Abstract
Central to any beneficial interaction is the capacity of partners to detect and respond to significant changes in the other. Recent studies of microbial mutualists show their close integration with host development, immune responses, and acclimation to a dynamic external environment. While the significance of microbial players is broadly appreciated, we are just beginning to understand the genetic, ecological, and physiological mechanisms that generate variation in symbiont functions, broadly termed “symbiont plasticity” here. Some possible mechanisms include shifts in symbiont community composition, genetic changes via DNA acquisition, gene expression fluctuations, and variation in symbiont densities. In this review, we examine mechanisms for plasticity in the exceptionally stable mutualisms between insects and bacterial endosymbionts. Despite the severe ecological and genomic constraints imposed by their specialized lifestyle, these bacteria retain the capacity to modulate functions depending on the particular requirements of the host. Focusing on the mutualism between Blochmannia and ants, we discuss the roles of gene expression fluctuations and shifts in bacterial densities in generating symbiont plasticity. This symbiont variation is best understood by considering ant colony as the host superorganism. In this eusocial host, the bacteria meet the needs of the colony and not necessarily the individual ants that house them.
Introduction
Symbiotic interactions profoundly affect the life history, physiology, and adaptation of all organisms. Among the most pervasive symbioses are those between a bacterial partner and a eukaryotic host. While the best-studied microbial associates are still disease-causing parasites, recent work illustrates the profound importance of commensal and mutualistic interactions, ranging from astonishingly diverse microbiota of gastrointestinal tracts, to simpler interactions with one or few symbiotic partners. By combining the biochemical functions of two domains of life, the establishment of bacterial-eukaryotic mutualisms often represent key evolutionary innovations (Margulis and Fester, 1991), allowing partners to thrive in niches and utilize resources that otherwise would be inaccessible.
In addition to their significance in evolutionary transitions, beneficial microbes are closely tied to host responses on physiological and ecological timescales. The function of symbiotic populations or communities can vary considerably, depending on host developmental stage (Chun et al., 2008), temperature (Baird et al., 2009), immune responses (Wen et al., 2008), resource availability (Zahran, 1999), host diet (Ley et al., 2008), or drug treatments (Dethlefsen et al., 2008). It is clear that microbes play important roles in mediating physiological changes across host life stages and environmental contexts. To understand how organisms develop, grow, maintain health, and respond appropriately to an ever-changing environment, we must consider the symbiotic players involved.
The role of symbiosis in the host's dynamic life history is a key but complex area of study. By what mechanisms do symbiont functions vary within hosts, including shifts in community composition, gene expression patterns, or symbiont densities? Understanding these mechanisms sets the stage to address the more difficult question of whether observed changes are meaningful to the host. That is, do changes in microbial symbionts reflect coordinated responses that benefit the host, selfish phenotypes in a stressed interaction, or neutral consequences of an environmental perturbation? The answers to these questions will surely depend on the age and integration of the association, the environmental variation encountered, and the genetic and ecological processes at work. By exploring symbiont plasticity in a variety of systems, we can begin to understand the range of possible responses of microbes to hosts and the general principles guiding those responses.
In any set of comparisons, the extremes can be especially informative. Among beneficial microbes, the long-term mutualists of insects engage in exceptionally stable and constrained interactions. As rare examples of intracellular mutualists, these bacteria are closely integrated into host development and nutritional physiology, reflecting long-term coevolution (Buchner, 1965). Like any organism, insects housing endosymbionts do not live in a static environment; hosts grow, age, and cope with environmental variation. In social insects, such as ants, the host colony develops and matures through the activities of different castes. What roles do insect symbionts play across these types of variation? Given their long evolutionary history in a constrained environment, these mutualists might lack certain mechanisms for genetic and functional variation found in more labile symbiotic interactions. On the other hand, functional plasticity may be all the more important in otherwise stable associations.
In this review, we explore the impact of symbiotic lifestyle on genetic variation and the ability of microbes to respond to distinct host life stages or environmental contexts. We cite examples from a range of insect endosymbionts, and focus on a long-term ant mutualist whose host might be considered the colony itself. Despite the severe evolutionary and genomic constraints imposed upon insect mutualists, recent work and our own data illustrate that the bacteria retain the capacity to modulate functions in response to particular requirements of the host.
Lifestyle Constraints in Insect Mutualists
Among multicellular organisms, insects as a group form exceptionally diverse associations with microbes, including viruses, protists, bacterial pathogens, and facultative bacterial endosymbionts that form labile associations. In addition, many insects possess bacterial mutualists that live exclusively within specialized host cells (bacteriocytes) and female reproductive organs. In contrast to intracellular pathogens and facultative secondary symbionts, these long-term primary endosymbionts are considered essential for host survival and reproduction (Buchner, 1965; Ishikawa, 2003). Primary mutualists often occur in insects that feed on nutritionally unbalanced diets, such as plant sap (e.g., aphids and psyllids), vertebrate blood (tsetse flies and lice), or grains (weevils). By supplying key nutrients missing in these diets, the bacteria allow insects to exploit food sources that would otherwise be inadequate (Baumann et al., 2000; Akman-Gündüz and Douglas, 2009). Unlike environmentally acquired symbionts, primary mutualists undergo maternal transmission to offspring. The congruence of host and bacterial phylogenies points to cospeciation for tens or hundreds of millions of years (e.g., Munson et al., 1991; Thao et al., 2000; Baumann and Baumann, 2005).
Interestingly, primary mutualists also occur in insects that feed on complex diets, such as ants in the tribe Camponotini and cockroaches (Buchner, 1965). These bacteria are thought to play more general nutritional roles involving nitrogen recycling and other functions. As a model to explore symbiont plasticity in a long-term mutualism, our labs focus on the ant mutualist called Candidatus Blochmannia (Blochman, 1882; Sauer et al., 2000). The bacteria are located in ant ovaries, consistent with maternal transmission to host offspring, and within bacteriocytes that are intercalated among enterocytes of the ant midgut (Buchner, 1965; Dasch, 1975; Sauer et al., 2002). Blochmannia has been studied most extensively in Camponotus, the second largest ant genus with ∼1000 species worldwide. Sometimes termed carpenter ants because many species excavate nests in wood, its nesting habits also include live trees and shrubs, within soil beneath rocks, and the rainforest canopy where Camponotus and other ant genera can comprise up to 94% of arthropods and 86% of the biomass (Davidson et al., 2003). Phylogenetic analyses indicate a single infection of Blochmannia at least before the divergence of three genera in the Camponitini >30 MYA and cospeciation since that time (Sameshima et al., 1999; Sauer et al., 2000; Degnan et al., 2004).
Stability of Symbiont Lineages
In interactions involving multiple symbionts, such as the microbiota of GI tracts, shifts in symbiont community profiles provide a potential mechanism for functional plasticity. By contrast, such shifts are not a major source of variation for most insect mutualists, due to their stability and exceptionally low genetic diversity. With some notable exceptions (see below), primary mutualists often consist of one bacterial lineage in a given host group. Moreover, the bacteria within a host individual are genetically homogeneous or very similar, due to continual diversity-purging population bottlenecks upon each inoculation of developing host eggs or embryos (Buchner, 1965; Mira and Moran, 2002).
