Abstract
Maintenance of specific heterochromatic domains is crucial for genome stability. In eukaryotic cells, a fraction of the tandem-repeated ribosomal RNA (rRNA) genes is organized in the heterochromatic structures. The principal determinant of rDNA silencing is the nucleolar remodelling complex, NoRC, that consists of TIP5 (TTF-1-interacting protein-5) and the ATPase SNF2h. Here we showed that TIP5 not only mediates the establishment of rDNA silencing but also the formation of perinucleolar heterochromatin that contains centric and pericentric repeats. Our data indicated that the TIP5-mediated heterochromatin is indispensable for stability of silent rRNA genes and of major and minor satellite repeats. Moreover, depletion of TIP5 impairs rDNA silencing, upregulates rDNA transcription levels and induces cell transformation. These findings point to a role of TIP5 in protecting genome stability and suggest that it can play a role in the cellular transformation process.
Keywords: cell proliferation, genomic stability, heterochromatin, NoRC, rDNA silencing
Introduction
Formation of specific heterochromatic domains is crucial for genome stability (Grewal and Jia, 2007; Peng and Karpen, 2008). This is exemplified by the heterochromatin structure of repetitive major satellite (pericentric) and minor satellite (centric) DNA sequences whose maintenance and accurate reproduction throughout multiple cell divisions represents a major challenge to ensure genome stability. In interphase, the centromeric heterochromatin is predominantly located either at the nuclear periphery or around the nucleolus (Haaf and Schmid, 1991; Pluta et al, 1995). The nucleolus is the subnuclear body where the tandemly repeated ribosomal RNA (rRNA) genes synthesize ribosomal RNA, the major components of the ribosome. In humans and apes, rRNA genes are located between the short arm and the satellite body of acrocentric chromosomes. Standard laboratory strains of mice, which are thought to have originated mainly from a European subspecies, Mus musculus domesticus, and partially from an Asian subspecies, M.m.musculus/molossinus, have rDNA clusters within the centromeric regions of chromosome-12, 15, 16, 18 and 19 (Dev et al, 1977; Davisson, 1989; Kurihara et al, 1994). However, the combinations of chromosomes that include rDNA have been shown to be highly polymorphic among individuals (Suzuki et al, 1990). Due to the linear proximity, centromeres of chromosomes bearing rDNA repeats associate with nucleoli. Notably, also chromosomes devoid of rRNA genes have their centromeres associated with the nucleolus at a frequency more than that expected for a random distribution (Carvalho et al, 2001 and references therein). The basis of this association probably relies on the linear proximity along the chromosome and on the repeated nature of DNA sequence, which provides multiple binding sites for specific proteins capable of forming multimeric complexes. In each cell, a fraction of rRNA genes is transcriptionally silent and organized in heterochromatic structures by epigenetic mechanisms (reviewed by Santoro, 2005). By contrast, the ‘active' euchromatic fraction represents rRNA genes competent for transcription whose activity is modulated according to the requirement of cell metabolism (Grummt, 2003; Moss et al, 2007). CpG-methylated silent rRNA genes were shown to assemble adjacent to the perinucleolar heterochromatin in mouse neuronal cells, suggesting an intricate relationship between these heterochromatic regions and silent rRNA copies (Akhmanova et al, 2000).
The presence of heterochromatic silent rDNA repeats raises question regarding their function, which could be to either dose the rRNA transcript levels and/or to affect the structure of other types of chromatin localized nearby. Here we show that the role of TIP5 (TTF-1-interacting protein-5), the key subunit of the NoRC (nucleolar remodelling complex), is not only restricted to the formation of heterochromatin at the rDNA repeats, but that it also extends its action in establishing the perinucleolar heterochromatin and repressive histone marks at major and minor satellite sequences. The results indicate that TIP5 is crucial to maintain the stability of silent rRNA genes and centric and pericentric repeats. We also show that cells depleted of TIP5 upregulate rDNA transcription levels, increase cellular proliferation rates and have a transformed phenotype. The results uncovered a role of TIP5 in protecting genome stability and suggest that deregulation of rDNA silencing can contribute to cellular transformation.
Results
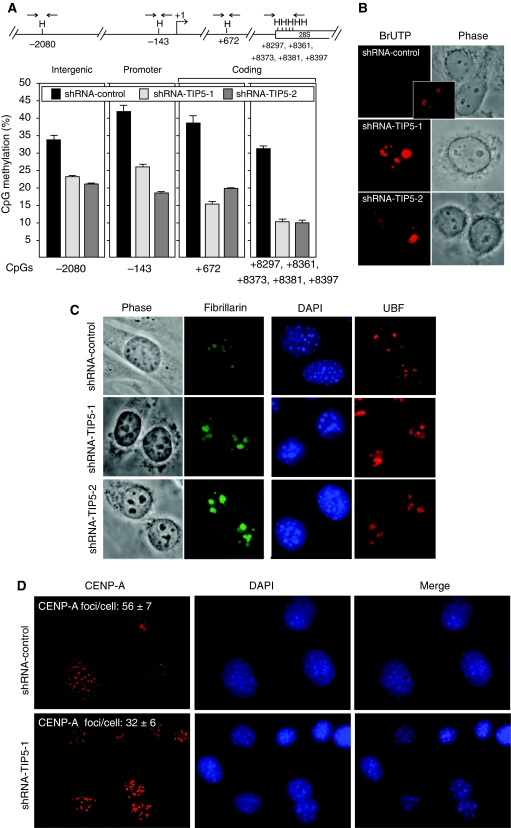
The principal determinant that establishes rDNA silencing is NoRC that consists of TIP5 and the ATPase SNF2h (Strohner et al, 2001; Santoro et al, 2002). NoRC binds to the rDNA promoter and represses rDNA transcription through recruitment of histone-modifying and DNA-methylating enzymes (Santoro et al, 2002; Zhou et al, 2002; Santoro and Grummt, 2005; Mayer et al, 2006). We have recently shown that depletion of TIP5, the NoRC subunit recruiting repressor complexes at the rDNA locus, affects rDNA silencing in mammalian cells (Santoro et al, 2009). Two cell lines (shRNA-TIP5-1 and shRNA-TIP5-2), derived from NIH3T3 cells, were established, each expressing a different shRNA sequence directed against TIP5 and both showing a reduction in TIP5 of about 80% when compared with a control cell line (Supplementary Figure S1A and B). Previous results showed that TIP5 binds to the rDNA promoter region and induces de novo methylation of these sequences (Santoro et al, 2002; Li et al, 2005). In NIH3T3 cells, about 40–50% of the rDNA promoter sequences are CpG-methylated. As shown in Figure 1A, rDNA CpG methylation levels were reduced over the entire rRNA gene in both shRNA-TIP5 cell lines when compared with that in control cells, underscoring the role of TIP5 in initiating local silencing events, which then spread over the whole rDNA unit (Figure 1A). Measurements of rRNA transcription by qRT–PCR and in vivo BrUTP incorporation showed higher levels of rRNA synthesis in both shRNA-TIP5 cell lines with respect to that in the control cell line (Supplementary Figure S1C and Figure 1B). Similarly, 45S pre-rRNA synthesis was enhanced in NIH3T3 cells 10 days after infection with a retrovirus expressing miRNA directed against TIP5 sequences (Supplementary Figure S1D). All these results indicate that depletion of TIP5 induces loss of rDNA silencing and enhances rRNA production.
