Abstract
Translocated in liposarcoma, Ewing's sarcoma and TATA-binding protein-associated factor 15 constitute an interesting and important family of proteins known as the TET proteins. The proteins function in several aspects of cell growth control, including multiple different steps in gene expression, and they are also found mutated in a number of specific diseases. For example, all contain domains for binding nucleic acids and have been shown to function in both RNA polymerase II-mediated transcription and pre-mRNA splicing, possibly connecting these two processes. Chromosomal translocations in human sarcomas result in a fusion of the amino terminus of these proteins, which contains a transcription activation domain, to the DNA-binding domain of a transcription factor. Although the fusion proteins have been characterized in a clinical environment, the function of the cognate full-length protein in normal cells is a more recent topic of study. The first part of this review will describe the TET proteins, followed by detailed descriptions of their multiple roles in cells. The final sections will examine changes that occur in gene regulation in cells expressing the fusion proteins. The clinical implications and treatment of sarcomas will not be addressed but have recently been reviewed.
Keywords: TLS/FUS, EWS, TAF 15, transcription, splicing, cancer, RNA-binding, arginine methylation, neuronal disease, fusion proteins, translocations
Introduction
The TET family of proteins consists of translocated in liposarcoma (TLS), Ewing's sarcoma (EWS) and TATA-binding protein-associated factor 15 (TAF15), and has various roles in gene expression. TLS, EWS and TAF15 are predominantly nuclear and are highly expressed in all human fetal and adult tissues examined (Zinszner et al., 1994; Morohoshi et al., 1996; Andersson et al., 2008). Each of these proteins contains an amino terminus rich in Gln, Gly, Ser and Tyr, and all three also contain a conserved RNA-binding domain (RBD), RGG regions that may affect RNA binding, and a Cys2–Cys2 zinc finger that may bind nucleic acids (Figure 1).
Figure 1.
Domain structure of TET and SARFH/Cabeza proteins. The activation domain (AD), RGG boxes, RBD, Cys2–Cys2 zinc finger (ZF) and sarcoma breakpoints (arrowheads) are depicted for TLS, EWS and TAF15. The RGG boxes, RBD and Cys2–Cys2 ZF are also depicted for SARFH/Cabeza, the D. melanogaster ortholog.
Both EWS and TLS (also called FUS) were originally discovered as a result of characteristic chromosomal translocations in EWS and myxoid liposarcoma, respectively. In the former, the chimeric protein consists of the amino terminus of EWS joined to the DNA-binding domain of the transcription factor FLI-1, whereas in the latter, the full-length CHOP protein replaces the carboxy-terminus of TLS (Delattre et al., 1992; Crozat et al., 1993; Rabbitts et al., 1993). TAF15 was originally found as a TAF in the general transcription initiation TFIID complex, and cloned both through biochemical methods and through its homology in the RBD to TLS and EWS (Bertolotti et al., 1996; Morohoshi et al., 1996; Tora, 2002). The TAF15 gene has subsequently been found translocated to the transcription factor CIZ in acute leukemia (Martini et al., 2002) and to the nuclear receptor CHN/TEC in extraskeletal myxoid chondrosarcoma (Attwooll et al., 1999; Panagopoulos et al., 1999; Sjogren et al., 1999), but these occur at a much lower frequency than translocations involving TLS and EWS. In all cases, the fusions are expressed under the control of the TET protein promoter.
The amino terminus of TET proteins can function like a transcriptional activation domain when fused to a DNA-binding domain (Zinszner et al., 1994; Bertolotti et al., 1999). Although the amino terminus of all TET proteins is enriched for Gln, Gly, Ser and Tyr residues, there are also some variations between the TET proteins. The amino terminus of EWS is rich in Pro and Thr and contains many copies of a hexapeptide repeat of consensus Ser-Tyr-Gly-Gln-Gln-Ser, with absolute conservation of Tyr at the second position and high conservation of Gln at the fourth position (Ng et al., 2007). This region is reminiscent of the heptapeptide repeats that constitute the carboxy-terminal domain (CTD) of the RNA polymerase (RNAP) II largest subunit, and the high Gln and Pro content is similar to activation domains of various transcription factors (Delattre et al., 1992). Computational modeling and synthetic constructs indicated that multiple Tyr residues, or at least an aromatic side chain since Phe can substitute, are required for transcription activation, and that the region is highly disordered, so structure is not a key determinant (Lee, 2007; Ng et al., 2007).
The 90-amino acid RBD in TET proteins folds into a structure with a sheet of four anti-parallel β-strands perpendicular to two α-helices (Burd and Dreyfuss, 1994). Within this domain, two short motifs, RNP-1 and RNP-2, directly contact RNA via hydrogen bonds and ring stacking. However, unlike other proteins with this type of RBD, TET proteins have an acidic residue at the second position and a Thr residue in the fourth position of RNP-1 as well as an unusually long loop after the first α-helix (Bertolotti et al., 1999), which may affect the structure of the protein since this region contributes to the hydrophobic core of the domain and to RNA-binding specificity or affinity. The RBD is the most conserved region within the TET protein family (Figure 2). Sequence-specific RNA binding by TET proteins has been examined by various groups. Early on, it was shown that EWS binds polyU and polyG sequences through its carboxy-terminal RGG domain (Ohno et al., 1994). Subsequent experiments showed that TLS also has affinity for polyG and polyU homoribopolymers, and in vitro systematic evolution of ligands by exponential enrichment (Tuerk and Gold, 1990) experiments suggested that the RBD and RGG motifs co-operate to bind GGUG RNA, but with relatively low affinity (Kd = 250 nM) (Lerga et al., 2001).
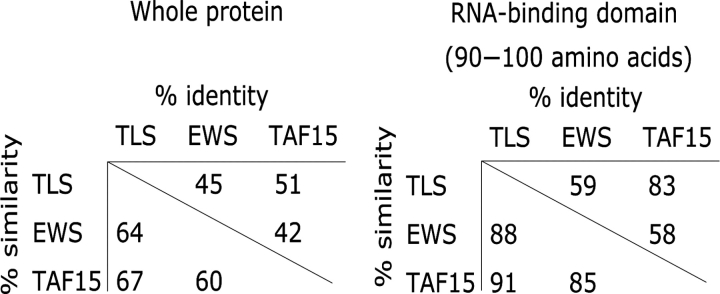
Figure 2.
TET proteins are highly related. Amino acid similarity and identity compared over the whole protein and within the RBD for TLS, EWS and TAF15.
In addition to the RBD, TET proteins contain other domains that bind nucleic acids. The zinc finger of TET proteins resembles those in ZIS, a splicing protein that contains two zinc fingers, a stretch of acidic amino acids and a Ser/Arg-rich (RS) domain (Ladomery and Dellaire, 2002). Limited proteolysis followed by MALDI-TOF and circular dichroism analyses demonstrated that both the RBD and zinc finger regions of TLS fold into protease-resistant structures, and NMR analysis suggested that the zinc finger may bind RNA (Iko et al., 2004). The carboxy-terminus of TLS also contains three RGG motifs that may increase RNA affinity of the RBD or zinc finger, and may also be the site of post-translational modifications that regulate RNA binding or protein–protein interactions (Burd and Dreyfuss, 1994). The carboxy-terminus of TAF15 contains ∼20 copies of perfect or degenerate Gly-Gly-Tyr-Gly-Gly-Asp-Arg repeats, and this region is likely to have a role in RNA binding as it encompasses many RGG boxes (Burd and Dreyfuss, 1994; Bertolotti et al., 1996; Morohoshi et al., 1996).
TET proteins also bind single-stranded DNA and possibly double-stranded DNA. TLS promotes D-loop formation, a process where a single strand of DNA invades and pairs with one of the strands in a double-stranded region of DNA, and this process is necessary for DNA repair and recombination (Baechtold et al., 1999; Bertrand et al., 1999). DNA binding is likely to occur through the Cys2–Cys2 zinc finger at the carboxy-terminus of the protein, a domain frequently found in transcription factors that is well known for binding nucleic acids, particularly DNA (Pieler and Theunissen, 1993). There are, however, reports of RBDs mediating sequence-specific DNA binding (Ding et al., 1999; Ladomery and Dellaire, 2002). Further analysis of each of the nucleic acid-binding domains is required to separate the specificity, affinity and function.
A Drosophila melanogaster ortholog was found to share homology with the RBD and the carboxy-terminus of TLS and EWS, and named SARFH (sarcoma-associated RNA-binding fly homolog) or Cabeza (Immanuel et al., 1995; Stolow and Haynes, 1995). This protein both binds RNA in vitro through its RBD and co-localizes with sites of active RNAP II transcription (Immanuel et al., 1995; Stolow and Haynes, 1995).
Regulation of TET proteins
TET proteins contain many sites for post-translational modifications, including phosphorylation and arginine methylation. Such modifications in or near the RBD and RGG motifs may affect RNA- and DNA-binding, interactions with other proteins, protein stability or subcellular localization (Burd and Dreyfuss, 1994).
TLS and EWS are the target of various Ser/Thr protein kinases. Ser 42 in TLS, but not in the TLS–CHOP fusion protein, is phosphorylated by the protein kinase ATM in response to double-strand breaks in DNA caused by ionizing radiation (Gardiner et al., 2008). EWS and EWS–FLI1 are phosphorylated at Thr 79 by the JNK and p38 families of protein kinases in response to DNA damage, and weakly phosphorylated by ERK in response to mitogens (Klevernic et al., 2008). TET proteins are also substrates for the PKC family: EWS is phosphorylated at Ser 266 and TLS at Ser 256 (Deloulme et al., 1997; Perrotti et al., 1998). Phosphorylation of Ser 256 protects TLS from proteolytic degradation by masking the binding site that targets TLS for proteasome-mediated degradation (Perrotti et al., 2000). Separately, the form of TLS that has homologous DNA pairing activity was also found to be phosphorylated in the zinc finger domain at Ser 439 (Guipaud et al., 2006). TLS and TAF15 are also substrates for Tyr phosphorylation. TLS is a target of the fibroblast growth factor receptor 1 kinase and a distinct fraction of TLS is phosphorylated on Tyr residues, and localized to the cytoplasm (Klint et al., 2004). Although Tyr phosphorylation of TLS results in cytoplasmic localization, TAF15 phosphorylation on Tyr residues by v-Src leads to increased transcriptional activation by TAF15 in the nucleus (Lee et al., 2004). The amino terminus of EWS is also post-translationally modified by O-linked-β-N-acetylglucosaminylation, which may overlap the sites of and thus prevent Ser/Thr phosphorylation (Bachmaier et al., 2009). These data indicate that phosphorylation of TET proteins regulates cellular localization, protein stability and function.
