Abstract
Cholesterol is distributed unevenly between different cellular membrane compartments, and the cholesterol content increases from the inner bilayers toward the plasma membrane. It has been suggested that this cholesterol gradient is important in the sorting of transmembrane proteins. Cholesterol has also been to shown play an important role in lateral organization of eukaryotic cell membranes. In this study the aim was to determine how transmembrane proteins influence the lateral distribution of cholesterol in phospholipid bilayers. Insight into this can be obtained by studying how cholesterol interacts with bilayer membranes of different composition in the presence of designed peptides that mimic the transmembrane helices of proteins. For this purpose we developed an assay in which the partitioning of the fluorescent cholesterol analog CTL between LUVs and mβCD can be measured. Comparison of how cholesterol and CTL partitioning between mβCD and phospholipid bilayers with different composition suggests that CTL sensed changes in bilayer composition similarly as cholesterol. Therefore, the results obtained with CTL can be used to understand cholesterol distribution in lipid bilayers. The effect of WALP23 on CTL partitioning between DMPC bilayers and mβCD was measured. From the results it was clear that WALP23 increased both the order in the bilayers (as seen from CTL and DPH anisotropy) and the affinity of the sterol for the bilayer in a concentration dependent way. Although WALP23 also increased the order in DLPC and POPC bilayers the effects on CTL partitioning was much smaller with these lipids. This indicates that proteins have the largest effect on sterol interactions with phospholipids that have longer and saturated acyl chains. KALP23 did not significantly affect the acyl chain order in the phospholipid bilayers, and inclusion of KALP23 into DMPC bilayers slightly decreased CTL partitioning into the bilayer. This shows that transmembrane proteins can both decrease and increase the affinity of sterols for the lipid bilayers surrounding proteins. This is likely to affect the sterol distribution within the bilayer and thereby the lateral organization in biomembranes.
Abbreviations used: CTL, cholestatrienol; DLPC, dilauroylphosphatidcholine; DMPC, dimyrisoylphosphatidylcholine; DPH, diphenylhexatriene; DTPC, ditridecanoylphosphatidylcholine; KX, molar fraction partitioning coefficient; LUV, large unilamellar vesicle; mβCD, methyl-β-cyclodextrin; POPC, 1-palmitoyl-2-oleoyl-phosphatidylcholine; PSM, palmitoylsphingomyelin
Introduction
Cholesterol is an essential component in the membranes of mammalian cells. An interesting property of cholesterol is that it together with phospholipids can form a so called liquid ordered phase, by ordering the acyl chains in fluid phospholipids. Thereby, cholesterol can induce liquid-liquid phase separation in bilayer membranes, i.e., cholesterol can influence the lateral organization in lipid bilayers.
How the lateral organization of a phospholipid bilayer is influenced by cholesterol depends on the phospholipid composition of the bilayer. Cholesterol has been shown to interact differently with different phospholipid types (1–3), and to prefere phospholipids with saturated acyl chains over those with unsaturated chains (see (4) and references within). Hence, cholesterol can facilitate lateral separation in complex bilayer membranes by favoring saturated acyl chains over unsaturated ones. For example, it has been observed that saturated sphingomyelin together with cholesterol can form ordered domains that laterally separate from disordered domains containing unsaturated phosphatidylcholine (5,6). As it is known that sphingomyelin and cholesterol have a similar distribution in cellular membranes it is thought that they are cosorted, and that sorting is linked to the formation of domains enriched in sphingolipids and cholesterol (reviewed in Holthuis et al. (7)).
Cholesterol has also been suggested to be involved in sorting and trafficking of transmembrane proteins (8). Sorting of transmembrane proteins between Golgi and the plasma membrane seems to depend, at least partially, on the length of the hydrophobic transmembrane segments in the proteins (9–11). Proteins with longer transmembrane helices are transported to the plasma membrane whereas those with shorter transmembrane helices stay in the inner membrane compartments. As it is known that cholesterol can increase the bilayer thickness and that the plasma membranes are thicker than Golgi membranes it has been suggested that the cholesterol gradient in the bilayers may drive the sorting of transmembrane proteins as the membrane components strive to minimize the hydrophobic mismatch (8) or due cholesterol induced changes to the bilayer material properties (12). This could mean that proteins with longer transmembrane helices are targeted to thicker membrane patches, i.e., cholesterol and sphingolipid enriched domains in the Golgi, leading to transport to the plasma membrane. Proteins with shorter transmembrane segments on the other hand could partition into the thinner cholesterol-poor domains.
It is well known that transmembrane proteins can affect the structure of the surrounding lipids (reviewed in de Planque and Killian (13), Lee (14), and Marsh (15)), but to what degree transmembrane proteins can influence the lateral organization in lipid bilayers remains unknown. Membrane proteins could increase the heterogeneity in cellular membranes e.g., by having different affinity for different lipids, and in fact it has been observed that several different membrane proteins preferentially interact with lipids that have a specific acyl chain length (16–20). As different proteins seem to have different acyl chain preferences, deriving from the effective hydrophobic length of their transmembrane segments, proteins should be considered as potential promoters of nanoscopic domain formation in cellular membranes.
If some proteins attract phospholipids with longer acyl chains, do the same proteins also attract cholesterol? Such attraction could be a result of the presence of long chain phospholipids at the surface of the protein or transmembrane proteins having an ordering effect on the surrounding lipids. This could result in the formation of nanoscopic domains around the proteins. Other proteins attracting short chain phospholipids could have the opposite effect, and expel cholesterol from the surrounding lipid environment.