Stable mutualisms involving multiple bacterial lineages occur in insects, but few are well characterized. The best studied is a dual, or coprimary, nutritional endosymbiosis in sharpshooters (Takiya et al., 2006). In addition, so-called secondary symbionts have infected mealybugs several times independently and coevolve with hosts, although over shorter time periods than the primary symbiont (Thao et al., 2002). Replacement of one mutualist lineage with a distinct microbial species has occurred in weevils (Conord et al., 2008) and aphids (Fukatsu and Ishikawa, 1996) and might play an important role over evolutionary timescales. Thus, changes in the profiles of mutualists are possible in a very limited sense, either due to shifts in the relative abundance of two mutualists, or lineage replacements over evolutionary time.
If we define the symbiotic community of insects more broadly, to also include diverse facultative associates, then shifts in the composition of a “super symbiotic system” (Koga et al., 2003) may be quite significant as the host responds to environmental conditions. These secondary symbionts are common in some insect groups, such as aphids, psyllids, and tsetse flies. The bacteria transfer frequently across species and help hosts cope with abiotic and biotic factors. For instance, aphid secondary symbionts increase tolerance to high temperatures (Montllor et al., 2002; Russell and Moran, 2006), reduce rates of fungal infection (Scarborough et al., 2005), confer resistance to parasitoids (Oliver et al., 2003; Moran et al., 2005b; Hansen et al., 2007; Degnan and Moran, 2008), and play a role in host plant preferences (Tsuchida et al., 2004). These bacteria might also complement the nutritional functions of primary mutualists. For example, secondary symbionts compensate for the loss of primary mutualists in the lab, presumably due to nutritional functions provided (Koga et al., 2003). In addition, tryptophan biosynthesis genes missing from the tiny (422 kb) genome of a Buchnera strain (Perez-Brocal et al., 2006) are encoded by the secondary symbiont that is abundant in the same aphid species, suggesting that both symbionts are necessary to provision this essential amino acid (Gosalbes et al., 2008). In this sense, facultative endosymbionts expand the potential for evolutionary innovation and may catalyze the formation of new, primary endosymbioses.
Lifestyle Constraints Reflected in the Genome
Genome degradation in insect mutualists
The constrained lifestyle of stable mutualists not only suppresses their genetic variation within hosts, but also has pervasive effects on patterns of DNA sequence and genome evolution (Moran, 1996; Andersson and Kurland, 1998; Moran et al., 2008). Across phylogenetically independent lineages, transitions to this lifestyle are coupled with apparent genetic degradation, including accelerated rates of molecular evolution, reduced genomic G + C content, as well as severe genome reduction (Fig. 1). This syndrome of genetic degradation may, in part, result from a reduction in effective population sizes (Ne), as these bacteria undergo successive bottlenecks when transmitted to host offspring (Andersson and Kurland, 1998; Funk et al., 2001; Abbot and Moran, 2002). According to the nearly neutral theory of evolution (Ohta, 1973), reduced Ne is expected to increase the rate of fixation of deleterious mutations by random genetic drift. Over time, the accumulation of deleterious mutations could negatively affect the fitness of the symbiont and its host.
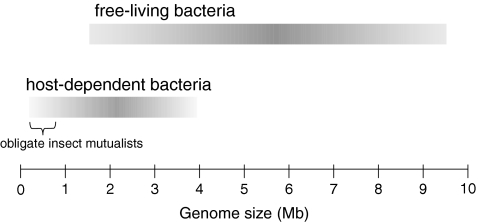
FIG. 1.
Genome size ranges for obligately host-associated and free-living bacterial species. Host-dependent bacteria typically undergo severe genome reduction. Primary mutualists of insects include the smallest bacterial genome known, the 160 kb chromosome of Carsonella associated with psyllids (Nakabachi et al., 2006).
Gene loss in insect mutualists is thought to reflect a combination of factors. Due to an underlying deletion bias in bacteria (Mira et al., 2001), genes persist only if selection favors their maintenance. Within a host cellular environment, genes encoding metabolic diversity and other functions may become redundant and lost under relaxed selection. In addition, selection to maintain genes will be less effective in these mutualists, due to reduced Ne. Fast nucleotide mutation rates and strong AT bias also drive gene disruption and loss. A recent population genomic study of the aphid mutualist, Buchnera, showed that replication errors within polyA tracts (abundant in AT-rich genomes) introduce frameshifts and trigger the process of gene erosion (Moran et al., 2009). Gene transfer to the host nucleus is also a very real possibility, as suggested by EST data for aphids (Nakabachi et al., 2005) and the absence of numerous critical functions in mutualist genomes, particularly the 160-kb genome of a psyllid symbiont (Nakabachi et al., 2006).
Despite severe gene loss in most functional categories, endosymbionts retain machinery for their own basic cellular processes (e.g., transcription, translation, and replication) and specific functions that benefit the host (e.g., amino acid or vitamin biosynthetic genes, as well as urease genes in Blochmannia of ants). Conspicuous losses in these mutualists include the absence of mobile DNA, the deletion of recA from Buchnera and Blochmannia, and the absence of the replication gene dnaA from Wigglesworthia of tsetse flies (Akman et al., 2002) and from Blochmannia of ants (Gil et al., 2003; Degnan et al., 2005). Relating to functional plasticity, mutualists have lost gene regulation functions to varying degrees (see Appendix). For a comparison of gene contents, we point the reader to recent reviews on this topic (Zientz et al., 2004; Moran et al., 2008; Feldhaar and Gross, 2009).
Genome stability in insect mutualists
Typically, bacterial evolution involves extensive genome fluidity, including lateral gene transfer, inversions, and translocations. Among parasitic microbes, horizontal transfer of pathogenicity islands and antibiotic resistance genes has indisputable significance. Likewise, genome fluidity generates diversity in many beneficial microbes. For example, the transfer of symbiotic loci on plasmids or genomic islands influences nitrogen-fixing rhizobia (Mergaert et al., 1997), and phage-mediated transfer of toxin genes shapes the defensive phenotype of aphid secondary symbionts (Degnan and Moran, 2008).
These major sources of genetic innovation for most bacteria—the acquisition of foreign DNA and shuffling of existing genes—is unavailable in at least some long-term mutualists. Instead, gene deletions are irreversible and may constrain the evolutionary potential of host and symbiont alike. Primary mutualists show no signs of horizontal gene transfer, with the notable exception of plasmid movement in some Buchnera lineages (Van Ham et al., 2000; Gil et al., 2006). For genera that have had more than one genome sequenced, the genome dynamics within the context of a long-term association can be evaluated. Such studies have found striking genome stability in Buchnera (Tamas et al., 2002) and Blochmannia (Degnan et al., 2005), implying no inversions or translocations during the tens of millions of year of evolution within hosts. Genome differences within a mutualist group are dominated by differential gene loss. This overall lack of gene acquisition or shuffling may reflect the loss of recombination genes and mobile DNA, combined with limited opportunities to encounter genetically distinct bacteria.
Gene Expression Variation
The increased accessibility of functional genomic approaches has opened new doors to explore gene expression in various symbiotic systems. Studies using microarrays, subtractive hybridization, EST sequencing, and meta-transcriptome analysis are illuminating the role of transcriptional plasticity in symbiotic interactions. As a starting point, many studies begin with the bacterial partner, for which genomic data are often available first. Ideally, expression analysis will include both partners in the symbiosis, to detect coordinated responses. The development of microarrays with host and symbiont loci is a promising avenue for this work (Wilson et al., 2006).
