Abstract
A surprisingly large and unrelated number of human tumors depend on sustained HEDGEHOG-GLI (HH-GLI) signaling for growth. This includes cancers of the skin, brain, colon, lungs, prostate, blood and pancreas among others. The basis of such commonality is not obvious. HH-GLI signaling has also been shown to be active in and required for cancer stem cell survival and expansion in different cancer types, and its activity is essential not only for tumor growth but also for recurrence and metastatic growth, two key medical problems. Here we review recent data on the role of HH-GLI signaling in cancer focusing on the role of the GLI code, the regulated combinatorial and cooperative function of repressive and activating forms of all Gli transcription factors, as a signaling nexus that integrates not only HH signals but also those of multiple tumor suppressors and oncogenes. Recent data support the view that the context-dependent regulation of the GLI code by oncogenes and tumor suppressors constitutes a basis for the widespread involvement of GLI1 in human cancers, representing a perversion of its normal role in the control of stem cell lineages during normal development and homeostasis.
Keywords: Gli, Hedgehog, cancer, stem cells, oncogenes, tumor suppressors, cancer stem cells
Introduction
A number of morphogenetic signaling pathways regulating developmental processes and organ homeostasis are critical players in tumorigenesis. Among them, Hedgehog-Gli (Hh-Gli) plays an important role in determining proper embryonic patterning and in controlling growth and cell fate during animal development. Similarly, in the adult, Hh-Gli signaling is involved in tissue maintenance and repair, regulating stem cell behavior in several instances. Importantly for the focus of this review, its aberrant activation drives tumorigenesis in humans and animals.
Secreted Hh ligands (Sonic, Indian and Desert Hhs in mice and humans) initiate signaling in receiving cells by binding and inactivating the 12-pass transmembrane receptor Patched1 (Ptc1), which relieves its catalytic inhibition of the 7-pass transmembrane protein Smoothened (Smo). Consequently, active Smo triggers an intracellular signaling cascade that enables activation and inhibits the formation of repressors of the Gli transcription factors (for example Riobó and Manning, 2007; Ruiz i Altaba et al., 2006, 2007; Aikin et al., 2008; Varjosalo and Taipale, 2008). Thus, Hh signaling regulates the function of the Gli proteins and their activation.
Recent evidence points to additional mechanisms of GLI modulation by oncogenes and tumor suppressors. In this review, we focus on how Hh and non-Hh inputs regulate the ‘GLI code’ (Ruiz i Altaba, 1997, 1998, 1999): the combinatorial and cooperative function of the GLI transcription factors. Rather than being comprehensive, here we selectively highlight recent advances in the integration of oncogenic and tumor suppressor pathways with HH-GLI signaling and how these inputs modulate the activity of the GLI code in cancer. We suggest that the context-dependent regulation of the GLI code by Hh and non-Hh signals is a driving force in human cancer likely taking place in cancer stem cells. The challenging view and general hypothesis of the GLI code as an integration nexus for numerous signaling inputs (Ruiz i Altaba et al., 2002, 2004, 2007) holds important implications for understanding how the major oncogenic pathways interact during tumorigenesis and for the development of more effective and focused anticancer therapies.
The GLI code
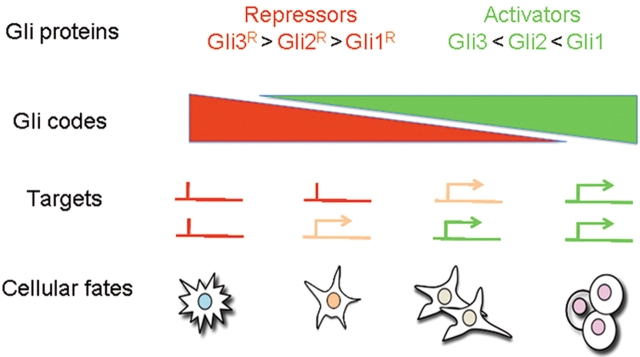
The Gli proteins, Gli1-3 in frogs, mice and humans, are zinc finger transcription factors that mediate transcriptional responses to Hh signaling (Figure 1). Generally, in the absence of Hh ligands, Gli1 is transcriptionally repressed, whereas Gli3 and possibly Gli2 are phosphorylated, recognized by the F-box protein β-TrCP and proteolytically processed to truncated repressor forms (Wang et al., 2000; Pan et al., 2006), with the consequent silencing of Hh-Gli targets. Gli2 and Gli3 can be also phosphorylated by Dyrk2, a kinase inducing their proteasome-dependent degradation (Varjosalo et al., 2008). In the presence of Hh ligands, the Gli code is modified: Gli1 is activated transcriptionally and the processing of Gli2 and Gli3 is inhibited, leading to the accumulation of their full-length forms and the activation of specific Hh-Gli target genes. Moreover, Gli function is also controlled by acetylation (Canettieri et al., 2010). The balance of the collective activator and repressor functions of these three transcription factors seems to determines the status of the Hh transcriptional program and ultimately the behavior for responding cells (Figures 1 and 2).
Figure 1.
The GLI code. Different combinations of GLI activator (in green) and repressor forms (in red), with different potencies, are proposed to activate different sets of target genes that result in specific cellular fates and proliferation rates. The diagram illustrates how different (combinatorial and quantitative) GLI codes give different cellular outcomes.
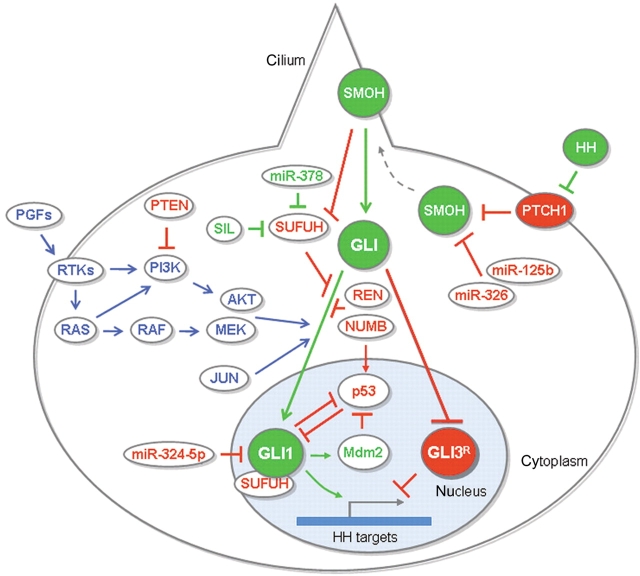
Figure 2.
Integration of oncogenic and tumor suppressor inputs by the GLI code in cancer. Upon inhibition of PTCH1 function by HH ligands, the repression on SMOH is released, SMOH moves into the primary cilium and activates downstream signaling by stabilizing activating full-length GLI proteins (GLI1) and blocking the production of GLI repressors (GLI3R). The mammalian GLI code includes three proteins. Generally, GLI1 is an activator although it exists in N′ and C′ Δ deleted activator and repressor forms, respectively, GLI2 has activator and C′ Δ repressor functions and GLI3 is a weak activator and its C′ Δ form is a strong repressor. Components of the classical HH pathway are in filled circles, in red for inhibitors and in green for activators. Positive and negative regulators of HH-GLI signaling are in unfilled circles, in blue for the PGF-RTK-RAS-RAF-MEK, PI3K-AKT and JUN pathways, in green for activators and in red for repressors. The color of the arrow is dictated by the final effect on the GLI code: red arrow for a final repressive effect, green arrow for a final activating effect on the GLI code. See text for further details.
The Gli proteins encode both activator and repressor functions. Like their fly homolog Cubitus interruptus (Ci) (Aza-Blanc et al., 1997), Gli2 and Gli3 possess an amino-terminal repressor domain and a carboxy-terminal activator domain flanking the central five zinc-finger DNA-binding domains. Gli1, however, lacks a similar amino-terminal repressor domain and while it exists in full-length, N′ Δ and C′ Δ forms, the latter with activator and repressor functions, respectively (Stecca and Ruiz i Altaba, 2009), it functions as the terminal and thus critical transcriptional activator of the Hh pathway. Its function is reinforced by a positive feedback loop as its transcription is induced by Hh signaling, making it, so far, the best reliable read-out of an active pathway (Lee et al., 1997; Bai et al., 2004), and it is consistently transcribed in Hh-responding cells. Gli2 can act as activator or repressor, whereas Gli3, which can exist and act as a weak activator, mostly functions as a repressor of transcription (Ruiz i Altaba, 1998, 1999; Sasaki et al., 1999; Aza-Blanc et al., 2000; Litingtung et al., 2002; Bai et al., 2004; Stamataki et al., 2005), mimicking the repressive function of Drosophila Ci (Aza-Blanc et al., 1997). The three Gli proteins operate together in responding cells to integrate intercellular Hh signaling and other inputs, and each can have positive or negative effects (Ruiz i Altaba, 1998; Nguyen et al., 2005; Tyurina et al., 2005).
The Gli proteins act in a context- and species-dependent manner. In the frog neural plate, Gli1 mediates the effects of Shh in the induction of floor plate differentiation, whereas both Gli2 and Gli3 inhibit this function (Lee et al., 1997), although whether they act positively or negatively is stage- and target-specific (Nguyen et al., 2005). In the spinal cord, Gli1 and Gli2 induce motor neurons, but Gli3 has an opposite function (Ruiz i Altaba, 1998). In mice, Gli1 seems to be dispensable for development (Park et al., 2000), whereas Gli2 and Gli3 have specific and partially overlapping functions. However, Gli1 can rescue the function of Gli2 (Bai and Joyner, 2001) and Gli1/Gli2 double mutants have stronger defects than the single Gli2 mutants (Park et al., 2000). In contrast, during floor plate development Gli2 appears to be the major player (Matise et al., 1998) and is required for initial Shh signaling (Bai et al., 2002). Gli3 has been proposed to act primarily as an inhibitor of Shh-Gli1. Loss of ventral spinal cord cell types seen in Shh mutant mice can be partially rescued in Shh/Gli3 double mutants (Litingtung and Chiang, 2000), suggesting that in this context Gli3 acts mainly as a repressor and Shh as an inhibitor of Gli3. However, an activator function of Gli3 is required for proper spinal cord patterning (Bai et al., 2004). The anterior hindbrain of Gli2 null embryos displays ventral patterning defects as severe as those observed in Shh null embryos suggesting that in the anterior hindbrain, unlike in the spinal cord, Gli3 cannot compensate for the loss of Gli2 activator function (Lebel et al., 2007). In zebrafish Gli1 is required for the development of the ventral central nervous system (Karlstrom et al., 2003) and is the main activator of Hh target genes (Tyurina et al., 2005).
In mice, Gli1 expression is dependent on Gli2 and/or Gli3-mediated transcription and it is induced by Hh ligands (Sasaki et al., 1999). However, degradation is also relevant for Gli1: two degradation sequences, degron N and degron C, mediate recognition by the β-TrCP E3 ubiquitin ligase to allow ubiquitination and degradation by the proteasome (Huntzicker et al., 2006). Gli1 can be also targeted for proteolysis by Itch, another E3 ubiquitin ligase (Di Marcotullio et al., 2006). Similarly, Ci/Gli can be degraded through the ubiquitin E3 ligase adaptors Roadkill and HIB/SPOP in an Hh-dependent manner (Kent et al., 2006; Zhang et al., 2006; Chen et al., 2009), the latter being mediated by multiple Ser/Thr-rich degrons (Zhang et al, 2009).