Methylation of arginine residues is a post-translational modification that occurs on many RNA-binding proteins and may affect protein–RNA and protein–protein interactions, protein stability or subcellular localization (Liu and Dreyfuss, 1995). Although methylation does not affect the positive charge on Arg residues, it does alter the steric properties of the side chain contacts to RNA or proteins and may decrease the hydrophilicity of a protein (Beyer et al., 1977). This modification is performed by protein arginine methyltransferases (PRMTs) and the structure of Arg allows for mono- and di-methylation. Type I PRMTs, including PRMT1 and PRMT3, monomethylate and asymmetrically dimethylate Arg residues, whereas type II PRMTs monomethylate and symmetrically dimethylate Arg residues. Dimethylation of hnRNPs accounts for the bulk of arginine methylation in the nucleus (Jobert et al., 2009a), occurs on ∼12% of Arg residues in hnRNPs (Liu and Dreyfuss, 1995) and targets hnRNPs for nuclear export (Stallcup, 2001).
TET proteins were found to be targets of PRMTs, and all three are substrates for PRMT1. TLS contains over 20 asymmetrically dimethylated Arg residues in the RGG motifs at the carboxy-terminus of the protein (Rappsilber et al., 2003). Similarly, EWS is extensively methylated in vitro and in vivo, and the majority of RGG sites are asymmetrically dimethylated, with rare cases of monomethylation and no instances of symmetric dimethylation (Belyanskaya et al., 2001; Pahlich et al., 2005). Such post-translational modification may alter cellular localization, including nuclear export of methylated EWS (Belyanskaya et al., 2003; Araya et al., 2005). TAF15 is also methylated on Arg residues in its RGG boxes by PRMT1, but in this case, the modification appears to be important for nuclear localization rather than nuclear export (Jobert et al., 2009a). Furthermore, cellular stress or lack of Arg methylation through knockdown of PRMT1 causes TAF15 to localize to stress granules (Jobert et al., 2009a). PRMT8, a type I PRMT that is located at the cell surface and predominantly expressed in brain tissue, binds the third RGG box in EWS (Pahlich et al., 2008), but the functional significance of this interaction is unclear. It should be noted that a brain-specific form of EWS, which results from alternative splicing, has been detected (Melot et al., 2001). The brain-specific form of EWS bound by PRMT8 may be monomethylated or asymmetrically dimethylated, opening the possibility for differential regulation of EWS in different cellular compartments and cell types.
Roles in transcription
TET proteins are likely to function in RNAP II transcription by interacting with TFIID and subunits of RNAP II itself. TFIID consists of TBP and a heterogeneous mixture of TAFs in stoichiometric or sub-stoichiometric ratios, and the various complexes may have different effects on basal or activated transcription (Brou et al., 1993). It is possible that the proteins associated with core TFIID components may affect promoter choice and recruitment of processing factors. Since each of the TET proteins co-purifies with a separate fraction of TFIID in a sub-stoichiometric ratio (Bertolotti et al., 1996, 1998), these related proteins may have distinct functions and may regulate different groups of genes.
TLS, EWS and TAF15 also associate directly with RNAP II (Bertolotti et al., 1996, 1998). EWS interacts with the Rbp3 subunit of RNAP II, whereas TAF15 interacts with Rbp3, Rbp5 and Rpb7 (Bertolotti et al., 1998). Another study indicated that the EWS amino terminal trans-activation domain also interacts with Rpb4 and Rpb7 subunits (Zhou and Lee, 2001). Rpb7 resembles the bacterial σ factor and in yeast may have a role in transcription regulation (Petermann et al., 1998). The amino terminus of EWS binds RNAP II and although this region is present in the fusion protein, it is unclear whether EWS–FLI1 is also able to do so. One study found that the EWS–FLI1 fusion protein does not interact with RNAP II (Bertolotti et al., 1998), whereas another group found that not only does EWS–FLI1 interact with RNAP II but also that overexpression of RPB7 increases EWS–FLI1, but not FLI1, transactivation (Petermann et al., 1998).
In addition to directly contacting general transcription factors and RNAP II, TET proteins may regulate transcription through contacting activators or repressors. TLS, but not EWS, interacts with the DNA-binding domain of various nuclear hormone receptors, without affecting the ability of the receptor to bind DNA response elements, suggesting that TLS may have a role in activating transcription of certain receptors under specific conditions (Powers et al., 1998). EWS binds various proteins containing a POU DNA-binding domain, including the transcriptional activator Oct4, which is expressed in embryonic stem and germ cells to maintain an undifferentiated totipotent state (Lee et al., 2005), and Brn3a, which is expressed in the developing and adult nervous systems to promote development of specific neuronal lineages (Thomas and Latchman, 2002).
The amino terminus of each TET protein was shown to act as a transcriptional activator when fused to a DNA-binding domain (Bailly et al., 1994; Zinszner et al., 1994; Bertolotti et al., 1999). In the case of EWS, however, transcription activation potential is decreased or even abolished in the full-length protein, indicating that the RBD and RGG boxes may regulate the activation domain in the normal protein (Li and Lee, 2000; Rossow and Janknecht, 2001; Alex and Lee, 2005). Although the EWS–FLI1 fusion protein activates transcription in vitro (Uren et al., 2004), others suggest that the in vitro system does not reflect in vivo events (Ng et al., 2009). EWS interacts with both the CREB-binding protein (CBP; Fujimura et al., 2001; Rossow and Janknecht, 2001; Araya et al., 2003) transcriptional co-activator and also the transcriptional repressor SF1 (Zhang et al., 1998), indicating that it may positively and negatively regulate transcription.
TLS can also repress transcription by RNAP III, which transcribes small structural and catalytic RNAs (A.Y.T. and J.L.M, submitted for publication). Repression occurs both in vitro and in vivo, and changes in levels of TLS protein in cells affect levels of RNAP III transcripts. This regulation is likely to occur through TLS binding to TBP and possibly affecting interactions with the general RNAP III transcription machinery. TLS joins a list of cancer-related factors, such as p53 and Rb, which can affect transcription by more than one RNAP.
Roles in splicing
Many experiments have suggested a role for TET proteins in pre-mRNA splicing. Early on, mass spectrometry identified TLS as hnRNP P2 in the H complex of proteins that assembles non-specifically and in an ATP-independent manner on pre-mRNAs in vitro (Calvio et al., 1995). More significantly, TLS was found to crosslink to the pre-mRNA 3′ splice site during the second step of splicing (Wu and Green, 1997) and was also found at the 5′ splice site in a large complex containing hyperphosphorylated RNAP II, U1 snRNP, p54nrb/PSF and transcription elongation factors P-TEFb, Tat-SF1 and TFIIF (Kameoka et al., 2004). Large-scale purification of functional spliceosomes and mass spectrometry analyses confirmed the presence of TET proteins in the spliceosome (Rappsilber et al., 2002; Zhou et al., 2002).
TET proteins bind various SR proteins and other known splicing factors. For example, the carboxy-terminus of TLS was shown to interact with SR proteins TASR (TLS-associated SR protein; also known as SRp38) and SC35 (Yang et al., 1998, 2000b; Lerga et al., 2001; Shin and Manley, 2002). In addition, TLS associates with SRp75 and SRm160 and with hnRNPs A1 and C1/C2 and PTB (Lerga et al., 2001; Meissner et al., 2003). The carboxy-termini of TLS and EWS were found to interact with YB-1, a splicing activator that may also have a role in mRNA packaging (Chansky et al., 2001).
Interactions between TET proteins and known splicing factors can affect patterns of alternative splicing. Transient transfection of TLS and reporter constructs in HeLa cells showed that TLS overexpression increased production of the 13S and 12S isoforms from an adenovirus E1A pre-mRNA, reflecting preferential use of downstream alternative 5′ splice sites (Yang et al., 1998). However, in an erythroleukemic cell line that lacks the transcription/splicing factor Spi-1, transient expression of TLS and the E1A reporter construct increased the use of the upstream 5′ splice site and thus production of the 9S isoform (Hallier et al., 1998). The differing results observed with TLS may reflect the presence or absence of Spi-1, which can also affect E1A splicing in these assays, and which binds directly to TLS (Hallier et al., 1998). These contrasting results highlight the difficulties in analyzing effects on splicing solely by transient overexpression assays. It will be important in the future to confirm and extend these results using other methods, such as RNAi-mediated knockdowns and in vitro splicing assays, to determine how TET proteins affect alternative splicing and to clarify what interactions are important in different cell types or conditions.
Connecting transcription and splicing
Transcription and splicing are likely to be connected by proteins with roles in both processes. Proteins and their various mechanisms of coordinating transcription and splicing are being characterized, and some may be gene-specific. It is possible that TET proteins connect transcription and splicing, since the amino terminus mediates interactions with RNAP II and the carboxy-terminus binds to splicing factors. TET proteins may recruit splicing factors to the RNAP II CTD, which co-ordinates pre-mRNA processing events (for review, see Hirose and Manley, 2000; Bentley, 2005; de Almeida and Carmo-Fonseca, 2008). As mentioned above, TLS was found as a component of in vitro transcription and splicing complexes (Kameoka et al., 2004), further suggesting that TLS may bridge these processes. Confirmation that TLS links transcription to splicing and elucidation of the mechanism by which this occurs would add to existing knowledge about the regulation of gene expression and may suggest how this protein family functions in disease. Furthermore, it is possible that each TET protein recruits different splicing factors to separate types of genes; such specificity could be achieved through TET proteins associating with distinct populations of TFIID.
In addition to the type of promoter and recruitment of specific splicing factors, transcription elongation rate may also affect alternative splicing (Kornblihtt et al., 2004) and splicing may affect mRNA transcription (Manley, 2002). Tat-SF1, a protein distantly related to the TET proteins, stimulates the Tat protein of HIV-1 and acts as a general transcription elongation factor (Zhou and Sharp, 1996; Li and Green, 1998). Tat-SF1 contains two tandem RRMs with homology to EWS and TLS and a highly acidic carboxy-terminus (Zhou and Sharp, 1996). Tat-SF1 recruits the CTD kinase P-TEFb to Tat bound to TAR RNA to stimulate transcription, and U snRNPs to elongating RNAP II to enhance splicing (Fong and Zhou, 2001).
TET proteins in other complexes
The TET proteins interact with a diverse range of proteins. In keeping with this, they may act to connect a number of processes within the cell, possibly by acting as a scaffold. For example, EWS has been shown to interact with a large number of other proteins using high-throughput mapping of protein–protein interactions (Rual et al., 2005). In addition, systematic analysis of proteins in a complex containing the RNase III enzyme Drosha, which processes microRNA precursors, identified EWS along with a number of other proteins involved in RNA binding (Gregory et al., 2004). The functional significance of EWS co-eluting with Drosha in this large complex remains to be determined, but it provides a possible link between the TET proteins and regulation of microRNAs.
