Abstract
The “channeling hypothesis” of DNA electrophoresis in sparse, ordered arrays of posts predicts that the DNA will move through the array relatively unhindered if (i) the spacing between the posts is larger than the DNA coil and (ii) the electric field lines are straight. We tested this hypothesis by studying the electrophoretic separation of a small plasmid DNA (pUC19, 2686 base pairs) and a large, linear DNA (λ-DNA, 48 500 base pairs) in a hexagonal array of 1 μm diameter posts with a pitch of 7 μm. At low electric field strengths, these DNAs are separated due to the long-lived, rope-over-pulley collisions of λ-DNA with the posts. The resolution is lost as the electric field increases due to the onset of channeling by the λ-DNA. Using a diffusive model, we show that channeling arises at low electric fields due to the finite size of the array. This channeling is not intrinsic to the system and is attenuated by increasing the size of the array. Higher electric fields lead to intrinsic channeling, which is attributed to the disparate time scales for a rope-over-pulley collision and transverse diffusion between collisions. The onset of channeling is a gradual process, in agreement with extant Brownian dynamics simulation data. Even at weak electric fields, the electrophoretic mobility of λ-DNA in the array is considerably higher than would be expected if the DNA frequently collided with the posts.
INTRODUCTION
Developing new methods to separate DNA by size is a long-standing goal in microfluidics. This body of work is motivated by the slow speed of pulsed-field gel electrophoresis, which can require hours to days to resolve large DNA by size. Perhaps the most well-studied microfluidic method to separate long DNA is the microfabricated post array.1 Prototype separations in these devices have resolved long DNA in the minute range using very dense arrays of nanoposts2, 3 or quasiordered arrays of self-assembled magnetic beads.4, 5
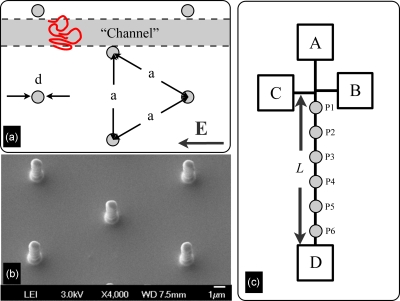
A number of Brownian dynamics simulations of DNA transport in post arrays6, 7, 8 suggested that the particular architecture of these devices2, 3, 4, 5 is crucial to their success. To focus our discussion, let us consider the idealized device depicted in Fig. 1a. The separation arm of length L contains a hexagonal array of small cylindrical posts with diameter d and center-to-center spacing a. Let us consider a DNA of nominal radius of gyration Rg moving through this array. In nanopost systems,2, 3 the gap between columns of the array, (a−d)∕2, is generally smaller than the equilibrium size of the DNA, 2Rg. As a result, the DNA is unable to completely relax in the interstices of the device and frequently interacts with the posts. If the gap is larger than the equilibrium size of the DNA, (a−d)∕2>2Rg, then there exists a “channel” between the columns of the array wherein the DNA can move unhindered. The aforementioned simulation data6, 7, 8 clearly demonstrate that if the electric field lines are straight, then the DNA will tend to move through this gap without colliding with the posts. We refer to this simulation result as the “channeling hypothesis.”
Figure 1.
(a) Schematic illustration of the channeling hypothesis. The separation device consists of a hexagonal array of total length L containing posts of diameter d with a center-to-center spacing a. If (a−d)∕2>2Rg, where Rg is the radius of gyration of the DNA, then there exists a channel for unhindered DNA transport through the array. (b) SEM image of the oxidized silicon post array used here. (c) Schematic of the experimental setup. Measurements of the fluorescence intensity are obtained at positions Pn located 2.5n mm along the separation arm.
