Abstract
Glial-guided neuronal migration is a key step in the development of laminar architecture of cortical regions of the mammalian brain. We previously reported that neuronal protein astrotactin (ASTN1) functions as a neuron-glial ligand during CNS glial-guided migration. Here, we identify a new Astn family member, Astn2, that is expressed at high levels in migrating, cerebellar granule neurons, along with Astn1, at developmental stages when glial-guided migration is ongoing. Biochemical and flow cytometry experiments show that ASTN2 forms a complex with ASTN1 and regulates surface expression of ASTN1. Live imaging of Venus-tagged ASTN1 in migrating cerebellar granule cells reveals the intracellular trafficking of ASTN1-Venus, with ASTN1-Venus accumulating in the forward aspect of the leading process where new sites of adhesion will form. Treatment of migrating neurons with Dynasore, a soluble noncompetitive inhibitor of Dynamin, rapidly arrests the migration of immature granule cells in a reversible manner, suggesting the critical importance of receptor trafficking to neuronal locomotion along Bergmann glial fibers in the developing cerebellum. Together, these findings suggest that ASTN2 regulates the levels of ASTN1 in the plasma membrane and that the release of neuronal adhesions to the glial fiber during neuronal locomotion involves the intracellular trafficking of ASTN1.
Introduction
The normal development of cortical regions of the mammalian brain depends on the migrations of neuronal precursors from germinal zones and assembly into neuronal laminae, where synaptic connections form (Rakic, 1978; Hatten, 1999). In higher vertebrates, radial glial fibers guide the migration of the principal output neurons of the cerebral cortex and the two major classes of neurons in the cerebellar cortex, the Purkinje neuron and the granule cell. Since migration disorders are associated with cortical malformations, mental retardation and epilepsy (Dobyns and Truwit, 1995; Barkovich et al., 1996; Ross and Walsh, 2001), a clearer understanding of the molecular control of CNS neuronal migration could be relevant to the diagnosis and treatment of human brain disorders.
The cerebellar cortex has long provided a model for glial-guided neuronal migration (Rakic, 1971). Live imaging studies of cerebellar granule neuron migration in vitro reveal distinct features of glial-guided migration, including the extension of a highly polarized leading process in the direction of migration (Edmondson and Hatten, 1987; Komuro and Rakic, 1998), assembly of an interstitial adhesion junction beneath the cell soma (Edmondson and Hatten, 1987; Gregory et al., 1988), localization of acto-myosin contractile motors in the proximal portion of the leading process and formation of a perinuclear cage of tubulin (Rivas and Hatten, 1995; Solecki et al., 2004). The migration cycle involves forward movement of the centrosome into the proximal portion of the leading process preceding translocation of the neuronal nucleus, the activation of acto-myosin motors located in the proximal aspect of the leading process (Solecki et al., 2009), and the release of the adhesion junction, initiating forward movement of the cell soma. Directed movements of the centrosome and the orientation of the leading process apparently set the direction of neuronal locomotion on glial fibers (Solecki et al., 2004; Bellion et al., 2005; Schaar and McConnell, 2005; Tsai et al., 2007; Umeshima et al., 2007).
The neuronal protein astrotactin (ASTN1) is a well studied receptor for glial-guided neuronal migration (Edmondson et al., 1988; Fishell and Hatten, 1991; Zheng et al., 1996; Adams et al., 2002). Other receptor systems that function in CNS migration include neuregulin, which binds to ErbB4 on the glial surface (Anton et al., 1997; Rio et al., 1997), and BDNF, which stimulates granule neuron migration (Borghesani et al., 2002). Although integrins function as adhesion receptors in a wide range of cell migrations (Ridley et al., 2003), genetic studies indicate that integrin-based adhesions are not essential for glial-guided neuronal migration (Fishell and Hatten, 1991; Belvindrah et al., 2007).
Astn2, a gene of unknown function that is homologous to Astn1, has recently been implicated in several common disorders of the nervous system, including attention deficit hyperactivity disorder (ADHD), autism and schizophrenia (Lesch et al., 2008; Vrijenhoek et al., 2008; Glessner et al., 2009). Here we show that Astn2 is abundant in migrating cerebellar granule neurons when glial-guided migration is ongoing. ASTN2 forms a complex with ASTN1 that regulates the polarized trafficking of ASTN1 during migration.
Materials and Methods
Construction of the full-length Astn2 mouse cDNA and expression vectors.
Astn2 cDNA fragments were identified by screening a P7 cerebellar cDNA library with a probe for the Astn1 EGF domain, and by PCR walking, using E17 brain first strand marathon ready cDNA (BD Biosciences) with the following primers: 5′-GTCTCCTTCTCTTTGTGCG-3′ and 5′-GGCGAGGTGGCATTGATC-3′. The identified Astn2 cDNA fragments were joined by restriction digest and cloned into the pGW1 and pRK5 expression vectors. To generate the pGW1-Astn2-Myc fusion, the C terminus of Astn2 was amplified using an antisense primer that contained the Myc coding sequence. This PCR product was swapped into the XhoI and SalI sites, replacing the untagged C-terminal region. To produce pCIG-Astn2-Venus and pCIG-Astn2-mCherry C-terminal fusions, the Venus and mCherry cDNAs were fused in frame with the 3′ end of Astn2 coding sequence by joining PCR. The resulting Astn2-Venus or -mCherry fusion inserts (XhoI/NotI) were subcloned along with the rest of the Astn2 cDNA (XmaI/XhoI) into pCIG expression vector by three-way ligation into the XmaI and NotI sites. To generate ASTN2 constructs that lacked either EGF, MP, or FN domains for coimmunoprecipitation experiments, the following primers were used: pRK5-Astn2-ΔEGF-Venus, 5′-GGATATCCTAATGCTCTCAGATGTCTG-3′, 5′-CCGGCCTTTGGTTTCCCC-3′; pRK5-Astn2-ΔMP-Venus, 5′-GGGTACCCCTACTGGTGTTCTGGTAAAGG-3′ and 5′-ACGCGTCGACGGATCCATGAAAGTCGTGC-3′; pRK5-Astn2-ΔFN-Venus, 5′-GCAGTCGCCGCCGCAGCAAAGGTTTAC-3′ and 5′-CGCTCGAGTCCAGTAAGAACTCATCAGTGC-3′. The ΔEGF, ΔMP and ΔFN products were inserted into EcoRV/XhoI, Acc651/SalI or EcoRV/XhoI digests of pRK5-Astn2, respectively. A C-terminal-Venus fusion protein was generated by joining PCR fragments.
We generated a C-terminal Myc-tagged version of Astn1, pGW1-Astn1-Myc, by PCR using an antisense primer that contained the Myc coding sequence as described above for pGW1-Astn2-Myc. To produce pCIG-Astn1-Venus, the Venus sequence was fused in frame with the 3′ end of the Astn1 coding sequence by joining PCR. The resulting Astn1-Venus fusion inserts (EcoRI/NotI) were subcloned, along with the rest of the Astn1 cDNA (XmaI/EcoRI), into the pCIG expression vector by three-way ligation into XmaI and NotI sites.
Northern blot analysis of Astn2 expression in developing brain.
RNA was extracted using Tri-Reagent (Molecular Research Center), separated on formaldehyde-agarose gels, and transferred onto Hybond-XL membrane (GE Healthcare). Northern blot hybridization was performed using a P32 labeled Astn2 probe corresponding to nucleotides 61-741 of the open reading frame of Astn2 in hybridization solution (6× SSPE, 5× Denhardt's, 0.5% SDS, and 50 mg of single stranded salmon sperm DNA) overnight. After washing, the membrane was exposed to film (Kodak Life Sciences), stripped in boiling 0.1% SDS and rehybridized with a 1.2 kb Gapdh riboprobe.
In situ hybridization.
P6, P10 and adult C57BL/6J mice were perfused with 4% paraformaldehyde, after which brains were removed by dissection, immersed in sucrose (30%, 4°C, overnight), embedded in Neg-50 (Richard-Allan Scientific) and sectioned (60 μm) with a Leica RM2265 microtome (Leica Microsystems). Hybridization and detection were performed as described previously (Schaeren-Wiemers and Gerfin-Moser, 1993), using the Astn2 probe (described above) or Astn1 probe (a 1749 base pair region derived from the 3′ UTR of the Astn1 gene). No staining was detected with a sense probe after a prolonged incubation period. Radioactive in situ hybridization was performed as described previously (Magdaleno et al., 2006).
Generation and purification of anti-ASTN2 antibodies.
The ASTN2 C-terminal peptide (KITCEEKMVSMARNTYGETKGR) was synthesized by the Rockefeller University Proteomics Resource Center and coupled to bovine thyroglobulin (Sigma-Aldrich). The coupled peptide was then used to immunize a female New Zealand White rabbit to generate an ASTN2-specific polyclonal antibody (Covance Inc.). The rabbit antiserum was affinity purified on a column containing ASTN2 peptide coupled to Affi-Gel-15 resin (Bio-Rad Laboratories). Bound antibodies were removed from the column with both low pH glycine and high salt MgCl2 and tested for specific immunoreactivity for ASTN2 by immunoblotting and immunostaining.
Cell lines and transfections.
HEK293T cells (American Type Culture Collection) were cultured at 37°C + 5% CO2 in DMEM (Invitrogen) + 10% fetal bovine serum, glutamine (4 mm), and penicillin-streptomycin (20 U/ml). For imaging and immunocytochemistry experiments, HEK293T cells were transiently transfected with Lipofectamine 2000 (Invitrogen) in poly-d-lysine- and Matrigel (BD Biosciences)-coated 8 well glass tissue culture chamber slides (BD Biosciences) as per the manufacturer's instructions. For immunoblotting of transfected cell lysate and live cell staining for flow cytometry, HEK293T cells were transiently transfected with Lipofectamine 2000 (Invitrogen) in 6 cm dishes (Corning Life Sciences) with 8 μg of DNA per construct. For coimmunoprecipitation experiments, 4 μg of the Astn1-Myc, control Venus, and control Myc constructs were cotransfected with 8 μg of Astn2-Venus construct. After 24–48 h, transiently transfected HEK293T cells were harvested for immunoblotting or processed for immunostaining.
Retrovirus production.