More recently, TAF15 was found associated with U1 snRNA, chromatin and RNA, in a complex distinct from the Sm-containing U1 snRNP that functions in splicing. Although the function of this particle has not been determined, the TAF15-U1 snRNA interaction increased after transcriptional inhibition and the complex localized to perinucleolar structures (Jobert et al., 2009b). These findings raise the possibility that TET proteins may regulate transcription and splicing through multiple mechanisms.
DNA repair and genomic stability
TET proteins have also been implicated in the DNA repair pathway. In vitro pairing on membrane (POM) assays demonstrated that TLS mediates annealing of complementary DNA strands to form D-loops, and this function is lost in TLS–CHOP (Baechtold et al., 1999; Bertrand et al., 1999). The POM blot followed by mass spectrometry revealed that all TET proteins are able to pair homologous DNA in vitro, and that this activity was specific to TET proteins and PSF, a splicing factor with domains similar to those in the TET proteins (Guipaud et al., 2006). Thus, TET proteins may have a role in DNA repair, especially at sites of active transcription or after cellular signaling via kinase cascades.
In an animal model, TLS knockout mice have high genomic instability due to chromosomal pairing defects and enhanced sensitivity to radiation (Hicks et al., 2000; Kuroda et al., 2000). The phenotype of TLS knockout mice depends on the genetic background: inbred TLS−/− mice showed defects in B-lymphocyte development, genomic instability and perinatal death (Hicks et al., 2000). Although outbred TLS−/− mice were able to survive until adulthood, they displayed defects in spermatogenesis and sensitivity to ionizing radiation (Kuroda et al., 2000). The c-ABL and ATM kinases are both activated by DNA damage and target TLS (Perrotti et al., 1998; Gardiner et al., 2008), so cells lacking TLS might not undergo appropriate DNA repair, resulting in genomic instability. DNA damage can also cause TLS to bind a non-coding RNA near the cyclin D1 promoter. This subsequently inhibits the histone acetyl transferase activity of CBP/p300 and results in transcriptional repression of the cyclin D1 gene (Wang et al., 2008). These data suggest that TLS could have multiple roles in DNA repair, including a link to the cell cycle through regulating transcription of cyclin D1.
Similarly, knockout of EWS in mice resulted in B-lymphocyte defects, decreased meiotic recombination, sensitivity to ionizing radiation, and pre-natal mortality of inbred mice and high rates of post-natal mortality in outbred strains (Li et al., 2007). Although EWS and TLS are quite similar, loss of EWS does not result in increased levels of TLS, and TLS has a role in pairing autosomes whereas EWS is involved with pairing the XY sex chromosomes during meiosis (Li et al., 2007). Morpholino-mediated knockdown of EWS orthologs in zebrafish resulted in mitotic defects during development, apoptosis of preneural cells and embryonic lethality, whereas siRNA-mediated knockdown of EWS in HeLa cells also led to mitotic defects and apoptosis (Azuma et al., 2007). Members of the TET family thus have distinct, non-redundant roles in meiosis and functional conservation between species, as demonstrated by studies using animal models.
Neuronal functions and disease
Although TET proteins are predominantly nuclear, TLS shuttles between the nucleus and the cytoplasm attached to RNA (Zinszner et al., 1997b) and thus may function in RNA transport. TLS was found in a complex with RNA-transporting proteins, translational regulators and mRNA, and this kinesin-associated granule transports RNA to the plus ends of microtubules, such as the dendrites of neurons (Kanai et al., 2004). TLS binds and is transported with the mRNA encoding Nd1-L, a protein that stabilizes actin, and thus might have a role in regulating actin organization in dendritic spines (Fujii and Takumi, 2005). Both microtubules and actin target RNA-bound TLS to dendritic granules, and this process is activated by metabotropic glutamate receptor 5 (mGluR5) excitation of synapses (Fujii et al., 2005). TLS also binds Sam68, an RNA-binding protein that functions in splicing but is also found in dendritic granules in neurons, and the N-methyl-d-aspartate receptor, which is involved in synaptic signaling (Belly et al., 2005). Taken together, these results indicate that TLS could affect mRNA transport with either actin or microtubules. This may alter dendritic structure after excitation and may affect long-term synaptic plasticity.
TLS has also been implicated in a form of familial amyotrophic lateral sclerosis (ALS), a fatal neurodegenerative disease. Genetic linkage analysis had previously implicated a region of chromosome 16 as the source of an autosomal dominant mutation. Very recently, two groups found that specific mutations in the highly conserved last 13 amino acids of TLS correlated with familial ALS but not sporadic ALS (Kwiatkowski et al., 2009; Vance et al., 2009). Single nucleotide changes were found that result in the substitution of a single amino acid, including missense mutations in each of the last five Arg residues of TLS in ALS patients, with the most common being an Arg to Cys substitution (R521C). These mutations resulted in increased cytoplasmic localization of TLS, apparently in an aggregated form, in the motor neurons of patients, as well as in transfected cells and rat cortical neurons and in cells expressing a fluorescently tagged version of the mutant TLS protein (Kwiatkowski et al., 2009; Vance et al., 2009). The carboxy-terminus is crucial for normal functioning of the protein in neurons, since the mutated Arg residues may be targets for dimethylation that could alter subcellular localization of TLS. The role of TLS in ALS remains to be characterized, and it is currently not clear whether ALS results from loss or alteration of TLS function, or from a toxic effect of the cytoplasmic aggregates.
Fusion proteins and sarcomas
TLS and EWS were discovered translocated in sarcomas. Sarcomas are aggressive cancers that occur in connective tissue and of mesodermal origin, possibly originating from mesenchymal progenitor cells and rarely appear in carcinomas of epithelial origin (Arvand and Denny, 2001). Sarcomas are relatively infrequent, accounting for under 10% of human cancers, and bone sarcomas tend to affect children and adolescents whereas soft tissue sarcomas such as liposarcoma are more common in adults (Riggi and Stamenkovic, 2007; Osuna and de Alava, 2009).
Translocation results in the TET protein promoter driving expression of the fusion protein. Decreased level of the full-length TET protein in cells containing the translocation is unlikely to be the cause of transformation given that mice heterozygous for either EWS or TLS are indistinguishable from their wild-type littermates (Kuroda et al., 2000; Li et al., 2007). Instead, introduction of the gain-of-function fusion protein into cells causes deregulated expression of target genes and alters the differentiation pattern of certain cell types (Aman, 1999; Martini et al., 2002). Fusion proteins target genes that permit cell growth and require other genetic alterations for tumor development (reviewed in Janknecht, 2005; Xia and Barr, 2005). Variations in chromosome breakpoints lead to fusion proteins with differing transcriptional activation strengths and abilities to transform cells, and these correlate with phenotype, tumor progression and patient prognosis (Zoubek et al., 1996; Lin et al., 1999; Gonzalez et al., 2007). Each type of sarcoma is caused by one or a small number of related fusion proteins, suggesting that the fusion proteins can only activate the oncogenic pathway in specific cell types.
The process of translocation has been studied and characteristic sequences at the fusion gene breakpoints recognized by specific DNA-binding proteins may have a role in recombination (Panagopoulos et al., 1997). Sequencing at the breakpoints and the presence of extra nucleotides indicate that certain proteins may be required and involved in the process of chromosomal breakage and re-ligation reactions. In the case of TLS–CHOP, the protein Translin, which is often found at recombination hotspots, binds to DNA sequences near some TLS and CHOP chromosome breakpoints (Kanoe et al., 1999; Hosaka et al., 2000; Yu and Hecht, 2008). The translocation breakpoints also contain binding sites for topoisomerase II, which creates staggered ends of double-stranded DNA and allows for the re-joining of DNA strands (Kanoe et al., 1999). Alu repeat sequences or an octamer similar to the bacterial Chi recombination site could also promote translocation, suggesting that an established pathway is involved in the process of recombination (Xiang et al., 2008).
Many variations to EWS–FLI1 and TLS–CHOP (Figure 3) have been documented in clinical literature and involve a TET protein fused to the DNA-binding domain or a full-length transcription factor (for a full list, see Oda and Tsuneyoshi, 2009; Osuna and de Alava, 2009). Clinical therapies are beyond the scope of this review but are discussed elsewhere (Olsen et al., 2002; Bernstein et al., 2006; Oda and Tsuneyoshi, 2009). In the following sections, we will address some of the mechanisms through which TLS–CHOP and EWS–FLI1 lead to cellular transformation. We focus on these two fusions as they are the most prevalent and best studied. The role of these fusion proteins has been studied both by introducing the fusion protein into normal cells and by knocking down expression of the fusion protein in sarcoma cells.
Figure 3.
Structure of TLS–CHOP and EWS–FLI1 fusion proteins. TLS and EWS are as described in Figure 1. CHOP contains a DNA-binding domain (DBD) and a Leucine Zipper (LZ) domain. Translocation breakpoint occurs upstream of the CHOP initiation site; TLS–CHOP fusion protein contains amino acids derived from sequences in the 5′ UTR of CHOP (hatched region). FLI1 contains an activation domain (AD) and an ETS DBD.
TLS–CHOP fusion protein
Myxoid liposarcoma displays a number of characteristic phenotypes, including morphology change to round nuclei in lipoblasts, intracellular lipid accumulation and induction of adipocyte-specific genes (Osuna and de Alava, 2009). Myxoid liposarcoma arises from translocation t(12;16)(q13:p11) and expression of the TLS–CHOP fusion protein. CHOP is a transcription factor that negatively regulates the CAAT/enhancer binding protein (C/EBP) family; this family includes genes that control adipogenesis (Zinszner et al., 1994). CHOP dimerizes and induces growth arrest in response to cellular stress. Although TLS–CHOP can dimerize, it does not result in growth arrest and acts in a dominant negative manner, possibly by competing for binding partners or inappropriately activating transcription (Barone et al., 1994). TLS–CHOP is more highly localized to the nucleus than TLS, and is not exported to the cytoplasm or located to the nucleolus when transcription is inhibited (Zinszner et al., 1994, 1997a), which allows for unregulated gene expression.
