Abstract
Mutations in p97, a major cytosolic AAA (ATPases associated with a variety of cellular activities) chaperone, cause inclusion body myopathy associated with Paget's disease of the bone and frontotemporal dementia (IBMPFD). IBMPFD mutants have single amino-acid substitutions at the interface between the N-terminal domain (N-domain) and the adjacent AAA domain (D1), resulting in a reduced affinity for ADP. The structures of p97 N–D1 fragments bearing IBMPFD mutations adopt an atypical N-domain conformation in the presence of Mg2+·ATPγS, which is reversible by ADP, showing for the first time the nucleotide-dependent conformational change of the N-domain. The transition from the ADP- to the ATPγS-bound state is accompanied by a loop-to-helix conversion in the N–D1 linker and by an apparent re-ordering in the N-terminal region of p97. X-ray scattering experiments suggest that wild-type p97 subunits undergo a similar nucleotide-dependent N-domain conformational change. We propose that IBMPFD mutations alter the timing of the transition between nucleotide states by destabilizing the ADP-bound form and consequently interfere with the interactions between the N-domains and their substrates.
Keywords: p97, VCP, IBMPFD, structure, conformational change
Introduction
P97/VCP is one of the most abundant cytosolic proteins and participates, often through interactions with different adaptor proteins, in numerous activities from homotypic membrane fusion, protein degradation, and transcriptional control to cell cycle regulation (for recent reviews see Woodman, 2003; Wang et al, 2004; Ye, 2006). Many p97 functions are linked to its ability to bind polyubiquitinated proteins or proteins that modulate ubiquitination (Wang et al, 2004; Ye, 2006). Cellular accumulation of polyubiquitinated proteins is correlated with mutations of p97 in patients suffering from the Inclusion Body Myopathy associated with Paget's disease of the bone and Frontotemporal Dementia (IBMPFD) and a range of other neurodegenerative disorders (Watts et al, 2004; Haubenberger et al, 2005; Schroder et al, 2005). P97 is a member of the type II AAA+ (extended family of ATPases Associated with a variety of cellular Activities) ATPase, which are characterized by the presence of two conserved AAA ATPase domains (Neuwald et al, 1999). Similar to other type II AAA+ proteins such as NSF and ClpA, p97 forms a homo-hexameric barrel and hydrolyses ATP to drive mechanical work necessary for its function.
Structurally, p97 consists of an N-terminal cofactor-binding domain (N-domain) followed by two AAA ATPase domains (D1 and D2) in tandem (Figure 1A). The first glimpse of p97 at high resolution was provided by the crystal structure of a truncated form of p97 consisting of just the N- and D1-domains (N–D1 from residues 21–463) (Zhang et al, 2000). In this structure, the N-domain (residues 21–184) contains two sub-domains: an N-terminal double ψ-barrel and a C-terminal four-stranded β-barrel. The D1-domain (residues 211–463) with bound ADP also comprises two sub-domains: a large N-terminal RecA-like domain with an α/β-fold and a smaller C-terminal α-helical bundle domain. The N–D1 linker (residues 185–210) lacks a regular secondary structure. The D1-domains form a hexameric ring and their N-domains radiate outwards and are roughly in the plane of the D1 ring. Hexameric ring formation is predominantly mediated through interactions between RecA-like sub-domains; the small α-helical sub-domains of D1 are located on the periphery of the D1 ring (Figure 1B and D). The full-length murine p97 structure was solved a few years later (DeLaBarre and Brunger, 2003; Huyton et al, 2003). In these structures, N- and D1-domains are identically arranged as in the N–D1 structure; the D2-domains form a larger ring, showing a substantial amount of flexibility.
Figure 1.
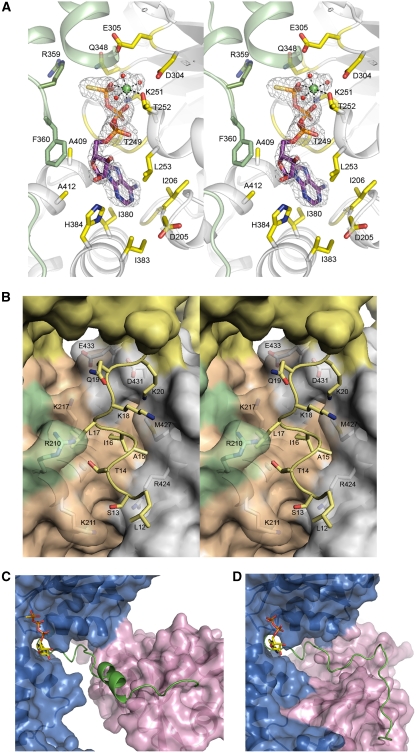
Structures of wild-type and mutant p97 N–D1 with bound ADP or ATPγS. Mutations, either identified in IBMPFD patients (yellow) or introduced in this work (R86A in red and R53A in green), are shown as ball-and-stick models. The N-domains are in magenta and the colours of the D1-domains are alternating in grey (cognate of N) and blue. (A) A schematic diagram for the domain arrangement of p97. (B) Mapping of various mutations to the Cα trace of the wild-type p97 hexamer with bound ADP (PDB:1E32) (Zhang et al, 2000). The structure is viewed down the six-fold axis from the D1- to D2-domain. (C) Stereoscopic rendition of a magnified portion in Figure 1B showing a detailed distribution of mutations in p97 with bound ADP. All mutation sites are labelled. (D) Mapping of various mutations to the Cα trace of the mutant (R155H) structure with bound Mg2+·ATPγS. (E) A stereo pair showing a detailed distribution of the mutation sites in the ATPγS-bound structure. (F) Superposition of the hexamers of the ATPγS and ADP forms. The two structures are portrayed in Cα traces with the N-domain from the ATPγS form in pink and that from the ADP form in magenta. Diameters and heights for the hexamers are given. (G) A stereo pair showing a portion of the superposition in Figure 1F. The superposition was based solely on the D1 RecA-like domain. The ADP form shows the N-domain in magenta, N–D1 linker in forest green, and D1-domain in dark blue, whereas the ATPγS form has its N-domain coloured in pink, N–D1 linker in light green, and D1-domain in light blue.
Interactions of p97 with ubiquitinated proteins are usually mediated through adaptor proteins (Rape et al, 2001; Meyer et al, 2002; Richly et al, 2005), but isolated N-domains or full-length p97 can also bind polyubiquitin chains directly (Dai and Li, 2001; Rape et al, 2001). Although N-domain is implicated in binding to an ubiquitin-like domain termed as UBX, which is present in many p97 cofactors (Schuberth and Buchberger, 2008), it is unclear whether or not the ubiquitin-binding site of an N-domain contributes directly to substrate recognition. Interestingly, crystallographic investigations in the presence of various forms of nucleotides invariably found ADP bound in the D1-domains and the N-domains adopted the in-plane conformation, yet different forms of nucleotides were found bound to the D2-domains in the full-length p97 structures (see review by Pye et al, 2006). Despite the lack of support from crystallographic studies, conflicting evidence on the direction of N-domain movement in response to different forms of nucleotides were reported by electron microscopy (EM) and small-angle X-ray scattering (SAXS) experiments (Rouiller et al, 2002; Davies et al, 2005). Nevertheless, the fact that ubiquitin binding was not observed in the presence of ADP (Dai and Li, 2001) suggests that a nucleotide-dependent conformational change is required to expose the binding site on the N-domain.
A number of single mutations in p97 have been associated with IBMPFD. The majority of these mutations were localized to the N-domain (Watts et al, 2004; Haubenberger et al, 2005; Hubbers et al, 2007; Djamshidian et al, 2009) at residues Arg155, Gly157, Arg159, Arg95, and Arg93, two were in the N–D1 linker region (Watts et al, 2004, 2007) at Arg191 and Leu198, and three others were found in the D1-domain (Watts et al, 2004, 2007) at Ala232, Thr262, and Asn387 (Table I). No mutations were found in the D2-domain. The most frequently observed mutation occurs at residue Arg155. All mutants seem to have normal cellular expression and slightly increased ATPase activity (Weihl et al, 2006; Halawani et al, 2009). It is, however, not yet understood how these single mutations in p97 affect its function, which in turn alters the cellular processing of polyubiquitinated substrates. Here, we present the biochemical and structural evidence to support our hypothesis that IBMPFD mutations may alter communication at the interface between N- and D1-domains. We observed that all IBMPFD mutations occur without exception at the N–D1 interface. Using X-ray crystallography, we show that N–D1 fragments of p97 bearing various IBMPFD mutations adopt an atypical N-domain conformation in the presence of Mg2+·ATPγS. This novel conformation can be reproduced by introducing a specific mutation that is not related to the IBMPFD mutations to the wild-type N–D1 fragment at the interface. We further show that IBMPFD mutants have a reduced binding affinity for ADP.
