Abstract
Objective
To investigate the role of mitochondrial modifiers in the development of deafness associated with 12S rRNA A1555G mutation.
Methods
Four Chinese families with nonsyndromic and aminoglycoside-induced deafness were studied by clinical and genetic evaluation, molecular and biochemical analyses of mitochondrial DNA (mtDNA).
Results
These families exhibited high penetrance and expressivity of hearing impairment. Penetrances of hearing loss in WZD31, WZD32, WZD33, and WZD34 pedigrees ranged from 50 to 67% and from 39 to 50%, respectively, when aminoglycoside-induced hearing loss was included or excluded. Matrilineal relatives in these families developed hearing loss at the average of 14, 13, 16, and 15 years of age, respectively, when aminoglycoside-induced deafness was excluded. Mutational analysis of entire mtDNA in these families showed the homoplasmic A1555G mutation and distinct sets of variants belonging to haplogroup B5b1. Of these, the tRNAThr G15927A mutation locates at the fourth base in the anticodon stem (conventional position 42) of tRNAThr. A guanine (G42) at this position of tRNAThr is highly conserved from bacteria to human mitochondria. The lower levels and altered electrophoretic mobility of tRNAThr were observed in cells carrying A1555G and G15927A mutations or only G15927A mutation but not cells carrying only A1555G mutation. The abolished base pairing (28C-42G) of this tRNAThr by the G15927A mutation caused a failure in tRNA metabolism, worsening the mitochondrial dysfunctions altered by the A1555G mutation.
Conclusion
The G15927A mutation has a potential modifier role in increasing the penetrance and expressivity of the deafness-associated 12S rRNA A1555G mutation in those Chinese pedigrees.
Keywords: aminoglycoside, Chinese, haplogroup, hearing loss, mitochondrial, modifier, mutation, ototoxicity, penetrance, ribosomal RNA, tRNA, variant
Introduction
Hearing loss is a very common congenital disorder affecting one in 1000 newborns. Hearing loss can be caused by hereditary and environmental factors including ototoxic drugs such as aminoglycoside antibiotics. Mutations in mitochondrial DNA (mtDNA) have been found to be one of the most important causes of sensorineural hearing loss in some countries [1,2]. Of these, the C1494T mutation in the 12S rRNA has been associated with both aminoglycoside induced and nonsyndromic hearing loss in some Chinese and Spanish pedigrees [3–5]. The 12S rRNA A1555G mutation, however, has been associated with both aminoglycoside-induced and nonsyndromic hearing loss in many families worldwide [6–16]. The administration of aminoglycosides induces or worsens hearing loss in these participants carrying the A1555G or C1494T mutation. In the absence of aminoglycosides, matrilineal relatives within and among families carrying the A1555G or C1494T mutation exhibited a wide range of penetrance, severity and age-of-onset in hearing loss [3,9,10,12,14]. Functional characterization of cell lines derived from matrilineal relatives of a large Arab–Israeli family or one large Chinese family demonstrated that the A1555G or C1494T mutation conferred mild mitochondrial dysfunction and sensitivity to aminoglycosides [3,17–20]. These findings strongly indicated that the A1555G and C1494T mutations by themselves are insufficient to produce the deafness phenotype. Therefore, other modifier factors including aminoglycosides, nuclear and mitochondrial genetic modifiers modulate the expressivity and penetrance of hearing loss associated with the A1555G or C1494T mutation [19–22].
These modifier factors, however, remain poorly defined. To investigate the role of mitochondrial genetic factors in the development of hearing loss, a systematic and extended mutational screening of mtDNA has been initiated in several cohorts of hearing-impaired participants. In the earlier studies, we showed the highly variable penetrance and expressivity of hearing loss in 40 Han Chinese families carrying the A1555G mutation [12–15,22–25]. Our data suggested that the tRNAGlu A14693G, tRNAThr T15908C, tRNAArg T10454C, tRNASer(UCN) G7444A, and tRNACys G5821A variants may influence the phenotypic manifestation of the A1555G mutation [15,22,24]. In this study, we characterized another four Han Chinese pedigrees carrying the A1555G mutation with high penetrance and expressivity of hearing loss. A wide range of penetrance and severity of hearing loss, however, was observed in the matrilineal relatives within and among these families. To assess the contribution that mtDNA variants make toward the phenotypic expression of the A1555G mutation, we performed PCR amplification of fragments spanning entire mtDNA and subsequent DNA sequence analysis in the matrilineal relatives of those families. Interestingly, the G15927A mutation, which locates at the fourth base in the anticodon stem, corresponding to conventional position 42 of the tRNAThr, is implicated to influence the penetrance and expressivity of hearing loss associated with the A1555G mutation. Functional significance of the G15927A mutation was evaluated by examining for the steady-state levels and aminoacylation capacities of mitochondrial tRNAThr, tRNALys, tRNALeu(CUN), tRNAGly, and tRNASer(AGY) using lymphoblastoid cell lines derived from an affected matrilineal relative carrying both A1555G and G15927A mutations, from an affected Chinese participant carrying only A1555G mutation, a hearing normal individual carrying only the G15927A mutation and from a married-in-control individual lacking those mtDNA mutations. Furthermore, it was implied that TRMU and GJB2 mutations modulated the phenotypic manifestation of hearing loss associated with the A1555G mutation [21,26]. To further examine the role of the TRMU and GJB2 genes in the phenotypic expression of the A1555G mutation, we performed a mutational analysis of the TRMU and GJB2 genes in the hearing-impaired and normal hearing participants of these families.
