Abstract
Sulphur is an essential element for plant growth and development as well as for defence against biotic and abiotic stresses. Increasing sulphate utilization efficiency (SUE) is an important issue for crop improvement. Little is known about the genetic determinants of sulphate utilization efficiency. No gain-of-function mutants with improved SUE have been reported to date. Here the isolation and characterization of two low-sulphur-tolerant mutants, sue3 and sue4 are reported using a high-throughput genetic screen where a ‘sulphur-free’ solid medium was devised to give the selection pressure necessary to suppress the growth of the wild-type seedlings. Both mutants showed improved tolerance to low sulphur conditions and well-developed root systems. The mutant phenotype of both sue3 and sue4 was specific to sulphate deficiency and the mutants displayed enhanced tolerance to heavy metal and oxidative stress. Genetic analysis revealed that sue3 was caused by a single recessive nuclear mutation while sue4 was caused by a single dominant nuclear mutation. The recessive locus in sue3 is the previously identified VirE2-interacting Protein 1. The dominant locus in sue4 is a function-unknown locus activated by the four enhancers on the T-DNA. The function of SUE3 and SUE4 in low sulphur tolerance was confirmed either by multiple mutant alleles or by recapitulation analysis. Taken together, our results demonstrate that this genetic screen is a reasonable approach to isolate Arabidopsis mutants with improved low sulphur tolerance and potentially with enhanced sulphate utilization efficiency. The two loci identified in sue3 and sue4 should assist in understanding the molecular mechanisms of low sulphur tolerance.
Keywords: Activation tagging, Arabidopsis, At1g43700, At3g55880, low-sulphur tolerance, root, sulphate utilization efficiency
Introduction
Sulphur is an essential element for all living organisms. In higher plants, sulphur plays important roles in plant growth and development as well as in plant defences against biotic and abiotic stresses although sulphur content accounts for only 0.1% of the total dry matter. Sulphate is the major form of inorganic sulphur in soil and is absorbed by plant roots and transported by a family of sulphate transporters (Takahashi et al., 1997; Yoshimoto et al., 2003; Kataoka et al., 2004a, b; Rouached et al., 2009). Sulphate is reduced in the chloroplasts and assimilated into cysteine, the first organic product of sulphate reduction in plants and a central component in sulphur metabolism (Leustek et al., 2000). Under normal growth conditions in Arabidopsis sulphate reduction takes place in plastids and cysteine synthesis occurs in plastids, mitochondria, and the cytosol (Heeg et al., 2008; Kopriva et al., 2009).
Plants accumulate little cysteine but maintain a high flux of the amino acid. Cysteine is the donor of reduced sulphur for the synthesis of methionine and other S-containing metabolites (Kopriva, 2006). Methionine is the precursor for S-adenosyl methionine (SAM), the precursor for ethylene (Burstenbinder et al., 2007), polyamines, and nicotinamine which is important for Fe nutrition in plants (Ling et al., 1999; Zuchi et al., 2009). Some of the intermediate metabolites of the sulphur metabolic pathway are precursors for a number of vitamins and the sulpholipids of chloroplasts. Glutathione plays many important roles in plants including defence against biotic (Parisy et al., 2007; Schlaeppi et al., 2008) and abiotic stresses (Noctor and Foyer, 1998; Vernoux et al., 2000; Xiang et al., 2001), regulation of stress-related gene expression (Ball et al., 2004; Maruyama-Nakashita et al., 2005), cell division (Vernoux et al., 2000; Henmi et al., 2005), and nodulation in symbiosis (Frendo et al., 2005). The synthesis of glutathione in higher plants is tightly regulated (Xiang and Oliver, 1998, 2002; Kopriva, 2006; Kopriva et al., 2009). Glutathione and S-methylmethionine are major forms of organic sulphur storage and transport in higher plants (Bourgis et al., 1999; Leustek et al., 2000).
Sulphate uptake and assimilation is generally believed to be demand-driven (Lappartient and Touraine, 1996; Lappartient et al., 1999) and co-ordinated with nitrogen metabolism (Kim et al., 1999). Sulphate content in Arabidopsis thaliana was reported to be related to APR2 in which a single-amino acid substitution decreased its enzyme activity leading to sulphate accumulation in the plant (Loudet et al., 2007). O-acetyl-L-serine (OAS) plays a regulatory role in the synthesis of cysteine by controlling the oligomerization of the cysteine synthase complex, thus co-ordinating between serine as the nitrogen source and sulphide as the sulphate assimilation intermediate (Leustek et al., 2000; Wirtz and Hell, 2007; Kopriva et al., 2009). OAS is also a positive regulator of sulphate uptake and assimilation (Hirai et al., 2003).
Sulphur deficiency is recognized as an increasingly important problem in agriculture, especially in developed countries where SO2 pollution has been significantly reduced. Associated with sulphur deficiency is an increase in some plant diseases (Bearchell et al., 2005). Plant sulphur nutrition not only affects crop yield but also quality (Blake-Kalff et al., 1998; Tabe et al., 2002; Chiaiese et al., 2004; Falk et al., 2007; Taylor et al., 2008). Therefore, increasing the sulphur utilization efficiency (SUE) of plants is becoming an important issue. SUE was described as ‘improved capture of resources, the accumulation of greater reserves of sulphur, and improved mechanisms for the remobilization of these reserves’ (Hawkesford, 2000). However, little is known about the genetic basis and molecular mechanisms underlying SUE. A genetic approach can be powerful in defining the basis of important agronomic traits as well as in basic research.
A mutant screen for increased sulphur utilization efficiency has not been reported. The major obstacle is the difficulty of establishing a sulphur level that can provide an effective selection pressure. Since nutrients and phytohormones are well known to regulate root development and cross-talk between nutrients, hormones, and reactive oxygen species play important roles in re-shaping root architecture (Signora et al., 2001; Kutz et al., 2002; Schachtman and Shin, 2006), mutants with improved SUE may be expected to have mutations in a wide range of genetic loci involved in phytohormone homeostasis and root architecture, in addition to sulphur assimilation and metabolism.
Other mutant screen strategies have been exploited to isolate different sulphur nutrient-related mutants. Arabidopsis mutants of a sulphate transporter were isolated by selecting for selenate tolerance (Shibagaki et al., 2002; Kassis et al., 2007). Sulphur-responsive mutants were isolated using the GFP reporter driven by a sulphate-responsive promoter and this has led to new findings about the mechanisms of the sulphur deficiency response (Maruyama-Nakashita et al., 2004, 2005, 2006; Ohkama-Ohtsu et al., 2004).
Genetic screens for mutants involved in plant responses to other nutrients have been successfully conducted. These included the phosphate accumulating mutants pho1 (Poirier et al., 1991) which led to the identification of PHO1 (Hamburger et al., 2002), pho2 (Delhaize and Randall, 1995), and other phosphate mutants (Chen et al., 2000; Chang et al., 2005; Sanchez-Calderon et al., 2006). Nitrogen-related mutants (Tsay et al., 1993; Yu et al., 2004), low potassium-insensitive mutants (Zhao et al., 2001) which led to the elucidation of K+ uptake regulated by a protein kinase (Xu et al., 2006), and iron nutrient-related mutants (Ling et al., 1996, 1999, 2002; Vert et al., 2002; Yuan et al., 2005; Ogo et al., 2007) are among the other mutants characterized. Existing crop germplasms also contain genetic variations that are valuable resources for plant nutrition research (Fang and Wu, 2001; Liao and Yan, 2001; Zhao et al., 2001). These studies demonstrate that genetic approaches will continue to be powerful tools in plant nutrition research.
Here a high-throughput genetic screen is reported where a ‘sulphur-free’ solid medium without added sulphur or toxic metals was devised that gives sufficient low sulphur selection and allows thousands of seeds to be screened on a single plate. The isolation and the characterization of two low-sulphur-tolerant mutants, sue3 (sulphur utilization efficiency) and sue4 validate the genetic screen as a feasible procedure for isolating gain-of-function mutants with potentially improved SUE. Both sue3 and sue4 displayed a well-developed root system under low-sulphur conditions and enhanced tolerance to heavy metal (cadmium) and oxidative stress (paraquat). Through molecular genetic analysis, the recessive mutation in sue3 was identified as the VirE2-interacting Protein 1 (VIP1) and the dominant mutation in sue4 was identified as a small unknown protein with four membrane spanning domains activated by the enhancers on T-DNA. Our results demonstrate that the genetic screen developed here is a reasonable approach to isolate Arabidopsis mutants with improved tolerance to low sulphur conditions and potentially with increased sulphur utilization efficiency. The two loci identified in sue3 and sue4 should assist understanding the pertinent molecular mechanisms involved in low sulphur tolerance.
Materials and methods
Arabidopsis growth
Arabidopsis thaliana ecotype Columbia (Col-0) was used throughout the study. Plants were grown in soil at 22 °C and with a 14 h photoperiod unless specified otherwise.
Generation of an activation tagging library
A large-scale activation tagging was carried out in the Columbia background using Agrobacterium strain C58C1 harbouring the pSKI015 plasmid as described by Weigel et al. (2000). About 55 000 independent transgenic plants were generated and T2 seeds were collected in 55 pools. Each pool consisted of approximately 1000 independent lines. These pools constituted the activation-tagging library which was later used for mutant screens.
Heavy metal and oxidative stress tolerance assay
For the heavy metal tolerance assay, seeds of the sue mutants and wild type were sterilized, sown on 1/2× MS medium supplemented with 0, 1, 10, 100 μM of CdCl2, and incubated at 22 °C constant temperature and 24 h light conditions. After 12 d, germination rates were determined.
The oxidative stress tolerance assay was conducted as above for heavy metal tolerance except that the 1/2× MS medium was supplemented with 0, 1, 2, and 3 μM paraquat (Sigma, USA). Seeds were able to germinate and cotyledons opened on the media. As the incubation continued the seedlings were bleached. The survival rate (percentage of green seedlings) was counted after 12 d.
Kinetic analysis of sulphate uptake
Sulphate uptake was measured using Na235SO4 as described by Maruyama-Nakashita et al. (2004) with slight modifications as liquid-cultured Arabidopsis seedlings were used. Seeds were germinated and cultured in 1/2× MS liquid medium for 2 weeks. Before the uptake experiments the culture medium was decanted and the seedlings were washed twice with deionized water.