If the channeling hypothesis is correct, then the successful separation of long DNA in magnetic bead arrays4, 5 is attributed to the inherent disorder of the system; the occasional misplaced post in these relatively sparse arrays halts any incipient channeling. Moreover, the channeling hypothesis implies that long DNA cannot be separated in a sparse, ordered post array. This corollary is unfortunate since sparse, ordered post arrays are easy to fabricate relative to nanopost arrays2, 3 and do not require the external magnet and flow control systems used for high-quality separations in magnetic bead arrays.5
In a recent publication,9 we tested the channeling hypothesis using λ-DNA [48 500 base pairs, Rg=0.7 μm (Ref. 10)] in a polydimethylsiloxane (PDMS) array with d=1.2 μm and a=3 μm. While Brownian dynamics simulations using a uniform electric field8, 9 predicted that this DNA should channel through the array when the electric field is strong, we observed frequent collisions with the posts at all electric field strengths. These collisions are due to the curvature of the electric field caused by the insulating posts and the symmetry of the array. Upon incorporating the details of the electric field into the Brownian dynamics simulation, we found that the simulated mobility agrees well with the experimental data.9
While our prior work refuted the channeling hypothesis for one particular array geometry, it did not eliminate the possibility of channeling in general. Rather, the latter work demonstrated that the assumption of uniform electric field lines is critical to the channeling hypothesis. If we can remove (or at least minimize) the electric field gradients, then we should observe channeling above some characteristic value of the electric field. In a hexagonal post array, the electric field gradients are controlled by two features: The physiochemical properties of the posts and the post density, d∕a. If the posts were ionically conductive, then the electric field lines would penetrate them. Unfortunately, it is not clear how to construct the system in Fig. 1a in a noninsulating material. In contrast, changing the post density is straightforward. As the parameter a∕d becomes large, the perturbation of the uniform electric field caused by the presence of a post is localized around that post. In the interstices, the electric field is relatively uniform. Indeed, for a finite-sized system, we must observe channeling in the limit d∕a→0. The extent of this perturbation is also a function of the post geometry. For example, the local electric field lines around a stack of magnetic beads would be different than a cylindrical obstacle. As a result, we would expect the channeling in an array of magnetic beads of diameter d to occur at a smaller value of a∕d than cylindrical obstacles with the same diameter.
In the present contribution, we present experimental data on the electrophoresis of λ-DNA in the silicon dioxide post array chip depicted in Fig. 1b. The large pore size in this system satisfies both of the criteria required for channeling of λ-DNA. To determine the onset of channeling, we also added a small, circular plasmid DNA (pUC19, 2686 base pairs). The latter molecule is too small to make a rope-over-pulley collision with the posts. The loss of separation resolution between the pUC19 and λ-DNA thereby serves as an unambiguous signal of complete channeling by the long DNA.
METHODS AND MATERIALS
The micropost array used for these experiments consists of a 50 μm wide, 16 mm long hexagonal array of 1 μm diameter posts with a center-to-center spacing of 7 μm. As seen in the schematic of Fig. 1c, the separation arm is integrated with a shifted-T injection using a 400 μm offset. To fabricate the chip, we first grew 100 nm of silicon dioxide on a [100] silicon wafer via thermal oxidation at 1200 °C for 1 h. We then spin coated the oxide layer with S1813 photoresist (Shipley) and transferred the negative image of the channel and post pattern into the resist by optical lithography. After a lift-off step using a 300 Å layer of chrome, we used a buffered oxide etch to create the positive image of the chip in the oxide layer. We then stripped the remaining chrome and etched the patterned wafer to a depth of 6 μm using a deep trench etcher (Plasma Therm SLR 770). The wafer was diced and the four reservoirs were opened using a diamond drill bit. To provide electrical insulation, we repeated the thermal oxidation step for 3 h to grow a 350 nm layer of oxide on the patterned wafer. A scanning electron microscopy (SEM) image of the finished post array is shown in Fig. 1b.
The assembled device consists of a silicon chip sandwiched between a 1×3 in.2 microscope slide and a 22×40 mm2 No. 1 coverslip. We drilled the microscope slide with access holes and glued it to the back side of the silicon chip. To bond the coverslip to the front side of the channel, we used a variant of the microfluidic stickers method.11 We first deposited a monolayer of tridecafluoro-1,1,2,2-tetrahydrooctyl-1-trichlorosilane (United Chemicals) on a flat piece of degassed, cured PDMS (Sylgard 184, Dow Corning) to aid in release from the optical adhesive. The coverslip was then cleaned in 1M NaOH and spin coated with a thin layer of NOA- 81 optical adhesive (Norland). The PDMS was placed on top of the liquid NOA-81 film and the system was exposed through the PDMS for 60 s using a high-intensity UV light (Daigger, 100 W, 365 nm). The oxygen-permeable PDMS prevents curing of a thin layer of NOA-81 in contact with the PDMS. This “sticky” coverslip was bonded immediately to the front side of the silicon chip by a 60 s UV exposure and 12 h of thermal curing at 55 °C. At the conclusion of the bonding, we glued four fittings (Upchurch 131 reservoirs) to the back side of the microscope slide to provide 120 μl reservoirs.