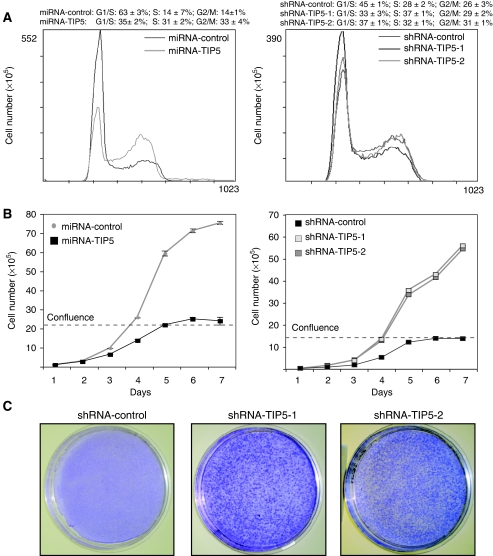
Figure 1.
Depletion of TIP5 impairs perinucleolar heterochromatin formation. (A) Depletion of TIP5 decreases CpG methylation at the entire rDNA repeat. A schema representing a single mouse rDNA repeat and the analysed HpaII (H) sites. The arrows represent the primers used to amplify the HpaII-digested DNA. The data represent the amounts of HpaII-resistant rDNA normalized to the total rDNA calculated by amplification with primers encompassing DNA sequences lacking HpaII sites and undigested DNA. The error bars indicate the s.d. of three independent experiments. (B) Depletion of TIP5 enhances rDNA transcription. rRNA transcripts were detected by in situ BrUTP incorporation after same exposure time. The inset shows a longer exposure of one control cell. (C) Depletion of TIP5 alters the number and size of nucleoli. Indirect immunofluorescence analysis of the nucleolar protein fibrillarin (left panel) or UBF (right panel) in shRNA-TIP5 and control cells. (D) Indirect immunofluorescence analysis of shRNA-control and shRNA-TIP5-1 cells using anti-CENP-A antibodies. The values represent the average number of CENP-A-stained foci of 50 cells scored at random.
TIP5 mediates the formation of perinucleolar heterochromatin
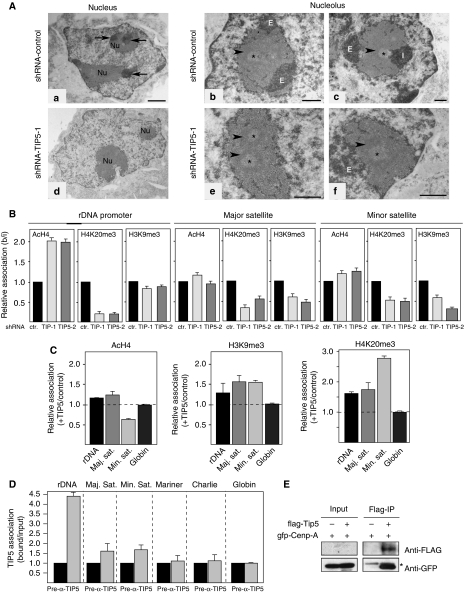
The nucleolus is the subnuclear body where rRNA is transcribed, processed and assembled into ribosomal subunits, and is a principal component of the nuclear architecture. Alterations in the nucleolar structure are often detected by changes in the levels of rRNA synthesis (Sirri et al, 2008). For example, as consequence of elevated nucleolar activities, cancer cells show enlarged nucleoli, which are commonly used by pathologists to identify tumour formation (White, 2005). To determine whether the nucleolar structure is affected in TIP5-depleted cells, we analysed the cellular localization of the nucleolar proteins fibrillarin or UBF (upstream binding factor) by immunofluorescence (Figure 1C). Statistical analyses of 100 shRNA-TIP5 and control cells selected at random showed that the nucleoli of shRNA-TIP5 cells diminished in number (shRNA-control cells: 6.09±1.47 nucleoli per cell; shRNA-TIP5 cells: 3.90±1.47 nucleoli per cell) and had enlarged structures (shRNA-control cells: 12.64±3.37 μm2/cell; shRNA-TIP5 cells: 21.19±6.55 μm2/cell), a characteristic indication of elevated rDNA transcription activities. Notably, TIP5-depleted cells showed an altered staining with DAPI (4,6-diamidino-2-phenylindole), a fluorochrome able to detect condensed heterochromatic loci (CC) formed by coalescence of centromeres in mouse interphase cells (reviewed by Maison and Almouzni, 2004). Whereas in control cells these CC are equally distributed all over the nucleus, in shRNA-TIP5 nuclei they are diminished in number and increased in size, indicating that structural changes occurred at these loci. Consistent with this, we detected structural alterations at the centromeric loci in interphase shRNA-TIP5 cells after immunostaining with antibodies against the core kinetochore CENP-A (Figure 1D) (reviewed by Black and Bassett, 2008). Although cellular amounts of CENP-A remain the same in both shRNA-control and shRNA-TIP5 cells (Supplementary Figure S2A), similar to the CC visualized by DAPI staining, the CENP-A-stained foci in TIP5-depleted cells were diminished in number (shRNA-control cells: 56±7 per cell; shRNA-TIP5 cells: 32±6 per cell) and increased in size. The heterochromatin of pericentric and centric regions were shown to localize also alongside the nucleolus (Carvalho et al, 2001; Guenatri et al, 2004 and Supplementary Figure S2B) and, in the specific case of mouse neuronal cells, even adjacent to methylated silent rRNA genes (Akhmanova et al, 2000). To analyse whether perinucleolar distribution of heterochromatin is affected in TIP5-depleted cells, we performed electron microscopic analysis (Figure 2A). The nucleolus is organized in three main structures: the fibrillar centre (FC), the dense fibrillar component (DFC) and the granular component (GC). Repressed rDNA genes are localized in the FC and initiation of rDNA transcription occurs at the FC–DFC boundary. The resulting pre-rRNA transcripts emerge into the DFC where they are cleaved and modified by the small nucleolar RNPs (snoRNPs) and processing enzymes (reviewed by Boisvert et al, 2007). Using a contrast specific for nucleic acids (Junéra et al, 1995), two kinds of heterochromatin (CC) with distinct positioning relative to the nucleolus were defined: intra-CC (ICC) in contact with the FC and extra-CC (ECC) at the nucleolar periphery (Figure 2A). In 13 shRNA-control and 14 shRNA-TIP5 cells selected at random, the number of ICC and ECC was analysed. As rDNA transcription is initiated at the periphery of the FC, most probably the ICC in contact with the FC corresponds to the rDNA-bearing chromosomes and the ECC to chromosomes not containing rDNA repeats. The number of ICC was 84% in control cells and 4% in shRNA-TIP5 cells, whereas the ECC was observed in 70 and 56% of control and shRNA-TIP5 cells, respectively. All these results indicated that depletion of TIP5 and reduction of rDNA silencing levels affects the nucleolar structure and the formation of condensed chromatin within and in close proximity of the nucleolus.
