Abstract
The mechanisms underlying hepatitis C virus (HCV) resistance to type 1 interferon (IFN) are not well understood. The recent development of a cell culture model of HCV infection allows analysis of host-virus interactions, including IFN resistance, in the context of the complete virus life cycle. In this report we show that HCV strongly triggers the phosphorylation of PKR and eIF2α as it expands in infected cells. In addition, HCV infection attenuates the induction of interferon-stimulated gene (ISG) protein expression despite normal induction of ISG mRNAs. We also show that ISG protein induction is restored to normal levels and the antiviral effect of IFN is enhanced when PKR expression is down-regulated by shRNA in IFN-treated infected cells. These results suggest that the ability of HCV to activate PKR may, paradoxically, be advantageous for the virus during an IFN response by preferentially suppressing the translation of ISGs.
Introduction
Hepatitis C virus (HCV) is a major human pathogen. Over 170 million people are chronically infected, many of whom will develop chronic liver disease and hepatocellular carcinoma (Alter and Seeff, 2000). There is no vaccine against HCV and the most widely used therapy, type I interferon (IFN) combined with ribavirin, is successful in only a fraction of chronically infected patients and it has toxic side effects (Patel and McHutchison, 2004).
HCV, the sole member of genus Hepacivirus within the Flaviviridae family (Maniloff, 1995), is an enveloped, single-stranded, positive-sense RNA virus (Choo et al., 1991). The HCV genome contains a long open reading frame (ORF) that encodes a single polyprotein of approximately 3000 amino acids (Choo et al., 1991). The ORF is flanked by 5’ and 3’ nontranslated regions (NTR) that contain essential sequences for RNA translation and replication (Friebe et al., 2005; Friebe et al., 2001; Honda et al., 1999). Polyprotein translation is driven by a highly structured internal ribosome entry site (IRES) located in the 5’ NTR (Honda et al., 1999). The polyprotein is co- and post-translationally processed by cellular and viral proteases leading to the expression of the structural (Core, E1 and E2) and non-structural proteins (p7, NS2, NS3, NS4A, NS4B, NS5A and NS5B) (Penin et al., 2004).
Type I interferons (IFNα/β) are produced in response to many virus infections and they induce a variety of IFN-stimulated genes (ISGs) (Goodbourn et al., 2000) some of which have antiviral activity (Samuel, 2001). Using the recently developed HCV JFH1 in vitro infection system (Lindenbach et al., 2005; Wakita et al., 2005; Zhong et al., 2005), we and others have shown that HCV efficiently blocks double-stranded RNA signaling by NS3/4A-dependent and -independent mechanisms (Cheng et al., 2006; Foy et al., 2005; Li et al., 2005), thereby preventing the production of type I IFN by the infected cell. Nevertheless, in vivo studies in experimentally infected chimpanzees (Hoofnagle, 2002; Su et al., 2002) and naturally HCV-infected humans (Alter and Seeff, 2000) have demonstrated that HCV infection strongly induces the expression of ISG mRNAs in the liver. However, HCV persists in the liver despite the induction of these ISGs (Alter and Seeff, 2000), raising the possibility that HCV can block the effector function of the ISGs in the infected cells.
The mechanisms underlying HCV resistance to IFN are not well understood. Previous attempts to answer these questions used systems, e.g. subgenomic replicons and viral protein over-expression, that reproduce only isolated aspects of the HCV viral cycle. Nonetheless, these studies yielded a list of candidate resistance mechanisms, including inhibition of Jak-STAT signaling by several HCV proteins, induction of interleukin 8 expression by NS5A, induction of SOCS-3 signaling by HCV core protein, transcriptional suppression of ISGs by HCV core protein and repression of PKR protein kinase by HCV NS5A and E2 proteins and by the IRES element of HCV (reviewed in Wohnsland et al., 2007).
The recently developed HCV cell culture infection system (Lindenbach et al., 2005; Wakita et al., 2005; Zhong et al., 2005) permits analysis of all the steps in the HCV life cycle, including its IFN resistance mechanisms, in a more physiological context. In this study, we tested the hypothesis that HCV evades the antiviral effect of IFN by blocking its effector functions downstream of the ISG mRNAs. We discovered that, although HCV does not block the IFN-induced ISG mRNA transcription, it strongly suppresses ISG protein expression and global cellular protein synthesis at the same time that it strongly induces the phosphorylation of PKR and eIF2α. Importantly, ISG protein expression is restored and the antiviral effect of IFN enhanced in PKR-down-regulated cells, suggesting that by inducing PKR phosphorylation HCV inhibits the production of antiviral ISG proteins in infected cells.
Results
HCV infected cells are less responsive to type I IFN than JFH-1 full-length stable replicon cells
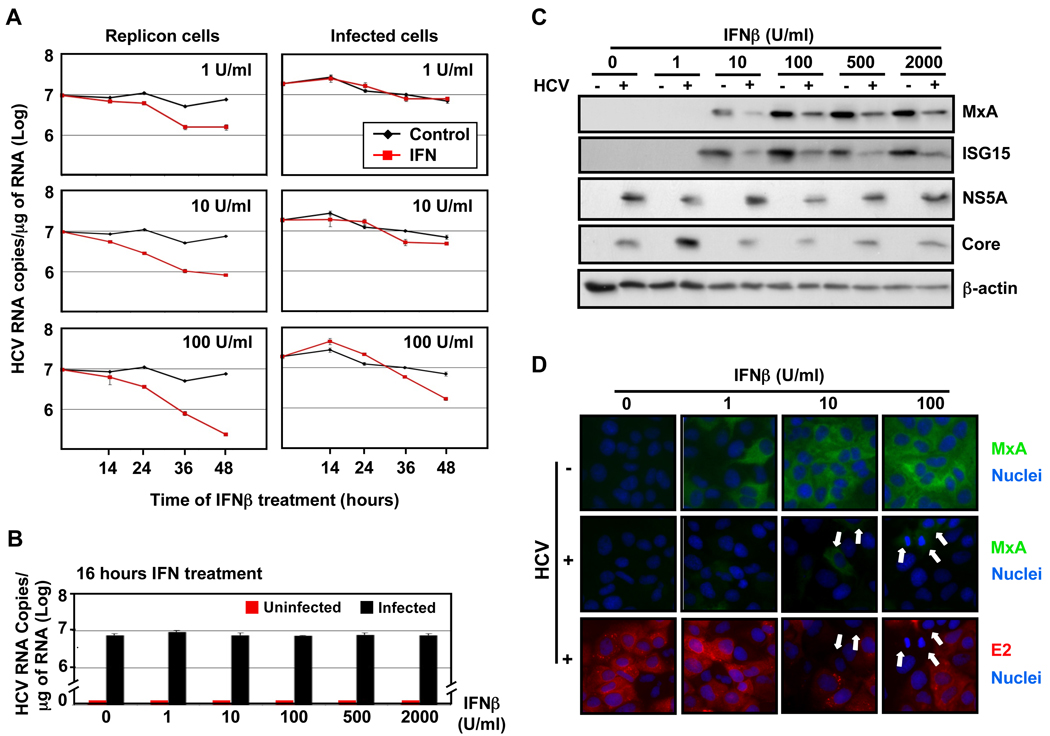
We examined the antiviral effect of type I IFN against HCV in JFH-1 full-length stable replicon cells and persistently infected cells by analyzing their intracellular HCV RNA content at various times after IFNβ treatment. As shown in Fig. 1A, HCV RNA levels were strongly reduced in JFH-1 stable replicon cells as early as 24 hours after administration of doses as low as 1 IU/ml of IFN. In contrast, the intracellular HCV RNA content of persistently infected cells was unchanged until 48 hour after administration of 100 IU/ml of IFN. These results confirm previous reports demonstrating the high susceptibility of HCV replicon systems to type I IFN treatment (Guo et al., 2001) and illustrate that HCV-infected cells are quite significantly more resistant to IFN than stable full-length replicon cells.
Fig. 1. Suppression of ISG protein induction in HCV-infected cells.
(A) Persistently infected Huh-7 cells and JFH-1 full-length stable replicon cells were treated with different doses of IFNβ as indicated in the figure, and at various times thereafter the intracellular HCV RNA and GAPDH mRNA (for normalization) were quantified by RT-qPCR. Data are represented as Mean ± AVEDEV; n=3. This experiment is representative of 2 independent experiments. (B–D) Huh-7 cells were infected with JFH-1 virus at moi=0.2 and at day 5 post-infection uninfected (−) and infected (+) cells were treated for 16 hours with different doses of IFNβ. (B) Quantification of the intracellular HCV RNA by RT-qPCR in total RNA isolated from HCV-infected or uninfected cells. GAPDH mRNA quantification was used for normalization. Data are represented as Mean ± AVEDEV; n=3. This experiment is representative of 3 independent experiments. (C) Analysis by Western-blotting of ISG (MxA and ISG15) and viral (NS5A and core) protein expression induced by IFNβ in cell extracts of HCV-infected or uninfected cells. β-actin expression was examined as protein loading control. Each sample represented a pool of three replicas. (D) Immunofluorescence analysis of MxA (in green) and viral E2 (in red) protein expression in uninfected and HCV-infected cells after IFNβ treatment. The nuclei were stained by Hoechst solution (blue). White arrows in HCV-infected panels point to cells with high MxA expression levels and with no or very low E2 staining. These images are representative of three different experiments.