Table 1. Structural characterization of p97 mutantsa.
| p97 mutations | Location in the structure with bound ADP | Location in structure with bound ATPγS | Closest residues in ADP form | Closest residues in ATPγS form | ||
|---|---|---|---|---|---|---|
| From D1 | From N | From D1 | From N | |||
| R53A | N–surf, exposedb | N–surf, exposedb | None | None | None | None |
| R86A | N–D1 int, buriedc | Exposedd | E261 | D204 | None | None |
| R93Ce | N–D1 int, buried | N–D1 int, buried | None | E195 | None | E194 |
| R95G/Ce | N–D1 int, buried | N–D1 int, buried | G263 | E196 | K386 | D204 |
| R155H/C/P/Le | N–D1 int, buried | Exposed | K386 | None | None | None |
| G157Re | N–D1 int, buried | Exposed | N443 | None | None | None |
| R159H/Ce | N–D1 int, buried | Exposed | A232 | I126 | None | E124 |
| R191Qe | N–D1 int, buried | N–D1 int, buried | None | E162,D121 | None | E162,E195 |
| L198We | N–D1 int, buried | N–D1 int, buried | None | V87,R86 | None | N90,R93 |
| A232Ee | N–D1 int, buried | Exposed | I437d | G125f | M442f | None |
| T262Ae | N–D1 int, buried | N–D1 int, buried | R225 | None | E221 | R64 |
| N387He | N–D1 int, buried | Exposed | None | R155,R159g | None | N199,R95 |
| aAll mutants form hexamers in solution. | ||||||
| b'N–surf, exposed' means that the mutation is located on the surface of the N-domain exposed to solvent. | ||||||
| c‘N–D1 int, buried' means that the mutation is located at the interface between the N- and D1-domains and is buried. | ||||||
| dThis residue is exposed to solvent. | ||||||
| eThose mutations were identified in patients with IBMPFD (Haubenberger et al, 2005; Hubbers et al, 2007; Djamshidian et al, 2009; Schroder et al, 2005; Watts et al, 2004; Watts et al, 2007; Kumar et al, 2010). | ||||||
| fInteracting with residues from a neighbouring subunit. | ||||||
| gInteracting with main chain atoms of the indicated residue. | ||||||
Results
IBMPFD mutations are located at the interface between the N- and D1-domains
By mapping the IBMPFD mutations to earlier available p97 structures (Zhang et al, 2000; DeLaBarre and Brunger, 2003; Huyton et al, 2003), we found that they cluster at the interface between the N- and D1-domains (N–D1 interface; Figure 1B; Table I), which includes the N–D1 linker. Specifically, Arg95, Arg155, Gly157, and Arg159 make contacts with residues from the D1-domain. Arg93 interacts with Glu195 of the N–D1 linker, which forms a large part of the interface. Side chains of Arg191 and Leu198 in the linker contact Glu162 and Val87, respectively, in the N-domain. The other three IBMPFD mutations at residues Ala232, Thr262, and Asn387 are located on the D1-domain side of the interface. Although Ala232 interacts with residues from the N-domain of a neighbouring subunit, Thr262 and Asn387 contact residues from the cognate N-domain. Biochemically, the full-length R155H, R155P, and A232E recombinant proteins exhibit elevated ATPase activities (Weihl et al, 2006; Halawani et al, 2009). Similar to the wild-type enzyme, mutant p97 invariably forms hexamers as determined by Blue-Native PAGE and gel filtration analysis (Ma and Xia, 2008; Halawani et al, 2009; Tang et al, 2009).
Structures of p97 mutants with bound Mg2+·ATPγS reveal a novel N-domain conformation
As all IBMPFD mutations are located in either the N-domain or the D1-domain, the N–D1 fragment of p97 (residues 1 to 481 plus a C-terminal hexahistidine tag) bearing either the R155H or R95G mutation was produced in large quantities and subjected to structural studies. The choice of using the N–D1 fragment was also based on the consideration of its subsequent crystallization. Arg155 is the most frequently observed position for mutations and Arg95 is in close proximity to another mutation, T262A, at the D1-domain. Crystals were obtained in the presence of Mg2+·ATPγS; they diffracted X-rays to better than 3 Å resolution (Table II). The crystals of the R155H and R95G mutants display the symmetries of the space groups R3 and P1, respectively.
Table 2. Statistics on the quality of diffraction data sets and refined atomic models.
| Mutant | R155H | R95G | R86A | R155H |
|---|---|---|---|---|
| Bound nucleotide | ATPγS | ATPγS | ATPγS | ADP |
| Data collection | ||||
| Space group | R3 | P1 | P1 | P212121 |
| Unit cell [a, b, c, Å] [α, β, γ, deg] | 134.2,134.2,182.9 90.0,90.0,120.0 | 92.76,103.3,107.7 97.7,91.9,89.7 | 90.89,102.6,107.2 97.5,90.6,91.5 | 146.5,170.7,256.9 90.0,90.0,90.0 |
| Resolution (last shell) (Å) | 50–2.15 (2.23–2.15) | 50–2.8 (2.90–2.80) | 50–2.85 (2.95–2.85) | 50–3.42 (3.51–3.42) |
| Rmergea (last shell) (%) | 5.7 (52.4)b | 5.0 (49.3) | 4.9 (50.2) | 10.4 (43.7) |
| Completeness (last shell) (%) | 99.5 (95.6) | 90.5 (71.5) | 91.8 (67.8) | 92.8 (66.1) |
| Total observations | 235 711 | 151 406 | 200 205 | 470 333 |
| Unique reflections | 66 557 | 87 240 | 85 163 | 76 375 |
| I/σ(I) (last shell) | 19.7 (1.72) | 12.3 (0.8) | 13.8 (1.02) | 7.2 (1.0) |
| Refinement statistics | ||||
| Resolution (Å) | 32–2.2 | 25–2.80 | 40–2.85 | 25–3.42 |
| Rfree (last shell) (%) | 19.3 (28.8)c | 27.3 (44.1) | 29.0 (39.2) | 26.3 (37.2) |
| Rwork (last shell) (%) | 17.1 (21.0) | 24.0 (39.1) | 25.8 (34.7) | 21.7 (33.3) |
| r.m.s.d. bond length (Å) | 0.012 | 0.021 | 0.020 | 0.022 |
| r.m.s.d. bond angle (deg) | 1.38 | 1.51 | 1.52 | 2.08 |
| Number of atoms | 7369 | 21 445 | 21 453 | 41 406 |
| Number of residues | 899 | 2706 | 2712 | 5244 |
| Number of solvent molecules | 263 | 121 | 93 | 0 |
| Number of ATPγS/ADP | 2 | 6 | 6 | 12 |
| Number of Mg2+ ions | 2 | 6 | 6 | 0 |
| Ramachandran analysis | ||||
| Most favoured (%) | 90.4 | 90.2 | 89.3 | 87.5 |
| Allowed (%) | 9.6 | 8.8 | 9.7 | 11.2 |
| Generously allowed (%) | 0 | 1.0 | 1.0 | 1.3 |
| Disallowed (%) | 0 | 0 | 0 | 0 |
| aRmerge is defined as ∑∣Ih,i–〈Ih〉∣/∑Ih,i, where Ih,i is the intensity for ith observation of a reflection with Miller index h, and 〈Ih〉 is the mean intensity for all measured Ihs and Friedel pairs. | ||||
| bValues in parentheses are for the highest resolution shells. | ||||
| cThe crystal is a merohedral twin relating h, k, l to h, –h, –k, –l. A consequence of a sharpened intensity distribution in cases of twinned crystals, the R-factors are lower than in cases of non-twinned crystals. | ||||
| Cell parameters are indicated in italics. | ||||
The structure of the p97 R155H mutant was solved by molecular replacement (MR), using the coordinates of wild-type p97 N–D1 as a phasing model (Zhang et al, 2000). However, the N- and D1-domains had to be searched for separately, indicating that the arrangement between the N- and D1-domains for this mutant in the presence of Mg2+·ATPγS differs from that of the wild type. There are two copies of mutant N–D1 protomers in the crystallographic asymmetric unit (AU) and the structure was successfully refined, converging to an Rfree and Rwork of 19.3 and 17.1%, respectively (Table II). The structure for the R95G mutant was successfully determined by MR, using the structure of the R155H mutant N–D1 as a search template, indicating that the two mutants with bound Mg2+·ATPγS have a similar or identical conformation. There exists a complete N–D1 hexamer in a crystallographic AU for this mutant. The structure was refined to an Rfree value of 27.3% and an Rwork value of 24.0% (Table II).