Among insect endosymbionts, gene expression began with the striking observation that symbionts produce almost exclusively one protein, called symbionin, which we now know is encoded by groEL (Ishikawa, 1984; Baumann et al., 1996). Since that time, we have learned that groEL is constitutively overexpressed in many other symbioses (Aksoy, 1995; Charles et al., 1997; Fares et al., 2004). This chaperonin may help to stabilize proteins that accumulate deleterious changes in the small populations of host-dependent microbes (Moran, 1996). More recently, functional genomic studies have illustrated reduced levels of gene expression differences in primary mutualists compared to Escherichia coli and other free-living bacteria. Studies in Buchnera shows high expression levels of certain chaperonins and amino acid biosynthetic genes, but very modest transcriptional responses when the host is exposed to heat shock (Wilcox et al., 2003) or changes in dietary amino acid content (Moran et al., 2005a). The loss of regulatory genes may limit the ability of long-term mutualists to alter transcriptional patterns in response to environmental fluctuations.
In the Blochmannia–Camponotus association, the ecology and physiology of hosts may be particularly variable and enhance selection for plasticity of endosymbiont functions. The bacterium must not only operate in different developmental stages, but also function in different castes and through the entire life cycle of an ant colony. Hymenopteran castes represent the most extreme morphological, biochemical, and behavioral variation ever documented among genetically equivalent organisms (Evans and Wheeler, 2001). Ants and their colonies tend to be especially long-lived, with workers typically living several months and the queens living several years to decades (Hölldobler and Wilson, 1990). As workers age, they change behaviorally and physiologically as they progress from working within nests to foraging. Two distinct types of workers, minors and majors, are biased to certain tasks and have physiologies to match. Queens are radically different than workers, as they are specialized to begin new colonies and have heavy nutritional demands to support egg production (Wheeler and Buck, 1995). The variety of food exchanges in ant colonies implies that the interaction of Blochmannia and an individual host will affect other individuals and the colony as a whole.
A key difference in the Blochmannia–ant mutualism in comparison to those in nonsocial insects is the reduced impact of symbiont loss to the individual host. Ant workers survive antibiotic treatment and in many respects appear to be normal. For example, elimination of Blochmannia with rifampicin had no effect on worker mortality (Feldhaar et al., 2007), and antibiotic treatment of other stages does not result in death (the authors' unpublished data). These patterns make sense if we consider the ant colony as a superorganism (Hölldobler and Wilson, 2009). The effects of symbionts on this superorganism involve nutrient levels that will be distributed to the colony members with the greatest nutrient demand. The symbionts meet the needs of the colony, not necessarily the individual ants that house them. Individuals can carry out resource-intensive processes above the level found in solitary insects, because the symbiont products synthesized in many individual ants can flow toward larvae or queens, greatly enhancing their performance. As predicted under this framework, the major difference between workers with versus without Blochmannia involved the ability of these workers to rear brood (Feldhaar et al., 2007).
Given this exceptional variation at the host level, there is much interest in mechanisms that underlie functional variation in Blochmannia across host life stages and castes. In a recent microarray study, Stoll et al. (2009) tracked changes in Blochmannia transcription across distinct developmental stages and worker ages for the host species Camponotus floridanus. Shifts in Blochmannia gene expression were modest and typically fell below a threefold change, although several (∼20) genes showed greater than fivefold variation in expression level. This overall reduction in expression fluctuations is similar to earlier findings in Buchnera, and might reflect the loss of many regulation genes in both endosymbionts (Wilcox et al., 2003; Moran et al., 2005a).
While insect mutualists are missing many gene regulation functions, they vary in the extent of this loss (Table 1). For example, Blochmannia of Camponotus pennsylvanicus (i.e., Blochmannia pennsylvanicus) diverged from Blochmannia floridanus ∼20 MYA and retains key regulatory genes that B. floridanus has lost. These distinct regulatory functions include hns, encoding the histone-like nucleoid-structuring protein (H-NS) that functions as a major bacterial chromatin component and can play an important role in global regulation; ispH involved in control of the stringent response that allows bacteria to survive conditions of amino acid starvation; hfq, a pleiotropic posttranscriptional regulator; mntR, a DNA-binding transcriptional repressor; and yfgB involved in posttranscriptional modification 23S rRNA. Having retained these regulatory capabilities, it is possible that B. pennsylvanicus shows a greater capacity for gene expression fluctuations, an hypothesis we are currently testing.
Symbiont Density Fluctuations
In symbionts with limited gene regulation capabilities, shifts in bacterial densities may offer an important mechanism for plasticity. Many studies have shown dramatic differences in the densities of primary mutualists within hosts, with numbers depending upon host developmental stage, sex, or age (Wolschin et al., 2004; Kono et al., 2008), host nutrition (Feldhaar et al., 2007; Wilkinson et al., 2007), and stressors such as predation (Mahadav et al., 2008).
It is often difficult to distinguish whether symbiont proliferation and decline represent adaptive responses that benefit the host, or alternatively, a breakdown in the normal regulation of symbiont numbers. In Camponotus, Blochmannia densities vary widely throughout development and show hints of an adaptive response. Symbionts reach their highest densities in pupae undergoing metamorphosis and in young imagines (Wolschin et al., 2004; Stoll et al., 2009). At these stages, high metabolic requirements include sclerotization and melanization of the cuticle and may require increased synthesis of amino acids (especially tyrosine), nitrogen recycling, and other symbiont nutritional functions. Further supporting the hypothesis that symbiont proliferation benefits hosts, the same host stages show a modest increase in the relative expression of symbiont nutritional functions (Zientz et al., 2004; Stoll et al., 2009). Interestingly, Blochmannia densities declined in workers fed a diet lacking essential amino acids (Feldhaar et al., 2007). While one might predict that increased symbiont load could help to compensate for dietary deficiencies, bacterial replication probably imposes some cost to the host. Apparently, workers deprived of essential nutrients lack the resources to support high symbiont densities. Notably, the unusual absence of dnaA in Blochmannia might provide the host some control over replication.
Transcriptional Slippage as a Potential Mechanism for Plasticity
Transcription in AT-rich endosymbionts may be influenced by the long, homopolymeric tracts (especially polyA regions) that are common in protein-coding genes. Such tracts are generally considered deleterious because they are prone to slippage errors during replication (which may disrupt the reading frame, as noted above). In addition, RNA polymerase errors along these tracts may introduce frameshifts in transcripts, producing a heterogeneous pool of mRNA transcripts with varied numbers of A's in the homopolymeric region. When this happens, some fraction of transcripts will contain frameshifts and fail to encode full-length proteins. An accurate picture of gene expression profiles in AT-rich symbionts will depend on understanding whether such transcriptional slippage is prevalent.
As a case study, Tamas et al. (2008) tested for transcriptional slippage in several Buchnera and Blochmannia genes that contain long polyA tracts. cDNA libraries showed variable tract lengths, indicating a high prevalence of RNA polymerase slippage. While transcriptional slippage typically disrupts the reading frame of an intact gene, it also restores the reading frame in transcripts of presumed pseudogenes. At genes that contain frameshifts in the genomic DNA, 12–50% of transcripts contained corrected reading frames that could potentially yield full-length proteins. Oddly, a surprising conservation of frameshifts within such genes suggests that they are maintained by selection over evolutionary time (Tamas et al., 2008).
