF ΔpcrA (CAL1175); F. wild type (CAL1173); G. ΔrecF (CAL1174); H. ΔrecF ΔpcrA +pcrB pcrA (CAL1148).
Under normal growth conditions, ICEBs1 is found stably integrated into a tRNA gene in B. subtilis (Fig. 1A). However, ICEBs1 is activated and can excise from the genome when cells experience DNA damage and activate the recA-dependent SOS response, or when the signaling protein RapI is activated in response to a high density of cells lacking ICEBs1 (Auchtung et al., 2005). Excision of ICEBs1 leads to the formation of a circular product and if suitable recipients are present, ICEBs1 can transfer from donor to recipient (Auchtung et al., 2005). Previous observations indicated that ICEBs1 might be capable of autonomous replication after excision. Under certain conditions, a single donor appeared to transfer ICEBs1 to more than one recipient (Auchtung et al., 2005). There was a small increase in ICEBs1 gene copy number after induction of the SOS response (J.D. Wang, ADG, unpublished results), and excision of ICEBs1 alters the subcellular location of the replication machinery (Berkmen et al., 2009).
Taking advantage of the ability to readily induce excision of ICEBs1 in the majority of cells in a population (Auchtung et al., 2005, Lee et al., 2007), we found that ICEBs1 undergoes plasmid-like autonomous replication after induction. ICEBs1 replication initiated at oriT and required the ICEBs1-encoded relaxase NicK. ICEBs1 replication also required the chromosomally-encoded helicase PcrA that is used by several rolling circle replicating (RCR) plasmids and did not require the helicase DnaC that is needed for chromosomal replication. By controlling excision and replication of ICEBs1, we were able to address the role of replication in ICEBs1 conjugation and stability. We found that replication in the donor is required for genetic stability of ICEBs1.
RESULTS
Autonomous replication of ICEBs1 following induction
The cell-cell signaling regulator RapI induces expression of ICEBs1 genes required for ICEBs1 excision and conjugation (Auchtung et al., 2005, Lee et al., 2007, Lee & Grossman, 2007, Berkmen et al., 2009, Bose* et al., 2008). Overproduction of RapI causes ICEBs1 to excise and form a circle, typically in >95% of cells (Auchtung et al., 2005, Lee et al., 2007). We found that expression of RapI caused ICEBs1 to replicate. We used DNA microarrays to measure the relative copy number of chromosomal and ICEBs1 genes with and without induction of ICEBs1. In cells carrying an excision-competent ICEBs1, the copy number of ICEBs1 genes increased ~2–5-fold one hour after induction of rapI (Fig. 1B, triangles). The ICEBs1 gene nicK had the highest copy number (4.6) and genes located rightward from nicK had progressively lower copy numbers (ydcS-yddD, copy numbers 4 to 3; yddF-yddM, copy numbers <3). Our results indicate that induction of ICEBs1 leads to its autonomous replication. Replication likely starts near nicK and proceeds rightward from nicK (Fig. 1A) around the ICEBs1 circle, sometimes terminating before completing a round of replication to the left of nicK.
The copy number of chromosomal genes adjacent to ICEBs1 also increased after induction of ICEBs1 (Fig. 1B). The increase was relatively modest (<1.5-fold) and extended far into the chromosomal region, especially on the right side. Replication of this region required induction with RapI and the presence of an adjacent replication-competent ICEBs1 (see below). We suspect that replication initiates in ICEBs1 in some cells before excision, and continues into the flanking chromosomal region.
Virtually nothing is known about the mechanisms and proteins involved in autonomous replication of ICEs, or the function of replication in their conjugation and stability. It has been generally assumed that ICEs do not replicate autonomously (e.g., Burrus & Waldor, 2004), although a couple of reports indicate this might not be the case for an ICE and ICE-like element that use a single-strand transfer mechanism (Ramsay et al., 2006, Wang et al., 2001). Therefore, we determined the mechanism of replication of ICEBs1 and the role of replication in its function.
ICEBs1 replication proceeds unidirectionally from a site in or near nicK
Analysis of an excision-defective ICEBs1 mutant confirmed that replication initiates in or near nicK and proceeds rightward. Excision of ICEBs1 requires site-specific recombination between attL and attR (Fig. 1A), and deletion of attR prevents excision (Lee et al., 2007). The copy number of nicK and genes to its right increased following induction of the excision-defective ICEBs1 ΔattR mutant (Fig. 1C, filled triangles). In contrast, there was little or no increase in the copy number of genes to the left of nicK in the ΔattR mutant (Fig. 1C, open triangles). The copy number of nicK and genes to its right was lower in the ΔattR mutant (3.3 to 2) than in the WT (4.6 to 3). For elements replicating unidirectionally, a complete round of replication is required to generate two intact origin regions, each capable of reinitiation. ICEBs1 cannot complete a round of replication without excision and the ΔattR mutant has only one replication-competent origin, likely explaining why the copy number of ICEBs1 genes is lower in the ΔattR strain than in the WT. We conclude that replication of genes to the left of nicK (int-ydcQ) requires excision and circularization of ICEBs1, replication is unidirectional, initiates in or near nicK and proceeds rightward, and excision is not needed for initiation of ICEBs1 replication. In addition, autonomous replication of ICEBs1 was much less processive than normal chromosomal replication, perhaps indicating differences in the replication proteins used (see below).
Replication of ICEBs1 initiates within oriT and requires nicK
The ICEBs1 conjugative relaxase encoded by nicK was required for ICEBs1 replication. NicK is a homolog of Rep DNA relaxases required for replication of RCR plasmids. ICEBs1 NicK makes a ssDNA break in a GC-rich inverted repeat in oriT, in the 5′ end of nicK, and is required for conjugative transfer of ICEBs1 from donor to recipient (Lee & Grossman, 2007). There was no increase in ICEBs1 gene copy number following induction of a ΔnicK mutant, nor was there any increase in copy number of the flanking chromosomal genes (Fig. 1D, ΔnicK). These results indicate that nicK is required for replication of ICEBs1 and the flanking chromosomal genes.
ICEBs1 replication initiated at or near nic, the site of nicking in oriT. Using quantitative real-time PCR (qRT-PCR), we measured the copy number on each side of nic 2 hours after induction of ICEBs1 (Fig. 2A) in both wild type and excision-defective ICEBs1. In the excision-defective (ΔattR) strain, the copy number of the region ~170 bp to the right of nic was 3.6 and the copy number ~30 bp to the left of nic was <1 (Fig. 2B). These results indicate that, in the ΔattR strain, the region to the left of nic did not replicate while that to the right of nic did. The qRT-PCR assays also showed that there is more replication of the region to the right of nic in the WT (copy number 9.3) than in the ΔattR mutant (copy number 3.6), confirming that replication of excised circular ICEBs1 is more productive than that of integrated ICEBs1. Based on these results, we conclude that the unidirectional replication of ICEBs1 requires NicK and initiates at nic in oriT. We suspect that oriT and NicK recruit replication proteins to ICEBs1 in a way analogous to that of a dsDNA origin and its cognate Rep protein of RCR plasmids.
Figure 2. Unidirectional replication of ICEBs1 initiates within oriT.
