Abstract
Fully-automated brain segmentation methods have not been widely adopted for clinical use because of issues related to reliability, accuracy, and limitations of delineation protocol. By combining the probabilistic-based FreeSurfer (FS) method with the Large Deformation Diffeomorphic Metric Mapping (LDDMM) based label propagation method, we are able to increase reliability and accuracy, and allow for flexibility in template choice. Our method uses the automated FreeSurfer subcortical labeling to provide a coarse to fine introduction of information in the LDDMM template-based segmentation resulting in a fully-automated subcortical brain segmentation method (FS+LDDMM).
One major advantage of the FS+LDDMM-based approach is that the automatically generated segmentations generated are inherently smooth, thus subsequent steps in shape analysis can directly follow without manual post-processing or loss of detail.
We have evaluated our new FS+LDDMM method on several databases containing a total of 50 subjects with different pathologies, scan sequences and manual delineation protocols for labeling the basal ganglia, thalamus, and hippocampus. In healthy controls we report Dice overlap measures of 0.81, 0.83, 0.74, 0.86 and 0.75 for the right caudate nucleus, putamen, pallidum, thalamus and hippocampus respectively. We also find statistically significant improvement of accuracy in FS+LDDMM over FreeSurfer for the caudate nucleus and putamen of Huntington’s disease and Tourette’s syndrome subjects, and the right hippocampus of Schizophrenia subjects.
Keywords: Computational Anatomy, Automated Segmentation, MR Imaging, FreeSurfer, Hippocampus, Basal Ganglia, Thalamus
1 Introduction
High-resolution structural magnetic resonance neuroimaging facilitates quantitative insight into normal brain structure and changes that occur in neuropsychiatric diseases such as Alzheimer’s, Parkinson’s, Huntington’s and schizophrenia among others. Accurate segmentation of subcortical nuclei such as the hippocampus, thalamus and basal ganglia influences the reliability and validity of subsequent volumetric and shape analyses. Even though it is closest to the gold standard, manual segmentation of entire datasets has become less desireable, or not feasible, comparing to accurate and reliable automated methods for the following reasons: 1) Databases now can contain upwards of hundreds, sometimes thousands cross-sectional and longitudinal MR images, and time required for training and actual segmentation is often significant – segmenting several structures may easily take several hours per scan. 2) Intra-rater reliability can be dificult to maintain for large databases segmented over weeks or months as “rater drift”, which is rater variation over time, becomes more significant (Spinks et al., 2002; Lacerda et al., 2003; Nugent III et al., 2007). Furthermore, studies involving multiple raters face the additional challenge of maintaining inter-rater reliability. 3) Finally, manual segmentations, even when iteratively performed using the transverse, coronal and sagittal views, usually result in jagged boundaries, which makes shape analysis dificult (see below).
The need for accurate, robust and cost-effective segmentation tools have led to the development of several automated or semi-automated tools for extracting and measuring anatomical shape and form, e.g. (Pitiot et al., 2004; Xia et al., 2007; Chupin et al., 2007; Yang and Duncan, 2004; Fischl et al., 2002, 2004; Hogan et al., 2000; Shen et al., 2002; Khan et al., 2005). These methods can be divided into the following broad categories:
Knowledge-driven methods make use of implicit or explicit anatomical knowledge to guide the segmentation.
Probabilistic-based methods treat segmentation as a classification problem and estimate the labeling that maximizes an a-posteriori probability given specific constraints.
Deformable template-based methods involve finding a geometric transformation from a pre-labeled template scan to the target scan, and propagating the labels with the same transformation to label the target brain.
Some recent knowledge-driven methods include Pitiot et al. (2004), which used expert-knowledge in the form of implicit training set statistics and explicit anatomical constraints to evolve deformable templates for each structure. More recently, Xia et al. (2007) applied a knowledge-driven approach to automatically segment the caudate nucleus by first delineating the lateral ventricles, then used shape and positional information to localize the boundaries. Chupin et al. (2007) used explicit knowledge to generate landmarks for guiding the competitive region growing of the hippocampus/amygdala complex. Although generally fully automated and fast, these knowledge-driven methods are specifically tailored and optimized for individual structures. Additionally, dificulties may be encountered if pathology, scan sequence, or manual delineation protocol differ from those that the method is designed for. Examples of probabilistic-based methods include Yang and Duncan (2004), which incorporated a level-set approach to their maximum a posteriori (MAP) estimation while constraining according to neighbouring structures. Recently, the FreeSurfer (FS) tool (Fischl et al., 2002, 2004) has been made available free for use in brain neuroanatomical analysis. Freesurfer’s subcortical processing pipeline uses a probabilistic approach to perform automated labeling of 37 brain structures, where each voxel in the MR image volume is classified using a probabilistic atlas generated by a training set of 41 manually labeled brains (surfer.nmr.mgh.harvard.edu/fswiki/AtlasSubjects). The procedure includes a neighborhood function to encode spatial information, a forward model of the MR scanner parameters to improve sequence-independence, and a nonlinear function to account for morphological differences between the atlas and the target brain. A key feature of FreeSurfer’s subcortical pipeline is that it is fully automated; manual correction steps are needed only for the cortical segmentation stages or for poor-quality MR scans due to high noise or movement where the initial Talairach normalization may fail. Note that we did not perform any manual correction steps for any scans we tested. However, voxel-wise labeling methods that employ probabilistic-atlases often involve averaging a training set which may cause some fine details to be lost when labeling voxels in a target scan. Also, similar to manual segmentations, voxel-wise labeling can also lead to non-smooth subcortical segmentations that may confound downstream shape analysis algorithms Wang et al. (2007a) due to “shape-noise”. Another limitation is with respect to the adaptability to differing protocols for defining subcortical shapes. Since the protocol for creating the atlas is fixed, it is not possible for individual groups to change it to adapt it to their own working protocol without recreating the training set.
Various intensity-based non-rigid registration tools for computing geometric transformations have been developed that are usable in deformable-template segmentation (Maintz and Viergever, 1998). However, issues such as tissue inhomogeneity, weakly-defined boundaries, or high variability between subjects present challenges for gray-scale registration methods alone, thus incorporating additional constraints such as corresponding landmarks can help to initialize the computation of matching transformation. It should be noted that several deformable-template methods exist which do not require landmark placement; in particular, Svarer et al. (2005) and Heckemann et al. (2006) segment subcortical structures with a high level of accuracy using multiple atlas propagation and label fusion.
Very high-dimensional registration methods can be viewed as desirable in the context of deformable-template segmentation since they can allow for displacements at a fine scale. However, a collection of previous work using very high-dimensional registration for segmenting subcortical structures in the brain (such as the basal ganglia, the hippocampus, the thalamus etc.) relied on manual placement of landmarks for initialization (Csernansky et al., 2004b; Wang et al., 2003; Hogan et al., 2000; Haller et al., 1997). For example, hippocampus mapping required the placement of 12 landmarks for global alignment and a further 22 local landmarks in each target scan (Haller et al., 1997). Shen et al. (2002) required landmarks to be placed on the boundaries of the hippocampi, but did not require correspondence between the two sets of landmarks. However, the number of landmarks required was very high, with at least 50 for each hippocampus.
Our previous work on caudate nuclei segmentation (Khan et al., 2005) involved placing landmarks on segmented ventricle surfaces to increase reliability by limiting degrees of freedom in landmark placement. These were subsequently used to initialize the computation of diffeomorphic transformations between the template and target scans using large deformation diffeomorphic mapping (LDDMM) (Beg et al., 2005) and the resulting maps were used to propagate the segmentation in the template to generate target subcortical segmentations.
Although the template-based segmentation methods discussed approach the accuracy of manual segmentations (Haller et al., 1997; Csernansky et al., 2000; Wang et al., 2007b), they may require substantial manual intervention and therefore are less attractive than fully-automated methods because of practicality and reliability issues specially in dealing with large databases. In this paper, we propose a new, fully-automated subcortical segmentation pipeline that uses the FreeSurfer subcortical segmentation to substitute for the landmark-based initialization in the diffeomorphic deformable template-based (i.e. LDDMM) segmentation, thereby eliminating the manual intervention step (i.e., landmark placement).