Dose-dependent sulphur uptake experiments were conducted in medium with the indicated concentration of sulphur, and every medium contained 10 μM Na235SO4 (2.06G Bq mmol−1, Amershan, UK). Time-dependent sulphur uptake experiments were conducted in liquid sulphur-free medium supplemented with 10 μM Na235SO4 (2.06 GBq mmol−1, Amershan, UK), After termination of sulphate uptake, seedlings were blotted dry with paper towels and the fresh weight measured before the plants were ground in deionized water and the radioactivity determined with a scintillation counter (Beckman LS1701).
Thiols and sulphur contents analysis
The mutants and the wild type were germinated on the MS or low-sulphur medium (75 μM sulphate) for 7 d. MS medium-germinated seedlings were transferred to soil and low-sulphur medium-germinated seedlings were transferred to sulphur-free hydroponic medium, and grown for 42 d before free sulphate quantification. Tissues were rinsed with deionized water and then dried at 60 °C. The dried tissues were weighed and ground in a mortar for free sulphate quantification.
The mutants and the wild type were germinated on the MS or low-sulphur medium (75 μM sulphate) for 4 d, then transferred to MS medium or sulphur-free medium and grown under the long-day photoperiod (16/8 h day/night) for 10 d. Tissues were rinsed with deionized water and then dried at 60 °C. The dried tissues were weighed and ground in a mortar for total sulphur quantification.
Total sulphur and free sulphate were quantified as described by Kolthoff (1969).
The mutants and the wild type grown in the liquid culture system with the long-day photoperiod (16/8 h) were used for GSH and Cys contents analysis as described by Xiang et al. (2001).
Genetic analysis
Backcrosses were made with the wild-type Arabidopsis thaliana Columbia ecotype and the F1 plants selfed. F1 and F2 seeds were assayed on sulphate-free medium for the mutant phenotype and the results analysed with χ2 test.
Identification of the T-DNA tagged locus
The single T-DNA insertion site in the mutant was cloned by TAIL-PCR (Liu et al., 1995) using three short arbitrary degenerate(AD) primers (AD1: 5′-NTCGA(G/C)T(A/T)T(G/C)G(A/T)GTT-3′, AD2: 5′-NGTCGA(G/C)(A/T)GANA(A/T)GAA-3′ and AD3: 5′-(A/T)GTGNAG(A/T)ANCANAGA-3′) and specific primer SK-LB3 (5′-TTGACCATCATACTCATTGCTG-3′) for the sue mutants, and positively identified by sequencing.
DNA gel blot analysis
Genomic DNA gel blot analysis for the mutant and the wild type was performed as described by Xiang et al. (1997) using the bar sequence as probe. RNA gel blot analysis was performed as described previously (Xiang and Oliver, 1998, 2002).
Genomic PCR screen for At1g43700 knockout mutant
T-DNA insertion lines Salk_0001014 was obtained from the ABRC and screened for homozygous progeny as described using specific primers (forward primer: 5′-GAGGAAGGTTCAGACACTTCAGA-3′; reverse primer: 5′-TACATCAAATATTGCAGCCCG-3′) and the T-DNA primer (LBb1) suggested by Alonso et al. (2003).
Recapitulation analysis
The At3G55880 cDNA was amplified by RT-PCR using specific primers (forward primer: 5′-GTACAAAAAAGCAGGCTGCATGGGTTTGATTAGCAAAGA-3′ and reverse primer: 5′-GTACAAGAAAGCTGGGTCTCAAAGTGAACTTACGGATT-3′), cloned into pDONR207, and subsequently shuttled into the expression binary vector pCB2004 (Lei, 2007). The construct was introduced into Agrobacterium tumefaciens C58C1, which was used to transform the Columbia wild type as described by Clough and Bent (1998). Low sulphate assays were performed for T2 transgenic plants harbouring an overexpression construct to reconfirm the At3g55880 gene function.
Over-expression of At3g55880 in tobacco for low sulphate tolerance assay
To generate transgenic tobacco, the above-mentioned pCB2004-At3g55880 construct was used to transform tobacco as previously described by Horsch et al. (1986). Primary transformants were tested positive by RT-PCR. Low sulphate tolerance assays were performed using T2 homozygous lines and the control (the wild type transformed with the empty vector pCB2004).
RT-PCR analysis
Total RNA was prepared from tissues indicated in the figures by the TRIZOL reagent (Invitrogen), and 1 μg of RNA from each sample was used for the reverse transcription reaction. Subsequently, 1 μl of the reverse transcription reaction was used as the template for PCR amplification. The PCR products were examined on a 0.8% agarose gel stained with ethidium bromide. The same RNA samples and primers were used for real-time PCR analysis that was performed and statistically analysed as described by Livak and Schmittgen (2001). SYBR green was used to monitor the kinetics of the PCR product in real-time RT-PCR. As an internal control, the tubulin transcript was used to quantify the relative transcript level of each target gene in each tissue type. RT-PCR was carried out using rTaq DNA polymerase and gene-specific primers. For SUE3, RT-PCR was performed to confirm the null expression of the disrupted locus At1g43700 with gene-specific primers: 5′-GAGGAAGGTTCAGACACTTCAGA-3′ and 5′-TACATCAAATATTGCAGCCCG-3′. For the RT-PCR analysis of At5g62200 and At5g62210, gene specific primers were used (At5g62200: 5′-ATGGCGTCCGTACGACTCTT-3′ and 5′-TTACAAGAGCAATGTGGTAC-3′; At5g62210: 5′-ATGGAGTGCTCTCTCTCATC-3′ and 5′-TCAAACAACCACAGCCGCAA-3′. For SUE4, RT-PCR was performed to confirm the over-expression of At3g55880 with gene-specific primers (At3g55870: 5′-ATGATCAAGAGGCTCCAAG-3′ and 5′-CTAGATTGTTGTATCACTT-3′, At3g55880: 5′-ATGGGTTTGATTAGCAAAGAA-3′ and 5′-TCAAAGTGAACTTACGGATTC-3′, At3g55890: 5′-GGTAGGGTTTTTATGGTTGATC-3′ and 5′-ACTTCTGGCTCTTTTCATGT-3′).
Results
Establishing a high-throughput genetic screen for mutants with improved tolerance to low sulphate
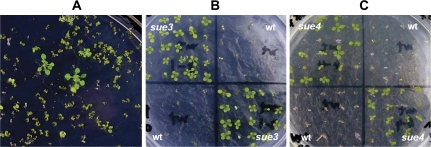
To meet the high-throughput criterion, the procedure must allow thousands of seeds to be screened on a single plate. It was found that, in order to suppress the wild-type seedling growth, it was necessary to set up a ‘sulphur-free’ growth condition. Therefore, a sulphur-free medium was developed that contained no added sulphur, which is described in the Supplementary data at JXB online. On this medium, the wild-type seeds were able to germinate, the cotyledons were able to open but arrested at the cotyledon stage. Although occasionally one pair of true leaves emerged, further growth was stopped (Fig. 1A). By contrast, the mutants continued to grow. The emergence of two to three pairs of true leaves and a rapidly elongating primary roots were also evident in the mutants (Fig. 1A). These characteristics were used as visual selection markers for our mutant screen. In addition, up to 3500 seeds could be screened in a single 150 mm plate, rendering the screen high-throughput. Therefore, a simple high-throughput genetic selection on sulphur-free agarose medium was capable of screening for Arabidopsis mutants with alterations in their growth in a sulphur-limited environment.
Fig. 1.
Arabidopsis mutants with improved low-sulphur tolerance isolated with a high-throughput genetic screen. (A) Primary screen. Seeds from the activation-tagging library were germinated on sulphur-free medium as described in the Materials and methods. Three putative mutants in the image taken at day 12 post-germination show continued growth as evidenced by 2–3 pairs of true leaves and long roots in contrast to the rest of the seedlings with arrested growth on the plate. (B) Secondary screen of sue3. To confirm the phenotype of the mutants from the primary screen, a secondary screen was conducted as described in the Materials and methods. The wild-type (wt) and the mutant seeds (sue3) were sown on sulphur-free medium. Continued growth was evident for the mutant. The image was recorded when seedlings were 10 d old. (C) Secondary screen of sue4. To confirm the phenotype of the mutants from the primary screen, a secondary screen was conducted as described in Materials and methods. The wildtype (wt) and the mutant seeds (sue4) were sown on sulphur-free medium. Continued growth was evident for the mutant. The image was recorded when seedlings were 10 d old. (This figure is available in colour at JXB online.)
Isolation of Arabidopsis mutants with improved tolerance to low sulphate
To facilitate the isolation of gain-of-function mutants with improved abiotic stress tolerance, an activation-tagging library of 55 000 independent lines was generated with the T-DNA mutagen pSKI015 as described by Weigel et al. (2000). This library was used for isolating mutants with improved tolerance to drought and salt (Gao and Xiang, 2008; Yu et al., 2008). A few low-nitrogen-tolerant Arabidopsis mutants were also isolated from this library (Yu et al., 2004).
Using the above-established conditions, the whole activation-tagging library was screened. Approximately 15 000 seeds were screened from each pool. The primary screen resulted in the isolation of 55 putative mutants that were rescued and grown to maturity and their seeds harvested. All the mutants were subjected to a secondary screen to confirm their mutant phenotype as demonstrated in Fig. 1B and C. After the secondary screen, only three gain-of-function mutants were confirmed and designated as sueN (sulphate utilization efficiency, N being a numeric). All these mutants were able to continue to grow after the cotyledon stage on the sulphur-free medium and showed similar phenotypes with the emergence of two to three pairs of true leaves. As a result, the biomass of the mutants was significantly higher than that of the wild type. The mutants sue3 and sue4 were characterized further.
The well-developed root system of sue3 and sue4
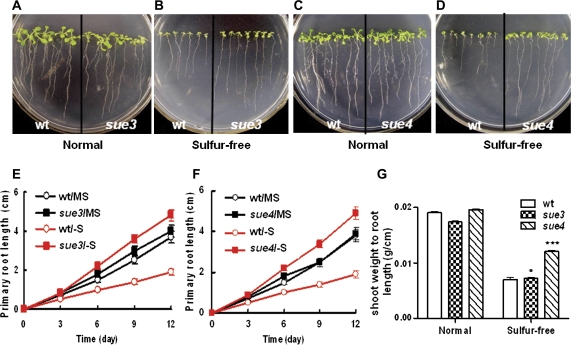
Besides the continued growth of shoots on sulphur-free medium, the mutants also showed faster root elongation. To display the root phenotype better, the mutants were germinated and grown vertically as shown in Fig. 2. On MS medium containing 1.5 mM sulphate, both mutants showed similar growth of roots and shoots to that of the wild type (Fig. 2A, C). However, on the sulphur-free medium, both mutants displayed a phenotype of longer primary root length (Fig. 2B, D). The primary roots of the mutants elongated much faster than those of the wild type. At day 12 after sowing, the primary root length of the mutants was about twice that of the wild type (Fig. 2E, F). These results suggest that the root elongation of the mutants were more responsive to the low-sulphur condition.
