Abstract
The HA of influenza virus is a paradigm for a transmembrane protein thought to be associated with membrane-rafts, liquid-ordered like nanodomains of the plasma membrane enriched in cholesterol, glycosphingolipids, and saturated phospholipids. Due to their submicron size in cells, rafts can not be visualized directly and raft-association of HA was hitherto analyzed by indirect methods. In this study, we have used GUVs and GPMVs, showing liquid disordered and liquid ordered domains, to directly visualize partition of HA by fluorescence microscopy. We show that HA is exclusively (GUVs) or predominantly (GPMVs) present in the liquid disordered domain, regardless of whether authentic HA or domains containing its raft targeting signals were reconstituted into model membranes. The preferential partition of HA into ld domains and the difference between lo partition in GUV and GPMV are discussed with respect to differences in packaging of lipids in membranes of model systems and living cells suggesting that physical properties of lipid domains in biological membranes are tightly regulated by protein-lipid interactions.
Abbreviations used: CFP, cyan fluorescent protein; CT, cytoplasmic tail; DRM, detergent resistant membrane; FLIM, fluorescence lifetime imaging microscopy; FRET, Förster resonance energy transfer; GPI-CFP, glycosylphosphatidylinositol fused to CFP; GPMVs, giant plasma membrane vesicles; GUVs, giant unilamellar vesicles; HA, hemagglutinin; ld, liquid-disordered; lo, liquid-ordered; PBS, phosphate buffered saline; TGN, trans-Golgi network; TMD, transmembrane domain; YFP, yellow fluorescent protein
Introduction
The assembly site of influenza virus is the apical plasma membrane of the host cell. Three viral transmembrane proteins, the HA, the neuraminidase, and the proton channel M2, as well as the matrix protein M1 and the eight viral RNA-protein complexes have to be recruited to the assembly site. Lipid domains of the plasma membrane have been suggested to play an important role for local enrichment of viral components. HA that mediates binding of the virus to the host cell and fusion with the target membrane (1) has been found to be enriched in DRM fractions (2–4). Typical lipid components of those fractions are saturated phospholipids, glycosphingolipids and cholesterol that are known to form lo domains in model systems (5). This has led to the hypothesis that lipid rafts (3,6,7), which resemble lo domains and are essentially composed of those lipids, might function as assembly site. This was supported by the observation that the lipid composition of the influenza virus envelope is more similar to that of a raft than of the overall plasma membrane (3,8). Although it has been shown that DRM fractions may not resemble the native state of lipid rafts (9,10), independent approaches to assess the lateral organization of HA supported a role of raft-like domains in virus maturation. Fluorescent and electron microscopy studies on fixed cells revealed a nonrandom distribution of HA, with clustering of HA in the plasma membrane of mammalian cells at length scales between 20 and 900 nm being also sensitive to cholesterol (4,11). In the plasma membrane of living cells, we found that the lateral distribution of fluorescent protein tagged HA is cholesterol dependent (12,13). For both full length HA (12) as well as a HA fragment containing essentially the transmembrane and cytoplasmic domain of HA (13) we found colocalization with well established raft markers such as GPI-CFP that disappeared on removal of cholesterol. Myristoylated/palmitoylated or double palmitoylated GPI-anchored proteins, here fused to CFP, are thought partition into rafts due to the higher degree of order in their saturated acyl chains anchoring them to the membrane (14,15). This is in line with a recent study, where fluorescence photoactivation localization microscopy of HA tagged with a photoactivatable green fluorescent protein showed cholesterol sensitive clusters of HA with a size ranging from 40 nm to several micrometers (16). However, because only clusters at the nanometer length scale, i.e., with the size of rafts (16), could be disintegrated by extraction of cholesterol the latter studies concluded clustering of HA was not exclusively dependent on cholesterol, but other mechanisms, e.g., association with the cytoskeleton, may be involved in lateral HA organization. Very recently, a proton NMR study on influenza virus and host cell membranes concluded that at physiological temperatures a major fraction of envelope lipids has ld properties and protein clustering during virus assembly cannot be explained only by lipid phase behavior (17).
To address these different conclusions we aimed at visualization of lipid domain partition of HA. However, lipid domains in the plasma membrane of living cells are typically of submicroscopic size and it is difficult to study the role of raft domains in local HA enrichment directly by light microscopy. Therefore, we have investigated the lateral organization of HA and HA fragments containing the membrane-targeting signals in various model systems forming lipid domains in the μm range. These domains can be detected easily by appropriate fluorescent lipid analogs that enrich preferentially either in the ld or in the lo domain. In one approach, fluorescent labeled full length HA and a fluorescent synthetic peptide corresponding to the HA transmembrane domain were reconstituted in GUVs of various lipid mixtures forming lipid domains. In a second approach, GPMVs, so called blebs (18), were generated from the plasma membrane of cells expressing full length HA or constructs of HA tagged with variants of the green fluorescent proteins. GPMVs have been shown to form lipid domains at low temperature (18). In all cases we found that HA and respective constructs were preferentially localized in the liquid-disordered domain. However, although HA was excluded entirely from the lo domains in GUVs of synthetic lipids, the results on GPMVs indicated that lo domains in biological membranes have properties that allow integration of HA. We presume lipid-lipid interaction to be an essential determinant for domain formation in model systems, whereas in the plasma membrane, and hence GPMVs, due to the high protein content protein-lipid interactions strongly modulate the physical properties of lipid domains at physiological temperatures.
Materials and Methods
Cloning of fluorescent proteins
HA-Cerulean
The HA gene from influenza virus strain A/FPV/Rostock/34 (H7N1) was inserted into the plasmid pECerulean (Clontech, Saint-Germain-en-Laye, France) (19) such that the resulting construct contains the amino acid sequence LRPEAPRARDPPVAT between the end of HA and the start of Cerulean. An uncleaved variant of HA containing glycine instead of arginine at position 339 at the cleavage site of HA1 (20) named HA-Cer was expressed. For more details see Engel et al. (12).
TMD-HA-YFP
The construct has been described by Scolari et al. (13). The sequence consisting of the HA TMD and CT as well as of 38 amino acid of the HA ectodomain proximal to the N-terminus of the TMD was amplified by polymerase chain reaction from the cDNA encoding the HA gene from influenza virus strain A/FPV/Rostock/34 (H7N1) (21). The sequence contains a glycosylation site. For more details see Scolari et al. (13).
Lipid domain marker
GPI-CFP has been provided by Keller et al. (22). Lyn-YFP consists of a myristoylated and palmitoylated peptide MGCIKSKRKDNLNDDE from the lyn-kinase fused to the N-terminus of the yellow-fluorescent protein.
All constructs contain the A206K mutation in the fluorescent proteins. This prevents their dimerization (23). Mutations were introduced following the manufacturer's protocol of the Quikchange site-directed mutagenesis kit (Stratagene, La Jolla, CA). For more details see Scolari et al. (13).