The regulation of the Gli proteins involves the function of several other modulators, including that of Suppressor of Fused (Sufu), Zic proteins and microRNAs. Sufu both sequesters the Gli proteins in the cytoplasm and represses transcription by recruiting the histone deacetylase complex SAP18-mSin3 (Kogerman et al., 1999; Cheng and Bishop, 2002). The inhibitory function of Sufu must be suspended during Hh pathway activation to induce a positive Gli code, but how this happens is not clear. It has been suggested that Smo activation inhibits Sufu function (Svärd et al., 2006) and that SCL/TAL interrupting locus (SIL), a cytoplasmic protein overexpressed in pancreatic adenocarcinoma, de-represses GLI1 from SUFUH (Kasai et al., 2008; Figure 2). In addition, Hh signaling itself can promote degradation of Sufu through the ubiquitin-proteasome system (Yue et al., 2009). Finally, Sufu has also been shown to recruit GSK3β for Gli3 for processing, yielding the potent and pan-dominant Gli3 C′ Δ repressor (Kise et al., 2009).
Gli proteins physically interact with the Zic transcription factors through their highly related five zinc finger domains and regulate each other in a context-dependent manner (Brewster et al., 1998; Koyabu et al., 2001; Mizugishi et al., 2001; Nguyen et al., 2005). In addition, recent studies show that Kif7, the mammalian homologue of Drosophila Costal2, physically interacts with the Gli proteins and can act both positively and negatively on Hh signaling (Cheung et al., 2009; Liem et al., 2009; Endoh-Yamagami et al., 2009). A recently identified serine/threonine kinase, ULK3, which shares some homology with Drosophila Fused protein, has been shown to enhance GLI1 and GLI2 transcriptional activity (Maloverjan et al., 2009). The conserved regulatory protein 14-3-3 has been described to bind all three Gli proteins, decreasing Hh signaling activity (Asaoka et al., 2009). Finally, recent reports suggest that Hh signaling can be regulated by specific microRNAs (miR). miR-125b, miR-324-5p and miR-326 functionally suppress Smo, and miR-324-5p also targets Gli1 in cerebellar granule cell precursors (GCPs) and human medulloblastoma cell lines (Ferretti et al., 2008). Moreover, the miR-17/92 cluster synergizes with Shh signaling in GCPs and medulloblastoma (Northcott et al., 2009; Uziel et al., 2009; Figure 2).
Different states of the Gli code are proposed to activate different or partially overlapping sets of target genes, resulting in distinct cellular responses (Figure 1). Gli activators and repressor targets are only partially known (Sasaki et al., 1997; Kasper et al., 2006; Clement et al., 2007; Vokes et al., 2007, 2008) and can respond to combinatorial and cooperative Gli activity (Nguyen et al., 2005). The outcome of Hh signaling varies according to the receiving cell type. Genes generally induced by Hh activity, such as Ptc1, Hip1 and Gli1, can trigger positive or negative feedbacks on the pathway, which modify the strength or duration of the Hh signal. Additional Gli targets include genes contributing to the regulation of proliferation and differentiation (e.g. CyclinD1 and D2, N-Myc, Wnts, PdgfRα, Igf2, FoxM1, Hes1) (Dahmane et al., 1997; Kenney and Rowitch, 2000; Mullor et al., 2001, Teh et al., 2002; Kenney et al., 2003; Ingram et al., 2008), survival (Bcl2) (Regl et al., 2004), self-renewal (Bmi1, Nanog) (Leung et al., 2004; Clement et al., 2007; Stecca and Ruiz i Altaba, 2009), angiogenesis (Vegf) (Pola et al., 2001), epithelial-mesenchymal transition (Snail1, Sip1, Elk1 and Msx2) (Li et al., 2006; Ohta et al., 2009; Varnat et al., 2009) and invasiveness (Osteopontin) (Das et al., 2009). Given the broad range of possible direct/indirect targets in different cellular contexts, it is not surprising that deregulation of Hh signaling can be pathogenic.
Integration of multiple signaling inputs by the GLI code in cancer
GLI1 was originally identified as an amplified gene in a human glioma (Kinzler et al., 1987). Germ line loss-of-function mutations in PTCH1 (Hahn et al., 1996; Johnson et al., 1996), which constitutively activate the HH pathway, were then found in patients with Gorlin's syndrome, a familiar condition with predisposition to basal cell carcinoma (BCC), medulloblastoma and rhabdomyosarcoma (Gorlin, 1995). Somatic misexpression of GLI1 was shown to directly induce skin hyperplasias or tumors in frog embryos and to mark human sporadic BCCs, indicating the presence of an active pathway in this most common human sporadic tumor type (Dahmane et al., 1997). Other studies also showed that the sustained activation of the Hh-Gli pathway in the mouse epidermis leads to BCC development through the misexpression of Shh (Oro et al., 1997), GLI1 (Nilsson et al., 2000) or Gli2 (Grachtchouk et al., 2000; Hutchin et al., 2005).
Hedgehog signaling regulates the growth of the external germinal layer of the cerebellum, which contains GCPs. Shh, produced by Purkinje neurons, induces GCP proliferation (Dahmane and Ruiz i Altaba, 1999; Wallace, 1999; Wechsler-Reya and Scott, 1999) and the differentiation of Bergmann glia (Dahmane and Ruiz i Altaba, 1999). Interestingly, Ptc1+/− mice and humans harboring inactivating mutations that abrogate PTCH1 function, develop medulloblastoma (Johnson et al., 1996; Hahn et al., 1996, 1998; Goodrich et al., 1997). These studies suggest that at least a subset of murine and human medulloblastomas arise from GCPs that maintain an inappropriately active SHH-GLI pathway. In this regard, it appears that medulloblastoma can initiate in multipotent stem cells or in GCPs in mice, but suggest that Hh-induced tumors must commit to a neuronal lineage in order to grow (Schuller et al., 2008; Yang et al., 2008).
The biological context of Hh-Gli signaling in the development of a particular cell type or tissue indicates that Hh-Gli activity has to be turned down after it has induced or promoted a particular developmental process. Such tight control of the Gli code, turning it towards a repressive state after the end of ligand-driven signaling, is disrupted in cancer. For instance, the inability to cease Shh signaling reception in GCPs in the cerebellum could lead to medulloblastoma development (Dahmane and Ruiz i Altaba, 1999; Wallace, 1999; Weschler-Reya and Scott, 1999; Ruiz i Altaba et al., 2002), as in Ptc1+/− mice (Goodrich et al., 1997; Hahn et al., 1998). Similarly, the failure to turn down the proliferative and self-renewing functions of the Gli code in an activating state in stem cells/precursors might lead to an abnormal proliferative response, provoking an expansion of their numbers thus leading to tumor formation (Clement et al., 2007; Bar et al., 2007; Ehtesham et al., 2007; Peacock et al., 2007; Dierks et al., 2008; Stecca and Ruiz i Altaba, 2009; Zhang et al., 2009).
What are then the mechanisms that contribute to the normal tight regulation of the Gli code? Other than structural alterations in pathway components such as SMOH or PTCH1, recent data point towards a context-dependent interplay between Hh-Gli signaling and oncogenic and tumor suppressor functions that modulate the activity of the Gli code. Here we highlight recent findings on the crosstalk of two major oncogenic signaling pathways—RAS-RAF-MEK, PI3K-AKT—and two major tumor suppressors—PTEN and p53—with the HH-GLI pathway.
Regulation of the Gli code by oncogenes
Discovery of the role of Gli2 in FGF signaling in the ventro-posterior mesoderm of frog embryos (Brewster et al., 2000) and the synergism of Hh and EGF signaling in neural stem cells (Palma and Ruiz i Altaba, 2004; Palma et al., 2005) allowed the proposal of a funnel hypothesis, by which multiple signaling inputs converge on and regulate the Gli code in development and cancer (Ruiz i Altaba et al., 2004; Figures 2 and 3). Recent data indeed suggest that the Gli code are regulated by multiple oncogenic signaling events, including peptide growth factors (PGF), receptor tyrosine kinases (RTKs), RAS, MEK, phosphoinositide-3 kinase (PI3K) and AKT (reviewed in Ruiz i Altaba et al., 2007). PGF activate RTKs, which in turn activate several signal-transduction cascades that include RAS-RAF-MEK and PI3K/AKT. These are key signaling pathways involved in the regulation of cell proliferation, survival and differentiation. Abnormal activation of these pathways commonly occurs in human cancers due to mutations at multiple levels (Wellbrock et al., 2004).
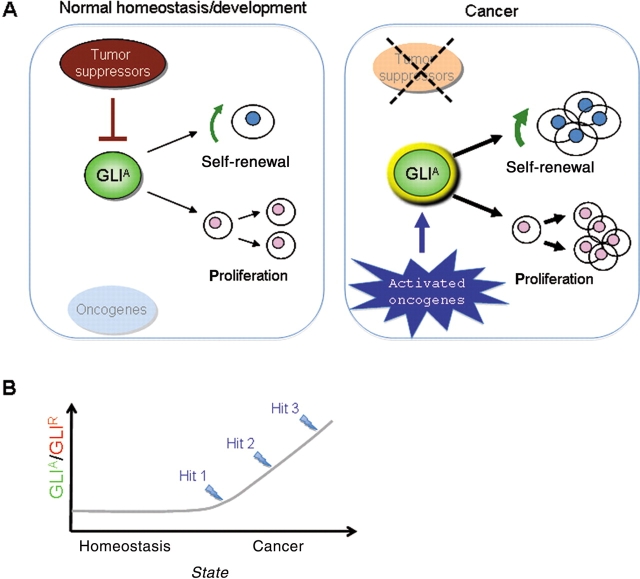
Figure 3.
Regulation of the GLI code by oncogenic and tumor suppressor inputs in stem cells and cancer. During development, HH signaling tightly promotes the formation of labile GLI activators. The action of tumor suppressors further restrains GLI positive function, which is boosted by converging pathways and inputs such as those triggered by oncogenic PGF-RTK signals. (A and B) Mutations or epigenetic changes that lead to the loss of tumor suppressors (for example p53, PTEN) and activation of oncogenic pathways (for example RAS-RAF-MEK, PI3K-AKT) could unlock the normally restricted proliferative and self-renewing activities of GLI activators leading to an abnormal expansion of cancer stem cells (Ruiz i Altaba et al., 2007).
An interplay between RAS-RAF-MEK and PI3K/AKT and HH-GLI signaling pathways is supported by multiple lines of evidence. For instance, oncogenic H- or N-RAS and AKT1 potentiate GLI1 function, by enhancing its transcriptional activity and nuclear localization, and counteracting its cytoplasmic retention by SUFUH in melanoma, gliomas and prostate cancer cells (Stecca et al., 2007). Furthermore, endogenous GLI1 activity requires endogenous AKT and MEK function (Stecca et al., 2007). Consistently, PI3K/AKT has a positive function on Hh-Gli signaling in NIH 3T3 cells (Riobó et al., 2006a) and HH and AKT synergize in tumor formation in zebrafish (Ju et al., 2009). Similarly, K-RAS suppresses GLI1 degradation in pancreatic cancer cells (Ji et al., 2007) and K-RAS-MEK-ERK cascade has a positive regulatory role in GLI transcriptional activity in gastric cancer (Seto et al., 2009). The RAS effectors responsible for GLI stimulation are likely to be MEK1 or ERK1/2, through GLI1 NH2-terminal domain, although GLI1 does not seem to be phosphorylated by ERK2 (Riobó et al., 2006b).