Participants and methods
Participants and audiological examinations
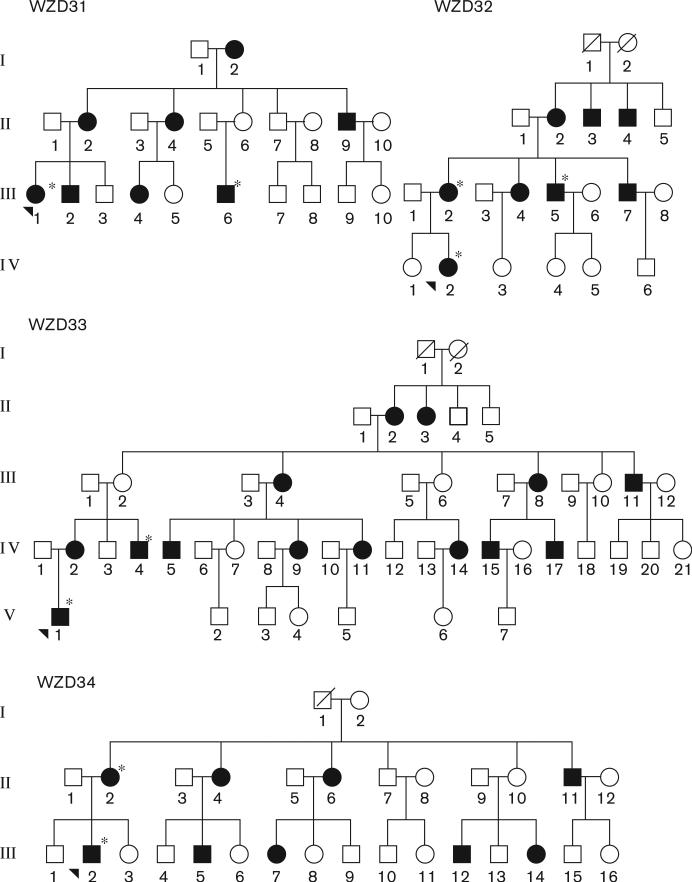
As the part of genetic screening program for hearing impairment, four Han Chinese families, as shown in Fig. 1, were ascertained through the Otology Clinic of Wenzhou Medical College. A comprehensive history and physical examination were performed to identify any syndromic findings, the history of the use of aminoglyco-sides, genetic factors related to the hearing impairment in members of these pedigrees. An age-appropriate audiological examination was performed and this examination included pure-tone audiometry and/or auditory brainstem response, immittance testing and distortion product otoacoustic emissions. The pure-tone audiometry was calculated from the sum of the audiometric thresholds at 500, 1000, 2000, 4000, and 8000 Hz. The severity of hearing impairment was classified into five grades: normal < 26 dB; mild = 26–40 dB; moderate = 41–70 dB; severe = 71–90 dB; and profound > 90 dB. Informed consent was obtained from participants before their participation in the study, in accordance with the Cincinnati Children's Hospital Medical Center Institutional Review Board and Ethics Committee of Wenzhou Medical College. The 262 control DNA used for screening for the presence of mtDNA variants were obtained from a panel of unaffected participants from Han Chinese ancestry.
Fig. 1.
Four Han Chinese pedigrees with aminoglycoside-induced and nonsyndromic hearing impairment. Hearing impaired individuals are indicated by filled symbols. Arrowhead denotes probands. Asterisks denote individuals who had a history of exposure to aminoglycosides.
Mutational analysis of mitochondrial genome
Genomic DNA was isolated from whole blood of participants using Puregene DNA Isolation kits (Gentra Systems, Minneapolis, Minnesota, USA). Participant's DNA fragments spanning the 12S rRNA gene were amplified by PCR using oligodeoxynucleotides corresponding to positions 618–635 and 1988–2007 [11]. For the detection of the A1555G mutation, the amplified segments were digested with a restriction enzyme BsmAI [11]. Equal amounts of various digested samples were then analyzed by electrophoresis through 1.5% agarose gel. The proportions of digested and undigested PCR product were determined by laser densitometry after ethidium bromide staining to determine if the A1555G mutation is in homoplasmy in these participants.
The entire mitochondrial genomes of four probands were PCR amplified in 24 overlapping fragments by use of sets of the light-strand and the heavy strand oligonucleotide primers, as described elsewhere [27]. Each fragment was purified and subsequently analyzed by direct sequencing in an ABI 3700 automated DNA sequencer using the Big Dye Terminator Cycle (Applied Bio-systems, Foster City, California, USA) sequencing reaction kit. The resultant sequence data were compared with the updated consensus Cambridge sequence (GenBank accession number: NC_001807) [28].
For the examination of the G15927A mutation, the first PCR segments (1122 bp) were amplified using genomic DNA as template and oligodeoxynucleotides corresponding to mtDNA at positions 14 856–15 978 to rule out the coamplification of possible nuclear pseudogenes [29]. Then, the second PCR product (138 bp) was amplified using the first PCR fragment as template and oligodeoxynucleotides corresponding to mtDNA at positions 15 822–15 960 and subsequently digested with restriction enzymes HpaII as the G15927A mutation abolished the site for this enzyme. Equal amounts of various digested samples were then analyzed by electrophoresis through 7% polyacrylamide gel. The proportions of digested and undigested PCR product were determined by laser densitometry after ethidium bromide staining to determine if the G15927A mutation is in homoplasmy in these participants. The allele frequency of G15927A variant was determined by PCR amplification using the genomic DNA derived from 262 Han Chinese controls and subsequent restriction enzyme analysis of PCR products, as described above.
Cell cultures
Lymphoblastoid cell lines were immortalized by transformation with the Epstein–Barr virus, as described elsewhere [30]. Cell lines derived from one affected individual (WZD32-III-7) carrying A1555G and G15927A mutations and a married-in-control (WZD32-III-1) in this Chinese family, an affected individual (WZD1-IV-2) from the other Chinese family carrying only A1555G mutation (Tang et al., 2007) [25] and a hearing normal individual (A4) carrying only the G15927A mutation were grown in RPMI 1640 medium (Invitrogen, Carlsbad, California, USA), supplemented with 10% fetal bovine serum.