Figure 2.
TIP5 mediates heterochromatin formation at major and minor satellites. (A) The distribution of the heterochromatin (CC) associated with the nucleoli of shRNA-control and shRNA-TIP5 cells. A general view of the nuclei (a, d) and the nucleoli (b, c, e, f). The contrast procedure reveals in dark the structures containing the nucleic acids, DNA or RNAs. (a) In the nucleus of an shRNA-control cell, the CC (arrows) are visible within the nucleolus (Nu) or at the nucleolar periphery. (d) In the nucleus of an shRNA-TIP5 cell, no CC are visible in or close to the nucleoli (Nu). (b, c) In shRNA-control nucleoli the CC are detected close (I, intra-CC) to the FC (*) or at the nucleolar periphery (E, extra-CC). (e, f) In shRNA-TIP5 nucleoli few CC are present (E). The arrowheads indicate the DFC (dense FC). Bar: (a, d)=1 μm; (b, c, e, f)=0.5 μm. (B) Depletion of TIP5 decreases repressive histone modification levels at the rDNA, major and minor satellite repeats. Quantitative ChIP analysis of cross-linked chromatin was precipitated with the indicated antibodies. The data are presented as the amounts of bound normalized to input and shRNA-control cell levels. The error bars indicate the s.d. of three independent experiments. (C) Overexpression of TIP5 modifies the heterochromatin of rDNA, major and minor satellites. The data are presented as a modified histone fold-change of NIH3T3 cells transiently transfected with TIP5-expression plasmids versus that in cells transfected with control plasmids. The error bars indicate the s.d. of two independent experiments. (D) ChIP showing association of TIP5 with a minor fraction of satellite repeats in NIH3T3 cells. The data are presented as the amounts of bound normalized to input and pre-immunoserum levels. The error bars indicate the s.d. of four independent experiments. (E) CENP-A interacts with TIP5 in vivo. HEK293T cells were co-transfected with GFP-tagged CENP-A plasmids in the presence and absence of pcDNA-FLAG-TIP5 and precipitated with anti-FLAG antibodies. Co-precipitated CENP-A was visualized on immunoblots using antibodies against GFP and FLAG. The signal indicated by the asterisk represents IgG. 10% of the lysate used for IP is shown (input). The low levels of FLAG-TIP5 in the input were below the detection limit.
TIP5 mediates heterochromatin at centric and pericentric repeats
The centric and pericentric domains consist of repetitive minor and major satellite DNA repeats, respectively. These sequences are enriched in nucleosomes containing histones H3 tri-methylated at Lys9 (H3K9me3; Peters et al, 2003). To analyse whether TIP5 affects the epigenetic features at major and minor satellite repeats, we measured histone modifications by quantitative ChIP analysis (Figure 2B). Consistent with the CpG methylation results (Figure 1A), depletion of TIP5 increased the amounts of acetylated histone H4 (AcH4, active histone mark) at the rDNA promoter and decreased the levels of K20 trimethylation of histone H4 (H4K20me3, repressive histone mark), underscoring the role of TIP5 in establishing rDNA silencing. In contrast, the levels of H3K9me3 were slightly reduced. This is consistent with previous results showing that H3K9me3 is also present at active rRNA genes (Yuan et al, 2007). Notably, the levels of H3K9me3 and H4K20me3 were drastically reduced at both major and minor satellite sequences, whereas levels of histone acetylation were only slightly changed. These results indicated that impairment of heterochromatin formation by TIP5 depletion is not only restricted to the rDNA locus but also occurs at the centric and pericentric sequences. To determine whether increase in the levels of TIP5 affects the formation of repressive chromatin at these repeats, we overexpressed TIP5 in NIH3T3 cells using a retroviral TIP5-expression vector. As shown in Figure 2C, when TIP5 was overexpressed, the levels of H3K9me3 and H4K20me3 increased at the rDNA, major and minor satellite repeats but not at the α-globin genes. Notably, minor satellite repeats showed the most drastic changes, including a 40% reduction in AcH4 levels. All these results indicated that TIP5 modulates the formation of repressive chromatin not only at the rDNA locus but also at the major and minor satellite repeats.
The results described so far suggest that TIP5 may bind to centric and pericentric repeats and establish heterochromatic structures using similar mechanisms used to silence the rDNA locus (Santoro et al, 2002; Zhou et al, 2002). Previous immunofluorescence studies showed that TIP5 localized exclusively within the nucleoli of NIH3T3 cells (Strohner et al, 2001). Colocalization of TIP5 with DAPI-stained heterochromatic loci, with the exception of those regions adjacent to the nucleolus, was never detected. To further assay whether TIP5 binds to centric and pericentric DNA, we performed ChIP assay. Consistent with previous results (Santoro et al, 2002), we found specific association of TIP5 with rRNA genes (Figure 2D). By contrast, the bound/input value of TIP5 immunoprecipitation (IP) with major and minor satellite repeats was much lower than that with rDNA sequences, although reproducibly higher when compared with a pre-immunoserum control IP and to a control α-globin gene and Mariner and Charlie transposon sequences. These results indicate that either TIP5 interacts with a minor fraction of centric–pericentric repeats or that this association is either weak and/or transient. To further examine the interaction of TIP5 with centric repeats, we analysed whether TIP5 associates with the core kinetochore protein CENP-A. After transfection of HEK293T cells with a plasmid expressing GFP-CENP-A with or without a FLAG-TIP5 expression vector, we detected anti-FLAG precipitated proteins on immunoblots using anti-GFP and anti-FLAG antibodies. As shown in Figure 2E, a significant amount of CENP-A was associated with TIP5. By contrast, no signal was detected when IP was performed in cells not expressing FLAG-TIP5. This result suggests that the interaction of TIP5 with centric repeats can be mediated by the association with the core kinetochore protein CENP-A.