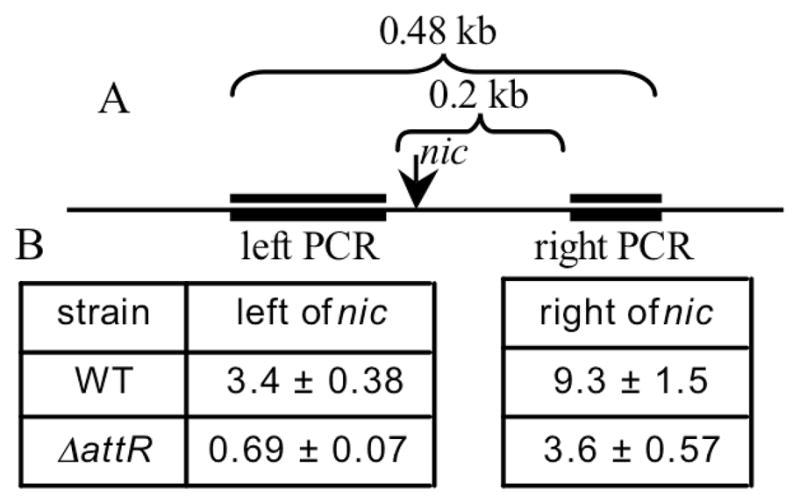
A. Schematic diagram of ICEBs1 oriT. The long horizontal line represents the 0.8 kb oriT region. The arrow indicates the site (nic) on the top strand of oriT that is nicked by NicK. The thick double lines depict the two regions of oriT assayed by qRT-PCR. Values above the brackets indicate the distance between the ends of the two regions assayed.
B. Copy number of sites flanking nic. Strains JMA168 (WT) and CAL108 (ΔattR) were grown in minimal D-glucose medium at 37°C and expression of RapI (Pspank(hy)-rapI) was induced by addition of IPTG for 2 hours. Cell lysates were prepared and assayed by qRT-PCR for the left PCR site, the right PCR site, and a control PCR site outside of ICEBs1 (ydbT). Copy number values were normalized to qRT-PCR assays of an uninduced cell lysate of IRN342. Data are the average and standard deviation from four independent biological replicates.
The catalytic subunit of DNA polymerase (PolC) and single strand DNA binding protein (Ssb) associate with replicating ICEBs1 in vivo
We monitored association of various proteins with ICEBs1 using crosslinking and immunoprecipitation followed by hybridization to DNA microarrays (ChIP-chip). Since ICEBs1 seems to replicate by a rolling circle mechanism, we tested for association of single strand DNA binding protein (Ssb) and the catalytic subunit of DNA polymerase (PolC) with replicating ICEBs1 (Fig. 3A, B). We used Ssb-GFP and PolC-GFP fusions and immunoprecipitated with anti-GFP antibodies. After induction, ICEBs1 DNA was significantly enriched in the immunoprecipitates, indicating that both Ssb-GFP (Fig. 3A) and PolC-GFP (Fig. 3B) were associated with replicating ICEBs1. For both Ssb-GFP and PolC-GFP, enrichment of ICEBs1 DNA was dependent on induction of ICEBs1, required addition of specific antibodies, and did not occur in cells without the GFP-tag (data not shown). These results indicate that Ssb and PolC are recruited to ICEBs1 after activation and associate with ICEBs1 DNA during replication. Chromosomal genes to the right of ICEBs1 were modestly enriched in the ChIP samples, indicating that, Ssb and PolC are involved in replication of both ICEBs1 and the flanking chromosomal genes.
Figure 3. DNA polymerase PolC, single-strand binding protein Ssb and alternative helicase PcrA associate with ICEBs1 during replication, but the replicative helicase DnaC does not.
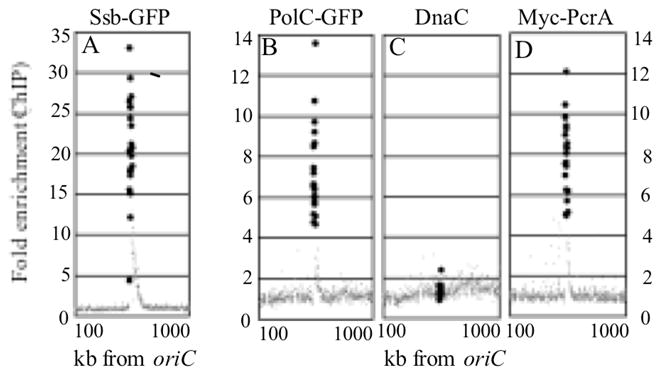
The association of various proteins with DNA was assessed by chromatin immunoprecipitation followed by hybridization to DNA microarray (ChIP-chip). Strains were grown in defined minimal L-arabinose medium at 30°C and expression of RapI (Pxyl-rapI) was induced by addition of D-xylose for 2 hours. Cells were fixed with formaldehyde and protein-DNA complexes were immunoprecipitated with antibodies to GFP (A, B) DnaC (C) or c-Myc (D). Cy-labeled samples were hybridized to DNA microarrays of spotted PCR products. Fold-enrichment of DNA by immunoprecipitation is plotted versus position in the chromosome. Data for genes from 100 to 1000 kb to the right of oriC are shown. Values for ICEBs1 genes are represented as large closed circles and non-ICEBs1 genes are dark gray dots. Data presented are from one representative experiment. A. Ssb-GFP (CAL635); B. PolC-GFP (CAL879); C. DnaC (CAL635); D. Myc-PcrA (CAL992).
PolC and the β-clamp (DnaN) of the B. subtilis replicative DNA polymerase are required for ICEBs1 replication
We found that the catalytic subunit PolC and the β-clamp DnaN (a.k.a. the processivity clamp) of the B. subtilis replicative polymerase were required for autonomous replication of ICEBs1. The replication inhibitor hydroxyphenylazo-uracil (HPUra) binds PolC that is associated with DNA and arrests replication (Brown, 1970). There was little or no increase in the copy number of nicK when ICEBs1 was induced shortly after treatment of cells with HPUra (Table 1, line 1). In contrast, similar induction of ICEBs1 without treatment with HPUra resulted in a copy number of ~6 for nicK (Table 1, line 2). In addition, in a dnaNts mutant, there was little or no increase in the copy number of nicK after induction of ICEBs1 at non-permissive temperature (47°C) (Table 1, line 3). Similar treatment of wild type (dnaN+) cells resulted in an increase in the copy number of nicK of ~9-fold (Table 1, line 4).
Table 1.
PolC and DnaN are required for replication of ICEBs1.
| line | Strain a | Treatment b | nicK copy number c |
|---|---|---|---|
| 1 | WT (Pxyl-rapI) | HPUra 5 min; xylose 2 hr | 1 ± 0.4 |
| 2 | WT (Pxyl-rapI) | xylose 2 hr | 6 ± 2 |
| 3 | dnaNts (Pspank(hy)-rapI) | 47°C 5 min; IPTG 90 min | 0.9 ± 0.1 |
| 4 | WT (Pspank(hy)-rapI) | 47°C 5 min; IPTG 90 min | 9 ± 3 |
| 5 | dnaCts (Pspank(hy)-rapI) | 47°C 5 min; IPTG 90 min | 27 ± 9 |
| 6 | dnaBts (Pxyl-rapI) | 47°C 1 hr; xylose 2 hr | 4 ± 1 |
| 7 | WT (Pxyl-rapI) | 47°C 1 hr; xylose 2 hr | 9 ± 2 |
Strains containing Pxyl-rapI were CAL874 (WT, lines 1, 2, 7) and CAL934 (dnaBts, line 6). Pspank(hy)-rapI strains were JMA168 (WT, line 4), CAL1161 (dnaNts, line 3), and CAL1035 (dnaCts, line 5).
Prior to the indicated treatment, all cells were grown at 30°C in defined minimal medium with L-arabinose as carbon source. Replication was blocked with HPUra (line 1) or shift of ts mutants to non-permissive temperature (lines 3, 5, 6) for the indicated amount of time, and then ICEBs1 was induced by induction of rapI with xylose or IPTG, as indicated. Full induction of ICEBs1 with Pxyl-rapI takes a bit longer than that with Pspank(hy).