Although speculative and difficult to test, it is possible that endosymbionts with reduced regulatory capabilities might exploit transcriptional slippage to alter the abundances of full-length proteins, achieving an indirect form of transcriptional plasticity. A DNA frameshift within a polyA tract could reduce (but thanks to transcriptional slippage, not eliminate) the production of intact transcripts and full-length proteins. Replication slippage that corrects the reading frame in the genomic DNA would be a very simple back-mutation that restores the higher expression level. In this sense, the persistence of error-prone tracts could offer a fortuitous avenue for plasticity. In any event, when quantifying transcription profiles in symbionts with homopolymeric tracts, it will be important to consider that the effective expression level of loci with polyA tracts depends on the prevalence of transcriptional slippage.
Conclusions
Symbioses vary in their complexity, from the highly diverse microbiota of GI tracts, to relatively simple, binary associations. These distinct lifestyles affect the genetic diversity of microbial symbionts and their capacity to respond to variation at the host level. At an extreme end of the spectrum, the specialized lifestyle of long-term insect mutualists severely constrains the genetic and metabolic diversity of these bacteria. Once established within a host group, these bacteria specialize at the expense of their abilities to acquire new functions or to shuffle existing genes. While one might imagine that gene expression variation could be especially important under such circumstances, these bacteria have lost many regulatory genes in the course of extensive genome reduction. As predicted, some species show only a modest capacity for transcriptional variation. Shifts in microbial densities may offer a primary mechanism to vary expression levels of symbiont functions.
In an ant–bacterial mutualism, the eusocial nature of the host may enhance symbiont plasticity, because the bacteria function within physiologically distinct castes and mediate exchanges among them. In the Blochmannia–Camponotus symbiosis, observed symbiont plasticity is best understood by considering the host colony as a superorganism, with the bacteria meeting the needs of the colony but not necessarily the individual ants that house them. For example, the symbiont is not essential to individual workers, but rather enhances this caste's contribution to colony growth.
Though exceptional in their constrained lifestyle, stable mutualists of insects illustrate general connections among symbiotic lifestyle, genome change, and mechanisms of plasticity. Challenges for future work will include distinguishing adaptive responses from neutral or even harmful perturbations in symbiont functions, and clarifying the actual signals that hosts and symbionts exchange to coordinate their responses.
Appendix
Appendix:
Regulation Genes Retained in Select Insect Mutualists
| Gene | Escherichia coli bnumber | Blochmannia pennsylvanicus | Blochmannia floridanus | Wigglesworthia brevipalpis | Buchnera–Acyrthosiphon pisum | Buchnera–Schizaphis graminum | Buchnera–Baizongia pistaciae | Function |
|---|---|---|---|---|---|---|---|---|
| alaS | b2697 | Bpen174 | Bfl168 | WGLp234 | BU403 | BUsg390 | Bbp364 | Alanyl-tRNA synthetase; transcriptional repressor |
| birA | b3973 | Bpen190 | Bfl184 | WGLp513 | – | – | – | Bifunctional biotin-[acetyl-CoA-carboxylase] ligase and transcriptional repressor |
| bolA | b0435 | – | – | – | BU473 | BUsg457 | Bbp418 | Transcriptional dual regulator |
| cgtA | b3183 | Bpen098 | Bfl095 | WGLp475 | BU389 | BUsg376 | Bbp352 | GTPase involved in DNA replication and ribosome assembly; transcriptional repressor |
| clpP | b0437 | Bpen253 | Bfl246 | WGLp155 | BU475 | BUsg459 | Bbp419 | Serine protease subunit; posttranscriptional regulator |
| cpxR | b3912 | – | – | WGLp538 | – | – | – | Transcriptional regulator in 2-component system |
| cspC | b1823 | Bpen462 | Bfl448 | – | BU322 | – | – | Predicted DNA-binding transcriptional regulator |
| cspE | b0623 | – | – | WGLp177 | BU489 | BUsg473 | Bbp433 | Transcription antiterminator and regulator of RNA stability |
| dksA | b0145 | Bpen154 | Bfl149 | – | BU198 | BUsg192 | Bbp184 | RNA polymerase-binding transcription factor |
| dnaA | b3702 | – | – | – | BU012 | BUsg012 | Bbp012 | Transcriptional dual regulator HU-alpha; (HU-2) |
| fis | b3261 | – | – | – | BU400 | BUsg387 | – | DNA-binding protein; a trans activator for transcription |
| flhC | b1891 | – | – | WGLp031 | – | – | – | Regulator of flagellar biosynthesis; transcription initiation factor |
| flhD | b1892 | – | – | WGLp030 | – | – | – | Regulator of flagellar biosynthesis; transcription initiation factor |
| fliA | b1922 | – | – | WGLp199 | – | – | – | Regulation of flagellar operons |
| frr | b0172 | Bpen282 | Bfl274 | WGLp389 | BU234 | BUsg228 | Bbp216 | Ribosome releasing factor |
| greA | b3181 | Bpen099 | Bfl096 | WGLp233 | BU384 | BUsg371 | Bbp347 | Transcription elongation factor |
| hflC | b4175 | Bpen085 | Bfl082 | WGLp186 | BU567 | BUsg547 | Bbp512 | Regulator of FtsH protease |
| hfq | b4172 | Bpen082 | – | WGLp184 | – | – | – | RNA-binding protein that affects many cellular processes |
| hha | b0460 | – | – | WGLp524 | – | – | – | Hemolysin expression modulating protein |
| hns | b1237 | Bpen448 | – | WGLp367 | BU272 | – | – | H-NS transcriptional dual regulator |
| hslK | b4174 | Bpen084 | Bfl081 | WGLp185 | BU568 | BUsg548 | Bbp513 | Regulator of FtsH protease |
| hslU | b3931 | – | – | WGLp277 | BU579 | BUsg558 | Bbp524 | ATPase component of the HslVU protease |
| hslV | b3932 | – | – | WGLp278 | BU578 | BUsg557 | Bbp523 | Peptidase component of the HslVU protease |
| hslX | b4173 | Bpen083 | Bfl080 | – | – | – | – | Putative GTPase; possible regulator of HflKC |
| hupA | b4000 | – | – | WGLp509 | BU032 | BUsg033 | Bbp033 | Transcriptional dual regulator HU-α (HU-2) |
| ihfA (himA) | b1712 | – | – | – | BU131 | BUsg123 | – | Integration host factor alpha |
| ihfB (himD) | b0912 | – | – | – | BU308 | BUsg298 | – | Integration host factor beta |
| ispH | b0029 | Bpen124 | – | – | – | – | – | Control of stringent response |
| lon | b0439 | Bpen307 | Bfl299 | WGLp153 | BU477 | BUsg461 | Bbp421 | ATP-dependent protease |
