Abstract
Background:
Osteosarcoma (OS) is the most common primary bone tumour in children and adolescents. Patients who respond poorly to chemotherapy have a higher risk of metastatic disease and 5-year survival rates of only 10–20%. Therefore, identifying molecular targets that are specific for OS, or more specifically, metastatic OS, will be critical to the development of new treatment strategies to improve patient outcomes.
Methods:
We performed a transcriptomic analysis of chemo-naive OS biopsies and non-malignant bone biopsies to identify differentially expressed genes specific to OS, which could provide insight into OS biology and chemoresistance.
Results:
Statistical analysis of the OS transcriptomes found differential expression of several metallothionein family members, as well as deregulation of genes involved in antigen presentation. Tumours also exhibited significantly increased expression of ID1 and profound down-regulation of S100A8, highlighting their potential as therapeutic targets for OS. Finally, we found a significant correlation between OS and impaired osteoclastogenesis and antigen-presenting activity. The reduced osteoclastogenesis and antigen-presenting activity were more profound in the chemoresistant OS samples.
Conclusion:
Our results indicate that OS displays gene signatures consistent with decreased antigen-presenting activity, enhanced chemoresistance, and impaired osteoclastogenesis. Moreover, these alterations are more pronounced in chemoresistant OS tumour samples.
Keywords: osteosarcoma, bone tumour, chemoresistance, RNA expression profile
Osteosarcoma (OS) is the most common primary malignant bone tumour in children and adolescents (Chou and Gorlick, 2006). Disease progression is marked by aggressive growth, local recurrence, and poor long-term survival rates because of the development of fatal pulmonary metastasis in up to 50% of patients (Wang, 2005). Chemotherapy over many weeks results in disease-free survival rates of 50–70% for patients with non-metastatic disease (Bacci et al, 1997; Marina et al, 2004). However, patients who have a poor response to chemotherapy have a higher risk of developing pulmonary metastases, which results in survival rates of <20% (Chou and Gorlick, 2006). To develop new-targeted therapies to treat OS will require knowledge of the specific defects associated with non-metastatic and metastatic disease. Tests to predict whether a patient will respond to current chemotherapeutics or develop metastases would enhance our ability to select appropriate treatment strategies. To this end, a number of studies of OS have generated gene expression signatures, which have provided insights into OS biology (Leonard et al, 2003; Baird et al, 2005) and chemoresistance (Ochi et al, 2004; Man et al, 2005; Mintz et al, 2005). However, these studies have focused on comparisons of chemosensitive vs chemoresistant, or metastatic vs non-metastatic disease. Studies comparing non-malignant bone vs OS tissue have not been earlier reported.
In this study, we compared the transcriptomes of chemo-naive OS biopsies, collected at the time of diagnosis, with samples of non-malignant bone. Statistical analysis of the expression profiles shows that osteosarcomas are characterised by an early deregulation of genes involved in drug resistance, tumour progression, antigen presentation, and osteoclastogenesis. Furthermore, in biopsies from patients who developed metastatic disease, these changes were significantly more pronounced. These data suggest that patient prognosis is determined early in tumour development and that enhancing antigen presentation or osteoclastogenesis may be of clinical value in treating OS.
Materials and Methods
Patient samples
Patients presented to the Oncology Clinic at the Princess Alexandra or at The Wesley Hospitals (Brisbane, Queensland, Australia). Tumour biopsies were collected at the time of initial diagnosis, before preoperative chemotherapy, with informed consent from patients/guardians and with approval from the relevant institutional Research Ethics Committees. Twenty-three biopsies were available and subjected to gene expression profiling analysis. Clinical data detailing response to chemotherapy was available for 22 out of 23 patients (Table 1). Patients were classified as good responders (R) if the tumours had ⩾90% tumour necrosis, or poor responders (N) if the tumours had <90% necrosis in response to preoperative chemotherapy (doxorubicin, 25 mg m–2 and cisplatin, 100 mg m–2) as determined by histologic examination at the time of definitive surgery (Salzer-Kuntschik et al, 1983). Non-malignant bone was collected with consent from five patients presenting for hip or knee replacement surgery.
Table 1. Clinical information for the osteosarcoma patients used in the study.
| Tumour ID | Age (years) | Gender | Site of tumour | Tumour necrosis (%) | Response |
|---|---|---|---|---|---|
| G3 | 15 | M | Proximal tibia | 100 | R |
| S4 | 29 | F | Sacrum | <5 | N |
| S5 | 27 | F | Proximal humerus | <50 | N |
| S7 | 39 | M | Pelvis | <50 | N |
| M8 | 24 | F | Distal femur | 25 | N |
| L9 | 17 | M | Tibia | 75 | N |
| W10 | 14 | M | Distal femur | 75 | N |
| A13 | 7 | F | Proximal tibia | 80 | N |
| M14 | 67 | M | Pelvis | 75 | N |
| S15 | 76 | F | Tibia | <90 | N |
| W16 | 15 | F | Proximal Humerus | <90 | N |
| O17 | 12 | M | Distal femur | 75 | N |
| M18 | 18 | M | 8th rib | 98 | R |
| K19 | 17 | M | Proximal tibia | 50 | N |
| E20 | 13 | F | Femur | 95 | R |
| M21 | 15 | M | Distal femur | 95 | R |
| W22 | 15 | F | Femur | 80 | N |
| G23 | 18 | M | Distal femur | 80 | N |
| F2BR | 18 | M | Proximal tibia | 80 | N |
| A3BR | 19 | F | Distal femur | 80 | N |
| V4BR | 16 | M | Calcaneum | <5 | N |
| M7BR | 14 | F | Distal femur | >92 | R |
| T1a | 37 | F | Proximal tibia | U | U |
Response to chemotherapy: R=good response; N=poor response; U=unknown.
This patient was used in the comparison between osteosarcoma and non-malignant bone, but not in the chemotherapy response study.
Microarray and data analysis
Extraction of RNA from cells and tumours was performed using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). Each OS patient sample was analysed in duplicate using dye swapping. Five non-malignant bone samples were analysed individually. Labelled reference RNA and labelled tumour RNA were combined before hybridisation to Agilent Whole Human Genome Oligo Microarrays (Agilent Technologies, Santa Clara, CA, USA). Arrays were scanned on an Agilent DNA Microarray Scanner G2505B (Agilent Technologies). The microarray data discussed in this work have been deposited in NCBI's Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO series accession number GSE19276. Data extraction was performed using ImaGene version 6.1 (BioDiscovery Inc, El Segundo, CA, USA). Statistical analysis of the data was performed with GeneSpring GX software versions 7.2 and 10.0.1 (Agilent Technologies). Details of the analysis are described in Supplementary Material.