Phenotypic interactions between RAS and GLI have been described in various mouse models. For instance, oncogenic K-RAS genetically cooperates with activated (truncated) Gli2 to induce undifferentiated pancreatic tumors (Pasca di Magliano et al., 2006; Nolan-Stevaux et al., 2009), H-RAS genetically cooperates with a specific Ptc1 allele to induce tumorigenesis (Wakabayashi et al., 2007), and Shh signaling enhances K-RAS-induced pancreatic tumors (Morton et al., 2007). Interestingly, mice expressing endogenous levels of mutant K-RAS in the pancreas show an activation of the Hh pathway, indicating the action of oncogenic RAS on the endogenous Hh-Gli pathway during tumor development (Hingorani et al., 2005). Importantly, mouse melanomas induced by activated N-RAS in an INK4a mutant background display an active pathway and require Hh-Gli function (Stecca et al., 2007). An additional report suggests that K-RAS regulates Smo-independent expression of Gli genes in mouse pancreatic ductal adenocarcinoma and that GLI1 is required for K-RAS-mediated transformation (Nolan-Stevaux et al., 2009).
A crosstalk between the EGFR-MEK/ERK and HH-GLI signaling pathways has been reported in human keratinocytes, both in normal conditions and in cancer. In normal keratinocytes EGFR signaling modulates HH-GLI target gene expression (Kasper et al., 2006), and during their transformation EGFR-MEK/ERK signaling induces activation of JUN, which cooperates with GLI1 and GLI2 (Schnidar et al., 2009). In turn, it has been shown that GLI1 and GLI2 directly regulate the expression of JUN (Laner-Plamberger et al., 2009).
The crosstalk between RAS/AKT and GLI1 might be more complex than expected, since it can have a different outcomes depending on the cellular type. For instance, oncogenic H-RAS promotes GLI1-mediated cancer cell proliferation but inhibits GLI1-mediated osteogenic differentiation of C3H10T1/2 cells (Stecca et al., 2007).
Another pathway that regulates the GLI code is triggered by TGFβ signaling (Dennler et al., 2007). The TGFβ superfamily comprises a group of growth factors involved in embryonic development, tissue repair and differentiation, exerting both positive and negative effects on cancer development (Derynck et al., 2001; Siegel and Massagué, 2003). Receptor activation leads to phosphorylation of cytoplasmic proteins of the Smad family, which translocate into the nucleus where they regulate target gene expression. Similar to RAS, TGFβ interacts with the Hh pathway downstream of Smo, activating the Gli code: in transgenic mice overexpressing TGFβ1 in the skin, Gli2 expression is induced in a Smad3-dependent manner (Dennler et al., 2007). These actions appear consistent with the binding of Smads to GLIs (Liu et al., 1998).
In addition, there is evidence that the Notch pathway can also affect the Gli code. Notch signaling, which is important in binary cell-fate decisions and inhibiting differentiation in many developmental systems (Artavanis-Tsakonas et al., 1999), can play as tumor suppressor or as oncogene depending on the context. In the mammalian skin, Notch1 deficiency increases the level of Gli2, causing the development of BCC-like tumors (Nicolas et al., 2003).
Recently, it has been shown that GLI1 is an important mediator of oncogenic EWS-FLI1 function (Joo et al., 2009), which is produced by a chromosomal translocation occurring in the majority of Ewing's sarcomas (Arvand and Denny, 2001).
Finally, it has been suggested that Nuclear Factor-kappaB (NF-κB), a key transcription factor that orchestrates numerous processes, including proliferation, apoptosis and inflammatory responses (Karin et al., 2002), contributes to Shh activation in pancreatic cancer cells (Nakashima et al., 2006; Kasperczyk et al., 2009). Additional oncogenes or oncogene mediators, such as β-catenin (Maeda et al., 2006; Ulloa et al., 2007) could also regulate the Gli code.
An unresolved issue is whether the primary targets of all these oncogenic pathways that affect the GLI code are the GLI proteins themselves or rather their regulators, such as SUFUH. Another issue is whether oncogene action requires a basal level of GLI expression or cells harboring oncogenes become tumorigenic after the further selection of events that activate the HH-GLI pathway. It has been reported that oncogenic RAS cannot transcriptionally activate GLI1 in human melanoma cells in the absence of HH signals, that sporadic NRASQ61K;Ink4a−/− mouse melanomas express and require Hh-Gli activity and that the transcriptional activity and nuclear translocation of GLI1 is regulated by oncogenic RAS/AKT (Stecca et al., 2007). Thus, the data so far support the idea that cells with an active HH-GLI pathway—commonly stem cells and precursors (see below)—are targets of oncogenic events.
Modulation of the Gli code by tumor suppressors
Several lines of evidence suggest that the Gli code can be tightly regulated by the action of tumor suppressors (Figure 2). For instance, the tumor suppressor PTEN (phosphatase and tensin homolog deleted on chromosome 10), which regulates neural stem cell behavior (Groszer et al., 2001) is frequently mutated, deleted or silenced in various human cancers (Teng et al., 1997), has been shown to negatively regulate the transcriptional activity of GLI1 in glioblastoma cells (Stecca et al., 2007).
Moreover, the tumor suppressor p53 negatively controls the activity of GLI1 in neural stem cells and human cancer cells. p53 inhibits GLI1-driven neural stem cell self-renewal, cell proliferation and tumor growth, by negatively regulating transcriptional activity, nuclear localization and levels of GLI1 protein (Stecca and Ruiz i Altaba, 2009). In turn, activation of the Hh pathway by constitutive activated Smo mutants, Shh ligand and overexpression of Gli1 and Gli2, inhibits the accumulation of p53, through the phosphorylation and activation of the p53 inhibitor Mdm2 (Abe et al., 2008; Stecca and Ruiz i Altaba, 2009). An essential role for the Hh signaling in tumorigenesis induced by loss of p53 is suggested by the finding that epidermal tumors induced by blockade of p53 in frog embryos (Wallingford et al., 1997) require endogenous Gli1 activity (Stecca and Ruiz i Altaba, 2009). The homeostatic balance between p53/GLI1 activities appears to be critical. In normal conditions the balance between p53 and GLI1 seems to be in favor of the former: p53 is expressed in neural stem cells where it regulates their self-renewal (Gil-Perotin et al., 2006; Meletis et al., 2006). In contrast, loss of p53, which is a common event in cancer progression, could free the normally restricted activities of GLI1, leading to expansion of cancer stem cells and their derived progenitors (Stecca and Ruiz i Altaba, 2009; Figure 3). Similarly, it has been recently shown that overexpression of Gli2 and p53 deficiency genetically cooperate in the progression from benign to malignant cartilage tumors in a mouse model, and have additive effects on negatively regulating chondrocyte apoptosis (Ho et al., 2009).
Another putative tumor suppressor gene, RENKCTD11, which maps to 17p13.2 near the p53 locus, has been reported to antagonize Hh signaling in the cerebellum. REN inhibits medulloblastoma growth by affecting Gli1 nuclear accumulation (Di Marcotullio et al., 2004). Conversely, REN knock-down enhances Hh signaling and proliferation in GCPs and impairs neuronal differentiation (Argenti et al., 2005), supporting a role for REN as an inhibitory signal required for restraining Hh-Gli activity and cessation of GCP expansion.
An additional factor suppressing Hh signaling in GCPs is Numb, an evolutionary conserved protein endowed with tumor suppressor properties that segregates asymmetrically determining self-renewal or differentiation fates during neural progenitor division (Rhyu et al., 1994). Numb targets Gli1 for proteasome degradation through the E3 ligase Itch (Di Marcotullio et al., 2007). The Numb-induced Itch-dependent proteolytic processing of Gli1 thus limits the extend and duration of Hh signaling during neural progenitor differentiation and its disruption could be an important event in brain tumorigenesis. With respect to the inhibitory effect of p53 and Numb on GLI1, it is interesting to note that Numb controls p53 activity as well (Colaluca et al., 2008).
Evidence for the integration of multiple signaling inputs by the GLI code in cancer stem cells
Hh-Gli signaling controls proliferation and self-renewal of neural stem cells in neurogenic niches of the brain, such as the embryonic neocortex, the subventricular zones of the lateral ventricles and the subgranular layer of the hippocampus (Lai et al., 2003; Machold et al., 2003; Palma and Ruiz i Altaba, 2004; Ahn and Joyner, 2005; Palma et al., 2005), and the level of GLI1 expression determines the number of neural stem cells in multiple brain regions (Stecca and Ruiz i Altaba, 2009). In humans, HH-GLI signaling controls self-renewal and tumorigenicity of cancer stem cells in glioma (Clement et al., 2007), as well as multiple myeloma (Peacock et al., 2007), myeloid leukemia (Dierks et al., 2008; Zhao et al., 2009) and colon cancer (Varnat et al., 2009).
Cancer stem cells are defined as tumor cells that self-renew, give rise to less undifferentiated progeny and induce tumorigenesis in immunocompromised mice. No marker or set of markers can unequivocally identify them as they are operationally defined (Ruiz i Altaba and Brand, 2009). Cancer stem cells are found in many tumor types (both solid and liquid) (Hamburger and Salmon, 1977; Lapidot et al., 1994), and could originate from stem cells that have lost the ability to regulate their self-renewal and proliferation or from a more differentiated population of progenitor cells that have acquired abilities to self-renew. In cancers dependent on sustained Hh-Gli pathway activity, cancer stem cells could derive from normal stem cells/early progenitors with basal Hh-Gli activity. In these ‘tumor initiating cells’ the accumulation of genetic hits, such as activation of oncogenes or inactivation of tumor suppressors, acquired during tumor progression, could shift the state of the Gli code to a hyperactive one (Ruiz i Altaba et al., 2004, 2007). Recent data lend to support this view: knock-down of p53 cooperates with Gli1 in controlling both neural stem self-renewal and tumor growth in vivo, and it increases the levels of GLI1 protein, with the consequent enhancement of the proliferation of human glioma stem cells (Stecca and Ruiz i Altaba, 2009). Likewise, the latter display an hyperactivated GLI code, with GLI1 and GLI2 highly expressed and required for their self-renewal and survival, whereas positive GLI3 does not seem to play a crucial role (Clement et al., 2007). Inhibition of SMOH induced a strengthening of negative modulators and repressive GLI proteins, leading to a GLI code switch towards a repressive state. We can thus suggest that p53 and PTEN deficiencies and accumulation of other genetic hits, such as RAS mutation and elevated AKT levels, favor the appearance and evolution of cancer stem cells with a progressively hyperactive GLI code states (Figure 3; Ruiz i Altaba et al., 2004; 2007).