TIP5 protects the stability of silent rDNA, centric and pericentric repeats
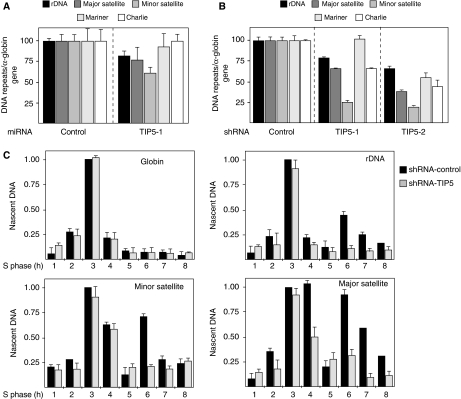
Repetitive DNA sequences, which constitute half of the genome in some organisms, often undergo homologous recombination, thus instigating genomic instability resulting from a gain or loss of DNA (Moazed, 2001; Grewal and Jia, 2007). Assembly of DNA repeats into a silent chromatin is generally thought to serve as mechanism ensuring repeat stability by limiting access to the recombination machinery. Consistent with this, in the budding yeast Saccharomyces cerevisiae, the stability of the rDNA repeats requires a Sir2-containing chromatin-silencing complex (Straight et al, 1999). Similarly, Drosophila Su(var)3-9 and RNAi mutants caused an increase in the amount of extrachromosomal circular rDNA, a typical result of rDNA recombination events (Peng and Karpen, 2007). The role of heterochromatic marks in preventing illicit DNA recombination events at repetitive sequences is also supported by data showing that the number of mouse minor satellite repeats decreased in Dnmt1-deficient cells (Jaco et al, 2008). To determine whether TIP5 protects genome stability, we depleted TIP5 in NIH3T3 cells for 10 days by miRNA, measured the amount of rDNA repeats by quantitative PCR and compared it to the levels of the α-globin gene (Figure 3A). In miRNA-TIP5 cells, we detected an about 20% reduction in the number of chromosomal rDNA copies as compared with that in the control cells, suggesting a role of TIP5 in preventing recombination events at the rDNA locus. Major and, more pronouncedly, minor satellite amounts were significantly reduced, indicating that as well these repeats are destabilized in the absence of TIP5. To determine whether this decrease in DNA levels was extended to other repeats, we analysed the amounts of the interspersed DNA transposons Mariner and Charlie. As shown in Figure 3A, both transposons were not substantially affected by TIP5 depletion, indicating that these are not the primary targets for TIP5. Consistent with these results, loss of rDNA, major and minor satellite repeats was also detected to a higher extent in shRNA-TIP5 stable cell lines (Figure 3B). However, in contrast to a short-time (10 days) TIP5 depletion, Charlie repeat levels diminished in both stable shRNA-TIP5 cells, whereas Mariner DNA levels decreased only in shRNA-TIP5-2, a likely effect of prolonged events of instability that the stable cell lines faced. All these results indicate that TIP5 protects the stability of rDNA, centric and pericentric repeats.
Figure 3.
TIP5 protects genome stability. (A) Depletion of TIP5 induces loss of rDNA, major and minor satellite repeats. qPCR of genomic DNA from NIH3T3 cells infected for 10 days with a retrovirus expressing miRNA-TIP5 and (B) from shRNA-TIP5 cells. The values were normalized to the amounts of α-globin genes and to control cells. The error indicate the s.d. of three independent experiments. (C) Depletion of TIP5 alters the replication timing profiles of rDNA, major and minor satellite repeats. Synchronized cells were pulse-labelled with BrdU in 1-h intervals and nascent DNA was immunoprecipitated using anti-BrU antibodies. To calibrate for DNA recovery during IP, BrdU-labelled E. coli DNA was added to the reactions. Nascent DNA was measured by qPCR. The values represent the amounts of immunoprecipitated DNA normalized to the amounts of BrdU-labelled β-lactamase gene. The error bars indicate the s.d. of two independent experiments.
A typical feature of the heterochromatin is that its replication occurs usually in the late S-phase. The replication timing is tightly regulated and correlates with the chromatin states (Goren and Cedar, 2003). Consistent with this, rRNA genes show a biphasic replication profile: active genes replicate early whereas silent replicate late in S-phase (Li et al, 2005). To determine whether depletion of TIP5 induces loss of either early and/or late replicating repeats, we performed anti-BrdU IPs of nascent DNA from S-phase-synchronized cells pulse-labelled with 5-bromodeoxyuridine (BrdU). In both shRNA-control and shRNA-TIP5 cells, S-phase progression took about 7–8 h to complete and replication of the early replicating α-globin gene occurred 3 h after entry into S-phase (Figure 3C and Supplementary Figure S3). As expected, rDNA from control cells replicated both in early and mid-late S-phase (3 and 6 h) (Figure 3C). By contrast, the amounts of late replicating silent rDNA decreased in shRNA-TIP5 cells. Notably, the levels of early replicating rRNA genes did not increase proportionally, suggesting that depletion of TIP5 induces loss of rDNA repeats that replicate in late S-phase. Similarly, the levels of both late replicating major and minor satellite repeats diminished in shRNA-TIP5 cells, whereas the levels of the early replicating fraction was not affected. All these results indicate that TIP5 protects the genome stability of repeats replicating in mid-late S-phase and suggest that TIP5 has a role in the duplication and inheritance of the chromatin state of rDNA, centric and pericentric repeats.
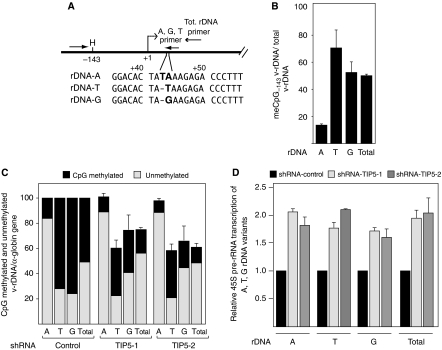
If TIP5 protects genome stability by establishing a heterochromatin at the rDNA repeats, depletion of TIP5 should instigate loss of silent CpG-methylated genes. To test this, we took advantage of the well-known presence of polymorphisms at the human and mouse rDNA sequences (Arnheim and Southern, 1977; Kominami et al, 1981). We tracked active and silent rRNA genes using a polymorphism that we found located at position +42/+43 (Figure 4A and Supplementary Figure S4A and B). We referred to these sequences as rDNA-A, rDNA-T and rDNA-G variants (v-rDNA). This polymorphism is conserved among mouse cells and tissues that we analysed so far (data not shown). To analyse the v-rDNA, we established polymorphism-specific rDNA amplifications that amplify specifically either rDNA-A or rDNA-G or rDNA-T sequences (Supplementary Figure S4C). NIH3T3 cells are characterized by a unique v-rDNA CpG methylation pattern that could not be found so far in other examined cell lines or tissues: about 85% of rDNA-A genes lack CpG methylation (i.e. active copies), whereas about 70 and 50% of rDNA-T and rDNA-G genes are CpG-methylated (i.e. silent copies) (Figure 4B and Supplementary Figure S4D). If the TIP5-mediated heterochromatin protects the stability of silent rRNA genes, the amounts of rDNA-A variants (85% unmethylated copies) should not be greatly affected by TIP5 depletion. To test this, we measured the amounts of v-rDNA in shRNA-TIP5 cells by quantitative PCR and compared it to the expression levels of the α-globin gene. As shown in Figure 4C, the levels of rDNA-G and rDNA-T genes decreased whereas rDNA-A copies were not affected, suggesting that stability of active variants is not influenced by TIP5. Notably, whereas the number of methylated silent genes decreases, the amounts of unmethylated rDNA-A, rDNA-T and total genes remain relatively unchanged, underscoring the role of TIP5-mediated rDNA heterochromatin in protecting the genome stability of silent rRNA genes.
Figure 4.