The aim of this study was to obtain new information about how transmembrane proteins can influence how cholesterol interacts with phospholipid bilayers. A convenient way to study this is to measure sterol partitioning between LUVs and mβCD (21–24). For this purpose, we developed and tested what we believe to be a new fluorescence-based method. The tests showed that the fluorescent cholesterol analog, CTL, responded similarly as cholesterol to changes in the lipid composition, i.e., the probe should also respond similarly to incorporation of proteins into the LUVs.
Model peptides designed to mimic the transmembrane helices of proteins were included to the lipid bilayers to give information about the influence of proteins. Peptide inclusion increased the affinity of CTL for the phospholipid bilayer, and the affect was decreased when the phospholipid acyl chains were shorter or unsaturated. In addition, the effect of peptides on the affinity of sterols for phospholipid bilayers was clearly dependent on the peptide structure. Hence, we conclude that sterols sense proteins in the bilayer and that based on the results in this study transmembrane proteins could affect the lateral organization in cellular membranes in a way that may be important for the sorting of lipids and proteins within the cell.
Experimental Procedures
Materials
All phospholipids were purchased from Avanti Polar Lipids (Alabaster, AL). PSM was purified from egg yolk sphingomyelin by reverse-phase high performance liquid chromatography (Supelco Discovery C18 column, dimensions 250 × 21.2 mm, 5 μm particle size) using methanol/water (95:5, volume ratio) as the eluent. DPH was obtained from Molecular Probes (Eugene, OR). CTL (cholesta-5,7,9(11)-trien-3-β-ol) was synthesized as described by Fischer et al. (25) and purified by reverse-phase high performance liquid chromatography with acetonitrile/methanol (7:3, vol/vol) as the eluent. The concentrations of the fluorophore stock solutions were determined based on their extinction coefficients (DPH: 88,000 cm−1M−1 at 350 nm in methanol; CTL: 11,250 cm−1M−1 at 324 nm in ethanol). Phospholipid stock solutions were prepared in hexane/2-propanol (3:2, volume ratio). The solutions were stored at −20°C and warmed to ambient temperature before use. The water used in all experiments was purified by reverse osmosis followed by passage through a Millipore UF Plus water purification system (Millipore, Billerica, MA), to yield a product with a resistivity of 18.2 MΩcm.
Partitioning of CTL between mβCD and phospholipid vesicles
Lipid vesicles for the partitioning studies were prepared by mixing phospholipids, CTL (2 mol %), and peptides in organic solvent. The solvent was then evaporated under a constant stream of nitrogen, after which the resulting film was rehydrated above the gel-to-fluid transition of the phospholipids in the samples. The rehydrated samples were then vortexed, and briefly bath-sonicated to form multilamellar vesicles. To create LUVs the multilamellar vesicles were then extruded through a membrane with 200 nm pores. The quality of the resulting LUVs was checked by measuring the vesicle size on a Malvern Zetasizer (Worcestershire, UK). For the partition assay 100 nmol lipids (LUVs) were portioned into 10 glass tubes. mβCD was added to nine of the tubes, after which the solutions were diluted with milli-Q water to a final phospholipid concentration of 40 μM. The final concentration of mβCD in the tubes was 0, 0.04, 0.08, 0.15, 0.25, 0.35, 0.50, 0.60, 0.80, and 1.0 mM.
The samples were then incubated 2 h at 37°C or overnight at 25°C, after which the steady-state anisotropy of CTL was measured (at 37°C or 25°C) on a PTI Quantamaster 1 (Photon Technology International, NJ) spectrofluorimeter operating in the T-format, with both the excitation and emission slits set to 5 nm. The samples were excited at 324 nm and the emission was measured at 390 nm.
The molar concentration of CTL in the LUVs in each sample was calculated from the measured anisotropies according to
| (1) |
where CCTL is the total concentration of CTL in the samples, rLUV is the anisotropy of CTL in the specific phospholipid bilayer, ri is the CTL anisotropy in the sample, and rCD is the anisotropy of CTL in the CTL-mβCD complex. The anisotropy of the CTL-mβCD complex was measured for a range of the CTL-mβCD ratios and was determined to 0.175 at 25°C and 0.170 at 37°C.
The molar fraction partition coefficient KX was calculated as described by Tsamaloukas et al. (22) based on the equation
| (2) |
where CL is the phospholipid concentration, CCD is the cyclodextrin concentration, is the cholesterol concentration in lipid bilayers, and is the concentration of cholesterol in complex with mβCD. The partition coefficients were calculated by plotting the calculated molar concentrations of CTL in the phospholipid bilayers against the mβCD concentration and fitting the obtained curves with the following equation
| (3) |
Determination of the DPH anisotropy in phospholipid bilayers
Samples were prepared by mixing phospholipids, DPH (0.5 mol %) and peptides (0–7 mol %) in organic solvent, after which the solvent was evaporated under a constant stream of nitrogen. The dry films were rehydrated in milli-Q water at a temperature above the gel-to-fluid transition of the phospholipids in the sample. The hydrated samples were then vortexed and bath sonicated briefly to obtain multilamellar vesicles, with a final lipid concentration of 50 μM. The anisotropy of DPH was measured at 25°C and 37°C using the same instrument as in CTL anisotropy measurements, with the slits at 5 nm. All samples were excited at 358 nm and the fluorescence emission was measured at 430 nm.
