Abstract
Mitochondria play a pivotal role in energy metabolism, programmed cell death and oxidative stress. Mutated mitochondrial DNA in diseased cells compromises the structure of key enzyme complexes and, therefore, mitochondrial function, which leads to a myriad of health-related conditions such as cancer, neurodegenerative diseases, diabetes and aging. Early detection of mitochondrial and metabolic anomalies is an essential step towards effective diagnoses and therapeutic intervention. Reduced nicotinamide adenine dinucleotide (NADH) and flavin adenine dinucleotide (FAD) play important roles in a wide range of cellular oxidation–reduction reactions. Importantly, NADH and FAD are naturally fluorescent, which allows noninvasive imaging of metabolic activities of living cells and tissues. Furthermore, NADH and FAD autofluorescence, which can be excited using distinct wavelengths for complementary imaging methods and is sensitive to protein binding and local environment. This article highlights recent developments concerning intracellular NADH and FAD as potential biomarkers for metabolic and mitochondrial activities.
Keywords: energy metabolism, flavin, flavin adenine dinucleotide, mitochondria, mitochondrial anomalies, reduced nicotinamide adenine dinucleotide
Mitochondrial function is essential to the life of all eukaryotic cells and human health by supporting the energy demands for all cellular processes [1]. In addition, mitochondria play a key mechanistic role in programmed cell death (apoptosis), free radical generation and oxidative stress (a hallmark of aging), and biomolecular sensing of glucose, oxygen and nitric oxide. As a result, it is not surprising that mitochondrial dysfunction is widely linked to a range of diseases and health problems such as cancer, Alzheimer’s disease and other neurodegenerative diseases [2–5]. Mitochondria have also been a target for developing therapeutic drugs, due to their role in cell survival and health related conditions [6]. For example, some anticancer agents are designed to stimulate proapoptotic mitochondrial events in tumor cells [7–9]. Mitochondrial anomalies dysregulate energy production, which supports many pathways of intermediary metabolism, and, therefore, have also attracted attention as another target for drug discovery and clinical intervention [10]. Mitochondrial dysfunction and oxidative stress have been associated with the majority of neurodegenerative diseases. As a result, the reduction of mitochondrial oxidative stress is another target for new therapeutic drugs aimed at either preventing or slowing down the progression of neurodegenerative disorders [11]. Uncoupling of the mitochondrial electron transport chain (ETC) has also been a pharmacological target for treating obesity [12]. 2,4 dinitrophenol has been used to induce some weight loss by enhancing energy expenditure via mitochondrial uncoupling. Efficient biomarkers and noninvasive techniques that would allow for real time monitoring of mitochondrial function would, therefore, provide a powerful tool towards effective diagnosis of health problems and an opportunity for early therapeutic intervention.
Decades after the pioneering work by Chance and colleagues [13–21], there is a resurgent interest in using intracellular coenzymes, such as reduced nicotinamide adenine dinucleotide (NADH) and flavin adenine dinucleotide (FAD) – see Figure 1 for chemical structure, as intrinsic biomarkers for metabolic activities and mitochondrial anomalies. For example, NADH and its oxidized form (NAD+) are involved in mitochondrial function, energy metabolism, calcium homeostasis, gene expression, oxidative stress, aging and apoptosis. Chance and Williams, in a series of seminal reports, demonstrated that the respiratory chain activities of isolated in vitro mitochondria correlate with the redox coenzymes in the ETC [17–20]. Mutated mitochondrial DNA in cancer, for example, results in an alteration of the conformations (and function) of NADH dehydrogenase (complex I) and cytochrome C (complex III) in the ETC, which leads to the generation of free radicals and apoptosis [22]. The reduced NADH phosphate (NAD[P]H) is involved in the reductive biosynthesis of fatty acids and steroids, antioxidation and oxidative stress, and the oxidized form, NADP+, participates in calcium homeostasis [23,24]. Some evidence suggests that intracellular NADH concentration is greater (up to tenfold, by some estimates) than that of NAD(P)H [24,25]. The sensitivity of the autofluorescence of these pyridine nucleotides to cell pathology remains inconsistent and may depend on the experimental techniques, conditions and cell lines [26,27]. Reduced FAD (FADH2) and FAD are another redox pair associated with respiration in all eukaryotic cells. Some data suggest that flavoproteins, such as lipoamide dehydrogenase (LipDH) and electron transfer flavoprotein, contribute significantly to the cellular autofluorescence [28,29]. FAD is largely associated with mitochondria and the oxidative phosphorylation pathway [30,31]. NAD(P)H, FADH2 and their oxidized counterparts are critical for a broad array of oxidation–reduction (redox) reactions in living cells [23,32,33]. In particular, the redox ratio (NAD+:NADH) allows for real-time monitoring of the metabolic state of a cell during pathophysiological changes.
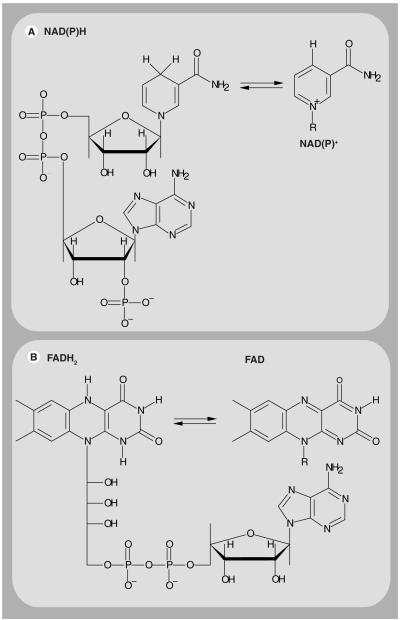
Figure 1. The chemical structure of NAD(P)H, FADH2 and their oxidized forms.
(A) The chemical structure of NAD(P)H is shown along with the corresponding nicotinamide ring of oxidized NAD(P)+ (right), which constitutes the reactive moiety that accepts a hydrogen ion and two electrons. (B) The chemical structure of FADH2 is also shown. The isoalloxazine ring is the reactive part of FAD (right) that is responsible for light absorption in the UV region with visible emission. Flavin mononucleotide has a similar chemical structure to FAD but it does not contain the ribose and adenine moieties (not shown). NAD(P)H and FAD are fluorescent while NAD(P)+ and FADH2 are not.FAD: Flavin adenine dinucleotide; FADH2: Reduced flavin adenine dinucleotide; NAD(P)+: Oxidized nicotinamide adenine dinucleotide phosphate; NAD(P)H: Reduced nicotinamide adenine dinucleotide phosphate.
Based on their functional role in cell biology, intracellular NADH and FAD have a diagnostic potential as natural biomarkers for cellular redox reactions, energy metabolism and mitochondrial anomalies under different pathophysiological conditions [34,35]. Importantly, these coenzymes are naturally fluorescent and, therefore, genuine, noninvasive imaging of metabolic activities can be carried out in living cells and tissues. The autofluorescence properties of NADH and FAD eliminate potential toxicity, nonspecific binding and interference with biomolecular functions that are associated with the use of exogenous dyes. In addition, the autofluorescence of these coenzymes can be excited using distinct illumination wavelengths, ranging from UV to infrared regions, for complementary imaging using one photon (1P) - and two photon (2P)-fluorescence microscopy, respectively. Finally, the NADH and FAD autofluorescence is sensitive to protein binding and local environment. When combined, these properties are essential for a useful biomarker such as NADH and FAD.
The objective of this article is to highlight recent development of these intracellular co enzymes as natural biomarkers for metabolic activities and mitochondrial anomalies. The structure and spectroscopic properties of these coenzymes is reviewed as a guide for experimental design and data interpretation. Recent studies on NADH and flavin are highlighted within the context of their biological function in metabolic activities and mitochondrial function. In addition, fluorescence based methods for monitoring cellular coenzymes are compared with conventional biochemical assays that require cell lysates. Finally, the potential and challenges associated with using these coenzymes as biomarkers is discussed and compared with conventional, exogenous markers for mitochondria. This article is not intended to be comprehensive. Rather, it is written to serve as biological, biomedical and technical resource for new researchers who may be interested in exploiting these natural biomarkers for diagnostic and mechanistic studies.
Coenzymes as biomarkers for metabolic activities
Glycolysis
In the cytoplasm, glycolysis involves ten steps in the following overall reaction [36]:
In anaerobic glycolysis, NAD+ is regenerated by pyruvate reduction to lactate, which is catalyzed by lactate dehydrogenase [37]. Under aerobic conditions, however, the high energy electrons from cytosolic NADH are shuttled, using the glycerol 3 phosphate and malate–aspartate shuttles, to the respiratory chain in the inner mitochondrial membrane [1,23,38]. Glycolysis also provides an alternative pathway for ATP production in eukaryotic cells under pathophysiological conditions caused by mitochondrial anomalies (as a result of cancer, for example). Therefore, monitoring the changes in the cytosolic NADH concentration can indicate the glycolytic rate under different physiological states. In addition, the correlation between intracellular glucose uptake and the cytosolic NADH concentration could establish this coenzyme as a natural biomarker for diabetes. Conventional biochemical methods have provided most of the current information regarding NADH concentration in cell lysates. However, these assays provide snap shots of the metabolic and redox state of cells and lack information regarding the morpho logical context [25,39–41]. By contrast, fluorescence microspectroscopy techniques are noninvasive, with the spatio–temporal resolution required to differentiate between physiological states, as well as of NADH compartmentalization (e.g., cytoplasm, mitochondria or nucleus).
A number of studies have demonstrated the sensitivity of intrinsic NADH autofluorescence (concentration) to cell physiology [42,43] and pathology [39]. Tilton etal. examined the effects of elevated glucose levels on glycolysis, sorbitol pathway activity and the cytosolic redox state of NADH:NAD+ [44]. In their model system of isolated blood free glomeruli from the kidneys of male, Sprague-Dawley rats, they found support for the hypothesis that metabolic imbalances, associated with a reduced ratio of cytosolic NADH:NAD+ (caused by increased glucose metabolism), play an important role in mediating glucose- and diabetes induced glomerular dysfunction. Following glucose metabolism by pancreatic β-cells, the increase in cytosolic ATP is the key signal for initiating insulin secretion by causing a blockade of ATP-dependent potassium channels.
Glucose metabolism in glycolysis is pivotal to glucose induced insulin secretion from pancreatic β-cells [45–47]. Dukes etal. investigated NADH production during glycolysis and its role in β-cell glucose signaling [45]. The induction of mitochondrial membrane depolarization leads to an elevation in cytosolic calcium and insulin secretion. Dukes etal. identified the critical metabolic step by which glucose initiates changes in ATP-sensitive potassium channel activity, membrane potential and calcium concentration in β-cells. Eto etal. also reported on the role of the NADH shuttle system in glucose induced activation of mitochondrial metabolism and insulin secretion [38]. In addition to the pyruvate byproduct, Eto etal. argued that additional glycolytic factors appear to be required for the generation of the mitochondrial signals that lead to insulin secretion. By inhibiting the NADH shuttles, glucose-induced increases in NADH autofluorescence were observed, while the mitochondrial membrane potential and ATP content were reduced along with the abolishment of glucoseinduced insulin secretion. These results support the hypothesis that the NADH shuttle couples glycolysis with the activation of mitochondrial energy metabolism to trigger insulin secretion. In animal diabetes models, the increased ratio of free cytosolic NADH:NAD+ is considered one of the earliest metabolic imbalances linked to increased blood flow in these tissues [46].
Piston and coworkers studied the redox signal, as a function of pyruvate, in intact pancreatic islets using flavoproteins and NAD(P)H autofluorescence microscopy [47,48]. As LipDH is in direct equilibrium with mitochondrial NADH, the authors used LipDH and NAD(P)H autofluorescence to assess the NAD(P)H production as a function of glucose dosage. They observed that glucose dose response is consistent with an increase in NAD(P)H. By contrast, the transient rise in NAD(P)H in response to pyruvate stimulation was not accompanied by a significant change in LipDH, which indicates that pyruvate raises cellular NAD(P)H without a significant increase in mitochondrial NADH and, therefore, fails to produce the ATP necessary for stimulating insulin secretion [47].
Mitochondrial role in energy metabolism
Inside the mitochondrial matrix, the tricarboxylic acid (TCA) cycle (also known as the Krebs cycle or citric acid cycle) eventually transforms acetyl-CoA, which is derived from glycolytic pyruvate into carbon dioxide, as summarized by the following equation [1,49]:
In addition to one ribonucleotide guanosine triphosphate, the TCA cycle produces three NADH molecules and one FADH2, which feed into the respiratory chain. The ETC and ATP synthase comprise the oxidative-phosphorylation pathway (Figure 2) in the impermeable inner membrane of mitochondria, where the majority of ATP is synthesized [23,50]. The respiratory chain consists of a series of coupled transmembrane enzymes, namely NADH dehydrogenase (complex I), succinate dehydrogenase (complex II), ubiquinol cytochrome C reductase (complex III), and cytochrome C oxidase (complex IV) [1,23,49], which serve as electron carriers. The protein subunits of these enzyme complexes are genetically encoded by nuclear (complex II) and mitochondrial DNA (complex I, III and IV). In the initial steps of the ETC, NADH and FADH2 are oxidized (i.e., oxidation reaction with NAD+ + H+ + 2e− for each NADH) triggering an electron transfer to complex I (or NADH–ubiquinone oxidoreductase), complex II (or succinate–ubiquinone oxidoreductase) and then complex III via coenzyme Q (also known as ubisemiqui none) [23,32,33]. Complex I also includes flavin mononucleotide (FMN) as an intermediate electron acceptor. The electron continues to complex IV with the help of cytochrome C as another electron carrier. Concomitant with the electron transfer between complexes I, III and IV is the formation of a proton gradient across the inner membrane. In addition, two water molecules are generated by complex IV for every oxygen molecule (i.e., reduction reaction with 0.5O2 + 2H+ + 2e− for each water molecule produced). The transmembrane proton gradient drives ADP phosphorylation via ATP synthase to produce the majority of ATP. The same electrochemical potential also regulates the rate of respiratory-chain activities and facilitates calcium ion transport across the inner membrane of mitochondria. The newly synthesized ATP is then transported out of the mitochondrial matrix via adenine nucleotide translocase with a concomitant, but opposite, migration of ADP [4]. Studies also demonstrated that accumulation of calcium into mitochondria regulates mitochondrial metabolism and causes a transient depolarization of the mitochondrial membrane potential, which causes a transient decline in ATP production [51,52].
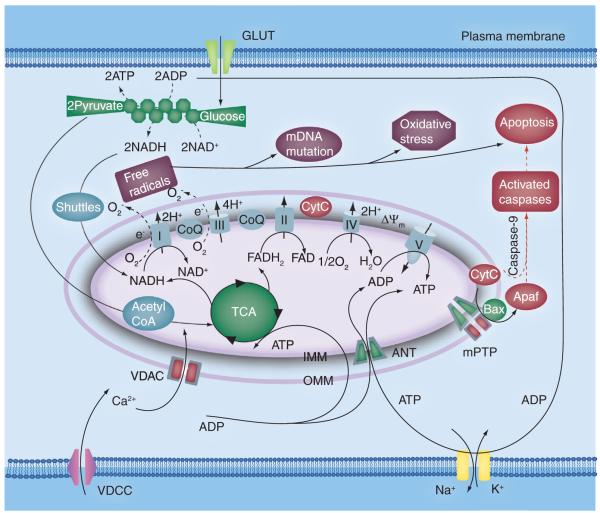
Figure 2. Mitochondrial role in energy metabolism, apoptosis and oxidative stress in eukaryotic cells.