To condition the assembled chip, we used a series of fluid exchanges by emptying reservoirs A–C and using a gravity head in reservoir D, following the notation of Fig. 1c. We first pumped purified water through the channels and then displaced the water with a 1 wt % solution of polyvinylpyrollidone (PVP) (molecular weight=1.3×106, Sigma) in tris-borate-EDTA (TBE) 5× buffer supplemented with 0.07 wt % ascorbic acid (Sigma-Aldrich) for 90 min. We found that the polymer coating was essential to avoid adsorption of DNA to the posts. The polymer coating should also suppress electro-osmotic flow, although there are reports that TBE 5× buffer alone is sufficient for suppression.12, 13 After the coating was complete, the excess PVP was flushed with a solution of TBE 5× and 0.07 wt % ascorbic acid for at least 12 h. To minimize evaporation, the reservoirs were covered in parafilm during the latter step.
Our experiments used linear λ-DNA (48 500 base pairs, New England Biolabs) and the circular pUC19 plasmid DNA (2686 base pairs, New England Biolabs). These DNAs were dyed with YOYO-1 (Invitrogen) at a ratio of one dye molecule:five base pairs. Although the commercial pUC19 plasmid consists of a mixture of supercoiled and open-circle topoisomers, we suspect that photonicking of the dyed DNA during our experiments leads to a dominantly open-circle state.14 The dyed stock solutions of λ-DNA and pUC19 were mixed and diluted with TBE 5× to a final concentration of 7.5 and 5 μg∕ml, respectively. For the control experiments with pUC19, the dye procedure was repeated to provide a stock concentration of 10 μg∕ml.
Prior to a given set of experiments, we first gently air dried the reservoirs and then filled reservoirs A, C, and D in Fig. 1c with 100 μl of TBE 5× buffer supplemented with 0.07 wt % ascorbic acid and 100 mM DL-dithiothreitol (Fluka) to scavenge oxygen. Reservoir B was filled with dyed DNA mixture. For the experiments using only pUC19, 100 μl of the pUC19 solution at 10 μg∕ml was added; for the remaining experiments, we added 100 μl of the DNA mixture. We mounted the chip on an inverted epifluorescence microscope (Leica DMI-4000) and observed the DNA using a 63×, 1.4 numerical aperture oil immersion objective. To obtain electropherograms, we used a small square field diaphragm setting to limit the field of view and focused the emitted light onto a photomultiplier tube (Hamamatsu). To obtain images, we used the full field of view and an electron-multiplying charge coupled device camera (Photometrics Cascade II). The experimental system is entirely automated and controlled by a custom LABVIEW script. The electric potentials were applied in each reservoir using a LabSmith HVS-1500 programmable power supply and platinum electrodes. We determined appropriate potentials for separation arm electric fields of 20–110 V∕cm by modeling the resistance of the arms of the chips as proportional to their length and computing the electric fields via Kirchoff’s law. Electropherograms were collected every 2.5 mm downstream from the injection point, as indicated in Fig. 1c.
To compute the resolution Rs, we attempted to fit the overall electropherogram with the sum of two Gaussian functions. If this approach was successful (Rs>0.5), we computed the resolution via the standard definition
| (1) |
where is the time corresponding to the maximum in the fitted Gaussian and σi is the variance of the Gaussian for species i. If we could not resolve the two peaks, we report the result as zero resolution.
All of the experimental data presented here were collected on a single chip. After approximately 40 injections, the fluid in the reservoirs was removed with a pipet and the chip was recoated with PVP following the protocol described above.
RESULTS
The key assumption in our experiments is that the pUC19 plasmid mobility represents the maximum mobility inside the post array. To test this assumption, we measured the mobility of pUC19 as a function of electric field using the multipoint method described previously.9 As seen in Fig. 2a, the mobility of pUC19 is essentially independent of the electric field. The average value of this mobility, , is similar to the free-solution mobility for DNA we obtained in our prior experiments in PDMS channels.9
Figure 2.
