Abstract
A number of meiosis-specific mRNAs are initially weakly transcribed, but then selectively removed during fission yeast mitotic growth. These mRNAs harbour a region termed DSR (determinant of selective removal), which is recognized by the YTH family RNA-binding protein Mmi1p. Mmi1p directs the destruction of these mRNAs in collaboration with nuclear exosomes. However, detailed molecular mechanisms underlying this process of selective mRNA elimination have remained elusive. In this study, we demonstrate the critical role of polyadenylation in this process. Two-hybrid and genetic screens revealed potential interactions between Mmi1p and proteins involved in polyadenylation. Additional investigations showed that destruction of DSR-containing mRNAs by exosomes required polyadenylation by a canonical poly(A) polymerase. The recruitment of Pab2p, a poly(A)-binding protein, to the poly(A) tail was also necessary for mRNA destruction. In cells undergoing vegetative growth, Mmi1p localized with exosomes, Pab2p, and components of the polyadenylation complex in several patchy structures in the nucleoplasm. These patches may represent the sites for degradation of meiosis-specific mRNAs with untimely expression.
Keywords: exosome, fission yeast, meiosis, mRNA degradation, polyadenylation
Introduction
Most eukaryotic cells have the genetic potential to carry out meiosis, but in reality only a limited population of cells, such as germ cells, actually performs meiotic division. As the untimely expression of meiotic genes during vegetative proliferation could be destructive, it is presumable that cells strictly separate the mitotic and meiotic cell division programmes. Consistent with this hypothesis, our previous analyses revealed that the fission yeast, Schizosaccharomyces pombe, possesses a mechanism for the elimination of meiosis-specific mRNAs from vegetatively growing cells (Harigaya et al, 2006). These mRNAs harbour a unique region, termed DSR (determinant for selective removal), which serves as a marker for mRNA degradation. Mmi1p was identified as a new type of RNA-binding protein that binds to the DSR region and directs transcript degradation. Furthermore, we also observed that Mei2p, the master regulator of meiosis in fission yeast (Watanabe and Yamamoto, 1994; Watanabe et al, 1997), sequesters Mmi1p in a nuclear dot structure (Mei2 dot) during meiotic prophase to prevent Mmi1p from directing the degradation of meiosis-specific mRNAs during this stage of cell division (Harigaya et al, 2006). The Mei2 dot was observed to be composed of Mei2p and meiRNA encoded by the sme2 gene, and to be attached to the sme2 locus on chromosome II (Watanabe et al, 1997; Yamashita et al, 1998; Shimada et al, 2003).
The transcription of meiotic genes does not stop completely during vegetative growth in fission yeast, and elimination of unnecessary meiosis-specific messages by Mmi1p seems to be physiologically indispensable, as growth is severely impaired if cells lose Mmi1p expression (Harigaya et al, 2006). The mRNA elimination system involving DSR and Mmi1p is likely to be the first example of a mechanism to selectively remove unnecessary mRNA species to maintain a certain cellular status. Hence, it is of great interest to clarify of the detailed mechanisms responsible for this selective elimination. Mmi1p is a relatively small protein of 488 amino acids with no obvious known features, other than a putative YTH family RNA-binding domain. Mmi1p also seems to cooperate with the exosome, a multi-subunit protein complex with nuclease activity (Mitchell et al, 1997; Allmang et al, 1999), to degrade mRNAs within the nucleus (Harigaya et al, 2006).
To gain further insight into the molecular mechanisms that underlie selective elimination of DSR-containing mRNAs, we set out to identify and characterize new components of this targeted degradation system. The analysis of the factors identified in the search delineates polyadenylation of the target mRNAs and subsequent recruitment of a poly(A)-binding protein to them as crucial steps in selective mRNA degradation.
Results
Identification of factors involved in 3′-end processing of mRNA, which participate in selective mRNA elimination
To search for factors that might cooperate with Mmi1p in facilitating selective elimination of meiosis-specific mRNAs, we carried out a genome-wide, yeast two-hybrid screen using Mmi1p as bait. Several candidates, including Rna15p and Pab2p, were identified as possible Mmi1p-interacting proteins in this analysis (Supplementary Figure S1). Rna15p (SPAC644.16) is an apparent orthologue of Saccharomyces cerevisiae RNA15, which is a subunit of the multi-subunit cleavage factor CF1A, a component of the polyadenylation complex (Minvielle-Sebastia et al, 1994; Kessler et al, 1996). Pab2p is a previously characterized nuclear poly(A)-binding protein (Perreault et al, 2007).
To identify factors that might be necessary to promote selective mRNA elimination, we also carried out a screen for mutations that could suppress meiotic arrest in the sme2Δ mutant, based on the observation that a reduction in Mmi1p activity suppressed this arrest (Harigaya et al, 2006). The sme2Δ mutant lacks meiRNA and cannot form the previously characterized Mei2 dot structures (Watanabe et al, 1997; Yamashita et al, 1998; Shimada et al, 2003), resulting in a failure to downregulate Mmi1p. In this screen, we observed that an insertion introduced in an intron of the pla1 gene, which encodes a poly(A) polymerase composing the polyadenylation complex (Ohnacker et al, 1996), could suppress sme2Δ (Supplementary Figure S2). This insertion (pla1-10) resulted in the production of a truncated form of Pla1p (Supplementary Figure S2).
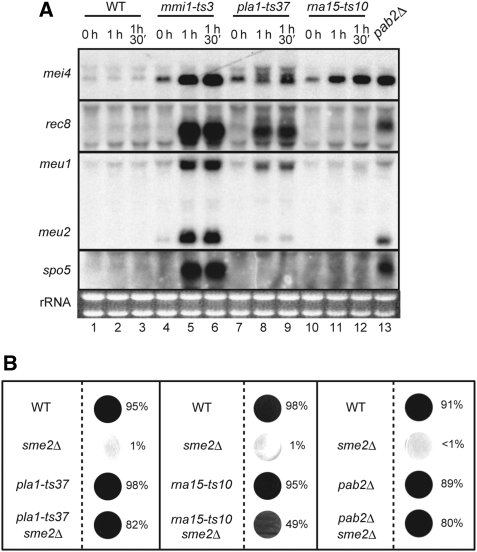
We confirmed the physical interaction of Mmi1p with Pla1p, Rna15p and Pab2p by immunoprecipitation and pull-down analyses, as shown in Supplementary Figure S3. Given these results, we tested whether the loss of function in pla1, rna15 or pab2 could affect expression of meiosis-specific transcripts in vegetative cells. As pla1 and rna15 are essential for cell growth, we isolated temperature-sensitive (ts) mutants of these two genes and used a viable pab2 deletion mutant in subsequent experiments. As shown in Figure 1A, cells of each mutant at least partially accumulated meiosis-specific mRNAs when the respective gene function was eliminated. However, the levels of mRNA accumulation in these mutants were generally lower than those observed in the mmi1-ts3 mutant. The pattern of affected mRNAs also seemed to vary to some extent according to the gene mutated, suggesting an underlying complexity of the whole system relevant to selective mRNA elimination.
