Abstract
Heptahelical protein 1 (HHP1) is a negative regulator in abscisic acid (ABA) and osmotic signalling in Arabidopsis. The physiological role of HHP1 was further investigated in this study using transgenic and knock-out plants. In HHP1::GUS transgenic mutants, GUS activity was found to be mainly expressed in the roots, vasculature, stomata, hydathodes, adhesion zones, and connection sites between septa and seeds, regions in which the regulation of turgor pressure is crucial. By measuring transpiration rate and stomatal closure, it was shown that the guard cells in the hhp1-1 mutant had a decreased sensitivity to drought and ABA stress compared with the WT or the c-hhp1-1 mutant, a complementation mutant of HHP1 expressing the HHP1 gene. The N-terminal fragment (amino acids 1–96) of HHP1 was found to interact with the transcription factor inducer of CBF expression-1 (ICE1) in yeast two-hybrid and bimolecular fluorescence complementation (BiFC) studies. The hhp1-1 mutant grown in soil showed hypersensitivity to cold stress with limited watering. The expression of two ICE1-regulated genes (CBF3 and MYB15) and several other cold stress-responsive genes (RD29A, KIN1, COR15A, and COR47) was less sensitive to cold stress in the hhp1-1 mutant than in the WT. These data suggest that HHP1 may function in the cross-talk between cold and osmotic signalling.
Keywords: Cold stress, crosstalk, osmotic stress, HHP1
Introduction
Plants use an interconnected signalling network to cope with the abiotic stresses of drought, high salt, and cold/low temperature (Chinnusamy et al., 2005; Yamaguchi-Shinozaki and Shinozaki, 2006). Stresses can occur at different growth stages during development and more than one stress can affect the plant simultaneously. Drought stress restricts the growth of plants due to the stress of osmosis (osmotic stress), leading to a lack of nutrients and reduced photosynthesis (Zhu, 2002). Salt stress leads to physiological drought and ion toxicity, which limit plant growth (Zhu, 2002). Osmotic stress is also brought about by chilling (low temperatures above freezing) and freezing temperatures (Thomashow, 1999). Osmotic stresses resulting from drought, high salt, and cold are transduced by plants by either an abscisic acid (ABA)-dependent or an ABA-independent signalling pathway (Shinozaki and Yamaguchi-Shinozaki, 2007). These pathways lead to the expression of some common downstream stress-responsive proteins, such as RD29A, RD29B, COR15A, COR47, KIN1, and ADH1 (Yamaguchi-Shinozaki and Shinozaki, 1993, 1994; Stockinger et al., 1997). These stress-responsive proteins can be classified into two categories, those functioning in stress tolerance and those involved in signal transduction. The proteins that are directly involved in tolerance include chaperones, late embryogenesis abundant proteins, osmotin, anti-freeze proteins, mRNA-binding proteins, water channel proteins, and several key enzymes involved in the biosynthesis of osmolytes, such as proline and sugar (Fowler and Thomashow, 2002; Kreps et al., 2002; Seki et al., 2002). The proteins involved in signal transduction and the regulation of gene expression include various transcription factors, which may act co-operatively (Yamaguchi-Shinozaki and Shinozaki, 2006; Chinnusamy et al., 2007). Due to the complex environment that plants face, it is expected that more signalling components involved in plant responses to abiotic stresses remain to be discovered. One candidate is heptahelical protein 1 (HHP1), which may function as a negative regulator in ABA and osmotic signalling (Chen et al., 2009).
HHP1 is a member of the HHP family in Arabidopsis that consists of at least five members HHP1, HHP2, HHP3, HHP4, and HHP5 (Hsieh and Goodman, 2005). HHP family proteins are homologous to PAQR family proteins, which include the membrane progestin receptor from fish, the adiponectin receptor from mouse, and YOL002c from yeast (Yamauchi et al., 2003; Zhu et al., 2003; Lyons et al., 2004; Tang et al., 2005). As shown in our previous study (Chen et al., 2009), HHP1 may be involved in stress sensitivity and act as a negative regulator in response to ABA and osmotic stress. The HHP1 T-DNA insertion mutant hhp1-1 shows a higher sensitivity to ABA and osmotic stress than the wild-type (WT), as shown by the germination rate and post-germination growth rate, and the induced expression of stress-responsive genes (RD29A, RD29B, ADH1, KIN1, COR15A, and COR47) is more sensitive to exogenous ABA and osmotic stress in the hhp1-1 mutant than in the WT. The hypersensitivity of the hhp1-1 mutant is reversed in c-hhp1-1, a complementation mutant of HHP1 expressing the HHP1 gene. These data show that mutation of HHP1 renders plants hypersensitive to ABA and osmotic stress and that HHP1 might be a negative regulator in ABA and osmotic signalling.
A gene network that can collect and interpret abiotic stresses, including drought, salt, and cold, has been described (Shinozaki et al., 2003; Yamaguchi-Shinozaki and Shinozaki, 2006; Chinnusamy et al., 2007; Shinozaki and Yamaguchi-Shinozaki, 2007; Tran et al., 2007). Central to this network are several transcription factors, including MYB2, NAC, ABF, DREB2, CBF, and ICE1 (Yamaguchi-Shinozaki and Shinozaki, 2006; Chinnusamy et al., 2007), which are regulated through ABA-independent and ABA-dependent pathways (Shinozaki and Yamaguchi-Shinozaki, 2007). It is expected that more signalling components will be found. ICE1 encodes a MYC-like basic helix-loop-helix transcription factor that regulates the expression of CBF3/DREB1A-controlled genes responsible for cold tolerance (Chinnusamy et al., 2003). The activity of ICE1 is regulated by two opposing processes, sumoylation by SIZ1, which activates ICE1, and ubiquitination by HOS1, which causes degradation of ICE1 (Dong et al., 2006; Miura et al., 2007).
Cold stress limits the normal growth development of plants directly by the inhibition of metabolic reactions and indirectly through cold-induced osmotic (chilling-induced inhibition of water uptake and freezing-induced cellular dehydration) and oxidative stresses (Chinnusamy et al., 2007). On exposure to low temperatures above freezing, plants can acquire freezing tolerance and this process is called cold acclimation (Chinnusamy et al., 2007). Cold-stress signalling in plants is considered to be ABA-independent, although HOS10 is speculated to regulate ABA-mediated cold acclimation (Zhu et al., 2005). ICE1 and C-repeat-binding factors (CBFs), also known as dehydration-responsive element-binding protein 1s (DREB1s), form a transcriptional cascade that is the best-characterized of the transcription factor pathways involved in cold stress-responsive mechanisms (Lee et al., 2005). This pathway controls an important regulon of cold stress-targeted genes, including several cold responsive (COR) genes that render plants cold tolerant (Lee et al., 2005). Under cold stress, CBF3 expression is activated by ICE1 (Chinnusamy et al., 2007). In addition to this direct induction of CBF3 expression, ICE1 appears to regulate MYB15 negatively, an upstream negative regulator of the CBF genes CBF1, CBF2, and CBF3 (Agarwal et al., 2006).
Although the abiotic stresses of drought, high salinity, and cold can lead to similar osmotic stress, it is thought that plants distinguish between complicated environmental changes through a complex signalling network that enables the plant to respond appropriately. The role of HHP1 in both cold and osmotic stress signalling is discussed.
Materials and methods
Plant materials and growth conditions
The Arabidopsis thaliana ecotype Columbia (Col-0) was used as the WT. The screening of the T-DNA insertion knockout mutant of HHP1 (hhp1-1), generation of the HHP1 complemented mutant of hhp1-1 (c-hhp1-1), and verification of the mutants have been described previously (Chen et al., 2009). Seeds were surface-sterilized with 1% sodium hypochlorite and 0.5% Tween 20 and washed with sterile water. Stratification was performed by plating seeds on 1/2 MS medium (Murashige and Skoog, 1962) containing 3% (w/v) sucrose and 0.8% phytagar and incubating them at 4 °C for 4 d. The plates were then transferred to a growth chamber at 22 °C and 50–60% relative humidity (RH) under long-day growth conditions (16/8 h light/darkness). After 10 d, the seedlings were transferred to soil and incubated at 22 °C and 50–60% RH under long-day growth conditions.
Dehydration and cold stress treatment on seedling plants
For dehydration treatment, 10-d-old (stage 1.04) WT seedlings grown on minimal medium (1/2 MS medium, no sucrose added) were transferred onto a filter paper in a covered Petri dish and subjected to dehydration in an environment of 70% RH, 22 °C, and 39 PAR units (photosynthetically active radiation units, μmol m−2 s−1) for 0 (control), 2, 5, 12, or 24 h. For cold treatment, 10-d-old (stage 1.04) WT seedlings grown on minimal medium were placed on ice in a freezing chamber set at 0–4 °C for 0 (control), 2, 5, 12, or 24 h, and ice chips were sprinkled on the seedlings during the cold treatment. The treated seedlings were then frozen in liquid nitrogen and stored at –80 °C for RNA extraction.