| metK | b2942 | Bpen259 | Bfl252 | WGLp316 | BU408 | BUsg393 | – | MetK S-adenosylmethionine synthetase monomer |
| metR | b3828 | – | – | – | – | BUsg030 | – | MetR transcriptional dual regulator |
| mntR | b0817 | Bpen403 | – | – | – | – | – | Putative toxin |
| nusA | b3169 | Bpen107 | Bfl103 | WGLp227 | BU378 | BUsg366 | Bbp341 | Transcription termination/antitermination L factor |
| ptsG | b1101 | – | – | – | BU356 | BUsg344 | Bbp326 | PTS system, glucose-specific IIBC component |
| putA | b1014 | – | – | WGLp434 | – | – | – | Proline dehydrogenase, P5C dehydrogenase |
| pyrI | b4244 | – | – | WGLp590 | BU370 | BUsg358 | – | Aspartate carbamoyltransferase, regulatory subunit |
| recA | b2699 | – | – | WGLp235 | – | – | – | DNA strand exchange and renaturation, DNA-dependent ATPase, DNA- and ATP-dependent coprotease |
| rho | b3783 | Bpen607 | Bfl586 | WGLp303 | BU596 | BUsg572 | Bbp538 | Transcription termination factor Rho monomer; polarity suppressor |
| rplA | b3984 | Bpen580 | Bfl560 | WGLp519 | BU037 | BUsg038 | Bbp038 | 50S ribosomal subunit protein L1; posttranscriptional regulation of ribosomal protein genes |
| rplD | b3319 | Bpen199 | Bfl192 | WGLp544 | BU523 | BUsg504 | Bbp466 | 50S ribosomal subunit protein L4; transcriptional and posttranscriptional regulation of ribosomal protein genes |
| rplJ | b3985 | Bpen579 | Bfl559 | WGLp520 | BU036 | BUsg037 | Bbp037 | 50S ribosomal subunit protein L10; posttranscriptional regulation of ribosomal protein genes |
| rplT | b1716 | Bpen365 | Bfl354 | WGLp084 | BU128 | BUsg120 | Bbp122 | 50S ribosomal subunit protein L20; autoregulates its own expression and that of L35 |
| rpmH | b3703 | Bpen014 | Bfl015 | WGLp014 | BU013 | BUsg013 | Bbp013 | 50S ribosomal subunit protein L34; inhibitor of the decarboxylases involved in polyamine biosynthesis |
| rpoD | b3067 | Bpen057 | Bfl056 | WGLp468 | BU055 | BUsg052 | Bbp052 | RNA polymerase, sigmaD factor; primary sigma factor during exponential growth |
| rpoH | b3461 | Bpen653 | Bfl626 | WGLp072 | BU025 | BUsg026 | Bbp027 | RNA polymerase, sigma(32) factor; regulation of proteins induced at high temperatures |
| rpsH | b3306 | Bpen212 | Bfl205 | WGLp557 | BU510 | BUsg491 | Bbp453 | 30S ribosomal subunit protein S8; posttranscriptional regulation of ribosomal protein genes |
| rpsJ | b3321 | Bpen197 | Bfl190 | WGLp542 | BU525 | BUsg506 | Bbp468 | 30S ribosomal subunit protein S10; role in transcription antitermination |
| rpsT | b0023 | Bpen120 | Bfl116 | WGLp296 | BU151 | BUsg144 | Bbp140 | 30S ribosomal subunit protein S20; inhibitor of the decarboxylases involved in polyamine biosynthesis |
| rseP (yaeL) | b0176 | Bpen286 | Bfl278 | WGLp385 | – | – | – | Cleaves RseA, thereby activating sigmaE-mediated stress response |
| slyA | b1642 | Bpen380 | Bfl369 | WGLp336 | – | – | – | Transcriptional regulator for cryptic hemolysin |
| sufB (ynhE) | b1683 | Bpen370 | Bfl359 | WGLp360 | – | – | – | SufBCD complex activates the cysteine desulfurase; secondary pathway of iron-sulfur cluster assembly |
| sufC (ynhD) | b1682 | Bpen371 | Bfl360 | WGLp359 | – | – | – | Component of SufBCD complex |
| sufD (ynhC) | b1681 | Bpen372 | Bfl361 | WGLp358 | – | – | – | Component of SufBCD complex |
| sufE | b1679 | Bpen374 | Bfl363 | WGLp356 | – | – | – | Sulfur acceptor that activates SufS cysteine desulfurase |
| suhB | b2533 | Bpen555 | Bfl535 | WGLp285 | BU285 | BUsg274 | Bbp264 | Enhances synthesis of sigma32 in mutant; extragenic suppressor |
| thrS | b1719 | Bpen362 | Bfl351 | WGLp081 | BU125 | BUsg117 | Bbp119 | Threonyl-tRNA synthetase; translation attenuation and efficiency |
| uspA | b3495 | – | – | – | – | – | Bbp530 | Universal stress global stress response regulator |
| ychA | b1214 | – | – | – | BU173 | BUsg167 | Bbp163 | Predicted transcriptional regulator |
| ychF | b1203 | Bpen354 | Bfl344 | WGLp351 | BU191 | BUsg185 | Bbp180 | Putative GTP-binding protein; translation attenuation and efficiency |
| ydgL (rsxA) | b1627 | – | – | – | BU113 | – | Bbp108 | Integral membrane protein of SoxR-reducing complex; posttranscriptional regulation |
| ydgM (rsxB) | b1628 | – | – | – | BU114 | BUsg106 | Bbp109 | Member of SoxR-reducing complex; posttranscriptional regulation |
| ydgN (rsxC) | b1629 | – | – | – | BU115 | – | Bbp110 | Member of SoxR-reducing complex; posttranscriptional regulation |
| ydgO (rsxD) | b1630 | – | – | – | BU116 | BUsg108 | Bbp111 | Integral membrane protein of SoxR-reducing complex; posttranscriptional regulation |
| ydgP (rsxG) | b1631 | – | – | – | BU117 | BUsg109 | Bbp112 | Member of SoxR-reducing complex; posttranscriptional regulation |
| ydgQ (rsxE) | b1632 | – | – | – | BU118 | BUsg110 | Bbp113 | Integral membrane protein of SoxR-reducing complex; posttranscriptional regulation |
| yhdO | b3244 | Bpen305 | Bfl297 | – | BU398 | BUsg385 | Bbp361 | Protease involved in Microcin maturation and sensitivity to the DNA gyrase inhibitor |
| yidZ | b3711 | Bpen038 | Bfl038 | – | – | – | – | Putative transcriptional regulator LysR-type |
| yjeA | b4155 | – | – | – | BU582 | – | Bbp528 | Putative regulator of pyruvate oxidase |
| yqeI | b2847 | Bpen401 | Bfl390 | WGLp467 | – | – | – | Predicted transcriptional regulator |
| yrbA | b3190 | Bpen046 | Bfl045 | WGLp328 | BU385 | BUsg372 | Bbp348 | Predicted DNA-binding transcriptional regulator |
| zur | b4046 | Bpen026 | Bfl026 | – | – | – | – | Putative transcriptional repressor |
| 443 regulation genes | absent from the six mutualists above | |||||||
These gamma-Proteobacterial endosymbionts are closely related to E. coli, and their gene contents are generally a subset of the E. coli genome. To avoid inconsistencies in annotation methods, we first identified E. coli orthologs in each mutualist using the reciprocal smallest distance algorithm (Wall et al., 2003). M. H. Serres kindly provided a list of E. coli genes that have been assigned to Regulation category in the MultiFun catalog of roles (Serres et al., 2004). In addition to the genes listed above, E. coli possesses numerous (443) regulation genes that are absent from the six mutualists considered here. Reflecting their multiple functions in the cell, many genes (including the loci listed here) have more than one functional designation within Multifun. The table lists the primary annotation ID for the orthologs in mutualists.
