Introduction
Molecular recognition in biological systems relies on the existence of specific attractive interactions between two partner molecules. Structure-based drug design seeks to identify and optimize such interactions between ligands and their host molecules, typically proteins, given their three-dimensional structures. This optimization process requires knowledge about interaction geometries and approximate affinity contributions of attractive interactions that can be gleaned from crystal structure and associated affinity data.
Here we compile and review the literature on molecular interactions as it pertains to medicinal chemistry through a combination of careful statistical analysis of the large body of publicly available X-ray structure data and experimental and theoretical studies of specific model systems. We attempt to extract key messages of practical value and complement references with our own searches of the CSDa,(1) and PDB databases.(2) The focus is on direct contacts between ligand and protein functional groups, and we restrict ourselves to those interactions that are most frequent in medicinal chemistry applications. Examples from supramolecular chemistry and quantum mechanical or molecular mechanics calculations are cited where they illustrate a specific point. The application of automated design processes is not covered nor is design of physicochemical properties of molecules such as permeability or solubility.
Throughout this article, we wish to raise the readers’ awareness that formulating rules for molecular interactions is only possible within certain boundaries. The combination of 3D structure analysis with binding free energies does not yield a complete understanding of the energetic contributions of individual interactions. The reasons for this are widely known but not always fully appreciated. While it would be desirable to associate observed interactions with energy terms, we have to accept that molecular interactions behave in a highly nonadditive fashion.3,4 The same interaction may be worth different amounts of free energy in different contexts, and it is very hard to find an objective frame of reference for an interaction, since any change of a molecular structure will have multiple effects. One can easily fall victim to confirmation bias, focusing on what one has observed before and building causal relationships on too few observations. In reality, the multiplicity of interactions present in a single protein−ligand complex is a compromise of attractive and repulsive interactions that is almost impossible to deconvolute. By focusing on observed interactions, one neglects a large part of the thermodynamic cycle represented by a binding free energy: solvation processes, long-range interactions, conformational changes. Also, crystal structure coordinates give misleadingly static views of interactions. In reality a macromolecular complex is not characterized by a single structure but by an ensemble of structures. Changes in the degrees of freedom of both partners during the binding event have a large impact on binding free energy.
The text is organized in the following way. The first section treats general aspects of molecular design: enthalpic and entropic components of binding free energy, flexibility, solvation, and the treatment of individual water molecules, as well as repulsive interactions. The second half of the article is devoted to specific types of interactions, beginning with hydrogen bonds, moving on to weaker polar interactions, and ending with lipophilic interactions between aliphatic and aromatic systems. We show many examples of structure−activity relationships; these are meant as helpful illustrations but individually can never confirm a rule.
General Design Aspects
Entropic and Enthalpic Components of Binding
Like any other spontaneous process, a noncovalent binding event takes place only when it is associated with a negative binding free energy (ΔG), which is the well-known sum of an enthalpic term (ΔH) and an entropic term (−TΔS). These terms may be of equal or opposite sign and thus lead to various thermodynamic signatures of a binding event, ranging from exothermic to entropy-driven. An increasing body of data from isothermal titration calorimetry (ITC) is available on the thermodynamic profiles for many complexes.5,6 Where crystal structure information exists as well, it is tempting to speculate about the link between thermodynamics and geometry of protein−ligand complexes. A rough correlation between the burial of apolar surface area and free energy could be derived,(5) but beyond that, practically useful relationships between structure and the components of free energy have remained elusive. This is not surprising, as both entropy and enthalpy terms obtained from calorimetric experiments contain solute and solvent contributions and thus cannot be interpreted on the basis of structural data alone. The direct experimental estimation of solvent effects has been attempted(7) but always requires additional assumptions. Only theoretical treatments allow a separation of these effects.(8) Thus, computer modeling can support the interpretation of experimental observations.
Furthermore, from an experimental point of view, such studies are very demanding: Error margins are significantly larger than the fitting errors usually reported(9) and depend on subtle details of experimental setup.(10) Entropy and enthalpy values are sensitive to conditions such as salt concentrations and choice of buffer. Protonation and deprotonation steps associated with the binding need to be understood in detail,(11) as significant differences in protonation states can occur even within congeneric series of ligands.(12)
Freire suggested that best-in-class drugs bind to their targets in an enthalpy-driven fashion.(13) His group established a comprehensive ITC data set on HIV protease inhibitors, concluding that second- and third-generation inhibitors bind through a strong enthalpy component. One may argue that this could be caused by a higher number of specific backbone interactions, partially replacing the larger lipophilic surface area of the first generation inhibitors, and that the larger reliance on backbone interactions could be the cause for the broader coverage of HIV protease mutants. Still, even in this data set small structural changes cause large difference in relative ΔH and TΔS contributions that are hard to explain structurally. Amprenavir and darunavir differ only in one cyclic ether moiety, but there is a drastic difference of about 10 kcal/mol in binding enthalpy between these two ligands.(14)
Since entropic and enthalpic components of binding are highly dependent on many system-specific properties, the practitioner has to conclude that optimizing for free energy is still the only viable approach to structure-based design. Perhaps the greatest advantage in the attempt to interpret components of ΔG is that it forces us to think about two fundamental topics in unprecedented detail: viewing protein−ligand complexes as flexible entities rather than fixed structures and the role of desolvation effects. These two topics will be discussed next.
Flexibility and Cooperativity
A discussion of the thermodynamics of ligand binding is not complete without mentioning the phenomenon of entropy−enthalpy compensation. The validity and generality of this phenomenon have been a contentious topic for many years. There is ample evidence of meaningless and spurious correlations between ΔH and TΔS, often due to far larger variations (sometimes through larger experimental errors) in ΔH than in ΔG.15,16 Also, compensation is no thermodynamic requirement:17,18 If changes in ΔH were always compensated by opposing changes in TΔS, optimization of binding affinities would hardly be possible.
Nevertheless, there seems to be a mechanism behind the compensation frequently observed(19) in host−guest chemistry(20) and in protein−ligand interactions.(21) In short, the tighter and more directed an interaction, the less entropically favorable it is. Bonding opposes motion, and motion opposes bonding.(22) However, the detailed nature of these compensatory mechanisms is highly system-dependent and the mechanisms do not obey a single functional form. As a consequence, noncovalent interactions can be positively cooperative; that is, the binding energy associated with their acting together is greater than the sum of the individual binding free energies. Detailed studies by Williams on glycopeptide antibiotics indicate that cooperativity may be caused by structural tightening upon introduction of additional interactions; interaction distances become shorter and enthalpically more favorable. Evidence for such effects also exists in protein−protein(23) and protein−ligand complexes. Streptavidin becomes better packed upon binding to biotin as evident from structural, mass spectrometry, and deuterium exchange NMR experiments.(24) Hunter has proposed an alternative model of cooperativity.(25) Flexible molecules may exist in a series of partially bound states, where not all interactions are made at the same time. Upon introduction of further interactions, the balance shifts toward more frequent formation of the interactions and less mobility. There is experimental evidence for both the structural tightening model(26) and the model of partially bound states(27) in different systems. Since proteins bind ligands in many geometric arrangements ranging from loosely surface-bound to deeply buried, it is likely that both are valid in different contexts. Ligands with increasingly longer linear and lipophilic side chains have been observed to bind with increasingly favorable entropies and increasingly unfavorable enthalpies against trypsin(28) and carbonic anhydrase.(29) Longer side chains retain more residual mobility and form less tight (or less frequent) interactions with the protein surface, trading enthalpy for entropy.
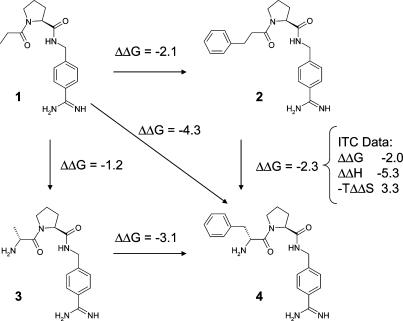
In an impressively comprehensive study on several series of thrombin inhibitors, the Klebe group has analyzed the sources of cooperativity between a lipophilic interaction and a hydrogen bond.30,31 The findings from this study can be summarized as follows: (i) In the presence of the amino substituent forming a hydrogen bond to the backbone of Gly216, the gain in binding free energy of a given P3 substituent is significantly larger. This is illustrated by the double functional group replacement cycle in Figure 1. (ii) Both B-factor analyses and MD studies confirm a decrease in residual motion of the P3 substituent in the presence of the hydrogen bond. MD studies also indicate that the average hydrogen bond distance is shorter with a P3 substituent attached. (iii) In accordance with this finding, the introduction of the hydrogen bond makes bonding significantly more exothermic, but the gain in enthalpy is largely compensated by an entropic disadvantage.
Figure 1.
Cooperativity of hydrogen bond formation and hydrophobic contacts in a set of thrombin inhibitors. Extension of the lipophilic side chain alone increases affinity by 2.1 kcal/mol. Addition of the amino group increases affinity by 1.2 kcal/mol. Cooperativity therefore amounts to 4.3 − 2.1 − 1.2 = 1.0 kcal/mol. Data from refs (30) and (31) were converted to kcal/mol and rounded to 1 decimal place.
What can we learn from this study? First, small changes in ΔG often mask large and mutually compensating changes in ΔH and TΔS. Focusing on ΔG in designing new molecules is certainly still the safest bet. Second, in medicinal chemistry we often rely on cooperative effects without even noticing, and yet in our minds we attempt to decompose binding free energies into additive elements. There is nothing wrong with this empirical approach as long as we keep in mind that it is primarily useful to teach us about the limits of additivity. The knowledge that specific interactions, in particular strongly directed ones like hydrogen bonds, rigidify a protein−ligand complex may help us to exploit cooperativity in a more rational fashion. The Klebe data should also make us think about the degree of mobility required in interactions with different parts of the protein. Flexible domains may require more flexible ligand moieties than highly ordered ones. The thermodynamic signature of a “good” ligand is not necessarily dominated by an enthalpic term. Our traditional focus on visible interactions and on the induced fit model has led to an overly enthalpic view of the world that neglects flexibility and cooperativity. It also neglects local solvation phenomena, whose details strongly affect the thermodynamic profile of a molecular recognition event. We should learn to incorporate these into our thinking, at least in a qualitative fashion.
Desolvation and the Hydrophobic Effect
The classic concept of the hydrophobic effect is as follows: A hydrophobic solute disrupts the structure of bulk water and decreases entropy because of stronger bonding and ordering of water molecules around the solute. Such disruptions can be minimized if nonpolar solute molecules aggregate. Water then forms one larger “cage” structure around the combined solutes, whose surface area will be smaller than the combined surface areas of isolated solutes. This maximizes the amount of free water and thus the entropy.
If this mechanism were the sole driving force for a protein−ligand interaction, all binding events involving hydrophobic partners would be entropy-driven. But spectroscopic evidence indicates that hydrogen bonds at hydrophobic surfaces are weaker(32) and water molecules more flexible(33) than originally assumed. In addition, already simple water models show that size and surface curvature of solutes have a dramatic impact on their solvation thermodynamics.(34) Complexation thermodynamics driven by enthalpic forces have been regularly observed in host−guest chemistry and have been dubbed the “nonclassical hydrophobic effect”.(35) Homans has carefully studied the ligand-binding thermodynamics of the mouse major urinary protein (MUP). The key to the extremely favorable enthalpy of binding of ligands to MUP seems to be the suboptimal hydration of the binding pocket in the unbound state.(36) The negative change in heat capacity upon binding, a hallmark of the hydrophobic effect, is also observed with MUP ligands. It is largely determined by ligand desolvation, with minor contribution from desolvation of the protein.(37) Similar observations have been made for hydrophobic cavities in other proteins.(38) It is quite likely that the synthetic hosts displaying an enthalpy-driven complexation behavior are also incompletely hydrated in the unbound state because they typically feature narrow lipophilic clefts not unlike that of MUP (see ref (39) for a recent example). Desolvation costs are also the likely cause why some ligands that seem to fit into a binding site cannot experimentally be confirmed to be inhibitors. An ITC study on the interactions between simple benzamidines and trypsin concludes that the unfavorable desolvation of the oxyanion hole area upon binding of p-tert-butylbenzamidinium causes the observed complete loss of binding affinity for this molecule.(28)
From these studies it becomes clear that carefully probing the hydration state of a protein binding pocket in the unbound state should be a standard element of structure-based design, as it will likely point out regions where most binding energy can be gained. Failure to match key hydrophobic areas with appropriate ligand moieties can have a severe impact on binding affinity. Figure 2 shows that an optimized factor Xa inhibitor is rendered virtually inactive when the key interacting group in the S4 pocket is replaced by hydrogen.(40) Not only does the isopropyl group optimally interact with the S4 pocket (see Hydrophobic Interactions) but its removal leads to a very unfavorably solvated state of the pocket41,42 that is likely the main reason for the dramatic loss of affinity.
Figure 2.

Binding mode of a factor Xa inhibitor from GSK.(40) The depicted compound (PDB code 2j4i) has a Ki of 1 nM. Replacing the isopropyl group (marked in red) by hydrogen reduces the affinity to 39 μM.
Structural Water
Any ligand binding event displaces water molecules from the binding site. In structure-based design, most of these are never explicitly considered because they are highly disordered and therefore rarely crystallographically observed.(43) Enzyme binding sites are in fact characterized by easily displaceable water, as shown by Ringe in her seminal work on transferring protein crystals to organic solvents.(44) Those water molecules that are observed need to be carefully analyzed; they might be replaceable or they might be considered as part of the protein structure.(45) Analyses of high-resolution crystal structures by Poornima and Dean showed that protein−ligand contacts are often mediated by water molecules situated in deep grooves in the binding site and forming multiple hydrogen bonds with both binding partners.(46) Relibase+, a database and query system for analyzing protein−ligand complexes, contains a module that allows a rigorous assessment of water structure in binding sites.(47)
The release of a water molecule from a rigid environment should be entropically favorable. In a classic contribution, Dunitz has estimated the upper limit of the entropy gain for transferring a water molecule from a protein to water to be 2 kcal/mol at room temperature.(48) To reach the 2 kcal/mol limit, a water molecule will have to be very tightly bound in a rigid protein structure. The entropy gain in releasing the water molecule will then be offset by a loss in enthalpy, and conversely, less tightly bound water molecules might approach or even exceed the entropy in bulk water. It has been observed that even rather tightly bound water molecules can retain a very high amount of residual mobility.49,50 Furthermore, protein flexibility may be significantly affected by water binding. Cases are known where proteins become more rigid(51) or more flexible(52) upon water binding. So how should one assess which water molecules are replaceable?
Various flavors of molecular dynamics and free energy calculations have been applied to study this problem. Essex et al., using thermodynamic integration calculations in Monte Carlo simulations, found that the calculated binding free energies for individual water molecules allowed a rough classification into those displaced by ligands and those conserved across protein−ligand complexes; the conserved ones are more tightly bound.(53) Li and Lazaridis computed the thermodynamics of water displacement in concanavalin A−carbohydrate complexes and concluded that “the final outcome of water displacement is sensitive to the details of the binding site and cannot be predicted by simple empirical rules”. The complexity of the contributions of specific water molecules to ligand binding has recently been re-emphasized in a comprehensive study by Jorgensen et al.(54)
Extensive computer simulations are not generally feasible in a fast-paced drug discovery environment, but simple geometric parameters describing the immediate protein environment of a crystallographic water molecule can serve as a useful guide to estimate whether it is displaceable by a ligand moiety.(55) In our hands, a simple geometric rank function based on the distances and angles of neighboring donor and acceptor atoms in the protein, developed by Kellogg and co-workers, has served as a practically useful metric.(56) Water molecules with high ranks should be regarded as part of the binding site.57,58 These experience three or more hydrogen bonds with the local environment and are usually located in buried polar cavities. Replacement of an optimally coordinated water molecule will require very high design precision because otherwise the enthalpy loss might be hard to regain. In contrast, displaceable water molecules often participate in a water network without tight binding to the protein. More “sloppiness” in the design is allowed in these cases.
A number of studies allow a direct comparison between a ligand binding to a structural water and a very close analogue displacing it. A nitrile substituent has successfully been used as an isostere of a water hydrogen-bonded to a pyridine-like acceptor nitrogen in EGFR kinase(59) and p38 MAP kinase inhibitors(60) as well as in inhibitors of scytalone dehydratase.(61) In all of these cases affinity was retained or even improved. In peptide inhibitors of iNOS,(62) an amide and an amine functionality were independently replaced by the respective aminoxides and yielded equipotent inhibitors replacing a water molecule bound by the heme carboxylates. In all of these cases no more than two hydrogen bonds are formed between protein and water, indicating nonideal coordination. Campiani et al. reported an instructive counterexample. They attempted to replace a water molecule in acetylcholinesterase by derivatives of huperzine and observed a dramatic loss in affinity.(63) The designed phenol ligands clearly would not reach the position of the water, which in addition is firmly coordinated by three hydrogen bonds to the protein.
The binding site of PDE10 contains two buried water molecules that are hydrogen-bonded to each other and to two and three protein residues, respectively (Figure 3). This high coordination suggests a large enthalpic penalty for the removal of any of these. We found that the water molecule with almost ideal tetrahedral coordination has not been displaced by any known PDE10 ligand, while 1 out of 15 different inhibitor series was able to expel the less tightly bound water molecule.
Figure 3.
Overlay of representatives of 15 different PDE10 inhibitor classes. Two water molecules are deeply buried in the binding site of the apo form and are hydrogen-bonded to two (left) and three (right) protein residues, respectively. Only the less tightly bound water molecule has been displaced by PDE10 inhibitors so far.
Wherever possible, a comparison between water placements in multiple independently solved crystal structures should be made to minimize errors during the modeling of water sites in the refinement process.(64) Strongly bound water molecules are often conserved across multiple crystal structures. Alternatively, water sites have been predicted computationally through MD simulations(65) or empirical functionals.(41) An increasingly important area of investigation is the analysis of entire water clusters. Understanding the hydration properties of subpockets within binding sites can be the key to understanding ligand binding, as exemplified by research on the OppA protein.66,67 Crystal structures of complexes between OppA and peptides of the general structure Lys-X-Lys reveal that certain water molecules adopt the same position independently of the nature of residue X. The strength of the interactions between these water molecules can help explain the binding affinities of the OppA−peptide complexes. tRNA-guanine transglycosylase is another protein in which a conserved water cluster has been carefully studied.68,69 Achieving a more general understanding of the properties of water clusters is a challenge for the future. For example, ring-shaped water clusters seem to be prominent in outer hydration spheres(70) and perhaps in purely hydrophobic binding pockets,(71) as they benefit from cooperativity of their hydrogen bonds.(72)
Repulsive Interactions
An important element of the design process is the analysis of repulsive interactions one may have artificially generated upon modifying experimentally determined coordinates of a complex in silico. This is even more critical if part of the protein is treated as flexible during a minimization step, since locally introduced strain can be absorbed by other parts of the structure and can be dissipated to such an extent that it becomes unrecognizable. It is therefore advisible to keep the protein rigid with the exception of only those parts that are known to adapt to ligands or to create alternative hypothetical low-energy states of a protein that are then kept fixed in the design process. The availability of multiple crystal structures of the same protein with different ligands is most useful here, as their overlays allow the identification of backbone movements and flexible side chains. Severe repulsive interactions are not observed in nature, so distances significantly below the shortest experimentally observed ones are to be avoided. General steric fit can be assessed through surface depictions of both binding partners. In addition, one may rely on general molecular mechanics terms or simple van der Waals distance criteria to analyze steric clashes. Conformations should be carefully checked; force field minimization often has the effect of achieving a good interaction at the expense of a strained torsion angle. Torsion angles are the softest conformation parameters and yet have the largest effects on geometry.(73)
Specific Intermolecular Interactions
Hydrogen bonds are by far the most important specific interactions in biological recognition processes. Geometries of hydrogen bonds follow strict rules, which were among the first to be extracted from crystal structure databases. Interactions between NH and carbonyl groups,(74) interactions of hydroxyl groups with carbonyl, ether, and ester groups,(75) and hydrogen bonds to aromatic heterocycles(76) were studied in detail. Systems like Isostar(77) and Superstar(78) have proven useful for visualizing such interaction preferences. The spatial distribution of intermolecular contacts between carbonyl groups and NH donors has been converted to energy maps, highlighting commonalities and differences between ester and amide acceptors.(79) This approach has also been applied to protein structures, where good agreement was found between statistical analysis and quantum mechanical calculations.(80) Here we will not exhaustively cover the topic of hydrogen bonding but will present key facts and concepts.