Quantitative real-time PCR analysis of HHP1 or cold stress-responsive gene expression profiles
Using the pine tree method, total RNA was isolated from Arabidopsis WT or hhp1-1 or c-hhp1-1 mutants that had undergone dehydration or cold treatment (Chang et al., 1993) and genomic DNA was removed using TURBO DNA-free kits (Ambion, USA). First-strand cDNA synthesized using SuperScript™ III reverse transcriptase (Invitrogen, USA) and an oligo(dT)15 primer was used as the template for real-time PCR using gene-specific primers designed using Primer Express 2.0 (Applied Biosystems). ACTIN2 was used as the internal control in the same cDNA sample. The gene-specific primers used to produce a single amplicon of about 70 bp were: HHP1 (5′-CCCCGTGGATGCAAAGAG-3′ and 5′-TGAGCCTCCTAAGAAAACGAAGA-3′), RD29A (5′-TGATCGATGCACCAGGCGTAAC-3′ and 5′-CCCTGGTGGAATAATTTCCTCCG-3′), KIN1 (5′-ATGCGAAAGATCAAACTCCCCAAA-3′ and 5′-TTCGGATCGACTTATGTATCGTGA-3′), COR15A (5′-CAGTGAAACCGCAGATACATTGGG-3′ and 5′-CGGCTTCTTTTCCTTTCTCCTCC-3′), COR47 (5′-GGAGTACAAGAACAACGTTCCCGA-3′ and 5′-TGTCGTCGCTGGTGATTCCTCT-3′), ICE1 (5′-TGGGATTGAGGTTTCTGGGTTGA-3′ and 5′-CTGAACACTCTCAGCCGCTTTACC-3′), MYB15 (5′-GCTATCATCAGCTTACACCAAATACTTG-3′ and 5′-GGTTCTTCCAGGCAGTTTTGC-3′), CBF1 (5′-CTGAAATGTTTGGCTCCGATTACG-3′ and 5′-TTCGGACAACTCGTGGCCAA-3′), CBF2 (5′-ATATGGATGAAGAGGCGATGTTGG-3′ and 5′-CGACGGTAAAAGCATCCCTTCG-3′), CBF3 (5′-CTGAAGCTGCGTTGGCGTTT-3′ and 5′-TCTCCTCCATGTCGAAGCCA-3′), and ACTIN2 (5′-TGTGGATCTCCAAGGCCGAGTA-3′ and 5′-CCCCAGCTTTTTAAGCCTTTGATC-3′). Real-time PCR was performed as described previously (Chen et al., 2009).
HHP1 promoter::GUS (HHP1::GUS) assay
The generation of the HHP1::GUS mutants used in the study and their screening and verification have been described previously (Chen et al., 2009). GUS staining assays were performed as described previously by Chen et al. (2009). GUS staining patterns were verified in eight independent homozygous T3 lines, and representative individuals from representative lines were chosen for photography. Seedlings were observed using a Leica MZ75 stereomicroscope and photographs were taken using an Olympus digital camera C-5050ZOOM (Olympus) and arranged using PhotoImpact version 8.0 (Ulead Systems).
Transpiration rate measurement under drought stress
To measure the transpiration rate under drought stress, detached fresh leaves were placed abaxial side up on open Petri dishes and weighed after different times (0, 10, 30, 60, 90, 120, 150, 180, 240, 300, 360, 450, and 1500 min) at room temperature. Leaves at similar developmental stages (third to fifth true rosette leaves) from 21-d-old soil-grown WT, hhp1-1, or c-hhp1-1 plants grown under long-day growth conditions were used. The weight of the detached leaf at 0 min was taken as 100% and the transpiration rate was measured in three independent experiments (15 detached leaves per experiment).
Stomatal closure measurement under ABA stress
To investigate the effect of ABA on stomatal aperture, a double-blind stomatal movement assay was performed (Kuhn et al., 2006; Pei et al., 1997). Detached 21-d-old rosette leaves of similar size were floated for 2.5 h on stomatal opening solution (50 mM KCl, 50 μM CaCl2, and 10 mM MES, pH 6.15), then 0, 1, 10, or 100 μM ABA was added to the solution for a further 2.5 h. The leaves were then briefly homogenized at 1600 rpm for 20 s in an homogenizer (SH-100 sample homogenizer, KURABO, Japan) and placed on a microscope slide. Stomatal apertures were observed and recorded using a light microscope (Olympus BX50 microscope with an Olympus DP70 digital camera). The width of the stomatal aperture and the height of the stomata were measured for those stomata surrounded by guard cells with an in focus inner edge of 16–22 μm. The stomatal aperture/height ratio was measured in three independent experiments (20–30 stomata per experiment).
Salt and drought tolerance assays on adult Arabidopsis plants
For salt or drought tolerance assays on the soil-grown plants, 7-d-old (stage 1.02) WT, hhp1-1, or c-hhp1-1 seedlings were transplanted to soil for 14 d under long-day growth conditions as above, they were then subjected to salt or drought by watering with ddH2O (control), 50, 100, or 200 mM NaCl or by withholding water for 14 d, and then the plants were removed from the salt or drought stress and received water. Three days after recommencing watering, the number of surviving plants was counted and photographed; the photographs shown are representative of those for plants in 12 independent pots, with three types of plants (WT, hhp1-1, and c-hhp1-1) per pot to minimize experimental variation. The entire test was repeated three times (12 pots/independent experiment).
Yeast two-hybrid (Y2H) screening and interaction assays
To understand how HHP1 works in the cell, Y2H experiments were performed to identify interacting partners of HHP1 in Arabidopsis using cDNA library CD4-30. The full-length coding sequence of HHP1 was amplified by sticky-end PCR using the primer pairs 5′-TATGGACCAAAATGGTCATAACGACGAAGCA-3′ (HHP1_Y 5′ primer_1) plus 5′-CTTAACAACCAACGTGGTCACGCCAGT-3′ (HHP1_Y 3′ primer_1) and 5′-TGGACCAAAATGGTCATAACGACGAA-3′ (HHP1_Y 5′ primer_2) plus 5′-GATCCTTAACAACCAACGTGGTCA-3′ (HHP1_Y 3′ primer_2) and cloned into pGBKT7 to generate pGBK-HHP1. The N-terminal fragment of HHP1 (nHHP1, encoding amino acids 1–96 of HHP1), used as bait in the Y2H screening, was generated by insertion of a stop codon by a single insertion of T at nucleotide position 292 of the HHP1 coding sequence in pGBK-HHP1 using site-directed mutagenesis (QuikChange II XL Site-Directed Mutagenesis Kit, Stratagene), generating construct pGBK-nHHP1. The Arabidopsis cDNA library (CD4-30) provided by the ABRC was constructed in Stratagene's HybriZap vector using mRNA isolated from Arabidopsis inflorescences, including inflorescence meristems, floral meristems, and floral buds up to about stages 8 or 9. The yeast strain AH109 was used for cDNA library screening, and yeast transformation was performed according to the manufacturer's manual (Clontech). Transformants were plated onto synthetic dropout (SD) medium lacking leucine, tryptophan, and histidine. Positive clones were re-plated on SD medium lacking leucine, tryptophan, histidine, and adenine, supplemented with X-α-gal, and the identities of the blue colonies recovered from the plate determined by DNA sequencing.
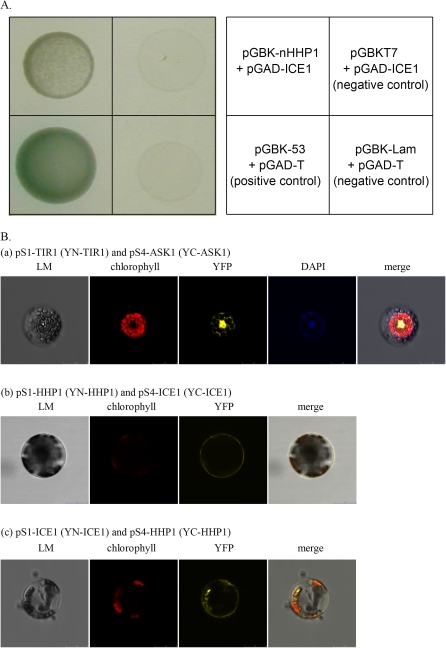
For Y2H interaction assays, the full-length coding sequence of ICE1 was amplified by PCR using the primer pairs 5′-GAATTCATGGGTCTTGACGGAAAC-3′ (ICE1 forward) and 5′-GAATTCTCAGATCATACCAGCATACC-3′ (ICE1 reverse), and cloned into pGADT7 to generate pGAD-ICE1. The interaction of the hybrid proteins was tested by co-transformation of yeast cells with pGBK-nHHP1 as bait and pGAD-ICE1 as prey. The transformants were plated onto SD medium lacking leucine and tryptophan. Positive clones were inoculated onto SD medium lacking leucine, tryptophan, histidine, and adenine, supplemented with X-α-gal. The plasmids extracted from positive clones were subjected to PCR analysis to determine whether they carried the desired insert DNA. The combinations of pGBKT7 plus pGAD-ICE1 or pGBK-Lam (human lamin C(66–230)) plus pGAD-T (SV40 large T-antigen(87–708)) were used as negative controls, while the combination of pGBK-53 (murine p53(72–390)) and pGAD-T was used as the positive control.