IFN-stimulated protein induction is reduced in HCV infected cells
To understand the basis of the relative resistance of infected cells to IFN, we examined the impact of HCV on the cellular response to type I IFN by analyzing the accumulation of IFN-induced antiviral effector proteins in infected and uninfected Huh-7 cells. Huh-7 cells were infected with JFH-1 at low multiplicity of infection (moi=0.2). Five days later, IFNβ was added and cellular extracts were analyzed for HCV RNA and for ISG (i.e. MxA and ISG15) and viral (i.e. NS5A and core) proteins 16 hours later by RT-qPCR and Western-blotting, respectively. As shown in Figs. 1B and 1C, IFNβ had little or no impact on intracellular HCV RNA and HCV core and NS5A protein levels during the 16 hours of the experiment. However, HCV-infected cells produced much less MxA and ISG15 protein than HCV-uninfected cells at all IFNβ doses studied (Fig. 1C). Similar results were obtained when Huh-7 (S. Fig. 1A) or Huh-7.5.1 (S. Fig. 1B) cells were infected at high multiplicity of infection and treated with IFNβ at 72 hours post-infection, or when persistently infected Huh-7 cells were treated with IFNβ (S. Fig. 1C) or IFNα (S. Fig. 1D). In contrast, ISG protein expression was strongly induced by IFNβ in both Con-1 (genotype 1b) and JFH-1 (genotype 2a) full-length stable replicon cells compared to their corresponding cured cells (S. Fig 1E) underlining the differential IFN sensitivity of replicon and infected cells. Collectively, these results indicate that HCV attenuates the expression of IFN inducible proteins in infected Huh-7 cells.
Next, we analyzed the differences in ISG protein accumulation observed in Fig. 1C and S. Fig. 1 at the single cell level by immunofluorescence of uninfected and HCV-infected IFNβ-treated Huh-7 cells. Cells were fixed and co-stained for MxA (in green) and viral E2 (in red) protein expression. As shown in Fig. 1D, all IFNβ–treated uninfected cells displayed homogeneous MxA staining while only rare cells in the IFNβ–treated infected cultures were MxA positive (white arrows in Fig. 1D), and these cells were HCV E2 negative. In contrast, the HCV E2-positive (i.e. infected) cells were MxA-negative. Collectively, these results confirm, at the single cell level, that ISG protein expression is attenuated in HCV-infected Huh-7 cells.
HCV does not block transcription or nucleo-cytoplasmic transport of ISG mRNAs
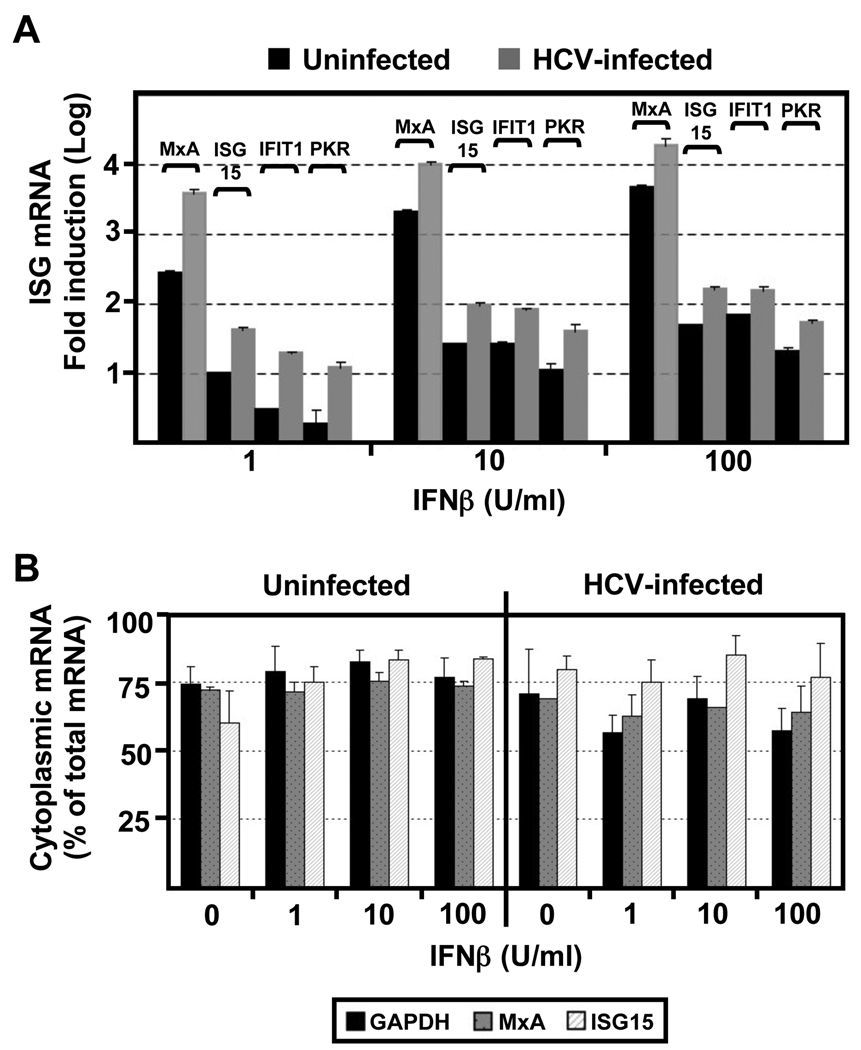
The foregoing results could reflect defects at the level of IFN signaling, ISG mRNA transcription, nucleo-cytoplasmic transport, stability, translation, or ISG protein stability. To determine if HCV blocks either IFN signaling or ISG mRNA transcription or stability, we treated HCV-infected and uninfected Huh-7 cells with different doses of IFNβ and monitored the ISG mRNA level 16 hours later by RT-qPCR. As shown in Fig. 2A, and consistent with our previous results (Cheng et al., 2006), all tested ISG mRNAs (MxA, ISG15, IFIT1 and PKR) were induced in HCV-infected cells at least as much as in uninfected cells, indicating that HCV does not suppress type 1 IFN signaling or ISG mRNA transcription or stability in infected Huh-7 cells. To determine if HCV blocks the nucleo-cytoplasmic transport of the ISG mRNAs, we compared the cytoplasmic and nuclear levels of MxA, ISG15 and GAPDH mRNA in uninfected and HCV-infected IFN-treated Huh-7 cells. As shown in Fig. 2B, HCV does not alter the nucleo-cytoplasmic distribution of these ISG mRNAs. Collectively, these results demonstrate that HCV attenuates the induction of ISG proteins post-transcriptionally in infected cells.
Fig. 2. The accumulation and nucleo-cytoplasmic distribution of ISG mRNAs is normal in HCV-infected cells.
Huh-7 cells were infected with JFH-1 virus at moi=0.2. At day 5 post-infection uninfected and HCV-infected cells were treated with different doses of IFNβ for 16 hours. (A) Quantification of ISG (MxA, ISG15, IFIT1 and PKR) mRNA levels by RT-qPCR in total RNA samples isolated from uninfected and HCV-infected cells. GAPDH mRNA quantification from the same samples was used for normalization. Results display the average fold induction for each ISG mRNA relative to its expression level in untreated uninfected Huh-7 cells (Mean ± AVEDEV; n=3). This experiment is representative of 3 independent experiments. (B) Quantification of MxA, ISG15 and GAPDH mRNAs in the nuclear and cytoplasmic fractions of uninfected and HCV-infected cells. Results displayed as percentage of cytoplasmic versus total mRNA content for each sample. Each bar in the graph corresponds to the average and the average deviation (Mean ± AVEDEV; n=3) of three replicas.