Modes of action of HEDGEHOG-GLI signaling in cancer
Apart from cancers harboring HH pathway activating mutations (e.g. BCC), which might be HH ligand independent, HH ligands appear to function in an autocrine manner in several human cancers (Ruiz i Altaba, 2008). This is the case of lung (Watkins et al., 2003; Yuan et al., 2007), pancreatic (Thayer et al., 2003; Feldmann et al., 2007), digestive tract (Berman et al., 2003) and prostate cancers (Karhadkar et al., 2004; Sanchez et al., 2004; Sheng et al., 2004), gliomas (Clement et al., 2007; Bar et al., 2007; Ehtesham et al., 2007), as well as melanomas (Stecca et al., 2007) and colon cancers (Varnat et al., 2009). In melanoma it has been shown that: (a) epithelial cells, but not the surrounding stroma, express SHH, GLI1 and PTCH1; (b) melanoma cells in vitro respond to HH pathway inhibition through cyclopamine (a SMOH antagonist) or siRNA treatment down-regulating the expression of GLI1 and PTCH1; (c) inhibition of SMOH with shRNAs in melanoma cells dramatically reduces tumor growth in an orthotopic xenograft model in vivo; (d) systemic cyclopamine treatment abolishes metastatic growth in the lungs of mice (Stecca et al., 2007). These effects are specific, since GLI1 epistatically rescues the inhibitory effect of cyclopamine on cell proliferation, and the latter mimics inhibition of SMOH via RNA interference (Clement et al., 2007; Stecca et al., 2007). This finding indicates that HH signaling functions in an autocrine manner and demonstrate that epithelial tumor cells require HH-GLI signaling.
Lately it has been proposed that tumor-derived HH induces a response only on adjacent stroma, including fibroblasts, endothelial cells and immune cells, which in turn provides growth and survival signals to the tumor cells (Yauch et al., 2008; Olive et al., 2009). These studies suggest that HH ligands fail to activate signaling in tumor epithelial cells, proposing a ligand-dependent activation of HH signaling in the stromal microenvironment. Inhibition of HH signaling using small molecules, anti-Hh antibody or genetic ablation of Smo in the mouse stroma results in growth inhibition in pancreatic and colorectal xenograft of human primary tumors and cell lines (Yauch et al., 2008). The paracrine requirement of Hh signaling in tumorigenesis has been noted also in B16F0 murine melanoma cells (Geng et al., 2007). Nevertheless, these results raise a number of important questions that need to be addressed (Ruiz i Altaba, 2008). For instance, the apparent lack of response to HH signaling in epithelial tumor cells in xenografts contrast with a wealth of data on human melanoma, brain, prostate, pancreatic, digestive tract and lung cancers (Dahmane et al., 2001; Berman et al., 2003, Thayer et al., 2003; Watkins et al., 2003; Karhadkar et al., 2004; Sanchez et al., 2004; Clement et al., 2007; Stecca et al., 2007; Yuan et al., 2007; Varnat et al., 2009). Furthermore, the finding that tumor-derived HH stimulates expression of Gli1 and Ptc1 in the infiltrating mouse stroma (as the human stroma transplanted in a primary graft is quickly replaced) but not in the tumor itself in pancreatic and colorectal xenografts (Yauch et al., 2008) is incongruous with the expression of SHH, GLI1 and PTCH1 found in human PSA+ prostate cancer epithelial cells, in MelanA+/MITF+ human melanoma cells (Sanchez et al., 2004; Stecca et al., 2007) and in Cytokeratin+ primary colon cancer epithelial cells (Varnat et al., 2009), but not in the surrounding stroma. We note, however, that the action of HH-GLI signaling may not only be context-dependent, but also species-specific. For instance, signaling in the rodent prostate appears to be exclusively from epithelium to stroma (Pu et al., 2004) whereas it appears to be autocrine in epithelial cells in humans (Sanchez et al., 2004; Varnat et al., 2009). Xenografts are chimeras and thus their signaling is hybrid in nature. Results from one species need to be taken with a grain (or a mountain) of salt when transposed into the context of another. Since the mode of action of HH-GLI signaling has important implication in the design of therapeutic antagonists, it will be important to further dissect the cellular and molecular mechanisms of HH-GLI action in human cancers.
Several lines of evidence suggest that in cancer stem cells derived from different tumor types HH-GLI acts in an autocrine manner. In glioma, CD133+ cancer stem cells express SHH, GLI1 and PTCH1 and their self-renewal, survival and tumorigenicity require SMOH and GLI1 activity, as shown by the inhibition with cyclopamine and RNA interference (Clement et al., 2007). CD44+/CD24−/low/Lin− putative breast cancer stem cells have higher levels of GLI1 and PTCH1 (Liu et al., 2006). Furthermore, cancer stem cells with HH pathway activity have also been identified in multiple myeloma (Peacock et al., 2007), and genetic studies in chronic myeloid leukemia (CML) cancer stem cells (Bcr-Abl-driven Lin−/Sca1+/c-Kit+) show that loss of Smo causes depletion of CML stem cells, whereas constitutive active Smo increases CML stem cell number and accelerates disease (Dierks et al., 2008; Zhao et al., 2009). Interestingly, pharmacological inhibition of Smo reduces not only the propagation of CML driven by wild-type BCR-ABL, but also the growth of imatinib-resistant mouse and human CML (Zhao et al., 2009). Finally, clonogenic CD133+ colon cancer stem cells also require HH-GLI activity (Varnat et al., 2009).
Collectively, the studies summarized above indicate that HH signaling regulates self-renewal and survival of cancer stem cells from different types of tumors, and suggest that HH pathway inhibition will have beneficial effects in vivo reducing tumor burden and metastatic growth. Indeed, systemic cyclopamine treatment of adult mice for a restricted period inhibits tumor growth and prevents recurrence without any major side effects (Sanchez and Ruiz i Altaba, 2005; Stecca et al., 2007; Varnat et al., 2009). They also provide a solid basis for new targeted therapies that include inhibition of HH-GLI function directly (for example Lauth et al., 2007; von Hoff et al., 2009), as well as indirectly and synergistically by blockade of cooperating oncogenic pathways and restoration of tumor suppressive functions.
Funding
Work from the authors' laboratory was supported by grants from the NIH, NSF, Oncosuisse, and from the Swissbridge, Leenards and Jeantet Foundations to A.R.A. B.S. is currently supported by an institutional grant from the Istituto Toscano Tumori.
Acknowledgements
We thank F. Varnat, C. Mas, A. Duquet, A. Lorente Trigos, I. Siegl-Cachedenier and all other Ruiz i Altaba lab members for discussion.
Conflict of interest: A.R.A. is an advisor to Phistem.
References
- Abe Y., Oda-Sato E., Tobiume K., Kawauchi K., Taya Y., Okamoto K., Oren M., Tanaka N. Hedgehog signaling overrides p53-mediated tumor suppression by activating Mdm2. Proc. Natl. Acad. Sci. USA. 2008;105:4838–4843. doi: 10.1073/pnas.0712216105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn S., Joyner A.L. In vivo analysis of quiescent adult neural stem cells responding to Sonic hedgehog. Nature. 2005;437:894–897. doi: 10.1038/nature03994. [DOI] [PubMed] [Google Scholar]
- Aikin R.A., Ayers K.L., Thérond P.P. The role of kinases in the Hedgehog signalling pathway. EMBO Rep. 2008;9:330–336. doi: 10.1038/embor.2008.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argenti B., Gallo R., Di Marcotullio L., Ferretti E., Napolitano M., Canterini S., De Smaele E., Greco A., Fiorenza M.T., Maroder M., et al. Hedgehog antagonist REN(KCTD11) regulates proliferation and apoptosis of developing granule cell progenitors. J. Neurosci. 2005;25:8338–8346. doi: 10.1523/JNEUROSCI.2438-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S., Rand M.D., Lake R.J. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- Arvand A., Denny C.T. Biology of EWS/ETS fusions in Ewing's family tumors. Oncogene. 2001;20:5747–5754. doi: 10.1038/sj.onc.1204598. [DOI] [PubMed] [Google Scholar]
- Asaoka Y., Kanai F., Ichimura T., Tateishi K., Tanaka Y., Ohta M., Seto M., Tada M., Ijichi H., Ikenoue T., et al. Identification of a suppressive mechanism for hedgehog signaling through a novel interaction of Gli with 14-3-3. J. Biol. Chem. 2010;285:4185–4194. doi: 10.1074/jbc.M109.038232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aza-Blanc P., Ramírez-Weber F.A., Laget M.P., Schwartz C., Kornberg T.B. Proteolysis that is inhibited by hedgehog targets Cubitus interruptus protein to the nucleus and converts it to a repressor. Cell. 1997;89:1043–1053. doi: 10.1016/s0092-8674(00)80292-5. [DOI] [PubMed] [Google Scholar]
- Aza-Blanc P., Lin H.Y., Ruiz i Altaba A., Kornberg T.B. Expression of the vertebrate Gli proteins in Drosophila reveals a distribution of activator and repressor activities. Development. 2000;127:4293–4301. doi: 10.1242/dev.127.19.4293. [DOI] [PubMed] [Google Scholar]
- Bai C.B., Joyner A.L. Gli1 can rescue the in vivo function of Gli2. Development. 2001;128:5161–5172. doi: 10.1242/dev.128.24.5161. [DOI] [PubMed] [Google Scholar]
- Bai C.B., Auerbach W., Lee J.S., Stephen D., Joyner A.L. Gli2, but not Gli1, is required for initial Shh signaling and ectopic activation of the Shh pathway. Development. 2002;129:4753–4761. doi: 10.1242/dev.129.20.4753. [DOI] [PubMed] [Google Scholar]
- Bai C.B., Stephen D., Joyner A.L. All mouse ventral spinal cord patterning by hedgehog is Gli dependent and involves an activator function of Gli3. Dev. Cell. 2004;6:103–115. doi: 10.1016/s1534-5807(03)00394-0. [DOI] [PubMed] [Google Scholar]