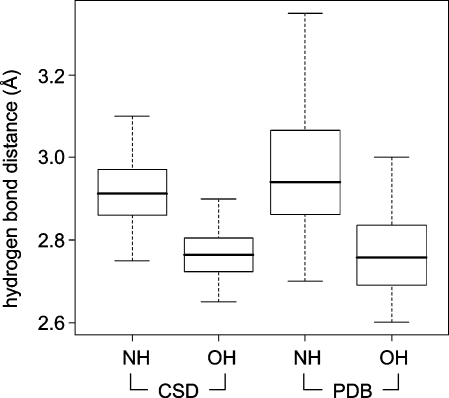
Frequency distributions of classical hydrogen bond distances give rise to sharp peaks, allowing one to distinguish small differences in median distances. In the CSD, hydrogen bonds between amide C=O and OH have a median distance of 2.75 Å. With NH donors, the median distance increases to about 2.9 Å. Within binding site regions of the PDB, the same median distances are observed. PDB distributions are significantly broader but still clearly separated from each other (Figure 4). In theory, this means that a hydrogen bond should be within about ±0.1 Å of its median observed distance. If this cannot be achieved in a relaxed state, the design should be modified. In practice, however, such rigid distance criteria can rarely be applied, since the adaptation of protein structure to a modified ligand is hard to predict. Furthermore, the coordinate precision of protein structures is much lower than that of small molecule crystal structures, and it varies considerably not only as a function of overall resolution.81,82
Figure 4.
Box plots (box region corresponding to the central 50% of the distribution, dotted lines extending to max 1.5 times this interval) of hydrogen bond length distributions with NH and OH as donors: (left) CSD statistics; (right) PDB statistics. In the PDB, the distributions around roughly the same mean are significantly broader.
We have analyzed the preferred geometries of a variety of acceptors and donors by searches in the CSD (see Figures S-1 and S-2 of Supporting Information). Even for the weakest acceptors (ether, sulfonamide), the median distance is shorter than the sum of van der Waals radii of the donor and acceptor heavy atom. Charged hydrogen bonds are considerably shorter than comparable uncharged ones. For example, the salt bridge between carboxylate and ammonium has a median distance of 2.79 Å. With a neutral amine as a counterpart, this distance increases to 2.83 Å. A hydrogen bond between amide carbonyl and amine is typically 2.9 Å long.
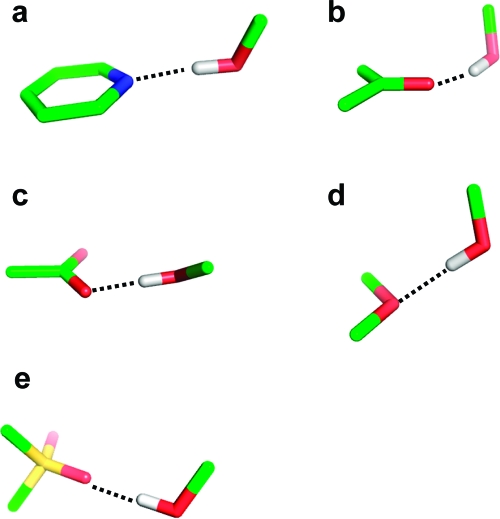
The angular preferences of hydrogen bonds are also quite pronounced. A number of typical geometries are shown in Figure 5. The angle donor−hydrogen···acceptor is generally above 150°. Only ethers, which are rather weak acceptors, show a somewhat higher variability of this angle. Hydrogen bonds are generally formed along the direction of the free electron pair of the acceptor atom. For example, the preferred angle C=O···H has its peak at 120°, corresponding to the lone pair direction of the carbonyl oxygen. This mnemonic rule does not hold for sulfonyl groups, where hydrogen bonds along the S=O axis are slightly preferred. For sulfonyl and carboxylate groups, one needs to further distinguish between syn and anti directions with respect to the second oxygen atom. The syn orientation is preferred over anti in both cases. The interaction with the syn lone pair is almost always an asymmetric one with the donor atom interacting with only one of the oxygen atoms.(83) The last geometric feature to be monitored is the deviation of the hydrogen bond from the acceptor plane in the case of acceptors comprising π systems (aromatic ring plane or the plane formed by carbonyl and its two substituent atoms). The deviation from the acceptor plane is generally below 25−30°.
Figure 5.
Schematic depiction of the most preferred geometries of hydrogen bond interactions with various types of acceptors: (a) pyridine nitrogen, (b) carbonyl oxygen, (c) carboxylic acid, (d) ether oxygen, (e) sulfonyl group.
While the preferred geometries of hydrogen bonds are easily defined, their contribution to affinity is highly context-dependent. Davis and Teague,(84) Kubinyi,(85) and Ladbury(86) have collected many illustrative examples. Hydrogen bonds always convey specificity to a recognition process but do not always add much binding free energy. Desolvation of the donor and the acceptor must occur for the hydrogen bond to form, such that the effects of hydration and hydrogen bond formation nearly cancel out.(87)
Thus, a primary question in molecular design should be which donors and acceptors need to be satisfied and not how more hydrogen bonds can be formed. Analyses of entire protein structures showed that both NH and CO groups form hydrogen bonds to a very large percentage,(88) in particular in high resolution crystal structures, and the remaining cases can usually be explained by artifacts of the crystal structures or inadequacies of the search methods.(89) The number of observed hydrogen bonds clearly increases with decreasing solvent accessibility.(90) There is evidence, however, that not satisfying a hydrogen bond donor in a protein, i.e., burying a donor in a desolvated state, has more drastic energetic consequences than not satisfying an acceptor. Homas et al. report a penalty of 4.3−5.3 kcal/mol for a hydroxyl group binding in a hydrophobic pocket, due to the cost of desolvation.(91) Relatively little loss of free energy is incurred upon removal of a hydrogen bond to a backbone carbonyl group. Bartlett and co-workers showed that replacements of NH by a methylene group hardly reduced the binding affinity of peptidomimetic thermolysin inhibitors, whereas the replacement of NH by oxygen allowed a dramatic loss in affinity by 4 kcal/mol.(92) The binding of acylated tripeptides and their analogues to vancomycin gives a similar picture: replacement of NH by methylene leads to a loss of 1.5 kcal in binding free energy, replacement by oxygen to a loss of 4.1 kcal/mol.(93) In a series of Chk1 inhibitors, a reduction in binding affinity of 1.4 kcal/mol was observed in changing a pyrrole to a furane ring where in both cases the heteroatoms point toward the Cys87 backbone carbonyl group. As in the previous two examples, this loss of affinity is probably mostly due to the introduction of a repulsive O···O interaction rather than due to the removal of the NH···O hydrogen bond.(94)
A look at the world of kinase inhibitors confirms this trend. The vast majority of kinase inhibitors with any direct hinge interactions form a hydrogen bond to the backbone NH situated at the center of the hinge strand. Where this is not the case, the backbone NH is usually solvated (see, for example, PDB code 1zz2 and ref (95)). We are aware of only a single ligand where the backbone NH has no good counterpart: 4,5,6,7-tetrabromobenzotriazole forms two halogen bonds to the flanking backbone carbonyl groups but no direct interaction with the NH group (PDB code 1p5e). In contrast, hinge binding ligands often leave one backbone carbonyl group in a desolvated state without a compensatory interaction. PDB entries 3blr and 3fmd may serve as examples.
So far, we have neglected the fact that hydrogen bonds can vary quite considerably in their intrinsic strength. To a first approximation, this might be justified because a stronger hydrogen bond also implies higher desolvation costs, so the net free energy gain of a stronger hydrogen bond might be minimal. However, the wide variation of the free energy changes associated with hydrogen bonds indicates that this picture is too simplistic. Just like solvent accessibility, the geometry of hydrogen bonds and the nature of neighboring atoms are key parameters determining this free energy change; hydrogen bond strengths should be carefully assessed as well.
The propensities of many functional groups to form hydrogen bonds have been experimentally determined and tabulated by Abraham(96) and more recently by Laurence in the form of the pKBHX scale, details of which have been reviewed in this journal.(97) Where experimental data are not available, acceptor strengths can be obtained from quantum chemical calculations.(98) Calculated acceptor strengths and those derived from IR spectroscopy generally correlate well with each other, indicating that the entropic component of hydrogen bond formation in water is relatively constant across acceptors.
What can be learned from such parameters? First, acceptor strengths are not directly related to proton basicities. The most basic center in a molecule is not necessarily the best acceptor. An appreciation of acceptor strengths can thus lead to a better understanding of desolvation effects and interaction preferences, for example, of more complex heterocycles. Second, although rarely reported, cases are known where acceptor (or donor) strengths correlate with affinity, leading to important insights into SAR.99,100 Finally, these values can help design molecules with better overall properties, for example, by modulating the nature of solubilizing groups. There is good general agreement between statistical likelihoods of hydrogen bond formation in the CSD and acceptor strengths. For example, nitrogen acceptors in aromatic heterocycles are more frequently observed to form hydrogen bonds in the CSD than oxygen acceptors,(101) in agreement with pKBHX data.
Our CSD searches also indicate a qualitative agreement between hydrogen bond frequencies and pKBHX acceptor strengths, as shown by the data in Table 1 for acyclic oxygen atoms as acceptors and for a selection of planar heterocycles. For the CSD searches, very generic substructures have been used, and so the results should be taken as a qualitative and average property of each functional group. In addition to the nature of the functional group, many factors influence acceptor strength. For example, cyclic amides (lactams) and cyclic ethers are significantly stronger acceptors than acyclic ones. Electron-donating substituents at the aromatic rings increase the acceptor strength, while electron-withdrawing ones decrease it. Aromatic ethers such as anisole are weaker acceptors than aliphatic ones. Because of its extremely low propensity to form hydrogen bonds, diphenyl ether might even be regarded as a bioisostere of diphenylmethane.
Table 1. Hydrogen Bond Acceptor Strengths (pKBHX Scale) and Frequencies Observed in the CSD for Selected Hydrogen Bonds Types.
| typical acceptor strength (pKBHX) | frequency of hydrogen bond to OH [%] | |
|---|---|---|
| Acyclic Oxygen Acceptors | ||
| amide | 2.2−2.6 | 48 |
| ketone | 1.1−1.2 | 39 |
| sulfone | 1.1 | 37 |
| sulfonamide | 1.0 | 30 |
| ether | 1.0−1.2 | 16 |
| Planar Heterocycles | ||
| N-alkylated imidazole N | 2.72 | 41 |
| pyridine N | 1.86 | 32 |
| oxazole N | 1.67 | 30 |
| pyrazine N | 0.92 | 25 |
| furane O | −0.4 | 0.5 |
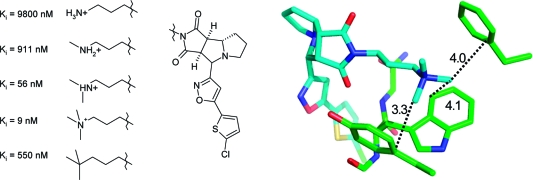
Only 30% of the sulfones and sulfonamides form hydrogen bonds. This raises the question of which type of interaction this functional group prefers. In the CSD, we find that about 80% of the SO2 groups are in proximity (3.3−3.9 Å) to an aliphatic carbon atom (in our queries we used any methylene unit −CH2− as a prototype hydrophobic group). In the PDB, 39% of the ligand sulfonyl groups are found to form a hydrogen bond with either a protein donor or a structural water molecule, while 74% are located in or close to van der Waals distance (3.3−3.9 Å) to an aliphatic group. Notably, of the sulfonyl groups situated in a hydrophobic environment in the PDB, only 36% are found to interact simultaneously as a hydrogen bond acceptor but 79% of the hydrogen-bonded sulfonyl groups are found to interact simultaneously with a hydrophobic group. These findings clearly indicate a dual character of the weakly polar sulfonyl groups as a hydrogen bond acceptor and as a hydrophobic group. An example of the types of hydrophobic environments that sulfonyl groups are found in is given in Figure 6, which depicts a section of the binding site of the ligand CRA-27934 within the cathepsin S cocrystal structure (PDB code 2fra). The sulfonyl group forms several van der Waals interactions with nonpolar atoms and forms weak hydrogen bonds with Cα-H donors. Further illustrative PDB examples include complexes of FK506 binding protein (1j4h) and phenylethanolamine N-methyltransferases (2onz). Hydrogen-bonded sulfonyl groups are prominent in cocrystal structures of matrix metalloproteinases (e.g., 3ehx).
Figure 6.
Closest interactions (distances in Å) formed by the sulfonyl oxygen atoms of a cathepsin S ligand within the active site (PDB code 2fra). Most side chains have been omitted for clarity.
One further aspect of hydrogen bonds requires the attention of the designer. Hydrogen bonds are rarely isolated interactions but are strongly influenced by additional polar atoms in the vicinity. Hydrogen bonds can mutually reinforce each other in networks. This is observed for aligned hydrogen bonds in protein secondary structures.(102) Calculations on biotin-bound streptavidin indicate that cooperative hydrogen bonding may be one source of the particular strength of this complex.(103) Ring-shaped water clusters owe their stability to cooperative binding.(72) The stacking of multiple β strands in amyloid fibrils has been, in part, ascribed to cooperative hydrogen bonding(104) in a way reminiscent of the association of urea molecules in nonpolar solvents.(105) Also, one should consider that branched hydrogen bonds are common in protein−ligand interfaces.(106) Hydrogen bonds with suboptimal geometries may be stabilized by additional partners that help to satisfy the total hydrogen bonding potential. Before labeling a particular interaction as “weak” because of its geometry, one should therefore assess its environment. It will be important to learn to recognize and to prospectively apply more complex hydrogen bonding patterns(107) that can be derived from crystal structure analysis. In this respect, materials science and supramolecular chemistry can fertilize the area of structure-based drug design, for example, through tools like the Cambridge Crystallographic Data Center’s Mercury.(108)
Neighboring acceptor and donor groups can also weaken hydrogen bonds. Originally formulated by Jorgensen,(109) the “secondary electrostatics hypothesis” was soon confirmed by Zimmerman(110) and is now widely accepted. Hydrogen bonds to model peptides in the extended (C5) conformer are weakened by the proximity of the neighboring carbonyl oxygen to the NH donor.(111)
While the hydrogen bond distances of salt bridges are shorter than those between neutral motifs, indicating stronger interactions, this does not necessarily translate into more favorable binding free energies. The contribution of charge-assisted protein−ligand hydrogen bonds to binding depends critically on the protein environment. Theoretical and experimental studies of solvent-exposed salt bridges reveal little free energy gain for the pairing of monovalent ions.(112) We are not aware of any example in which the formation of charge-assisted hydrogen bonds on the surface of the binding site of a complex has led to significant affinity improvements. This can be rationalized with the high solvation free energy of solvent exposed charged protein residues, which needs to be compensated by ligand binding. In contrast, a number of SAR examples exists in which charge-assisted hydrogen bonds are crucial binding hot spots. Common to these is that the interacting protein motif is at least partially buried and held in position by other interactions to surrounding residues. A textbook example is the critical salt bridge between Asp189 at the bottom of the S1 pocket of trypsin-like serine proteases with benzamidine inhibitors, where removal of the amidine group or reduction of basicity strongly reduces activity. More recently, it was found that the introduction of a 2-amino group in lin-benzoguanine as a ligand of tRNA-guanine transglycosylase leads to a 50-fold lower Ki.(113) The modified ligand forms a cooperative charge-assisted hydrogen bonding network involving a part of the protein backbone and a more remote glutamate side chain (PDB code 2z7k). In a series of zanamivir analogues binding to neuraminidase, it was found that a positively charged amino group was roughly 30-fold more active than a neutral hydroxyl substituent.(114) This substituent bridges two Glu and Asp side chains with formal negative charges providing additional electrostatic stabilization (PDB code 1bji). Because of the long-range character of charged interactions, not only the direct protein−ligand contacts but also the influence of more distant protein charges should be considered when designing charge-assisted hydrogen bonds.
Weak Hydrogen Bonds
During recent years, an increasing amount of attention has been paid to weaker hydrogen bond interactions. This increased attention is a double edge sword: It sharpens the eye for interactions that previously went unnoticed, but it also increases the risk of overinterpretation. Numerous accounts of weak hydrogen bonds fall in the latter category,115,116 as they try to ascribe molecular recognition to selected observed interactions. Nevertheless, weak hydrogen bonds often do contribute to protein−ligand binding in a subtly directional fashion, and it is important to understand that they are, at the very least, not repulsive. The nature of weak hydrogen bonds has been extensively analyzed and reviewed by Desiraju.106,117 Here we will only briefly discuss the nature of a few representative cases.
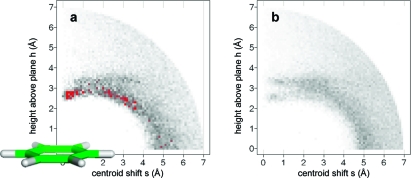
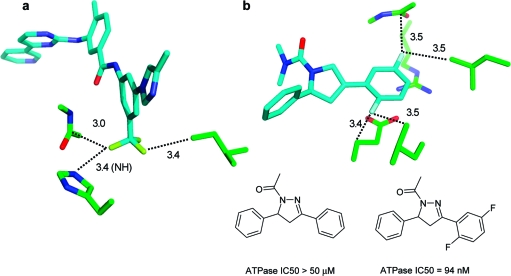
Aromatic rings can act as acceptors of hydrogen bonds. Tsuzuki et al. have calculated the gas phase interaction between benzene and ammonia to be 2.2 kcal/mol, only slightly more attractive than that between benzene and methane (1.5 kcal/mol).(118) In the most stable orientation, one NH vector is oriented perpendicularly to the ring plane at the benzene ring center. Likewise, 2-pyridone and benzene adopt a T-shaped NH···π hydrogen-bonded arrangement according to calculations and spectroscopic analysis of the supersonic-jet-cooled dimer.(119) In proteins, interactions between NH donors and aromatic side chains are observed rarely and usually at long distances (>3.5 Å). Clearly, an aromatic ring can only be a “reserve” acceptor for a strong donor. Tyr, Phe, and in particular Trp side chains interact more frequently with polarized CH groups as donors.120,121 There are few clear-cut cases where ligand phenyl rings accept hydrogen bonds from protein amide NH groups, and it is unlikely that these interactions are the root cause of affinity gains. Figure 7 shows two examples with reported NH···π contacts. Addition of a substituted phenyl ring to a weak Chk1 kinase ligand (Figure 7a) increased affinity from from 8.5 to 0.026 μM.(122) The additional phenyl ring forms multiple interactions, including a contact to a valine side chain, and displaces several water molecules. One of these had been coordinated to the backbone NH below. Another NH···π interaction reported for a PDE10 complex(123) structure in reality does not exist: The glutamine side chain forms two classical hydrogen bonds to water and a tyrosine and thus fully satisfies the hydrogen bonding potential of the NH2 unit (Figure 7b). While CSD statistics clearly indicate a preference of aromatic CH groups to be oriented above phenyl ring planes, there is no such preference for NH and OH groups. Figure 8 shows radial density plots of the distance above the aromatic plane of query hydrogen atoms versus their distance from the centroid in the plane (centroid shift). These two coordinates project the three-dimensional distribution of query atoms around the phenyl ring onto two dimensions with the centroid as the origin, facilitating visual inspection (details on the query setup and calculation of the radial distribution plots are found in Materials and Methods). Hydrogen atoms in XCH units polarized by neighboring heteroatoms (X = O, N) have a clear preference for interactions above the ring plane, whereas NH and OH groups rarely undergo π hydrogen bonds. The NH···π interaction itself might be stronger than the CH···π interaction, but this energy gain is overcompensated by a higher desolvation cost, since a strong donor will form significantly better interactions with a stronger acceptor than with a phenyl ring.
Figure 7.