Copy number was calculated relative to uninduced IRN342 (no rapI) controls (average of 2) grown at the same temperatures with the addition of xylose (30°C) or IPTG (47°C). Data are the average and standard deviation from two to four independent experiments. Samples from strains with Pxyl-rapI were hybridized to DNA microarrays of spotted PCR products. Samples from strains with Pspank(hy)-rapI were hybridized to DNA microarrays of spotted oligonucleotides. The difference is simply due to the arrays that were available at the time of the experiments.
The defects in replication were not a secondary effect of a possible defect in nicking in oriT. Primer extension assays showed that the ICEBs1 oriT was nicked in the dnaNts mutant and in the presence of HPUra (data not shown). Defects in excision can cause a reduction in replication, as discussed above, and ICEBs1 excision frequencies were reduced by HPUra treatment and by the dnaNts mutation (24% and 47% excision, respectively). However, these reductions in excision could not account for the blocks in replication, as excision is not needed for replication of the genes to the right of oriT, including nicK (Fig. 1C). We conclude that both PolC and the β-clamp of the chromosomal replicative DNA polymerase are required for replication of ICEBs1.
The B. subtilis replicative helicase, DnaC, is not required for ICEBs1 replication and does not associate with replicating ICEBs1 in vivo
We found that the replicative helicase in B. subtilis, DnaC, is not required for autonomous replication of ICEBs1 and is not associated with replicating ICEBs1 DNA. We tested for effects of DnaC helicase activity on ICEBs1 replication using two temperature-sensitive mutations, one in dnaC and one in dnaB. dnaB encodes a component of the helicase loader and is required for DnaC to assemble at oriC and at stalled replication forks (Bruand et al., 2001, Velten et al., 2003). dnaCts mutations cause a rapid and almost immediate block in replication elongation after shift to non-permissive temperature (Karamata & Gross, 1970). In contrast, at non-permissive temperature, dnaBts mutations cause a block in replication initiation and replication does not stop until ongoing rounds have finished (Karamata & Gross, 1970).
To test for a role of the replicative helicase in replication of ICEBs1, we shifted dnaCts and dnaBts mutants to the non-permissive temperature for 5 or 60 minutes, respectively. Maintaining the cells at the non-permissive temperature, we then induced ICEBs1 by overexpression of RapI. The dnaCts mutation did not prevent ICEBs1 replication (Table 1, line 5). If anything, ICEBs1 replicated more efficiently in this mutant than in the wild type under similar conditions (Table 1, line 4). We do not know what causes the more efficient replication in the dnaCts mutant, but it might be due to SOS induction in the mutant at the non-permissive temperature or perhaps indirect effects on machinery involved in ICEBs1 replication. ICEBs1 also replicated in the dnaBts mutant. The copy number of nicK was ~4 (Table 1, line 6), and was less than that in the comparably treated wild type strain in which the copy number of nicK was ~9 (Table 1, line 7). The reduced replication of ICEBs1 in the dnaBts mutant is likely due to a decreased excision frequency (12.5%). We were also unable to detect association of DnaC with replicating ICEBs1 DNA using ChIP-chip with antibodies to DnaC (Fig. 3C). The same antibodies are able to detect helicase at oriC in ChIP experiments (W.K. Smits, A. I. Goranov, ADG, unpublished results). Based on these results, we conclude that neither the replicative helicase DnaC, nor the helicase loader protein DnaB, are required for autonomous replication of ICEBs1, and that DnaC is not detectably associated with autonomously replicating ICEBs1.
The helicase PcrA associates with replicating ICEBs1
We found that the chromosomally-encoded helicase PcrA associates with replicating ICEBs1 DNA. PcrA is required for replication of RCR plasmids in B. subtilis and other Gram-positive bacteria (Khan, 2005). Since ICEBs1 replication has features similar to that of RCR plasmids and DnaC is not required and none of the ICEBs1 genes are predicted to encode a helicase, we decided to test PcrA. We made a myc-pcrA fusion and used anti-Myc monoclonal antibodies to immunoprecipitate Myc-PcrA. We found that ICEBs1 DNA was highly enriched in the anti-Myc-PcrA immunoprecipitates (Fig. 3D). Enrichment of ICEBs1 DNA was dependent on RapI-overexpression and did not occur if cells did not contain the Myc-tagged PcrA (data not shown). These results indicate that, like PolC and Ssb, PcrA is recruited to ICEBs1 after induction and associates with ICEBs1 DNA during replication. Chromosomal genes to the right of ICEBs1 were modestly enriched in the Myc-PcrA immunoprecipitates, indicating that, in addition to mediating the replication of ICEBs1, PcrA may also mediate the replication of flanking chromosomal genes.
PcrA is required for ICEBs1 replication and mating
pcrA was required for autonomous replication of ICEBs1. pcrA is essential, even though it is not required for replication of the B. subtilis chromosome (Petit et al., 1998). PcrA appears to be needed to disassemble chromosomal recombination complexes that can block replication (Petit & Ehrlich, 2002, Anand et al., 2007), and the lethality of pcrA null mutants is suppressed by null mutations in genes of the recFOR recombination pathway. We constructed a pcrA null mutation in a recF null mutant. After induction, there was no detectable replication of ICEBs1 in the pcrA recF double mutant (Fig. 1E). ICEBs1 did replicate after similar induction in wild type (Fig. 1F) and a recF single mutant (Fig. 1G). Ectopic expression of pcrA (and the upstream uncharacterized gene pcrB) from its native promoter, at a heterologous chromosomal location, restored ICEBs1 replication in the pcrA recF double mutant (Fig. 1H), indicating that the defect in ICEBs1 replication in the double mutant was due to loss of pcrA and not a polar effect on the downstream genes ligA (DNA ligase) and yerH (unknown). The defect in replication in the pcrA recF double mutant was not due to a defect in nicking of ICEBs1 in oriT since primer extension assays showed that ICEBs1 was nicked at the same frequency in the double mutant as in the recF single mutant (data not shown). There was a modest reduction of ICEBs1 excision in the pcrA recF double mutant (54% excision), but this cannot account for the block in replication. Based on these results, we conclude that the helicase PcrA is required for autonomous replication of ICEBs1. PcrA function in ICEBs1 replication is most likely analogous to its function in plasmid replication.
pcrA was required for ICEBs1 conjugation. We mixed different ICEBs1 donor cells with potential recipients and measured mating efficiencies (Fig. 4A). In the conditions used, wild type and recF mutant donors had approximately 3% and 8% mating efficiencies (transconjugants per donor), respectively (Fig. 4A). In contrast, the mating efficiency of the pcrA recF mutant donor was <0.0001% (Fig. 4A). This mating defect was due to loss of pcrA and not a polar effect on the downstream genes as it was fully complemented by ectopic expression of pcrA (Fig. 4A). Based on these results, we conclude that the helicase PcrA is required in donor cells for conjugative transfer of ICEBs1. This requirement could reflect a requirement for replication of ICEBs1 in mating, or a requirement for the activity of PcrA independently of ICEBs1 replication.
Figure 4. Effects of mutations in pcrA, dnaC, and dnaN on ICEBs1 mating efficiency.