Structures of individual domains of all Mg2+·ATPγS-bound mutants are identical to those of wild type with bound ADP, as shown by their pair-wise alignments (Supplementary Table S1). Regardless of variations in space group symmetries and in mutations, all N–D1 fragments with bound Mg2+·ATPγS have virtually identical structures, as shown by the small root-mean-square (r.m.s.) deviations in structure superposition and by the large number of superimposed residues. Moreover, these point mutations have little effect on the overall structure of the domain in which they reside.
When the structures of the hexameric D1 rings of mutants with Mg2+·ATPγS bound were aligned with the ADP-bound wild-type p97 N–D1, the RecA-like sub-domains of D1 rings superimpose nicely, whereas the positions and orientations for the D1 α-helical sub-domains and the N-domains deviate significantly (Supplementary Figure S1). In particular, the N-domains display dramatic conformational changes by swinging upwards by 12.5 Å (Up-conformation; Figure 1C, E; Supplementary Figure S3), as measured from the distance travelled by the centres of gravity of the N-domain (residues 22–190) between the two nucleotide-binding states. This movement of the N-domain includes a large rotation of 93° (Supplementary Figure S3), which results in a significant increase in the height of the N–D1 from 43 to 62 Å, but only marginal expansion of the diameter of the hexameric ring from 162 to 164 Å (Figure 1F).
ADP-bound mutant p97 displays a Down-conformation of the N-domain resembling wild-type p97
To determine whether the source of the novel Up-conformation was the mutation in p97 or the presence of Mg2+·ATPγS bound in the active site, we crystallized the N–D1 fragment of the R155H mutant in the presence of ADP. The ADP-bound crystals diffracted X-rays to ∼3.4 Å resolution (Table II) and belong to the orthorhombic space group P212121. To avoid potential phase bias in MR phasing with respect to the position of the N-domain and to the bound nucleotide, the phasing model contained only one D1-domain without nucleotides. Electron densities for N-domains and bound nucleotide moieties were revealed by difference Fourier calculations. A complete model for the ADP-bound N–D1 fragment of the R155H mutant was built and refined (Table II).
In a crystallographic AU, two hexameric rings stack face-to-face with a small shift in positions of their respective ring axes and are slightly rotated with respect to each other, producing inter-digitating N-domains (Supplementary Figure S2A). The N-domain displays a conformation nearly co-planar with the D1 hexamer ring, which is similar to the wild-type p97 with ADP bound to the D1-domain. Indeed, structure alignment between the mutant and wild-type molecules shows an r.m.s. deviation of 1.034 Å for 433 residues (Supplementary Table S1); hence this conformation is referred to as the Down-conformation. The crystal-packing environment is only compatible with the N-domains taking the Down-conformation (Supplementary Figure S2B). The difference Fourier density at each of the 12 nucleotide-binding sites has the unmistakable shape of a bound ADP molecule (Supplementary Figure S2C). This result establishes the interdependency between the conformation of the N-domain and the nucleotide state in the D1-domain for IBMPFD mutants. Moreover, a mutant p97 hexamer is able to push N-domains of all its protomers into identical conformations in response to the presence of different forms of nucleotides.
X-ray scattering shows nucleotide-dependent conformational changes of wild-type and mutant N–D1 fragments in solution
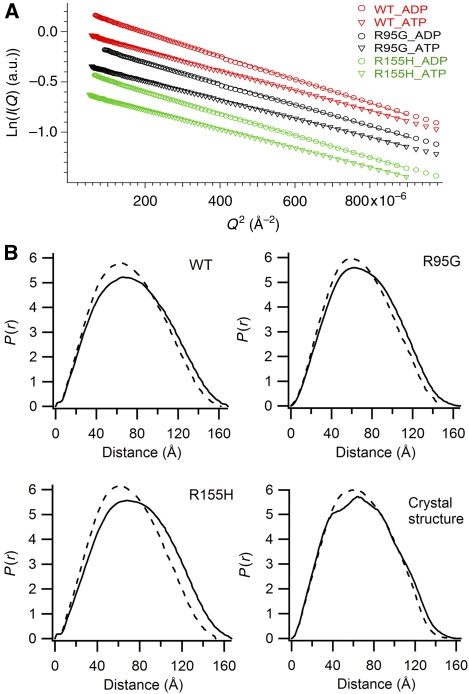
To show that the conformational changes of p97 seen in crystal structures could also happen in solution, wild-type and mutant N–D1 fragments were studied in the presence of ATPγS or ADP by X-ray scattering experiments. Scattering data sets covering different resolution ranges for each sample were scaled together and merged, yielding intensity data over the reciprocal Q range from 0.005 to 0.7 Å−1 (1200 and 9 Å in real space). The radii of gyration (Rg), determined from Guinier analysis (Glatter and Kratky, 1982), are consistently 3–5 Å smaller for the ATPγS-bound N–D1 fragment compared with the ADP-bound form, which is in agreement with the calculated values from crystal structures (Table III; Figure 2A). The smaller Rg values indicate the presence of more spherical molecules in solution for the N–D1 in the ATPγS form. Importantly, wild-type p97 N–D1 also displays a smaller Rg in response to the presence of ATPγS in solution, indicating that the wild-type p97 undergoes a similar conformational change, although this change may limit to only a few subunits in a p97 hexamer. The conformational change of N–D1 in solution can also be shown by the distance distribution functions, p(r), obtained by Fourier transformation of scattering intensities (Svergun, 1992), in which a significant shift in the distribution towards shorter vectors was observed for the ATPγS-bound N–D1 fragments (Figure 2B), again indicating a more spherical molecule. This shift in p(r) is most obvious at vector lengths beyond 90 Å, consistent with the large-scale N-domain conformational change. Furthermore, calculated changes in the distribution function (Figure 2B) based on crystal structures are in agreement with the experimentally obtained distribution functions, suggesting that the crystallographically observed differences in the conformation of N-domain exist in solution not only for p97 mutants, but also for wild-type p97.
Table 3. Radius of gyration (Rg) measured by SAXS for wild-type and mutant p97 N–D1 fragments in solutiona.
| Protein | Bound nucleotide | Rg (Å)b |
|---|---|---|
| ND1-WT | ADP | 58.51±0.02 |
| ATPγS | 55.30±0.02 | |
| ND1-R95G | ADP | 55.45±0.03 |
| ATPγS | 52.26±0.03 | |
| ND1-R155H | ADP | 56.35±0.03 |
| ATPγS | 53.27±0.03 | |
| aCalculated Rg values were based on Rg=∑miRi2/∑mi, where mi is atomic weight for atom i and Ri is the distance from the centre of mass of the protein to atom i. The Rg values for ATPγS and ADP bound p97 N–D1 fragments are 51.6 and 52.9 Å, respectively. These calculations were made in the absence of solvent molecules. | ||
| bRg values were obtained from low Q Guinier plot (Glater and Kratky, 1982). | ||
Figure 2.
Nucleotide-binding-associated conformational changes in solution observed by X-ray scattering experiments. (A) Guinier plots (Guinier and Fournet, 1955) for the wild-type and mutant p97 N–D1 fragments in the presence of 0.1 mM ATPγS or ADP. Wild-type data are in red, R95G and R155H mutants are in black and green, respectively. Open circles are for ADP and open triangles for ATPγS. (B) Distance distribution functions, p(r), normalized to a common total probability, with standard error of the data in Å propagated through the inverse Fourier transform implemented in GNOM (Svergun, 1992) for the wild-type and mutant N–D1 fragments. The calculated distribution (Glatter, 1980) is given in the last panel based on crystal structure in the absence of bound-solvent molecules. The dashed and solid lines are for ATPγS and ADP, respectively.
Nucleotide-binding environment in the presence of ATPγS or ADP
In contrast to the earlier observation of invariably bound ADP in D1-domains (Zhang et al, 2000; Huyton et al, 2003; DeLaBarre and Brunger, 2003, 2005), unbiased difference Fourier maps showed unambiguously the bound ATPγS molecules in D1 subunits of both mutant crystals obtained in the presence of ATPγS (Figure 3A). When Cα atoms of the wild type and mutant N–D1 were superimposed, the adenosine moieties of the bound nucleotides align very well and the immediate environment around the adenosine moiety shows little change. Contributing to the adenosine-binding pocket are residues from the N–D1 linker (Asp205, Val206, and Gly207), the Walker A motif (Thr249, Gly250, and Leu253), the small α-helical bundle domain (Ile380, Ile383, Gly408, Ala409, His384, and Ala412), and residue Phe360 from a neighbouring subunit (Figure 3A). In contrast, the phosphate groups in the alignment between ADP and ATPγS forms deviate, even though the same set of residues are involved in contacting the α- and β-phosphates in both the wild-type and mutant structures, including the conserved Walker A residues Gly248, Thr249, Gly250, Lys251, Thr252, and Leu253. The γ-phosphate in the ATPγS structure is stabilized by the ionic interaction with a magnesium ion (Mg2+), by hydrogen bonds with Gln348 and Lys251, by Arg359, an Arg finger residue from a neighbouring subunit, and by two water molecules associated with the Mg2+ ion.