Mitochondrial tRNA Northern analysis
Total mitochondrial RNA were obtained using TOTALLY RNA kit (Ambion, Austin, Texas, USA) from mitochondria isolated from lymphoblastoid cell lines (~4.0 × 108 cells), as described previously [21,31]. Five microgram of total mitochondrial RNA were electrophoresed through a 10% polyacrylamide/7 mol/l urea gel in Tris–borate–EDTA buffer (after heating the sample at 65°C for 10 min), and then electroblotted onto a positively charged nylon membrane (Roche, Basel, Switzerland) for the hybridization analysis with oligodeoxynucleotide probes. For the detection of tRNAThr, tRNALeu(CUN), tRNALys, tRNAGly, and tRNASer(AGY), the following nonradioactive digoxigenin (DIG)-labeled oligodeoxynucleotides specific for each RNA were used: 5′-CCTTGGAAAAAG GTTTTCATCTC C-3′ (tRNAThr); 5′-TACTTTTATTTGGAGTTGCACC-3′ [tRNALeu(CUN)]; 5′-TCACTGTAAAGAGGTGTTGGTTCTCTTAATCTT-3′ (tRNALys); 5′-TACTCTTTTTTGAATGTTGTC-3′ (tRNAGly); 5′-TTAGCAGTTCTTGTGAGCTTTCTC-3′ [tRNASer(AGY)] [28]. DIG-labeled oligodeoxynucleotides were generated by using DIG oligonucleotide Tailing kit (Roche). The hybridization was carried out as detailed elsewhere [31]. Quantification of density in each band was made as detailed previously [32].
Mitochondrial tRNA aminoacylation analysis
Total mitochondrial RNAs were isolated as above but under acid condition. The total mitochondrial RNA (2 mg) was electrophoresed at 41C through an acid 10% polyacrylamide/7 mol/l urea gel in a 0.1 mol/l sodium acetate (pH 5.0) to separate the charged and uncharged tRNA, as detailed elsewhere [33,34]. Then RNAs were electroblotted onto a positively charged membrane (Roche) and hybridized sequentially with the specific tRNA probes as above.
Genotyping analysis of TRMU gene
The genotyping for the TRMU A10S variant in subjects from three pedigrees was PCR-amplified for exon 1 and was followed by digestion the 467-pb segment with the restriction enzyme Bsp1286I. The forward and reverse primers for exon 1 are 5′-ACAGCGCAGAAGAAGAGCAGT-3′ and 5′-ACAACGCCACGACGGACG-3′, respectively. The Bsp1286I-digested products were analyzed on 1.5% agarose gels [21]. The PCR segments were subsequently analyzed by direct sequencing in an ABI 3700 automated DNA sequencer using the Big Dye Terminator Cycle sequencing reaction kit. The resultant sequence data were compared with the TRMU genomic sequence (GenBank accession number: AF448221) [21].
Mutational analysis of GJB2 gene
The DNA fragments spanning the entire coding region of GJB2 gene were amplified by PCR using the following oligodeoxynucleotides: forward-5′TATGACACTCCCCAGCACAG3′ and reverse-5′GGGCAATGCTTAAACTGGC3′. PCR amplification and subsequent sequencing analysis were performed as detailed elsewhere [11]. The results were compared with the wild-type GJB2 sequence (GenBank accession number: M86849) to identify the mutations.
Results
Clinical and genetic evaluations of four Chinese pedigrees
Four Han Chinese participants were diagnosed as aminoglycoside ototoxicity by the Otology Clinics at the Wenzhou Medical College. Mutational analysis of the 12S rRNA gene revealed that four participants harbored the A1555G mutation (data not shown). A comprehensive history and physical examination as well as audiological examination were performed to identify any syndromic findings, the history of the use of aminoglycosides, genetic factors related to the hearing impairment in all available members of these families. In fact, comprehensive family medical histories of those probands and other members of these Chinese families showed no other clinical abnormalities, including diabetes, muscular diseases, visual dysfunction, and neurological disorders. The restriction enzyme digestion and subsequent electrophoresis of available members in three pedigrees indicated that the A1555G mutation was indeed present in homoplasmy in matrilineal relatives but not other members of these families (data not shown).
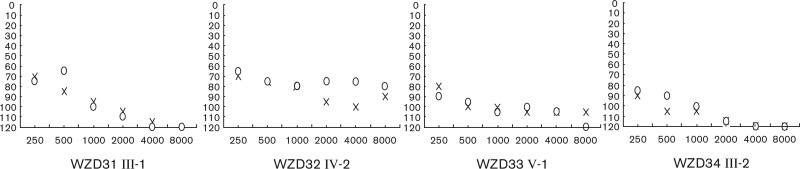
Of these, the proband (III-1) of WZD31 family received gentamycin for high fever at the age of 3 years. She exhibited bilateral hearing impairment 1 month later after the drug administration. As illustrated in Fig. 2, she had profound hearing impairment (98 dB at both right and left ears, with a slope-shaped pattern). Of other members in this family, III-6 exhibited profound hearing loss after the administration of gentamycin at the age of 2 years. Of other matrilineal relatives, six participants exhibited hearing deficit, ranging from severe (I-2, II-4, III-2), to moderate (II-2, II-9, III-4), whereas others had normal hearing.
Fig. 2.
Air conduction audiogram of four affected participants with the A1555G mutation. X, left ear; O, right ear.
In the family WZD32, the proband IV-2 was a 17-year-old girl. She was treated with gentamycin for high fever at the age of 2 years. As shown in Fig. 2, she had bilateral severe hearing loss (75 dB at right ear, 84 dB at left ear; a flat-shaped pattern). Two matrilineal relatives (III-2, III-5) had profound hearing loss after administration of aminoglycosides. Other four matrilineal relatives without exposure to aminoglycosides suffer from hearing impairment as the sole clinical symptom, ranged from profound (III-7), to severe (II-2), to moderate (II-3, II-4, III-4) to normal hearing.
In family WZD33, the proband V-1, as shown in Fig. 2, was evaluated as profound hearing loss (102 dB at right ear, 99 dB at left ear; a slope-shaped pattern) by the Otology Clinic of Wenzhou Medical College at the age of 12 years. He developed hearing loss at the age of 2 years after administration of gentamycin for high fever. Of other family members, IV-4 exhibited severe hearing loss after an administration with gentamycin at the age of 10 years. Other matrilineal relatives, who did not have a history of exposure to aminoglycosides, had hearing loss, varying from profound (III-4, IV-15, IV-17), to severe (II-2, II-3, III-8, III-11, IV-11), to moderate (IV-2, IV-5, IV-9) and to mild (IV-14), and to normal hearing of others.