Transfection and treatment of cells
CHO-K1 cells (American Type Culture Collection) were maintained in DMEM with 10% FBS (Invitrogen, Karlsruhe, Germany) and 1% penicillin-streptomycin at 37°C and 5% CO2. Cells were grown to ∼80% confluence on 35-mm culture dishes with glass bottoms (MatTek, Ashland, MA), transfected using Lipofectamine 2000 (Invitrogen, Karlsruhe, Germany) following the manufacturer's protocol 24 h before carrying out the experiments. Metabolic labeling, immunoprecipitation with 2.5 μL anti-green fluorescent protein antibodies (Invitrogen), SDS-PAGE, and fluorography were carried out as described in Veit et al. (21). Endoglycosidase-H and PNGase-F digestions of immunoprecipitated samples were carried out as described by the manufacturer (New England BioLabs, Frankfurt am Main, Germany).
Preparation of cells and giant plasma membrane vesicles
GPMVs were prepared according to Sengupta et al. (14) and Baumgart et al. (18) from confluent CHO-K1 cells 24 h after transfection with HA-Cer, TMD-HA-YFP, or GPI-CFP. Cells grown in flasks were washed twice with GPMV buffer (2 mM CaCl2, 10 mM Hepes, 0.15 M NaCl, pH 7.4). GPMV buffer (1.5 mL) containing 25 mM formaldehyde and 2 mM dithiothreitol were added and flasks were incubated for 1 h at 37°C shaking slowly (60–80 cycles/min). After incubation, detached GPMVs were decanted gently into a conical glass tube. GPMVs were allowed to sediment for 30 min at 4°C. For microscopy, 30 μL of GPMVs were labeled with a 20-μM R18 (Octadecylrhodamine-B-chloride; Invitrogen) solution. Images of the equatorial plane of the GPMVs were taken at 10°C.
Peptide synthesis
Rh-TMD, containing 28 amino acid residues of the TMD of HA (strain Japan/305/57, H2; Rh-TMD: Rh-βA-ILAIYATVAGSLSLAIMMAGISFWMCSNKKK) was synthesized as described recently by Nikolaus et al. (24).
Preparation of GUVs
Phospholipids and 1-palmitoyl-2-[6-[(7-nitro-2-1,3-benzoxadiazol-4-yl)amino]hexanoyl]-sn-glycero-3-phosphocholine (C6-NBD-PC) were obtained from Avanti Polar Lipids (Birmingham, AL), cholesterol from Sigma-Aldrich (Taufkirchen, Germany). Lipids were used without further purification. Viral lipids were isolated from influenza virus X31 or A/PR/8/34 according to (25).
GUVs were prepared from lipid films dried on indium tin oxide-coated glass slides (Präzisions Glas and Optik GmbH, Iserlohn, Germany) by electroswelling according to Angelova et al. (26). Rh-TMD was incorporated as described by Nikolaus et al. (24).
Labeling and isolation of HA
Influenza A virus strain X31 was grown and purified as described in Krumbiegel et al. (27). Virus protein (20 mg) was incubated for 2 h at RT (25°C) in the dark with TAMRA (5/6-Carboxytetramethylrhodamine; emp Biotech, Berlin, Germany), added from a 5 mM DMSO stock to molar excess of 10 over HA. Uncoupled TAMRA was removed by washing labeled virus three times in PBS (pH 7.4) and harvested by centrifugation at 52,000 × g. The pellet was solubilized by adding 500 μL PBS, pH 7.4, containing 28% (w/v) octylglycoside (Alexis, Lausen, Switzerland) and gently shaken on ice in the dark for 1 h (28). After centrifugation for 1 h (100,000 × g, 4°C), the HA containing supernatant was purified by affinity chromatography on ricin A agarose (Ricinus communis lectin; Sigma-Aldrich). To remove detergent and galactose, 1 vol sample was dialyzed against 500 vol PBS, pH 7.4. The purity of rhodamine-labeled HA (Rh-HA) was verified by SDS-PAGE, fluorography, and Western blot.
Reconstitution of HA in GUVs
Two different protocols for reconstitution were used.
Protocol 1
Proteoliposomes were prepared according to Papadopulos et al. (29). Triton X-100 solubilized labeled HA was mixed with a Triton X-100 solubilized lipid mixture of 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC)/N-Stearoyl-D-erythro-Sphingosylphosphorylcholine (SSM)/cholesterol (Chol) (1:1:1; molar ratio) with 1 mol % C6-NBD-PC at a lipid/protein ratio of 20:1 (w/w) and incubated for 1 h. To remove detergent and to generate proteoliposomes 1 g of SM2 BioBeads (Bio-Rad, Munich, Germany) per 70 mg of Triton X-100 were added and rotated at 4°C. After 12 h the same amount of BioBeads was added for another 4 h. The turbid suspension was withdrawn carefully from the beads and washed twice with PBS and collected by centrifugation (55,000 × g, 1 h, 4°C) to remove remaining detergent.
GUVs with reconstituted viral protein were generated from preformed proteoliposomes according to Girard et al. (30). Proteoliposomes were diluted with distilled water to 0.4–0.8 mg/mL lipid, 0.02 g sucrose/g lipid was added to protect proteins during the following dehydration (31), and 50 μL of this suspension was deposited onto each indium tin oxide slide. Partial dehydration was achieved by placing the slides in a sealed chamber containing a saturated NaCl solution overnight. From here, formation of GUVs follows the electroformation method described above.
Protocol 2
For direct reconstitution of purified rhodamine-labeled HA into GUVs, films from 100 nmol lipids (DOPC/SSM/Chol at the molar ratio of 36:36:28, 0.7 mol % C6-NBD-PC or X31 lipids, 0.7 mol % C6-NBD-PC) were first dried on titanium plates. Subsequently, 0.6 nmol Rh-HA in 50 μL PBS were spotted in little droplets and the lipid-HA film was dehydrated using a vacuum desiccator at 500 mbar for 1 h. Afterward GUVs were generated by electroswelling.
Confocal imaging
Confocal images of the equatorial plane of GUVs and GPMVs were taken with an inverted confocal laser scanning microscope FluoView 1000 (Olympus, Hamburg, Germany) with a 60× (N.A. 1.35) oil-immersion objective at various temperatures using a custom built temperature chamber. Cerulean and CFP were excited at 440 nm using a laser diode and detected in the range of 460–490 nm. YFP was excited at 515 nm using an Argon ion laser and detected in the range of 535–575 nm. TAMRA and NBD were excited with the 559-nm laser line of a He-Ne laser (Melles Griot, Bensheim, Germany) and the 488-nm laser line of an Argon ion laser (Melles Griot), respectively. The emission of TAMRA and NBD was recorded from 560 to 670 nm and 500 to 530 nm, respectively. R18 was excited with the 559 nm laser (DM 405/488/559/635) and emission was detected from 560 to 670 nm.