Results
CTL partitioning between mβCD and phospholipid bilayers
The affinity of sterols for lipid bilayers has been measured successfully previously by measuring how sterols partition between cyclodextrins and lipid bilayers (21–24,26). In the majority of these studies, 3H-cholesterol was used as a probe, and in two studies cholesterol partitioning was measured using isothermal titration calorimetry. A draw back in all the methods used previously is that rather large quantities of material is needed, and that relatively high cholesterol concentrations were used to keep the phospholipid concentrations lower. In this study, we wanted to minimize material use, and especially lower the sterol concentration below domain forming concentrations. Therefore we developed what we believe to be a new fluorescence-based method to determine the affinity of sterols for lipid bilayers with different composition. CTL was chosen as the probe, because it has been shown to be a good analog of cholesterol (27). To avoid formation of sterol rich domains that could complicate the interpretation of the results the amount of sterol in the bilayers were set to 2 mol %.
To calculate partition coefficients the fraction of CTL in LUVs and in complex with mβCD needed to be determined. If the CTL anisotropy in lipid bilayers is different from that of CTL in the cyclodextrin complexes this parameter could be used for this purpose. Therefore, we first measured the anisotropy in CTL mβCD complexes. Complexes were prepared by rehydrating dry CTL films in 0.04, 0.25, 0.5, or 1.0 mM mβCD solutions, with a final CTL concentration of 0.8 μM. The anisotropy of CTL in these complexes was measured and as seen in the results shown in Fig. 1 A the anisotropy was ∼0.175 in the complexes, irrespective of the total CLT-mβCD ratios in the solutions. As the anisotropy of CTL in all studied phospholipid bilayers have been significantly higher than this, we could distinguish between CTL in bilayers and in cyclodextrin complexes by measuring the anisotropy. Next we prepared POPC LUVs containing 2 mol % CTL and prepared a series of samples with 40 mM POPC LUVs and 0–1 mM mβCD. To insure that equilibrium was reached the samples were incubated overnight at 25°C before the fluorescence measurements were carried out. When the anisotropy of CTL in these samples was measured at 25°C a clear relation between mβCD concentration and anisotropy could be seen (Fig. 1 A). As the mβCD concentration increased, the anisotropy decreased toward the anisotropy observed in the CTL-mβCD complexes. This indicated the CTL was being removed from the lipid bilayers by mβCD.
Figure 1.

Representative data that describes the partition method. (A) The anisotropy of CTL in POPC bilayers and in mβCD complexes was measured as a function of mβCD concentration at 25°C. (B) Using Eq. 1 the concentration of CTL in lipid bilayers can be calculated from the anisotropy data and by fitting the data with Eq. 3 the partition coefficient and the stoichiometry of the mβCD-CTL complexes are obtained.
The recorded anisotropy data could then be used to calculate the fraction of CTL in the LUVs in the presence of varying mβCD concentrations using Eq. 1. The resulting data is shown in Fig. 1 B. To obtain the molar fraction partition coefficients for CTL partitioning between LUVs and mβCD, and the stoichiometry of the CTL-mβCD complexes the data in Fig. 1 B was fitted with Eq. 3. As is clear from the figure, the data fit well with this model for the partitioning. The stoichiometry of the CTL-mβCD complexes was 2 according to the fit, in agreement with what has been shown previously for cholesterol-mβCD complexes (22). The partition coefficient was 6.5 mM, which was much lower than has been observed for cholesterol (22,23), but is in agreement with the previous observation that CTL is removed much faster both from pure sterol and phospholipid-sterol monolayers by β-cyclodextrin than cholesterol (28).
Lipid effects on CTL partitioning
To further test the method and to learn more about what determines the affinity of sterols for phospholipid bilayers we tested how CTL partitioning between LUVs and mβCD was influenced by the lipid composition in the LUVs. Because previous studies offered data on cholesterol partitioning between POPC/PSM LUVs and mβCD, we also studied this system with our method to be able to compare how CTL and cholesterol interactions with phospholipid bilayers are affected by changes in the lipid composition.
LUVs of POPC and 0, 33, or 50 mol % PSM were prepared and CTL partitioning was studied at 25°C and 37°C. The results from measurements at 37°C are shown in Fig. S2 and Fig. S3 in the Supporting Material. As expected, the addition of PSM increased the affinity of CTL for the lipid bilayers both at 25°C and 37°C. Hence, it seems that CTL responds to changes in bilayer composition similarly as cholesterol.
To further test how the affinity of the sterol is affected by lipid bilayer composition, we measured CTL partitioning between mβCD and DLPC, DTPC, and DMPC bilayers at 25°C and 37°C, i.e., in fluid bilayers composed of saturated acyl chains. The results from the experiments at 37°C are shown in Fig. S2. The results show that the affinity of CTL for phospholipid bilayers depend on bilayer thickness as KX increased at both temperatures with increasing acyl chain length in the phospholipids.
How cholesterol interacts with phospholipids has been shown to correlate well with the chain order in the lipid bilayers (23). To see whether CTL partitioning has a similar correlation on chain order we measured DPH anisotropy in all different lipid compositions used in the partition studies. Fig. S3 shows the correlation between KX and the anisotropy reported by DPH at both 25°C and 37°C, and clearly there is a fairly good correlation between KX and DPH anisotropy. A bonus from using CTL in the partition assay is the recorded anisotropy values for the sterol in different phospholipid environments. Hence, it is possible to evaluate also how the order of the sterol in different bilayers correlates with KX. From Fig. S3, it is clear that KX also correlates with the anisotropy of the sterol, i.e., the more ordered the bilayer surrounding the sterol the higher its affinity for the bilayer.