Bimolecular fluorescence complementation (BiFC) and confocal imaging
In this study, the protein–protein interaction between HHP1, nHHP1, or ΔnHHP1 (bait protein) and ICE1 (prey protein) was verified by BiFC analysis in planta. The coding sequences of HHP1, ΔnHHP1, and ICE1 were amplified by sticky-end PCR using the primer pairs 5′-CATGGACCAAAATGGTCATAACGACGAA-3′ (HHP1_B 5′ primer_1) plus 5′-GTACCACAACCAACGTGGTCAC-3′ (HHP1_B 3′ primer_1) and 5′-GTACCATGGACCAAAATGGTCATAACGA-3′ (HHP1_B 5′ primer_2) plus 5′-CACAACCAACGTGGTCACGCCA-3′ (HHP1_B 3′ primer_2) for HHP1; 5′-CCTCCATGTTTGGACACATTTGATTGGTT-3′ (ΔnHHP1_B 5′ primer_1) plus 5′-CACAACCAACGTGGTCACGCCA-3′ (HHP1_B 3′ primer_2) and 5′-GTACCCTCAATGTTTGGACACATTTGA-3′ (ΔnHHP1_B 5′ primer_2) plus 5′-GATCCACAACCAACGTGGTCAC-3′ (ΔnHHP1_B 3′ primer _2) for ΔnHHP1; 5′-GATCCATGGGTCTTGACGGAAACA-3′ (ICE1_B 5′ primer_1) plus 5′-CTCAGATCATACCAGCATACCCTG-3′ (ICE1_ B 3′ primer_1) and 5′-CATGGGTCTTGACGGAAACAATGG-3′ (ICE1_B 5′ primer_2) plus 5′-GATCCTCAGATCATACCAGCATACC-3′ (ICE1_B 3′ primer_2) for ICE1, and were cloned into pSAT1-nEYFP-C1 or pSAT4-cEYFP-C1-B to fuse HHP1, ΔnHHP1, or ICE1 to the C-terminus of nEYFP (YN) and to the C-terminus of cEYFP (YC); these constructs were named pS1-HHP1, pS4-HHP1, pS1-ΔnHHP1, pS4-ΔnHHP1, pS1-ICE1, and pS4-ICE1. The N-terminal fragment of HHP1 (nHHP1) encoding amino acids 1–96 of HHP1 was generated by insertion of a stop codon by a single insertion of T at nucleotide position 292 of the HHP1 CDS in pS1-HHP1 or pS4-HHP1 using site-directed mutagenesis (QuikChange II XL Site-Directed Mutagenesis Kit, Stratagene) and these constructs were named pS1-nHHP1 and pS4-nHHP1, respectively. In the BiFC experiment, pS1-TIR1 (transport inhibitor response 1) plus pS4-ASK1 (Arabidopsis Skp1-like protein) (positive control), pS1-bait protein plus pS4-prey protein, or pS1-prey protein plus pS4-bait protein were co-transformed into Arabidopsis mesophyll protoplasts and the transformed protoplasts incubated at 25 °C for 16 h in the dark before confocal imaging. Isolation of Arabidopsis mesophyll protoplasts and polyethylene glycol (PEG) transformation were performed as described previously (Yoo et al., 2007) with some modifications. Well-expanded leaves from 3–4-week-old WT Arabidopsis plants grown under short-day growth conditions (12 h light and 12 h darkness) were cut into 0.5–1 mm strips using a fresh sharp razor blade and the strips transferred into enzyme solution (20 mM MES, pH 5.7, 1% (w/v) cellulase R10, 0.25% (w/v) macerozyme R10, 0.4 M mannitol, 20 mM KCl, 10 mM CaCl2, 5 mM β-mercaptoethanol, and 0.1% BSA), placed in a vacuum desiccator for 30 min, then digested in the dark without shaking for 3 h at 25 °C. The enzyme solution containing protoplasts was filtered through a 70 μm nylon mesh into a 50 ml Falcon tube and the filtrate centrifuged at 100 g for 2 min at room temperature. The protoplasts were washed using cold W5 solution (2 mM MES, pH 5.7, 154 mM NaCl, 125 mM CaCl2, 5 mM KCl, and 5 mM glucose) and resuspended in cold W5 solution and kept on ice for 30 min. The number of protoplasts was counted under the microscope using a haemocytometer, then the W5 solution was removed by centrifugation at 100 g for 2 min at room temperature and the protoplasts resuspended at 1× 106 ml−1 in MMG (4 mM MES, pH 5.7, 0.4 M mannitol, and 15 mM MgCl2) and kept at room temperature. Twenty microlitres of DNA (20–40 μg of plasmid DNA) was added to a 1.5 ml microfuge tube, followed by 200 μl of protoplasts, and the suspension gently mixed, then 220 μl of 40% PEG solution was added and the suspension mixed completely by gently tapping the tube. The transformation mixture was incubated at room temperature for 7 min, then was diluted with 880 μl of W5 solution at room temperature and mixed well by gently inverting the tube to stop the transformation process. The PEG solution was removed by centrifugation at 100 g for 2 min at room temperature and the protoplasts gently re-suspended in 1 ml of W5 solution and incubated at 25 °C for 12–16 h before observation.
Confocal imaging was performed using a Leica TCS SP5 confocal spectral microscope imaging system with an argon blue laser at 488 nm, a 500 nm beamsplitter for excitation, and the spectral detector set between 515 nm and 540 nm.
Chilling and cold tolerance assay
To determine the chilling (4 °C) tolerance of Arabidopsis seedlings, germinated WT, hhp1-1, or c-hhp1-1 seedlings grown on 1/2 MS medium containing 1.5% sucrose and 0.8% phytagar were incubated at 4±1 °C with white light (20.8–23.4 PAR units) under long-day conditions. After 6 weeks, the survival rate of the 42-d-old seedlings was visually determined in three independent experiments (60 seedlings per experiment).
The cold (below 0 °C) tolerance assay was performed on Arabidopsis seedlings grown on 1/2 MS medium as described previously (Agarwal et al., 2006; Miura et al., 2007; Yoo et al., 2007) with some modifications. To determine the cold (below 0 °C) tolerance of Arabidopsis seedlings grown on medium, 10-d-old (stage 1.04) WT, hhp1-1, or c-hhp1-1 seedlings grown on 1/2 MS medium containing 1.5% sucrose and 0.8% phytagar under continuous light (32.4–45.5 PAR units) were cold acclimated at 4±1 °C under continuous light (20.8–23.4 PAR units) for 4 d, then were transferred onto ice in a freezing chamber (MIR-154, SANYO, Japan) set to 0 °C in the dark for 16 h, then the chamber was programmed to cool at 1 °C h−1. The seedlings were treated with cold stress at freezing temperatures (–2, –4, –6, –8, or –10 °C) for 2 h, then incubated at 4±1 °C in the dark for 12 h before being transferred to 22 °C under continuous light (32.4–45.5 PAR units). After 2 d, the survival rate of the seedlings was determined visually in three independent experiments (60 seedlings per experiment).
The cold (below 0 °C) tolerance assay was performed on soil-grown Arabidopsis seedlings as described previously (Yoo et al., 2007) with some modifications. To determine the cold tolerance of soil-grown Arabidopsis seedlings, 21-d-old WT, hhp1-1, or c-hhp1-1 plants grown in soil under continuous light (32.4–45.5 PAR units) were cold acclimated at 4±1 °C for 1 h, then were transferred into a freezing chamber (MIR-154, SANYO, Japan) set to 0 °C in the dark for 1 h, and ice chips were sprinkled over them. The chamber was then programmed to cool at 4 °C h−1. The plants were treated with cold stress at freezing temperatures (–3, –5, –7, or –10 °C) for 1 h, then incubated at 4±1 °C in the dark for 1 d, and transferred to 22 °C under continuous light (32.4–45.5 PAR units). After 7 d, the survival rate of the seedlings was visually determined in three independent experiments (60 seedlings per experiment).
Results
Expression profile of HHP1 at different growth stages and in different organs by histochemical β-glucuronidase (GUS) analysis
Significant HHP1 expression is seen in the reproductive organs (Hsieh and Goodman, 2005), and the expression profile of HHP1 shows two peaks at stages 1.04 and 6.50 using quantitative real-time PCR (data not shown) or public microarray data (Genevestigator database, https://www.genevestigator.com). To study further when and where HHP1 is expressed, homozygous HHP1::GUS T3 mutants harbouring the HHP1::GUS transgene were analysed using histochemical GUS activity staining (Fig. 1).