Global cellular protein synthesis is reduced in IFN-treated HCV-infected cells
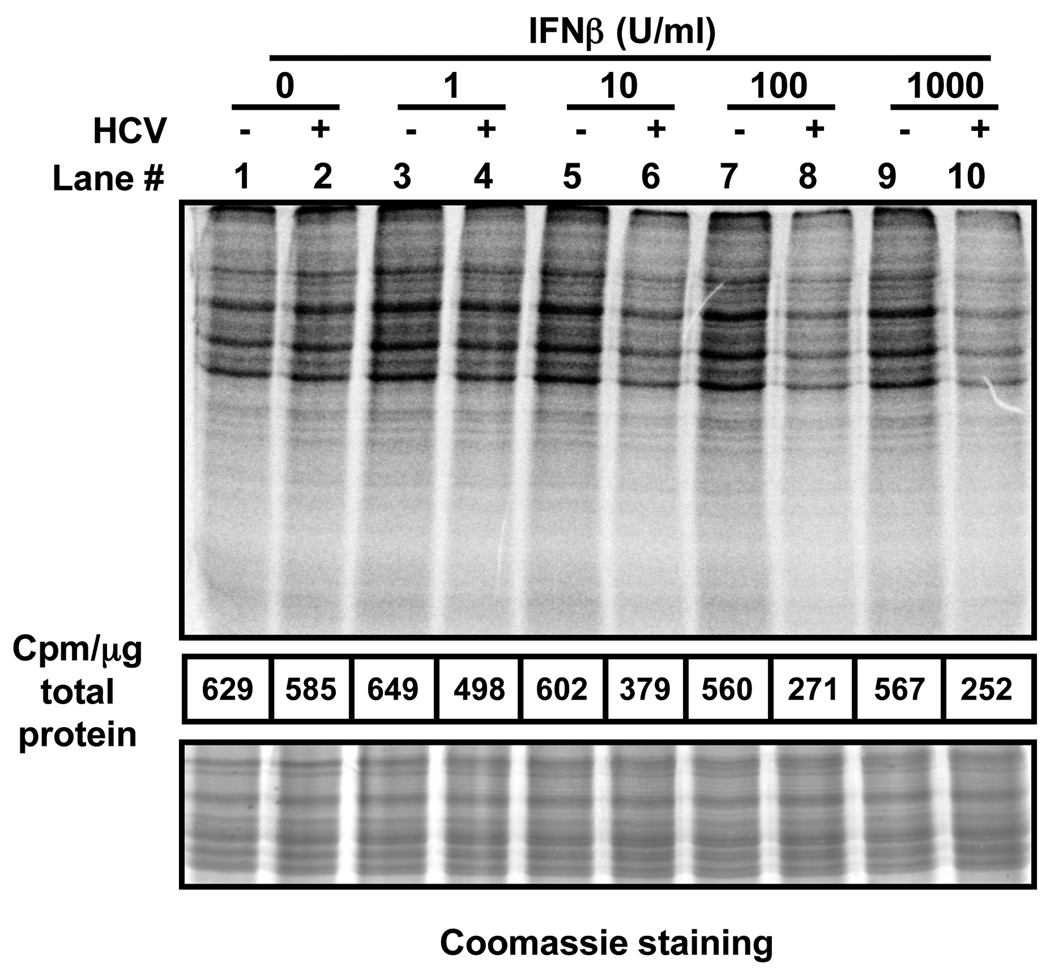
The lack of an effect on the steady state content of ISG mRNA (Fig. 2) suggests that HCV could block the induction of ISG proteins (Fig. 1) at the translational level. To test that hypothesis, metabolic pulse labeling experiments were carried out. As described in Fig. 1, Huh-7 cells were inoculated with JFH-1 at low multiplicity of infection (moi=0.2) and 5 days later, infected and uninfected cells were treated with different concentrations of IFNβ. Sixteen hours later, the cells were pulse-labeled for 1h with 35S-Met and cellular extracts were analyzed by electrophoresis and phosphorimaging. As shown in Fig. 3, HCV had little or no effect on global protein synthesis (compare lanes 1 and 2). Similarly, IFNβ had no effect in uninfected Huh-7 (compare lane 1 with lanes 3, 5, 7 and 9). However, cellular protein synthesis was clearly reduced in the IFN-treated HCV-infected cells (compare lane 2 with lanes 6, 8 and 10) and this was confirmed by quantitation of cellular protein radioactivity after methanol-chloroform precipitation of the samples (middle panel). Similar results were obtained when IFNα was used instead of IFNβ (S. Fig. 2). Time of addition experiments revealed that the impact of IFN on global protein synthesis in HCV infected cells increased as a function of time and the extent of HCV infection (S. Table 1). In addition, these experiments also revealed that HCV infection itself had a slight negative impact on general protein synthesis in the absence of IFN. Collectively, these results suggest that HCV slightly reduces de novo protein synthesis in infected Huh-7 cells and this effect is greatly enhanced in the presence of IFN. The mechanism responsible for these interesting results was investigated in the following experiments.
Fig. 3. Global cellular protein synthesis is inhibited in IFNβ-treated HCV-infected cells.
Huh-7 cells were infected with JFH-1 virus at moi=0.2. At day 5 post-infection uninfected (−) and HCV-infected (+) cells were treated with different doses of IFNβ for 16 hours, after which the cultures were metabolically labeled for 1 hour with 35S-Met. Protein extracts prepared in RIPA buffer were quantified by BCA. Equivalent amounts from each sample were analyzed by SDS-PAGE, Coomassie staining (bottom panel) and autoradiography (top panel). The figure is representative of 2 independent experiments with 2 replicas per sample. The radioactivity present after methanol-chloroform (4:1) precipitation of each sample was quantified and the results were displayed as counts per minute (cpm) per microgram of total protein (middle panel).
HCV infection triggers PKR and eIF2α protein phosphorylation
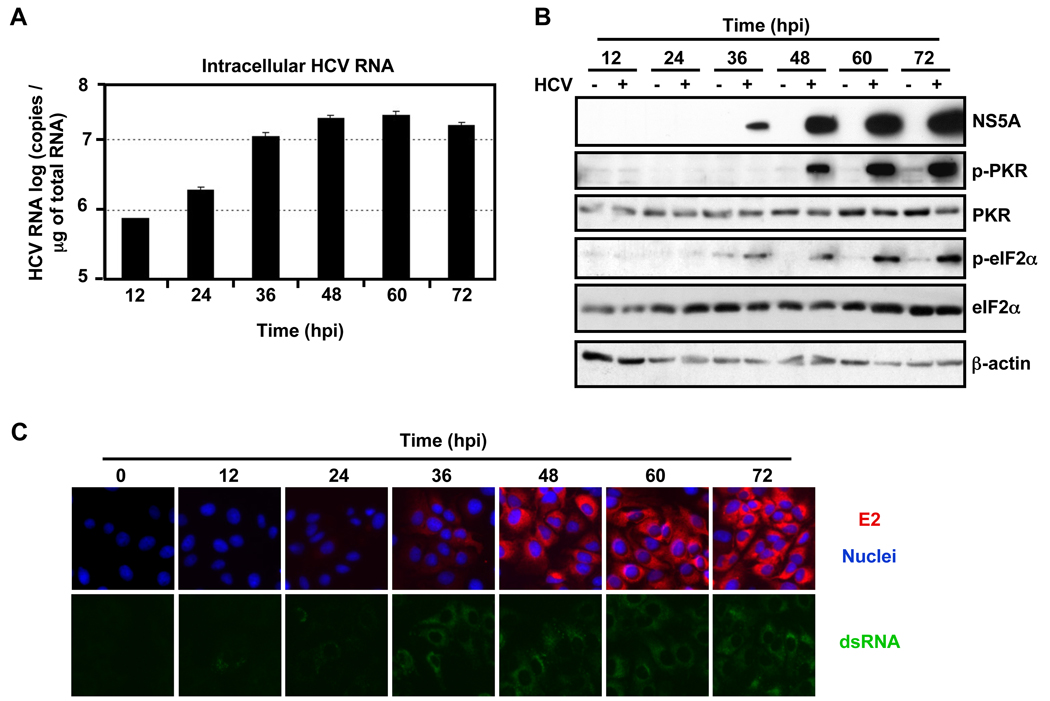
Cellular protein synthesis is tightly regulated at the level of initiation, elongation and termination. The eukaryotic initiation factor 2 (eIF2) is a key regulator of translation initiation in multiple settings, including viral infection (Hershey, 1991). When eIF2α is phosphorylated by kinases such as PKR, PERK, GCN2, and HRI, translation initiation and, therefore, protein synthesis is inhibited (reviewed in Garcia et al., 2006). Kang et al have recently reported that transfection of replication competent HCV JFH-1 RNA induces PKR and eIF2α phosphorylation (Kang et al., 2009). Given the inhibition of protein synthesis we observed during HCV infection (S. Table 1) and in IFN treated HCV-infected cells (compare lanes 6, 8 and 10 to lanes 5, 7 and 9 in Fig. 3), we examined the phosphorylation status of eIF2α and its kinases during HCV infection. Huh-7 cells were infected at a high multiplicity of infection (moi=3) and cellular extracts were analyzed by Western-blotting at different times after infection. As shown in Fig. 4A, intracellular HCV RNA expanded rapidly during the infection and it was accompanied by a corresponding increase in viral E2 (Fig. 4C), NS5A (Fig. 4B) and core (S. Fig. 3A) protein accumulation which, collectively, reflect active viral replication via double-stranded RNA (dsRNA) replicative intermediates (Fig. 4C). Interestingly, HCV infection strongly triggered the phosphorylation of the dsRNA-dependent protein kinase (PKR) and eIF2α proteins (Fig. 4B), but not PERK or GCN2 (S. Fig. 3B), suggesting that PKR is responsible for the observed eIF2α phosphorylation. In addition, PKR and eIF2α proteins were also highly phosphorylated in HCV-infected Huh-7.5.1 cells and in Huh-7 cells that were treated with neutralizing IFNα/β receptor antibodies, suggesting that HCV-induced PKR phosphorylation does not reflect the induction type I IFN induction in the infected cells (S. Fig. 3C and 3D).
Fig. 4. HCV infection triggers the phosphorylation of PKR and eIF2α proteins.
Huh-7 cells were infected (+) or not (−) with JFH-1 d183 virus at moi=3. (A) At the indicated time points post-inoculation, total RNA was isolated from HCV-infected or uninfected cells and the intracellular HCV RNA levels were quantified by RT-qPCR. GAPDH mRNA quantification was used for normalization. Data are represented as Mean ± AVEDEV; n=3. (B) At the indicated time points cellular extracts were prepared in RIPA buffer, their protein content was quantified by BCA and the accumulation of cellular eIF2α , phospho-eIF2α (p-eIF2α), PKR, phospho-PKR (p-PKR) and viral NS5A proteins was analyzed by Western-blotting.β-actin expression was examined as protein loading control. The results displayed are representative of 3 independent experiments. (C) Cultures infected in parallel were fixed at the indicated time points and processed for immunofluorescence for the detection of viral E2 (in red) or dsRNA (in green). The nuclei were stained by Hoechst solution (blue).