In conclusion, the results from studies of CTL affinity for different phospholipid bilayers shows that CTL senses phospholipid structure similarly to cholesterol, suggesting that results obtained with the probe can be used to evaluate cholesterol interactions with phospholipids.
The effect of peptides on CTL partitioning
To be able to correlate the results from the partitioning assay with the acyl chain order in the lipid bilayers with and without peptides we measured chain order in the samples. The effect of peptides on the acyl chain order in lipid bilayers was determined by measuring the anisotropy of two probes, CTL and DPH, in DMPC bilayers containing 0–7 mol % peptide (Fig. 2). Inclusion of WALP23 led to an increased order in the bilayers as seen from both the CTL (Fig. 2 A) and DPH (Fig. 2 B) anisotropy. The effect of KALP23 was much smaller, but also for this peptide the anisotropy of both probes was increased slightly with increasing peptide concentration.
Figure 2.
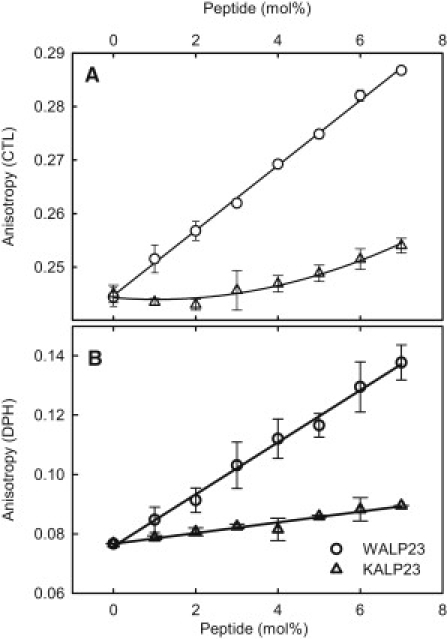
Effect of peptide inclusion on the acyl chain order in the bilayers. The steady-state anisotropy of (A) CTL and (B) DPH was measured in DMPC bilayers with and without peptides at 37°C.
Having determined the ordering effect of the peptide, the next step was to determine how this effect influenced sterol affinity for the DMPC bilayer. This was done using the same approach as for pure lipid systems, except that 0–7 mol % of peptide was added to the DMPC bilayers. After the samples had been incubated 2 h at 37°C, the equilibrium distribution of CTL between LUVs and mβCD was determined from CTL anisotropy measurements. First the effect of WALP23 was studied. Representative data are shown in Fig. 3 A, and as can be seen in the figure an increasing amount of WALP23 in the bilayers led to less efflux of CTL from the bilayers. The calculated partition coefficients are shown in Fig. 3 B. As the figure shows, KX increased with increasing WALP23 concentration, reaching a plateau at ∼5 mol % WALP23. This indicates that WALP23 addition increased the affinity of sterols for the bilayer.
Figure 3.
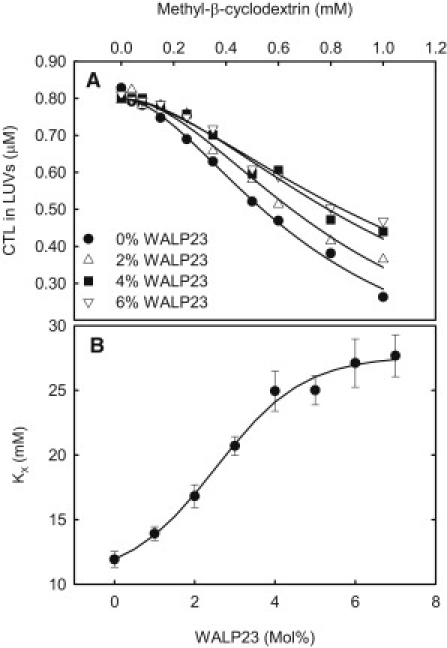
Influence of WALP23 on sterol partitioning between DMPC bilayers and mβCD at 37°C. (A) Representative data showing the effect of WALP23 on the partitioning of CTL between DMPC bilayer and mβCD. (B) Summary of the calculated partition coefficients.
To get further information about how WALP23 influences the sterol affinity for phospholipid bilayers similar experiments were carried out in DLPC and POPC bilayers. The results from these experiments showed that the influence of WALP23 (2 or 5 mol %) on KX depended on the phospholipid composition in the bilayer (Fig. 4). In both DLPC and POPC bilayers the addition of WALP23 lead to an increased acyl chain order as seen from DPH and CTL anisotropy (Fig. 4 and Fig. S4). KX increased on inclusion of the peptide, i.e., CTL bound stronger to the bilayer in the presence of the peptide. However, the effect was much smaller than with DMPC bilayers. Furthermore, there was a smaller effect of increased acyl chain order, as probed with DPH anisotropy, on KX in DLPC and POPC bilayers than in DMPC bilayers. This suggests that to what degree proteins affect sterol-phospholipid interactions depends on phospholipid structure.
Figure 4.
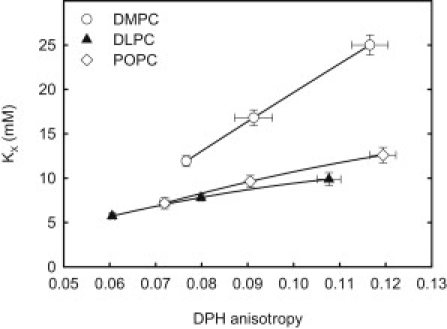
Relationship between measured partition coefficients and the acyl chain order in WALP23 containing bilayers. The anisotropy of DPH and partitioning of CTL was measured in DMPC, DLPC, and POPC bilayers in the presence of 0, 2, or 5 mol % WALP23 at 37°C.