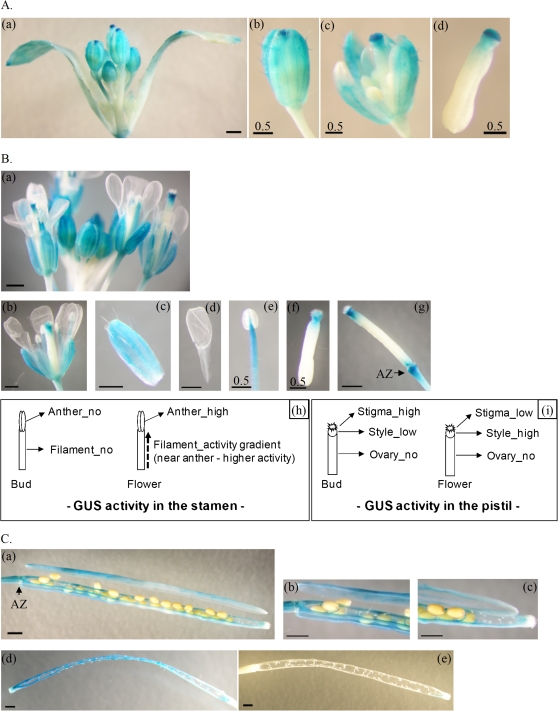
Fig. 1.
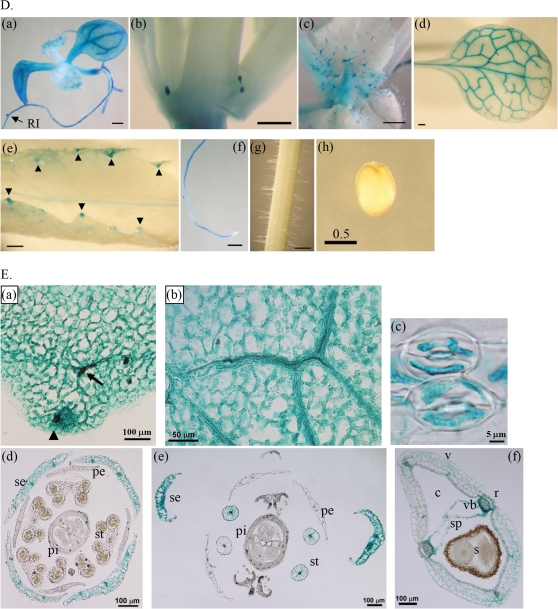
Expression profiles of HHP1 at different growth stages and in different organs or tissues in transgenic HHP1::GUS Arabidopsis mutants as demonstrated by histochemical analyses of GUS. Histochemical GUS activity was examined in buds (A), flowers (B), and siliques (C) during reproductive growth (stages 6 and 8), and in individual organs or tissues of the plant (D) or parafilm sections of rosette leaves and reproductive organs (E). (A–C) GUS expression in the intact bud cluster (Aa) and its dissected parts (Ab–d), in the whole flower cluster (Ba) and its dissected sepal (Bc), petal (Bd), stamen (Be), pistil (Bf), and intact pistil (Bg), or in the intact silique before (Ca), during (Cd), or after (Ce) seed scattering and in the dissected parts before seed scattering (Cb, c). The intensity of GUS activity in the buds or flowers is summarized in two simple diagrams (Bh, i). The GUS staining patterns shown are representative of those for eight independent homozygous transgenic HHP1::GUS T3 lines. The arrowheads and arrows indicate hydathodes and vasculature, respectively. Scale bars correspond to 1 mm, except in Ab–d, Be, f, and Dh, where they correspond to 0.5 mm. Abbreviations: AZ, adhesion zone; c, carpel; pe, petal; pi, pistil; r, replum; RI, lateral root initiation site; s, seed; se, sepal; sp, septum; st, stamen; v, valve; vb, vascular bundle.
Changes in GUS expression pattern were observed during the development of the reproductive organs (buds, flowers, and siliques) (Fig. 1A–C, Ed–f). In the buds, GUS activity was clearly seen in trichomes, the veins of sepals, and the stigmata of pistils, but not in the stamens and petals (Fig. 1A, Ed). High GUS expression was seen in the stigma of the pistil in mature buds (Fig. 1Ac, d). In the flowers, GUS activity was predominantly present in the pistils (Fig. 1Bf, g), stamens (Fig. 1Be, Ee), adhesion zones (Fig. 1Bg), and veins of sepals (Fig. 1Bc, Ee), but not in the petals (Fig. 1Bd). As the bud developed to flower, GUS activity changed in both the stamen and pistil (Fig. 1B). In the stamen, the activity changed from low to high and was centred on the anther (Fig. 1Bb, Ee). In the pistil, GUS activity changed from high in the stigma/low in the style to low in the stigma/high in the style (Fig. 1Bf, g, i) The adhesion zones of older flowers showed higher GUS activity than those in younger flowers (Fig. 1A, B). The intensity of GUS activity changes during the transition from buds to flowers is summarized schematically in Fig. 1Bh and Bi. In the siliques, GUS activity was present in the adhesion zones, septa, veins of carpels, and connection sites between septa and seeds, while seeds, pedicels, and remains of styles showed little or no GUS activity (Fig. 1Ca–c, Ef). When seed pods became brown and shattered, the septa and the remaining connection sites between the septa and seeds showed high GUS activity (Fig. 1Cd). After seed scattering and withering of the siliques, the remains of the siliques showed no GUS activity (Fig. 1Ce). Taken together, these results show that GUS activity varies throughout the entire development of the reproductive organ (Fig. 1A–C).
In summary, the high GUS activity in the lateral root initiation site (Fig. 1Da), the vasculature of rosette leaves and reproductive organs (Fig. 1Dd, Ea, b, d–f), the hydathodes (Fig. 1De, Ea), the guard cells of stomata (Fig. 1Ec), the adhesion zones (Fig. 1Bg, Ca), and the connection sites between septa and seeds (Fig. 1Ca–d) suggests that HHP1 plays an important role in the regulation of turgor pressure in response to osmotic stress. These results agree with those of a previous study that indicated that HHP1 may be a regulator of ABA and osmotic signalling (Chen et al., 2009).
HHP1 expression is increased by drought stress, but is not affected by cold stress
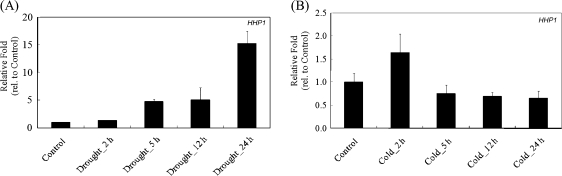
The HHP1 expression profiles of 10-d-old (stage 1.04) WT Arabidopsis seedlings left untreated (control) or subjected for different times to one of two osmotic stresses, drought or cold, were analysed by real-time PCR. ACT2 was used to normalize HHP1 expression and the results are presented as the fold expression relative to that in the untreated WT (control). HHP1 expression was significantly increased 5-fold (5 h and 12 h) to 15-fold (24 h) by drought stress (Fig. 2A), while cold stress had no significant effect (Fig. 2B).
Fig. 2.
Expression profiles of HHP1 in response to drought or cold stresses. HHP1 expression in 10-d-old (stage 1.04) WT Arabidopsis seedlings without (control) or with drought (A) or cold (B) treatment for the indicated time analysed by real-time PCR. The data are the mean ±standard error of the mean (SEM) for three independent amplification reactions and are representative of two independent biological replicates, each consisting of 10–15 seedlings.
The hhp1-1 mutant shows lower sensitivity of guard cells to drought and ABA
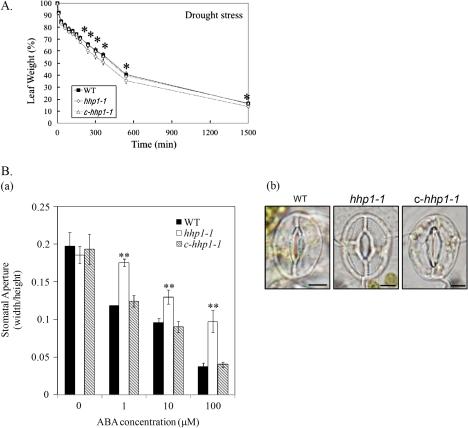
In plants, the guard cells integrate signals about water availability, exogenous CO2, hormones, light, and other environmental conditions to regulate the size of the stomatal aperture. Under drought stress, the stomata in the leaf epidermis are closed to prevent water being lost through transpiration. ABA acts as an endogenous anti-transpirant to decrease the rate of water loss through stomatal pores (Wang and Song, 2008). Our results above showed high GUS activity in the guard cells of stomata (Fig. 1Ec). In addition, HHP1 expression is significantly increased by ABA (Chen et al., 2009) or drought stress (Fig. 2A). It was therefore of interest to know whether HHP1 played a role in the regulation of the stomatal aperture in the response to drought stress. The transpiration rate and stomatal closure were analysed in WT, hhp1-1, and c-hhp1-1 plants subjected to drought stress or ABA treatment. To measure the transpiration rate under drought stress, fresh leaves were detached from 21-d-old soil-grown WT, hhp1-1, and c-hhp1-1 plants and were weighed at different time points after detachment. Stomatal closure is a key ABA-controlled process that determines the rate of transpiration under water-deficit conditions (Leung and Giraudat, 1998). Under drought stress, the loss in weight of detached rosette leaves from hhp1-1 transgenic plants was significantly greater than that for rosette leaves from WT and c-hhp1-1 plants (Fig. 3A), indicating that stomata closure in hhp1-1 plants might be less sensitive to water deficit than in the WT and c-hhp1-1. To determine whether this decreased sensitivity of the hhp1-1 mutant to drought was mediated through ABA, fresh leaves were detached from 21-d-old soil-grown WT, hhp1-1, and c-hhp1-1 plants and treated with ABA, followed by stomatal aperture measurement. As shown in Fig. 3B, ABA-induced stomatal closure was decreased and less significant in hhp1-1 leaves than in WT and c-hhp1-1 leaves, implying that the guard cells of the hhp1-1 mutant have an impaired response to ABA. Taken together, these results show that HHP1 acts as a positive regulator of ABA-mediated stomatal closure under exogenous water deficit stress.