In glycolysis, an alternative pathway for producing ATP under mitochondrial dysfunction, two ATP, two NAD+ and two pyruvate molecules are generated for each glucose molecule consumed via a series of ten catalyzed reactions. The high-energy electrons of glycolytic NAD+ are shuttled (via the malate–aspirate and glycerol-3-phosphate shuttles) into the mitochondria, while the final destination of the pyruvate molecules is the tricarboxylic acid cycle in the mitochondrial matrix. Oxidative phosphorylation involves a series of coupled enzyme complexes (I–IV) in the electron transport chain to generate a proton gradient across the inner mitochondrial membrane that activates the ATP synthase (complex V), which provide the majority of cellular ATP. In addition, the free electrons in the electron transport chain (especially from complex I and III) generate singlet oxygen species, which cause oxidative stress and the loss of ΔΨm in the absence of antioxidant defense mechanisms. The release of CytC from the inner mitochondrial membrane triggers apoptosis ANT: Adenine nucleotide translocator; CoQ: Coenzyme Q; CytC: Cytochrome C; FAD: Flavin adenine dinucleotide; FADH2: Reduced flavin adenine dinucleotide; IMM: Inner mitochondrial membrane; mPTP: Mitochondrial permeability transition pore; NADH: Reduced nicotinamide adenine dinucleotide; OMM: Outer mitochondrial membrane; TCA: Tricarboxylic acid; VDAC: Voltage-dependent anion channel; VDCC: Voltage-dependent calcium channel.
The pioneering work of Chance highlighted the potential of NADH and FAD autofluo rescence, under UV excitation, as biomarkers for monitoring the respiratory chain activities [13,15,16]. Recently, Bird etal. investigated the functional response of the intracellular reduction:oxidation ratio in normal human breast cells (MCF10A) to confluence, serum starvation and potassium cyanide poisoning [43]. Using 2P fluorescence lifetime imaging (FLIM), the authors reported a significant decrease in the fluorescence lifetime of both free and protein-bound NADH, as well as the contribution of protein-bound NADH, as cells progress towards confluency. Bird etal. demon strated that treating the cells with potassium cyanide treatment or serum starvation yielded similar changes. Their results suggest that the ratio of free:protein bound NADH is related to changes in the NADH:NAD+ ratio.
Care must be taken, however, in using only the fluorescence lifetime to quantify the free and enzyme bound NADH in cells, since the fluorescence of both free and enzyme-bound NADH in solution decays as a multiexponential, which makes the cellular data interpretation rather challenging. By contrast, the rotational diffusion directly depends on the size of the rotating biomolecule and its surroundings. Vishwasrao etal. demonstrated that the NADH autofluorescence in brain slices isolated from the hippocampus of a young mouse, correlates with the metabolic state transition from normoxia to hypoxia [42]. Using 2P fluorescence micro spectroscopy techniques, the authors monitored the changes in NADH concentration, conformation (free vs enzyme bound) and local environment as a function of oxygen content in neuronal tissues. Similar measurements were carried out to examine the sensitivity of intracellular NADH to cell pathophysiology and ETC enzymatic inhibition using potassium cyanide in breast cancer and normal cells [53]. These studies demonstrated an increase in the overall NADH autofluorescence (and, therefore, concentration) upon the interruption of the respiratory-chain activities using either potassium cyanide or hypoxia.
Reduced FAD (nonfluorescent) and FAD (fluorescent) are coenzymes associated with the respiratory activities in all eukaryotic cells. Recently, it was demonstrated that the autofluo rescence intensity of flavin and flavoproteins is reduced after treatment with the mito chondrial inhibitor cyanide in pancreatic islet [48], rat cardiac myocytes [29,54] and guinea pig ventricular myocytes [55]. As a result, real-time monitoring of mitochondrial NADH and FAD will provide a ratiometric measure of energy respiration as well as the redox state of a cell under certain physiological conditions [29]. The ratiometric measure of both coenzymes also requires a noninvasive technique with enough spatial resolution to distinguish between cytosolic and mitochondrial NADH. Luckily, FAD and FMN are strictly localized within the mitochondria. The compartmentalization of NADH and FAD makes these natural biomarkers a more powerful means to monitor, in real time, the metabolic activities in cells and tissues.
Mitochondria-mediated oxidative stress & apoptosis
Oxidative stress
Under physiological conditions, effective antioxidant defense mechanisms regulate the level of reactive oxygen species (ROS), which are highly reactive, oxygen-containing molecules, such as hydrogen peroxide, nitric oxide and hydroxyl ions, produced in small quantities during natural metabolic reactions. Under pathological conditions, antioxidant defense mechanisms fail, which leads to an elevated level of ROS and, therefore, oxidative stress. Mitochondria are a major source of ROS and free radicals even in the presence of antioxidant defense mechanisms. The unpaired electrons generated in respiration Figure 2, particularly by complex I and (to lesser degree) complex III, produce superoxide ions after reacting with oxygen. An elevated level of ROS and oxidative stress has harmful effects on cells, caused by lipid peroxidation, DNA damage and inhibition of the mitochondrial respiratory chain, leading to numerous health-related conditions such as aging. A common pathogenic mechanism for neurodegenerative diseases, such as Alzheimer’ s disease and Parkinson’ s disease, involve the aggregation and disposition of misfolded proteins, which leads to progressive degradation of the CNS [56]. Evidence suggests that brain amyloid β which is commonly associated with Alzheimer’ s disease, is linked mechanisti cally to an increased level of oxidative stress [57] and apoptosis. Oxidative stress also appears to provide a critical link between environmental and genetic risk factors in the pathogenesis of Parkinson’ s disease [58]. Epidemiologic studies suggest that pesticides and other environmental toxins that inhibit complex I are involved in the pathogenesis of Parkinson’ s disease [59]. Fat induced increase in oxidative stress is an important mechanism of obesity associated metabolic syndrome, both in humans and mice [60], and may be a useful therapeutic target. Furukawa etal. demonstrated that increased ROS production in the adipose tissue of obese mice is accompanied by an elevated expression of NAD(P)H oxidase and a decreased expression of antioxidative enzymes (e.g., superoxide dismutase, catalase and glutathione peroxidase) [60]. Elevated fatty acids levels increased the oxidative stress via NAD(P)H oxidase activation, while oxidative stress caused dysregulation of fat-derived hormone production in cultured adipocytes. Furukawa etal. observed that inhibiting NAD(P)H oxidase reduced ROS production in adipose tissue and improved diabetes, hyperlipidemia and hepatic steatosis in obese mice. The generation of ROS also plays a role in the cytotoxicity of adaphostin, a potential anticancer drug undergoing preclinical testing [61]. Alcohol also promotes ROS generation in human or animal models through different mechanisms [62,63]. As a result, ROS production and oxidative stress in liver cells trigger the development of alcoholic liver disease [64,65].
Changes in the NAD+:NADH ratio, mitochondrial function and ATP production are associated with oxidative stress, independent of the underlying mechanisms. Therefore, it would be advantageous to monitor the oxidative stress state of a cell by using coenzymes such as NADH and FAD as natural biomarkers. For example, the sensitivity of intracellular NADH autofluorescence in yeast cells has recently been investigated as a function of hydrogen peroxide and peroxonitrite treatment [66]. Different levels of hydrogen peroxide and peroxonitrite have been demonstrated to initiate necrosis, apoptosis and reversible injury in yeast cells. Concurrently, the level of intracellular NADH increased at the beginning of the apoptotic process and then decreased continuously until cell death. The time course of these intracellular NADH changes was attributed to the response of the mitochondrial redox state to oxidative stress [66]. The results indicate that different pathways of oxidative injury in yeast cells trigger different responses in the mitochondrial redox state. Yang etal. also investigated the sensitivity of the FAD:NAD(P)H ratio and NAD(P)H conformation in HepG2 cells to ROS, which was induced by cadmium chloride exposure [67]. Using confocal spectroscopy and time-resolved fluorescence measurements, the results suggested that changes in NAD(P)H metabolism precedes the increase in ROS production and cellular oxidative stress.
Programmed cell death
Natural programmed cell death (apoptosis), which is tightly regulated by a cascade of bio chemical reactions, is an evolutionarily conserved suicide process essential for embryonic development, homeostasis of adult tissues and organs, and the destruction of virally infected, injured or DNA-damaged cells [68,69]. Cells undergoing apoptosis would exhibit morphological characteristics, such as plasma membrane blebbing, cell shrinking, fragmentation and nuclear chromatin condensation. Apoptosis is mediated by extrinsic (or death receptor) pathways, which are beyond the scope of this article, and intrinsic (or mitochondrial) pathways [70]. In the mito chondrial pathway for apoptosis, the inner mitochondrial transmembrane potential (ΔΨm) [71] is usually lost and mitochondrial release of cytochrome C, which activates apoptotic activating factor-1, eventually triggers caspase activation. A potential mechanism for cytochrome C release from the mitochondria is the swelling and rupture of the outer membrane of mitochondria upon the opening of the mitochondrial permeability transition pores (mPTP), which include the adenine trans locator (adenine nucleotide translocase) and the voltage-dependent anion channel at the inner and outer membranes, respectively [65]. A number of effectors (e.g., calcium and ADP/ATP concentrations and the membrane potential of mitochondria) influence the mPTP activation. Under pathological conditions of cellular calcium overload, particularly in association with oxidative stress, mitochon drial calcium uptake may trigger events that lead to apoptosis [51,52]. In the presence of ATP and cytochrome C, two different domains of apoptotic activating factor-1 and procaspase-9 interact to form the apoptosome. The down stream caspases are, in turn, activated by the apoptosome-associated caspase-9 (Figure 2). Mitochondria also regulate apoptosis by releasing apoptosis promoting factors into the cytosol. Enhancement of the rate of apoptosis is associated with viral and bacterial infections, neurodegenerative disorder and toxin-induced diseases [72]. Some evidence suggests that cancer chemopreventive agents might prevent cancer by suppressing growth or inducing apoptosis in precancerous cells [73,74].
The concentration of intracellular NAD(P)H is sensitive to the onset of apoptosis in several model systems [75–79]. For example, Petit etal. investigated the correlation between the oxidation/depletion of NAD(P)H and apoptosis in Jurkat T cells [77]. High-performance liquid chromatography and UV-excited NAD(P)H fluorescence measurements indicate oxidation and depletion of NAD(P)H prior to the onset of apoptosis. In addition, cytofluorimetric multi parameter ana lysis and cell sorting experiments indicate a close temporal relationship between NAD(P)H oxidation/depletion and the dissipation of ΔΨm. The observed NAD(P)H depletion preceded other apoptosis induced changes, such as enhanced superoxide generation, loss of cytosolic potassium, decreased cytoplasmic pH, nuclear DNA fragmentation, cell shrinkage and loss of viability [77]. The authors also reported a correlation among the activation of caspases (9 and 3), the appearance of superoxide anions in the mitochondria, and NAD(P)H concentration. Their findings suggest a functional relationship between NAD(P)H depletion, the membrane potential loss prior to caspase 3 activation and changes in the redox state of the cell. Other studies have demonstrated that calcium- and superoxide induced mitochondrial swelling triggered the release of cytochrome C and increased caspase activity from isolated mitochondria [79]. 1P autofluorescence of NAD(P)H, FAD and their redox ratio in primary human foreskin keratinocytes is also sensitive to the early stages of cisplatin treatment (an apoptosis promoting agent) [80]. Recently, Wang etal. also demonstrated that the 2P-autofluorescence lifetime imaging of NADH can be used to monitor mitochondria mediated apoptosis after staurosporine treatment and hydrogen peroxide-induced necrosis in live HeLa cells and 143B osteosarcoma [81].
Transcriptional role of nuclear NADH
Carboxyl terminal binding protein (CtBP) is a corepressor that is involved in the transcription pathways during development, cell cycle regulation and transformation [82,83]. Zhang et al. observed that binding of CtBP to cellular and viral transcriptional repressors is regulated by nuclear NAD+ and NADH [82]. Comparing the total 2P-autofluorescence intensity to a standard curve of free NAD(P)H in solution, Zhang etal. estimated there to be 113 μM of NAD(P)H in the nucleus of cultured Cos-7 cells. Agents capable of increasing NADH levels stimulate binding of CtBP to its partners in vivo and potentiate CtBP mediated repression. The results of this study support the hypothesis that monitoring changes in the nuclear NAD+:NADH ratio would allow CtBP to serve as a redoxsensor for transcription [82]. NAD+ has also emerged as a putative metabolic regulator of transcription. A possible mechanism involves silent information regulator 2, which is a family of protein deacetylases (sirtuins) that play an important role in regulating transcription in a NAD+ dependent manner [84,85]. NAD+ metabolism and sirtuins are also involved in the mechanisms associated with cell survival under conditions of oxidative stress and toxicity [86].
Mitochondria dysfunction in human diseases
It is well established that mitochondrial anomalies correlate with a wide range of health problems, such as cancer, neurodegenerative diseases, diabetes, cardiomyopathy and aging [5,32,87–90]. As a result, biomarkers that are sensitive to mitochondrial activities would enhance early diagnosis of potential health problems and, therefore, therapeutic intervention [89].
Cancer
In addition to the six known hallmarks of cancer (self sufficiency in growth signals, insensitivity to growth-inhibitory [antigrowth] signals, evasion of programmed cell death, limit less replicative potential, sustained angiogenesis and metastasis [91]), bioenergetics is another feature of cancer cells that was first discovered by Otto Warburg [37,92]. Warburg hypothesized that cancer development may originate from the impairment of mitochondrial respiration with a parallel increase in the cellular glycolysis [92]. Mitochondria in cancer cells exhibit both phenotypic and genotypic differences, compared with normal cells [32,93]. Studies have also demonstrated that mitochondrial DNA, cholesterol content and lipid types in the inner membrane, and protein activities (especially the cytochrome C oxidase) are usually skewed in cancer cells [88,94]. High levels of free radicals generated by mitochondria are also found in cancer cells [95].
Some studies have demonstrated an elevated autofluorescence of intracellular NADH in different cancer cell lines under UV illumination [39,96]. Skala etal. investigated the sensitivity of intracellular NADH in hamster cheek pouch epithelial cells to treatment with dimethylbenz[α] anthracene as a model of oral carcinogenesis [97]. Using 2P FLIM, the fast and slow autofluorescence lifetime components were assigned to free and protein bound NADH, respectively. Skala etal. concluded that the decrease in protein bound NADH fluorescence lifetime with dysplasia is caused by a shift from oxidative phosphorylation to glycolysis, which is consistent with the predictions of neoplastic metabolism [98]. They also demonstrated that both low- and high-grade precancers could be discriminated from normal tissue based on the lifetime of protein-bound NADH. In another report [98], the same group monitored the cellular NADH and FAD autofluorescence lifetime to identify metabolic fingerprints of living cells at the earliest stages of cancer development. The redox ratio was significantly decreased in the less differentiated basal epithelial cells compared with the more mature cells in the superficial layer of the normal stratified squamous epithelium, indicating an increase in metabolic activity in cells with an increased nuclear:cytoplasmic ratio.