Recombinant, ecotropic, replication-incompetent retroviruses were produced as previously described (Tomoda et al., 1999; Solecki et al., 2001). Briefly, HEK293T cells were cotransfected with a retroviral construct and pCL-Eco, an ecotropic packaging construct. Twenty-four hours after transfection, medium was replaced with granule cell medium lacking glucose and the resulting culture supernatant containing retroviruses was harvested 24 and 48 h later. The retrovirus supernatant was then supplemented with glucose (6 mm) and filtered with a 0.22 μm filter (Millipore Corp.).
Subcellular fractionation experiments.
Whole brain was dissected from C57BL/6J mouse embryos on embryonic day 10 (E10), E12, E14, and E16; cerebellar tissue was dissected from neonatal mice on the day of birth (P0), P2, P4, P6, P10, P14 or adult (P56), transferred to immunoprecipitation buffer (IPB), [PBS containing 1 mm EDTA, 1 mm EGTA, and protease inhibitors (Roche Applied Science)] and homogenized using a PT 1200CL Polytron (Kinematica) and Dounce homogenizer. The tissue homogenate was centrifuged in a bench top microcentrifuge at 4°C at top speed for 15 min to pellet insoluble/nuclear material, and the soluble fraction was used for further analysis. For cell fractionation experiments, the P10 cerebella homogenate soluble fraction was fractionated by ultracentrifugation in a TLA-45 rotor (Beckman Coulter) at 100,000 × g for 50 min at 4°C. The crude membrane fraction (pellet) was resuspended in homogenization buffer. To separate the membrane fraction into integral and peripheral membrane fractions, a portion of the membrane pellet was resuspended in 0.1 m Na2Co3 pH 11.5, 1 mg/ml saponin, 5 mm EDTA, and protease inhibitors, incubated on ice 30 min, and separated by ultracentrifugation at 100,000 × g for 1 h. The peripheral membrane fraction (supernatant) was collected and the integral membrane fraction (pellet) was resuspended in homogenization buffer. Protein concentrations were determined as described by Bradford (1976).
SDS-PAGE and immunoblotting.
Transiently transfected HEK293T cells were lysed in Triton IPB (IPB + 1% Triton X-100), and solubilized for 20 min (4°C). The cell lysate was then centrifuged at top speed for 15 min in a bench top microcentrifuge at 4°C. For coimmunoprecipitation experiments, a portion of the supernatant was reserved for input loading control, and the remaining 500 μl of the soluble cell lysate was incubated with Protein A/G beads (EMD Biosciences) and 2 μg of either anti-ASTN1 or rabbit anti-GFP antibodies (Invitrogen) for 3 h at 4°C, washed once with Triton IPB, two times with Triton IPB + 500 mm NaCl, then twice with IPB, and eluted in 30 μl of sample buffer. For the coimmunoprecipitations to test calcium dependence, one set of transiently transfected cells was lysed in IPB containing 1 mm CaCl2, 0.5 mm MgCl2 (in the absence of EDTA and EGTA). Protein samples were boiled for 5 min and separated on 7.5 or 10% SDS-polyacrylamide electrophoresis gels. Membranes were blocked in 5% nonfat milk in TBS-Tween and proteins were detected by primary antibody incubation for 1 h at room temperature in blocking buffer using the following antibodies: rabbit anti-ASTN2 (1:200), rabbit anti-ASTN1 (1:200), anti-ERBB4 (1:400, Santa Cruz Biotechnology), anti-MMP9 (1:500, Torrey Pines Biolabs), mouse anti-GAPDH (1:1000, Millipore Bioscience Research Reagents), mouse anti-c-Myc (1:50, EMD Biosciences), mouse anti-JL8 GFP (1:2000, Clontech Laboratories). Subsequently, membranes were incubated for 1 h with a horseradish peroxidase (HRP)-conjugated secondary antibody using the following antibodies: Sheep-anti-mouse IgG (1:10,000, Jackson ImmunoResearch Laboratories) or Donkey-anti-rabbit IgG-HRP (1:50,000, GE Healthcare). HRP-antibody signals were detected with enhanced chemiluminescence reagents, ECL or ECL+ (GE Healthcare), and membranes were exposed to Biomax film (Kodak Bioscience). In some experiments, blots were stripped in a Tris buffer containing 2% SDS and 0.7% β–mercaptoethanol, blocked and reprobed as described.
Preparation of cerebellar cell cultures, immunostaining, retroviral infection, and nucleofection.
Suspensions of cerebellar cells, purified granule cell progenitors (GCPs) and glial cells were prepared from P5–P7 C57BL/6J mice (Jackson Laboratory) as described previously (Hatten, 1985). For immunostaining experiments, the cells were plated in 16 well LabTek glass tissue culture chamber slides (Nalge Nunc) coated with low concentrations of poly-d-lysine (0.001–0.01 mg/ml) at a density of 175,000–225,000 cells/well. In some experiments, expression vectors encoding fluorescently labeled ASTN1, ASTN2 or clathrin light chain proteins were introduced into GCPs by retroviral infection or nucleofection. For retroviral constructs, cells were infected overnight with 0.5 ml of pCX retrovirus containing Venus. For nucleofection, we used the Amaxa Mouse Neuron Nucleofection kit (Lonza) as per the manufacturer's instructions and program A30 to deliver 30 μg of pCIG-Astn1-Venus and 30 μg of pCIG-Astn2-mCherry. For live cell imaging experiments, we plated cells electroporated with ASTN1-Venus and ASTN2-mCherry at a final cell density of 106 cells/dish in MatTek dishes pretreated with low concentrations of poly-d-lysine (0.001–0.01 mg/ml) as described previously (Edmondson and Hatten, 1987). In control experiments, we immunostained cultures with antibodies to GFAP to confirm that neurons were migrating on a glial substrate. Images were acquired using a Zeiss Axiovert 200M microscope, equipped with a PerkinElmer Wallac UltraView spinning disc confocal scan head (PerkinElmer) and Orca ER cooled CCD camera (Hamamatsu Photonics). Cells were imaged for 20–80 min and 9 μm z-stacks (7 sections per stack) were taken every 1–3.25 min. Images were processed and analyzed using MetaMorph software (Universal Imaging Corp.).
Inhibition of endocytosis with Dynasore, a soluble noncompetitive inhibitor of Dynamin.
To test the effect of Dynasore (Sigma-Aldrich) on granule cell migration in vitro, we plated cells infected with 0.5 ml of pCX Venus retrovirus suspension for 48 h. After imaging neuronal migration for 1 h, cells were treated with dimethyl sulfoxide [DMSO, 100 μl in 100 ml of BME supplemented as described previously (Solecki et al., 2004) (Sigma-Aldrich)] or Dynasore [80 μm final concentration; 80 mm stock solution prepared in DMSO and diluted 1:1000 in BME supplemented as described previously (Solecki et al., 2004)] for 30 min (35.5°C), after which images of migrating neurons were acquired at a time lapse of 5 min [5 μm z-stacks (3–6 sections per stack)], using a Zeiss microscope fitted with a PerkinElmer Ultraview Vox spinning disc confocal laser scan head (PerkinElmer) and Volocity acquisition software (PerkinElmer). To remove the Dynasore, we washed the cells three times with granule cell medium (35.5°C), and imaged migrating neurons as above for 1 h.
To examine the effect of Dynasore on granule cell migration in vivo, we prepared organotypic slice cultures as previously described (Solecki et al., 2001). To label a subset of the GCP population with a fluorescent marker, we introduced Venus by retroviral infection, incubating the slice cultures with pCX-Venus retroviral supernatant (0.5 ml/well) overnight. Twenty-four hours later, we added Dynasore (40 or 80 μm final concentration as above) to some of the cultures and incubated the slices 48 h (35.5°C). To assay the effect of Dynasore on GCP migration, the organotypic slice cultures were processed for immunohistochemistry with antibodies against EGFP and TUJ1, as described previously (Solecki et al., 2001) and with DRAQ5 (Cell Signaling Technology) to visualize nuclei.
Apoptosis assay.
To assay for apoptosis, organotypic slice cultures were incubated with the pCX retrovirus containing Venus cDNA and treated with DMSO or with DMSO and Dynasore (40 or 80 μm final concentration) as above, then were processed for immunohistochemistry as described below and stained with sheep anti-GFP and rabbit anti-caspase 3 antibodies (Lossi et al., 1998). The number of cells that were double-labeled with anti-GFP and anti-caspase 3 antibodies were counted per unit area (Adams et al., 2002).
Immunocytochemistry.
Unless otherwise noted, cultured cells and organotypic slice cultures were fixed with 4% paraformaldehyde, blocked in 10% normal goat serum/PBS, and permeabilized in 0.05–0.1% Triton X-100/PBS. Primary antibodies were used at the following concentrations: rabbit anti-GFP antibodies (1:2000, Invitrogen), sheep anti-GFP antibodies (1:150, Biogenesis UK), mouse anti-TUJ1 antibodies (1:1000), rabbit anti-ASTN2 (1:200, Covance), rabbit anti-ASTN1 (1:200, Covance), mouse anti-c-Myc (1:50, EMD Biosciences), rabbit anti-caspase 3 (1:500, R&D Systems), mouse anti-TAG-1 IgM (1:2, 4D7), mouse anti-GFAP (1:500, Sigma). Secondary antibodies (Invitrogen): goat anti-rabbit IgG-488 Alexa Fluor, goat anti-rabbit IgG-555 Alexa Fluor, goat anti-mouse IgG-488 Alexa Fluor, goat anti-mouse IgG-555 Alexa Fluor, goat anti-chicken IgG-488 Alexa Fluor, were used at 1:500. DRAQ5 was used per the manufacturer's instructions. For live cell staining, we washed transfected HEK293T cell cultures once with warm culture media and immunostained with rabbit anti-GFP antibodies (1:2000, Invitrogen) for 10 min at 37°C + 5% CO2. Cultures were then washed with cold media, fixed with 4% paraformaldehyde on ice for 15 min to prevent antibody internalization, and incubated with secondary antibodies. Slides were imaged using a Radiance 2000 confocal laser-scanning microscope (Carl Zeiss).
Flow cytometry.