Fluorescence microscopy and FLIM
FLIM images were taken as described previously (32). In short, a confocal laser scanning microscope with an inverted IX81 fluorescence microscope equipped with a Fluoview 1000 scanhead (Olympus, Tokyo, Japan) was used. For excitation, a pulsed picosecond diode laser was used with a wavelength of 468 nm. Fluorescence was collected for ∼100 s with the same objective and selected with a 540/40 bandpass filter. The average photon count rate was ∼2–4 × 104 counts/s. Fluorescence was detected with an avalanche photo diode using the method of time-correlated single photon counting.
For analysis, the lipid bilayer of the GUVs or GPMVs was selected. Fluorescence decay curves were independently fitted for ld and lo domains as a sum of two exponential terms:
where F(t) is the fluorescence intensity at time t and αi is a preexponential factor representing the fractional contribution to the time-resolved decay of the component with a lifetime τi such that Σiαi = 1. The decay parameters were recovered using a nonlinear least-squares iterative fitting procedure based on the Marquardt algorithm. For the calculation of lifetime histograms the fitting procedure was carried out for every pixel of a selected region of interest.
Results
In Fig. 1 an overview on the various constructs used for studying lateral partition among lo and ld domains of GUVs and GPMVs is shown. GUVs either consist of a mixture of DOPC, SM, and cholesterol or of lipids extracted from influenza virus. GPMVs were derived from CHO-K1 cells (see Material and Methods).
Figure 1.
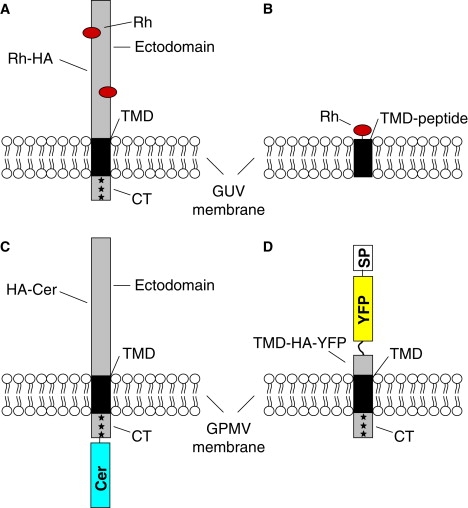
Overview on HA constructs and synthetic peptides. (A and B) Rhodamine (TAMRA) labeled full length HA (A, Rh-HA) or synthetic peptide (TMD-peptide) corresponding to the transmembrane domain of influenza HA (B, Rh-TMD) were incorporated into GUVs of different lipid compositions. (C and D) Full length HA tagged C-terminally with Cer (C, HA-Cer) or a HA fragment linked N-terminally to YFP and signal peptide (SP) (D, TMD-HA-YFP) were expressed in the plasma membrane of CHO-K1 cells. GPMVs were generated subsequently and used for confocal scanning microscopy. Rh, TAMRA. ∗Palmitoylation site.
Lateral organization of reconstituted HA in GUVs
Two different reconstitution protocols lead to visualization of fluorescent HA in GUV membranes (Fig. 2). In Protocol 1 (Fig. 2, A and B), HA from X31 Influenza virus labeled with TAMRA (Rh-HA) was reconstituted in GUVs according to the protocol adapted from (29,30) (see Protocol 1, Material and Methods). To this end, HA was reconstituted into proteoliposomes from which GUVs were generated. In Protocol 2, reconstituted GUVs were prepared by applying purified Rh-HA directly onto preformed lipid films (see Protocol 2, Material and Methods). With both protocols, similar results were obtained (Fig. 2). Whereas the Rh-HA was homogenously distributed in pure DOPC GUVs (not shown), we observed a lateral inhomogeneous organization of Rh-HA in lipid domain forming GUVs consisting of DOPC/SSM/Chol (1:1:1, molar ratio) (Fig. 2 A) or of DOPC/SSM/Chol (1:1:0.8, molar ratio) (not shown). As shown, Rh-HA colocalizes with the green fluorescent lipid analog C6-NBD-PC that is known to enrich in ld domains (33). To mimic a more native lipid environment, Rh-HA was reconstituted into GUVs prepared from virus lipid extracts from A/PR/8/34 (Fig. 2 B) or X31(Fig. 2, C and D, at 25 and 37°C, respectively). Those GUVs also form rather large ld and lo domains as visualized by C6-NBD-PC (Fig. 2, B–D). Again, a preference of HA for the ld domain was observed. When using lipid extracts from X31 enrichment of HA in ld domains was even more pronounced in comparison to lipids from A/PR/8/34.
Figure 2.
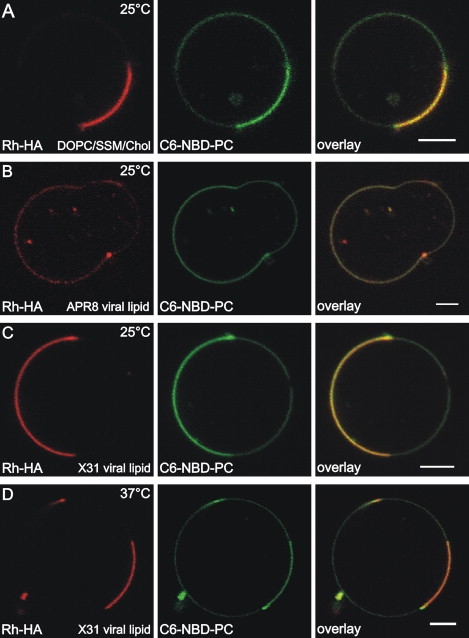
Lateral organization of full length HA in domain forming GUVs. Rhodamine (TAMRA)-labeled full length HA (Rh-HA) was incorporated into GUVs made from (A) DOPC/SSM/Chol (1:1:1, molar ratio), (B) influenza virus lipid extracts of A/PR/8/34, or (C and D) X31. Rh-HA was reconstituted according to Protocol 1 using proteoliposomes (A and B) or following Protocol 2 (C and D) (for details see Material and Methods and Results). Rh-HA, rhodamine fluorescence (red); C6-NBD-PC fluorescence (green) as marker for liquid-disordered domains; overlay. Scale bar = 5 μm. Images were taken at (A–C) 25°C or (D) 37°C.
Lateral organization of the TMD-peptide of HA in GUVs
To clarify whether any conformational change of the HA ectodomain due to extraction and reconstitution may affect partition behavior between lipid domains, we studied the lateral organization of the tetramethylrhodamine labeled peptide Rh-TMD (Fig. 1 B) corresponding to the HA transmembrane domain sequence. The peptide was incorporated into membranes during GUV preparation (1 mol %, see Material and Methods). The partition behavior of Rh-TMD was very similar to that of intact full-length HA as judged by comparison with C6-NBD-PC distribution (see above): a homogenous distribution in pure DOPC vesicles (not shown) and a strong partition into the ld domain of GUVs consisting either of DOPC/SSM/Chol (1:1:1) or of virus lipid extracts (A/PR/8/34) at 25°C (Fig. 3), and also at 10°C and 37°C (not shown).
Figure 3.