Fig. 2.
Primary root elongation on MS and sulphur-free medium :wild type versus mutants. Seeds of the wild type and the mutant were germinated on the MS or sulphur-free medium and the plates were placed vertically. The image was recorded when seedlings were 12 d old. (A) The wild type (wt) versus sue3 on MS medium. (B) The wild type (wt) versus sue3 on sulphur-free medium. (C) Growth curves of primary roots. The primary root length was measured every 3 d for the wild type and sue3 grown on MS as in (A) (wt/MS versus sue3/MS) and sulphur-free medium as in (B) (wt/–S versus sue3/–S). Values represent the mean of >30 plants and error bars represent SEM. (D) The wild type (wt) versus sue4 on MS medium. (E) The wild type (wt) versus sue4 on sulphur-free medium. (F) Growth curves of primary roots. The primary root length was measured every 3 d for the wild type and sue4 grown on MS as in (A) (wt/MS versus sue4/MS) and sulphur-free medium as in (B) (wt/–S versus sue4/–S). Values represent the mean of >30 plants and error bars represent SEM. (G) Shoot weight to root length. The primary root length was measured as in (C) and (F), and the fresh weight of shoot was measured after 12 d. Values represent the mean of >30 plants and error bars represent SEM. **P <0.01, ***P <0.001. (This figure is available in colour at JXB online.)
The sue3 and sue4 mutants had a more established primary root system than the wild type, which might contribute to the better growth of mutants on sulphur-free medium. Therefore, shoot weight to primary root length was measured. At day 12 after sowing, the ratio of shoot weight to primary root length of sue4 was about twice that of the wild type on the sulphur-free medium (Fig. 2G), while sue3 was about 50% higher than the wild type. On the normal medium no significant differences were observed between the mutants and the wild type (Fig. 2G).
The mutant phenotype is specific for sulphur deficiency.
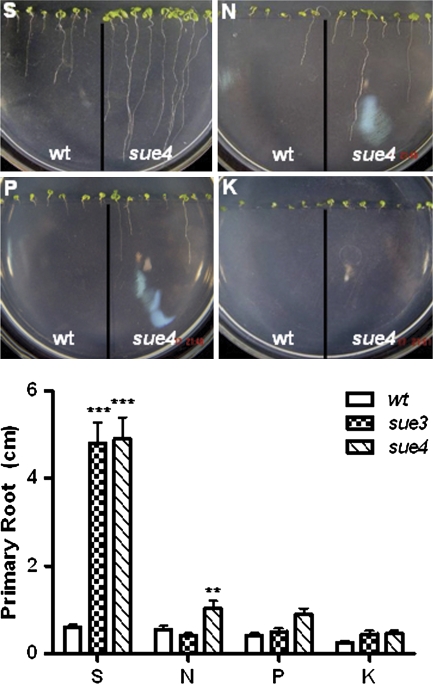
To confirm whether the mutant phenotype is specific for sulphur deficiency, root elongation was examined on nitrogen-, phosphate-, and potassium-deficient medium. Figure 3 shows that the increased elongation rate of the primary roots in the mutants was specific for sulphur-deficiency. On the N-, P-, and K-deficient medium, root elongation of both wild type and mutants was greatly inhibited. However, under N-deficiency sue4 roots grew significantly longer than sue3 and the wild type. However, under P- and K-deficiency medium, the difference of primary root elongation was not statistically significant between the wild type and mutants. Both mutants showed a significantly faster root growth rate on the S-deficient medium, indicating that the root elongation of the mutants is a specific response to sulphur deficiency.
Fig. 3.
Mutant phenotype is specific for sulphate deficiency. The specificity to nutrient deficiency was assayed for the mutants on the medium with specific nutrient deficiency (N, nitrogen; P, phosphate; K, potassium; S, sulphur) as described in the Materials and methods. The seeds of the mutant sue3 and sue4 and the wild type (wt) were sown on the same plate, germinated vertically, and grown for 12 d before primary root length was measured. Values represent the mean of >30 plants and error bars represent SEM. *0.05< P <0.01, **P <0.01, ***P <0.001. (This figure is available in colour at JXB online.)
No growth difference under normal sulphur conditions between the mutants and the wild type
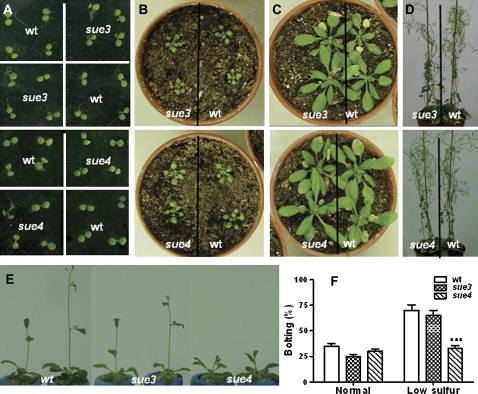
Under normal sulphur conditions (750 μM sulphate on 1/2× MS), no obvious difference was observed between the mutants and the wild type throughout their life cycle (Fig. 4A, D). However, in low-sulphur hydroponic culture with 75 μM sulphate, flowering was delayed in sue4 compared with that in the wild type. The wild type and sue3 showed a bolting frequency of 70% while sue4 only 33% at the same time after growth in low-sulphur hydroponic culture for 4 weeks (Fig. 4E, F). No significant difference was observed between sue3 and the wild type. Both mutants did not show any significant difference in flowering timing in normal-sulphur hydroponic culture (liquid 1/2× MS medium) (Fig. 4F).
Fig. 4.
Growth behaviour of sue3, sue4, and the wild type under normal conditions and hydroponic culture. (A) 8-d-old seedlings grown on MS medium, sue3 versus wt (upper) and sue4 versus wt (lower). (B) 2-week-old plants grown in soil, sue3 versus wt (upper) and sue4 versus wt (lower). (C) 4-week-old plants grown in soil, sue3 versus wt (upper) and sue4 versus wt (lower). (D) 7-week-old plants grown in soil, sue3 versus wt (upper) and sue4 versus wt (lower). (E) After 3 weeks grown in hydroponics system with sulphate concentration of 75 μM, the wild type and the sue3 began to bolt, while sue4 was delayed. (F) Bolting frequency under low sulphur stress. The wild type (wt) and sue4 seeds were sown on 1/2× MS medium (750 μM sulphate) and low sulphur medium (75 μM sulphate) for 7 d, and the contents in the low sulphur medium is the same as in the 1/2× MS medium except the sulphur concentration. Then seedlings were translated in hydroponics system containing 1/2× MS medium and low sulphur medium for 3 weeks. Values represent the mean of >20 seedlings. Error bars represent SEM. *0.05< P <0.01, **P <0.01, ***P <0.001. (This figure is available in colour at JXB online.)
The mutants show improved tolerance to heavy metal and oxidative stress
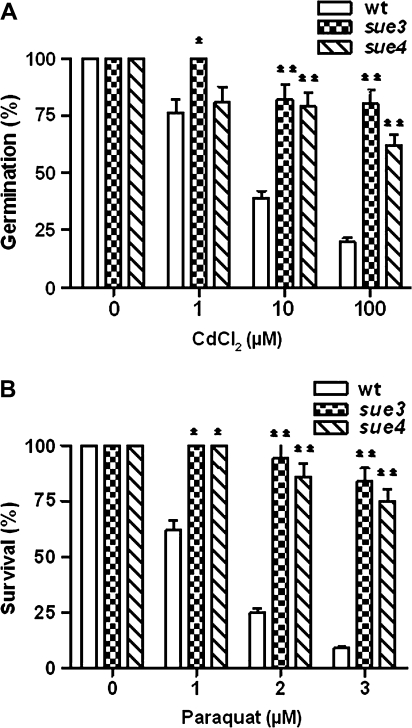
The sulphur nutrient level is directly linked to heavy metal and oxidative stress tolerance (Noctor and Foyer, 1998). To find out whether the mutant has improved tolerance to heavy metal and oxidative stress, they were assayed for growth on 1/2× MS medium supplemented with heavy metal CdCl2 or paraquat. The results in Fig. 5A show that the mutants had significantly higher germination frequency in the presence of various concentrations of CdCl2. Within the range of CdCl2 concentrations tested, the higher the CdCl2 concentration the greater the difference between the wild type and the mutant. At 100 μM CdCl2, the mutant showed a germination frequency of more than 60% while wild-type control only 20%. This suggests that the tolerance to heavy metal stress was significantly improved in the sue3 and sue4 mutants.
Fig. 5.
Improved tolerance of the mutants to heavy metal and oxidative stress. (A) Germination frequency under heavy metal stress. The wild type (wt) and mutant (sue3 and sue4) seeds were sown on 1/2× MS medium (750 μM sulphate) containing heavy metal CdCl2 of indicated concentrations. For each treatment, 25 seeds of each mutant and the wild type were sown on the same plate. Four replica plates were used. Germination frequency was scored 12 d after sowing. Values represent the mean of >100 seeds and error bars represent SEM. *P <0.01, **P <0.001. (B) Germination frequency under oxidative stress. The wild-type and mutant seeds were germinated on 1/2× MS medium supplemented with paraquat of indicated concentrations. For each treatment, 25 seeds of each mutant and the wild type were sown on the same plate and four replica plates were used. Germination frequencies were scored 12 d after sowing. Values represent the mean of 100 plants and error bars represent SEM. *P <0.01, **P <0.001.
The germination frequency under oxidative stress was assayed by germinating seeds on the 1/2× MS medium containing 0, 1, 2, and 3 μM paraquat. The results in Fig. 5B show that wild-type seedlings were more rapidly bleached and their germination frequency decreased as paraquat concentration increased in the medium. By contrast, the germination frequency of both mutants was still above 70% under the same conditions. These results indicate that the tolerance of the mutants to oxidative stress was significantly improved compared with the wild type.