In the family WZD34, the proband III-2 came to otology clinic of Wenzhou Medical College at the age of 15 years. He received gentamycin for high fever at the age of 4 years. As illustrated in Fig. 2, he suffered from profound hearing loss (105 dB at right ear, 109 dB at left ear, with a slope-shaped pattern). His mother (II-2) exhibited profound hearing loss after administration of gentamycin. Other matrilineal relatives, who were not exposed to aminoglycosides, had hearing loss, ranging from severe (II-4), to moderate (II-3, II-11, III-5, III-12), to mild (II-6, III-7, III-14), to normal hearing.
Mutational analysis of mitochondrial genome
To assess the role of mtDNA variants in the phenotypic expression of the A1555G mutation, we performed a PCR amplification of fragments spanning entire mitochondrial genome and subsequent DNA sequence analysis in four probands. In addition to the identical A1555G mutation, as shown in Table 1, these participants exhibited distinct sets of mtDNA polymorphism. We failed to detect the presence of the secondary deafness-associated tRNAGlu A14693G, tRNAThr T15908C, tRNAArg T10454C, tRNASer(UCN) G7444A, and tRNACys G5821A variants in these probands. Of other nucleotide changes, there are 30 variants in the noncoding region D-loop, five known variants in 12S rRNA gene, two variants in the 16S rRNA gene, one novel variant (G5744A) in the noncoding L-strand origin, one known variant (G15927A) in the tRNAThr gene, the previously identified CO2/tRNALys intergenic 9 bp deletion corresponding to mtDNA at positions 8271–8279 [14], 15 known and six novel silent variants in the protein encoding genes as well as 10 missense mutations (one novel and nine known) in the protein encoding genes (http://www.mitomap.org) [35]. These missense mutations are the G7269A (V456M) in the CO1 gene, the G8584A (A20T) and A8860G (T112A) in A6 gene, the A10398G (T114A) in the ND3 gene, the C12084T (S442F) in the ND4 gene, the A12361G (T8A) in the ND5 gene, and the C14766T (T7I), A15326G (T194A), A15662G (I306 V), and A15851G (I369 V) in the cyto b gene.
Table 1.
mtDNA variants in four Han Chinese families with hearing loss
| Gene | Position | Replacement | Conservation (H/B/M/X)a | CRS | WZD31 | WZD32 | WZD33 | WZD34 | Previously reportedb |
|---|---|---|---|---|---|---|---|---|---|
| D-Loop | 73 | A to G | A | G | G | G | G | Yes | |
| 103 | G to A | G | A | A | A | Yes | |||
| 131 | T to C | T | C | Yes | |||||
| 200 | A to G | A | G | Yes | |||||
| 204 | T to C | A | C | C | Yes | ||||
| 207 | G to A | G | A | Yes | |||||
| 263 | A to G | A | G | G | G | G | Yes | ||
| 297 | A to C | A | C | No | |||||
| 303 | C to CC | C | CC | Yes | |||||
| 310 | T to CTC/TC | T | TC | TC | CTC | TC | Yes | ||
| 481 | C to T | C | T | No | |||||
| 514 | C Del | C | C del | Yes | |||||
| 515 | A Del | A | A del | Yes | |||||
| 523 | A Del | A | A del | A del | A del | Yes | |||
| 524 | C Del | C | C del | C del | C del | Yes | |||
| 16111 | C to T | C | T | T | Yes | ||||
| 16140 | T to C | T | C | C | C | C | Yes | ||
| 16182 | A to C | A | C | Yes | |||||
| 16183 | A to C | A | C | C | C | C | Yes | ||
| 16189 | T to CC/C | T | CC | CC | C | CC | Yes | ||
| 16194 | A to C | A | C | Yes | |||||
| 16195 | T to A | T | A | Yes | |||||
| 16196 | G to TG | G | TG | Yes | |||||
| 16234 | C to T | C | T | T | Yes | ||||
| 16243 | T to C | T | C | C | C | C | Yes | ||
| 16291 | C to T | C | T | Yes | |||||
| 16344 | C to T | C | T | Yes | |||||
| 16355 | C to T | C | T | Yes | |||||
| 16463 | A to G | A | G | G | Yes | ||||
| 16519 | T to C | T | C | C | C | Yes | |||
| 12S rRNA | 709 | G to A | G/G/A/ | G | A | A | A | Yes | |
| 750 | A to G | A/G/G/ | A | G | Yes | ||||
| 961 | C to CC | T/A/T/A | C | CC | CC | Yes | |||
| 1438 | A to G | A/A/A/G | A | G | G | G | G | Yes | |
| 1555 | A to G | A/A/A/A | A | G | G | G | G | Yes | |
| 1598 | G to A | G/A/T/T | G | A | A | A | A | Yes | |
| 16S rRNA | 2706 | A to G | A/G/A/A | A | G | G | G | G | Yes |
| 3107 | C Del | C/T/T/T | C | C Del | C Del | C Del | Yes | ||
| ND1 | 4161 | C to T | C | T | No | ||||
| ND2 | 4769 | A to G | A | G | G | G | G | Yes | |
| 4895 | A to G | A | G | G | Yes | ||||
| 4985 | G to A | G | A | A | Yes | ||||
| NC4 | 5744 | G to A | G | A | No | ||||
| CO1 | 6446 | G to A | G | A | Yes | ||||
| 7028 | C to T | C | T | T | T | Yes | |||
| 7269 | G to A (Val to Met) | V/M/M/I | G | A | No | ||||
| NC7 | 8271-79 | 9-bp del | Del | Del | Del | Yes | |||
| ATPase6 | 8584 | G to A (Ala to Thr) | A/V/V/I | G | A | A | A | A | Yes |
| 8784 | A to G | A | G | Yes | |||||
| 8829 | C to T | C | T | T | T | Yes | |||
| 8860 | A to G (Thr to Ala) | T/A/A/T | A | G | G | G | Yes | ||
| CO3 | 9947 | G to A | G | A | Yes | ||||
| 9950 | T to C | T | C | C | C | Yes | |||
| ND3 | 10398 | A to G (Thr to Ala) | T/T/T/A | A | G | G | G | G | Yes |
| ND4 | 11101 | A to G | A | G | No | ||||
| 11335 | T to C | T | C | C | Yes | ||||
| 11437 | T to C | T | C | No | |||||
| 11719 | G to A | G | A | A | A | A | Yes | ||
| 11893 | A to G | A | G | Yes | |||||
| 12084 | C to T (Ser to Phe) | S/L/I/I | C | T | Yes | ||||
| ND5 | 12361 | A to G (Thr to Ala) | T/S/I/M | A | G | G | G | G | Yes |
| 14148 | A to G | A | G | No | |||||
| ND6 | 14560 | G to A | G | A | A | No | |||
| Cyt b | 14766 | T to C (Thr to Ile) | T/S/I/S | T | C | Yes | |||
| 15223 | C to T | C | T | T | T | T | No | ||
| 15326 | A to G (Thr to Ala) | T/M/I/I | A | G | G | G | G | Yes | |
| 15508 | C to T | C | T | T | T | T | Yes | ||
| 15662 | A to G (Ile to Val) | I/L/F/L | A | G | G | G | G | Yes | |
| 15734 | G to A | G | A | Yes | |||||
| 15850 | T to C | T | C | C | Yes | ||||
| 15851 | A to G (Ile to Val) | I/A/S/M | A | G | G | G | Yes | ||
| tRNA | 15927 | G to A | G/G/G/G | G | A | A | A | A | Yes |
CRS, consensus Cambridge sequence; mtDNA, mitochondrial DNA.