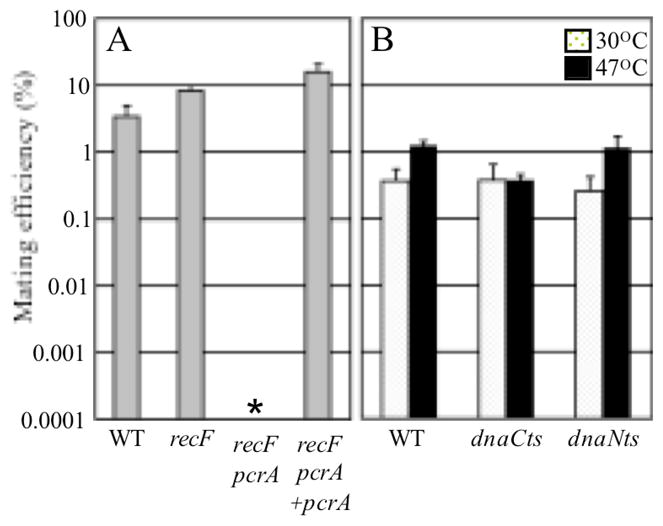
Efficiencies of transfer of ICEBs1 (kanamycin-resistant) from the indicated donor strains into the recipient CAL419 (streptomycin-resistant) were calculated from the number of kanamycin-resistant, streptomycin-resistant trans-conjugants per initial donor X 100%.
A. Wild type (CAL1173), recF (CAL1174), recF pcrA (CAL1175), and recF pcrA +pcrA (CAL1148) donors were grown in minimal L-arabinose medium at 30°C and expression of RapI (Pxyl-rapI) was induced by addition of xylose for 2 hours. Mating mixtures were incubated at 37°C for 3 hours. Gray bars show the average and standard deviation from two independent mating assays. The asterisk indicates that the mating efficiency of the recF pcrA double mutant was <0.0001%.
B. Wild type (JMA168), dnaCts (CAL1035), and dnaNts (CAL1161) donors were grown in minimal L-arabinose medium at 30°C. A portion was shifted to 47°C for 5 minutes, and then IPTG was added to all donor cultures at 30°C and 47°C for 90 minutes to induce expression of RapI (Pspank(hy)-rapI). Mating mixtures were incubated at 30°C or 47°C for 3 hours. Speckled (30°C) and black (47°C) bars show the average and standard deviation from two to four independent mating experiments.
Replication is not required for ICEBs1 mating
DNA replication of a donor element might play a direct role in conjugative DNA transfer if leading-strand DNA synthesis (dependent on a helicase) from the nicked oriT is required to unwind the DNA template and generate a single DNA strand for transfer (Lanka & Wilkins, 1995, Llosa et al., 2002). Alternatively, a helicase could promote DNA unwinding and conjugation independently of replication (Lanka & Wilkins, 1995). Since replication is required for plasmid maintenance, determining whether or not replication of a plasmid is required for conjugative transfer is complicated (e.g., Meyer, 2009). However, since ICEBs1 is maintained as an integrated element, and its excision and autonomous replication are not obligatory, we could test whether or not autonomous replication is required to produce a single strand template for transfer.
Horizontal transfer of ICEBs1 appeared to be independent of autonomous replication in the donor. We blocked replication of ICEBs1 (and the chromosome) in a dnaNts mutant. As a control, we compared the mating efficiency to that of a dnaCts mutant in which chromosomal, but not ICEBs1, replication was blocked. We grew the mutants and wild type strains at permissive temperature (30°C), shifted a portion of each culture to non-permissive temperature (47°C) to inactivate the temperature-sensitive replication components, then induced ICEBs1 by over-production of RapI. The wild type donor had mating efficiencies of 0.4% and 1% at 30°C and 47°C, respectively (Fig. 4B). The dnaCts donor had mating efficiencies of 0.4% both at 30°C and 47°C (Fig. 4B), indicating that replication of the donor chromosome is not required for ICEBs1 mating. The dnaNts donor had mating efficiencies of 0.3% and 1% at 30°C and 47°C, respectively (Fig. 4B). These efficiencies were indistinguishable from those of the wild type, indicating that replication of ICEBs1 in the donor was not required for mating. To verify that ICEBs1 did not replicate in the dnaNts cells under the conditions used for mating, we measured ICEBs1 replication using genomic microarrays in donor cells subjected to a mock mating procedure. We found that ICEBs1 replicated in mock treated WT donor cells incubated at 47°C (nicK copy number 6.7), whereas ICEBs1 did not replicate in identically treated dnaNts donor cells (nicK copy number 0.84). The simplest interpretation of these results is that replication of ICEBs1 is not required for ICEBs1 mating. We conclude that the loss of ICEBs1 mating in the pcrA null mutant is most likely due to a requirement for the unwinding activity of the PcrA helicase and not due to the defect in ICEBs1 replication.
Replication is required for stability of ICEBs1 after induction
We determined the effects of replication of ICEBs1 on its stability. If ICEBs1 re-integrates shortly after excision, then autonomous replication of ICEBs1 should have a relatively small effect on its stability as ICEBs1 would resume being replicated with the chromosome. However, if re-integration is delayed, occurring several generations after excision, then replication might be important for the maintenance of ICEBs1. We found that after induction, autonomous replication of extrachromosomal ICEBs1 allowed for its stable maintenance and re-integration in a population of growing and dividing cells.
Before induction, all of the replication-competent (ICEBs1 nicK+) and replication-defective (ICEBs1 ΔnicK) cells tested (>150 for each strain) were resistant to kanamycin, indicating the presence of ICEBs1. In addition, PCR analysis indicated that >99% of cells from each strain contained attL, the junction between the left end of integrated ICEBs1 and the chromosome (Fig. 5A), and <0.1% contained attB, the empty chromosomal attachment site, indicating that <1 cell in 103 had lost ICEBs1 before induction (Fig. 5B, C).
Figure 5. NicK-dependent replication is required for stability of ICEBs1 after excision.
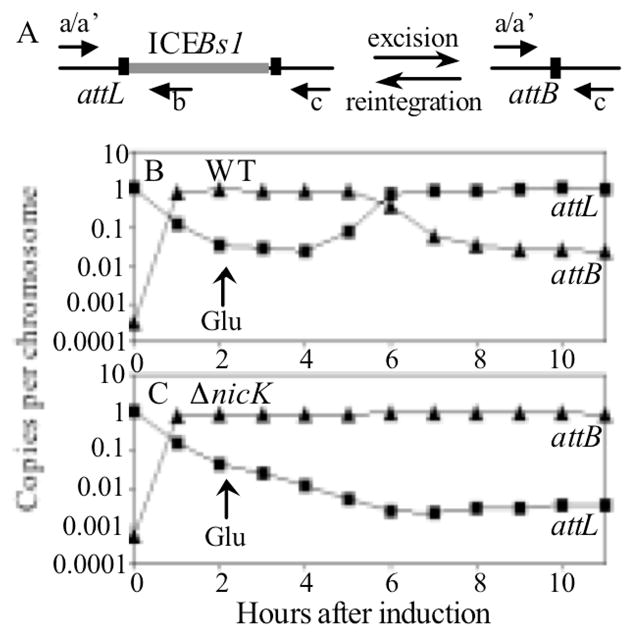
A. Schematic for detecting attL, the junction between the chromosome and the left end of ICEBs1, and attB, the empty attachment site in the bacterial chromosome formed after excision of ICEBs1. ICEBs1 is indicated by the gray line in the drawing on the left and is not shown after excision on the right. qRT-PCR using primer pairs a and b or a′ and c detected attL and attB, respectively.
B, C. Replication-competent wild type, CAL874 (B), and replication-defective ΔnicK, CAL1215 (C), strains were grown in minimal L-arabinose medium at 37°C and expression of Pxyl-rapI was induced during exponential growth (OD600 ~0.2) by addition of D-xylose for 2 hours. Expression of Pxyl-rapI was then repressed by removal of xylose and resuspension (and dilution) into minimal D-glucose medium at an OD600 of ~0.025. Samples were taken at the indicated times, covering ~7 doublings as cells entered stationary phase, and qRT-PCR was used to determine the copy number of attL and attB relative to that of a nearby gene (ydbT) outside of ICEBs1. The relative copy number is plotted as a function of time after addition of xylose to induce ICEBs1. The arrow at 2 hours indicates the shift to glucose (Glu) to repress expression of Pxyl-rapI. Data presented are from one representative experiment.