(a) Mobility of pUC19 as a function of the electric field. Each data point represents the average of three to five data points. (b) Images of pUC19 and λ-DNA under an electric field of 20 V/cm. The images were taken at the same location in the post array (10 mm downstream) with an offset of Δt=25 s (enhanced online). (c) Representative electropherograms obtained 15 mm downstream under electric fields of 20, 40, 60, 80, and 100 V/cm. Each electropherogram corresponds to an individual experiment. The fluorescence intensity for each electropherogram has been rescaled to a maximum intensity of unity.
We then proceeded to determine whether the λ-DNA will begin to move with the same mobility as the pUC19 as the electric field increases. There are two simple ways to address this question: (i) Perform experiments with λ-DNA exclusively and compare the resulting mobility to the data in Fig. 2a or (ii) inject a mixture of λ-DNA and pUC19 and measure the separation resolution. We chose option (ii) because it allowed us to minimize the run-to-run (and day-to-day) changes in the measured mobility due to changes in buffer pH, random perturbations to the reservoir pressures, and other experimental uncertainties. While the latter factors cause the separation resolution to vary somewhat from run to run, we will see shortly that the presence of a single peak in a given run indicates that the λ-DNA and pUC19 possess similar mobilities under the particular experimental conditions of that run. We define the point where we can no longer distinguish two peaks in the electropherogram as the channeling limit.
Figure 2c presents several representative electropherograms obtained at the end of our array (15 mm) at different electric fields. At the lowest electric field of 20 V∕cm, two peaks are clearly resolved. Even at a shorter distance of 10 mm, the videomicroscopy data presented in Fig. 2b (enhanced online) definitively identify the peaks. The first wave of DNA to enter the viewing window clearly consists of a large number of small, freely moving DNA, corresponding to pUC19. In contrast, the latter images feature a lower number of bright molecules, corresponding to λ-DNA. These large DNAs also occasionally undergo rope-over-pulley collisions at 20 V∕cm.
The mobility of the pUC19 appearing in Fig. 2c is about 10% lower than in Fig. 2a. Some of these differences arises from deconvolving the peak in Fig. 2c into two Gaussians. Since the bands are not base line resolved, the overlap between the pUC19 and λ-DNA peaks can make the deconvolved pUC19 “band” appear to be slower. The fitting error should only reduce the mobility measurement by a few percent, which is insufficient to explain the mobility difference between the mixture and the individual species.
We envision two possible reasons for the change in mobility between Figs. 2a, 2c: (i) A difference in the background flow between experiments that could be due to a breakdown in the PVP coating and ensuing electro-osmotic flow or (ii) pUC19∕λ-DNA interactions in the mixture that would not be present when these species are run individually. We tested these hypotheses with our 20 V∕cm data since the DNAs are relatively well resolved by the end of the channel under this electric field. We used our fitting routine to deconvolve the electrophoregrams obtained at 7.5, 10.0, 12.5, and 15.0 mm downstream from the injection point for a number of different injections of DNA. We did not attempt the fitting at the first two detection points (2.5 and 5.0 mm) since the electropherogram is essentially one peak at these short distances. To test for day-to-day variability, we used electropherograms obtained on two separate days. Between the two days, the chip was cleaned following the protocol described in Sec. 2.
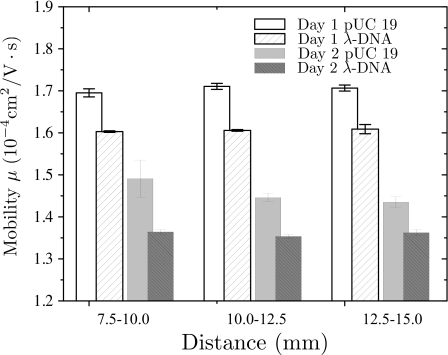
Figure 3 presents the mobility of pUC19 and λ-DNA over the last three 2.5 mm long sections of our channel. These data are averaged over multiple experiments conducted with the same electric field. While the mobility difference remains the same between days, the absolute mobilities vary. Moreover, the mobility is relatively constant within a given day. These data are consistent with a change in the background electro-osmotic flow on different days, which would affect each species in a uniform manner. These data further support our decision to measure the channeling effect with a mixture, rather than individual species.