Figure 1.
Components of the polyadenylation complex and the poly(A)-binding protein Pab2p contribute to the elimination of DSR-containing mRNAs. (A) JY450 (WT), JV564 (mmi1-ts3), JT452 (pla1-ts37) and JT453 (rna15-ts10) cells were cultured at 25°C in YE liquid medium and then shifted to 37°C. After the indicated times, total RNA was extracted from each sample and processed for northern blot analysis (lanes 1–12). For JT454 (pab2Δ), total RNA was extracted from cells cultured at 30°C (lane 13). A total of 5μg of RNA was loaded in each lane. (B) Suppression of sme2Δ by pla1-ts37, rna15-ts10 and pab2Δ. JY450 (WT), JZ464 (sme2Δ), JT452 (pla1-ts37), JT453 (rna15-ts10), JT455 (pla1-ts37 sme2Δ) and JT456 (rna15-ts10 sme2Δ) cells were cultured in YE medium at 25°C and spotted onto an SSA plate. Incubation was continued at 25°C for 4 days, and then cells were stained with iodine and the sporulation efficiency of each cell line was calculated. JT454 (pab2Δ) and JT457 (pab2Δ sme2Δ) cells and control JY450 and JZ464 cells were cultured at 30°C and after 2 days the sporulation efficiency was measured at 30°C.
We then evaluated whether the mutations that we created in pla1, rna15 and pab2 could suppress meiotic arrest in sme2Δ cells. As shown in Figure 1B, the mutants suppressed the sme2Δ phenotype. To our surprise, the pla1-ts37 and rna15-ts10 mutations could suppress the sme2Δ phenotype at 25°C (Figure 1B), although strains carrying these mutations exhibited no obvious growth retardation at this temperature (data not shown). As pla1-10, the original insertion mutation that suppressed sme2Δ, did not seem to retard vegetative growth, a defect in the polyadenylation complex might have a more significant impact on meiotic transcripts than on mitotic transcripts. Overall, the observed genetic interactions suggested that polyadenylation might contribute to the selective elimination of meiosis-specific mRNAs in mitotic cells.
Excessive polyadenylation of meiosis-specific mRNAs generated in an exosome mutant
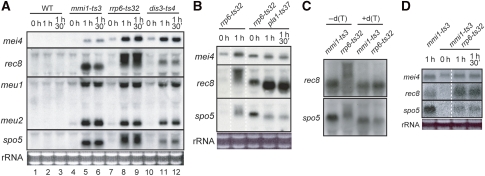
We have previously shown that Mmi1p could physically interact with Rrp6p (Harigaya et al, 2006), which is a nucleus-specific subunit of the exosome (Briggs et al, 1998; Burkard and Butler 2000). Furthermore, a defect in Rrp6p was as effective in suppressing the sme2Δ phenotype as a reduction in Mmi1p activity, suggesting that exosomes were likely to eliminate meiosis-specific mRNAs within the cell nucleus (Harigaya et al, 2006). We next examined the expression of meiosis-specific transcripts in the ts exosome mutant cell lines, rrp6 and dis3; the latter of which encodes a core subunit of the exosome (Ohkura et al, 1988; Dziembowski et al, 2007; Murakami et al, 2007). As shown in Figure 2A, the transcripts of five meiotic genes, namely mei4, rec8, meu1, meu2 and spo5, were detected in both rrp6-ts32 and dis3-ts4 cells shifted to the restrictive temperature in growth medium (lanes 8, 9, 11 and 12) at levels that were comparable to those observed in mmi1-ts3 cells (lanes 5 and 6). These transcripts were never induced by a temperature shift in wild-type cells, indicating that nuclear exosomes are required for the elimination of meiosis-specific mRNAs during vegetative growth (lanes 2 and 3; Harigaya et al, 2006).
Figure 2.
DSR-containing mRNAs suffer Pla1p-dependent excessive polyadenylation in the rrp6-ts32 strain at the restrictive temperature. (A) Northern blot analysis of the expression of meiosis-specific genes mei4, rec8, meu1, meu2 and spo5 in exosome mutants. Cells were cultured in YE liquid medium at 25°C, shifted to the restrictive temperature 37°C and then sampled for RNA extraction after incubation for the indicated times. Lanes 1–3, JY450 (WT; wild-type); lanes 4–6, JV564 (mmi1-ts3); lanes 7–9, JT432 (rrp6-ts32); and lanes 10–12, JT449 (dis3-ts4). Total RNA (5 μg) was loaded in each lane. (B) JT432 (rrp6-ts32) and JT458 (rrp6-ts32 pla1-ts37) cells were cultured at 25°C, and then shifted to 37°C. At the indicated times, total RNA was extracted from each sample and processed for northern blot analysis as in (A). (C) Northern blots of RNase H-treated rec8 and spo5 transcripts derived from growing mutant strains as indicated. RNase H treatment was performed in the absence (lanes 1 and 2) and presence (lanes 3 and 4) of poly(dT). (D) JV564 (mmi1-ts3) and JT496 (mmi1-ts3 rrp6-ts32) cells were cultured at 25°C, and then shifted to 37°C. At the indicated times, total RNA was extracted from each sample and processed for northern blot analysis as in (A).
In the exosome mutants, we noted that some ectopically expressed, meiosis-specific transcripts appeared to be smeary and larger than the natural transcripts produced during meiosis or those expressed in the mmi1-ts3 mutant. For example, rec8 transcripts accumulated in rrp6-ts32 cells were extremely larger, spo5 transcripts were substantially larger and mei4 transcripts were slightly larger in comparison with natural transcripts (Figure 2A). In the experiments described below, we tested whether the increase in the size of these transcripts might result from excessive polyadenylation.
First, we analysed rec8, spo5 and mei4 transcripts produced in the absence of active canonical poly(A) polymerase. As shown in Figure 2B, the increase in transcript size observed in the rrp6-ts32 mutant was almost completely lost in the rrp6-ts32 pla1-ts37 double mutant, supporting the idea that the increase was caused by polyadenylation. Notably, the double mutant accumulated a considerable quantity of meiosis-specific transcripts at the permissive temperature 25°C, implying that the rrp6-ts32 and the pla1-ts37 alleles were both partially defective at this temperature, although each single mutant showed no obvious growth defect. The confirmation that the rrp6-ts32 allele was not fully functional at 25°C was based on the observation that the rrp6-ts32 pab2Δ double mutant suffered severe growth retardation at this temperature (data not shown).