Fig. 3.
The hhp1-1 mutant shows lower sensitivity of guard cells to drought and ABA. (A) To examine the transpiration rate under drought stress, fresh leaves from 21-d-old soil-grown WT, hhp1-1, and c-hhp1-1 plants grown under long-day conditions were excised and weighed at various time points after detachment and the weight of the detached leaf at 0 min taken as 100%. The values are the mean ±SEM for three independent experiments, with approximately 15 detached leaves per experiment. (B) Effects of ABA on stomatal aperture in WT, hhp1-1, and c-hhp1-1 plants. Detached fresh leaves of similar size and age from 21-d-old soil-grown WT, hhp1-1, and c-hhp1-1 plants grown under long-day conditions were floated on stomatal opening solution for 2.5 h, then were treated with 0, 1, 10, or 100 μM ABA for 2.5 h. The stomatal aperture was then measured (a) and photographs were taken of stomata in detached leaves from WT, hhp1-1, and c-hhp1-1 plants treated with 100 μM ABA (b). The values in B(a) are the mean width/height ratio ±SEM for three independent experiments, with approximately 20–30 stomata per experiment. The scale bars in B(b) correspond to 5 μm. In (A) and (Ba), the asterisks indicate the level of significance of differences between the WT and hhp1-1 or c-hhp1-1 plants under the same growth conditions. (*P <0.05; **P <0.01, Student's t test). (This figure is available in colour at JXB online.)
Proteins interacting with HHP1 in a yeast two-hybrid (Y2H) system assay
To understand how HHPl works in the cell, Y2H experiments were performed to identify interacting partners of HHP1 in Arabidopsis using cDNA library CD4-30. When the full-length HHP1 protein (construct pGBK-HHP1) was used as bait, no positive (survival) clone was found on SD medium lacking leucine, tryptophan, and histidine (data not shown). This suggested that HHP1 may be a membrane-associated protein that cannot enter the nucleus, a necessary condition in the Y2H system. HHP1 is predicted to have an N-terminal segment of 96 amino acids, a C-terminal segment of six amino acids, and six short loops ranging from five to 20 amino acids, connecting TMs. Based on transmembrane domain prediction using the hidden Markov model (TMHMM) (Hsieh and Goodman, 2005), HHP1 adopts an N-terminus-inside topology. Although these data need to be verified in further experiments, the N-terminal fragment of HHP1 was used as bait in another Y2H screening.
Using the N-terminal region of HHP1 (construct pGBK-nHHP1) as bait, 12 positive clones with signals of various intensities were identified in the initial screening of 2.7×105 transformants. The prey plasmids from the 12 clones were isolated and sequenced. Two encoded ZFHD1 (at1g69600) and one encoded ICE1 (at3g26744). The full-length coding sequence for each clone was amplified from a cDNA library and cloned into pGAD. The full-length prey plasmids were co-transformed into yeast with pGBK-nHHP1 and the cells plated on plates lacking Leu, Trp, His, and Ade, supplemented with X-α-gal. Only two of the transformants, harboring ICE1 (Fig. 4A) or ZFHD1 (data not shown), respectively, showed high β-galactosidase activity. No self activation was found for pGAD-ICE1 (Fig. 4A). It is interesting that both ICE1 and ZFHD1 participate in the signalling networks of cold or osmotic stresses (Yamaguchi-Shinozaki and Shinozaki, 2006). Protein–protein interaction between HHP1 and ICE1 was further studied using BiFC analyses, as detailed below, while the putative interaction between HHP1 and ZFHD1 is currently under investigation.
Fig. 4.
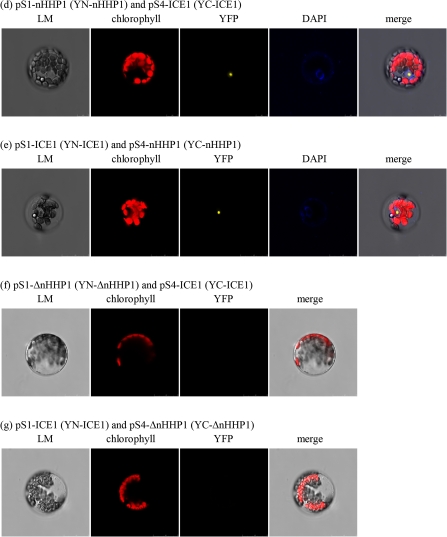
Protein-protein interaction between HHP1 and ICE1 verified by Y2H (A) or BiFC (B). (A) AH109 yeast cells were transformed with plasmids pGBK-nHHP1 plus pGAD-ICE1, pGBKT7 plus pGAD-ICE1 (negative control), pGBK-53 plus pGAD-T (positive control), or pGBK-Lam plus pGAD-T (negative control) and inoculated on SD medium lacking leucine, tryptophan, histidine, and adenine, supplemented with X-α-gal. (B) pS1-TIR1 plus pS4-ASK1 (a), pS1-HHP1 plus pS4-ICE1 (b), pS1-ICE1 plus pS4-HHP1 (c), pS1-nHHP1 plus pS4-ICE1 (d), pS1-ICE1 plus pS4-nHHP1 (e), pS1-ΔnHHP1 plus pS4-ICE1 (f), or pS1-ICE1 plus pS4-ΔnHHP1 (g) were transformed into Arabidopsis mesophyll protoplasts by PEG transformation and YFP fluorescence observed by confocal spectral microscopy.
Protein–protein interaction between HHP1 and ICE1 verified by BiFC analyses
Y2H experiments (Fig. 4A) revealed that protein–protein interaction occurred between ICE1 and the N-terminus of HHP1. BiFC analysis was used to detect and confirm this interaction in planta (Fig. 4B). To examine the protein–protein interaction between full-length HHP1 and ICE1, pS1-HHP1 (YN-HHP1) plus pS4-ICE1 (YC-ICE1) or pS1-ICE1 (YN-ICE1) plus pS4-HHP1 (YC-HHP1) were co-transformed into Arabidopsis mesophyll protoplasts by PEG transformation and YFP fluorescence examined using confocal spectral microscopy. Co-transformation with pS1-TIR1 (YN-TIR1) and pS4-ASK1 (YC-ASK1) was used as the positive control (Citovsky et al., 2006) and gave strong YFP fluorescence in the cytosol and nucleus of the transformed protoplasts (Fig. 4Ba). On co-expression of YN-HHP1 and YC-ICE1 (Fig. 4Bb) or YN-ICE1 and YC-HHP1 (Fig. 4Bc), YFP fluorescence was detected predominantly in the plasma membrane in the transformed protoplast, showing that HHP1 interacts with ICE1 in the plasma membrane. To determine which domain of HHP1 interacted with ICE1, Arabidopsis mesophyll protoplasts were co-transformed with pS1-nHHP1 (YN-nHHP1), coding for the N-terminal 96 amino acid fragment of HHP1, plus pS4-ICE1 (YC-ICE1) (Fig. 4Bd), pS1-ICE1 (YN-ICE1) plus pS4-nHHP1 (YC-nHHP1) (Fig. 4Be), pS1-ΔnHHP1 (YN-ΔnHHP1, harbouring HHP1 lacking the N-terminal 96 amino acids, i.e. containing the seven transmembrane regions and the C-terminal domain) plus pS4-ICE1 (YC-ICE1) (Fig. 4Bf), or pS1-ICE1 (YN-ICE1) plus pS4-ΔnHHP1 (YC-ΔnHHP1) (Fig. 4Bg). The results showed that nHHP1 (the N-terminal domain) interacted with ICE1 in the nucleus (Fig. 4Bd, e), while ΔnHHP1 (the 7TM part of HHP1) was unable to interact with ICE1 (Fig. 4Bf, g). It is interesting that the YFP fluorescence in cells co-expressing nHHP1 and ICE1 was inside the nucleus (Fig. 4Bd, e), implying that the protein complex of nHHP1 and ICE1 may move into a specific site in the nucleus. In summary, protein–protein interaction was seen between HHP1 and ICE1, and the N-terminus of HHP1 was responsible for its interaction with ICE1 (Fig. 4).