Structures referred to as NH···π interactions in the literature. (a) Chk1 kinase ligands. The orange structure has a Ki of 8.5 μM (PDB code 2c3l), and the green structure has a Ki of 0.026 μM (PDB code 2c3k).(122) The additional phenyl ring displaces several water molecules, one of which was coordinated to the backbone NH below. The shortest NH···phenyl distance in the green structure is 3.4 Å. (b) A PDE10 complex structure with a reported NH···π contact(123) where in reality the glutamine side chain forms two classical hydrogen bonds to water and a tyrosine. Distances are in Å.
Figure 8.
Radial distribution of hydrogen atoms around a phenyl ring (CSD statistics): (a) hydrogen bound to sp2 carbon flanked by one or two heteroatoms (N, O); (b) hydrogen bound to O or N. Queries were set up as described in Materials and Methods. The phenyl ring in (a) is drawn roughly to scale and should serve as an interpretation aid for the in-plane (s) and above-plane distances (h). Darker gray corresponds to higher density; peaks above a numerical value of 70 are colored red.
Interactions between CF and polar hydrogen atoms HX (where X = O, N) frequently occur in the PDB and CSD, even if such interactions cannot be classified as strong hydrogen bonds.(124) We have observed a thrombin inhibitor to change its binding mode upon fluorination of an aryl ring, such that a CF···HN interaction is formed.(125) In another study on factor VIIa inhibitors, a fluorinated phenyl ring was shown to act as an isostere of a pyridine.(126) An increase of affinity from 455 to 68 nM was observed in sitagliptin analogues binding to DPP-IV when going from 3,4-difluorinated to 2,4,5-trifluorinated triazolopiperazines.(127) The additional ortho-F forms interactions at 3.2 Å distance with NH2 groups of Asn and Arg side chains (PDB code 1x70).
The weaker a hydrogen bond donor, the longer is the hydrogen bond distance and the broader is the distribution of the observed distances in crystallographic databases. This is illustrated in Figure 9 for the donor series OH, NH, acetylene CH, and CH in six-membered aromatic rings. The larger variation in median distances should not be overlooked in analyzing and designing structures. The most prominent weak donor is the CH group. In particular in kinase inhibitors, C−H···O interactions are frequently observed.(128) They play an equally important role in stabilizing planar conformations of linked heterocyclic systems. A team at Vertex working on GSK3 inhibitors has published an instructive study on the interplay between steric repulsion and classical and weak hydrogen bonds.(129) Often, CH groups bound to N or O atoms in aromatic heterocycles act as donors. The Cα-H unit of proteins is also a weak donor. Calculations indicate that Cα-H···O=C interactions are about one-half the strength of an NH···O=C hydrogen bond.(130) Analysis of peptide X-ray structures show that the Cα-H unit can substitute for stronger donors.(131) A close Cα-H···F interaction has been observed in a thrombin−ligand complex where the introduction of the fluorine atom led to a 5-fold affinity increase.(132) Finally, cation−π interactions, discussed further below, might be regarded as hydrogen-bonded systems, since it is often not the cationic center itself, but an electron-deficient alkyl substituent, that forms the direct contact. Protonated histidines can also act as strong CH donors.(133)
Figure 9.
Box plots of hydrogen bond length distributions for the interaction between weak and strong donors and amide carbonyl oxygen as acceptor (CSD statistics). An increase in median hydrogen bond length and in breadth of the distribution is observed for decreasing donor strength.
Halogen Bonds
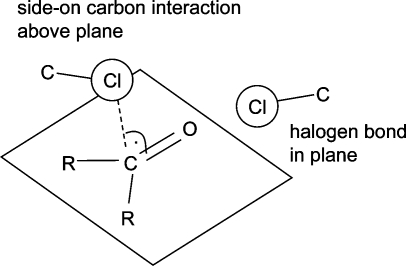
In an analysis of crystal structure data published in 1986, Ramasubbu et al. concluded that “the halogen X in a C−X bond is capable of significant interactions with electrophiles, nucleophiles, and other halogens. The electrophiles approach X of the C−X “side on”, nearly normal to C−X, and the nucleophiles nearly “head-on” and behind the C−X bond.”(134) This observation, valid for the halogens Cl, Br, and I, is an ideal introduction to the following two sections on halogen bonds and multipolar interactions.
In contrast to fluorine, the heavier halogens have unique electronic properties when bound to aryl or electron withdrawing alkyl groups. They show an anisotropy of electron density distribution with a positive area (σ-hole) of electrostatic potential opposite the C−X bond.(135) In a molecular orbital framework, the origin of the σ hole can be explained from the fact that the three pairs of unshared electrons follow an approximate s2p2p2 configuration forming a belt of negative charge around its central region, whereas the third p orbital along the C−X axis is distorted toward carbon, forming the C−X single bond. This leads to attractive interactions between C−X moieties and carbonyl groups or other classical H-bond acceptors, with a preference for linear C−X···B arrangements. For example, distances below van der Waals radius (3.3 Å) between carbon-bound chlorine and sp2-hybridized oxygen almost exclusively occur with linear C−Cl···O geometries.(136) The strength of halogen bonds increases with the size of the halogen atom. Because of its mostly electrostatic nature, it also depends on the electronegativity of the carbon substituents in the C−X partner and on the electron density of the binding partner.137,138 Halogen bonds are significantly weaker than hydrogen bonds. Ab initio calculations on formaldehyde−halobenzene dimers give gas phase interaction energies below 2.5 kcal/mol, corresponding to the strength of CH···O hydrogen bonds.(139) This is in accord with observations that halogen bonds involving iodine can displace less conventional hydrogen bond donors such as the C−H imine moiety.(140) QM/MM calculations on ligand−enzyme complexes(141) cannot be expected to give meaningful values because halogen substituents typically form multiple interactions besides a halogen bond and because desolvation effects are not accounted for.
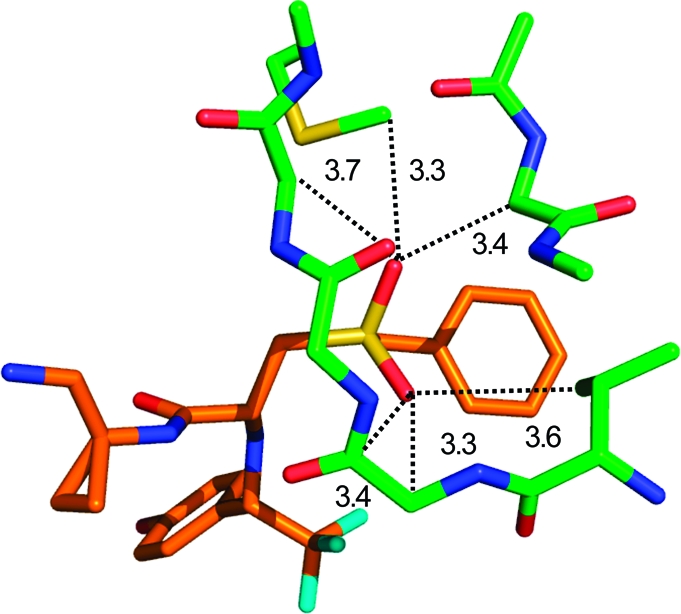
A large amount of structural and SAR data proves the existence of halogen bonds both in small molecule complexes and in protein−ligand complexes.142,143 Typically, replacement of hydrogen by iodine leads to the largest affinity differences. A 200-fold affinity gain from H to I has been observed in a series of adenosine kinase inhibitors(144) (PDB code 1lij), whereas Cl only led to a 34-fold affinity increase. A 100-fold increase in binding affinity was observed in a class of HDM2 inhibitors. Figure 10 shows that the closest contact between iodine and protein is formed by a carbonyl oxygen, whereas other van der Waals contacts are significantly longer. Note that iodine could be replaced by an acetylene substituent, leading to 4-fold lower affinity.(145) Acetylene and iodine substituents are comparable in length, and the affinity data suggest again that a halogen bond to iodine is comparable in strength to a weak hydrogen bond. A 300-fold affinity difference upon iodine substitution was observed in a series of HIV RT inhibitors.(146) In this series, the carbonyl oxygen of Tyr188 forms a halogen bond with an iodine subsituent. In a high resolution structure, this carbonyl oxygen is observed to bend slightly toward the iodine, although not enough to abolish a β-sheet hydrogen bond with the Tyr181 nitrogen.(147) In the PDB it is not uncommon to find carbonyl groups forming both a halogen bond and a hydrogen bond. Calculations on model systems indicate that energies of halogen bonds to amide carbonyl groups are largely independent from simultaneously formed hydrogen bonds.(148) Affinity gains by iodine substitution drop with longer I···O distances, as observed, for example, in a series of BACE inhibitors(149) (PDB code 2iqg), where the affinity gain was only 25-fold.
Figure 10.
Iodine bond to a backbone carbonyl group in a HDM2 p53 domain crystal structure(145) (PDB code 1t4e, distances in Å).
In spite of the fact that halogen bonds formed by chlorine are usually much weaker, they can play a unique role in some complexes. The broad spectrum bactericide triclosan inhibits the FabI (enoyl reductase) component of bacterial fatty acid synthesis.(150) Complex structures with several enoyl reductases have been solved, and all show the same binding mode of triclosan. One chlorine is situated at the outer rim of the active site and interacts almost linearly at a distance of 3.3−3.5 Å with a backbone C=O. Strikingly, an extensive synthetic program directed at replacing this chlorine atom by both lipophilic and hydrogen bonding substituents did not lead to compounds with higher potency against P. falciparum enoyl reductase.(151)
We conclude that halogen bonds are weak but specific interactions that can lead to clear gains in binding affinity. They belong to the arsenal of structure-based design tools just like weak hydrogen bonds and, like these, have the advantage that they are associated with a lower desolvation cost than classical hydrogen bonds.
Orthogonal Multipolar Interactions
This particular interaction motif, characterized by a close orthogonal contact between two dipolar functional groups, has only recently received detailed attention.(152) Note that in a completely orthogonal arrangement between two dipoles, the actual dipole contribution to interaction energy is zero such that higher order electrostatic and dispersion terms must be responsible for the attractiveness of the interaction. The disappearance of the dipole term may turn a repulsive electrostatic interaction into an attractive one. Manas et al. provide an instructive example involving a nitrile interacting with divalent sulfur.(153)
Fluorine substituents are sometimes found at short distances (3.0−3.7 Å) and orthogonal to carbonyl carbon atoms. This arrangement is not the most frequent fluorine interaction observed in the PDB.(154) Investigations of a model system in chemical double mutant cycles confirmed the weakly attractive nature of the orthogonal C−F···C=O interaction, with a contribution in binding free enthalpy in apolar environments of ΔΔG = −0.2 to −0.3 kcal/mol. Multiple SAR examples have shown that increases in binding affinity can be obtained when this interaction is present.(155) Elastase is known to bind peptidic inhibitors with trifluoracetyl groups much more strongly than those containing acetyl groups,(156) and the corresponding X-ray structures feature three orthogonal CF···C=O contacts.(157) A similar effect has been observed in the SAR of abl kinase inhibitors. In the active site pocket, the CF3 group of nilotinib forms one interaction orthogonal to an amide besides two further contacts to the imidazole NH of a histidine as well as an isoleucine side chain(158) (Figure 11a). The CF3 derivative is over 5-fold more active than the corresponding methyl derivative in an autophosphorylation assay.(159) Many classes of p38 MAP kinase inhibitors typically contain a phenyl ring in the lipophilic back pocket of the ATP binding site situated behind the gatekeeper residue.(125) A Merck team working on kinesin spindle protein(160) reports a dramatic boost in affinity upon fluorine substitution (Figure 11b), again with one of the two fluorine atoms interacting orthogonally with an amide bond. It should not be assumed that this increase in binding free energy can be ascribed to the interactions formed by fluorine only. Large components might be due to changes in residual mobility and desolvation. Still, the examples show the value of fluorine scans and in particular its targeted use in “fluorophilic” pockets.
Figure 11.
(a) Fluorine interactions in the complex between nilotinib and abl kinase (PDB code 3cs9).(159) (b) Kinesin spindle protein structure (2fl6) and activity data of two closely related inhibitor structures.(160)Distances are in Å.
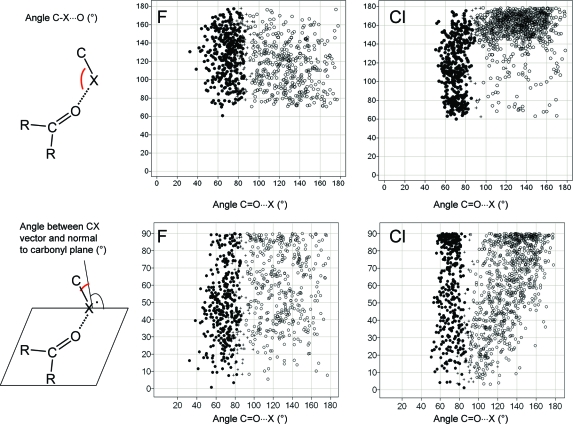
Since halogens can interact both with the oxygen and with the carbon atoms of carbonyl groups, forming halogen bonds and multipolar interactions, respectively, it is instructive to look at the relative propensities of both interactions in the same context. Figure 12 shows results of CSD searches between carbonyl groups and halogens, where either the halogen−carbon or the halogen−oxygen distance is below van der Waals contact. Fluorine does not have an orientation preference, and the distinction between close contacts to carbonyl C and O seems somewhat arbitrary. CF bonds approach carbonyl centers from any direction. For Cl, the scatter plot divides into two distinct distributions for halogen bonds and multipolar interactions. Both types of contacts occur roughly with the same frequency. Chlorine does form multipolar interactions with carbonyl groups as fluorine does but shows a tendency for the C−Cl bond to be parallel rather than orthogonal to the amide plane, a consequence of the anisotropic distribution of electron density around the Cl atom. For clarity, Figure 13 shows the two preferred orientations of C−Cl vectors with respect to carbonyl groups. We have repeated these searches for bromine and iodine and find qualitatively similar distributions as for chlorine, with an increasing shift toward more halogen bonds and fewer side-on interactions. Analogous distributions extracted from the PDB are so much more scattered that a clear orientation pattern is not recognizable anymore (data not shown).
Figure 12.
Occurrence of close contacts between F (left) and Cl (right) atoms and carbonyl groups in the CSD. Scatter plots show two different angle distributions. Points are categorized by their distance to the carbon and oxygen atoms of the C=O unit: (●) close contact between halogen and carbonyl C (<3.3 Å for F, <3.5 Å for Cl); (○) close contact between halogen and carbonyl O (<3.1 Å for F, <3.3 Å for Cl); (+) both close contact criteria satisfied.
Figure 13.
Preferred interaction geometries between chlorine and carbonyl groups derived from the CSD queries in Figure 12.
Halogens and Aromatic Rings
Various groups have analyzed crystallographic data on interactions between aromatic rings and halogen substituents, arriving at partially contradictory conclusions. On the basis of CSD searches, Schneider et al. observe a preference for CH···halogen interactions but often with significant van der Waals contacts to the π system, and they do not explicitly distinguish between fluorine and the heavier halogens.(161) A similar CSD study by Prasanna and Guru Row even concludes that the propensity for the formation of CX···π interactions is higher in the case of fluorine than other halogens,(162) in stark contrast to a study of fluorine interactions in the PDB.(154) Yet another study focused exclusively on interactions between fluorine and hydrogen bond donors, including very weak ones.(163) More recently, the attractive nature of interactions between heavier halogen substituents and aromatic rings has been emphasized, in particular in the context of serine proteases(164) and reviews on halogen bonding.(143) Unfortunately, the cited studies are not suitable to derive a clear picture of the orientation preferences and interaction energies of halogens around aromatic rings, since they suffer from one or more technical deficiencies: a focus on one type of interaction instead of a consideration of the relative importance of all alternatives, the lack of differentiation between different moieties (e.g., halogen bound to aliphatic vs aromatic carbon), and the analysis of database searches by means of uncorrected one-dimensional frequency diagrams or scatter plots instead of probability densities.
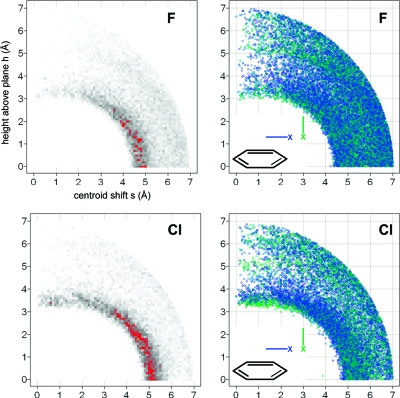
The query results depicted in Figure 14 give a more holistic picture. The radial distribution plots of both F and Cl have in common that the probability density reaches a maximum in the plane of the aryl ring, peaking at a centroid-to-halogen distance of 4.6−4.8 Å for F and of 4.9−5.2 Å for Cl. Thus, both Cl and F prefer, by a wide margin, interactions with CH over those with the π system. Gas phase calculations(165) and analyses of PDB structures(154) confirm this trend. Desiraju classifies CH···halogen interactions as “very weak” hydrogen bonds.(166) In a crystallographic study of fluorinated benzenes, he observed H···F contacts below van der Waals distance (∼2.6 Å) with increased fluorine content of the aryl ring, i.e., with increased hydrogen acidity. Also, short distances were only observed when CH···F angles approach linearity. In our larger data set we observe this trend as well: Distances below ∼2.6 Å are only observed with CH···F angles above 120° (Figure S-3), indicative of a weak hydrogen bond character of this interaction.
Figure 14.
Radial distribution of fluorine atoms (top) and chlorine atoms (bottom) around phenyl rings (CSD statistics). Darker gray corresponds to higher density; peaks above a numerical value of 90 are colored red. Scatter plots of all hits for fluorine and chlorine (right-hand side) are colored by the angle between the phenyl plane and the C−X vector from blue (0°, in plane) to green (90°, orthogonal).
Figure 14 (right-hand side) also shows scatter plots of the individual occurrences of halogen···phenyl interactions, colored by the angle between the C−X vector and the normal to the aromatic plane. The hydrogen bond interactions do not show a dependence on the orientation of the C−X vector. There is, however, a clear trend for the C−Cl vector to be perpendicular to the aromatic plane at the closest observed distances from the centroid (3.2−3.4 Å) and directly above the plane. This coincides with a minor second maximum in the distribution function of Cl. Thus, a minor fraction of Cl atoms do form a halogen bond-like interaction with phenyl rings, with the σ-hole facing the π system. This trend is slightly more pronounced for Br, again with a peak clearly below van der Waals radius at 3.5 Å (Figure S-6). Note that the searches in Figure 14 included halogen atoms bound to sp3 carbon only. Supporting Information Figures S-4 to S-6 give an overview of all halogen interactions with phenyl rings, including halogens bound to aromatic rings. The difference in distribution between halogens bound to aliphatic and aromatic carbon clearly indicates that the driving force for the larger fraction of halogens situated above the ring plane is the stacking interaction between the two aryl rings and not the halogen interaction itself. Overall, the picture emerges that the orthogonal interaction between the heavier halogens and aromatic rings is attractive, as also confirmed by quantum chemical calculations in the gas phase,164,167 but that its energetic contribution to ligand binding in water has been overrated because of the high binding affinity gains observed with ligands containing chlorinated aromatic moieties as factor Xa and thrombin inhibitors. Depending on the nature of the ligand, the difference between a methyl group and Cl at the bottom of the S1 pocket can vary between 6-fold168,169 and 10-fold.(164) Desolvation effects are an unlikely cause of this difference.(169) However, the chlorine and methyl substituents form a significant number of additional contacts besides interacting with the tyrosine side chain that need to be taken into account in a full analysis. In particular for thrombin, the closest contact is often the formation of a halogen bond with a main chain carbonyl group (see, for example, PDB entries 1ta6 and 2jh5) and not with the tyrosine phenyl ring. Finally, chemical double mutant experiments in CDCl3 gave repulsive values for all three halogens Br, Cl, and F interacting with the π faces of a phenyl ring. Fluorine had the most pronounced repulsive effect of about 0.7 kcal/mol.(170) These results are consistent with the crystallographic data and the use of a solvent less polar than water.