Depletion of TIP5 induces loss of CpG methylated, silent rDNA repeats. (A) A schema representing rDNA polymorphisms at +43/+44 (A, G and T sequences). The arrows represent the primers used to specifically amplify v-rDNA. (B) The CpG methylation profile of the v-rDNA promoter region in NIH3T3 cells. Polymorphism-specific qPCR. The data represent the amounts of HpaII-resistant v-rDNA normalized to the corresponding total v-rDNA calculated by amplifications using primers encompassing v-DNA sequences lacking HpaII sites and undigested DNA. The error bars indicate the s.d. of three independent experiments. (C) TIP5 mediates the stability of silent rRNA genes. Polymorphism-specific qPCR of v-rDNA from shRNA-TIP5 and control cells. The data were normalized to the amounts of α-globin genes and to control cells. Silent, methylated rDNA represents the HpaII-resistent fraction relative to v-rDNA amounts. The HpaII-digested fraction corresponds to active genes, lacking CpG methylation. The error bars indicate the s.d. of three independent experiments. (D) Depletion of TIP5 enhances the transcription of active rRNA genes. rRNA transcripts originating from v-rDNA variants were measured by qRT–PCR. The data are presented as the amounts of v-rRNA transcripts normalized to GAPDH mRNA levels and to control cells. The error bars indicate the s.d. of four independent experiments.
Changes in the amounts of silent and active rRNA genes is generally discussed as a mechanism that cells use to modulate and dose rRNA transcript levels (reviewed by Santoro, 2005 and McStay, 2006). However, until now, there was no satisfying correlation between the levels of synthesized 45S pre-rRNA transcripts and the number of rRNA genes (French et al, 2003). In agreement with these results, the data described so far indicate that rDNA transcription is upregulated in TIP5-depleted cells (Figure 1B and Supplementary Figures S1C and D), although the number of unmethylated, active genes is the same as that in the control cells (Figure 4C). These results suggest that enhancement of rDNA transcription in TIP5-depleted cells does not depend on the number of active genes. To further investigate this point, we compared the levels of rRNA transcripts synthesized by each class of rDNA variants. As shown in Figure 4D and Supplementary Figure S4D, all the variants, including the rDNA-A genes whose copy number was not affected by depletion of TIP5, transcribed at higher levels. These results strengthen the view that rDNA transcription is preferentially modulated by altering the transcriptional activity of each gene and not by altering the number of genes. Moreover, the data imply that TIP5 and the levels of rDNA silencing influence and modulate the transcription rate of active rRNA genes.
Depletion of TIP5 induces cellular transformation
As genome instability and elevated rDNA transcription are typical features of cancer cells, we tested whether TIP5 can contribute to cellular transformation. As shown in Figure 5A, the population of both shRNA-TIP5 and miRNA-TIP5 cells decreased in the G1/S and accumulated in the S and G2-phases. Consistent with these results, shRNA TIP5 cells showed increased incorporation of BrdU into nascent DNA and higher levels of cyclin-A (Supplementary Figures S5A and B). Comparison of the proliferation rates indicated that shRNA-TIP5 and miRNA-TIP5 cells proliferate faster than control cells, suggesting a functional link between TIP5 and control of cell growth and proliferation (Figure 5B). Notably, TIP5-depleted cells showed a transformed phenotype, continuing to proliferate beyond confluence, forming cellular foci and peeling off the culture surface in large mass (Figures 5B and C). A similar phenotype was obtained with NIH3T3 cells transformed with known oncoproteins like Ras (Tognon et al, 1998). All these results indicate that depletion of TIP5 can contribute to cellular transformation and strengthen the intimate link between rDNA transcription, genome instability and cancer.
Figure 5.
Depletion of TIP5 induces cellular transformation. (A) FACS analysis of miRNA and shRNA-TIP5 cells. Data were quantified from two independent experiments. (B) The growth curve of miRNA-TIP5 and shRNA-TIP5 and control cells. Cellular confluence was reached at about day 5. Similar results were obtained in three independent experiments performed in triplicates. The error bar values of shRNA cells are hidden by symbols. (C) The transforming activity of TIP5 depletion. Cells were plated at low density on a 10-cm-diameter plate. After 14 days (8 days after confluence) cells were stained with methylene blue.
Discussion
Our study provides insights into the role of TIP5 and rDNA silencing in mammalian cells. The results indicate that both depletion and overexpression of TIP5 affect heterochromatin formation at rDNA, centric and pericentric repeats, implying an intimate relationship that links TIP5 with rDNA silencing and formation of centromeric heterochromatin. In mouse and human chromosomes, the rRNA genes are positioned very close to the centromeres (Henderson et al, 1974; Elsevier and Ruddle, 1975) and centromeres of chromosomes bearing rDNA repeats associate with the nucleolus (Carvalho et al, 2001). Importantly, silent methylated rRNA genes were found localized in proximity to the centromeric heterochromatin in mouse neuronal cells (Akhmanova et al, 2000). According to our data, such spatial and linear closeness may allow TIP5, bound to silent rRNA genes, to interact with centric repeats and to aid in establishing heterochromatic structures using similar mechanisms as used to silence the rDNA locus (Santoro et al, 2002; Zhou et al, 2002) (Figures 6A and B). Although our ChIP data showed that this interaction is probably weak and transient, the association of TIP5 with the centromeric protein CENP-A suggested that this interaction indeed takes place. Alternatively, the repressive chromatin of silent rDNA copies may affect the centric and pericentric heterochromatin either by spreading mechanisms or by creating a nucleolar/perinucleolar compartment enriched in chromatin repressor complexes. In both cases, decrease of rDNA silencing after TIP5 depletion would affect the spreading of heterochromatin and reduce the levels of repressor complexes within and nearby the nucleolus. Notably, a role of the perinucleolar compartment in mediating the incorporation of repressive chromatin factors was recently discussed for the establishment of the inactive X-chromosome that contacts the nucleolus during mid-to-late S-phase to faithfully duplicate its epigenetic character (Zhang et al, 2007). Future studies will investigate whether formation of inactive X-chromatin at the nucleolar periphery is also a process that may depend on the levels of rDNA silencing and TIP5.
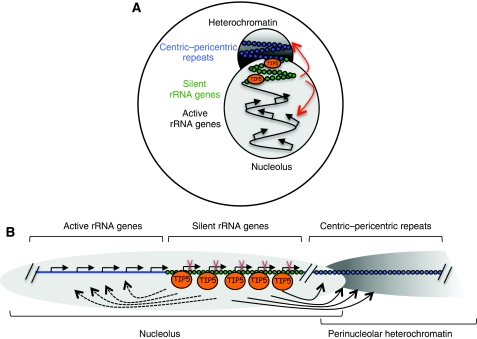
Figure 6.
TIP5 mediates the heterochromatin at the nucleolar/perinucleolar associated chromatin. A model showing the role of TIP5 in establishing heterochromatin at regions located adjacent to the nucleolus. The cellular (A) and linear (B) distribution of active/silent rRNA genes and centromeric heterochromatin within the nucleolus and at the perinuclear periphery. In a transient association model, TIP5 interacts transiently and/or weakly with nearby localized chromatin domains (centric-pericentric repeats) from its stable binding sites (silent rRNA genes). Alternatively, spread of heterochromatin from silent rRNA genes or formation of nucleolar/perinucleolar compartment enriched in chromatin repressor complexes can affect the perinucleolar heterochromatin. In this model, it is also proposed that TIP5 and silent rRNA copies have a role in mediating the transcriptional activity of active rRNA genes as suggested by the results shown in Figure 4D.