Sulphate uptake, non-protein thiols, total sulphur, and free sulphate content in the mutants versus the wild type
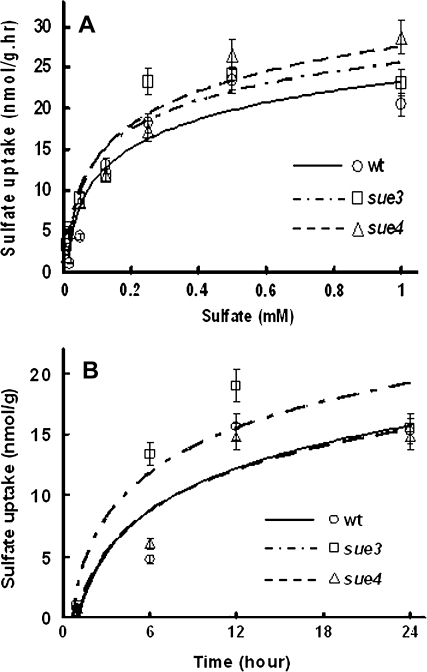
Considering that sulphate uptake may be one of the major determinants of sulphate utilization efficiency, sulphate uptake experiments were conducted as described in the Materials and methods using Na235SO4. The kinetics of sulphur uptake are shown in Fig. 6. The results of dose-dependent sulphate uptake experiments (with different sulphate concentrations) in Fig. 6A indicate that the mutants had the same Vmax values as the wild type, and the results of time-dependent sulphate uptake experiments in Fig. 6B indicate that the mutant sue3 had a significantly higher rate of sulphate uptake than the wild type at low sulphate levels (10 μM) while sue4 had a rate similar to the wild type.
Fig. 6.
Kinetics of sulphate uptake. (A) Dose-dependent sulphur uptake. Na235SO4 uptake experiments were conducted in medium with the indicated concentration of sulphate, and every medium contented 10 μM Na235SO4. Uptake was allowed for 6 h before samples were prepared for scintillation counting as described in the Materials and methods. Values represent the mean of three experiments and error bars represent SEM. (B) Time-dependent sulphur uptake. Sulphate uptake experiments were conducted in sulphur-free medium supplemented with 10 μM Na235SO4. Uptake was allowed for the time indicated before samples were prepared for scintillation counting as described in the Materials and methods. Values represent the mean of three experiments and error bars represent SEM.
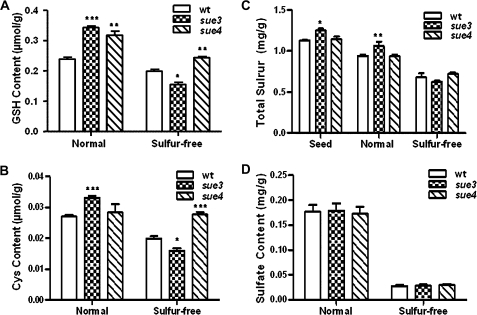
To determine whether sulphate metabolism is changed in the sue mutants, the non-protein thiols cysteine and GSH were quantified using HPLC as described by Xiang and Oliver (1998) as well as free sulphate and total sulphur content as described by Kolthoff (1969) for the mutants and the wild-type plants. Figure 7A and B show that under normal sulphur conditions, the GSH content was significantly higher in sue mutants than in the wild type. After being shifted to sulphur-free medium for 48 h, the GSH content was significantly higher in sue4 while that in sue3 was significantly lower than the wild type. The cysteine content showed a similar result to GSH except under normal conditions when no difference was observed between sue4 and the wild type.
Fig. 7.
Non-protein thiols, total sulphur, and free sulphate content in the mutants versus the wild type. (A, B) 7-d-old sue4 and the wild type were transplanted from MS hydroponic medium (Normal) to sulphate-free hydroponic medium (Sulphur-free) for 48 h before tissue extracts were prepared for quantification of GSH and cysteine. Three replicate experiments were conducted for each treatment. Error bars represent SEM. *0.05<P <0.01, **P <0.01, ***P <0.001. (C) Total sulphur content of seedlings and seeds. Seeds of the wild type and the mutants were germinated on the MS or low-sulphur medium (75 μM sulphate) for 4 d, then transplanted into MS medium (Normal) or sulphur-free medium (Sulphur-free) for 10 d before quantifying total sulphur content. Air-dry wild-type and mutant seeds were directly quantified for total sulphur content (Seed). For each treatment, three replica experiments were used and error bars represent SEM. *0.05<P <0.01, **P <0.01, ***P <0.001. (D) Free sulphate content. Seeds of the wild type and the mutants were germinated on the MS or low-sulphur medium (75 μM sulphate) for 7 d, respectively. MS medium-germinated seedlings were transferred to soil and grew for 42 d before free sulphate quantification (Normal). Low-sulphur medium-germinated seedlings were transferred to sulphur-free hydroponic medium and grew for 42 d before free sulphate quantification (Sulphur-free). For each treatment, three replica experiments were used and error bars represent SEM. *0.05< P < 0.01, **P <0.01, ***P <0.001.
Figure 7C shows that total sulphur after acid hydrolysis was not significantly different between sue4 and the wild type under both normal conditions and after sulphur-free stress. However, sue3 had a significantly higher total sulphur content than the wild type under normal conditions while after sulphur-free stress total sulphur content in the sue3 dropped to the level in the wild type. Total sulphur content in seeds of sue3 was significantly higher than that of the wild type and sue4 while no difference was observed between sue4 and the wild type.
Free sulphate content was not significantly different between the mutants and the wild type (Fig. 7D) under either normal sulphur conditions (soil-grown) or sulphur-free stress (hydroponic culture). As expected, the free sulphate level was greatly reduced for all under the sulphur-free stress compared with that under normal conditions. This is also true for non-protein thiols and total sulphur except for the cysteine level in sue4 under sulphur-free conditions which was similar to that under normal conditions.
Genetic analysis of sue3 and sue4
To reveal the nature of the mutations in both mutants, crosses were made for genetic analysis. The wild type as female parent was crossed with the homozygous mutant as the pollen donor. F1 plants were selfed to obtain F2 seeds. Both F1 and F2 seeds were assayed for growth on the sulphur-free medium.
For mutant sue3, all 24 F1 seeds were sensitive to low sulphur levels. Among the F2 seeds that resulted from F1 selfing, 59 seeds were low-sulphur-tolerant and 161 were low-sulphur-sensitive. A chi square analysis showed that this did not deviate significantly from a 1:3 (tolerant to sensitive) segregation ratio (χ2=0.388, P >0.1). These results indicate that the sue3 mutant was caused by a single recessive nuclear locus. The recessive nature of the mutation implies that the tagged gene was probably disrupted by the T-DNA insertion.
For the mutant sue4, 360 F1 seeds were low-sulphur-tolerant and 19 were low-sulphur-sensitive. The segregation was close to the expected 379 to 0 ratio (χ2=0.953, P >0.1). The 19 sensitive seeds might result from non-viable seeds. Among the F2 seeds that resulted from F1 selfing, 140 seeds were low-sulphur-tolerant and 49 were low-sulphur-sensitive. A chi square analysis showed that this did not deviate significantly from a 3:1 (tolerant to sensitive) segregation ratio (χ2=0.492, P >0.1). These results indicate that the sue4 mutant was caused by a single dominant locus. Moreover, the mutant phenotype co-segregated with herbicide resistance (data not shown). The dominance of the mutation suggests that the tagged gene is activated by the enhancers on the T-DNA.
Identification of T-DNA tagged loci in the mutants
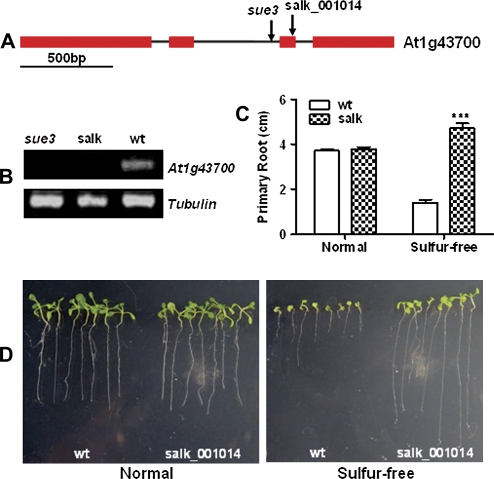
By using TAIL-PCR (Liu et al., 1995), the T-DNA insertion junction of sue3 was amplified and sequenced, three copies of T-DNA were identified, one was inserted around 382 bp upstream of the ATG codon of At5g62200, the other two copies were inserted at the same location in inverted repeats in the second intron of the At1g43700 (see Supplementary Fig. 1A at JXB online). RT-PCR analysis indicated that there were no significant changes of transcript level of At5g62200 and At5g62210 between the wild type and sue3 (see Supplementary Fig. 1B at JXB online), indicating that the T-DNA insertion did not affect the expression of these two genes. However, At1g43700 was completely disrupted (Fig. 8), consistent with the genetic analysis results. The full length of At1g43700 is 2011 bp, with four exons and three introns (Fig. 8A). At1g43700 is the previously identified VirE2-interacting Protein1 (VIP1), which mediates nuclear translocation of VirE2 (Tzfira et al., 2001; Li et al., 2005).
Fig. 8.
Multiple mutant allele analysis of low sulphur tolerance for the At1g43700 locus. (A) Illustration of At1g43700 gene structure and location of T-DNA insertions. The arrows indicate the positions of the T-DNA insertions within the sue3 and Salk_001014 (Col-0 background). At1g43700 are represented by exons (red rectangles) and introns (black lines). (B) Confirmation of the knockout of At1g43700 in Salk_001014 (salk) and sue3 by RT-PCR. Tubulin was used as loading control. (C) Primary root length of Salk_001014 versus wt. For each treatment, 10 seeds of each line were vertically germinated and grown on the same normal sulphate (Normal) or sulphur-free (Sulphur-free) medium for 12 d before primary root length was measured. Three replicate plates were used and error bars represent SEM. *0.05< P <0.01, **P <0.01, ***P <0.001. (D) A typical result of root elongation experiment in (C), showing wild-type seedlings (wt) and Salk_001014 vertically grown on normal and sulphur-free medium for 12 d. (This figure is available in colour at JXB online.)
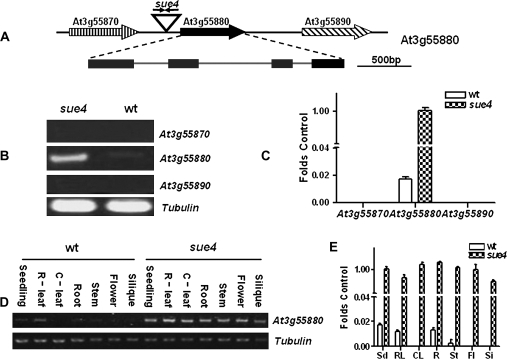
In sue4, two copies of T-DNA were also detected by Southern blot and identified via TAIL-PCR and sequencing (data not shown). It was found that the two copies were inserted in inverted repeats at the same position that is 1758 bp from the stop codon of At3g55870 and 930 bp from the ATG codon of At3g55880. The At3g55880 locus comprises four exons and three introns (Fig. 9A) and is predicted to encode an unknown small membrane protein with four membrane-spanning domains. Based on the dominance nature of the mutation from our genetic analysis, the results of real-time RT-PCR analysis in Fig. 9B and C indicated that At3g55880 was activated while its neighbouring genes At3g55870 and At3g55890 were not. In the wild type, At3g55880 is expressed at low levels in seedling and rosette leaves, roots, and inflorescence stems of mature plants. In sharp contrast, At3g55880 is constitutively expressed at high levels (Fig. 9E, F).