For flow cytometry experiments, HEK293T cell cultures were harvested in 1 mm EDTA-PBS 48 h after transfection. To visualize surface ASTN1 or ASTN2, we performed live cell staining with rabbit anti-GFP antibodies (1:20,000 in culture medium, Invitrogen) for 10 min at 37°C + 5% CO2, washed the cells with cold culture media, cold PBS containing 10% normal goat serum (NGS), and then incubated in a single cell suspension with goat anti-rabbit IgG-647 Alexa Fluor (1:5000, Invitrogen) for 25 min at 4°C. After washing with PBS containing 10% NGS at 4°C, the cells were resuspended in 1 ml of PBS. Propidium iodide (100 ng/ml, Sigma-Aldrich) was added for dead cell exclusion and cells were incubated on ice for 15 min before flow cytometry.
Flow cytometry analysis was performed on the FACSort Analyzer (BD Biosciences) using CellQuest Pro 5.2 software (BD Biosciences). Detectors, compensation levels, and acquisition gates were set based on nontransfected and single color controls and a total of 20,000 cells were analyzed per condition. The resulting data were analyzed using FlowJo 8.3.3 (Tree Star Inc.). The analysis included a gating for live cells (propidium iodide-negative) and cell size, and the resulting cell population was presented in a dot plot as FL4 (Alexa 647 signal on y-axis) versus FL1 (Venus/EYFP signal on x-axis). Quadrants were fitted based on background signal and identical quadrants were applied to each condition in the experiment to allow for comparisons. The lower right quadrant values represent single/transfection (Venus/EYFP)-positive cells, whereas upper right quadrant values represent double-positive/surface labeled cells (Venus/EYFP expression positive and Alexa 647 live stain positive) in each experimental condition. In addition, results were graphically displayed as the average percentage of surface labeled cells.
Results
Bioinformatics, identification and cloning of Astn2, and sequence comparison with Astn1
To identify genes related to Mus musculus (mouse) Astn1, full-length cDNA sequence of Astn1 and its translated protein sequences were used as queries to BLAST search the NIH mouse and Homo sapiens (human) cDNA databases. Identified mouse sequences were confirmed in the Celera Genomics Database. Like Astn1, Astn2 is a vertebrate-specific gene, with putative homologs in Homo sapiens (human), Pan troglodytes (chimp), and Canis familiaris (dog). In mouse, Astn2 is located in the qC1 region of chromosome 4 (UCSC Genome Browser, http://genome.ucsc.edu), contains 24 exons, has two splice forms, and spans 1023.681 kb (Fig. 1 A). RT-PCR analysis demonstrates that both Astn2 splice forms are detected in cerebellar granule neurons (data not shown).
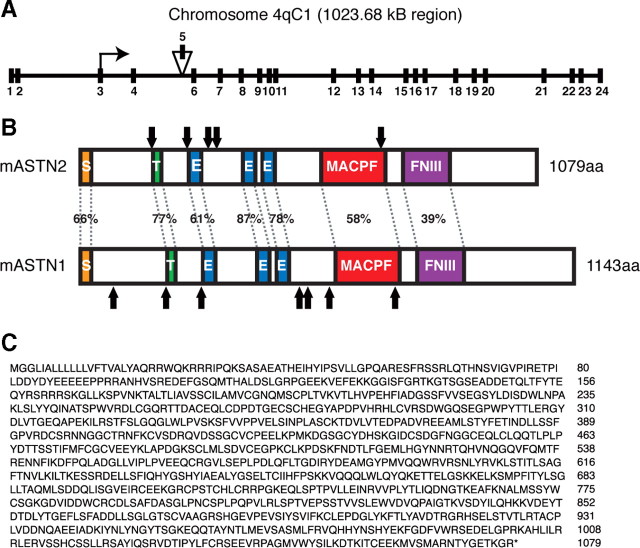
Figure 1.
Astn2 genomic and protein structure. A, Schematic showing genomic structure of Astn2 on mouse chromosome 4qC1. Astn2 contains 24 exons and spans 1023.68 kb. Isoform a lacks exon 5 (156 nt). B, Schematic illustrating homology between mouse ASTN2 and ASTN1, arrows indicate N-linked glycosylation sites. ASTN2 contains a signal sequence (S), a transmembrane domain (T), three EGF repeats (E), a MAC/Perforin domain (MACPF), and a fibronectin III domain (FNIII). C, ASTN2 isoform a amino acid sequence.
The mouse EST database contained a partial sequence with the first three EGF repeats of Astn2 and a P7 mouse brain cDNA library screen generated a partial cDNA. A search of the TIGR Gene Indices (now the Dana Farber Gene Index Project, http://compbio.dfci.harvard.edu/tgi/) identified additional upstream gene sequence (∼750 nt), and the upstream region of the Astn2 cDNA was generated by 5′RACE and PCR walking using E17 brain cDNA as a template. The newly identified Astn2 sequence yielded a longer open reading frame containing a putative start site that would produce a protein of 1079 aa (Fig. 1 C). Although additional upstream methionines were identified, we concluded that the methionine indicated is the functional start site based on the location of a conserved signal sequence (Fig. 1 B), the size of the ASTN2 protein detected in cerebellum by Western blot analysis using an antibody specific to ASTN2, as well as the ability to in vitro translate a protein of this size, using this methionine (data not shown). Using this start site, there are two isoforms of Astn2, a larger splice variant, isoform b, that expresses a protein of 1132 aa, and a shorter variant, isoform a, that lacks exon 5 (Fig. 1 A) and expresses a protein of 1079 aa. In this study, isoform a was used, as the alternatively spliced exon did not contain any conserved domains. Sequence analysis revealed that the ASTN2 protein contains a signal sequence, transmembrane domain, three epidermal growth factor-like (EGF) repeats, a MAC/Perforin (MACPF, MP) domain, a fibronectin type III (FNIII, FN) domain, and five N-linked glycosylation sites (Fig. 1 B), similar to ASTN1. ASTN1 and ASTN2 are 48% identical and 65% homologous overall, with individual domain homologies ranging from 39 to 87% (Fig. 1 B).
Astn2 is expressed in neurons in the developing and adult brain
Northern blot and in situ analyses demonstrate that Astn2 is expressed in embryonic, postnatal and adult brain (Fig. 2). Notably, Astn2 is most highly expressed in the cerebellum at each time point. In situ analysis illustrates that Astn2 is expressed primarily in the cerebellum, but is also expressed in the cortex, olfactory bulb, hindbrain, and dentate gyrus of the hippocampus at P6, P10, and adult stages (Fig. 2 B,E,H). Between P6 and P10, when granule cell migration occurs in the cerebellum, Astn2 is expressed in granule cell progenitors (GCPs) in the external germinal layer (EGL), in GCPs migrating along Bergmann glial cells in the molecular layer, in the internal granule layer (IGL), and in Purkinje neurons (Fig. 2 C,F,N). In adult mouse cerebellum, Astn2 transcripts are detected in postmitotic neurons in the adult cerebellum (Fig. 2 I). Radioactive in situ hybridization demonstrates that the expression level of Astn1 is significantly lower than that of Astn2 in the developing cerebellum (supplemental Fig. 1, available at www.jneurosci.org as supplemental material).
Figure 2.
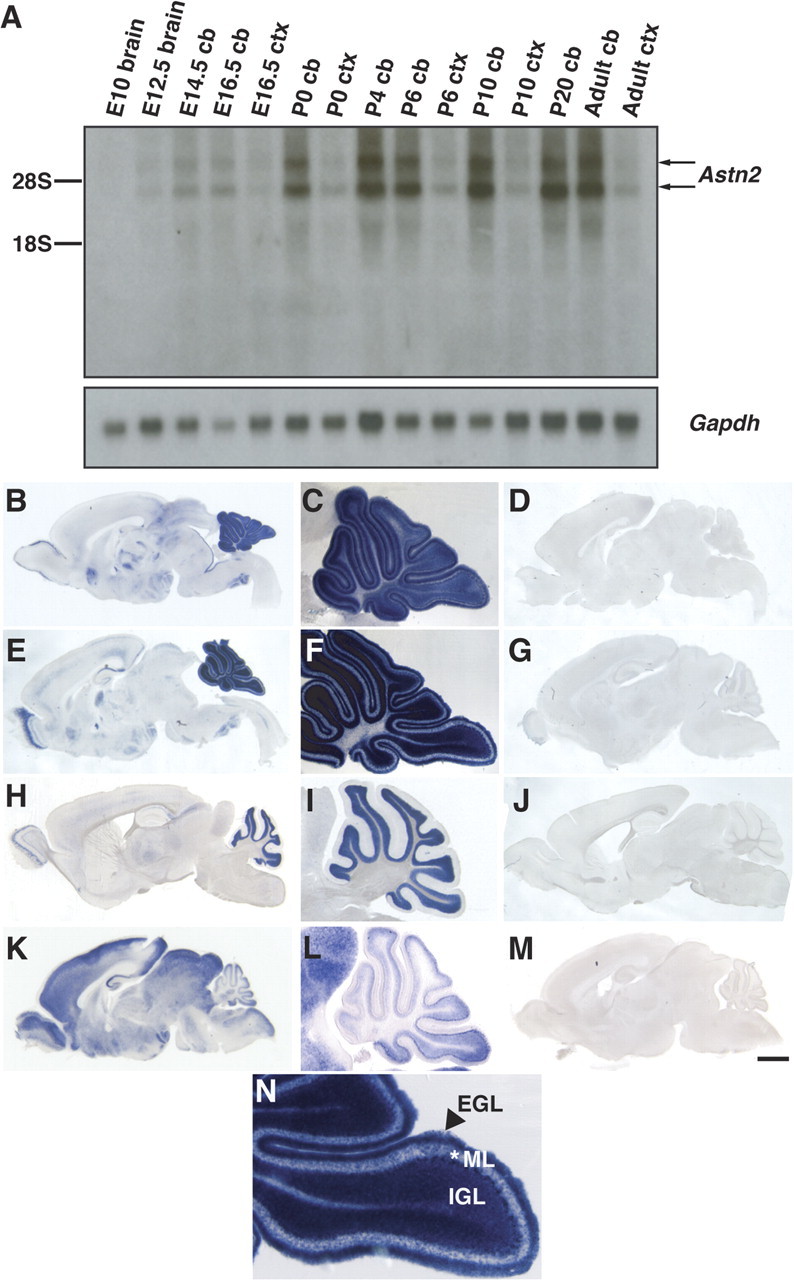
Astn2 is expressed in the developing and adult brain. A, Northern blot of Astn2 from brain, cerebellum (cb), or cortex (ctx) RNA. Astn2 expression is enriched in the cerebellum; Gapdh control is shown in the bottom panel. Northern blotting reveals that there are two Astn2 transcripts (4.2 and 5.8 kb), which is likely due to alternative splicing in an untranslated region. B–J, N, In situ hybridization of sagittal brain sections reveals that Astn2 is expressed in the cerebellum, cortex, olfactory bulb, hindbrain, and hippocampus at P6 (B), P10 (E), and adult (H) stages. Magnification (5×) of the cerebellum reveals that at P6 (C) and P10 (F, N), Astn2 is expressed in granule neurons in the EGL, IGL, and the molecular layer (ML), and Purkinje neurons. At adult stages (H, I), Astn2 is localized to Purkinje neurons and granule neurons in the IGL. K–M, In situ hybridization of Astn1 at P6. D, G, J, M, Corresponding sense controls. Scale bar: (in M) B, D, K, M, 1000 μm; C, F, I, L, 400 μm; E, G, 1200 μm; H, J, 1500 μm; N, 200 μm.