Our data show that TIP5 is involved in maintaining genome stability. In the yeast S. cerevisiae, recruitment of the nucleolar protein complexes RENT (regulator of nucleolar silencing and telophase exit) and Cohibin to rDNA suppresses unequal recombination at the rDNA repeats (Mekhail et al, 2008 and references herein). This suppression is seemingly linked to the ability of these complexes to induce rDNA silencing. Until now, the repetitive nature of the rRNA genes represented a limit in determining which rRNA genes (active or silent) undergo instability in the absence of these chromatin repressor complexes. By tracking rRNA genes with polymorphic variations, we showed that TIP5-mediated heterochromatin formation specifically protects silent rRNA genes from illicit recombination events whereas active genes are not affected. Similarly, our data showed that the stability of major and minor satellite sequences depends on TIP5. Loss of rDNA, major and minor satellite repeats is restricted to sequences replicating in the mid-late S-phase. This is consistent with previous results showing that TIP5 binds to rRNA genes, which replicate in the second half of S-phase (Li et al, 2005). Furthermore, it suggests that TIP5 is involved in the heterochromatin formation of centric repeats replicating in mid-late S-phase. As formation and maintenance of heterochromatic structures at these repeats are crucial for genome stability (Peters et al, 2001), our results show that the TIP5-mediated heterochromatin has an important role in protecting the genome from inappropriate chromosomal rearrangements.
Cells in the absence of TIP5 proliferated beyond confluence and had a transformed phenotype, a likely result of the genome instability that we detected in TIP5-depleted cells. Loss of genome stability is known to be a principal molecular step in cancer formation, contributing importantly to rapid selection of clonal cell populations that are able to overcome the various environmental challenges that arise during carcinogenic progression. In addition, in cancer cells, rDNA transcription is enhanced, contributing to increased production of ribosomes and protein synthesis of the rapidly proliferating tumours (Ruggero and Pandolfi, 2003; White, 2005). Disruption in one or more of the steps that control protein biosynthesis has been associated with alterations in the cell cycle and regulation of cell growth (White, 2005). Consistent with this, we have recently shown that depletion of TIP5 and impairment of rDNA silencing enhances ribosome synthesis and increases protein production (Santoro et al, 2009). A lower content of rDNA methylation was reported for several tumours (Qu et al, 1999; Shiraishi et al, 1999; Ghoshal et al, 2004), strengthening the notion of the role of CpG methylation in repressing rDNA transcription (Santoro and Grummt, 2001). Moreover, rDNA CpG methylation levels were found to be higher in ovarian cancer patients with long progression survival as compared with that in patients with short survival, an indication that rDNA silencing levels may influence cell growth properties essential for active tumour proliferation and tumour aggressiveness (Powell et al, 2002). Surprisingly, upregulation of rDNA transcription in TIP5-depleted cells does not depend on the de-repression of silent genes. Whereas the amount of silent genes decreases in these cells, the number of unmethylated active genes is not affected. Consistent with this, a specific class of rDNA variants (rDNA-A) synthesized higher rRNA transcript levels after TIP5 depletion, although the majority of these genes are active and their stability is not affected by TIP5. It seems, therefore, that TIP5 and/or presence of heterochromatic silent repeats indirectly affects the transcription rate of active genes, probably by enriching the nucleolar compartment of the chromatin repressor complexes. However, we cannot exclude the possibility that upregulation of rDNA transcription is a consequence of genome instability that caused the acquisition of aberrant mechanisms of rDNA transcriptional regulation, thus representing an advantage for the elevated protein synthesis necessary for high proliferative rates.
Although it remains to be estimated to which extent the genome instability or enhancement of rDNA transcription in TIP5-depleted cells contributed to the transformed phenotype, our results provide evidences that the TIP5-mediated heterochromatin has a crucial role in protecting genome stability and regulating rDNA transcription, thus contributing to the cellular transformation process.
Materials and methods
Stable and transient TIP5 knockdown
NIH3T3 cells were stably transfected with plasmids expressing shRNA-TIP5-1 (5′-GGACGATAAAGCAAAGATGTTCAAGAGACATCTTTGCTTTATCGTCC) and shRNA-TIP5-2 (5′-GCAGCCCAGGGAAACTAGATTCAAGAGATCTAGTTTCCCTGGGCTGC) sequences under the control of the H1 promoter. Plasmids expressing control miRNA or miRNA targeting TIP5 (5′-GATCAGCCGCAAACTCCTCTGAGTTTTGGCCACTGACTGACTCAGAGGATTGCGGCTGAT) were constructed according to the Block-iT Pol II miR RNAi system (Invitrogen). Infections were performed according to manufacturer's instructions. Cells were analysed 10 days after infection.
Transcription and ChIP analysis
45S pre-rRNA transcription was measured by qRT–PCR in accordance with the standard procedure using the Universal Master mix (Diagenode). The primer sequences used to detect 45S pre-rRNA and GAPDH were as reported by Santoro and Grummt (2005). The rDNA transcription levels were normalized to GAPDH mRNA levels. ChIP analysis was performed as previously described (Santoro et al, 2002). rDNA, major and minor satellite sequences were amplified with previously reported primers (Santoro et al, 2002; Martens et al, 2005). rDNA methylation was measured as previously described (Santoro et al, 2002).
Indirect immunofluorescence
In vivo BrUTP incorporation was performed as previously described (Grob et al, 2009). To detect fibrillarin and UBF, cells were fixed in methanol for 20 min at −20°C, air-dried for 5 min and rehydrated with PBS for 5 min. Incubations with antibodies were performed as previously described (Strohner et al, 2001). DNA was stained with DAPI (Molecular Probes). The area of nucleoli was quantified using ImageJ software (NIH).
Electron microscopy
The DNAs and RNAs were contrasted with uranyl after methylation and acetylation of the amino and carboxyl groups as described by Junéra et al (1995). Briefly, cell pellets were fixed in glutaraldehyde, incubated in methanol and acetic anhydride, and embedded in Epon. The sections were contrasted by uranyl acetate for 60 min at RT. The specificity of the contrast was verified on ribosomes.
Immunoprecipitation
To monitor the interaction of TIP5 and CENP-A in vivo, we transfected HEK293T cells with expression vectors encoding the respective proteins (pcDNA-Flag-Tip5 and GFP-CENP-A (gift from K Sullivan)). After 48 h, we lysed the cells in a lysis buffer (50 mM Tris–HCl (pH 7.8), 150 mM KCl, 5 mM MgCl2, 5 mM EDTA, 20% glycerol, 0.1% NP-40, 0.1 mM PMSF, proteinase inhibitor cocktail (Roche)) at 4°C for 30 min. The cleared lysate was subjected to IP overnight at 4°C using an immobilized antibody against FLAG (anti-FLAG M2 affinity gel; Sigma). The precipitates were washed three times with a buffer containing 150 mM KCl, separated on either 6 or 12% SDS–polyacrylamide gels and analysed on western blots using anti-FLAG M2 (Sigma) and anti-GFP (Roche) antibodies.