Since HCV triggered PKR and eIF2α phosphorylation, it was not surprising that global cellular protein synthesis slightly decreased during HCV infection (S. Table 1). Nonetheless, the HCV core (S. Fig. 3A), E2 and NS5A (Fig. 4C and 4B respectively) protein content increased progressively in the infected cells despite the reduction in cellular protein synthesis, indicating that HCV protein synthesis is relatively insensitive to the inhibitory effects of phosphorylated eIF2α , as previously reported (Robert et al., 2006; Shimoike et al., 2009; Terenin et al., 2008).
HCV increases PKR and eIF2α phosphorylation in IFN-treated cells
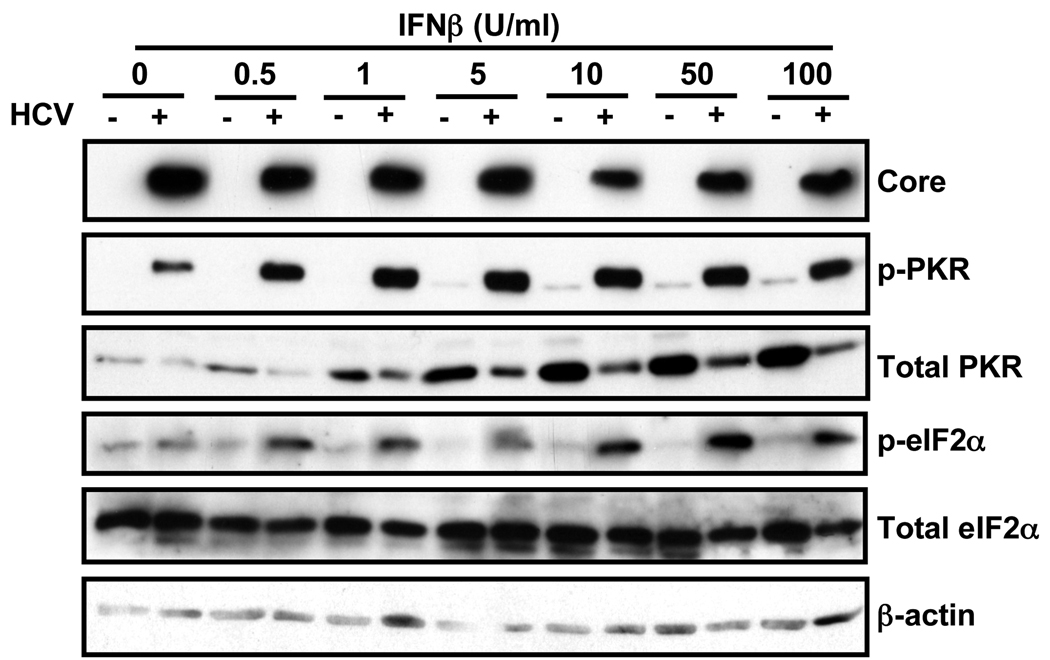
Although PKR expression is induced by IFN, its activation requires binding to dsRNA and subsequent autophosphorylation (Thomis and Samuel, 1993). To determine if the inhibition of global protein synthesis we observed in IFN-treated HCV-infected cells (Fig. 3) was due to hyperphosphorylation of PKR and eIF2α, uninfected and HCV-infected Huh-7 cells were treated with a wide range of IFNβ concentrations for 16 hours at which time cellular extracts were prepared and the levels of viral core protein and cellular eIF2α and PKR proteins and their phosphorylated forms were determined by Western-blotting. As shown in Fig. 5, IFN had little or no effect on viral core and total eIF2α protein levels. In contrast, IFN strongly induced the accumulation of PKR protein, a well-known ISG (Samuel, 1991), in uninfected cells. Importantly, PKR protein induction was greatly attenuated in the infected cells, similar to the attenuated induction of MxA and ISG15 protein accumulation shown in Fig. 1. Nonetheless, reflecting the dsRNA content of the infected cells, PKR- and, therefore, eIF2α-phosphorylation were much more strongly induced in IFN-treated HCV-infected cells than in uninfected cells.
Fig. 5. HCV infection strongly enhances phosphorylation of PKR and eIF2α proteins in IFNβ-treated cells.
Huh-7 cells were infected with JFH-1 virus at moi=0.2. At day 5 post-infection uninfected (−) and infected (+) cells were treated for 16 hours with different doses of IFNβ. Total cellular protein in extracts prepared in RIPA buffer was quantified by BCA. Equivalent amounts from each sample were subjected to Western-blot analysis for the detection of viral core and cellular eIF2α, p-eIF2α, PKR and p-PKR proteins. β-actin expression was examined as protein loading control. The figure is representative of 2 independent experiments with 3 replicas per sample.
HCV infection kinetics are unaffected by down-regulation of PKR expression
As shown in Fig. 4 and S. Fig. 3, HCV protein and RNA expansion proceeded in parallel with the phosphorylation of PKR and eIF2α in infected cells despite the fact that total protein and ISG protein synthesis were suppressed (S. Table 1). To confirm the relative insensitivity of HCV to translational regulation by phosphorylated PKR, we examined the impact of PKR down-regulation on the kinetics and magnitude of HCV infection. We first generated Huh-7 cells in which PKR expression was down-regulated by transduction with lentivirus vectors that express short hairpin RNAs (shRNAs) that specifically target PKR. As controls, naïve Huh-7 cells and Huh-7 cells that were co-transduced with lentivirus vectors that express GFP and shRNAs specific against GFP were used. After verifying that total PKR and GFP protein content was down-regulated (S. Fig. 4A), naïve, PKR- or GFP- down-regulated Huh-7 cells were infected with JFH-1 virus at high multiplicity of infection (moi=5) and the extracellular infectivity and intracellular HCV RNA content were determined at various times thereafter. The kinetics of both extracellular infectivity (Fig. 6A) and intracellular HCV RNA (S. Fig. 4B) in PKR down-regulated cells were virtually that same as in GFP- down- regulated or naïve Huh-7 cells. These kinetic results indicate that HCV infection is not altered by reduced levels of PKR, indicating that HCV is resistant to the translational inhibitory effects of the phosphorylated forms of PKR and eIF2α that it induces during infection.
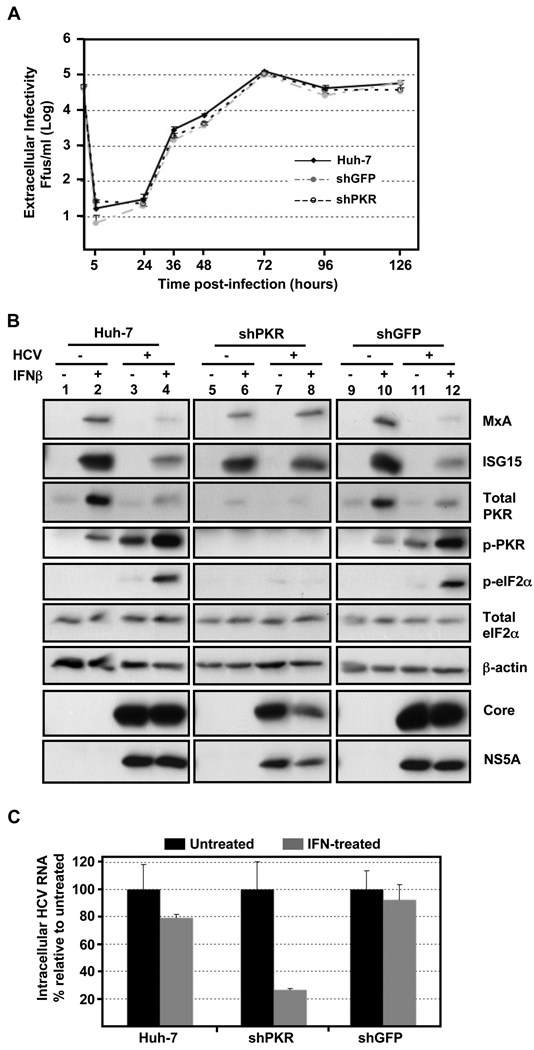
Fig. 6. PKR downregulation does not affect HCV infection kinetics, restores HCV-induced ISG protein suppression and enhances the antiviral effect of IFN.
Huh-7 cells were transduced with lentiviral vectors expressing GFP and shRNAs against GFP or PKR. (A) PKR-, GFP-down-regulated and Huh-7 cells, used as controls, were infected with JFH-1 d183 virus at moi=5 and the extracellular infectivity was determined at various times thereafter. Data are represented as Mean ± AVEDEV; n=3. The graph is representative of 3 independent experiments. (B) Transduced and control cells were infected (+) or not (−) with JFH-1 d183 virus at moi=5. At 70 hpi the cultures were treated (+) or not (−) with 100 U/ml of IFNβ for 20 hours, after which cellular extracts were prepared in RIPA buffer. Equivalent amounts of total protein were analyzed by Western-blotting for core, NS5A, MxA, ISG15, PKR, p-PKR, eIF2α and p-eIF2α proteins. β-actin expression was examined as protein loading control. (C) The HCV RNA present in samples generated in parallel was extracted and analyzed by RT-qPCR. GAPDH mRNA quantification of the same samples was used for normalization. IFN-treated samples are represented by grey bars and untreated controls by black bars. The results are displayed as Mean ± AVEDEV; n=3. Panels are representative of 3 independent experiments with 3 replicas per sample.