Expression profiles of cold stress-responsive genes under cold stress
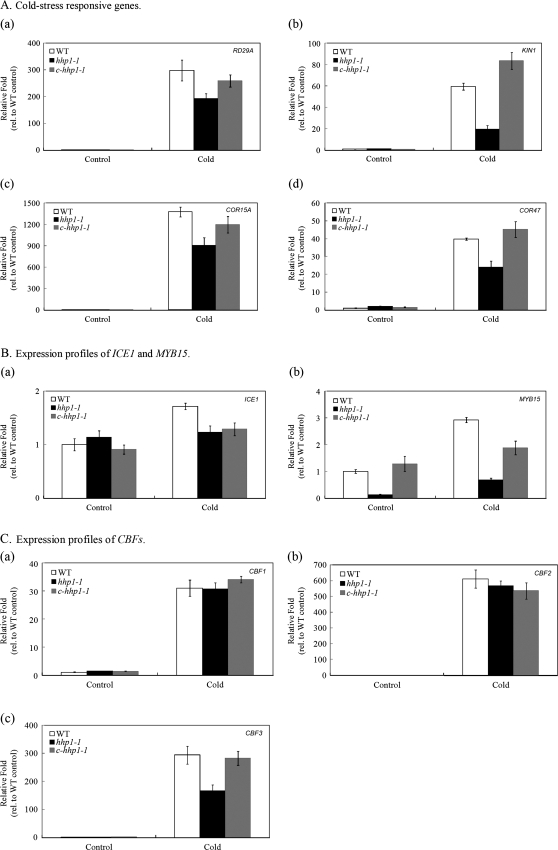
In order to elucidate the relationship between HHP1 and ICE1 further, the expression profiles of the cold stress-responsive genes RD29A, KIN1, COR15A, and COR47 (Fig. 5A), of ICE1 and MYB15, a known protein–protein interaction pair (Fig. 5B), and of CBF1, CBF2, and CBF3, transcription factors induced by cold stress (Fig. 5C) (Xiong et al., 2002; Zhu et al., 2005, 2007; Chinnusamy et al., 2007; Miura et al., 2007) in 10-d-old (stage 1.04) WT, hhp1-1 and c-hhp1-1 seedlings grown on minimal medium and exposed to cold stress for 16 h were determined by real-time PCR. ACT2 was used to normalize the expression of the target genes and the results are presented as the fold expression relative to that of untreated WT (control). Without cold treatment, the expression of most cold stress-responsive genes was low in all three types of seedlings. Compared with WT and c-hhp1-1 seedlings, the cold-induced expression of RD29A, KIN1, COR15A, COR47 (Fig. 5A), MYB15 (Fig. 5Bb), and CBF3 (Fig. 5Cc) was repressed in hhp1-1 seedlings (it should be noted that the scale in 5B is much smaller than in 5A and C). This indicates that HHP1 may be an upstream activator in the cold-signalling pathway. Furthermore, since the induced expression of MYB15 and CBF3 in response to cold stress was significantly repressed in hhp1-1 seedlings compared with WT and c-hhp1-1 seedlings, this suggests that HHP1 may be an activator in the induction of CBF3 and MYB15 by cold stress and be involved in the ICE1-mediated signalling pathway in response to cold stress. The cold stress signalling pathway is interconnected with the osmotic stress signalling pathways (Xiong et al., 2002; Yamaguchi-Shinozaki and Shinozaki, 2005, 2006) and the authors speculated that HHP1 may act as a cross-talk point.
Fig. 5.
Expression profiles of cold stress-responsive genes. The expression of RD29A (Aa), KIN1 (Ab), COR15A (Ac), COR47 (Ad), ICE1 (Ba), MYB15 (Bb), CBF1 (Ca), CBF2 (Cb), or CBF3 (Cc) mRNAs in 10-d-old (stage 1.04) WT, hhp1-1, and c-hhp1-1 seedlings grown on minimal medium after cold treatment (0–4 °C) for 16 h was analysed by real-time PCR. The data are the mean ±SEM for three independent amplification reactions and are representative of two independent biological replicates, each consisting of 10–15 seedlings.
Adult hhp1-1 plants show a similar tolerance to salt or drought stresses to the WT
Real-time PCR results showed that HHP1 expression was significantly increased by salt stress (Chen et al., 2009) or drought stress (Fig. 2). Furthermore, HHP1 may contribute to osmotic stress sensitivity and play a negative regulatory role in seed germination and early growth in Arabidopsis (Chen et al., 2009). However, these results were derived using juvenile Arabidopsis seedlings. To elucidate the relationship between HHP1 and salt or drought stress further, salt or drought stress tolerance was examined in adult WT, hhp1-1, or c-hhp1-1 plants treated with different concentrations of NaCl or drought stress. Under conditions of salt stress (Fig. 6B-D) or drought stress (Fig. 6E), no significant difference was seen between hhp1-1 plants and either WT or c-hhp1-1 plants in the tolerance assay. These results suggest that HHP1 may play a less important role in tolerance to exogenous salt and drought stress in adult plants, although the guard cells in hhp1-1 plants showed lower sensitivity to drought or ABA (Fig. 3).
Fig. 6.
No significant difference is seen between adult hhp1-1 plants and adult WT or c-hhp1-1 plants in tolerance to salt or drought stress. For the salt or drought tolerance assay, 21-d-old adult soil-grown WT, hhp1-1, and c-hhp1-1 plants grown under long-day conditions were subjected to salt (B-D) or drought (E) by watering with water (A, control) or 50 mM (B), 100 mM (C), or 200 mM NaCl (D) or withholding water (E) for 14 d. The plants were then removed from the salt or drought stress and received water for 3 d, then the number of surviving plants was counted. The numbers 1, 2, and 3 represent WT, hhp1-1, and c-hhp1-1 plants, respectively. Photographs were taken on day 38 when the inflorescence was excised and were taken in top view. The photographs are representative of 12 independent pots, with the three types of plant (WT, hhp1-1, and c-hhp1-1) grown in the same pot to minimize experimental variations. The entire test was repeated three times, using 12 plants each time. (This figure is available in colour at JXB online.)
Under conditions of water deficit, the hhp1-1 mutant shows lower tolerance to cold stress
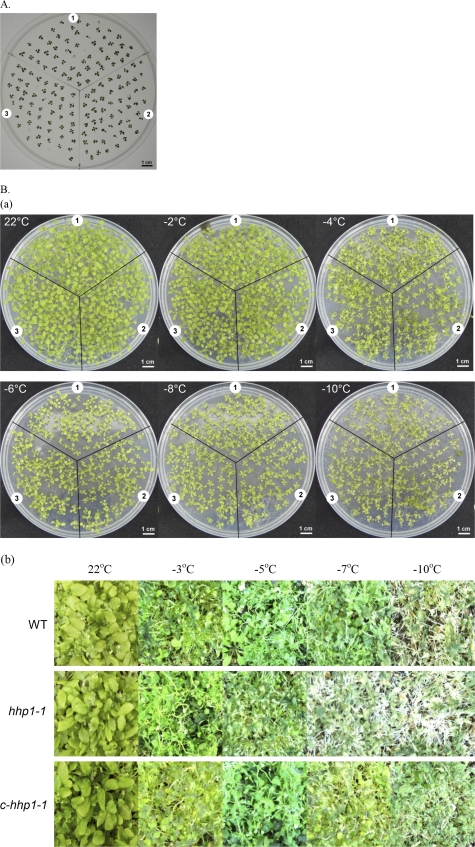
Our results showed that HHP1, acting as an activator in the induction of CBF3 and MYB15 expression by cold stress, interacts with ICE1 and participates in the ICE1-mediated signalling pathway in response to cold stress (Figs 4, 5), although cold stress had no significant effect on HHP1 expression (Fig. 2). The induced expression of cold stress-responsive genes, such as COR15A or COR47, in the hhp1-1 mutant was less sensitive to cold stress (Fig. 5), indicating that HHP1 may participate in tolerance to cold stress in Arabidopsis. In order to elucidate further whether HHP1 was involved in cold stress sensitivity, chilling or cold stress tolerance was examined in WT, hhp1-1, or c-hhp1-1 plants treated with low or freezing temperatures. Under both chilling (4 °C) and cold stress (–2 to –10 °C) conditions, no significant difference in tolerance was seen between hhp1-1 (sector 2), WT (sector 1), and c-hhp1-1 (sector 3) plants grown on medium (Fig. 7A, Ba). However, in plants grown in soil under water-deficit conditions, the survival rate of the hhp1-1 plants was lower than that of the WT and c-hhp1-1 plants at temperatures of –7 °C and –10 °C (Fig. 7Bb). These results suggest that HHP1 may be involved in tolerance to a combination of cold and drought stresses, although mutation of HHP1 did not affect the tolerance of plants to drought (Fig. 6), chilling (Fig. 7A), or cold stress (Fig. 7Ba).
