Abstract
Co-stimulation by the chemokine, SDF-1/CXCL12, has been shown to increase the amount of IL-10 secreted by TCR-stimulated human T cells, however, the molecular mechanisms of this response are unknown. Knowledge of this signaling pathway may be useful since extensive evidence indicates that deficient IL-10 secretion promotes autoimmunity. The human IL-10 locus is highly polymorphic. We report here that SDF-1 co-stimulates IL-10 secretion from T cells containing all three of the most common human IL-10 promoter haplotypes that are identified by SNPs at -1082, -819, and -592 bp (numbering is relative to the transcription start site). We further show that SDF-1 primarily co-stimulates IL-10 secretion by a diverse population of CD45RA- (“memory”) phenotype T cells that includes cells expressing the presumed regulatory T cell marker, Foxp3. To address the molecular mechanisms of this response, we showed that SDF-1 co-stimulates the transcriptional activities in normal human T cells of reporter plasmids containing 1.1 kb of all three of the common IL-10 promoter haplotypes. IL-10 promoter activity was ablated by mutating two non-polymorphic binding sites for the AP-1 transcription factor, and chromatin immunoprecipitation assays of primary human T cells revealed that SDF-1 co-stimulation enhances AP-1 binding to both of these sites. Together, these results delineate the molecular mechanisms responsible for SDF-1 co-stimulation of T cell IL-10 secretion. Since it is preserved among several human haplotypes and in diverse T cell populations including Foxp3+ T cells, this pathway of IL-10 regulation may represent a key mechanism for modulating expression of this important immunoregulatory cytokine.
Keywords: T cells, chemokines, cytokines, transcription factors, gene regulation
INTRODUCTION
IL-10 potently inhibits inflammatory immune reactions in both humans and mice, primarily by inhibiting the immune activation of monocytes, dendritic cells, and macrophages, and their secretion of inflammatory mediators. For example, IL-10 profoundly inhibits monocyte expression of MHC Class II, ICAM-1, B7.1, B7.2, and TLR4, thereby limiting their ability to act as antigen presenting cells (APCs) to promote the activation of Th1-type inflammatory T cells (1). IL-10 also critically limits the initiation, duration, and associated pathology of inflammatory immune responses by limiting the secretion of many chemokines and pro-inflammatory cytokines. IL-10 thereby ameliorates sepsis, endotoxemia, pancreatitis, uveitis, and hepatitis in mouse models (1). Interestingly, mice with the IL-10 gene deleted only from their T cells develop gut inflammation with immune features similar to human inflammatory bowel disease, even though gut macrophages and other cell types in these animals expressed IL-10 (2). T cell-derived IL-10 also plays important roles in inhibiting allergy and asthma (3). Therapies aimed at increasing T cell secretion of IL-10 may therefore hold promise for ameliorating autoimmunity. Unfortunately, the molecular mechanisms responsible for regulating IL-10 secretion by T cells are only poorly understood.
Stromal Cell-Derived Factor-1 (SDF-1; CXCL12) is a member of the superfamily of chemokines that regulate the growth and migration of multiple cell types (4–6). CXCR4 is the receptor for SDF-1 and is expressed by most T cell subsets. SDF-1/CXCR4 signaling results in T cell adhesion (7), chemotaxis (8), and gene expression important for regulating cell-cycle progression and apoptosis (9). SDF-1 critically modulates T cell immune functions. For example, SDF-1/CXCR4 signal transduction contributes to regulating lymphocyte development (10) and homeostasis (11), and both SDF-1 and CXCR4 are critical for (9,12,13) regulating T cell infection by the human immunodeficiency virus type-1 (HIV-1) (14,15). SDF-1/CXCR4 signaling also induces the expression of multiple genes, via mechanisms including activation of the MEK-1/ERK Mitogen-Activated Protein (MAP) kinase pathway which regulates downstream transcription factors including AP-1 (9,16–18). Nevertheless, the immunological role(s) of SDF-1 regulation of gene expression are, as yet, incompletely understood.
Mapping of the human IL-10 promoter has revealed multiple predicted transcription factor binding sites and two microsatellite regions that are likely to regulate IL-10 gene transcription (19–23). Interestingly, IL-10 promoter polymorphisms among humans affect levels of IL-10 secretion and susceptibility to autoimmune diseases and related immune conditions (20–25). These polymorphisms include forty-six single-nucleotide polymorphisms (SNPs) and multiple variations within two microsatellites. Thirty-eight of the published polymorphisms are located in the 5′ flanking and presumed promoter region of the IL-10 gene, suggesting that they affect IL-10 expression. In particular, three linked SNPs located within 1.1 kb of the transcription start site characterize three haplotypes that we shall refer to here as ATA, ACC and GCC, that are defined by G or A at -1082 bp, C or T at -819 bp, and C or A at -592 bp (numbering is relative to the transcription start site). The ATA, ACC, and GCC haplotypes also contain other linked polymorphisms, including other SNPs and variations in two microsatellite CA repeat regions located −4 kb and −1 kb (summarized in (23)). The ATA, ACC, and GCC haplotypes are particularly common: each is present at least heterozygously in 20 – 33 % of Caucasian individuals, and the ATA haplotype is present in up to 70 % of Asian sub-populations (23,25).
The haplotypes are therefore likely to have been preserved and disseminated among human populations via selective pressure. Consistent with this idea, the ATA, ACC, and GCC haplotypes are associated with low, medium, and high levels of IL-10 secretion, respectively (25,26). In contrast to the higher-IL-10-expressing ACC and GCC haplotypes, the lower-IL-10-expressing ATA haplotype is epidemiologically associated with the development of autoimmune diseases, including Sjogren’s syndrome, rheumatoid arthritis, systemic lupus erthematosus, and psoriasis (27,28). ATA is also associated with other immune-related conditions, including increased risk of graft rejection after renal or cardiac transplantation (28), increased risk of HIV-1 infection and faster progression to AIDS (29), and decreased risk of graft-vs.-host-disease and death after HLA-identical hematopoietic cell transplantation (23,26). Strikingly, the range of IL-10 secretion levels associated with the haplotypes is relatively small: GCC+ individuals secrete on the average only 2 - 3 -fold more IL-10 than ATA+ individuals (25,26). Thus, even small differences in IL-10 secretion evidently can significantly impact the development of immune-related diseases including autoimmunity.
Below, we show that the chemokine, SDF-1/CXCL12, potently co-stimulates IL-10 secretion by human memory T cell subsets. In addition, we address the molecular mechanisms responsible for this co-stimulation by isolating and characterizing in primary human T cells SDF-1 and TCR stimulation effects on ATA, ACC, GCC human IL-10 promoter constructs. These results significantly enhance understanding of the molecular mechanisms responsible for regulating IL-10 secretion by T cells. By establishing that SDF-1 is a potent regulator of T cell IL-10 secretion these results in addition suggest a critical role for SDF-1 in preventing the development of the diverse autoimmune diseases and immune disregulations that are linked to IL-10 deficiency.
MATERIALS AND METHODS
Cells
Human PBMC were isolated from the defibrinated peripheral blood of healthy volunteers using a ficoll gradient and were used for experiments following 12–24 hr of culture at 37 °C. Blood was obtained and used with informed consent and approval by the Mayo Institutional Review Board. Typical PBMC preparations contained 75–80 % CD3+ T lymphocytes (70–75 % CD4+ and 25–30 % CD8+), and essentially 100 % of CD3+ cells expressed CXCR4. T cells were purified from buffy coats using the RosetteSep Human T cell enrichment cocktail according to the manufacturer’s instructions (Stem Cell Technologies, Canada). Where indicated, additional purification of T cell subsets utilized the CD4 and CD8 Multisort kits from Miltenyi Biotech (Auburn, California). The purified CD4+ or CD8+ T cell preparations were divided into two parts and further separated into naïve vs. memory cells as follows: CD45RA or CD45RO beads were added and CD45RA- (memory) or CD45RO- (naïve) cells were isolated by negative-selection. Purities of the isolated cell populations were determined by flow cytometry to be 97–99 % for CD4+CD45RO- and CD4+CD45RA- cells, and ≥ 88% for CD8+CD45RO-and CD8+CD45RA- cells.