- Bar E.E., Chaudhry A., Lin A., Fan X., Schreck K., Matsui W., Piccirillo S., Vescovi A.L., DiMeco F., Olivi A., et al. Cyclopamine-mediated hedgehog pathway inhibition depletes stem-like cancer cells in glioblastoma. Stem Cells. 2007;25:2524–2533. doi: 10.1634/stemcells.2007-0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman D.M., Karhadkar S.S., Maitra A., Montes De Oca R., Gerstenblith M.R., Briggs K., Parker A.R., Shimada Y., Eshleman J.R., Watkins D.N., et al. Widespread requirement for Hedgehog ligand stimulation in growth of digestive tract tumours. Nature. 2003;425:846–851. doi: 10.1038/nature01972. [DOI] [PubMed] [Google Scholar]
- Brewster R., Lee J., Ruiz i Altaba A. Gli/Zic factors pattern the neural plate by defining domains of cell differentiation. Nature. 1998;393:579–583. doi: 10.1038/31242. [DOI] [PubMed] [Google Scholar]
- Brewster R., Mullor J.L., Ruiz i Altaba A. Gli2 functions in FGF signaling during antero-posterior patterning. Development. 2000;127:4395–4405. doi: 10.1242/dev.127.20.4395. [DOI] [PubMed] [Google Scholar]
- Canettieri G., Di Marcotullio L., Greco A., Coni S., Antonucci L., Infante P., Pietrosanti L., De Smaele E., Ferretti E., Miele, et al. Histone deacetylase and Cullin3-RENKCTD11 ubiquitin ligase interplay regulates hedgehog signaling through Gli acetylation. Nat. Cell Biol. 2010;12:132–142. doi: 10.1038/ncb2013. [DOI] [PubMed] [Google Scholar]
- Chen M.H., Wilson C.W., Li Y.J., Law K.K., Lu C.S., Gacayan R., Zhang X., Hui C.C., Chuang P.T. Cilium-independent regulation of Gli protein function by Sufu in Hedgehog signaling is evolutionarily conserved. Genes Dev. 2009;23:1910–1928. doi: 10.1101/gad.1794109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S.Y., Bishop J.M. Suppressor of Fused represses Gli-mediated transcription by recruiting the SAP18-mSin3 corepressor complex. Proc. Natl. Acad. Sci. USA. 2002;99:5442–5447. doi: 10.1073/pnas.082096999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung H.O., Zhang X., Ribeiro A., Mo R., Makino S., Puviindran V., Law K.K., Briscoe J., Hui C.C. The kinesin protein Kif7 is a critical regulator of Gli transcription factors in mammalian hedgehog signaling. Sci. Signal. 2009;2 doi: 10.1126/scisignal.2000405. ra29. [DOI] [PubMed] [Google Scholar]
- Clement V., Sanchez P., de Tribolet N., Radovanovic I., Ruiz i Altaba A. HEDGEHOG-GLI1 signaling regulates human glioma growth, cancer stem cell self-renewal, and tumorigenicity. Curr. Biol. 2007;17:165–172. doi: 10.1016/j.cub.2006.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colaluca I.N., Tosoni D., Nuciforo P., Senic-Matuglia F., Galimberti V., Viale G., Pece S., Di Fiore PP. NUMB controls p53 tumour suppressor activity. Nature. 2008;451:76–80. doi: 10.1038/nature06412. [DOI] [PubMed] [Google Scholar]
- Dahmane N., Ruiz i Altaba A. Sonic hedgehog regulates the growth and patterning of the cerebellum. Development. 1999;126:3089–3100. doi: 10.1242/dev.126.14.3089. [DOI] [PubMed] [Google Scholar]
- Dahmane N., Lee J., Robins P., Heller P., Ruiz i Altaba A. Activation of the transcription factor Gli1 and the Sonic hedgehog signalling pathway in skin tumours. Nature. 1997;389:876–881. doi: 10.1038/39918. [DOI] [PubMed] [Google Scholar]
- Dahmane N., Sánchez P., Gitton Y., Palma V., Sun T., Beyna M., Weiner H., Ruiz i Altaba A. The Sonic Hedgehog-Gli pathway regulates dorsal brain growth and tumorigenesis. Development. 2001;128:5201–5212. doi: 10.1242/dev.128.24.5201. [DOI] [PubMed] [Google Scholar]
- Das S., Harris L.G., Metge B.J., Liu S., Riker A.I., Samant R.S., Shevde L.A. The hedgehog pathway transcription factor, GLI1 promotes malignant behavior of cancer cells by upregulating Osteopontin. J. Biol. Chem. 2009;284:22888–22897. doi: 10.1074/jbc.M109.021949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennler S., André J., Alexaki I., Li A., Magnaldo T., ten Dijke P., Wang X.J., Verrecchia F., Mauviel A. Induction of sonic hedgehog mediators by transforming growth factor-beta: Smad3-dependent activation of Gli2 and Gli1 expression in vitro and in vivo. Cancer Res. 2007;67:6981–6986. doi: 10.1158/0008-5472.CAN-07-0491. [DOI] [PubMed] [Google Scholar]
- Derynck R., Akhurst R.J., Balmain A. TGF-beta signaling in tumor suppression and cancer progression. Nat. Genet. 2001;29:117–129. doi: 10.1038/ng1001-117. [DOI] [PubMed] [Google Scholar]
- Di Marcotullio L., Ferretti E., De Smaele E., Argenti B., Mincione C., Zazzeroni F., Gallo R., Masuelli L., Napolitano M., Maroder M., et al. REN (KCTD11) is a suppressor of Hedgehog signaling and is deleted in human medulloblastoma. Proc. Natl. Acad. Sci. USA. 2004;101:10833–10838. doi: 10.1073/pnas.0400690101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Marcotullio L., Ferretti E., Greco A., De Smaele E., Po A., Sico M.A., Alimandi M., Giannini G., Maroder M., Screpanti I., et al. Numb is a suppressor of Hedgehog signalling and targets Gli1 for Itch-dependent ubiquitination. Nat. Cell Biol. 2006;8:1415–1423. doi: 10.1038/ncb1510. [DOI] [PubMed] [Google Scholar]
- Di Marcotullio L., Ferretti E., Greco A., De Smaele E., Screpanti I., Gulino A. Multiple ubiquitin-dependent processing pathways regulate hedgehog/gli signaling: implications for cell development and tumorigenesis. Cell Cycle. 2007;6:390–393. doi: 10.4161/cc.6.4.3809. [DOI] [PubMed] [Google Scholar]
- Dierks C., Beigi R., Guo G.R., Zirlik K., Stegert M.R., Manley P., Trussell C., Schmitt-Graeff A., Landwerlin K., Veelken H., et al. Expansion of Bcr-Abl-positive leukemic stem cells is dependent on Hedgehog pathway activation. Cancer Cell. 2008;14:238–249. doi: 10.1016/j.ccr.2008.08.003. [DOI] [PubMed] [Google Scholar]
- Ehtesham M., Sarangi A., Valadez J.G., Chanthaphaychith S., Becher M.W., Abel T.W., Thompson R.C., Cooper M.K. Ligand-dependent activation of the hedgehog pathway in glioma progenitor cells. Oncogene. 2007;26:5752–5761. doi: 10.1038/sj.onc.1210359. [DOI] [PubMed] [Google Scholar]
- Endoh-Yamagami S., Evangelista M., Wilson D., Wen X., Theunissen J.W., Phamluong K., Davis M., Scales S.J., Solloway M.J., de Sauvage F.J., et al. The mammalian Cos2 homolog Kif7 plays an essential role in modulating Hh signal transduction during development. Curr. Biol. 2009;9:1320–1326. doi: 10.1016/j.cub.2009.06.046. [DOI] [PubMed] [Google Scholar]
- Feldmann G., Dhara S., Fendrich V., Bedja D., Beaty R., Mullendore M., Karikari C., Alvarez H., Iacobuzio-Donahue C., Jimeno A., et al. Blockade of hedgehog signaling inhibits pancreatic cancer invasion and metastases: a new paradigm for combination therapy in solid cancers. Cancer Res. 2007;67:2187–2196. doi: 10.1158/0008-5472.CAN-06-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti E., De Smaele E., Miele E., Laneve P., Po A., Pelloni M., Paganelli A., Di Marcotullio L., Caffarelli E., Screpanti I., et al. Concerted microRNA control of Hedgehog signalling in cerebellar neuronal progenitor and tumour cells. EMBO J. 2008;27:2616–2627. doi: 10.1038/emboj.2008.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng L., Cuneo K.C., Cooper M.K., Wang H., Sekhar K., Fu A., Hallahan D.E. Hedgehog signaling in the murine melanoma microenvironment. Angiogenesis. 2007;10:259–267. doi: 10.1007/s10456-007-9078-9. [DOI] [PubMed] [Google Scholar]
- Gil-Perotin S., Marin-Husstege M., Li J., Soriano-Navarro M., Zindy F., Roussel M.F., Garcia-Verdugo J.M., Casaccia-Bonnefil P. Loss of p53 induces changes in the behavior of subventricular zone cells: implication for the genesis of glial tumors. J. Neurosci. 2006;26:1107–1116. doi: 10.1523/JNEUROSCI.3970-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich L.V., Milenković L., Higgins K.M., Scott M.P. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science. 1997;277:1109–1113. doi: 10.1126/science.277.5329.1109. [DOI] [PubMed] [Google Scholar]
- Gorlin R.J. Nevoid basal cell carcinoma syndrome. Dermatol. Clin. 1995;13:113–125. [PubMed] [Google Scholar]
- Grachtchouk M., Mo R., Yu S., Zhang X., Sasaki H., Hui C.C., Dlugosz A.A. Basal cell carcinomas in mice overexpressing Gli2 in skin. Nat. Genet. 2000;24:216–217. doi: 10.1038/73417. [DOI] [PubMed] [Google Scholar]
- Groszer M., Erickson R., Scripture-Adams D.D., Lesche R., Trumpp A., Zack J.A., Kornblum H.I., Liu X., Wu H. Negative regulation of neural stem/progenitor cell proliferation by the Pten tumor suppressor gene in vivo. Science. 2001;294:2186–2189. doi: 10.1126/science.1065518. [DOI] [PubMed] [Google Scholar]
- Hahn H., Wicking C., Zaphiropoulous P.G., Gailani M.R., Shanley S., Chidambaram A., Vorechovsky I., Holmberg E., Unden A.B., Gillies S., et al. Mutations of the human homolog of Drosophila patched in the nevoid basal cell carcinoma syndrome. Cell. 1996;85:841–851. doi: 10.1016/s0092-8674(00)81268-4. [DOI] [PubMed] [Google Scholar]
- Hahn H., Wojnowski L., Zimmer A.M., Hall J., Miller G., Zimmer A. Rhabdomyosarcomas and radiation hypersensitivity in a mouse model of Gorlin syndrome. Nat. Med. 1998;4:619–622. doi: 10.1038/nm0598-619. [DOI] [PubMed] [Google Scholar]
- Hamburger A.W., Salmon S.E. Primary bioassay of human tumor stem cells. Science. 1977;197:461–463. doi: 10.1126/science.560061. [DOI] [PubMed] [Google Scholar]
- Hingorani S.R., Wang L., Multani A.S., Combs C., Deramaudt T.B., Hruban R.H., Rustgi A.K., Chang S., Tuveson D.A. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. 2005;7:469–483. doi: 10.1016/j.ccr.2005.04.023. [DOI] [PubMed] [Google Scholar]
- Ho L., Stojanovski A., Whetstone H., Wei Q.X., Mau E., Wunder J.S., Alman B. Gli2 and p53 cooperate to regulate IGFBP-3-mediated chondrocyte apoptosis in the progression from benign to malignant cartilage tumors. Cancer Cell. 2009;16:126–136. doi: 10.1016/j.ccr.2009.05.013. [DOI] [PubMed] [Google Scholar]