To estimate the length of poly(A) additions, poly(A) tracts were removed by RNase H digestion in the presence of oligo(dT) (Hausen and Stein, 1970). After digestion, both rec8 and spo5 transcripts yielded a single, non-smeary band in gel electrophoresis of a size that was close to that observed for fragments derived from mmi1-ts3 cells (Figure 2C). These results strongly suggest that rec8 and spo5 transcripts that have escaped from destruction by exosomes are likely to exhibit various extents of excessive polyadenylation.
Interestingly, excessive polyadenylation of rec8, spo5 and mei4 transcripts, as was seen in the rrp6-ts32 mutant, did not occur in the mmi1-ts3 rrp6-ts32 double mutant (Figure 2D). This suggested that Mmi1p was responsible for the excessive polyadenylation observed in rrp6-ts32 cells, and that the polyadenylation reaction involving Mmi1p might be unusual either quantitatively or qualitatively.
Meanwhile, slow migrating bands were also observed for mei4 mRNA produced in pla1-ts37 cells shifted to the restrictive temperature, though their intensity was moderate (Figure 1, lanes 8 and 9). They turned out not to represent excessive polyadenylation. Transcripts in these bands were not homogeneous with regard to the 3′ end, but they generally carried a shorter poly(A) tail and a longer 3′ UTR stretch, which apparently resulted from a failure in cleavage at the proper polyadenylation site (data not shown).
Polyadenylation is required for the selective elimination of meiosis-specific mRNAs
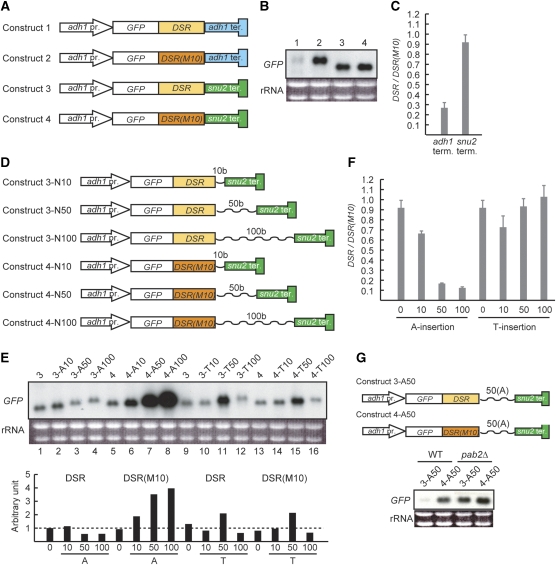
To establish the significance of polyadenylation for the elimination of meiosis-specific transcripts, we devised a system for transcribing a DSR-containing reporter gene with or without the addition of a poly(A) tail. The reporter constructs used are shown schematically in Figure 3A. The parental construct (Construct 1) was comprised of the constitutive adh1 promoter, the jellyfish green fluorescent protein (GFP) open reading frame (ORF), the DSR region derived from spo5 and the terminator region of adh1. The transcripts produced from Construct 1 were barely detectable in mitotic cells (Figure 3B, lane 1). To confirm that DSR-dependent selective elimination was functional for Construct 1, we substituted the DSR sequence with a non-functional sequence termed DSR-M10 that had six nucleotide substitutions (data not shown). The resultant construct (Construct 2) escaped degradation, resulting in transcript accumulation within mitotic cells (Figure 3B, lane 2). We then proceeded to examine the effect of a poly(A) tail in this system. To terminate transcripts with no poly(A) addition, we replaced the adh1 terminator in Construct 1 with the terminator of the snu2 gene, which encodes a small nuclear RNA (Brennwald et al, 1988; Zhou et al, 1999). Interestingly, this construct (Construct 3) produced detectable amounts of transcripts in mitotic cells, which were nearly as much as its counterpart with DSR-M10 instead of a functional DSR (Construct 4; Figure 3B, lanes 3 and 4). Data were quantified by measuring the intensity of each band in Figure 3B and then calculating the ratio between the counterparts bearing DSR and DSR(M10) (Figure 3C). These results suggest that a poly(A) tail is necessary for targeting DSR-containing mRNAs for selective elimination.
Figure 3.
A poly(A) tail is required for the elimination of DSR-containing RNA. (A) Schematic illustration of the reporter constructs used in (B). A detailed explanation of each construct is provided in the text. adh1 pr., the promoter for the adh1 gene; GFP, the ORF for jellyfish green fluorescent protein; DSR, the DSR sequence of the spo5 gene; DSR(M10), a defective form of DSR; adh1 ter., the terminator for the adh1 gene; and snu2 ter., the terminator for the snu2 gene. (B) Constructs 1–4, as shown in (A), were integrated at the lys1 locus of the parental strain JY333. The resulting four strains were cultured in YE at 30°C. Total RNA was extracted from each culture and analysed by northern blot analysis. RNA (5 μg) was loaded in each lane and the GFP sequence was used as the probe. (C) Comparison of the band intensity in (B) between lanes 1 and 2 (adh1 term.) and lanes 3 and 4 (snu2 term.). (D) Schematic illustration of the reporter constructs used in (E). Either a poly(dA) tract or a poly(dT) tract was inserted after the DSR sequence in Construct 3 and Construct 4. The tract length was 10, 50 or 100 bases. (E) Constructs carrying a poly(dA) or poly(dT) tract, as shown in (D), were integrated at the lys1 locus of JY333. The resulting strains were examined for GFP expression by northern blot analysis as described in (B). The intensity of each band was measured and normalized by the amount of rRNA as the loading control, which is graphically shown under the northern blot panel. (F) Comparison of the band intensity in (E) between lanes 1 and 5 (A-0), lanes 2 and 6 (A-10), lanes 3 and 7 (A-50), lanes 4 and 8 (A-100), lanes 9 and 13 (T-0), lanes 10 and 14 (T-10), lanes 11 and 15 (T-50) and lanes 12 and 16 (T-100). (G) Evaluation of the necessity of Pab2p for the elimination of DSR-containing RNA with an artificial poly(A) tract. The expression of GFP was monitored by northern blot analysis in wild-type (JY450) and pab2Δ (JT454) cells carrying either Construct 3-A50 or 4-A50.