Yu and Heikal developed an autofluorescence dynamics imaging assay to quantify the concentration and conformation of NADH in normal breast (Hs578Bst) and cancer (Hs578T) cell lines [53]. The authors estimated the intracellular NADH level in the breast cancer cell model to be almost twice that of their nontransformed counterpart. For the first time, Yu and Heikal observed a 2P-autofluorescence associated anisotropy of intracellular NADH at the single-cell level, which directly indicates the presence of free and enzyme-bound NADH at equilibrium with distinct fluorescence properties and molecular sizes [53]. Chemotherapeutic agents have been demonstrated to enhance apoptosis by disrupting the inner membrane potential of mitochondria, enhancing the release of cytochrome C and activating caspases [72]. Kirkpatrick etal. used endogenous NADH, FAD and tryptophan fluorescence spectroscopy to measure metabolic changes in response to treatment with cancer chemopreventive agents (namely, N 4 [hydroxyphenyl]retinamide) in ovarian and bladder cancer cell lines [74]. Their results suggest that the NADH signal changed in a similar dose dependent manner, in all cells, but started at different baseline levels. Toxic doses of photodynamic therapy have been demonstrated to cause in vitro and in vivo damage, which is concomitant with a reduction of intracellular NADH autofluorescence and mitochondrial activities relative to that of controls.
Neurodegenerative diseases
Mitochondria are critical regulators of programmed cell death, mitochondrial DNA mutations and the level of free radicals, which are key features of age related neurodegenerative diseases such as Alzheimer’ s, Parkinson’ s and Huntington s disease [99,100]. Mitochondrial dysfunction, amyloid l mediated processes, transition metal accumulation and genetic factors are major sources of free radicals. A better understanding of the redox imbalance will lead to a better understanding of the pathogenesis of neurodegenerative diseases and, ultimately, novel therapeutic approaches.
The concentrations of intracellular NADH and flavin are good biomarkers of mitochondrial function and mitochondrial anomalies. As a result, these coenzymes have a diagnostic potential for neurodegenerative diseases. For example, in Alzheimer s disease, amyloid-β peptide aggregates in the extracellular space to form senile plaques. Recently, Webb and coworkers used 2P-autofluorescence and second-harmonic generation to characterize the intrinsic emissions in ex vivo acute brain slices from transgenic mouse models with Alzheimer’ s disease [101]. Their results demonstrated that senile plaques exhibit autofluorescence with a distinct emission spectrum, as well as weakly generated second harmonic from the dendritic microtubule arrays near senile plaques.
Diabetes
Diabetes is a leading cause of specific microvascular diseases, such as blindness, renal failure, nerve damage and diabetes-accelerated athero sclerosis [102]. Glycolysis and mitochondria mediated glucose metabolism are essential for glucose induced insulin secretion by pancreatic β-cells [103,104]. The high energy electrons from the glycolysis generated NADH are transferred to mitochondria through the NADH shuttles. Inhibition of these shuttles leads to an increase in the NADH autofluorescence level. Concomitantly, the mitochondrial membrane potential and ATP content were reduced when glucose-induced insulin secretion was abrogated. Molecular mechanisms that are implicated in glucose mediated vascular damage involve hyperglycemia-induced overproduction of superoxide by the mitochondrial ETC [102].
As a result, quantitative imaging of cytosolic and mitochondrial NADH can be used as a biomarker of glycolytic rate, mitochondrial function and insulin secretion. For example, Noda etal. demonstrated the connection between the metabolic activities of ETC and TCA enzymes and their impact on insulin secretion using a murine, pancreatic β-cell line model with suppressed transcription of mitochondrial DNA [105]. The authors evaluated 2P-autofluorescence of NADH, insulin secretion, the membrane potential and the ATP and ADP content in this β-cell model as a function of ethidium bromide, an inhibitor of mito chondrial DNA transcription. The results of the study by Noda etal. suggest that reduced expression of the mitochondrial ETC enzymes causes NADH accumulation in the β-cells while facilitating anaerobic glucose metabolism. Glucose induced insulin secretion and membrane potential were also lost upon treatment with ethidium bromide.
Experimental approaches to interrogate intracellular coenzymes
Conventional biochemical methods
Traditional biochemical techniques have been useful for estimating cellular levels of NADH [106,107] and flavin [108]. For example, enzymatic methods [39], cycling assays [109,110] and high performance liquid chromatography analyses [111] have been used to determine the concentration of NADH in cells and tissues. A NAD+:NADH assay kit that is commercially available, with a sensitivity of approximately 0.2 μM, is based on an alcohol-dehydrogenase cycling reaction. Umemura and Kimura developed a single extraction procedure and a spectrophotometric assay (an enzymatic cycling assay using a fluorescent marker) to determine the concentration of oxidized and reduced NAD in cell monolayers [112]. Recently, a capillary electro phoresis method was developed, using an enzymatic cycling reaction to determine levels of both NAD+ and NADH at the single cell level and with a single run [113]. Another assay, using bacterial luciferase with a liquid scintillation spectrometer, has also been used to determine subpico molar concentrations of NADH and FMN [114]. These conventional biochemical assays provide an accurate and sensitive measure of the intrinsic metabolic co factors. In addition, some of these methods are capable of distinguishing between NAD(P)H and NADH, which is not possible using spectro scopic techniques owing to the similarity in their spectral properties. However, despite these advantages these methods require destroying the cells or tissues and, therefore, only provide snapshots of the metabolic activities in the absence of any morphological context.
Steady state, 1P spectroscopy
Absorption
Free NADH and NAD+ absorb a maximum of approximately 260 and 340 nm, respectively, in an aqueous buffer (pH 7.1), with an extinction coefficient of approximately 6.22 × 103 cm−1 M−1 at 340-nm absorption for the reduced form. These absorption properties are similar to those of free NAD(P)H and NAD(P)+, a disadvantage for spectroscopy-based assays used to differentiate between the metabolic activities associated with either NADH or NAD(P)H. The corresponding FAD absorption (Figure 3) is red shifted with two bands at 376 and 450 nm (with an extinction coefficient of approximately 11,300 cm−1 M−1 at 450 nm absorption) in addition to the 270-nm band assigned for adenine absorption [115]. While the absorption spectrum of NADH is insensitive to enzyme (e.g., mitochondria malate dehydro genase) titration, the FAD moiety in LipDH reveals a slight red shift of the absorption bands (362 and 456 nm). With knowledge of the extinction coefficient (ε), the relative NADH:FAD concentration (c) can be calculated (A = εcl) using the measured absorbance (A) of NADH (at ~340 nm) and FAD (at ~450 nm) in a cuvette (length [l]). Chance was the first to use a double beam spectro photometer to monitor intra cellular NADH in the absorption spectrum [116]. However, such UV-visible absorption spectra are usually broad with some degree of overlapping, which may undermine the specificity of the absorbing biomolecule identity. In addition, absorption spectroscopy would require the knowledge of the sample thickness and the influence of cellular environment on the extinction coefficient of a given biomolecule.
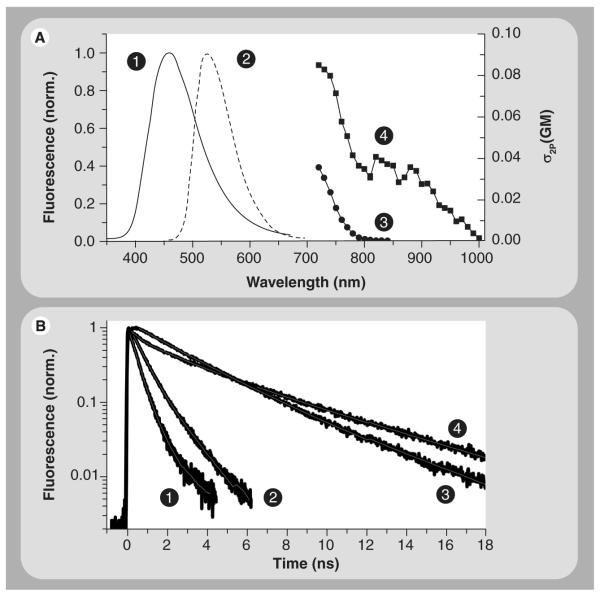
Figure 3. Fluorescence lifetime, two-photon excitation cross-section and emission spectra of NADH and FAD.
(A) The one-photon-fluorescence spectrum of NADH (1) peaks at approximately 458 nm as compared with approximately 528 nm emission of FAD in a buffered solution. Two-photon-excitation crosssection spectra of NADH (3) and FAD (4) are shown in GM units (1 GM = 10−50 cm4.s.photon−1.molecule−1) [29]. These results indicate that both NADH and FAD can be excited nonlinearly using 730 nm, but only FAD can be excited when λ is approximately 850–950 nm (4). As a result, the two photon-excitation wavelength, as well as the detection filters, can be used to separate the contribution of intracellular NADH and FAD autofluorescence. (B) Time-resolved fluorescence NADH (1) and FAD (3) decays as multiexponential with distinct time constants [29,42,53] and sensitivity to protein binding (mitochondria malate dehydrogenase (2); lipoamide dehydrogenase (4)). As a result, fluorescence lifetime can be used as a contrasting observable to differentiate between these two coenzymes using fluorescence lifetime imaging techniques.FAD: Flavin adenine dinucleotide; NADH: Reduced nicotinamide adenine dinucleotide.
Emission
The fluorescence emission of free NADH in aqueous buffer with pH 7.4 is maximal at approximately 458 nm, which is sensitive to both viscosity and enzyme titration. For example, NADH emission peaks at approximately 436 nm (i.e., 22 nm blue shift) when fully bound with mitochondria malate dehydrogenase. In contrast to NADH, NAD+ is not fluorescent. These fluorescence properties also mimic those of NAD(P)H and NADP+ in aqueous solution. By comparison, free FAD emits around 528 nm in buffer at room temperature with a slight blue shift in protein environment [117], as well as in the presence of 80% (by volume) of glycerol. For example, the fluorescence emission of LipDH is blue shifted by approximately 13 nm with respect to that of free FAD emission [115]. As a result, the autofluorescence of NAD(P)H (458 nm) and FAD (~528 nm) can be detected independently following selective 1P excitation at 340 and 450 nm, respectively (Figure 3A).
Fluorescence spectrophotometry has been used for over half a century to monitor in vitro and in vivo autofluorescence both for basic research and in a clinical setting. Duysens and Amesz were the first to report the autofluorescence spectra of NADH in intact cells using fluorescence spectrophotometry [118]. Similar measurements followed on various biological systems, such as human blood sample [119], human breast normal and cancer cell lines [26], Chinese hamster ovary cells, rat neurons, normal- and SV-40-transformed human keratinocytes [120], bovine oligodendroglia, mink and murine fibroblast [121], and isolated rat hepatocytes [122] under different metabolic conditions. The relative levels of the metabolic cofactors in these models were assessed under different physiological and pathophysiological conditions. In addition, in vivo autofluorescence spectrophotometry has also been developed to detect intrinsic NADH and/or flavoprotein signals in rabbit models [123], the brain and kidney of rats [14], murine muscle tissues [124] and squamous cell carcinoma [96]. Furthermore, NADH fluorescence has already been used clinically [125]. Fluorescence spectrophotometry is cost effective, easy to perform and can be performed in intact live cells and tissues. However, an absolute concentration cannot be obtained from this assay owing to the sensitivity of these cofactors to the local cellular environment. In addition, these measurements are usually conducted on cell ensembles in a cuvette and, therefore, lack the spatial resolution to determine the concentration of intra cellular coenzymes in different compartments of living cells.
1P-autofluorescence microscopy
In conventional 1P-microscopy, the sample is usually illuminated by UV light to excite intracellular NADH or FAD, based on their absorption spectra, and an autofluorescence image is captured under a physiological condition. This approach has been used for imaging intracellular coenzymes in a variety of biological models, such as contracting xenopus skeletal muscle fibers [126], hippocampal slices [127] and pancreatic islet β-cells [48]. Mitochondrial activities, reflected by changes in cellular autofluorescence, were monitored upon manipulation of physiological conditions such as response to oxygen availability, electrical stimulation and glucose application. For example, NAD(P)H and FAD autofluorescence as a marker of neuronal activation in CA1 of murine hippocampal slices under different electrical, glutamate iontophoresis, bath-applied kainate stimuli [127]. The results demonstrate that the NAD(P)H transients in mature slices do not reflect neuronal calcium dynamics. Furthermore, the study demonstrates that NADH and FAD signals are sensitive indicators of both the spatial and temporal characteristics of postsynaptic neuronal activation [127].
Reinert etal. exploited NADH and flavoproteins autofluorescence, excited by UV light from a 100-W mercury–xenon lamp, to directly monitor the in vivo neuronal activity in the cere bellar cortex of the ketamine/xylazine anesthetized mouse under electrical surface stimulation [128,129]. The authors demonstrated that the majority of the autofluorescence signal is attributed to activating the postsynaptic targets of the parallel fibers. Blocking mitochondrial respiration with sodium cyanide or inactivation of flavoproteins with diphenyleneiodonium substantially reduced the autofluorescence signal, which was attributed to the oxidation–reduction of flavoproteins. Reinert etal. also ruled out the effects of changes in hemoglobin oxygenation or blood flow. However, traditional UV excitation of NADH and flavoproteins suffers from problems such as scattering, extended photobleaching, smaller penetration depth and possible photodamage to samples [130,131]. In addition, the UV light used for intrinsic NADH imaging may cause photodamage to cellular DNA [132].
2P-autofluorescence microscopy
Two photon microscopy is based on nonlinear excitation of molecules using a high numerical aperture objective to tightly focus infrared laser pulses [133–137], which overcome some of the previously mentioned challenges associated with UV excitation. For example, 2P-microscopy provides high spatial resolution owing to the nonlinear excitation and, thus, the confocal pinhole (descanning) is not required, which minimizes the loss of fluorescence photons. Such inherent resolution also reduces the overall phototoxicity (or photodamage), especially when used properly, and allows for localized photochemistry. The infrared laser pulses used in 2P microscopy also enhance the penetration depth into biological tissues while reducing laser scattering detection. The 2P excitation cross section spectra of NADH [29,138,139] and flavin [29] in phosphate buffered saline (pH 7.5) are demonstrated in Figure 3 [29,140]. Based on these spectral profiles, selective 2P-excitation of NAD(P)H and FAD becomes possible using 730 nm (Figure 4A) [29,140] and 900 nm (Figure 4B), respectively [29]. Nichols etal. assessed the extent of 2P-induced photodamage and its effect on DNA synthesis in rat basophilic leukemia cells during NADH imaging [141].
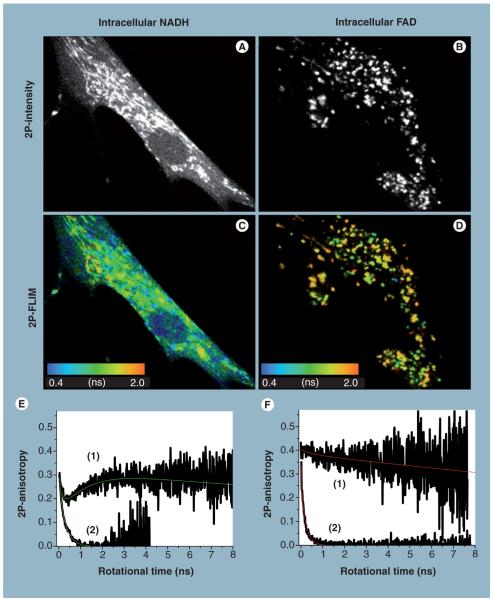
Figure 4. Two-photon autofluorescence intensity, lifetime and anisotropy of intracellular NADH and flavin in living cells.