An anti-ASTN2-specific antibody was generated to assay protein expression (supplemental Fig. 2, available at www.jneurosci.org as supplemental material). Western blot analysis of mouse brain and cerebellar lysates at different developmental stages showed low levels of ASTN2 expression during embryonic developmental stages and high levels of expression during the first 2 weeks postnatal, the stage of cerebellar development when granule neurons undergo glial-guided migration (Fig. 3 A). In addition, low levels of ASTN2 were observed in lysates of adult mouse brain. Among tissue lysates we analyzed at P7, ASTN2 levels were highest in brain, eye, heart, and lung, with low levels of protein detected in the liver and kidney, and minimal levels detected in the spleen and skin (data not shown). Thus, the spatiotemporal pattern of ASTN2 is developmentally regulated, with peak levels of ASTN2 protein expression in postnatal brain.
Figure 3.
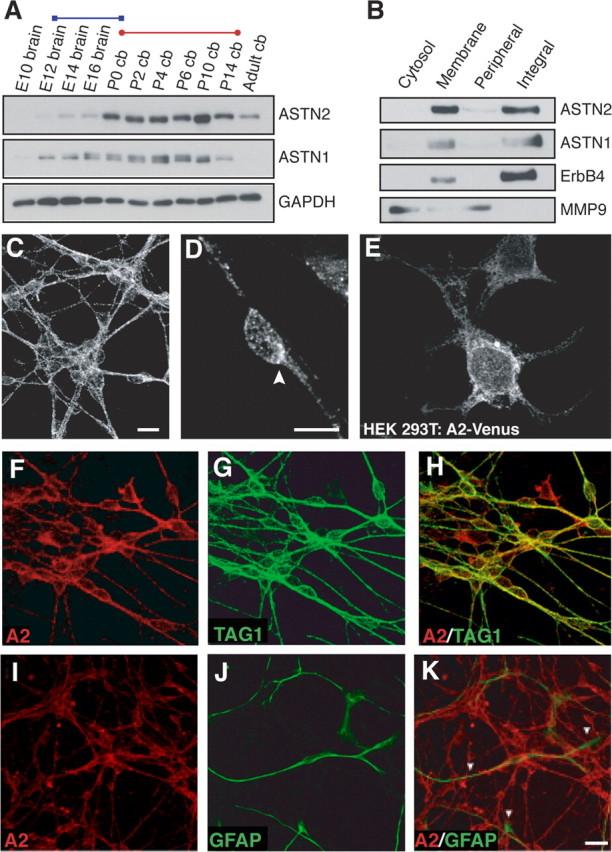
ASTN2 is an integral membrane protein expressed in cerebellar neurons. A, Expression of ASTN2 and ASTN1 in the brain is developmentally regulated. ASTN2 protein is expressed in the embryonic, postnatal, and adult brain, and peak expression occurs during the radial migration of the granule neurons into the IGL (red bar), whereas ASTN1 expression overlaps with both the tangential migration of the granule cell precursors in the cerebellar anlage (blue bar) and the migration of granule neurons (red bar). GAPDH immunostaining was performed as a loading control. B, Cell fractionation experiments show that ASTN2 and ASTN1 are enriched in membranes and are integral membrane proteins. Immunoblotting was performed for integral membrane (ErbB4) and peripheral membrane (MMP9) proteins as controls. C, D, Immunocytochemistry reveals ASTN2 is localized in a punctate pattern in cerebellar granule neurons with an accumulation of protein at one pole of the cell (arrowhead in D). E, Venus-tagged ASTN2 (A2-Venus) is localized in a punctate pattern in HEK293T cells. F–K, Immunocytochemistry shows that ASTN2 (A2) is expressed in cerebellar granule neurons, but not astroglia. The anti-ASTN2 antibody double labels Tag-1-positive cerebellar granule neurons (H), but not GFAP-positive astroglia (K). Scale bars: (in C) C, E–H, 10 μm; (in D) D, 5 μm; (in K) I–K, 20 μm. cb, Cerebellum.
ASTN2 is an integral membrane protein expressed in cerebellar granule neurons
To examine whether ASTN2, like ASTN1, is a membrane protein, we used differential centrifugation to separate plasma membrane and organelle fractions from lysates of purified cerebellar granule cells and examined the amount of ASTN1 and ASTN2 in each fraction by Western blot analysis. Cell fractionation experiments demonstrate that ASTN1 and ASTN2 are enriched in membranes and fractionate with integral membrane proteins (Fig. 3 B). Thus, ASTN2 like ASTN1 is apparently an integral membrane protein.
Immunostaining of cerebellar granule neurons in vitro with anti-ASTN2 antibodies revealed a punctate staining pattern along cell membranes and in the cytoplasm (Fig. 3 C,D). Staining was evident in the cell soma and along neuronal processes with an accumulation of ASTN2 protein frequently appearing at one pole of the cell (Fig. 3 D, arrowhead) in neurons with a migratory profile. A punctate distribution of ASTN2 was also seen in HEK293T cells that over expressed Venus-tagged ASTN2 (Fig. 3 E). In contrast to polarized, migrating granule cells, we did not observe a concentrated zone of ASTN2 in HEK293T cells. These findings suggest that ASTN2 is expressed on cell membranes and in cytoplasmic vesicles and polarizes in the direction of movement in migrating neurons.
To examine whether ASTN2 is expressed in neurons and glial cells, we immunostained cultures of cerebellar granule cells and glia with antibodies against ASTN2. In cocultures of cerebellar granule neurons and astroglial cells, anti-ASTN2 antibody labeled granule neurons, identified by their expression of the granule cell marker TAG-1 (Fig. 3 F–H), but not glial cells, identified by their expression of the glial marker GFAP (Fig. 3 I–K). Similar results were obtained by immunostaining tissue sections of P6 mouse cerebellum with antibodies against ASTN2 and GFAP (data not shown). These results were confirmed by RT-PCR analysis of purified cerebellar granule neurons and purified glia (data not shown). Together, these results show that ASTN2 is a developmentally regulated neuronal membrane protein that is highly expressed in granule neurons of the cerebellum.
Interaction of ASTN1 and ASTN2
The overlapping spatiotemporal pattern of expression of ASTN1 and ASTN2 raised the possibility that the two proteins interact. To assay whether ASTN1 binds ASTN2, we performed coimmunoprecipitation experiments in HEK293T cells with tagged versions of the ASTN proteins. Venus-tagged ASTN2 coimmunoprecipitated with Myc-tagged ASTN1 (Fig. 4 B, top panel, left lane), indicating that ASTN1 interacts with ASTN2 in vitro. To define the domain of ASTN2 that binds ASTN1, we performed coimmunoprecipitation experiments with ASTN2 domain deletion constructs (Fig. 4 A). The ASTN2-ΔEGF-Venus, ASTN2-ΔMP-Venus, and ASTN2-ΔFN-Venus deletion proteins all coimmunoprecipitate with Myc-tagged ASTN1, suggesting that ASTN1:ASTN2 binding does not involve a single conserved domain of the ASTN2 protein (Fig. 4 B).
Figure 4.
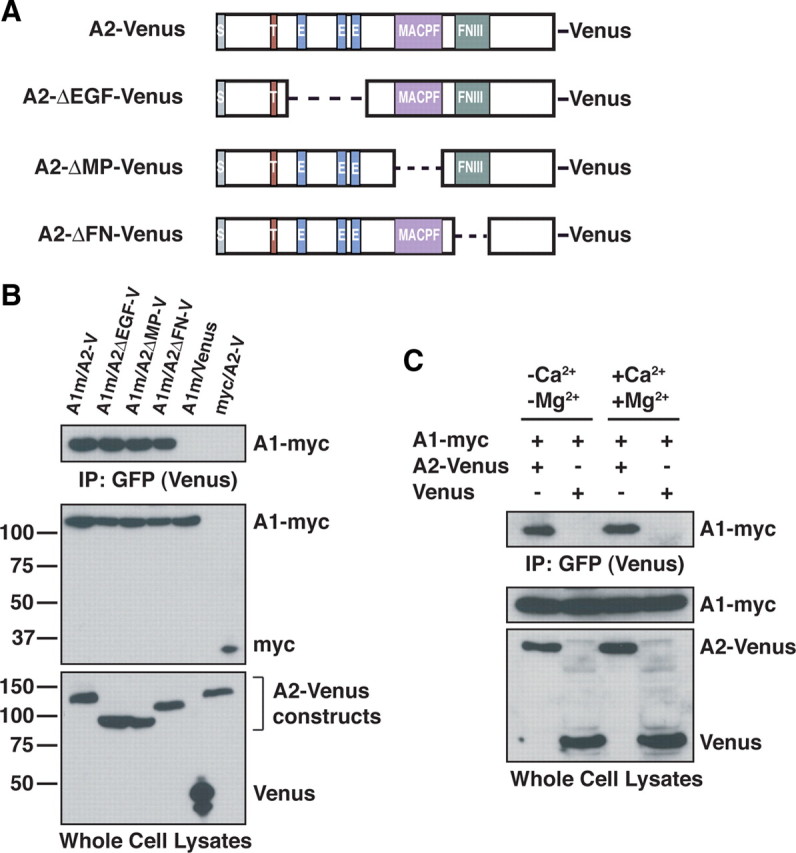
ASTN2 interacts with ASTN1. A, Schematic representation of Venus-tagged ASTN2 domain deletions. Dashed lines represent deleted regions. B, Coimmunoprecipitation reveals Venus-tagged ASTN2 (A2-V and A2-Venus) interacts with Myc-tagged ASTN1 (A1m and A1-myc). Coimmunoprecipitation with Venus-tagged ASTN2 constructs lacking the EGF (A2ΔEGF-V), MACPF (A2ΔMP-V), and FNIII (A2ΔFN-V) domains show that the ASTN2/ASTN1 interaction does not depend on any individual conserved domain. Anti-GFP antibody was used to immunoprecipitate and immunoblot Venus-tagged proteins. Anti-Myc and Anti-GFP (to recognize Venus) antibodies were used for immunoblotting. C, Coimmunoprecipitation demonstrates that the ASTN1/ASTN2 interaction is calcium independent.