Cell culture and stimulation
Cells were cultured in Medium A (RPMI 1640 supplemented with 10 % FCS, 10 mM Hepes pH 7.4, 2 mM L-glutamine, 1 mM sodium pyruvate, and 2 μM 2-ME) at 1 × 106 - 2 × 106 cells/ml. Stimulations were performed at 37 °C and SDF-1α (R&D Systems, Minneapolis, MN) was used at 5 × 10−8 M. TCR ligation was achieved by cross-linking anti-TCR mAbs: for reporter assays, chromatin immunoprecipitation assays, and cytokine production assays, the TCR was ligated via plate-bound 1 μg/ml anti-CD3 mAb (OKT3, Ortho Biotech, Bridewater, NJ); for biochemical assays, the TCR was ligated via 1 μg/ml soluble OKT3 mAb that was then cross-linked with 0.1 mg/ml goat anti-mouse IgG (Sigma, St. Louis, MO). Where indicated, cells were pretreated for 90 min with either 50 μM PD098059 (Sigma, St. Louis, MO) or an equivalent amount of vehicle (DMSO) as a control.
Assays of IL-10 and marker expression by human T cells
Cells were stimulated as indicated, then assayed. Stimulation times were 24 hr for both IL-10 intracellular cytokine staining and ELISA. Cell-surface staining of CD4, CD3, CD8, CD69, CCR7, and CXCR3 and intracellular staining of IL-10 were performed using the Cytoperm/Cytofix intracellular cytokine staining kit (BD Pharmingen, San Diego, CA). The kit was used according to the manufacture’s directions, except that cells were treated with 4X GolgiPlug for 6 hr prior to staining. Intracellular Foxp3 staining was performed using the Foxp3 staining set from eBioscience (San Diego, CA). Supernatant IL-10 was assayed using an ELISA kit (BD Pharmingen, San Diego, CA).
IL-10 promoter expression constructs
DNA sequence −1111 bp to +1 bp from the human IL-10 promoter (numbering is relative to the transcription start site) was amplified by PCR (primers available upon request) from normal human blood and subcloned into the luciferase vector, pGL4 (Promega, Madison, WI). Site-directed mutagenesis of the GCC promoter construct was performed using the Quikchange II XL kit (Stratagene, La Jolla, CA) to change the GGGTCA AP-1 site at −775 bp to GTTCAG and the CCAATCA AP-1 site at −202 bp to CAGGCAG. After subcloning and/or mutagenesis, the identity of each construct was confirmed by DNA sequencing.
Transient transfections and assays of activities of IL-10 promoter constructs in primary human T cells
Purified human T cells (3 × 107) were mixed with 100 μg of the indicated expression plasmid. To compare different transfections done the same day, the cells were additionally transfected with 40 ng of pRL-TK (Promega, Madison, WI), which encodes a Renilla luciferase. Cells were transfected by electroporation using a BTX Electro-square-porator model T820 (BTX Inc., San Diego, CA) as described (30), except that 350 volts were used. Typical transfection efficiencies were 10 – 20 %. Transfected cells were cultured in Medium A for 2.5 hr and then stimulated for 18 hr, as indicated. Luciferase activities derived from both experimental and control pRL-TK plasmids were measured in the same samples using the Dual-Luciferase assay kit (Promega, Madison, WI) and Renilla luciferase values were used for normalization. Results shown are presented as Fold-Increases as compared to unstimulated samples in the same experiment.
Assay of active, phosphorylated ERK
Cells were stimulated as indicated, then whole cell lysates were assayed for active ERK phosphorylated on Thr-202 and Tyr-204 after SDS-PAGE and immunoblotting with phospho-specific anti- p44/p42 ERK1/ERK2 (Thr-202/Tyr-204; Cell Signaling Technology, Beverly, MA). Total ERK2 protein was determined as a control by stripping bound antibodies off the same blots and re-immunoblotting with p44 ERK kinase antiserum (Santa Cruz Biotechnology, Inc., Santa Cruz, CA).
Assay of IL-10 mRNA
Cells were stimulated as indicated. RNA was isolated using Trizol (Invitrogen, Carlsbad, CA), and IL-10 and actin mRNA was assayed using the Thermoscript RT-PCR System (Invitrogen) according to manufacturer’s directions. Briefly, each reaction utilized 2 μg of total RNA that was subjected cDNA synthesis, 10 % of the cDNA reaction was then subjected to 35 cycles of PCR at denaturing, annealing, and elongation temperatures of 94 °C, 55 °C, and 72 °C, respectively, in an i-Cycler PCR machine (BioRad, Hercules, CA). The primers used were CTCTGTTGCCTGGTCCTCCTG and GTCCTCCAGCAAGGACTCC for IL-10 mRNA, and ATGTTTGAGACCTTCAACAC and ACGTCACACTTCATGATGGA for actin mRNA.
Chromatin Immunoprecipitation Assay
Purified T cells (2 × 106 cells per test) were stimulated for 12 hr as indicated. Chromatin immunoprecipitation assays were performed on lysed cells using a ChIP Assay kit (Upstate, Lake Placid, New York). ChIP assays were performed according to the manufacturer’s instructions, except that the sonicated suspension was pre-cleared for 3 hr at 4 °C before incubation with 1μg of either c-Fos rabbit polyclonal antisera (Santa Cruz Biotech, Santa Cruz, CA) or normal rabbit control polyclonal IgG (R&D systems, Minneapolis, MN) overnight at 4 °C, and the salmon sperm DNA/protein A agarose slurry was blocked for 30 min at room temperature in 5 % BSA/PBS before incubation with the immune complexes for 3 hr at 4 °C. Sonicated DNA was between 200 and 1000 bp in length. Each sample was subjected to thirty cycles of PCR at denaturing, annealing, and elongation temperatures of 94 °C, 55 °C, and 72 °C, respectively, using an i-Cycler PCR machine (BioRad, Hercules, CA). The primers used to amplify the −775 bp AP-1 site were GTGCTGGAGATGGTGTACAGTAGG and GGTAAGAGTAGTCTGCACTTGCTG and the primers used to amplify the −202 bp AP-1 site were CCTAGGAACACGCGAATGAGAACC and GCTTAGAGCTCCTCCTTCTCTAACC. As a control, 0.12 % of each input DNA sample prior to immunoprecipitation was assayed by PCR amplification under identical conditions of the −202 bp AP-1 site (“input”).
RESULTS
SDF-1 co-stimulates IL-10 secretion by human T cells of both GCC+ and GCC− genomic haplotypes
We analyzed SDF-1 co-stimulation of IL-10 secretion by T cells from different individuals. Human PBMC were isolated from normal blood donors, stimulated in vitro with immobilized anti-TCR-CD3 mAb ± SDF-1, and assayed for IL-10 production. SDF-1 co-stimulation increased the number of T cells producing IL-10 protein, as shown by intracellular cytokine staining of CD4+ CD3+ cells (Fig. 1A). SDF-1 co-stimulation also increased the number of T cells producing IL-10 protein among CD8+ CD3+ T cells (Fig. 1A and data not shown). SDF-1 co-stimulation of IL-10-expressing T cells was consistently observed among different blood donors (Fig. 1B). The percentage of IL-10-expressing T cells induced after CD3 mAb alone or CD3 + SDF-1 stimulation varied among individual donors (Fig. 1B), therefore, we asked if IL-10 promoter haplotype influenced the ability of SDF-1 to co-stimulate T cell IL-10 expression. The GCC haplotype has previously been associated with higher IL-10 levels (25,26), therefore, genomic DNA sequencing of the -1082 bp SNP site was used to classify blood donors as either GCC+ or GCC−. All individuals who lacked the GCC haplotype were confirmed by DNA sequencing to contain either ATA and/or ACC haplotypes (data not shown). SDF-1 co-stimulation enhanced the amount of IL-10 secreted by all T cells, including individuals both with and without the −1082 bp G SNP (p < 0.05) (Fig. 1C). In contrast to SDF-1 co-stimulation, SDF-1 alone had no effect on T cell IL-10 production or secretion as measured either by intracellular cytokine staining or ELISA (Fig. 1A, 1B and data not shown). Together, the results in this section indicate that SDF-1 co-stimulates IL-10 production and secretion by human T cells, via a mechanism independent of the presence or absence of the GCC IL-10 promoter haplotype.