The oncogenic transformation in human liposarcoma has been attributed to uncontrolled transcriptional activation by TLS–CHOP and interference with adipocytic differentiation to promote liposarcoma development. TLS–CHOP-containing cells express the neural-specific genes Nexin and Neuronatin, as well as the RET proto-oncogene, at higher levels than they are expressed in normal fat tissue (Thelin-Jarnum et al., 1999). TLS–CHOP activates the DOL54 gene, which is a secreted protein that can promote tumorigenicity (Kuroda et al., 1999). In addition, TLS–CHOP halts C/EBP-mediated activation of the PPARγ adipocyte differentiation cascade (Perez-Mancera et al., 2007, 2008) to promote cell proliferation and liposarcoma development.
Significantly, the TLS–CHOP fusion protein is only able to direct tumor development in certain cell types: despite expressing TLS–CHOP in all cells, a transgenic mouse model only develops liposarcomas (Perez-Losada et al., 2000a). Overexpression of CHOP alone is not enough to transform cells in culture or induce tumors in mice (Zinszner et al., 1994, Perez-Losada et al., 2000b). Thus, while transcriptional activation conferred by the amino terminus of TLS is required, unregulated expression of TLS–CHOP is only able to activate the liposarcoma tumorigenesis pathway in adipocytes. Similarly, in patients, each type of translocation (and the resulting fusion protein) corresponds to a specific type of sarcoma, suggesting that unregulated expression of target genes can only drive tumor formation in certain cell types.
EWS–FLI1 fusion protein
EWS typically arises in bones as a result of translocation t(11;22)(q24;q12) joining the EWS and FLI1 genes (Delattre et al., 1992). The EWS–FLI1 fusion protein is found in ∼85% of the cases, whereas EWS fusion to other transcription factors, including ERG, ETV-1, E1AF, ATF-1 and WT-1, comprise the remainder (Oda and Tsuneyoshi, 2009). The ETS family, which includes FLI1, ERG, ETV-1 and E1AF, is a large group of transcription factors found only in metazoans and contains a characteristic DNA-binding domain that recognizes GGAA or GGAT sequences (for a review, see Oikawa and Yamada, 2003). EWS fusions to other transcription factors, including ATF-1 and WT-1, have been found but are infrequent, and are reviewed elsewhere (Gerald and Haber, 2005; Riggi et al., 2007). The amino terminus of FLI1 contains a weak transcription activation domain, which is replaced by the stronger EWS activation domain in the fusion protein. EWS–FLI1, like TLS–CHOP and unlike EWS, is predominantly nuclear with very little cytoplasmic localization (Yang et al., 2000a).
The breakpoints in EWS are not homogeneous but are instead clustered within a small region of EWS and spread over a larger region in the FLI1 gene. Although it is near the EWS breakpoint and could be included in-frame in the fusion protein, the RBD of EWS was not detected during examination of a large number of tumors containing EWS–FLI1 or EWS–ERG fusion proteins, suggesting that the RBD may decrease or inhibit the oncogenic potential of the fusion protein (Zucman et al., 1993). The particular variant of the EWS–FLI1 fusion correlates with patient prognosis, reflecting different transcriptional activation potentials of the resulting fusion proteins (de Alava et al., 1998; Lin et al., 1999; Aryee et al., 2000). Indeed, the transcriptional activation and metastatic activity of different isoforms of EWS–FLI1 in a mouse model correlated with disease progression in human patients (Gonzalez et al., 2007). Furthermore, duplication or loss of certain chromosome regions, which is frequent in Ewing's sarcoma patients, also correlates with disease progression and clinical outcome (Savola et al., 2009).
Introduction of EWS–FLI1 into a cultured cell line confers changes typical of transformation: alterations in cell morphology, an increased proliferation rate and anchorage-independent growth. Expression of EWS–FLI1 leads to growth arrest in primary cells (Lessnick et al., 2002) but to growth and inhibition of differentiation in transformed cells (Eliazer et al., 2003) and prevents apoptosis in immortalized 3T3 cells (Yi et al., 1997). Many of these changes are the result of de-regulated gene expression by the fusion protein. Both FLI1 and EWS–FLI1 bind the same sequences, due to the ETS domain of FLI1, but EWS–FLI1 drives stronger activation of target genes due to its interaction with RNAP II through the EWS amino terminus (Bailly et al., 1994; Yang et al., 2000a). However, EWS–FLI1 proteins with mutations in the DNA-binding domain were still able to transform cells (Jaishankar et al., 1999; Welford et al., 2001), indicating that some of the oncogenic effects are independent of DNA binding.
EWS–FLI1 affects a range of genes to alter gene expression and cause cellular transformation (for reviews, see Janknecht, 2005; Kovar, 2005; Riggi and Stamenkovic, 2007). EWS–FLI1, but not FLI1 alone, rapidly up-regulates transcription of the gene encoding the SH2 domain-containing adapter protein EAT-2, and this is correlated with transformation (Thompson et al., 1996). EWS–FLI1 also up-regulates expression of the morphogenic gene manic fringe (MFNG), which has a role in somatic development and can cause tumorigenesis when overexpressed (May et al., 1997). EWS–FLI1 binds to the promoter of the telomerase reverse transcriptase (TERT) gene and recruits CBP/p300 to activate TERT transcription (Takahashi et al., 2003), and an elevated level of TERT is important for continued cell division and a characteristic of many cancers. EWS–FLI1 also represses the tumor suppressor TGF-β type II receptor to relieve growth inhibition (Hahm et al., 1999). Depleting EWS–FLI1 protein in EWS-derived cells caused decreased tumorigenicity in vivo and reduced growth and cell cycle arrest in vitro, partly by alleviating repression of the Rb tumor suppressor and decreasing cyclin D1 and CDK2 levels (Tanaka et al., 1997; Chansky et al., 2004; Prieur et al., 2004; Smith et al., 2006; Hu et al., 2008). Similarly, examination of global changes in Ewing's sarcoma cell lines indicated that EWS–FLI alters cell cycle and proliferation genes to down-regulate differentiation (Kauer et al., 2009).
To find genes regulated by EWS–FLI1 but not FLI1, cells overexpressing these proteins were analyzed by cDNA microarray. EWS–FLI1 binds the promoter region and up-regulates expression of the Id2 (inhibitor of DNA binding 2) gene (Nishimori et al., 2002). Id2 is a helix-loop-helix protein that has a dominant negative repressive effect on basic HLH transcription factors and can affect cell differentiation and proliferation (Nishimori et al., 2002). Similarly, EWS–FLI1, but not FLI1, binds to the promoter and up-regulates the expression of DAX1, a nuclear receptor (Mendiola et al., 2006). Further gene expression analysis suggests that the DAX1 pathway regulates ∼10% of EWS–FLI1 target genes, especially those involved in G1 to S cell cycle progression (Garcia-Aragoncillo et al., 2008). DAX1 is thus an important direct target of EWS–FLI1 and indicates a pathway that is important for proliferative growth of sarcoma cells. Taken together, EWS–FLI1 represses key tumor suppressors and activates a range of genes to promote transformation in cells and tumorigenesis in vivo.
Conclusions
The TET proteins are involved in a wide range of cellular processes and tightly regulated to ensure appropriate localization and activity. The domains in these proteins and early experiments suggested a role in RNAP II transcription and splicing, possibly coupling these processes, but subsequent studies have shown that TET proteins are also involved in a wide range of other processes, including repressing RNAP III transcription, DNA repair and RNA transport in neurons. The mechanism of TET protein activity, regulation of their cellular localization and the domains of the proteins involved in various processes remain to be clarified, especially in the recent discovery that implicates TLS in familial ALS.
The amino terminus of TET proteins is also found fused to transcription factors, and these oncogenic fusion proteins in sarcomas are responsible for inappropriate transcriptional activation of target genes. Many therapies are in development, including knocking down the levels of fusion proteins, and further knowledge of the pathways that are disrupted by the fusion proteins will aid both the understanding of sarcoma development and the identification of other possible drug targets.
Funding
A.Y.T. was partially funded by a Postgraduate Scholarship from the Natural Sciences and Engineering Research Council of Canada and related work in the lab of J.L.M. was supported by grants from the National Institutes of Health.
