Abstract
Actinin-associated LIM protein (ALP) and Enigma are two subfamilies of Postsynaptic density 95, discs large and zonula occludens-1 (PDZ)–Lin-11, Isl1 and Mec-3 (LIM) domain containing proteins. ALP family members have one PDZ and one LIM domain, whereas Enigma proteins contain one PDZ and three LIM domains. Four ALP and three Enigma proteins have been identified in mammals, each having multiple splice variants and unique expression patterns. Functionally, these proteins bind through their PDZ domains to α-actinin and bind through their LIM domains or other internal protein interaction domains to other proteins, including signaling molecules. ALP and Enigma proteins have been implicated in cardiac and skeletal muscle structure, function and disease, neuronal function, bipolar disorder, tumor growth, platelet and epithelial cell motility and bone formation. This review will focus on recent advances in the biological roles of ALP/Enigma PDZ–LIM domain proteins in cardiac muscle and provide insights into mechanisms by which mutations in these proteins are related to human cardiac disease.
Keywords: PDZ, LIM, cypher, ZASP, cardiomyopathy, Z-disc
Introduction
Postsynaptic density 95, discs large and zonula occludens-1 (PDZ) and Lin-11, Isl1 and Mec-3 (LIM) domains, two common motifs that mediate protein–protein interactions, have been found to be of fundamental importance for multiple biological processes. The PDZ domain is a protein interaction motif consisting of 80–100 amino acid residues with a highly conserved four residue GLGF sequence within the domain (Ponting and Phillips, 1995). PDZ domains are often found to bind to short peptide motifs at the C-terminus of partner proteins, although recent data suggest other internal PDZ binding motifs (Jemth and Gianni, 2007). PDZ domain containing proteins are involved in a wide range of cellular processes, including maintenance of epithelial cell polarity and morphology, the organization of postsynaptic density and the regulation of membrane protein activity and trafficking (Harris and Lim, 2001; Long et al., 2003; Jani and Schock, 2007; Maday et al., 2008).
The LIM domain is a conserved zinc finger motif with the cysteine-rich consensus sequence CX2CX16–23HX2CX2CX2CX16–21CX2(C/H/D) (Gill, 1995). LIM domains have been identified in a variety of proteins. Despite data regarding the well-defined interaction of LIM-domain containing proteins and partner proteins, no consensus motifs for LIM domain binding have been identified as yet. Diverse biological roles of LIM domain containing proteins include regulation of actin structure and dynamics, neuronal path finding, integrin-dependent adhesion and signaling, cell-fate determination and tissue-specific gene expression (Kadrmas and Beckerle, 2004; Jani and Schock, 2007; Schaffar et al., 2008).
Since identification of the first PDZ–LIM domain protein, Enigma (Wu and Gill, 1994), 10 PDZ–LIM domain proteins have been identified in mammals. On the basis of similarities in protein structure, PDZ–LIM proteins have been divided into four subfamilies: Actinin-associated LIM protein (ALP) [ALP, CLP36, reversion-induced LIM (RIL) and Mystique], Enigma [Enigma, Enigma homologue protein (ENH) and Cypher/ZASP], LIM kinases (LMK1 and LMK2) and LMO7 (te Velthuis and Bagowski, 2007). Genes of the PDZ–LIM domain family have been reviewed recently (te Velthuis and Bagowski, 2007). In the current review, we will focus on structure and function of the ALP and Enigma subfamily proteins and their involvement in cardiac development, function and disease.
ALP subfamily proteins
Four proteins: ALP (PDLIM3), CLP36 (CLIM1, Elfin, PDLIM1), RIL (PDLIM4) and Mystique (SLIM, PDLIM2) have been classified into the ALP subfamily. Each member of the subfamily contains an N-terminal PDZ domain and a C-terminal LIM domain. α-Actinin-associated LIM protein (ALP) was first identified as a 39 kDa protein in rat skeletal muscle with a slightly shorter isoform in heart (Xia et al., 1997). ALP includes both an N-terminal PDZ domain, which interacts with the α-actinin-2, and a C-terminal LIM domain. Thus, ALP shares a high degree of homology with previously characterized CLP-36 (Wang et al., 1995) and RIL (Kiess et al., 1995) proteins, suggesting a novel protein family containing an N-terminal PDZ domain and one C-terminal LIM motif (Xia et al., 1997).
CLP36 (36 kDa C-terminal LIM domain protein) protein was first cloned from rat hepatocytes and contains a conserved C-terminal LIM domain. CLP36 shares 45.1% amino acid identity with RIL (Wang et al., 1995). Studies with the human ortholog of rat CLP36, hCLIM1 (Kotaka et al., 1999) and the mouse ortholog of rat CLP36, Elfin (Kotaka et al., 2001), revealed that CLP36/hCLIM1/Elfin also contain a PDZ domain at their N-termini. Further, studies revealed that CLP36 co-localizes with α-actinin-2 at the Z-lines in myocardium (Kotaka et al., 1999, 2000). In addition, CLP36 binds to α-actinin-1 and associates with F-actin filaments and stress fibers in human activated platelets and endothelial cells (Bauer et al., 2000). These data are consistent with previous findings in mouse epithelial cells, in which CLP36 was found to localize to actin stress fibers. This subcellular localization is dependent upon the CLP36 PDZ domain and its association with the non-muscle forms of α-actinin, α-actinin-1 and α-actinin-4 (Vallenius et al., 2000). These data suggest that CLP36 might be involved in not only the maintenance of sarcomeres in muscle cells, but also in actin stress fiber-mediated cellular processes, such as cell shape, migration, polarity and cytokinesis in non-muscle cells.
RIL protein, identified initially in rat fibroblasts (Kiess et al., 1995) and later in human lymphocytes (Szpirer et al., 1996; Bashirova et al., 1998), is the smallest protein in the ALP subfamily and is highly conserved in a wide range of species. RIL interacts with α-actinin (Torrado et al., 2004; Vallenius et al., 2004) via its PDZ domain and with PTP-BL, a submembranous protein tyrosine phosphatase, via its C-terminus (van den Berk et al., 2004). Moreover, RIL interacts with the AMPA glutamate receptor in dendritic spines through the C-terminal LIM domain (Schulz et al., 2004).
Mystique is the most recently identified member of the ALP protein family (Torrado et al., 2004; Loughran et al., 2005). Mystique also interacts with α-actinin (Torrado et al., 2004; Vallenius et al., 2004). The LIM domain of Mystique is necessary for the suppression of anchorage-independent growth (Loughran et al., 2005). Moreover, Mystique functions as an ubiquitin E3 ligase acting on STAT proteins to cause their proteosome-mediated degradation (Tanaka et al., 2005).