PKR downregulation restores ISG protein expression in HCV-infected cells and enhances the antiviral effect of IFN
In view of the foregoing, we hypothesized that, by activating PKR, HCV triggers the phosphorylation of eIF2α and, thereby, inhibits the synthesis of interferon-stimulated antiviral proteins. To demonstrate that HCV inhibits ISG protein synthesis by inducing PKR activation, Huh-7 cells were transduced with the same lentiviruses described in Fig. 6A and PKR- or GFP- down-regulated and control Huh-7 cells were infected at high multiplicity of infection (moi=5) six days later. At the peak of the infection (70 hpi), the cells were treated with 100 U/ml of IFNβ for 20 hours when cellular extracts were prepared for protein analysis by Western-blotting. As shown in Fig. 6B, MxA, ISG15 and PKR protein expression was strongly induced by IFN in non-transduced or GFP shRNA-transduced, HCV-uninfected Huh-7 cells (lanes 2 and 10, respectively), in contrast to HCV infected cells in which they were much less strongly induced (lanes 4 and 12), reflecting the corresponding phosphorylation of PKR and eIF2α proteins. As expected, neither PKR, phospho-PKR or phospho-eIF2α was detectable under any conditions in the PKR down-regulated cells (lanes 9–12). In the absence of PKR, however, the magnitude of IFN-stimulated MxA and ISG15 protein expression in HCV-infected cells was restored to the level in uninfected cells (compare lanes 8 and 6, respectively) without a corresponding change in mRNA (data not shown), suggesting that the HCV-induced PKR and eIF2α phosphorylation was responsible for the suppression of ISG protein accumulation shown in Fig. 1 and Fig 6B (lanes 4 and 12). Interestingly, while the level of viral core and NS5A proteins was not altered by IFN in Huh-7 and GFP down-regulated cells, both viral proteins were suppressed in IFN-treated PKR down-regulated cells suggesting that the antiviral effect of IFN is increased in cells with reduced levels of PKR.
To further investigate the role of PKR downregulation on the antiviral effect of IFN infected GFP-, PKR-down-regulated and Huh-7 cells were treated with 100 U/ml of IFN and intracellular HCV RNA levels were determined by RT-qPCR 20 hours later. As expected from the low moi infection experiments shown in Fig. 1A, IFN treatment had minimal (10–20%) impact on the HCV RNA content of naïve and GFP shRNA-transduced Huh-7 cells (Fig. 6C). In contrast, IFN treatment reduced the HCV RNA content of PKR deficient cells by ~70–80%, suggesting that the increased ISG protein expression in those cells (Fig. 6B, lane 12) mediated the antiviral effects of IFN.
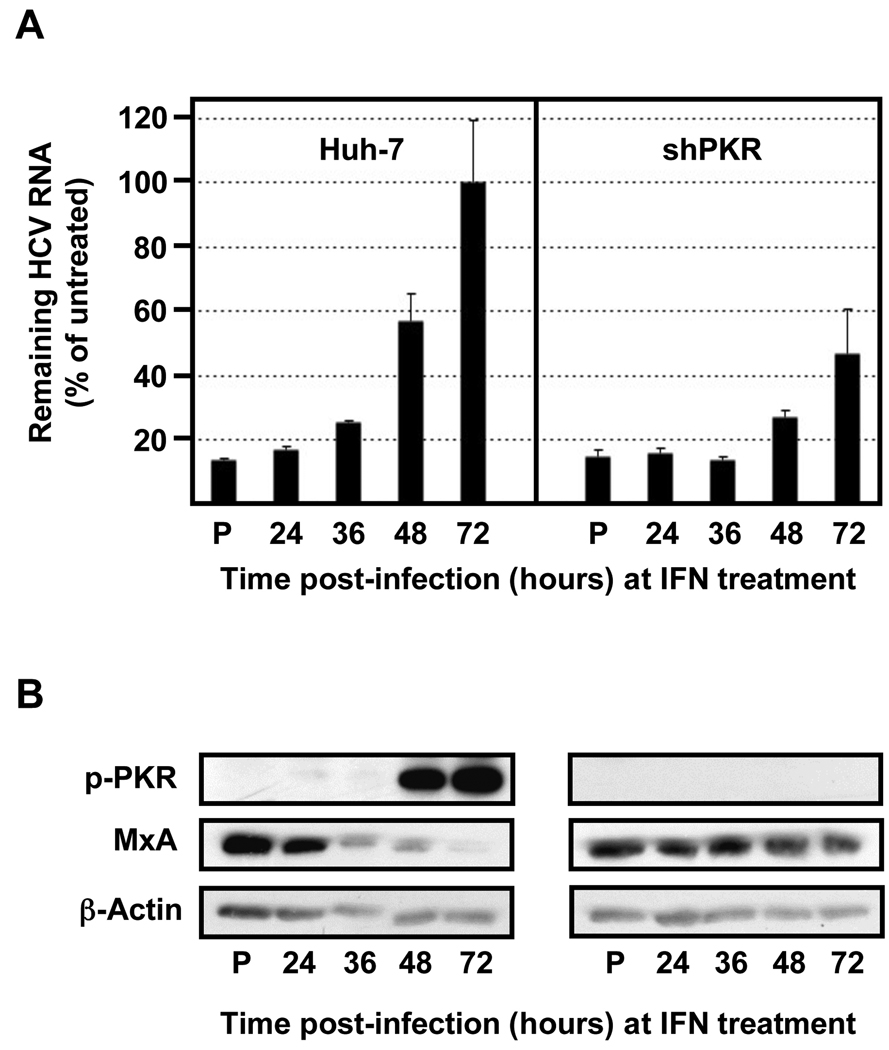
Interestingly, as shown in Fig. 7, IFN time of addition experiments in infected PKR down-regulated and control Huh-7 cells revealed a direct correlation between the degree of PKR phosphorylation and HCV resistance to IFN and an inverse correlation between PKR phosphorylation and MxA protein accumulation, used as a read out for ISG protein expression. Collectively, these results suggest that HCV-induced PKR phosphorylation is responsible for the reduced levels of ISG proteins in infected cells and for the resistance of HCV to the antiviral effects of IFN.
Fig. 7. The HCV resistance to IFN correlates with PKR phosphorylation and is inversely related to ISG protein expression.
Huh-7 cells were transduced with lentiviral vectors expressing shRNAs against PKR, as indicated in Figure 6. Transduced and control cells were pre-treated with 100 U/ml of IFN (P) or remain untreated (the rest). After 16 hours all the cells were infected with JFH-1 d183 virus at moi=3 and at the indicated times post infection cells were treated with 100 U/ml of IFNβ for 20 hours, after which cellular extracts were prepared for RNA and protein analysis. (A) The HCV RNA present in those samples was analyzed by RT-qPCR. The quantification of GAPDH mRNA was used for normalization. Data are displayed as percentage of remaining HCV RNA in IFN-treated samples relative to that in the untreated ones at each time point. Bars in the graph represent the Mean ± AVEDEV; n=3. The graph is representative of 2 independent experiments with 3 replicas per sample. (B) Equivalent amounts of total protein from samples generated in parallel were analyzed by Western-blotting for MxA and p-PKR protein detection. β-actin expression was examined as protein loading control. Panels are representative of 2 independent experiments.
Discussion
One of the main characteristics of HCV is its tendency to persist despite the massive type 1 IFN stimulated gene (ISG) expression response it induces in the liver (Alter and Seeff, 2000; Hoofnagle, 2002; Lanford et al., 2007; Su et al., 2002). This is consistent with the fact that IFN monotherapy is ineffective in the vast majority of HCV-infected patients (Deutsch and Hadziyannis, 2008; McHutchison et al., 1998). Moreover, a large proportion of patients are also resistant to treatment with massive doses of pegylated interferon plus ribavirin (Fried et al., 2002; Hadziyannis et al., 2004; McHutchison et al., 1998; Miglioresi et al., 2003; Sjogren et al., 2007). A desire to understand the molecular basis for the resistance of HCV to the interferon stimulated antiviral response it induces in the infected liver motivated the experiments described in this report.
In this study, we compared the response of HCV-infected and -uninfected Huh-7 cells to type 1 interferon in the in vitro JFH-1 infection system in order to understand the mechanism(s) whereby HCV resists IFN. We showed that IFN strongly induces ISG mRNA transcription in HCV-infected cells, reaching even higher levels than in uninfected cells, indicating that HCV does not block IFN signaling. These results differ from the negative impact of individually over-expressed HCV proteins on IFN-induced Jak-Stat pathway activation and ISG mRNA transcription (reviewed in Wohnsland et al., 2007), perhaps reflecting the more physiological protein expression levels achieved in the context of the complete viral life cycle in our studies. However, the current results confirm previous observations showing comparable induction of the IFN-stimulated response element (ISRE)-promoter in HCV-infected and -uninfected cells (Cheng et al., 2006), and they are consistent with the strong ISG mRNA induction observed in HCV-infected livers from naturally infected humans (Alter and Seeff, 2000) and experimentally infected chimpanzees (Hoofnagle, 2002; Lanford et al., 2007; Su et al., 2002).