Fig. 1. SDF-1 co-stimulates IL-10 secretion by human T cells of both GCC+ and GCC− genomic haplotypes.
A) Normal human PBMC were purified from peripheral blood, then stimulated in vitro for 24 hr with 1 μg/ml immobilized CD3 mAb + SDF-1 (5 × 10−8 M). IL-10 expression was determined by intracellular cytokine staining. Results shown are gated on CD3+ T cells. B) Results of analyzing multiple PBMC donors as in A). C) Normal human T cells were purified and stimulated as in A), then secreted IL-10 was determined by ELISA in triplicate (S.D. for all data points were ≤ 10 %). The results from different donors are grouped according to a haplotype-characteristic SNP at −1082 relative to the IL-10 transcription start site: donors in which the GCC haplotype is present are at left, donors in which the GCC haplotype is absent are at right. Lines connect results from the same donor.
SDF-1 co-stimulates activity of GCC, ACC, and ATA IL-10 promoter haplotype constructs expressed in human T cells
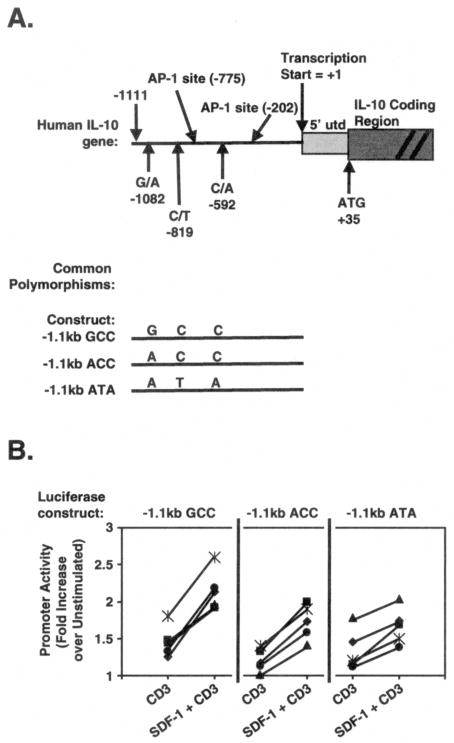
We next addressed the molecular mechanisms responsible for SDF-1 co-stimulation of T cell IL-10 secretion. Since we found no effect of SDF-1/CXCR4 signaling on mRNA stability regulated by the IL-10 3′-untranslated region (31,32) (data not shown), we analyzed SDF-1 co-stimulation of IL-10 promoter activity. 1.1 kb 5′ of the human IL-10 transcription start site contains the three SNPs that define the GCC, ACC, and ATA haplotypes discussed in this report, as well as several predicted transcription factor binding sites (Fig. 2A, top). We therefore subcloned this region from GCC, ACC, and ATA human haplotypes into a luciferase reporter plasmid vector, creating haplotype luciferase reporter constructs (Fig. 2A, bottom). The reporter plasmids were transiently transfected into normal human T cells, and luciferase was assayed and compared after in vitro stimulation of the T cells with TCR mAbs +/−SDF-1 (Fig. 2B). All three IL-10 promoter haplotype constructs responded with increased transcriptional activity in response to CD3 mAb stimulation of the TCR (for each construct, p < 0.05; n = 5). Moreover, SDF-1 co-stimulation enhanced the activity of all three haplotypes of the IL-10 promoter luciferase constructs (for each construct, p < 0.05; n = 5). Thus, all three of the most common human polymorphic IL-10 promoter haplotypes respond to TCR stimulation by increasing transcriptional activity, and SDF-1 further co-stimulates the TCR-dependent transcriptional activity of all three haplotypes.
Fig. 2. SDF-1 co-stimulates activity of GCC, ACC, and ATA IL-10 promoter haplotype constructs expressed in human T cells.
A) Top, cartoon of the proximal human IL-10 promoter region, showing SNPs referred to in the text and the putative AP-1 transcription factor binding sites. Bottom, cartoons showing the 1.1 kb fragments of the IL-10 promoters of the most common haplotypes (ATA, ACC, and GCC) that were subcloned into a luciferase reporter plasmid to create IL-10 promoter haplotype constructs. B) The IL-10 promoter luciferase constructs containing the indicated haplotypes were transiently-transfected into purified human T cells. Cells were stimulated in vitro for 18 hr with 1 μg/ml immobilized CD3 mAb ± SDF-1 (5 × 10−8 M) and assayed for luciferase activity. Fold-increases in promoter activity were calculated as compared to unstimulated samples in the same experiment, and the results of multiple independent experiments were then normalized so that the fold-increase of the unmutated, unstimulated construct in each experiment = 1. The results of multiple experiments performed on different days are shown; lines connect results from the same experiment; each point denotes the mean of 2 independent determinations; y-axes denote the fold-increase in luciferase activity as compared to unstimulated cells analyzed in the same experiment.
SDF-1 co-stimulates the MEK-1/ERK pathway in human T cells, and both IL-10 mRNA accumulation and IL-10 secretion by human T cells requires MEK-1 activity
Both the TCR and SDF-1 signaling activate the MEK-1/ERK pathway in human T cells (9,16–18). We therefore asked if SDF-1 co-stimulates TCR-mediated activation of this pathway. PBMC T cells were stimulated with either SDF-1 or cross-linked anti-TCR (CD3) mAb, lysed and assayed for active, phosphorylated ERK. The results indicate that both SDF-1 and CD3 mAb stimulated ERK activation, and that ERK activation was enhanced by co-stimulation with both SDF-1 and CD3 mAb (Fig. 3A, upper gel). The same gel was stripped and reprobed with antiserum recognizing total ERK2, as a loading control (Fig. 3A, lower gel). Thus, SDF-1 co-stimulation enhances TCR-mediated ERK activation. We next asked if the MEK-1/ERK pathway is required for T cell IL-10 secretion. For this purpose we used the MEK-1 inhibitor drug, PD098059. PD098059 pretreatment abrogated IL-10 mRNA accumulation after either TCR or TCR+SDF-1 stimulation of normal human T cells (Fig. 3B). The same MEK-1 inhibitor also inhibited TCR and TCR+SDF-1 -mediated stimulation of IL-10 protein secretion by human T cells (Fig. 3C). Thus, SDF-1 co-stimulates the MEK-1/ERK pathway in human T cells, and both IL-10 mRNA accumulation and IL-10 secretion by stimulated human T cells is sensitive to upstream blockade of the MEK-1/ERK pathway.
Fig. 3. SDF-1 co-stimulates the MEK-1/ERK pathway in human T cells, and both IL-10 mRNA accumulation and IL-10 secretion by human T cells requires MEK-1 activity.
A) Cells were stimulated with 1 μg/ml cross-linked CD3 mAb ± SDF-1 (5 × 10−8 M) for 12 min as indicated, then active, phosphorylated ERK was assayed by SDS-PAGE and immunoblotting with specific antisera (P-ERK; upper gel). As a control, the same membrane was stripped and reprobed with antisera recognizing total ERK2 (lower gel). B) Normal human T cells were pretreated with either vehicle (DMSO) or the MEK-1 inhibitor, PD098059 (PD), then stimulated as in Fig. 1A. Cells were then lysed and IL-10 mRNA levels were assayed by RT-PCR (upper gel). As a control, actin mRNA levels of the same samples were similarly assayed by RT-PCR (lower gel). C) Normal human T cells were pretreated with either vehicle (DMSO) or the MEK-1 inhibitor, PD098059 (PD), then stimulated as in Fig. 1A. Secreted IL-10 was assayed by ELISA as in Fig. 1C. The bar graph shows mean IL-10 levels ± S.D. of three independent determinations. Results in A) – C) are each representative of 3 independent experiments.