2 Method
Let images be represented by functions I: Ω → ℝ, where Ω ∈ ℝ3 is the domain of the 3D MR volume. The goal is to find the geometric transformation ϕj: Ω → Ω such that each target image Ij, j = 1 ··· N is registered accurately to the template I0; i.e. minimizing an appropriate metric such as , where ||||L2 is the L2 norm in the space of functions I. Let the operator Ψ represent the process of segmentation of an image manually Ψman (I) or via Freesurfer ΨFS (I). In all such cases, the segmentation yields a labeling Ψ: Ω → ℤ that labels each voxel of the image with an integer label for the subcortical structure to which the voxel belongs. Given the segmentation of a particular structure in the template I0, the corresponding structure in target space can be computed by transforming the manual template segmentation, .
2.1 FreeSurfer Labeling
The first step in the FS+LDDMM pipeline is the generation of the FreeSurfer subcortical labels as demonstrated in Fischl et al. (2002, 2004). FreeSurfer generates a 37 labels of the brain (Fischl et al., 2002), ΨFS (I), that includes 18 labels of subcortical structures and cerebro-spinal fluid (CSF) used by FS+LDDMM. Manual correction in FreeSurfer is not required in our work since we are restricted to subcortical structures; manual edits are required for cortical segmentation in FreeSurfer which is not the focus of this subcortical method.
In short, the FreeSurfer pipeline consistes of five stages: an affine registration with Talairach space, an initial volumetric labeling, bias field correction, nonlinear alignment to the Talairach space, and a final labeling of the volume. For a full description of the FreeSurfer processing steps, please refer to Fischl et al. (2002) and Fischl et al. (2004).
2.2 Region of Interest Generation
The LDDMM geometric transformation is computed on a region of interest (ROI) bounding the structures of interest and not on the whole brain; this is essential as global whole brain mappings using gradient methods are prone to trapping in local minima, thus restriction to a region of interest has a better likelihood of meeting the assumption that the images being registered can be mapped with an invertible transformation, a key to LDDMM computation.
The first step in generating the ROI sub-volumes is the coarse registration of the target image, Ij, to the template image, I0, centered around each subcortical structure of interest (SOI). This ensures that subcortical structures of interest such as the left or the right hippocampus or basal ganglia are in gross alignment with the corresponding structures in the template I0 before defining the boundaries of each ROI. Computing a separate affine transformation for each structure group, using the labels provided by FS, significantly improves the outcome compared to using a single transformation for the whole brain. The affine transformation matrix is found by minimizing the cost function
using standard gradient descent with the translation initialized by the center of mass of each image. The target MR image Ij and FreeSurfer labels ΨFS(Ij) are now transformed with this structure-specific affine transformation T to generate images T(Ij) and T(ΨFS(Ij) which are in gross alignment with I0.
A rectangular bounding box ρSOI: Ω → {0, 1} is defined on the template image I0 to be centered around the structure of interest using the extent of the labels of the structure of interest in the template. In addition to the extent of the template structures, we impose an allowance of 8 voxels in each direction to account for any mis-alignment that is likely to occur. We have found this allowance to be sufficient for all our test cases. This bounding box is then used to cut a sub-volume ROI ρSOI · I0 in the template and ρSOI · Ij in the target MR image. The corresponding FreeSurfer labels ρSOI · ΨFS (I0) and ρSOI · T(ΨFS(Ij)) are also transformed to sub-volumes.
For generation of all the stuctures tested on the same brain, four ROI’s would need to be defined as 1.) left caudate, left putamen, left nucleus accumbens, left pallidum, left thalamus, 2.) right caudate, right putamen, right nucleus accumbens, right pallidum, right thalamus, 3.) left hippocampus, 4.) right hippocampus. Figure 1 depicts the ROI generation process for a single ROI, in this case the right basal ganglia (ROI 2.).
Fig. 1.
Procedure for generating a region of interest (ROI) for a template (I0) and a target (Ij) MR image. The first step involves finding the affine transformation, T, which minimizes the mean-square error between the FreeSurfer structures of interest (SOI) of the template, , and the target, . After applying T to the target images, a bounding box, ρSOI, is defined based on the extents of the FreeSurfer template SOI,
2.3 Histogram-based Intensity Normalization
Inside the ROI defined for each structure, we perform a variant of histogram matching to ensure homogeneity of corresponding tissue type intensities between the images. Prior to this, the MR image intensities are rescaled by linearly mapping the range between the 0.5 and 99.5 percentile intensities to the full image intensity range. Now, given the Freesurfer segmentations of cerebro-spinal fluid (CSF), of gray matter (GM) and of white matter (WM) compartments in the brain, we define histogram landmarks as the median image intensity in each of these structures in the ROI. We find the piece-wise linear intensity transform that aligns these histogram landmarks to produce the histogram landmark-matched target MR image, HLM (ρSOI · T(Ij)). Essentially this intensity normalization step is a specialization of the intensity scale standardization by Nyul et al. (2000) where we assume knowledge of the tissue intensity distributions.
2.4 LDDMM-based Diffeomorphic Registration
LDDMM (Beg et al., 2005) generates a diffeomorphic transformations by minimizing the following energ functional:
| (1) |
where vt is a time-dependent vector field that is integrated to find the mapping, ϕ, and I0 and I1 are the template and target images respectively. The mapping, ϕ: Ω → Ω, is smooth and has a smooth inverse, thus anatomy is mapped consistently, without fusions or tears, while preserving smoothness of anatomical features. In this paper, we will denote LDDMM registration as a function, LDDMM: (I0, I1) ↦ ϕ, which takes two input images and outputs the optimal diffeomorphic map between the sub-volumes.
In keeping with a multi-resolution coarse-to-fine strategy, a three stage procedure for computing the optimal diffeomorphic transformation was developed, where at each stage, additional anatomical information is added into the optimization process, starting with binary segmentations, followed by smoothed MR sub-volume images, and then eventually by the unsmoothed MR sub-volume image to provide texture for the final mapping. Each step thus designed helps guide the optimization away from potential local minima as the subsequent matching stages initialize with the optimal velocity vector field and map ϕ computed at the previous stage.
In the first stage, LDDMM registration is performed using the FreeSurfer cerebro-spinal fluid (CSF) labels, or equivalently, the portion of the ventricles in the subvolume giving
Ventricles are a good choice for performing gross first-level mapping for subcortical nuclei; in the case that ventricles are considerably different in size, as is often the case in diseased states, then the larger ventricles have been properly registered.
In the second stage, the MRI sub-volume ROI convolved with a Gaussian Gσ mask of size (3×3×3, and standard deviation σ = 0.5) is used with the initial flow taken from the optimal flow found in the first stage ϕ(1). Hence, at the second stage, we get:
At the third stage, smoothing is removed and the original MR sub-volume ROI’s are mapped, with the mapping initialized with the optimal from the previous step ϕ(2):
After the multi-stage mapping, the expert manual segmentation given in the template space I0 is propagated to the target ROI space using the final LD-DMM transformation ϕ(3), followed by the inverse affine transformation to the target Ij whole brain space, thus generating the final automated labeling of the structures of interest:
The resulting automated target labels can then be thresholded if required and converted to binary images as interpolation has made them continuous through the course of the procedure. We threshold at the mid-intensity (e.g. 128 for 256-level images) to obtain binary segmentations prior to computation of binary image similarity metrics. Surface models for each segmentation are generated as iso-surfaces at the mid-intensity; the vertices of the surface models are used in the surface distance metrics.
2.5 Comparison Metrics
Accuracy and reliability of our FS+LDDMM automated segmentations are computed quantitatively through the use of several comparison metrics to manual and FreeSurfer segmentations. To compare the spatial similarity of the contours, we used the following metrics:
-
Dice Similarity Coefficient (DSC)
(2) where V(A) and V(B) are the volumes of segmented images A and B, where it is assumed A and B are binary segmentations. Perfect spatial correspondence between the two binary images will result in DSC = 1, whereas no corresponce will result in DSC = 0.