The effect of KALP23 on CTL partitioning between LUVs and mβCD was studied next. This was done in the same way as with WALP23 and representative data is shown in Fig. 5 A. The calculated partition coefficients are shown in Fig. 5 B. As can be seen from the figure, the addition of KALP23 did not increase the sterol's affinity for the bilayer. Instead, KALP23 inclusion led to decreased CTL partitioning into phospholipid bilayers. By doing sucrose gradient centrifugation experiments, as described by Killian et al. (29), we insured that this was not due to nonhomogenous incorporation of KALP23 in DMPC bilayers (results not shown). Hence, we can conclude that addition of peptides, mimicking the transmembrane helices in proteins, to phospholipid bilayers can have either a positive or a negative effect on the affinity of sterols for the bilayers.
Figure 5.
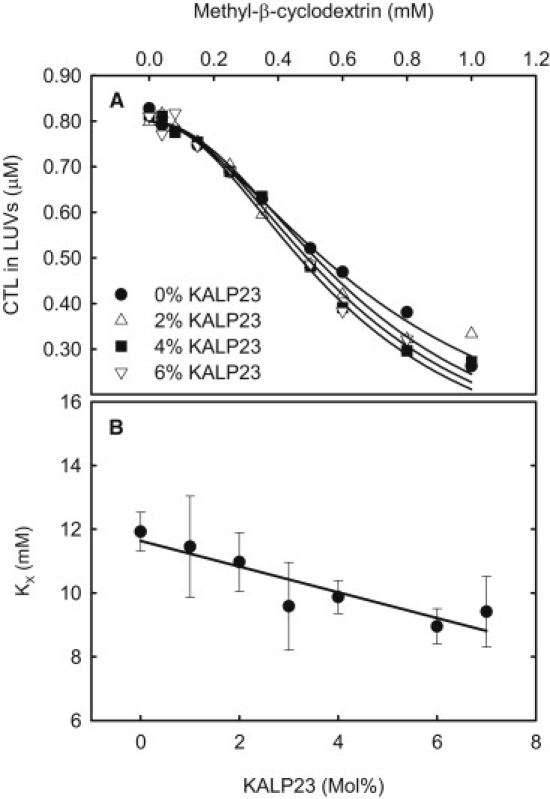
Influence of KALP23 on sterol partitioning between DMPC bilayers and mβCD at 37°C. (A) Representative data showing the effect of KALP23 on the partitioning of CTL between DMPC bilayer and mβCD. (B) Summary of the calculated partition coefficients.
Discussion
Cholesterol interactions with phospholipid bilayers
CTL has been shown to be the fluorescent cholesterol analog that mimics cholesterol best (27). For example it was reported that CTL orders the surrounding lipid acyl chains and interacts with phospholipids similarly as cholesterol. Therefore, CTL has been used in studies of cholesterol enriched domains (30,31). In this study, we have further characterized CTL-phospholipid interactions by measuring the equilibrium portioning of the probe between phospholipid vesicles and mβCD. The first apparent difference between cholesterol and CTL that we observed was that CTL partitioned more into mβCD than cholesterol. For example, KX at 37°C with pure POPC bilayers was 6.5 mM for CTL whereas it has been reported to be 30–47 mM for cholesterol (21–23). This finding was not surprising because it has been shown that rate of CTL efflux from monolayers is markedly higher than that of cholesterol (28). We think that this effect is mostly due to differences in sterol-mβCD interactions, and not so much dependent on sterol-phospholipid interactions.
To compare how CTL and cholesterol interacts with phospholipids one must compare how the sterol partitioning behavior is affected by changes in phospholipid composition. For this purpose, we studied how CTL partitioning was affected by inclusion of PSM into POPC bilayers. It has been reported previously that the addition of 50 mol % PSM to POPC bilayers increases KX for cholesterol three to four times at 37°C (21,24). For CTL the KX was increased approximately six times by PSM addition at the same temperature. The higher relative partition coefficient could mean that CTL has a higher affinity for PSM containing bilayers than cholesterol, but we think that the increase was due to a markedly lower sterol concentration (2 mol %) in the bilayers than in the cholesterol studies (15–30 mol %). This presumption is based on observations by Tsamaloukas and co-workers regarding sterol content and partitioning behavior (24). Addition of 33 mol % PSM to POPC bilayers increased KX for CTL 4 times. This is closely matching what previously has been observed for cholesterol in the same systems (23). Further, we confirmed that CTL, like cholesterol, has an increased affinity for phospholipid bilayers with increasing acyl chain order. In conclusion, we consider CTL to mimic the membrane properties of cholesterol well enough that CTL data can be used to make predictions regarding cholesterol partition between lipid bilayers and mβCD.
Effect of transmembrane peptides on sterol partitioning
It is well known that cholesterol likes lipids that form ordered bilayers. We showed recently that the affinity of cholesterol for phospholipid bilayers is more or less directly dependent on the acyl chain order in lipid bilayers (23). In this study, we showed that CTL affinity for bilayers also depends on the acyl chain order as seen from DPH anisotropy (Fig. S2). Similar results were also obtained in a recent molecular dynamics study of cholesterol interactions with phospholipid bilayers (32). In this study, the area per lipid correlates fairly well with the free energy barriers for cholesterol desorption.