PCR analysis
For validation, 11 genes were selected at random from the 20 most highly differentially expressed molecules between non-malignant bone and OS. For PCR, 2 μg of non-malignant bone or OS tumour biopsy RNA was reverse transcribed with BioScript (Bioline, Sydney, Australia); PCR was performed using Taq DNA polymerase with ThermoPol II buffer (New England Biolabs, Ipswich, MA, USA) at an annealing temperature of 53–55°C for 30 cycles on a ThermoHybaid PxE0.2 (Thermo Scientific, Waltham, MA, USA). Primers were as follows: ID1 (forward 5′-CGGATCTGAGGGAGAACAAG-3′ and reverse 5′-CTGAGAAGCACCAAACGTGA-3′), PRDX4 (forward 5′-GAGGACTTGGGCCAATAAGG-3′ and reverse 5′-TTCACTACCAGGTTTCCAGC-3′), TPM2 (forward 5′-CGAGAGTAAATGTGGGGACC-3′ and reverse 5′-TAAAGGATGAAGCCAGTGCC-3′), MT1E (forward 5′-TGCTTGTTCGTCTCACTGG-3′ and reverse 5′-AAAGAAATGCAGCAAATGGC-3′), FKBP9 (forward 5′-TACCTGAAAACTGTGAGCGG-3′ and reverse 5′-GTTCATCTGGTTTGGCTTCC-3′), S100A13 (forward 5′-ACCTTATGACCTGTCAGCCC-3′ and reverse 5′-CCGAGTCCTGATTCACATCC-3′), S100A8 (forward 5′-TGGGCATCATGTTGACCGAGCTG-3′ and reverse 5′-GCCACGCCCATCTTTATCACCAGA-3′), CTSG (forward 5′-CGCATCTTCGGTTCCTACG-3′ and reverse 5′-GCTTCTCATTGTTGTCCTTATCC-3′), VWA5B2 (forward 5′-TACTCGGGAGCTACTCTTCC-3′ and reverse 5′-CATATGGCTGTGTCAGAGGG-3′), AZU1 (forward 5′-AGCATCAGGTCGTTCAGGTT-3′ and reverse 5′-CAGAATCAAGGCAGGCACTTC-3′), PFC (forward 5′-GCTCTGTCACCTGCTCCAA-3′ and reverse 5′-GCGGCTTCGTGTCTCCTTA-3′).
Results
Gene expression profiling of OS vs non-malignant bone
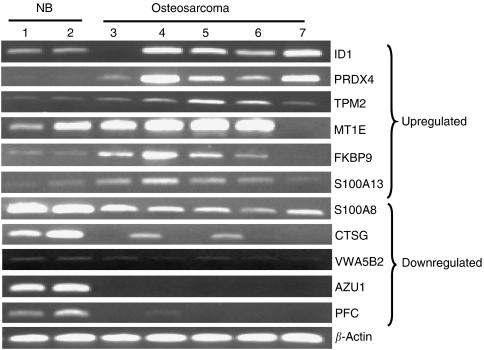
We compared gene expression in 23 OS biopsies and 5 non-malignant bone samples. Our analysis yielded a suite of 305 differentially expressed genes (two-fold or greater, P<0.05) between OS and non-malignant bone, of which 206 were annotated (Supplementary Table 1). Table 2 lists the 10 most highly up and 10 most highly downregulated genes, 11 of which were selected at random and their differential expression confirmed by PCR in two non-malignant bone samples and in five randomly selected OS tumour biopsies (Figure 1). Of the 36 upregulated genes, 47% were associated with cellular growth and proliferation (e.g. ID1, ANXA2, BTG3, MT2A, ITGB1, NDUFAF2). The most well-represented family of genes within this group was the metallothionein family, linked to intrinsic and acquired drug resistance (Cherian et al, 2003). Seven members of this family, MT1E, MT1H, MT1X, MT2A, MT1B, MT1G, and MT1L, were upregulated in our OS samples, and three were among the 10 most highly upregulated genes (Table 2). The inhibitor of DNA binding 1, ID1 (+4.07, P=0.003), peroxiredoxin 4, PRDX4 (+3.63, P=0.007), S100 calcium-binding protein A13, S100A13 (+2.66, P=0.009), annexin 2, ANXA2 (+2.62, P=0.003), and destrin, DSTN (+2.50, P=0.001), earlier reported as positive regulators of angiogenesis, tumour progression and invasion (Fong et al, 2003; Landriscina et al, 2006; Estornes et al, 2007; Lee et al, 2008; Mussunoor and Murray, 2008; Iwatsuki et al, 2009) were all induced in OS samples (Table 2).
Table 2. Osteosarcoma vs non-malignant bone.
| Probe ID | Symbol | Description and accession number | Fold change | P-value |
|---|---|---|---|---|
| A_23_P252306 | ID1 | Inhibitor of DNA binding 1, dominant-negative helix-loop-helix protein [NM_002165] | 4.07 | 0.00276 |
| A_23_P114232 | PRDX4 | Peroxiredoxin 4 [NM_006406] | 3.63 | 0.00664 |
| A_23_P216501 | TPM2 | Tropomyosin 2 (β) [NM_213674] | 3.30 | 0.00237 |
| A_23_P206724 | MT1E | Metallothionein 1E [NM_175617] | 3.11 | 0.00661 |
| A_23_P334709 | FKBP9 | FK506-binding protein 9, 63 kDa [NM_007270] | 3.08 | 0.0111 |
| A_23_P163782 | MT1H | Metallothionein 1H [NM_005951] | 3.02 | 0.0169 |
| A_24_P125096 | MT1X | Metallothionein 1X [NM_005952] | 2.72 | 0.0316 |
| A_23_P372874 | S100A13 | S100 calcium-binding protein A13 [NM_005979] | 2.66 | 0.00955 |
| A_32_P94798 | ANXA2 | Annexin A2 [NM_001002857] | 2.62 | 0.00359 |
| A_23_P408095 | DSTN | Destrin (actin depolymerising factor) [NM_001011546] | 2.50 | 0.00140 |
| A_23_P434809 | S100A8 | S100 calcium-binding protein A8 (calgranulin A) [NM_002964] | −100.00 | 0.00549 |
| A_23_P37856 | HBA1 | Homo sapiens haemoglobin, α 1 (HBA1), mRNA [NM_000558] | −50.00 | 4.26E−10 |
| A_23_P140384 | CTSG | Cathepsin G [NM_001911] | −16.67 | 0.0222 |
| A_23_P80867 | VWA5B2 | von Willebrand factor A domain containing 5B2 [AL834499] | −11.11 | 0.0222 |
| A_23_P153741 | AZU1 | Azurocidin 1 (cationic antimicrobial protein 37) [NM_001700] | −6.67 | 0.0485 |
| A_23_P22444 | CFP | Properdin P factor, complement [NM_002621] | −6.25 | 0.0496 |
| A_23_P208866 | GMFG | Glia maturation factor, γ [NM_004877] | −5.88 | 0.00748 |
| A_24_P207195 | IRX3 | Iroquois homeobox protein 3 [NM_024336] | −5.88 | 0.00263 |
| A_23_P403886 | GLYAT | Glycine-N-acyltransferase [NM_005838] | −5.56 | 0.00391 |
| A_23_P156708 | TNXB | Tenascin XB [NM_019105] | −5.56 | 0.00351 |
Top 10 upregulated and 10 downregulated genes between osteosarcoma biopsies and non-malignant bone samples.
Figure 1.
Validation of highly differentially expressed genes in osteosarcoma vs non-malignant bone. Eleven genes were selected at random from Table 1 and validated by PCR in two non-malignant bone samples (lanes 1 and 2), and five randomly selected osteosarcoma patients (lanes 3–7). The results are shown in groups of genes upregulated and downregulated in osteosarcoma compared with non-malignant bone.
Analysis of the 170 downregulated genes found a large number associated with the inflammatory (26%) and cell-mediated (31%) immune response, as well as with antigen presentation (24%). Ingenuity pathway analysis (IPA) (Ingenuity Systems, Mountain View, California, USA) identified the antigen-presentation pathway as downregulated in OS with HLA-C, HLA-DOA, HLA-DPB1, HLA-DPA1, and HLA-E all expressed 2.17- to 3.45-fold lower in the lesions than in non-malignant bone (P=0.002–0.04). However, the most highly differentially expressed gene was the S100 calcium-binding protein A8 (S100A8), a marker of a number of inflammatory conditions (Zreiqat et al, 2007), which was downregulated 100-fold in OS (P=0.005) (Table 2). Also downregulated were cathepsin G (CTSG; −16.67, P=0.02) and azurocidin 1 (AZU1; −6.67, P=0.05), which regulate monocyte/macrophage function and chemotaxis in inflammatory conditions (Pereira, 1995; Miyata et al, 2007).