The above analysis of various halogen interactions highlights the fact that halogen atoms are not merely lipophilic appendages to fill hydrophobic cavities. In designing ligands, we should be more aware of the detailed nature of lipophilic pockets to optimally exploit the weak directional nature of halogen interactions. The typical “good” halogen environment consists of multiple C−H···X contacts. The more polarized (acidic) these groups are, the stronger the interaction is, up to the point where desolvation effects start to have a detrimental effect. Because of its high electron density and low polarizability, fluorine prefers dipolar interactions more strongly than the other halogens.(171) The difference between typical fluorophilic and fluorophobic environments has become particularly apparent in a study on neprilysin inhibitors by the Diederich group.(172) Any fluorine substituent at a phenyl ring in the S1′ pocket led to a decrease in affinity, a result explained by electrostatically unfavorable close contacts of organic fluorine with the negatively polarized π-surfaces of surrounding aromatic amino acid side chains. In contrast, fluorination of the benzimidazole moiety of the ligand led to an increase in binding affinity. This ring system is located in a plane with three surrounding guanidinium groups of arginine side chains.
Hydrophobic Interactions
Many studies have shown that the single best structural parameter correlating with binding affinity is the amount of hydrophobic surface buried upon ligand binding. It holds for diverse sets of protein−ligand complexes,(173) for free energies obtained from ITC measurements,(5) and for protein−protein interactions.(174) On the basis of oil/water partitioning experiments, the magnitude of the hydrophobic effect was estimated to be around 30 cal/(mol·Å2),(175) which is the equivalent of 0.7 kcal/mol or a 3.5-fold increase in binding constant for a methyl group. Essentially all empirical scoring functions used in docking and de novo design build on similar relationships, and already the visualization of matching molecular surfaces can be a useful tool in design.
This is, of course, a highly simplified view. First, we have seen above that details of (de)solvation and cooperative effects govern free energy gains through hydrophobic interactions. Second, optimal filling of a hydrophobic pocket is often achieved long before the van der Waals limit is reached. For synthetic host−guest complexes, it has been empirically established that optimal binding is observed when the guest occupies about 55% of the volume of the host.(176) The Diederich laboratory has investigated a series of plasmepsin II inhibitors with a wide variety of flexible alkyl chains binding into an induced lipophilic tunnel and could confirm the validity of the “55% rule” for this system.(177) These findings stress the importance of residual flexibility in binding events. Finally, we have seen above in the discussions on halogen interactions and weak hydrogen bonds that a simple distinction between “polar” and “unpolar” groups is not useful; there is a gray zone of weakly polar and directed interactions in between.
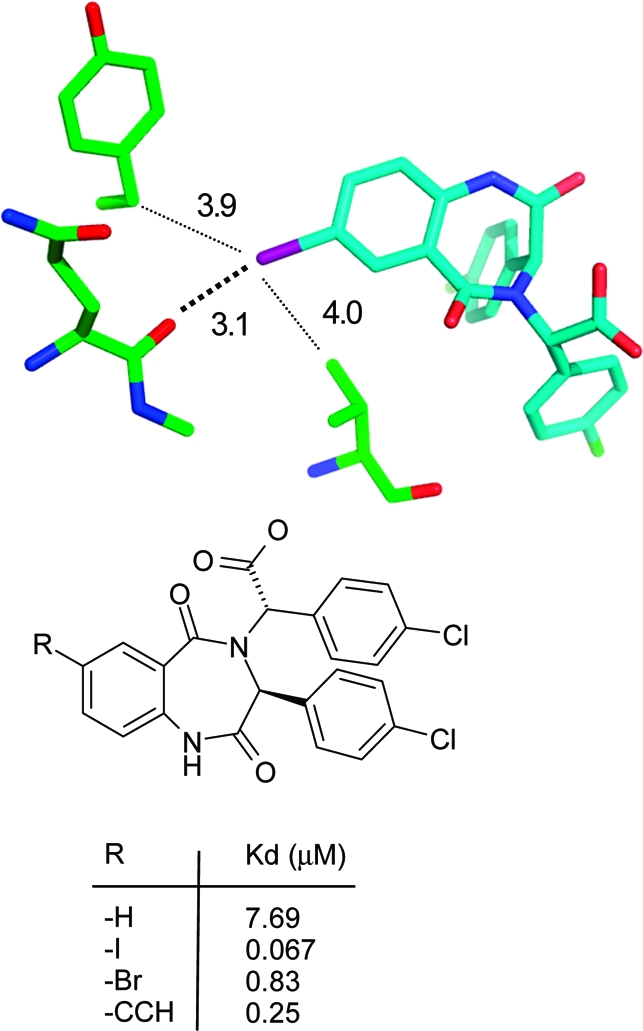
Particularly large gains in binding free energy of several kcal/mol per heavy atom can be obtained when a lipophilic protein pocket is optimally occupied by nonpolar ligand atoms.178,179 An instructive example stems from the optimization of interactions in the S1 specificity pocket of the serine protease DPP-IV which is composed of several hydrophobic Val, Trp, and Tyr side chains. Substitution of the meta-phenyl hydrogen atom by a −CH2F moiety resulted in a 400-fold increase in binding in a series of aminobenzoquinolizine inhibitors.(180) Figure 15 illustrates the excellent fit of this substituent to the asymmetric S1 pocket, with five short hydrophobic contacts to the surrounding side chain atoms. The 9-fold difference in Kd between a methyl and a monofluoromethyl group can be attributed to the slightly bigger volume enabling a tighter fit and the roughly 2-fold higher interaction strength of fluorine compared to hydrogen. This example emphasizes the potential rewards that can be gained from the fine-tuning of hydrophobic shape complementarity.
Figure 15.
X-ray complex crystal structure of human DPP-IV with an aminobenzoquinolizine inhibitor (R = CH2F) and affinity data for three derivatives.(180) The closest hydrophobic protein contacts of the CH2F moiety (distances are less than the sum of van der Waals radius + 0.5 Å) are displayed (PDB code 3kwj).
As pointed out above, a significant amount of the large affinity gains imparted by hydrophobic interactions in narrow pockets is due to poor solvation of the pocket in the unbound state. Simulations on the hydration structure of different binding sites suggest that the buried water molecules are very poorly hydrogen-bonded.42,181 Recent experimental studies on cavity hydration indicate that small, completely apolar cavities might even be completely empty rather than occupied by a single water molecule.(50)
Aryl−Aryl and Alkyl−Aryl Interactions
Aryl rings deserve special consideration in the context of hydrophobic interactions. Interactions with aryl-containing amino acids like Trp, Phe, Tyr, and His are ubiquitous in proteins,182−184 and they often expose their aromatic side chain to the binding site. The special shape and electronic properties of aromatic rings, which give rise to large polarizabilities and a considerable quadrupole moment, result in a set of preferred interaction geometries. For interactions between two π systems, the T-shaped edge-to-face and the parallel-displaced stacking arrangement are predominant. High-level quantum mechanical calculations of the dimerization energy of benzene predict these two arrangements to be isoenergetic (De = −2.5 kcal/mol),(185) with an absolute value in good quantitative agreement with experimental results (De = −1.6 to −2.4 kcal/mol).186,187 In protein structures, the parallel-displaced geometry is found somewhat more frequently than the T-shaped arrangement.(188)
Introduction of substituents or insertion of heteroatoms into aromatic rings influences the relative propensities for edge-to-face vs parallel-displaced arrangement. Some guidelines for the optimization of aryl interactions of a given geometry are summarized here, taken mainly from the comprehensive review of Meyer et al.:(35) Stacking arrangements of an electron-poor and an electron-rich aromatic ring profit from charge transfer. Stacking between electron-deficient rings is generally preferred over stacking of electron-rich ones. The preferred orientation of heteroaromatic rings is significantly affected by the alignment of positive and negative partial charges and molecular dipoles. Consideration of the detailed distribution of atomic charges and molecular electrostatic potentials is warranted here. The usefulness of electrostatic potential maps for the assessment of interaction strengths with aryl rings has been pointed out.(189) It should be noted, however, that such maps should not be interpreted as only reflecting local electron density. Through-space substituent effects can dominate.(190)
The attractiveness of edge-to-face interactions can be increased when the interacting hydrogen atom is rendered more acidic, most effectively by introducing strongly electron-withdrawing substituents in ortho- and/or para-position. High-level ab initio calculations confirm this trend:(191) The T-shaped interaction between benzene as a donor and fluorobenzene as the acceptor is ∼0.3 kcal/mol weaker than that of the benzene dimer. With reverted roles (fluorobenzene para-H as the donor), the interaction becomes ∼0.6 kcal/mol stronger relative to the benzene dimer.
Investigations of model systems led to the conclusion that aliphatic−aromatic and aromatic−aromatic edge-to-face contacts provide similar levels of stabilization.(192) For T-shaped aliphatic−aromatic interactions there is computational and experimental evidence that the interaction energy increases with increasing acidity of the interacting CH unit. High-level ab initio calculations of dimer dissociation energies between benzene and ethane, ethylene, and acetylene, respectively, yield a clear correlation in which the more acidic sp-hybridized acetylene (−2.8 kcal/mol) is about 1 kcal/mol more tightly bound than the sp3-hybridized ethane (−1.8 kcal/mol).(193) Database mining in CSD and PDB for interactions of phenyl rings with methyl groups provides further support (Figure 16). While methyl groups connected to another sp3-hybridized carbon atom (Figure 16b) show a rather broad range of interaction geometries, the distribution of methyl groups bound to electronegative atoms is much more biased toward edge-to-face interactions (Figure 16a). This behavior is qualitatively reproduced in protein structures (Figure 16c). The stronger electrostatic interaction and directionality arising from electron-withdrawing substituents have also been found by model calculations on fluorinated alcohols.(194)
Figure 16.
Radial distribution of carbon atoms around phenyl rings: (a) CH3 bound to sp3 C (CSD statistics); (b) CH3 bound to O or N (CSD statistics); (c) CH3 bound to O or N (PDB statistics). Darker gray corresponds to higher density; peaks above a numerical value of 90 are colored red.
Edge-to-face and π−π stacking interactions are not limited to (hetero)aromatic rings but can also be exploited with H-bonding arrays such as guanidinium−carboxylate ion pairs of Arg and Glu/Asp side chains or with the π faces of amide bonds. These motifs require hydrogen bonding interactions within the π-plane but are rather apolar and highly polarizable in a perpendicular direction. Interaction energies of intramolecular amide stacking were found to be competitive with nearest-neighbor hydrogen bonding in IR spectroscopic measurements of small model peptides.(195) In our own searches, we find the typical distance between the amide and aromatic plane to be in the rather broad range of 3.2−3.7 Å.
Several SAR examples from medicinal chemistry programs illustrate the points made above. The S4 binding pocket of the serine protease factor Xa is composed of the side chains of Tyr, Phe, and Trp and provides an “aromatic box” with opportunities for both edge-to-face and stacking interactions. The example in Figure 2 above clearly illustrates the strong gains in binding affinity that can be obtained by filling this pocket. Similarly, in a series of aminothiazole inhibitors, a more than 300-fold gain in binding affinity could be achieved by inserting a 3-fluorophenylpyridone substituent into this pocket.(196) Both ligand aryl rings interact with the surrounding aromatic side chains. In the same target, a more than 60-fold lower IC50 value could be obtained by introducing a carbonyl group adjacent to the nitrogen atom of a morpholine ring.(168) As illustrated in Figure 17, the additional C=O motif does not interact directly with the protein but exerts its beneficial effect through polarization of the ring CH2 on top of Trp (C−Trp distance: 3.5 Å), thereby increasing the CH2···π interaction as well as through conformational preorganization of a perpendicular arrangement to the adjacent phenyl ring. The beneficial effect of acidifying CH groups on top of an edge-to-face oriented phenyl ring can also be seen in a series of 2-anilinothiazolone steroid dehydrogenase inhibitors.(197) Introduction of a F-substituent adjacent to two aromatic CH groups in contact with a Tyr ring increases binding from IC50 = 110 nM to IC50 = 17 nM (PDB code 2rbe). Enhanced van der Waals contacts of the fluorine atom with a neighboring alanine side chain might also contribute. Finally, a drastic improvement in IC50 for inhibition of p38 MAP kinase was observed when inserting an oxygen atom between an amide and a methyl group (Figure 18).(198) The resulting methyl hydroxamate sits on top of a Phe aromatic ring and interacts additionally with a leucine side chain and through a weak hydrogen bond with a backbone carbonyl group. Acidification of the methyl protons through the attached hydroxamate certainly enhances the interactions with the Phe ring and the carbonyl acceptor. Similar to chlorine substituents, acidified CH groups can engage in both lipophilic and weakly polar interactions.
Figure 17.
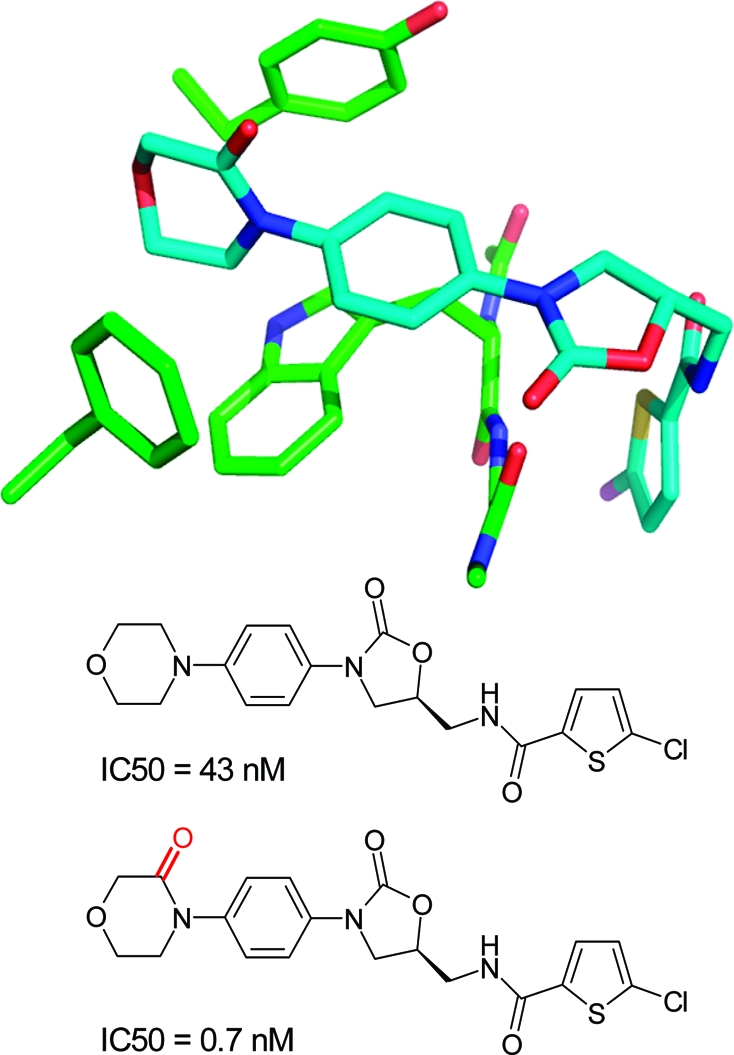
Structure and IC50 values(168) of factor Xa inhibitors (PDB code 2w26, distances in Å).
Figure 18.
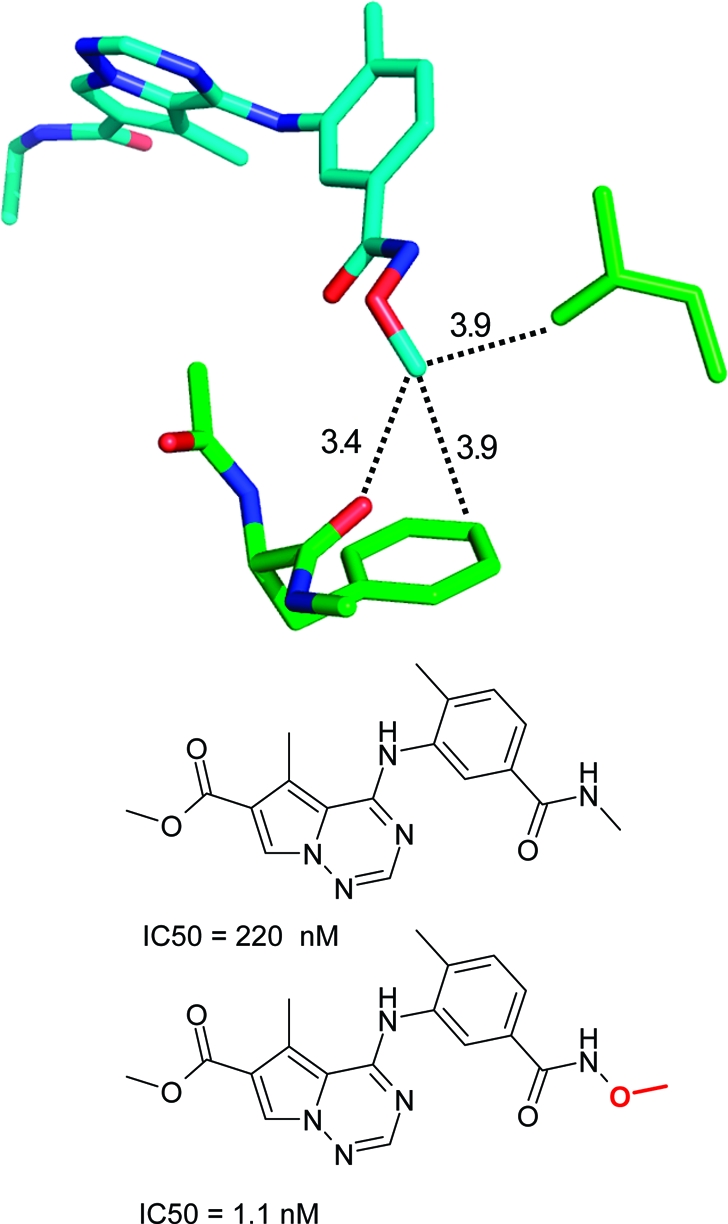
Effect of a methyl group on the IC50 of a p38 MAP kinase inhibitor(198) (PDB code 2rg5).
Cation−π Interactions
Cation−π interactions have been extensively studied in protein structures. Gallivan and Dougherty found that cation−π interations are rarely buried(199) and that arginine side chains are more likely to form such interactions than lysine. Among the aromatic side chains, tryptophan is featured most prominently.(200) Through a series of mutation studies, the interaction strength between a buried Trp and a Lys, Arg, or His side chain has been determined to range from −0.8 to −0.5 kcal/mol.(201) With increasing solvent exposure of the partners, the interaction energy dropped significantly. These values are consistent with several earlier studies performed with biological and synthetic receptor systems (summarized in ref (35)). An instructive example is the complex structure of periplasmic lysine-, arginine-, and ornithine-binding protein (LAO). LAO binds lysine and arginine with the same affinity of about 15 nM.(202) The cationic centers of the amino acids ligands are sandwiched between the aromatic rings of two Phe residues at about 3.6 Å distance (PDB codes 1laf and 1lst). In addition, several hydrogen bonds are formed between the guanidine and ammonium groups of the ligands and surrounding residues.
Arginine stacking interactions have rarely been utilized proactively. Detailed thermodynamic studies of galectin and lectin ligand complexes have been performed.203,204 A group at Vernalis found arginine stacking interactions to be an important factor in a series of 70 kDa heat shock protein inhibitors,(205) an example of which has adorned the cover of this journal in 2009. Arginine stacking plays a role in nucleic acid binding sites. In particular adenine−arginine pair interactions are conserved within several protein families.(206)
The interaction between alkylammonium ions and aromatic rings can be seen as a special class of alkyl−aryl interaction. We have seen above that methyl groups favorably interact with the π face of aromatic rings when bound to an electronegative atom. A (formally) positively charged nitrogen is a particularly strong electronegative substituent, and therefore, the direct interaction of an alkylated ammonium group with an aromatic ring leads to a strongly attractive interaction. Ammonium groups in medicinal chemistry are rarely permanently charged quaternary ions, so there is at least one proton on the nitrogen atom whose interactions need to be considered in the design process. Figure S-7 shows examples illustrating these effects. Carbamoylcholine binds to acetylcholine binding protein through a number of short contacts between the N-Me groups and Trp and Tyr side chains in the protein. Nicotine forms similar contacts with a different set of rings (Trp and the lower Tyr residue, contacts not drawn in Figure S-7a). In addition, the NH group of the nicotine cation forms a hydrogen bond to a backbone carbonyl group.(207) The marketed acetylcholinesterase inhibitor donepezil binds to its target in a similar fashion, the NH group being solvated by a structural water molecule.(208) The strong desolvation component of cationic interactions is highlighted particularly well in a recent study on factor Xa inhibitors by the Diederich group(209) (Figure 19). Stepwise methylation of an ammonium substituent binding to the S4 pocket increased the binding affinity by 3 orders of magnitude. This leads to the conclusion that the desolvation cost of ammonium NH groups by far outweighs the energy gain of a direct NH··π interaction, even if it is intrinsically attractive.(210) This is in agreement with studies on a β-hairpin model, where an enhancement of a cation−π interaction of about 0.2−0.3 kcal/mol per additional methyl group on a lysine was found.(211)
Figure 19.