-
L1 Error
(3) where ΨM denotes the manual segmentation, and ΨA denotes the automated segmentation and ||f||L1 = Σx∈Ω|f(x)|. The L1 error is a voxel-wise measure of the intensity difference between two images, or segmentations in this case, normalized by the sum total of intensities in the manual segmentation. When binary segmentations are used, the L1 error becomes a normalized overlap score, the advantage is that binary segmentations are not required for this metric.
-
Symmetrized Hausdorff Distance
is the directed Hausdorff distance, where d(a, b) is the euclidean distance between two points on two different surfaces. To symmetrize this metric, we use the following:
(4) The Hausdorff distance gives an upper bound on the mismatch between the contours of the segmentations.
-
Symmetrized Mean Surface Distance
is the directed mean surface distance, and is symmetrized similarly with:
(5) The mean surface distance expresses on average the error between the two segmentation contours.
To determine whether the FS+LDDMM procedure generates segmentations that are more accurate than the FreeSurfer segmentations used for initialization, we performed paired t-tests on all the comparative metrics, i.e. the Dice similarity coefficient (DSC), L1 error, symmetrized Hausdorff distance and the symmetrized mean surface distance; we report the significance (p-value) of a mean difference.
3 Materials
We have used five different MR databases to validate the FS+LDDMM automated segmentations of various subcortical structures against expert manual segmentations (“gold-standard”). Specifically, we have looked at the basal ganglia in Huntington’s Disease, Tourette’s Syndrome and control subjects, the hippocampus in schizophrenia and Alzheimer’s Disease subjects, and the thalamus in control subjects. These datasets were used because they represented a variety of MR scanning parameters, subcortical structures and diseases. In addition, the datasets have also been used in previously published work validating landmark-initialized diffeomorphic image matching (Haller et al., 1997; Csernansky et al., 2000, 2004a; Wang et al., 2007b). The Huntington’s Disease (HD) dataset consisted of 16 subjects (7 male, 9 female), mean age 37 (SD=11) years, possessing the HD gene but not yet diagnosed with the disease. Images were acquired on a 1.5T GE Genesis Signa scanner using a SPGR sequence (TR=18ms,TE=3ms,N=2,flip angle=20°) with an axial orientation, image dimensions 256×256×124 and voxel dimensions 0.9375×0.9375×1.5 mm. The left and right caudate nucleus and putamen were manually outlined following the protocol used by Aylward et al. (2004). Note that the manual segmentation protocol used for this dataset differs from that of FreeSurfer. First, the scans are aligned along the axial plane passing through the anterior-commisure and posterior-commisure (AC-PC) and perpendicular to the inter-hemispheric fissure. The caudate and putamen are then outlined beginning with the most inferior slice where the caudate and putamen are clearly seperated by the internal capsule, and continuing in the superior direction. Thus, axial slices of the caudate and putamen inferior to the initial slice are included in the FreeSurfer training set, but are not included in the manual segmentations; the two will still be compared despite the protocol differences. One subject was randomly chosen as the template to generate the FS+LDDMM caudate and putamen segmentations for the remaining 15 subjects.
The Tourette syndrome (TS) dataset consisted of five subjects diagnosed with TS, and five age-matched healthy controls (Wang et al., 2007b). Images were acquired on a 1.5T Siemens Sonata scanner using an MPRAGE sequence (TR=9.7ms, TE=4ms, flip angle=12°, t=6.5min), with image dimensions 256 × 256 × 128, and voxel dimensions 1 × 1 × 1.25 mm. The right caudate nucleus, putamen, globus pallidus and nucleus accumbens were manually outlined according to the definitions detailed by Wang et al. (2007b). The template used for this dataset was also from Wang et al. (2007b), averaged from seven T1 acquisitions on a healthy comparison subject and with manually outlined basal ganglia structures.
The schizophrenia dataset was made up of five schizophrenic subjects, and five healthy controls matched in pairs according to age and parental socioeconomic status (Haller et al., 1997). Images were acquired using an MPRAGE sequence (TR=10ms, TE=4ms, TI=300ms, flip angle=8°, t=6min10sec) with a sagittal orientation, image dimensions 256 × 256 × 160 and voxel dimensions 1 × 1 × 1.25 mm. The right hippocampus in each scan was manually outlined according to the procedure detailed by Haller et al. (1997). An additional healthy control subject was used as a template to generate the FS+LDDMM segmentations.
The Alzheimers Disease (AD) dataset consisted of five elderly subjects diagnosed with dementia of Alzheimers type (DAT) with a clinical dementia rating (CDR) of 0.5, and five elderly control subjects with a CDR score of 0 (Csernansky et al., 2000). Images were acquired on a 1.5T Siemens Magnetom SP-4000 scanner using an MPRAGE sequence (TR=10ms, TE=4ms, N=1, t=11.0min), with image dimensions 160×256×256 and voxel dimensions 1×1×1 mm. The right hippocampus in each scan was manually outlined according to the procedure detailed by Haller et al. (1997) and Csernansky et al. (2000). A separate elderly control subject was used as a template to generate the FS+LDDMM segmentations of the right hippocampi for the ten subjects.
Note that the hippocampi manual outlines used for these datasets differ slightly from the CMA protocol used by FreeSurfer; the CMA protocol includes the fimbria, the strip of white matter superior to the hippocampus and inferior to the lateral ventricle, whereas this region is left out in our datasets. Finally, to test thalamus segmentation, we used a dataset consisting of four healthy controls, chosen randomly from the comparison set used by Csernansky et al. (2004a). These images were acquired using a turbo-fast, low-angle shots sequence (TR=20ms, TE=5.4ms, N=1, flip angle=30°, t=13.5min) with image dimensions 256 × 256 × 256 and voxel dimensions 1 × 1 × 1 mm. The same template used to segment the Tourette’s Syndrome dataset was used for these subjects as well.
Table 1 summarizes the vital information of all the datasets used.
Table 1.
Summary of data-sets tested reporting subject information as the number of subjects, the male/female distribution, the mean age with std. dev. or range, the disease state, the structures of interest, and reporting scan parameters as the scan sequence name, the rise time and echo time in ms, and number of acquisitions.
| Subject Information | Scan Parameters | ||||||
|---|---|---|---|---|---|---|---|
| N | Sex | Age | Dis. State | Structures | Sequence | TR/TE/N | |
| HD | 16 | 7/9 | 37 ± 11 | Pre-sym. HD | L. Caud. | SPGR | 18/3/2 |
| R. Caud. | |||||||
| L. Put. | |||||||
| R. Put. | |||||||
| TS | 5 | 3/2 | 36.5 ± 9.81 | Tourette’s | R. Caud. | MPRAGE | 9.7/4/1 |
| R. Put. | |||||||
| R. Pall. | |||||||
| R. N. Acc. | |||||||
| 5 | 3/2 | 34.8 ± 14.0 | Healthy | see above | see above | see above | |
| Schiz. | 5 | 5/0 | 33.0 (23–44) | Schiz. | R. Hippo. | MPRAGE | 10/4/1 |
| 5 | 5/0 | 32.4 (24–42) | Healthy | R. Hippo. | see above | see above | |
| AD | 5 | 3/2 | 74.0 ± 4.8 | CDR 0.5 | R. Hippo. | MPRAGE | 10/4/1 |
| 5 | 3/2 | 74.2 ± 5.2 | CDR 0 | R. Hippo. | see above | see above | |
| Thal. | 4 | 2/2 | 31.2 ± 11.9 | Healthy | R. Thal. | MPRAGE | 20/5.4/1 |
4 Results
Compilation of the aforementioned metrics and statistics for each dataset can be seen in Tables 2, 3, 4, 5, 6, 7, 8 and 9. An increase in spatial overlap (DSC) with the manual “gold standard” for the FS+LDDMM over the FreeSurfer segmentations can be seen for the majority of structures tested, with statistically significant improvement shown as well. Similarly, the L1 Error metrics are lower for FS+LDDMM segmentations which also indicates they are more similar to the manual segmentations than FreeSurfer. Note that for smaller structures, such as the nucleus accumbens and globus pallidus, overlap measures report relatively higher error since discrepencies on the boundaries of the segmentation are more significant due to the small structural volume. Therefore, examining the surface distances (Hausdorff distance, mean surface distance) leads to more meaningful comparisons between structures as only accuracy of the segmentation boundaries is taken into account. Our tabulated results show a decrease FS+LDDMM surface distances over FreeSurfer thus the FS+LDDMM segmentation boundaries follow the manual “gold standard” boundaries closer than FreeSurfer.