The 2P-autofluorescence intensity imaging of endogenous NADH (A) and FAD (B) in HTB125 cells were recorded using 740 and 900 nm excitations, respectively, under different magnifications to examine their distribution throughout a single cell. The corresponding 2P-autofluorescence lifetime imaging reveals that the average autofluorescence lifetime of NADH (C) is relatively faster than that of FAD (D), as shown in the color bar. Furthermore, these intensity and lifetime images indicate a heterogeneous concentration, conformation and surrounding environment of these coenzymes. Time-resolved associated anisotropy of intracellular NADH (E), curve (1), provides direct evidence of the presence of two populations of free and enzyme-bound species at equilibrium [42,53]. Associate anisotropy decays are indicative of two species with distinctive sizes and autofluorescence properties (e.g., quantum yield or lifetime). As a point of reference, the anisotropy of free NADH in a buffer exhibits simple decay (E), curve (1), with a fast rotational time. By contrast, time-resolved autofluorescence anisotropy of intracellular FAD (F), curve (1), indicates a mostly enzyme-bound with a much slower rotational time than free FAD in a buffer (F), curve (2). These combined results demonstrate the potential of an integrated experimental approach [53,157] towards conducting biochemical analyses on living cells and tissues, without the need for their destruction as required with conventional biochemical techniques. 2P: Two-photon; FAD: Flavin adenine dinucleotide; FLIM: Fluorescence lifetime imaging; NADH: Reduced nicotinamide adenine dinucleotide.
Two-photon fluorescence microscopy of intrinsic NADH has been used to monitor energy metabolism in macrophages, pancreatic islet cells, skeletal muscle cells [142–144], brain slices [42,139], cardiomyocytes [29], human breast normal and cancer cells [53], cochlea [145] and murine skeletal muscle in vivo [146]. In addition, the redox and metabolic states of a cell can be determined using ratiometric measure of FAD and NADH autofluorescence [29,98]. Rocheleau etal. used 2P-excitation of NADH and 1P-excitation of intrinsic flavin to measure the β-cell redox potential (i.e., NADH:LipDH ratio) in pancreatic islets [48]. Kasischke etal. resolved metabolic signatures in astrocyte processes and neurons deep in brain tissue slices using 2P-autofluorescence functional imaging of NADH [139]. The results support the hypothesis for neurometabolic coupling in which early oxidative metabolism in neurons is eventually sustained by late activation of the astrocyte–neuron-lactate shuttle. Christie etal. developed a thinned skull preparation in the Alzheimer’ s disease mouse model that allows for imaging of senile plaques over several months using 2P-microscopy imaging of thioflavine S stained senile plaques in the Tg2576 transgenic mouse model of Alzheimer’ s disease [147]. Their results indicate that thioflavine S-positive plaques appear and then stabilize, supporting a dynamic feedback model of plaque growth in this Alzheimer’ s in vivo model.
The 2P autofluorescence of single cardiac cells has been assigned to intrinsic flavoproteins based on excitation (750, 800 and 900 nm), inhibition of oxidative phosphorylation [29] and a spectral unmixing algorithm [54]. The limited number of publications on 2P-microscopy imaging of cellular flavin reflect its experimental limitations, namely a low excitation cross-section and an abysmal fluorescence quantum yield (0.03) [148], which is attributed to dynamic quenching by adenine [148,149], van der Waals interactions with tryptophan residues and/or the sulfur atoms in cysteine residues [150,151]. The actual concentrations and distribution of FAD in live cells remains uncertain, especially under different physiological conditions. Using fluorescence assays, Brolin and Agren reported concentrations of FAD (113–291 μmol/kg dry weight) and FMN (9–139 μmol/kg dry weight) in pancreatic islets and other organs from obese hyperglycemic mice [152].
Autofluorescence lifetime imaging microscopy
In contrast to intensity-based microscopy, the excited state fluorescence lifetime is extremely sensitive to both the molecular conformation and surrounding environment of a fluorophore. Moreover, the fluorescence lifetime is also independent of the fluorophore concentration under typical physiological conditions. Time-correlated single photon counting (TCSPC) technique is routinely used for time-domain measurements of fluorescence lifetime [153,154]. For example, the fluorescence of free NADH in a pH7.4 buffer decays as a biexponential (Figure 3B, curve 1) with an average lifetime of 395 ± 20 ps at room temperature [42,53]. In the presence of enzymes, the NADH fluorescence decay remains multi exponential (Figure 3B, curve 2), but the average lifetime increases significantly upon binding [42,53]. By comparison, the fluorescence of free FAD in a pH7.4 buffer decays as a biexponential (Figure 3B, curve 3) with τ1 = 2.57 ns (a3 = 0.71) and τ2 = 4.42 ns (a2 = 0.29), with an average lifetime of 3.13 ns [Yu Q, Heikal AA, Unpublished Data]. When bound to LipDH, the flavin fluo rescence decays as a triple exponential (Figure 3B, curve 4) with τ1 = 268 ps (a1 = 0.34), τ2 = 2.17 ns (a2 = 0.26), τ3 = 5.3 ns (a3 = 0.39), with an estimate average lifetime of 2.75 ns [Yu Q, Heikal AA, Unpublished Data]. The distinctive fluorescence-decay parameters of intracellular NADH and flavin allow for a facile separation of these coenzymes and their conformational states (e.g., free vs enzyme bound) in cellular environment.
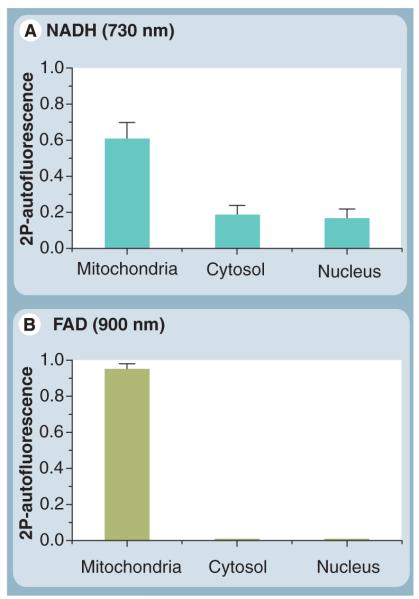
The FLIM technique implements the TCSPC principle in imaging mode [155], and is becoming a powerful tool for biological and biomedical studies. Recently, Yu and Heikal used an integrated fluorescence microspectroscopy approach to measure the NADH content in breast cancer and normal cells [53]. Representative 2P-autofluorescence intensity and lifetime images of intrinsic NADH (Figures 4A & C, excited at 730 nm) are distinct from those of flavin autofluorescence (Figures 4B & D, excited at 900 nm) in normal breast cells (HTB125). The 2P-autofluorescence intensity (Figure 4A) depends on the concentration of coenzymes throughout the cell as compared with the autofluorescence lifetime, which is sensitive to their conformation and microenvironment. On average, the autofluorescence lifetime of intracellular flavin is longer than that of NADH through out the cell. In addition, pixel to pixel analyses of high spatial resolution FLIM images yield a quantitative measure of mitochondrial, cytosolic and nuclear population of native NADH (Figure 5A). Comparative analyses of FAD images also reveal dominant compartmentalization of flavin in the mitocondria (Figure 5B). A number of FLIM studies of intracellular NAD(P)H autofluorescence have been reported on a range of cell lines such as dermal fibroblasts from artificial skin constructs [156]
Figure 5. Pixel-to-pixel analysis of intracellular NADH and FAD two-photon autofluorescence intensity at the single-cell level.
(A) Under 730-nm excitation, pixel-to-pixel ana lysis (binning: 3) reveals that approximately 61% of intracellular NADH is localized in mitochondria (oxidative phosphorylation and tricarboxylic acid cycle), while 19% of the population could be found in the cytoplasm (glycolysis). In addition, approximately 17% of native NADH seems to exist in the nucleus (transcriptional pathways [82]), after the background signal was subtracted from the real autofluorescence signal. (B) Intracellular flavin, however, is localized mainly in the mitochondria (>95%) when monitored at 900-nm excitation. 2P: Two-photon; FAD: Flavin adenine dinucleotide; NADH: Reduced nicotinamide adenine dinucleotide.
Recently, the in vivo 2P-FLIM measurements of FAD were reported in normal and precancerous epithelial mucosa tissues in hamsters [98]. In the study by Skala etal., FAD fluorescence lifetime under 890-nm excitation was fit with two species, with the longer lifetime component assigned to free FAD and the shorter component attributed to protein bound FAD [98]. The authors observed a decreased contribution of protein-bound FAD in the high-grade precancer epithelia as estimated by changes in the autofluorescence lifetime [98]. By contrast, flavin exists predominantly in mitochondria (Figure 6B) with negligible concentration in the cytosol and nucleus. Using these 2P-autofluorescence intensity and lifetime images, an imaging processing algorithm [157] was developed to convert the intracellular autofluorescence into NADH concentration in living cells [53]. The analyses of these measurements, carried out on a calibrated microscope, indicate that the concentration of intracellular flavin (144 ± 3.8 μM, n = 6) [Yu Q, Heikal AA, Unpublished Data] is relatively lower than that of NADH [53]. Since the 2P-excitation cross section of LipDH bound FAD is approximately ten times larger than that of free FAD [29], used as a reference, the average concentration of flavin is approximately 14 μM in HTB125 cells.
Owing to the multiexponential decay of the fluorescence of free NADH, FAD and their enzyme bound complexes in buffer, care must be taken during data interpretation of the autofluorescence lifetime measurements in living cells and tissues. It is not an easy task to assign different fluorescence decay components in a FLIM image to specific molecular conformation (e.g., free vs enzyme bound), excited state processes (e.g., charge transfer, energy transfer or isomerization) or environmental effects.
2P-autofluorescence polarization anisotropy
Using the same TCSPC principle as FLIM, time resolved fluorescence anisotropy measurements can be carried out to directly quantify the rotation dynamics of a fluorophore, which depend on the molecular size, binding with macro molecules, energy transfer and the viscosity of the surrounding environment [153]. Following the excitation of a fluorophore, using polarized laser pulses, molecules in their excited state tumble (or rotate), causing fluorescence depolarization. The parallel and perpendicular polarizations (with respect to the excitation laser polarization) of fluorescence intensity decays are then used to calculate the anisotropy decay constant (or rotational time). As a result, the anisotropy based approach offers the most direct and sensitive discrimination between free and enzyme bound NADH [42,53].
Vishwasrao etal. reported a global ana lysis of associated anisotropy decays of intrinsic NADH autofluorescence in brain hippocampal tissue, isolated from Sprague Dawley rats, in response to the metabolic transition from normoxia and hypoxia [42]. Using this sensitive and noninvasive approach, the authors were able to directly quantify the free and enzyme-bound population fractions of intracellular NADH in neural tissue for the first time. Their results also indicate that the NADH response to hypoxic inhibition of energy metabolism is more complicated than a simple increase in NADH concentration. In sub sequent studies on cultured cells, Yu and Heikal, using a similar flourescence anisotropy approach, observed, for the first time, that intracellular NADH autofluorescence also exhibits an associated anisotropy (Figure 4e, curve 1) at the single cell level [53]. Their results provided direct evidence for the equilibrated existence of free and enzyme bound populations of cellular NADH, since each conformation exhibits distinct fluorescence and rotational dynamics. By contrast, time-resolved autofluorescence anisotropy of intracellular flavin exhibits biexponential anisotropy (Figure 4F, curve 1) with a dominant slow rotational component that indicates a mostly enzyme-bound coenzyme [Yu Q, Heikal AA, Unpublished Data]. The rotational time of flavin in cells is much slower than that of free FAD in buffer at room temperature (Figure 4F, curve 2). The fluorescence based assay of the concentration of coenzymes and their conformation will enhance our ability to precisely quantify the role of NADH and FAD in vivo in metabolic activities, and to develop an accurate picture of their biochemistry and, ultimately, cellular energy metabolism.
Commercially available, extrinsic markers
Conventional labels are chemically designed to maximize their fluorescence quantum yield and to minimize their spectral (absorption and emission) overlap with the endogenous bio molecules, such as tryptophan, collagen, NADH and FAD, in biological samples. Using simple labeling protocols, these fluorescent markers allow for monitoring the spatial and temporal changes of mitochondrial structure, function and membrane potential under different pathophysiological conditions [158,159].
Rhodamine-123 is a lipophilic, photostable fluorophore with a high affinity for mitochondria and with minimal cell toxicity. Rhodamine-123 has been used for membrane potential studies [71,159–161] owing to its its high extinction coefficient (ε ~7.5 × 104 cm−1 mol−1) and fluorescence (~525 nm) quantum yield (~0.9) [71,162]. MitoTracker© derivatives (Invitrogen) are another class of mitochondrial markers, which contain a thiol reactive chloromethyl moiety that appears to be responsible for their high affinity for mitochondria. These mitochondrial markers exhibit a wide range of spectral properties (e.g., absorption: 490–644 nm and emission: 516–665 nm) that allows for experimental design flexibility and avoids the natural autofluorescence of cells. The fluorescence of another mitochondria label, JC 1 (5,5′,6,6′ tetrachloro 1,1′,3,3′ tetraethylbenzimidazolylcarbocyanine iodide), is sensitive to both its concentration and the mitochondrial membrane potential [163]. The JC 1 emission, which is green (529 nm) for a monomeric confor mation and at low membrane potential, becomes red (~590 nm) at a high membrane potential due to aggregate formation. JC-1 has also been used to monitor mitochondrial changes upon poisoning, uncoupling, anoxia [164] and apoptosis [165,166]. Tetramethylrhodamine methylester has been used to assess the concentration of mitochondria in neurons as a function of ATP demands and/or calcium buffering [167]. The results led to the hypothesis that neurons locally regulate the mitochondrial metabolic state. Genetically encoded fluorescent proteins also provide labeling specificity, large extinction coefficients, high fluorescent quantum yield, pH sensitivity and a wide range of mutations for multicolor labeling [168–173]. Mahajan etal., used Förster resonance energy transfer to demonstrate a direct interaction between Bcl 2 and Bax in individual mitochondria, labeled with green and blue fluorescent proteins, respectively [174], and their role in regulating apoptosis. Llopis etal., also used green fluorescent protein mutants to measure the local pH in the mitochondrial matrix and other cellular compartments [175] in both HeLa cells (pH 7.98) and rat neonatal cardiomyocytes (pH 7.91). Partikian etal. measured the diffusion of green fluorescent proteins in the mitochondrial matrix of fibroblast, liver, skeletal muscle and epithelial cell lines [176], which is three- to four times slower than its diffusion in water.
In oxidative stress studies, reduced dichlorofluorescein (DCFH)-diacetate has typically been used as a substrate for fluorescence based assays that measure the relative amounts of intracellular ROS [177]. This nonfluorescent substrate is taken up by the cell via passive diffusion and hydrolyzed by intracellular esterases to form DCFH, which is nonfluorescent. Upon encountering oxidants (e.g., hydrogen peroxide, nitric oxide and peroxynitrite oxidation), DCFH is oxidized to DCF, which is fluorescent. Oxidative stress in the cells can then be estimated by quantifying the DCF fluorescence intensity.