Model peptide studies have shown that on positive hydrophobic mismatch, i.e., when the hydrophobic length of transmembrane helices exceed the hydrophobic thickness of phospholipid bilayers, the acyl chains in lipids next to the peptides are stretched to minimize the hydrophobic mismatch (33). This indicates that membrane proteins can influence the chain order in lipid bilayers, and perhaps thereby influence the cholesterol distribution within the bilayer. In this study, we studied how inclusion of two transmembrane peptides, WALP23 and KALP23, influenced the affinity of CTL for the lipid bilayers. Both WALP23 and KALP23 affected the chain order (as seen from CTL and DPH anisotropy) as could be expected based on previous studies (34). WALP23 clearly increased the chain order in the bilayers whereas KALP23 had only a small ordering effect on the acyl chain order (Fig. 2).
Addition of WALP23 to DMPC bilayers clearly increased the affinity of CTL for the bilayers (Fig. 3). The affinity increased with peptide concentration and reached a plateau at ∼5 mol % WALP23. As with the peptide free bilayers, there was a clear correlation between the chain order in the bilayers and the sterol affinity for the bilayer up to 5 mol % WALP23. At higher WALP23 concentrations the partition coefficients did not increase although the order in the bilayer was increased up to the highest used peptide concentration according to both CTL and DPH anisotropy. It has been estimated that the first shell of phospholipids surrounding a WALP23 peptide is ∼10 lipids/monolayer (33). Therefore, all phospholipids in the bilayer are likely to be in contact with WALP23 with 5 mol % of the peptide in the bilayers. Hence, it seems that the maximum effect of WALP23 on the CTL partitioning is reached at a concentration at which all DMPC molecules are in contact with the peptide i.e., when there is at most a single shell of lipids between the peptide transmembrane segments.
In POPC and DLPC bilayers the addition of WALP23 also increased the acyl chain order and the affinity of CTL for the bilayers (Fig. 4). In both POPC and DLPC bilayers the effect of WALP23 on the chain order (as seen from DPH anisotropy) was similar to that in DMPC however the effect on KX was small compared to that in DMPC bilayers (Fig. 4). We conclude that if the chains are too short or unsaturated the CTL (and cholesterol) will not interact more much favorably with the lipids although WALP23 orders and stretches the chains. This is in agreement with the general view that cholesterol interacts favorably with phospholipids that have long saturated acyl chains.
Addition of KALP23 to DMPC bilayers decreased the affinity of CTL for the bilayers although the peptide at least at higher concentrations had a small ordering effect on the bilayers (Fig. 5). Although KALP23 is built up of the same number of amino acids as WALP23, it has been shown that it has a shorter effective hydrophobic length than WALP23 (35). It was estimated that KALP23 has an effective hydrophobic length in between a WALP16 and a WALP19, i.e., a 6–10 Å shorter than WALP23. The hydrophobic length of KALP23 is estimated to be a close match with the hydrophobic thickness of fluid DMPC bilayers (36). KALP23 has also been shown induced cubic phase in di-16:1-PE that forms bilayers that are slightly thicker than DMPC (35). This indicates that there is negative hydrophobic mismatch between KALP23 and di-16:1-PE. As the lysine side chains are thought to prefer a position no deeper in the bilayer than the lipid phosphate region (34) it is possible that there may be a slight negative hydrophobic mismatch between KALP23 and fluid DMPC bilayers. Because there should be a positive hydrophobic mismatch between WALP23 and fluid DMPC bilayer we think that the fact that WALP23 and KALP23 had the opposite effect on KX is related to the difference in effective hydrophobic length of the peptides.
Biological implication
The sorting of transmembrane proteins within cells have been indicated to be dependent on the cholesterol gradient in cellular membranes (8). It is believed that the membrane thickening effect of cholesterol can be an important factor that guides proteins to their destination, and cholesterol is also known to form so called membrane rafts together with sphingolipids. These rafts have been indicated to have a role in lipid trafficking in cells (37).We think colocalization of proteins and cholesterol in cell membranes could be an important step in membrane trafficking. The fact that peptides mimicking proteins transmembrane segments can increase sterol affinity for a bilayer, or decrease it, indicates that protein can attract or expel cholesterol from the phospholipid environment in which they are embedded. An important factor seems to be how proteins influence the chain order, and thereby the thickness of the surrounding lipid bilayer. As proteins that are destined for the plasma membrane on average have longer transmembrane helices than those that reside in the inner membrane compartments (8) we predict that these proteins may have an ordering effect on the Golgi membranes, and thereby may attract cholesterol. It has been predicted that proteins with larger cross-sectional size, i.e., several transmembrane helices, have a larger effect on the surrounding lipid bilayer (38). Hence, such proteins may also affect sterol affinity for bilayers even more than single-spanning helices. The results in this study also showed that proteins modulate the affinity of sterols the most for phospholipids with longer saturated acyl chains. Therefore, it seems that proteins interactions with phospholipids can favor formation of lateral domains enriched in saturated lipids, like sphingomyelin, and cholesterol. This could lead to a segregation of lipids and protein in the bilayer plane that can be an initial step in the sorting of the membrane components.
Conclusions
In this study, we developed what we believe to be a new method that can be used to measure sterol affinity for phospholipid bilayers. The method uses the fluorescent cholesterol analog CTL and we showed that the probe interacted similarly with phospholipids as cholesterol. This method was then used to study how peptides mimicking the transmembrane helices of proteins affect the affinity of sterols for phospholipid bilayers. We showed that peptides could both increase and reduce sterol affinity for phospholipid bilayers, and that the degree to which peptides affected sterol affinity also depended on the lipid composing in the bilayers. The fact that the sterol affinity for the bilayers was altered by peptides suggests that proteins can affect the lateral organization of cholesterol in cell membrane. It is likely that this influence of proteins on lipid organization can play a role in the trafficking of lipids and proteins within cellular membranes.