Conservation of amino acid for polypeptides or nucleotide for RNAs in human (H), bovine (B), mouse (M), and Xenopus laevis (X).
See the online mitochondrial genome database http://www.mitomap.org
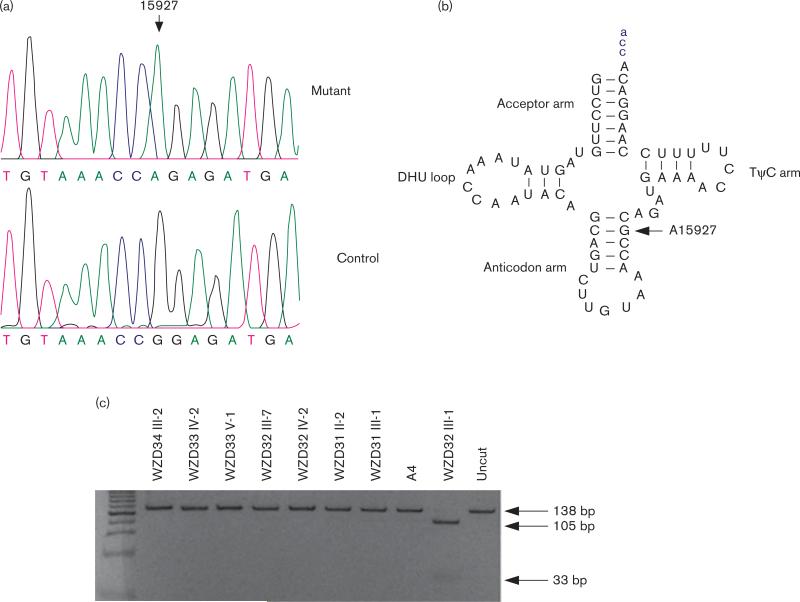
These variants in RNAs and polypeptides were further evaluated by phylogenetic analysis of these variants and sequences from other organisms including mouse [36], bovine [37], and Xenopus laevis [38]. Variants in the polypeptides showed no evolutionary conservation. Of RNA variants, the G-to-A transition at position 15927 (G15927A) in the tRNAThr gene of all probands (Fig. 3a) is of special interest. The G15927A mutation, as shown in Fig. 3b, locates at the fourth base in the anticodon stem, corresponding to conventional position 42 of the tRNAThr [39]. In fact, a guanine at this position is a highly conserved base in sequenced threonine tRNA from bacteria to human mitochondria [40]. It is anticipated that the G15927A mutation disrupts a very conservative base pairing (28C-42G) on the anticodon stem of this tRNAThr, thus causing a failure in tRNA metabolism. To determine whether the G15927A mutation is homoplasmic, the fragments spanning the tRNAThr gene were PCR amplified and subsequently digested with HpaII. As shown in Fig. 3c, there was no detectable wild-type DNA in all available matrilineal relatives, indicating that the G15927A variant was homoplasmic in these matrilineal relatives. In addition, allele frequency analysis showed that two of the 262 Chinese control participants carried this G15927A variant.
Fig. 3.
Identification and qualification of the G15927A mutation in the mitochondrial tRNAThr gene. (a) Partial sequence chromatograms of tRNAThr gene from an affected individual (WZD32-III-2) and a married-in-control (WZD32-III-1). An arrow indicates the location of the base changes at position 15927. (b) The location of the G15927A mutation in the mitochondrial tRNAThr. Cloverleaf structure of human mitochondrial tRNAThr is derived from the study of Florentz et al. [39]. Arrow indicates the position of the G15927A mutation. (c) Quantification of G15927A mutation in the tRNAThr gene of mutants and controls derived from the Chinese families. PCR products around the G15927A mutation were digested with HpaII and analyzed by electrophoresis in a 7% polyacrylamide gel stained with ethidium bromide.