Conventional fluorescent labels are designed to report primarily on a specific aspect of mitochondria (e.g., structure or membrane potential), from which other physiological processes might be extrapolated. Nonspecific binding of these markers may also undermine the potential significance for a given observable readout. In addition, these extrinsic markers may interfere with the biological function of their cellular targets and must be controlled for. Finally, the fluorescence properties of some of these extrinsic labels can be less sensitive to changes in their intermolecular interactions and local environment.
Conclusion & future perspective
The intracellular coenzymes NAD(P)H and FAD are integral to a complex network of oxidation–reduction reactions and metabolic activities essential for the survival of mammalian cells and human health. Mitochondrial anomalies or mutations of the catalytic enzymes participating in these functions can have deleterious effects on cellular processes, which would lead to numerous health problems. These coenzymes are naturally fluorescent and, therefore, allow for true, noninvasive, imaging of metabolic activities in living cells and tissues. By contrast, exogenous fluorescence dyes can be toxic and may suffer from nonspecific binding as well as interference with biomolecular functions. In addition, the fluorescence of these coenzymes can be excited using different illumination wavelengths, ranging from UV to infrared regions, for complementary imaging using 1P- and 2P-fluorescence microscopy, respectively. Importantly, the fluorescence properties of NADH and FAD are sensitive to protein binding and local environment.
The concentrations and distributions of these naturally fluorescent coenzymes correlate with the metabolic and physiological states of cells and tissues, both in vitro as well as in vivo. Unlike extrinsic fluorescent markers, which must be added to cells or tissues, cellular co enzymes are nontoxic and unlikely to interfere with the diagnostic readout because they are natural participants in redox reactions and cellular metabolic activities. Flavin, which is almost exclusively associated with mitochondria and the oxidative phosphorylation pathway [30,31], exhibits a relatively higher fluorescence quantum efficiency than NADH. Based on the 2P-excitation cross-section, flavin can be selectively excited at approx imately 900 nm, which is suitable for deep-tissue imaging without the excitation spectral overlap. Ratiometric monitoring of metabolic activities can be achieved with simultaneous detection of intracellular NAD(P)H and flavin autofluorescence. The main challenge with flavin autofluorescence, however, is the relatively low cellular concentration of flavin and flavoproteins compared with NAD(P)H [148,149]. Collectively, the fluorescence properties and functional roles of NAD(P)H and FAD are the trademarks of a good biomarker.
Conventional biochemical methods are very useful in quantifying the cellular contents of biomolecules, such as NADH and FAD. These methods inherently require the destruction of cells (or tissues) and, therefore, yield snapshots of the metabolic and redox state of cells without the morphological context. By contrast, fluorescence microspectroscopy techniques are noninvasive with the spatiotemporal resolution required to differentiate between physiological states using NADH and flavin autofluorescence. Traditional cuvette based studies of these cellular coenzymes lack both the spatial information and real time monitoring of cellular response to metabolic state modulation. As a result, integrated fluorescence microspectroscopy and multiparametric approaches hold great potential for biological and biomedical research using intracellular coenzymes as natural biomarkers. When combined with these noninvasive experimental approaches, these natural biomarkers can report, in real time, on the spatiotemporal changes of the cell physiology.
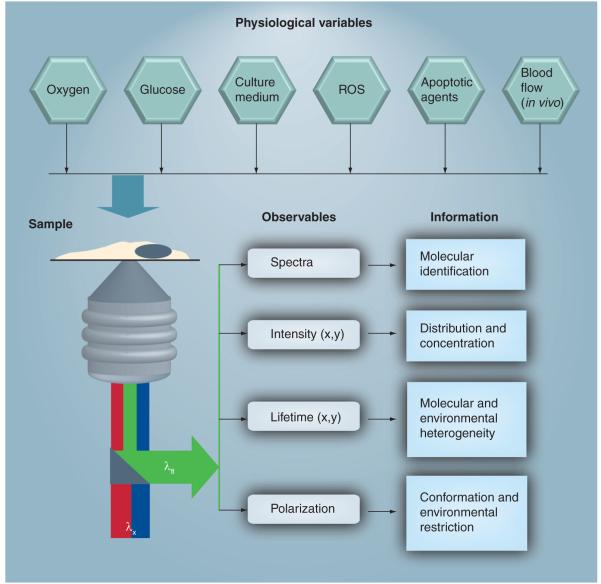
Despite the advantages of these natural biomarkers for biological research and biomedical/clinical applications, some inherent challenges remain. First, the spatial distribution of NADH concentration requires noninvasive, quantitative techniques with high spatial resolution to differentiate between cytosolic (glycolysis), mitochondrial (oxidative phosphorylation and TCA cycle) and nuclear (transcription) coenzymes. Second, an ability to selectively monitor the concentration of these coenzymes is essential for a meaningful readout without bias caused by possible photodamage. Third, the fact that these coenzymes are involved in a range of metabolic activities and biochemical reactions can complicate data interpretation. Fourth, the concentration of these coenzymes is sensitive to many physiological conditions, such as oxygen, in vivo blood flow, enzymatic mutations, the presence of other metabolites and chemical stimulation. As a result, a multiparameteric approach [92] becomes critical in monitoring one pathophysiological readout while fixing the others. Figure 6 illustrates this multi parametric approach from the autofluorescence microspectroscopy perspective of intracellular NAD(P)H and FAD. Fifth, in vivo imaging is essential for clinical diagnostics to realize the full potential of these coenzymes as natural biomarkers for numerous health problems including cancer, neurodegenerative diseases, diabetes and aging [178,179]. Recent developments in multi photon microscopy [136,180–184] and endo scopy [183,185] makes these very promising techniques for such clinical applications. The infrared laser pulses used in multiphoton microscopy can penetrate deeper into tissues than UV or visible lasers owing to the inherent tissue scattering and absorption associated with linear excitation. (By comparison with mammalian cell studies, there are limited studies on intracellular coenzymes in plant cells [186,187], which should be considered as an opportunity to exploit their characteristics as biomarkers.) To date, studies have mainly been qualitative with a focus on the diagnostic potential of these intracellular coenzymes. It is time to shift our effort towards more quantitative and mechanistic information for further advancement of cell biology, biomedicine and therapeutic targets, especially with the development of sophisticated microspectroscopic techniques.
Figure 6. Multiparametric approach for noninvasive imaging of intracellular coenzymes autofluorescence in living cells or tissues.
From the sample perspective, a range of physiological parameters (e.g., oxygen, glucose content, temperature, culture medium, reactive oxygen species and apoptotic agents for in vitro studies as compared with blood flow associated with in vivo imaging) has to be controlled for meaningful interpretation. The coenzyme of choice will be determined by the biological and medical hypothesis to be tested and will dictate the experimental design, such as excitation wavelength (λx), readout fluorescence (λfl) variables and information to be gained. Confocal one-photon (blue) or two-photon (red) microscopy are used to selectively excite reduced nicotinamide adenine dinucleotide (NADH) and flavin adenine dinucleotide with high spatial (x,y) and temporal resolution of their autofluorescence intensity. Spectral resolution of the autofluorescence emission, using a microscope-compatible spectrofluorimeter, can be used as a coenzyme fingerprint for their identification and protein binding state. Autofluorescence lifetime imaging enables us to assess the conformation (free vs protein bound) and environmental heterogeneity of intracellular NADH and flavin (i.e., how the cellular environment may influence their fluorescence quantum yield). In addition, these fluorescence lifetime and intensity images, recorded simultaneously on a calibrated microscope, can be used to construct concentration images of endogenous NADH and flavin at the single-cell level [53,157]. For a direct assessment of the size, conformation and environmental restriction of these coenzymes, time-resolved fluorescence anisotropy images can be used at the single-cell [53] or tissue level [42]. Simultaneous differential interference contrast imaging (not shown) also allow for monitoring changes in cell morphology.ROS: Reactive oxygen species.
Executive summary.
Coenzymes as biomarkers for metabolic activities
The ratio of cytosolic reduced nicotinamide adenine dinucleotide (NADH):oxidized nicotinamide adenine dinucleotide (NAD+) plays an important role in mediating glucose- and diabetes-induced glomerular dysfunction.
Inhibiting the NADH shuttles leads to glucose-induced increases in NADH autofluorescence and a reduction in ΔΨm, ATP content and glucose-induced insulin secretion.
Pyruvate stimulation increases cytosolic NADH phosphate without significant changes in mitochondrial NADH and ATP production needed for stimulating insulin secretion.
NADH and flavin autofluorescence (concentration) are sensitive to the respiratory-chain response to inhibitors (e.g., hypoxia and potassium cyanide), uncoupling (carbonyl cyanide-p-trifluoromethoxyphenylhydrazone), serum starvation and cell confluency.
In the absence of effective antioxidant defense mechanisms, elevated levels of reactive oxygen species and oxidative stress trigger changes in energy respiration, redox imbalance, elevated expression of NADH phosphate oxidase and a reduced expression of antioxidative enzymes.
The concentration of NADH is sensitive to the onset of mitochondria-mediated apoptosis and the dissipation of ΔΨm.
Nuclear NAD+ and NADH regulate carboxyl-terminal binding protein binding to cellular and viral transcription repressors.
Mitochondrial dysfunction in human diseases
Mitochondrial anomalies are associated with numerous health problems such as cancer, neurodegenerative diseases, Type 2 diabetes and aging.
Cancer cells exhibit an elevated level of NADH and a much reduced intracellular flavin concentration.
Low (mild-to-moderate dysplasia) and high (severe dysplasia and carcinoma in situ) grade precancers can be discriminated from normal tissues using NADH conformation (free vs enzyme bound).
Intracellular NADH phosphate autofluorescence in both normal and cancerous breast cells exhibits, associated anisotropy decays, which is direct evidence for the equilibrated presence of free and protein-bound NADH.
Senile plaques exhibit autofluorescence with a distinct emission spectrum in ex vivo and acute brain slices from Alzheimer’s diseased transgenic mouse model.
Inhibition of mitochondrial DNA transcription causes an increase in NADH autofluorescence and a reduction of the glucose-induced insulin secretion and ΔΨm in murine pancreatic β-cells.
Experimental approaches for interrogating intracellular coenzymes
Biochemical assays provide an accurate measure of intracellular coenzymes. Owing to their requirement for cell destruction, these assays provide snapshots of the metabolic activities without the morphological context.
Absorption-based concentration estimates are hindered by the spectral overlap and the knowledge of environmental effects on the extinction coefficient, the sample thickness and averaging over cell ensemble.
Fluorescence (one- or two-photon) microscopy allow for noninvasive monitoring of intracellular distribution of coenzymes and, therefore, the metabolic activities in living cells and tissues.
Fluorescence lifetime and polarization imaging microscopy allows for a noninvasive approach for quantifying the concentration and free-to-protein-bound fraction of intracellular coenzymes in living cells and tissues.
Commercially available, extrinsic markers
There are several commercially available markers for mitochondria (e.g., rhodamine-123, MitoTrackers©, JC-1, TMRM and green fluorescent protein) and oxidative stress (e.g., dichlorofluorescin-diacetate).
These markers allow for monitoring different aspects of mitochondria, such as shape and membrane potential.
However, these dyes may cause problems, such as nonspecific binding, toxicity and possible interference with the function of biological targets.
Conclusion
NADH and reduced flavin adenine dinucleotide play important roles in a wide range of cellular oxidation–reduction reactions.
NADH and flavin adenine dinucleotide are naturally fluorescent with distinct excitation and detection wavelengths.
The autoflourescence of these coenzymes is sensitive to protein binding and the local environment.
Fluorescence methods are not applicable to nonfluorescent species, such as NAD(P)+ and reduced flavin adenine dinucleotide.
When combined with noninvasive, quantitative methodologies, intracellular coenzymes can serve as natural biomarkers for a myriad of metabolic activities and mitochondrial anomalies.
Acknowledgements
The author would like to thank Q Yu (Pennsylvania State University, Department of Bioengineering, PA, USA) for her help in obtaining some of the experimental data used in Figure 4, KM Krise (Pennsylvania State University, Department of Chemistry) for his help in preparing Figure 1, and ED Sheets (University of Minnesota-Duluth, Duluth, MN, USA) for helpful comments.
This work was partially supported by the NIH (AG030949) and the National Science Foundation (MCB0718741).
Footnotes
Financial & competing interests disclosure The author has no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Bibliography
Papers of special note have been highlighted as:
▪ of considerable interest
- 1.Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Molecular Biology of the Cell. 4th Edition Garland Science; NY, USA: 2002. [Google Scholar]
- 2.Christen Y. Oxidative stress and Alzheimer disease. Am. J. Clin. Nutr. 2000;71(2):621S–629S. doi: 10.1093/ajcn/71.2.621s. [DOI] [PubMed] [Google Scholar]
- 3.Jacquard C, Trioulier Y, Cosker F, et al. Brain mitochondrial defects amplify intracellular [Ca2+] rise and neurodegeneration but not Ca2+ entry during NMDA receptor activation. FASEB J. 2006;20(7):1021–1023. doi: 10.1096/fj.05-5085fje. [DOI] [PubMed] [Google Scholar]
- 4.Duchen MR. Mitochondria in health and disease: perspectives on a new mitochondrial biology. Mol. Aspects Med. 2004;25(4):365–451. doi: 10.1016/j.mam.2004.03.001. ▪▪ Insightful perspective on mitochondria in health, diseases and basic biology.