Figure 3.
Mobility of pUC19 and λ-DNA at 20 V∕cm over the indicated 2.5 mm long intervals downstream from the injection point. The data are at the average of three runs each day.
The data in Fig. 3 do not support pUC19∕λ-DNA interactions. We would expect that the pUC19∕λ-DNA interactions, if present, would decrease as these DNAs move down the channel and separate into two bands. As a result, the pUC19 mobility data in Fig. 3 should increase with the measurement interval. Moreover, since these DNAs are relatively well resolved at the final detection point, we would further expect that the mobility over the last interval would be similar to that in Fig. 2a. However, this is not the case. Within the experimental error, the mobility data in Fig. 3 are independent of the position in the channel. Thus, our data do not support a lowered mobility of the mixture caused by pUC19∕λ-DNA interactions.
The electrophoretic mobility of the slower λ-DNA second peak in Fig. 2c at 20 V∕cm, obtained from the Gaussian fit, is 1.64×10−4 cm2∕V s. If the pUC19 peak represents the limiting electrophoretic mobility, then the λ-DNA moves at 94% of the maximum possible velocity. Although we can still resolve these two DNAs, the relative electrophoretic mobility of the λ-DNA is much larger than the ∼75%–80% value obtained in a denser array that led to frequent collisions with the posts.9 Thus, although we do not see “complete” channeling of the λ-DNA, it is clear that the posts offer little impediment to the motion even at the lowest electric fields used here. This claim is verified by the videomicroscopy data in Fig. 2b (enhanced online).
The area under the slower peak is also greater than the area under the faster peak. Although this is consistent with the relative mass concentrations of λ-DNA and pUC19 loaded into the reservoirs, the area under the curve is not a reliable method for peak identification in our experimental system. If a λ-DNA collides with a post and makes a rope-over-pulley collision during its passage through the detection window, it will contribute more integrated fluorescence intensity than an unimpeded pUC19 molecule. The latter effect is offset to some extent by an injection bias toward pUC19. The electric field in the large reservoir is relatively weak compared to the electric field in the microchannels so the injected DNAs (which were located near the reservoir exit) need to be replenished by diffusion inside the reservoir between injections. The smaller pUC19 diffuses more quickly toward the sink at the entrance to the injection arm and thus, tends to be injected at a higher concentration.
As the electric field increases, we observe a gradual decrease in the resolution of the peaks. Several representative electropherograms are presented in Fig. 2c. To permit a more quantitative analysis, we obtained a number of electropherograms between 20 and 110 V∕cm at 10 V∕cm intervals. We computed the separation resolution by first fitting the electropherogram with the sum of two Gaussians and then computing the resolution with Eq. 1. When the two peaks strongly overlap, as is the case in Fig. 2c at 100 V∕cm, we simply state that there is no resolution. While our fitting program will produce a resolution value for these high electric fields, the algorithm is a six parameter fit and the uncertainty in the computed resolution is large. We chose Rs=0.5 as the cutoff, following Gidding’s guideline for detecting two peaks.15
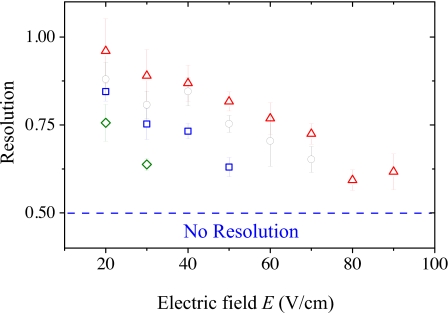
The separation resolution depends on the dynamics inside the post array and the finite size (and DNA distribution) in the injection plug. Owing to the latter, we never observed two peaks in the electropherogram until we reached the third detection position at 7.5 mm. Figure 4 presents the separation resolution data obtained from this point onward as a function of the electric field. The resolution increases with the distance downstream from the injection point and decreases monotonically with the electric field strength until a regime where the resolution cannot be measured.
Figure 4.
Separation resolution at different electric fields at 7.5 mm (green diamonds), 10 mm (blue squares), 12.5 mm (black circles), and 15 mm (red triangles). If two peaks could not be resolved by the fitting program (Rs<0.5), the resolution is reported as zero. Each data point represents the average of six to ten electropherograms.