Example of cation−π interactions: a factor Xa inhibitor scaffold with systematically varied S4 pocket side chains (PDB code 2bok, distances in Å).(209)
Interactions Formed by Sulfur
Divalent sulfur is a highly versatile atom. It is characterized by the ability to interact both with electron-poor and with electron-rich functional groups. Electron-rich ones tend to approach divalent sulfur along the extension of the C−S bond (σ* direction), while electron-poor ones tend to approach it along the direction of the lone pairs.(212) An example is the pair C=O···SC2, where the carbonyl oxygen acts as the electron-rich partner. Attractive carbonyl−sulfur interactions can have a strong influence on conformational equilibria.(73)
Methionine side chains are both lipophilic and flexible. Both properties make them good targets in design, for example, in the binding sites of nuclear hormone receptors.(213) How do methionine side chains interact with aryl rings? Ab initio calculations have shed some light on the nature of these interactions. When benzene and either methanethiol(214) or H2S(215) are used as model systems, the lowest energy arrangement is that of an SH···π hydrogen bond, which may serve as a model for cysteine interactions only.(215) Arrangements in which the sulfur lone pairs point at the aromatic ring face are avoided. With dimethyl sulfide as a model system to represent the methionine side chain,(216) the most stable geometry is one in which the methyl groups interact with the π system, analogous to the interactions between methoxy groups and phenyl rings described above. Direct S···π interactions could not be reproduced.
Methionine···aryl interactions are quite frequent in protein structures. Pal and Chakrabarti found methionine as frequently as aromatic amino acids in the environment of the tryptophan indole ring (distance between ring carbon and S of <4.0 Å).(217) Out of 1276 methionine residues, 9% were found to be in contact with an aromatic edge and 8% with an aromatic face. Imai et al. found that CH···S interactions account for the majority of all methionine sulfur interactions (∼40%) in the PDB. Close contacts between methionine sulfur and π systems occurred with a frequency of about 22%,(121) including electron-poor π systems such as amides. In the CSD, Zauhar et al. found a pronounced preference for thioethers to form CH···S interactions in the plane of the aryl ring.(218)Our searches agree with these findings. In fact, the trend that sulfur atoms of thioethers are preferentially found in the phenyl ring plane and not above it is also visible in the corresponding PDB query (Figure S-8). Overall, it seems that with regard to phenyl ring interactions, divalent sulfur behaves like a weakly negatively polarized atom not unlike chlorine.
The binding sites of adenine rings (as part of enzyme substrates or cofactors) repeatedly feature methionine side chains. Often, the C−S−C fragment is positioned in a coplanar fashion above the purine ring at van der Waals distance (∼4.0 Å). This arrangement is exemplified by the complex of catechol-O-methyltransferase with its cofactor S-adenosylmethionine. The cofactor is sandwiched between Met 91 and His 142 (Figure 20). The distance between the sulfur atom and the closest ring atom is 3.6 Å. It has not been studied in detail whether this type of arrangement is predefined by the (often highly conserved) adenine binding pockets or whether it is due to the electronic nature of the adenine ring system. There is a second common geometric arrangement between adenine and methionine side chains in which the terminal methyl group forms the interaction as found in the ab initio model studies of dimethyl sulfide and benzene.(216) This geometry is also observed with other nucleobases, for example, in the complex between herpes simplex virus type 1 thymidine kinase and thymidine,(219) and it is confirmed as the preferred interaction geometry between phenyl rings and sulfur-bound methyl groups in the CSD (Figure S-8).
Figure 20.
Interaction between methionine and adenine in the complex between S-adenosylmethionine and catechol-O-methyltranferase (PDB code 2cl5, distances in Å).
Summary of Typical Interaction Distances
It is our experience that successful interactive structure-based design as well as a meaningful analysis of experimental or calculated structures requires an intimate knowledge of the typical interaction geometries, in particular of the typical distances involved. We therefore conclude by listing typical interaction distance ranges in a separate table for reference purposes (Table 2). It lists distances for specific interactions discussed in the previous paragraphs, plus distance ranges of several further frequently occurring interactions. For interactions involving aromatic rings, we avoid distances to centroids and list atom−atom distances instead because these can be readily measured with most molecular modeling software packages. All distance ranges were derived by the same method (see Materials and Methods) from CSD data.
Table 2. Summary of Typical Interaction Distances of Selected Noncovalent Interactionsa.
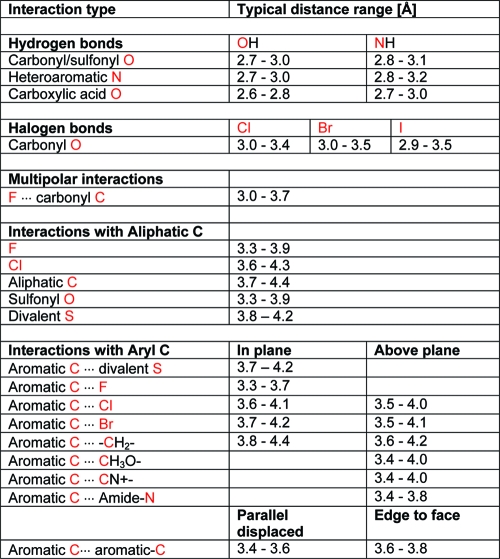 |
Distance values are given as the lower and upper 90% percentiles of the corresponding histogram peak extracted from CSD searches. Interactions are formed with the atoms highlighted in red. In the case of aryl carbon atoms, the closest distances observed were chosen.
Conclusions
We have shown how an understanding of typical noncovalent protein−ligand interactions can arise from the synergistic use of crystal structure database searching, structures, and association properties of biomolecular and synthetic receptor−ligand systems and various computational models. Awareness of geometric preferences of a specific interaction enables its recognition and application in different contexts. Although a clear decomposition of the energetic contributions of individual interactions is, strictly speaking, impossible, approximate relative propensities can be derived. Keeping in mind the big picture, i.e., the likelihood of alternative interactions any particular group can undergo, helps to avoid the mistake of overemphasizing any particular interaction. In this sense, there is still a lot to be learned from crystal structure databases. The results of such studies should become textbook knowledge but could also very well serve as a basis for improved knowledge-based force fields on the basis of functional groups rather than pairwise atom contacts. Structure-based design can certainly benefit from neighboring disciplines, in particular the analysis of supramolecular synthons in crystal engineering and the inverse design processes applied in protein engineering, both of which may foster an increased understanding of larger recurring patterns beyond the concept of functional group interactions.
It is striking that the multitude of different interaction types reviewed here has not yet become part of crystal structure visualization and molecular design programs. Visualization often does not go beyond the concepts of hydrogen bonds and van der Waals contacts. We have implemented these nonclassical types of interactions in our version of the macromolecular crystal structure database and visualization system, complemented by a flagging of unfavorable interactions such as unsatisfied hydrogen bond donors and acceptors in a lipophilic environment. Most of these options are now available in the form of PyMol scripts as part of Proasis2.(220) Already such simple enhancements have led to a better understanding of protein−ligand interactions at play and were well received by medicinal chemists. In the short term, we see possibilities for further improvements in a better visual representation of poorly solvated binding pockets, which offer potentially large affinity gains. Local dipolar interactions have been touched upon in this article. Beyond that, helix dipoles can be very strong but, to our knowledge, have not been proactively used in the design process. Such secondary structure elements can, for instance, be visualized in Relibase+.(47)
The increased availability of structural information has led to a great amount of detailed knowledge, but it has also overly focused our attention on “visible interactions”. We need to become more aware of the fact that many effects not explicitly present in a receptor−ligand complex structure (solvation/desolvation effects, changes in mobility, cooperativity) play a large and sometimes dominant role. More holistic conceptual models such as the one proposed by Hunter(87) could increase the awareness of solvent effects; similar concepts for entropy might be hard to develop. An increased appreciation of entropic aspects can perhaps only grow on the basis of detailed investigations of individual model systems and accurate thermodynamic data. Studies that carefully probe the energetic contributions of individual subpockets or ligand components, such as the decomposition of known ligands into fragments, can provide a significant amount of insight and should become routine practice in medicinal chemistry.
Finally, as the level of confidence in structure-based design has increased, it is important to emphasize that any modeled binding mode remains a model until it is experimentally validated. Sadly, it has become common practice today to mingle models and reality. We close with a call for a more careful assessment of modeled structures. No binding modes from docking or molecular dynamics calculations should go unchallenged. The knowledge base for such scrutiny is large and is waiting for its application.
Materials and Methods
All statistical data on small molecule crystal structures described in this article were derived from searches in the CSD, version 5.30 (November 2008), with the ConQuest 1.11 program.(221) Unless noted otherwise, the following general search flags were set: R factor of ≤0.10, “3D coordinates determined”, “not disordered”, “no ions”, “no errors”, “not polymeric”, and “only organic.” Protein−ligand interactions were analyzed by extracting binding sites in SD format(222) from Proasis2,(220) a curated version of the PDB, and generating a separate database in CSD format with the program PreQuest.(223) Binding sites were extracted around all HET entries, except metals and commonly found small ions, in the PDB as of December 8, 2008. Those with less than 5 or more than 100 ligand atoms were removed, excluding in particular large peptidic ligands. Multiple occurrences of the same ligand in one PDB structure were counted as separate entries. This database contains a total of 77 378 binding sites derived from 25 096 PDB entries. Queries were run in identical manner in the CSD and the PDB ligand database with one exception: For CSD searches, explicit hydrogen definitions were used, whereas the absence of hydrogen atoms in the PDB ligands required the use of implicit hydrogen counts on carbon.
Where preferred interaction distances are quoted in the text, they were derived in the following manner: Distance histograms were generated from counts divided by 4πr2, where r is the interaction distance, in order to account for the increasing volume available at larger distances. Histograms were plotted in 0.05 Å bins. The peaks in the distance histograms were visually located and characterized by their median and the lower and upper 90% percentiles (i.e., the range containing 80% of the hits). These are the range values given in the text and in Table 2. Angle distributions were treated in a similar fashion. Angles between geometric objects (atoms, planes) were normalized by the “cone correction” (1/sin(angle)).(79)
To derive frequencies of occurrence of noncovalent interactions, CSD entries containing the interacting fragments exactly once were compiled. The frequency of occurrence is defined by the fraction of entries that form the interaction within the geometric limits defined earlier.
Maps for interactions between functional groups and phenyl rings were created by measuring, for a query atom X, the distance above the phenyl ring plane h and the distance from the centroid to the query atom r (Figure 21). From these parameters, the in-plane distance between X and the centroid was calculated (centroid shift s). A limit of 7 Å was used for the centroid−X distance. To avoid spurious mapping of interactions “behind” the phenyl ring, queries were limited to a half sphere by limiting the angle C−H1−X to ≤90°. Plots of h vs s effectively project the space of this half sphere to two dimensions. Radial distribution plots were generated by counting hits in 0.1 Å × 0.1 Å distance bins (h vs s) and scaled by 1/(2πr2), where r is the distance between the centroid and the query atom X. In addition, all bins were scaled by the total number of counts.
Figure 21.
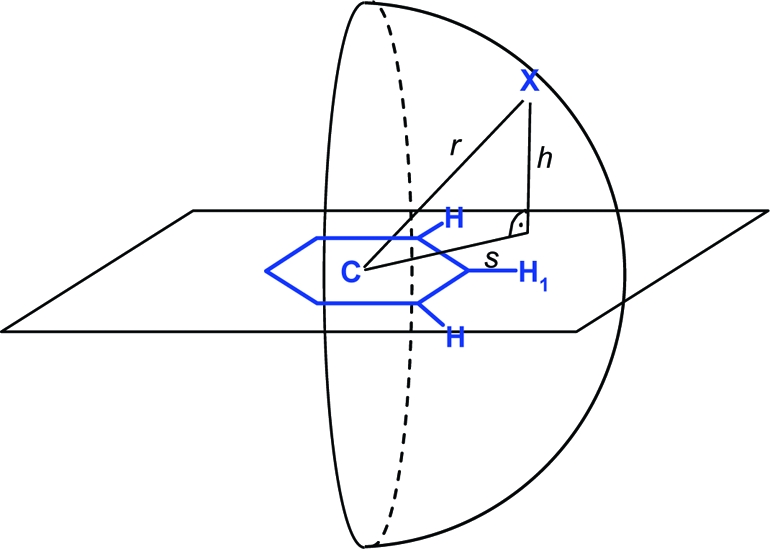
Query setup used for searching interactions between query atoms X and phenyl rings.
All depictions of chemical structures in this article have been generated with PyMOL.(224)
Acknowledgments
We thank Julian Fuchs for support in compiling interaction statistics and Johannes Hermann, François Diederich, and Klaus Müller for many valuable discussions. We also thank two anonymous referees for many constructively critical comments and additional references.
Biographies
Caterina Bissantz earned her Pharm.D. (1998) from University of Freiburg, Germany, her M.Sc. (1999) from Purdue University, West Lafayette, IN, under the supervision of Dr. Don Bergstrom in the field of synthetic medicinal chemistry, and her Ph.D. (2002) from ETH Zurich, Switzerland, under the supervision of Prof. Gerd Folkers and Dr. Didier Rognan in the field of virtual screening and GPCR modeling. In 2002, she joined Roche, where she is a member of the Molecular Design Group. Her special areas of interest are structure-based and GPCR drug design. She has coauthored more than 40 papers and patents.
Bernd Kuhn received his M.S. in Chemistry (1994) from the University of Karlsruhe, Germany, and Ph.D. (1998) from the Swiss Federal Institute of Technology, Lausanne, Switzerland, under the supervision of Prof. Tom Rizzo. Following postdoctoral studies as a DAAD fellow in macromolecular simulations with Prof. Peter Kollman at the University of California, San Francisco, he joined the start-up company Prospect Genomics in 2000. In 2001 he moved to F. Hoffmann-La Roche, Basel, Switzerland, where he is a member of the Molecular Design group with a focus on structure-based computational methods and their application to drug design projects. His scientific contributions include more than 90 papers and patents.
Martin Stahl received his Ph.D. in Theoretical Chemistry from the University of Marburg, Germany, under the supervision of Prof. G. Frenking. He joined Roche in 1997 and initially worked on structure-based design projects at the Basel and Palo Alto sites. In 2001, he was appointed head of molecular design in Basel. Since 2005, he has been a section head in medicinal chemistry. A member of the global chemistry leadership team, he is responsible for the Roche-wide coordination of molecular design and cheminformatics work. He has helped to develop and apply multiple novel methods in computational chemistry and is a fervent advocate of joint interactive design sessions between chemists and molecular designers. Martin has coauthored over 60 scientific articles and is an Advisory Board Member of Journal of Medicinal Chemistry.
Supporting Information Available
Additional figures of CSD statistics and PDB examples referenced in the text by the prefix “S-”. This material is available free of charge via the Internet at http://pubs.acs.org.
Footnotes
Abbreviations: CSD, Cambridge Structural Database; PDB, Protein Data Bank; ITC, isothermal titration calorimetry; MD, molecular dynamics; MUP, mouse major urinary protein; EGFR, epidermal growth factor receptor; MAP, mitogen-activated protein; iNOS, inducible nitric oxide synthetase; PDE10, phosphodiesterase 10; OppA, oligopeptide-binding; DPP-IV, dipeptidylpeptidase IV; LAO, lysine-, arginine-, ornithine-binding protein.
Supplementary Material
References
- Allen F. H. The Cambridge Structural Database: a quarter of a million crystal structures and rising. Acta Crystallogr. 2002, B58, 380–388. [DOI] [PubMed] [Google Scholar]
- Bernstein F. C.; Koetzle T. E.; Williams G. J. B.; Meyer J., E. F.; Brice M. D.; Rodgers J. R.; Kennard O.; Shimanouchi T.; Tasumi M. The protein data bank: a computer-based archival file for macromolecular structures. J. Mol. Biol. 1977, 112, 535–542; http://www.rcsb.org/pdb/. [DOI] [PubMed] [Google Scholar]
- Mark A. E.; van Gunsteren W. F. Decomposition of the free energy of a system in terms of specific interactions. Implications for theoretical and experimental studies. J. Mol. Biol. 1994, 240, 167–176. [DOI] [PubMed] [Google Scholar]
- Dill K. A. Additivity principles in biochemistry. J. Biol. Chem. 1997, 272, 701–704. [DOI] [PubMed] [Google Scholar]
- Olsson T. S. G.; Williams M. A.; Pitt W. R.; Ladbury J. E. The thermodynamics of protein−ligand interaction and solvation: insights for ligand design. J. Mol. Biol. 2008, 384, 1002–1017. [DOI] [PubMed] [Google Scholar]
- http://scorpio.biophysics.ismb.lon.ac.uk/scorpio.html (accessed March 12, 2010).