- 5.Duchen MR. Roles of mitochondria in health and disease. Diabetes. 2004;53(Suppl. 1):S96–S102. doi: 10.2337/diabetes.53.2007.s96. [DOI] [PubMed] [Google Scholar]
- 6.Armstrong JS. Mitochondria: a target for cancer therapy. Br. J. Phamcol. 2006;147:239–248. doi: 10.1038/sj.bjp.0706556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.E Flescher. Jasmonates in cancer therapy. Cancer Lett. 2007;245:1–10. doi: 10.1016/j.canlet.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 8.Neuzil J, Dong LF, Ramnathapuram L. Vitamin E analogues as a novel group of mitocans: anti-cancer agents that act by targeting mitochondria. Mol. Asp. Med. 2007;28:607–645. doi: 10.1016/j.mam.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 9.Kagan VE, Bayir A, Bayir H, et al. Mitochondria targeted disruptors and inhibitors of cytochrome C/cardiolipin peroxidase complexes: a new strategy in anti apoptotic drug discovery. Mol. Nutr. Food. Res. 2009;53(1):104–114. doi: 10.1002/mnfr.200700402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Howell N, Taylor SW, Fahy E, Murphy A, Ghosh SS. Restoring energy in a power crisis: mitochondrial targets for drug development. Drug Discov. Today Targets. 2003;2(5):208–216. [Google Scholar]
- 11.Szeto HH. Mitochondria-targeted peptide antioxidants: novel neuroprotective agents. AAPS J. 2006;8(3):E521–E531. doi: 10.1208/aapsj080362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harper JA, Dickinson K, Brand MD. Mitochondrial uncoupling as a target for drug development for the treatment of obesity. Obesity Rev. 2001;2(4):255–265. doi: 10.1046/j.1467-789x.2001.00043.x. [DOI] [PubMed] [Google Scholar]
- 13.Chance B. Pyridine nucleotide as an indicator of the oxygen requirements for energy-linked functions of Mitochondria. Circ. Res. 1976;5(Suppl. 1):131I–138I. [PubMed] [Google Scholar]
- 14.Chance B, Cohen P, Jobsis F, Schoener B. Intracellular oxidation–reduction states in vivo. Science. 1962;137:499–508. doi: 10.1126/science.137.3529.499. [DOI] [PubMed] [Google Scholar]
- 15.Chance B, Jamieson D, Coles H. Energy-linked pyridine nucleotide reduction: inhibitory effects of hyperbaric oxygen in vitro and in vivo. Nature. 1965;206(981):257–263. doi: 10.1038/206257a0. [DOI] [PubMed] [Google Scholar]
- 16.Chance B, Legallais V, Schoener B. Metabolically linked changes in fluorescence emission spectra of cortex of rat brain, kidney and adrenal gland. Nature. 1962;195:1073–1075. doi: 10.1038/1951073a0. [DOI] [PubMed] [Google Scholar]
- 17.Chance B, Williams GR. Respiratory enzymes in oxidative phosphorylation. II. Difference spectra. J. Biol. Chem. 1955;217(1):395–407. [PubMed] [Google Scholar]
- 18.Chance B, Williams GR. Respiratory enzymes in oxidative phosphorylation. I. Kinetics of oxygen utilization. J. Biol. Chem. 1955;217(1):383–393. [PubMed] [Google Scholar]
- 19.Chance B, Williams GR. Respiratory enzymes in oxidative phosphorylation. III. The steady state. J. Biol. Chem. 1955;217(1):409–427. [PubMed] [Google Scholar]
- 20.Chance B, Williams GR. Respiratory enzymes in oxidative phosphorylation. IV. The respiratory chain. J. Biol. Chem. 1955;217(1):429–438. [PubMed] [Google Scholar]
- 21.Chance B, Williams GR, Holmes WF. Higgins J: Respiratory enzymes in oxidative phosphorylation. V. A mechanism for oxidative phosphorylation. J. Biol. Chem. 1955;217(1):439–451. [PubMed] [Google Scholar]
- 22.Jones JB, Song JJ, Hempen PM, Parmigiani G, Hruban RH, Kern SE. Detection of mitochondrial DNA mutations in pancreatic cancer offers a “mass” ive advantage over detection of nuclear DNA mutations. Cancer Res. 2001;61(4):1299–1304. [PubMed] [Google Scholar]
- 23.Stryer L. Biochemistry. 4th Edition WH Freeman and Co.; NY, USA: 1999. [Google Scholar]
- 24.Ying W. NAD+/NADH and NADP+/NADPH in cellular functions and cell death: regulation and biological consequences. Antioxid. Redox Signal. 2008;10(2):179–206. doi: 10.1089/ars.2007.1672. [DOI] [PubMed] [Google Scholar]
- 25.Klaidman LK, Leung AC, Adams JD., Jr High performance liquid chromatography ana lysis of oxidized and reduced pyridine dinucleotides in specific brain regions. Anal. Biochem. 1995;228(2):312–317. doi: 10.1006/abio.1995.1356. [DOI] [PubMed] [Google Scholar]
- 26.Glassman WS, Steinberg M, Alfano RR. Time resolved and steady state fluorescence spectroscopy from normal and malignant cultured human breast cell lines. Lasers Life Sci. 1994;6(2):91–98. [Google Scholar]
- 27.Palmer GM, Keely PJ, Breslin TM. Ramanujam N: Autofluorescence spectroscopy of normal and malignant human breast cell lines. Photochem. Photobiol. 2003;78(5):462–469. doi: 10.1562/0031-8655(2003)078<0462:asonam>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 28.Kunz WS, Kunz W. Contribution of different enzymes to flavoprotein fluorescence of isolated rat liver mitochondria. Biochim. Biophys. Acta. 1985;841(3):237–246. doi: 10.1016/0304-4165(85)90064-9. [DOI] [PubMed] [Google Scholar]
- 29.Huang S, Heikal AA, Webb WW. Two photon fluorescence spectroscopy and microscopy of NAD(P)H and flavoprotein. Biophys. J. 2002;82:2811–2825. doi: 10.1016/S0006-3495(02)75621-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koke JR, Wylie W, Wills M. Sensitivity of flavoprotein fluorescence to oxidative state in single isolated heart cells. Cytobios. 1981;32(127–128):139–145. [PubMed] [Google Scholar]
- 31.Scholz R, Thurman RG, Williamson JR, Chance B, Bucher T. Flavin and pyridine nucleotide oxidation–reduction changes in perfused rat liver. I. Anoxia and subcellular localization of fluorescent flavoproteins. J. Biol. Chem. 1969;244(9):2317–2324. [PubMed] [Google Scholar]
- 32.Modica Napolitano JS, Singh KK. Mitochondrial dysfunction in cancer. Mitochondrion. 2004;4:755–762. doi: 10.1016/j.mito.2004.07.027. [DOI] [PubMed] [Google Scholar]
- 33.Scheffler IE. Mitochondria. Wiley-Liss; NY, USA: 1999. [Google Scholar]
- 34.Benson RC, Meyer RA, Zaruba ME. McKhann GM: Cellular autofluorescence – is it due to flavins? J. Histochem. Cytochem. 1979;27(1):44–48. doi: 10.1177/27.1.438504. [DOI] [PubMed] [Google Scholar]
- 35.Müller F. Chemistry and Biochemistry of Flavoenzymes. CRC Press; FL, USA: 1991. [Google Scholar]
- 36.Berg M, Tymoczko JL, Stryer L. Biochemistry. Freeman; NY, USA: 1995. [Google Scholar]
- 37.Warburg O. On the origin of cancer cells. Science. 1956;123(3191):309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 38.Eto K, Tsubamoto Y, Terauchi Y, et al. Role of NADH shuttle system in glucose induced activation of mitochondrial metabolism and insulin secretion. Science. 1999;283(5404):981–985. doi: 10.1126/science.283.5404.981. [DOI] [PubMed] [Google Scholar]
- 39.Uppal A, Gupta PK. Measurements of NADH concentration in normal and malignant human tissues from breast and oral cavity. Biotechnol. Appl. Biochem. 2003;37:45–50. doi: 10.1042/ba20020052. [DOI] [PubMed] [Google Scholar]
- 40.Liu ZH, Cai RX, Wang J. Current Development in the Determination of Intracellular NADH Level. Springer; NY, USA: 2006. [Google Scholar]
- 41.Wang W. NAD+/NADH and NADP+/NADPH in cellular functions and cell death: regulation and biological consequences. Antioxid. Redox Signal. 2008;10(2):179–206. doi: 10.1089/ars.2007.1672. [DOI] [PubMed] [Google Scholar]
- 42.Vishwasrao HD, Heikal AA, Kasischke KA, Webb WW. Conformational dependence of intracellular NADH on metabolic state revealed by associated fluorescence anisotropy. J. Biol. Chem. 2005;280:25119–25126. doi: 10.1074/jbc.M502475200. ▪▪ Original paper that revealed an associated anisotropy behavior of intracellular reduced nicotinamide adenine dinucleotide in brain slices as a direct measure of free and protein-bound coenzyme in response to hypoxia.
- 43.Bird DK, Yan L, Vrotsos KM, et al. Metabolic mapping of MCF 10A human breast cells via multiphoton fluorescence lifetime imaging of the coenzyme NADH. Cancer Res. 2005;65:8766–8773. doi: 10.1158/0008-5472.CAN-04-3922. [DOI] [PubMed] [Google Scholar]
- 44.Tilton RG, Baier LD, Harlow JE, Smith SR, Ostrow E, Williamson JR. Diabetes-induced glomerular dysfunction: links to a more reduced cytosolic ratio of NADH/NAD+ Kidney Int. 1992;41:778–788. doi: 10.1038/ki.1992.121. [DOI] [PubMed] [Google Scholar]
- 45.Dukes ID, McIntyre MS, Mertz RJ, et al. Dependence on NADH produced during glycolysis for β-cell glucose signaling. J. Biol. Chem. 1994;269(15):10979–10982. [PubMed] [Google Scholar]
- 46.Ido Y, Kilo C, Williamson JR. Cytosolic NADH/NAD+, free radicals, and vascular dysfunction in early diabetes mellitus. Diabetologia. 1997;40:S115–S117. doi: 10.1007/s001250051422. [DOI] [PubMed] [Google Scholar]
- 47.Rocheleau JV, Head WS, Piston DW. Quantitative NAD(P)H/flavoprotein autofluorescence imaging reveals metabolic mechanisms of pancreatic islet pyruvate response. J. Biol. Chem. 2004;279:31780–31787. doi: 10.1074/jbc.M314005200. [DOI] [PubMed] [Google Scholar]
- 48.Rocheleau J, Head WS, Piston D. Two photon NAD(P)H and one-photon flavoprotein autofluorescence imaging to examine the metabolic mechanisms of pancreatic islet β-cell function. Microsc. Microanal. 2003;9:218–219. [Google Scholar]
- 49.Alberts B, Bray D, Johnson A, et al. Essential Cell Biology. Garland Publishing Inc.; NY, USA: 2003. [Google Scholar]
- 50.Scheffler IE. Mitochondria. 2nd Edition Wiley Liss; NY, USA: 2007. [Google Scholar]
- 51.Hajnoczky G, Csordas G, Das S, et al. Mitochondrial calcium signaling and cell death: approaches for assessing the role of mitochondrial Ca2+ uptake in apoptosis. Cell Calcium. 2006;40(5–6):553–560. doi: 10.1016/j.ceca.2006.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Walsh C, Barrow S, Voronina S, Chvanov M, Petersen OH, Tepikin A. Modulation of calcium signalling by mitochondria. Biochim. Biophys. Acta. 2009;1787(11):1374–1382. doi: 10.1016/j.bbabio.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 53.Yu Q, Heikal AA. Two photon autofluorescence dynamics imaging reveals sensitivity of intracellular NADH concentration and conformation to cell physiology at the single-cell level. J. Photochem. Photobiol. B. 2009;95(1):46–57. doi: 10.1016/j.jphotobiol.2008.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chorvat D, Jr, Kirchnerova J, Cagalinec M, Smolka J, Mateasik A, Chorvatova A. Spectral unmixing of flavin autofluorescence components in cardiac myocytes. Biophys. J. 2005;89(6):L55–L57. doi: 10.1529/biophysj.105.073866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Romashko DN, Marban E, O’Rourke B. Subcellular metabolic transients and mitochondrial redox waves in heart cells. Proc. Natl Acad. Sci. USA. 1998;95(4):1618–1623. doi: 10.1073/pnas.95.4.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Skovronsky DM, Lee VMY, Trojanowski JQ. Neurodegenerative diseases: new concepts of pathogenesis and their therapeutic implications. Annu. Rev. Pathol. Mech. Dis. 2006;1:151–170. doi: 10.1146/annurev.pathol.1.110304.100113. [DOI] [PubMed] [Google Scholar]
- 57.Smith MA, Rottkamp CA, Nunomura A, Raina AK, Perry G. Oxidative stress in Alzheimer’s disease. Biochim. Biophys. Acta. 2000;1502:139–144. doi: 10.1016/s0925-4439(00)00040-5. [DOI] [PubMed] [Google Scholar]
- 58.Ischiropoulos H, Beckman JS. Oxidative stress and nitration in neurodegeneration: cause, effect, or association? J. Clin. Invest. 2003;111:163–169. doi: 10.1172/JCI17638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sherer TB, Betarbet R, Greenamyre JT. Environment, mitochondria, and Parkinson’ s disease. Neuroscientist. 2002;8:192–197. doi: 10.1177/1073858402008003004. [DOI] [PubMed] [Google Scholar]
- 60.Furukawa S, Fujita T, Shimabukuro M, et al. Increased oxidative stress in obesity and its impact on metabolic syndrome. J. Clin. Invest. 2004;114(12):1752–1761. doi: 10.1172/JCI21625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Le SB, Hailer MK, Buhrow S, et al. Inhibition of mitochondrial respiration as a source of adaphostin induced reactive oxygen species and cytotoxicity. J. Biol. Chem. 2007;282(12):8860–8872. doi: 10.1074/jbc.M611777200. [DOI] [PubMed] [Google Scholar]
- 62.Koch OR, Pani G, Borrello S, et al. Oxidative stress and antioxidant defenses in ethanol-induced cell injury. Mol. Aspects Med. 2004;25(1–2):191–198. doi: 10.1016/j.mam.2004.02.019. [DOI] [PubMed] [Google Scholar]
- 63.Bailey SM, Cunningham CC. Contribution of mitochondria to oxidative stress associated with alcoholic liver disease. Free Radic. Biol. Med. 2002;32(1):11–16. doi: 10.1016/s0891-5849(01)00769-9. [DOI] [PubMed] [Google Scholar]
- 64.Zhou Z, Wang L, Song Z, Lambert JC, McClain CJ, Kang YJ. A critical involvement of oxidative stress in acute alcohol induced hepatic TNF α-production. Am. J. Pathol. 2003;163:1137–1146. doi: 10.1016/s0002-9440(10)63473-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Adachi M, Ishii H. Role of mitochondria in alcoholic liver injury. Free Radic. Biol. Med. 2002;32(6):487–491. doi: 10.1016/s0891-5849(02)00740-2. [DOI] [PubMed] [Google Scholar]
- 66.Liang J, Wu WL, Liu ZH, Meib YJ, Cai RX, Shen P. Study the oxidative injury of yeast cells by NADH autofluorescence. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2007;67(2):355–359. doi: 10.1016/j.saa.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 67.Yang MS, Li D, Lin T, Zheng JJ, Zheng W, Qu JY. Increase in intracellular free/bound NAD(P)H as a cause of Cd-induced oxidative stress in the Hep G2 cells. Toxicology. 2008;247(1):6–10. doi: 10.1016/j.tox.2008.01.021. [DOI] [PubMed] [Google Scholar]
- 68.Elmore S. Apoptosis: a review of programmed cell death. Toxicol. Pathol. 2007;35(4):495–516. doi: 10.1080/01926230701320337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lockshin RA, Zakeri Z. Programmed cell death and apoptosis: origins of the theory. Nat. Rev. Mol. Biol. 2001;2:545–550. doi: 10.1038/35080097. [DOI] [PubMed] [Google Scholar]
- 70.Crow MT, Mani K, Nam YJ, Kitsis RN. The mitochondrial death pathway and cardiac myocyte apoptosis. Circulation Res. 2004;95:957–970. doi: 10.1161/01.RES.0000148632.35500.d9. [DOI] [PubMed] [Google Scholar]
- 71.Chen LB. Mitochondrial membrane potential in living cells. Annu. Rev. Cell Biol. 1988;4:155–181. doi: 10.1146/annurev.cb.04.110188.001103. [DOI] [PubMed] [Google Scholar]
- 72.Vermeulen K, Van Bockstaele DR, Berneman ZN. Apoptosis: mechanisms and relevance in cancer. Ann. Hematol. 2005;84:627–639. doi: 10.1007/s00277-005-1065-x. [DOI] [PubMed] [Google Scholar]
- 73.Brewer MA, Utzinger U, Li Y, et al. Fluorescence spectroscopy as a biomarker in a cell culture and in a nonhuman primate model for ovarian cancer chemopreventive agents. J. Biomed. Optics. 2002;7(1):20–26. doi: 10.1117/1.1427672. [DOI] [PubMed] [Google Scholar]
- 74.Kirkpatrick ND, Zou C, Brewer MA, Brands WR, Drezek RA, Utzinger U. Endogenous fluorescence spectroscopy of cell suspensions for chemopreventive drug monitoring. Photochem. Photobiol. 2005;81(1):125–134. doi: 10.1562/2004-08-09-RA-267. [DOI] [PubMed] [Google Scholar]
- 75.Nieminen AL, Byrne AM, Herman B, Lemasters JJ. Mitochondrial permeability transition induced by t-buOOH:NAD(P)H and reactive oxygen species. Am. J. Physiol. Cell Physiol. 1997;272:C1286–C1294. doi: 10.1152/ajpcell.1997.272.4.C1286. [DOI] [PubMed] [Google Scholar]
- 76.Shinho A, Matsuda M, Handa J, Chance B. Poor recovery of mitochondrial redox state in CA1 after transient forebrain ischemia in gerbils. Stroke. 1998;29:2421–2424. doi: 10.1161/01.str.29.11.2421. [DOI] [PubMed] [Google Scholar]
- 77.Petit PX, Gendron M-C, Schrantz N, et al. Oxidation of pyridine nucleotides during Fas and ceramide induced apoptosis in Jurkat cells: correlation with changes in mitochondria, glutathione depletion, intracellular acidification and caspase 3 activation. Biochem. J. 2001;353:357–367. [PMC free article] [PubMed] [Google Scholar]
- 78.Rouhanizadeh M, Hwang J, Clempus RE, et al. Oxidized-1-palmitoyl-2-arachidonoyl sn-glycero-3-phosphorylcholine induces vascular endothelial superoxide production: implication of NADPH oxidase. Free Radic. Biol. Med. 2005;39(11):1512–1522. doi: 10.1016/j.freeradbiomed.2005.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Galindo MF, Jordán J, González García C, Cena V. Reactive oxygen species induce swelling and cytochrome C release but not transmembrane depolarization in isolated rat brain mitochondria. Br. J. Pharmacol. 2003;139:797–804. doi: 10.1038/sj.bjp.0705309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Levitt JM, Baldwin A, Papadakis A, et al. Intrinsic fluorescence and redox changes associated with apoptosis of primary human epithelial cells. J. Biomed. Opt. 2006;11(6):064012. doi: 10.1117/1.2401149. [DOI] [PubMed] [Google Scholar]
- 81.Wang H-W, Gukassyan V, Chen C-T, et al. Differentiation of apoptosis from necrosis by dynamic changes of reduced nicotinamide adenine dinucleotide fluorescence lifetime in live cells. J. Biomed. Opt. 2008;13:054011. doi: 10.1117/1.2975831. [DOI] [PubMed] [Google Scholar]
- 82.Zhang Q, Piston DW, Goodman RH. Regulation of corepressor function by nuclear NADH. Science. 2002;295(8):1895–1897. doi: 10.1126/science.1069300. ▪▪ Original paper describing the role of nuclear NADH in regulating corepressor function.