To determine whether artificial addition of a poly(A) tract could stimulate the elimination of DSR-containing transcripts, we inserted poly(dA) sequences of various lengths between the DSR and the snu2 terminator on Construct 3, as is schematically shown in Figure 3D (Construct 3-A10, 10 nucleotides; Construct 3-A50, 50 nucleotides; and Construct 3-A100, 100 nucleotides). Corresponding constructs were also prepared from Construct 4, which carried DSR-M10 (Constructs 4-A10, 4-A50 and 4-A100; Figure 3D). Although the transcripts produced in this system did not carry a poly(A) tail at their very 3′ end, the presence of a poly(A) tract seemed to perform a stabilizing function for the transcripts of derivatives of Construct 4; more transcripts accumulated within the cell as the poly(A) tract became longer (4-A10 through 4-A100; Figure 3E, lanes 5–8). However, this effect was not observed for derivatives of Construct 3, which carried the DSR. In these cells, the amount of transcripts did not increase, but rather decreased, as the poly(A) tract became longer (3-A10 through 3-A100; Figure 3E, lanes 1–4). This difference was obvious when the ratio between the counterparts with and without the functional DSR was plotted (Figure 3F).
To confirm the specificity of poly(A), we prepared analogues of the above constructs by substituting poly(dA) with poly(dT) to produce Constructs 3-T10, 3-T50, 3-T100, 4-T10, 4-T50 and 4-T100 (Figure 3D). Transcript abundance was comparable between 3-T10 and 4-T10, between 3-T50 and 4-T50 and between 3-T100 and 4-T100 (Figure 3E and F), although the T50 constructs exhibited abundance nearly twice as much as the others for reasons unknown. These data indicate that DSR-dependent, selective mRNA elimination does not operate using a poly(U) tract. Thus, it seemed that a poly(A) tract normally stabilizes the transcripts in this system, but the presence of a DSR sequence antagonizes this stabilization.
As shown in Figure 1A, Pab2p function was required for the degradation of endogenous DSR-containing mRNAs. This necessity for degradation was confirmed using the artificial reporter constructs Construct 3-A50 and Construct 4-A50. The former, which harboured a functional DSR, underwent transcript elimination in growing wild-type cells. However, both constructs accumulated transcripts in growing pab2Δ cells, indicating that Pab2p was necessary for the elimination of transcripts derived from Construct 3-A50 (Figure 3G). Thus, an important function of a poly(A) tract in selective mRNA elimination may be facilitating the recruitment of Pab2p to the transcript targeted for degradation.
The TRAMP complex is unlikely to be involved in selective elimination of meiotic mRNAs
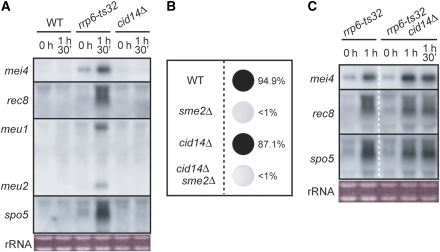
There have been reports of polyadenylation activating the degradation of certain RNA species by exosomes. The best-studied case in this regard may be the system involving the TRAMP protein complex (LaCava et al, 2005; Vanacova et al, 2005; Wyers et al, 2005). The TRAMP complex has poly(A) polymerase activity and polyadenylates various substrates, including rRNA, tRNA, snRNA and snoRNA, before substrate degradation. We therefore addressed whether the TRAMP complex was relevant to the selective elimination of meiosis-specific mRNAs. In fission yeast, Cid14p is most probably the catalytic subunit of TRAMP responsible for poly(A) polymerization (Win et al, 2006; Bühler et al, 2007). We examined the expression of meiosis-specific transcripts in viable, vegetatively growing cid14Δ cells. As shown in Figure 4A, the accumulation of meiosis-specific transcripts was not observed in these cells, which was in contrast to the results for rrp6-ts32 cells, which were used as a control. Furthermore, cid14Δ did not suppress meiotic arrest in sme2Δ cells, unlike what occurred with a defect in mmi1 or rrp6 (Figure 4B). These observations suggest that the TRAMP complex is unlikely to have a major role in DSR-dependent, selective mRNA elimination.
Figure 4.
Cid14p is most likely not involved in the Mmi1-dependent selective elimination of the meiotic mRNAs (A) JY450 (WT), JT432 (rrp6-ts32) and JT654 (cid14Δ) cells were cultured at 25°C in YE medium, and then shifted to 37°C. At the indicated times, total RNA was extracted from each sample and processed for northern blot analysis. (B) JY450 (WT), JZ464 (sme2Δ), JT654 (cid14Δ) and JT655 (cid14Δ sme2Δ) cells were cultured in YE medium at 30°C and spotted onto an SPA plate. Incubation was continued at 30°C for 1 day, and then cells were stained with iodine and the sporulation efficiency was calculated. (C) JT432 (rrp6-ts32) and JT656 (rrp6-ts32 cid14Δ) cells were cultured at 25°C in YE medium, and then shifted to 37°C. At the indicated times, total RNA was extracted from each sample and processed for northern blot analysis.
We also tested whether Cid14p was relevant to the excessive polyadenylation of meiosis-specific transcripts in rrp6-ts32 cells. The rrp6-ts32 mutant and rrp6-ts32 cid14Δ double mutant exhibited quite similar transcript patterns (Figure 4C), indicating that the contribution of Cid14p to excessive polyadenylation, if any, is probably much smaller than the contributions of Pla1p. However, as the size of the transcripts seemed to be slightly reduced in the double mutant, we cannot completely dismiss the possibility that Cid14p may have a minor influence on this selective elimination system.
Subcellular localization of the factors involved in the selective elimination
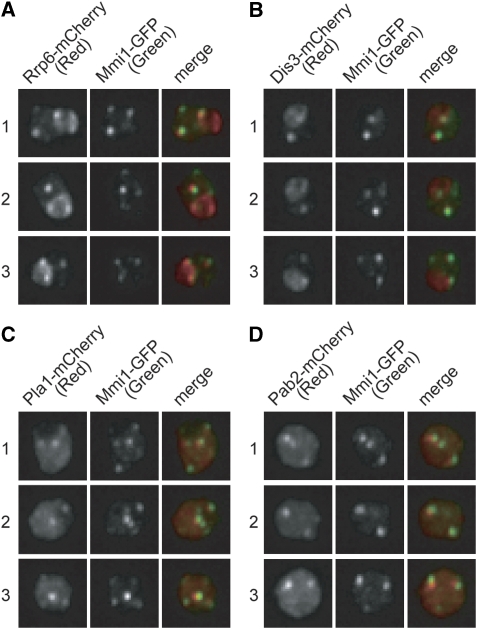
We have previously reported that Mmi1p forms several patchy structures in the nucleus of growing cells (Harigaya et al, 2006). To determine whether the factors cooperating with Mmi1p might also localize to these patches, Rrp6p, Dis3p, Pla1p and Pab2p were tagged with a fluorescent marker using expression constructs. The majority of Rrp6p and Dis3p proteins, which compose the exosome, was localized rather evenly in the nucleolus and also formed bright patches in the nucleoplasm. These patches showed co-localization with Mmi1p (Figure 5A and B).