Unexpectedly, the current results demonstrate that not only IFNβ- but also IFNα-induced ISG protein expression is attenuated in HCV-infected cells. Importantly, IFN time of addition experiments revealed that maximal suppression of ISG protein accumulation was achieved when IFN was added at the peak of HCV infection rather than early in the infection, as previously reported (Erickson et al., 2008; Kang et al., 2009), presumably because early IFN addition inhibits HCV infection sufficiently to prevent the induction of PKR phosphorylation. These results suggest that a minimum level of HCV replication is needed to trigger PKR phosphorylation and suppression of ISG protein expression.
Since our results indicate that HCV does not block transcription or nucleo-cytoplasmic transport of ISG mRNAs we performed pulse-labeling experiments to determine if HCV inhibits the translation or the stability of ISG proteins. Pulse labeling experiments demonstrated that HCV slightly inhibits global cellular protein synthesis, that this inhibition is greatly enhanced in IFN-treated cells (both IFNβ and IFNα) and that both effects occur in parallel with the strong phosphorylation of PKR and eIF2α in HCV infected cells. In agreement with these results, Kang et al. have recently shown that electroporation of a replication-competent HCV JFH-1 RNA genome triggered the phosphorylation of PKR and eIF2α proteins in Huh-7 cells (Kang et al., 2009). In contrast to these findings, others have shown that the HCV NS5A and E2 proteins inhibit PKR activation (Gale et al., 1998; Gale et al., 1997; Taylor et al., 1999) and mediate IFN-resistance in subgenomic replicon systems (Sumpter et al., 2004). We suggest that these discrepancies probably reflect intrinsic differences of the experimental systems used in each study (e.g. genotype 1a and 1b replicons vs genotype 2a infectious virus), since we showed that the induction of ISG proteins by IFN treatment is not suppressed in Con-1 and JFH-1 full-length stable replicon cells. We suggest that the results imply that replicon models do not represent the whole spectra of viral-host interactions that occur during HCV infection or that they reflect the elimination of phospho-PKR-inhibitable clones during the replicon clonal selection process. Nonetheless, the results reported herein don’t dispute the PKR-inactivating potential of NS5A and E2. Indeed, the ability of HCV to induce PKR phosphorylation in our system might have been even stronger in the absence of the attenuating potential of NS5A and E2. They do, however, argue against the hypothesis that NS5A and E2 function as viral evasion proteins since our results show that suppression of PKR phosphorylation enhances the antiviral effect of IFN against HCV, at least in the current JFH1 cell culture infection system.
PKR activation, through eIF2α phosphorylation and the inhibition of translation, has been shown to inhibit a variety of other viral infections and to mediate some of the antiviral effects of IFN (Garcia et al., 2006; Samuel, 2001). To counteract this antiviral effect many RNA viruses inhibit the activation of PKR (Garcia et al., 2006). In contrast, HCV, and Sindbis virus (a member of the alphavirus family) strongly activate PKR and eIF2α phosphorylation, but have evolved eIF2α-independent mechanisms to initiate translation (Robert et al., 2006; Shimoike et al., 2009; Terenin et al., 2008; Ventoso et al., 2006). Consistent with these observations, our results indicate that HCV expansion is unaffected by reduced levels of PKR, since downregulation of PKR did not alter significantly HCV infection kinetics.
Importantly, IFN-mediated ISG protein expression was restored if PKR protein production and, thus, PKR and eIF2α phosphorylation was suppressed in HCV infected cells. ISG protein restoration correlated with an increase in the antiviral effect of IFN, suggesting that the antiviral effects of IFN against HCV are dependent on the expression of antiviral ISGs other than PKR. The fact that HCV-IRES translation has been shown to be relatively resistant to eIF2α phosphorylation (Shimoike et al., 2009; Terenin et al., 2008) and to the antiviral effect of IFN (Koev et al., 2002; Terenin et al., 2008) compared to other viral IRES and cap-dependent translation (Koev et al., 2002; Terenin et al., 2008), makes the activation of a classical antiviral factor, i.e. PKR, paradoxically a potential survival mechanism for HCV since it blocks the expression of antiviral proteins induced by IFN. This new concept could have important therapeutic implications since the administration of PKR inhibitors that would prevent eIF2α phosphorylation, could enhance the effectiveness of IFN-based therapy by increasing the accumulation of antiviral ISG proteins.
Clinical studies have shown that patients with high HCV load are less responsive to IFN therapy than patients with lower viral load (Fried et al., 2002; Manns et al., 2001). This is consistent with our observation that the magnitude of ISG protein induction is inversely related to the HCV content of IFN-treated cells. If hepatic PKR and eIF2α phosphorylation also reflects the magnitude of HCV viral load in vivo, one would expect the expression of antiviral ISG proteins to be attenuated in those patients. This hypothesis could be tested by analysis of the ISG protein content of HCV-infected hepatocytes in interferon-treated HCV infected patients and chimpanzees in the future. It may also explain why massive doses of interferon are required to terminate HCV infection in vivo and why most treated patients don’t respond to interferon therapy even when it is combined with ribavirin.
In summary, the current results indicate that HCV strongly induces PKR phosphorylation but it appears to be resistant to translational inhibition by PKR. In contrast, HCV strongly suppresses the ability of IFN to induce ISG proteins by inducing hyper-phosphorylation of PKR in the presence of IFN. Since phosphorylated PKR triggers translational suppression of cellular mRNAs but has little or no impact on the kinetics or magnitude of HCV infection, our results suggest that, by inducing PKR phosphorylation, HCV suppresses the translation of potentially antiviral ISGs that would otherwise threaten its survival. These findings may explain how HCV can thrive in the liver in the face of a robust intrahepatic ISG mRNA response, i.e. by suppressing translation of ISG proteins.
Experimental Procedures
Biological materials and reagents
The origin of Huh-7 (Zhong et al., 2005), Huh-7.5.1 (Zhong et al., 2005), Huh-7.5.1 subclone 2 (clone 2) and the full-length JFH-1 stable replicon cell line (Gastaminza et al., 2008) and HEK-293T (Graham et al., 1977) cells have been described previously. All cells were maintained in DMEM (Cellgro; Mediatech, Herndon, VA) supplemented with 10% FBS (Cellgro), 10 mM HEPES (Invitrogen, Carlsbad, CA), 100 units/ml penicillin, 100 mg/ml streptomycin, and 2 mM L-glutamine (Invitrogen) in 5% CO2 at 37C. The plasmid containing the JFH-1 (Kato et al., 2003) genome was kindly provided by T. Wakita. Lentiviral vectors encoding short hairpin RNAs (shRNAs) targeting PKR and GFP were commercially available (Sigma Aldrich, St. Louis, MO). Vectors encoding compatible packaging proteins and VSV-G were provided by Inder Verma (Salk Institute, La Jolla, CA). Rabbit polyclonal antibodies for the detection of MxA, ISG15, PKR and eIF2α proteins were purchased form Santa Cruz Biotechnology (Santa Cruz, CA). The rabbit phospho-specific antibody against PKR (pT446) was from Epitomics (Burlingame, CA), while phospho-eIF2α (Ser51), -PERK (Thr980), and -GCN2 (Thr898) were from Cell Signaling Technology (Danvers, MA). For detection of dsRNA (K1 antibody from English & Scientific Consulting Bt, Hungary), β-actin (Sigma Aldrich) and viral core protein (Affinity BioReagents, Golden, CO) mouse monoclonal antibodies were used. The recombinant human IgG anti-E2 and the MS5 anti-NS5A antibodies were provided by D. Burton (The Scripps Research Institute) and M. Houghton (Chiron), respectively. The recombinant human IFNβ-1a and IFNα-2a were purchased from PBL InterferonSource (Piscataway, NJ) and Thapsigargin from Sigma Aldrich. Protease and phosphatase inhibitors were obtained from Roche (Indianapolis, IN).
Virus techniques
The original JFH-1 virus was generated by transfection of an in vitro transcribed full-length JFH-1 HCV RNA into Huh-7 cells as previously described (Zhong et al., 2005). JFH-1 and high titer virus stocks of JFH-1 day 183 virus (Zhong et al., 2006) were produced by inoculation of Huh-7 or highly susceptible Huh-7.5.1 subclone 2 cells, respectively, at moi=0.01 as described before (Gastaminza et al., 2008).
Protein analyses
For protein labeling, cultures were washed twice with warm PBS and labeled for 1h at various times post-infection or after IFN treatment with 35S-met to a final concentration of 200 µCi/ml. Total extracts were prepared in RIPA buffer (150 mM NaCl, 50 mM Tris pH=8, 1% NP-40, 0.5% Deoxycholate, 1% SDS) and after protein quantification by BCA, equivalent amounts of total protein per sample were processed by SDS-PAGE and autoradiography. For the radioactivity quantification, equivalent amounts of total protein were subjected to methanol:chloroform (4:1) precipitation. Precipitates were resuspended in RIPA buffer and after addition of 4 volumes of MicroSpint20 (PerkinElmer, Waltham, MA) the radioactivity was measured in a TopCount-NXT Scintillation Counter (PerkinElmer). The results were expressed as counts per minute per microgram of total protein.