- 83.Chen S, Whetstine JR, Ghosh S, et al. The conserved NAD(H) dependent corepressor CTBP-1 regulates Caenorhabditis elegans life span. Proc. Natl Acad. Sci. USA. 2009;106:1496–1501. doi: 10.1073/pnas.0802674106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lin S-J, Guarente L. Nicotinamide adenine dinucleotide, a metabolic regulator of transcription, longevity and disease. Curr. Opin. Cell Biol. 2003;15(2):241–246. doi: 10.1016/s0955-0674(03)00006-1. [DOI] [PubMed] [Google Scholar]
- 85.Buck SW, Gallo CM, Smith JS. Diversity in the Sir2 family of protein deacetylases. J. Leukoc. Biol. 2004;75:939–950. doi: 10.1189/jlb.0903424. [DOI] [PubMed] [Google Scholar]
- 86.Yang T, Sauve AA. NAD metabolism and sirtuins: metabolic regulation of protein deacetylation in stress and toxicity. AAPS J. 2006;8(4):E632–E643. doi: 10.1208/aapsj080472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Singh KK. Mitochondrial dysfunction is a common phenotype in aging and cancer. Ann. NY Acad. Sci. 2004;1019:260–264. doi: 10.1196/annals.1297.043. [DOI] [PubMed] [Google Scholar]
- 88.Modica-Napolitano JS, Singh KK. Mitochondria as targets for detection and treatment of cancer. Expert Rev. Mol. Med. 2002;4(9):1–19. doi: 10.1017/S1462399402004453. [DOI] [PubMed] [Google Scholar]
- 89.Modica-Napolitano JS, Kulawiec M. Singh KK: Mitochondria and human cancer. Curr. Mol. Med. 2007;7(1):121–131. doi: 10.2174/156652407779940495. [DOI] [PubMed] [Google Scholar]
- 90.Lu B. Mitochondrial dynamics and neurodegeneration. Curr. Neurol. Neurosci. Rep. 2009;9(3):212–219. doi: 10.1007/s11910-009-0032-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 92.Mayevsky A. Mitochondrial function and energy metabolism in cancer cells: past overview and future perspectives. Mitochondrion. 2009;9:165–179. doi: 10.1016/j.mito.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 93.Pederson PL. Tumor mitochondria and the bioenergetics of cancer cells. Prog. Exp. Tumor Res. 1978;22:198–274. doi: 10.1159/000401202. [DOI] [PubMed] [Google Scholar]
- 94.Chang LO, Schnaitman CA, Morris HP. Comparison of the mitochondrial membrane proteins in rat liver and hepatomas. Cancer Res. 1971;31(2):108–113. [PubMed] [Google Scholar]
- 95.Ishii T, Yasuda K, Akatsuka A, Hino O, Hartman PS, Ishii N. A mutation in the SDHC gene of complex II increases oxidative stress, resulting in apoptosis and tumorigenesis. Cancer Res. 2005;65(1):203–209. [PubMed] [Google Scholar]
- 96.Villette S, Pigaglio-Deshayes S, Vever-Bizet C, Validire P, Bourg-Heckly G. Ultraviolet induced autofluorescence characterization of normal and tumoral esophageal epithelium cells with quantitation of NAD(P)H. Photochem. Photobiol. Sci. 2006;5:483–492. doi: 10.1039/b514801d. [DOI] [PubMed] [Google Scholar]
- 97.Skala MC, Riching KM, Gendron-Fitzpatrick A, et al. In vivo multiphoton microscopy of NADH and FAD redox states, fluorescence lifetimes, and cellular morphology in precancerous epithelia. Proc. Natl Acad. Sci. USA. 2007;104(49):19494–19499. doi: 10.1073/pnas.0708425104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Skala MC, Riching KM, Bird DK, et al. In vivo multiphoton fluorescence lifetime imaging of protein-bound and free nicotinamide adenine dinucleotide in normal and precancerous epithelia. J. Biomed. Opt. 2007;12(2):024014. doi: 10.1117/1.2717503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- 100.Krishnan KJ, Reeve AK, Turnbull DM. Do mitochondrial DNA mutations have a role in neurodegenerative disease? Biochem. Soc. Trans. 2007;35:1232–1235. doi: 10.1042/BST0351232. [DOI] [PubMed] [Google Scholar]
- 101.Kwan AC, Duff K, Gouras GK, Webb WW. Optical visualization of Alzheimer s pathology via multiphoton excited intrinsic fluorescence and second harmonic generation. Optics Express. 2009;17(5):3679–3689. doi: 10.1364/oe.17.003679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Brownlee M. Biochemistry and molecular cell biology of diabetic complications. Nature. 2001;414:813–820. doi: 10.1038/414813a. [DOI] [PubMed] [Google Scholar]
- 103.Wiederkehr A, Wollheim CB. Impact of mitochondrial calcium on the coupling of metabolism to insulin secretion in the pancreatic β-cell. Cell Calcium. 2008;44(1):64–76. doi: 10.1016/j.ceca.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 104.Wollheim CB, Maechler P. β-cell mitochondria and insulin secretion: messenger role of nucleotides and metabolites. Diabetes. 2002;51(1):S37–S42. doi: 10.2337/diabetes.51.2007.s37. [DOI] [PubMed] [Google Scholar]
- 105.Noda M, Yamashita S, Takahashi N, et al. Switch to anaerobic glucose metabolism with NADH accumulation in the β-cell model of mitochondrial diabetes: characteristics of β-HC9 cells deficient in mitochondrial DNA transcription. J. Biol. Chem. 2002;277(44):41817–41826. doi: 10.1074/jbc.M207690200. [DOI] [PubMed] [Google Scholar]
- 106.Gore M, Ibbott F, McIlwain H. The cozymase of mammalian brain. Biochem. J. 1950;47(1):121–127. doi: 10.1042/bj0470121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sporty JL, Kabir MM, Turteltaub KW, Ognibene T, Lin SJ, Bench G. Single sample extraction protocol for the quantification of NAD and NADH redox states in Saccharomyces cerevisiae. J. Sep. Sci. 2008;31(18):3202–3211. doi: 10.1002/jssc.200800238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Britz-McKibbin P, Markuszewski MJ, Iyanagi T, Matsuda K, Nishioka T, Terabe S. Picomolar ana lysis of flavins in biological samples by dynamic pH junction sweeping capillary electrophoresis with laser-induced fluorescence detection. Anal. Biochem. 2003;313(1):89–96. doi: 10.1016/s0003-2697(02)00510-9. [DOI] [PubMed] [Google Scholar]
- 109.Giblin FJ, Reddy VN. Pyridine nucleotides in ocular tissues as determined by the cycling assay. Exp. Eye Res. 1980;31(5):601–609. doi: 10.1016/s0014-4835(80)80019-4. [DOI] [PubMed] [Google Scholar]
- 110.Matsumura H, Miyachi S. Cycling assay for nicotinamide adenine dinucleotides. Methods Enzymol. 1980;69:465–470. [Google Scholar]
- 111.Klaidman LK, Leung AC, Adams JD., Jr High-performance liquid chromatography ana lysis of oxidized and reduced pyridine dinucleotides in specific brain regions. Anal. Biochem. 1995;228(2):312–317. doi: 10.1006/abio.1995.1356. [DOI] [PubMed] [Google Scholar]
- 112.Umemura K, Kimura H. Determination of oxidized and reduced nicotinamide adenine dinucleotide in cell monolayers using a single extraction procedure and a spectrophotometric assay. Anal. Biochem. 2005;338(1):131–135. doi: 10.1016/j.ab.2004.10.020. [DOI] [PubMed] [Google Scholar]
- 113.Xie W, Xu A, Yeung ES. Determination of NAD+ and NADH in a single cell under hydrogen peroxide stress by capillary electrophoresis. Anal. Chem. 2009;81(3):1280–1284. doi: 10.1021/ac802249m. [DOI] [PubMed] [Google Scholar]
- 114.Stanley PE. Determination of subpicomole levels of NADH and FMN using bacterial luciferase and the liquid scintillation spectrometer. Anal. Biochem. 1971;39(2):441–453. doi: 10.1016/0003-2697(71)90434-9. [DOI] [PubMed] [Google Scholar]
- 115.Winstead JA, Moss SA. γ-irradiated flavin adenine dinucleotide: a d-amino acid oxidase inhibitor. Radiat. Res. 1972;52(3):520–527. [PubMed] [Google Scholar]
- 116.Chance B. Spectrophotometry of intracellular respiratory pigments. Science. 1954;120(3124):767–775. doi: 10.1126/science.120.3124.767. [DOI] [PubMed] [Google Scholar]
- 117.Chorvat D, Jr, Chorvatova A. Spectrally resolved time-correlated single photon counting: a novel approach for characterization of endogenous fluorescence in isolated cardiac myocytes. Eur. Biophys. J. 2006;36:73–83. doi: 10.1007/s00249-006-0104-4. [DOI] [PubMed] [Google Scholar]
- 118.Duysens LN, Amesz J. Fluorescence spectrophotometry of reduced phosphopyridine nucleotide in intact cells in the near ultraviolet and visible region. Biochim. Biophys. Acta. 1957;24(1):19–26. doi: 10.1016/0006-3002(57)90141-5. [DOI] [PubMed] [Google Scholar]
- 119.Uppal A, Ghosh N, Datta A, Gupta PK. Fluorimetric estimation of the concentration of NADH from human blood samples. Biotechnol. Appl. Biochem. 2005;41(Pt 1):43–47. doi: 10.1042/BA20030197. [DOI] [PubMed] [Google Scholar]
- 120.Papadopoulos AJ, Zhadin NN, Steinberg ML, Alfano RR. Fluorescence spectroscopy of normal, SV40-transformed human keratinocytes, and carcinoma cells. Cancer Biochem. Biophys. 1999;17:13–23. [PubMed] [Google Scholar]
- 121.Benson RC, Meyer RA, Zaruba ME, McKhann GM. Cellular autofluorescence – is it due to flavins? J. Histochem. Cytochem. 1979;27(1):44–48. doi: 10.1177/27.1.438504. [DOI] [PubMed] [Google Scholar]
- 122.Croce AC, Ferrigno A, Vairetti M, Bertone R, Freitasa I, Bottirolia G. Autofluorescence properties of isolated rat hepatocytes under different metabolic conditions. Photochem. Photobiol. Sci. 2004;3(10):920–926. doi: 10.1039/b407358d. [DOI] [PubMed] [Google Scholar]
- 123.Ranji M, Kanemoto S, Matsubara M, et al. Fluorescence spectroscopy and imaging of myocardial apoptosis. J. Biomed. Opt. 2006;11(6):064036. doi: 10.1117/1.2400701. [DOI] [PubMed] [Google Scholar]
- 124.Pogue BW, Pitts JD, Mycek MA, et al. In vivo NADH fluorescence monitoring as an assay for cellular damage in photodynamic therapy. Photochem. Photobiol. 2001;74(6):817–824. doi: 10.1562/0031-8655(2001)074<0817:ivnfma>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 125.Mayevsky A, Chance B. Oxidation–reduction states of NADH in vivo: from animals to clinical use. Mitochondrion. 2007;7(5):330–339. doi: 10.1016/j.mito.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 126.Hogan MC, Stary CM, Balaban RS, Combs CA. NAD(P)H fluorescence imaging of mitochondrial metabolism in contracting Xenopus skeletal muscle fibers: effect of oxygen availability. J. Appl. Physiol. 2005;98(4):1420–1426. doi: 10.1152/japplphysiol.00849.2004. [DOI] [PubMed] [Google Scholar]
- 127.Shuttleworth CW, Brennan AM, Connor JA. NAD(P)H fluorescence imaging of postsynaptic neuronal activation in murine hippocampal slices. J. Neurosci. 2003;23(8):3196–3208. doi: 10.1523/JNEUROSCI.23-08-03196.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Reinert KC, Dunbar RL, Gao W, Chen G, Ebner TJ. Flavoprotein autofluorescence imaging of neuronal activation in the cerebellar cortex in vivo. J. Neurophysiol. 2004;92:199–211. doi: 10.1152/jn.01275.2003. [DOI] [PubMed] [Google Scholar]
- 129.Reinert KC, Gao W, Chen G, Ebner TJ. Flavoprotein autofluorescence imaging in the cerebellar cortex in vivo. J. Neurosci. Res. 2007;85(15):3221–3232. doi: 10.1002/jnr.21348. [DOI] [PubMed] [Google Scholar]
- 130.Williams RM, Piston DW, Webb WW. Two-photon molecular excitation provides intrinsic 3-dimensional resolution for laser-based microscopy and microphotochemistry. FASEB J. 1994;8:804–813. doi: 10.1096/fasebj.8.11.8070629. [DOI] [PubMed] [Google Scholar]
- 131.Gniadecki R, Thorn T, Vicanova J, Petersen A, Wulf HC. Role of mitochondria in ultraviolet-induced oxidative stress. J. Cell Biochem. 2000;80:216–222. doi: 10.1002/1097-4644(20010201)80:2<216::aid-jcb100>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 132.Lisby S, Gniadecki R, Wulf HC. UV-induced DNA damage in human keratinocytes: quantitation and correlation with long-term survival. Exp. Dermatol. 2005;14:349–355. doi: 10.1111/j.0906-6705.2005.00282.x. [DOI] [PubMed] [Google Scholar]
- 133.Bhawalkar JD, Shih A, Pan SJ, et al. Two-photon laser scanning fluorescence microscopy – from a fluorophore and specimen perspective. Bioimaging. 1996;4:168–178. [Google Scholar]
- 134.Centonze VE, White JG. Multiphoton excitation provides optical sections from deeper within scattering specimens that confocal imaging. Biophys. J. 1998;75:2015–2024. doi: 10.1016/S0006-3495(98)77643-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Denk W, Strickler JH, Webb WW. Two-photon laser scanning fluorescence microscopy. Science. 1990;245:73–76. doi: 10.1126/science.2321027. [DOI] [PubMed] [Google Scholar]
- 136.Masters BR, So PTC. Antecedents of two-photon excitation laser scanning microscopy. Microsc. Res. Tech. 2004;63:3–11. doi: 10.1002/jemt.10418. [DOI] [PubMed] [Google Scholar]