Supporting Material
Four figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(10)00550-3.
Supporting Material
Acknowledgments
The authors thank J. Peter Slotte and Bodil Westerlund for fruitful discussion and J. Peter Slotte for access to excellent laboratory facilities.
This study was supported by the Academy of Finland, the Sigrid Juselius Foundation, Medicinska Understödsföreningen Liv och Hälsa R.F., the Magnus Ehrnrooth Foundation, and the Otto Malm Foundation.
References
- 1.Demel R.A., Jansen J.W., van Deenen L.L. The preferential interaction of cholesterol with different classes of phospholipids. Biochim. Biophys. Acta. 1977;465:1–10. doi: 10.1016/0005-2736(77)90350-9. [DOI] [PubMed] [Google Scholar]
- 2.van Dijck P.W.M. Negatively charged phospholipids and their position in the cholesterol affinity sequence. Biochim. Biophys. Acta. 1979;555:89–101. doi: 10.1016/0005-2736(79)90074-9. [DOI] [PubMed] [Google Scholar]
- 3.Thomas P.D., Poznansky M.J. Cholesterol transfer between lipid vesicles. Effect of phospholipids and gangliosides. Biochem. J. 1988;251:55–61. doi: 10.1042/bj2510055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ohvo-Rekilä H., Ramstedt B., Slotte J.P. Cholesterol interactions with phospholipids in membranes. Prog. Lipid Res. 2002;41:66–97. doi: 10.1016/s0163-7827(01)00020-0. [DOI] [PubMed] [Google Scholar]
- 5.de Almeida R.F., Fedorov A., Prieto M. Sphingomyelin/phosphatidylcholine/cholesterol phase diagram: boundaries and composition of lipid rafts. Biophys. J. 2003;85:2406–2416. doi: 10.1016/s0006-3495(03)74664-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Veatch S.L., Keller S.L. Miscibility phase diagrams of giant vesicles containing sphingomyelin. Phys. Rev. Lett. 2005;94:148101. doi: 10.1103/PhysRevLett.94.148101. [DOI] [PubMed] [Google Scholar]
- 7.Holthuis J.C., van Meer G., Huitema K. Lipid microdomains, lipid translocation and the organization of intracellular membrane transport (Review) Mol. Membr. Biol. 2003;20:231–241. (Review) [PubMed] [Google Scholar]
- 8.Bretscher M.S., Munro S. Cholesterol and the Golgi apparatus. Science. 1993;261:1280–1281. doi: 10.1126/science.8362242. [DOI] [PubMed] [Google Scholar]
- 9.Cole N.B., Ellenberg J., Lippincott-Schwartz J. Retrograde transport of Golgi-localized proteins to the ER. J. Cell Biol. 1998;140:1–15. doi: 10.1083/jcb.140.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Munro S. Sequences within and adjacent to the transmembrane segment of alpha-2,6-sialyltransferase specify Golgi retention. EMBO J. 1991;10:3577–3588. doi: 10.1002/j.1460-2075.1991.tb04924.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Masibay A.S., Balaji P.V., Qasba P.K. Mutational analysis of the Golgi retention signal of bovine beta-1,4-galactosyltransferase. J. Biol. Chem. 1993;268:9908–9916. [PubMed] [Google Scholar]
- 12.Lundbaek J.A., Andersen O.S., Nielsen C. Cholesterol-induced protein sorting: an analysis of energetic feasibility. Biophys. J. 2003;84:2080–2089. doi: 10.1016/S0006-3495(03)75015-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Planque M.R., Killian J.A. Protein-lipid interactions studied with designed transmembrane peptides: role of hydrophobic matching and interfacial anchoring. Mol. Membr. Biol. 2003;20:271–284. doi: 10.1080/09687680310001605352. [DOI] [PubMed] [Google Scholar]
- 14.Lee A.G. How lipids and proteins interact in a membrane: a molecular approach. Mol. Biosyst. 2005;1:203–212. doi: 10.1039/b504527d. [DOI] [PubMed] [Google Scholar]
- 15.Marsh D. Protein modulation of lipids, and vice-versa, in membranes. Biochim. Biophys. Acta. 2008;1778:1545–1575. doi: 10.1016/j.bbamem.2008.01.015. [DOI] [PubMed] [Google Scholar]
- 16.Williamson I.M., Alvis S.J., Lee A.G. Interactions of phospholipids with the potassium channel KcsA. Biophys. J. 2002;83:2026–2038. doi: 10.1016/S0006-3495(02)73964-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O'Keeffe A.H., East J.M., Lee A.G. Selectivity in lipid binding to the bacterial outer membrane protein OmpF. Biophys. J. 2000;79:2066–2074. doi: 10.1016/S0006-3495(00)76454-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lehtonen J.Y., Kinnunen P.K. Evidence for phospholipid microdomain formation in liquid crystalline liposomes reconstituted with Escherichia coli lactose permease. Biophys. J. 1997;72:1247–1257. doi: 10.1016/S0006-3495(97)78771-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fernandes F., Loura L.M., Hemminga M.A. Dependence of M13 major coat protein oligomerization and lateral segregation on bilayer composition. Biophys. J. 2003;85:2430–2441. doi: 10.1016/s0006-3495(03)74666-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fernandes F., Loura L.M., Prieto M. Quantification of protein-lipid selectivity using FRET: application to the M13 major coat protein. Biophys. J. 2004;87:344–352. doi: 10.1529/biophysj.104.040337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niu