- Chervenak M. C.; Toone E. J. A direct measure of the contribution of solvent reorganization to the enthalpy of binding. J. Am. Chem. Soc. 1994, 116, 10533–10539. [Google Scholar]
- Gilson M. K.; Zhou H.-X. Calculation of protein-ligand binding affinities. Annu. Rev. Biophys. Biomol. Struct. 2007, 36, 21–42. [DOI] [PubMed] [Google Scholar]
- Myszka D. G.; Abdiche Y. N.; Arisaka F.; Byron O.; Eisenstein E.; Hensley P.; Thomson J. A.; Lombardo C. R.; Schwarz F.; Stafford W.; Doyle M. L. The ABRF-MIRG’02 study: assembly state, thermodynamic, and kinetic analysis of an enzyme/inhibitor interaction. J. Biomol. Tech. 2003, 14, 247–269. [PMC free article] [PubMed] [Google Scholar]
- Tellinghuisen J. Calibration in isothermal titration calorimetry: heat and cell volume from heat of dilution of NaCl(aq). Anal. Biochem. 2007, 360, 47–55. [DOI] [PubMed] [Google Scholar]
- Klebe G.; Dullweber F.; Boehm H.-J.. Thermodynamic Models of Drug−Receptor Interactions: A General Introduction. In Drug−Receptor Thermodynamics: Introduction and Applications; Raffa R. B., Ed.; Wiley: Chichester, U.K., 2001; pp 83−103. [Google Scholar]
- Dullweber F.; Stubbs M. T.; Musil D.; Stürzebecher J.; Klebe G. Factorising ligand affinity: a combined thermodynamic and crystallographic study of trypsin and thrombin inhibition. J. Mol. Biol. 2001, 313, 593–614. [DOI] [PubMed] [Google Scholar]
- Freire E.A Thermodynamic Guide to Affinity Optimization of Drug Candidates. In Protein Reviews; Waksman G., Ed.; Springer: New York, 2005; Vol. 3, pp 291−307. [Google Scholar]
- Freire E. Do enthalpy and entropy distinguish first in class from best in class?. Drug Discovery Today 2008, 13, 869–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exner O. Entropy−enthalpy compensation and anticompensation: solvation and ligand binding. Chem. Commun. 2000, 1655–1656. [Google Scholar]
- Sharp K. Entropy−enthalpy compensation: fact or artifact?. Protein Sci. 2001, 10, 661–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallicchio E.; Kubo M. M.; Levy R. M. Entropy−enthalpy compensation in solvation and ligand binding revisited. J. Am. Chem. Soc. 1998, 120, 4526–4527. [Google Scholar]
- Ford D. M. Enthalpy−entropy compensation is not a general feature of weak association. J. Am. Chem. Soc. 2005, 127, 16167–16170. [DOI] [PubMed] [Google Scholar]
- Dunitz J. D. Win some, lose some: enthalpy−entropy compensation in weak molecular interactions. Chem. Biol. 1995, 2, 709–712. [DOI] [PubMed] [Google Scholar]
- Houk K. N.; Leach A. G.; Kim S. P.; Zhang X. Binding affinities of host−guest, protein−ligand and protein−transition state complexes. Angew. Chem., Int. Ed. 2003, 42, 4872–4897. [DOI] [PubMed] [Google Scholar]
- Whitesides G. M.; Krishnamurthy V. M. Designing ligands to bind proteins. Q. Rev. Biophys. 2005, 38, 385–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams D. H.; Stephens E.; O’Brien D. P.; Zhou M. Understanding noncovalent interactions: ligand binding energy and catalytic efficiency from ligand-induced reductions in motion within receptors and enzymes. Angew. Chem., Int. Ed. 2004, 43, 6596–6616. [DOI] [PubMed] [Google Scholar]
- Frederick K. K.; Marlow M. S.; Valentine K. G.; Wand J. Conformational entropy in molecular recognition by proteins. Nature 2007, 448, 325–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams D. H.; Stephens E.; Zhou M. Ligand binding energy and catalytic efficiency from improved packing within receptors and enzymes. J. Mol. Biol. 2003, 329, 389–399. [DOI] [PubMed] [Google Scholar]
- Hunter C. A.; Tomas S. Cooperativity, partially bound states, and enthalpy−entropy compensation. Chem. Biol. 2003, 10, 1023–1032. [DOI] [PubMed] [Google Scholar]
- Williams D. H.; Davies N. L.; Koivisto J. J. Importance of structural tightening, as opposed to partially bound states, in the determination of chemical shift changes at noncovalently bonded interfaces. J. Am. Chem. Soc. 2004, 126, 14267–14272. [DOI] [PubMed] [Google Scholar]
- Chekmeneva E.; Hunter C. A.; Packer M. J.; Turega S. M. Evidence for partially bound states in cooperative molecular recognition interfaces. J. Am. Chem. Soc. 2008, 130, 17718–17725. [DOI] [PubMed] [Google Scholar]
- Talhout R.; Villa A.; Mark A. E.; Engberts J. B. F. N. Understanding binding affinity: a combined isothermal titration calorimetry/molecular dynamics study of the binding of a series of hydrophobically modified benzamidinium chloride inhibitors to trypsin. J. Am. Chem. Soc. 2003, 125, 10570–10579. [DOI] [PubMed] [Google Scholar]
- Krishnamurthy V. M.; Bohall B. R.; Semetey V.; Whitesides G. M. The paradoxical thermodynamic basis for the interaction of ethylene glycol, glycine, and sarcosine chains with bovine carbonic anhydrase II: an unexpected manifestation of enthalpy/entropy compensation. J. Am. Chem. Soc. 2006, 128, 5802–5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muley L.; Baum B.; Smolinski M.; Freindorf M.; Heine A.; Klebe G.; Hangauer D. Enhancement of hydrophobic interactions and hydrogen bond strength by cooperativity: synthesis, modeling and MD-simulations of a congeneric series of thrombin inhibitors. J. Med. Chem. 2010, 53, 2126–2135. [DOI] [PubMed] [Google Scholar]
- Baum B.; Muley L.; Smolinski M.; Heine A.; Hangauer D.; Klebe G.. Non-additivity of functional group contributions in protein−ligand binding: a comprehensive study by crystallography and isothermal titration calorimetry. J. Mol. Biol. 2010, DOI: 10.1016/j.jmb.2010.02.007. [DOI] [PubMed] [Google Scholar]
- Scatena L. F.; Brown M. G.; Richmond G. L. Water at hydrophobic surfaces: weak hydrogen bonding and strong orientation effects. Science 2001, 292, 908–912. [DOI] [PubMed] [Google Scholar]
- Finney J. L.; Soper A. K. Solvent structure and perturbations in solutions of chemical and biological importance. Chem. Soc. Rev. 1994, 23, 1–10. [Google Scholar]
- Dill K. A.; Truskett T. M.; Vlachy V.; Hribar-Lee B. Modeling water, the hydrophobic effect, and ion solvation. Annu. Rev. Biophys. Biomol. Struct. 2005, 34, 173–199. [DOI] [PubMed] [Google Scholar]
- Meyer E. A.; Castellano R. K.; Diederich F. Interactions with aromatic rings in chemical and biological recognition. Angew. Chem., Int. Ed. 2003, 42, 1210–1250. [DOI] [PubMed] [Google Scholar]
- Homans S. W. Water, water everywhere—except where it matters?. Drug Discovery Today 2007, 12, 534–539. [DOI] [PubMed] [Google Scholar]
- Syme N. R.; Dennis C.; Phillips S. E. V.; Homans S. W. Origin of heat capacity changes in a “nonclassical” hydrophobic interaction. ChemBioChem 2007, 8, 1509–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berne B. J.; Weeks J. D.; Zhou R. Dewetting and hydrophobic interaction in physical and biological systems. Annu. Rev. Phys. Chem. 2009, 60, 85–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaerner F.-G.; Kahlert B.; Nellesen A.; Zienau J.; Ochsenfeld C.; Schrader T. Molecular tweezer and clip in aqueous solution: unexpected self-assembly, powerful host-guest complex formation, quantum chemical 1H NMR shift calculation. J. Am. Chem. Soc. 2006, 128, 4831–4841. [DOI] [PubMed] [Google Scholar]
- Young R. J.; Campbell M.; Borthwick A. D.; Brown D.; Burns-Kurtis C. L.; Chan C.; Convery M. A.; Crowe M. C.; Dayal S.; Diallo H.; Kelly H. A.; King N. P.; Kleanthous S.; Mason A. M.; Mordaunt J. E.; Patel C.; Pateman A. J.; Senger S.; Shah G. P.; Smith P. W.; Watson N. S.; Weston H. E.; Zhou P. Structure- and property-based design of factor Xa inhibitors: pyrrolidin-2-ones with acyclic alanyl amides as P4 motifs. Bioorg. Med. Chem. Lett. 2006, 16, 5953–5957. [DOI] [PubMed] [Google Scholar]
- Abel R.; Young T.; Farid R.; Berne B. J.; Friesner R. A. Role of the active-site solvent in the thermodynamics of factor Xa ligand binding. J. Am. Chem. Soc. 2008, 130, 2817–2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young T.; Abel R.; Kim B.; Berne B. J.; Friesner R. A. Motifs for molecular recognition exploiting hydrophobic enclosure in protein−ligand binding. Proc. Natl. Acad. Sci. U.S.A. 2007, 104, 808–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt M.; Park B. H. Water: now you see it, now you don’t. Structure 1993, 1, 223–226. [DOI] [PubMed] [Google Scholar]
- Ringe D.; Mattos C. Analysis of the binding surfaces of proteins. Med. Res. Rev. 1999, 19, 321–331. [DOI] [PubMed] [Google Scholar]
- Ladbury J. E. Just add water! The effect of water on the specificity of protein−ligand binding sites and its potential application to drug design. Chem. Biol. 1996, 3, 973–980. [DOI] [PubMed] [Google Scholar]
- Poornima C. S.; Dean P. M. Hydration in drug design. 2. Influence of local site surface shape on water binding. J. Comput.-Aided Mol. Des. 1995, 9, 513–520. [DOI] [PubMed] [Google Scholar]
- http://www.ccdc.cam.ac.uk/products/life_sciences/relibase/ (accessed March 12, 2010).
- Dunitz J. D. The entropic cost of bound water in crystals and biomolecules. Science 1994, 264, 670. [DOI] [PubMed] [Google Scholar]
- Denisov V. P.; Venu K.; Peters J.; Hoerlein H. D.; Halle B. Orientational disorder and entropy of water in protein cavities. J. Phys. Chem. B 1997, 101, 9380–9389. [Google Scholar]
- Matthews B. W.; Liu L. A review about nothing: are apolar cavities in proteins really empty?. Protein Sci. 2009, 18, 494–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y.; Ratner M. A.; Jarrold M. F. One water molecule stiffens a protein. J. Am. Chem. Soc. 2000, 122, 2950–2951. [Google Scholar]
- Fischer S.; Verma C. S. Binding of buried structural water increases the flexibility of proteins. Proc. Natl. Acad. Sci. U.S.A. 1999, 96, 9613–9615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barillari C.; Taylor J.; Viner R.; Essex J. W. Classification of water molecules in protein binding sites. J. Am. Chem. Soc. 2007, 129, 2577–2587. [DOI] [PubMed] [Google Scholar]
- Michel J.; Tirado-Rives J.; Jorgensen W. L. Energetics of displacing water molecules from protein binding sites: consequences for ligand optimization. J. Am. Chem. Soc. 2009, 131, 15403–15411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Sosa A. T.; Mancera R. L.; Dean P. M. WaterScore: a novel method for distinguishing between bound and displaceable water molecules in the crystal structure of the binding site of protein-ligand complexes. J. Mol. Model. 2003, 9, 172–182. [DOI] [PubMed] [Google Scholar]
- Amadasi A.; Surface J. A.; Spyrakis F.; Cozzini P.; Mozzarelli A.; Kellogg G. E. Robust classification of “relevant” water molecules in putative protein binding sites. J. Med. Chem. 2008, 51, 1063–1067. [DOI] [PubMed] [Google Scholar]
- Boehm H.-J.; Boehringer M.; Bur D.; Gmuender H.; Huber W.; Klaus W.; Kostrewa D.; Kuehne H.; Luebbers T.; Meunier-Keller N.; Mueller F. Novel inhibitors of DNA gyrase: 3D structure based biased needle screening, hit validation by biophysical methods, and 3D guided optimization. A promising alternative to random screening. J. Med. Chem. 2000, 43, 2664–2674. [DOI] [PubMed] [Google Scholar]
- Garcia-Sosa A. T.; Firth-Clark S.; Mancera R. L. Including tightly-bound water molecules in de novo drug design. Exemplification through the in silico generation of poly(ADP-ribose)polymerase ligands. J. Chem. Inf. Model. 2005, 45, 624–633. [DOI] [PubMed] [Google Scholar]
- Wissner A.; Berger D. M.; Boschelli D. H.; Floyd M. B. Jr.; Greenberger L. M.; Gruber B. C.; Johnson B. D.; Mamuya N.; Nilakantan R.; Reich M. F.; Shen R.; Tsou H.-R.; Upeslacis E.; Wang Y. F.; Fei B. W.; Ye F.; Zhang N. 4-Anilino-6,7-dialkoxyquinoline-3-carbonitrile Inhibitors of epidermal growth factor receptor kinase and their bioisosteric relationship to the 4-anilino-6,7-dialkoxyquinazoline inhibitors. J. Med. Chem. 2000, 43, 3244. [DOI] [PubMed] [Google Scholar]
- Liu C.; Wrobleski S. T.; Lin J.; Ahmed G.; Metzger A.; Wityak J.; Gillooly K. M.; Shuster D. J.; McIntyre K. W.; Pitt S.; Shen D. R.; Zhang R. F.; Zhang H.; Doweyko A. M.; Diller D.; Henderson I.; Barrish J. C.; Dodd J. H.; Schieven G. L.; Leftheris K. 5-Cyanopyrimidine derivatives as a novel class of potent, selective, and orally active inhibitors of p38.alpha. MAP kinase. J. Med. Chem. 2005, 48, 6261–6270. [DOI] [PubMed] [Google Scholar]
- Chen J. M.; Xu S. L.; Wawrzak Z.; Basarab G. S.; Jordan D. B. Structure-based design of potent inhibitors of scytalone dehydratase: displacement of a water molecule from the active site. Biochemistry 1998, 37, 17735–17744. [DOI] [PubMed] [Google Scholar]
- Seo J.; Igarashi J.; Li H.; Martasek P.; Roman L. J.; Poulos T. L.; Silverman R. B. Structure-based design and synthesis of Nω-nitro-l-arginine-containing peptidomimetics as selective inhibitors of neuronal nitric oxide synthase. Displacement of the heme structural water. J. Med. Chem. 2007, 50, 2089–2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campiani G.; Kozlkowskl A. P.; Wang S.; Ming L.; Naccl V.; Saxena A.; Doctor B. P. Synthesis and anticholinesterase activity of huperzine A analogues containing phenol and catechol replacements for the pyridone ring. Bioorg. Med. Chem. Lett. 1998, 8, 1413–1418. [DOI] [PubMed] [Google Scholar]
- Karplus P. A.; Faerman C. Ordered water in macromolecular structure. Curr. Opin. Struct. Biol. 1994, 4, 770–776. [Google Scholar]
- Henchman R. H.; McCammon J. A. Extracting hydration sites around proteins from explicit water simulations. J. Comput. Chem. 2002, 23, 861–869. [DOI] [PubMed] [Google Scholar]
- Tame J. R. H.; Sleigh S. H.; Wilkinson A. J.; Ladbury J. E. The role of water in sequence-independent ligand binding by an oligopeptide transporter protein. Nat. Struct. Biol. 1996, 3, 998–1001. [DOI] [PubMed] [Google Scholar]
- Davies T. G.; Hubbard R. E.; Tame J. R. H. Relating structure to thermodynamics: the crystal structures and binding affinity of eight OppA−peptide complexes. Protein Sci. 1999, 8, 1432–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritschel T.; Kohler P. C.; Neudert G.; Heine A.; Diederich F.; Klebe G. How to replace the residual solvation shell of polar active site residues to achieve nanomolar inhibition of tRNA-guanine transglycosylase. ChemMedChem 2009, 4, 2012–2023. [DOI] [PubMed] [Google Scholar]
- Stengl B.; Meyer E. A.; Heine A.; Brenk R.; Diederich F.; Klebe G. Crystal structures of tRNA-guanine transglycosylase (TGT) in complex with novel and potent inhibitors unravel pronounced induced-fit adaptations and suggest dimer formation upon substrate binding. J. Mol. Biol. 2007, 370, 492–511. [DOI] [PubMed] [Google Scholar]
- Lee J.; Kim S.-H. Water polygons in high-resolution protein crystal structures. Protein Sci. 2009, 18, 1370–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaitheeswaran S.; Yin H.; Rasaiah J. C.; Hummer G. Water clusters in nonpolar cavities. Proc. Natl. Acad. Sci. U.S.A. 2004, 101, 17002–17005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xantheas S. S. Cooperativity and hydrogen bonding network in water clusters. Chem. Phys. 2000, 258, 225–231. [Google Scholar]
- Brameld K. A.; Kuhn B.; Reuter D. C.; Stahl M. Small molecule conformational preferences derived from crystal structure data. A medicinal chemistry focused analysis. J. Chem. Inf. Model. 2008, 48, 1–24. [DOI] [PubMed] [Google Scholar]
- Taylor R.; Kennard O.; Versichel W. Geometry of the imino-carbonyl (N−H···O:C) hydrogen bond. 1. Lone-pair directionality. J. Am. Chem. Soc. 1983, 105, 5761–5766. [Google Scholar]
- Lommerse J. P. M.; Price S. L.; Taylor R. Hydrogen bonding of carbonyl, ether, and ester oxygen atoms with alkanol hydroxyl groups. J. Comput. Chem. 1997, 18, 757–774. [Google Scholar]
- Nobeli I.; Price S. L.; Lommerse J. P. M.; Taylor R. Hydrogen bonding properties of oxygen and nitrogen acceptors in aromatic heterocycles. J. Comput. Chem. 1997, 18, 2060–2074. [Google Scholar]
- Bruno I. J.; Cole J. C.; Lommerse J. P. M.; Rowland R. S.; Taylor R.; Verdonk M. L. IsoStar: a library of information about nonbonded interactions. J. Comput.-Aided Mol. Des. 1997, 11, 525–537. [DOI] [PubMed] [Google Scholar]
- Boer D. R.; Kroon J.; Cole J. C.; Smith B.; Verdonk M. L. SuperStar: comparison of CSD and PDB-based interaction fields as a basis for the prediction of protein−ligand interactions. J. Mol. Biol. 2001, 313, 275–287. [DOI] [PubMed] [Google Scholar]
- Grzybowski B. A.; Ishchenko A. V.; DeWitte R. S.; Whitesides G. M.; Shaknovich E. I. Development of a knowledge-based potential for crystals of small organic molecules: calculation of energy surfaces for C=O···H−N hydrogen bonds. J. Phys. Chem. B 2000, 104, 7293–7298. [Google Scholar]
- Morozov A. V.; Kortemme T.; Tsemekhman K.; Baker D. Close agreement between the orientation dependence of hydrogen bonds observed in protein structures and quantum mechanical calculations. Proc. Natl. Acad. Sci. U.S.A. 2004, 101, 6946–6951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis A. M.; Teague S. J.; Kleywegt G. J. Application and limitations of X-ray crystallographic data in structure-based ligand and drug design. Angew. Chem., Int. Ed. 2003, 42, 2718–2736. [DOI] [PubMed] [Google Scholar]
- DePristo M. A.; Bakker P. I. W.; Blundell T. L. Heterogeneity and inaccuracy in protein structures solved by X-ray crystallography. Structure 2004, 12, 831–838. [DOI] [PubMed] [Google Scholar]
- Görbitz C. H.; Etter M. C. Hydrogen bonds to carboxylate groups. The question of three-center interactions. J. Chem. Soc., Perkin Trans. 2 1992, 131–135. [Google Scholar]
- Davis A. M.; Teague S. J. Hydrogen bonding, hydrophobic interactions, and failure of the rigid receptor hypothesis. Angew. Chem., Int. Ed. 1999, 38, 736–749. [DOI] [PubMed] [Google Scholar]
- Kubinyi H.Hydrogen Bonding, the Last Mystery in Drug Design? In Pharmacokinetic Optimization in Drug Research; Testa B., van de Waterbeemd H., Folkers G., Guy R., Eds.; Wiley-VCH: Weinheim, Germany, 2001; pp 513−524. [Google Scholar]
- Williams M. A.; Ladbury J. E. Hydrogen bonds in protein−ligand complexes. Methods Princ. Med. Chem. 2003, 19, 137–161. [Google Scholar]
- Hunter C. A. Quantifying intermolecular interactions: guidelines for the molecular recognition toolbox. Angew. Chem., Int. Ed. 2004, 43, 5310–5324. [DOI] [PubMed] [Google Scholar]
- McDonald I. K.; Thornton J. M. Satisfying hydrogen bonding potential in proteins. J. Mol. Biol. 1994, 238, 777–793. [DOI] [PubMed] [Google Scholar]