Figure 3.
Changes in the structure of N–D1 on binding of ATPγS. (A) Stereoscopic pair showing details of the ATPγS-binding environment. The nucleotide-binding site is located at the subunit interface. One subunit is coloured in green and the other in grey. Residues making contact with bound ATPγS are drawn as sticks and are labelled. The ATPγS molecule is shown as a stick model with carbon in purple, oxygen in red, nitrogen in blue, phosphorous in magenta, and sulphur in yellow. The ATPγS molecule is enclosed in a difference electron density cage in grey, contoured at the 2.5σ level. The Mg2+ ion is shown as a green ball with the three coordinating water molecules in red. (B) Stereo pair showing the ordering of an N-terminal segment from Leu12 to Lys20 in the ATPγS form and its environment. The re-ordered residues are depicted as stick models in yellow and their environment is shown as a semi-transparent molecular surface with all interacting residues rendered as stick models with labels. The yellow surface is the N-domain; orange and green surfaces are the cognate D1-domain and N–D1 linker, respectively. The grey surface is from an adjacent D1-domain. (C) Conformation of the N–D1 linker in the presence of ATPγS. The N–D1 linker is shown as a ribbon model in green; N- and D1-domains are shown as molecular surfaces in magenta and in blue, respectively. The bound ATPγS is shown as a stick model. (D) Conformation of the N–D1 linker in the presence of ADP. The N–D1 linker is shown as a coil in green. The D1-domain is similarly orientated as in (C). The bound ADP is shown as a stick model.
Also new in the mutant structures of p97 is the presence of a Mg2+ ion in the nucleotide-binding site of every subunit. The Mg2+ ion is at the centre of an octahedral mer-triaquo complex with the additional three oxo ligands coming from the side chain of the highly conserved Thr252 and from the β- and γ-phosphates (Figure 3A). The acidic residues of the DEXX sequence (Asp304 and Glu305) in the Walker B motif make hydrogen bonds with two of the water molecules in the Mg2+ coordination sphere and, additionally, Asp304 stabilizes Thr252. As expected, most of the changes in the nucleotide-binding environment are a consequence of the introduction of the γ-phosphate.
Structural differences between ADP and ATPγS-bound N–D1
Prominent structural changes on ATPγS binding are present at several levels. With ATPγS bound, the N-terminal residues from Leu9 to Lys20 become ordered in all non-crystallographic symmetry (NCS)-related subunits of the mutant structures. This is true for both R3 and P1 space groups and is, therefore, not attributable to crystal lattice contacts. Rather, it is conceivably a consequence of the large conformational change of the N-domain, bringing the earlier disordered N-terminal peptide segment in contact with the D1-domain (Figure 3B). Specifically, the ordered N-terminal peptide stretch, which, in the ADP form, was disordered and pointed away from the D1-domain, now reaches into a shallow, hydrophilic valley formed between the first helix of the cognate D1-domain (α4) and the helix α12 from a neighbouring D1-domain (Figure 3B).
A pronounced change in secondary structure on ATPγS binding was the appearance of a three-turn α-helix (Arg191–Asp200) in the linker between the N- and D1-domains (Figure 3C and D). In contrast to the random coil conformation in the ADP-bound structure, well-defined electron density representing helical secondary structure became visible in composite-omit maps for a portion of the linker in the ATPγS-bound structures. The same helical conformation of the N–D1 linker was observed in both space groups and in all NCS-related subunits, suggesting that it was induced by ATPγS binding or by the N-domain conformational switch. Secondary structure prediction for the linker pointed at a high helical propensity for residues in the sequence between Arg191 and Asp200, coinciding with our crystallographic observation. Therefore, the coil-to-helix transition in the N–D1 linkers is reminiscent of a contracted spring pulling the N-domains out of the planar conformation on ATPγS binding.
Movements of secondary structural elements and of entire sub-domains induced by ATPγS binding were easily observed on global alignment of N–D1 hexamers. In ADP-bound structures, the six RecA-like sub-domains of the D1 hexamer associate with each other to form the centre ring with the six smaller α-helical sub-domains attached to the periphery of the ring. Superposition of the ATPγS-bound hexamer to this ring shows well-aligned hexameric RecA-like domains (Supplementary Figure S1), but results in significant displacements of the peripheral α-sub-domains, in addition to the conformational changes in the N-domains described above (Figure 1F and G). Optimal alignment of the RecA-like sub-domains of the ADP-bound wild type and the ATPγS-bound mutants shows well superimposed β-sheets; however, the first three helices of the D1-domain, α4, α5, and α6, and the loop between helices α9 and α10 are offset (Figure 1G). In the ATPγS-bound state, helix α4 connects directly to the C-terminus of the N–D1 linker, α5 moves laterally by about 7 Å, and α6, preceding the Walker A motif, translates vertically and laterally by 3 and 5 Å, respectively. In addition, secondary structure elements in the small α-helical sub-domains of D1 are out of alignment, moving in the opposite direction of the N-domains (Figure 1G); the only exception is the first helix, which connects to the RecA-like domain.
Changes in the nucleotide-binding affinity of mutant p97 N–D1 fragments
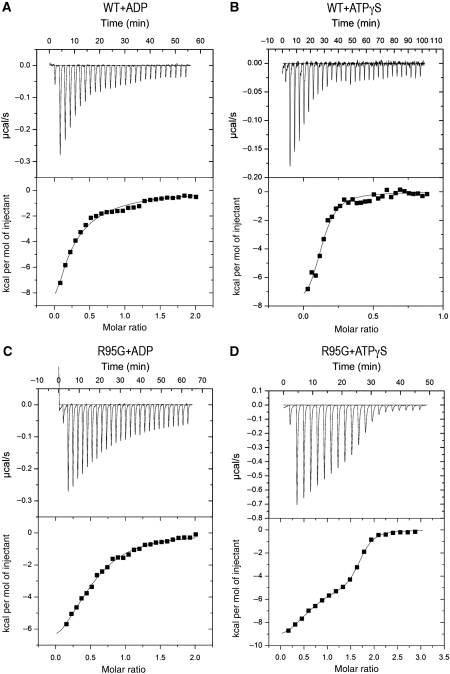
Despite the apparent preference of wild-type p97 for binding ADP in the D1-domains in both forms (full-length and the N–D1 fragment), our crystallographic analysis of IBMPFD mutants suggested that their nucleotide-binding tendency in D1 was altered, as both ATPγS and ADP forms of mutant p97 can be crystallized. To test this hypothesis, we used isothermal titration calorimetry (ITC), which allows simultaneous measurements of nucleotide-binding affinities and stoichiometry (O'Brien et al, 2001). For the wild-type N–D1, we determined a dissociation constant (Kd) of 0.88 μM towards ADP (Table IV) with a stoichiometry of 0.35, suggesting that only two out of six sites are available for binding, which is consistent with reported values (Briggs et al, 2008). In contrast, mutant p97 N–D1 fragments displayed reduced binding affinities for ADP and the level of reduction is site dependent (Table IV). For example, the R155H mutant showed a maximum reduction with a Kd of 4.25 μM. Notably, the changes in the binding stoichiometry are correlated with the changes in binding affinities for the mutants (Table IV).
Table 4. Dissociation constants (Kd) and binding stoichiometry (N) of wild-type and mutant p97 N–D1 fragments for ATPγS and ADP determined by ITCa.
| N–D1 p97 | ATPγS | ADP | ||
|---|---|---|---|---|
| Kd (μM)b | N | Kd (μM) | N | |
| Wild type | 0.89±0.28 | 0.12±0.01 | 0.88±0.18 | 0.35±0.06 |
| R53A | 0.62±0.18 | 0.20±0.02 | 0.95±0.13 | 0.34±0.02 |
| R86A | 0.06±0.03 | 0.42±0.09 | 1.85±0.09 | 0.54±0.07 |
| R95G | 0.13±0.02 | 0.56±0.01 | 2.27±0.11 | 0.62±0.08 |
| R155H | 0.13±0.01 | 0.61±0.01 | 4.25±0.54 | 0.72±0.18 |
| aAll numbers shown are averages of three independent experiments. | ||||
| bITC with ATPγS for R86A, R95G, and R155H mutants showed biphasic titration curves and data were fitted with a two-site model. The Kd values for those mutants are derived from fitting to the first phase. | ||||
Consistent with earlier findings (Briggs et al, 2008), wild-type p97 showed an apparent Kd value for ATPγS of 0.89 μM, similar to that for ADP. Unexpectedly, the titration profiles with ATPγS for mutants were biphasic and can only be fitted to a two-site model (Figure 4). The apparent Kd values for the high-affinity site were well determined and close to 0.1 μM for all mutants (Table IV), whereas those for the low-affinity site were associated with significant errors (data not shown). Again, mutant p97 displayed higher stoichiometry than wild type in the ATPγS titration experiments.