Two hours after induction (addition of xylose to induce expression of Pxyl-rapI), ICEBs1 had excised from >90% of the cells of both wild type and the nicK mutant, as >90% of cells contained attB and <10% contained attL (Fig. 5BC). At this point, we repressed expression of Pxyl-rapI by removal of xylose and addition of glucose.
Several hours after repression by glucose, we observed that the WT replication-competent ICEBs1 began to re-integrate into attB, as the percentage of cells that had attL increased while those with attB decreased (Fig. 5B). By 6–9 hours after repression of Pxyl-rapI, the percentage of cells with the integrated WT ICEBs1 (attL) leveled off at ~95%, and only ~5% still had attB (Fig. 5B). In stark contrast, >98% of cells with the replication-defective ICEBs1 continued to have an empty attB long after repression of Pxyl-rapI (Fig. 5C). The copy number of the replication-defective ICEBs1 circular form had decreased in these cells to <0.002 copies/cell, as determined by quantitation of attICEBs1 (described in Auchtung et al., 2005), while that of the replication-competent ICEBs1 was ~0.4 copies/cell. Consistent with the PCR results, 7–8 hours after repression, the replication-competent ICEBs1 was present in >95% of cells as judged by resistance to kanamycin, while the non-replicating ICEBs1 was present in <0.1% of cells. We also used a pcrA recF double mutant to block replication of ICEBs1. As with the replication-defective nicK mutant, ICEBs1 (nicK+) was unstable after excision in the pcrA recF double mutant, but stable in the recF single mutant (data not shown). Together, our results indicate that autonomous replication is critical for maintenance of ICEBs1 in a population of cells, and that reintegration, at least under the conditions tested, begins several generations after removal of the inducing signal.
DISCUSSION
We found that ICEBs1 replicates autonomously by a plasmid-like rolling circle mechanism. Replication initiates at the ICEBs1 origin of transfer and requires the element-encoded DNA relaxase NicK. Replication also requires the catalytic subunit of the host DNA polymerase PolC, the host processivity clamp DnaN, and the host-encoded helicase PcrA. ICEBs1 replication does not require the host replicative helicase DnaC. Autonomous replication of ICEBs1 is important for its stable maintenance and re-integration in a population of growing cells, but does not appear to be required for mating.
Rolling circle replication and conjugation
Rolling circle replication initiates when a DNA relaxase nicks the double strand origin of replication and covalently attaches to the 5′-end of the nicked DNA (Khan, 2005). It has been proposed that conjugation evolved from adaptation of a protein secretion system to transport an RCR DNA relaxase and the attached DNA strand (Llosa et al., 2002). Consistent with this idea, oriT and NicK are required for both conjugative transfer (Lee & Grossman, 2007) and unidirectional replication of ICEBs1. Furthermore, PcrA, which is required for replication of RCR plasmids in B. subtilis and other Gram-positive bacteria (Khan, 2005), is required for replication and conjugation of ICEBs1. PcrA is recruited to the nicked origin of RCR plasmids by direct interaction with the cognate Rep DNA relaxase and unwinds the DNA template for leading strand synthesis from the nicked 3′-end (reviewed in Khan, 2005). NicK is homologous to Rep DNA relaxases and is likely involved in recruiting PcrA to the nicked ICEBs1 template. PcrA likely unwinds the double-stranded ICEBs1 template to generate the single-strand DNA substrate needed for transfer through the ICEBs1 mating pore.
Early studies of E. coli F and other conjugative plasmids demonstrated that there is conjugation-dependent replication in donor cells (Ohki & Tomizawa, 1968, Rupp & Ihler, 1968). Since then, a direct role for DNA replication in conjugation has remained an intriguing possibility, especially for RCR conjugative plasmids (Lanka & Wilkins, 1995, Llosa et al., 2002). Although some experiments indicate that replication is not required for conjugation (Sarathy & Siddiqi, 1973, Kingsman & Willetts, 1978), others demonstrate the difficulty of totally abolishing replication of conjugative plasmids (Meyer, 2009).
We found that B. subtilis DnaN is required for replication of ICEBs1 but is not required for conjugation. Therefore, we propose that DNA replication is not required for ICEBs1 conjugation and that the B. subtilis PcrA helicase, not rolling circle replication, mediates unwinding and generation of a single-strand of ICEBs1 DNA for conjugation. It is theoretically possible that ICEBs1 is able to replicate in a subpopulation of the dnaNts mutant cells at the non-permissive temperature, that we would not be able to detect this, and that these cells would be the donors for mating. This seems highly unlikely as the mating efficiency of this subpopulation would have to be considerably higher than what we typically observe for WT cells under the conditions tested.
Autonomous replication of integrative and conjugative elements
Similar to ICEBs1, the 501.8 kb ICEMlSymR7A element of Mesorhizobium loti R7A may have a bi-functional oriT and DNA relaxase that allows it to replicate autonomously and be stably maintained in growing cells (Ramsay et al., 2006). Autonomous replication may be a common, if not universal, feature of ICEs that is partly derived from the evolutionary relationship between conjugal relaxases and replication relaxases. It may be difficult to detect replication of other ICEs since many excise at very low frequencies and may only replicate transiently. However, sensitive and quantitative PCR-based methods might be used to detect the unique junctions formed in the chromosome (attB) and in the circularized element (attP) after excision. An increase in the copy number of attP vs attB would indicate that the excised ICE replicates more frequently than the chromosome (AB & ADG, unpublished results).
A regulatory switch controls the mode of ICEBs1 replication
ICEs appear to be comprised of discrete functional modules (Toussaint & Merlin, 2002, Burrus et al., 2002), and typically excise from the host chromosome to form a circle before conjugative transfer. ICEBs1 contains a phage-like module required for integration, excision, and regulation (Auchtung et al., 2007, Lee et al., 2007, Bose* et al., 2008), a plasmid-like module involved in DNA replication and transfer (Lee & Grossman, 2007, Berkmen et al., 2009), and a cell-cell signaling module (Auchtung et al., 2005). The particular combination of modules present in ICEBs1 allows it to switch from an integrated element to a replicating plasmid and back. This conversion is controlled by a phage-like proteolytic regulatory switch. In one state, the repressor ImmR accumulates and prevents expression of excisionase and ICEBs1 replication and transfer functions, maintaining ICEBs1 in its integrated form. Induction of ICEBs1 causes the second state by triggering the degradation of ImmR, thereby de-repressing ICEBs1 gene expression (Bose* et al., 2008), leading to excision and autonomous replication. DNA damage or the presence of a high density of neighboring cells that lack ICEBs1 independently activate this regulatory switch (Auchtung et al., 2005, Bose* et al., 2008). We found that the kinetics of reintegration of ICEBs1 after removal of an inducing signal are slow. The delay in re-integration is likely due to the time it takes to deplete the excisionase and re-establish repression. Autonomous replication of ICEBs1 contributes to its overall stability within a population. Since many ICEs are related to conjugative plasmids, and excise from the host chromosome prior to conjugative transfer, we suspect that this switch between an integrated passively replicating state and the extra-chromosomal autonomously replicating state is widespread among ICEs.