Alignment analysis of the ALP family of proteins revealed a novel ALP subfamily-specific 34-amino-acid motif termed ALP-like motif (AM), which contains a putative consensus protein kinase C (PKC) phosphorylation site and two α-helices (Te Velthuis et al., 2007). These data further support the notion that ALP family proteins may serve as signaling molecules and may be regulated by PKC-dependent signaling.
Enigma subfamily proteins
The Enigma subfamily is comprised of three members: Enigma, ENH and Cypher (mouse)/ZASP (human). These subfamily members contain a single PDZ domain at the N-terminus and three LIM domains at the C-terminus. Enigma was initially characterized in humans as a novel 457-amino-acid protein containing three LIM domains at the C-terminus, with one of the LIM domains interacting with the insulin receptor (Wu and Gill, 1994). The LIM domains of Enigma bind to the receptor tyrosine kinase Ret and the adaptor protein APS, and thus are implicated in signal transduction processes such as mitogenic activity, insulin related actin organization and glucose metabolism (Durick et al., 1998; Borrello et al., 2002; Barres et al., 2005, 2006).
ENH protein, the second Enigma subfamily member, was identified in rat brain (Kuroda et al., 1996). Enigma's three C-terminal LIM domains interact with PKC both in vivo and in vitro (Kuroda et al., 1996). A 596-amino-acid human ortholog of rat ENH protein was identified (Ueki et al., 1999). ENH interacts with α-actinin at the Z-line via its PDZ domain (Ueki et al., 1999; Nakagawa et al., 2000; Niederlander et al., 2004).
Cypher was cloned and identified from mice as a striated muscle restricted PDZ/LIM domain protein which colocalizes with α-actinin-2 at the Z-line (Zhou et al., 1999). The human ortholog of Cypher, ZASP was independently identified in heart and skeletal muscle (Faulkner et al., 1999). A third group isolated Cypher from mice and named the protein Oracle (Passier et al., 2000).
Alignment and experimental analysis of the ALP/ZASP family of proteins revealed a ZASP-like motif (ZM motif) in Cypher/ZASP, ALP and CLP36 (Klaavuniemi et al., 2004; Klaavuniemi and Ylanne, 2006; Te Velthuis et al., 2007). The ZM motif interacts with the spectrin repeats of α-actinin (Klaavuniemi et al., 2004; Klaavuniemi and Ylanne, 2006).
The ALP/Enigma subfamily proteins contain one PDZ domain and one or three LIM domains. Structural analysis of the PDZ domain of ALP/Enigma proteins demonstrated that it is a classical class I PDZ domain as previously suggested by its interaction with the C-terminal region of α-actinin-2 (Au et al., 2004). Recently, it has been shown that the PDZ domain of ALP/Enigma proteins also interacts with Myotilin and Calsarcin/FATZ C-terminal motifs, which have the characteristics of class III PDZ-binding domain sequences (von Nandelstadh et al., 2009a; Zheng et al., 2009). LIM domain(s) of ALP/Enigma subfamily proteins interact with different domains in various proteins, particularly in signaling factors. This common structural feature supports the notion that ALP/Enigma proteins serve as adaptor proteins, where the PDZ domain tethers the protein to the cytoskeleton and the LIM domain or additional internal domains, recruit signaling proteins to implement corresponding functions.
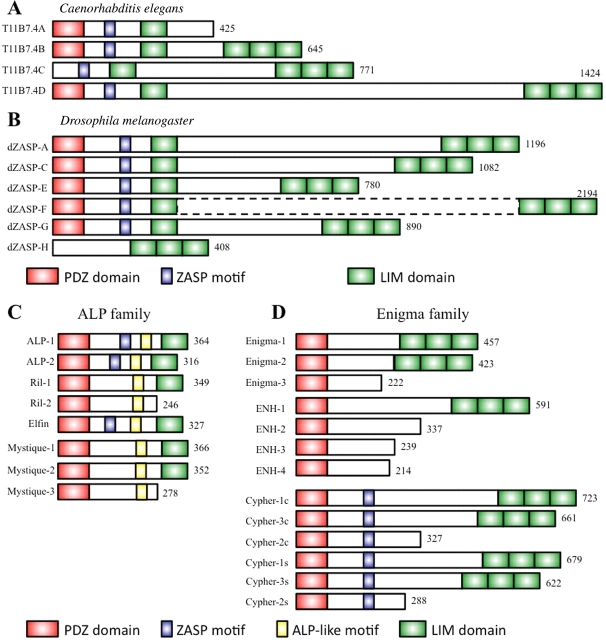
Intriguingly, Caenorhabditis elegans has a single gene orthologous to alp/enigma, termed alp-1, which encodes ALP and Enigma-like molecules (McKeown et al., 2006; Lecroisey et al., 2007) (Figure 1A). Similarly, Drosophila melanogaster has a single ortholog of alp/enigma (dzasp) which gives rise to ALP and Enigma-like molecules (Machuca-Tzili et al., 2006; Jani and Schock, 2007). Database analysis of Flybase (http://flybase.org) reveals six transcript isoforms for this factor (Figure 1B). However, recent expression studies of larval development have characterized 10 transcripts and an additional six, previously unidentified, exons (Benna et al., 2009).
Figure 1.
Characterization of ALP/Enigma protein family in invertebrates (A and B) and vertebrate (C and D). (A) The C. elegans T11B7.4 gene encodes four splice isoforms. T11B7.4a is similar to ALP family proteins in vertebrate containing one PDZ and one LIM domain. T11B7.4b and d are similar to Enigma family proteins in vertebrate containing one PDZ and four LIM domains. T11B7.4c does not have the PDZ domain. (B) The Drosophila homolog of human ZASP has six identified splice isoforms. (C) In vertebrate, there are four genes (ALP, CLP36, Mystique and RIL) which encode ALP family proteins, and multiple splice isoforms. (D) In vertebrate, there are three genes (Enigma, ENH and Cypher/ZASP) for Enigma subfamily with multiple splice isoforms. ZM is a conserved motif in Cypher/ZASP, ALP, CLP36 and Drosophila ZASP. ZM motif interacts with the rod-region of α-actinin-2. AM is a conserved motif in ALP subfamily. AM contains a potential PKC phosphorylation site.
Splice isoforms in ALP and Enigma families
Several studies have examined alternative splice isoforms and their functions in ALP and Enigma sub-families. Each member of the ALP-subfamily varies in the number of splice isoforms. Studies of ALP in mammals and chicken have isolated two isoforms detected in heart and smooth muscle cells with the molecular weight of 36 and 40 kDa, respectively, which result from alternative splicing (Pomies et al., 1999; Andersen et al., 2004; Klaavuniemi et al., 2004). Both of these transcripts contain the PDZ and LIM domain (Figure 1C).