Table 2.
Huntington’s Disease Metrics: Compilation of overlap metrics (DSC, L1 Error), and symmetrized surface distances (Hausdorff Distance, Mean Surface Distance) for the pre-symptomatic Huntington’s Disease dataset, consisting of 16 subjects, one being chosen arbitrarily as a template image. Results for the left and right caudate nucleus, and left and right putamen are shown as the mean ± standard deviation and range for the 15 computed segmentations. Paired t-tests were used to detect a statistically significant difference in the metrics between the two methods, with p-values reported for each structure and metric
| Structure | Method | DSC | L1 Error | Haus. Dist. | Mean Dist. |
|---|---|---|---|---|---|
| L. Caud. | FreeSurfer | 0.80 ± 0.03 (0.69, 0.83) | 0.20 ± 0.04 (0.17, 0.30) | 6.74 ± 2.62 (3.19, 12.76) | 0.92 ± 0.23 (0.67, 1.47) |
| FS+LDDMM | 0.84 ± 0.01 (0.82, 0.87) | 0.15 ± 0.01 (0.13, 0.18) | 3.81 ± 0.72 (2.61, 5.34) | 0.65 ± 0.09 (0.51, 0.87) | |
| p-value | 2.75 × 10−5 | 1.86 × 10−5 | 5.95 × 10−4 | 2.30 × 10−4 | |
| L. Put. | FreeSurfer | 0.77 ± 0.02 (0.72, 0.81) | 0.25 ± 0.04 (0.21, 0.35) | 4.85 ± 0.86 (3.53, 6.78) | 1.16 ± 0.16 (0.93, 1.40) |
| FS+LDDMM | 0.84 ± 0.03 (0.76, 0.89) | 0.15 ± 0.02 (0.11, 0.21) | 3.26 ± 0.81 (2.34, 5.71) | 0.70 ± 0.15 (0.54, 1.17) | |
| p-value | 1.79 × 10−6 | 1.08 × 10−7 | 4.25 × 10−7 | 1.07 × 10−7 | |
| R. Caud. | FreeSurfer | 0.77 ± 0.02 (0.74, 0.80) | 0.22 ± 0.03 (0.18, 0.29) | 10.95 ± 13.94 (3.37, 60.94) | 3.82 ± 11.10 (0.75, 45.41) |
| FS+LDDMM | 0.81 ± 0.04 (0.69, 0.86) | 0.19 ± 0.05 (0.14, 0.34) | 6.88 ± 3.49 (4.41, 15.40) | 0.92 ± 0.34 (0.67, 1.94) | |
| p-value | 8.17 × 10−4 | 4.31 × 10−3 | 0.27 | 0.31 | |
| R. Put. | FreeSurfer | 0.78 ± 0.05 (0.62, 0.83) | 0.25 ± 0.09 (0.18, 0.53) | 4.49 ± 1.16 (3.01, 6.92) | 1.12 ± 0.31 (0.75, 1.97) |
| FS+LDDMM | 0.85 ± 0.02 (0.78, 0.87) | 0.15 ± 0.03 (0.13, 0.24) | 3.28 ± 0.85 (2.36, 5.09) | 0.68 ± 0.14 (0.58, 1.16) | |
| p-value | 1.14 × 10−5 | 1.62 × 10−5 | 3.83 × 10−4 | 1.10 × 10−6 | |
Table 3.
Tourette’s Syndrome Metrics: Compilation of overlap metrics (DSC, L1 Error), and symmetrized surface distances (Hausdorff Distance, Mean Surface Distance) for the 5 healthy control subjects. Results for the right caudate nucleus (R. Caud.), right nucleus accumbens (R. N. Acc.), right globus pallidus (R. Pall.) and right putamen (R. Put.) are shown as the mean ± standard deviation and range for the 5 computed segmentations. Paired t-tests were used to detect a statistically significant difference in the metrics between the two methods, with p-values reported for each structure and metric
| Structure | Method | DSC | L1 Error | Haus. Dist. | Mean Dist. |
|---|---|---|---|---|---|
| R. Caud. | FreeSurfer | 0.76 ± 0.02 (0.74, 0.78) | 0.23 ± 0.02 (0.21, 0.26) | 5.81 ± 3.56 (3.10, 12.01) | 0.98 ± 0.10 (0.90, 1.10) |
| FS+LDDMM | 0.81 ± 0.02 (0.79, 0.83) | 0.17 ± 0.02 (0.16, 0.19) | 11.12 ± 5.54 (4.86, 17.31) | 0.92 ± 0.22 (0.67, 1.20) | |
| p-value | 0.014 | 0.0083 | 0.21 | 0.63 | |
| R. N. Acc. | FreeSurfer | 0.60 ± 0.06 (0.51, 0.67) | 0.47 ± 0.11 (0.37, 0.63) | 4.37 ± 1.13 (3.02, 6.09) | 1.29 ± 0.41 (0.96, 1.91) |
| FS+LDDMM | 0.54 ± 0.05 (0.50, 0.63) | 0.43 ± 0.09 (0.34, 0.57) | 4.00 ± 1.08 (2.40, 5.33) | 1.24 ± 0.29 (0.90, 1.69) | |
| p-value | 0.021 | 0.094 | 0.56 | 0.57 | |
| R. Pall. | FreeSurfer | 0.71 ± 0.06 (0.62, 0.77) | 0.32 ± 0.07 (0.24, 0.41) | 4.64 ± 0.98 (3.17, 5.62) | 1.21 ± 0.21 (0.89, 1.41) |
| FS+LDDMM | 0.74 ± 0.06 (0.65, 0.81) | 0.26 ± 0.06 (0.21, 0.37) | 3.55 ± 0.97 (2.45, 5.03) | 1.02 ± 0.19 (0.84, 1.29) | |
| p-value | 0.22 | 0.12 | 0.012 | 0.15 | |
| R. Put. | FreeSurfer | 0.79 ± 0.01 (0.78, 0.81) | 0.21 ± 0.02 (0.18, 0.23) | 4.53 ± 1.64 (2.48, 6.97) | 0.99 ± 0.12 (0.78, 1.10) |
| FS+LDDMM | 0.83 ± 0.02 (0.81, 0.86) | 0.16 ± 0.02 (0.14, 0.17) | 4.77 ± 1.41 (2.54, 6.15) | 0.84 ± 0.09 (0.74, 0.98) | |
| p-value | 0.020 | 0.0098 | 0.60 | 0.064 | |
Table 4.