In prior studies, we showed that ASTN1-mediated binding of granule cell membranes to glial cells is calcium independent (Stitt and Hatten, 1990). To determine whether the binding of ASTN1 to ASTN2 is calcium dependent, we performed coimmunoprecipitations of ectopically expressed Myc-tagged ASTN1 and Venus-tagged ASTN2 in HEK293T cells in the presence and absence of calcium and magnesium. The results of these experiments demonstrate that the interaction between ASTN1 and ASTN2 is calcium independent (Fig. 4 C).
ASTN1 and ASTN2 colocalize with endosomal markers and Clathrin light chain
To examine whether ASTN proteins colocalize with endosomes, we coexpressed ASTN1 and ASTN2-mCherry with GFP-Endo (RhoB fusion), a marker of early and late endosomes, in HEK293T cells and visualized expression of transfected proteins by indirect immunofluorescence (ASTN1 and GFP-Endo) or by direct visualization of the fluorophore-conjugated fusion protein (ASTN2-mCherry). In single, optical sections of fixed cells, both ASTN1 and ASTN2 are expressed in a subset of RhoB-positive endosomes, as well as in vesicles that are not RhoB positive (Fig. 5 A). Coexpression of ASTN1 and ASTN2 with GFP-Endo did not alter colocalization of either protein with RhoB-positive endosomes (data not shown).
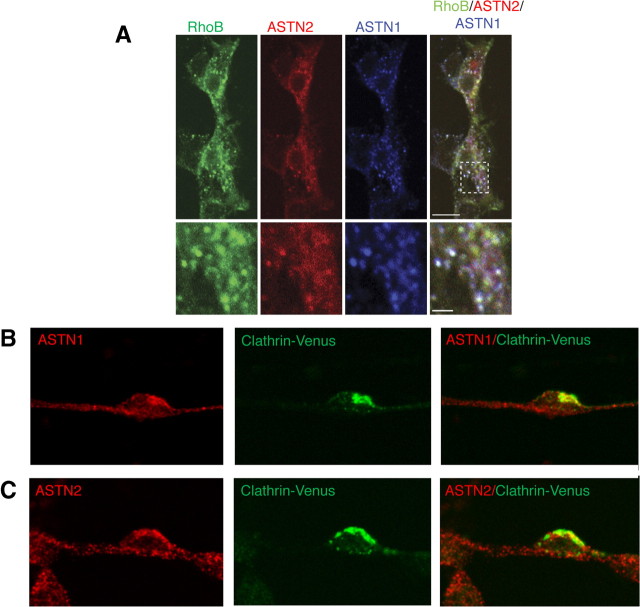
Figure 5.
ASTN1 and ASTN2 colocalize with endosomal markers. A, Coexpression of ASTN1 and ASTN2-mCherry with the endosomal marker GFP-Endo (RhoB) in HEK293T cells demonstrates that the ASTN proteins partially colocalize with early and late endosomes. B, C, Endogenous ASTN1 and ASTN2 proteins colocalize with Venus-tagged Clathrin light chain (Clathrin-Venus, expressed via retrovirus infection) in migrating cerebellar granule neurons. Images shown represent single optical confocal sections. Scale bars: (in A) A, 10 μm; B, C, 5 μm.
Having demonstrated that ectopically expressed ASTN proteins localize to endosomes in transfected cells, we next investigated whether endogenous ASTN1 and ASTN2 localize to endosomes in cerebellar granule neurons. To examine whether ASTN1 and ASTN2 colocalize with coated vesicles in living granule neurons, we used retroviral constructs to express Venus-tagged Clathrin light chain and immunostained with anti-ASTN1 or anti-ASTN2 antibodies to localize endogenous ASTN proteins. By confocal microscopy, ASTN1 and ASTN2 colocalize with Clathrin light chain-Venus in migrating cerebellar granule cells (Fig. 5 B,C). We observed a speckled localization pattern for the two ASTN proteins and Clathrin light chain-Venus, with an accumulation of all three proteins in the base of the leading process and in the edge of the migrating neuron opposite the interstitial adhesion junction with the glial fiber (data not shown).
Trafficking of ASTN1 and ASTN2 in migrating cerebellar granule cells
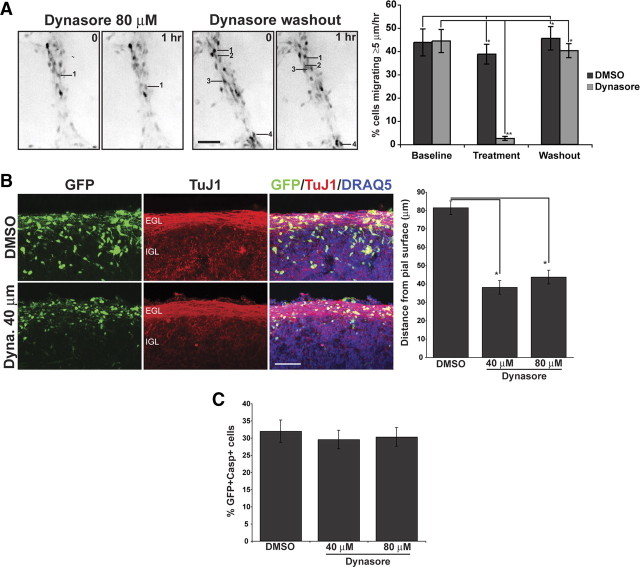
The colocalization of ASTN1 and ASTN2 with Clathrin light chain in migrating granule neurons suggested a role for receptor trafficking in neuronal migration. As clathrin-dependent endocytosis requires the GTPase Dynamin, we used a soluble noncompetitive inhibitor of Dynamin, Dynasore (Macia et al., 2006), to determine whether receptor trafficking plays a role in glial-guided neuronal migration. In vitro treatment of migrating granule cell precursors with Dynasore rapidly decreased the percentage of migrating granule neurons over the course of an hour, with complete recovery within 1 h of washout of the inhibitor (Fig. 6 A). Similar results were obtained when we treated acute organotypic cultures of P6 cerebellum. Treatment with Dynasore for 48 h significantly inhibited migration of cerebellar granule neurons from the EGL into the IGL (Fig. 6 B). Immunostaining with anti-GFP and anti-caspase 3 antibodies did not reveal a change in the number of caspase 3-positive cells in organotypic slices treated with Dynasore (Fig. 6 C) suggesting that the inhibitory effect of Dynasore on glial-guided migration was not due to increased cell death. These studies suggest that inhibiting receptor trafficking by treatment with Dynasore rapidly and reversibly inhibited granule neuron migration on glial fibers.
Figure 6.
The Dynamin inhibitor Dynasore inhibits glial-guided neuronal migration. A, A granule cell culture migration assay demonstrates that the effects of a soluble noncompetitive inhibitor of Dynamin, Dynasore, on neuronal migration are reversible. Dissociated cerebellar granule neurons, labeled with a Venus encoding retrovirus, were imaged for three 60 min periods; vehicle (DMSO) or DMSO and either 40 or 80 μm Dynasore were added during the second imaging period. To remove the Dynasore, we washed the cells three times with granule cell medium, and imaged migrating neurons as above for 1 h. The images in A are inverted for better resolution and show migrating granule neurons labeled with Venus. In this study, we selected neurons with the features of actively migrating neurons, i.e., an elongated cell soma that was apposed to and flattened against Bergmann-like glial fibers. Migrating neurons are labeled by number in the 0 h panel and their final position is indicated by the same number in the 1 h panel. The number of cells that migrated >5 μm/h was counted and expressed as a percentage of total labeled cells.(*p <0.001, **p <0.0001).B, Cerebellar slices transfected with Venus-expressing retrovirus were treated with Dynasore (Dyna.) at 40 or 80 μm concentration, or with control vehicle (DMSO). TuJ1 staining reveals the parallel fiber layer of differentiated granule neurons in the inner EGL. In slices incubated with Dynasore, Venus-expressing neurons seem to arrest in the EGL compared with DMSO control cells. DRAQ5 was used to counterstain nuclei. Quantitation of neurons in organotypic cerebellar slices shows that neurons remain significantly closer to the pial surface in the presence of 40 or 80 μm Dynasore, relative to controls (*p < 0.001). C, In slices double immunostained with antibodies to GFP and caspase 3, 48 h incubation with Dynasore did not significantly alter the percentage of GFP-positive cells that also were caspase 3 positive. Significance was assessed via Student's t test; values are mean ± SEM (*p < 0.001). Scale bars, 50 μm.
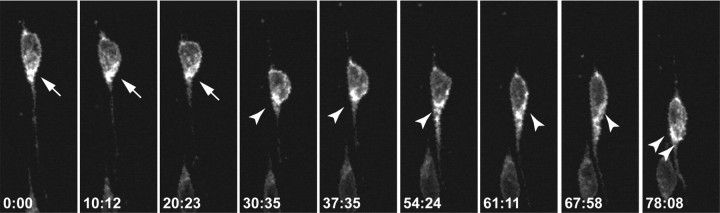
To visualize the dynamics of ASTN1 in live, migrating neurons, we expressed ASTN1-Venus in cerebellar granule cells in vitro. Live imaging revealed dynamic changes in the localization of ASTN1-Venus protein during migration. As the neuron moved, ASTN1-Venus accumulated at the anterior pole of the neuronal soma and in the base of the leading process during migration (Fig. 7, supplemental Movie 1, available at www.jneurosci.org as supplemental material). When the neuron-glial adhesion site is released, just before forward movement of the neuron along the glial fiber, ASTN1-Venus accumulates in the base of the leading process (Fig. 7, arrows). As the neuron moves forward, ASTN1-Venus moves into the leading process (Fig. 7, arrowheads). The forward flow of ASTN1-Venus protein down the leading process continues during the stationary phase of the movement cycle, resulting in an accumulation of ASTN1-Venus at the front of the neuronal soma (double arrowheads) after the cell soma glides forward on the glial guide. The localization of ASTN1-Venus at the front of the cell soma is consistent with the formation of a new adhesion site between the neuron and glial fiber.