Figure 4.
Calorimetric titration of wild-type and mutant p97 N–D1 with ATPγS or ADP. (A) ITC for wild-type p97 with ADP. Raw data was obtained from 27 automatic injections of 10 μl of 100 μM ADP solution (first injection 2 μl) into 1.4236 ml of 10 μM p97 solution at 25°C (top panel) and fitted (bottom panel, continuous line) to a one-site model. (B) ITC for wild-type p97 with ATPγS; 30 injections of 8 μl of 100 μM ATPγS solution were made into 20 μM p97 solution (top panel). The data was fitted to a one-site model (bottom panel). (C) ITC for the R95G mutant with ADP; 27 injections of 10 μl of 100 μM ADP solution were made into 10 μM mutant p97 solution (top panel). The resulting heat data was fitted to a one-site model (bottom panel). (D) ITC for the R95G mutant with ATPγS; 30 injections of 10 μl of 100 μM ATPγS solution were made into 10 μM mutant p97 solution (top panel). The data was fitted to a two-site model (bottom panel).
Molecular signature of IBMPFD can be reproduced by the R86A mutation introduced into the wild-type N–D1 interface
On the basis of our crystallographic and biochemical analysis, the structural and biochemical hallmarks of IBMPFD mutants are the less constrained position of N-domains and the changes in relative affinity for ATPγS and ADP at the D1 nucleotide-binding site. To verify that these characteristics are a consequence of unfavourable interaction by IBMPFD mutations at the N–D1 interface, we introduced a mutation, R86A, to the wild-type N–D1 fragment, which was not associated with known IBMPFD mutations, but which we predicted would disrupt major interactions of the N-domain with residues Asp204 and Asp205 in N–D1 linker at the interface (Figure 1; Table I). The R86A mutant crystallizes in the presence of Mg2+·ATPγS (Table II) with its N-domain in the Up-conformation. Its biphasic ITC profile for ATPγS and an altered nucleotide-binding affinity (Table IV) are consistent with the characteristics of a typical IBMPFD mutant (Table I). As a control, we introduced the mutation, R53A, which alters a residue located on the surface, but distant from the N–D1 interface. The R53A mutant had properties similar to those of the wild-type enzyme (Figure 1; Tables I and IV).
Discussion
Conformational states of the N-domain are correlated with the nucleotide-binding states in the D1-domain
Perhaps the single most important discovery of our crystallographic work is the interdependency between the nucleotide-binding states of the D1-domain and the conformational changes of the N-domain in p97 mutants. The question is whether this observation can be extended to the wild-type p97 or other type II AAA ATPases such as Escherichia coli ClpA. Although an equivalent to the Down-conformation for the N-domain was observed in the crystal structure of ClpA with bound ADP (Guo et al, 2002) and while that for the ATP-bound form has yet to be obtained, it is generally accepted that the conformation of the N-domain in ClpA is rather mobile (Ishikawa et al, 2004). Consistent with this view, the crystal structure of Thermus thermophilus ClpB, another type II AAA ATPase in E. coli, showed two distinct conformations of the N-domains, whereas all D1-domains feature bound AMP-PNP (Lee et al, 2003). Similar conclusion for a flexible N-domain in p97 was drawn based on missing densities in EM analyses (Rouiller et al, 2002). However, conflicting observations on the movement of N-domain in response to ATP binding were reported. For example, a downwards movement of N-domains positioned below the D1 ring in response to ATP binding was observed by SAXS (Davies et al, 2005). In contrast, cryo-EM at 24 Å resolution in the presence of AMP-PNP suggested an upwards repositioning of the N-domain (Beuron et al, 2003). Furthermore, cryo-EM also observed nucleotide-dependent changes in the N-domain of p97 in complex with p47 (Beuron et al, 2006). In spite of these observations, crystallographic experiments with wild-type p97 have not provided evidence for or against nucleotide-dependent changes in the N-domain conformation (Zhang et al, 2000; Huyton et al, 2003; DeLaBarre and Brunger, 2003, 2005). Our high-resolution crystallographic data correlated, for the first time, the movement of the N-domain in p97 with the ATP/ADP states in the D1-domain of a type II AAA protein. Although the initial observation was made on IBMPFD mutants, it does extend to the wild type as shown by solution SAXS experiments, in which similar changes in vector distributions and in Rg values were taking place in response to different forms of nucleotides irrespective of whether wild-type p97 or mutants were used. Thus, we surmise that the N-domains of wild-type p97 undergo the same nucleotide-dependent conformational changes. However, unlike mutant p97 whose six N-domains are capable of taking the Up-conformation uniformly at high ATPγS concentrations, the wild-type p97 seems to have only a few subunits undergoing N-domain conformational changes, leading to asymmetry in the conformations of N-domains in hexameric p97. This asymmetric distribution of the N-domain conformations of wild-type p97 in the presence of ATPγS may be the reason for its unsuccessful crystallization.
Changes in the interaction at the N–D1 interface on ATP–ADP transition
As a result of ATPγS binding, the hexameric N–D1 fragments gain 10 510 Å2 accessible surface area compared with the ADP-bound form (Supplementary Table S2), suggesting loss of interactions among different domains and subunits. During the ADP to ATPγS transition, a major reduction (679 Å2 per subunit) in contacts between N- and D1-domains comes from changes in interaction with cognate subunits. However, there is a gain of 164 Å2 per subunit for the N-domain in interaction with the D1-domain of a neighbouring subunit. As a result, the Up-conformation for the N-domain in the ATPγS-bound form is conceivably less stable than the Down-conformation of the ADP-bound form.
The N-domain conformational change is accompanied not only by a coil-to-helix transition in the N–D1 linker, but also by an extensive rearrangement of interactions. Table I shows the changes in interacting partners for residues that bear IBMPFD mutations before and after the conformational transition. Among the 10 listed IBMPFD mutations, all are buried in the N–D1 interface in the ADP-bound form, whereas only five are buried in the ATPγS-bound form. This observation is consistent with the amount of buried surface area (BSA) lost in the ADP to ATPγS transition and suggests that IBMPFD mutations affect the stability of the N-domain conformation more in the ADP-bound form. It is likely that mutations of other residues at the interface, as exemplified by the R86A mutation, would also result in p97 malfunction similar to those observed in the disease-associated mutants.
Functional defect in IBMPFD mutants is the result of altered nucleotide binding in the D1-domain
In addition to having a lower ATPase activity with respect to the D2-domain, the D1-domain of the wild-type p97 has a large amount of pre-bound ADP, as shown by ITC (Table IV), heat, or urea denaturation experiments (Davies et al, 2005; Briggs et al, 2008). The pre-bound ADP is difficult to exchange, even though the apparent binding affinities for ADP or ATPγS were reported to be rather similar (DeLaBarre and Brunger, 2003; Davies et al, 2005; Briggs et al, 2008). Consistent with this, wild-type p97 can only be crystallized with ADP bound to the D1-domain. Notably, not all subunits in a p97 hexamer are pre-occupied with ADP and the stoichiometry of the pre-bound ADP in purified wild-type p97 is in the range of 0.5–0.8. In this work, we confirmed these findings on the wild-type p97 N–D1 fragment. Mutant p97 N–D1 fragments behave rather differently: they can be crystallized in both ADP- and ATPγS-bound forms with characteristic N-domain conformations associated with either nucleotide form bound at the D1-domain. Their D1 sites more easily exchange nucleotides, as evidenced by a reduced affinity for ADP and a lowered level of pre-bound ADP (Table IV). Furthermore, mutant p97 N–D1 fragments display biphasic ITC profiles titrating with ATPγS and have higher apparent binding affinities for ATPγS.
A possible interpretation of these data is that IBMPFD mutations introduce a defect in the nucleotide-binding environment of the D1 site. As the N–D1 interface in the Down-conformation is in close proximity to the nucleotide-binding site, it is conceivable that the ADP-bound form is more affected by the mutations than the ATP-bound form when the N-domain is in the Up-conformation. As most mutations lead to changes in charged interactions (Table I), the influence on the D1 nucleotide-binding site is most likely electrostatic in nature. Consequently, the nucleotide-binding affinity at the D1 site must be dependent on the N-domain conformation. Indeed, the measurement of altered ADP-binding affinity by ITC for mutants support this conclusion.