- Huntzicker E.G, Estay I.S., Zhen H., Lokteva L.A., Jackson P.K., Oro A.E. Dual degradation signals control Gli protein stability and tumor formation. Genes Dev. 2006;20:276–281. doi: 10.1101/gad.1380906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchin M.E., Kariapper M.S., Grachtchouk M., Wang A., Wei L., Cummings D., Liu J., Michael L.E., Glick A., Dlugosz A.A. Sustained Hedgehog signaling is required for basal cell carcinoma proliferation and survival: conditional skin tumorigenesis recapitulates the hair growth cycle. Genes Dev. 2005;19:214–223. doi: 10.1101/gad.1258705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram W.J., McCue K.I., Tran T.H., Hallahan A.R., Wainwright B.J. Sonic Hedgehog regulates Hes1 through a novel mechanism that is independent of canonical Notch pathway signalling. Oncogene. 2008;27:1489–1500. doi: 10.1038/sj.onc.1210767. [DOI] [PubMed] [Google Scholar]
- Ji Z., Mei F.C., Xie J., Cheng X. Oncogenic KRAS activates hedgehog signaling pathway in pancreatic cancer cells. J. Biol. Chem. 2007;282:14048–14055. doi: 10.1074/jbc.M611089200. [DOI] [PubMed] [Google Scholar]
- Johnson R.L., Rothman A.L., Xie J., Goodrich L.V., Bare J.W., Bonifas J.M., Quinn A.G., Myers R.M., Cox D.R., Epstein E.H., et al. Human homolog of patched, a candidate gene for the basal cell nevus syndrome. Science. 1996;272:1668–1671. doi: 10.1126/science.272.5268.1668. [DOI] [PubMed] [Google Scholar]
- Joo J., Christensen L., Warner K., States L., Kang H.G., Vo K., Lawlor E.R., May W.A. GLI1 is a central mediator of EWS/FLI1 signaling in Ewing tumors. PLoS One. 2009;4:e7608. doi: 10.1371/journal.pone.0007608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju B., Spitsbergen J., Eden C.J., Taylor M.R., Chen W. Co-activation of hedgehog and AKT pathways promote tumorigenesis in zebrafish. Mol. Cancer. 2009;8:40. doi: 10.1186/1476-4598-8-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karhadkar S.S., Bova G.S., Abdallah N., Dhara S., Gardner D., Maitra A., Isaacs J.T., Berman D.M., Beachy P.A. Hedgehog signalling in prostate regeneration, neoplasia and metastasis. Nature. 2004;431:707–712. doi: 10.1038/nature02962. [DOI] [PubMed] [Google Scholar]
- Karin M., Cao Y., Greten F.R., Li Z.W. NF-KB in cancer: from innocent bystander to major culprit. Nat. Rev. Cancer. 2002;2:301–310. doi: 10.1038/nrc780. [DOI] [PubMed] [Google Scholar]
- Karlstrom R.O., Tyurina O.V., Kawakami A., Nishioka N., Talbot W.S., Sasaki H., Schier A.F. Genetic analysis of zebrafish gli1 and gli2 reveals divergent requirements for gli genes in vertebrate development. Development. 2003;130:1549–1564. doi: 10.1242/dev.00364. [DOI] [PubMed] [Google Scholar]
- Kasai K., Inaguma S., Yoneyama A., Yoshikawa K., Ikeda H. SCL/TAL1 interrupting locus derepresses GLI1 from the negative control of suppressor-of-fused in pancreatic cancer cell. Cancer Res. 2008;68:7723–7729. doi: 10.1158/0008-5472.CAN-07-6661. [DOI] [PubMed] [Google Scholar]
- Kasper M., Schnidar H., Neill G.W., Hanneder M., Klingler S., Blaas L., Schmid C., Hauser-Kronberger C., Regl G., Philpott M.P., et al. Selective modulation of Hedgehog/GLI target gene expression by epidermal growth factor signaling in human keratinocytes. Mol. Cell. Biol. 2006;26:6283–6298. doi: 10.1128/MCB.02317-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasperczyk H., Baumann B., Debatin K.M., Fulda S. Characterization of sonic hedgehog as a novel NF-kappaB target gene that promotes NF-kappaB-mediated apoptosis resistance and tumor growth in vivo. FASEB J. 2009;23:21–33. doi: 10.1096/fj.08-111096. [DOI] [PubMed] [Google Scholar]
- Kenney A.M., Rowitch D.H. Sonic hedgehog promotes G(1) cyclin expression and sustained cell cycle progression in mammalian neuronal precursors. Mol. Cell. Biol. 2000;20:9055–9067. doi: 10.1128/mcb.20.23.9055-9067.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney A.M., Cole M.D., Rowitch D.H. Nmyc upregulation by sonic hedgehog signaling promotes proliferation in developing cerebellar granule neuron precursors. Development. 2003;130:15–28. doi: 10.1242/dev.00182. [DOI] [PubMed] [Google Scholar]
- Kent D., Bush E.W., Hooper J.E. Roadkill attenuates Hedgehog responses through degradation of Cubitus interruptus. Development. 2006;133:2001–2010. doi: 10.1242/dev.02370. [DOI] [PubMed] [Google Scholar]
- Kinzler K.W., Bigner S.H., Bigner D.D., Trent J.M., Law M.L., O'Brien S.J., Wong A.J., Vogelstein B. Identification of an amplified, highly expressed gene in a human glioma. Science. 1987;236:70–73. doi: 10.1126/science.3563490. [DOI] [PubMed] [Google Scholar]
- Kise Y., Morinaka A., Teglund S., Miki H. Sufu recruits GSK3beta for efficient processing of Gli3. Biochem. Biophys. Res. Commun. 2009;387:569–574. doi: 10.1016/j.bbrc.2009.07.087. [DOI] [PubMed] [Google Scholar]
- Kogerman P., Grimm T., Kogerman L., Krause D., Undén A.B., Sandstedt B., Toftgård R., Zaphiropoulos P.G. Mammalian suppressor-of-fused modulates nuclear-cytoplasmic shuttling of Gli-1. Nat. Cell Biol. 1999;1:312–319. doi: 10.1038/13031. [DOI] [PubMed] [Google Scholar]
- Koyabu Y., Nakata K., Mizugishi K., Aruga J., Mikoshiba K. Physical and functional interactions between Zinc and Gli proteins. J. Biol. Chem. 2001;276:6889–6892. doi: 10.1074/jbc.C000773200. [DOI] [PubMed] [Google Scholar]
- Lai K., Kaspar B.K., Gage F.H., Schaffer D.V. Sonic hedgehog regulates adult neural progenitor proliferation in vitro and in vivo. Nat. Neurosci. 2003;6:21–27. doi: 10.1038/nn983. [DOI] [PubMed] [Google Scholar]
- Laner-Plamberger S., Kaser A., Paulischta M., Hauser-Kronberger C., Eichberger T., Frischauf A.M. Cooperation between GLI and JUN enhances transcription of JUN and selected GLI target genes. Oncogene. 2009;28:1639–1651. doi: 10.1038/onc.2009.10. [DOI] [PubMed] [Google Scholar]
- Lapidot T., Sirard C., Vormoor J., Murdoch B., Hoang T., Caceres-Cortes J., Minden M., Paterson B., Caligiuri M.A., Dick J.E. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367:645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- Lauth M., Bergström A., Shimokawa T., Toftgård R. Inhibition of GLI-mediated transcription and tumor cell growth by small-molecule antagonists. Proc. Natl. Acad. Sci. USA. 2007;104:8455–8460. doi: 10.1073/pnas.0609699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebel M., Mo R., Shimamura K., Hui C.C. Gli2 and Gli3 play distinct roles in the dorsoventral patterning of the mouse hindbrain. Dev. Biol. 2007;302:345–55. doi: 10.1016/j.ydbio.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Lee J., Platt K.A., Censullo P., Ruiz i Altaba A. Gli1 is a target of Sonic hedgehog that induces ventral neural tube development. Development. 1997;124:2537–2552. doi: 10.1242/dev.124.13.2537. [DOI] [PubMed] [Google Scholar]
- Leung C., Lingbeek M., Shakhova O., Liu J., Tanger E., Saremaslani P., Van Lohuizen M., Marino S. Bmi1 is essential for cerebellar development and is overexpressed in human medulloblastomas. Nature. 2004;428:337–341. doi: 10.1038/nature02385. [DOI] [PubMed] [Google Scholar]
- Li X., Deng W., Nail C.D., Bailey S.K., Kraus M.H., Ruppert J.M., Lobo-Ruppert S.M. Snail induction is an early response to Gli1 that determines the efficiency of epithelial transformation. Oncogene. 2006;25:609–621. doi: 10.1038/sj.onc.1209077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liem K.F., Jr., He M., Ocbina P.J., Anderson K.V. Mouse Kif7/Costal2 is a cilia-associated protein that regulates Sonic hedgehog signaling. Proc. Natl. Acad. Sci. USA. 2009;106:13377–13382. doi: 10.1073/pnas.0906944106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litingtung Y., Chiang C. Specification of ventral neuron types is mediated by an antagonistic interaction between Shh and Gli3. Nat. Neurosci. 2000;3:979–985. doi: 10.1038/79916. [DOI] [PubMed] [Google Scholar]
- Litingtung Y., Dahn R.D., Li Y., Fallon J.F., Chiang C. Shh and Gli3 are dispensable for limb skeleton formation but regulate digit number and identity. Nature. 2002;418:979–983. doi: 10.1038/nature01033. [DOI] [PubMed] [Google Scholar]
- Liu F., Massagué J., Ruiz i Altaba A. Carboxy-terminally truncated Gli3 proteins associate with Smads. Nat. Genet. 1998;20:325–326. doi: 10.1038/3793. [DOI] [PubMed] [Google Scholar]
- Liu S., Dontu G., Mantle I.D., Patel S., Ahn N.S., Jackson K.W., Suri P., Wicha M.S. Hedgehog signaling and Bmi-1 regulate self-renewal of normal and malignant human mammary stem cells. Cancer Res. 2006;66:6063–6071. doi: 10.1158/0008-5472.CAN-06-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machold R., Hayashi S., Rutlin M., Muzumdar M.D., Nery S., Corbin J.G., Gritli-Linde A., Dellovade T., Porter J.A., Rubin L.L., et al. Sonic hedgehog is required for progenitor cell maintenance in telencephalic stem cell niches. Neuron. 2003;39:937–950. doi: 10.1016/s0896-6273(03)00561-0. [DOI] [PubMed] [Google Scholar]
- Maeda O., Kondo M., Fujita T., Usami N., Fukui T., Shimokata K., Ando T., Goto H., Sekido Y. Enhancement of GLI1-transcriptional activity by beta-catenin in human cancer cells. Oncol. Rep. 2006;16:91–96. [PubMed] [Google Scholar]
- Maloverjan A., Piirsoo M., Michelson P., Kogerman P., Osterlund T. Identification of a novel serine/threonine kinase ULK3 as a positive regulator of Hedgehog pathway. Exp. Cell Res. 2010;316:627–637. doi: 10.1016/j.yexcr.2009.10.018. [DOI] [PubMed] [Google Scholar]
- Matise M.P., Epstein D.J., Park H.L., Platt K.A., Joyner A.L. Gli2 is required for induction of floor plate and adjacent cells, but not most ventral neurons in the mouse central nervous system. Development. 1998;125:2759–2770. doi: 10.1242/dev.125.15.2759. [DOI] [PubMed] [Google Scholar]
- Meletis K., Wirta V., Hede S.M., Nistér M., Lundeberg J., Frisén J. p53 suppresses the self-renewal of adult neural stem cells. Development. 2006;133:363–369. doi: 10.1242/dev.02208. [DOI] [PubMed] [Google Scholar]