Tourette’s Syndrome Metrics: Compilation of overlap metrics (DSC, L1 Error), and symmetrized surface distances (Hausdorff Distance, Mean Surface Distance) for the 5 Tourette’s syndrome patients. Results for the right caudate nucleus (R. Caud.), right nucleus accumbens (R. N. Acc.), right globus pallidus (R. Pall.) and right putamen (R. Put.) are shown as the mean ± standard deviation and range for the 5 computed segmentations. Paired t-tests were used to detect a statistically significant difference in the metrics between the two methods, with p-values reported for each structure and metric
| Structure | Method | DSC | L1 Error | Haus. Dist. | Mean Dist. |
|---|---|---|---|---|---|
| R. Caud. | FreeSurfer | 0.73 ± 0.04 (0.68, 0.77) | 0.26 ± 0.03 (0.23, 0.29) | 9.16 ± 7.70 (3.79, 20.56) | 1.09 ± 0.19 (0.93, 1.34) |
| FS+LDDMM | 0.78 ± 0.02 (0.75, 0.80) | 0.20 ± 0.02 (0.18, 0.23) | 12.08 ± 7.43 (3.26, 18.82) | 1.02 ± 0.24 (0.75, 1.27) | |
| p-value | 0.10 | 0.071 | 0.67 | 0.72 | |
| R. N. Acc. | FreeSurfer | 0.57 ± 0.11 (0.46, 0.70) | 0.47 ± 0.13 (0.32, 0.61) | 3.73 ± 1.34 (2.67, 5.61) | 1.16 ± 0.43 (0.66, 1.65) |
| FS+LDDMM | 0.60 ± 0.03 (0.56, 0.62) | 0.39 ± 0.06 (0.34, 0.48) | 3.85 ± 0.73 (3.02, 4.67) | 1.03 ± 0.20 (0.85, 1.29) | |
| p-value | 0.67 | 0.25 | 0.88 | 0.50 | |
| R. Pall. | FreeSurfer | 0.73 ± 0.06 (0.67, 0.81) | 0.27 ± 0.06 (0.19, 0.31) | 4.33 ± 1.35 (2.40, 5.56) | 1.10 ± 0.28 (0.71, 1.31) |
| FS+LDDMM | 0.78 ± 0.05 (0.72, 0.83) | 0.22 ± 0.04 (0.19, 0.26) | 2.99 ± 0.94 (2.34, 4.37) | 0.96 ± 0.12 (0.81, 1.08) | |
| p-value | 0.10 | 0.067 | 0.16 | 0.18 | |
| R. Put. | FreeSurfer | 0.80 ± 0.05 (0.73, 0.85) | 0.20 ± 0.05 (0.15, 0.27) | 4.72 ± 1.33 (3.00, 6.24) | 0.94 ± 0.24 (0.68, 1.26) |
| FS+LDDMM | 0.83 ± 0.03 (0.79, 0.87) | 0.16 ± 0.03 (0.13, 0.19) | 4.27 ± 0.73 (3.26, 4.80) | 0.78 ± 0.08 (0.67, 0.87) | |
| p-value | 0.055 | 0.039 | 0.45 | 0.14 | |
Table 5.
Schizophrenia Metrics: Compilation of overlap metrics (DSC, L1 Error), and symmetrized surface distances (Hausdorff Distance, Mean Surface Distance) for the 5 healthy control subjects. Results for the right hippocampus (R. Hip.) are shown as the mean ± standard deviation for the 5 computed segmentations. Paired t-tests were used to detect a statistically significant difference in the metrics between the two methods, with p-values reported for each metric
| Structure | Method | DSC | L1 Error | Haus. Dist. | Mean Dist. |
|---|---|---|---|---|---|
| R. Hip. | FreeSurfer | 0.73 ± 0.03 (0.69, 0.76) | 0.32 ± 0.04 (0.29, 0.39) | 5.44 ± 0.91 (4.29, 6.53) | 1.25 ± 0.15 (1.02, 1.43) |
| FS+LDDMM | 0.75 ± 0.03 (0.70, 0.77) | 0.25 ± 0.04 (0.21, 0.29) | 5.76 ± 1.37 (3.64, 6.90) | 1.03 ± 0.17 (0.78, 1.21) | |
| p-value | 0.25 | 0.0070 | 0.56 | 0.047 |
Table 6.
Schizophrenia Metrics: Compilation of overlap metrics (DSC, L1 Error), and symmetrized surface distances (Hausdorff Distance, Mean Surface Distance) for the 5 schizophrenia patients. Results for the right hippocampus (R. Hip.) are shown as the mean ± standard deviation for the 5 computed segmentations. Paired t-tests were used to detect a statistically significant difference in the metrics between the two methods, with p-values reported for each metric
| Structure | Method | DSC | L1 Error | Haus. Dist. | Mean Dist. |
|---|---|---|---|---|---|
| R. Hip. | FreeSurfer | 0.72 ± 0.04 (0.67, 0.77) | 0.33 ± 0.05 (0.27, 0.39) | 6.29 ± 2.00 (3.45, 8.65) | 1.26 ± 0.25 (0.91, 1.53) |
| FS+LDDMM | 0.77 ± 0.03 (0.72, 0.81) | 0.22 ± 0.03 (0.18, 0.27) | 3.75 ± 0.94 (2.91, 5.30) | 0.86 ± 0.12 (0.71, 0.97) | |
| p-value | 0.056 | 0.0039 | 0.011 | 0.0089 |
Table 7.
Alzheimer’s Disease Metrics: Compilation of overlap metrics (DSC, L1 Error), and symmetrized surface distances (Hausdorff Distance, Mean Surface Distance) for the 5 healthy control (CDR 0) subjects. Results for the right hippocampus (R. Hip.) are shown as the mean ± standard deviation and range for the 5 computed segmentations. Paired t-tests were used to detect a statistically significant difference in the metrics between the two methods, with p-values reported for each metric
| Structure | Method | DSC | L1 Error | Haus. Dist. | Mean Dist. |
|---|---|---|---|---|---|
| R. Hip. | FreeSurfer | 0.69 ± 0.07 (0.61, 0.75) | 0.40 ± 0.11 (0.29, 0.55) | 7.24 ± 2.27 (4.63, 10.22) | 1.56 ± 0.51 (1.17, 2.19) |
| FS+LDDMM | 0.73 ± 0.03 (0.70, 0.77) | 0.35 ± 0.06 (0.27, 0.41) | 6.50 ± 2.67 (3.55, 10.78) | 1.52 ± 0.41 (1.14, 2.14) | |
| p-value | 0.20 | 0.23 | 0.16 | 0.74 |
Table 8.
Alzheimer’s Disease Metrics: Compilation of overlap metrics (DSC, L1 Error), and symmetrized surface distances (Hausdorff Distance, Mean Surface Distance) for the 5 Alzheimer’s (CDR 0.5) patients. Results for the right hippocampus (R. Hip.) are shown as the mean ± standard deviation and range for the 5 computed segmentations. Paired t-tests were used to detect a statistically significant difference in the metrics between the two methods, with p-values reported for each metric
| Structure | Method | DSC | L1 Error | Haus. Dist. | Mean Dist. |
|---|---|---|---|---|---|
| R. Hip. | FreeSurfer | 0.70 ± 0.03 (0.66, 0.75) | 0.37 ± 0.04 (0.31, 0.43) | 5.20 ± 1.05 (4.11, 6.71) | 1.22 ± 0.13 (1.12, 1.44) |
| FS+LDDMM | 0.72 ± 0.04 (0.67, 0.78) | 0.35 ± 0.09 (0.26, 0.47) | 4.61 ± 1.17 (3.06, 6.29) | 1.23 ± 0.22 (1.00, 1.57) | |
| p-value | 0.31 | 0.56 | 0.23 | 0.85 |
Table 9.