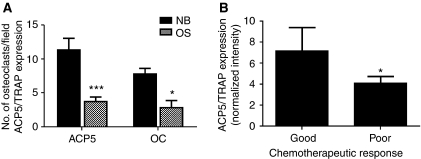
The OS biopsies also revealed a transcriptomic signature characteristic of reduced osteoclastogenesis. ID1 is an inhibitor of osteoclast differentiation (Lee et al, 2006) and was induced four-fold in OS biopsies. Similarly, there was a 100-fold down-regulation of S100A8, which is highly expressed in osteoclasts (Zreiqat et al, 2007), and significantly lower expression of another 13 genes associated with negative regulation of osteoclast differentiation/function, or indicating diminished osteoclast presence or activity. These genes included von Willebrand factor A domain, 5B2 (VWA5B2; −11.11, P=0.02), FGR, a member of the Src family of protein tyrosine kinases (−5.56, P=0.02), TYRO protein kinase-binding protein (TYROBP; −5.26, P=0.04), the Rac small GTPase RAC2 (−4.76, P=0.01), RelA/p65 (−4.55, P=0.005), MYC (−3.70, P=2.77 × 10−16), signal regulatory protein α (SIRPα/SIRPA; −2.94, P=0.006), tartrate-resistant acid phosphatase (ACP5/TRAP, −2.86, P=0.005), BCL2 (−2.5-fold, P=0.005), high-mobility group box 1 (HMGB1; −2.33, P=0.002), V-ATPase (ATP6V0D1; −2.27, P=0.04), leukotriene B4 receptor (LTB4R; −2.22, P=0.03), and gelsolin (GSN, −2.08, P=0.04). To see whether the reduction in ACP5/TRAP gene expression correlated with a decrease in the number of osteoclasts in OS, we performed immunohistochemistry on FFPE sections of OS biopsies and of non-malignant bone with a monoclonal antibody to ACP5/TRAP. We found a 2.5-fold decrease in the number of osteoclasts in OS biopsies compared with non-malignant bone, which correlated with the observed 2.3-fold decrease in ACP5/TRAP gene expression (Figure 2A). Furthermore, the decrease in ACP5/TRAP expression was significantly more marked in the biopsies of patients, which showed a poor response to chemotherapy treatment than in those who exhibited a good response (Figure 2B), suggesting that a reduction in osteoclastogenesis is not only associated with OS in general, but also with chemoresistance.
Figure 2.
Impaired osteoclastogenesis in osteosarcoma correlates with chemoresistance. (A) Osteoclast number is expressed as the average number of osteoclasts per × 20 field in immunohistochemically stained FFPE sections from osteosarcoma biopsies (OS) and non-malignant bone (NB). The ACP5 expression as measured by microarray analysis and expressed as normalised intensity. ***P<0.0001, *P=0.0491. Bars: mean+s.e.m. (B) ACP5 expression in good and poor responders, as measured by microarray analysis and expressed as normalised intensity. *P=0.0428. Bars: mean+s.e.m.
Gene expression profiling of good responders vs poor responders
Osteosarcomas are inherently drug-resistant tumours (Chou and Gorlick, 2006), and, therefore, the most commonly used predictor of disease outcome is a patient's initial response to chemotherapy. Unfortunately, this response cannot be assessed at the time of presentation. To specifically search for genes that could be predictive of chemotherapeutic response and drug resistance at the time of diagnosis, patients were divided into good (n=5) and poor (n=17) responders using the criteria already described. A set of 123 genes was found to be significantly differentially expressed (P<0.05) between the two groups, of which 61 were annotated (Table 3). Most of these genes (94%) were upregulated in the good responders and were associated with cellular development, growth, and proliferation, suggesting that good responders may have tumours that are more proliferative and may, therefore, be more sensitive to the effects of chemotherapy. Among the most highly upregulated genes in good responders was thymosin β 10 (TSMB10, +5.34-fold, P=0.017), which had been identified within the suite of differentially expressed genes between good and poor responders in an earlier study of OS tumour biopsies (Ochi et al, 2004).
Table 3. Good response vs poor response.
| Genbank | Synonym | Common name | Fold change | P-value |
|---|---|---|---|---|
| NM_021103 | TMSB10 | Thymosin, β 10 | 5.34 | 0.0171 |
| NM_000582 | SPP1 | Secreted phosphoprotein 1 (osteopontin, bone sialoprotein I, early T-lymphocyte activation 1) | 4.82 | 0.0171 |
| NM_006332 | IFI30 | Interferon, γ-inducible protein 30 | 4.36 | 0.0224 |
| NM_002952 | RPS2 | Ribosomal protein S2 | 3.70 | 0.0487 |
| NM_005514 | HLA-B | Major histocompatibility complex, class I, B | 3.56 | 0.0487 |
| NM_000455 | STK11 | Serine/threonine kinase 11 (Peutz–Jeghers syndrome) | 3.49 | 0.0249 |
| NM_147780 | CTSB | Cathepsin B | 3.48 | 0.0355 |
| NM_153461 | IL17RC | Interleukin 17 receptor C | 3.39 | 0.0415 |
| NM_002032 | FTH1 | Ferritin, heavy polypeptide 1 | 3.32 | 0.0355 |
| NM_005507 | CFL1 | Cofilin 1 (non-muscle) | 3.29 | 0.0251 |
| NM_002116 | HLA-A | Major histocompatibility complex, class I, A | 3.17 | 0.0379 |
| NM_003332 | TYROBP | TYRO protein tyrosine kinase-binding protein | 3.16 | 0.0487 |
| NM_001022 | RPS19 | Ribosomal protein S19 | 3.16 | 0.023 |
| NM_053275 | RPLP0 | Ribosomal protein, large, P0 | 3.14 | 0.0439 |
| NM_000992 | RPL29 | Ribosomal protein L29 a.k.a. heparin/eparan sulphate interacting protein (HIP) | 3.11 | 0.0497 |
| NM_002117 | HLA-C | Major histocompatibility complex, class I, C | 2.99 | 0.0401 |
| NR_002205 | FTHL12 | Ferritin, heavy polypeptide-like 12 | 2.95 | 0.0392 |
| NM_001404 | EEF1G | Eukaryotic translation elongation factor 1 γ | 2.93 | 0.0443 |
| NM_001014 | RPS10 | Ribosomal protein S10 | 2.83 | 0.0171 |
| NM_001003 | RPLP1 | Ribosomal protein, large, P1 | 2.81 | 0.0397 |
| NM_003295 | TPT1 | Tumour protein, translationally controlled 1 | 2.66 | 0.0365 |
| BC031631 | CFLP1 | Cofilin pseudogene 1 | 2.65 | 0.0232 |
| NM_006435 | IFITM2 | Interferon induced transmembrane protein 2 (1-8D) | 2.61 | 0.0489 |
| NM_002948 | RPL15 | Ribosomal protein L15 | 2.60 | 0.0497 |
| NM_001020 | RPS16 | Ribosomal protein S16 | 2.58 | 0.0355 |
| NM_000982 | RPL21 | Ribosomal protein L21 | 2.56 | 0.0489 |
| NM_178230 | COAS2 | Cyclophilin-LC | 2.51 | 0.0444 |
| NM_000979 | RPL18 | Ribosomal protein L18 | 2.50 | 0.0455 |