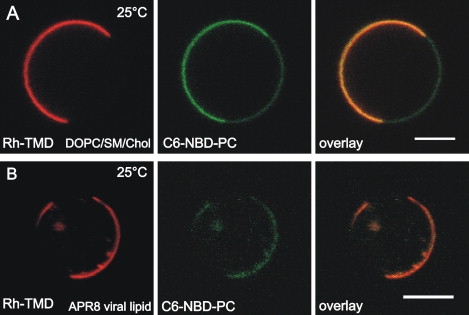
Lateral organization of TMD-peptide in domain forming GUVs. Rhodamine (TAMRA) labeled synthetic peptide (Rh-TMD) corresponding to the transmembrane domain of influenza virus hemagglutinin was incorporated into GUVs made from (A) DOPC/SSM/Chol (1:1:1, molar ratio) or from (B) influenza virus lipid extracts. Rh-TMD, rhodamine fluorescence (red); C6-NBD-PC fluorescence (green) as marker for liquid-disordered domains; overlay. Scale bar = 5 μm. Images were taken at 25°C.
Lateral organization of fluorescent HA in plasma membrane blebs
Lipid domain specific partition of HA has been essentially studied in the plasma membrane of mammalian cells expressing HA (see Introduction). However, lipid rafts in those membranes are of dynamic nanoscale size and cannot be visualized directly in living cells. Therefore, we have taken advantage of a method published recently for generating large blebs from the plasma membrane of mammalian cells (18). Based on partition of lipid raft markers, those GPMVs were found to form large lipid domains at lower temperature (10°C).
We applied this method to cells transiently expressing either full length HA or a membrane anchoring fragment derived from the same HA (Fig. 1C and D). The full length HA (HA-Cer) was tagged at its CT (C-terminus) with the fluorescent protein Cerulean, a variant of CFP with improved quantum yield and a higher excitation coefficient (19). The membrane anchored fragment (TMD-HA-YFP) consists of the TMD and the CT of the protein as well as a short sequence of the HA ectodomain. Essentially, the HA ectodomain was replaced by the YFP. In contrast to HA-Cer, this construct circumvents tagging of the cytoplasmic tail that may interfere with its role in lateral organization. Indeed, previous studies indicated that mutations in the TMD and the CT of HA reduce association with detergent-resistant fractions (2,4,8,34). As we have already shown (13), endoglycosidase digestion shows that TMD-HA-YFP acquires Endo-H resistant carbohydrates like full length HA, a modification that occurs during the transport of plasma membrane proteins through the Golgi complex (35). Likewise, labeling of HA-Cer and of TMD-HA-YFP with 3[H]-palmitate confirmed their palmitoylation (12,13).
To avoid interference of fluorescence of Cer, CFP, or YFP and the fluorescence of the lipid domain marker C6-NBD-PC, we have used the red fluorescent lipid-like octadecylrhodamine B chloride (R18) that is known to partition into the ld domain (36). A micrometer-scaled segregation of fluid phase domains was induced in GPMVs by lowering the temperature (10°C) as described previously (36). As shown in Fig. 4, apart from GPMVs with a homogenous distribution of R18 we observed domain forming vesicles indicated by the heterogeneous distribution of R18 at 10°C. At 25°C we could not detect visible lipid domain either in GPMVs or in the plasma membrane of CHO-K1 cells. To characterize further the R18 enriched domain at low temperature, we generated blebs from CHO-K1 cells expressing GPI-CFP on the outer plasma membrane leaflet. This construct is known to enrich in lo domains and has been often used as a raft marker (22). Indeed, GPI-CFP was excluded from R18 containing domains (Fig. 4 A).
Figure 4.
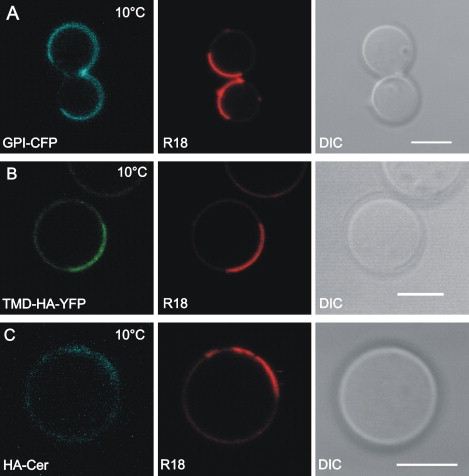
Lateral arrangement of HA-Cer and TMD-HA-YFP in GPMVs. GPMVs were derived from CHO-K1 cells expressing (A) GPI-CFP, (B) TMD-HA-YFP, or (C) HA-Cer in the plasma membrane. Confocal images were taken after cooling samples to 10°C. R18 fluorescence (red) as marker for liquid-disordered domains. DIC, differential interference contrast image. Scale bar = 5 μm.
TMD-HA-YFP was found to partition preferentially to the ld domain in GPMVs as indicated by colocalization with R18 (Fig. 4 B). We also found HA-Cer enriched in ld domains (Fig. 4 C). Note that the differences in partition of TMD-HA-YFP and HA-Cer between lo and ld domains is less pronounced compared to full length HA and TMD-peptide of HA in GUVs prepared from synthetic or virus lipids. A quantitative analysis of the fluorescence intensities is provided in Table 1 giving the ratio of fluorescence intensity in the lo and ld domain in percent for each of the studied systems. On average the ratio is ten times higher in GPMV (0.266) as compared to the GUV system (0.027). At 25°C or 37°C neither clustering for HA-Cer and TMD-HA-YFP nor lipid domain formation (see above) was observed.
Table 1.
Quantitative fluorescence intensity analysis of HA and peptide partitioning between lo and ld domains in GUVs and GPMVs
| Vesicle |
GUV |
GPMV |
||||||
|---|---|---|---|---|---|---|---|---|
| Protein | Rh-HA | Rh-TMD | TMD-HA-YFP | HA-Cer | ||||
| Figure | 2 A | 2 B | 2 C | 2 D | 3 A | 3 B | 4 B | 4 C |
| Lipids | DOPC/SSM/Chol | APR8 | X31∗ | X31† | DOPC/SSM/Chol | APR8 | ||
| Ratio | 0.020 | 0.126 | 0.001 | 0.003 | 0.006 | 0.005 | 0.185 | 0.347 |
| ±SE | 0.027 | 0.016 | 0.004 | 0.006 | 0.016 | 0.018 | 0.023 | 0.057 |
| n | 8 | 7 | 6 | 8 | 11 | 7 | 12 | 6 |
Table shows the ratio of the fluorescence intensity of the HA protein or peptide in the lo and ld domains, respectively. Average fluorescence intensities were determined at the membrane of the two distinct domains using Olympus FluoView 1000 software (Olympus, Hamburg, Germany). Average background fluorescence was subtracted before calculating the ratio of the fluorescence intensity (lo/ld). Note, the ratio refers to the fluorescence intensity but not to the ratio of amount of HA protein (or peptide) in the two domains. n, number of vesicles.
At 25°C.
At 37°C.