- 137.Zipfel WR, Williams RM, Webb WW. Nonlinear magic: multiphoton microscopy in the biosciences. Nat. Biotechnol. 2003;21(11):1369–1377. doi: 10.1038/nbt899. [DOI] [PubMed] [Google Scholar]
- 138.Xu C, Webb WW. Measurement of two-photon excitation cross sections of molecular fluorophores with data from 690 to 1050 nm. J. Opt. Soc. Am. B. 1996;13(3):481–491. [Google Scholar]
- 139.Kasischke KA, Vishwasrao HD, Fisher PJ, Zipfel WR, Webb WW. Neural activity triggers neuronal oxidative metabolism followed by astrocytic glycolysis. Science. 2004;305(5680):99–103. doi: 10.1126/science.1096485. [DOI] [PubMed] [Google Scholar]
- 140.Zipfel WR, Williams RM, Christie R, Nikitin AY, Hyman BT, Webb WW. Livetissue intrinsic emission microscopy using multiphoton-excited native fluorescence and second harmonic generation. Proc. Natl Acad. Sci. USA. 2003;100(12):7075–7080. doi: 10.1073/pnas.0832308100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Nichols MG, Barth EE, Nichols JA. Reduction in DNA synthesis during two-photon microscopy of intrinsic reduced nicotinamide adenine dinucleotide fluorescence. Photochem. Photobiol. 2005;81(2):259–269. doi: 10.1562/2004-08-05-RA-263. [DOI] [PubMed] [Google Scholar]
- 142.Perriott LM, Kono T, Whitesell RR, et al. Gluocose uptake and metabolism by cultured human skeletal muscle cells: rate-limiting steps. Am. J. Physiol. Endocrinol. Metab. 2001;281:E72–E80. doi: 10.1152/ajpendo.2001.281.1.E72. [DOI] [PubMed] [Google Scholar]
- 143.Kable EPW, Kiemer AK. Non-invasive live-cell measurement of changes in macrophage NAD(P)H by two-photon microscopy. Immunol. Lett. 2005;96:33–38. doi: 10.1016/j.imlet.2003.12.013. [DOI] [PubMed] [Google Scholar]
- 144.Bennett BD, Jetton TL, Ying G, Magnuson MA, Piston DW. Quantitative subcellular imaging of glucose metabolism within intact pancreatic islets. J. Biol. Chem. 1996;271:3647–3651. doi: 10.1074/jbc.271.7.3647. [DOI] [PubMed] [Google Scholar]
- 145.Tiede LM, Rocha-Sanchez SM, Hallworth R, Nichols MG, Beisel K. Determination of hair cell metabolic state in isolated cochlear preparations by two-photon microscopy. J. Biomed. Opt. 2007;12(2):021004. doi: 10.1117/1.2714777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Rothstein EC, Carroll S, Combs CA, Jobsis PD, Balaban RS. Skeletal muscle NAD(P)H two-photon fluorescence microscopy in vivo: topology and optical inner filters. Biophys. J. 2005;88:2165–2176. doi: 10.1529/biophysj.104.053165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Christie RH, Bacskai BJ, Zipfel WR, et al. Growth arrest of individual senile plaques in a model of Alzheimer’s disease observed by in vivo multiphoton microscopy. J. Neurosci. 2001;21:858–864. doi: 10.1523/JNEUROSCI.21-03-00858.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Weber G. Fluorescence of riboflavin and flavin-adenine dinucleotide. Biochem. J. 1950;47(1):114–121. doi: 10.1042/bj0470114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Visser AJ. Kinetics of stacking interactions in flavin adenine dinucleotide from timeresolved flavin fluorescence. Photochem. Photobiol. 1984;40(6):703–706. doi: 10.1111/j.1751-1097.1984.tb04640.x. [DOI] [PubMed] [Google Scholar]
- 150.de Kok A, Visser AJ. Flavin binding site differences between lipoamide dehydrogenase and glutathione reductase as revealed by static and time-resolved flavin fluorescence. FEBS Lett. 1987;218(1):135–138. doi: 10.1016/0014-5793(87)81033-5. [DOI] [PubMed] [Google Scholar]
- 151.Digris AV, Shakoun VV, Novikov EG, van Hoek A, Claiborne A, Visser AJ. Thermal stability of a flavoprotein assessed from associative analysis of polarized time-resolved fluorescence spectroscopy. Eur. Biophys. J. 1999;28:526–531. doi: 10.1007/s002490050235. [DOI] [PubMed] [Google Scholar]
- 152.Brolin SE, Agren A. Assay of flavin nucleotides in pancreatic islets by a differential fluorimetric technique. Biochem. J. 1977;163(1):159–162. doi: 10.1042/bj1630159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Lakowicz JR. Principles of Fluorescence Spectroscopy. 3rd Edition Springer; NY, USA: 2006. [Google Scholar]
- 154.O’Connor DV, Phillips D. Time-Correlated Single-Photon Counting. Academic Press; London, UK: 1984. [Google Scholar]
- 155.Becker W. Advanced Time-Correlated Single-Photon Counting Techniques. Springer; NY, USA: 2005. [Google Scholar]
- 156.Niesner R, Peker B, Schlüsche P, Gericke K-H. Noniterative biexponential fluorescence lifetime imaging in the investigation of cellular metabolism by means of NAD(P)H aut ofluorescence. Chem. Phys. Chem. 2004;5:1141–1149. doi: 10.1002/cphc.200400066. [DOI] [PubMed] [Google Scholar]
- 157.Yu Q, Proia M, Heikal AA. Integrated biophotonics approach for noninvasive and multiscale studies of biomolecular and cellular biophysics. J. Biomed. Opt. 2008;13(4):041315. doi: 10.1117/1.2952297. [DOI] [PubMed] [Google Scholar]
- 158.Fink C, Morgan F, Loew LM. Intracellular fluorescent probe concentrations by confocal microscopy. Biophys. J. 1998;75(4):1648–1658. doi: 10.1016/S0006-3495(98)77607-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Huang M, Camara AK, Stowe DF, Qi F, Beard DA. Mitochondrial inner membrane electrophysiology assessed by rhodamine-123 transport and fluorescence. Ann. Biomed. Eng. 2007;35(7):1276–1285. doi: 10.1007/s10439-007-9265-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Johnson LV, Walsh ML, Chen LB. Localization of mitochondria in living cells with rhodamine 123. Proc. Natl Acad. Sci. USA. 1980;77(2):990–994. doi: 10.1073/pnas.77.2.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Scaduto RC, Jr, Grotyohann LW. Measurement of mitochondrial membrane potential using fluorescent rhodamine derivatives. Biophys. J. 1999;76(Pt 1):469–477. doi: 10.1016/S0006-3495(99)77214-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kubin RF, Fletcher AN. Fluoresecence quantum yields of some rhodamine dyes. J. Lumin. 1982;27:455–462. [Google Scholar]
- 163.Sureau F, Moreau F, Millot JM, et al. Microspectrofluorometry of the protonation state of ellipticine, an antitumor alkaloid, in single cells. Biophys. J. 1993;65(5):1767–1774. doi: 10.1016/S0006-3495(93)81273-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Duchen MR, McGuinness O, Brown LA, Crompton M. On the involvement of a cyclosporine A sensitive mitochondrial pore in myocardial reperfusion injury. Cardiovasc. Res. 1993;27(10):1790–1794. doi: 10.1093/cvr/27.10.1790. [DOI] [PubMed] [Google Scholar]
- 165.Ankarcrona M, Dypbukt JM, Bonfoco E, et al. Glutamate-induced neuronal death: a succession of necrosis or apoptosis depending on mitochondrial function. 1995. pp. 961–973. [DOI] [PubMed]
- 166.Cossarizza A, Kalashnikova G, Grassilli E, et al. Mitochondrial modifications during rat thymocyte apoptosis: a study at the single cell level. Exp. Cell Res. 1994;214(1):323–330. doi: 10.1006/excr.1994.1264. [DOI] [PubMed] [Google Scholar]
- 167.Verburg J, Hollenbeck PJ. Mitochondrial membrane potential in axons increases with local nerve growth factor or semaphorin signaling. J. Neurosci. 2008;38(33):8306–8315. doi: 10.1523/JNEUROSCI.2614-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Tsien RY. The green fluorescent protein. Ann. Rev. Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 169.Misteli T, Spector DL. Applications of the green fluorescent protein in cell biology and biotechnology. Nat. Biotechnol. 1997;15:961–964. doi: 10.1038/nbt1097-961. [DOI] [PubMed] [Google Scholar]
- 170.Zimmer M. Green fluorescent protein (GFP): applications, structure, and related photophysical behavior. Chem. Rev. 2002;102(3):759–782. doi: 10.1021/cr010142r. [DOI] [PubMed] [Google Scholar]
- 171.Billinton N, Knight AW. Seeing the wood through the trees: a review of techniques for distinguishing green fluorescent protein from endogenous autofluorescence. Anal. Biochem. 2001;291(2):175–197. doi: 10.1006/abio.2000.5006. [DOI] [PubMed] [Google Scholar]
- 172.Heikal AA, Hess ST, Baird GS, Tsien RY, Webb WW. Molecular spectroscopy and dynamics of intrinsically fluorescent proteins: coral red (dsRed) and yellow (citrine) Proc. Natl Acad. Sci. USA. 2000;97(22):11996–12001. doi: 10.1073/pnas.97.22.11996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Hess ST, Heikal AA, Webb WW. Fluorescence photoconversion kinetics in novel green luorescent protein pH sensors (pHluorins) J. Phys. Chem. B. 2004;108:10138–10148. [Google Scholar]
- 174.Mahajan NP, Linder K, Berry G, Gordon GW, Heim R, Herman B. Bcl-2 and Bax interactions in mitochondria probed with green fluorescent protein and fluorescence resonance energy transfer. Nat. Biotechnol. 1998;16:547–552. doi: 10.1038/nbt0698-547. [DOI] [PubMed] [Google Scholar]
- 175.Llopis J, McCaffery JM, Miyawaki A, Farquhar MG, Tsien RY. Measurement of cytosolic, mitochondrial, and Golgi pH in single living cells with green fluorescent proteins. Proc. Natl Acad. Sci. USA. 1998;95(12):6803–6808. doi: 10.1073/pnas.95.12.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Partikian A, ölveczky B, Swaminathan R, Li Y, Verkman AS. Rapid diffusion of greenfluorescent protein in the mitochondrial matrix. J. Cell Biol. 1998;140(4):821–829. doi: 10.1083/jcb.140.4.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Vaquero EC, Edderkaoui M, Pandol SJ, Gukovsky I, Gukovskaya AS. Reactive oxygen species produced by NAD(P)H oxidase inhibit apoptosis in pancreatic cancer cells. J. Biol. Chem. 2004;279:34643–34654. doi: 10.1074/jbc.M400078200. [DOI] [PubMed] [Google Scholar]
- 178.Mayevsky A, Rogatsky GG. Mitochondrial function in vivo evaluated by NADH fluorescence: from animal models to human studies. Am. J. Physiol. Cell. Physiol. 2007;292:C615–C640. doi: 10.1152/ajpcell.00249.2006. ▪▪ Detailed review of NADH use to monitorin vivo mitochondrial function from animal models to human.
- 179.Mayevsky A, Barbiro-Michaely E. Use of NADH fluorescence to determine mitochondrial function in vivo. Int. J. Biochem. Cell Biol. 2009;41(10):1977–1988. doi: 10.1016/j.biocel.2009.03.012. [DOI] [PubMed] [Google Scholar]
- 180.Masters BR, So PTC. Confocal microscopy and multi-photon excitation microscopy of human skin in vivo. Opt. Express. 2001;8(1):1–10. doi: 10.1364/oe.8.000002. [DOI] [PubMed] [Google Scholar]
- 181.Brecht M, Fee MS, Garaschuk O, et al. Novel approaches to monitor and manipulate single neurons in vivo. J. Neurosci. 2004;24(42):9223–9227. doi: 10.1523/JNEUROSCI.3344-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Helmchen F, Denk W. Deep tissue two-photon microscopy. Nat. Methods. 2005;2:932–940. doi: 10.1038/nmeth818. [DOI] [PubMed] [Google Scholar]
- 183.Jung JC, Mehta AD, Aksay E, Stepnoski R, Schnitzer MJ. In vivo mammalian brain imaging using one- and two-photon fluorescence microendoscopy. J. Neurophysiol. 2004;92:3121–3133. doi: 10.1152/jn.00234.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Lin SX, Maxfield FR. Fluorescence imaging in living animals. Focus on uptake and trafficking of fluorescent conjugates of folic acid in intact kidney determined using intravital two-photon microscopy. Am. J. Physiol. Cell Physiol. 2004;287(2):C257–C259. doi: 10.1152/ajpcell.00235.2004. [DOI] [PubMed] [Google Scholar]
- 185.Atkinson RJ, Shorthouse AJ, Hurlstone DP. Novel colorectal endoscopic in vivo imaging and resection practice: a short practice guide for interventional endoscopists. Tech. Coloproctol. 2007;11(1):7–16. doi: 10.1007/s10151-007-0319-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Kasimova MR, Grigiene J, Krab K, et al. The free NADH concentration is kept constant in plant mitochondria under different metabolic conditions. Plant Cell. 2006;18:688–698. doi: 10.1105/tpc.105.039354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Sandoval FJ, Zhang Y, Roje S. Flavin nucleotide metabolism in plants: monofunctional enzymes synthesize FAD plastids. J. Biol. Chem. 2008;283(45):30890–30900. doi: 10.1074/jbc.M803416200. [DOI] [PMC free article] [PubMed] [Google Scholar]