From Eq. 1, there are two mechanisms for the loss of resolution. In the first, the band broadening becomes so severe that although the two peaks may be separated, they cannot be detected easily. Referring back to Fig. 2c, we see that the peaks actually become sharper as the electric field increases. This behavior is expected in a finish-line system; the time for a band of fixed width to cross the detector decreases as the electric field increases. Rather, the loss of resolution is due to the fact that the difference between the peak times, , becomes small. We can conclude that the λ-DNA electrophoretic mobility approaches the pUC19 electrophoretic mobility as the electric field increases.
DISCUSSION
Ogston sieving of pUC19
Before we proceed to discuss the channeling of λ-DNA, it is useful to briefly comment on the factors contributing to the pUC19 electrophoretic mobility. Since the pUC19 is too short to form a rope-over-pulley, we would not expect any long-lived interactions between the pUC19 and the posts. However, the geometry of our post array bears a remarkable similarity to the lattice models used by Slater and co-workers for gel electrophoresis (see, for example, Ref. 16). As a result, it is tempting to assume that the pUC19 electrophoretic mobility is governed by something akin to Ogston sieving [or, more precisely, the Ogston-Morris-Rodbard-Chrambach (OMRC) model]. However, the electric fields used in our experiments are too strong for Ogston sieving to be a valid model. Indeed, even the lowest electric field videomicroscopy data appearing in Fig. 2b (enhanced online) show that the transport is dominated by convection. In the context of the Ogston sieving model of Slater and co-workers, we note that the curvature of the field lines around our large obstacles (in conjunction with the strong electric field) make the “rejected moves” used in the lattice model specious.16 Rather, we would expect that the DNAs are convected smoothly around the obstacle with minimal delay. Note that this argument is only valid for large fields, as the lattice model gives essentially the same electrophoretic mobility for conductive and insulating obstacles in the low-field limit.17
Even if Ogston sieving was the dominant transport mechanism for the small pUC19 DNA, it would have a little effect on the averaged electrophoretic mobility in the post array because the fractional volume, f, available to the DNA is very high in our dilute post system. To be quantitative, if we neglect the size of the plasmid DNA, then the fractional volume in a hexagonal array is given by
| (2) |
The OMRC model assumes that the electrophoretic mobility in this artificial “gel” is μ=μ0f, where μ0 is the free-solution electrophoretic mobility.18 For our system, d∕a=1∕7 and the corresponding fractional volume is f=0.98. The difference between the free-solution electrophoretic and the Ogston sieving electrophoretic mobilities is thus within the experimental error. Owing to the strong electric field and the high fractional volume, we can safely assume that the pUC19 peak represents a DNA molecule moving through the array with minimal interactions with the posts.
Channeling in a finite-length array
Our results for the loss of resolution between λ-DNA and pUC19 can be understood in the context of a simple model for the channeling process.9 As we established above, the pUC19 hardly interacts with the posts and moves with a field-independent electrophoretic mobility. Let us assume that this maximum electrophoretic mobility inside the array is the free-solution electrophoretic mobility and denote its value as μ0. In contrast to the pUC19, the λ-DNA will tend to make nc rope-over-pulley collisions with the posts before reaching some distance L inside the array. The nominal duration of a collision is τc. Owing to the straight electric field lines in our dilute array, we assume that the collisions arise from transverse diffusion to the edges of the channel between the posts. As a result, the time between collisions, τd, is independent of the electric field and the distance traveled between collisions, lc=μ0Eτd, scales linearly with the electric field. At some distance L inside the post array, the nominal number of collisions experience by the λ-DNA is
| (3) |
Once the DNA has experienced many collisions, nc⪢1, then we assume that it has adequately sampled the collision time distribution. At this point, τc represents a reasonable approximation for the averaged collision time and the average electrophoretic mobility of the λ-DNA is
| (4) |
Note that if we assume τd=a2∕D and τc=L∕μ0E for a DNA of contour length L, Eq. 4 reduces to the result appearing in Ref. 9.
If the DNAs have only made a few collisions, then the average electrophoretic mobility is not stationary in time. Rather, this mobility will change each time the DNA collides because these collisions make a non-neglible contribution to the total residence time in the array. To aid in the remainder of our discussion, let us denote by the characteristic number of collisions after which the average electrophoretic mobility is essentially independent of time. Thus, Eq. 4 is valid for .