Owing to the presence of pre-bound ADP, isothermal titration with ATPγS is effectively only a measurement of apparent binding affinity (Table IV) (Zhang and Zhang, 1998). What IBMPFD mutations do to the protein is to alter the interaction at the N–D1 interface, reduce its affinity for ADP (∼4 μM), and the amount of pre-bound ADP, leading to a less constrained Down-conformation even with ADP bound at the D1 site. Indeed, IBMPFD mutations lead to the increase in the apparent affinity for ATPγS (∼0.1 μM; Table IV). Owing to the lowered affinity for ADP in the mutants, any pre-bound ADP is now more likely to be exchanged and the ITC profiles for ATPγS become biphasic with the first phase representing the titration of empty sites followed by the second phase, in which ATPγS competitively displaces pre-bound ADP molecules.
Implications for p97 ATPase cycle in the D1-domain
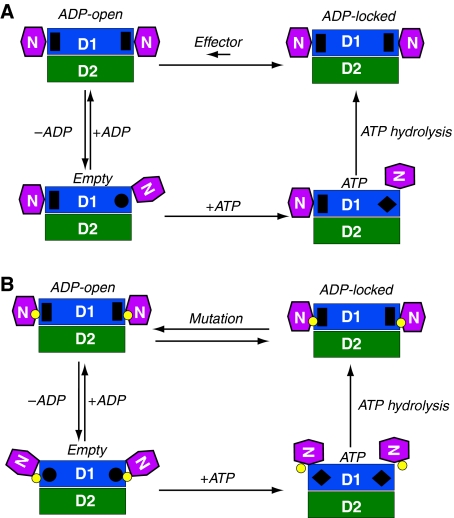
A model for the ATP cycle in the D1-domain emerges from integrating structural and biochemical data of wild-type and mutant p97 (Figure 5). In this model, we propose four nucleotide-binding states for the D1-domain and assume that the state of each protomer is independent of the states of other protomers, as evidence has yet to be obtained for supporting a coordinated action among different subunits in a wild-type p97 hexamer. First, there is an ‘ATP' state, with ATP bound and the N-domain in the Up-conformation. Crystallographic and X-ray scattering experiments support the existence of this state in subunits of both mutant and wild-type p97. One should keep in mind that because of non-exchangeable, pre-bound ADP in a wild-type p97 hexamer, not all subunits would bring their N-domains in the Up-conformation even with an excess of ATP in solution. We, therefore, hypothesize that there is an ‘ADP-locked' state, with non-exchangeable, pre-bound ADP at the D1 site and the N-domain in the Down-conformation. This state seems to be important for wild-type p97 function and the pre-bound ADP is particularly difficult to change. The structure of the N–D1 fragment of wild-type p97 may represent this conformation. In a third state, termed as ‘ADP-open', ADP is bound, but exchangeable. This state was observed for mutant p97 by its biphasic ITC titration profile and is presumably in equilibration with the ADP-locked state. The structure of R155H with bound ADP represents this conformation. The fourth state is the ‘Empty' state, with nucleotide-binding sites unoccupied and the N-domain in an unknown position.
Figure 5.
Mechanistic model for the control of N-domain movement and implication for IBMPFD. Schematic diagram for the control of the N-domain conformation in (A) the wild-type and (B) IBMPFD mutant p97 N–D1 fragment. The N-, D1-, and D2-domains are in magenta, blue, and green, respectively, and as labelled. The small yellow circles between N- and D1-domains represent positions of mutations. Four states are defined for each nucleotide-binding site in D1: Empty state, ATP state, ADP-locked, and ADP-open, as labelled. Each protomer is assumed to operate independently. The stimuli for changes in D1 nucleotide state may come either from the N-domain or from ATP hydrolysis of the D2-domain. In the ADP-locked state, the N-domains are in Down-conformation with a pre-bound ADP shown as a black rectangle. This ADP-locked state has been observed crystallographically in wild-type p97. In the ATP state, the N-domains of hexameric wild-type p97 could be either in an Up-conformation with bound ATP shown as a black diamond or in a Down-conformation in an ADP-locked state, whereas the N-domains of mutants adopt only the Up-conformation as observed in this study. On the basis of available structural and biochemical information, we introduce two additional conformational states. The Empty state has the N-domain conformation undefined and the nucleotide-binding site shown as a black circle. The ADP-open state also has an N-domain conformation similar to that of Down-conformation as determined by the crystal structure of R155H mutant with bound ADP, which is also shown as a black rectangle. Bound ADP can only be exchanged through the ADP-open state. In wild-type p97, the equilibration between ADP-open and ADP-locked favours heavily the latter and is presumably regulated by effectors such as p47 or ATP hydrolysis in D2-domain. In IBMPFD mutants, the tight control between ADP-open and ADP-locked is disrupted and the equilibration is now favouring the ADP-open state.
The difference between the wild type and mutants, however, lies in the transition between the ADP-locked state and the ADP-open state. We propose that in the wild-type protein, this transition is tightly controlled and characterized by the asymmetry in nucleotide-binding states in D1-domains of different subunits, resulting in a low concentration of the ADP-open state (Figure 5A), whereas in IBMPFD mutants, this control mechanism is altered, leading to a high concentration of subunits in the ADP-open state (Figure 5B). Consequently, p97 mutants undergo a uniform N-domain conformational change in response to high concentrations of ATPγS, allowing successful crystallization. In contrast, the asymmetry in N-domain conformation in wild-type p97 produces a non-uniform protein preparation, resisting crystallization. We speculate that in wild-type p97, the control for the transition between ADP-locked and ADP-open state in D1 could be achieved in two ways: (1) the binding of adaptor proteins to the N-domain and (2) the hydrolysis of ATP in D2-domain. The N-domain was shown to display an influence on the ATPase activities of both N–D1 and the full-length p97 orthologue VAT, as the N-domain-deleted p97 mutants have higher ATPase activities (Tang et al, unpublished; Gerega et al, 2005). The binding of adaptor protein p47 to the N-domain was shown to inhibit the ATPase activity of p97 (Meyer et al, 1998). The communications between D1 and D2 were also known to exist for p97 and other type II AAA proteins. For example, it was shown that the absence of D2-domain inhibits the nucleotide exchange activity in D1 (Davies et al, 2005). The yeast Hsp104, another type II AAA protein, displays cooperative kinetics and inter-domain communication for its two ATPase domains (Hattendorf and Lindquist, 2002). However, the exact details of these potential control mechanisms for the switching of D1 nucleotide states remain elusive.
The unique properties exhibited by IBMPFD mutants in comparison with wild-type p97 give indications on how wild-type enzyme may function. For example, the differences in D1-domain ADP-binding affinity and in nucleotide exchangeability observed in our study and others suggest that in a wild-type p97 hexamer, the asymmetry in nucleotide-binding states of D1-domains may be important for function and the N-domains from different subunits are unlikely to undergo a concerted Up- or Down-conformational change. However, between the two possible mechanisms for a non-concerted N-domain movement, existing data seem insufficient to distinguish a random from a sequential conformational change of the N-domains. Importantly, the fact that all IBMPFD mutations in either the N- or D1-domain are able to induce the same changes in conformation and thermodynamic properties in p97 suggests that it is the conformational change in the N-domain, not the mutation per se, that is important for the control of D1 ATPase activity.
Materials and methods
Cloning, expression, and purification of p97 N–D1 fragments of wild type and mutants
Protein expression and purification of wild-type and mutant p97 N–D1 fragments is detailed elsewhere (Tang et al, 2009). Briefly, the expression plasmid pQE-p97 for the wild-type full-length p97 was generated by inserting a PCR-amplified product (accession number: BC110913, Thermo Scientific, Huntsville, AL) into a pQE60 expression vector (Qiagen, Valencia, CA) at the BamHI/BglII sites. The expression plasmid pQE-p97ND1 encoding Met1 to Gly481 (N–D1 fragment) was then generated by deletion mutagenesis, removing residues 482 to 806 from the plasmid pQE-p97, using the QuikChange® Site-Directed Mutagenesis kit (Stratagene, La Jolla, CA). The wild-type construct contains a hexahistidine tag at its C-terminus. All mutant constructs were made based on this wild-type plasmid by using the QuikChange kit with appropriate primers. The proteins were expressed in E. coli strain M15 (QIAexpression system, Qiagen). After centrifugation, the E. coli cell pellet was suspended in buffer A (25 mM Tris–HCl, pH 7.6, 0.3 mM NaCl, 1 mM DTT) and lysed by sonication. After the removal of cell debris, the supernatant was incubated with Ni-NTA resin (Qiagen) pre-equilibrated with buffer A for 1 h at 4oC. The Ni-NTA resin was washed and bound protein eluted in steps with buffer A supplemented with different concentrations of imidazole (50, 100, or 250 mM).