- Fleming P. J.; Rose G. D. Do all backbone polar groups in proteins form hydrogen bonds?. Protein Sci. 2005, 14, 1911–1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efimov A. V.; Brazhnikov E. V. Relationship between intramolecular hydrogen bonding and solvent accessibility of side-chain donors and acceptors in proteins. FEBS Lett. 2003, 554, 389–393. [DOI] [PubMed] [Google Scholar]
- Barratt E.; Bronowska A.; Vondrasek J.; Cerny J.; Bingham R.; Phillips S.; Homans S. W. Thermodynamic penalty arising from burial of a ligand polar group within a hydrophobic pocket of a protein receptor. J. Mol. Biol. 2006, 362, 994–1003. [DOI] [PubMed] [Google Scholar]
- Morgan B. P.; Scholtz J. M.; Ballinger M. D.; Zipkin I. D.; Bartlett P. A. Differential binding energy: a detailed evaluation of the influence of hydrogen bonding and hydrophobic groups on the inhibition of thermolysin by phosphorus-containing inhibitors. J. Am. Chem. Soc. 1991, 113, 297–301. [Google Scholar]
- McComas C. C.; Crowley B. M.; Boger D. L. Partitioning the loss in vancomycin binding affinity for d-Ala-d-Lac into lost H-bond and repulsive lone pair contributions. J. Am. Chem. Soc. 2003, 125, 9314–9315. [DOI] [PubMed] [Google Scholar]
- Foloppe N.; Fisher L. M.; Howes R.; Kierstan P.; Potter A.; Robertson A. G. S.; Surgenor A. E. Structure-based design of novel Chk1 inhibitors: insights into hydrogen bonding and protein−ligand affinity. J. Med. Chem. 2005, 48, 4332–4345. [DOI] [PubMed] [Google Scholar]
- Leftheris K.; Ahmed G.; Chan R.; Dyckman A. J.; Hussain Z.; Ho K.; Hynes J. Jr.; Letourneau J.; Li W.; Lin S.; Metzger A.; Moriarty K. J.; Riviello C.; Shimshock Y.; Wen J.; Wityak J.; Wrobleski S. T.; Wu H.; Wu J.; Desai M.; Gillooly K. M.; Lin T. H.; Loo D.; McIntyre K. W.; Pitt S.; Shen D. R.; Shuster D. J.; Zhang R.; Diller D.; Doweyko A.; Sack J.; Baldwin J.; Barrish J.; Dodd J.; Henderson I.; Kanner S.; Schieven G. L.; Webb M. The discovery of orally active triaminotriazine aniline amides as inhibitors of p38 MAP kinase. J. Med. Chem. 2004, 47, 6283–6291. [DOI] [PubMed] [Google Scholar]
- Abraham M. H.; Platts J. A. Hydrogen bond structural group constants. J. Org. Chem. 2001, 66, 3484–3491. [DOI] [PubMed] [Google Scholar]
- Laurence C.; Brameld K. A.; Graton J.; Le Questel J.-Y.; Renault E. The pKBHX database: toward a better understanding of hydrogen-bond basicity for medicinal chemists. J. Med. Chem. 2009, 52, 4073–4086. [DOI] [PubMed] [Google Scholar]
- a Hagelin H.; Murray J. S.; Brinck T.; Berthelot M.; Politzer P. Family-independent relationships between computed molecular surface quantities and solute hydrogen bond acidity/basicity and solute-induced methanol O−H infrared frequency shifts. Can. J. Chem. 1995, 73, 483–488. [Google Scholar]; b Gancia E.; Montana J. G.; Manallack D. T. Theoretical hydrogen bonding parameters for drug design. J. Mol. Graph. Modell. 2001, 19, 349–362. [DOI] [PubMed] [Google Scholar]; c Hao M.-H. Theoretical calculation of hydrogen-bonding strength for drug molecules. J. Chem. Theory Comput. 2007, 2, 863–872. [DOI] [PubMed] [Google Scholar]; d Kenny P. W. Hydrogen bonding, electrostatic potential, and molecular design. J. Chem. Inf. Model. 2009, 49, 1234–1244. [DOI] [PubMed] [Google Scholar]; e Nocker M.; Handschuh S.; Tautermann C.; Liedl K. R. Theoretical Prediction of Hydrogen Bond Strength for use in Molecular Modeling. J. Chem. Inf. Model. 2009, 49, 2067–2076. [DOI] [PubMed] [Google Scholar]
- Bingham A. H.; Davenport R. J.; Gowers L.; Knight R. L.; Lowe C.; Owen D. A.; Parry D. M.; Pitt W. R. A novel series of potent and selective IKK2 inhibitors. Bioorg. Med. Chem. Lett. 2004, 14, 409–412. [DOI] [PubMed] [Google Scholar]
- Morris J. J.; Hughes L. R.; Glen A. T.; Taylor P. J. Non-steroidal antiandrogens. Design of novel compounds based on an infrared study of the dominant conformation and hydrogen-bonding properties of a series of anilide antiandrogens. J. Med. Chem. 1991, 34, 447–455. [DOI] [PubMed] [Google Scholar]
- Nobeli I.; Price S. L.; Lommerse J. P. M.; Taylor R. Hydrogen bonding properties of oxygen and nitrogen acceptors in aromatic heterocycles. J. Comput. Chem. 1997, 18, 2060–2074. [Google Scholar]
- Koch O.; Bocola M.; Klebe G. Cooperative effects in hydrogen-bonding of protein secondary structure elements: a systematic analysis of crystal data using Secbase. Proteins 2005, 61, 310–317. [DOI] [PubMed] [Google Scholar]
- DeChancie J.; Houk K. N. The origins of femtomolar protein−ligand binding: hydrogen-bond cooperativity and desolvation energetics in the biotin-(strept)avidin binding site. J. Am. Chem. Soc. 2007, 129, 5419–5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsemekhman K.; Goldschmidt L.; Eisenberg D.; Baker D. Cooperative hydrogen bonding in amyloid formations. Protein Sci. 2007, 16, 761–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jadzyn J.; Zywucki B. Molecular structure of hydrogen bonded N,N′-diethylurea in nonpolar solvents. J. Mol. Struct. 1987, 158, 293–300. [Google Scholar]
- Panigrahi S. K.; Desiraju G. R. Strong and weak hydrogen bonds in the protein−ligand interface. Proteins 2007, 67, 128–141. [DOI] [PubMed] [Google Scholar]
- Etter M. C. Encoding and decoding hydrogen-bond patterns of organic compounds. Acc. Chem. Res. 1990, 23, 120–126. [Google Scholar]
- Macrae C. F.; Bruno I. J.; Chisholm J. A.; Edgington P. R.; McCabe P.; Pidcock E.; Rodriguez-Monge L.; Taylor R.; van de Streek J.; Wood P. A. Mercury CSD 2.0: new features for the visualization and investigation of crystal structures. J. Appl. Crystallogr. 2008, 41, 466–470. [Google Scholar]
- Jorgensen W. L.; Pranata J. Importance of secondary interactions in triply hydrogen bonded complexes: guanine−cytosine vs uracil-2,6-diaminopyridine. J. Am. Chem. Soc. 1990, 112, 2008–2010. [Google Scholar]
- Zimmerman S. C.; Murray T. J. New supramolecular architectures using hydrogen bonding. Philos. Trans. R. Soc. London, Ser. A 1993, 345, 49–56. [Google Scholar]
- Scheiner S.; Kar T. Underlying source of the relation between polypeptide conformation and strength of NH···O hydrogen bonds. J. Mol. Struct. 2007, 844−845, 166–172. [Google Scholar]
- Luo R.; David L.; Hung H.; Devaney J.; Gilson M. K. Strength of solvent-exposed salt-bridges. J. Phys. Chem. B 1999, 103, 727–736. [Google Scholar]
- Hoertner S. R.; Ritschel T.; Stengl B.; Kramer C.; Schweizer W. B.; Wagner B.; Kansy M.; Klebe G.; Diederich F. Potent inhibitors of tRNA-guanine transglycosylase, an enzyme linked to the pathogenicity of the Shigella bacterium: charge-assisted hydrogen bonding. Angew. Chem., Int. Ed. 2007, 46, 8266–8269. [DOI] [PubMed] [Google Scholar]
- Taylor N. R.; Cleasby A.; Singh O.; Skarzynski T.; Wonacott A. J.; Smith P. W.; Sollis S. L.; Howes P. D.; Cherry P. C.; Bethell R.; Colman P.; Varghese J. Dihydropyrancarboxamides related to zanamivir: a new series of inhibitors of influenza virus sialidases. 2. Crystallographic and molecular modeling study of complexes of 4-amino-4H-pyran-6-carboxamides and sialidase from influenza virus types A and B. J. Med. Chem. 1998, 41, 798–807. [DOI] [PubMed] [Google Scholar]
- Klaholz B. P.; Moras D. C−HO hydrogen bonds in the nuclear receptor RAR gamma: a potential tool for drug selectivity. Structure 2002, 10, 1197–1204. [DOI] [PubMed] [Google Scholar]
- Tóth G.; Bowers S. G.; Truong A. P.; Probst G. The role and significance of unconventional hydrogen bonds in small molecule recognition by biological receptors of pharmaceuctical relevance. Curr. Pharm. Des. 2007, 13, 3476–3493. [DOI] [PubMed] [Google Scholar]
- Desiraju G. R.; Sarkel S. N−H···O, O−H···O, and C−H···O hydrogen bonds in protein−ligand complexes: strong and weak interactions in molecular recognition. Proteins 2004, 54, 247–259. [DOI] [PubMed] [Google Scholar]
- Tsuzuki S.; Honda K.; Uchimaru T.; Mikami M.; Tanaba K. Origin of the attraction and directionality of the NH/π interaction: comparison with OH/pi and CH/pi interactions. J. Am. Chem. Soc. 2000, 122, 11450–11458. [Google Scholar]
- Ottiger P.; Pfaffen C.; Leist R.; Leutwyler S.; Bachorz R. A.; Klopper W. Strong N−H···pi hydrogen bonding in amide−benzene interactions. J. Phys. Chem. B 2009, 113, 2937–2943. [DOI] [PubMed] [Google Scholar]
- Steiner T.; Koellner G. Hydrogen bonds with pi-acceptors in proteins: frequencies and role in stabilizing local 3D structures. J. Mol. Biol. 2001, 305, 535–557. [DOI] [PubMed] [Google Scholar]
- Imai Y. N.; Inoue Y.; Yamamoto Y. Propensities of polar and aromatic amino acids in noncanonocal interactions: nonbonded contacts analysis of protein−ligand complexes in crystal structures. J. Med. Chem. 2007, 50, 1189–1196. [DOI] [PubMed] [Google Scholar]
- Foloppe N.; Fisher L. M.; Francis G.; Howes R.; Kierstan P.; Potter A. Identification of a buried pocket for potent and selective inhibition of Chk1: prediction and verification. Bioorg. Med. Chem. Lett. 2002, 14, 1792–1804. [DOI] [PubMed] [Google Scholar]
- Verhoest P. R.; Chapin D. S.; Corman M.; Fonseca K.; Harms J. F.; Hou X.; Marr E. S.; Menniti F. S.; Nelson F.; O’Connor R.; Pandit J.; Proulx-LaFrance C.; Schmidt A. W.; Schmidt C. J.; Suiciak J. A.; Liras S. Discovery of a novel class of phosphodiesterase 10A inhibitors and identification of clinical candidate 2-[4-(1-methyl-4-pyridin-4-yl-1H-pyrazol-3-yl)-phenoxymethyl]-quinoline (PF-2545920)(I) for the treatment of schizophrenia. J. Med. Chem. 2009, 52, 5188–5196. [DOI] [PubMed] [Google Scholar]
- Dunitz J. D.; Taylor R. Organic fluorine hardly ever accepts hydrogen bonds. Chem.—Eur. J. 1997, 3, 89–98. [Google Scholar]
- Böhm H.-J.; Banner D.; Bendels S.; Kansy M.; Kuhn B.; Müller K.; Obst-Sander U.; Stahl M. Fluorine in medicinal chemistry. ChemBioChem 2004, 5, 637–643. [DOI] [PubMed] [Google Scholar]
- a Parlow J. J.; Kurumbail R. G.; Stegeman R. A.; Stevens A. M.; Stallings W. C.; South M. S. Synthesis and X-ray crystal structures of substituted fluorobenzene and benzoquinone inhibitors of the tissue factor VIIa complex. Bioorg. Med. Chem. Lett. 2003, 13, 3721–3725. [DOI] [PubMed] [Google Scholar]; b Parlow J. J.; Kurumbail R. G.; Stegeman R. A.; Stevens A. M.; Stallings W. C.; South M. S. Design, synthesis, and crystal structure of selective 2-pyridone tissue factor VIIa inhibitors. J. Med. Chem. 2003, 46, 4696–4701. [DOI] [PubMed] [Google Scholar]
- Kim D.; Wang L.; Beconi M.; Eiermann G. J.; Fisher M. H.; He H.; Hickey G. J.; Kowalchick J. E.; Leiting B.; Lyons K.; Marsilio F.; McCann M. E.; Patel R. A.; Petrov A.; Scapin G.; Patel S. B.; Roy R. S.; Wu J. K.; Wyvratt M. J.; Zhang B. B.; Zhu L.; Thornberry N. A.; Weber A. E. (2R)-4-Oxo-4-[3-(trifluoromethyl)-5,6-dihydro[1,2,4]triazolo[4,3-a]pyrazin-7(8H)-yl]-1-(2,4,5-trifluorophenyl)butan-2-amine: a potent, orally active dipeptidyl peptidase IV inhibitor for the treatment of type 2 diabetes. J. Med. Chem. 2005, 48, 141–151. [DOI] [PubMed] [Google Scholar]
- Pierce A. C.; Sandretto K. L.; Bemis G. W. Kinase inhibitors and the case for CH···O hydrogen bonds in protein−ligand binding. Proteins 2002, 49, 567–576. [DOI] [PubMed] [Google Scholar]
- Pierce A. C.; ter Haar E.; Binch H. M.; Kay D. P.; Patel S. R.; Li P. CHO and CHN hydrogen bonds in ligand design: a novel quinazolin-4-ylthiazol-2-ylamine protein kinase inhibitor. J. Med. Chem. 2005, 48, 1278–1281. [DOI] [PubMed] [Google Scholar]
- Vargas R.; Garza J.; Dixon D. A.; Hay B. P. How strong is the Calpha-H···O=C hydrogen bond?. J. Am. Chem. Soc. 2000, 122, 4750–4755. [Google Scholar]
- Steiner T. Water molecules which apparently accept no hydrogen bonds are systematically involved in C−H···C interactions. Acta Crystallogr. 1995, D51, 93–97. [DOI] [PubMed] [Google Scholar]
- Olsen J. A.; Banner D. W.; Seiler P.; Sander U. O.; D’Arcy A.; Stihle M.; Mueller K.; Diederich F. A fluorine scan of thrombin inhibitors to map the fluorophilicity/fluorophobicity of an enzyme active site: evidence for C−F···C=O interactions. Angew. Chem., Int. Ed. 2003, 42, 2507–2511. [DOI] [PubMed] [Google Scholar]
- Chakrabarti P.; Bhattacharyya R. Geometry of nonbonded interactions involving planar groups in proteins. Prog. Biophys. Mol. Biol. 2007, 95, 83–137. [DOI] [PubMed] [Google Scholar]
- Ramasubbu N.; Parthasarathy R.; Murray-Rust P. Angular preferences of intermolecular forces around halogen centers: preferred directions of approach of electrophiles and nucleophiles around the carbon−halogen bond. J. Am. Chem. Soc. 1986, 108, 4308–4314. [Google Scholar]
- Clark T.; Hennemann M.; Murray J. S.; Politzer P. Halogen bonding: the sigma hole. J. Mol. Model. 2007, 13, 291–296. [DOI] [PubMed] [Google Scholar]
- Lommerse J. P. M.; Stone A. J.; Taylor R.; Allen F. H. The nature and geometry of intermolecular interactions between halogens and oxygen or nitrogen. J. Am. Chem. Soc. 1996, 118, 3108–3116. [Google Scholar]
- Glaser R.; Chen N.; Wu H.; Knotts N.; Kaupp M. 13C NMR study of halogen bonding of haloarenes: measurement of solvent effects and theoretical analyses. J. Am. Chem. Soc. 2004, 126, 4412–4419. [DOI] [PubMed] [Google Scholar]
- Sarwar M. G.; Dragisic B.; Salsberg L. J.; Gouliaras C.; Taylor M. S. Thermodynamics of halogen bonding in solution: substituent, structural, and solvent effects. J. Am. Chem. Soc. 2010, 132, 1646–1653. [DOI] [PubMed] [Google Scholar]
- Riley K. E.; Merz K. M. Jr. Insights into the strength and origin of halogen bonding: the halobenzene−formaldehyde dimer. J. Phys. Chem. A 2007, 111, 1688–1694. [DOI] [PubMed] [Google Scholar]
- Aakeroey C. B.; Fasulo M.; Schultheiss N.; Desper J.; Moore C. Structural competition between hydrogen bonds and halogen bonds. J. Am. Chem. Soc. 2007, 129, 13772–13773. [DOI] [PubMed] [Google Scholar]
- Lu Y.; Shi T.; Wang Y.; Yang H.; Yan X.; Luo X.; Jiang H.; Zhu W. Halogen bonding—a novel interaction for rational drug design?. J. Med. Chem. 2009, 52, 2854–2862. [DOI] [PubMed] [Google Scholar]
- Auffinger P.; Hays F. A.; Westhof E.; Ho P. S. Halogen bonds in biological molecules. Proc. Natl. Acad. Sci. U.S.A. 2004, 101, 16789–16794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metrangolo P.; Neukirch H.; Pilati T.; Resnati G. Halogen bonding based recognition processes: a world parallel to hydrogen bonding. Acc. Chem. Res. 2005, 38, 386–395. [DOI] [PubMed] [Google Scholar]
- Iltzsch M. H.; Uber S. S.; Tankersley K. O.; el Kouni M. H. Structure−activity relationship for the binding of nucleoside ligands to adenosine kinase from Toxoplasma gondii. Biochem. Pharmacol. 1995, 49, 1501–1512. [DOI] [PubMed] [Google Scholar]
- Parks D. J.; LaFrance L. V.; Calvo R. R.; Milkiewicz K. L.; Gupta V.; Lattanze J.; Ramachandren K.; Carver T. E.; Petrella E. C.; Cummings M. D.; Maguire D.; Grasberger B. L.; Lu T. 1,4-Benzodiazepine-2,5-diones as small molecule antagonists of the HDM2-p53 interaction: discovery and SAR. Bioorg. Med. Chem. Lett. 2005, 15, 765–770. [DOI] [PubMed] [Google Scholar]
- Benjahad A.; Guillemont J.; Andries K.; Nguyen C. H.; Grierson D. S. 3-Iodo-4-phenoxypyridinones (IOPY’s), a new family of highly potent non-nucleoside inhibitors of HIV-1 reverse transcriptase. Bioorg. Med. Chem. Lett. 2003, 13, 4309–4312. [DOI] [PubMed] [Google Scholar]
- Himmel D. M.; Das K.; Clark A. D. Jr.; Hughes S. H.; Benjahad A.; Oumouch S.; Guillemont J.; Coupa S.; Poncelet A.; Csoka I.; Meyer C.; Andries K.; Nguyen C. H.; Grierson D. S.; Arnold E. Crystal structures for HIV-1 reverse transcriptase in complexes with three pyridinone derivatives: a new class of non-nucleoside inhibitors effective against a broad range of drug-resistant strains. J. Med. Chem. 2005, 48, 7582–7591. [DOI] [PubMed] [Google Scholar]
- Regier Voth A.; Khuu P.; Oishi K.; Ho P. S. Halogen bonds as orthogonal molecular interactions to hydrogen bonds. Nat. Chem. 2009, 1, 74–79. [DOI] [PubMed] [Google Scholar]
- Maillard M. C.; Hom R. K.; Benson T. E.; Moon J. B.; Mamo S.; Bienkowski M.; Tomasselli A. G.; Woods D. D.; Prince D. B.; Paddock D. J.; Emmons T. L.; Tucker J. A.; Dappen M. S.; Brogley L.; Thorsett E. D.; Jewett N.; Sinha S.; John V. Design, synthesis, and crystal structure of hydroxyethyl secondary amine-based peptidomimetic inhibitors of human β-secretase. J. Med. Chem. 2007, 50, 776–781. [DOI] [PubMed] [Google Scholar]
- Heath R. J.; Yu Y.-T.; Shapiro M. A.; Olson E.; Rock C. O. Broad spectrum antimicrobial biocides target the FabI component of fatty acid synthesis. J. Biol. Chem. 1998, 273, 30316–30320. [DOI] [PubMed] [Google Scholar]
- Freundlich J. S.; Anderson J. W.; Sarantakis D.; Shieh H.-M.; Yu M.; Valderramos J.-C.; Lucumi E.; Kuo M.; Jacobs W. R.; Fidock D. A.; Schiehser G. A.; Jacobus D. P.; Sacchettini J. C. Synthesis, biological activity, and X-ray crystal structural analysis of diaryl ether inhibitors of malarial enoyl acyl carrier protein reductase. Part 1: 4′-Substituted triclosan derivatives. Bioorg. Med. Chem. Lett. 2005, 15, 5247–5252. [DOI] [PubMed] [Google Scholar]
- Paulini R.; Müller K.; Diederich F. Orthogonal multipolar interactions in structural chemistry and biology. Angew. Chem., Int. Ed. 2005, 44, 1788–1805. [DOI] [PubMed] [Google Scholar]
- Manas E. S.; Unwalla R. J.; Xu Z. B.; Malamas M. S.; Miller C. P.; Harris H. A.; Hsiao C.; Akopian T.; Hum W.-T.; Malakian K.; Wolfrom S.; Bapat A.; Bhat R. A.; Stahl M. L.; Somers W. S.; Alvarez J. C. Structure-based design of estrogen receptor-β selective ligands. J. Am. Chem. Soc. 2004, 126, 15106–15119. [DOI] [PubMed] [Google Scholar]
- Zhou P.; Zou J.; Tian F.; Shang Z. Fluorine bonding. How does it work in protein−ligand interactions?. J. Chem. Inf. Model. 2009, 49, 2344–2355. [DOI] [PubMed] [Google Scholar]
- Müller K.; Faeh C.; Diederich F. Fluorine in pharmaceuticals: looking beyond intuition. Science 2007, 317, 1881–1886. [DOI] [PubMed] [Google Scholar]
- Dimicoli J. L.; Bieth J.; Lhoste J. M. Trifluoroacetylated peptides as substrates and inhibitors of elastase: a nuclear magnetic resonance study. Biochemistry 1976, 15, 2230–2236. [DOI] [PubMed] [Google Scholar]
- Hughes D. L.; Sieker L. C.; Bieth J.; Dimicoli J. L. Crystallographic study of the binding of a trifluoroacetyl dipeptide anilide inhibitor with elastase. J. Mol. Biol. 1982, 162, 645–658. [DOI] [PubMed] [Google Scholar]
- Cowan-Jacob S. W.; G. F.; Floersheimer A.; Furet P.; Liebetanz J.; Rummel G.; Rheinberger P.; Centeleghe M.; D. F.; Manley P. W. Structural biology contributions to the discovery of drugs to treat chronic myelogenous leukaemia. Acta Crystallogr. 2007, D63, 80–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowan-Jacob S. Personal communication in 2009.