Fig. 7.
Tolerance assay under chilling or cold stress conditions. (A) For the chilling tolerance assay, the plants were grown at 4 °C for 6 weeks after germination at 22 °C and the survival rate determined. (B) For the cold tolerance assay on plants grown on medium (a), 10-d-old seedlings were cold acclimated at 4 °C for 4 d, placed on ice in a freezing chamber at 0 °C for 16 h, treated with cold stress at –2, –4, –6, –8, or –10 °C for 2 h, and the survival rate was scored visually after 2 d. For the cold-tolerance assay on soil-grown plants (b), 21-d-old seedlings were treated at 4 °C for 1 h, transferred to a freezing chamber at 0 °C for 1 h, treated with cold stress at –3, –5, –7, or –10 °C for 1 h, and the survival rate scored visually after 7 d. Numbers 1, 2, and 3 in (A) and (B) represent the WT, hhp1-1, and c-hhp1-1 plants, respectively. The photograph in (A) was taken on day 42, those in (Ba) on day 17, and those in (Bb) on day 28. The photographs are representative of three independent experiments, with approximately 60 seedlings per experiment.
Discussion
HHP1 is involved in drought and cold stress responses
HHP1 has been shown to be involved in ABA-mediated osmotic stress signalling and to act as a negative regulator (Chen et al., 2009). To examine further the role of HHP1 in the signalling network for abiotic stresses, including drought, salinity, and cold, phenotypic analysis related to drought or cold signalling was carried out. Measurement of the transpiration rate and stomatal aperture showed that HHP1 acted as a positive regulator in ABA-mediated guard cell control to regulate stomatal closure under conditions of exogenous water deficit stress (Fig. 3). hhp1-1 plants showed a decreased sensitivity of guard cells to drought compared to WT and c-hhp1-1 plants (Fig. 3). However, this slight difference did not result in a significant difference in tolerance to salinity or drought in the adult plant (Fig. 6). It is possible that HHP1 is not major player in drought and salinity tolerance, which require many other factors in the signalling networks (Zhang et al., 2004).
The most important finding in this study was the discovery that HHP1 can interact with ICE1, a key regulator in freezing tolerance (Chinnusamy et al., 2003). The protein–protein interaction between HHP1 and ICE1 placed HHP1 in the ICE1-mediated cold stress signalling pathway. ICE1 encodes a MYC-like bHLH transcriptional activator, which binds to the CBF3 promoter. Transgenic plants overexpressing ICE1 do not show increased CBF3 expression at warm temperatures, but, at low temperatures, show higher expression of CBF3 and of two cold responsive genes, RD29A and COR15A, which are under the control of CBF3 (Chinnusamy et al., 2003). This suggests that a cold-induced modification of ICE1 is required for it to function in cold signalling. In this study, the ICE1-regulated gene CBF3 was expressed at a lower level in the hhp1-1 mutant (Fig. 5C), supporting the idea that HHP1 is involved in cold stress signalling. HHP1 may play an important part in ABA-regulated osmotic stress sensitivity (Chen et al., 2009). On the other hand, ICE1-mediated cold stress signalling is mainly viewed as an ABA-independent pathway (Yamaguchi-Shinozaki and Shinozaki, 2005), although the role of ABA in cold stress-responsive gene expression is not clear (Yamaguchi-Shinozaki and Shinozaki, 2006). Our finding that HHP1 interacts with ICE1 suggests a novel regulatory mechanism for ICE1. Since HHP1 expression was induced by drought, but not by cold (Fig. 2), it is likely that the drought-induced accumulation of HHP1 positively regulates ICE1 function in cold signalling. As shown in Fig. 7, the hhp1-1 mutant showed a decreased tolerance to the combination of dehydration and cold stresses. This demonstrates that HHP1 may contribute to tolerance to a specified stress condition of drought plus cold, which is often encountered by plants during winter.
The expression of CBF3, but not that of CBF1 and CBF2, was less sensitive to cold in the hhp1-1 mutant (Fig. 5). This supports the idea that HHP1 can regulate the function of ICE1 through protein–protein interaction, since ICE1 is found to bind directly to the promoter region of CBF3 (Chinnusamy et al., 2003). It is known that the expression of CBF1 or CBF2 is not regulated through ICE1 (Yamaguchi-Shinozaki and Shinozaki, 2006). At least four other protein homologues of HHP1 (HHP2, HHP3, HHP4, and HHP5) are present in Arabidopsis (Hsieh and Goodman, 2005). It would be of interest to examine whether other HHP proteins are involved in the regulation of other CBFs through protein–protein interactions.
The detailed action of HHP1 requires further investigation
The phenotype of reduced tolerance to cold/drought of the hhp1-1 mutant reported here may represent only one of the functions of HHP1. First, not all possible degrees of drought or cold that a plant may encounter were tested. Second, analysis of the HHP1::GUS mutants showed that HHP1 was expressed in the roots, vasculature, stomata, hydathodes, adhesion zones, and the connection sites between septa and seeds (Fig. 1), sites at which the turgor pressure is regulated. This tissue- and stage-specific expression pattern may reflect functions other than the phenotype of cold/drought tolerance.
The two proteins interacting with HHP1, ICE1 (Fig. 4) and ZFHD1 (data not shown), participate in the ABA-independent signalling pathway in response, respectively, to cold or osmotic stress (Chinnusamy et al., 2007; Yamaguchi-Shinozaki and Shinozaki, 2006). The possible interaction between HHP1 and ZFHD1 is currently under investigation. The localization of HHP1 in the plasma membrane might not be compatible with the function of ICE1, a nucleus-targeted transcription factor and a candidate HHP1 interaction partner. One possibility is that ICE1 dissociates from HHP1 when necessary, or, alternatively, that the N-terminal fragment of HHP1 may be released by controlled proteolysis, as seen with membrane-associated transcription factors, such as those in the NAC protein family (Kim et al., 2007). The detailed physiological mechanism by which HHP1 affects ICE1 function requires further investigation.
It was recently reported that ICE1 plays a role in the development of stomata (Kanaoka et al., 2008) and this finding enables a link to be established between environmental responses and the formation of stomata. This is very interesting, since HHP1 expression, as revealed by GUS analysis, was clearly seen in the stomata (Fig. 1E), although the stomata in hhp1-1 plants appeared normal (Fig. 3Bb). The detailed mechanism of how HHP1 operates in physiological terms requires further investigation.
HHP1 may function as a regulator of cross-talk between various osmotic stress signalling pathways in Arabidopsis
Based on microarray data, Seki et al. (2002) reported that expression of more than 50% of the drought-inducible genes (around 300 in total) is induced by high salinity, whereas cold stress only results in expression of 10%. By contrast, expression of the DREB1/CBF genes is induced by cold stress, but not by drought stress (Liu et al., 1998). This suggests that independent pathways exist for cold and drought signalling. Originally, ICE1 was placed in an ABA-independent pathway in the gene network responding to osmotic stress and cold stress signalling (Chinnusamy et al., 2007). In our previous study (Chen et al., 2009), it was shown that the induced expression of HHP1 in response to high-salinity or drought is mediated by the ABA-dependent pathway. In the present study, HHP1 was found to affect the major ABA-independent cold signalling pathway through its interaction with ICE1. Our data, therefore, support the idea that HHP1 may act as a cross-talk point between cold stress and drought stress signalling.
HHP1 is likely to be an upstream positive regulator of the drought and cold stress signalling pathways. It is interesting to note that it plays a negative regulatory role in seed germination and early growth (Chen et al., 2009), but acted as a positive regulator of ABA-mediated stomatal closure in response to water-deficit stress (Fig. 3) and cold stress (Figs 5, 7). It is possible that HHP1 may play different roles in different organs (or cells) and at different growth stages under water deficit stress in Arabidopsis.
Acknowledgments
We thank Dr Jong-Ching Su and Dr Ming-Hsiun Hsieh for helpful discussion. We are grateful to Dr Shiang-Jiuun Chen and Ms Yi-Chun Chuang of TC5 Bio-Image Tools, Technology Commons, College of Life Science, NTU, for help with the confocal laser scanning microscopy. This project was supported by the National Science Council, Taiwan (NSC 97-2313-B-002-011-MY3).