SDF-1 co-stimulates activity of the human IL-10 promoter in T cells by enhancing binding of the AP-1 transcription factor to two non-polymorphic binding sites located at −775 bp and −202 bp
We next addressed the mechanism by which SDF-1 co-stimulation of the MEK-1/ERK pathway leads to enhanced IL-10 promoter activity. Activation of the MEK-1/ERK pathway frequently leads to activation of downstream AP-1-dependent transcriptional activity (9,33,34), and we recently showed that SDF-1 selectively co-stimulates TCR-mediated AP-1 transcriptional activity (17). Since the human IL-10 promoter contains two putative AP-1 binding sites located approximately at −775 bp and −202 bp in all three common haplotypes of the IL-10 promoter (Fig. 2A), we hypothesized that AP-1 might mediate SDF-1 co-stimulatory effects on IL-10 gene transcription in T cells. To test this idea, we first asked if the activity of the IL-10 promoter constructs in human T cells requires the two putative AP-1 sites. Mutagenesis of both AP-1 sites of the IL-10 promoter construct significantly impaired both TCR and TCR+SDF-1 -mediated activation of the IL-10 promoter in T cells (Fig. 4A), suggesting a critical role for AP-1 in regulating activity of the IL-10 promoter in T cells. Consistent with this idea, the transcriptional activity in T cells of the un-mutated IL-10 promoter constructs was abrogated by the MEK-1 inhibitor drug, PD098059 (data not shown).
Fig. 4. SDF-1 co-stimulates activity of the human IL-10 promoter in T cells by enhancing binding of the AP-1 transcription factor to two non-polymorphic binding sites located at −775 bp and −202 bp.
A) Either the GCC haplotype IL-10 promoter luciferase construct or this construct with both putative AP-1 sites mutated (see Fig. 2A) was transiently-transfected into purified human T cells. The cells were then stimulated as indicated and assayed for luciferase activity as in Fig. 2B. Fold-increases in promoter activity were calculated as compared to unstimulated samples in the same experiment, and the results of multiple independent experiments were then normalized so that the fold-increase of the unmutated, unstimulated construct in each experiment = 10. B) Normal human T cells of GCC/GCC genotype were stimulated as in Fig. 1A for 12 hr. The presence of the AP-1 subunit, c-Fos, bound to two sites of the endogenous IL-10 promoter was then detected by chromatin immunoprecipitation (ChIP) assay. Chromatin immunoprecipitation of the SDF-1 + CD3 sample was also performed with control IgG (far right lane). The upper and middle gels denote c-Fos binding to the AP-1 sites located at approximately −775 and −202 bp of the human IL-10 promoter (numbering is relative to the transcription start site). The lower gel shows a control PCR showing the amount of IL-10 promoter DNA present in each sample prior to chromatin immunoprecipitation (“input”). The results shown are representative of 2 independent experiments using different donors.
To further address whether SDF-1 co-stimulates IL-10 promoter activity by enhancing AP-1 binding to the IL-10 promoter, we employed a chromatin immunoprecipitation (ChIP) assay. The most common transactivating form of AP-1 is composed of one c-Fos and one c-Jun subunit (35,36). Purified human T cells were stimulated in vitro and c-Fos-specific antibodies were used to immunoprecipitate AP-1 together with bound genomic DNA. PCR characterization of precipitated DNA revealed that, as compared to unstimulated cells or cells stimulated with either TCR or SDF-1 alone, TCR+SDF-1 stimulation enhanced c-Fos binding to both the −775 bp AP-1 binding site and the −202 AP-1 binding site of the endogenous IL-10 promoter (Fig. 4B, upper and middle gels). In contrast to the results obtained from using c-Fos-specific antibodies for chromatin immunoprecipitation, IL-10 promoter DNA was not specifically immunoprecipitated by non-specific control IgG (Fig. 4B, far right lane). Moreover, all samples contained similar amounts of IL-10 genomic DNA prior to immunoprecipitation (Fig. 4B, lower gel, “input”). Together, the results in this section indicate that SDF-1 co-stimulation enhances AP-1 binding to two sites of the human IL-10 promoter, and that this event is required for the promoter’s transcriptional activity in T cells.
SDF-1 co-stimulates IL-10 secretion by a subset of human CD4+ and CD8+ memory-phenotype T cells
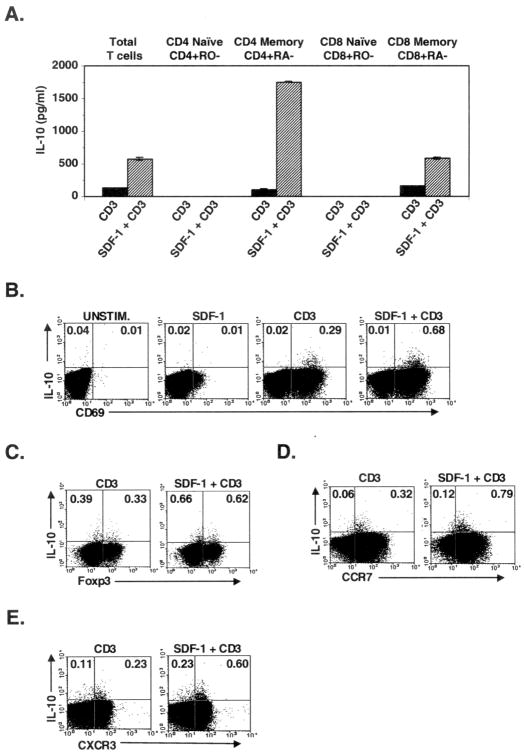
To gain a better understanding of the immunological role of SDF-1 co-stimulation of T cell IL-10 secretion, we characterized T cell subsets that respond to SDF-1 co-stimulation with enhanced IL-10 production and secretion. Several T cell subsets have been previously shown to produce IL-10, including Th2 effector cells and various regulatory and memory T cell subsets (1,37–39). We therefore purified human peripheral blood T cells according to their expression of CD4, CD8, and the naive/memory markers, CD45RA and CD45RO. Purified T cells were stimulated in vitro and assayed for secreted IL-10. Compared to naive-phenotype (CD45-RO-negative) T cells, SDF-1 potently co-stimulated IL-10 secretion by both CD4+ and CD8+ memory-phenotype (CD45-RA-negative) T cells (Fig. 5A). The effect of SDF-1 was particularly potent on CD4+CD45-RA-negative T cells: SDF-1 enhanced CD3-mediated IL-10 secretion by over 10-fold in 4 out of 5 donors (data not shown). Similar results were obtained using intracellular cytokine staining to detect IL-10-producing T cells (data not shown). Thus, SDF-1 co-stimulates IL-10 secretion by human CD4+ and CD8+ memory-phenotype T cells, particularly the CD4+ memory-phenotype T cells. Cell-surface and other FACS analysis was then used together with intracellular cytokine staining to further characterize the CD4+ memory-phenotype T cells that express IL-10 after SDF-1 co-stimulation. Interestingly, CD4+ memory T cells that responded to SDF-1 co-stimulation with enhanced IL-10 production included cells both positive and negative for the transcription factor Foxp3 that has been associated with regulatory T cells (Fig. 5C). In contrast to Foxp3 staining, all IL-10+ CD4+ memory-phenotype T cells were positive for the activation marker, CD69 (Fig. 5B). Similarly, all IL-10+ cells were positive for the activation marker, CD25 (data not shown). Thus, the CD4+ memory-phenotype T cells that respond to SDF-1 co-stimulation with enhanced IL-10 production appear to include both Foxp3+ regulatory and other types of memory-phenotype T cells. In addition to regulatory T cells, memory-phenotype T cell populations contain both “central” and “peripheral” memory T-cells that are defined via expression or non-expression of the CCR7 chemokine receptor, respectively (40). Most IL-10-secreting cells expressed CCR7, however, SDF-1 co-stimulated IL-10 production by both CCR7+ and CCR7− subsets of the CD4+ memory-phenotype T cells (Fig. 5D). Thus, SDF-1 appears to co-stimulate IL-10 secretion by a subset of both central and peripheral memory T cells. Finally, we assayed expression of the CXCR3 chemokine receptor by SDF-1-co-stimulated IL-10-expressing CD4+ memory-phenotype T cells. CXCR3 expression has been associated with both activation and tissue-homing effector and memory T cell subsets (41–43). Most IL-10-secreting cells expressed CXCR3, although SDF-1 co-stimulated IL-10 expression of CD4+ memory-phenotype T cells which were also CXCR3- (Fig. 5E). Together, these results indicate that SDF-1 is capable of co-stimulating IL-10 secretion by diverse T cell subsets within the population characterized by expression of the previously-activated/”memory” cell-surface CD45 markers, including presumed regulatory Foxp3+ T cells, both central and peripheral memory T cells, and CXCR3+ memory T cells.