- Mizugishi K., Aruga J., Nakata K., Mikoshiba K. Molecular properties of Zic proteins as transcriptional regulators and their relationship to GLI proteins. J. Biol. Chem. 2001;276:2180–2188. doi: 10.1074/jbc.M004430200. [DOI] [PubMed] [Google Scholar]
- Morton J.P., Mongeau M.E., Klimstra D.S., Morris J.P., Lee Y.C., Kawaguchi Y., Wright C.V., Hebrok M., Lewis B.C. Sonic hedgehog acts at multiple stages during pancreatic tumorigenesis. Proc. Natl. Acad. Sci. USA. 2007;104:5103–5108. doi: 10.1073/pnas.0701158104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullor J.L., Dahmane N., Sun T., Ruiz i Altaba A. Wnt signals are targets and mediators of Gli function. Curr. Biol. 2001;11:769–773. doi: 10.1016/s0960-9822(01)00229-9. [DOI] [PubMed] [Google Scholar]
- Nakashima H., Nakamura M., Yamaguchi H., Yamanaka N., Akiyoshi T., Koga K., Yamaguchi K., Tsuneyoshi M., Tanaka M., Katano M. Nuclear factor-kappaB contributes to hedgehog signaling pathway activation through sonic hedgehog induction in pancreatic cancer. Cancer Res. 2006;66:7041–7049. doi: 10.1158/0008-5472.CAN-05-4588. [DOI] [PubMed] [Google Scholar]
- Nguyen V., Chokas A.L., Stecca B., Ruiz i Altaba A. Cooperative requirement of the Gli proteins in neurogenesis. Development. 2005;132:3267–3279. doi: 10.1242/dev.01905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas M., Wolfer A., Raj K., Kummer J.A., Mill P., van Noort M., Hui C.C., Clevers H., Dotto G.P., Radtke F. Notch1 functions as a tumor suppressor in mouse skin. Nat. Genet. 2003;33:416–421. doi: 10.1038/ng1099. [DOI] [PubMed] [Google Scholar]
- Nilsson M., Unden A.B., Krause D., Malmqwist U., Raza K., Zaphiropoulos P.G., Toftgard R. Induction of basal cell carcinomas and trichoepitheliomas in mice overexpressing GLI-1. Proc. Natl. Acad. Sci. USA. 2000;97:3438–3443. doi: 10.1073/pnas.050467397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan-Stevaux O., Lau J., Truitt M.L., Chu G.C., Hebrok M., Fernández-Zapico M.E., Hanahan D. GLI1 is regulated through Smoothened-independent mechanisms in neoplastic pancreatic ducts and mediates PDAC cell survival and transformation. Genes Dev. 2009;23:24–36. doi: 10.1101/gad.1753809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northcott P.A., Fernandez-L A., Hagan J.P., Ellison D.W., Grajkowska W., Gillespie Y., Grundy R., Van Meter T., Rutka J.T., Croce C.M., et al. The miR-17/92 polycistron is up-regulated in sonic hedgehog-driven medulloblastomas and induced by N-myc in sonic hedgehog-treated cerebellar neural precursors. Cancer Res. 2009;69:3249–3255. doi: 10.1158/0008-5472.CAN-08-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta H., Aoyagi K., Fukaya M., Danjoh I., Ohta A., Isohata N., Saeki N., Taniguchi H., Sakamoto H., Shimoda T., et al. Cross talk between hedgehog and epithelial-mesenchymal transition pathways in gastric pit cells and in diffuse-type gastric cancers. Br. J. Cancer. 2009;100:389–398. doi: 10.1038/sj.bjc.6604846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olive K.P., Jacobetz M.A., Davidson C.J., Gopinathan A., McIntyre D., Honess D., Madhu B., Goldgraben M.A., Caldwell M.E., Allard D., et al. Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science. 2009;324:1457–1461. doi: 10.1126/science.1171362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oro A.E., Higgins K.M., Hu Z., Bonifas J.M., Epstein E.H., Jr., Scott M.P. Basal cell carcinomas in mice overexpressing sonic hedgehog. Science. 1997;276:817–821. doi: 10.1126/science.276.5313.817. [DOI] [PubMed] [Google Scholar]
- Palma V., Ruiz i Altaba A. Hedgehog-GLI signaling regulates the behavior of cells with stem cell properties in the developing neocortex. Development. 2004;131:337–345. doi: 10.1242/dev.00930. [DOI] [PubMed] [Google Scholar]
- Palma V., Lim D.A., Dahmane N., Sánchez P., Brionne T.C., Herzberg C.D., Gitton Y., Carleton A., Alvarez-Buylla A., Ruiz i Altaba A. Sonic hedgehog controls stem cell behavior in the postnatal and adult brain. Development. 2005;132:335–344. doi: 10.1242/dev.01567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y., Bai C.B., Joyner A.L., Wang B. Sonic hedgehog signaling regulates Gli2 transcriptional activity by suppressing its processing and degradation. Mol. Cell. Biol. 2006;26:3365–3377. doi: 10.1128/MCB.26.9.3365-3377.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H.L., Bai C., Platt K.A., Matise M.P., Beeghly A., Hui C.C., Nakashima M., Joyner A.L. Mouse Gli1 mutants are viable but have defects in SHH signaling in combination with a Gli2 mutation. Development. 2000;127:1593–1605. doi: 10.1242/dev.127.8.1593. [DOI] [PubMed] [Google Scholar]
- Pasca di Magliano M., Sekine S., Ermilov A., Ferris J., Dlugosz A.A., Hebrok M. Hedgehog/Ras interactions regulate early stages of pancreatic cancer. Genes Dev. 2006;20:3161–3173. doi: 10.1101/gad.1470806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock C.D., Wang Q., Gesell G.S., Corcoran-Schwartz I.M., Jones E., Kim J., Devereux W.L., Rhodes J.T., Huff C.A., Beachy P.A., et al. Hedgehog signaling maintains a tumor stem cell compartment in multiple myeloma. Proc. Natl. Acad. Sci. USA. 2007;104:4048–4053. doi: 10.1073/pnas.0611682104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pola R., Ling L.E., Silver M., Corbley M.J., Kearney M., Blake Pepinsky R., Shapiro R., Taylor F.R., Baker D.P., Asahara T., et al. The morphogen Sonic hedgehog is an indirect angiogenic agent upregulating two families of angiogenic growth factors. Nat. Med. 2001;7:706–711. doi: 10.1038/89083. [DOI] [PubMed] [Google Scholar]
- Pu Y., Huang L., Prins G.S. Sonic hedgehog-patched Gli signaling in the developing rat prostate gland: lobe-specific suppression by neonatal estrogens reduces ductal growth and branching. Dev. Biol. 2004;273:257–275. doi: 10.1016/j.ydbio.2004.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regl G., Kasper M., Schnidar H., Eichberger T., Neill G.W., Philpott M.P., Esterbauer H., Hauser-Kronberger C., Frischauf A.M., Aberger F. Activation of the BCL2 promoter in response to Hedgehog/GLI signal transduction is predominantly mediated by GLI2. Cancer Res. 2004;64:7724–7731. doi: 10.1158/0008-5472.CAN-04-1085. [DOI] [PubMed] [Google Scholar]
- Rhyu M.S., Jan L.Y., Jan Y.N. Asymmetric distribution of numb protein during division of the sensory organ precursor cell confers distinct fates to daughter cells. Cell. 1994;76:477–491. doi: 10.1016/0092-8674(94)90112-0. [DOI] [PubMed] [Google Scholar]
- Riobó N.A., Manning D.R. Pathways of signal transduction employed by vertebrate Hedgehogs. Biochem. J. 2007;40:369–379. doi: 10.1042/BJ20061723. [DOI] [PubMed] [Google Scholar]
- Riobó N.A., Lu K., Ai X., Haines G.M., Emerson C.P., Jr. Phosphoinositide 3-kinase and Akt are essential for Sonic Hedgehog signaling. Proc. Natl. Acad. Sci. USA. 2006a;103:4505–4510. doi: 10.1073/pnas.0504337103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riobó N.A., Haines G.M., Emerson C.P., Jr. Protein kinase C-delta and mitogen-activated protein/extracellular signal-regulated kinase-1 control GLI activation in hedgehog signaling. Cancer Res. 2006b;66:839–845. doi: 10.1158/0008-5472.CAN-05-2539. [DOI] [PubMed] [Google Scholar]
- Ruiz i Altaba A. Catching a Gli-mpse of Hedgehog. Cell. 1997;90:193–196. doi: 10.1016/s0092-8674(00)80325-6. [DOI] [PubMed] [Google Scholar]
- Ruiz i Altaba A. Combinatorial Gli gene function in floor plate and neuronal inductions by Sonic hedgehog. Development. 1998;125:2203–2212. doi: 10.1242/dev.125.12.2203. [DOI] [PubMed] [Google Scholar]
- Ruiz i Altaba A. Gli proteins encode context-dependent positive and negative functions: implications for development and disease. Development. 1999;126:3205–3216. doi: 10.1242/dev.126.14.3205. [DOI] [PubMed] [Google Scholar]
- Ruiz i Altaba A. Hedgehog-Gli Signaling in Human Disease. Austin, TX: Landes Bioscience, Springer; 2006. [Google Scholar]
- Ruiz i Altaba A. Therapeutic inhibition of Hedgehog-GLI signaling in cancer: epithelial, stromal, or stem cell targets? Cancer Cell. 2008;14:281–283. doi: 10.1016/j.ccr.2008.09.007. [DOI] [PubMed] [Google Scholar]
- Ruiz i Altaba A., Brand A.H. Entity versus property: tracking the nature, genesis and role of stem cells in cancer. Conference on stem cells and cancer. EMBO Rep. 2009;10:832–836. doi: 10.1038/embor.2009.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz i Altaba A., Sánchez P., Dahmane N. Gli and hedgehog in cancer: tumours, embryos and stem cells. Nat. Rev. Cancer. 2002;2:361–372. doi: 10.1038/nrc796. [DOI] [PubMed] [Google Scholar]
- Ruiz i Altaba A., Stecca B., Sánchez P. Hedgehog-Gli signaling in brain tumors: stem cells and paradevelopmental programs in cancer. Cancer Lett. 2004;204:145–157. doi: 10.1016/S0304-3835(03)00451-8. [DOI] [PubMed] [Google Scholar]
- Ruiz i Altaba A., Mas C., Stecca B. The Gli code: an information nexus regulating cell fate, stemness and cancer. Trends Cell Biol. 2007;17:438–447. doi: 10.1016/j.tcb.2007.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez P., Ruiz i Altaba A. In vivo inhibition of endogenous brain tumors through systemic interference of Hedgehog signaling in mice. Mech. Dev. 2005;122:223–230. doi: 10.1016/j.mod.2004.10.002. [DOI] [PubMed] [Google Scholar]
- Sanchez P., Hernández A.M., Stecca B., Kahler A.J., DeGueme A.M., Barrett A., Beyna M., Datta M.W., Datta S., Ruiz i Altaba A. Inhibition of prostate cancer proliferation by interference with SONIC HEDGEHOG-GLI1 signaling. Proc. Natl. Acad. Sci. USA. 2004;101:12561–12566. doi: 10.1073/pnas.0404956101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki H., Hui C., Nakafuku M., Kondoh H. A binding site for Gli proteins is essential for HNF-3beta floor plate enhancer activity in transgenics and can respond to Shh in vitro. Development. 1997;124:1313–1322. doi: 10.1242/dev.124.7.1313. [DOI] [PubMed] [Google Scholar]