References
- Alex D., Lee K.A. RGG-boxes of the EWS oncoprotein repress a range of transcriptional activation domains. Nucleic Acids Res. 2005;33:1323–1331. doi: 10.1093/nar/gki270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aman P. Fusion genes in solid tumors. Semin. Cancer Biol. 1999;9:303–318. doi: 10.1006/scbi.1999.0130. [DOI] [PubMed] [Google Scholar]
- Andersson M.K., Stahlberg A., Arvidsson Y., Olofsson A., Semb H., Stenman G., Nilsson O., Aman P. The multifunctional FUS, EWS and TAF15 proto-oncoproteins show cell type-specific expression patterns and involvement in cell spreading and stress response. BMC Cell Biol. 2008;9:37. doi: 10.1186/1471-2121-9-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araya N., Hirota K., Shimamoto Y., Miyagishi M., Yoshida E., Ishida J., Kaneko S., Kaneko M., Nakajima T., Fukamizu A. Cooperative interaction of EWS with CREB-binding protein selectively activates hepatocyte nuclear factor 4-mediated transcription. J. Biol. Chem. 2003;278:5427–5432. doi: 10.1074/jbc.M210234200. [DOI] [PubMed] [Google Scholar]
- Araya N., Hiraga H., Kako K., Arao Y., Kato S., Fukamizu A. Transcriptional down-regulation through nuclear exclusion of EWS methylated by PRMT1. Biochem. Biophys. Res. Commun. 2005;329:653–660. doi: 10.1016/j.bbrc.2005.02.018. [DOI] [PubMed] [Google Scholar]
- Arvand A., Denny C.T. Biology of EWS/ETS fusions in Ewing's family tumors. Oncogene. 2001;20:5747–5754. doi: 10.1038/sj.onc.1204598. [DOI] [PubMed] [Google Scholar]
- Aryee D.N., Sommergruber W., Muehlbacher K., Dockhorn-Dworniczak B., Zoubek A., Kovar H. Variability in gene expression patterns of Ewing tumor cell lines differing in EWS–FLI1 fusion type. Lab. Invest. 2000;80:1833–1844. doi: 10.1038/labinvest.3780194. [DOI] [PubMed] [Google Scholar]
- Attwooll C., Tariq M., Harris M., Coyne J.D., Telford N., Varley J.M. Identification of a novel fusion gene involving hTAFII68 and CHN from a t(9;17)(q22;q11.2) translocation in an extraskeletal myxoid chondrosarcoma. Oncogene. 1999;18:7599–7601. doi: 10.1038/sj.onc.1203156. [DOI] [PubMed] [Google Scholar]
- Azuma M., Embree L.J., Sabaawy H., Hickstein D.D. Ewing sarcoma protein ewsr1 maintains mitotic integrity and proneural cell survival in the zebrafish embryo. PLoS One. 2007;2:e979. doi: 10.1371/journal.pone.0000979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmaier R., Aryee D.N., Jug G., Kauer M., Kreppel M., Lee K.A., Kovar H. O-GlcNAcylation is involved in the transcriptional activity of EWS–FLI1 in Ewing's sarcoma. Oncogene. 2009;28:1280–1284. doi: 10.1038/onc.2008.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baechtold H., Kuroda M., Sok J., Ron D., Lopez B.S., Akhmedov A.T. Human 75-kDa DNA-pairing protein is identical to the pro-oncoprotein TLS/FUS and is able to promote D-loop formation. J. Biol. Chem. 1999;274:34337–34342. doi: 10.1074/jbc.274.48.34337. [DOI] [PubMed] [Google Scholar]
- Bailly R.A., Bosselut R., Zucman J., Cormier F., Delattre O., Roussel M., Thomas G., Ghysdael J. DNA-binding and transcriptional activation properties of the EWS-FLI-1 fusion protein resulting from the t(11;22) translocation in Ewing sarcoma. Mol. Cell. Biol. 1994;14:3230–3241. doi: 10.1128/mcb.14.5.3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barone M.V., Crozat A., Tabaee A., Philipson L., Ron D. CHOP (GADD153) and its oncogenic variant, TLS–CHOP, have opposing effects on the induction of G1/S arrest. Genes Dev. 1994;8:453–464. doi: 10.1101/gad.8.4.453. [DOI] [PubMed] [Google Scholar]
- Belly A., Moreau-Gachelin F., Sadoul R., Goldberg Y. Delocalization of the multifunctional RNA splicing factor TLS/FUS in hippocampal neurones: exclusion from the nucleus and accumulation in dendritic granules and spine heads. Neurosci. Lett. 2005;379:152–157. doi: 10.1016/j.neulet.2004.12.071. [DOI] [PubMed] [Google Scholar]
- Belyanskaya L.L., Gehrig P.M., Gehring H. Exposure on cell surface and extensive arginine methylation of Ewing sarcoma (EWS) protein. J. Biol. Chem. 2001;276:18681–18687. doi: 10.1074/jbc.M011446200. [DOI] [PubMed] [Google Scholar]
- Belyanskaya L.L., Delattre O., Gehring H. Expression and subcellular localization of Ewing sarcoma (EWS) protein is affected by the methylation process. Exp. Cell Res. 2003;288:374–381. doi: 10.1016/s0014-4827(03)00221-0. [DOI] [PubMed] [Google Scholar]
- Bentley D.L. Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr. Opin. Cell Biol. 2005;17:251–256. doi: 10.1016/j.ceb.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Bernstein M., Kovar H., Paulussen M., Randall R.L., Schuck A., Teot L.A., Juergens H. Ewing's sarcoma family of tumors: current management. Oncologist. 2006;11:503–519. doi: 10.1634/theoncologist.11-5-503. [DOI] [PubMed] [Google Scholar]
- Bertolotti A., Lutz Y., Heard D.J., Chambon P., Tora L. hTAF(II)68, a novel RNA/ssDNA-binding protein with homology to the pro-oncoproteins TLS/FUS and EWS is associated with both TFIID and RNA polymerase II. EMBO J. 1996;15:5022–5031. [PMC free article] [PubMed] [Google Scholar]
- Bertolotti A., Melot T., Acker J., Vigneron M., Delattre O., Tora L. EWS, but not EWS-FLI-1, is associated with both TFIID and RNA polymerase II: interactions between two members of the TET family, EWS and hTAFII68, and subunits of TFIID and RNA polymerase II complexes. Mol. Cell. Biol. 1998;18:1489–1497. doi: 10.1128/mcb.18.3.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolotti A., Bell B., Tora L. The N-terminal domain of human TAFII68 displays transactivation and oncogenic properties. Oncogene. 1999;18:8000–8010. doi: 10.1038/sj.onc.1203207. [DOI] [PubMed] [Google Scholar]
- Bertrand P., Akhmedov A.T., Delacote F., Durrbach A., Lopez B.S. Human POMp75 is identified as the pro-oncoprotein TLS/FUS: both POMp75 and POMp100 DNA homologous pairing activities are associated to cell proliferation. Oncogene. 1999;18:4515–4521. doi: 10.1038/sj.onc.1203048. [DOI] [PubMed] [Google Scholar]
- Beyer A.L., Christensen M.E., Walker B.W., LeStourgeon W.M. Identification and characterization of the packaging proteins of core 40S hnRNP particles. Cell. 1977;11:127–138. doi: 10.1016/0092-8674(77)90323-3. [DOI] [PubMed] [Google Scholar]
- Brou C., Chaudhary S., Davidson I., Lutz Y., Wu J., Egly J.M., Tora L., Chambon P. Distinct TFIID complexes mediate the effect of different transcriptional activators. EMBO J. 1993;12:489–499. doi: 10.1002/j.1460-2075.1993.tb05681.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd C.G., Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- Calvio C., Neubauer G., Mann M., Lamond A.I. Identification of hnRNP P2 as TLS/FUS using electrospray mass spectrometry. RNA. 1995;1:724–733. [PMC free article] [PubMed] [Google Scholar]
- Chansky H.A., Hu M., Hickstein D.D., Yang L. Oncogenic TLS/ERG and EWS/Fli-1 fusion proteins inhibit RNA splicing mediated by YB-1 protein. Cancer Res. 2001;61:3586–3590. [PubMed] [Google Scholar]
- Chansky H.A., Barahmand-Pour F., Mei Q., Kahn-Farooqi W., Zielinska-Kwiatkowska A., Blackburn M., Chansky K., Conrad E.U., 3rd, Bruckner J.D., Greenlee T.K., et al. Targeting of EWS/FLI-1 by RNA interference attenuates the tumor phenotype of Ewing's sarcoma cells in vitro. J. Orthop. Res. 2004;22:910–917. doi: 10.1016/j.orthres.2003.12.008. [DOI] [PubMed] [Google Scholar]
- Crozat A., Aman P., Mandahl N., Ron D. Fusion of CHOP to a novel RNA-binding protein in human myxoid liposarcoma. Nature. 1993;363:640–644. doi: 10.1038/363640a0. [DOI] [PubMed] [Google Scholar]
- de Alava E., Kawai A., Healey J.H., Fligman I., Meyers P.A., Huvos A.G., Gerald W.L., Jhanwar S.C., Argani P., Antonescu C.R., et al. EWS–FLI1 fusion transcript structure is an independent determinant of prognosis in Ewing's sarcoma. J. Clin. Oncol. 1998;16:1248–1255. doi: 10.1200/JCO.1998.16.4.1248. [DOI] [PubMed] [Google Scholar]
- de Almeida S.F., Carmo-Fonseca M. The CTD role in cotranscriptional RNA processing and surveillance. FEBS Lett. 2008;582:1971–1976. doi: 10.1016/j.febslet.2008.04.019. [DOI] [PubMed] [Google Scholar]
- Delattre O., Zucman J., Plougastel B., Desmaze C., Melot T., Peter M., Kovar H., Joubert I., de Jong P., Rouleau G., et al. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature. 1992;359:162–165. doi: 10.1038/359162a0. [DOI] [PubMed] [Google Scholar]
- Deloulme J.C., Prichard L., Delattre O., Storm D.R. The prooncoprotein EWS binds calmodulin and is phosphorylated by protein kinase C through an IQ domain. J. Biol. Chem. 1997;272:27369–27377. doi: 10.1074/jbc.272.43.27369. [DOI] [PubMed] [Google Scholar]
- Ding J., Hayashi M.K., Zhang Y., Manche L., Krainer A.R., Xu R.M. Crystal structure of the two-RRM domain of hnRNP A1 (UP1) complexed with single-stranded telomeric DNA. Genes Dev. 1999;13:1102–1115. doi: 10.1101/gad.13.9.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliazer S., Spencer J., Ye D., Olson E., Ilaria R.L., Jr. Alteration of mesodermal cell differentiation by EWS/FLI-1, the oncogene implicated in Ewing's sarcoma. Mol. Cell. Biol. 2003;23:482–492. doi: 10.1128/MCB.23.2.482-492.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong Y.W., Zhou Q. Stimulatory effect of splicing factors on transcriptional elongation. Nature. 2001;414:929–933. doi: 10.1038/414929a. [DOI] [PubMed] [Google Scholar]
- Fujii R., Takumi T. TLS facilitates transport of mRNA encoding an actin-stabilizing protein to dendritic spines. J. Cell Sci. 2005;118:5755–5765. doi: 10.1242/jcs.02692. [DOI] [PubMed] [Google Scholar]