EXPERIMENTAL PROCEDURES
Bacterial strains and growth conditions
Strains were constructed by natural transformation and grown in LB or defined minimal medium (Harwood & Cutting, 1990). All strains (Table 2) contain pheA1 trpC2, except for CAL1161, which is trp+ (Berkmen & Grossman, 2007). Most ICEBs1 strains contained a kanamycin-resistance cassette {Δ(rapI-phrI)342::kan} (Auchtung et al., 2005). ICEBs1 was induced by over-expression of rapI from a xylose-inducible promoter using amyE::{(Pxyl-rapI), spc} (Berkmen et al., 2009) or lacA::{(Pxyl-rapI), tet}, or from an IPTG-inducible promoter using amyE::{(Pspank(hy)-rapI), spc} (Auchtung et al., 2005). Temperature-sensitive replication mutations included: dnaB134(ts) (Mendelson & Gross, 1967), linked to zhb83::Tn917(mls) (Rokop et al., 2004); dnaC30(ts) (Karamata & Gross, 1970) linked to mls between yybS and cotF (Lin & Grossman, 1998); and dnaN5(ts) (Karamata & Gross, 1970).
Table 2.
Bacillus subtilis strains used.
| Strain | Relevant genotype (reference) |
|---|---|
| CAL108 | Δ(rapI-phrI)342::kan ΔattR100::tet amyE::{(Pspank(hy)-rapI) spc} (Lee et al., 2007) |
| CAL306 | Δ(rapI-phrI)342::kan ΔnicK306 amyE::{(Pspank(hy)-rapI) spc} (Lee & Grossman, 2007) |
| CAL419 | ICEBs10 str-84 comK::cat (Auchtung et al., 2005) |
| CAL635 | Δ(rapI-phrI)342::kan amyE::{(Pxyl-rapI) spc} lacA::{rpsF (ssb-mgfpmut2) tet} |
| CAL874 | Δ(rapI-phrI)342::kan amyE::{(Pxyl-rapI) spc} |
| CAL879 | Δ(rapI-phrI)342::kan amyE::{(Pxyl-rapI) spc} {(polC::polC-gfpmut2) cat} |
| CAL934 | Δ(rapI-phrI)342::kan amyE::{(Pxyl-rapI) spc} dnaB134(ts) zhb83::Tn917(mls) |
| CAL992 | Δ(rapI-phrI)342::kan amyE::{(Pxyl-rapI) spc} pcrA::{Pspank-(myc-pcrA), cat} |
| CAL1035 | Δ(rapI-phrI)342::kan amyE::{(Pspank(hy)-rapI) spc} dnaC30(ts) Δ449::mls |
| CAL1148 | Δ(rapI-phrI)342::kan lacA::{(Pxyl-rapI) tet} ΔrecF::spc ΔpcrA::cat thrC::{(pcrB-pcrA)1126 mls} |
| CAL1161 | Δ(rapI-phrI)342::kan amyE::{(Pspank(hy)-rapI) spc} dnaN5(ts) (ypjG-hepT)122 |
| CAL1173 | Δ(rapI-phrI)342::kan lacA::{(Pxyl-rapI) tet} amyE::{(Pspank(hy)) spc} thrC::cat |
| CAL1174 | Δ(rapI-phrI)342::kan lacA::{(Pxyl-rapI) tet} ΔrecF::spc thrC::cat |
| CAL1175 | Δ(rapI-phrI)342::kan lacA::{(Pxyl-rapI) tet} ΔrecF::spc ΔpcrA1021::cat thrC::phl |
| CAL1215 | Δ(rapI-phrI)342::kan ΔnicK306 amyE::{(Pxyl-rapI) spc} |
| IRN342 | Δ(rapI-phrI)342::kan (Auchtung et al., 2005) |
| JMA168 | Δ(rapI-phrI)342::kan amyE::{(Pspank(hy)-rapI) spc} (Auchtung et al., 2005) |
| JMA222 | ICEBs10 (Auchtung et al., 2005) |
polC-gfpmut2 (cat) is a single cross-over insertion of pCAL862 at polC and is the only functional copy of polC in the cell and is expressed from the endogenous promoter. It is analogous to the polC-gfpmut2 constructs described previously (Lemon & Grossman, 1998, Berkmen & Grossman, 2006) except for the antibiotic marker.
ssb-mGFPmut2 is expressed from lacA::{(rpsF ssb-mgfpmut2) tet} in cells that also contain wild type ssb (Berkmen & Grossman, 2006).
The alleles amyE::{Pspank(hy), spc} (empty vector) (Auchtung et al., 2005), thrC1165::cat and thrC1167::phl (phleomycin) were used as controls for antibiotic resistance and threonine auxotrophy in mating experiments (Fig. 4). thrC1165::cat and thrC1167::phl were constructed using long-flanking homology PCR (Wach, 1996).
We deleted the entire pcrA open reading frame, except for the last 44 codons, and inserted a chloramphenicol-inducible cat lacking its transcription terminator, using long-flanking homology PCR. We introduced ΔpcrA1021::cat into strains carrying ΔrecF::spc, (Sciochetti et al., 2001). Growth of the ΔpcrA ΔrecF mutant requires chloramphenicol, likely due to chloramphenicol-dependent transcription of ligA from the upstream cat. ligA is downstream from pcrA and encodes DNA ligase.
For complementation of ΔpcrA, we cloned pcrA, the upstream gene pcrB and another 341 bp upstream including the endogenous promoter, into a thrC vector to generate pCAL1126. This plasmid was used to introduce thrC::{(pcrB pcrA) mls}1126 into the chromosome.
Myc-tagged PcrA was expressed from Pspank-myc-pcrA (cat), a single crossover insertion of pCAL984 into pcrA that fuses a 3xMyc epitope tag to the N-terminal end of PcrA. The myc-pcrA fusion is expressed from the IPTG-inducible promoter Pspank with an optimized ribosome-binding site and is the only functional copy of pcrA.
Preparation of DNA for analysis of ICEBs1 and chromosome replication
Cells growing in minimal medium were mixed with an equal volume of cold (−20°C) methanol, harvested by centrifugation, and cell pellets were frozen at −80°C. Cells from 5 ml of culture (OD600~1) were resuspended in 0.3 ml of A buffer (Lin & Grossman, 1998) containing 1 mg/ml lysozyme, lysed by incubation at 37°C for 30 min, followed by room temperature additions of 3 μl 100 mg/ml RNase A for 5 min, 0.3 ml 2 x IP buffer (Lin & Grossman, 1998) for 5 min, then addition of 15 μl 10% SDS and 3 μl Proteinase K (Qiagen, stock solution ≥600 mAU/ml) at 37°C for 30 min. Cell lysates were extracted twice, once with phenol:chloroform:isoamyl alcohol (1:1:48) and once with chloroform alone. Genomic DNA was precipitated from 0.5 ml of extracted lysate by addition of 15 μl of 5M NaCl and 1.25 ml of cold (−20°C) ethanol and resuspended in 10mM Tris-HCl 0.1mM EDTA pH 8. For microarray analysis, genomic DNA was digested with HaeIII, run on MinElute PCR Purification columns, washed twice with PB, once with PE (Qiagen), then eluted with water.
To verify that both ssDNA and dsDNA were recovered, we added purified 32P-labelled ssDNA or dsDNA fragments to cell suspensions and monitored 32P levels at each step of purification. There was no difference in the recovery of a 7.3 kb fragment of ICEBs1 (ydcS-yddG′), whether single-stranded or double-stranded.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) of DNA bound to epitope-tagged and untagged proteins was done essential as described (Lin & Grossman, 1998, Breier & Grossman, 2007). Tests using unfixed cell lysates were conducted to determine the amount of antibodies needed to immunoprecipitate each epitope-tagged or untagged protein from cell lysates.