For immunofluorescence experiments, Huh-7 cells were either infected with JFH-1 as indicated or maintained uninfected as controls. After 5 days cells were treated with different doses of IFNβ for 16 hours. At this point the cultures were washed with PBS and fixed for 20 min with 4% paraformaldehyde at room temperature for further processing by immunofluorescence as previously described (Zhong et al., 2005). Images were obtained using a Zeiss fluorescence microscope.
For Western-blot analysis, cell extracts were prepared in RIPA buffer supplemented with protease and phosphatase inhibitors and after protein quantification by BCA equivalent amounts of total protein for each sample were separated by SDS-PAGE and transferred to Immobilon membranes. The membranes were blocked for 1h at room temperature with PBS-5% milk and incubated with the primary antibodies diluted in 0.5% milk-0.1% Tween 20 in PBS at room temperature, for varying periods of time, depending on each antibody. After washing 4 times for 15 min with 0.1% Tween 20 in PBS, the membranes were incubated with a dilution of goat-anti-rabbit or anti mouse IgG conjugated to horseradish peroxidase in 0.5% milk-0.1% Tween 20 in PBS. After washing 4 times for 15 min with 0.1% Tween 20 in PBS, membranes were developed using the SuperSignal-West-Pico or -Femto substrates purchased from Thermo Scientific (Rockford, IL).
RNA analyses
Total RNA was extracted from the cells using the guanidinium isothiocyanate extraction method (Zhong et al., 2005) after adding 20 µg of glycogen (Roche) per sample as a carrier. ISG (MxA, ISG15, PKR and IFIT1), HCV and GAPDH (as control) RNA levels were measured by reverse transcription real-time quantitative PCR (RT-qPCR) as described previously (Zhong et al., 2005) using the primers listed in Supplemental Data.
For nucleo-cytoplasmic fractionation, cell cultures were resuspended in isotonic buffer (150 mM NaCl-1.5 mM MgCl2-10 mM Tris HCl-0.25% NP40-RNase inhibitors-1mM DTT, pH 8.5) and the nuclei were separated by centrifugation for 5 min at 3000 rpm at 4C. After an additional wash in the same buffer the supernatants were pooled as cytoplasmic fraction and the pellet was used as nuclear fraction. The extraction of RNA from the nuclear fraction was carried out as indicated above for total RNA. RNA from the cytoplasmic fractions was isolated by incubation with proteinase K (1 µg/ml) in 100 mM NaCl-50 mM Tris-HCl-5 mM EDTA-0.5% SDS, pH7.5 buffer for 1h at 37C, followed by extraction with acid phenol-chloroform and recovered by ethanol precipitation. Quantification of MxA, ISG15 and GAPDH mRNA content in nuclear and cytoplasmic fractions was carried out as described above.
Huh-7 transduction with lentiviruses expressing PKR or GFP shRNAs
Vesicular stomatitis virus glycoprotein (VSV-G)-pseudotyped lentiviral particles were produced in HEK-293T cells by co-transfection of lentiviral vectors encoding shRNAs (targeting PKR or GFP) or GFP protein together with plasmids encoding compatible packaging proteins and VSV-G, as described previously (Dreux et al., 2007). 48 hours post-transfection, cell supernatants were collected, filtered through 0.45 µm filters and used to transduce Huh-7 cells. Out of 5 different PKR shRNAs (Sigma), #5 was selected because of its efficacy in down-regulating PKR protein in the absence of cytotoxic effect (measured by the classical MTT assay). Its sequence is available in the Supplemental Data. The shRNA vector specific for GFP was commercially available as Mission eGFP shRNA Control Vector (Sigma).
Supplementary Material
Acknowledgements
We are grateful to Takaji Wakita (National Institute of Infectious Diseases, Tokyo, Japan) for providing the infectious JFH-1 and full-length JFH-1 replicon clones, Ralf Bartenschlager (University of Heidelberg, Heidelberg, Germany) for the full-length SfiI Con-1 replicon clone, Dennis Burton (The Scripps Research Institute, La Jolla, CA) for the recombinant human IgG anti-E2, Michael Houghton (Chiron) for the MS5 anti-NS5A antibody, and Inder Verma (Salk Institute, La Jolla, CA) for lentiviral plasmids. We are grateful to Bryan Boyd, Josan Chung and Christina Whitten for excellent technical assistance. We are grateful to Stefan Wieland, Pablo Gastaminza, Marlene Dreux, Ken Takahasi and Vladimir Kravchenko for helpful discussions, advice and critically reading the manuscript. Urtzi Garaigorta is a fellow from The Irvington Institute Fellowship Program of the Cancer Research Institute. This work was supported by grant R01-AI079043 from the NIH. This is manuscript number 20244 from the Scripps Research Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alter HJ, Seeff LB. Recovery, persistence, and sequelae in hepatitis C virus infection: a perspective on long-term outcome. Semin Liver Dis. 2000;20:17–35. doi: 10.1055/s-2000-9505. [DOI] [PubMed] [Google Scholar]
- Cheng G, Zhong J, Chisari FV. Inhibition of dsRNA-induced signaling in hepatitis C virus-infected cells by NS3 protease-dependent and -independent mechanisms. Proc Natl Acad Sci U S A. 2006;103:8499–8504. doi: 10.1073/pnas.0602957103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo QL, Richman KH, Han JH, Berger K, Lee C, Dong C, Gallegos C, Coit D, Medina-Selby R, Barr PJ, et al. Genetic organization and diversity of the hepatitis C virus. Proc Natl Acad Sci U S A. 1991;88:2451–2455. doi: 10.1073/pnas.88.6.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutsch M, Hadziyannis SJ. Old and emerging therapies in chronic hepatitis C: an update. J Viral Hepat. 2008;15:2–11. doi: 10.1111/j.1365-2893.2007.00887.x. [DOI] [PubMed] [Google Scholar]
- Dreux M, Boson B, Ricard-Blum S, Molle J, Lavillette D, Bartosch B, Pecheur EI, Cosset FL. The exchangeable apolipoprotein ApoC-I promotes membrane fusion of hepatitis C virus. J Biol Chem. 2007;282:32357–32369. doi: 10.1074/jbc.M705358200. [DOI] [PubMed] [Google Scholar]
- Erickson AK, Seiwert S, Gale M., Jr Antiviral potency analysis and functional comparison of consensus interferon, interferon-alpha2a and pegylated interferon-alpha2b against hepatitis C virus infection. Antivir Ther. 2008;13:851–862. [PMC free article] [PubMed] [Google Scholar]
- Foy E, Li K, Sumpter R, Jr, Loo YM, Johnson CL, Wang C, Fish PM, Yoneyama M, Fujita T, Lemon SM, et al. Control of antiviral defenses through hepatitis C virus disruption of retinoic acid-inducible gene-I signaling. Proc Natl Acad Sci U S A. 2005;102:2986–2991. doi: 10.1073/pnas.0408707102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friebe P, Boudet J, Simorre JP, Bartenschlager R. Kissing-loop interaction in the 3' end of the hepatitis C virus genome essential for RNA replication. J Virol. 2005;79:380–392. doi: 10.1128/JVI.79.1.380-392.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friebe P, Lohmann V, Krieger N, Bartenschlager R. Sequences in the 5' nontranslated region of hepatitis C virus required for RNA replication. J Virol. 2001;75:12047–12057. doi: 10.1128/JVI.75.24.12047-12057.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried MW, Shiffman ML, Reddy KR, Smith C, Marinos G, Goncales FL, Jr, Haussinger D, Diago M, Carosi G, Dhumeaux D, et al. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N Engl J Med. 2002;347:975–982. doi: 10.1056/NEJMoa020047. [DOI] [PubMed] [Google Scholar]
- Gale M, Jr, Blakely CM, Kwieciszewski B, Tan SL, Dossett M, Tang NM, Korth MJ, Polyak SJ, Gretch DR, Katze MG. Control of PKR protein kinase by hepatitis C virus nonstructural 5A protein: molecular mechanisms of kinase regulation. Mol Cell Biol. 1998;18:5208–5218. doi: 10.1128/mcb.18.9.5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale MJ, Jr, Korth MJ, Tang NM, Tan SL, Hopkins DA, Dever TE, Polyak SJ, Gretch DR, Katze MG. Evidence that hepatitis C virus resistance to interferon is mediated through repression of the PKR protein kinase by the nonstructural 5A protein. Virology. 1997;230:217–227. doi: 10.1006/viro.1997.8493. [DOI] [PubMed] [Google Scholar]
- Garcia MA, Gil J, Ventoso I, Guerra S, Domingo E, Rivas C, Esteban M. Impact of protein kinase PKR in cell biology: from antiviral to antiproliferative action. Microbiol Mol Biol Rev. 2006;70:1032–1060. doi: 10.1128/MMBR.00027-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gastaminza P, Cheng G, Wieland S, Zhong J, Liao W, Chisari FV. Cellular determinants of hepatitis C virus assembly, maturation, degradation, and secretion. J Virol. 2008;82:2120–2129. doi: 10.1128/JVI.02053-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodbourn S, Didcock L, Randall RE. Interferons: cell signalling, immune modulation, antiviral response and virus countermeasures. J Gen Virol. 2000;81:2341–2364. doi: 10.1099/0022-1317-81-10-2341. [DOI] [PubMed] [Google Scholar]