Replication timing
Cells were synchronized at the G1/S phase as previously reported (Li et al, 2005). The synchronized cells were pulse-labelled (30 min) with 30 μM 5′-BrdU in 1-h intervals. Nascent DNA was isolated, purified and measured by qPCR. To calibrate DNA recovery, BrdU-labelled Escherichia coli DNA was added to the reactions before IPs. Semi-quantitative PCRs were normalized to the amounts of β-lactamase calculated by qPCR.
DNA copy number
Repetitive DNA sequences were quantitatively amplified from logarithmic dilutions of genomic DNA using previously reported primer sequences (Santoro et al, 2002; Martens et al, 2005). The data were normalized to the amounts of the α-globin gene.
Polymorphism-specific PCR
The following primers pairs were used to specifically amplify the v-rDNA variants: rDNA-A (+63/+42) Rev: TAAATCGAAAGGGTCTCTTT; rDNA-T (+62/+41) Rev: TAAATCGAAAGGGTCTCTTA; rDNA-G (+62/+41) Rev: TAAATCGAAAGGGTCTCTTC; total rDNA (+87/+66) Rev: TAGGCTGGACAAGCAAAACAG; total rDNA (+1/+20) For: ACTGACACGCTGTCCTTTCC; total rDNA (−165/−145) For: GACCAGTTGTTCCTTTGAGG.
Antibodies
Anti-TIP5 antibodies were purchased from Diagenode. Anti-acetylated histone H4, anti-H3K9me3 and H4K20me3 were obtained from Upstate. Anti-UBF and anti-cyclin-A antibodies were from Santa Cruz Biotechnology. A previously characterized human autoimmune serum with specificity against fibrillarin was used (Sirri et al, 2002).
Supplementary Material
Acknowledgments
We thank K Sullivan for providing the GFP-CENP-A plasmid and Mabel San Roman for her help in electron microscopy analysis. This work was supported by the ETH Zurich (RS, MF), the Swiss National Science Foundation (SNF) (RS, CG), the 7th EC Framework Program (PERSIST), the Centre National de la Recherche Scientifique and the Association pour la Recherche sur le Cancer (VS, DH-V).
Footnotes
The authors declare that they have no conflict of interest.
References
- Akhmanova A, Verkerk T, Langeveld A, Grosveld F, Galiart N (2000) Characterisation of transcriptionally active and inactive chromatin domains in neurons. J Cell Sci 24: 4463–4474 [DOI] [PubMed] [Google Scholar]
- Arnheim N, Southern EM (1977) Heterogeneity of the ribosomal genes in mice and men. Cell 11: 363–370 [DOI] [PubMed] [Google Scholar]
- Black BE, Bassett EA (2008) The histone variant CENP-A and centromere specification. Curr Opin Cell Biol 20: 91–100 [DOI] [PubMed] [Google Scholar]
- Boisvert FM, van Koningsbruggen S, Navascués J, Lamond AI (2007) The multifunctional nucleolus. Nat Rev Mol Cell Biol 8: 574–585 [DOI] [PubMed] [Google Scholar]
- Carvalho C, Pereira HM, Ferreira J, Pina C, Mendonca D, Rosa AC, Carmo-Fonseca M (2001) Chromosomal G-dark bands determine the spatial organization of centromeric heterochromatin in the nucleus. Mol Cell Biol 12: 3563–3572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davisson MT (1989) Centromeric heterochromatin variants. In Genetic Variants and Strains of the Laboratory Mouse, Lyon MF, Searle AG (eds), pp 146–166. Oxford: Oxford University Press
- Dev VG, Tantravahi R, Miller DA, Miller OJ (1977) Nucleolus organizers in Mus musculus subspecies and in the RAG mouse cell line. Genetics 86: 389–398 [PMC free article] [PubMed] [Google Scholar]
- Elsevier SM, Ruddle FH (1975) Location of genes coding for 18S and 28S ribosomal RNA within the genome of Mus musculus. Chromosoma 52: 219–228 [DOI] [PubMed] [Google Scholar]
- French SL, Osheim YN, Cioci F, Nomura M, Beyer AL (2003) In exponentially growing Saccharomyces cerevisiae cells, rRNA synthesis is determined by the summed RNA polymerase I loading rate rather than by the number of active genes. Mol Cell Biol 23: 1558–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoshal K, Majumder S, Datta J, Motiwala T, Bai S, Sharma SM, Frankel W, Jacob ST (2004) Role of human ribosomal RNA (rRNA) promoter methylation and of methyl-CpG-binding protein MBD2 in the suppression of rRNA gene expression. J Biol Chem 279: 6783–6793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goren A, Cedar H (2003) Replicating by the clock. Nat Rev Mol Cell Biol 4: 25–32 [DOI] [PubMed] [Google Scholar]
- Grewal SI, Jia S (2007) Heterochromatin revisited. Nat Rev Genet 8: 35–46 [DOI] [PubMed] [Google Scholar]
- Grob A, Roussel P, Wright JE, McStay B, Hernandez-Verdun D, Sirri V (2009) Involvement of SIRT7 in resumption of rDNA transcription at the exit from mitosis. J Cell Sci 122: 489–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grummt I (2003) Life on a planet of its own: regulation of RNA polymerase I transcription in the nucleolus. Genes Dev 17: 1691–1702 [DOI] [PubMed] [Google Scholar]
- Guenatri M, Bailly D, Maison C, Almouzni G (2004) Mouse centric and pericentric satellite repeats from distinct functional heterochromatin. J Cell Biol 166: 493–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haaf T, Schmid M (1991) Chromosome topology in mammalian interphase nuclei. Exp Cell Res 192: 325–332 [DOI] [PubMed] [Google Scholar]
- Henderson AS, Eicher EM, Yu MT, Atwood KC (1974) The chromosomal location of ribosomal DNA in the mouse. Chromosoma 49: 155–160 [DOI] [PubMed] [Google Scholar]
- Jaco I, Canela A, Vera E, Blasco MA (2008) Centromere mitotic recombination in mammalian cells. J Cell Biol 181: 885–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junéra HR, Masson C, Geraud G, Hernandez-Verdun D (1995) The three-dimensional organization of ribosomal genes and the architecture of the nucleoli vary with G1, S and G2 phases. J Cell Sci 108: 3427–3441 [DOI] [PubMed] [Google Scholar]
- Kominami R, Urano Y, Mishima Y, Muramatsu M (1981) Organization of ribosomal RNA gene repeats of the mouse. Nucleic Acids Res 9: 3219–3233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara Y, Suh DS, Suzuki H, Moriwaki K (1994) Chromosomal locations of Ag-NORs and clusters of ribosomal DNA in laboratory strains of mice. Mamm Genome 5: 225–228 [DOI] [PubMed] [Google Scholar]