Healthy Control Metrics: Compilation of overlap metrics (DSC, L1 Error), and symmetrized surface distances (Hausdorff Distance, Mean Surface Distance) for the healthy control dataset, consisting of 4 subjects. Results for the right thalamus (R. Thal.) are shown as the mean ± standard deviation and range for the 4 computed segmentations. Paired t-tests were used to detect a statistically significant difference in the metrics between the two methods, with p-values reported for each metric
| Structure | Method | DSC | L1 Error | Haus. Dist. | Mean Dist. |
|---|---|---|---|---|---|
| R. Thal. | FreeSurfer | 0.85 ± 0.01 (0.84, 0.86) | 0.16 ± 0.01 (0.14, 0.17) | 7.32 ± 1.68 (5.99, 9.78) | 1.16 ± 0.12 (1.03, 1.31) |
| FS+LDDMM | 0.86 ± 0.03 (0.82, 0.88) | 0.14 ± 0.03 (0.12, 0.19) | 4.71 ± 1.59 (2.92, 6.54) | 1.10 ± 0.26 (0.87, 1.46) | |
| p-value | 0.38 | 0.31 | 0.19 | 0.67 |
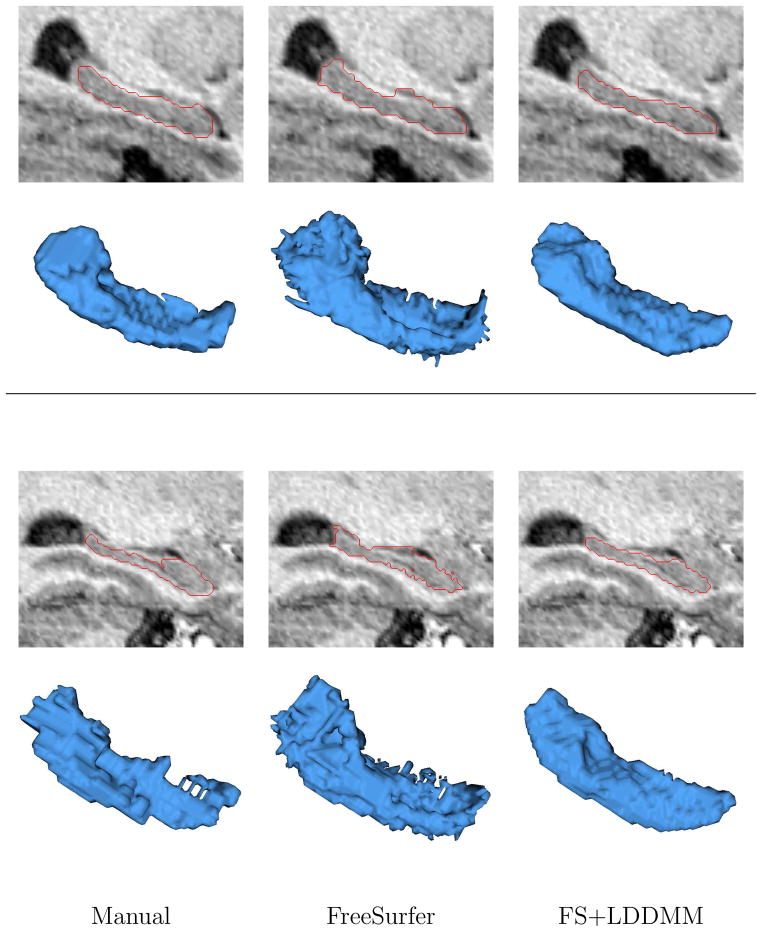
Figures 2, 3 and 4 show overlays and surface visualizations of the manual, FreeSurfer, and FS+LDDMM outlines for representative subjects from the Huntington’s Disease, Tourette’s Syndrome and schizophrenia datasets respectively. Smoothness and accuracy in the FS+LDDMM segmentations can clearly be seen over the FreeSurfer segmentations used for initialization. Furthermore, even the smoothness of the manual segmentations is less than desired for shape analysis.
Fig. 2.
Comparison of caudate and putamen outlines from a manual rater, FreeSurfer, and FS+LDDMM (left to right) on a subject from the presymptomatic Huntington’s disease dataset. The first two rows are MR overlays of an axial slice on the first row, and a coronal slice on the second row. The last two rows are surface renderings showing a medial sagittal view of the left structures on the third row, and the right structures on the fourth row (caudate - yellow, putamen - purple). Here it can be seen that the anatomical definitions in the manual outlines are different than the FreeSurfer atlas; FS+LDDMM overcomes this barrier by using a cohort template from the same manual rater. Also note the jagged inaccuracies in the FreeSurfer putamen labeling, a by-product of it’s voxel-based segmentation approach
Fig. 3.
Right hemisphere basal ganglia segmentations of a Tourette’s Syndrome subject shown in a coronal slice (top), surfaces in a medial sagittal view (middle), and surfaces in a lateral sagittal view (bottom). Structures shown are the right caudate (yellow), right putamen (purple), right nucleus accumbens (blue), and right globus pallidus (orange). The manual, FreeSurfer, and FS+LDDMM segmentations (left to right) show how the use of LDDMM refines the structural boundaries and produces segmentations that vary smoothly
Fig. 4.
Hippocampal outlines and surface renderings of a diseased subject (top), and a healthy subject (bottom) from the schizophrenia dataset. Manual, FreeSurfer and FS+LDDMM segmentations (left to right) are shown for each subject as a sagittal overlay, and a 3D surface rendering. Several surface irregularities can be seen on the FreeSurfer hippocampal surfaces which would make further shape analysis difficult without smoothing and loss of detail. The manual tracings also lack smoothness since these segmentations are delineated on 2D-slices
The FreeSurfer subcortical segmentations were computed with version 3.0.5 and were run on 2.4 Ghz AMD Opteron workstations, with processing time for each subject in the range of 10 to 15 hours. Note that the FreeSurfer processing only needs to be performed once per subject regardless of the number of ROI’s being computed. Image registration with LDDMM was performed using flows discretized to 20 timesteps on an SGI Altix 3700 (64 Itanium CPU’s, 64-bit, 64 GB RAM) using 8-processors for each ROI. LDDMM run-times for each ROI were 89.3 ± 36.3 minutes (mean ± standard deviation) with a range of (13.5,179.0) minutes.
5 Discussion
In this paper, we proposed a new, fully automated, diffeomorphic deformable-template pipeline, i.e., FS+LDDMM, for segmentating subcortical structures. The fusion of the probabilistic voxel-based classification method, FreeSurfer, and the deformable template-based LDDMM segmentation procedure overcomes the limitations in each individual strategy. In addition, as each method is improved independently, the improvements will be inherited by FS+LDDMM. The method is also very attractive to those already using the FreeSurfer processing and analysis pipeline, as the increase in computational effort becomes marginal. We have demonstrated proof of concept of this fully automated procedure and have shown results for 7 different subcortical structures (caudate nucleus, putamen, globus pallidus, nucleus accumbens, thalamus and hippocampus).
Even though we have demonstrated the FS+LDDMM method with a single, random, scan as the template, this method is suitable for a number of choices for a template. Rohlfing et al. (2004) outlined three additional scenarios: averaged template, a representative individual who is part of the study as template, and multiple subjects employed as template with subsequent decision fusion based final classification.
The advantages of using an averaged template is it can encompass the inter-subject variability within the group. However, it is possible that the averaging procedure will create an image that is blurry or may not truly be the average subject in the group. In addition, creating the averaged template requires a set of labeled training images, which demands a great deal of manual effort to construct. This is akin to the probabilistic atlas approach employed by FreeSurfer or the maximum probability maps by Hammers et al. (2003).
Selecting a subject that is most similar to the target datasets to be the template circumvenes the possible blurry averaging issue while reducing inter-subject variability. However, it does require a set of pre-labeled subjects as potential candidates and a computational step to determine similarity (usually through non-rigid registration) between each target and each potential template.
Using multiple subjects as template involves performing the label-propagation for all templates, then combining the results using methods such as probability maps or decision fusion classification. Although this method has been shown to be the most accurate (Rohlfing et al., 2004), it also requires the most resources.