Fig. 9.
At3g55880 locus is activated in sue4 mutant. (A) Illustration of At3g55880 locus in sue4. Two copies of T-DNA were inserted in an inverted orientation at position 20597, 1758 bp downstream from the stop codon of At3g55870 and 930 bp upstream from the ATG codon of At3g55880. (B) RT-PCR analysis of transcript levels of the neighbouring genes. Transcript levels for At3g55870, At3g55880, and At3g55890 were compared in the wild type and the sue4 mutant. The experiment was repeated three times, and a typical result is presented. (C) Real-time RT-PCR analysis of transcript levels of the neighbouring genes. Using the same samples and primers as in (B), real-time RT-PCR was performed for 30 cycles. The relative transcript level was obtained as folds of the tubulin transcript level, which was used as the internal control. Values represent the mean of three experiments and error bars represent SEM. (D) RT-PCR analysis of the expression patterns of At3g55880 in the mutant and the wild type. RNA was isolated from seedlings (Sd), roots (R), rosette leaves (R-leaf), cauline leaves (C-leaf), inflorescence stem (St), flowers (Fl), and siliques (Si) of the wild type and sue4 plants, respectively. Tubulin was used as a loading control. The experiment was repeated three times, and a typical result is presented. (E) Real-time RT-PCR analysis of the expression patterns of At3g55880 in the mutant and the wild type. Using the same samples and primers as in (D), real-time RT-PCR was performed for 30 cycles. The relative transcript level was obtained as folds of the tubulin transcript level, which was used as the internal control. Values represent the mean of three experiments and error bars represent SEM.
As the phenotype of sue mutants seems to be specific to sulphur deprivation, it is important to determine whether SUE3 and SUE4 are regulated by sulphur. The results of RT-PCR analysis indicate that neither At1g43700 nor At3g55880 was responsive to low-sulphur stress (see Supplementary Fig. 1D at JXB online).
Multiple mutant allele analysis for low-sulphate tolerance of sue3
To confirm the gene-to-trait relationship in sue3, one Salk T-DNA insertion line Salk_001014 was obtained. The position of the T-DNA insertion is shown in Fig. 8A. In order to analyse the low sulphate tolerance of Salk_001014, the homozygous lines were isolated first with genomic PCR as described by Alonso et al. (2003). The homozygous plants were identified (see Supplementary Fig. 1C at JXB online). The disruption of At1g43700 in sue3 and Salk_001014 was confirmed by RT-PCR analysis. The results shown in Fig. 8B indicate that both sue3 and Salk_001014 had undetectable transcript levels of At1g43700.
The homozygous mutant of Salk_001014 was subsequently assayed for low-sulphur-tolerance phenotypes. Figure 8C and D show the markedly elongated primary roots of the knockout mutant under sulphur-free conditions compared with that of the wild type, while no difference in root elongation was observed under normal sulphur conditions between the mutant and the wild type. These results confirm that loss-of-At1g43700 caused the low-sulphur-tolerance phenotype in sue3.
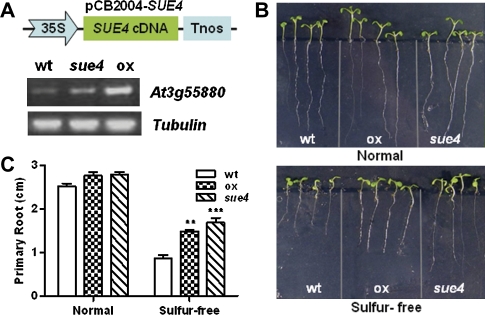
Recapitulation of sue4 phenotype in the wild type by overexpressing the tagged gene At3G55880
To confirm that the low-sulphur-tolerance phenotype was conferred by the activated expression of At3g55880, the cDNA of At3g55880 was cloned into the binary expression vector pCB2004 for generating transgenic plants. The cDNA was under the control of the 35S promoter. Homozygous lines (T2 generation) were confirmed for the transgene over-expression (Fig. 10A). Results of the low-sulphur tolerance assay experiments are shown in Fig. 10B and C. Under normal sulphur conditions (1/2 MS), there was no significant difference in root development between the pCB2004-SUE4 transgenic line, sue4, and the wild type. However, under sulphur-free conditions, the primary root of the transgenic line and sue4 was significantly longer than that of the wild type. This result confirms that over-expression of the At3g55880 confers the improved low-sulphur tolerance in sue4.
Fig. 10.
Recapitulation of sue4 phenotype in the wild type by overexpressing At3g55880 cDNA. (A) Illustration of the overexpression construct pCB2004-SUE4 and verification of the overexpression transgenic line (ox) via RT-PCR analysis. Tubulin was used as loading control. (B) Root elongation assay. The wild type (wt), sue4, and the ox seeds were vertically germinated and grown for 12 d on 1/2× MS medium and sulphur-free medium (–S) before primary root length was measured. (C) Primary root length after 12 d growth as in (B). Values represent the mean of >30 plants and error bars represent SEM. *0.05 < P <0.01, **P <0.01, ***P <0.001. (This figure is available in colour at JXB online.)
Constitutively overexpressing At3g55880 confers low-sulphur tolerance in transgenic tobacco
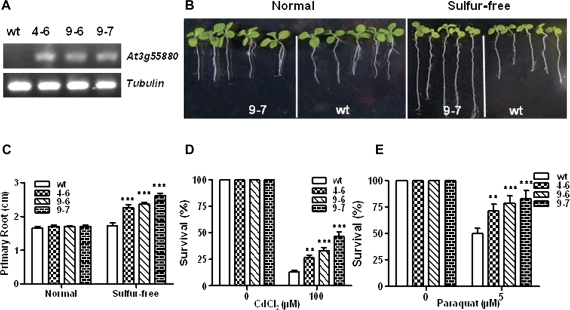
The same pCB2004-SUE4 construct was transferred into tobacco. Three transgenic lines 4-6, 9-6, and 9-7 were confirmed by RT-PCR analysis (Fig. 11A) and assayed for low-sulphur tolerance. The transgenic lines displayed a more elongated primary root under sulphur-free conditions compared with the wild type (Fig. 11B, C). The representative line 9-7 is shown in Fig. 11B. The tolerance to oxidative and heavy metal stress was also improved in the transgenic lines compared with the wild type (Fig. 11D, E). These results with transgenic tobacco not only further confirm that activated expression of SUE4 causes the low-sulphur-tolerance phenotype, but also demonstrate the potential of this gene to improve low-sulphur-tolerance of crops.
Fig. 11.
Overexpression of SUE4 in tobacco recapitulates sue4-like phenotype. (A) Verification of overexpression transgenic lines in tobacco via RT-PCR analysis. Tubulin was used as loading control. (B) Root elongation assay. The pCB2004-SUE4 was used to generate transgenic tobacco plants. The overexpression line 4-6, 9-6, 9-7, and the wild type were assayed for primary root elongation on 1/2× MS medium (normal) and sulphur-free medium. Primary root length was measured at day 20 after sowing. (C) Primary root length after 20 d growth as in (A). Values represent the mean of >50 plants and error bars represent SEM. *0.05 < P <0.01, **P <0.01, ***P <0.001. (D) Survival ratio under oxidative stress. The wild type, 4–6, and 9–7 were co-planted on MS medium and MS containing 5 μM paraquat. Values represent the mean of >50 plants and error bars represent SEM. *0.05 <P <0.01, **P <0.01, ***P <0.001. (E) Survival ratio under Cd stress. The wild type, 4–6, and 9–7 were co-planted on MS medium and MS containing 100 μM CdCl2 in hydroponic system. Values represent the mean =20 plants and error bars represent SEM. *0.05 <P < 0.01, **P <0.01, ***P <0.001. (This figure is available in colour at JXB online.)
Discussion
An ideal genetic screen for sulphur nutrient utilization mutants must meet two criteria. First, a reproducible and experimentally feasible sulphur deficiency condition must be established and, second, a high-throughput process needs to be designed. A modified hydroponic culture has proved ideal for limiting nutrient levels and was used for isolating low-nitrogen tolerant mutants of Arabidopsis (Yu et al., 2004). That set-up, however, does not meet the high-throughput requirement. This genetic screen developed for isolating mutants with improved tolerance to low sulphate growth conditions meets both criteria. Compared with the other screens for nutrient defect mutants, the key to this screen is to restrict sulphur in the medium to a sulphur-free level. Under nitrate stress, plants may respond by changing root architecture, root-to-shoot ratio, and seed germination rate. Therefore, various nitrogen stress-responsive mutants were isolated (Alboresi et al., 2005; Forde and Walch-Liu, 2009; Wang et al., 2009). However, the demand for sulphur by plants is relatively low compared with other macronutrients, making it hard for sulphur stress-responsive mutants.
With this genetic screen, three mutants with improved growth under low sulphur stress have been successfully isolated, thus demonstrating the feasibility of the screen. The establishment of this genetic screen should facilitate the isolation of more low sulphur-tolerant mutants in the future, an important step towards understanding the genetic basis of sulphur utilization efficiency.
The mutants displayed continued growth after germination and a well-developed root system under the sulphur-free conditions. The development of root systems usually reflects the ability of roots to adjust their growth to environmental factors. One of the factors is the nutrient availability. Nitrogen and sulphur often limit plant growth (Kutz et al., 2002; Potters et al., 2007) and cause changes in auxin homeostasis, which may lead to the increased root growth in order to capture the nutrients from the environment (Kutz et al., 2002; Potters et al., 2007). On the other hand, sulphur utilization is also an important factor for plant growth (Hawkesford, 2000).Thus the mutants must have enhanced sulphur utilization or enhanced sulphur-absorbing capability which, in part, might be contributed by the well-developed root system. In addition to the improved root system, the capacity in sue3 of sulphate uptake as revealed by the kinetic analysis should be beneficial to the mutant.