Acknowledgments
We are grateful to Margaret McFall-Ngai for organizing the 2008 ASM Beneficial Microbes meeting, and thank all of the conference participants for lively discussion. We thank two anonymous reviewers for their suggestions that improved the manuscript, and thank Norman Buck for comments on the DNA regulation table. This work was supported by grants from National Science Foundation (D.E.W. and J.J.W.) and from the National Institutes of Health (J.J.W.).
Disclosure Statement
No competing financial interests exist.
References
- Abbot P. Moran N.A. Extremely low levels of genetic polymorphism in endosymbionts (Buchnera) of aphids (Pemphigus) Mol Ecol. 2002;11:2649–2660. doi: 10.1046/j.1365-294x.2002.01646.x. [DOI] [PubMed] [Google Scholar]
- Akman L. Yamashita A. Watanabe H. Oshima K. Shiba T. Hattori M. Aksoy S. Genome sequence of the endocellular obligate symbiont of tsetse flies, Wigglesworthia glossinidia. Nat Genet. 2002;32:402–407. doi: 10.1038/ng986. [DOI] [PubMed] [Google Scholar]
- Akman-Gündüz E. Douglas A. Symbiotic bacteria enable insect to use a nutritionally inadequate diet. Proc Biol Sci. 2009;276:987–991. doi: 10.1098/rspb.2008.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aksoy S. Molecular analysis of the endosymbionts of tsetse flies: 16S rDNA locus and over-expression of a chaperonin. Insect Mol Biol. 1995;4:23–29. doi: 10.1111/j.1365-2583.1995.tb00004.x. [DOI] [PubMed] [Google Scholar]
- Andersson S.G.E. Kurland C.G. Reductive evolution of resident genomes. Trends Microbiol. 1998;6:263–268. doi: 10.1016/s0966-842x(98)01312-2. [DOI] [PubMed] [Google Scholar]
- Baird A.H. Bhagooli R. Ralph P.J. Takahashi S. Coral bleaching: the role of the host. Trends Ecol Evol. 2009;24:16–20. doi: 10.1016/j.tree.2008.09.005. [DOI] [PubMed] [Google Scholar]
- Baumann L. Baumann P. Cospeciation between the primary endosymbionts of mealybugs and their hosts. Curr Microbiol. 2005;50:84–87. doi: 10.1007/s00284-004-4437-x. [DOI] [PubMed] [Google Scholar]
- Baumann P. Baumann L. Clark M.A. Levels of Buchnera aphidicola chaperonin GroEL during growth of the aphid Schizaphis graminum. Curr Microbiol. 1996;32:279–285. [Google Scholar]
- Baumann P. Moran N. Baumann L. Bacteriocyte-associated endosymbionts of insects. In: Dworkin M., editor. The Prokaryotes: A Handbook on the Biology of Bacteria; Ecophysiology, Isolation, Identification, Applications. Springer-Verlag; New York: 2000. pp. 403–438. [Google Scholar]
- Blochman F. Über das vorkommen von bakterienähnlichen gebilden in den geweben und eiern verschiedener insekten. Zentbl Bakteriol. 1882;11:234–240. [Google Scholar]
- Buchner P. Endosymbiosis of Animals with Plant Microorganisms. Interscience Publishers; New York: 1965. [Google Scholar]
- Charles H. Heddi A. Guillaud J. Nardon C. Nardon P. A molecular aspect of symbiotic interactions between the weevil Sitophilus oryzae and its endosymbiotic bacteria: over-expression of a chaperonin. Biochem Biophys Res Commun. 1997;239:769–774. doi: 10.1006/bbrc.1997.7552. [DOI] [PubMed] [Google Scholar]
- Chun C.K. Troll J.V. Koroleva I. Brown B. Manzella L. Snir E. Almabrazi H. Scheetz T.E. Bonaldo Mde F. Casavant T.L. Soares M.B. Ruby E.G. McFall-Ngai M.J. Effects of colonization, luminescence, and autoinducer on host transcription during development of the Squid-Vibrio association. Proc Natl Acad Sci USA. 2008;105:11323–11328. doi: 10.1073/pnas.0802369105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conord C. Despres L. Vallier A. Balmand S. Miquel C. Zundel S. Lemperiere G. Heddi A. Long-term evolutionary stability of bacterial endosymbiosis in Curculionoidea: additional evidence of symbiont replacement in the Dryophthoridae family. Mol Biol Evol. 2008;25:859–868. doi: 10.1093/molbev/msn027. [DOI] [PubMed] [Google Scholar]
- Dasch G.A. Morphological and Molecular Studies on Intracellular Bacterial Symbiotes of Insects. Yale University; New Haven, CT: 1975. p. 329. [Google Scholar]
- Davidson D.W. Cook S.C. Snelling R.R. Chua T.H. Explaining the abundance of ants in lowland tropical rainforest canopies. Science. 2003;300:969–972. doi: 10.1126/science.1082074. [DOI] [PubMed] [Google Scholar]
- Degnan P. Lazarus A. Brock C. Wernegreen J. Host-symbiont stability and fast evolutionary rates in an ant-bacterium association: cospeciation of Camponotus species and their endosymbionts, Candidatus Blochmannia. Syst Biol. 2004;53:95–110. doi: 10.1080/10635150490264842. [DOI] [PubMed] [Google Scholar]
- Degnan P.H. Lazarus A.B. Wernegreen J.J. Genome sequence of Blochmannia pennsylvanicus indicates parallel evolutionary trends among bacterial mutualists of insects. Genome Res. 2005;15:1023–1033. doi: 10.1101/gr.3771305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnan P.H. Moran N.A. Evolutionary genetics of a defensive facultative symbiont of insects: exchange of toxin-encoding bacteriophage. Mol Ecol. 2008;17:916–929. doi: 10.1111/j.1365-294X.2007.03616.x. [DOI] [PubMed] [Google Scholar]
- Dethlefsen L. Huse S. Sogin M.L. Relman D.A. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans J.D. Wheeler D.E. Expression profiles during honeybee caste determination. Genome Biol. 2001;2 doi: 10.1186/gb-2000-2-1-research0001. RESEARCH0001 Epub 2000 Dec 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fares M.A. Moya A. Barrio E. GroEL and the maintenance of bacterial endosymbiosis. Trends Genet. 2004;20:413–416. doi: 10.1016/j.tig.2004.07.001. [DOI] [PubMed] [Google Scholar]
- Feldhaar H. Gross R. Insects as hosts for mutualistic bacteria. Int J Med Microbiol. 2009;299:1–8. doi: 10.1016/j.ijmm.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldhaar H. Straka J. Krischke M. Berthold K. Stoll S. Mueller M.J. Gross R. Nutritional upgrading for omnivorous carpenter ants by the endosymbiont Blochmannia. BMC Biol. 2007;5:48. doi: 10.1186/1741-7007-5-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukatsu T. Ishikawa H. Phylogenetic position of yeast-like symbiont of Hamiltonaphis styraci (Homoptera, Aphididae) based on 18S rDNA sequence. Insect Biochem Mol Biol. 1996;26:383–388. doi: 10.1016/0965-1748(95)00105-0. [DOI] [PubMed] [Google Scholar]