- Fujii R., Okabe S., Urushido T., Inoue K., Yoshimura A., Tachibana T., Nishikawa T., Hicks G.G., Takumi T. The RNA binding protein TLS is translocated to dendritic spines by mGluR5 activation and regulates spine morphology. Curr. Biol. 2005;15:587–593. doi: 10.1016/j.cub.2005.01.058. [DOI] [PubMed] [Google Scholar]
- Fujimura Y., Siddique H., Lee L., Rao V.N., Reddy E.S. EWS-ATF-1 chimeric protein in soft tissue clear cell sarcoma associates with CREB-binding protein and interferes with p53-mediated trans-activation function. Oncogene. 2001;20:6653–6659. doi: 10.1038/sj.onc.1204684. [DOI] [PubMed] [Google Scholar]
- Garcia-Aragoncillo E., Carrillo J., Lalli E., Agra N., Gomez-Lopez G., Pestana A., Alonso J. DAX1, a direct target of EWS/FLI1 oncoprotein, is a principal regulator of cell-cycle progression in Ewing's tumor cells. Oncogene. 2008;27:6034–6043. doi: 10.1038/onc.2008.203. [DOI] [PubMed] [Google Scholar]
- Gardiner M., Toth R., Vandermoere F., Morrice N.A., Rouse J. Identification and characterization of FUS/TLS as a new target of ATM. Biochem. J. 2008;415:297–307. doi: 10.1042/BJ20081135. [DOI] [PubMed] [Google Scholar]
- Gerald W.L., Haber D.A. The EWS-WT1 gene fusion in desmoplastic small round cell tumor. Semin. Cancer Biol. 2005;15:197–205. doi: 10.1016/j.semcancer.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Gonzalez I., Vicent S., de Alava E., Lecanda F. EWS/FLI-1 oncoprotein subtypes impose different requirements for transformation and metastatic activity in a murine model. J. Mol. Med. 2007;85:1015–1029. doi: 10.1007/s00109-007-0202-5. [DOI] [PubMed] [Google Scholar]
- Gregory R.I., Yan K.P., Amuthan G., Chendrimada T., Doratotaj B., Cooch N., Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- Guipaud O., Guillonneau F., Labas V., Praseuth D., Rossier J., Lopez B., Bertrand P. An in vitro enzymatic assay coupled to proteomics analysis reveals a new DNA processing activity for Ewing sarcoma and TAF(II)68 proteins. Proteomics. 2006;6:5962–5972. doi: 10.1002/pmic.200600259. [DOI] [PubMed] [Google Scholar]
- Hahm K.B., Cho K., Lee C., Im Y.H., Chang J., Choi S.G., Sorensen P.H., Thiele C.J., Kim S.J. Repression of the gene encoding the TGF-beta type II receptor is a major target of the EWS–FLI1 oncoprotein. Nat. Genet. 1999;23:222–227. doi: 10.1038/13854. [DOI] [PubMed] [Google Scholar]
- Hallier M., Lerga A., Barnache S., Tavitian A., Moreau-Gachelin F. The transcription factor Spi-1/PU.1 interacts with the potential splicing factor TLS. J. Biol. Chem. 1998;273:4838–4842. doi: 10.1074/jbc.273.9.4838. [DOI] [PubMed] [Google Scholar]
- Hicks G.G., Singh N., Nashabi A., Mai S., Bozek G., Klewes L., Arapovic D., White E.K., Koury M.J., Oltz E.M., et al. Fus deficiency in mice results in defective B-lymphocyte development and activation, high levels of chromosomal instability and perinatal death. Nat. Genet. 2000;24:175–179. doi: 10.1038/72842. [DOI] [PubMed] [Google Scholar]
- Hirose Y., Manley J.L. RNA polymerase II and the integration of nuclear events. Genes Dev. 2000;14:1415–1429. [PubMed] [Google Scholar]
- Hosaka T., Kanoe H., Nakayama T., Murakami H., Yamamoto H., Nakamata T., Tsuboyama T., Oka M., Kasai M., Sasaki M.S., et al. Translin binds to the sequences adjacent to the breakpoints of the TLS and CHOP genes in liposarcomas with translocation t(12;6) Oncogene. 2000;19:5821–5825. doi: 10.1038/sj.onc.1203943. [DOI] [PubMed] [Google Scholar]
- Hu H.M., Zielinska-Kwiatkowska A., Munro K., Wilcox J., Wu D.Y., Yang L., Chansky H.A. EWS/FLI1 suppresses retinoblastoma protein function and senescence in Ewing's sarcoma cells. J. Orthop. Res. 2008;26:886–893. doi: 10.1002/jor.20597. [DOI] [PubMed] [Google Scholar]
- Iko Y., Kodama T.S., Kasai N., Oyama T., Morita E.H., Muto T., Okumura M., Fujii R., Takumi T., Tate S., et al. Domain architectures and characterization of an RNA-binding protein, TLS. J. Biol. Chem. 2004;279:44834–44840. doi: 10.1074/jbc.M408552200. [DOI] [PubMed] [Google Scholar]
- Immanuel D., Zinszner H., Ron D. Association of SARFH (sarcoma-associated RNA-binding fly homolog) with regions of chromatin transcribed by RNA polymerase II. Mol. Cell. Biol. 1995;15:4562–4571. doi: 10.1128/mcb.15.8.4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaishankar S., Zhang J., Roussel M.F., Baker S.J. Transforming activity of EWS/FLI is not strictly dependent upon DNA-binding activity. Oncogene. 1999;18:5592–5597. doi: 10.1038/sj.onc.1202940. [DOI] [PubMed] [Google Scholar]
- Janknecht R. EWS-ETS oncoproteins: the linchpins of Ewing tumors. Gene. 2005;363:1–14. doi: 10.1016/j.gene.2005.08.007. [DOI] [PubMed] [Google Scholar]
- Jobert L., Argentini M., Tora L. PRMT1 mediated methylation of TAF15 is required for its positive gene regulatory function. Exp. Cell Res. 2009a;315:1273–1286. doi: 10.1016/j.yexcr.2008.12.008. [DOI] [PubMed] [Google Scholar]
- Jobert L., Pinzon N., Van Herreweghe E., Jady B.E., Guialis A., Kiss T., Tora L. Human U1 snRNA forms a new chromatin-associated snRNP with TAF15. EMBO Rep. 2009b;10:494–500. doi: 10.1038/embor.2009.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kameoka S., Duque P., Konarska M.M. p54(nrb) associates with the 5′ splice site within large transcription/splicing complexes. EMBO J. 2004;23:1782–1791. doi: 10.1038/sj.emboj.7600187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai Y., Dohmae N., Hirokawa N. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron. 2004;43:513–525. doi: 10.1016/j.neuron.2004.07.022. [DOI] [PubMed] [Google Scholar]
- Kanoe H., Nakayama T., Hosaka T., Murakami H., Yamamoto H., Nakashima Y., Tsuboyama T., Nakamura T., Ron D., Sasaki M.S., et al. Characteristics of genomic breakpoints in TLS–CHOP translocations in liposarcomas suggest the involvement of Translin and topoisomerase II in the process of translocation. Oncogene. 1999;18:721–729. doi: 10.1038/sj.onc.1202364. [DOI] [PubMed] [Google Scholar]
- Kauer M., Ban J., Kofler R., Walker B., Davis S., Meltzer P., Kovar H. A molecular function map of Ewing's sarcoma. PLoS One. 2009;4:e5415. doi: 10.1371/journal.pone.0005415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klevernic I.V., Morton S., Davis R.J., Cohen P. Phosphorylation of EWS and EWS–Fli1 in response to DNA damage. Biochem. J. 2008;418:625–634. doi: 10.1042/BJ20082097. [DOI] [PubMed] [Google Scholar]
- Klint P., Hellman U., Wernstedt C., Aman P., Ron D., Claesson-Welsh L. Translocated in liposarcoma (TLS) is a substrate for fibroblast growth factor receptor-1. Cell Signal. 2004;16:515–520. doi: 10.1016/j.cellsig.2003.09.007. [DOI] [PubMed] [Google Scholar]
- Kornblihtt A.R., de la Mata M., Fededa J.P., Munoz M.J., Nogues G. Multiple links between transcription and splicing. RNA. 2004;10:1489–1498. doi: 10.1261/rna.7100104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovar H. Context matters: the hen or egg problem in Ewing's sarcoma. Semin. Cancer Biol. 2005;15:189–196. doi: 10.1016/j.semcancer.2005.01.004. [DOI] [PubMed] [Google Scholar]
- Kuroda M., Wang X., Sok J., Yin Y., Chung P., Giannotti J.W., Jacobs K.A., Fitz L.J., Murtha-Riel P., Turner K.J., et al. Induction of a secreted protein by the myxoid liposarcoma oncogene. Proc. Natl. Acad. Sci. USA. 1999;96:5025–5030. doi: 10.1073/pnas.96.9.5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda M., Sok J., Webb L., Baechtold H., Urano F., Yin Y., Chung P., de Rooij D.G., Akhmedov A., Ashley T., et al. Male sterility and enhanced radiation sensitivity in TLS(−/−) mice. EMBO J. 2000;19:453–462. doi: 10.1093/emboj/19.3.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski T.J., Jr., Bosco D.A., Leclerc A.L., Tamrazian E., Vanderburg C.R., Russ C., Davis A., Gilchrist J., Kasarskis E.J., Munsat T., et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–1208. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- Ladomery M., Dellaire G. Multifunctional zinc finger proteins in development and disease. Ann. Hum. Genet. 2002;66:331–342. doi: 10.1017/S0003480002001215. [DOI] [PubMed] [Google Scholar]
- Lee K.A. Ewings family oncoproteins: drunk, disorderly and in search of partners. Cell Res. 2007;17:286–288. doi: 10.1038/cr.2007.22. [DOI] [PubMed] [Google Scholar]
- Lee H.J., Kim S., Pelletier J., Kim J. Stimulation of hTAFII68 (NTD)-mediated transactivation by v-Src. FEBS Lett. 2004;564:188–198. doi: 10.1016/S0014-5793(04)00314-X. [DOI] [PubMed] [Google Scholar]
- Lee J., Rhee B.K., Bae G.Y., Han Y.M., Kim J. Stimulation of Oct-4 activity by Ewing's sarcoma protein. Stem Cells. 2005;23:738–751. doi: 10.1634/stemcells.2004-0375. [DOI] [PubMed] [Google Scholar]
- Lerga A., Hallier M., Delva L., Orvain C., Gallais I., Marie J., Moreau-Gachelin F. Identification of an RNA binding specificity for the potential splicing factor TLS. J. Biol. Chem. 2001;276:6807–6816. doi: 10.1074/jbc.M008304200. [DOI] [PubMed] [Google Scholar]
- Lessnick S.L., Dacwag C.S., Golub T.R. The Ewing's sarcoma oncoprotein EWS/FLI induces a p53-dependent growth arrest in primary human fibroblasts. Cancer Cell. 2002;1:393–401. doi: 10.1016/s1535-6108(02)00056-9. [DOI] [PubMed] [Google Scholar]
- Li K.K., Lee K.A. Transcriptional activation by the Ewing's sarcoma (EWS) oncogene can be cis-repressed by the EWS RNA-binding domain. J. Biol. Chem. 2000;275:23053–23058. doi: 10.1074/jbc.M002961200. [DOI] [PubMed] [Google Scholar]