The model proposed here permits two types of channeling behavior. In the first type, the λ-DNAs have simply failed to make enough collisions to reach the asymptotic limit . In the most extreme limit, the DNA will, on average, have made less than one collision. This type of channeling can be attenuated by increasing the length of the separation channel. In the second type of channeling, the collision time becomes very small compared to the diffusion time (τc⪡τd). In this case, the channeling is intrinsic and cannot be alleviated by increasing the length of the separation channel. Both types of channeling can also occur simultaneously; it is possible to have only a few collisions and for those collisions to contribute little to the residence time in the array.
Let us begin by considering the first type of channeling. If we assume the initial position of the DNA is uniformly distributed inside the channel, then the first-passage time to hit one of the boundaries is τd=a2∕12D.19 In the latter, D=0.47 μm2∕s is the molecular diffusion coefficient of λ-DNA.10 The corresponding diffusion time is 8.7 s. Based on the electropherograms presented in Fig. 2c, we begin to have substantial resolution around 200 s, indicating that . Note that for all of the electropherograms in Fig. 2c, the λ-DNA is still moving much more quickly than if it made frequent collisions.9 Thus, we are being very strict in our definition of channeling.
We could also imagine that an unhooking DNA molecule should release closer to one side of the channel. In this case, the time between collisions should be computed from a first-passage time analysis with an initial position biased toward one side of the interval.19 The latter model would reduce the diffusion time by an O (1) constant, but it would not affect our general conclusions.
If the λ-DNA channeling for a given electric field strength is the result of the finite length of the array, we should again resolve the peaks if we increase the post array length. Indeed, this is exactly what we observe in Fig. 4. From Eq. 3, we see that the post array length L required to reach the required number of collisions, , should scale linearly with the electric field. The latter scaling is consistent with the data reported in Fig. 4; as we move the observation point 2.5 mm further downstream, the electric field corresponding to the loss of separation resolution shifts up by 20 V∕cm. We shall not place much emphasis on the slope of this line or its zero residual since both quantities would be affected by changing our definition of no resolution or decreasing the step size of the electric field between experiments.
The dependence of the resolution on distance at a fixed electric field provides further support for our claim that the channeling is due to the finite size of the array. Explicitly, the standard resolution model in separation sciences predicts that the resolution should scale with distance like .15 This model assumes, among other things, that the average electrophoretic mobility and dispersivity of each species are stationary. In the context of our post array system, this assumption corresponds to the DNA making many collisions with the posts before reaching the detection point. The data in Fig. 4 do not follow this scaling law. Indeed, the scaling of the resolution with length varies with the electric field. We posit that the failure of the data in Fig. 4 to obey the standard scaling law is due to the low number of collisions and the concomitant nonstationary electrophoretic mobility and dispersivity for λ-DNA.
The loss of resolution at E=80 V∕cm and L=12.5 mm seems to be close to the onset of intrinsic channeling for λ-DNA in our array. Although the resolution returns when we increase the detection distance from 12.5 to 15 mm, it is very close to the cutoff value. In the context our diffusion model, we are now approaching the second regime of channeling where the collisions no longer make a significant contribution to the residence time in the array. To provide quantitative estimate, let us model the average collision time for a DNA of contour length L as20
| (5) |
The stained contour length of λ-DNA is approximately 20.5 μm.21 If we assume that μ0 is the averaged electrophoretic mobility from Fig. 2a, then the collision time is approximately 200 ms. There is thus a clear separation between the diffusion and collision time scales at this point.
CONCLUDING REMARKS
In the present contribution, we have experimentally confirmed the channeling hypothesis for long DNA electrophoresis in a sparse, ordered post array. The uniform electric field in the interstices of our very sparse (but electrically insulating) post array is the key to the realization of channeling since we can minimize the convective steering of DNA by the curved field lines present in more dense systems. At low electric fields, channeling arises from the finite number of collisions with the posts. Upon increasing the length of the array, this type of channeling can be alleviated. In contrast, channeling is intrinsic to the system at high electric fields because the collisions make a small contribution to the total residence time in the array. The onset of the channeling behavior is gradual, in accord with previously published Brownian dynamics simulation data for a related system.8
ACKNOWLEDGMENTS
This work was supported by a CAREER award from the National Science Foundation (Grant No. CBET-0642794) and the David and Lucile Packard Foundation. We are grateful to Alyssa Terry for assistance in device fabrication and Daniel W. Olson for acquiring some of the data presented here. Portions of this work were performed in the University of Minnesota NanoFabrication Center, which receives partial support from the NSF through the NNIN.