Figure 7.
Live imaging of ASTN1-Venus dynamics in a migrating cerebellar granule neuron. Purified granule neurons were electroporated with Venus-tagged ASTN1 and mCherry-tagged ASTN2, and Venus-tagged ASTN1 was imaged throughout a migration cycle. Live imaging revealed dynamic changes in the localization of ASTN1-Venus protein during migration. As the neuron moved, ASTN1-Venus accumulated at the anterior pole of the neuronal soma and in the base of the leading process during migration (arrow). At the beginning of the migration cycle, when the neuron adheres to the glial fiber, ASTN1-Venus levels were high in the base of the cell soma. The forward flow of ASTN1-Venus protein continued during the stationary phase of the movement cycle, resulting in an accumulation of ASTN1-Venus at the front of the neuronal soma. In the third step of the cycle, with the release of the interstitial junction, ASTN1-Venus accumulated in the proximal domain of the leading process where a new adhesion site will form (double arrowheads).
ASTN2 regulates the surface localization of ASTN1
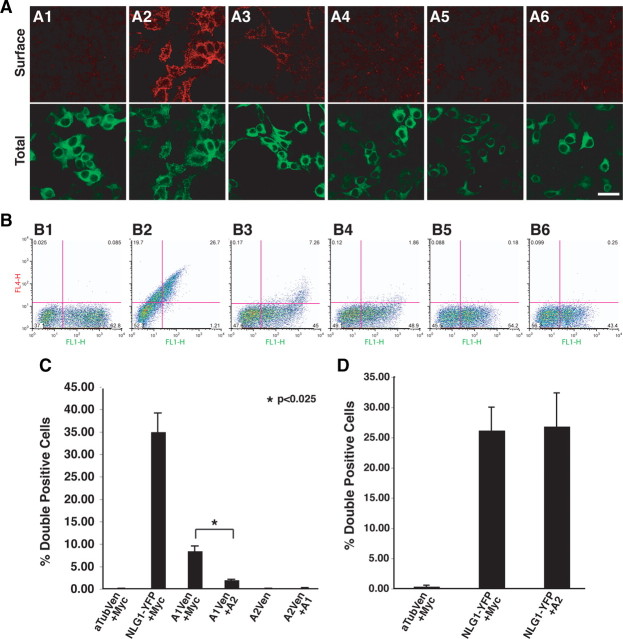
Previous studies illustrated that ASTN1 is exposed on the cell surface of cerebellar granule neurons (Fishell and Hatten, 1991; Zheng et al., 1996), and functions in the formation of adherence junctions during cerebellar granule cell migration along Bergmann glial fibers. To localize the C terminus of ASTN1 and of ASTN2, we transfected HEK293T cells with Venus-tagged α-tubulin (Fig. 8 A1), a cytoplasmic protein, neuroligin-1-EYFP (NLG1-EYFP), an integral membrane protein (Fig. 8 A2) (Scheiffele et al., 2000), C-terminal Venus-tagged ASTN1 (Fig. 8 A3) and C-terminal Venus-tagged ASTN2 (Fig. 8 A5). To visualize surface expression of these fluorescently tagged proteins, we used a live staining protocol incubating the cells with anti-GFP antibodies before fixation, and with an Alexa Fluor 555-conjugated secondary antibody after fixation (Fig. 8 A, top row). In addition to visualizing the surface expression of the fusion proteins by indirect immunofluorescence, we also directly imaged fluorescently tagged α-tubulin, NLG1, ASTN1 and ASTN2 in fixed cells (Fig. 8 A, bottom row). These experiments showed surface expression of NLG1 (Fig. 8 A2) and the C terminus of ASTN1 (Fig. 8 A3), but not of α-Tubulin (Fig. 8 A1) or the C terminus of ASTN2 (Fig. 8 A5). Coexpression of ASTN1 did not lead to an increase in surface localization of ASTN2 (Fig. 8 A6).
Figure 8.
Coexpression with ASTN2 reduces the cell surface localization of ASTN1. HEK293T cells were transfected with a Venus or EYFP fusion protein, and a second construct (Myc, ASTN2, or ASTN1). A, The top row shows a live cell stain using a rabbit anti-GFP primary and an Alexa Fluor 555-conjugated secondary antibody indicating expression of the Venus/EYFP-conjugated protein on the surface of the cell; while the bottom row shows the total cell expression of the fusion protein. A1, α-Tubulin-Venus and Myc; A2, neuroligin-EYFP and Myc; A3, ASTN1-Venus and Myc; A4, ASTN1-Venus and ASTN2; A5, ASTN2-Venus and Myc; A6, ASTN2-Venus and ASTN1. In the absence of ASTN2, Venus-tagged ASTN1 localizes on the cell surface (A3). Live cell surface staining of transfected HEK293T cells demonstrates that Venus-tagged ASTN1 is no longer exposed on the cell surface when coexpressed with ASTN2 (A4). These experiments also demonstrated that ASTN2-Venus is not exposed on the cell surface in the absence (A5, B5) or presence (A6, B6) of ASTN1. B, For the flow cytometry experiments, an AlexaFluor-647 secondary antibody is substituted for the AlexaFluor-555 antibody, and the data are presented as dot plots of surface labeling with the axes as FL1-H (Venus/EYFP signal on x-axis) versus FL4-H (Alexa 647 signal on y-axis). Lower right quadrant values represent singly labeled Venus/EYFP-positive cells, and upper right quadrant values represent doubly labeled Venus/EYFP and Alexa Fluor 647 live stain-positive cells. B1, α-Tubulin-Venus and Myc; B2, neuroligin-EYFP and Myc; B3, ASTN1-Venus and Myc; B4, ASTN1-Venus and ASTN2; B5, ASTN2-Venus and Myc; B6, ASTN2-Venus and ASTN1. C, Quantitation of surface labeling from B. Average surface labeling from three experiments was calculated and Student's t test was performed to determine significance. Values are mean ± SEM. D, Flow cytometry analysis reveals that coexpression of ASTN2 does not alter the surface localization of EYFP-tagged NLG1. Graphs represents average surface labeling from three experiments and significance was assessed via Student's t test; values are mean ± SEM. αTub, α-Tubulin; A1, ASTN1; A2, ASTN2; Ven, Venus. Scale bar, 20 μm.
We then used flow cytometry to quantitate the amount of ASTN2 exposed on the cell surface in the presence or absence of ASTN1. Flow cytometry analysis revealed that the C terminus of ASTN1 (Fig. 8 B3) and NLG1 (Fig. 8 B2) were expressed on the cell surface of HEK293T cells. In contrast, the C terminus of ASTN2 was not detected on the cell surface of HEK293T cells, in the presence (Fig. 8 B6) or absence of ASTN1 (Fig. 8 B5). Quantitation of the flow cytometry results indicated that in the absence of ASTN1, the C terminus of ASTN2 was detected on the surface of 0.063 ± 0.006% of the cells (Fig. 8 C, mean ± SEM). In the presence of ASTN1, ASTN2 was detected in 0.222 ± 0.066% of the cells (Fig. 8 C, mean ± SEM). Live staining of purified cerebellar granule cells confirmed that endogenous ASTN2 is not exposed on the cell surface (data not shown).
We next examined whether ASTN2 affects the surface localization of ASTN1-Venus in HEK293T cells. Live cell immunostaining revealed a discrete, speckled cell surface-staining pattern in the cells cotransfected with ASTN1-Venus and Myc control vectors (Fig. 8 A3), demonstrating that the C terminus of ASTN1 is exposed on the cell surface. Interestingly, when ASTN1-Venus was coexpressed with ASTN2 (Fig. 8 A4) the intensity of surface labeling decreased to levels that approached those of the negative control (Fig. 8 A1). Quantitation of ASTN1-Venus levels in the presence and absence of ASTN2 by flow cytometry (Fig. 8 B,C) demonstrated that coexpression of ASTN1-Venus with ASTN2 decreased the levels of ASTN1-Venus surface localization fourfold, from 8.457 ± 1.075% of cells with surface labeling to 2.017 ± 0.115% with surface labeling (mean ± SEM). The change in ASTN1-Venus cell surface localization was significant (Student's t test, p = 0.021, n = 3). To examine whether ASTN2 reduces the surface expression of NLG1, we coexpressed ASTN2 and NLG1-EYFP in HEK293T cells and measured surface expression of NLG1-EYFP by flow cytometry. The results of these experiments showed no significant change in the cell surface localization of NLG1-EYFP in cells that coexpressed ASTN2 (Student's t test p = 0.932, n = 3) (Fig. 8 D).
To examine whether the surface levels of ASTN1 depended on endocytosis, we compared the surface localization of ASTN1-Venus in HEK293T cells treated with Dynasore, a soluble noncompetitive inhibitor of Dynamin, with cells treated with DMSO alone (vehicle). The results of these experiments showed that Dynasore treatment increased the number of HEK293T cells that expressed ASTN1-Venus on the cell surface twofold (supplemental Fig. 3, available at www.jneurosci.org as supplemental material). ASTN1 surface expression increased from 6.57 ± 0.04% of control cells treated with DMSO alone to 11.1 ± 0.17% (mean ± SEM) of cells treated with DMSO and Dynasore. These findings support the idea that surface localization of ASTN1-Venus is regulated by clathrin-mediated receptor endocytosis.
Discussion
The present study shows that Astn2 is abundant in cerebellar granule neurons, along with Astn1, at developmental stages when glial-guided migration is ongoing. Surprisingly, biochemical studies and flow cytometry experiments demonstrate that the function of the two highly homologous Astn gene family members is not redundant. ASTN1 is exposed on the cell surface and, as demonstrated previously (Stitt and Hatten, 1990; Zheng et al., 1996), provides a ligand for neuron-glial adhesion during migration. The present study shows that ASTN2 forms a complex with ASTN1, which regulates the surface expression of ASTN1. Live imaging experiments show that forward movement of the neuron involves the cycling of ASTN1-Venus-containing vesicles and accumulation of ASTN1-Venus labeling in the forward aspect of the soma and the proximal aspect of the leading process where new adhesions will form. Together, these findings suggest that receptor trafficking dictates the polarized distribution of ASTN1 on the surface of the migrating neuron in the direction of movement.