Physical properties of lipid domains in GUVs and GPMVs
To characterize and compare lo and ld domains of GUVs and GPMVs, we applied fluorescence lifetime imaging of the fluorescent lipid analog C6-NBD-PC. Recently, we have shown that the NBD fluorescence lifetime of C6-NBD-PC depends on the lipid composition and, in particular, on lipid-dependent physical properties of the bilayer (32). C6-NBD-PC showed a double exponential fluorescence decay: a shorter lifetime τ1 of ∼2–3 ns originating from the red edge excitation shift of NBD, and a longer lifetime τ2 sensitive to the lipid domain. The contribution of τ1 is <5%. The lifetime τ2 is significantly longer in the lo domain in comparison to the ld domain as shown in Fig. 5 A for DOPC/SSM/Chol (1:1:1) GUVs at 25°C. In accordance with previous data the average lifetime τ2 in the ld was around 7 ns and in the lo domains around 12 ns, respectively (32). A similar pronounced difference of τ2 between both domains was observed for GUVs prepared from virus lipids (Fig. 5 B). However, for GPMVs the difference between τ2 in ld and lo domains was much smaller. Although the lifetime of C6-NBD-PC in ld domains of GPMVs was shifted slightly to higher values, the lifetime in the lo domain was significantly shorter even at 10°C in comparison to the lo domain of GUVs (Fig. 5, B and C).
Figure 5.
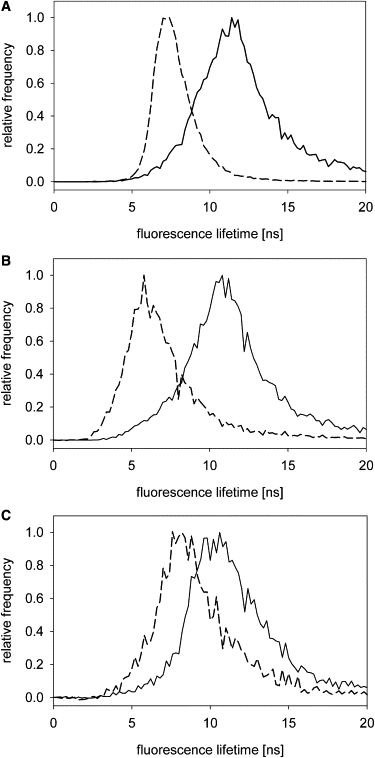
Fluorescence lifetime of C6-NBD-PC GUVs and GPMVs. C6-NBD-PC showed a double exponential fluorescence decay. Only the longer lifetime (τ2) sensitive to the lipid domain is shown. (A) Lifetime histograms of C6-NBD-PC in GUVs prepared from DOPC/SSM/Chol mixtures (1:1:1 mol/mol/mol) for ld (dashed line) and lo (solid line) domains at 25°C. (B) Lifetime histograms of C6-NBD-PC in GUVs prepared from total lipids isolated from influenza A/PR/8/34 for ld (dashed line) and lo (solid line) domains at 10°C. (C) Lifetime histograms of C6-NBD-PC in GPMVs prepared from CHO-K1 cells for ld (dashed line) and lo (solid line) domains at 10°C. Histograms were normalized by setting the maximum to 1.
Discussion
Influenza virus hemagglutinin is considered a typical example for integral membrane proteins that strongly enrich in rafts thought to resemble the lo domains observed in model membranes. However, direct visualization of HA partition into such domains of mammalian cell plasma membranes expressing the virus protein has not been reported. In this study, we have used model systems that are known to form lipid domains in the microscopic range easily detectable by fluorescent lipid analogs with a preference either for the lo or ld domain. In one system, either full length HA or the TMD-peptide of HA was reconstituted into GUVs prepared from a mixture of well defined raft-lipids or from lipids extracted from the viral envelope. As another model system, GPMVs were obtained from cell plasma membranes, thus containing a large variety of lipids and, apart from the protein of interest—HA-Cer or TMD-HA-YFP—other membrane proteins. For all studied examples a strong preference for the ld domain was observed. However, the preference of full length HA for the ld domain was less pronounced in GPMVs, which mimic biological membranes much better than GUVs as we will discuss below.
Full length HA was almost exclusively localized in the ld domain in GUVs either consisting of a well defined composition of synthetic lipids or viral lipid extracts. This extreme partition was observed not only at 25°C but also at 37°C. A similar pronounced preference for the ld domain was found for the TMD-peptide of HA indicating that partition can be determined sufficiently by the transmembrane domain. The enrichment of the TMD-peptide in the ld domain is unexpected when comparing the thickness of the bilayer with that of the TMD. The thickness of the hydrophobic part of a DOPC/SSM/Chol (1:1:1) bilayer is 36 Å in the lo and 27 Å in the ld phase (37). The Rh-TMD peptide is essentially of α-helical structure (J. Nikolaus and A. Herrmann, unpublished results). Considering 1.5 Å per amino acid in an α-helical conformation, the hydrophobic stretch of the TMD-peptide (25 aa) is 37.5 Å. Hence, one would expect partition of the TMD-peptide in the lo phase where it faces only a small positive mismatch of 1.5 Å. Surprisingly, despite a positive mismatch of 10.5 Å, TMD-peptides are incorporated into the liquid disordered phase. However, this behavior is consistent with recent studies on domain partition of various membrane proteins and peptides in model membranes (38–44). Vidal and McIntosh (39) and McIntosh et al. (45) studied lateral sorting of model transmembrane peptides of different hydrophobic length—23 or 29 amino acids—between lo and ld domains by confocal fluorescence microscopy at 4°C and 37°C. These transmembrane domains match the hydrocarbon thickness of ld and lo domains, respectively. At both temperatures the peptides were primarily localized in the ld domain independent of hydrophobic matching. Distinct physical properties favor the incorporation of TMDs into the ld phase. For example, lo domains have a higher bilayer compressibility, which for SM/cholesterol (1:1, molar ratio) is approximately nine times that of a phosphatidylcholine bilayer (46), and higher bending moduli compared to the nonraft domains (47). That is, lo domains are more tightly packed as confirmed by the significantly higher fluorescence lifetime of C6-NBD-PC in comparison to the ld domain (see Fig. 5) (32). Hence, more energy is required to separate adjacent lipid molecules to enable incorporation of TMDs into the bilayer than in ld domains, which are of lower cohesive energies (39). Thus, it is energetically favorable for TMDs to partition into the ld domain. At present we do not know how the TMD-peptide of HA responds to the hydrophobic mismatch that occurs in the ld domain. Tilting of peptides might be one possibility to match the hydrophobic thickness of the bilayer avoiding exposure of hydrophobic side chains to polar environment, which is energetically unfavorable (48,49). However, using polarized ATR-FTIR it could be shown that the TMD of HA in DMPC membranes having a hydrophobic thickness of ∼21 Å is in an α-helical conformation and orientated almost parallel to the membrane normal (50).