RIL has two splice isoforms in human fetal brain (Bashirova et al., 1998). These two transcripts include one major transcript that contains seven exons. The minor transcript lacks the sixth exon and consequently its encoded protein lacks the LIM domain (Figure 1C).
Three alternative splice isoforms have been identified for Mystique in humans (Loughran et al., 2005). The largest of these three, Mystique1, contains a LIM domain at the C-terminus. Mystique2, although smaller, also contains the LIM domain. Mystique3, the smallest of the three isoforms, lacks the C-terminal LIM domain. Only one murine transcript has been identified for Mystique which encodes a protein sharing ∼77% homology with human Mystique2 (Loughran et al., 2005; Figure 1C).
In contrast to ALP, RIL and Mystique, only a single transcript of human and mouse CLP36 has been identified to date (Kotaka et al., 2000, 2001; Figure 1C).
Similar to the ALP subfamily, Enigma subfamily members have multiple splice variants. Early studies in rat led to the isolation of an Enigma transcript that was shown to be involved in bone formation (Boden et al., 1998). Analysis of mouse Enigma gene also revealed a single transcript (Pola et al., 2004). In contrast to these rodent species, three Enigma splice variants have been identified in humans. The hEnigma variant one encodes a 457-amino-acid protein containing one PDZ domain and three LIM domains. Similarly, the hEnigma variant two encodes a 423-amino-acid protein that contains one PDZ domain and three LIM domains. However, the third splice variant hEnigma variant three encodes a 222-amino-acid protein that contains a single PDZ domain, but lacks any LIM domain (Pola et al., 2004; Figure 1D).
For ENH, one isoform and three splice isoforms have been identified from rat and mouse, respectively (Nakagawa et al., 2000; Moser and White, 2006). Mouse ENH isoform 1 (mENH-1) encodes a full-length 591-amino-acid protein containing one PDZ and three LIM domains. Two smaller transcripts were termed mouse isoforms 2 and 3 (mENH-2 and mENH-3). mENH-2 and mENH-3 encode proteins of 337-amino-acid and 239-amino-acid, respectively, both lacking the three LIM domains. Analysis of human ENH transcripts revealed three transcripts (hENH-1, -2, -3) similar to those in mouse, and a fourth transcript hENH-4, encoding a protein of 214-amino-acid lacking the three LIM domains (Niederlander et al., 2004; Fig. 1D).
In mouse, six splice variants of Cypher have been characterized, which fall into two classes, one specific to cardiac and the other predominant in skeletal muscle (Huang et al., 2003). These isoforms include short (Cypher2c, 2s) and long (Cypher1c, 1s, 3c, 3s) subtypes within both cardiac and skeletal muscle. The mouse cypher gene contains 17 exons. Cardiac-specific isoforms contain amino acids encoded by exon 4 and skeletal muscle predominant isoforms contain amino acids encoded by exons 5–7. Exon 10 is the last exon of both Cypher2 isoforms and exon 12 is differentially spliced to generate Cypher1 (included) or Cypher3 (excluded) isoforms. An N-terminal PDZ domain is common in both short and long isoforms, whereas three LIM domains are unique to long isoforms (Fig. 1D). Four human splice variants of Cypher/ZASP have been identified, with one long and one short isoforms specific to cardiac or predominant in skeletal muscle (Faulkner et al., 1999; Vatta et al., 2003; also see NCBI database).
Functional roles of ALP subfamily proteins in heart
ALP subfamily proteins are found in heart. ALP is expressed at highest levels in skeletal and cardiac muscle (Xia et al., 1997). Ablation of ALP in mice leads to a form of selective right ventricle (RV) cardiomyopathy without showing obvious alterations in skeletal muscle (Jo et al., 2001; Pashmforoush et al., 2001). Moreover, ALP-deficiency leads to a decrease in trabeculation, irreversible chamber dilation and dysmorphogenesis of the embryonic RV, suggesting that ALP plays an essential role in the development of RV chamber (Pashmforoush et al., 2001). In vitro studies with intact myocardium reveal co-localization of ALP with both α-actinin and β-catenin at the intercalated disc (Pashmforoush et al., 2001). Furthermore, these studies found that ALP enhances the ability of α-actinin to crosslink actin filaments, thus suggesting ALP might serve as a genetic modifier of the response of embryonic ventricular muscle to biomechanical stress that accompanies exposure to normal workload in utero (Pashmforoush et al., 2001). ALP-deficient mice also exhibit altered regional RV function and abrogate hypertrophic remodeling in response to hypoxic stress (Lorenzen-Schmidt et al., 2005).
CLP36 is highly expressed in heart and is present in many other tissues including lung, liver, spleen and blood. CLP36 has been implicated in many processes including hypoxia and regulation of actin stress fibers (Wang et al., 1995; Kotaka et al., 1999; Bauer et al., 2000; Vallenius and Makela, 2002). In situ hybridization in mouse embryos reveals that CLP36 is expressed early in heart development and is present throughout the developing organ (Kotaka et al., 2000). However, the function of CLP36 in heart remains to be determined.
The other two ALP subfamily members, RIL and Mystique are expressed in a wide variety of tissues. RIL plays a role in tumor cell growth and neuronal signaling (Schulz et al., 2004; Iida et al., 2009), whereas Mystique may function in epithelial cell migration (Loughran et al., 2005). To date, there has been no evidence defining the function of these two ALP subfamily proteins in heart.
Functional roles of Enigma subfamily proteins in heart
The role of Cypher/ZASP in heart has been investigated in a diverse set of species, including human, mouse, zebrafish and fly (Faulkner et al., 1999; Zhou et al., 1999, 2001; Huang et al., 2003; Vatta et al., 2003; van der Meer et al., 2006; Jani and Schock, 2007; Arimura et al., 2009; Zheng et al., 2009; von Nandelstadh et al., 2009b). Ablation of Cypher in mice causes perinatal lethality within 1 week, accompanied by striated muscle failure and congenital cardiomyopathy (Zhou et al., 2001). Given the relatively normal sarcomerogenesis in non-contracting embryonic diaphragm muscle in Cypher-deficient mice, it was concluded that Cypher is required for maintenance of Z-line structure during muscle contraction, but not required for Z-line assembly. Deletion of Cypher in the postnatal heart causes severe dilated cardiomyopathy and premature death within 5 months, with gradually disrupted Z-lines, further supporting the hypothesis that Cypher is required for maintenance of Z-line structure (Zheng et al., 2009).