| NM_001022 | RPS19 | Ribosomal protein S19 | 2.47 | 0.0392 |
| NM_003295 | TPT1 | Tumour protein, translationally controlled 1 | 2.47 | 0.0439 |
| BC034271 | FANCC | Fanconi anaemia, complementation group C | 2.45 | 0.0214 |
| NM_001997 | FAU | Finkel–Biskis–Reilly murine sarcoma virus (FBR-MuSV) ubiquitously expressed (fox derived); RPS30 | 2.45 | 0.043 |
| NM_001006 | RPS3A | Ribosomal protein S3A | 2.45 | 0.0497 |
| AY358369 | SIGLEC5 | Sialic acid-binding Ig-like lectin 5 (CD170) | 2.45 | 0.0489 |
| NM_181468 | ITGB4BP | Integrin β 4-binding protein | 2.40 | 0.0444 |
| NM_000990 | RPL27A | Ribosomal protein L27a | 2.39 | 0.0392 |
| NM_002489 | NDUFA4 | NADH dehydrogenase (ubiquinone) 1 α subcomplex, 4, 9 kDa | 2.32 | 0.0439 |
| AK098605 | FMN2 | Formin 2 | 2.32 | 0.0443 |
| NM_001019 | RPS15A | Ribosomal protein S15a | 2.32 | 0.0401 |
| NM_002107 | H3F3A | H3 histone, family 3A | 2.30 | 0.0303 |
| NM_005620 | S100A11 | S100 calcium-binding protein A11 (calgizzarin) | 2.30 | 0.0487 |
| NM_006013 | RPL10 | Ribosomal protein L10 | 2.28 | 0.0489 |
| NM_005009 | NME4 | Non-metastatic cells 4, protein expressed in | 2.27 | 0.0365 |
| NM_006886 | ATP5E | ATP synthase, H+ transporting, mitochondrial F1 complex, å subunit | 2.23 | 0.0171 |
| NM_001008741 | LOC388817 | Peptidylprolyl isomerase A-like | 2.23 | 0.0444 |
| NM_032828 | ZNF587 | Zinc-finger protein 587 | 2.22 | 0.0487 |
| NM_015933 | HSPC016 | Hypothetical protein HSPC016 | 2.22 | 0.0258 |
| NM_024040 | CUEDC2 | CUE domain containing 2 | 2.19 | 0.0214 |
| NM_006808 | SEC61B | Sec61 β subunit | 2.18 | 0.0465 |
| NM_002406 | MGAT1 | Mannosyl (α-1,3-)-glycoprotein β-1,2-N-acetylglucosaminyltransferase | 2.14 | 0.0415 |
| NM_002797 | PSMB5 | Proteasome (prosome, macropain) subunit, β type, 5 | 2.13 | 0.0171 |
| NM_001021 | RPS17 | Ribosomal protein S17 | 2.10 | 0.0489 |
| NM_000182 | HADHA | Hydroxyacyl-coenzyme A dehydrogenase/3-ketoacyl-coenzyme A thiolase/enoyl-coenzyme A hydratase (trifunctional protein), α subunit | 2.07 | 0.0392 |
| NM_012067 | AKR7A3 | Aldo-keto reductase family 7, member A3 (aflatoxin aldehyde reductase) | 2.06 | 0.0224 |
| XM_376787 | RPS26P10 | Ribosomal protein S26 pseudogene 10 | 2.06 | 0.043 |
| NM_005340 | HINT1 | Histidine triad nucleotide-binding protein 1 | 2.04 | 0.0444 |
| NM_145893 | A2BP1 | Ataxin 2-binding protein 1 | −2.44 | 0.0357 |
| NM_015503 | SH2B1 | SH2-B adaptor protein | −2.33 | 0.0487 |
| NM_178033 | CYP4X1 | Cytochrome P450, family 4, subfamily X, polypeptide 1 | −2.08 | 0.0487 |
| NM_003893 | LDB1 | LIM domain-binding 1 | −2.04 | 0.0355 |
| CR749256 | XRCC2 | X-ray repair complementing defective repair in Chinese hamster cells 2 | −2.00 | 0.0357 |
Genes differentially expressed between biopsies of good responders and poor responders. The list shows 61 annotated genes from the original list of 123 genes.
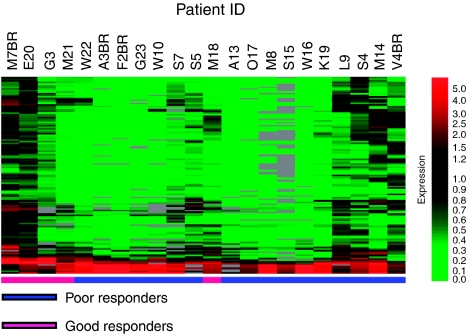
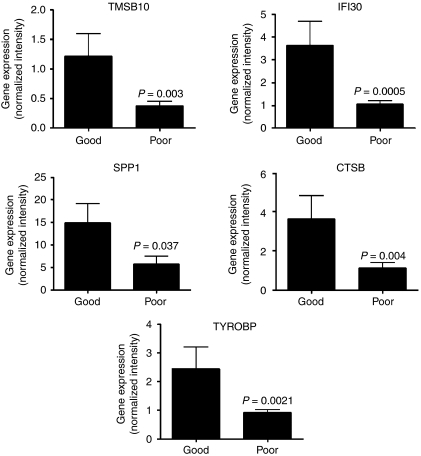
To test the suitability of this set of 123 genes to separate between good and poor responders, we performed unsupervised hierarchical clustering of the data. With the exception of one patient (M18), the gene set was able to clearly separate 21 out of 22 (>95%) OS patients on the basis of their response to chemotherapy (Figure 3). We examined the possibility that some of these genes could serve as individual predictors of chemotherapeutic response. We selected individual genes from the list based on their ⩾three-fold expression (Table 3), and looked at their levels of expression in individual patients. Of the selected genes, only TMSB10, SPP1, CTSB, TYROBP/DAP12, and IFI30 showed significant (P<0.05) differential expression between patients in the two groups, with IFI30 (P=0.0005) showing the most significant difference (Figure 4).
Figure 3.
Unsupervised clustering of 123 differentially expressed genes between good responders and poor responders. Genes were selected based on normalised data values that were greater or less in 5 good responders than those in 17 poor responders by a factor of two-fold, with statistically significant differences when grouped by ‘response’ according to a parametric test Welch t-test (P-value cutoff 0.05). Multiple testing correction was applied (Benjamini and Hochberg false discovery rate). Level of expression – lowest (light green), highest (bright red).
Figure 4.
Expression of highly differentially expressed genes between good and poor responders. Expression, measured as normalised spot intensity, in each of the 5 good responders and 17 poor responders of 6 highly differentially expressed genes between the two groups. P-value (ANOVA, Welch T-test, Benjamini and Hochberg false discovery rate). Bars: s.e.m.
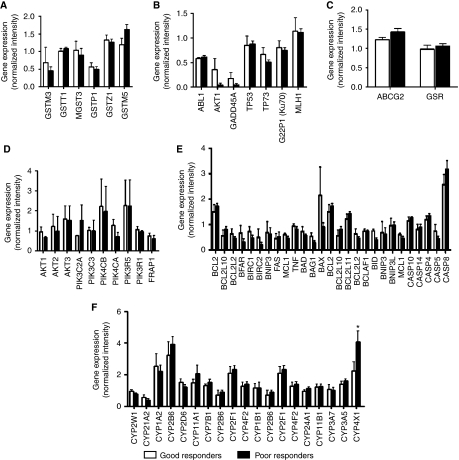
One of the major obstacles to effective treatment of OS patients is intrinsic or acquired resistance to the cytotoxic effects of anticancer agents. The mechanism dictating this resistance in OS is still unknown, but may involve a number of gene families, which mediate detoxification, increased efflux from the cell, and increased DNA repair (Chou and Gorlick, 2006). Therefore, understanding the mechanism of drug resistance and identifying genes that are involved may lead to new therapies that could improve survival. The cytochrome P450 family of enzymes functions in the detoxification of anticancer drugs (Simpson, 1997). As our signature found CYP4X1 differentially expressed between good and poor responders, we looked at the mRNA expression levels in each patient of CYP4X1 and other cytochrome P450 family members (Figure 5F). Of the 50 or more P450 genes present on the array, only 19 had detectable expression levels in our tumour biopsies, and of these, only CYP4X1 had significant differential expression associated with chemotherapeutic response. However, given that CYP4X1 is an orphan P450 protein with no assigned biological function (Stark et al, 2008), and that it was downregulated in good responders, its function in OS drug response remains unclear.