Figure 5.
Factors involved in selective elimination of DSR-containing mRNA form patchy structures in the nucleoplasm. (A–D) GFP-tagged Mmi1p (Mmi1–GFP) was co-expressed with either (A) Rrp6–mCherry, (B) Dis3–mCherry, (C) Pla1–mCherry or (D) Pab2–mCherry from the respective endogenous promoters. Three independent cells (numbered 1–3) were examined for each combination. An image of the nuclear region, stacked along the z-axis, is shown.
Pla1p was localized evenly in the nucleoplasm, occasionally forming patchy foci (Supplementary Figure S4). Most of these foci seemed to coincide with the Mmi1p patches (Figure 5C). Pab2p localized to the nucleolus and the nucleoplasm and formed patchy foci in the latter. Again, these foci seemed to coincide with the Mmi1p patches (Figure 5D). On the basis of these observations, we propose that the machinery for the selective meiotic mRNA elimination is localized to several patchy multi-protein structures in the nucleoplasm, which include the following components: Mmi1p, exosomes, canonical poly(A) polymerase, a poly(A)-binding protein and possibly some additional factors. Similar analyses have shown that Rna15p also co-localizes with Mmi1p (data not shown). Cid14p is reported to be enriched in the nucleolus, but it does not seem to form patchy structures in the nucleus (Win et al, 2006). These data may provide additional evidence that the TRAMP complex is unlikely to participate in this Mmi1p-dependent mRNA elimination system.
Discussion
In this study, we have demonstrated that the acquisition of a poly(A) tail is crucial for mRNA degradation by exosomes in the DSR/Mmi1p-dependent selective elimination system. An important role for the poly(A)-binding protein, Pab2p, in the system has been also demonstrated.
A polyadenylation-mediated RNA degradation system has been well established in bacteria and was the subject of a recent review (Richards et al, 2008). In Escherichia coli, mRNA stability is established through a stem-loop structure at the 3′ end of the mRNA. When an mRNA is to be degraded, it is cleaved by endoribonuclease RNase E, the major component of the degradosome. This cleavage provides a signal for the polyadenylation of the resulting fragments involving the recruitment of poly(A) polymerase I (PAP I) to the fragments, most likely as a result of its interaction with the degradosome. Poly(A) polymerase I adds a poly(A) tail to the fragments, which in turn promotes the binding of polynucleotide phosphorylase (PNPase), another component of the degradosome. On binding to the poly(A) tail, PNPase degrades the RNA fragment in cooperation with the helicase RhlB, which is also a component of the degradosome. It should be noted that polyadenylation is now established as having a much more crucial role in bacterial RNA quality control than was previously anticipated. Thus, the basic scheme for selective elimination of meiosis-specific mRNAs in fission yeast seems to have many similarities to this bacterial system, except that the bacterial system represents a general degradation mechanism involving most mRNAs with no apparent strong sequence specificity for the mRNA target.
By contrast, polyadenylation is thought to facilitate the function of mRNAs in eukaryotic cells through mRNA stabilization, promotion of cytoplasmic export and stimulation of translation. Thus, contrary to prokaryotes, eukaryotes use mRNA polyadenylation for the prevention of transcript degradation. However, recent studies have provided examples whereby polyadenylation activates the degradation of certain eukaryotic RNA species by exosomes. In this regard, of particular interest is the TRAMP complex (LaCava et al, 2005; Vanacova et al, 2005; Wyers et al, 2005), which has poly(A) polymerase activity and polyadenylates various substrates, including rRNA, tRNA, snRNA and snoRNA before their destruction. For example, TRAMP polyadenylates tRNAs that are not methylated properly and converts them to degradation substrates for exosomes, thereby contributing to the quality control of a cell's tRNA population (Kadaba et al, 2004). Recent studies have also indicated that polyadenylation of yeast cryptic unstable transcripts (CUTs) by TRAMP directs them to degradation by nuclear exosomes (Wyers et al, 2005). Our analyses, however, suggest that the TRAMP complex is not directly relevant to the selective elimination of meiosis-specific mRNAs. Rather, this selective elimination system apparently uses the general polyadenylation complex, which includes Pla1p and Rna15p, to polyadenylate transcripts targeted for degradation.
We also describe the unexpected observation that mutation of pla1-ts37 or rna15-ts10 could suppress sme2Δ at the permissive temperature for vegetative growth. In our original screening, we isolated an insertion mutation in pla1, which permitted vegetative growth but suppressed meiotic arrest in the sme2Δ strain. These observations indicate that a small flaw in the polyadenylation system that does not severely affect mRNA production can effectively block the Mmi1p-dependent selective elimination mechanism.
A recent study has shown that the fission yeast pab2Δ strain accumulates excessively polyadenylated snoRNAs, although it does not seem to affect mRNAs (Lemay et al, 2010). The authors proposed that Pab2p promotes poly(A) tail trimming from pre-snoRNAs through nuclear exosome recruitment. Whether this observation is directly relevant to the function of Pab2p in selective elimination remains unclear, but it is interesting to note that Pab2p apparently activates exosomes in both systems.
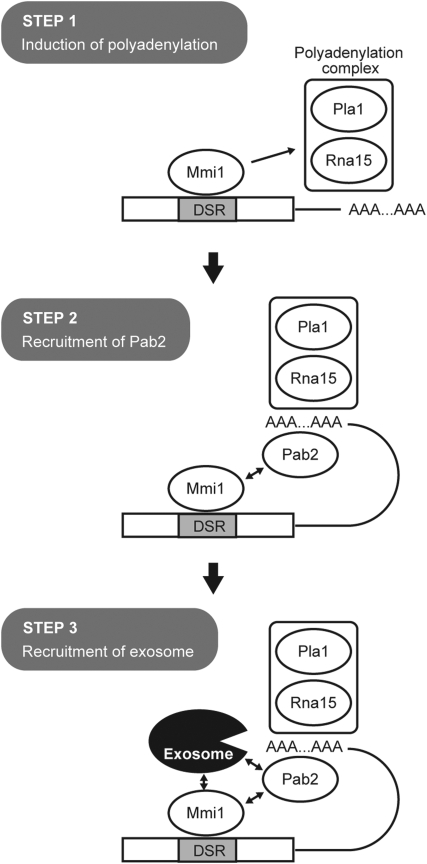
The important question of how eukaryotic cells determine whether polyadenylated RNAs associated with nearly the same protein complex are targeted to the exosome for degradation or to the cytoplasm for function remains unanswered (Libri, 2010). Although the data obtained in this study are not yet sufficient to establish a clear mechanistic scheme for the selective elimination of mRNA, we would like to remark the followings, with a sketchy diagram comprising three steps (Figure 6).