Mitochondrial tRNA analysis
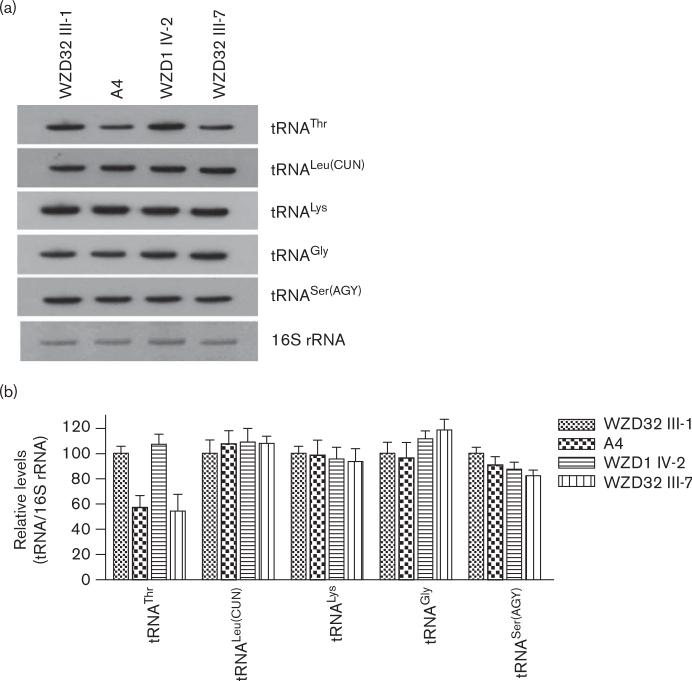
To examine whether the G15927A mutation affects the stability of the tRNAThr, we determined the steady state level of the tRNAThr determined by isolating total mitochondrial RNA from lymphoblastoid cell lines, separating them on a 10% polyacrylamide, 7-mol/l urea gel, electroblotting and hybridizing with a nonradioactive DIG-labeled oligodeoxynucleotide probe specific for tRNAThr. After the blots were stripped, the DIG-labeled probes, including tRNALeu(CUN), tRNALys, and tRNASer(AGY) as representatives of the whole H-strand transcription unit and tRNAGly derived from the L-strand transcription unit [32], were hybridized with the same blots for normalization purposes. As shown in Fig. 4a, the amount of tRNAThr in the mutant cell line derived from affected individual (WZD32-III-7) carrying A1555G and G15927A mutations and a hearing normal individual (A4) carrying only the G15927A mutation were markedly decreased, compared with those in the cell line derived from the married-in-control (WZD32-III-1) lacking those mtDNA mutations. For comparison, the average levels of tRNAThr, tRNALeu(CUN), tRNALys, tRNASer(AGY), and tRNAGly in various control or mutant cell lines were normalized to the average levels in the same cell line for 16S rRNA, respectively. As shown in Fig. 4b, the average levels of tRNAThr in the cell lines derived from WZD32-III-7 and A4 were 55 and 57% of the control after normalization to 16S rRNA, whereas the average levels of tRNALeu(CUN), tRNALys, tRNASer(AGY), and tRNAGly these two cell lines were comparable with those of the control after normalization to 16S rRNA. On the contrary, the levels of tRNAThr, tRNALeu(CUN), tRNALys, tRNASer(AGY), and tRNAGly in one cell line derived from an affected individual (WZD1-IV-2) carrying only A1555G mutation were comparable with those in the cell line derived from the married-in-control (WZD32-III-1).
Fig. 4.
Northern blot analysis of mitochondrial tRNA. (a) Equal amounts (5 μg) of total mitochondrial RNA from various cell lines were electrophoresed through a denaturing polyacrylamide gel, electroblotted and hybridized with digoxigenin (DIG)-labeled oligonucleotide probes specific for the tRNAThr. The blots were then stripped and rehybridized with DIG-labeled tRNALeu(CUN), tRNASer(AGY), tRNALys, and tRNAGly, respectively. (b) Quantification of mitochondrial tRNA levels. Average relative tRNAThr, content per cell, normalized to the average content per cell of 16S rRNA in cells derived from an affected individual (WZD32-III-7) carrying A1555G and G15927A mutations, a hearing normal individual (A4) carrying only the G15927A mutation, an individual (WZD1-IV-2) carrying only A1555G mutation, a married-in-control (WZD32-III-1) lacking those mitochondrial DNA mutations. The values for the latter are expressed as percentages of the average values for the control cell line (WZD32-III-1). The calculations were based on three independent determinations of each tRNA content in each cell line and three determinations of the content of reference RNA marker in each cell line.
Analysis of aminoacylation of mitochondrial tRNA
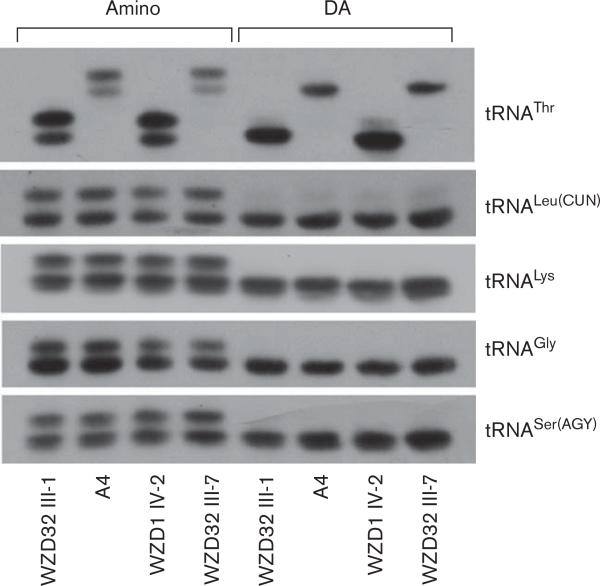
To test whether the G15927A mutation affects the aminoacylation of tRNAs, the aminoacylation capacity of tRNAThr, tRNALeu(CUN), tRNALys, tRNASer(AGY), and tRNAGly in wild-type and mutant cell lines were examined by the use of electrophoresis in an acid polyacrylamide/urea gel system to separate uncharged tRNA species from the corresponding charged tRNA [33], electroblotting and hybridizing with tRNA probes, as above. As shown in Fig. 5, the upper band represented the charged tRNA, and the lower band was uncharged tRNA. Electrophoretic patterns showed that either charged or uncharged tRNAThr in cell lines carrying the G15927A mutation migrated much slower than those of cell lines lacking this variant. However, there were no obvious differences in electrophoretic mobility of tRNALeu(CUN), tRNALys, tRNASer(AGY), and tRNAGly between the cell line carrying the G15927A variant and cell lines lacking this variant. To determine whether the lower band is uncharged tRNA and upper band is charged tRNA, a sample of mt tRNA, deacylated by heating 10 min at 60°C at pH 8.3 [33], was run in parallel. As shown in Fig. 3, the deacylated samples gave all cases a single hybridization band comigrating with lower band of the acid-purified tRNA samples. The proportions of aminoacylation of the tRNAs in cell line derived from the married-in-control individual (WZD32-III-1) lacking these mutations were 58, 42, 45, 45, and 39% in the tRNAThr, tRNALeu(CUN), tRNALys, tRNASer(AGY), and tRNAGly, respectively. The levels of other three cell lines were comparable with those of the married-in-control individual (WZD32-III-1).