Figure 5.
Expression of genes classically associated with chemotherapeutic response. (A) Glutathione S-transferase family, (B) DNA damage response, (C) drug transport, (D) survival, (E) apoptosis, and (F) CYP family. *P<0.05. Bars: mean+s.e.m.
Other enzyme families responsible for resistance to many chemotherapeutic agents include the glutathione-S-transferases (GSTs) (Ekhart et al, 2009) and the ATP-binding cassette (ABC) transporters (Sharom, 2008). We, therefore, compared the expression of GST and ABC family members between good and poor responders. Only five GSTs were expressed in our samples, but none significantly (Figure 5A). Similarly, only one ABC transporter, ABCG2, was found expressed in our samples, but its levels were not significantly different between the two groups (Figure 5C). Moreover, we found no significant difference in the expression of other genes involved in DNA damage response, drug metabolism, apoptosis, or survival (Figures 5B, D and E). Taken together, our data indicate that genes classically associated with multi-drug resistance do not correlate with chemotherapeutic response in OS.
Discussion
Despite intensive multi-agent chemotherapy, OS remains an aggressive, highly metastatic, and relatively drug-resistant tumour with poor long-term survival rates (Chou and Gorlick, 2006). The mechanisms behind metastasis and chemoresistance in OS are not well understood, but are likely to be due to the innate biology of metastatic and chemoresistant lesions (Gorlick and Meyers, 2003). Therefore, understanding the basic tumour biology is central to understanding OS pathogenesis and chemoresistance. In this study, we used chemo-naive OS biopsies and, for the first time, compared their transcriptomes to those of non-malignant bone. We identified a unique gene signature showing increased expression of genes associated with tumour progression and drug resistance, and decreased expression of genes associated with antigen presentation and osteoclastogenesis in all OS lesions. In addition, tumours that were chemoresistant were characterised by more pronounced inhibition of osteoclastogenesis markers and antigen-presenting activity than tumours that were chemosensitive. We noted no significant difference in the expression of genes classically associated with drug resistance between chemosensitive and chemoresistant tumours.
This study identified ID1 as a potentially important molecule in the regulation of many of the characteristics of OS. Increased expression of ID1 in the OS biopsies has been shown to be involved in the proliferation, survival, angiogenesis, metastasis (Ling et al, 2006), and formation of a permissive metastatic niche in other cancer types (Lyden et al, 1999), and may be similarly involved in OS. Recently, ID1 has been identified as a novel inhibitor of PTEN and p53, leading to AKT-dependent activation of the canonical Wnt-signalling pathway (Lee et al, 2009). The Wnt-signalling pathway has earlier been shown to be important to the regulation of OS progression (Hoang et al, 2004; Geryk-Hall and Hughes, 2009; Kansara et al, 2009). Thus, there is evidence that the overexpression of ID1 in OS may be causally involved in the growth, survival, and metastatic behaviour of OS. If true, then targeting of the Akt and Wnt pathways could be of value in OS (Geryk-Hall and Hughes, 2009).
Mintz et al (2005) earlier reported that genes involved in osteoclast differentiation and function in OS were mostly associated with a poor chemotherapeutic response. We now report that osteoclast numbers are decreased in all OS lesions. Thus, the loss of osteoclasts in OS may be involved in OS metastasis, although the mechanism by which OS induce osteoclast loss is unknown. An earlier transcriptomic study in seven OS biopsies (Patino-Garcia et al, 2009) showed up-regulation of EBF2, a known transcription factor for osteoprotegerin (OPG), a major negative regulator of osteoclastogenesis (Khosla, 2001). In contrast, our study identified the up-regulation of ID1 in OS. As ID1 is a negative regulator of osteoclast differentiation (Lee et al, 2006), it is possible that the overexpression of ID1 in OS lesions may provide an explanation for OS-induced osteoclast loss. Our microarray analysis of primary OS lesions also indicates that S100A8 was profoundly reduced in OS lesions (P=0.005; fold decrease=100). S100A8 is highly expressed in osteoclasts in which it functions as a chemotactic-signalling molecule involved in the coupling of osteoclast and osteoblast activity (Zreiqat et al, 2007). In addition, decreased expression of 15 other genes involved in osteoclast development and function (Pereira et al, 1990; Garcia et al, 1996; Lowell et al, 1996; Chertov et al, 1997; Chellaiah et al, 2000; Duong et al, 2000; Battaglino et al, 2002; Lundberg et al, 2007; Shahbazi et al, 2007; Kawano et al, 2008; Zhou et al, 2008; Soehnlein and Lindbom, 2009) was observed in the OS biopsies. Among these was CTSG, which is necessary for the recruitment of osteoclast precursors (Wilson et al, 2009b) and for the activation of MMP9, which in turn activates TGFβ to enhance osteoclast activity (Wilson et al, 2009a); TYROBP/DAP12, which is essential for RANK signalling and osteoclast multinucleation and differentiation (Humphrey et al, 2004; Mocsai et al, 2004), and the NFkB subunit, RelA/p65, which promotes osteoclastogenesis by inhibiting JNK-mediated osteoclast apoptosis (Vaira et al, 2008; Soysa and Alles, 2009). We also observed decreased expression of osteoclast cellular components such as ACP5/TRAP, a classic marker of mature and active osteoclasts (Hayman, 2008) and ATP6V0D1, which is found in the osteoclast membrane and critical for its resorptive activity (Xu et al, 2007). Furthermore, the decreased expression of osteoclast cellular components correlated with a decrease in the number of osteoclast cell counts in immunohistochemically stained sections of OS compared with non-malignant bone. Thus, we provide evidence that OS lesions are associated with a reduction in osteoclasts.
The down-regulation of osteoclast differentiation in OS was accompanied by down-regulation of five MHC Class I and II genes belonging to the antigen-presentation pathway and of HMGB1. The correlation between MHC Class I and II deficiencies in human tumours and metastatic potential/reduced survival rates in patients is well documented (Meissner et al, 2005; Chamuleau et al, 2006; Ramnath et al, 2006; Seliger, 2008), whereas the interaction of HMGB1 with Toll-like receptor 4 on dendritic cells has been shown to be essential for tumour antigen processing and presentation (Apetoh et al, 2007). Interestingly, we observed a significant increase in the expression of five genes with overlapping functions in antigen presentation and osteoclastogenesis in the good responder group – IFI30 (P=0.0005), TYROBP/DAP12 (P=0.002), TMSB10 (P=0.003), CTSB (P=0.004), and SPP1 (P=0.037). The IFI30 expression, in melanoma (Goldstein et al, 2008) and squamous cell carcinoma (Wenzel et al, 2008), has been shown to enhance antigen presentation and to activate CTSB (Goldstein et al, 2008), which is required for osteoclast fusion during differentiation (McMichael et al, 2009). In addition, TYROBP/DAP12 is associated with active innate immune responses (Lanier, 2009) and, together with SPP1, also has a crucial function in osteoclast differentiation (Humphrey et al, 2004; Mocsai et al, 2004; Dalla-Torre et al, 2006; Inui et al, 2009). Furthermore, the expression of SPP1 in OS lesions has been correlated with improved overall survival (Dalla-Torre et al, 2006). Although there is some evidence connecting TMSB10 expression to carcinogenesis (Lee et al, 2001; Sardi et al, 2002; Alldinger et al, 2005), this is the first report linking TMSB10 expression to good chemotherapeutic response in OS. In addition, good responders had significantly higher levels of ACP5/TRAP gene expression than poor responders. Thus, these results are the first to show a possible association between antigen presentation and osteoclastogenesis in the biology of OS tumours and in their chemotherapeutic response.