Figure 6.
A putative model for the cooperation of Mmi1p, the polyadenylation complex, Pab2p and the exosome in the induction of selective elimination of a DSR-containing mRNA. In Step 1, Mmi1p bound to the DSR sequence on the transcript interacts with a conventional polyadenylation complex and promotes polyadenylation of apparently unusual nature. In Step 2, the poly(A)-binding protein Pab2p is recruited to the produced poly(A) tail. The affinity of Mmi1p for Pab2p may facilitate this recruitment. In Step 3, Pab2p, likely in collaboration with Mmi1p, recruits a nuclear exosome to the target transcript, which then digests it from the 3′ end.
We speculate that the physical affinity of Mmi1p for the polyadenylation complex, on top of its affinity for the nuclear exosome, may contribute significantly to gather exosomes to DSR-containing mRNAs. As mentioned in the Results section, Mmi1p seems to direct polyadenylation that is unusual either quantitatively or qualitatively, which might represent the major function of Mmi1p in the selective elimination (Figure 6, Step 1). The produced poly(A) tail, together with Mmi1p, probably recruits Pab2p (Step 2). Then, Pab2p, together with Mmi1p, may recruit a nuclear exosome to the transcript (Step 3).
We speculate further that Mmi1p and Pab2p may alter the fate of mRNAs through kinetic changes, slowing down their export from the nucleus and targeting them to a default exosome pathway, for instance. This possibility was previously hypothesized with regard to the putative role of Pab2p in the processing of pre-snoRNAs, under the naming of a ‘take the money and run' strategy (Libri, 2010). This type of mechanism may be compatible with the observation that a modest flaw in the polyadenylation complex led to the suppression of selective elimination.
Intracellular localization analyses have shown that the factors necessary for selective mRNA elimination, including Mmi1p, Pla1p, Pab2p and exosomes, are assembled together into several patchy structures in the nucleoplasm of growing cells. One simple speculation is that these patches are the sites for selective degradation of meiosis-specific mRNAs. However, the number and position of these patches do not seem to be constant. Our preliminary analyses suggest that they are associated with neither major meiotic genes nor heterochromatin regions, although some of them do seem to show preferential localization on the periphery of the nucleolus. Thus, the nature of these patchy structures remains to be established. In this regard, it is noteworthy that several foci enriched with poly(A) RNA have been observed at the periphery of the nucleolar region in fission yeast mutants defective in nuclear mRNA export (Mizuki et al, 2007). Whether the patchy structures with Mmi1p localization have any relevance to the general control of mRNA quality, transport and metabolism remains an intriguing question.
Materials and methods
Yeast strains and growth media
Table I provides a summary of the yeast strains used in this study. The general genetic procedures for S. pombe have been described previously (Gutz 1974; Moreno et al, 1991). The pab2Δ mutant was produced by the direct chromosomal integration method (Bähler et al, 1998). The production of various proteins tagged with a fluorescent marker was also performed using this method. The temperature-sensitive alleles of rrp6, dis3, pla1 and rna15 used in this study were produced essentially as described previously for the mmi1 ts alleles (Harigaya et al, 2006 and Supplementary data therein). Briefly, we first constructed strains carrying a kanamycin cassette downstream of the respective gene on the chromosome. Using genomic DNA extracted from each strain as a template, we amplified DNA fragments that covered from roughly 0.5 kb upstream of the respective ORF to roughly 0.5 kb downstream of the kanamycin cassette by an error-prone PCR method (Zhou et al, 1991). Wild-type cells (JY450) were transformed with the resulting DNA fragments, which carried no annotated neighbouring gene, and transformants resistant to G418 (indicating the gain of the kanamycin cassette) were isolated at 25°C. They were replica-plated at 37°C, and strains showing temperature-sensitive growth were picked. The introduction of each ts mutation to the proper gene was confirmed by complementation tests.
Table 1. S. pombe strains used in this study.
| Strain | Genotype | Source |
|---|---|---|
| JY333 | h− ade6-M216 leu1 | Laboratory stock |
| JY450 | h90 ade6-M216 leu1 | Laboratory stock |
| JZ464 | h90 sme2∷ura4 ade6-M216 leu1 ura4-D18 | Laboratory stock |
| JV564 | h− mmi1-ts3-(3HA)≪kan ade6-M216 leu1 | Laboratory stock |
| JV838 | h90 pab2-GFP≪kanr ade6-M216 leu1 | This study |
| JT3 | h90 pla1-GFP≪kanr ade6-M216 leu1 | This study |
| JT432 | h90 rrp6-32-GFP≪kan ade6-M216 leu1 | This study |
| JT449 | h90 dis3-4≪kan ade6-M216 leu1 | This study |