Fig. 5.
In-vivo aminoacylation assays for mitochondrial tRNA. Equal amounts (2 μg) of total mitochondrial RNA purified from various cell lines under acid conditions were treated with electrophoresis at 4°C through an acid (pH 5.1) 10% polyacrylamide/7 mol/l urea gel, electroblotted onto a positively charged nylon membrane, and hybridized with digoxigenin (DIG)-labeled oligonucleotide probes specific for mitochondrial tRNAThr. Samples were also deacylated (DA) by heating 10 min at 60°C at pH 8.3. The blots were then stripped and rehybridized with DIG-labeled probes for tRNALeu(CUN), tRNASer(AGY), tRNALys, and tRNAGly, respectively.
Mutational analysis of TRMU gene
Our earlier study showed that the TRMU A10S mutation modulated the phenotypic manifestation of the A1555G mutation in the Israeli/European pedigrees [21]. To assess whether the TRMU A10S variant also plays a role in phenotypic expression of the A1555G mutation in these Chinese families, we carried out a mutational screening of exon 1 in TRMU gene in three affected matrilineal relatives of each pedigree who did not have a history of exposure to aminoglycosides. We failed to detect any variant in TRMU exon 1 in these affected matrilineal relatives of these Chinese pedigrees.
Mutational analysis of GJB2 gene
To examine the role of GJB2 gene in phenotypic expression of the A1555G mutation, we performed the mutational screening of GJB2 gene in three affected matrilineal relatives of each pedigree who did not have a history of exposure to aminoglycosides. None of variants in GJB2 gene was found in these affected matrilineal relatives of these Chinese pedigree. Indeed, the absence of variant in the GJB2 gene in those participants with hearing impairment indicates that the GJB2 gene may not be a modifier of the phenotypic effects of the A1555G mutation in those participants.
Discussion
In this study, we investigated the role of modifiers in the phenotypic manifestation of the A1555G mutation in four Chinese pedigrees with aminoglycoside-induced and nonsyndromic hearing loss. Hearing loss as a sole clinical phenotype was only present in the maternal lineage of those pedigrees. These Chinese families exhibit much higher penetrance and expressivities of hearing loss than in other 27 Chinese pedigrees carrying the A1555G mutation [14,23,25]. As shown in Table 2, the penetrances of hearing loss (affected matrilineal relatives/total matrilineal relatives) in WZD31, WZD32, WZD33, and WZD34 pedigrees were 66.7, 66.7, 51.9, and 50%, respectively, when aminoglycoside-induced deafness was included. When the effect of aminoglycosides was excluded, the penetrances of hearing loss in these pedigrees were 50, 40.7, 44.4, and 39%, respectively. The penetrances of hearing loss in these Chinese pedigrees are comparable with the previously identified five Chinese pedigrees with a high penetrance of hearing loss, with the average of 54 and 31.7%, respectively, when aminoglycoside-induced deafness was included or excluded [15,22,24]. In contrast, the average penetrances of hearing loss in the other 27 pedigrees carrying the A1555G mutation were 9.3 and 1.7%, respectively, when aminoglycoside-induced deafness was included or excluded [14,23,25]. Furthermore, as shown in Table 2, when aminoglycoside-induced deafness was excluded, matrilineal relatives in these four Chinese families developed hearing loss at the average of 14, 13, 16, and 15 years of age, respectively, whereas the average age-of-onset for hearing loss were from 18 to 21 years from matrilineal relatives of 11 Chinese pedigrees carrying the A1555G mutation but lacking secondary mtDNA mutations, respectively [14,25]. Thus, matrilineal relatives in these four families had earlier age-onset of hearing loss than those in the other 11 Chinese families carrying the A1555G mutation. To assess whether the differences in penetrance and age-at-onset of hearing loss without aminoglycosides differ based on the presence and absence of additional mtDNA variant, a statistical analysis was performed by the unpaired, two-tailed Student's t-test contained in Microsoft Excel. The penetrances and age-at-onset of hearing loss among Chinese pedigrees carrying the additional mtDNA variants showed significantly higher than those pedigrees lacking significant mtDNA variants, when aminoglycoside-induced deafness was excluded (P < 0.0001 and P = 0.001).
Table 2.
Summary of clinical and molecular data for 21 Chinese families carrying the A1555G mutation
| Pedigree | Number of matrilineal relatives | Penetrance (including the use of drugs) (%)a | Penetrance (excluding the use of drugs) (%) | Average age-at-onset (excluding the use of drugs) | Second mtDNA mutations | mtDNA haplogroup |
|---|---|---|---|---|---|---|
| WZD31 | 12 | 66.7 | 50 | 14 | tRNAThr G15927A | B5b1 |
| WZD32 | 12 | 66.7 | 41.7 | 13 | tRNAThr G15927A | B5b1 |
| WZD33 | 27 | 51.9 | 44.4 | 16 | tRNAThr G15927A | B5b1 |
| WZD34 | 19 | 50 | 39 | 15 | tRNAThr G15927A | B5b1 |
| WZD1 [25] | 17 | 5.9 | 0 | None | B4C1C1 | |
| WZD2 | 21 | 9.5 | 4.7 | 19 | None | D4a |
| WZD3 | 16 | 12.5 | 0 | None | D5a2 | |
| WZD4 | 24 | 29.2 | 16.7 | 20 | None | F1a1 |
| WZD5 | 31 | 3.2 | 0 | None | D52b | |
| WZD6 | 8 | 25 | 12.5 | 19 | None | D4b2b |
| WZD7 | 30 | 10 | 3.3 | 21 | None | D5a2 |
| BJ101 [14] | 14 | 5 | 0 | None | F3 | |
| BJ102 | 13 | 13 | 8 | 18 | None | N9a1 |
| BJ103 | 20 | 5 | 0 | None | D4a | |
| BJ104 | 11 | 4 | 0 | None | D4b2b | |
| BJ105 [15] | 15 | 67 | 45 | tRNACys G5821A | F3 | |
| BJ106 [24] | 18 | 33 | 33 | None | M7b | |
| BJ107 [22] | 34 | 35 | 24 | tRNAThr T15908C | N | |
| BJ108 | 16 | 63 | 38 | tRNAGlu A14693G | F | |
| BJ109 | 9 | 67 | 44 | tRNAArg T10454C | M | |
| BJ110 [14] | 17 | 59 | 5.9 | CO1/tRNASer(UCN) G7444A | D4a |
mtDNA, mitochondrial DNA.