Targeted gene knock-down of zebrafish cypher using antisense morpholinos leads to deformation of somites, dilation of the pericardium and thinning of the cardiac ventricular wall (van der Meer et al., 2006). Deletion of dCypher/ZASP in Drosophila results in a lack of striations and Z-lines in larvae, suggesting that dCypher/ZASP is necessary for Z-line assembly and filament organization (Jani and Schock, 2007; Benna et al., 2009). However, dCypher/ZASP is the only ALP/Enigma family protein in Drosophila, in contrast to the multiple ALP/Enigma family proteins in mammalian species. Therefore, it is possible that the multiple ALP/Enigma family members in mammals may have redundant roles.
In heart, Cypher/ZASP plays a structural role, primarily mediated through its interaction with cytoskeletal Z-line proteins (Sheikh et al., 2007). In addition, there is increasing evidence that Cypher/ZASP also performs signaling functions. Studies in both human and mouse reveal that Cypher/ZASP interacts with and directs PKC to the Z-line, where PKC phosphorylates downstream targets, including Cypher. Mutations in Cypher alter its affinity for PKC (Zhou et al., 1999; Arimura et al., 2004). These data suggest that PKC via its association with Cypher is involved in cardiac function. Cypher also interacts with Calsarcin/FATZ/Myozenin, a Z-line associated protein (Frey and Olson, 2002), although specific domains essential for this interaction were not identified. We recently reported that Cypher interacts with the C-termini of Calsarcin-1 and Myotilin, another Z-line associated protein, through its PDZ domain (Zheng et al., 2009). In an independent study, von Nandelstadh et al. (2009a) also demonstrated that PDZ domains of Cypher/ZASP, ALP, CLP-36 and RIL interact with C-termini of Myotilin and Calsarcin. However, the functional role of these interactions in heart needs further investigation.
In contrast to the predominant expression and function of Cypher in striated muscle, Enigma and ENH are expressed in multiple tissues, such as skeletal muscle, heart, bone and brain (Ueki et al., 1999; Niederlander et al., 2004). Enigma is involved in distinct functions including bone metabolism (Liu et al., 2002). In addition, Enigma is found at the Z-line and I-band in skeletal muscle (Guy et al., 1999). Currently, a functional role for Enigma in skeletal muscle and heart has not been determined.
Positive associations have been made between ENH variants and schizophrenic patients (Kato, 2007). It has been reported that ENH interacts with PKC epsilon and CaV2.2, forming a PKC epsilon-ENH-Ca2+ channel macromolecular complex that specifically modulates N-type Ca2+ channels (Maeno-Hikichi et al., 2003; Chen et al., 2006). However, Stanley and coworkers recently observed that ENH did not co-immunoprecipitate with CaV2.2 in rat brain lysates, calling these initial data into question (Gardezi et al., 2009). It has also been shown that ENH1 interacts with protein kinase D1 (PKD1) via its LIM domains and forms a complex with PKD1 and the alpha1C subunit of cardiac L-type voltage-gated calcium channel in rat neonatal cardiomyocytes (Maturana et al., 2008). The authors also reported that silencing of ENH1 with RNAi in rat neonatal cardiomyocytes prevented the binding of PKD1 to alpha1C and inhibited the alpha-adrenergic-induced increase of L-type calcium currents (Maturana et al., 2008).
Human cardiomyopathy resulting from mutations in ALP/Enigma proteins
Mutations in genes for cytoskeletal proteins localized at Z-lines in cardiac and skeletal muscle have been linked to the development of cardiomyopathy and skeletal myopathy (Frank et al., 2006; Selcen and Carpen, 2008). All ALP/Enigma PDZ–LIM domain proteins have been observed to interact with α-actinin via their PDZ domain, thereby serving as components of sarcomeric complexes at the Z-line. Indeed, deficiencies of ALP and Cypher have been linked to cardiac dysfunction in animal models as described above. However, only mutations in Cypher/ZASP have been shown to cause different forms of cardiac myopathy such as hypertrophic and dilated cardiomyopathy, left ventricular non-compaction (Vatta et al., 2003; Arimura et al., 2004; Xing et al., 2006; Sheikh et al., 2007) and skeletal muscle myopathy in humans (Selcen and Engel, 2005).
In human studies, many identified Cypher/ZASP mutations localize to the internal region, named ZASP/Cypher-like motif (Vatta et al., 2003; Klaavuniemi and Ylanne, 2006). Although evidence shows that the internal ZASP/Cypher-like motif also interacts with the spectrin repeats of α-actinin in cardiac muscle, no alterations in localization or stability of α-actinin have been found in cardiac cells with Cypher/ZASP mutations (Zhou et al., 2001; Klaavuniemi and Ylanne, 2006). Thus, the molecular mechanism by which mutations in Cypher result in cardiomyopathy and skeletal myopathy remains to be determined. However, the D626N mutation in the third LIM domain has been found to change the affinity of Cypher/ZASP with PKC, suggesting that the signaling function of Cypher/ZASP may also contribute to the cardiomyopathy phenotype (Arimura et al., 2004).
It has been reported recently that Phosphoglucomutase 1 (PGM1), a metabolic enzyme involved in glycolysis and gluconeogenesis, interacts with Cypher/ZASP at domains encoded by exons 4 and 10 of the Cypher/ZASP gene (Arimura et al., 2009). These studies also showed that two Cypher/ZASP mutations in exon 4 (S189L and T206I) and one mutation in exon 10 (I345M) have reduced binding to PGM1. In addition, binding of endogenous PGM1 and Cypher/ZASP is found to be enhanced by stress in cultured rat neonatal cardiomyocytes (Arimura et al., 2009). These data suggest that Cypher/ZASP may anchor PGM1 to the Z-disc under stress conditions and that impaired binding of PGM1 to Cypher/ZASP may contribute to the pathogenesis of cardiomyopathy in patients with relevant Cypher/ZASP mutations.
Concluding remarks
ALP/Enigma subfamily proteins are defined by an N-terminal PDZ domain and one or three C-terminal LIM domains. ALP/Enigma subfamily proteins interact with α-actinin via their PDZ domains to regulate or organize cytoskeletal structure. ALP/Enigma members also bind to multiple signaling proteins, including protein kinases and protein phosphatases, through their LIM domains or other domains to mediate distinct biological functions.