DNA microarrays for analysis of DNA synthesis and ChIP-chip
Chromosomal DNA or DNA from ChIP samples was labeled and hybridized to spotted DNA microarrays as described (Breier & Grossman, 2007). Two types of microarrays were used, those prepared from PCR products representing > 99% of the open reading frames in the B. subtilis genome and 295 intergenic regions (Breier & Grossman, 2007), and those prepared from a set of synthetic oligonucleotides designed to correspond to all annotated B. subtilis ORFs (Auchtung et al., 2005).
HaeIII digested genomic DNA samples were labeled with Cy5 and mixed with a reference genomic DNA labeled with Cy3, as described previously (Wang et al., 2007). By comparing the signal from the Cy5 sample to the Cy3 reference from replicate sets of test samples to that of similarly labeled uninduced IRN342 control samples, we determined the copy number of each gene in the genomic sample relative to the uninduced control.
ChIP DNA samples were labeled with Cy5 and mixed with the corresponding total DNA sample labeled with Cy3. The ratio of Cy5 ChIP to Cy3 total signals indicated the fold enrichment of genes due to immunoprecipitation. The raw Cy5 and Cy3 signals from the genomic microarrays and the ChIP microarrays were normalized as previously described (Breier & Grossman, 2007).
Quantitative PCR assays for ICEBs1 excision and replication
qRT-PCR was used to measure ICEBs1 excision and replication from purified DNA samples or from crude cell lysates. In the latter case, cells were lysed by suspension in water containing 0.05 mg/ml of lysozyme, incubation at 37°C for 30 minutes followed by heating at 105°C for 15 minutes, cooling to 4°C for 5 minutes, heating again to 105°C for 3 minutes, then finally cooling again to 4°C. The cell lysates were centrifuged for 10 minutes at room temperature and the cleared supernatant was assayed by qRT-PCR. We used the LightCycler 480 Real-Time PCR system with Syber Green detection reagents (Roche).
ICEBs1 excision and re-integration were measured using primer pair AB023-ABO24, to quantitate the left-hand integrated ICEBs1-chromosomal junction (attL), and primer pair CLO261–CLO262, to quantitate the unoccupied attachment site (attB) that is formed by excision. Primer pair CLO284-CLO285 was used to quantitate a control chromosomal region outside of ICEBs1 in ydbT, which is located 15 kb to the left of attL. Standard curves for attL and ydbT were generated using genomic DNA from uninduced cells of IRN342 in which ICEBs1 is integrated in single copy in the chromosome. Standard curves for attB and ydbT were also generated using genomic DNA from JMA222, an ICEBs1-cured strain that simulates 100% excision.
Replication of ICEBs1 regions to the left and right of the nic site in oriT were measured using primer pairs CLO274-CLO275 (left) and CLO280-CLO281 (right). Standard curves for these ICEBs1 sites and the ydbT control site were generated using genomic DNA from uninduced cells of IRN342, described above. Primer sequences are available upon request.
ICEBs1 nicking and mating assays
Nicking of ICEBs1 at oriT was assayed using purified genomic DNA digested with HaeIII (see above) in a primer extension assay with 32P-labelled primer CLO76, essentially as described (Lee & Grossman, 2007). The frequency of nicking in each sample was calculated by quantitating the amount of primer extension products terminating at the nicked site and at the HaeIII restriction site ~50 bp farther upstream in ydcQ. Primer extension into ydcQ would only occur if an oriT template is not nicked by NicK.
ICEBs1 mating assays were conducted using donor cells activated by expression of rapI from an IPTG-inducible promoter Pspank(hy) or from a xylose-inducible promoter Pxyl, essentially as described (Lee et al., 2007).
Acknowledgments
We thank M. Berkmen, P. Piggot and D. Rudner for gifts of strains, and G. Wright for providing HPUra. We thank M. Berkmen, A. Breier, M. Buttner, A. Salyers and W.K. Smits for helpful discussions, and M. Berkmen for comments on the manuscript. This work was supported, in part, by NIH grant GM50895.
REFERECES
- Anand SP, Zheng H, Bianco PR, Leuba SH, Khan SA. DNA helicase activity of PcrA is not required for the displacement of RecA protein from DNA or inhibition of RecA-mediated strand exchange. J Bacteriol. 2007;189:4502–4509. doi: 10.1128/JB.00376-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auchtung JM, Lee CA, Garrison KL, Grossman AD. Identification and characterization of the immunity repressor (ImmR) that controls the mobile genetic element ICEBs1 of Bacillus subtilis. Mol Microbiol. 2007;64:1515–1528. doi: 10.1111/j.1365-2958.2007.05748.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auchtung JM, Lee CA, Monson RE, Lehman AP, Grossman AD. Regulation of a Bacillus subtilis mobile genetic element by intercellular signaling and the global DNA damage response. Proc Natl Acad Sci U S A. 2005;102:12554–12559. doi: 10.1073/pnas.0505835102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkmen MB, Grossman AD. Spatial and temporal organization of the Bacillus subtilis replication cycle. Mol Microbiol. 2006;62:57–71. doi: 10.1111/j.1365-2958.2006.05356.x. [DOI] [PubMed] [Google Scholar]
- Berkmen MB, Grossman AD. Subcellular positioning of the origin region of the Bacillus subtilis chromosome is independent of sequences within oriC, the site of replication initiation, and the replication initiator DnaA. Mol Microbiol. 2007;63:150–165. doi: 10.1111/j.1365-2958.2006.05505.x. [DOI] [PubMed] [Google Scholar]
- Berkmen MB, Lee CA, Loveday EK, Grossman AD. Polar positioning of a conjugation protein from the integrative and conjugative element ICEBs1 of Bacillus subtilis. J Bacteriol. 2009 doi: 10.1128/JB.00860-09. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Bose* B, Auchtung* JM, Lee CA, Grossman AD. A conserved anti-repressor controls horizontal gene transfer by proteolysis. Mol Microbiol. 2008;70:570–582. doi: 10.1111/j.1365-2958.2008.06414.x. co-first authors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breier AM, Grossman AD. Whole-genome analysis of the chromosome partitioning and sporulation protein Spo0J (ParB) reveals spreading and origin-distal sites on the Bacillus subtilis chromosome. Mol Microbiol. 2007;64:703–718. doi: 10.1111/j.1365-2958.2007.05690.x. [DOI] [PubMed] [Google Scholar]
- Brown NC. 6-(p-hydroxyphenylazo)-uracil: a selective inhibitor of host DNA replication in phage-infected Bacillus subtilis. Proc Natl Acad Sci U S A. 1970;67:1454–1461. doi: 10.1073/pnas.67.3.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruand C, Farache M, McGovern S, Ehrlich SD, Polard P. DnaB, DnaD and DnaI proteins are components of the Bacillus subtilis replication restart primosome. Mol Microbiol. 2001;42:245–255. doi: 10.1046/j.1365-2958.2001.02631.x. [DOI] [PubMed] [Google Scholar]
- Burrus V, Pavlovic G, Decaris B, Guedon G. The ICESt1 element of Streptococcus thermophilus belongs to a large family of integrative and conjugative elements that exchange modules and change their specificity of integration. Plasmid. 2002;48:77–97. doi: 10.1016/s0147-619x(02)00102-6. [DOI] [PubMed] [Google Scholar]