- Funk D.J. Wernegreen J.J. Moran N.A. Intraspecific variation in symbiont genomes: bottlenecks and the aphid-Buchnera association. Genetics. 2001;157:477–489. doi: 10.1093/genetics/157.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil R. Sabater-Munoz B. Perez-Brocal V. Silva F.J. Latorre A. Plasmids in the aphid endosymbiont Buchnera aphidicola with the smallest genomes: a puzzling evolutionary story. Gene. 2006;370:17–25. doi: 10.1016/j.gene.2005.10.043. [DOI] [PubMed] [Google Scholar]
- Gil R. Silva F.J. Zientz E. Delmotte F. Gonzalez-Candelas F. Latorre A. Rausell C. Kamerbeek J. Gadau J. Hölldobler B. van Ham R.C.H.J. Gross R. Moya A. The genome sequence of Blochmannia floridanus: comparative analysis of reduced genomes. Proc Natl Acad Sci USA. 2003;100:9388–9393. doi: 10.1073/pnas.1533499100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosalbes M.J. Lamelas A. Moya A. Latorre A. The striking case of tryptophan provision in the cedar aphid Cinara cedri. J Bacteriol. 2008;190:6026–6029. doi: 10.1128/JB.00525-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen A.K. Jeong G. Paine T.D. Stouthamer R. Frequency of secondary symbiont infection in an invasive psyllid relates to parasitism pressure on a geographic scale in California. Appl Environ Microbiol. 2007;73:7531–7535. doi: 10.1128/AEM.01672-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hölldobler B. Wilson E.O. The Ants. Belknap Press of Harvard University Press; Cambridge: 1990. [Google Scholar]
- Hölldobler B. Wilson E.O. The Superorganism. W.W. Norton & Company; New York: 2009. [Google Scholar]
- Ishikawa H. Characterization of the protein species synthesized in vivo and in vitro by an aphid endosymbiont. Insect Biochem. 1984;14:417–425. [Google Scholar]
- Ishikawa H. Insect symbiosis: an introduction. In Insect Symbiosis. K. Bourtzis and T.A. Miller, eds. CRC Press; Boca Raton: 2003. pp. 1–21. [Google Scholar]
- Koga R. Tsuchida T. Fukatsu T. Changing partners in an obligate symbiosis: a facultative endosymbiont can compensate for loss of the essential endosymbiont Buchnera in an aphid. Proc Biol Sci. 2003;270:2543–2550. doi: 10.1098/rspb.2003.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono M. Koga R. Shimada M. Fukatsu T. Infection dynamics of coexisting beta- and gamma-proteobacteria in the nested endosymbiotic system of mealybugs. Appl Environ Microbiol. 2008;74:4175–4184. doi: 10.1128/AEM.00250-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley R.E. Lozupone C.A. Hamady M. Knight R. Gordon J.I. Worlds within worlds: evolution of the vertebrate gut microbiota. Nat Rev Microbiol. 2008;6:776–788. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadav A. Gerling D. Gottlieb Y. Czosnek H. Ghanim M. Parasitization by the wasp Eretmocerus mundus induces transcription of genes related to immune response and symbiotic bacteria proliferation in the whitefly Bemisia tabaci. BMC Genomics. 2008;9:342. doi: 10.1186/1471-2164-9-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulis L. Fester R. Symbiosis As a Source of Evolutionary Innovation. MIT Press; Cambridge: 1991. [PubMed] [Google Scholar]
- Mergaert P. Van Montagu M. Holsters M. Molecular mechanisms of Nod factor diversity. Mol Microbiol. 1997;25:811–817. doi: 10.1111/j.1365-2958.1997.mmi526.x. [DOI] [PubMed] [Google Scholar]
- Mira A. Moran N.A. Estimating population size and transmission bottlenecks in maternally transmitted endosymbiotic bacteria. Microb Ecol. 2002;44:137–143. doi: 10.1007/s00248-002-0012-9. [DOI] [PubMed] [Google Scholar]
- Mira A. Ochman H. Moran N.A. Deletional bias and the evolution of bacterial genomes. Trends Genet. 2001;17:589–596. doi: 10.1016/s0168-9525(01)02447-7. [DOI] [PubMed] [Google Scholar]
- Montllor C. Maxmen A. Purcell A. Facultative bacterial endosymbionts benefit pea aphids Acyrthosiphon pisum under heat stress. Ecol Entomol. 2002;27:189–195. [Google Scholar]
- Moran N.A. Accelerated evolution and Muller's rachet in endosymbiotic bacteria. Proc Natl Acad Sci USA. 1996;93:2873–2878. doi: 10.1073/pnas.93.7.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran N.A. Dunbar H.E. Wilcox J.L. Regulation of transcription in a reduced bacterial genome: nutrient-provisioning genes of the obligate symbiont Buchnera aphidicola. J Bacteriol. 2005a;187:4229–4237. doi: 10.1128/JB.187.12.4229-4237.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran N.A. McCutcheon J.P. Nakabachi A. Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet. 2008;42:165–190. doi: 10.1146/annurev.genet.41.110306.130119. [DOI] [PubMed] [Google Scholar]
- Moran N.A. McLaughlin H.J. Sorek R. The dynamics and time scale of ongoing genomic erosion in symbiotic bacteria. Science. 2009;323:379–382. doi: 10.1126/science.1167140. [DOI] [PubMed] [Google Scholar]
- Moran N.A. Russell J.A. Koga R. Fukatsu T. Evolutionary relationships of three new species of Enterobacteriaceae living as symbionts of aphids and other insects. Appl Environ Microbiol. 2005b;71:3302–3310. doi: 10.1128/AEM.71.6.3302-3310.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munson M.A. Baumann P. Clark M.A. Baumann L. Moran N.A. Voegtlin D.J. Campbell B.C. Evidence for the establishment of aphid-Eubacterium endosymbiosis in an ancestor of four aphid families. J Bacteriol. 1991;173:6321–6324. doi: 10.1128/jb.173.20.6321-6324.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabachi A. Shigenobu S. Sakazume N. Shiraki T. Hayashizaki Y. Carninci P. Ishikawa H. Kudo T. Fukatsu T. Transcriptome analysis of the aphid bacteriocyte, the symbiotic host cell that harbors an endocellular mutualistic bacterium, Buchnera. Proc Natl Acad Sci USA. 2005;102:5477–5482. doi: 10.1073/pnas.0409034102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabachi A. Yamashita A. Toh H. Ishikawa H. Dunbar H.E. Moran N.A. Hattori M. The 160-kilobase genome of the bacterial endosymbiont Carsonella. Science. 2006;314:267. doi: 10.1126/science.1134196. [DOI] [PubMed] [Google Scholar]
- Ohta T. Slightly deleterious mutant substitutions in evolution. Nature. 1973;246:96–98. doi: 10.1038/246096a0. [DOI] [PubMed] [Google Scholar]