- Li X.Y., Green M.R. The HIV-1 Tat cellular coactivator Tat-SF1 is a general transcription elongation factor. Genes Dev. 1998;12:2992–2996. doi: 10.1101/gad.12.19.2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Watford W., Li C., Parmelee A., Bryant M.A., Deng C., O'Shea J., Lee S.B. Ewing sarcoma gene EWS is essential for meiosis and B lymphocyte development. J. Clin. Invest. 2007;117:1314–1323. doi: 10.1172/JCI31222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin P.P., Brody R.I., Hamelin A.C., Bradner J.E., Healey J.H., Ladanyi M. Differential transactivation by alternative EWS–FLI1 fusion proteins correlates with clinical heterogeneity in Ewing's sarcoma. Cancer Res. 1999;59:1428–1432. [PubMed] [Google Scholar]
- Liu Q., Dreyfuss G. In vivo and in vitro arginine methylation of RNA-binding proteins. Mol. Cell. Biol. 1995;15:2800–2808. doi: 10.1128/mcb.15.5.2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley J.L. Nuclear coupling: RNA processing reaches back to transcription. Nat. Struct. Biol. 2002;9:790–791. doi: 10.1038/nsb1102-790. [DOI] [PubMed] [Google Scholar]
- Martini A., La Starza R., Janssen H., Bilhou-Nabera C., Corveleyn A., Somers R., Aventin A., Foa R., Hagemeijer A., Mecucci C., et al. Recurrent rearrangement of the Ewing's sarcoma gene, EWSR1, or its homologue, TAF15, with the transcription factor CIZ/NMP4 in acute leukemia. Cancer Res. 2002;62:5408–5412. [PubMed] [Google Scholar]
- May W.A., Arvand A., Thompson A.D., Braun B.S., Wright M., Denny C.T. EWS/FLI1-induced manic fringe renders NIH 3T3 cells tumorigenic. Nat. Genet. 1997;17:495–497. doi: 10.1038/ng1297-495. [DOI] [PubMed] [Google Scholar]
- Meissner M., Lopato S., Gotzmann J., Sauermann G., Barta A. Proto-oncoprotein TLS/FUS is associated to the nuclear matrix and complexed with splicing factors PTB, SRm160, and SR proteins. Exp. Cell Res. 2003;283:184–195. doi: 10.1016/s0014-4827(02)00046-0. [DOI] [PubMed] [Google Scholar]
- Melot T., Dauphinot L., Sevenet N., Radvanyi F., Delattre O. Characterization of a new brain-specific isoform of the EWS oncoprotein. Eur. J. Biochem. 2001;268:3483–3489. doi: 10.1046/j.1432-1327.2001.02251.x. [DOI] [PubMed] [Google Scholar]
- Mendiola M., Carrillo J., Garcia E., Lalli E., Hernandez T., de Alava E., Tirode F., Delattre O., Garcia-Miguel P., Lopez-Barea F., et al. The orphan nuclear receptor DAX1 is up-regulated by the EWS/FLI1 oncoprotein and is highly expressed in Ewing tumors. Int. J. Cancer. 2006;118:1381–1389. doi: 10.1002/ijc.21578. [DOI] [PubMed] [Google Scholar]
- Morohoshi F., Arai K., Takahashi E.I., Tanigami A., Ohki M. Cloning and mapping of a human RBP56 gene encoding a putative RNA binding protein similar to FUS/TLS and EWS proteins. Genomics. 1996;38:51–57. doi: 10.1006/geno.1996.0591. [DOI] [PubMed] [Google Scholar]
- Ng K.P., Potikyan G., Savene R.O., Denny C.T., Uversky V.N., Lee K.A. Multiple aromatic side chains within a disordered structure are critical for transcription and transforming activity of EWS family oncoproteins. Proc. Natl Acad. Sci. USA. 2007;104:479–484. doi: 10.1073/pnas.0607007104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng K.P., Li K.K., Lee K.A. In vitro activity of the EWS oncogene transcriptional activation domain. Biochemistry. 2009;48:2849–2857. doi: 10.1021/bi802366h. [DOI] [PubMed] [Google Scholar]
- Nishimori H., Sasaki Y., Yoshida K., Irifune H., Zembutsu H., Tanaka T., Aoyama T., Hosaka T., Kawaguchi S., Wada T., et al. The Id2 gene is a novel target of transcriptional activation by EWS-ETS fusion proteins in Ewing family tumors. Oncogene. 2002;21:8302–8309. doi: 10.1038/sj.onc.1206025. [DOI] [PubMed] [Google Scholar]
- Oda Y., Tsuneyoshi M. Recent advances in the molecular pathology of soft tissue sarcoma: implications for diagnosis, patient prognosis, and molecular target therapy in the future. Cancer Sci. 2009;100:200–208. doi: 10.1111/j.1349-7006.2008.01024.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno T., Ouchida M., Lee L., Gatalica Z., Rao V.N., Reddy E.S. The EWS gene, involved in Ewing family of tumors, malignant melanoma of soft parts and desmoplastic small round cell tumors, codes for an RNA binding protein with novel regulatory domains. Oncogene. 1994;9:3087–3097. [PubMed] [Google Scholar]
- Oikawa T., Yamada T. Molecular biology of the Ets family of transcription factors. Gene. 2003;303:11–34. doi: 10.1016/s0378-1119(02)01156-3. [DOI] [PubMed] [Google Scholar]
- Olsen R.J., Tarantolo S.R., Hinrichs S.H. Molecular approaches to sarcoma therapy. Sarcoma. 2002;6:27–42. doi: 10.1080/13577140220127530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osuna D., de Alava E. Molecular pathology of sarcomas. Rev. Recent Clin. Trials. 2009;4:12–26. doi: 10.2174/157488709787047585. [DOI] [PubMed] [Google Scholar]
- Pahlich S., Bschir K., Chiavi C., Belyanskaya L., Gehring H. Different methylation characteristics of protein arginine methyltransferase 1 and 3 toward the Ewing sarcoma protein and a peptide. Proteins. 2005;61:164–175. doi: 10.1002/prot.20579. [DOI] [PubMed] [Google Scholar]
- Pahlich S., Zakaryan R.P., Gehring H. Identification of proteins interacting with protein arginine methyltransferase 8: the Ewing sarcoma (EWS) protein binds independent of its methylation state. Proteins. 2008;72:1125–1137. doi: 10.1002/prot.22004. [DOI] [PubMed] [Google Scholar]
- Panagopoulos I., Lassen C., Isaksson M., Mitelman F., Mandahl N., Aman P. Characteristic sequence motifs at the breakpoints of the hybrid genes FUS/CHOP, EWS/CHOP and FUS/ERG in myxoid liposarcoma and acute myeloid leukemia. Oncogene. 1997;15:1357–1362. doi: 10.1038/sj.onc.1201281. [DOI] [PubMed] [Google Scholar]
- Panagopoulos I., Mencinger M., Dietrich C.U., Bjerkehagen B., Saeter G., Mertens F., Mandahl N., Heim S. Fusion of the RBP56 and CHN genes in extraskeletal myxoid chondrosarcomas with translocation t(9;17)(q22;q11) Oncogene. 1999;18:7594–7598. doi: 10.1038/sj.onc.1203155. [DOI] [PubMed] [Google Scholar]
- Perez-Losada J., Pintado B., Gutierrez-Adan A., Flores T., Banares-Gonzalez B., del Campo J.C., Martin-Martin J.F., Battaner E., Sanchez-Garcia I. The chimeric FUS/TLS–CHOP fusion protein specifically induces liposarcomas in transgenic mice. Oncogene. 2000a;19:2413–2422. doi: 10.1038/sj.onc.1203572. [DOI] [PubMed] [Google Scholar]
- Perez-Losada J., Sanchez-Martin M., Rodriguez-Garcia M.A., Perez-Mancera P.A., Pintado B., Flores T., Battaner E., Sanchez-Garcia I. Liposarcoma initiated by FUS/TLS–CHOP: the FUS/TLS domain plays a critical role in the pathogenesis of liposarcoma. Oncogene. 2000b;19:6015–6022. doi: 10.1038/sj.onc.1204018. [DOI] [PubMed] [Google Scholar]
- Perez-Mancera P.A., Vicente-Duenas C., Gonzalez-Herrero I., Sanchez-Martin M., Flores-Corral T., Sanchez-Garcia I. Fat-specific FUS-DDIT3-transgenic mice establish PPARgamma inactivation is required to liposarcoma development. Carcinogenesis. 2007;28:2069–2073. doi: 10.1093/carcin/bgm107. [DOI] [PubMed] [Google Scholar]
- Perez-Mancera P.A., Bermejo-Rodriguez C., Sanchez-Martin M., Abollo-Jimenez F., Pintado B., Sanchez-Garcia I. FUS-DDIT3 prevents the development of adipocytic precursors in liposarcoma by repressing PPARgamma and C/EBPalpha and activating eIF4E. PLoS One. 2008;3:e2569. doi: 10.1371/journal.pone.0002569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrotti D., Bonatti S., Trotta R., Martinez R., Skorski T., Salomoni P., Grassilli E., Lozzo R.V., Cooper D.R., Calabretta B. TLS/FUS, a pro-oncogene involved in multiple chromosomal translocations, is a novel regulator of BCR/ABL-mediated leukemogenesis. EMBO J. 1998;17:4442–4455. doi: 10.1093/emboj/17.15.4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrotti D., Iervolino A., Cesi V., Cirinna M., Lombardini S., Grassilli E., Bonatti S., Claudio P.P., Calabretta B. BCR-ABL prevents c-jun-mediated and proteasome-dependent FUS (TLS) proteolysis through a protein kinase CbetaII-dependent pathway. Mol. Cell. Biol. 2000;20:6159–6169. doi: 10.1128/mcb.20.16.6159-6169.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petermann R., Mossier B.M., Aryee D.N., Khazak V., Golemis E.A., Kovar H. Oncogenic EWS–Fli1 interacts with hsRPB7, a subunit of human RNA polymerase II. Oncogene. 1998;17:603–610. doi: 10.1038/sj.onc.1201964. [DOI] [PubMed] [Google Scholar]
- Pieler T., Theunissen O. TFIIIA: nine fingers—three hands? Trends Biochem. Sci. 1993;18:226–230. doi: 10.1016/0968-0004(93)90194-r. [DOI] [PubMed] [Google Scholar]
- Powers C.A., Mathur M., Raaka B.M., Ron D., Samuels H.H. TLS (translocated-in-liposarcoma) is a high-affinity interactor for steroid, thyroid hormone, and retinoid receptors. Mol. Endocrinol. 1998;12:4–18. doi: 10.1210/mend.12.1.0043. [DOI] [PubMed] [Google Scholar]
- Prieur A., Tirode F., Cohen P., Delattre O. EWS/FLI-1 silencing and gene profiling of Ewing cells reveal downstream oncogenic pathways and a crucial role for repression of insulin-like growth factor binding protein 3. Mol. Cell. Biol. 2004;24:7275–7283. doi: 10.1128/MCB.24.16.7275-7283.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabbitts T.H., Forster A., Larson R., Nathan P. Fusion of the dominant negative transcription regulator CHOP with a novel gene FUS by translocation t(12;16) in malignant liposarcoma. Nat. Genet. 1993;4:175–180. doi: 10.1038/ng0693-175. [DOI] [PubMed] [Google Scholar]