- Sasaki H., Nishizaki Y., Hui C., Nakafuku M., Kondoh H. Regulation of Gli2 and Gli3 activities by an amino-terminal repression domain: implication of Gli2 and Gli3 as primary mediators of Shh signaling. Development. 1999;126:3915–3924. doi: 10.1242/dev.126.17.3915. [DOI] [PubMed] [Google Scholar]
- Schnidar H., Eberl M., Klingler S., Mangelberger D., Kasper M., Hauser-Kronberger C., Regl G., Kroismayr R., Moriggl R., Sibilia M., et al. Epidermal growth factor receptor signaling synergizes with Hedgehog/GLI in oncogenic transformation via activation of the MEK/ERK/JUN pathway. Cancer Res. 2009;69:1284–1292. doi: 10.1158/0008-5472.CAN-08-2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüller U., Heine V.M., Mao J., Kho A.T., Dillon A.K., Han Y.G., Huillard E., Sun T., Ligon A.H., Qian Y., et al. Acquisition of granule neuron precursor identity is a critical determinant of progenitor cell competence to form Shh-induced medulloblastoma. Cancer Cell. 2008;14:123–134. doi: 10.1016/j.ccr.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto M., Ohta M., Asaoka Y., Ikenoue T., Tada M., Miyabayashi K., Mohri D., Tanaka Y., Ijichi H., Tateishi K., et al. Regulation of the hedgehog signaling by the mitogen-activated protein kinase cascade in gastric cancer. Mol. Carcinog. 2009;48:703–712. doi: 10.1002/mc.20516. [DOI] [PubMed] [Google Scholar]
- Sheng T., Li C., Zhang X., Chi S., He N., Chen K., McCormick F., Gatalica Z., Xie J. Activation of the hedgehog pathway in advanced prostate cancer. Mol. Cancer. 2004;3:29. doi: 10.1186/1476-4598-3-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel P.M., Massagué J. Cytostatic and apoptotic actions of TGF-beta in homeostasis and cancer. Nat. Rev. Cancer. 2003;3:807–821. doi: 10.1038/nrc1208. [DOI] [PubMed] [Google Scholar]
- Stamataki D., Ulloa F., Tsoni S.V., Mynett A., Briscoe J. A gradient of Gli activity mediates graded Sonic Hedgehog signaling in the neural tube. Genes Dev. 2005;19:626–641. doi: 10.1101/gad.325905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecca B., Ruiz i Altaba A. A GLI1-p53 inhibitory loop controls neural stem cell and tumour cell numbers. EMBO J. 2009;28:663–676. doi: 10.1038/emboj.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecca B., Mas C., Clement V., Zbinden M., Correa R., Piguet V., Beermann F., Ruiz i Altaba A. Melanomas require HEDGEHOG-GLI signaling regulated by interactions between GLI1 and the RAS-MEK/AKT pathways. Proc. Natl. Acad. Sci. USA. 2007;104:5895–5900. doi: 10.1073/pnas.0700776104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svärd J., Heby-Henricson K., Persson-Lek M., Rozell B., Lauth M., Bergström A., Ericson J., Toftgård R., Teglund S. Genetic elimination of suppressor of fused reveals an essential repressor function in the mammalian Hedgehog signaling pathway. Dev. Cell. 2006;10:187–197. doi: 10.1016/j.devcel.2005.12.013. [DOI] [PubMed] [Google Scholar]
- Teh M.T., Wong S.T., Neill G.W., Ghali L.R., Philpott M.P., Quinn A.G. FOXM1 is a downstream target of Gli1 in basal cell carcinomas. Cancer Res. 2002;62:4773–4780. [PubMed] [Google Scholar]
- Teng D.H., Hu R., Lin H., Davis T., Iliev D., Frye C., Swedlund B., Hansen K.L., Vinson V.L., Gumpper K.L., et al. MMAC1/PTEN mutations in primary tumor specimens and tumor cell lines. Cancer Res. 1997;57:5221–5225. [PubMed] [Google Scholar]
- Thayer S.P., di Magliano M.P., Heiser P.W., Nielsen C.M., Roberts D.J., Lauwers G.Y., Qi P., Gysin S., Fernández-del Castillo C., Yajnik V., et al. Hedgehog is an early and late mediator of pancreatic cancer tumorigenesis. Nature. 2003;425:851–856. doi: 10.1038/nature02009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyurina O.V., Guner B., Popova E., Feng J., Schier A.F., Kohtz J.D., Karlstrom R.O. Zebrafish Gli3 functions as both an activator and a repressor in Hedgehog signaling. Dev. Biol. 2005;277:537–556. doi: 10.1016/j.ydbio.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Ulloa F., Itasaki N., Briscoe J. Inhibitory Gli3 activity negatively regulates Wnt/beta-catenin signaling. Curr. Biol. 2007;17:545–550. doi: 10.1016/j.cub.2007.01.062. [DOI] [PubMed] [Google Scholar]
- Uziel T., Karginov F.V., Xie S., Parker J.S., Wang Y.D., Gajjar A., He L., Ellison D., Gilbertson R.J., Hannon G., et al. The miR-17 ∼ 92 cluster collaborates with the Sonic Hedgehog pathway in medulloblastoma. Proc. Natl. Acad. Sci. USA. 2009;106:2812–2817. doi: 10.1073/pnas.0809579106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varjosalo M., Taipale J. Hedgehog: functions and mechanisms. Genes Dev. 2008;22:2454–2472. doi: 10.1101/gad.1693608. [DOI] [PubMed] [Google Scholar]
- Varjosalo M., Björklund M., Cheng F., Syvänen H., Kivioja T., Kilpinen S., Sun Z., Kallioniemi O., Stunnenberg H.G., He W.W., et al. Application of active and kinase-deficient kinome collection for identification of kinases regulating hedgehog signaling. Cell. 2008;133:537–548. doi: 10.1016/j.cell.2008.02.047. [DOI] [PubMed] [Google Scholar]
- Varnat F., Duquet A., Malerba M., Zbinden M., Mas C., Gervaz P., Ruiz i Altaba A. Human colon cancer epithelial cells harbour active HEDGEHOG-GLI signalling that is essential for tumour growth, recurrence, metastasis and stem cell survival and expansion. EMBO Mol. Med. 2009;1:338–351. doi: 10.1002/emmm.200900039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vokes S.A., Ji H., McCuine S., Tenzen T., Giles S., Zhong S., Longabaugh W.J., Davidson E.H., Wong W.H., McMahon A.P. Genomic characterization of Gli-activator targets in sonic hedgehog-mediated neural patterning. Development. 2007;134:1977–1989. doi: 10.1242/dev.001966. [DOI] [PubMed] [Google Scholar]
- Vokes S.A., Ji H., Wong W.H., McMahon A.P. A genome-scale analysis of the cis-regulatory circuitry underlying sonic hedgehog-mediated patterning of the mammalian limb. Genes Dev. 2008;22:2651–2663. doi: 10.1101/gad.1693008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Hoff D.D., LoRusso P.M., Rudin C.M., Reddy J.C., Yauch R.L., Tibes R., Weiss G.J., Borad M.J., Hann C.L., Brahmer J.R., et al. Inhibition of the hedgehog pathway in advanced basal-cell carcinoma. N. Engl. J. Med. 2009;361:1164–1172. doi: 10.1056/NEJMoa0905360. [DOI] [PubMed] [Google Scholar]
- Wakabayashi Y., Mao J.H., Brown K., Girardi M., Balmain A. Promotion of Hras-induced squamous carcinomas by a polymorphic variant of the Patched gene in FVB mice. Nature. 2007;445:761–765. doi: 10.1038/nature05489. [DOI] [PubMed] [Google Scholar]
- Wallace V.A. Purkinje-cell-derived Sonic hedgehog regulates granule neuron precursor cell proliferation in the developing mouse cerebellum. Curr. Biol. 1999;9:445–448. doi: 10.1016/s0960-9822(99)80195-x. [DOI] [PubMed] [Google Scholar]
- Wallingford J.B., Seufertm D.W., Virta V.C., Vize P.D. p53 activity is essential for normal development in Xenopus. Curr. Biol. 1997;7:747–757. doi: 10.1016/s0960-9822(06)00333-2. [DOI] [PubMed] [Google Scholar]
- Wang B., Fallon J.F., Beachy P.A. Hedgehog-regulated processing of Gli3 produces an anterior/posterior repressor gradient in the developing vertebrate limb. Cell. 2000;100:423–434. doi: 10.1016/s0092-8674(00)80678-9. [DOI] [PubMed] [Google Scholar]
- Watkins D.N., Berman D.M., Burkholder S.G., Wang B, Beachy P.A., Baylin S.B. Hedgehog signalling within airway epithelial progenitors and in small-cell lung cancer. Nature. 2003;422:313–317. doi: 10.1038/nature01493. [DOI] [PubMed] [Google Scholar]
- Wechsler-Reya R.J., Scott M.P. Control of neuronal precursor proliferation in the cerebellum by Sonic Hedgehog. Neuron. 1999;22:103–114. doi: 10.1016/s0896-6273(00)80682-0. [DOI] [PubMed] [Google Scholar]
- Wellbrock C., Karasarides M., Marais R. The RAF proteins take centre stage. Nat. Rev. Mol. Cell Biol. 2004;5:875–885. doi: 10.1038/nrm1498. [DOI] [PubMed] [Google Scholar]
- Yang Z.J., Ellis T., Markant S.L., Read T.A., Kessler J.D., Bourboulas M., Schüller U., Machold R., Fishell G., Rowitch D.H., et al. Medulloblastoma can be initiated by deletion of Patched in lineage-restricted progenitors or stem cells. Cancer Cell. 2008;14:135–145. doi: 10.1016/j.ccr.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yauch R.L., Gould S.E., Scales S.J., Tang T., Tian H., Ahn C.P., Marshall D., Fu L., Januario T., Kallop D., et al. A paracrine requirement for hedgehog signalling in cancer. Nature. 2008;455:406–410. doi: 10.1038/nature07275. [DOI] [PubMed] [Google Scholar]
- Yuan Z., Goetz J.A., Singh S., Ogden S.K., Petty W.J., Black C.C., Memoli V.A., Dmitrovsky E., Robbins D.J. Frequent requirement of hedgehog signaling in non-small cell lung carcinoma. Oncogene. 2007;26:1046–1055. doi: 10.1038/sj.onc.1209860. [DOI] [PubMed] [Google Scholar]
- Yue S., Chen Y., Cheng S.Y. Hedgehog signaling promotes the degradation of tumor suppressor Sufu through the ubiquitin-proteasome pathway. Oncogene. 2009;28:492–499. doi: 10.1038/onc.2008.403. [DOI] [PubMed] [Google Scholar]
- Zhang Q., Shi Q., Chen Y., Yue T., Li S., Wang B., Jiang J. Multiple Ser/Thr-rich degrons mediate the degradation of Ci/Gli by the Cul3-HIB/SPOP E3 ubiquitin ligase. Proc. Natl. Acad. Sci. USA. 2009;106:21191–21196. doi: 10.1073/pnas.0912008106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Zhang L., Wang B., Ou C.Y., Chien C.T., Jiang J. A hedgehog-induced BTB protein modulates hedgehog signaling by degrading Ci/Gli transcription factor. Dev. Cell. 2006;10:719–729. doi: 10.1016/j.devcel.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Zhao C., Chen A., Jamieson C.H., Fereshteh M., Abrahamsson A., Blum J., Kwon H.Y., Kim J., Chute J.P., Rizzieri D., et al. Hedgehog signalling is essential for maintenance of cancer stem cells in myeloid leukemia. Nature. 2009;458:776–779. doi: 10.1038/nature07737. [DOI] [PMC free article] [PubMed] [Google Scholar]