Fig. 5. SDF-1 co-stimulates IL-10 secretion by a subset of human CD4+ and CD8+ memory-phenotype T cells.
Purified human T cells were separated into CD4, CD8, CD45RA and CD45RO populations then stimulated in vitro as in Fig. 1A. A) Secreted IL-10 was assayed by ELISA as in Fig. 1C. Each bar shows the mean of 2 determinations ± range. B–E) Purified CD4+ memory-phenotype T cells stimulated as in Fig. 5A were further characterized by intracellular cytokine staining for IL-10 expression together with staining for either cell-surface CD69, intracellular Foxp3, or cell-surface CCR7 or CXCR3. Results in A) – E) are each typical of 3–5 independent experiments using different donors.
DISCUSSION
Despite extensive genetic evidence linking particular human IL-10 promoter polymorphisms to various autoimmune diseases and immune conditions (19–23), the molecular mechanisms that regulate activity of the human IL-10 promoter in T cells have until now been only poorly understood. Moreover, despite previous reports describing SDF-1 co-stimulation of IL-10 secretion by human T cells (17,44,45), neither the particular T cell subset(s) targetted by SDF-1, nor any signaling pathway(s) or molecular mechanisms responsible for this effect of SDF-1, have been defined. Knowledge about these mechanisms becomes particularly urgent to acquire as epidemiological evidence is accumulating that small, genetically-determined differences in IL-10 secretion can significantly alter human susceptibility to certain autoimmune diseases and autoimmune conditions including Sjogren’s syndrome, rheumatoid arthritis, and graft-vs.-host disease (20–25). In addition, understanding the regulation of IL-10 secretion by T cells is of particular importance since experimental evidence indicates that T cell-derived IL-10, in contrast to IL-10 secreted by other cell types, is critical for inhibiting the development of autoimmunity (2) and the airway inflammation associated with allergy and asthma (3). We therefore investigated the molecular mechanisms responsible for SDF-1 co-stimulation of human T cell IL-10 secretion. In addition, we compared the effects of SDF-1 co-stimulation on common human T cell IL-10 locus haplotypes and investigated the subpopulation(s) of T cells in which IL-10 production is regulated by SDF-1.
Our results indicate that SDF-1 co-stimulates IL-10 secretion in human T cells via a mechanism in which SDF-1 enhances TCR-mediated activation of the MEK-1/ERK signaling pathway, thereby leading to enhanced AP-1 production, enhanced AP-1 binding to the IL-10 promoter, and enhanced IL-10 gene transcription. Moreover, we show that the co-stimulatory effects of SDF-1 on IL-10 promoter activity and T cell secretion of IL-10 are haplotype-independent, occurring similarly in all three of the most common haplotypes of the polymorphic IL-10 promoter. Finally, we show that SDF-1 co-stimulates IL-10 secretion by several T cell populations that are characterized by cell-surface memory-marker phenotypes, including T cells both expressing and not expressing the putative regulatory T cell marker, Foxp3. Together, our results represent a novel and important advance in the understanding of the molecular and genetic regulation of the human IL-10 promoter in T cells. The signaling pathway defined here may in addition represent a mechanism that could be usefully modulated therapeutically in order to regulate T cell IL-10 secretion and related immunopathologies.
No previous studies have determined if SDF-1 co-stimulates IL-10 in a haplotype -dependent or -independent manner. We found that SDF-1 co-stimulated IL-10 secretion by human T cells from all donors tested, whether or not the donor was positive for the previously-reported high-expressing IL-10 promoter haplotype that is identified by the -1082 G SNP (25,26). To more clearly define the effects of SDF-1 co-stimulation on IL-10 promoter activity, and the impact of different haplotypes on this effect of SDF-1, we isolated and cloned into luciferase analysis plasmids the three most common haplotypes of IL-10 promoters from human donors that are defined by three linked SNPs of ATA, ACC, and GCC. In normal human T cells, TCR stimulation increased the promoter activities of all three haplotypes, and SDF-1 co-stimulation significantly further enhanced the promoter activities of all three haplotypes. Thus, SDF-1 co-stimulates T cell IL-10 secretion in a similar manner in the three most common polymorphic variants of the human IL-10 promoter.
The above results suggested that SDF-1 co-stimulation of human IL-10 promoter activity is mediated via a non-polymorphic transcription factor binding site of the promoter. Several of our results indicate that SDF-1 co-stimulates T cell IL-10 gene transcription and IL-10 secretion by enhancing TCR-mediated activation of the MEK-1-ERK pathway and downstream AP-1, which then binds the IL-10 promoter at a non-polymorphic site and enhances its transcriptional activity. First, we recently showed that SDF-1/CXCR4 signals in T cells via the TCR, and that this novel pathway is required for SDF-1 to stimulate prolonged ERK activation and robust activation of AP-1-dependent transcriptional activity (16,17). SDF-1 is relatively unique among several chemokines in its ability to stimulate ERK activation in T cells for a prolonged period of time (18), suggesting that SDF-1-mediated ERK activation subserves a special biological role. Indeed, while blockade of MEK-1/ERK activation has no effect on SDF-1-directed chemotaxis (46), MEK-1/ERK activity is required for SDF-1 to induce the expression of multiple genes involved in signal transduction and for the prevention of apoptosis in a T cell line (9). ERK phosphorylates and regulates the functions of transcription factors (47,48), thereby regulating gene expression. Second, we showed that SDF-1 co-stimulates TCR-mediated activation of the MEK-1-ERK pathway, and that blockade of this pathway by a MEK-1 inhibitor drug abrogates both IL-10 mRNA accumulation and IL-10 secretion by stimulated T cells. Third, we showed that mutation of two non-polymorphic consensus AP-1 binding sites within the IL-10 promoter disrupts activation of this promoter in T cells. Fourth, we showed using a chromatin immunoprecipitation assay that SDF-1 + TCR stimulation of human T cells enhances the binding of c-Fos-containing AP-1 transcription factors to the IL-10 promoter. Together, these results provide strong support for the idea that the mechanism by which SDF-1 co-stimulates T cell IL-10 secretion is by enhancing TCR-mediated activation of the MEK-1/ERK signaling pathway, which leads to enhanced AP-1 production, enhanced AP-1 binding to the IL-10 promoter, and enhanced IL-10 gene transcription. Similar to the classic situation of CD28 co-stimulation of the IL-2 promoter, our results also indicate that activation of the ERK/AP-1 pathway by SDF-1 is not by itself sufficient to induce IL-10 secretion. Thus, in addition to activation of the ERK/AP-1 pathway, other regulatory signals induced in response to anti-CD3/TCR stimulation are likely to be required for IL-10 secretion.