- Cox C. D.; Breslin M. J.; Mariano B. J.; Coleman P. J.; Buser C. A.; Walsh E. S.; Hamilton K.; Huber H. E.; Kohl N. E.; Torrent M.; Yan Y.; Kuo L. C.; Hartman G. D. Kinesin spindle protein (KSP) inhibitors. Part 1: The discovery of 3,5-diaryl-4,5-dihydropyrazoles as potent and selective inhibitors of the mitotic kinesin KSP. Bioorg. Med. Chem. Lett. 2005, 15, 2041–2045. [DOI] [PubMed] [Google Scholar]
- Swierczynski D.; Luboradzki R.; Dolgonos G.; Lipkowski J.; Schneider H.-J. Non-covalent interactions of organic halogen compounds with aromatic systems: analyses of crystal structure data. Eur. J. Org. Chem. 2005, 1172–1177. [Google Scholar]
- Prasanna M. D.; Row T. N. G. C−halogen···pi interactions and their influence on molecular conformation and crystal packing: a database study. Cryst. Eng. 2000, 3, 135–154. [Google Scholar]
- Carosati E.; Sciabola S.; Cruciani G. Hydrogen bonding interactions of covalently bonded fluorine atoms: from crystallographic data to a new angular function in the GRID force field. J. Med. Chem. 2004, 47, 5114–5125. [DOI] [PubMed] [Google Scholar]
- Matter H.; Nazare M.; Guessregen S.; Will D. W.; Schreuder H.; Bauer A.; Urmann M.; Ritter K.; Wagner M.; Wehner V. Evidence for C−Cl/C−Br···pi interactions as an important contribution to protein−ligand binding affinity. Angew. Chem., Int. Ed. 2009, 48, 2911–2916. [DOI] [PubMed] [Google Scholar]
- Di Magno S. G.; Sun H. The strength of weak interactions: aromatic fluorine in drug design. Curr. Top. Med. Chem. 2006, 6, 1473–1482. [DOI] [PubMed] [Google Scholar]
- Desiraju G. R. Hydrogen bridges in crystal engineering: interactions without borders. Acc. Chem. Res. 2002, 35, 565–573. [DOI] [PubMed] [Google Scholar]
- Imai Y. N.; Inoue Y.; Nakanishi I.; Kitaura K. Cl−π interactions in protein−ligand complexes. Protein Sci. 2008, 17, 1129–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roehrig S.; Straub A.; Pohlmann J.; Lampe T.; Pernerstorfer J.; Schlemmer K.-H.; Reinemer P.; Perzborn E. Discovery of the novel antithrombotic agent 5-chloro-N-({(5S)-2-oxo-3-[4-(3-oxomorpholin-4-yl)phenyl]-1,3-oxazolidin-5-yl}methyl)thiophene-2-carboxamide (BAY 59-7939): an oral, direct factor Xa inhibitor. J. Med. Chem. 2005, 48, 5900–5908. [DOI] [PubMed] [Google Scholar]
- Baum B.; Mohamed M.; Zayed M.; Gerlach C.; Heine A.; Hangauer D.; Klebe G. More than a simple lipophilic contact: a detailed thermodynamic analysis of nonbasic residues in the S1 pocket of thrombin. J. Mol. Biol. 2009, 390, 56–69. [DOI] [PubMed] [Google Scholar]
- Adams H.; Cockroft S. L.; Guardigli C.; Hunter C. A.; Lawson K. R.; Perkins J.; Spey S. E.; Urch C. J.; Ford R. Experimental measurement of noncovalent interactions between halogens and aromatic rings. ChemBioChem 2004, 5, 657–665. [DOI] [PubMed] [Google Scholar]
- Dunitz J. D. Organic fluorine: odd man out. ChemBioChem 2004, 5, 614–621. [DOI] [PubMed] [Google Scholar]
- Morgenthaler M.; Aebi J. D.; Grüninger F.; Mona D.; Wagner B.; Kansy M.; Diederich F. A fluorine scan of non-peptidic inhibitors of neprilysin: fluorophobic and fluorophilic regions in an enzyme active site. J. Fluorine Chem. 2008, 129, 852–856. [Google Scholar]
- Boehm H.-J.; Klebe G. What can we learn from molecular recognition in protein−ligand complexes for the design of new drugs?. Angew. Chem., Int. Ed. 1996, 35, 2588–2614. [Google Scholar]
- Vallone B.; Miele A. E.; Vecchini P.; Chiancone E.; Brunori M. Free energy of burying hydrophobic residues in the interface between protein subunits. Proc. Natl. Acad. Sci. U.S.A. 1998, 95, 6103–6107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southall N. T.; Dill K. A.; Haymet A. D. J. A view of the hydrophobic effect. J. Phys. Chem. B 2002, 106, 521–533. [Google Scholar]
- Mecozzi S.; Rebek J. The 55% solution: a formula for molecular recognition in the liquid state. Chem.—Eur. J. 1998, 4, 1016–1022. [Google Scholar]
- Zurcher M.; Diederich F. Structure-based drug design: exploring the proper filling of apolar pockets at enzyme active sites. J. Org. Chem. 2008, 73, 4345–4361. [DOI] [PubMed] [Google Scholar]
- Kuhn B.; Hennig M.; Mattei P. Molecular recognition of ligands in dipeptidyl peptidase IV. Curr. Top. Med. Chem. 2007, 7, 609–619. [DOI] [PubMed] [Google Scholar]
- Peters J.-U.; Weber S.; Kritter S.; Weiss P.; Wallier A.; Boehringer M.; Hennig M.; Kuhn B.; Loeffler B.-M. Aminomethylpyrimidines as novel DPP-IV inhibitors: a 100 000-fold activity increase by optimization of aromatic substituents. Bioorg. Med. Chem. Lett. 2004, 14, 1491–1493. [DOI] [PubMed] [Google Scholar]
- Boehringer M.; Fischer H.; Hennig M.; Hunziker D.; Huwyler J.; Kuhn B.; Loeffler B. M.; Luebbers T.; Mattei P.; Narquizian R.; Sebokova E.; Sprecher U.; Wessel H. P. Aryl- and heteroaryl-substituted aminobenzo[a]quinolizines as dipeptidyl peptidase IV inhibitors. Bioorg. Med. Chem. Lett. 2010, 20, 1106–1108. [DOI] [PubMed] [Google Scholar]
- Carey C.; Cheng Y. K.; Rossky P. J. Hydration structure of the α-chymotrypsin substrate binding pocket: the impact of constrained geometry. Chem. Phys. 2000, 258, 415–425. [Google Scholar]
- Brandl M.; Weiss M. S.; Jabs A.; Suehnel J.; Hilgenfeld R. CH···pi-interactions in proteins. J. Mol. Biol. 2001, 307, 357–377. [DOI] [PubMed] [Google Scholar]
- Burley S. K.; Petsko G. A. Aromatic−aromatic interaction: a mechanism of protein structure-stabilization. Science 1985, 229, 23–28. [DOI] [PubMed] [Google Scholar]
- Burley S. K.; Petsko G. A. Weakly polar interactions in proteins. Adv. Protein Chem. 1988, 39, 125–89. [DOI] [PubMed] [Google Scholar]
- Tsuzuki S.; Honda K.; Uchimaru T.; Mikami M.; Tanabe K. Origin of attraction and directionality of the pi/pi interaction: model chemistry calculations of benzene dimer interaction. J. Am. Chem. Soc. 2002, 124, 104–112. [DOI] [PubMed] [Google Scholar]
- Grover J. R.; Walters E. A.; Hui E. T. Dissociation energies of the benzene dimer and dimer cation. J. Phys. Chem. 1987, 91, 3233–3237. [Google Scholar]
- Krause H.; Ernstberger B.; Neusser H. J. Binding energies of small benzene clusters. Chem. Phys. Lett. 1991, 184, 411–417. [Google Scholar]
- McGaughey G. B.; Gagné M.; Rappé A. K. Pi-stacking interactions—alive and well in proteins. J. Biol. Chem. 1998, 273, 15458–15463. [DOI] [PubMed] [Google Scholar]
- Mecozzi S.; West A. P. Jr.; Dougherty D. A. Cation−pi interactions in aromatics of biological and medicinal interest: electrostatic potential surfaces as a useful qualitative guide. Proc. Natl. Acad. Sci. U.S.A. 1996, 93, 10566–10571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler S. E.; Houk K. N. Through-space effects of substituents dominate molecular electrostatic potentials of substituted arenes. J. Chem. Theory Comput. 2009, 5, 2301–2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinnokrot M. O.; Sherrill C. D. Substituent effects in pi−pi interactions: sandwich and T-shaped configurations. J. Am. Chem. Soc. 2004, 126, 7690–7697. [DOI] [PubMed] [Google Scholar]
- Turk J. A.; Smithrud D. B. Synthesis and physical properties of protein core mimetics. J. Org. Chem. 2001, 66, 8328–8335. [DOI] [PubMed] [Google Scholar]
- Tsuzuki S.; Honda K.; Uchimaru T.; Mikami M.; Tanabe K. The magnitude of the CH/pi interaction between benzene and some model hydrocarbons. J. Am. Chem. Soc. 2000, 122, 3746–3753. [Google Scholar]
- Tsuzuki S.; Uchimaru T. Magnitude and physical origin of intermolecular interactions of aromatic molecules: recent progress of computational studies. Curr. Org. Chem. 2006, 10, 745–762. [Google Scholar]
- James W. H. III; Muller C. W.; Buchanan E. G.; Nix M. G. D.; Guo L.; Roskop L.; Gordon M. S.; Slipchenko L. V.; Gellman S. H.; Zwier T. S. Intramolecular amide stacking and its competition with hydrogen bonding in a small foldamer. J. Am. Chem. Soc. 2009, 131, 14243–14245. [DOI] [PubMed] [Google Scholar]
- Zbinden K. G.; Anselm L.; Banner D. W.; Benz J.; Blasco F.; Decoret G.; Himber J.; Kuhn B.; Panday N.; Ricklin F.; Risch P.; Schlatter D.; Stahl M.; Thomi S.; Unger R.; Haap W. Design of novel aminopyrrolidine factor Xa inhibitors from a screening hit. Eur. J. Med. Chem. 2009, 44, 2787–2795. [DOI] [PubMed] [Google Scholar]
- Yuan C.; St. Jean D. J.; Liu Q.; Cai L.; Li A.; Han N.; Moniz G.; Askew B.; Hungate R. W.; Johansson L.; Tedenborg L.; Pyring D.; Williams M.; Hale C.; Chen M.; Cupples R.; Zhang J.; Jordan S.; Bartberger M. D.; Sun Y.; Emery M.; Wang M.; Fotsch C. The discovery of 2-anilinothiazolones as 11b-HSD1 inhibitors. Bioorg. Med. Chem. Lett. 2007, 17, 6056–6061. [DOI] [PubMed] [Google Scholar]
- Hynes J. Jr.; Dyckman A. J.; Lin S.; Wrobleski S. T.; Wu H.; Gillooly K. M.; Kanner S. B.; Lonial H.; Loo D.; McIntyre K. W.; Pitt S.; Shen D. R.; Shuster D. J.; Yang X.; Zhang R.; Behnia K.; Zhang H.; Marathe P. H.; Doweyko A. M.; Tokarski J. S.; Sack J. S.; Pokross M.; Kiefer S. E.; Newitt J. A.; Barrish J. C.; Dodd J.; Schieven G. L.; Leftheris K. Design, synthesis, and anti-inflammatory properties of orally active 4-(phenylamino)-pyrrolo[2,1-f][1,2,4]triazine p38a mitogen-activated protein kinase inhibitors. J. Med. Chem. 2008, 51, 4–16. [DOI] [PubMed] [Google Scholar]
- Gallivan J. P.; Dougherty D. A. A computational study of cation−pi interactions vs salt bridges in aqueous media: implications for protein engineering. J. Am. Chem. Soc. 2000, 122, 870–874. [Google Scholar]
- Gallivan J. P.; Dougherty D. A. Cation−pi nteractions in structural biology. Proc. Natl. Acad. Sci. U.S.A. 1999, 96, 9459–9464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry B. W.; Elvekrog M. M.; Tommos C. Environmental modulation of protein cation−.pi. interactions. J. Am. Chem. Soc. 2007, 129, 5308–5309. [DOI] [PubMed] [Google Scholar]
- Oh B.-H.; Ames G. F.-L.; Kim S.-H. Structural basis for multiple ligand specificity of the periplasmic lysine-, arginine-, ornithine-binding protein. J. Biol. Chem. 1994, 269, 26323–26330. [PubMed] [Google Scholar]
- Soerme P.; Arnoux P.; Kahl-Knutsson B.; Leffler H.; Rini J. M.; Nilsson U. J. Structural and thermodynamic studies on cation−π interactions in lectin−ligand complexes: high-affinity galectin-3 inhibitors through fine-tuning of an arginine−arene interaction. J. Am. Chem. Soc. 2005, 127, 1737–1743. [DOI] [PubMed] [Google Scholar]
- Cumpstey I.; Salomonsson E.; Sundin A.; Leffler H.; Nilsson U. J. Double affinity amplification of galectin−ligand interactions through arginine−arene interactions: synthetic, thermodynamic, and computational studies with aromatic diamido thiodigalactosides. Chem.—Eur. J. 2008, 14, 4233–4245. [DOI] [PubMed] [Google Scholar]
- Williamson D. S.; Borgognoni J.; Clay A.; Daniels Z.; Dokurno P.; Drysdale M. J.; Foloppe N.; Francis G. L.; Graham C. J.; Howes R.; Macias A. T.; Murray J. B.; Parsons R.; Shaw T.; Surgenor A. E.; Terry L.; Wang Y.; Wood M.; Massey A. J. Novel adenosine-derived inhibitors of 70 kDa heat shock protein, discovered through structure-based design. J. Med. Chem. 2009, 52, 1510–1513. [DOI] [PubMed] [Google Scholar]
- Biot C.; Buisine E.; Kwasigroch J.-M.; Wintjens R.; Rooman M. Probing the energetic and structural role of amino acid/nucleobase cation−π interactions in protein−ligand complexes. J. Biol. Chem. 2002, 277, 40816–40822. [DOI] [PubMed] [Google Scholar]
- Celie P. H. N.; van Rossum-Fikkert S. E.; van Dijk W. J.; Brejc K.; Smit A. B.; Sizma T. K. Nicotine and carbamylcholine binding to nicotinic acetylcholine receptors as studied in AChBP crystal structures. Neuron 2004, 41, 907–914. [DOI] [PubMed] [Google Scholar]
- Kryger G.; Silman I.; Sussman J. L. Structure of acetylcholinesterase complexed with E2020 (Aricept): implications for the design of new anti-Alzheimer drugs. Structure 1999, 7, 297–307. [DOI] [PubMed] [Google Scholar]
- Salonen L. M.; Bucher C.; Banner D. W.; Haap W.; Mary J.-L.; Benz J.; Kuster O.; Seiler P.; Schweizer W. B.; Diederich F. Cation−pi interactions at the active site of factor Xa: dramatic enhancement upon stepwise N-alkylation of ammonium ions. Angew. Chem., Int. Ed. 2009, 48, 811–814. [DOI] [PubMed] [Google Scholar]
- Mo Y.; Subramanian G.; Gao J.; Ferguson D. M. Cation−pi interactions: an energy decomposition analysis and its implication in delta-opioid receptor−ligand binding. J. Am. Chem. Soc. 2002, 124, 4832–4837. [DOI] [PubMed] [Google Scholar]
- Hughes R. M.; Benshoff M. L.; Waters M. L. Effects of chain length and N-methylation on a cation−.pi. interaction in a .beta.-hairpin peptide. Chem.—Eur. J. 2007, 13, 5753–5764. [DOI] [PubMed] [Google Scholar]
- Rosenfield R. E.; Parthasarathy R.; Dunitz J. D. Directional preferences of nonbonded atomic contacts with divalent sulfur. 1. Electrophiles and nuclephiles. J. Am. Chem. Soc. 1977, 99, 4860–4862. [Google Scholar]
- Grether U.; Benardeau A.; Benz J.; Binggeli A.; Blum D.; Hilpert H.; Kuhn B.; Marki H. P.; Meyer M.; Mohr P.; Puntener K.; Raab S.; Ruf A.; Schlatter D. Design and biological evaluation of novel, balanced dual PPAR alpha/gamma agonists. ChemMedChem 2009, 4, 951–956. [DOI] [PubMed] [Google Scholar]
- Duan G. L.; Smith V. H.; Weaver D. F. Characterization of aromatic-thiol pi-type hydrogen bonding and phenylalanine−cysteine side chain interactions through ab initio calculations and protein database analyses. J. Biomol. Struct. Dyn. 2001, 19, 115–128.11565843 [Google Scholar]
- Ringer A. L.; Senenko A.; Sherrill C. D. Models of S/pi interactions in protein structures: Comparison of the H2S−benzene complex with PDB data. Protein Sci. 2007, 16, 2216–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pranata J. Sulfur−aromatic interactions: a computational study of the dimethyl sulfide−benzene complex. Bioorg. Chem. 1997, 25, 213–219. [Google Scholar]
- Pal D.; Chakrabarti P. Non-hydrogen bond interactions involving the methionine sulfur atom. J. Biomol. Struct. Dyn. 2001, 19, 115–128. [DOI] [PubMed] [Google Scholar]
- Zauhar R. J.; Colbert C. L.; Morgan R. S.; Welsh W. J. Evidence for a strong sulfur−aromatic interaction derived from crystallographic data. Biopolymers 2000, 53, 233–248. [DOI] [PubMed] [Google Scholar]
- Pilger B. D.; Perozzo R.; Alber F.; Wurth C.; Folkers G.; Scapozza L. Substrate diversity of herpes simplex virus thymidine kinase. J. Biol. Chem. 1999, 274, 31967–31973. [DOI] [PubMed] [Google Scholar]
- http://www.desertsci.com/ (accessed March 12, 2010).
- Bruno I. J.; Cole J. C.; Edgington P. R.; Kessler M.; Macrae C. F.; McCabe P.; Pearson J.; Taylor R. New software for searching the Cambridge Structural Database and visualizing crystal structures. Acta Crystallogr. 2002, B58, 389–397. [DOI] [PubMed] [Google Scholar]
- Dalby A.; Nourse J. G.; Hounshell W. D.; Gushurst A. K. I.; Grier D. L. Description of several chemical structure file formats used by computer programs developed at Molecular Design Ltd. J. Chem. Inf. Comput. Sci. 1992, 32, 244–255. [Google Scholar]
- http://www.ccdc.cam.ac.uk/products/csd_system/prequest/ (accessed March 12, 2010).
- DeLano W. L.The PyMOL Molecular Graphics System; DeLano Scientific: Palo Alto, CA; http://www.pymol.org (accessed March 12, 2010). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.