Our results confirm and extend previous studies showing that in lipid model systems the cohesive bilayer material properties determine the process of lateral sorting of transmembrane peptides (39,45) and even of full length integral membrane proteins. The strong enrichment of wild-type HA in the ld domain may be rather surprising, in particular, because reconstituted HA contains the typical raft targeting signal, such as palmitoylation and specific hydrophobic amino acids in its TMD. Thus, any signal that sorts HA into rafts of living cells (2,34) and an unfavorable hydrophobic mismatch are not relevant for the partition of the TMD-peptide of HA and wild-type HA in GUVs.
Although we also observed a strong preference of full length HA-Cer and TMD-HA-YFP for the ld domain in GPMVs derived from the plasma membrane of CHO-K1 cells, their preference was less pronounced compared to full length HA and TMD-peptide of HA in GUVs prepared from synthetic or virus lipids. This indicates that lo domains of GPMVs may differ in their physical properties to that in GUVs. Indeed, lifetime of C6-NBD-PC shows that packing of lipid domains in GPMVs is different from that of GUVs. In particular, the differences of lipid packing between ld and lo domains are much less pronounced for GPMVs (Fig. 5 C) in comparison to GUVs prepared from synthetic or virus lipids (Fig. 5, A and B). This is in agreement with our observations for GPMVs from HeLa cells (32) and data of Kaiser et al. on plasma membrane spheres from the human epithelial carcinoma cell line A431 (51). We suppose that due to the much higher membrane protein content of GPMVs, protein-lipid interactions may interfere with a very tight packing of lo domain (51–54) as found in GUVs. This would reduce the energy barrier for intercalation of membrane proteins thus explaining the partial presence of HA in lo domains of GPMVs. Furthermore, differences of partition between GPMVs and GUVs might also be due to lipid asymmetry in GPMVs affecting domain formation and properties. Although a partial loss of bilayer asymmetry on formation of GPMVs compared to the cell membrane has been observed (51,55), GUV do not show asymmetry at all.
We surmise a similar situation for the organization of virus envelope lipid phase. In the influenza virus envelope the cholesterol fraction is much higher than that of the host cell membranes (3,8). Despite the high cholesterol content a recent NMR study on plasma membrane—serving as a budding site for influenza viruses—and also on the viral envelope indicated that the physical state of those membranes at physiological temperature has ld like properties (17). Presumably, protein-lipid interactions interfere with the formation of tightly packed lo domains. This may not only prevent segregation of membrane proteins such as HA away from lo domains, but may even facilitate partition in those lipid phases. However, it remains to identify the underlying mechanism of cholesterol enrichment in the virus envelope. Polozov et al. (17) suggested the virus enriches itself in specific lipids such as cholesterol. In particular, palmitoylation of HA may ensure a recruitment of saturated lipids and cholesterol. Either HAs with such lipid environment associate and form smaller clusters. Oligomerization of the virus matrix protein M1 and interaction of M1 with the CT of HA during virus assembly cross-linking HAs may lead to membrane patches enriched in cholesterol that eventually form the virus budding site. Lateral cross-linking of membrane proteins and glycosphingolipids has been shown to trigger coalescence of nanoscale lipid heterogeneity into larger stabilized raft domains (15,56) that play a functional role for uptake and endocytosis of proteins (57), and viruses (58), and for cell signaling (59,60).
In summary, our studies with model membranes clearly show that HA does not partition into tightly packed lo domains. To explain cholesterol dependent clustering of HA spikes at the plasma membrane visualized by immuno-EM (11,61) and FPALM (16), and clustering of HA with raft-markers observed by FLIM-FRET (12,13) in HA-transfected cells, we conclude that protein-lipid interactions reduce lipid packing of lo domains in biological membranes enabling recruitment of proteins such as HA to those domains. This would be in line with the view that physical properties of lipid domains in biological membranes are tightly regulated by protein-lipid interactions (54,62). Besides differences in lipid packing, cellular and model membranes are also distinguished by the absence of cytoskeletal components in the latter. Recent reports indicated that microfilaments, especially cortical actin, might affect the formation, dynamics and maintenance of membrane-rafts in living cells (63,64). However, other studies found submicroscopic membrane heterogeneity to be independent of actin (65). Hence, future studies are required to unravel whether the absence of HA in lo domains is also due to the lack of microfilaments in GUVs and GPMVs.
Acknowledgments
Note added in proof: During the publication process of our manuscript the work of Johnson et al. (2010) was published reporting on a variable phase partitioning of HA in GPMVs from HeLa cell into either ld or lo phases or a nonpreferential partitioning:
Johnson, S.A., B. M. Stinson, …, T. Baumgart. 2010. Temperature dependent phase behavior and protein partitioning in giant plasma membrane vesicles. Biochim. Biophys. Acta. 1798:1427–1435.
We thank Gudrun Habermann and Sabine Schiller for technical support.
The work was supported by the Deutsche Forschungsgemeinschaft (SPP 1175, SFB 740, and TP C3 to A.H. and M.V.), the European Union (MRTN-CT-2004-005330 and MEST-CT-2004-007931 to A.P.P. and S.S.), and the Leibniz Graduate School of Molecular Biophysics (J.N.).
Footnotes
Elisa Bayraktarov's present address is Department of Biogeochemistry, Max Planck Institute for Marine Microbiology, Bremen, Germany.