This study revealed two interesting findings relating to OS chemosensitivity. First, we found that OS displayed a gene signature that was consistent with a chemoresistant phenotype. Second, genes associated with classical drug resistance were not overrepresented in chemoresistant OS lesions. These conclusions are supported by the increased expression of seven metallothionein family members in the OS biopsies. The function of metallothioneins in drug resistance has been well documented (Cherian et al, 2003; Theocharis et al, 2004; Surowiak et al, 2007), and an earlier transcriptomic study of OS reported up-regulation of MTIG and MT1L in poorly responsive tumours (Mintz et al, 2005). In this study, we found no correlation between their level of expression and response, a finding that is supported by others (Uozaki et al, 1997; Shnyder et al, 1998). Moreover, we found no correlation between good and poor responses and the expression of molecules classically associated with chemoresistance, such as GSTs and ABC transporters, or DNA damage response, apoptosis, drug metabolism, and survival genes. These results suggest that drug resistance may be a global characteristic of all osteosarcomas and that drug resistance, in chemoresistant lesions, is likely to be mediated by novel pathways.
Acknowledgments
This work was supported by funds from the Cancer Collaborative Group, Princess Alexandra Hospital, and grants from the Wesley Research Institute (#2006/10), The Australian National Health and Medical Research Council (#569689), and the Queensland Cancer Fund (#401542). We thank Ms Lani Hardy for technical assistance, Dr Emma Woodhall, Princess Alexandra Hospital Tumour Bank, and Mr Michael Doyle, Sullivan & Nicolaides Pathology, for the provision of patient tissue, Dr Steve Bouralexis, St Vincent's Institute, and Associate Professor Andreas Evdokiou, University of Adelaide, for osteosarcoma cell lines, and Dr Glen Boyle, Queensland Institute for Medical Research, for assistance with statistical analysis. We gratefully acknowledge the assistance of those who donated tissue for this study.
Footnotes
Supplementary Information accompanies the paper on British Journal of Cancer website (http://www.nature.com/bjc)
Supplementary Material
References
- Alldinger I, Dittert D, Peiper M, Fusco A, Chiappetta G, Staub E, Lohr M, Jesnowski R, Baretton G, Ockert D, Saeger HD, Grutzmann R, Pilarsky C (2005) Gene expression analysis of pancreatic cell lines reveals genes overexpressed in pancreatic cancer. Pancreatology 5: 370–379 [DOI] [PubMed] [Google Scholar]
- Apetoh L, Ghiringhelli F, Tesniere A, Obeid M, Ortiz C, Criollo A, Mignot G, Maiuri MC, Ullrich E, Saulnier P, Yang H, Amigorena S, Ryffel B, Barrat FJ, Saftig P, Levi F, Lidereau R, Nogues C, Mira JP, Chompret A, Joulin V, Clavel-Chapelon F, Bourhis J, Andre F, Delaloge S, Tursz T, Kroemer G, Zitvogel L (2007) Toll-like receptor 4-dependent contribution of the immune system to anticancer chemotherapy and radiotherapy. Nat Med 13: 1050–1059 [DOI] [PubMed] [Google Scholar]
- Bacci G, Mercuri M, Briccoli A, Ferrari S, Bertoni F, Donati D, Monti C, Zanoni A, Forni C, Manfrini M (1997) Osteogenic sarcoma of the extremity with detectable lung metastases at presentation. Results of treatment of 23 patients with chemotherapy followed by simultaneous resection of primary and metastatic lesions. Cancer 79: 245–254 [PubMed] [Google Scholar]
- Baird K, Davis S, Antonescu CR, Harper UL, Walker RL, Chen Y, Glatfelter AA, Duray PH, Meltzer PS (2005) Gene expression profiling of human sarcomas: insights into sarcoma biology. Cancer Res 65: 9226–9235 [DOI] [PubMed] [Google Scholar]
- Battaglino R, Kim D, Fu J, Vaage B, Fu XY, Stashenko P (2002) c-myc is required for osteoclast differentiation. J Bone Miner Res 17: 763–773 [DOI] [PubMed] [Google Scholar]
- Chamuleau ME, Ossenkoppele GJ, van de Loosdrecht AA (2006) MHC class II molecules in tumour immunology: prognostic marker and target for immune modulation. Immunobiology 211: 619–625 [DOI] [PubMed] [Google Scholar]
- Chellaiah M, Kizer N, Silva M, Alvarez U, Kwiatkowski D, Hruska KA (2000) Gelsolin deficiency blocks podosome assembly and produces increased bone mass and strength. J Cell Biol 148: 665–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherian MG, Jayasurya A, Bay BH (2003) Metallothioneins in human tumors and potential roles in carcinogenesis. Mutat Res 533: 201–209 [DOI] [PubMed] [Google Scholar]
- Chertov O, Ueda H, Xu LL, Tani K, Murphy WJ, Wang JM, Howard OM, Sayers TJ, Oppenheim JJ (1997) Identification of human neutrophil-derived cathepsin G and azurocidin/CAP37 as chemoattractants for mononuclear cells and neutrophils. J Exp Med 186: 739–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou AJ, Gorlick R (2006) Chemotherapy resistance in osteosarcoma: current challenges and future directions. Expert Rev Anticancer Ther 6: 1075–1085 [DOI] [PubMed] [Google Scholar]
- Dalla-Torre CA, Yoshimoto M, Lee CH, Joshua AM, de Toledo SR, Petrilli AS, Andrade JA, Chilton-MacNeill S, Zielenska M, Squire JA (2006) Effects of THBS3, SPARC and SPP1 expression on biological behavior and survival in patients with osteosarcoma. BMC Cancer 6: 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duong LT, Lakkakorpi P, Nakamura I, Rodan GA (2000) Integrins and signaling in osteoclast function. Matrix Biol 19: 97–105 [DOI] [PubMed] [Google Scholar]
- Ekhart C, Rodenhuis S, Smits PH, Beijnen JH, Huitema AD (2009) An overview of the relations between polymorphisms in drug metabolising enzymes and drug transporters and survival after cancer drug treatment. Cancer Treat Rev 35: 18–31 [DOI] [PubMed] [Google Scholar]
- Estornes Y, Gay F, Gevrey JC, Navoizat S, Nejjari M, Scoazec JY, Chayvialle JA, Saurin JC, Abello J (2007) Differential involvement of destrin and cofilin-1 in the control of invasive properties of Isreco1 human colon cancer cells. Int J Cancer 121: 2162–2171 [DOI] [PubMed] [Google Scholar]
- Fong S, Itahana Y, Sumida T, Singh J, Coppe JP, Liu Y, Richards PC, Bennington JL, Lee NM, Debs RJ, Desprez PY (2003) Id-1 as a molecular target in therapy for breast cancer cell invasion and metastasis. Proc Natl Acad Sci USA 100: 13543–13548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia C, Boyce BF, Gilles J, Dallas M, Qiao M, Mundy GR, Bonewald LF (1996) Leukotriene B4 stimulates osteoclastic bone resorption both in vitro and in vivo. J Bone Miner Res 11: 1619–1627 [DOI] [PubMed] [Google Scholar]