- Graham FL, Smiley J, Russell WC, Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol. 1977;36:59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- Guo JT, Bichko VV, Seeger C. Effect of alpha interferon on the hepatitis C virus replicon. J Virol. 2001;75:8516–8523. doi: 10.1128/JVI.75.18.8516-8523.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadziyannis SJ, Sette H, Jr, Morgan TR, Balan V, Diago M, Marcellin P, Ramadori G, Bodenheimer H, Jr, Bernstein D, Rizzetto M, et al. Peginterferon-alpha2a and ribavirin combination therapy in chronic hepatitis C: a randomized study of treatment duration and ribavirin dose. Ann Intern Med. 2004;140:346–355. doi: 10.7326/0003-4819-140-5-200403020-00010. [DOI] [PubMed] [Google Scholar]
- Hershey JW. Translational control in mammalian cells. Annu Rev Biochem. 1991;60:717–755. doi: 10.1146/annurev.bi.60.070191.003441. [DOI] [PubMed] [Google Scholar]
- Honda M, Beard MR, Ping LH, Lemon SM. A phylogenetically conserved stem-loop structure at the 5' border of the internal ribosome entry site of hepatitis C virus is required for cap-independent viral translation. J Virol. 1999;73:1165–1174. doi: 10.1128/jvi.73.2.1165-1174.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoofnagle JH. Course and outcome of hepatitis C. Hepatology. 2002;36:S21–S29. doi: 10.1053/jhep.2002.36227. [DOI] [PubMed] [Google Scholar]
- Kang JI, Kwon SN, Park SH, Kim YK, Choi SY, Kim JP, Ahn BY. PKR protein kinase is activated by hepatitis C virus and inhibits viral replication through translational control. Virus Res. 2009;142:51–56. doi: 10.1016/j.virusres.2009.01.007. [DOI] [PubMed] [Google Scholar]
- Kato T, Date T, Miyamoto M, Furusaka A, Tokushige K, Mizokami M, Wakita T. Efficient replication of the genotype 2a hepatitis C virus subgenomic replicon. Gastroenterology. 2003;125:1808–1817. doi: 10.1053/j.gastro.2003.09.023. [DOI] [PubMed] [Google Scholar]
- Koev G, Duncan RF, Lai MM. Hepatitis C virus IRES-dependent translation is insensitive to an eIF2alpha-independent mechanism of inhibition by interferon in hepatocyte cell lines. Virology. 2002;297:195–202. doi: 10.1006/viro.2002.1455. [DOI] [PubMed] [Google Scholar]
- Lanford RE, Guerra B, Bigger CB, Lee H, Chavez D, Brasky KM. Lack of response to exogenous interferon-alpha in the liver of chimpanzees chronically infected with hepatitis C virus. Hepatology. 2007;46:999–1008. doi: 10.1002/hep.21776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Foy E, Ferreon JC, Nakamura M, Ferreon AC, Ikeda M, Ray SC, Gale M, Jr, Lemon SM. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proc Natl Acad Sci U S A. 2005;102:2992–2997. doi: 10.1073/pnas.0408824102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbach BD, Evans MJ, Syder AJ, Wolk B, Tellinghuisen TL, Liu CC, Maruyama T, Hynes RO, Burton DR, McKeating JA, et al. Complete replication of hepatitis C virus in cell culture. Science. 2005;309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- Maniloff J. Identification and classification of viruses that have not been propagated. Arch Virol. 1995;140:1515–1520. doi: 10.1007/BF01322679. [DOI] [PubMed] [Google Scholar]
- Manns MP, McHutchison JG, Gordon SC, Rustgi VK, Shiffman M, Reindollar R, Goodman ZD, Koury K, Ling M, Albrecht JK. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet. 2001;358:958–965. doi: 10.1016/s0140-6736(01)06102-5. [DOI] [PubMed] [Google Scholar]
- McHutchison JG, Gordon SC, Schiff ER, Shiffman ML, Lee WM, Rustgi VK, Goodman ZD, Ling MH, Cort S, Albrecht JK Hepatitis Interventional Therapy Group. Interferon alfa-2b alone or in combination with ribavirin as initial treatment for chronic hepatitis C. N Engl J Med. 1998;339:1485–1492. doi: 10.1056/NEJM199811193392101. [DOI] [PubMed] [Google Scholar]
- Miglioresi L, Bacosi M, Russo F, Patrizi F, Saccenti P, Ursitti A, Angelis AD, Ricci GL. Consensus interferon versus interferon-alpha 2b plus ribavirin in patients with relapsing HCV infection. Hepatol Res. 2003;27:253–259. doi: 10.1016/s1386-6346(03)00269-9. [DOI] [PubMed] [Google Scholar]
- Patel K, McHutchison JG. Initial treatment for chronic hepatitis C: current therapies and their optimal dosing and duration. Cleve Clin J Med. 2004;71 Suppl 3:S8–S12. doi: 10.3949/ccjm.71.suppl_3.s8. [DOI] [PubMed] [Google Scholar]
- Penin F, Dubuisson J, Rey FA, Moradpour D, Pawlotsky JM. Structural biology of hepatitis C virus. Hepatology. 2004;39:5–19. doi: 10.1002/hep.20032. [DOI] [PubMed] [Google Scholar]
- Robert F, Kapp LD, Khan SN, Acker MG, Kolitz S, Kazemi S, Kaufman RJ, Merrick WC, Koromilas AE, Lorsch JR, et al. Initiation of protein synthesis by hepatitis C virus is refractory to reduced eIF2.GTP.Met-tRNA(i)(Met) ternary complex availability. Mol Biol Cell. 2006;17:4632–4644. doi: 10.1091/mbc.E06-06-0478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel CE. Antiviral actions of interferon. Interferon-regulated cellular proteins and their surprisingly selective antiviral activities. Virology. 1991;183:1–11. doi: 10.1016/0042-6822(91)90112-o. [DOI] [PubMed] [Google Scholar]
- Samuel CE. Antiviral actions of interferons. Clin Microbiol Rev. 2001;14:778–809. doi: 10.1128/CMR.14.4.778-809.2001. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoike T, McKenna SA, Lindhout DA, Puglisi JD. Translational insensitivity to potent activation of PKR by HCV IRES RNA. Antiviral Res. 2009;83:228–237. doi: 10.1016/j.antiviral.2009.05.004. [DOI] [PubMed] [Google Scholar]
- Sjogren MH, Sjogren R, Jr, Lyons MF, Ryan M, Santoro J, Smith C, Reddy KR, Bonkovsky H, Huntley B, Faris-Young S. Antiviral response of HCV genotype 1 to consensus interferon and ribavirin versus pegylated interferon and ribavirin. Dig Dis Sci. 2007;52:1540–1547. doi: 10.1007/s10620-007-9757-9. [DOI] [PubMed] [Google Scholar]
- Su AI, Pezacki JP, Wodicka L, Brideau AD, Supekova L, Thimme R, Wieland S, Bukh J, Purcell RH, Schultz PG, et al. Genomic analysis of the host response to hepatitis C virus infection. Proc Natl Acad Sci U S A. 2002;99:15669–15674. doi: 10.1073/pnas.202608199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumpter R, Jr, Wang C, Foy E, Loo YM, Gale M., Jr Viral evolution and interferon resistance of hepatitis C virus RNA replication in a cell culture model. J Virol. 2004;78:11591–11604. doi: 10.1128/JVI.78.21.11591-11604.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor DR, Shi ST, Romano PR, Barber GN, Lai MM. Inhibition of the interferon-inducible protein kinase PKR by HCV E2 protein. Science. 1999;285:107–110. doi: 10.1126/science.285.5424.107. [DOI] [PubMed] [Google Scholar]
- Terenin IM, Dmitriev SE, Andreev DE, Shatsky IN. Eukaryotic translation initiation machinery can operate in a bacterial-like mode without eIF2. Nat Struct Mol Biol. 2008;15:836–841. doi: 10.1038/nsmb.1445. [DOI] [PubMed] [Google Scholar]
- Thomis DC, Samuel CE. Mechanism of interferon action: evidence for intermolecular autophosphorylation and autoactivation of the interferon-induced, RNA-dependent protein kinase PKR. J Virol. 1993;67:7695–7700. doi: 10.1128/jvi.67.12.7695-7700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventoso I, Sanz MA, Molina S, Berlanga JJ, Carrasco L, Esteban M. Translational resistance of late alphavirus mRNA to eIF2alpha phosphorylation: a strategy to overcome the antiviral effect of protein kinase PKR. Genes Dev. 2006;20:87–100. doi: 10.1101/gad.357006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Krausslich HG, Mizokami M, et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005;11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohnsland A, Hofmann WP, Sarrazin C. Viral determinants of resistance to treatment in patients with hepatitis C. Clin Microbiol Rev. 2007;20:23–38. doi: 10.1128/CMR.00010-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, Wieland SF, Uprichard SL, Wakita T, Chisari FV. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci U S A. 2005;102:9294–9299. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong J, Gastaminza P, Chung J, Stamataki Z, Isogawa M, Cheng G, McKeating JA, Chisari FV. Persistent hepatitis C virus infection in vitro: coevolution of virus and host. J Virol. 2006;80:11082–11093. doi: 10.1128/JVI.01307-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.