| JT451 | h90 sme2∷ura4 dis3-4≪kan ade6-M216 leu1 | This study |
| JT452 | h90 pla1-37-GFP≪kan ade6-M216 leu1 | This study |
| JT453 | h90 rna15-10-GFP≪kan ade6-M216 leu1 | This study |
| JT454 | h90 pab2∷kan ade6-M216 leu1 | This study |
| JT455 | h90 sme2∷ura4 pla1-37-GFP≪kan ade6-M216 leu1 | This study |
| JT456 | h90 sme2∷ura4 rna15-10-GFP≪kan ade6-M216 leu1 | This study |
| JT457 | h90 sme2∷ura4 pab2∷kan ade6-M216 leu1 | This study |
| JT458 | h− rrp6-32-GFP≪kan pla1-37-GFP≪kan ade6-M216 leu1 | This study |
| JT459 | h− lys1∷adh1-GFP-DSR-adh ter.≪kan ade6-M216 leu1 | This study |
| JT460 | h− lys1∷adh1-GFP-DSR(M10)-adh ter.≪kan ade6-M216 leu1 | This study |
| JT461 | h− lys1∷adh1-GFP-DSR-snu2 ter.≪kan ade6-M216 leu1 | This study |
| JT462 | h− lys1∷adh1-GFP-DSR(M10)-snu2 ter.≪kan ade6-M216 leu1 | This study |
| JT463 | h− lys1∷adh1-GFP-DSR-10(A)-snu2 ter.≪kan ade6-M216 leu1 | This study |
| JT464 | h− lys1∷adh1-GFP-DSR-50(A)-snu2 ter.≪kan ade6-M216 leu1 | This study |
| JT465 | h− lys1∷adh1-GFP-DSR-100(A)-snu2 ter.≪kan ade6-M216 leu1 | This study |
| JT466 | h− lys1∷adh1-GFP-DSR(M10)-10(A)-snu2 ter.≪kan ade6-M216 leu1 | This study |
| JT467 | h− lys1∷adh1-GFP-DSR(M10)-50(A)-snu2 ter.≪kan ade6-M216 leu1 | This study |
| JT468 | h− lys1∷adh1-GFP-DSR(M10)-100(A)-snu2 ter.≪kan ade6-M216 leu1 | This study |
| JT469 | h− lys1∷adh1-GFP-DSR-10(T)-snu2 ter.≪kan ade6-M216 leu1 | This study |
| JT470 | h− lys1∷adh1-GFP-DSR-50(T)-snu2 ter.≪kan ade6-M216 leu1 | This study |
| JT471 | h− lys1∷adh1-GFP-DSR-100(T)-snu2 ter.≪kan ade6-M216 leu1 | This study |
| JT472 | h− lys1∷adh1-GFP-DSR(M10)-10(T)-snu2 ter.≪kan ade6-M216 leu1 | This study |
| JT473 | h− lys1∷adh1-GFP-DSR(M10)-50(T)-snu2 ter.≪kan ade6-M216 leu1 | This study |
| JT474 | h− lys1∷adh1-GFP-DSR(M10)-100(T)-snu2 ter.≪kan ade6-M216 leu1 | This study |
| JT475 | h90or− lys1∷adh1-GFP-DSR-50(A)-snu2 ter.≪kan pab2∷hyg ade6-M216 leu1 | This study |
| JT476 | h90or− lys1∷adh1-GFP-DSR(M10)-50(A)-snu2 ter.≪kan pab2∷hyg ade6-M216 leu1 | This study |
| JT485 | h90 LEU2>>mmi1(pro)-GFP-mmi1 rrp6-cherry≪hyg ade6-M216 leu1 | This study |
| JT486 | h90 LEU2>>mmi1(pro)-GFP-mmi1 dis3-cherry≪hyg ade6-M216 leu1 | This study |
| JT487 | h90 LEU2>>mmi1(pro)-GFP-mmi1 pla1-cherry≪hyg ade6-M216 leu1 | This study |
| JT488 | h90 LEU2>>mmi1(pro)-GFP-mmi1 pab2-cherry≪hyg ade6-M216 leu1 | This study |
| JT496 | h90or− mmi1-ts3-(3HA)≪kanr rrp6-ts32-GFP≪kanr ade6-M216 leu1 | This study |
| JT654 | h90 cid14∷natr ade6-M216 leu1 | Danesh Moazed |
| JT655 | h90 cid14∷natr sme2∷ura4 ade6-M216 leu1 | This study |
| JT656 | h90 rrp6-ts32-GFP≪kanr cid14∷natr ade6-M216 leu1 | This study |
| JT657 | h90 grn1-CFP≪kanr rrp6-mCherry≪hygr ade6-M210 leu1 | This study |
| JT658 | h90 grn1-CFP≪kanr pla1-mCherry≪hygr ade6-M216 leu1 | This study |
| JT682 | h90 rna15-GFP≪kanr ade6-M216 leu1 | This study |
Northern blot analysis
S. pombe cells growing exponentially in YE medium were sampled and total RNA was prepared from each sample. Northern blot analysis was performed essentially according to a standard protocol (Thomas, 1980), with modifications as described previously (Watanabe et al, 1988). A PCR-amplified fragment covering the entire ORF of a specific gene was used as the DNA probe to detect each transcript.
RNase H treatment
A volume of 8 μl of a RNA solution containing 20 μg total RNA was mixed with 2 μl of a specific oligonucleotide solution (100 pmol/μl), which contained an oligonucleotide complementary to the transcript of either mei4, or rec8 or spo5. A volume of 4 μl of oligo(dT) solution (100 pmol/μl) or DDW was then added to this mix. The final mixture was heated at 85°C for 5 min, incubated at 42°C for 10 min and then left at room temperature for 10 min. A volume of 20 μl of 10 × buffer (200 mM Tris–HCl (pH7.5), 20 mM KCl, 1 mM EDTA and 1 mM DTT), 20 μl of 100 mM MgCl2 and 1 μl of RNase H (TAKARA) were added to each mixture and digestion was carried out at 37°C for 30 min.
Microscopy
An Axioplan II microscope (Zeiss) and the Deltavision/SoftwoRx system (Olympus and Applied Precision) were used for fluorescence microscopy. For Figure 5, images consist of 10 optical sections along the z-axis taken at 0.4-μm intervals. The images were then deconvolved and merged into a single projection.
Supplementary Material
Acknowledgments
We thank D Moazed for providing the cid14Δ strain and T Andoh, T Tani and Y Hirose for helpful discussion. This study was supported by Grants-in-Aid for Scientific Research (S) to MY and (C) to AY from JSPS, and by a Grant-in-Aid for Scientific Research on Innovative Areas to AY from MEXT of Japan. The study was also supported in part by the MEXT Global COE Program (Integrative Life Science Based on the Study of Biosignaling Mechanisms). SY is the recipient of a JSPS Research Fellowship for Young Scientists (DC).
Footnotes
The authors declare that they have no conflict of interest.