Although all ALP/Enigma proteins are expressed in the heart, specific cardiac functions have been found only for ALP, ENH and Cypher/ZASP. In addition, with the exception of CLP36, each ALP/Enigma family protein has different splice isoforms. It will be important to investigate the potential role of these proteins and their splice isoforms in cardiac development, function and disease and to determine whether different members have overlapping/redundant roles in this context. Ablation of ALP or Cypher/ZASP in mice results in abnormal cardiac function. Furthermore, mutations of Cypher/ZASP have been linked with different forms of cardiomyopathy and skeletal myopathy (Sheikh et al., 2007). Molecular mechanisms by which Cypher/ZASP mutations lead to cardiomyopathy have not yet been fully elucidated. Studies with genetic animal models and other approaches may provide clues and help to develop therapeutic strategies for these diseases.
Funding
Work cited from the author's laboratory was supported by the National Institutes of Health, American Heart Association, Muscular Dystrophy Association and Children's Cardiomyopathy Foundation to J.C. Funding to M.Z. is provided by National Science Foundation of China (30770870 and 30971062) and National Key Basic Research Program of China (2007CB512100).
Conflict of interest: none declared.
Acknowledgements
The authors would like to thank Drs Angela Peter, Robert Lyon and Valeria Mezzano for their critical reading of the manuscript.
References
- Andersen O., Ostbye T.K., Gabestad I., Nielsen C., Bardal T., Galloway T.F. Molecular characterization of a PDZ–LIM protein in Atlantic salmon (Salmo salar): a fish ortholog of the alpha-actinin-associated LIM-protein (ALP) J. Muscle Res. Cell Motil. 2004;25:61–68. doi: 10.1023/b:jure.0000021363.07313.75. [DOI] [PubMed] [Google Scholar]
- Arimura T., Hayashi T., Terada H., Lee S.Y., Zhou Q., Takahashi M., Ueda K., Nouchi T., Hohda S., Shibutani M., et al. A Cypher/ZASP mutation associated with dilated cardiomyopathy alters the binding affinity to protein kinase C. J. Biol. Chem. 2004;279:6746–6752. doi: 10.1074/jbc.M311849200. [DOI] [PubMed] [Google Scholar]
- Arimura T., Inagaki N., Hayashi T., Shichi D., Sato A., Hinohara K., Vatta M., Towbin J.A., Chikamori T., Yamashina A., et al. Impaired binding of ZASP/Cypher with phosphoglucomutase 1 is associated with dilated cardiomyopathy. Cardiovasc. Res. 2009;83:80–88. doi: 10.1093/cvr/cvp119. [DOI] [PubMed] [Google Scholar]
- Au Y., Atkinson R.A., Guerrini R., Kelly G., Joseph C., Martin S.R., Muskett F.W., Pallavicini A., Faulkner G., Pastore A. Solution structure of ZASP PDZ domain; implications for sarcomere ultrastructure and enigma family redundancy. Structure. 2004;12:611–622. doi: 10.1016/j.str.2004.02.019. [DOI] [PubMed] [Google Scholar]
- Barres R., Gonzalez T., Le Marchand-Brustel Y., Tanti J.F. The interaction between the adaptor protein APS and Enigma is involved in actin organisation. Exp. Cell Res. 2005;308:334–344. doi: 10.1016/j.yexcr.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Barres R., Gremeaux T., Gual P., Gonzalez T., Gugenheim J., Tran A., Le Marchand-Brustel Y., Tanti J.F. Enigma interacts with adaptor protein with PH and SH2 domains to control insulin-induced actin cytoskeleton remodeling and glucose transporter 4 translocation. Mol. Endocrinol. 2006;20:2864–2875. doi: 10.1210/me.2005-0455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashirova A.A., Markelov M.L., Shlykova T.V., Levshenkova E.V., Alibaeva R.A., Frolova E.I. The human RIL gene: mapping to human chromosome 5q31.1, genomic organization and alternative transcripts. Gene. 1998;210:239–245. doi: 10.1016/s0378-1119(98)00080-8. [DOI] [PubMed] [Google Scholar]
- Bauer K., Kratzer M., Otte M., de Quintana K.L., Hagmann J., Arnold G.J., Eckerskorn C., Lottspeich F., Siess W. Human CLP36, a PDZ-domain and LIM-domain protein, binds to alpha-actinin-1 and associates with actin filaments and stress fibers in activated platelets and endothelial cells. Blood. 2000;96:4236–4245. [PubMed] [Google Scholar]
- Benna C., Peron S., Rizzo G., Faulkner G., Megighian A., Perini G., Tognon G., Valle G., Reggiani C., Costa R., et al. Post-transcriptional silencing of the Drosophila homolog of human ZASP: a molecular and functional analysis. Cell Tissue Res. 2009;337:463–476. doi: 10.1007/s00441-009-0813-y. [DOI] [PubMed] [Google Scholar]
- Boden S.D., Liu Y., Hair G.A., Helms J.A., Hu D., Racine M., Nanes M.S., Titus L. LMP-1, a LIM-domain protein, mediates BMP-6 effects on bone formation. Endocrinology. 1998;139:5125–5134. doi: 10.1210/endo.139.12.6392. [DOI] [PubMed] [Google Scholar]
- Borrello M.G., Mercalli E., Perego C., Degl'Innocenti D., Ghizzoni S., Arighi E., Eroini B., Rizzetti M.G., Pierotti M.A. Differential interaction of Enigma protein with the two RET isoforms. Biochem. Biophys. Res. Commun. 2002;296:515–522. doi: 10.1016/s0006-291x(02)00886-0. [DOI] [PubMed] [Google Scholar]
- Chen Y., Lai M., Maeno-Hikichi Y., Zhang J.F. Essential role of the LIM domain in the formation of the PKCepsilon-ENH-N-type Ca2+ channel complex. Cell Signal. 2006;18:215–224. doi: 10.1016/j.cellsig.2005.04.007. [DOI] [PubMed] [Google Scholar]
- Durick K., Gill G.N., Taylor S.S. Shc and Enigma are both required for mitogenic signaling by Ret/ptc2. Mol. Cell. Biol. 1998;18:2298–2308. doi: 10.1128/mcb.18.4.2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner G., Pallavicini A., Formentin E., Comelli A., Ievolella C., Trevisan S., Bortoletto G., Scannapieco P., Salamon M., Mouly V., et al. ZASP: a new Z-band alternatively spliced PDZ-motif protein. J. Cell Biol. 1999;146:465–475. doi: 10.1083/jcb.146.2.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank D., Kuhn C., Katus H.A., Frey N. The sarcomeric Z-disc: a nodal point in signalling and disease. J. Mol. Med. 2006;84:446–468. doi: 10.1007/s00109-005-0033-1. (Berlin, Germany) [DOI] [PubMed] [Google Scholar]