Crystallization and X-ray diffraction data collection
Before crystallization, protein solutions were supplemented with DTT, MgCl2, and ATPγS or ADP to a final concentration of 5, 40, and 4 mM, respectively. Crystallization experiments were performed using the hanging drop vapour diffusion technique, after mixing equal volumes of proteins with reservoir solutions. The R155H mutant (7 mg/ml) was set up for crystallization with a reservoir solution containing 100 mM citrate buffer pH 5.6, 6% benzamidine, 7% PEG3350, and 20% glycerol. Crystals of the R95G mutant were obtained by mixing the protein (5 mg/ml) with a reservoir solution consisting of 100 mM NaCl, 100 mM citrate pH 5.7, 13.5% PEG3350, and 20% glycerol. The R86A mutant (5 mg/ml) was crystallized with a well solution containing 100 mM NaCl, 4% benzamidine, 100 mM citrate, pH 5.8, 16.5% PEG3350, and 20% glycerol. Crystals ranging in size between 0.1 and 0.2 mm were obtained in 1–3 weeks at 15°C. The R155H mutant with ADP bound was crystallized by mixing the protein (7 mg/ml) in a 1:1 ratio with a reservoir solution containing 100 mM citrate buffer, pH 5.8, 300 mM NaCl, 2.5% benzamidine, 13.8% PEG3350, and 5% glycerol.
Crystals of p97 mutants were cryo-protected before X-ray diffraction experiments, which were carried out at 100K at the SER-CAT (ID-22) beamline of the advanced photon source (APS) at the Argonne National Lab (ANL). Diffraction images were collected with a Mar225 CCD detector and processed and scaled with the HKL2000 package (Otwinowski and Minor, 1997).
Structure determination and analysis of the N–D1 mutants
Crystals of the R155H mutant with bound Mg2+·ATPγS have the symmetry of the space group R3. There are two copies of N–D1 in a crystallographic AU. The structure was determined by MR with the program Phaser (Storoni et al, 2004), using templates derived from the wild-type N–D1 structure (PDB:1E32) (Zhang et al, 2000). Atomic models were examined and revised manually with the graphical modelling programme O (Jones et al, 1991) and refined with Refmac with NCS restraints (Murshudov et al, 1997).
Both the R95G and R86A mutants in ATPγS-bound form crystallized in the triclinic space group P1. Six copies of the p97 N–D1 protomer are present in the unit cell and the structures were solved with Phaser, using the R155H mutant structure as a search model. The structures were subsequently refined using Refmac with NCS restraints.
Crystals of the R155H mutant with ADP bound belong to the symmetry of the space group P212121. Twelve copies of p97 N–D1 are found in one AU in two hexamers. The structure was determined by MR with Phaser using the wild-type N–D1 structure (PDB:1E32) as a search model and refined using Refmac with NCS restraints. Statistics on the qualities of diffraction data sets and refined structures are given in Table II. Structure alignments were performed in O; accessible and buried surface area calculations were performed with Areaimol (Lee and Richards, 1971) in CCP4.
Structural analysis of wild-type and mutant p97 N–D1 fragments by solution X-ray scattering
Before data collection, protein samples were dialysed overnight at 4°C against 20 mM Tris, pH 8.0, 2 mM MgCl2, and 5% glycerol, in the presence of either 0.1 mM ADP or ATPγS. For various N–D1 fragments, protein concentrations of 0.5 and 2.0 mg/ml were used for the SAXS experiments and a concentration of 11 mg/ml was used for the wide-angle X-ray scattering experiments. SAXS experiments were conducted at the BIO-CAT (ID-18) beamline at APS, ANL. All buffer and sample solutions were filtered (0.1 μm filter) and subsequently pumped into a 1.5 mm diameter quartz capillary connected to a MicroLab® 500 series syringe pump (Hamilton Company, Reno, NV) with a flow rate of 12.5 μl/s. Samples were exposed to a focused X-ray beam with a wavelength of 1.033 Å and a flux of 2 × 1013 photons/s for 1 s. A Mar165 CCD detector placed 2304 mm and 169 mm from the sample, respectively, for small- and wide-angle data collections was used for measuring 2D scattering patterns. For each sample, 15 scattering images with 1 s exposure were collected. The scattering profiles were obtained by circularly integrating intensities over the image using a program written in Igor Pro (http://www.wavemetrics.com) provided by the beamline. After manual rejection of outliers, the scattering profiles from the same sample were averaged and the background scattering from the buffer solution was subtracted. The quality of the data was examined and the initial radius of gyration (Rg) was obtained from the Guinier plot at low resolution (Guinier and Fournet, 1955). Individual scattering profile curves for a given nucleotide-bound state that were collected at different protein concentrations and over different scattering angle ranges were scaled and merged together in GNOM (Svergun, 1992) to yield a low-noise composite curve covering a wide angular range. The vector distribution functions, p(r), calculated in GNOM by indirect Fourier transform from the scattering curve, were used to estimate the maximal vector length of p97 in solution.
Measurement of nucleotide-binding affinity by ITC
ITC experiments were performed using VP-ITC titration microcalorimeter (GE Healthcare, Piscataway, New Jersey). All titrations were performed in a buffer containing 50 mM Tris–HCl, pH 8.0, 2 mM MgCl2, and 5% glycerol at 25°C. Protein concentrations were determined photometrically at 280 nm using a calculated molar extinction coefficient of 21 430 M−1 cm−1. Protein solutions (10–20 μM) were dialysed against the same buffer overnight, which was also used for preparing ligand solutions (100–200 μM). Raw data in the form of the incremental heat per mole of added ligand for the titration were fitted by non-linear least squares to either one-site or two-site models using ORIGIN7 software. All experiments were performed at least three times.
Coordinates
Atomic coordinates of the refined structures have been deposited in the Protein Data Bank (http://www.pdb.org) with the pdb codes 3HU1 for the p97-ND1-R95G mutant, 3HU2 for the p97-ND1-R86A mutant, and 3HU3 for the p97-ND1-R155H mutant, respectively. All mutant structures are bound with ATPγS.
Supplementary Material
Acknowledgments
We thank the staff of the SER-CAT beamline at APS, ANL for their assistance with data collection. This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. We acknowledge the Natural Science Foundation of China (Program 30628006) for its support to the Tongji University, China and to DX. Our special thanks go to Drs. Susan Gottesman, Michael Maurizi, and Yihong Ye for critical reading of the manuscript. We also thank George Leiman for editorial assistance.
Footnotes
The authors declare that they have no conflict of interest.