References
- Allmang C, Kufel J, Chanfreau G, Mitchell P, Petfalski E, Tollervey D (1999) Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J 18: 5399–5410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bähler J, Wu JQ, Longtine MS, Shah NG, McKenzie A III, Steever AB, Wach A, Philippsen P, Pringle JR (1998) Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14: 943–951 [DOI] [PubMed] [Google Scholar]
- Brennwald P, Porter G, Wise JA (1988) U2 small nuclear RNA is remarkably conserved between Schizosaccharomyces pombe and mammals. Mol Cell Biol 8: 5575–5580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs MW, Burkard KT, Butler JS (1998) Rrp6p, the yeast homologue of the human PM-Scl 100-kDa autoantigen, is essential for efficient 5.8 S rRNA 3′ end formation. J Biol Chem 273: 13255–13263 [DOI] [PubMed] [Google Scholar]
- Bühler M, Haas W, Gygi SP, Moazed D (2007) RNAi-dependent and -independent RNA turnover mechanisms contribute to heterochromatic gene silencing. Cell 129: 707–721 [DOI] [PubMed] [Google Scholar]
- Burkard KT, Butler JS (2000) A nuclear 3′–5′ exonuclease involved in mRNA degradation interacts with Poly(A) polymerase and the hnRNA protein Npl3p. Mol Cell Biol 20: 604–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziembowski A, Lorentzen E, Conti E, Seraphin B (2007) A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat Struct Mol Biol 14: 15–22 [DOI] [PubMed] [Google Scholar]
- Gutz H, Heslot H, Leupold U, Loprieno N (1974) Schizosaccharomyces pombe. In Handbook of Genetics, King RC (ed), Vol. 1. New York, NY: Plenum Press [Google Scholar]
- Harigaya Y, Tanaka H, Yamanaka S, Tanaka K, Watanabe Y, Tsutsumi C, Chikashige Y, Hiraoka Y, Yamashita A, Yamamoto M (2006) Selective elimination of messenger RNA prevents an incidence of untimely meiosis. Nature 442: 45–50 [DOI] [PubMed] [Google Scholar]
- Hausen P, Stein H (1970) Ribonuclease H. An enzyme degrading the RNA moiety of DNA–RNA hybrids. Eur J Biochem 14: 278–283 [DOI] [PubMed] [Google Scholar]
- Kadaba S, Krueger A, Trice T, Krecic AM, Hinnebusch AG, Anderson J (2004) Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev 18: 1227–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler MM, Zhao J, Moore CL (1996) Purification of the Saccharomyces cerevisiae cleavage/polyadenylation factor I. Separation into two components that are required for both cleavage and polyadenylation of mRNA 3′ ends. J Biol Chem 271: 27167–27175 [DOI] [PubMed] [Google Scholar]
- LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D (2005) RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 121: 713–724 [DOI] [PubMed] [Google Scholar]
- Lemay JF, D'Amours A, Lemieux C, Lackner DH, St-Sauveur VG, Bähler J, Bachand F (2010) The nuclear poly(A)-binding protein interacts with the exosome to promote synthesis of noncoding small nucleolar RNAs. Mol Cell 37: 34–45 [DOI] [PubMed] [Google Scholar]
- Libri D (2010) Nuclear poly(A)-binding proteins and nuclear degradation: take the mRNA and run? Mol Cell 37: 3–5 [DOI] [PubMed] [Google Scholar]
- Minvielle-Sebastia L, Preker PJ, Keller W (1994) RNA14 and RNA15 proteins as components of a yeast pre-mRNA 3′-end processing factor. Science 266: 1702–1705 [DOI] [PubMed] [Google Scholar]
- Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D (1997) The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′ → 5′ exoribonucleases. Cell 91: 457–466 [DOI] [PubMed] [Google Scholar]
- Mizuki F, Namiki T, Sato H, Furukawa H, Matsusaka T, Ohshima Y, Ishibashi R, Andoh T, Tani T (2007) Participation of XPB/Ptr8p, a component of TFIIH, in nucleocytoplasmic transport of mRNA in fission yeast. Genes Cells 12: 35–47 [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P (1991) Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol 194: 795–823 [DOI] [PubMed] [Google Scholar]
- Murakami H, Goto DB, Toda T, Chen ES, Grewal SI, Martienssen RA, Yanagida M (2007) Ribonuclease activity of Dis3 is required for mitotic progression and provides a possible link between heterochromatin and kinetochore function. PLoS ONE 2: e317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkura H, Adachi Y, Kinoshita N, Niwa O, Toda T, Yanagida M (1988) Cold-sensitive and caffeine-supersensitive mutants of the Schizosaccharomyces pombe dis genes implicated in sister chromatid separation during mitosis. EMBO J 7: 1465–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnacker M, Minvielle-Sebastia L, Keller W (1996) The Schizosaccharomyces pombe pla1 gene encodes a poly(A) polymerase and can functionally replace its Saccharomyces cerevisiae homologue. Nucleic Acids Res 24: 2585–2591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perreault A, Lemieux C, Bachand F (2007) Regulation of the nuclear poly(A)-binding protein by arginine methylation in fission yeast. J Biol Chem 282: 7552–7562 [DOI] [PubMed] [Google Scholar]
- Richards J, Sundermeier T, Svetlanov A, Karzai AW (2008) Quality control of bacterial mRNA decoding and decay. Biochim Biophys Acta 1779: 574–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T, Yamashita A, Yamamoto M (2003) The fission yeast meiotic regulator Mei2p forms a dot structure in the horse-tail nucleus in association with the sme2 locus on chromosome II. Mol Biol Cell 14: 2461–2469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas PS (1980) Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci USA 77: 5201–5205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanacova S, Wolf J, Martin G, Blank D, Dettwiler S, Friedlein A, Langen H, Keith G, Keller W (2005) A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol 3: e189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Iino Y, Furuhata K, Shimoda C, Yamamoto M (1988) The S. pombe mei2 gene encoding a crucial molecule for commitment to meiosis is under the regulation of cAMP. EMBO J 7: 761–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Shinozaki-Yabana S, Chikashige Y, Hiraoka Y, Yamamoto M (1997) Phosphorylation of RNA-binding protein controls cell cycle switch from mitotic to meiotic in fission yeast. Nature 386: 187–190 [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Yamamoto M (1994) S. pombe mei2+ encodes an RNA-binding protein essential for premeiotic DNA synthesis and meiosis I, which cooperates with a novel RNA species meiRNA. Cell 78: 487–498 [DOI] [PubMed] [Google Scholar]
- Win TZ, Draper S, Read RL, Pearce J, Norbury CJ, Wang SW (2006) Requirement of fission yeast Cid14 in polyadenylation of rRNAs. Mol Cell Biol 26: 1710–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, Boulay J, Regnault B, Devaux F, Namane A, Seraphin B, Libri D, Jacquier A (2005) Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 121: 725–737 [DOI] [PubMed] [Google Scholar]
- Yamashita A, Watanabe Y, Nukina N, Yamamoto M (1998) RNA-assisted nuclear transport of the meiotic regulator Mei2p in fission yeast. Cell 95: 115–123 [DOI] [PubMed] [Google Scholar]
- Zhou D, Frendewey D, Lobo Ruppert SM (1999) Pac1p, an RNase III homolog, is required for formation of the 3′ end of U2 snRNA in Schizosaccharomyces pombe. RNA 5: 1083–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou YH, Zhang XP, Ebright RH (1991) Random mutagenesis of gene-sized DNA molecules by use of PCR with Taq DNA polymerase. Nucleic Acids Res 19: 6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.