- Rappsilber J., Ryder U., Lamond A.I., Mann M. Large-scale proteomic analysis of the human spliceosome. Genome Res. 2002;12:1231–1245. doi: 10.1101/gr.473902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J., Friesen W.J., Paushkin S., Dreyfuss G., Mann M. Detection of arginine dimethylated peptides by parallel precursor ion scanning mass spectrometry in positive ion mode. Anal. Chem. 2003;75:3107–3114. doi: 10.1021/ac026283q. [DOI] [PubMed] [Google Scholar]
- Riggi N., Cironi L., Suva M.L., Stamenkovic I. Sarcomas: genetics, signalling, and cellular origins. Part 1: the fellowship of TET. J. Pathol. 2007;213:4–20. doi: 10.1002/path.2209. [DOI] [PubMed] [Google Scholar]
- Riggi N., Stamenkovic I. The biology of Ewing sarcoma. Cancer Lett. 2007;254:1–10. doi: 10.1016/j.canlet.2006.12.009. [DOI] [PubMed] [Google Scholar]
- Rossow K.L., Janknecht R. The Ewing's sarcoma gene product functions as a transcriptional activator. Cancer Res. 2001;61:2690–2695. [PubMed] [Google Scholar]
- Rual J.F., Venkatesan K., Hao T., Hirozane-Kishikawa T., Dricot A., Li N., Berriz G.F., Gibbons F.D., Dreze M., Ayivi-Guedehoussou N., et al. Towards a proteome-scale map of the human protein–protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- Savola S., Klami A., Tripathi A., Niini T., Serra M., Picci P., Kaski S., Zambelli D., Scotlandi K., Knuutila S. Combined use of expression and CGH arrays pinpoints novel candidate genes in Ewing sarcoma family of tumors. BMC Cancer. 2009;9:17. doi: 10.1186/1471-2407-9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin C., Manley J.L. The SR protein SRp38 represses splicing in M phase cells. Cell. 2002;111:407–417. doi: 10.1016/s0092-8674(02)01038-3. [DOI] [PubMed] [Google Scholar]
- Sjogren H., Meis-Kindblom J., Kindblom L.G., Aman P., Stenman G. Fusion of the EWS-related gene TAF2N to TEC in extraskeletal myxoid chondrosarcoma. Cancer Res. 1999;59:5064–5067. [PubMed] [Google Scholar]
- Smith R., Owen L.A., Trem D.J., Wong J.S., Whangbo J.S., Golub T.R., Lessnick S.L. Expression profiling of EWS/FLI identifies NKX2.2 as a critical target gene in Ewing's sarcoma. Cancer Cell. 2006;9:405–416. doi: 10.1016/j.ccr.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Stallcup M.R. Role of protein methylation in chromatin remodeling and transcriptional regulation. Oncogene. 2001;20:3014–3020. doi: 10.1038/sj.onc.1204325. [DOI] [PubMed] [Google Scholar]
- Stolow D.T., Haynes S.R. Cabeza, a Drosophila gene encoding a novel RNA binding protein, shares homology with EWS and TLS, two genes involved in human sarcoma formation. Nucleic Acids Res. 1995;23:835–843. doi: 10.1093/nar/23.5.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi A., Higashino F., Aoyagi M., Yoshida K., Itoh M., Kyo S., Ohno T., Taira T., Ariga H., Nakajima K., et al. EWS/ETS fusions activate telomerase in Ewing's tumors. Cancer Res. 2003;63:8338–8344. [PubMed] [Google Scholar]
- Tanaka K., Iwakuma T., Harimaya K., Sato H., Iwamoto Y. EWS–Fli1 antisense oligodeoxynucleotide inhibits proliferation of human Ewing's sarcoma and primitive neuroectodermal tumor cells. J. Clin. Invest. 1997;99:239–247. doi: 10.1172/JCI119152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelin-Jarnum S., Lassen C., Panagopoulos I., Mandahl N., Aman P. Identification of genes differentially expressed in TLS–CHOP carrying myxoid liposarcomas. Int. J. Cancer. 1999;83:30–33. doi: 10.1002/(sici)1097-0215(19990924)83:1<30::aid-ijc6>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- Thomas G.R., Latchman D.S. The pro-oncoprotein EWS (Ewing's Sarcoma protein) interacts with the Brn-3a POU transcription factor and inhibits its ability to activate transcription. Cancer Biol. Ther. 2002;1:428–432. doi: 10.4161/cbt.1.4.23. [DOI] [PubMed] [Google Scholar]
- Thompson A.D., Braun B.S., Arvand A., Stewart S.D., May W.A., Chen E., Korenberg J., Denny C. EAT-2 is a novel SH2 domain containing protein that is up regulated by Ewing's sarcoma EWS/FLI1 fusion gene. Oncogene. 1996;13:2649–2658. [PubMed] [Google Scholar]
- Tora L. A unified nomenclature for TATA box binding protein (TBP)-associated factors (TAFs) involved in RNA polymerase II transcription. Genes Dev. 2002;16:673–675. doi: 10.1101/gad.976402. [DOI] [PubMed] [Google Scholar]
- Tuerk C., Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- Uren A., Tcherkasskaya O., Toretsky J.A. Recombinant EWS–FLI1 oncoprotein activates transcription. Biochemistry. 2004;43:13579–13589. doi: 10.1021/bi048776q. [DOI] [PubMed] [Google Scholar]
- Vance C., Rogelj B., Hortobagyi T., De Vos K.J., Nishimura A.L., Sreedharan J., Hu X., Smith B., Ruddy D., Wright P., et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Arai S., Song X., Reichart D., Du K., Pascual G., Tempst P., Rosenfeld M.G., Glass C.K., Kurokawa R. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;454:126–130. doi: 10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welford S.M., Hebert S.P., Deneen B., Arvand A., Denny C.T. DNA binding domain-independent pathways are involved in EWS/FLI1-mediated oncogenesis. J. Biol. Chem. 2001;276:41977–41984. doi: 10.1074/jbc.M106757200. [DOI] [PubMed] [Google Scholar]
- Wu S., Green M.R. Identification of a human protein that recognizes the 3′ splice site during the second step of pre-mRNA splicing. EMBO J. 1997;16:4421–4432. doi: 10.1093/emboj/16.14.4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S.J., Barr F.G. Chromosome translocations in sarcomas and the emergence of oncogenic transcription factors. Eur. J. Cancer. 2005;41:2513–2527. doi: 10.1016/j.ejca.2005.08.003. [DOI] [PubMed] [Google Scholar]
- Xiang H., Wang J., Hisaoka M., Zhu X. Characteristic sequence motifs located at the genomic breakpoints of the translocation t(12;16) and t(12;22) in myxoid liposarcoma. Pathology. 2008;40:547–552. doi: 10.1080/00313020802320424. [DOI] [PubMed] [Google Scholar]
- Yang L., Embree L.J., Tsai S., Hickstein D.D. Oncoprotein TLS interacts with serine-arginine proteins involved in RNA splicing. J. Biol. Chem. 1998;273:27761–27764. doi: 10.1074/jbc.273.43.27761. [DOI] [PubMed] [Google Scholar]
- Yang L., Chansky H.A., Hickstein D.D. EWS.Fli-1 fusion protein interacts with hyperphosphorylated RNA polymerase II and interferes with serine–arginine protein-mediated RNA splicing. J. Biol. Chem. 2000a;275:37612–37618. doi: 10.1074/jbc.M005739200. [DOI] [PubMed] [Google Scholar]
- Yang L., Embree L.J., Hickstein D.D. TLS-ERG leukemia fusion protein inhibits RNA splicing mediated by serine-arginine proteins. Mol. Cell. Biol. 2000b;20:3345–3354. doi: 10.1128/mcb.20.10.3345-3354.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi H., Fujimura Y., Ouchida M., Prasad D.D., Rao V.N., Reddy E.S. Inhibition of apoptosis by normal and aberrant Fli-1 and erg proteins involved in human solid tumors and leukemias. Oncogene. 1997;14:1259–1268. doi: 10.1038/sj.onc.1201099. [DOI] [PubMed] [Google Scholar]
- Yu Z., Hecht N.B. The DNA/RNA-binding protein, translin, binds microRNA122a and increases its in vivo stability. J. Androl. 2008;29:572–579. doi: 10.2164/jandrol.108.005090. [DOI] [PubMed] [Google Scholar]
- Zhang D., Paley A.J., Childs G. The transcriptional repressor ZFM1 interacts with and modulates the ability of EWS to activate transcription. J. Biol. Chem. 1998;273:18086–18091. doi: 10.1074/jbc.273.29.18086. [DOI] [PubMed] [Google Scholar]
- Zhou H., Lee K.A. An hsRPB4/7-dependent yeast assay for trans-activation by the EWS oncogene. Oncogene. 2001;20:1519–1524. doi: 10.1038/sj.onc.1204135. [DOI] [PubMed] [Google Scholar]
- Zhou Q., Sharp P.A. Tat-SF1: cofactor for stimulation of transcriptional elongation by HIV-1 Tat. Science. 1996;274:605–610. doi: 10.1126/science.274.5287.605. [DOI] [PubMed] [Google Scholar]
- Zhou Z., Licklider L.J., Gygi S.P., Reed R. Comprehensive proteomic analysis of the human spliceosome. Nature. 2002;419:182–185. doi: 10.1038/nature01031. [DOI] [PubMed] [Google Scholar]
- Zinszner H., Albalat R., Ron D. A novel effector domain from the RNA-binding protein TLS or EWS is required for oncogenic transformation by CHOP. Genes Dev. 1994;8:2513–2526. doi: 10.1101/gad.8.21.2513. [DOI] [PubMed] [Google Scholar]
- Zinszner H., Immanuel D., Yin Y., Liang F.X., Ron D. A topogenic role for the oncogenic N-terminus of TLS: nucleolar localization when transcription is inhibited. Oncogene. 1997a;14:451–461. doi: 10.1038/sj.onc.1200854. [DOI] [PubMed] [Google Scholar]
- Zinszner H., Sok J., Immanuel D., Yin Y., Ron D. TLS (FUS) binds RNA in vivo and engages in nucleo-cytoplasmic shuttling. J. Cell Sci. 1997b;110:1741–1750. doi: 10.1242/jcs.110.15.1741. (Pt 15) [DOI] [PubMed] [Google Scholar]
- Zoubek A., Dockhorn-Dworniczak B., Delattre O., Christiansen H., Niggli F., Gatterer-Menz I., Smith T.L., Jurgens H., Gadner H., Kovar H. Does expression of different EWS chimeric transcripts define clinically distinct risk groups of Ewing tumor patients? J. Clin. Oncol. 1996;14:1245–1251. doi: 10.1200/JCO.1996.14.4.1245. [DOI] [PubMed] [Google Scholar]
- Zucman J., Melot T., Desmaze C., Ghysdael J., Plougastel B., Peter M., Zucker J.M., Triche T.J., Sheer D., Turc-Carel C., et al. Combinatorial generation of variable fusion proteins in the Ewing family of tumours. EMBO J. 1993;12:4481–4487. doi: 10.1002/j.1460-2075.1993.tb06137.x. [DOI] [PMC free article] [PubMed] [Google Scholar]