This is the first study to indicate the importance of AP-1 in regulating IL-10 promoter activity in human T cells. Basal promoter elements have previously been shown to exist near the IL-10 promoter TATA-box (49). As for many inducible promoters, an Sp1 binding site at approximately −80 bp relative to the transcription start site is essential for IL-10 promoter activity in T cells and macrophages of both humans and mice (50–52). Analysis of the human IL-10 promoter in the human T lymphoma cell line HuT78 identified and characterized roles for NF-κB in its regulation (53), and a STAT3 site located at −120 bp was shown to be involved in LPS-induced activation of the human IL-10 promoter in a B cell line (54). A SMAD binding site previously characterized as mediating IL-10 secretion in T cells in response to TGF-β is unlikely to function in human T cells since it is not present in the human IL-10 promoter (55). In contrast to this SMAD binding site, AP-1 regulation of the IL-10 promoter in T cells is likely to be conserved between mouse and humans, since the mouse IL-10 promoter also contains an AP-1 binding site that was shown to be functional in a mouse Th2 cell clone (56). The ATA, ACC, and GCC haplotypes have been associated with low, medium, and high levels of IL-10 secretion, respectively (25,26). However, none of these previously-reported linked polymorphisms, nor any other polymorphisms of the IL-10 promoter described in the literature, specifically target either AP-1 site.
Our results further indicate that SDF-1 co-stimulates IL-10 secretion by several different subsets of previously-activated T cells. SDF-1 co-stimulated both central and peripheral memory T cells defined as CD45RA- and either CCR7+ or CCR7−, as well as CD45RA- T cells that are CXCR3+ and may therefore be capable of migrating into inflamed tissues. SDF-1 also appears to co-stimulate IL-10 secretion by CD4+CD45RA- T cells that are both positive and negative for the presumed regulatory T cell marker, Foxp3. SDF-1 co-stimulation of T cell IL-10 secretion may therefore have the potential to broadly antagonize inflammatory responses, both in the lymph nodes and within inflamed tissues.
The biological functions of many chemokine receptors including CXCR4 are often assumed to be derived primarily from their ability to regulate cellular migration. Thus, SDF-1/CXCR4 signaling has been proposed to play a homeostatic role in regulating the migration patterns of immune and other cell types (10,11). Here, we show that SDF-1/CXCR4 signaling also significantly regulates T cell gene expression of the critical immune regulatory cytokine, IL-10. SDF-1/CXCR4 signaling therefore appears to contribute to immune homeostasis via multiple critical mechanisms, including both immune cell migration and regulation of IL-10 gene expression. While IL-10 levels have not been specifically examined in mice or humans either completely or partially deficient in expression of SDF-1 and/or CXCR4, it is therefore possible that some phenotypes and conditions associated with mutation of these genes might be exacerbated by changes in SDF-1-dependent IL-10 secretion in addition to changes in SDF-1-dependent cellular migration. For example, mice completely deficient in either SDF-1 or CXCR4 die perinatally from multiple developmental defects in the brain, gut, and bone marrow (10). Humans bearing a relatively common human SDF-1 polymorphism (801A) that is thought to decrease SDF-1 expression (57) are at higher risk for certain conditions that might theoretically be exacerbated by low IL-10 levels, including transmitting HIV-1 maternally (58), developing early onset Type I diabetes after a shorter lag time (59), and developing lung cancer (60). In addition, rare mutations in CXCR4 that truncate CXCR4 and enhance some SDF-1-dependent signals are linked to WHIM syndrome, which is characterized by extensive Human Papilloma Virus infection in addition to other pathologies (61). Thus, SDF-1/CXCR4 signaling may contribute to immune homeostasis via multiple critical mechanisms, including both immune cell migration and regulation of IL-10 gene expression.
Acknowledgments
We are grateful to J. Tarara and the Mayo Flow Cytometry Core Facility for providing expert technical assistance and to L. Pease, R. Bram, S. Kauffman, and P. Leibson for comments on the manuscript.
Footnotes
This work was supported in part by the philanthropy of Barbara Lipps to the Mayo Clinic and by N.I.H. RO1 Grant #GM59763 (to K.E.H).
References
- 1.Groux H, Cottrez F. The complex role of IL-10 in autoimmunity. J Autoimmunity. 2003;20:281–285. doi: 10.1016/s0896-8411(03)00044-1. [DOI] [PubMed] [Google Scholar]
- 2.Roers A, Siewe L, Strittmatter E, Deckert M, Schluter D, Stenzel W, Gruber AD, Krieg T, Rajewsky L, Muller W. T cell-specific inactivation of the IL-10 gene in mice results in enhanced T cell responses but normal innate responses to lipopolysaccharide or skin irritation. J Exp Med. 2004;200:1289–1297. doi: 10.1084/jem.20041789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hawrylowicz CM, O’Garra A. Potential role of interleukin-10-secreting regulatory T cells in allergy and asthma. Nat Rev Immunol. 2005;5:271–283. doi: 10.1038/nri1589. [DOI] [PubMed] [Google Scholar]
- 4.Ansel KM, Cyster JG. Chemokines in lymphopoiesis and lymphoid organ development. Curr Opin Immunol. 2001;13:172–179. doi: 10.1016/s0952-7915(00)00201-6. [DOI] [PubMed] [Google Scholar]
- 5.Campbell JJ, Butcher EC. Chemokines in tissue-specific and microenvironment-specific lymphocyte homing. Curr Opin Immunol. 2000;12:336–341. doi: 10.1016/s0952-7915(00)00096-0. [DOI] [PubMed] [Google Scholar]
- 6.Murphy PM, Baggiolini M, Charo IF, Hebert CA, Horuk R, Matsushima K, Miller LH, Oppenheim JJ, Power CA. International union of pharmacology XXII. Nomenclature for chemokine receptors. Pharmacological Reviews. 2000;52:145–176. [PubMed] [Google Scholar]
- 7.Laudanna C, Kim JY, Constantin G, Butcher EC. Rapid leukocyte integrin activation by chemokines. Immunol Rev. 2002;186:37–46. doi: 10.1034/j.1600-065x.2002.18604.x. [DOI] [PubMed] [Google Scholar]
- 8.Phillips R, Ager A. Activation of pertussis toxin-sensitive CXCL12 (SDF-1) receptors mediates transendothelial migration of T lymphocytes across lymph node high endothelial cells. Eur J Immunol. 2002;32:837–847. doi: 10.1002/1521-4141(200203)32:3<837::AID-IMMU837>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 9.Suzuki Y, Rahman M, Mitsuya H. Diverse transcriptional response of CD4+ T cells to Stromal cell-derived factor (SDF)-1: cell survival promotion and priming effects of SDF-1 on CD4+ T cells. J Immunol. 2001;167:3064–3073. doi: 10.4049/jimmunol.167.6.3064. [DOI] [PubMed] [Google Scholar]