- Geryk-Hall M, Hughes DP (2009) Critical signaling pathways in bone sarcoma: candidates for therapeutic interventions. Curr Oncol Rep 11: 446–453 [DOI] [PubMed] [Google Scholar]
- Goldstein OG, Hajiaghamohseni LM, Amria S, Sundaram K, Reddy SV, Haque A (2008) Gamma-IFN-inducible-lysosomal thiol reductase modulates acidic proteases and HLA class II antigen processing in melanoma. Cancer Immunol Immunother 57: 1461–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlick R, Meyers PA (2003) Osteosarcoma necrosis following chemotherapy: innate biology versus treatment-specific. J Pediatr Hematol Oncol 25: 840–841 [DOI] [PubMed] [Google Scholar]
- Hayman AR (2008) Tartrate-resistant acid phosphatase (TRAP) and the osteoclast/immune cell dichotomy. Autoimmunity 41: 218–223 [DOI] [PubMed] [Google Scholar]
- Hoang BH, Kubo T, Healey JH, Yang R, Nathan SS, Kolb EA, Mazza B, Meyers PA, Gorlick R (2004) Dickkopf 3 inhibits invasion and motility of Saos-2 osteosarcoma cells by modulating the Wnt-beta-catenin pathway. Cancer Res 64: 2734–2739 [DOI] [PubMed] [Google Scholar]
- Humphrey MB, Ogasawara K, Yao W, Spusta SC, Daws MR, Lane NE, Lanier LL, Nakamura MC (2004) The signaling adapter protein DAP12 regulates multinucleation during osteoclast development. J Bone Miner Res 19: 224–234 [DOI] [PubMed] [Google Scholar]
- Inui M, Kikuchi Y, Aoki N, Endo S, Maeda T, Sugahara-Tobinai A, Fujimura S, Nakamura A, Kumanogoh A, Colonna M, Takai T (2009) Signal adaptor DAP10 associates with MDL-1 and triggers osteoclastogenesis in cooperation with DAP12. Proc Natl Acad Sci USA 106: 4816–4821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwatsuki M, Fukagawa T, Mimori K, Nakanishi H, Ito S, Ishii H, Yokobori T, Sasako M, Baba H, Mori M (2009) Bone marrow and peripheral blood expression of ID1 in human gastric carcinoma patients is a bona fide indicator of lymph node and peritoneal metastasis. Br J Cancer 100: 1937–1942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kansara M, Tsang M, Kodjabachian L, Sims NA, Trivett MK, Ehrich M, Dobrovic A, Slavin J, Choong PF, Simmons PJ, Dawid IB, Thomas DM (2009) Wnt inhibitory factor 1 is epigenetically silenced in human osteosarcoma, and targeted disruption accelerates osteosarcomagenesis in mice. J Clin Invest 119: 837–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano T, Troiano N, Adams DJ, Wu JJ, Sun BH, Insogna K (2008) The anabolic response to parathyroid hormone is augmented in Rac2 knockout mice. Endocrinology 149: 4009–4015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khosla S (2001) Minireview: the OPG/RANKL/RANK system. Endocrinology 142: 5050–5055 [DOI] [PubMed] [Google Scholar]
- Landriscina M, Schinzari G, Di Leonardo G, Quirino M, Cassano A, D’Argento E, Lauriola L, Scerrati M, Prudovsky I, Barone C (2006) S100A13, a new marker of angiogenesis in human astrocytic gliomas. J Neurooncol 80: 251–259 [DOI] [PubMed] [Google Scholar]
- Lanier LL (2009) DAP10- and DAP12-associated receptors in innate immunity. Immunol Rev 227: 150–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Kim K, Kim JH, Jin HM, Choi HK, Lee SH, Kook H, Kim KK, Yokota Y, Lee SY, Choi Y, Kim N (2006) Id helix-loop-helix proteins negatively regulate TRANCE-mediated osteoclast differentiation. Blood 107: 2686–2693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Kang MB, Jang SH, Qian T, Kim HJ, Kim CH, Kim Y, Kong G (2009) Id-1 activates Akt-mediated Wnt signaling and p27(Kip1) phosphorylation through PTEN inhibition. Oncogene 28: 824–831 [DOI] [PubMed] [Google Scholar]
- Lee SH, Zhang W, Choi JJ, Cho YS, Oh SH, Kim JW, Hu L, Xu J, Liu J, Lee JH (2001) Overexpression of the thymosin beta-10 gene in human ovarian cancer cells disrupts F-actin stress fiber and leads to apoptosis. Oncogene 20: 6700–6706 [DOI] [PubMed] [Google Scholar]
- Lee SU, Rhee M, Min YK, Kim SH (2008) Involvement of peroxiredoxin IV in the 16alpha-hydroxyestrone-induced proliferation of human MCF-7 breast cancer cells. Cell Biol Int 32: 401–405 [DOI] [PubMed] [Google Scholar]
- Leonard P, Sharp T, Henderson S, Hewitt D, Pringle J, Sandison A, Goodship A, Whelan J, Boshoff C (2003) Gene expression array profile of human osteosarcoma. Br J Cancer 89: 2284–2288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling MT, Wang X, Zhang X, Wong YC (2006) The multiple roles of Id-1 in cancer progression. Differentiation 74: 481–487 [DOI] [PubMed] [Google Scholar]
- Lowell CA, Niwa M, Soriano P, Varmus HE (1996) Deficiency of the Hck and Src tyrosine kinases results in extreme levels of extramedullary hematopoiesis. Blood 87: 1780–1792 [PubMed] [Google Scholar]
- Lundberg P, Koskinen C, Baldock PA, Lothgren H, Stenberg A, Lerner UH, Oldenborg PA (2007) Osteoclast formation is strongly reduced both in vivo and in vitro in the absence of CD47/SIRPalpha-interaction. Biochem Biophys Res Commun 352: 444–448 [DOI] [PubMed] [Google Scholar]
- Lyden D, Young AZ, Zagzag D, Yan W, Gerald W, O’Reilly R, Bader BL, Hynes RO, Zhuang Y, Manova K, Benezra R (1999) Id1 and Id3 are required for neurogenesis, angiogenesis and vascularization of tumour xenografts. Nature 401: 670–677 [DOI] [PubMed] [Google Scholar]
- Man TK, Chintagumpala M, Visvanathan J, Shen J, Perlaky L, Hicks J, Johnson M, Davino N, Murray J, Helman L, Meyer W, Triche T, Wong KK, Lau CC (2005) Expression profiles of osteosarcoma that can predict response to chemotherapy. Cancer Res 65: 8142–8150 [DOI] [PubMed] [Google Scholar]
- Marina N, Gebhardt M, Teot L, Gorlick R (2004) Biology and therapeutic advances for pediatric osteosarcoma. Oncologist 9: 422–441 [DOI] [PubMed] [Google Scholar]
- McMichael BK, Wysolmerski RB, Lee BS (2009) Regulated proteolysis of nonmuscle myosin IIA stimulates osteoclast fusion. J Biol Chem 284: 12266–12275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner M, Reichert TE, Kunkel M, Gooding W, Whiteside TL, Ferrone S, Seliger B (2005) Defects in the human leukocyte antigen class I antigen processing machinery in head and neck squamous cell carcinoma: association with clinical outcome. Clin Cancer Res 11: 2552–2560 [DOI] [PubMed] [Google Scholar]
- Mintz MB, Sowers R, Brown KM, Hilmer SC, Mazza B, Huvos AG, Meyers PA, Lafleur B, McDonough WS, Henry MM, Ramsey KE, Antonescu CR, Chen W, Healey JH, Daluski A, Berens ME, Macdonald TJ, Gorlick R, Stephan DA (2005) An expression signature classifies chemotherapy-resistant pediatric osteosarcoma. Cancer Res 65: 1748–1754 [DOI] [PubMed] [Google Scholar]