- Frey N., Olson E.N. Calsarcin-3, a novel skeletal muscle-specific member of the calsarcin family, interacts with multiple Z-disc proteins. J. Biol. Chem. 2002;277:13998–14004. doi: 10.1074/jbc.M200712200. [DOI] [PubMed] [Google Scholar]
- Gardezi S.R., Weber A.M., Li Q., Wong F.K., Stanley E.F. PDLIM5 is not a neuronal CaV2.2 adaptor protein. Nat. Neurosci. 2009;12:957–958. doi: 10.1038/nn0809-957a. author reply 958. [DOI] [PubMed] [Google Scholar]
- Gill G.N. The enigma of LIM domains. Structure. 1995;3:1285–1289. doi: 10.1016/s0969-2126(01)00265-9. [DOI] [PubMed] [Google Scholar]
- Guy P.M., Kenny D.A., Gill G.N. The PDZ domain of the LIM protein enigma binds to beta-tropomyosin. Mol. Biol. Cell. 1999;10:1973–1984. doi: 10.1091/mbc.10.6.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris B.Z., Lim W.A. Mechanism and role of PDZ domains in signaling complex assembly. J. Cell Sci. 2001;114:3219–3231. doi: 10.1242/jcs.114.18.3219. [DOI] [PubMed] [Google Scholar]
- Huang C., Zhou Q., Liang P., Hollander M.S., Sheikh F., Li X., Greaser M., Shelton G.D., Evans S., Chen J. Characterization and in vivo functional analysis of splice variants of cypher. J. Biol. Chem. 2003;278:7360–7365. doi: 10.1074/jbc.M211875200. [DOI] [PubMed] [Google Scholar]
- Iida Y., Matsuzaki T., Morishima T., Sasano H., Asai K., Sobue K., Takata K. Localization of reversion-induced LIM protein (RIL) in the rat central nervous system. Acta Histochem. Cytochem. 2009;42:9–14. doi: 10.1267/ahc.08038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jani K., Schock F. Zasp is required for the assembly of functional integrin adhesion sites. J. Cell Biol. 2007;179:1583–1597. doi: 10.1083/jcb.200707045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemth P., Gianni S. PDZ domains: folding and binding. Biochemistry. 2007;46:8701–8708. doi: 10.1021/bi7008618. [DOI] [PubMed] [Google Scholar]
- Jo K., Rutten B., Bunn R.C., Bredt D.S. Actinin-associated LIM protein-deficient mice maintain normal development and structure of skeletal muscle. Mol. Cell. Biol. 2001;21:1682–1687. doi: 10.1128/MCB.21.5.1682-1687.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadrmas J.L., Beckerle M.C. The LIM domain: from the cytoskeleton to the nucleus. Nat. Rev. Mol. Cell Biol. 2004;5:920–931. doi: 10.1038/nrm1499. [DOI] [PubMed] [Google Scholar]
- Kato T. Molecular genetics of bipolar disorder and depression. Psychiatry Clin. Neurosci. 2007;61:3–19. doi: 10.1111/j.1440-1819.2007.01604.x. [DOI] [PubMed] [Google Scholar]
- Kiess M., Scharm B., Aguzzi A., Hajnal A., Klemenz R., Schwarte-Waldhoff I., Schafer R. Expression of ril, a novel LIM domain gene, is down-regulated in Hras-transformed cells and restored in phenotypic revertants. Oncogene. 1995;10:61–68. [PubMed] [Google Scholar]
- Klaavuniemi T., Ylanne J. Zasp/Cypher internal ZM-motif containing fragments are sufficient to co-localize with alpha-actinin—analysis of patient mutations. Exp. Cell Res. 2006;312:1299–1311. doi: 10.1016/j.yexcr.2005.12.036. [DOI] [PubMed] [Google Scholar]
- Klaavuniemi T., Kelloniemi A., Ylanne J. The ZASP-like motif in actinin-associated LIM protein is required for interaction with the alpha-actinin rod and for targeting to the muscle Z-line. J. Biol. Chem. 2004;279:26402–26410. doi: 10.1074/jbc.M401871200. [DOI] [PubMed] [Google Scholar]
- Kotaka M., Ngai S.M., Carcia-Barcelo M., Tsui S.K.W., Fung K.P., Lee C.Y., Waye M.M.Y. Characterization of the human 36-kDa carboxyl terminal LIM domain protein (hCLIM1) J. Cell. Biochem. 1999;72:279–285. doi: 10.1002/(sici)1097-4644(19990201)72:2<279::aid-jcb12>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- Kotaka M., Kostin S., Ngai S., Chan K., Lau Y., Lee S.M., Li H., Ng E.K., Schaper J., Tsui S.K., et al. Interaction of hCLIM1, an enigma family protein, with alpha-actinin 2. J. Cell. Biochem. 2000;78:558–565. doi: 10.1002/1097-4644(20000915)78:4<558::aid-jcb5>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- Kotaka M., Lau Y.M., Cheung K.K., Lee S.M., Li H.Y., Chan W.Y., Fung K.P., Lee C.Y., Waye M.M., Tsui S.K. Elfin is expressed during early heart development. J. Cell. Biochem. 2001;83:463–472. doi: 10.1002/jcb.1244. [DOI] [PubMed] [Google Scholar]
- Kuroda S., Tokunaga C., Kiyohara Y., Higuchi O., Konishi H., Mizuno K., Gill G.N., Kikkawa U. Protein–protein interaction of zinc finger LIM domains with protein kinase C. J. Biol. Chem. 1996;271:31029–31032. doi: 10.1074/jbc.271.49.31029. [DOI] [PubMed] [Google Scholar]
- Lecroisey C., Segalat L., Gieseler K. The C. elegans dense body: anchoring and signaling structure of the muscle. J. Muscle Res. Cell Motil. 2007;28:79–87. doi: 10.1007/s10974-007-9104-y. [DOI] [PubMed] [Google Scholar]
- Liu Y., Hair G.A., Boden S.D., Viggeswarapu M., Titus L. Overexpressed LIM mineralization proteins do not require LIM domains to induce bone. J. Bone Miner. Res. 2002;17:406–414. doi: 10.1359/jbmr.2002.17.3.406. [DOI] [PubMed] [Google Scholar]
- Long J.F., Tochio H., Wang P., Fan J.S., Sala C., Niethammer M., Sheng M., Zhang M. Supramodular structure and synergistic target binding of the N-terminal tandem PDZ domains of PSD-95. J. Mol. Biol. 2003;327:203–214. doi: 10.1016/s0022-2836(03)00113-x. [DOI] [PubMed] [Google Scholar]