Under normal sulphur conditions, no significant difference was observed in root development and growth between the wild type and the mutants. This might be expected considering the tight regulation of sulphate uptake and assimilation that is demand-driven (Lappartient et al., 1999). Low sulphur conditions are known to stimulate root elongation (Shibagaki et al., 2002) while the sulphur-free condition suppresses wild-type root elongation and seedling growth as demonstrated in this study. Therefore, it is reasonable to speculate that the mutations might have altered low-sulphur signal sensing and stimulated root development under sulphur-free conditions.
Sulphur is an essential macroelement in plant. Sulphur deficiency often results in decreased crop yields and quality, but not the death of the plant. Under sulphur deficiency, several pathways in plant metabolism may cross-influence as networks (Hirai et al., 2003; Maruyama-Nakashita et al., 2003; Hirai and Saito, 2004; Nikiforova et al., 2006). More than 2000 genes in Arabidopsis were significantly altered in expression by sulphur deficiency (Nikiforova et al., 2003) as well as the alterations in the metabolism of sulphur-containing metabolites such as SAM, cysteine, and glutathione (Nikiforova et al., 2005).
To defend against abiotic stresses, glutathione plays many important roles in plants (Noctor and Foyer, 1998; Vernoux et al., 2000; Xiang et al., 2001). Usually, the abiotic stresses induce an initial decrease in the glutathione pool size in plants. Plants quickly respond by replenishing glutathione by the increased synthesis of glutathione (Xiang and Oliver, 1998) For example, under heavy metal stress, the GSH level is consumed for the biosynthesis of PCs, heavy metal-binding peptides which are involved in heavy metal tolerance and sequestration (Cobbett and Goldsbrough, 2002). The level of GSH in sue4 was unchanged under sulphur deficiency, while the levels of GSH in sue3 and the wild type were reduced. Normally, as a major form of organic sulphur storage, the level of GSH would be reduced under conditions of sulphur starvation. However, sue4 maintained a steady level of GSH level compared with the wild type. Similar results have been reported for a selenium-binding protein SBP1 whose expression responds to cellular sulphur and GSH demand. Overexpression of SBP1 clearly enhanced the tolerance to the heavy metal in the wild type with the affected GSH level. However, overexpression of SBP1 in cad2-1 (Howden et al., 1995), a cadmium-sensitive, glutathione-deficient mutant, did not affect GSH content while the mutant also has the phenotype of heavy metal tolerance (Dutilleul et al., 2008; Hugouvieux et al., 2009). In the mutants of the OAS-TL gene family, cysteine and glutathione levels were reduced only in roots of BSAS2:2 while both were reduced in roots and leaves of BSAS1:1 plants (Watanabe et al., 2008; Kopriva et al., 2009).
The level of GSH in both sue3 and sue4 was significantly higher than that in the wild type on the normal medium. Correlated with the results of the root to shoot ratio, sulphate uptake, and the total sulphur content, these results suggested that the mutant sue3 may have a higher capacity and a higher rate of sulphate uptake while sue4 may have higher sulphate utilization efficiency compared with the wild type.
Further study on the identification of the tagged gene in sue3 and sue4 mutants showed that sue3 was caused by the disruption of At1g43700 by the T-DNA insertion. Multiple mutant allele analysis for low sulphur tolerance of sue3 confirmed that loss-of-At1g43700 resulted in the phenotype of sue3. SUE3 is the previously identified VIP1 involved in the nuclear import of VirE2 in the T-DNA transfer process (Tzfira et al., 2001; Li et al., 2005). How VIP1 senses low-sulphur signals and regulates root system architecture under low-sulphur condition is not clear at present.
The dominant mutation in SUE4 was a consequence of activated expression of At3g55880 by the enhancers on the T-DNA. The activated expression of the At3g55880 in sue4 and recapitulation in wild-type Arabidopsis as well as in tobacco confirm that it was the altered expression pattern of SUE4, i.e. constitutive overexpression that caused the low-sulphur-tolerance phenotype. SUE4 is a small protein with four trans-membrane domains. Its function is currently unknown. It is not immediately obvious how this small protein works to increase low-sulphur tolerance and improve root system architecture under low-sulphur conditions. Considering the root phenotype of sue4 mutant, the dominant mutation might have perturbed auxin homeostasis which plays a dominant role in shaping root architecture. Likewise, loss-of-SUE3 might also somehow perturb auxin homeostasis in the sue3 mutant. The exact underlying mechanisms await further investigation.
Taken together, a high-throughput genetic screen has been reported for isolating mutants with improved low-sulphur tolerance and potentially with enhanced SUE. Its application was demonstrated by screening an Arabidopsis activation-tagging library, and confirmed by multiple mutant allele analysis and functional recapitulation experiments. Further functional analysis of the tagged genes (At1g43700 and At3g55880) should shed light on the genetic basis and molecular mechanisms of SUE. Moreover, these mutants may be useful materials for transcriptomic and metabolomic analysis to reveal transcriptional and metabolic networks as reported (Maruyama-Nakashita et al., 2003; Nikiforova et al., 2004; Tohge et al., 2005; Hirai et al., 2007; Yonekura-Sakakibara et al., 2008).
Supplementary data
Supplementary data can be found at JXB online.
Supplementary data. A detailed description of the genetic screen for low-sulphur-tolerance mutants.
Supplementary Table S1. Composition of stock solutions for sulphur-containing and sulfur-free medium.
Supplementary Table S2. Composition of stock solutions for MS medium.
Supplementary Fig. S1. T-DNA-tagged loci in sue3 and isolation of Salk_001014 homozygous lines.
Supplementary Material
Acknowledgments
This work was supported by grants from the NNSFC (90917004, 30770189, 30471038). The authors thank ABRC for providing the Arabidopsis mutants.
References
- Alboresi A, Gestin C, Leydecker MT, Bedu M, Meyer C, Truong HN. Nitrate, a signal relieving seed dormancy in Arabidopsis. Plant, Cell and Environment. 2005;28:500–512. doi: 10.1111/j.1365-3040.2005.01292.x. [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- Ball L, Accotto GP, Bechtold U, et al. Evidence for a direct link between glutathione biosynthesis and stress defence gene expression in Arabidopsis. The Plant Cell. 2004;16:2448–2462. doi: 10.1105/tpc.104.022608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bearchell SJ, Fraaije BA, Shaw MW, Fitt BD. Wheat archive links long-term fungal pathogen population dynamics to air pollution. Proceedings of the National Academy of Sciences, USA. 2005;102:5438–5442. doi: 10.1073/pnas.0501596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake-Kalff MM, Harrison KR, Hawkesford MJ, Zhao FJ, McGrath SP. Distribution of sulphur within oilseed rape leaves in response to sulphur deficiency during vegetative growth. Plant Physiology. 1998;118:1337–1344. doi: 10.1104/pp.118.4.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgis F, Roje S, Nuccio ML, et al. S-methylmethionine plays a major role in phloem sulphur transport and is synthesized by a novel type of methyltransferase. The Plant Cell. 1999;11:1485–1498. doi: 10.1105/tpc.11.8.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burstenbinder K, Rzewuski G, Wirtz M, Hell R, Sauter M. The role of methionine recycling for ethylene synthesis in Arabidopsis. The Plant Journal. 2007;49:238–249. doi: 10.1111/j.1365-313X.2006.02942.x. [DOI] [PubMed] [Google Scholar]
- Chang S, Shu H, Qin G, Wu Y. A new Arabidopsis phosphate-sensing mutants screening method. Chinese Agricultural Science Bulletin. 2005;21:202–204. [Google Scholar]
- Chen DL, Delatorre CA, Bakker A, Abel S. Conditional identification of phosphate-starvation-response mutants in Arabidopsis thaliana. Planta. 2000;211:13–22. doi: 10.1007/s004250000271. [DOI] [PubMed] [Google Scholar]
- Chiaiese P, Ohkama-Ohtsu N, Molvig L, Godfree R, Dove H, Hocart C, Fujiwara T, Higgins TJ, Tabe LM. Sulphur and nitrogen nutrition influence the response of chickpea seeds to an added, transgenic sink for organic sulphur. Journal of Experimental Botany. 2004;55:1889–1901. doi: 10.1093/jxb/erh198. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Cobbett C, Goldsbrough P. Phytochelatins and metallothioneins: roles in heavy metal detoxification and homeostasis. Annual Review of Plant Biology. 2002;53:159–182. doi: 10.1146/annurev.arplant.53.100301.135154. [DOI] [PubMed] [Google Scholar]
- Delhaize E, Randall PJ. Characterization of a phosphate-accumulator mutant of Arabidopsis thaliana. Plant Physiology. 1995;107:207–213. doi: 10.1104/pp.107.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutilleul C, Jourdain A, Bourguignon J, Hugouvieux V. The Arabidopsis putative selenium-binding protein family: expression study and characterization of SBP1 as a potential new player in cadmium detoxification processes. Plant Physiology. 2008;147:239–251. doi: 10.1104/pp.107.114033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk KL, Tokuhisa JG, Gershenzon J. The effect of sulphur nutrition on plant glucosinolate content: physiology and molecular mechanisms. Plant Biology. 2007;9:573–581. doi: 10.1055/s-2007-965431. [DOI] [PubMed] [Google Scholar]
- Fang P, Wu P. QTLs of nitrogen uptake and utilization efficiency in rice. Plant Nutrition and Fertilizer Science. 2001;7:159–165. [Google Scholar]
- Forde BG, Walch-Liu P. Nitrate and glutamate as environmental cues for behavioural responses in plant roots. Plant, Cell and Environment. 2009;32:682–693. doi: 10.1111/j.1365-3040.2008.01927.x. [DOI] [PubMed] [Google Scholar]