- Oliver K.M. Russell J.A. Moran N.A. Hunter M.S. Facultative bacterial symbionts in aphids confer resistance to parasitic wasps. Proc Natl Acad Sci USA. 2003;100:1803–1807. doi: 10.1073/pnas.0335320100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Brocal V. Gil R. Ramos S. Lamelas A. Postigo M. Michelena J.M. Silva F.J. Moya A. Latorre A. A small microbial genome: the end of a long symbiotic relationship? Science. 2006;314:312–313. doi: 10.1126/science.1130441. [DOI] [PubMed] [Google Scholar]
- Russell J.A. Moran N.A. Costs and benefits of symbiont infection in aphids: variation among symbionts and across temperatures. Proc Biol Sci. 2006;273:603–610. doi: 10.1098/rspb.2005.3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sameshima S. Hasegawa E. Kitade O. Minaka N. Matsumoto T. Phylogenetic comparison of endosymbionts with their host ants based on molecular evidence. Zool Sci. 1999;16:993–1000. [Google Scholar]
- Sauer C. Stackebrandt E. Gadau J. Hölldobler B. Gross R. Systematic relationships and cospeciation of bacterial endosymbionts and their carpenter ant host species: proposal of the new taxon Candidatus Blochmannia gen. nov. Int J Syst Evol Microbiol. 2000;50 Pt 5:1877–1886. doi: 10.1099/00207713-50-5-1877. [DOI] [PubMed] [Google Scholar]
- Sauer C. Dudaczek D. Hölldobler B. Gross R. Tissue localization of the endosymbiotic bacterium “Candidatus Blochmannia floridanus” in adults and larvae of the carpenter ant Camponotus floridanus. Appl Environ Microbiol. 2002;68:4187–4193. doi: 10.1128/AEM.68.9.4187-4193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarborough C.L. Ferrari J. Godfray H.C. Aphid protected from pathogen by endosymbiont. Science. 2005;310:1781. doi: 10.1126/science.1120180. [DOI] [PubMed] [Google Scholar]
- Serres M.H. Goswami S. Riley M. GenProtEC: an updated and improved analysis of functions of Escherichia coli K-12 proteins. Nucleic Acids Res. 2004;32:D300–D302. doi: 10.1093/nar/gkh087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoll S. Feldhaar H. Gross R. Transcriptional profiling of the endosymbiont Blochmannia floridanus during different developmental stages of its holometabolous ant host. Environ Microbiol. 2009;11:877–888. doi: 10.1111/j.1462-2920.2008.01808.x. [DOI] [PubMed] [Google Scholar]
- Takiya D.M. Tran P.L. Dietrich C.H. Moran N.A. Co-cladogenesis spanning three phyla: leafhoppers (Insecta: Hemiptera: Cicadellidae) and their dual bacterial symbionts. Mol Ecol. 2006;15:4175–4191. doi: 10.1111/j.1365-294X.2006.03071.x. [DOI] [PubMed] [Google Scholar]
- Tamas I. Klasson L. Canback B. Naslund A.K. Eriksson A.S. Wernegreen J.J. Sandstrom J.P. Moran N.A. Andersson S.G.E. 50 million years of genomic stasis in endosymbiotic bacteria. Science. 2002;296:2376–2379. doi: 10.1126/science.1071278. [DOI] [PubMed] [Google Scholar]
- Tamas I. Wernegreen J.J. Nystedt B. Kauppinen S.N. Darby A.C. Gomez-Valero L. Lundin D. Poole A.M. Andersson S.G.E. Endosymbiont gene functions impaired and rescued by polymerase infidelity at poly(A) tracts. Proc Natl Acad Sci USA. 2008;105:14934–14939. doi: 10.1073/pnas.0806554105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thao M.L. Gullan P.J. Baumann P. Secondary (gamma-Proteobacteria) endosymbionts infect the primary (beta-Proteobacteria) endosymbionts of mealybugs multiple times and coevolve with their hosts. Appl Environ Microbiol. 2002;68:3190–3197. doi: 10.1128/AEM.68.7.3190-3197.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thao M.L. Moran N.A. Abbot P. Brennan E.B. Burckhardt D.H. Baumann P. Cospeciation of psyllids and their primary prokaryotic endosymbionts. Appl Environ Microbiol. 2000;66:2898–2905. doi: 10.1128/aem.66.7.2898-2905.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchida T. Koga R. Fukatsu T. Host plant specialization governed by facultative symbiont. Science. 2004;303:1989. doi: 10.1126/science.1094611. [DOI] [PubMed] [Google Scholar]
- van Ham R.C. Gonzalez-Candelas F. Silva F.J. Sabater B. Moya A. Latorre A. Postsymbiotic plasmid acquisition and evolution of the repA1-replicon in Buchnera aphidicola. Proc Natl Acad Sci USA. 2000;97:10855–10860. doi: 10.1073/pnas.180310197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall D.P. Fraser H.B. Hirsh A.E. Detecting putative orthologs. Bioinformatics. 2003;19:1710–1711. doi: 10.1093/bioinformatics/btg213. [DOI] [PubMed] [Google Scholar]
- Wen L. Ley R.E. Volchkov P.Y. Stranges P.B. Avanesyan L. Stonebraker A.C. Hu C. Wong F.S. Szot G.L. Bluestone J.A. Gordon J.I. Chervonsky A.V. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455:1109–1113. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler D.E. Buck N.A. Storage proteins in ants during development and colony founding. J Insect Physiol. 1995;41:885–894. [Google Scholar]
- Wilcox J.L. Dunbar H.E. Wolfinger R.D. Moran N.A. Consequences of reductive evolution for gene expression in an obligate endosymbiont. Mol Microbiol. 2003;48:1491–1500. doi: 10.1046/j.1365-2958.2003.03522.x. [DOI] [PubMed] [Google Scholar]
- Wilkinson T.L. Koga R. Fukatsu T. Role of host nutrition in symbiont regulation: impact of dietary nitrogen on proliferation of obligate and facultative bacterial endosymbionts of the pea aphid Acyrthosiphon pisum. Appl Environ Microbiol. 2007;73:1362–1366. doi: 10.1128/AEM.01211-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A.C. Dunbar H.E. Davis G.K. Hunter W.B. Stern D.L. Moran N.A. A dual-genome microarray for the pea aphid, Acyrthosiphon pisum, and its obligate bacterial symbiont, Buchnera aphidicola. BMC Genomics. 2006;7:50. doi: 10.1186/1471-2164-7-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolschin F. Hölldobler B. Gross R. Zientz E. Replication of the endosymbiotic bacterium Blochmannia floridanus is correlated with the developmental and reproductive stages of its ant host. Appl Environ Microbiol. 2004;70:4096–4102. doi: 10.1128/AEM.70.7.4096-4102.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahran H.H. Rhizobium-legume symbiosis and nitrogen fixation under severe conditions and in an arid climate. Microbiol Mol Biol Rev. 1999;63:968–989. doi: 10.1128/mmbr.63.4.968-989.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zientz E. Dandekar T. Gross R. Metabolic interdependence of obligate intracellular bacteria and their insect hosts. Microbiol Mol Biol Rev. 2004;68:745–770. doi: 10.1128/MMBR.68.4.745-770.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]