- Lorenzen-Schmidt I., McCulloch A.D., Omens J.H. Deficiency of actinin-associated LIM protein alters regional right ventricular function and hypertrophic remodeling. Ann. Biomed. Eng. 2005;33:888–896. doi: 10.1007/s10439-005-3604-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughran G., Healy N.C., Kiely P.A., Huigsloot M., Kedersha N.L., O'Connor R. Mystique is a new insulin-like growth factor-l-regulated PDZ–LIM domain protein that promotes cell attachment and migration and suppresses anchorage-independent growth. Mol. Biol. Cell. 2005;16:1811–1822. doi: 10.1091/mbc.E04-12-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machuca-Tzili L., Thorpe H., Robinson T.E., Sewry C., Brook J.D. Flies deficient in Muscleblind protein model features of myotonic dystrophy with altered splice forms of Z-band associated transcripts. Hum. Genet. 2006;120:487–499. doi: 10.1007/s00439-006-0228-8. [DOI] [PubMed] [Google Scholar]
- Maday S., Anderson E., Chang H.C., Shorter J., Satoh A., Sfakianos J., Folsch H., Anderson J.M., Walther Z., Mellman I. A PDZ-binding motif controls basolateral targeting of syndecan-1 along the biosynthetic pathway in polarized epithelial cells. Traffic. 2008;9:1915–1924. doi: 10.1111/j.1600-0854.2008.00805.x. (Copenhagen, Denmark) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeno-Hikichi Y., Chang S.H., Matsumura K., Lai M.Z., Lin H., Nakagawa N., Kuroda S., Zhang J.F. A PKC epsilon-ENH-channel complex specifically modulates N-type Ca2+ channels. Nat. Neurosci. 2003;6:468–475. doi: 10.1038/nn1041. [DOI] [PubMed] [Google Scholar]
- Maturana A.D., Walchli S., Iwata M., Ryser S., Van Lint J., Hoshijima M., Schlegel W., Ikeda Y., Tanizawa K., Kuroda S. Enigma homolog 1 scaffolds protein kinase D1 to regulate the activity of the cardiac L-type voltage-gated calcium channel. Cardiovasc. Res. 2008;78:458–465. doi: 10.1093/cvr/cvn052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeown C.R., Han H.F., Beckerle M.C. Molecular characterization of the Caenorhabditis elegans ALP/Enigma gene alp-1. Dev. Dyn. 2006;235:530–538. doi: 10.1002/dvdy.20633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser K., White F.M. Phosphoproteomic analysis of rat liver by high capacity IMAC and LC-MS/MS. J. Proteome Res. 2006;5:98–104. doi: 10.1021/pr0503073. [DOI] [PubMed] [Google Scholar]
- Nakagawa N., Hoshijima M., Oyasu M., Saito N., Tanizawa K., Kuroda S. ENH, containing PDZ and LIM domains, heart/skeletal muscle-specific protein, associates with cytoskeletal proteins through the PDZ domain. Biochem. Biophys. Res. Commun. 2000;272:505–512. doi: 10.1006/bbrc.2000.2787. [DOI] [PubMed] [Google Scholar]
- Niederlander N., Fayein N.A., Auffray C., Pomies P. Characterization of a new human isoform of the enigma homolog family specifically expressed in skeletal muscle. Biochem. Biophys. Res. Commun. 2004;325:1304–1311. doi: 10.1016/j.bbrc.2004.10.178. [DOI] [PubMed] [Google Scholar]
- Pashmforoush M., Pomies P., Peterson K.L., Kubalak S., Ross J., Jr., Hefti A., Aebi U., Beckerle M.C., Chien K.R. Adult mice deficient in actinin-associated LIM-domain protein reveal a developmental pathway for right ventricular cardiomyopathy. Nat. Med. 2001;7:591–597. doi: 10.1038/87920. [DOI] [PubMed] [Google Scholar]
- Passier R., Richardson J.A., Olson E.N. Oracle, a novel PDZ–LIM domain protein expressed in heart and skeletal muscle. Mech. Dev. 2000;92:277–284. doi: 10.1016/s0925-4773(99)00330-5. [DOI] [PubMed] [Google Scholar]
- Pola E., Gao W., Zhou Y., Pola R., Lattanzi W., Sfeir C., Gambotto A., Robbins P.D. Efficient bone formation by gene transfer of human LIM mineralization protein-3. Gene Ther. 2004;11:683–693. doi: 10.1038/sj.gt.3302207. [DOI] [PubMed] [Google Scholar]
- Pomies P., Macalma T., Beckerle M.C. Purification and characterization of an alpha-actinin-binding PDZ–LIM protein that is up-regulated during muscle differentiation. J. Biol. Chem. 1999;274:29242–29250. doi: 10.1074/jbc.274.41.29242. [DOI] [PubMed] [Google Scholar]
- Ponting C.P., Phillips C. DHR domains in syntrophins, neuronal NO synthases and other intracellular proteins. Trends Biochem. Sci. 1995;20:102–103. doi: 10.1016/s0968-0004(00)88973-2. [DOI] [PubMed] [Google Scholar]
- Schaffar G., Taniguchi J., Brodbeck T., Meyer A.H., Schmidt M., Yamashita T., Mueller B.K. LIM-only protein 4 interacts directly with the repulsive guidance molecule A receptor. Neogenin. J. Neurochem. 2008;107:418–431. doi: 10.1111/j.1471-4159.2008.05621.x. [DOI] [PubMed] [Google Scholar]
- Schulz T.W., Nakagawa T., Licznerski P., Pawlak V., Kolleker A., Rozov A., Kim J., Dittgen T., Kohr G., Sheng M., et al. Actin/alpha-actinin-dependent transport of AMPA receptors in dendritic spines: role of the PDZ–LIM protein RIL. J. Neurosci. 2004;24:8584–8594. doi: 10.1523/JNEUROSCI.2100-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selcen D., Carpen O. The Z-disk diseases. Adv. Exp. Med. Biol. 2008;642:116–130. doi: 10.1007/978-0-387-84847-1_10. [DOI] [PubMed] [Google Scholar]
- Selcen D., Engel A.G. Mutations in ZASP define a novel form of muscular dystrophy in humans. Ann. Neurol. 2005;57:269–276. doi: 10.1002/ana.20376. [DOI] [PubMed] [Google Scholar]
- Sheikh F., Bang M.L., Lange S., Chen J. “Z”eroing in on the role of Cypher in striated muscle function, signaling, and human disease. Trends Cardiovasc. Med. 2007;17:258–262. doi: 10.1016/j.tcm.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szpirer C., Szpirer J., Riviere M., Hajnal A., Kiess M., Scharm B., Schafer R. Chromosomal assignment of three rat and human H-rev genes, putative tumor suppressors, down-regulated in malignantly HRAS-transformed cells. Mamm. Genome. 1996;7:701–703. doi: 10.1007/s003359900211. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Soriano M.A., Grusby M.J. SLIM is a nuclear ubiquitin E3 ligase that negatively regulates STAT signaling. Immunity. 2005;22:729–736. doi: 10.1016/j.immuni.2005.04.008. [DOI] [PubMed] [Google Scholar]