- Frendo P, Harrison J, Norman C, Hernandez Jimenez MJ, Van de Sype G, Gilabert A, Puppo A. Glutathione and homoglutathione play a critical role in the nodulation process of Medicago truncatula. Molecular Plant–Microbe Interactions. 2005;18:254–259. doi: 10.1094/MPMI-18-0254. [DOI] [PubMed] [Google Scholar]
- Gao L, Xiang CB. The genetic locus At1g73660 encodes a putative MAPKKK and negatively regulates salt tolerance in Arabidopsis. Plant Molecular Biology. 2008;67:125–134. doi: 10.1007/s11103-008-9306-8. [DOI] [PubMed] [Google Scholar]
- Hamburger D, Rezzonico E, MacDonald-Comber Petetot J, Somerville C, Poirier Y. Identification and characterization of the Arabidopsis PHO1 gene involved in phosphate loading to the xylem. The Plant Cell. 2002;14:889–902. doi: 10.1105/tpc.000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkesford MJ. Plant responses to sulphur deficiency and the genetic manipulation of sulphate transporters to improve S-utilization efficiency. Journal of Experimental Botany. 2000;51:131–138. [PubMed] [Google Scholar]
- Heeg C, Kruse C, Jost R, Gutensohn M, Ruppert T, Wirtz M, Hell R. Analysis of the Arabidopsis O-acetylserine(thiol)lyase gene family demonstrates compartment-specific differences in the regulation of cysteine synthesis. The Plant Cell. 2008;20:168–185. doi: 10.1105/tpc.107.056747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henmi K, Demura T, Tsuboi S, Fukuda H, Iwabuchi M, Ogawa K. Change in the redox state of glutathione regulates differentiation of tracheary elements in Zinnia cells and Arabidopsis roots. Plant Cell Physiology. 2005;46:1757–1765. doi: 10.1093/pcp/pci198. [DOI] [PubMed] [Google Scholar]
- Hirai MY, Saito K. Post-genomics approaches for the elucidation of plant adaptive mechanisms to sulphur deficiency. Journal of Experimental Botany. 2004;55:1871–1879. doi: 10.1093/jxb/erh184. [DOI] [PubMed] [Google Scholar]
- Hirai MY, Fujiwara T, Awazuhara M, Kimura T, Noji M, Saito K. Global expression profiling of sulphur-starved Arabidopsis by DNA macroarray reveals the role of O-acetyl-l-serine as a general regulator of gene expression in response to sulphur nutrition. The Plant Journal. 2003;33:651–663. doi: 10.1046/j.1365-313x.2003.01658.x. [DOI] [PubMed] [Google Scholar]
- Hirai MY, Sugiyama K, Sawada Y, et al. Omics-based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. Proceedings of the National Academy of Sciences, USA. 2007;104:6478–6483. doi: 10.1073/pnas.0611629104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsch RB, Klee HJ, Stachel S, Winans SC, Nester EW, Rogers SG, Fraley RT. Analysis of Agrobacterium tumefaciens virulence mutants in leaf discs. Proceedings of the National Academy of Sciences, USA. 1986;83:2571–2575. doi: 10.1073/pnas.83.8.2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howden R, Andersen CR, Goldsbrough PB, Cobbett CS. A cadmium-sensitive, glutathione-deficient mutant of Arabidopsis thaliana. Plant Physiology. 1995;107:1067–1073. doi: 10.1104/pp.107.4.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugouvieux V, Dutilleul C, Jourdain A, Reynaud F, Lopez V, Bourguignon J. Arabidopsis putative selenium-binding protein1 expression is tightly linked to cellular sulphur demand and can reduce sensitivity to stresses requiring glutathione for tolerance. Plant Physiology. 2009;151:768–781. doi: 10.1104/pp.109.144808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassis EE, Cathala N, Rouached H, Fourcroy P, Berthomieu P, Terry N, Davidian JC. Characterization of a selenate-resistant Arabidopsis mutant. Root growth as a potential target for selenate toxicity. Plant Physiology. 2007;143:1231–1241. doi: 10.1104/pp.106.091462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka T, Hayashi N, Yamaya T, Takahashi H. Root-to-shoot transport of sulphate in Arabidopsis. Evidence for the role of SULTR3;5 as a component of low-affinity sulphate transport system in the root vasculature. Plant Physiology. 2004a;136:4198–4204. doi: 10.1104/pp.104.045625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka T, Watanabe-Takahashi A, Hayashi N, Ohnishi M, Mimura T, Buchner P, Hawkesford MJ, Yamaya T, Takahashi H. Vacuolar sulphate transporters are essential determinants controlling internal distribution of sulphate in Arabidopsis. The Plant Cell. 2004b;16:2693–2704. doi: 10.1105/tpc.104.023960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Hirai MY, Hayashi H, Chino M, Naito S, Fujiwara T. Role of O-acetyl-l-serine in the coordinated regulation of the expression of a soybean seed storage-protein gene by sulphur and nitrogen nutrition. Planta. 1999;209:282–289. doi: 10.1007/s004250050634. [DOI] [PubMed] [Google Scholar]
- Kolthoff IM, Sandell EB, Meehan EJ, Bruckenstein S. Quantitative chemical analysis. London: Macmillan; 1969. [Google Scholar]
- Kopriva S. Regulation of sulphate assimilation in Arabidopsis and beyond. Annals of Botany. 2006;97:479–495. doi: 10.1093/aob/mcl006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopriva S, Mugford SG, Matthewman C, Koprivova A. Plant sulphate assimilation genes: redundancy versus specialization. Plant Cell Reports. 2009;28:1769–1780. doi: 10.1007/s00299-009-0793-0. [DOI] [PubMed] [Google Scholar]
- Kutz A, Muller A, Hennig P, Kaiser WM, Piotrowski M, Weiler EW. A role for nitrilase 3 in the regulation of root morphology in sulphur-starving Arabidopsis thaliana. The Plant Journal. 2002;30:95–106. doi: 10.1046/j.1365-313x.2002.01271.x. [DOI] [PubMed] [Google Scholar]
- Lappartient AG, Touraine B. Demand-driven control of root atp sulphurylase activity and uptake in intact canola (the role of phloem-translocated glutathione) Plant Physiology. 1996;111:147–157. doi: 10.1104/pp.111.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappartient AG, Vidmar JJ, Leustek T, Glass AD, Touraine B. Inter-organ signaling in plants: regulation of ATP sulphurylase and sulphate transporter genes expression in roots mediated by phloem-translocated compound. The Plant Journal. 1999;18:89–95. doi: 10.1046/j.1365-313x.1999.00416.x. [DOI] [PubMed] [Google Scholar]
- Leustek T, Martin MN, Bick JA, Davies JP. Pathways and regulation of sulphur metabolism revealed through molecular and genetic studies. Annual Review of Plant Physiology and Plant Molecular Biology. 2000;51:141–165. doi: 10.1146/annurev.arplant.51.1.141. [DOI] [PubMed] [Google Scholar]
- Li Y, Dhankher OP, Carreira L, Balish RS, Meagher RB. Arsenic and mercury tolerance and cadmium sensitivity in Arabidopsis plants expressing bacterial γ-glutamylcysteine synthetase. Environmental Toxicology and Chemistry. 2005;24:1376–1386. doi: 10.1897/04-340r.1. [DOI] [PubMed] [Google Scholar]
- Liao H, Yan XL. Genotypic variation in root morphological characteristics of common bean in relation to phosphorus efficiency. Journal of Integrative Plant Biology. 2001;43:1161–1166. [Google Scholar]
- Ling HQ, Pich A, Scholz G, Ganal MW. Genetic analysis of two tomato mutants affected in the regulation of iron metabolism. Molecular and General Genetics. 1996;252:87–92. doi: 10.1007/BF02173208. [DOI] [PubMed] [Google Scholar]
- Ling HQ, Koch G, Baumlein H, Ganal MW. Map-based cloning of chloronerva, a gene involved in iron uptake of higher plants encoding nicotianamine synthase. Proceedings of the National Academy of Sciences, USA. 1999;96:7098–7103. doi: 10.1073/pnas.96.12.7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling HQ, Bauer P, Bereczky Z, Keller B, Ganal M. The tomato fer gene encoding a bHLH protein controls iron-uptake responses in roots. Proceedings of the National Academy of Sciences, USA. 2002;99:13938–13943. doi: 10.1073/pnas.212448699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YG, Mitsukawa N, Oosumi T, Whittier RF. Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. The Plant Journal. 1995;8:457–463. doi: 10.1046/j.1365-313x.1995.08030457.x. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Loudet O, Saliba-Colombani V, Camilleri C, Calenge F, Gaudon V, Koprivova A, North KA, Kopriva S, Daniel-Vedele F. Natural variation for sulphate content in Arabidopsis thaliana is highly controlled by APR2. Nature Genetics. 2007;39:896–900. doi: 10.1038/ng2050. [DOI] [PubMed] [Google Scholar]
- Maruyama-Nakashita A, Nakamura Y, Yamaya T, Takahashi H. A novel regulatory pathway of sulphate uptake in Arabidopsis roots: implication of CRE1/WOL/AHK4-mediated cytokinin-dependent regulation. The Plant Journal. 2004;38:779–789. doi: 10.1111/j.1365-313X.2004.02079.x. [DOI] [PubMed] [Google Scholar]
- Maruyama-Nakashita A, Inoue E, Watanabe-Takahashi A, Yamaya T, Takahashi H. Transcriptome profiling of sulphur-responsive genes in Arabidopsis reveals global effects of sulphur nutrition on multiple metabolic pathways. Plant Physiology. 2003;132:597–605. doi: 10.1104/pp.102.019802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama-Nakashita A, Nakamura Y, Tohge T, Saito K, Takahashi H. Arabidopsis SLIM1 is a central transcriptional regulator of plant sulphur response and metabolism. The Plant Cell. 2006;18:3235–3251. doi: 10.1105/tpc.106.046458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama-Nakashita A, Nakamura Y, Watanabe-Takahashi A, Inoue E, Yamaya T, Takahashi H. Identification of a novel cis-acting element conferring sulphur deficiency response in Arabidopsis roots. The Plant Journal. 2005;42:305–314. doi: 10.1111/j.1365-313X.2005.02363.x. [DOI] [PubMed] [Google Scholar]
- Nikiforova VJ, Bielecka M, Gakiere B, Krueger S, Rinder J, Kempa S, Morcuende R, Scheible WR, Hesse H, Hoefgen R. Effect of sulphur availability on the integrity of amino acid biosynthesis in plants. Amino Acids. 2006;30:173–183. doi: 10.1007/s00726-005-0251-4. [DOI] [PubMed] [Google Scholar]
- Nikiforova V, Freitag J, Kempa S, Adamik M, Hesse H, Hoefgen R. Transcriptome analysis of sulphur depletion in Arabidopsis thaliana: interlacing of biosynthetic pathways provides response specificity. The Plant Journal. 2003;33:633–650. doi: 10.1046/j.1365-313x.2003.01657.x. [DOI] [PubMed] [Google Scholar]
- Nikiforova VJ, Gakiere B, Kempa S, Adamik M, Willmitzer L, Hesse H, Hoefgen R. Towards dissecting nutrient metabolism in plants: a systems biology case study on sulphur metabolism. Journal of Experimental Botany. 2004;55:1861–1870. doi: 10.1093/jxb/erh177. [DOI] [PubMed] [Google Scholar]