It is possible to include any of the aforementioned template selection strategies within the current framework without dramatically increasing computational costs, since the FreeSurfer initialization computation remains the same for each above strategy. Evaluation of the choice of template is being conducted on a large dataset, and it is beyond the scope of this paper. The intent of the current manuscript is to demonstrate proof of concept and applicability to various sub-cortical structures. Looking at recently published segmentation methods, FS+LDDMM demonstrates comparable levels of accuracy as indicated by spatial overlap: Pitiot et al. (2004) reported an average mean surface distance of 1.6 mm for the caudate nucleus; our mean surface distances ranged from 0.65 mm to 1.01 mm. Zhou and Rajapakse (2005) reported spatial overlaps (DSC) of 0.81, 0.84 and 0.83 for the caudate, putamen, and thalamus respectively, and Amini et al. (2004) reported a spatial overlap (DSC) of 0.88 for the thalamus, figures which are very close in magnitude with our results (DSC = 0.81, 0.83 and 0.86 for the caudate, putamen, and thalamus in healthy controls). Methods which are tailored for the segmentation of a specific structure (Xia et al., 2007; Chupin et al., 2007) are likely to achieve higher spatial overlap, such as those presented by Xia et al. (2007) (DSC = 0.873 ± 0.0234 for the caudate nucleus), however, this knowledge-driven method may be unlikely to achieve the same results on other datasets or pathologies and is usable only on the caudate nucleus. The hippocampus and amygdala segmentation method by Chupin et al. (2007) has been shown to perform equally well on diseased subjects (DSC=0.84), but the disadvantage with this method is it requires some manual interaction to place seed points and define a bounding box and is thus not a fully automated method. Another knowledge-driven approach by (Barra and Boire, 2001) uses fuzzy maps for segmentation and reports higher spatial overlaps (V(A ∩ B)/V(A) = 0.84, 0.88, 0.89 for the caudate, putamen and thalamus), but may require re-definition of the maps when used on atrophic structures, limiting applicability of the method. Methods incorporating decision fusion into multiple template-based label-propagation such as those by (Heckemann et al., 2006) (DSC = 0.89/0.90, 0.72/0.70, 0.89/0.90, 0.91/0.90, 0.80/0.80 and 0.83/0.81 for the left/right caudate, nucleus accumbens, putamen, thalamus, pallidum, and hippocampus) and (Hammers et al., 2007) (DSC = 0.83 and 0.76 for una3ected and atrophic hippocampi) report very high spatial overlap, however as discussed above, the manual labeling of the multiple templates required for this method is a trade-off and may not always be worthwhile. Note that it is possible to make use of multiple subject propagation and decision fusion in the FS+LDDMM framework to improve accuracy as well, an option that will be fully explored in the future.
The visualizations shown in Figures 2, 3 and 4 illustrate the stark difference in smoothness between manual, FreeSurfer and FS+LDDMM segmentations. The irregular boundary “shape-noise” observed in manual and FreeSurfer segmentations are not present in the FS+LDDMM segmentation because the latter is the transformation of the template expert manual segmentation via smooth diffeomorphic maps. Smooth segmentations allows shape analysis without additional smoothing, a step that would have involved loss of structural details.
As large research databases containing hundreds of brain scans become available, the FS+LDDMM fully automated method not only can be used to generate accurate segmentations for volume and shape analyses, but also addresses the issue that various research groups may follow anatomical definitions that are different from the FreeSurfer atlas. This issue arose in our HD dataset segmentations, where the manual outlines of the caudate and putamen differ from the FreeSurfer protocols as can be seen in Figure 2. To use FreeSurfer alone, a new atlas would have to be created with labels adhering to this anatomical definition protocol; a task which involves the manual labeling of several structures in many training-set scans to re-generate the probabilistic atlas. The issue is easier dealt with in FS+LDDMM since it can propagate a single template, defined using the desired anatomical protocol, and label each target structure accordingly; the initialization using the FreeSurfer labeling is still valid here since both template and target initialization labels are from FreeSurfer and thus are defined similarly.
Although the FS+LDDMM procedure has only been tested on data from four neuropsychiatric diseases, the robustness of the method and its initializers lends itself to applicability to subjects with different diseases. Firstly, because the deformable template can be chosen from the group of subjects requiring segmentation, scanner- and patient-specific variability may be reduced. Secondly, by initializing the LDDMM registration with the FreeSurfer labels helps account for potential morphological differences among the subjects; initial mapping of the cerebro-spinal fluid in the Huntington’s Disease subjects solves the issue of variable-sized lateral ventricles which would otherwise be problematic.
The benefits of automation come at a computational cost as run-times for our FS+LDDMM method are on the order of several hours if no parallelization is used. The FreeSurfer subcortical labeling can take between 10–15 hours on a single processor, and the LDDMM runs on the subcortical ROI’s can take up to a few hours as well, however, this cost is only computing time; it is straightforward to process the entire database without any manual intervention. With the advent of large scale distributed computational infrastructure such as the Biomedical Informatics Research Network (BIRN,www.nbirn.net) and the TeraGrid (www.teragrid.org), and the trend of increasing core counts in the face of declining hardware costs, computationally intensive processing is not ultimately prohibitive and therefore holds considerable promise for computational anatomy of MR brain images.
One limitation in the presented method is a lack of testing on high field strength (3T) MR scanner data. Although currently many existing large datasets, such as those produced by ADNI (Alzheimer’s Disease Neuroimaging Initiative, http://www.loni.uclas.edu/ADNI/predominantly use 1.5T scanners, it will be desirable to test images collected on 3T scanners to test effects of field inhomogeneity. A further limitation is that the method is designed to only segment subcortical structures; cortical segmentation is beyond the scope of this work.
Diffeomorphic transformations are not the universal solution to the segmentation problem, but since corresponding regions in the brain likely have the same structures, the use of bijective mappings is warranted. Crum et al. (2004) compares various non-rigid registration methods in neuroimaging applications and concludes that a high-dimensional diffeomorphic viscous fluid method was outperformed by a B-splines method. However, the fluid registration used did not incorporate the same degree of initialization that our method utilizes, particularly the lateral ventricle initialization which Crum et al. (2004) shows to be problematic. We believe that with sufficient levels of initialization, high-dimensional diffeomorphic transformations, like those generated by LDDMM, can lead to higher levels of accuracy.
Acknowledgments
The authors acknowledge the support of the following grants: NIH P50 MH071616, NSERC 31-611387, CHRP 751115 and Pacific Alzheimer Research Foundation 869294. Ali Khan was supported by NSERC PGS-M scholarship. The authors would also like to thank Bruce Fischl for initial discussions that have led to this work.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Ali R. Khan, Email: akhanf@sfu.ca.
Lei Wang, Email: lei@wustl.edu.
Mirza Faisal Beg, Email: mfbeg@ensc.sfu.ca.
References
- Amini L, Amini L, Soltanian-Zadeh H, Lucas C, Gity M. Automatic segmentation of thalamus from brain MRI integrating fuzzy clustering and dynamic contours. IEEE Trans Bio-Med Eng. 2004;51 (5):800–811. doi: 10.1109/TBME.2004.826654. [DOI] [PubMed] [Google Scholar]