- Burrus V, Waldor MK. Shaping bacterial genomes with integrative and conjugative elements. Res Microbiol. 2004;155:376–386. doi: 10.1016/j.resmic.2004.01.012. [DOI] [PubMed] [Google Scholar]
- Frost LS, Leplae R, Summers AO, Toussaint A. Mobile genetic elements: the agents of open source evolution. Nat Rev Microbiol. 2005;3:722–732. doi: 10.1038/nrmicro1235. [DOI] [PubMed] [Google Scholar]
- Harwood CR, Cutting SM. Molecular Biological Methods for Bacillus. John Wiley & Sons; Chichester: 1990. [Google Scholar]
- Karamata D, Gross JD. Isolation and genetic analysis of temperature-sensitive mutants of B. subtilis defective in DNA synthesis. Mol Gen Genet. 1970;108:277–287. doi: 10.1007/BF00283358. [DOI] [PubMed] [Google Scholar]
- Keeling PJ, Palmer JD. Horizontal gene transfer in eukaryotic evolution. Nat Rev Genet. 2008;9:605–618. doi: 10.1038/nrg2386. [DOI] [PubMed] [Google Scholar]
- Khan SA. Plasmid rolling-circle replication: highlights of two decades of research. Plasmid. 2005;53:126–136. doi: 10.1016/j.plasmid.2004.12.008. [DOI] [PubMed] [Google Scholar]
- Kingsman A, Willetts N. The requirements for conjugal DNA synthesis in the donor strain during Flac transfer. J Mol Biol. 1978;122:287–300. doi: 10.1016/0022-2836(78)90191-2. [DOI] [PubMed] [Google Scholar]
- Lanka E, Wilkins BM. DNA processing reactions in bacterial conjugation. Annu Rev Biochem. 1995;64:141–169. doi: 10.1146/annurev.bi.64.070195.001041. [DOI] [PubMed] [Google Scholar]
- Lee CA, Auchtung JM, Monson RE, Grossman AD. Identification and characterization of int (integrase), xis (excisionase) and chromosomal attachment sites of the integrative and conjugative element ICEBs1 of Bacillus subtilis. Mol Microbiol. 2007;66:1356–1369. doi: 10.1111/j.1365-2958.2007.06000.x. [DOI] [PubMed] [Google Scholar]
- Lee CA, Grossman AD. Identification of the origin of transfer (oriT) and DNA relaxase required for conjugation of the integrative and conjugative element ICEBs1 of Bacillus subtilis. J Bacteriol. 2007;189:7254–7261. doi: 10.1128/JB.00932-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemon KP, Grossman AD. Localization of bacterial DNA polymerase: evidence for a factory model of replication. Science. 1998;282:1516–1519. doi: 10.1126/science.282.5393.1516. [DOI] [PubMed] [Google Scholar]
- Lin DC, Grossman AD. Identification and characterization of a bacterial chromosome partitioning site. Cell. 1998;92:675–685. doi: 10.1016/s0092-8674(00)81135-6. [DOI] [PubMed] [Google Scholar]
- Llosa M, Gomis-Ruth FX, Coll M, de la Cruz F. Bacterial conjugation: a two-step mechanism for DNA transport. Mol Microbiol. 2002;45:1–8. doi: 10.1046/j.1365-2958.2002.03014.x. [DOI] [PubMed] [Google Scholar]
- Mendelson NH, Gross JD. Characterization of a temperature-sensitive mutant of Bacillus subtilis defective in deoxyribonucleic acid replication. J Bacteriol. 1967;94:1603–1608. doi: 10.1128/jb.94.5.1603-1608.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer R. Replication and conjugative mobilization of broad host-range IncQ plasmids. Plasmid. 2009;62:57–70. doi: 10.1016/j.plasmid.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- Ohki M, Tomizawa J. Asymmetric transfer of DNA strands in bacterial conjugation. Cold Spring Harb Symp Quant Biol. 1968;33:651–658. doi: 10.1101/sqb.1968.033.01.074. [DOI] [PubMed] [Google Scholar]
- Osborn AM, Boltner D. When phage, plasmids, and transposons collide: genomic islands, and conjugative- and mobilizable-transposons as a mosaic continuum. Plasmid. 2002;48:202–212. doi: 10.1016/s0147-619x(02)00117-8. [DOI] [PubMed] [Google Scholar]
- Petit MA, Dervyn E, Rose M, Entian KD, McGovern S, Ehrlich SD, Bruand C. PcrA is an essential DNA helicase of Bacillus subtilis fulfilling functions both in repair and rolling-circle replication. Mol Microbiol. 1998;29:261–273. doi: 10.1046/j.1365-2958.1998.00927.x. [DOI] [PubMed] [Google Scholar]
- Petit MA, Ehrlich D. Essential bacterial helicases that counteract the toxicity of recombination proteins. EMBO J. 2002;21:3137–3147. doi: 10.1093/emboj/cdf317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay JP, Sullivan JT, Stuart GS, Lamont IL, Ronson CW. Excision and transfer of the Mesorhizobium loti R7A symbiosis island requires an integrase IntS, a novel recombination directionality factor RdfS, and a putative relaxase RlxS. Mol Microbiol. 2006;62:723–734. doi: 10.1111/j.1365-2958.2006.05396.x. [DOI] [PubMed] [Google Scholar]
- Rokop ME, Auchtung JM, Grossman AD. Control of DNA replication initiation by recruitment of an essential initiation protein to the membrane of Bacillus subtilis. Mol Microbiol. 2004;52:1757–1767. doi: 10.1111/j.1365-2958.2004.04091.x. [DOI] [PubMed] [Google Scholar]
- Rupp WD, Ihler G. Strand selection during bacterial mating. Cold Spring Harb Symp Quant Biol. 1968;33:647–650. doi: 10.1101/sqb.1968.033.01.073. [DOI] [PubMed] [Google Scholar]
- Sarathy PV, Siddiqi O. DNA synthesis during bacterial conjugation. II. Is DNA replication in the Hfr obligatory for chromosome transfer? J Mol Biol. 1973;78:443–451. doi: 10.1016/0022-2836(73)90467-1. [DOI] [PubMed] [Google Scholar]
- Sciochetti SA, Blakely GW, Piggot PJ. Growth phase variation in cell and nucleoid morphology in a Bacillus subtilis recA mutant. J Bacteriol. 2001;183:2963–2968. doi: 10.1128/JB.183.9.2963-2968.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- te Poele EM, Bolhuis H, Dijkhuizen L. Actinomycete integrative and conjugative elements. Antonie Van Leeuwenhoek. 2008;94:127–143. doi: 10.1007/s10482-008-9255-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toussaint A, Merlin C. Mobile elements as a combination of functional modules. Plasmid. 2002;47:26–35. doi: 10.1006/plas.2001.1552. [DOI] [PubMed] [Google Scholar]
- Velten M, McGovern S, Marsin S, Ehrlich SD, Noirot P, Polard P. A two-protein strategy for the functional loading of a cellular replicative DNA helicase. Mol Cell. 2003;11:1009–1020. doi: 10.1016/s1097-2765(03)00130-8. [DOI] [PubMed] [Google Scholar]
- Wach A. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast. 1996;12:259–265. doi: 10.1002/(SICI)1097-0061(19960315)12:3%3C259::AID-YEA901%3E3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Wang J, Wang GR, Shoemaker NB, Salyers AA. Production of two proteins encoded by the Bacteroides mobilizable transposon NBU1 correlates with time-dependent accumulation of the excised NBu1 circular form. J Bacteriol. 2001;183:6335–6343. doi: 10.1128/JB.183.21.6335-6343.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JD, Sanders GM, Grossman AD. Nutritional Control of Elongation of DNA Replication by (p)ppGpp. Cell. 2007;128:865–875. doi: 10.1016/j.cell.2006.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters VL, Guiney DG. Processes at the nick region link conjugation, T-DNA transfer and rolling circle replication. Mol Microbiol. 1993;9:1123–1130. doi: 10.1111/j.1365-2958.1993.tb01242.x. [DOI] [PubMed] [Google Scholar]