- Nikiforova VJ, Kopka J, Tolstikov V, Fiehn O, Hopkins L, Hawkesford MJ, Hesse H, Hoefgen R. Systems rebalancing of metabolism in response to sulphur deprivation, as revealed by metabolome analysis of Arabidopsis plants. Plant Physiology. 2005;138:304–318. doi: 10.1104/pp.104.053793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor G, Foyer CH. ASCORBATE AND GLUTATHIONE: keeping active oxygen under control. Annual Review of Plant Physiology and Plant Molecular Biology. 1998;49:249–279. doi: 10.1146/annurev.arplant.49.1.249. [DOI] [PubMed] [Google Scholar]
- Ogo Y, Itai RN, Nakanishi H, Kobayashi T, Takahashi M, Mori S, Nishizawa NK. The rice bHLH protein OsIRO2 is an essential regulator of the genes involved in Fe uptake under Fe-deficient conditions. The Plant Journal. 2007;51:366–377. doi: 10.1111/j.1365-313X.2007.03149.x. [DOI] [PubMed] [Google Scholar]
- Ohkama-Ohtsu N, Kasajima I, Fujiwara T, Naito S. Isolation and characterization of an Arabidopsis mutant that overaccumulates O-acetyl-l-ser. Plant Physiology. 2004;136:3209–3222. doi: 10.1104/pp.104.047068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisy V, Poinssot B, Owsianowski L, Buchala A, Glazebrook J, Mauch F. Identification of PAD2 as a gamma-glutamylcysteine synthetase highlights the importance of glutathione in disease resistance of Arabidopsis. The Plant Journal. 2007;49:159–172. doi: 10.1111/j.1365-313X.2006.02938.x. [DOI] [PubMed] [Google Scholar]
- Poirier Y, Thoma S, Somerville C, Schiefelbein J. Mutant of Arabidopsis deficient in xylem loading of phosphate. Plant Physiology. 1991;97:1087–1093. doi: 10.1104/pp.97.3.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potters G, Pasternak TP, Guisez Y, Palme KJ, Jansen MA. Stress-induced morphogenic responses: growing out of trouble? Trends in Plant Science. 2007;12:98–105. doi: 10.1016/j.tplants.2007.01.004. [DOI] [PubMed] [Google Scholar]
- Rouached H, Secco D, Arpat AB. Getting the most sulphate from soil: regulation of sulphate uptake transporters in Arabidopsis. Journal of Plant Physiology. 2009;166:893–902. doi: 10.1016/j.jplph.2009.02.016. [DOI] [PubMed] [Google Scholar]
- Sanchez-Calderon L, Lopez-Bucio J, Chacon-Lopez A, Gutierrez-Ortega A, Hernandez-Abreu E, Herrera-Estrella L. Characterization of low phosphorus insensitive mutants reveals a crosstalk between low phosphorus-induced determinate root development and the activation of genes involved in the adaptation of Arabidopsis to phosphorus deficiency. Plant Physiology. 2006;140:879–889. doi: 10.1104/pp.105.073825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachtman DP, Shin R. Nutrient sensing and signaling: NPKS. Annual Review of Plant Biology. 2006;58:115–136. doi: 10.1146/annurev.arplant.58.032806.103750. [DOI] [PubMed] [Google Scholar]
- Schlaeppi K, Bodenhausen N, Buchala A, Mauch F, Reymond P. The glutathione-deficient mutant pad2-1 accumulates lower amounts of glucosinolates and is more susceptible to the insect herbivore Spodoptera littoralis. The Plant Journal. 2008;55:774–786. doi: 10.1111/j.1365-313X.2008.03545.x. [DOI] [PubMed] [Google Scholar]
- Shibagaki N, Rose A, McDermott JP, Fujiwara T, Hayashi H, Yoneyama T, Davies JP. Selenate-resistant mutants of Arabidopsis thaliana identify Sultr1;2, a sulphate transporter required for efficient transport of sulphate into roots. The Plant Journal. 2002;29:475–486. doi: 10.1046/j.0960-7412.2001.01232.x. [DOI] [PubMed] [Google Scholar]
- Signora L, De Smet I, Foyer CH, Zhang H. ABA plays a central role in mediating the regulatory effects of nitrate on root branching in Arabidopsis. The Plant Journal. 2001;28:655–662. doi: 10.1046/j.1365-313x.2001.01185.x. [DOI] [PubMed] [Google Scholar]
- Tabe L, Hagan N, Higgins TJ. Plasticity of seed protein composition in response to nitrogen and sulphur availability. Current Opinion in Plant Biology. 2002;5:212–217. doi: 10.1016/s1369-5266(02)00252-2. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Yamazaki M, Sasakura N, Watanabe A, Leustek T, Engler JA, Engler G, Van Montagu M, Saito K. Regulation of sulphur assimilation in higher plants: a sulphate transporter induced in sulphate-starved roots plays a central role in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA. 1997;94:11102–11107. doi: 10.1073/pnas.94.20.11102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor M, Chapman R, Beyaert R, Hernandez-Sebastia C, Marsolais F. Seed storage protein deficiency improves sulphur amino acid content in common bean (Phaseolus vulgaris L.): redirection of sulphur from gamma-glutamyl- S-methyl-cysteine. Journal of Agriculture and Food Chemistry. 2008;56:5647–5654. doi: 10.1021/jf800787y. [DOI] [PubMed] [Google Scholar]
- Tohge T, Nishiyama Y, Hirai MY, et al. Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. The Plant Journal. 2005;42:218–235. doi: 10.1111/j.1365-313X.2005.02371.x. [DOI] [PubMed] [Google Scholar]
- Tsay YF, Schroeder JI, Feldmann KA, Crawford NM. The herbicide sensitivity gene CHL1 of Arabidopsis encodes a nitrate-inducible nitrate transporter. Cell. 1993;72:705–713. doi: 10.1016/0092-8674(93)90399-b. [DOI] [PubMed] [Google Scholar]
- Tzfira T, Vaidya M, Citovsky V. VIP1, an Arabidopsis protein that interacts with Agrobacterium VirE2, is involved in VirE2 nuclear import and Agrobacterium infectivity. EMBO Journal. 2001;20:3596–3607. doi: 10.1093/emboj/20.13.3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernoux T, Wilson RC, Seeley KA, et al. The ROOT MERISTEMLESS1/CADMIUM SENSITIVE2 gene defines a glutathione-dependent pathway involved in initiation and maintenance of cell division during postembryonic root development. The Plant Cell. 2000;12:97–110. doi: 10.1105/tpc.12.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert G, Grotz N, Dedaldechamp F, Gaymard F, Guerinot ML, Briat JF, Curie C. IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. The Plant Cell. 2002;14:1223–1233. doi: 10.1105/tpc.001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Xing X, Wang Y, Tran A, Crawford NM. A genetic screen for nitrate regulatory mutants captures the nitrate transporter gene NRT1.1. Plant Physiology. 2009;151:472–478. doi: 10.1104/pp.109.140434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Kusano M, Oikawa A, Fukushima A, Noji M, Saito K. Physiological roles of the beta-substituted alanine synthase gene family in Arabidopsis. Plant Physiology. 2008;146:310–320. doi: 10.1104/pp.107.106831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Ahn JH, Blazquez MA, et al. Activation tagging in Arabidopsis. Plant Physiology. 2000;122:1003–1013. doi: 10.1104/pp.122.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtz M, Hell R. Dominant-negative modification reveals the regulatory function of the multimeric cysteine synthase protein complex in transgenic tobacco. The Plant Cell. 2007;19:625–639. doi: 10.1105/tpc.106.043125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang C, Oliver DJ. Glutathione metabolic genes coordinately respond to heavy metals and jasmonic acid in Arabidopsis. The Plant Cell. 1998;10:1539–1550. doi: 10.1105/tpc.10.9.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang C, Oliver DJ. Multilevel regulation of glutathione homeostasis in higher plants. In: Pessarakli M, editor. Handbook of plant and crop physiology. New York: Marcel Dekker Inc; 2002. pp. 539–548. [Google Scholar]
- Xiang C, Miao Z, Lam E. DNA-binding properties, genomic organization and expression pattern of TGA6, a new member of the TGA family of bZIP transcription factors in Arabidopsis thaliana. Plant Molecular Biology. 1997;34:403–415. doi: 10.1023/a:1005873500238. [DOI] [PubMed] [Google Scholar]
- Xiang C, Werner BL, Christensen EM, Oliver DJ. The biological functions of glutathione revisited in arabidopsis transgenic plants with altered glutathione levels. Plant Physiology. 2001;126:564–574. doi: 10.1104/pp.126.2.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Li HD, Chen LQ, Wang Y, Liu LL, He L, Wu WH. A protein kinase, interacting with two calcineurin B-like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell. 2006;125:1347–1360. doi: 10.1016/j.cell.2006.06.011. [DOI] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K, Tohge T, Matsuda F, Nakabayashi R, Takayama H, Niida R, Watanabe-Takahashi A, Inoue E, Saito K. Comprehensive flavonol profiling and transcriptome coexpression analysis leading to decoding gene-metabolite correlations in Arabidopsis. The Plant Cell. 2008;20:2160–2176. doi: 10.1105/tpc.108.058040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto N, Inoue E, Saito K, Yamaya T, Takahashi H. Phloem-localizing sulphate transporter, Sultr1;3, mediates re-distribution of sulphur from source to sink organs in Arabidopsis. Plant Physiology. 2003;131:1511–1517. doi: 10.1104/pp.014712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Chen X, Hong YY, Wang Y, Xu P, Ke SD, Liu HY, Zhu JK, Oliver DJ, Xiang CB. Activated expression of an Arabidopsis HD-START protein confers drought tolerance with improved root system and reduced stomatal density. The Plant Cell. 2008;20:1134–1151. doi: 10.1105/tpc.108.058263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu XM, Fang P, Xiang CB. Screening of low-nitrogen tolerant mutants of Arabidopsis thaliana. Plant Nutrition and Fertilizer Science. 2004;10:441–443. [Google Scholar]
- Yuan YX, Zhang J, Wang DW, Ling HQ. AtbHLH29 of Arabidopsis thaliana is a functional ortholog of tomato FER involved in controlling iron acquisition in strategy I plants. Cell Research. 2005;15:613–621. doi: 10.1038/sj.cr.7290331. [DOI] [PubMed] [Google Scholar]
- Zhao SQ, Lin C, Yang XH, Wu WH. Isolation and genetic analysis of Arabidopsis mutants with low-K+ tolerance. Journal of Integrative Plant Biology. 2001;43:105–107. [Google Scholar]
- Zuchi S, Cesco S, Varanini Z, Pinton R, Astolfi S. Sulphur deprivation limits Fe-deficiency responses in tomato plants. Planta. 2009;230:85–94. doi: 10.1007/s00425-009-0919-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.