References
- Volkmuth W. D. and Austin R. H., Nature (London) 358, 600 (1992). 10.1038/358600a0 [DOI] [PubMed] [Google Scholar]
- Kaji N., Tezuka Y., Takamura Y., Ueda M., Nishimoto T., Nakanishi H., Horiike Y., and Baba Y., Anal. Chem. 76, 15 (2004). 10.1021/ac030303m [DOI] [PubMed] [Google Scholar]
- Shi J., Fang A. P., Malaquin L., Pepin A., Decanini D., Viovy J. -L., and Chen Y., Appl. Phys. Lett. 91, 153114 (2007). 10.1063/1.2793616 [DOI] [Google Scholar]
- Doyle P. S., Bibette J., Bancaud A., and Viovy J. -L., Science 295, 2237 (2002). 10.1126/science.1068420 [DOI] [PubMed] [Google Scholar]
- Minc N., Fütterer C., Dorfman K. D., Bancaud A., Gosse C., Goubault C., and Viovy J. -L., Anal. Chem. 76, 3770 (2004). 10.1021/ac035246b [DOI] [PubMed] [Google Scholar]
- Patel P. D. and Shaqfeh E. S. G., J. Chem. Phys. 118, 2941 (2003). 10.1063/1.1532729 [DOI] [Google Scholar]
- Mohan A. and Doyle P. S., Phys. Rev. E 76, 040903(R) (2007). 10.1103/PhysRevE.76.040903 [DOI] [PubMed] [Google Scholar]
- Mohan A. and Doyle P. S., Macromolecules 40, 8794 (2007). 10.1021/ma071354e [DOI] [Google Scholar]
- Ou J., Cho J., Olson D. W., and Dorfman K. D., Phys. Rev. E 79, 061904 (2009). 10.1103/PhysRevE.79.061904 [DOI] [PubMed] [Google Scholar]
- Smith D. E., Perkins T. T., and Chu S., Macromolecules 29, 1372 (1996). 10.1021/ma951455p [DOI] [Google Scholar]
- Bartolo D., Degre G., Nghe P., and Studer V., Lab Chip 8, 274 (2008). 10.1039/b712368j [DOI] [PubMed] [Google Scholar]
- Han J. and Craighead H. G., J. Vac. Sci. Technol. A 17, 2142 (1999). 10.1116/1.581740 [DOI] [Google Scholar]
- Kaji N., Oki A., Ogawa R., Takamura Y., Nishimoto T., Nakanishi H., Horiike Y., Tokeshi M., and Baba Y., Isr. J. Chem. 47, 161 (2007). 10.1560/IJC.47.2.161 [DOI] [Google Scholar]
- Cole K. D., Gaigalas A., and Akerman B., Electrophoresis 27, 4396 (2006). 10.1002/elps.200600347 [DOI] [PubMed] [Google Scholar]
- Giddings J. C., Unified Separation Science (Wiley, New York, 1991). [Google Scholar]
- Slater G. W. and Guo H. L., Electrophoresis 17, 977 (1996). 10.1002/elps.1150170604 [DOI] [PubMed] [Google Scholar]
- Mercier J. -F., Tessier F., and Slater G. W., Electrophoresis 22, 2631 (2001). [DOI] [PubMed] [Google Scholar]
- Rodbard D. and Chrambach A., Proc. Natl. Acad. Sci. U.S.A. 65, 970 (1970). 10.1073/pnas.65.4.970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redner S., A Guide to First-Passage Processes (Cambridge University Press, Cambridge, 2001). [Google Scholar]
- Nixon G. I. and Slater G. W., Phys. Rev. E 50, 5033 (1994). 10.1103/PhysRevE.50.5033 [DOI] [PubMed] [Google Scholar]
- Randall G. C. and Doyle P. S., Macromolecules 38, 2410 (2005). 10.1021/ma048073g [DOI] [Google Scholar]