- te Velthuis A.J., Bagowski C.P. PDZ and LIM domain-encoding genes: molecular interactions and their role in development. ScientificWorldJournal. 2007;7:1470–1492. doi: 10.1100/tsw.2007.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Te Velthuis A.J., Isogai T., Gerrits L., Bagowski C.P. Insights into the molecular evolution of the PDZ/LIM family and identification of a novel conserved protein motif. 2007;2:e189. doi: 10.1371/journal.pone.0000189. PLoS One. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrado M., Senatorov V.V., Trivedi R., Fariss R.N., Tomarev S.I. Pdlim2, a novel PDZ–LIM domain protein, interacts with alpha-actinins and filamin A. Invest. Ophthalmol. Vis. Sci. 2004;45:3955–3963. doi: 10.1167/iovs.04-0721. [DOI] [PubMed] [Google Scholar]
- Ueki N., Seki N., Yano K., Masuho Y., Saito T., Muramatsu M. Isolation, tissue expression, and chromosomal assignment of a human LIM protein gene, showing homology to rat Enigma homologue (ENH) J. Hum. Genet. 1999;44:256–260. doi: 10.1007/s100380050155. [DOI] [PubMed] [Google Scholar]
- Vallenius T., Makela T.P. Clik1: a novel kinase targeted to actin stress fibers by the CLP-36 PDZ–LIM protein. J. Cell Sci. 2002;115:2067–2073. doi: 10.1242/jcs.115.10.2067. [DOI] [PubMed] [Google Scholar]
- Vallenius T., Luukko K., Makela T.P. CLP-36 PDZ–LIM protein associates with nonmuscle alpha-actinin-1 and alpha-actinin-4. J. Biol. Chem. 2000;275:11100–11105. doi: 10.1074/jbc.275.15.11100. [DOI] [PubMed] [Google Scholar]
- Vallenius T., Scharm B., Vesikansa A., Luukko K., Schafer R., Makela T.P. The PDZ–LIM protein RIL modulates actin stress fiber turnover and enhances the association of alpha-actinin with F-actin. Exp. Cell Res. 2004;293:117–128. doi: 10.1016/j.yexcr.2003.09.004. [DOI] [PubMed] [Google Scholar]
- van den Berk L.C., van Ham M.A., te Lindert M.M., Walma T., Aelen J., Vuister G.W., Hendriks W.J. The interaction of PTP-BL PDZ domains with RIL: an enigmatic role for the RIL LIM domain. Mol. Biol. Rep. 2004;31:203–215. doi: 10.1007/s11033-005-1407-8. [DOI] [PubMed] [Google Scholar]
- van der Meer D.L., Marques I.J., Leito J.T., Besser J., Bakkers J., Schoonheere E., Bagowski C.P. Zebrafish cypher is important for somite formation and heart development. Dev. Biol. 2006;299:356–372. doi: 10.1016/j.ydbio.2006.07.032. [DOI] [PubMed] [Google Scholar]
- Vatta M., Mohapatra B., Jimenez S., Sanchez X., Faulkner G., Perles Z., Sinagra G., Lin J.H., Vu T.M., Zhou Q., et al. Mutations in Cypher/ZASP in patients with dilated cardiomyopathy and left ventricular non-compaction. J. Am. Coll. Cardiol. 2003;42:2014–2027. doi: 10.1016/j.jacc.2003.10.021. [DOI] [PubMed] [Google Scholar]
- von Nandelstadh P., Ismail M., Gardin C., Suila H., Zara I., Belgrano A., Valle G., Carpen O., Faulkner G. A class III PDZ binding motif in the myotilin and FATZ families binds Enigma family proteins: a common link for Z-disc myopathies. Mol. Cell. Biol. 2009a;29:822–834. doi: 10.1128/MCB.01454-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Nandelstadh P., Ismail M., Gardin C., Suila H., Zara I., Belgrano A., Valle G., Carpen O., Faulkner G. A class III PDZ binding motif in the myotilin and FATZ families binds enigma family proteins: a common link for Z-disc myopathies. Mol. Cell. Biol. 2009b;29:822–834. doi: 10.1128/MCB.01454-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Harrison-Shostak D.C., Lemasters J.J., Herman B. Cloning of a rat cDNA encoding a novel LIM domain protein with high homology to rat RIL. Gene. 1995;165:267–271. doi: 10.1016/0378-1119(95)00542-e. [DOI] [PubMed] [Google Scholar]
- Wu R.Y., Gill G.N. LIM domain recognition of a tyrosine-containing tight turn. J. Biol. Chem. 1994;269:25085–25090. [PubMed] [Google Scholar]
- Xia H., Winokur S.T., Kuo W.L., Altherr M.R., Bredt D.S. Actinin-associated LIM protein: identification of a domain interaction between PDZ and spectrin-like repeat motifs. J. Cell Biol. 1997;139:507–515. doi: 10.1083/jcb.139.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y., Ichida F., Matsuoka T., Isobe T., Ikemoto Y., Higaki T., Tsuji T., Haneda N., Kuwabara A., Chen R., et al. Genetic analysis in patients with left ventricular noncompaction and evidence for genetic heterogeneity. Mol. Genet. Metab. 2006;88:71–77. doi: 10.1016/j.ymgme.2005.11.009. [DOI] [PubMed] [Google Scholar]
- Zheng M., Cheng H., Li X., Zhang J., Cui L., Ouyang K., Han L., Zhao T., Gu Y., Dalton N.D., et al. Cardiac-specific ablation of Cypher leads to a severe form of dilated cardiomyopathy with premature death. Hum. Mol. Genet. 2009;18:701–713. doi: 10.1093/hmg/ddn400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Chu P.H., Huang C., Cheng C.F., Martone M.E., Knoll G., Shelton G.D., Evans S., Chen J. Ablation of cypher, a PDZ–LIM domain Z-line protein, causes a severe form of congenital myopathy. J. Cell Biol. 2001;155:605–612. doi: 10.1083/jcb.200107092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Ruiz-Lozano P., Martone M.E., Chen J. Cypher, a striated muscle-restricted PDZ and LIM domain-containing protein, binds to alpha-actinin-2 and protein kinase C. J. Biol. Chem. 1999;274:19807–19813. doi: 10.1074/jbc.274.28.19807. [DOI] [PubMed] [Google Scholar]