The redistribution of ASTN1 from the site of adhesion beneath the cell soma to a forward site in the cell soma and base of the leading process, seen in live imaging of Venus-tagged ASTN1, is consistent with the cyclic formation and release of interstitial adhesions during the cycles of neuronal locomotion. It is also consistent with our earlier studies, using correlated video microscopy and electron microscopy of live, migrating granule cells, which showed the polarization of the Golgi apparatus, centrosome and associated microtubule networks in the direction of migration (Rakic, 1971; Edmondson and Hatten, 1987; Gregory et al., 1988), the movement of vesicles in living, migrating neurons (Edmondson and Hatten, 1987) and coated endo/exocytic figures and coated vesicles along the interstitial densities in migrating neurons (Gregory et al., 1988).
The conclusion that ASTN1 undergoes internalization and recycling during glial-guided migration is consistent with a general role for adhesion receptor internalization and recycling in growth cone motility and cell migration (Kamiguchi et al., 1998; Kamiguchi and Lemmon, 2000; Keleman et al., 2005), and with the recent studies of Rakic suggesting a role for the conserved exocyst protein complex (TerBush et al., 1996) in CNS neuronal migration (Letinic et al., 2009). It is also consistent with cell migrations that involve integrin-based adhesions, as a number of recent studies provide evidence that integrin endocytosis and recycling occurs at the front of a migrating cell (Caswell et al., 2009). Together, these findings support the emerging idea that receptor trafficking provides a mechanism to dictate the polarized distribution of adhesion receptors on the cell surface of migrating cells (Nishimura and Kaibuchi, 2007; Sigismund et al., 2008; Caswell et al., 2009).
Given the high degree of homology between ASTN1 and ASTN2, and the function of ASTN1 as a neuronal ligand for neuron-glial adhesion, we were surprised by the results of live antibody staining experiments showing that the C terminus of ASTN2 is not exposed on the cell surface. The results of the present study indicate that ASTN2 regulates the expression of ASTN1 on the cell surface, which functions in neuron-glial binding and facilitates neuronal migration (Stitt and Hatten, 1990; Fishell and Hatten, 1991; Zheng et al., 1996; Adams et al., 2002). Our results suggest that ASTN2 regulates the surface levels of ASTN1 and raises the possibility that ASTN2 may regulate the surface expression of other proteins. The epithelial membrane protein-2 (EMP2) provides another example of a protein that affects the membrane localization of other proteins. EMP2 associates with β1 integrins, reciprocally regulates the expression α6β1 to α5β1 on the cell surface, and controls the expression of GPI-linked proteins on the cell surface (Wadehra et al., 2002). Although we did not observe an effect of ASTN2 expression on the surface expression levels of the synaptic protein neuroligin-1, it will be important to determine whether ASTN2 interacts with other receptor systems. It will also be of interest to examine whether ASTN1/ASTN2 complexes regulate signaling events, especially downstream signaling pathways that are important for endosomal transport, such as Rho GTPase signaling pathways. Recent studies show that activation of RhoA-ROCK (Rho-associated coiled coil-containing protein kinase) via ENDO180 in endosomes located in the rear of the cell promote the release of adhesions in the back of the cell during migration (Sturge et al., 2006). Our results suggest a key role for ASTN2 in modulating the level of the neuron-glial receptor ASTN1 in the neuronal membrane and raise the question as to whether there are other potential functions for ASTN2.
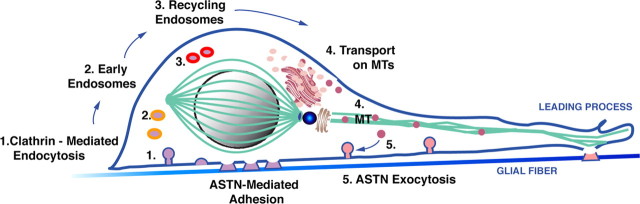
Our results on ASTN1 protein dynamics during migration in live imaging experiments suggest a new model for migration that involves receptor internalization and recycling (Fig. 9). During movement cycles, clathrin-mediated endocytosis releases ASTN-based adhesions beneath the cell soma (step 1) allowing the neuron to glide forward along the glial fiber. The accumulation of ASTN1 we observed at the front edge of the migrating neuron in the proximal portion of the leading process suggests that receptor dynamics dictate the direction of migration, as a new adhesion site (pink) will form in the area in which ASTN1 accumulation is observed. New adhesion sites (pink) are created, in part, by the recycling of ASTN1 receptors from the rear of the cell through intracellular endosomes (orange, bright pink; steps 2–3) transported via microtubules (green; step 4), and exocytosed into the plasma membrane (step 5). This model is supported by our observation that ASTN1 colocalizes with the Clathrin light chain and that treatment of cells with Dynasore, a soluble noncompetitive inhibitor of Dynamin, a large GTPase involved in the scission of membrane vesicles during clathrin-mediated endocytosis, rapidly arrests the migration of immature granule cells in a reversible manner. Elucidating the dependence of ASTN1 internalization and recycling on clathrin-dependent mechanisms, as well as clathrin-independent pathways, and defining mechanisms that regulate ASTN1 trafficking will provide further insight into the control of the saltatory adhesion and locomotion of neurons along glial fibers.
Figure 9.
Model for endocytosis and receptor recycling in glial-guided neuronal migration. Integrated model for neuronal migration demonstrates that Par6α signaling at the neuronal centrosome (blue) functions to maintain integrity of the perinuclear tubulin cage (green) and drives somal translocation. At the beginning of the movement cycle, the ASTN-mediated interstitial adhesion beneath the neuronal cell soma (step 1, purple circles) is disassembled and released via clathrin-mediated endocytosis, after which the neuron glides along the glial fiber. During movement, ASTN receptor recycling continues as ASTN1-containing vesicles traffic through early (step 2, orange circles) and late endosomes (step 3, bright pink circles), through intracellular compartments, including the trans-Golgi network (step 4, pink circles), before being transported along microtubules (green) and inserted onto the surface membrane via exocytosis (step 5, pink circles), to generate new adhesion sites at the front of the migrating neuron.
The present study suggests a unique and surprising role for ASTN2 in regulating surface levels of the neuron-glial receptor ASTN1. It remains to be determined whether ASTN2 regulates the surface expression of other proteins or receptors, whether the endocytosis and trafficking of ASTN1 activates downstream signaling pathways and whether these or other pathways are modulated by ASTN2. Recent human genetic studies associating mutations in Astn2 with neurological deficits such as ADHD (Lesch et al., 2008), autism (Glessner et al., 2009) and schizophrenia (Vrijenhoek et al., 2008) underscore the importance of understanding the role of ASTN2 in brain development and CNS disease.
Footnotes
This work was supported by National Institutes of Health–National Institute of Neurological Disorders and Stroke Grant 2RO1 NS015429-27 (M.E.H.) and National Research Service Award Training Grant GM07524 (P.M.W.). We gratefully acknowledge the critical contributions of Drs. Jee Hae Kim, Toshifumo Tomoda, and Thomas Curran, and thank Drs. David Solecki and Eve-Ellen Govek for critical comments on the manuscript. Dr. Jee Hae Kim generated the anti-ASTN1 antibody used in this work, Dr. Toshifumo Tomoda did the original BLAST searches for Astn2, and Dr. Thomas Curran performed the radioactive in situ analysis of Astn1 and Astn2. Dr. Franck Polleux generously provided pCIG2-iresEGFP and pCIG, Dr. Peter Scheiffele provided pNICE-neuroligin-1-EYFP, and Dr. David Solecki provided pMSCXβ-Venus-α-tubulin and pCX-I-BSR-Venus Clathrin light chain.