References
- 1.White J., Helenius A., Gething M.J. Hemagglutinin of influenza virus expressed from a cloned gene promotes membrane fusion. Nature. 1982;300:658–659. doi: 10.1038/300658a0. [DOI] [PubMed] [Google Scholar]
- 2.Scheiffele P., Roth M.G., Simons K. Interaction of influenza virus hemagglutinin with sphingolipid-cholesterol membrane domains via its transmembrane domain. EMBO J. 1997;16:5501–5508. doi: 10.1093/emboj/16.18.5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scheiffele P., Rietveld A., Simons K. Influenza viruses select ordered lipid domains during budding from the plasma membrane. J. Biol. Chem. 1999;274:2038–2044. doi: 10.1074/jbc.274.4.2038. [DOI] [PubMed] [Google Scholar]
- 4.Takeda M., Leser G.P., Lamb R.A. Influenza virus hemagglutinin concentrates in lipid raft microdomains for efficient viral fusion. Proc. Natl. Acad. Sci. USA. 2003;100:14610–14617. doi: 10.1073/pnas.2235620100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Silvius J.R. Role of cholesterol in lipid raft formation: lessons from lipid model systems. Biochim. Biophys. Acta. 2003;1610:174–183. doi: 10.1016/s0005-2736(03)00016-6. [DOI] [PubMed] [Google Scholar]
- 6.Simons K., Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 7.Brown D.A., London E. Structure and function of sphingolipid- and cholesterol-rich membrane rafts. J. Biol. Chem. 2000;275:17221–17224. doi: 10.1074/jbc.R000005200. [DOI] [PubMed] [Google Scholar]
- 8.Zhang J., Pekosz A., Lamb R.A. Influenza virus assembly and lipid raft microdomains: a role for the cytoplasmic tails of the spike glycoproteins. J. Virol. 2000;74:4634–4644. doi: 10.1128/jvi.74.10.4634-4644.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heerklotz H. Triton promotes domain formation in lipid raft mixtures. Biophys. J. 2002;83:2693–2701. doi: 10.1016/S0006-3495(02)75278-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Munro S. Lipid rafts: elusive or illusive? Cell. 2003;115:377–388. doi: 10.1016/s0092-8674(03)00882-1. [DOI] [PubMed] [Google Scholar]
- 11.Hess S.T., Kumar M., Zimmerberg J. Quantitative electron microscopy and fluorescence spectroscopy of the membrane distribution of influenza hemagglutinin. J. Cell Biol. 2005;169:965–976. doi: 10.1083/jcb.200412058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Engel S., Scolari S., Veit M. FLIM-FRET and FRAP reveal association of influenza virus hemagglutinin with membrane rafts. Biochem. J. 2010;425:567–573. doi: 10.1042/BJ20091388. [DOI] [PubMed] [Google Scholar]
- 13.Scolari S., Engel S., Herrmann A. Lateral distribution of the transmembrane domain of influenza virus hemagglutinin revealed by time-resolved fluorescence imaging. J. Biol. Chem. 2009;284:15708–15716. doi: 10.1074/jbc.M900437200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sengupta P., Hammond A., Baird B. Structural determinants for partitioning of lipids and proteins between coexisting fluid phases in giant plasma membrane vesicles. Biochim. Biophys. Acta. 2008;1778:20–32. doi: 10.1016/j.bbamem.2007.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schroeder R., London E., Brown D. Interactions between saturated acyl chains confer detergent resistance on lipids and glycosylphosphatidylinositol (GPI)-anchored proteins: GPI-anchored proteins in liposomes and cells show similar behavior. Proc. Natl. Acad. Sci. USA. 1994;91:12130–12134. doi: 10.1073/pnas.91.25.12130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hess S.T., Gould T.J., Zimmerberg J. Dynamic clustered distribution of hemagglutinin resolved at 40 nm in living cell membranes discriminates between raft theories. Proc. Natl. Acad. Sci. USA. 2007;104:17370–17375. doi: 10.1073/pnas.0708066104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Polozov I.V., Bezrukov L., Zimmerberg J. Progressive ordering with decreasing temperature of the phospholipids of influenza virus. Nat. Chem. Biol. 2008;4:248–255. doi: 10.1038/nchembio.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baumgart T., Hammond A.T., Webb W.W. Large-scale fluid/fluid phase separation of proteins and lipids in giant plasma membrane vesicles. Proc. Natl. Acad. Sci. USA. 2007;104:3165–3170. doi: 10.1073/pnas.0611357104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rizzo M.A., Springer G.H., Piston D.W. An improved cyan fluorescent protein variant useful for FRET. Nat. Biotechnol. 2004;22:445–449. doi: 10.1038/nbt945. [DOI] [PubMed] [Google Scholar]
- 20.Vey M., Orlich M., Garten W. Hemagglutinin activation of pathogenic avian influenza viruses of serotype H7 requires the protease recognition motif R-X-K/R-R. Virology. 1992;188:408–413. doi: 10.1016/0042-6822(92)90775-K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Veit M., Kretzschmar E., Rott R. Site-specific mutagenesis identifies three cysteine residues in the cytoplasmic tail as acylation sites of influenza virus hemagglutinin. J. Virol. 1991;65:2491–2500. doi: 10.1128/jvi.65.5.2491-2500.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keller P., Toomre D., Simons K. Multicolor imaging of post-Golgi sorting and trafficking in live cells. Nat. Cell Biol. 2001;3:140–149. doi: 10.1038/35055042. [DOI] [PubMed] [Google Scholar]
- 23.Zacharias D.A., Violin J.D., Tsien R.Y. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 2002;296:913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
- 24.Nikolaus J., Stöckl M., Herrmann A. Direct visualization of large and protein-free hemifusion diaphragms. Biophys. J. 2010;98:1192–1199. doi: 10.1016/j.bpj.2009.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bligh E.G., Dyer W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 26.Angelova M.I., Dimitrov D.S. Liposome electroformation. Faraday Discuss. Chem. Soc. 1986;81:303–311. [Google Scholar]
- 27.Krumbiegel M., Herrmann A., Blumenthal R. Kinetics of the low pH-induced conformational changes and fusogenic activity of influenza hemagglutinin. Biophys. J. 1994;67:2355–2360. doi: 10.1016/S0006-3495(94)80721-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Böttcher C., Ludwig K., Stark H. Structure of influenza hemagglutinin at neutral and at fusogenic pH by electron cryo-microscopy. FEBS Lett. 1999;463:255–259. doi: 10.1016/s0014-5793(99)01475-1. [DOI] [PubMed] [Google Scholar]
- 29.Papadopulos A., Vehring S., Herrmann A. Flippase activity detected with unlabeled lipids by shape changes of giant unilamellar vesicles. J. Biol. Chem. 2007;282:15559–15568. doi: 10.1074/jbc.M604740200. [DOI] [PubMed] [Google Scholar]