- 10.Zou YR, Kottmann AH, Kuroda M, Taniuchi I, Littman DR. Function of the chemokine receptor CXCR4 in haematopoiesis and in cerebellar development. Nature. 1998;393:595–599. doi: 10.1038/31269. [DOI] [PubMed] [Google Scholar]
- 11.Sawada S, Gowrishankar K, Kitamura R, Suzuki M, Suzuki G, Tahara S, Koito A. Disturbed CD4+ T cell homeostasis and in vitro HIV-1 susceptibility in transgenic mice expressing T cell line-tropic HIV-1 receptors. J Exp Med. 1998;187:1439–1449. doi: 10.1084/jem.187.9.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gonzalo JA, Lloyd CM, Peled A, Delaney T, Coyle AJ, Gutierrez-Ramos JC. Critical involvement of the chemotactic axis CXCR4/stromal cell-derived factor-1α in the inflammatory component of allergic airway disease. J Immunol. 2000;165:499–508. doi: 10.4049/jimmunol.165.1.499. [DOI] [PubMed] [Google Scholar]
- 13.Matin K, Salam MA, Akhter J, Hanada N, Senpuku H. Role of stromal-cell derived factor-1 in the development of autoimmune diseases in non-obese diabetic mice. Immunology. 2002;107:222–232. doi: 10.1046/j.1365-2567.2002.01478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Verani A, Lusso P. Chemokines as natural HIV antagonists. Curr Mol Med. 2002;2:691–702. doi: 10.2174/1566524023361862. [DOI] [PubMed] [Google Scholar]
- 15.Feng Y, Broder CC, Kennedy PE, Berger EA. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 16.Kremer KN, Humphreys TD, Kumar A, Qian NX, Hedin KE. Distinct role of ZAP-70 and Src Homology 2 domain-containing leukocyte protein of 76 kDa in the prolonged activation of Extracellular Signal-regulated protein kinase by the Stromal-Cell-derived Factor 1-α/CXCL12 chemokine. J Immunol. 2003;171:360–367. doi: 10.4049/jimmunol.171.1.360. [DOI] [PubMed] [Google Scholar]
- 17.Kumar A, Humphreys TD, Kremer KN, Bramati PS, Bradfield L, Edgar CE, Hedin KE. CXCR4 physically associates with the T Cell Receptor to signal in T cells. Immunity. 2006;25:213–224. doi: 10.1016/j.immuni.2006.06.015. [DOI] [PubMed] [Google Scholar]
- 18.Tilton B, Ho L, Oberlin E, Loetscher P, Baleux F, Clark-Lewis I, Thelen M. Signal transduction by CXC chemokine receptor 4: stromal cell-derived factor 1 stimulates prolonged protein kinase B and extracellular signal-regulated kinase 2 activation in T lymphocytes. J Exp Med. 2000;192:313–324. doi: 10.1084/jem.192.3.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim JM, Brannan CI, Copeland NG, Jenkins NA, Khan TA, Moore KW. Structure of the mouse IL-10 gene and chromosomal localization of the mouse and human genes. J Immunol. 1992;148:3618–3623. [PubMed] [Google Scholar]
- 20.Kube D, Platzer C, von Knethen A, Straub H, Bohlen H, Hafner M, Tesch H. Isolation of the human IL-10 promoter: Characterization of the promoter activity in Burkitt’s Lymphoma cell lines. Cytokine. 1995;7:1–7. doi: 10.1006/cyto.1995.1001. [DOI] [PubMed] [Google Scholar]
- 21.Eskdale J, Kube D, Tesch H, Gallagher G. Mapping of the human IL-10 gene and further characterization of the 5′ flanking sequence. Immunogenetics. 1997;46:120–128. doi: 10.1007/s002510050250. [DOI] [PubMed] [Google Scholar]
- 22.Kube D, Rieth H, Eskdale J, Kremsner PG, Gallagher G. Structural characterisation of the distal 5′ flanking region of the human IL-10 gene. Genes and Immunity. 2001;2:181–190. doi: 10.1038/sj.gene.6363750. [DOI] [PubMed] [Google Scholar]
- 23.Lin MT, Storer B, Martin PJ, Tseng LH, Gooley T, Chen PJ, Hansen JA. Relation of an IL-10 promoter polymorphism to graft-versus-host disease and survival after hematopoietic-cell transplantation. New Engl J Med. 2003;349:2201–2210. doi: 10.1056/NEJMoa022060. [DOI] [PubMed] [Google Scholar]
- 24.Howell WM. Cytokine polymorphisms, cancer susceptibility, and prognosis. ASHI Quarterly First Quarter. 2005;2005:10–15. [Google Scholar]
- 25.Eskdale J, Gallagher G, Verweij CL, Keijsers V, Westendorp RGJ, Huizinga TWJ. IL-10 secretion in relation to human IL-10 locus haplotypes. Proc Natl Acad Sci USA. 1998;95:9465–9470. doi: 10.1073/pnas.95.16.9465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoffman SC, Stanley EM, Cox ED, Craighead N, DiMercurio BS, Koziol DE, Harlan DM, Kirk AD, Blair PJ. Association of cytokine polymorphic inheritance and in vitro cytokine production in anti-CD3/CD28-stimulated peripheral blood lymphocytes. Transplantion. 2001;72:1444–1450. doi: 10.1097/00007890-200110270-00019. [DOI] [PubMed] [Google Scholar]
- 27.Bidwell J, Keen L, Gallagher G, Kimberly R, Huizinga T, McDermott MF, Oksenberg J, McNicholl J, Pociot F, Hardt C, D’Alfonso S. Cytokine gene polymorphism in human disease: on-line databases. Genes and Immunity. 1999;1:3–19. doi: 10.1038/sj.gene.6363645. [DOI] [PubMed] [Google Scholar]
- 28.Haukim N, Bidwell J, Smith AJP, Keen L, Gallagher G, Kimberly R, Huizinga T, McDermott DH, Oksenberg J, McNicholl J, Pociot F, Hardt C, D’Alfonso S. Cytokine gene polymorphism in human disease: on-line databases, Supplement 2. Genes and Immunity. 2002;3:313–330. doi: 10.1038/sj.gene.6363881. [DOI] [PubMed] [Google Scholar]
- 29.Shin HD, Winkler C, Stephens JC, Bream J, Young H, Goedert JJ, O’Brien TR, Vlahov D, Buchbinder S, Giorgi J, Rinaldo C, Donfield S, Willoughby A, O’Brien SJ, Smith MW. Genetic restriction of HIV-1 pathogenesis to AIDS by promoter alleles of IL-10. Proc Natl Acad Sci USA. 2000;97:14467–14472. doi: 10.1073/pnas.97.26.14467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bell MP, Huntoon C, Graham D, McKean DJ. The analysis of costimulatory receptor signaling cascades in normal T lymphocytes using in vitro gene transfer and reporter gene analysis. Nature Medicine. 2001;10:1155–1158. doi: 10.1038/nm1001-1155. [DOI] [PubMed] [Google Scholar]
- 31.Powell MJ, Thampson SAJ, Tone Y, Waldmann H, Tone M. Posttranscriptional regulation of IL-10 gene expression through sequences in the 3′-untranslated region. J Immunol. 2000;165:292–296. doi: 10.4049/jimmunol.165.1.292. [DOI] [PubMed] [Google Scholar]
- 32.Winzen R, Kracht M, Ritter B, Wilhelm A, Chen CYA, Shyu AB, Muller M, Gaestel M, Resch K, Holtmann H. The p38 MAP kinase pathway signals for cytokine-induced mRNA stabilization via MAP kinase-activated protein kinase 2 and an AU-rich region targeted mechanism. EMBO J. 1999;18:4969–4980. doi: 10.1093/emboj/18.18.4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vaudry D, Stork PJ, Lazarovici P, Eiden LE. Signaling pathways for PC12 cell differentiation: making the right connections. Science. 2002;296:1648–1649. doi: 10.1126/science.1071552. [DOI] [PubMed] [Google Scholar]
- 34.Harada T, Morooka T, Ogawa S, Nishida E. ERK induces p35, a neuron-specific activator of CDK5, through induction of Egr1. Nature Cell Biol. 2001;3:453–459. doi: 10.1038/35074516. [DOI] [PubMed] [Google Scholar]