References
- Adams NC, Tomoda T, Cooper M, Dietz G, Hatten ME. Mice that lack astrotactin have slowed neuronal migration. Development. 2002;129:965–972. doi: 10.1242/dev.129.4.965. [DOI] [PubMed] [Google Scholar]
- Anton ES, Marchionni MA, Lee KF, Rakic P. Role of GGF/neuregulin signaling in interactions between migrating neurons and radial glia in the developing cerebral cortex. Development. 1997;124:3501–3510. doi: 10.1242/dev.124.18.3501. [DOI] [PubMed] [Google Scholar]
- Barkovich AJ, Kuzniecky RI, Dobyns WB, Jackson GD, Becker LE, Evrard P. Malformations of the cortical development. Neuropediatrics. 1996;27:59–63. doi: 10.1055/s-2007-973750. [DOI] [PubMed] [Google Scholar]
- Bellion A, Baudoin JP, Alvarez C, Bornens M, Métin C. Nucleokinesis in tangentially migrating neurons comprises two alternating phases: forward migration of the Golgi/centrosome associated with centrosome splitting and myosin contraction at the rear. J Neurosci. 2005;25:5691–5699. doi: 10.1523/JNEUROSCI.1030-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belvindrah R, Graus-Porta D, Goebbels S, Nave KA, Müller U. Beta1 integrins in radial glia but not in migrating neurons are essential for the formation of cell layers in the cerebral cortex. J Neurosci. 2007;27:13854–13865. doi: 10.1523/JNEUROSCI.4494-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghesani PR, Peyrin JM, Klein R, Rubin J, Carter AR, Schwartz PM, Luster A, Corfas G, Segal RA. BDNF stimulates migration of cerebellar granule cells. Development. 2002;129:1435–1442. doi: 10.1242/dev.129.6.1435. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Caswell PT, Vadrevu S, Norman JC. Integrins: masters and slaves of endocytic transport. Nat Rev Mol Cell Biol. 2009;10:843–853. doi: 10.1038/nrm2799. [DOI] [PubMed] [Google Scholar]
- Dobyns WB, Truwit CL. Lissencephaly and other malformations of cortical development: 1995 update. Neuropediatrics. 1995;26:132–147. doi: 10.1055/s-2007-979744. [DOI] [PubMed] [Google Scholar]
- Edmondson JC, Hatten ME. Glial-guided granule neuron migration in vitro: a high-resolution time-lapse video microscopic study. J Neurosci. 1987;7:1928–1934. doi: 10.1523/JNEUROSCI.07-06-01928.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmondson JC, Liem RK, Kuster JE, Hatten ME. Astrotactin: a novel neuronal cell surface antigen that mediates neuron-astroglial interactions in cerebellar microcultures. J Cell Biol. 1988;106:505–517. doi: 10.1083/jcb.106.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishell G, Hatten ME. Astrotactin provides a receptor system for CNS neuronal migration. Development. 1991;113:755–765. doi: 10.1242/dev.113.3.755. [DOI] [PubMed] [Google Scholar]
- Glessner JT, Wang K, Cai G, Korvatska O, Kim CE, Wood S, Zhang H, Estes A, Brune CW, Bradfield JP, Imielinski M, Frackelton EC, Reichert J, Crawford EL, Munson J, Sleiman PM, Chiavacci R, Annaiah K, Thomas K, Hou C, et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009;459:569–573. doi: 10.1038/nature07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory WA, Edmondson JC, Hatten ME, Mason CA. Cytology and neuron-glial apposition of migrating cerebellar granule cells in vitro . J Neurosci. 1988;8:1728–1738. doi: 10.1523/JNEUROSCI.08-05-01728.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatten ME. Neuronal regulation of astroglial morphology and proliferation in vitro. J Cell Biol. 1985;100:384–396. doi: 10.1083/jcb.100.2.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatten ME. Central nervous system migration. Annu Rev Neurosci. 1999;22:511–539. doi: 10.1146/annurev.neuro.22.1.511. [DOI] [PubMed] [Google Scholar]
- Kamiguchi H, Lemmon V. Recycling of the cell adhesion molecule L1 in axonal growth cones. J Neurosci. 2000;20:3676–3686. doi: 10.1523/JNEUROSCI.20-10-03676.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiguchi H, Long KE, Pendergast M, Schaefer AW, Rapoport I, Kirchhausen T, Lemmon V. The neural cell adhesion molecule L1 interacts with the AP-2 adaptor and is endocytosed via the clathrin-mediated pathway. J Neurosci. 1998;18:5311–5321. doi: 10.1523/JNEUROSCI.18-14-05311.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keleman K, Ribeiro C, Dickson BJ. Comm function in commissural axon guidance: cell-autonomous sorting of Robo in vivo. Nat Neurosci. 2005;8:156–163. doi: 10.1038/nn1388. [DOI] [PubMed] [Google Scholar]
- Komuro H, Rakic P. Distinct modes of neuronal migration in different domains of developing cerebellar cortex. J Neurosci. 1998;18:1478–1490. doi: 10.1523/JNEUROSCI.18-04-01478.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesch KP, Timmesfeld N, Renner TJ, Halperin R, Röser C, Nguyen TT, Craig DW, Romanos J, Heine M, Meyer J, Freitag C, Warnke A, Romanos M, Schäfer H, Walitza S, Reif A, Stephan DA, Jacob C. Molecular genetics of adult ADHD: converging evidence from genome-wide association and extended pedigree linkage studies. J Neural Transm. 2008;115:1573–1585. doi: 10.1007/s00702-008-0119-3. [DOI] [PubMed] [Google Scholar]
- Letinic K, Sebastian R, Toomre D, Rakic P. Exocyst is involved in polarized cell migration and cerebral cortical development. Proc Natl Acad Sci U S A. 2009;106:11342–11347. doi: 10.1073/pnas.0904244106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lossi L, Zagzag D, Greco MA, Merighi A. Apoptosis of undifferentiated progenitors and granule cell precursors in the postnatal human cerebellar cortex correlates with expression of BCL-2, ICE, and CPP32 proteins. J Comp Neurol. 1998;399:359–372. [PubMed] [Google Scholar]
- Macia E, Ehrlich M, Massol R, Boucrot E, Brunner C, Kirchhausen T. Dynasore, a cell-permeable inhibitor of dynamin. Dev Cell. 2006;10:839–850. doi: 10.1016/j.devcel.2006.04.002. [DOI] [PubMed] [Google Scholar]
- Magdaleno S, Jensen P, Brumwell CL, Seal A, Lehman K, Asbury A, Cheung T, Cornelius T, Batten DM, Eden C, Norland SM, Rice DS, Dosooye N, Shakya S, Mehta P, Curran T. BGEM: an in situ hybridization database of gene expression in the embryonic and adult mouse nervous system. PLoS Biol. 2006;4:e86. doi: 10.1371/journal.pbio.0040086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura T, Kaibuchi K. Numb controls integrin endocytosis for directional cell migration with aPKC and PAR-3. Dev Cell. 2007;13:15–28. doi: 10.1016/j.devcel.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Rakic P. Neuron-glia relationship during granule cell migration in developing cerebellar cortex. A Golgi and electronmicroscopic study in Macacus Rhesus. J Comp Neurol. 1971;141:283–312. doi: 10.1002/cne.901410303. [DOI] [PubMed] [Google Scholar]
- Rakic P. Neuronal migration and contact guidance in the primate telencephalon. Postgrad Med J. 1978;54(Suppl 1):25–40. [PubMed] [Google Scholar]
- Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT, Horwitz AR. Cell migration: integrating signals from front to back. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- Rio C, Rieff HI, Qi P, Khurana TS, Corfas G. Neuregulin and erbB receptors play a critical role in neuronal migration. Neuron. 1997;19:39–50. doi: 10.1016/s0896-6273(00)80346-3. [DOI] [PubMed] [Google Scholar]
- Rivas RJ, Hatten ME. Motility and cytoskeletal organization of migrating cerebellar granule neurons. J Neurosci. 1995;15:981–989. doi: 10.1523/JNEUROSCI.15-02-00981.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross ME, Walsh CA. Human brain malformations and their lessons for neuronal migration. Annu Rev Neurosci. 2001;24:1041–1070. doi: 10.1146/annurev.neuro.24.1.1041. [DOI] [PubMed] [Google Scholar]
- Schaar BT, McConnell SK. Cytoskeletal coordination during neuronal migration. Proc Natl Acad Sci U S A. 2005;102:13652–13657. doi: 10.1073/pnas.0506008102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeren-Wiemers N, Gerfin-Moser A. A single protocol to detect transcripts of various types and expression levels in neural tissue and cultured cells: in situ hybridization using digoxigenin-labelled cRNA probes. Histochemistry. 1993;100:431–440. doi: 10.1007/BF00267823. [DOI] [PubMed] [Google Scholar]
- Scheiffele P, Fan J, Choih J, Fetter R, Serafini T. Neuroligin expressed in nonneuronal cells triggers presynaptic development in contacting axons. Cell. 2000;101:657–669. doi: 10.1016/s0092-8674(00)80877-6. [DOI] [PubMed] [Google Scholar]
- Sigismund S, Argenzio E, Tosoni D, Cavallaro E, Polo S, Di Fiore PP. Clathrin-mediated internalization is essential for sustained EGFR signaling but dispensable for degradation. Dev Cell. 2008;15:209–219. doi: 10.1016/j.devcel.2008.06.012. [DOI] [PubMed] [Google Scholar]
- Solecki DJ, Liu XL, Tomoda T, Fang Y, Hatten ME. Activated Notch2 signaling inhibits differentiation of cerebellar granule neuron precursors by maintaining proliferation. Neuron. 2001;31:557–568. doi: 10.1016/s0896-6273(01)00395-6. [DOI] [PubMed] [Google Scholar]
- Solecki DJ, Model L, Gaetz J, Kapoor TM, Hatten ME. Par6alpha signaling controls glial-guided neuronal migration. Nat Neurosci. 2004;7:1195–1203. doi: 10.1038/nn1332. [DOI] [PubMed] [Google Scholar]
- Solecki DJ, Trivedi N, Govek EE, Kerekes RA, Gleason SS, Hatten ME. Myosin II motors and F-actin dynamics drive the coordinated movement of the centrosome and soma during CNS glial-guided neuronal migration. Neuron. 2009;63:63–80. doi: 10.1016/j.neuron.2009.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitt TN, Hatten ME. Antibodies that recognize astrotactin block granule neuron binding to astroglia. Neuron. 1990;5:639–649. doi: 10.1016/0896-6273(90)90218-5. [DOI] [PubMed] [Google Scholar]
- Sturge J, Wienke D, Isacke CM. Endosomes generate localized Rho-ROCK-MLC2-based contractile signals via Endo180 to promote adhesion disassembly. J Cell Biol. 2006;175:337–347. doi: 10.1083/jcb.200602125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TerBush DR, Maurice T, Roth D, Novick P. The Exocyst is a multiprotein complex required for exocytosis in Saccharomyces cerevisiae . EMBO J. 1996;15:6483–6494. [PMC free article] [PubMed] [Google Scholar]
- Tomoda T, Bhatt RS, Kuroyanagi H, Shirasawa T, Hatten ME. A mouse serine/threonine kinase homologous to C. elegans UNC51 functions in parallel fiber formation of cerebellar granule neurons. Neuron. 1999;24:833–846. doi: 10.1016/s0896-6273(00)81031-4. [DOI] [PubMed] [Google Scholar]
- Tsai JW, Bremner KH, Vallee RB. Dual subcellular roles for LIS1 and dynein in radial neuronal migration in live brain tissue. Nat Neurosci. 2007;10:970–979. doi: 10.1038/nn1934. [DOI] [PubMed] [Google Scholar]
- Umeshima H, Hirano T, Kengaku M. Microtubule-based nuclear movement occurs independently of centrosome positioning in migrating neurons. Proc Natl Acad Sci U S A. 2007;104:16182–16187. doi: 10.1073/pnas.0708047104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrijenhoek T, Buizer-Voskamp JE, van der Stelt I, Strengman E, Sabatti C, Geurts van Kessel A, Brunner HG, Ophoff RA, Veltman JA. Recurrent CNVs disrupt three candidate genes in schizophrenia patients. Am J Hum Genet. 2008;83:504–510. doi: 10.1016/j.ajhg.2008.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadehra M, Iyer R, Goodglick L, Braun J. The tetraspan protein epithelial membrane protein-2 interacts with beta1 integrins and regulates adhesion. J Biol Chem. 2002;277:41094–41100. doi: 10.1074/jbc.M206868200. [DOI] [PubMed] [Google Scholar]
- Zheng C, Heintz N, Hatten ME. CNS gene encoding astrotactin, which supports neuronal migration along glial fibers. Science. 1996;272:417–419. doi: 10.1126/science.272.5260.417. [DOI] [PubMed] [Google Scholar]