- 30.Girard P., Pécréaux J., Bassereau P. A new method for the reconstitution of membrane proteins into giant unilamellar vesicles. Biophys. J. 2004;87:419–429. doi: 10.1529/biophysj.104.040360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Doeven M.K., Folgering J.H., Poolman B. Distribution, lateral mobility and function of membrane proteins incorporated into giant unilamellar vesicles. Biophys. J. 2005;88:1134–1142. doi: 10.1529/biophysj.104.053413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stöckl M., Plazzo A.P., Herrmann A. Detection of lipid domains in model and cell membranes by fluorescence lifetime imaging microscopy of fluorescent lipid analogues. J. Biol. Chem. 2008;283:30828–30837. doi: 10.1074/jbc.M801418200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shaw J.E., Epand R.F., Yip C.M. Correlated fluorescence-atomic force microscopy of membrane domains: structure of fluorescence probes determines lipid localization. Biophys. J. 2006;90:2170–2178. doi: 10.1529/biophysj.105.073510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin S., Naim H.Y., Roth M.G. Mutations in the middle of the transmembrane domain reverse the polarity of transport of the influenza virus hemagglutinin in MDCK epithelial cells. J. Cell Biol. 1998;142:51–57. doi: 10.1083/jcb.142.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Copeland C.S., Doms R.W., Helenius A. Assembly of influenza hemagglutinin trimers and its role in intracellular transport. J. Cell Biol. 1986;103:1179–1191. doi: 10.1083/jcb.103.4.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baumgart T., Hunt G., Feigenson G.W. Fluorescence probe partitioning between Lo/Ld phases in lipid membranes. Biochim. Biophys. Acta. 2007;1768:2182–2194. doi: 10.1016/j.bbamem.2007.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gandhavadi M., Allende D., McIntosh T.J. Structure, composition, and peptide binding properties of detergent soluble bilayers and detergent resistant rafts. Biophys. J. 2002;82:1469–1482. doi: 10.1016/S0006-3495(02)75501-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fastenberg M.E., Shogomori H., London E. Exclusion of a transmembrane-type peptide from ordered-lipid domains (rafts) detected by fluorescence quenching: extension of quenching analysis to account for the effects of domain size and domain boundaries. Biochemistry. 2003;42:12376–12390. doi: 10.1021/bi034718d. [DOI] [PubMed] [Google Scholar]
- 39.Vidal A., McIntosh T.J. Transbilayer peptide sorting between raft and nonraft bilayers: comparisons of detergent extraction and confocal microscopy. Biophys. J. 2005;89:1102–1108. doi: 10.1529/biophysj.105.062380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bacia K., Schuette C.G., Schwille P. SNAREs prefer liquid-disordered over “raft” (liquid-ordered) domains when reconstituted into giant unilamellar vesicles. J. Biol. Chem. 2004;279:37951–37955. doi: 10.1074/jbc.M407020200. [DOI] [PubMed] [Google Scholar]
- 41.Shogomori H., Hammond A.T., Brown D.A. Palmitoylation and intracellular domain interactions both contribute to raft targeting of linker for activation of T cells. J. Biol. Chem. 2005;280:18931–18942. doi: 10.1074/jbc.M500247200. [DOI] [PubMed] [Google Scholar]
- 42.Almeida P.F., Pokorny A., Hinderliter A. Thermodynamics of membrane domains. Biochim. Biophys. Acta. 2005;1720:1–13. doi: 10.1016/j.bbamem.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 43.Hammond A.T., Heberle F.A., Feigenson G.W. Crosslinking a lipid raft component triggers liquid ordered-liquid disordered phase separation in model plasma membranes. Proc. Natl. Acad. Sci. USA. 2005;102:6320–6325. doi: 10.1073/pnas.0405654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kalvodova L., Kahya N., Simons K. Lipids as modulators of proteolytic activity of BACE: involvement of cholesterol, glycosphingolipids, and anionic phospholipids in vitro. J. Biol. Chem. 2005;280:36815–36823. doi: 10.1074/jbc.M504484200. [DOI] [PubMed] [Google Scholar]
- 45.McIntosh T.J., Vidal A., Simon S.A. Sorting of lipids and transmembrane peptides between detergent-soluble bilayers and detergent-resistant rafts. Biophys. J. 2003;85:1656–1666. doi: 10.1016/S0006-3495(03)74595-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McIntosh T.J., Simon S.A., Huang C.H. Structure and cohesive properties of sphingomyelin/cholesterol bilayers. Biochemistry. 1992;31:2012–2020. doi: 10.1021/bi00122a017. [DOI] [PubMed] [Google Scholar]
- 47.Lundbaek J.A., Andersen O.S., Nielsen C. Cholesterol-induced protein sorting: an analysis of energetic feasibility. Biophys. J. 2003;84:2080–2089. doi: 10.1016/S0006-3495(03)75015-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mouritsen O.G., Bloom M. Mattress model of lipid-protein interactions in membranes. Biophys. J. 1984;46:141–153. doi: 10.1016/S0006-3495(84)84007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Killian J.A. Synthetic peptides as models for intrinsic membrane proteins. FEBS Lett. 2003;555:134–138. doi: 10.1016/s0014-5793(03)01154-2. [DOI] [PubMed] [Google Scholar]
- 50.Tatulian S.A., Tamm L.K. Secondary structure, orientation, oligomerization, and lipid interactions of the transmembrane domain of influenza hemagglutinin. Biochemistry. 2000;39:496–507. doi: 10.1021/bi991594p. [DOI] [PubMed] [Google Scholar]
- 51.Kaiser H.J., Lingwood D., Simons K. Order of lipid phases in model and plasma membranes. Proc. Natl. Acad. Sci. USA. 2009;106:16645–16650. doi: 10.1073/pnas.0908987106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jacobson K., Mouritsen O.G., Anderson R.G. Lipid rafts: at a crossroad between cell biology and physics. Nat. Cell Biol. 2007;9:7–14. doi: 10.1038/ncb0107-7. [DOI] [PubMed] [Google Scholar]
- 53.Hancock J.F. Lipid rafts: contentious only from simplistic standpoints. Nat. Rev. Mol. Cell Biol. 2006;7:456–462. doi: 10.1038/nrm1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lingwood D., Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010;327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- 55.Honerkamp-Smith A.R., Veatch S.L., Keller S.L. An introduction to critical points for biophysicists; observations of compositional heterogeneity in lipid membranes. Biochim. Biophys. Acta. 2009;1788:53–63. doi: 10.1016/j.bbamem.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kahya N., Brown D.A., Schwille P. Raft partitioning and dynamic behavior of human placental alkaline phosphatase in giant unilamellar vesicles. Biochemistry. 2005;44:7479–7489. doi: 10.1021/bi047429d. [DOI] [PubMed] [Google Scholar]
- 57.Römer W., Berland L., Johannes L. Shiga toxin induces tubular membrane invaginations for its uptake into cells. Nature. 2007;450:670–675. doi: 10.1038/nature05996. [DOI] [PubMed] [Google Scholar]
- 58.Ewers H., Romer W., Johannes L. GM1 structure determines SV40-induced membrane invagination and infection. Nat. Cell Biol. 2010;12(sup 11–12):11–18. doi: 10.1038/ncb1999. [DOI] [PubMed] [Google Scholar]
- 59.Sohn H.W., Tolar P., Pierce S.K. Membrane heterogeneities in the formation of B cell receptor-Lyn kinase microclusters and the immune synapse. J. Cell Biol. 2008;182:367–379. doi: 10.1083/jcb.200802007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zech T., Ejsing C.S., Harder T. Accumulation of raft lipids in T-cell plasma membrane domains engaged in TCR signaling. EMBO J. 2009;28:466–476. doi: 10.1038/emboj.2009.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Leser G.P., Lamb R.A. Influenza virus assembly and budding in raft-derived microdomains: a quantitative analysis of the surface distribution of HA, NA and M2 proteins. Virology. 2005;342:215–227. doi: 10.1016/j.virol.2005.09.049. [DOI] [PubMed] [Google Scholar]
- 62.Ge M., Gidwani A., Freed J.H. Ordered and disordered phases coexist in plasma membrane vesicles of RBL-2H3 mast cells. An ESR study. Biophys. J. 2003;85:1278–1288. doi: 10.1016/S0006-3495(03)74563-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Suzuki K.G., Fujiwara T.K., Kusumi A. GPI-anchored receptor clusters transiently recruit Lyn and G alpha for temporary cluster immobilization and Lyn activation: single-molecule tracking study 1. J. Cell Biol. 2007;177:717–730. doi: 10.1083/jcb.200609174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Goswami D., Gowrishankar K., Mayor S. Nanoclusters of GPI-anchored proteins are formed by cortical actin-driven activity. Cell. 2008;135:1085–1097. doi: 10.1016/j.cell.2008.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lasserre R., Guo X.J., He H.T. Raft nanodomains contribute to Akt/PKB plasma membrane recruitment and activation. Nat. Chem. Biol. 2008;4:538–547. doi: 10.1038/nchembio.103. [DOI] [PubMed] [Google Scholar]