- Li J, Santoro R, Koberna K, Grummt I (2005) The chromatin remodeling complex NoRC controls replication timing of rRNA genes. EMBO J 24: 120–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maison C, Almouzni G (2004) HP1 and the dynamics of heterochromatin maintenance. Nat Rev Mol Cell Biol 5: 296–304 [DOI] [PubMed] [Google Scholar]
- Martens JH, O'Sullivan RJ, Braunschweig U, Opravil S, Radolf M, Steinlein P, Jenuwein T (2005) The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO J 24: 800–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer C, Schmitz KM, Li J, Grummt I, Santoro R (2006) Intergenic transcripts regulate the epigenetic state of rRNA genes. Mol Cell 22: 351–361 [DOI] [PubMed] [Google Scholar]
- McStay B (2006) Nucleolar dominance: a model for rRNA gene silencing. Genes Dev 20: 1207–1214 [DOI] [PubMed] [Google Scholar]
- Mekhail K, Seebacher J, Gygi SP, Moazed D (2008) Role for perinuclear chromosome tethering in maintenance of genome stability. Nature 456: 667–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D (2001) Common themes in mechanisms of gene silencing. Mol Cell 8: 489–498 [DOI] [PubMed] [Google Scholar]
- Moss T, Langlois F, Gagnon-Kugler T, Stefanovsky V (2007) A housekeeper with power of attorney: the rRNA genes in ribosome biogenesis. Cell Mol Life Sci 64: 29–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng JC, Karpen GH (2007) H3K9 methylation and RNA interference regulate nucleolar organization and repeated DNA stability. Nat Cell Biol 9: 25–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng JC, Karpen GH (2008) Epigenetic regulation of heterochromatic DNA stability. Curr Opin Genet Dev 18: 204–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters AH, Kubicek S, Mechtler K, O'Sullivan RJ, Derijck AA, Perez-Burgos L, Kohlmaier A, Opravil S, Tachibana M, Shinkai Y, Martens JH, Jenuwein T (2003) Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell 12: 1577–1589 [DOI] [PubMed] [Google Scholar]
- Peters AH, O'Carroll D, Scherthan H, Mechtler K, Sauer S, Schofer C, Weipoltshammer K, Pagani M, Lachner M, Kohlmaier A, Opravil S, Doyle M, Sibilia M, Jenuwein T (2001) Loss of the Suv39 h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107: 323–337 [DOI] [PubMed] [Google Scholar]
- Pluta AF, Mackay AM, Ainsztein AM, Goldberg IG, Earnshaw WC (1995) The centromere: hub of chromosomal activities. Science 270: 1591–1594 [DOI] [PubMed] [Google Scholar]
- Powell MA, Mutch DG, Rader JS, Herzog TJ, Huang TH, Goodfellow PJ (2002) Ribosomal DNA methylation in patients with endometrial carcinoma: an independent prognostic marker. Cancer 94: 2941–2952 [DOI] [PubMed] [Google Scholar]
- Qu GZ, Grundy PE, Narayan A, Ehrlich M (1999) Frequent hypomethylation in Wilms tumors of pericentromeric DNA in chromosomes 1 and 16. Cancer Genet Cytogenet 109: 34–39 [DOI] [PubMed] [Google Scholar]
- Ruggero D, Pandolfi PP (2003) Does the ribosome translate cancer? Nat Rev Cancer 3: 179–192 [DOI] [PubMed] [Google Scholar]
- Santoro R (2005) The silence of the ribosomal RNA genes. Cell Mol Life Sci 62: 2067–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro R, Grummt I (2001) Molecular mechanisms mediating methylation-dependent silencing of ribosomal gene transcription. Mol Cell 8: 719–725 [DOI] [PubMed] [Google Scholar]
- Santoro R, Grummt I (2005) Epigenetic mechanism of rRNA gene silencing: temporal order of NoRC-mediated histone modification, chromatin remodeling, and DNA methylation. Mol Cell Biol 25: 2539–2546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro R, Li J, Grummt I (2002) The nucleolar remodeling complex NoRC mediates heterochromatin formation and silencing of ribosomal gene transcription. Nat Genet 32: 393–396 [DOI] [PubMed] [Google Scholar]
- Santoro R, Lienemann P, Fussenegger M (2009) Epigenetic engineering of ribosomal RNA genes enhances protein production. PLoS One 4: e6653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraishi M, Sekiguchi A, Chuu YH, Sekiya T (1999) Tight interaction between densely methylated DNA fragments and the methyl-CpG binding domain of the rat MeCP2 protein attached to a solid support. Biol Chem 380: 1127–1131 [DOI] [PubMed] [Google Scholar]
- Sirri V, Hernandez-Verdun D, Roussel P (2002) Cyclin-dependent kinases govern formation and maintenance of the nucleolus. J Cell Biol 156: 969–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirri V, Urcuqui-Inchima S, Roussel P, Hernandez-Verdun D (2008) Nucleolus: the fascinating nuclear body. Histochem Cell Biol 129: 13–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight AF, Shou W, Dowd GJ, Turck CW, Deshaies RJ, Johnson AD, Moazed D (1999) Net1, a Sir2-associated nucleolar protein required for rDNA silencing and nucleolar integrity. Cell 97: 245–256 [DOI] [PubMed] [Google Scholar]
- Strohner R, Nemeth A, Jansa P, Hofmann-Rohrer U, Santoro R, Langst G, Grummt I (2001) NoRC—a novel member of mammalian ISWI-containing chromatin remodeling machines. EMBO J 20: 4892–4900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Kurihara Y, Kanehisa T, Moriwaki K (1990) Variation in the distribution of silver-staining nucleolar organizer regions on the chromosomes of the wild mouse, Mus musculus. Mol Biol Evol 7: 271–282 [DOI] [PubMed] [Google Scholar]
- Tognon CE, Kirk HE, Passmore LA, Whitehead IP, Der CJ, Kay RJ (1998) Regulation of RasGRP via a phorbol ester-responsive C1 domain. Mol Cell Biol 18: 6995–7008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RJ (2005) RNA polymerases I and III, growth control and cancer. Nat Rev Mol Cell Biol 6: 69–78 [DOI] [PubMed] [Google Scholar]
- Yuan X, Feng W, Imhof A, Grummt I, Zhou Y (2007) Activation of RNA polymerase I transcription by cockayne syndrome group B protein and histone methyltransferase G9a. Mol Cell 27: 585–595 [DOI] [PubMed] [Google Scholar]
- Zhang LF, Huynh KD, Lee JT (2007) Perinucleolar targeting of the inactive X during S phase: evidence for a role in the maintenance of silencing. Cell 129: 693–706 [DOI] [PubMed] [Google Scholar]
- Zhou Y, Santoro R, Grummt I (2002) The chromatin remodeling complex NoRC targets HDAC1 to the ribosomal gene promoter and represses RNA polymerase I transcription. EMBO J 21: 4632–4640 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.