- Miyata J, Tani K, Sato K, Otsuka S, Urata T, Lkhagvaa B, Furukawa C, Sano N, Sone S (2007) Cathepsin G: the significance in rheumatoid arthritis as a monocyte chemoattractant. Rheumatol Int 27: 375–382 [DOI] [PubMed] [Google Scholar]
- Mocsai A, Humphrey MB, Van Ziffle JA, Hu Y, Burghardt A, Spusta SC, Majumdar S, Lanier LL, Lowell CA, Nakamura MC (2004) The immunomodulatory adapter proteins DAP12 and Fc receptor gamma-chain (FcRgamma) regulate development of functional osteoclasts through the Syk tyrosine kinase. Proc Natl Acad Sci USA 101: 6158–6163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mussunoor S, Murray GI (2008) The role of annexins in tumour development and progression. J Pathol 216: 131–140 [DOI] [PubMed] [Google Scholar]
- Ochi K, Daigo Y, Katagiri T, Nagayama S, Tsunoda T, Myoui A, Naka N, Araki N, Kudawara I, Ieguchi M, Toyama Y, Toguchida J, Yoshikawa H, Nakamura Y (2004) Prediction of response to neoadjuvant chemotherapy for osteosarcoma by gene-expression profiles. Int J Oncol 24: 647–655 [PubMed] [Google Scholar]
- Patino-Garcia A, Zalacain M, Folio C, Zandueta C, Sierrasesumaga L, San Julian M, Toledo G, De Las Rivas J, Lecanda F (2009) Profiling of chemonaive osteosarcoma and paired-normal cells identifies EBF2 as a mediator of osteoprotegerin inhibition to tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis. Clin Cancer Res 15: 5082–5091 [DOI] [PubMed] [Google Scholar]
- Pereira HA (1995) CAP37, a neutrophil-derived multifunctional inflammatory mediator. J Leukoc Biol 57: 805–812 [DOI] [PubMed] [Google Scholar]
- Pereira HA, Shafer WM, Pohl J, Martin LE, Spitznagel JK (1990) CAP37, a human neutrophil-derived chemotactic factor with monocyte specific activity. J Clin Invest 85: 1468–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramnath N, Tan D, Li Q, Hylander BL, Bogner P, Ryes L, Ferrone S (2006) Is downregulation of MHC class I antigen expression in human non-small cell lung cancer associated with prolonged survival? Cancer Immunol Immunother 55: 891–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzer-Kuntschik M, Delling G, Beron G, Sigmund R (1983) Morphological grades of regression in osteosarcoma after polychemotherapy – study COSS 80. J Cancer Res Clin Oncol 106: 21–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardi I, Tintori V, Marchi C, Veltroni M, Lippi A, Tucci F, Tamburini A, Bernini G, Faulkner L (2002) Molecular profiling of high-risk neuroblastoma by cDNA array. Int J Mol Med 9: 541–545 [PubMed] [Google Scholar]
- Seliger B (2008) Different regulation of MHC class I antigen processing components in human tumors. J Immunotoxicol 5: 361–367 [DOI] [PubMed] [Google Scholar]
- Shahbazi S, Lenting PJ, Fribourg C, Terraube V, Denis CV, Christophe OD (2007) Characterization of the interaction between von Willebrand factor and osteoprotegerin. J Thromb Haemost 5: 1956–1962 [DOI] [PubMed] [Google Scholar]
- Sharom FJ (2008) ABC multidrug transporters: structure, function and role in chemoresistance. Pharmacogenomics 9: 105–127 [DOI] [PubMed] [Google Scholar]
- Shnyder SD, Hayes AJ, Pringle J, Archer CW (1998) P-glycoprotein and metallothionein expression and resistance to chemotherapy in osteosarcoma. Br J Cancer 78: 757–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson AE (1997) The cytochrome P450 4 (CYP4) family. Gen Pharmacol 28: 351–359 [DOI] [PubMed] [Google Scholar]
- Soehnlein O, Lindbom L (2009) Neutrophil-derived azurocidin alarms the immune system. J Leukoc Biol 85: 344–351 [DOI] [PubMed] [Google Scholar]
- Soysa NS, Alles N (2009) NF-kappaB functions in osteoclasts. Biochem Biophys Res Commun 378: 1–5 [DOI] [PubMed] [Google Scholar]
- Stark K, Dostalek M, Guengerich FP (2008) Expression and purification of orphan cytochrome P450 4X1 and oxidation of anandamide. FEBS J 275: 3706–3717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surowiak P, Materna V, Maciejczyk A, Pudelko M, Markwitz E, Spaczynski M, Dietel M, Zabel M, Lage H (2007) Nuclear metallothionein expression correlates with cisplatin resistance of ovarian cancer cells and poor clinical outcome. Virchows Arch 450: 279–285 [DOI] [PubMed] [Google Scholar]
- Theocharis SE, Margeli AP, Klijanienko JT, Kouraklis GP (2004) Metallothionein expression in human neoplasia. Histopathology 45: 103–118 [DOI] [PubMed] [Google Scholar]
- Uozaki H, Horiuchi H, Ishida T, Iijima T, Imamura T, Machinami R (1997) Overexpression of resistance-related proteins (metallothioneins, glutathione-S-transferase pi, heat shock protein 27, and lung resistance-related protein) in osteosarcoma. Relationship with poor prognosis. Cancer 79: 2336–2344 [DOI] [PubMed] [Google Scholar]
- Vaira S, Alhawagri M, Anwisye I, Kitaura H, Faccio R, Novack DV (2008) RelA/p65 promotes osteoclast differentiation by blocking a RANKL-induced apoptotic JNK pathway in mice. J Clin Invest 118: 2088–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LL (2005) Biology of osteogenic sarcoma. Cancer J 11: 294–305 [DOI] [PubMed] [Google Scholar]
- Wenzel J, Tomiuk S, Zahn S, Kusters D, Vahsen A, Wiechert A, Mikus S, Birth M, Scheler M, von Bubnoff D, Baron JM, Merk HF, Mauch C, Krieg T, Bieber T, Bosio A, Hofmann K, Tuting T, Peters B (2008) Transcriptional profiling identifies an interferon-associated host immune response in invasive squamous cell carcinoma of the skin. Int J Cancer 123: 2605–2615 [DOI] [PubMed] [Google Scholar]
- Wilson TJ, Nannuru KC, Singh RK (2009a) Cathepsin G-mediated activation of pro-matrix metalloproteinase 9 at the tumor-bone interface promotes transforming growth factor-{beta} signaling and bone destruction. Mol Cancer Res 7(8): 1224–1233 [DOI] [PubMed] [Google Scholar]
- Wilson TJ, Nannuru KC, Singh RK (2009b) Cathepsin G recruits osteoclast precursors via proteolytic activation of protease-activated receptor-1. Cancer Res 69: 3188–3195 [DOI] [PubMed] [Google Scholar]
- Xu J, Cheng T, Feng HT, Pavlos NJ, Zheng MH (2007) Structure and function of V-ATPases in osteoclasts: potential therapeutic targets for the treatment of osteolysis. Histol Histopathol 22: 443–454 [DOI] [PubMed] [Google Scholar]
- Zhou Z, Han JY, Xi CX, Xie JX, Feng X, Wang CY, Mei L, Xiong WC (2008) HMGB1 regulates RANKL-induced osteoclastogenesis in a manner dependent on RAGE. J Bone Miner Res 23: 1084–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zreiqat H, Howlett CR, Gronthos S, Hume D, Geczy CL (2007) S100A8/S100A9 and their association with cartilage and bone. J Mol Histol 38: 381–391 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.