References
- Beuron F, Dreveny I, Yuan X, Pye VE, McKeown C, Briggs LC, Cliff MJ, Kaneko Y, Wallis R, Isaacson RL, Ladbury JE, Matthews SJ, Kondo H, Zhang X, Freemont PS (2006) Conformational changes in the AAA ATPase p97-p47 adaptor complex. EMBO J 25: 1967–1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuron F, Flynn TC, Ma J, Kondo H, Zhang X, Freemont PS (2003) Motions and negative cooperativity between p97 domains revealed by cryo-electron microscopy and quantised elastic deformational model. J Mol Biol 327: 619–629 [DOI] [PubMed] [Google Scholar]
- Briggs LC, Baldwin GS, Miyata N, Kondo H, Zhang X, Freemont PS (2008) Analysis of nucleotide binding to P97 reveals the properties of a tandem AAA hexameric ATPase. J Biol Chem 283: 13745–13752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai RM, Li CC (2001) Valosin-containg protein is a multi-ubiquitin chain-targeting factor required in ubiquitin-proteasome degradation. Nat Cell Biol 3: 740–744 [DOI] [PubMed] [Google Scholar]
- Davies JM, Tsuruta H, May AP, Weis WI (2005) Conformational changes of p97 during nucleotide hydrolysis determined by small-angle X-ray scattering. Structure 13: 183–195 [DOI] [PubMed] [Google Scholar]
- DeLaBarre B, Brunger AT (2003) Complete structure of p97/valosin-containing protein reveals communication between nucleotide domains. Nat Struc Biol 10: 856–863 [DOI] [PubMed] [Google Scholar]
- DeLaBarre B, Brunger AT (2005) Nucleotide dependent motion and mechanism of action of p97/VCP. J Mol Biol 347: 437–452 [DOI] [PubMed] [Google Scholar]
- Djamshidian A, Schaefer J, Haubenberger D, Stogmann E, Zimprich F, Auff E, Zimprich A (2009) A novel mutation in the VCP gene (G157R) in a german family with inclusion-body myopathy with paget disease of bone and frontotemporal dementia. Muscle Nerve 39: 389–391 [DOI] [PubMed] [Google Scholar]
- Gerega A, Rockel B, Peters J, Tamura T, Baumeister W, Zwickl P (2005) VAT, the thermoplasma homolog of mammalian p97/VCP, is an N domain-regulated protein unfoldase. J Biol Chem 280: 42856–42862 [DOI] [PubMed] [Google Scholar]
- Glatter O (1980) Computation of distance distribution function and scattering function of models for small angle scattering experiments. Acta Phys Austriaca 52: 243–256 [Google Scholar]
- Glatter O, Kratky O (1982) Small-Angle X-ray Scattering. London, UK: Acedamic Press [Google Scholar]
- Guinier A, Fournet G (1955) Small Angle Scattering of X-rays. New York, USA: Wiley Press [Google Scholar]
- Guo F, Maurizi MR, Esser L, Xia D (2002) Crystal structure of ClpA, an HSP100 chaperone and regulator of ClpAP protease. J Biol Chem 277: 46743–46752 [DOI] [PubMed] [Google Scholar]
- Halawani D, LeBlanc AC, Rouiller I, Michnick SW, Servant MJ, Latterich M (2009) Hereditary inclusion body myopathy-linked p97/VCP mutations in the NH2 domain and the D1 ring modulate p97/VCP ATPase activity and D2 ring conformation. Mol Cell Biol 29: 4484–4494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haubenberger D, Bittner RE, Rauch-Shorny S, Zimprich F, Mannhalter C, Wagner L, Mineva I, Vass K, Auff E, Zimprich A (2005) Inclusion body myopathy and Paget disease is linked to a novel mutation in the VCP gene. Neurology 65: 1304–1305 [DOI] [PubMed] [Google Scholar]
- Hattendorf DA, Lindquist SL (2002) Cooperative kinetics of both Hsp104 ATPase domains and interdomain communication revealed by AAA sensor-1 mutants. EMBO J 21: 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbers CU, Clemen CS, Kesper K, Boddrich A, Hofmann A, Kamarainen O, Tolksdorf K, Stumpf M, Reichelt J, Roth U, Krause S, Watts G, Kimonis V, Wattjes MP, Reimann J, Thal DR, Biermann K, Evert BO, Lochmüller H, Wanker EE et al. (2007) Pathological consequences of VCP mutations on human striated muscle. Brain 130: 381–393 [DOI] [PubMed] [Google Scholar]
- Huyton T, Pye VE, Briggs LC, Flynn TC, Beuron F, Kondo H, Ma J, Zhang X, Freemont PS (2003) The crystal structure of murine p97/VCP at 3.6 Å. J Struct Biol 144: 337–348 [DOI] [PubMed] [Google Scholar]
- Ishikawa T, Maurizi MR, Steven AC (2004) The N-terminal substrate-binding domain of ClpA unfoldase is highly mobile and extends axially from the distal surface of ClpAP protease. J Struct Biol 146: 180–188 [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan ZW, Kjeldgaard M (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A 47: 110–119 [DOI] [PubMed] [Google Scholar]
- Kumar KR, Needham M, Mina K, Davis M, Brewer J, Staples C, Ng K, Sue CM, Mastaglia FL (2010) Two Australian families with inclusion-body myopathy, Paget's disease of bone and frontotemporal dementia: novel clinical and genetic findings. Neuromuscul Disord 20: 330–334 [DOI] [PubMed] [Google Scholar]
- Lee B, Richards FM (1971) The interpretation of protein structures: estimation of static accessibility. J Mol Biol 55: 379–400 [DOI] [PubMed] [Google Scholar]
- Lee S, Sowa ME, Watanabe Y, Sigler PB, Chiu W, Yoshida M, Tsai TF (2003) The structure of ClpB: a molecular chaperone that rescues proteins from an aggregated state. Cell 115: 229–240 [DOI] [PubMed] [Google Scholar]
- Ma J, Xia D (2008) The use of blue native PAGE in the evaluation of membrane protein aggregation states for crystallization. J Appl Cryst 41: 1150–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer HH, Kondo H, Warren G (1998) The p47 co-factor regulates the ATPase activity of the membrane fusion protein, p97. FEBS Lett 437: 255–257 [DOI] [PubMed] [Google Scholar]
- Meyer HH, Wang Y, Warren G (2002) Direct binding of ubiquitin conjugates by the mammalian p97 adaptor complexes, p47 and Ufd1-Npl4. EMBO J 21: 5645–5652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Neuwald AF, Aravind L, Spouge JL, Koonin EV (1999) AAA+: a class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res 9: 27–43 [PubMed] [Google Scholar]
- O'Brien R, Ladbury JE, Chowdhry BZ (2001) Isothermal ittration calorimetry of biomolecules. In Protein-Ligand Interaction: Hydrodynamics and Calorimetry: A Practical Approach, Harding SE, Chowdhry BZ (eds) pp 263–274. Oxford: Oxford University Press [Google Scholar]
- Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Pye VE, Dreveny I, Briggs LC, Sands C, Beuron F, Zhang X, Freemont PS (2006) Going through the motions: the ATPase cycle of p97. J Struct Biol 156: 12–28 [DOI] [PubMed] [Google Scholar]
- Rape M, Hoppe T, Gorr I, Kalocay M, Richly H, Jentsch S (2001) Mobilization of processed, membrane-tethered SPT23 transcription factor by CDC48(UFD1/NPL4), a ubiquitin-selective chaperone. Cell 107: 667–677 [DOI] [PubMed] [Google Scholar]
- Richly H, Rape M, Braun S, Rumpf S, Hoege C, Jentsch S (2005) A series of ubiquitin binding factors connects CDC48/p97 to substrate multiubiquitylation and proteasomal targeting. Cell 120: 73–84 [DOI] [PubMed] [Google Scholar]
- Rouiller I, DeLaBarre B, May AP, Weis WI, Brunger AT, Milligan RA, Wilson-Kubalek EM (2002) Conformational changes of the multifunction p97 AAA ATPase during its ATPase cycle. Nat Struct Biol 9: 950–957 [DOI] [PubMed] [Google Scholar]
- Schroder R, Watts GD, Mehta SG, Evert BO, Broich P, Fliessbach K, Pauls K, Hans VH, Kimonis V, Thal DR (2005) Mutant valosin-containing protein causes a novel type of frontotemporal dementia. Ann Neurol 5: 457–461 [DOI] [PubMed] [Google Scholar]
- Schuberth C, Buchberger A. (2008) UBX domain proteins: major regulators of the AAA ATPase Cdc48/p97. Cell Mol Life Sci 65: 2360–2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storoni LC, McCoy AJ, Read RJ (2004) Likelihood-enhanced fast rotation functions. Acta Cryst D 60: 432–438 [DOI] [PubMed] [Google Scholar]
- Svergun DI (1992) Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J Appl Crystallogr 25: 495–503 [Google Scholar]
- Tang WK, Li DY, Esser L, Xia D (2009) Purification, crystallization and preliminary X-ray diffraction of disease-related mutants of p97. Acta Cryst F 65: 1166–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Song C, Li C (2004) Molecular perspectives on p97-VCP: progress in understanding its structure and diverse biological functions. J Struct Biol 146: 44–57 [DOI] [PubMed] [Google Scholar]
- Watts G, Thomasova D, Ramdeen S, Fulchiero E, Mehta S, Drachman D, Weihl C, Jamrozik Z, Kwiecinski H, Kaminska A, Kimonis V (2007) Novel VCP mutations in inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia. Clin Genet 72: 420–426 [DOI] [PubMed] [Google Scholar]
- Watts GD, Wymer J, Kovach MJ, Mehta SG, Mumm S, Darvish D, Pestronk A, Whyte MP, Kimonis VE (2004) Inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia is caused by mutant valosin-containing protein. Nat Genet 36: 377–381 [DOI] [PubMed] [Google Scholar]
- Weihl CC, Dalal S, Pestronk A, Hanson PI (2006) Inclusion body myopathy-associated mutations in p97/VCP impair endoplasmic reticulum-associated degradation. Hum Mol Genet 15: 189–199 [DOI] [PubMed] [Google Scholar]
- Woodman PG (2003) p97, a protein coping with multiple identities. J Cell Sci 116: 4283–4290 [DOI] [PubMed] [Google Scholar]
- Ye Y (2006) Diverse functions with a common regulator: ubiquitin takes command of an AAA ATPase. J Struct Biol 156: 29–40 [DOI] [PubMed] [Google Scholar]
- Zhang X, Shaw A, Bates PA, Newman RH, Gowen B, Orlova E, Gorman MA, Kondo H, Dokurno P, Lally J, Leonard G, Meyer H, van Heel M, Freemont PS (2000) Structure of the AAA ATPase p97. Mol Cell 6: 1473–1484 [DOI] [PubMed] [Google Scholar]
- Zhang YL, Zhang ZY (1998) Low-affinity binding determined by titration calorimetry using a high-affinity coupling ligand: a thermodynamic study of ligand binding to protein tyrosine phosphatase 1B. Anal Biochem 261: 139–148 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.