- 35.Murphy LO, Smith S, Chen RH, Fingar DC, Blenis J. Molecular interpretation of ERK signal duration by immediate early gene products. Nature Cell Biol. 2002;4:556–564. doi: 10.1038/ncb822. [DOI] [PubMed] [Google Scholar]
- 36.Treisman R. Ternary complex factors: growth factor regulated transcriptional activators. Curr Opin Genetics & Develop. 1994;4:96–101. doi: 10.1016/0959-437x(94)90097-3. [DOI] [PubMed] [Google Scholar]
- 37.Moore KW, Malefyt RDW, Coffman RL, O’Garra A. IL-10 and the IL-10 receptor. Annu Rev Immunol. 2001;19:683–765. doi: 10.1146/annurev.immunol.19.1.683. [DOI] [PubMed] [Google Scholar]
- 38.Yssel H, Malefyt RDW, Roncarolo MG, Abrams JS, Lahesmaa R, Spits H, deVries JE. IL-10 is produced by subsets of human CD4+ T cell clones and peripheral blood T cells. J Immunol. 1992;149:2378–2384. [PubMed] [Google Scholar]
- 39.Gouma G, Strober W. The immunological and genetic basis of inflammatory bowel disease. Nature Reviews Immunology. 2003;3:521–533. doi: 10.1038/nri1132. [DOI] [PubMed] [Google Scholar]
- 40.Sallusto F, Lanzavecchia A. Exploring pathways for memory T cell generation. J Clin Invest. 2001;108:805–806. doi: 10.1172/JCI14005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Loetscher M, Loetscher P, Brass N, Meese E, Moser B. Lymphocyte-specific chemokine receptor CXCR3: regulation, chemokine binding, and gene localization. Eur J Immunol. 1998;28:3696–3705. doi: 10.1002/(SICI)1521-4141(199811)28:11<3696::AID-IMMU3696>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 42.Hancock WW, Gao W, Faia K, Csizmadia V. Chemokines and their receptors in allograft rejection. Curr Opin Immunol. 2000;12:511–516. doi: 10.1016/s0952-7915(00)00130-8. [DOI] [PubMed] [Google Scholar]
- 43.Lazzeri E, Romagnani P. CXCR3-binding chemokines: novel multifunctional therapeutic targets. Curr Drug Targets- Immune, Endocrine, & Metabolic Disorders. 2005;5:109–118. doi: 10.2174/1568008053174723. [DOI] [PubMed] [Google Scholar]
- 44.Nanki T, Lipsky PE. Cutting Edge: stromal cell-derived factor-1 is a costimulator for CD4+ T cell activation. J Immunol. 2000;164:5010–5014. doi: 10.4049/jimmunol.164.10.5010. [DOI] [PubMed] [Google Scholar]
- 45.Molon B, Gri G, Bettella M, Gomez-Mouton C, Lanzavecchia A, Martinez-A C, Manes S, Viola A. T cell costimulation by chemokine receptors. Nature Immunol. 2005;6:465–471. doi: 10.1038/ni1191. [DOI] [PubMed] [Google Scholar]
- 46.Cherla RP, Ganju RK. Stromal cell-derived factor 1α-induced chemotaxis in T cells is mediated by nitric oxide signaling pathways. J Immunol. 2001;166:3067–3074. doi: 10.4049/jimmunol.166.5.3067. [DOI] [PubMed] [Google Scholar]
- 47.Marshall CJ. MAP kinase kinase kinase, MAP kinase kinase, and MAP kinase. Curr Opin Genetics & Develop. 1994;4:82–89. doi: 10.1016/0959-437x(94)90095-7. [DOI] [PubMed] [Google Scholar]
- 48.Chang L, Karin M. Mammalian MAP kinase signalling cascades. Nature. 2001;410:37–40. doi: 10.1038/35065000. [DOI] [PubMed] [Google Scholar]
- 49.Brenner S, Prosch S, Schenke-Layland K, Riese U, Gausmann U, Platzer C. cAMP-induced IL-10 promoter activation depends on CCAAT/Enhancer-Binding Protein expression and monocytic differentiation. J Biol Chem. 2003;278:5597–5604. doi: 10.1074/jbc.M207448200. [DOI] [PubMed] [Google Scholar]
- 50.Tone M, Powell MJ, Ton Y, Thompson SAJ, Waldmann H. IL-10 gene expression is controlled by the transcription factors Sp1 and Sp3. J Immunol. 2000;165:286–291. doi: 10.4049/jimmunol.165.1.286. [DOI] [PubMed] [Google Scholar]
- 51.Brightbill HD, Plevy SE, Modlin RL, Smale ST. A prominent role for Sp1 during lipopolysaccharide-mediated induction of the IL-10 promoter in macrophages. J Immunol. 2000;164:1940–1951. doi: 10.4049/jimmunol.164.4.1940. [DOI] [PubMed] [Google Scholar]
- 52.Ma W, Lim W, Gee K, Aucoin S, Nandan D, Kozlowski M, Diaz-Mitoma F, Kumar A. The p38 mitogen-activated kinase pathway regulates the human IL-10 promoter via the activation of Sp1 transcription factor in lipopolysaccharide-stimulated human macrophages. J Biol Chem. 2001;276:13664–13674. doi: 10.1074/jbc.M011157200. [DOI] [PubMed] [Google Scholar]
- 53.Mori N, Prager D. Activation of the IL-10 gene in the human T lymphoma line HuT 78: identification and characterization of NF-kappaB binding sites in the regulatory region of the IL-10 gene. Eur J Haematol. 1997;59:162–170. doi: 10.1111/j.1600-0609.1997.tb00970.x. [DOI] [PubMed] [Google Scholar]
- 54.Benkahrt EM, Siedlar M, Wedel A, Werner T, Ziegler-Heitbrock HWL. Role of STAT3 in lipopolysaccharide-induced IL-10 gene expression. J Immunol. 2000;165:1612–1617. doi: 10.4049/jimmunol.165.3.1612. [DOI] [PubMed] [Google Scholar]
- 55.Kitani A, Fuss I, Nakamura K, Kumaki F, Usui T, Strober W. Transforming growth factor (TGF)-beta-1-producing regulatory T cells induce SMAD-mediated IL-10 secretion that facilitates coordinated immunoregulatory activity and amelioration of TGF-beta-1-mediated fibrosis. J Exp Med. 2003;198:1179–1188. doi: 10.1084/jem.20030917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang ZY, Sato H, Kusam S, Sehra S, Toney LM, Dent AL. Regulation of IL-10 gene expression in Th2 cells by Jun proteins. J Immunol. 2005;174:2098–2105. doi: 10.4049/jimmunol.174.4.2098. [DOI] [PubMed] [Google Scholar]
- 57.Dommange F, Cartron G, Espanel C, Gallay N, Domenech J, Benboubker L, Ohresser M, Colombat P, Binet C, Watier H, Herault O. CXCL12 polymorphism and malignant cell dissemination/tissue infiltration in acute myeloid leukemia. FASEB J. 2006;20:1913–1915. doi: 10.1096/fj.05-5667fje. [DOI] [PubMed] [Google Scholar]
- 58.John GC, Rousseau C, Dong T, Rowland-Jones S, Nduati R, Mbori-Ngacha D, Rostron T, Kreiss JK, Richardson BA, Overbaugh J. Maternal SDF1 3′ A polymorphism is associated with increased perinatal Human Immunodeficiency Virus type 1 transmission. J Virol. 2000;74:5736–5739. doi: 10.1128/jvi.74.12.5736-5739.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dubois-Laforgue D, Hendel H, Caillat-Zucman S, Zagury JF, Winkler C, Boitard C, Timsit J. A common Stromal Cell-Derived Factor-1 chemokine gene variant is associated with the early onset of Type 1 diabetes. Diabetes. 2001;50:1121–1213. doi: 10.2337/diabetes.50.5.1211. [DOI] [PubMed] [Google Scholar]
- 60.Razmkhah M, Doroudchi M, Ghayumi SMA, Erfani N, Ghaderi A. Stromal cell-derived factor-1 (SDF-1) gene and susceptibility of Iranian patients with lung cancer. Lung Cancer. 2005;49:311–315. doi: 10.1016/j.lungcan.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 61.Hernandez PA, Gorlin RJ, Lukens JN, Taniuchi S, Bohinjec J, Francois F, Klotman ME, Diaz GA. Mutations in the chemokine receptor gene CXCR4 are associated with WHIM syndrome, a combined immunodeficiency disease. Nature Genetics. 2003;34:70–74. doi: 10.1038/ng1149. [DOI] [PubMed] [Google Scholar]