References
- Agarwal M, Hao Y, Kapoor A, Dong C-H, Fujii H, Zheng X, Zhu J- K. A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. Journal of Biological Chemistry. 2006;281:37636–37645. doi: 10.1074/jbc.M605895200. [DOI] [PubMed] [Google Scholar]
- Chang S, Puryear J, Cairney J. A simple and efficient method for isolating RNA from pine trees. Plant Molecular Biology Reporter. 1993;11:113–116. [Google Scholar]
- Chen C-c, Liang C-s, Kao A-l, Yang C-c. HHP1 is involved in osmotic stress sensitivity in Arabidopsis. Journal of Experimental Botany. 2009;60:1589–1604. doi: 10.1093/jxb/erp039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V, Jagendorf A, Zhu J- K. Understanding and improving salt tolerance in plants. Crop Science. 2005;45:437–448. [Google Scholar]
- Chinnusamy V, Ohta M, Kanrar S, Lee B-h, Hong X, Agarwal M, Zhu J-K. ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes and Development. 2003;17:1043–1054. doi: 10.1101/gad.1077503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V, Zhu J, Zhu J-K. Cold stress regulation of gene expression in plants. Trends in Plant Science. 2007;12:444–451. doi: 10.1016/j.tplants.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Citovsky V, Lee L-Y, Vyas S, Glick E, Chen M-H, Vainstein A, Gafni Y, Gelvin SB, Tzfira T. Subcellular localization of interacting proteins by bimolecular fluorescence complementation in planta. Journal of Molecular Biology. 2006;362:1120–1131. doi: 10.1016/j.jmb.2006.08.017. [DOI] [PubMed] [Google Scholar]
- Dong C-H, Agarwal M, Zhang Y, Xie Q, Zhu J- K. The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proceedings of the National Academy of Sciences, USA. 2006;103:8281–8286. doi: 10.1073/pnas.0602874103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler S, Thomashow MF. Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. The Plant Cell. 2002;14:1675–1690. doi: 10.1105/tpc.003483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh MH, Goodman HM. A novel gene family in Arabidopsis encoding putative heptahelical transmembrane proteins homologous to human adiponectin receptors and progestin receptors. Journal of Experimental Botany. 2005;56:3137–3147. doi: 10.1093/jxb/eri311. [DOI] [PubMed] [Google Scholar]
- Kanaoka MM, Pillitteri LJ, Fujii H, Yoshida Y, Bogenschutz NL, Takabayashi J, Zhu JK, Torii KU. SCREAM/ICE1 and SCREAM2 specify three cell-state transitional steps leading to arabidopsis stomatal differentiation. The Plant Cell. 2008;20:1775–1785. doi: 10.1105/tpc.108.060848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S-Y, Kim S-G, Kim Y-S, Seo PJ, Bae M, Yoon H-K, Park C- M. Exploring membrane-associated NAC transcription factors in Arabidopsis: implications for membrane biology in genome regulation. Nucleic Acids Research. 2007;35:203–213. doi: 10.1093/nar/gkl1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreps JA, Wu Y, Chang H-S, Zhu T, Wang X, Harper JF. Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiology. 2002;130:2129–2141. doi: 10.1104/pp.008532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn JM, Boisson-Dernier A, Dizon MB, Maktabi MH, Schroeder JI. The protein phosphatase AtPP2CA negatively regulates abscisic acid signal transduction in Arabidopsis, and effects of abh1 on AtPP2CA mRNA. Plant Physiology. 2006;140:127–139. doi: 10.1104/pp.105.070318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B-H, Henderson DA, Zhu J-K. The Arabidopsis cold-responsive transcriptome and its regulation by ICE1. The Plant Cell. 2005;17:3155–3175. doi: 10.1105/tpc.105.035568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung J, Giraudat J. Abscisic acid signal transduction. Annual Review of Plant Physiology and Plant Molecular Biology. 1998;49:199–222. doi: 10.1146/annurev.arplant.49.1.199. [DOI] [PubMed] [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. The Plant Cell. 1998;10:1391–1406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons TJ, Villa NY, Regalla LM, Kupchak BR, Vagstad A, Eide DJ. Metalloregulation of yeast membrane steroid receptor homologs. Proceedings of the National Academy of Sciences, USA. 2004;101:5506–5511. doi: 10.1073/pnas.0306324101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Jin JB, Lee J, Yoo CY, Stirm V, Miura T, Ashworth EN, Bressan RA, Yun D-J, Hasegawa PM. SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. The Plant Cell. 2007;19:1403–1414. doi: 10.1105/tpc.106.048397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiologia Plantarum. 1962;15:473–497. [Google Scholar]
- Pei ZM, Kuchitsu K, Ward JM, Schwarz M, Schroeder JI. Differential abscisic acid regulation of guard cell slow anion channels in Arabidopsis wild-type and abi1 and abi2 mutants. The Plant Cell. 1997;9:409–423. doi: 10.1105/tpc.9.3.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Ishida J, et al. Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. The Plant Journal. 2002;31:279–292. doi: 10.1046/j.1365-313x.2002.01359.x. [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. Gene networks involved in drought stress response and tolerance. Journal of Experimental Botany. 2007;58:221–227. doi: 10.1093/jxb/erl164. [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K, Seki M. Regulatory network of gene expression in the drought and cold stress responses. Current Opinion in Plant Biology. 2003;6:410–417. doi: 10.1016/s1369-5266(03)00092-x. [DOI] [PubMed] [Google Scholar]
- Stockinger E, Gilmour S, Thomashow M. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proceedings of the National Academy of Sciences, USA. 1997;94:1035–1040. doi: 10.1073/pnas.94.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang YT, Tianhua H, Matthew A, Bryan B, Jessica MB, Peter CE, Walter DF. PAQR proteins: a novel membrane receptor family defined by an Ancient7-transmembrane pass motif. Journal of Molecular Evolution. 2005;61:372–380. doi: 10.1007/s00239-004-0375-2. [DOI] [PubMed] [Google Scholar]
- Thomashow MF. PLANT COLD ACCLIMATION: freezing tolerance genes and regulatory mechanisms. Annual Review of Plant Physiology and Plant Molecular Biology. 1999;50:571–599. doi: 10.1146/annurev.arplant.50.1.571. [DOI] [PubMed] [Google Scholar]
- Tran LS, Nakashima K, Shinozaki K, Yamaguchi-Shinozaki K. Plant gene networks in osmotic stress response: from genes to regulatory networks. Methods in Enzymology. 2007;428:109–128. doi: 10.1016/S0076-6879(07)28006-1. [DOI] [PubMed] [Google Scholar]
- Wang P, Song CP. Guard-cell signalling for hydrogen peroxide and abscisic acid. New Phytologist. 2008;178:703–718. doi: 10.1111/j.1469-8137.2008.02431.x. [DOI] [PubMed] [Google Scholar]
- Xiong L, Schumaker KS, Zhu JK. Cell signalling during cold, drought, and salt stress. The Plant Cell. 2002;14:S165–S183. doi: 10.1105/tpc.000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. Characterization of the expression of a desiccation-responsive rd29 gene of Arabidopsis thaliana and analysis of its promoter in transgenic plants. Molecular and General Genetics. 1993;236:331–340. doi: 10.1007/BF00277130. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. The Plant Cell. 1994;6:251–264. doi: 10.1105/tpc.6.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends in Plant Science. 2005;10:88–94. doi: 10.1016/j.tplants.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annual Review of Plant Biology. 2006;57:781–803. doi: 10.1146/annurev.arplant.57.032905.105444. [DOI] [PubMed] [Google Scholar]
- Yamauchi T, Kamon J, Ito Y, et al. Cloning of adiponectin receptors that mediate antidiabetic metabolic effects. Nature. 2003;423:762–769. doi: 10.1038/nature01705. [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nature Protocols. 2007;2:1565–1572. doi: 10.1038/nprot.2007.199. [DOI] [PubMed] [Google Scholar]
- Zhang JZ, Creelman RA, Zhu J- K. From laboratory to field. Using information from Arabidopsis to engineer salt, cold, and drought tolerance in crops. Plant Physiology. 2004;135:615–621. doi: 10.1104/pp.104.040295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Dong C-H, Zhu J-K. Interplay between cold-responsive gene regulation, metabolism and RNA processing during plant cold acclimation. Current Opinion in Plant Biology. 2007;10:290–295. doi: 10.1016/j.pbi.2007.04.010. [DOI] [PubMed] [Google Scholar]
- Zhu J, Verslues PE, Zheng X, et al. HOS10 encodes an R2R3-type MYB transcription factor essential for cold acclimation in plants. Proceedings of the National Academy of Sciences, USA. 2005;102:9966–9971. doi: 10.1073/pnas.0503960102. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhu JK. Salt and drought stress signal transduction in plants. Annual Review of Plant Biology. 2002;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Rice CD, Pang Y, Pace M, Thomas P. Cloning, expression, and characterization of a membrane progestin receptor and evidence it is an intermediary in meiotic maturation of fish oocytes. Proceedings of the National Academy of Sciences, USA. 2003;100:2231–2236. doi: 10.1073/pnas.0336132100. [DOI] [PMC free article] [PubMed] [Google Scholar]