S.L., Litman B.J. Determination of membrane cholesterol partition coefficient using a lipid vesicle-cyclodextrin binary system: effect of phospholipid acyl chain unsaturation and headgroup composition. Biophys. J. 2002;83:3408–3415. doi: 10.1016/S0006-3495(02)75340-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsamaloukas A., Szadkowska H., Heerklotz H. Interactions of cholesterol with lipid membranes and cyclodextrin characterized by calorimetry. Biophys. J. 2005;89:1109–1119. doi: 10.1529/biophysj.105.061846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Halling K.K., Ramstedt B., Nyholm T.K. Cholesterol interactions with fluid-phase phospholipids: effect on the lateral organization of the bilayer. Biophys. J. 2008;95:3861–3871. doi: 10.1529/biophysj.108.133744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsamaloukas A., Szadkowska H., Heerklotz H. Thermodynamic comparison of the interactions of cholesterol with unsaturated phospholipid and sphingomyelins. Biophys. J. 2006;90:4479–4487. doi: 10.1529/biophysj.105.080127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fischer R.T., Stephenson F.A., Schroeder F. delta 5,7,9(11)-Cholestatrien-3 beta-ol: a fluorescent cholesterol analogue. Chem. Phys. Lipids. 1984;36:1–14. doi: 10.1016/0009-3084(84)90086-0. [DOI] [PubMed] [Google Scholar]
- 26.Leventis R., Silvius J.R. Use of cyclodextrins to monitor transbilayer movement and differential lipid affinities of cholesterol. Biophys. J. 2001;81:2257–2267. doi: 10.1016/S0006-3495(01)75873-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scheidt H.A., Muller P., Huster D. The potential of fluorescent and spin-labeled steroid analogs to mimic natural cholesterol. J. Biol. Chem. 2003;278:45563–45569. doi: 10.1074/jbc.M303567200. [DOI] [PubMed] [Google Scholar]
- 28.Ohvo-Rekilä H., Akerlund B., Slotte J.P. Cyclodextrin-catalyzed extraction of fluorescent sterols from monolayer membranes and small unilamellar vesicles. Chem. Phys. Lipids. 2000;105:167–178. doi: 10.1016/s0009-3084(00)00122-5. [DOI] [PubMed] [Google Scholar]
- 29.Killian J.A., Salemink I., Greathouse D.V. Induction of nonbilayer structures in diacylphosphatidylcholine model membranes by transmembrane alpha-helical peptides: importance of hydrophobic mismatch and proposed role of tryptophans. Biochemistry. 1996;35:1037–1045. doi: 10.1021/bi9519258. [DOI] [PubMed] [Google Scholar]
- 30.Alanko S.M., Halling K.K., Ramstedt B. Displacement of sterols from sterol/sphingomyelin domains in fluid bilayer membranes by competing molecules. Biochim. Biophys. Acta. 2005;1715:111–121. doi: 10.1016/j.bbamem.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 31.Björkqvist Y.J., Nyholm T.K., Ramstedt B. Domain formation and stability in complex lipid bilayers as reported by cholestatrienol. Biophys. J. 2005;88:4054–4063. doi: 10.1529/biophysj.104.054718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bennett W.F., MacCallum J.L., Tieleman D.P. Molecular view of cholesterol flip-flop and chemical potential in different membrane environments. J. Am. Chem. Soc. 2009;131:12714–12720. doi: 10.1021/ja903529f. [DOI] [PubMed] [Google Scholar]
- 33.de Planque M.R., Greathouse D.V., Killian J.A. Influence of lipid/peptide hydrophobic mismatch on the thickness of diacylphosphatidylcholine bilayers. A 2H NMR and ESR study using designed transmembrane alpha-helical peptides and gramicidin A. Biochemistry. 1998;37:9333–9345. doi: 10.1021/bi980233r. [DOI] [PubMed] [Google Scholar]
- 34.de Planque M.R., Kruijtzer J.A., Killian J.A. Different membrane anchoring positions of tryptophan and lysine in synthetic transmembrane alpha-helical peptides. J. Biol. Chem. 1999;274:20839–20846. doi: 10.1074/jbc.274.30.20839. [DOI] [PubMed] [Google Scholar]
- 35.Strandberg E., Morein S., Killian J.A. Lipid dependence of membrane anchoring properties and snorkeling behavior of aromatic and charged residues in transmembrane peptides. Biochemistry. 2002;41:7190–7198. doi: 10.1021/bi012047i. [DOI] [PubMed] [Google Scholar]
- 36.Kandasamy S.K., Larson R.G. Molecular dynamics simulations of model trans-membrane peptides in lipid bilayers: a systematic investigation of hydrophobic mismatch. Biophys. J. 2006;90:2326–2343. doi: 10.1529/biophysj.105.073395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klemm R.W., Ejsing C.S., Simons K. Segregation of sphingolipids and sterols during formation of secretory vesicles at the trans-Golgi network. J. Cell Biol. 2009;185:601–612. doi: 10.1083/jcb.200901145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Venturoli M., Smit B., Sperotto M.M. Simulation studies of protein-induced bilayer deformations, and lipid-induced protein tilting, on a mesoscopic model for lipid bilayers with embedded proteins. Biophys. J. 2005;88:1778–1798. doi: 10.1529/biophysj.104.050849. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