- Aylward E, Sparks B, Field K, Yallapragada V, Shpritz, Rosenblatt A, Brandt J, Gourley L, Liang K, Zhou H, Margolis R, Ross C. Onset and rate of striatal atrophy in preclinical Huntington’s disease. Neurology. 2004 July;63 (1):66–72. doi: 10.1212/01.wnl.0000132965.14653.d1. [DOI] [PubMed] [Google Scholar]
- Barra V, Boire JY. Automatic segmentation of subcortical brain structures in MR images using information fusion. IEEE Trans Med Imaging. 2001;20 (7):549–558. doi: 10.1109/42.932740. [DOI] [PubMed] [Google Scholar]
- Beg MF, Miller MI, Trouvé A, Younes L. Computing large deformation metric mappings via geodesic flows of diffeomorphisms. Int J Comput Vision. 2005 February;61 (2):139–157. [Google Scholar]
- Chupin M, Mukuna-Bantumbakulu AR, Hasboun D, Bardinet E, Baillet S, Kinkingnéhun S, Lemieux L, Dubois B, Garnerob L. Anatomically constrained region deformation for the automated segmentation of the hippocampus and the amygdala: Method and validation on controls and patients with Alzheimer’s disease. Neuroimage. 2007;34:995–1019. doi: 10.1016/j.neuroimage.2006.10.035. [DOI] [PubMed] [Google Scholar]
- Crum WR, Rueckert D, Jenkinson M, Kennedy D, Smith SM. A framework for detailed objective comparison of non-rigid registration algorithms in neuroimaging. MICCAI. 2004:679–686. [Google Scholar]
- Csernansky JG, Schindler MK, Splinter NR, Wang L, Gado M, Selemon LD, Rastogi-Cruz D, Posener JA, Thompson PA, Miller MI. Abnormalities of thalamic volume and shape in Schizophrenia. Am J Psychiat. 2004a;161:896–902. doi: 10.1176/appi.ajp.161.5.896. [DOI] [PubMed] [Google Scholar]
- Csernansky JG, Wang L, Joshi SC, Tilak Ratnanather J, Miller MI. Computational anatomy and neuropsychiatric disease: probabilistic assessment of variation and statistical inference of group difference, hemispheric asymmetry, and time-dependent change. Neuroimage. 2004b;23 (Supplement 1):S56–S68. doi: 10.1016/j.neuroimage.2004.07.025. [DOI] [PubMed] [Google Scholar]
- Csernansky J, Wang L, Joshi S, Miller J, Gado M, Kido D, McKeel D, Morris J, Miller M. Early DAT is distinguished from aging by high-dimensional mapping of the hippocampus. Neurology. 2000 December;55 (1):1636–1643. doi: 10.1212/wnl.55.11.1636. [DOI] [PubMed] [Google Scholar]
- Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, van der Kouwe A, Killiany R, Kennedy D, Klaveness S, Montillo A, Makris N, Rosen B, Dale1 AM. Whole brain segmentation: Automated labeling of neuroanatomical structures in the human brain. Neuron. 2002 January;33:341–355. doi: 10.1016/s0896-6273(02)00569-x. [DOI] [PubMed] [Google Scholar]
- Fischl B, Salat DH, van der Kouwe AJ, Makris N, Segonne F, Quinn BT, Dale AM. Sequence-independent segmentation of magnetic resonance images. Neuroimage. 2004;23 (Supplement 1):S69–S84. doi: 10.1016/j.neuroimage.2004.07.016. [DOI] [PubMed] [Google Scholar]
- Haller JW, Banerjee A, Christensen GE, Gado M, Joshi S, Miller MI, Sheline Y, Vannier MW, Csernansky JG. Three-dimensional hippocampal MR morphometry with high-dimensional transformation of a neuroanatomic atlas. Radiology. 1997;202:504–510. doi: 10.1148/radiology.202.2.9015081. [DOI] [PubMed] [Google Scholar]
- Hammers A, Allom R, Koepp MJ, Free SL, Myers R, Lemieux L, Mitchell TN, Brooks DJ, Duncan JS. Three-dimensional maximum probability atlas of the human brain, with particular reference to the temporal lobe. Hum Brain Mapp. 2003;19 (4):224–247. doi: 10.1002/hbm.10123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammers A, Heckemann R, Koepp MJ, Duncan JS, Hajnal JV, Rueckert D, Aljabar P. Automatic detection and quantifica-tion of hippocampal atrophy on MRI in temporal lobe epilepsy: A proof-of-principle study. Neuroimage. 2007 May;36 (1):38–47. doi: 10.1016/j.neuroimage.2007.02.031. [DOI] [PubMed] [Google Scholar]
- Heckemann RA, Hajnal JV, Aljabar P, Rueckert D, Hammers A. Automatic anatomical brain MRI segmentation combining label propagation and decision fusion. Neuroimage. 2006;33:115–126. doi: 10.1016/j.neuroimage.2006.05.061. [DOI] [PubMed] [Google Scholar]
- Hogan RE, Mark KE, Wang L, Joshi S, Miller MI, Bucholz RD. Mesial temporal sclerosis and temporal lobe epilepsy: MR imaging deformation-based segmentation of the hippocampus in five patients. Radiology. 2000;216:291–297. doi: 10.1148/radiology.216.1.r00jl41291. [DOI] [PubMed] [Google Scholar]
- Khan A, Aylward E, Barta P, Miller MI, Beg MF. Semiautomated basal ganglia segmentation using large deformation diffeomorphic metric mapping. MICCAI. 2005:238–245. doi: 10.1007/11566465_30. [DOI] [PubMed] [Google Scholar]
- Lacerda ALT, Hardan AY, Yorbik O, Keshavan MS. Measurement of the orbitofrontal cortex: a validation study of a new method. Neuroimage. 2003 Jul;19 (3):665–673. doi: 10.1016/s1053-8119(03)00137-x. [DOI] [PubMed] [Google Scholar]
- Maintz J, Viergever M. A survey of medical image registration. Med Image Anal. 1998;2 (1):1–36. doi: 10.1016/s1361-8415(01)80026-8. [DOI] [PubMed] [Google Scholar]
- Nugent TF, III, Herman DH, Ordonez A, Greenstein D, Hayashi KM, Lenane M, Clasen L, Jung D, Toga AW, Giedd JN, Rapoport JL, Thompson PM, Gogtay N. Dynamic mapping of hippocampal development in childhood onset schizophrenia. Schizophr Res. 2007 Feb;90 (1–3):62–70. doi: 10.1016/j.schres.2006.10.014. [DOI] [PubMed] [Google Scholar]
- Nyul LG, Udupa JK, Zhang X. New variants of a method of MRI scale standardization. IEEE Trans Med Imaging. 2000 February;19 (2):143–150. doi: 10.1109/42.836373. [DOI] [PubMed] [Google Scholar]
- Pitiot A, Delingette H, Thompson P, Ayache N. Expert knowledge guided segmentation system for brain MRI. Neuroimage. 2004;23 (Supplement 1):S85–S96. doi: 10.1016/j.neuroimage.2004.07.040. [DOI] [PubMed] [Google Scholar]
- Rohlfing T, Brandt R, Menzel R, Jr, CRM Evaluation of atlas selection strategies for atlas-based image segmentation with application to confocal microscopy images of bee brains. Neuroimage. 2004;21:1428–1442. doi: 10.1016/j.neuroimage.2003.11.010. [DOI] [PubMed] [Google Scholar]
- Shen D, Moffat S, Resnick S, Davatziko C. Measuring size and shape of the hippocampus in MR images using a deformable shape model. Neuroimage. 2002 February;15:422–434. doi: 10.1006/nimg.2001.0987. [DOI] [PubMed] [Google Scholar]
- Spinks R, Magnotta VA, Andreasen NC, Albright KC, Ziebell S, Nopoulos P, Cassell M. Manual and automated measurement of the whole thalamus and mediodorsal nucleus using magnetic resonance imaging. Neuroimage. 2002 Oct;17 (2):631–642. [PubMed] [Google Scholar]
- Svarer C, Madsen K, Hasselbalch SG, Pinborg LH, Haugbol S, Frok-jaer VG, Holm S, Paulson OB, Knudsen GM. MR-based automatic delineation of volumes of interest in human brain PET images using probability maps. Neuroimage. 2005 Feb;24 (4):969–979. doi: 10.1016/j.neuroimage.2004.10.017. [DOI] [PubMed] [Google Scholar]
- Wang L, Beg F, Ratnanather T, Ceritoglu C, Younes L, Morris J, Csernansky J, Miller M. Large deformation diffeomorphism and momentum based hippocampal shape discrimination in dementia of the Alzheimer type. IEEE Trans Med Imaging. 2007a;26 (4):462–470. doi: 10.1109/TMI.2005.853923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Lee DY, Bailey E, Hartlein JM, Gado MH, Miller MI, Black KJ. Validity of large-deformation high dimensional brain mapping of the basal ganglia in adults with Tourette syndrome. Psychiat Res-NeuroIm. 2007b;154:181–190. doi: 10.1016/j.pscychresns.2006.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Swank JS, Glick IE, Gado MH, Miller MI, Morris JC, Csernansky JG. Changes in hippocampal volume and shape across time distinguish dementia of the Alzheimer type from healthy aging. Neuroimage. 2003 Oct;20 (2):667–682. doi: 10.1016/S1053-8119(03)00361-6. [DOI] [PubMed] [Google Scholar]
- Xia Y, Bettinger K, Shen L, Reiss AL. Automatic segmentation of the caudate nucleus from human brain MR images. IEEE Trans Med Imaging. 2007 April;26 (4):509–517. doi: 10.1109/TMI.2006.891481. [DOI] [PubMed] [Google Scholar]
- Yang J, Duncan JS. 3D image segmentation of deformable objects with joint shape-intensity prior models using level sets. Med Image Anal. 2004;8:285–294. doi: 10.1016/j.media.2004.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Rajapakse JC. Segmentation of subcortical brain structures using fuzzy templates. Neuroimage. 2005;28:915–924. doi: 10.1016/j.neuroimage.2005.06.037. [DOI] [PubMed] [Google Scholar]