Affected matrilineal relatives/total matrilineal relatives.
The phenotypic variability of matrilineal relatives within and among these Chinese families indicated the involvement of modifier factors such as aminoglycosides, nuclear and mitochondrial genetic modifiers in the phenotypic manifestation of the A1555G mutation. Eight matrilineal relatives of these pedigrees exhibited severe or profound hearing loss after an administration with aminoglycosides, demonstrating the modifier role of aminoglycosides in deafness expression. The absence of TRMU A10S variant and GJB2 mutation ruled out their involvement in the phenotypic expression of the A1555G mutation in these Chinese families, similar to other Chinese families carrying the A1555G mutation [22,25]. Hence, other nuclear modifier genes may contribute to the phenotypic variability in these Chinese families. Furthermore, the secondary mtDNA mutations may act as modifiers influencing the phenotypic expression of the primary mtDNA mutations. The haplogroup J specific variants T4216C and G13708A may increase the penetrance of vision loss associated ND4 G11778A mutation [41] or hearing loss associated with tRNASer(UCN) A7445G mutation [32], whereas the haplogroup L1b specific variants ND1 T3308C and tRNAAla T5655C likely caused higher penetrance of deafness in an African pedigree than Japanese and French families carrying the T7511C mutation [31,42]. Furthermore, variants tRNAGlu A14693G, tRNAThr T15908C, tRNAArg T10454C, tRNASer(UCN) G7444A, and tRNACys G5821A may contribute to higher penetrance of hearing loss in five Chinese pedigrees carrying the A1555G mutation [15,22,24].
Mitochondrial genomes in these four Chinese pedigrees harbored the identical A1555G and G15927A mutations as well as distinct sets of mtDNA variants, belonging to Eastern Asian haplogroup B5b1 [43]. Of mtDNA variants, the 961insC mutation may influence the phenotypic manifestation of hearing loss in pedigrees WZD33 and WZD34, as in the case of a Chinese pedigree carrying both A1555G and 961insC mutations [12]. The homo-plasmic G15927A variant (conventional position 42 of tRNA), however, occurred at a highly evolutionarily conserved nucleotide of tRNAThr [39,40]. In fact, the G15927A variant disrupted a very conservative base pairing (28C-42G) on the anticodon stem of this tRNAThr. An abolished base pair at the same position in tRNAIle by the A4300G mutation associated with hypertrophic cardiomyopathy altered this tRNA metabolism [44]. Thus, the alteration of the tertiary structure of the tRNAThr by the G15927A mutation may lead to a failure in this tRNA metabolism. Approximately 45 and 43% reductions in the level of tRNAThr were observed in cells carrying both A1555G and G15927A mutations and cells carrying only the G15927A mutation, respectively, as compared with control cells lacking those mutations, while there was no significant reduction in the level of tRNAThr in cells carrying only A1555G mutation. This level of total tRNAThr in mutant cells is, however, above a proposed threshold, which is 30% of the control level of tRNA, to support a normal rate of mitochondrial translation [31,32]. Furthermore, an altered electrophoretic mobility of tRNAThr was observed in cell lines carrying the G15927A mutation, as compared with those lacking this variant. In contrast, the G15927A mutation did not affect the aminoacylation efficiency of tRNAThr. The lower level and altered electrophoretic mobility of tRNAThr in cells carrying the G15927A mutation most probably results from the alteration of the tertiary structure of this tRNA by the G15927A mutation. The homoplasmic form, mild biochemical defect and presence of two of 262 Han Chinese controls indicated that the G15927A mutation itself is insufficient to produce a clinical phenotype. A failure in mitochondrial tRNA metabolism, caused by the G15927A mutation, may impair mitochondrial protein synthesis, thereby worsening mitochondrial dysfunctions altered by the A1555G mutation. Therefore, the G15927A mutation may have a potential modifier role in increasing the penetrance and expressivity of the deafness-associated 12S rRNA A1555G mutation in these Chinese pedigrees.
Acknowledgements
This study was supported by Public Health Service Grants RO1DC05230 and RO1DC07696 from the National Institute on Deafness and Other Communication Disorders, and grants from National Basic Research Priorities Program of China 2004CCA02200, Ministry of Public Heath of Zhejiang Province 2006A100 and Ministry of Science and Technology of Zhejiang Province 2007G50G2090026 and 2007C13021 to M.X.G.
References
- 1.Fischel-Ghodsian N. Genetic factors in aminoglycoside toxicity. Pharmacogenomics. 2005;6:27–36. doi: 10.1517/14622416.6.1.27. [DOI] [PubMed] [Google Scholar]
- 2.Guan MX. Prevalence of mitochondrial 12S rRNA mutations associated with aminoglycoside ototoxicity. Voltra Rev. 2005;105:211–237. doi: 10.1016/j.mito.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 3.Zhao H, Li R, Wang Q, Yan Q, Deng JH, Han D, et al. Maternally inherited aminoglycoside-induced and non-syndromic deafness is associated with the novel C1494T mutation in the mitochondrial 12S rRNA gene in a large Chinese family. Am J Hum Genet. 2004;74:139–152. doi: 10.1086/381133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen J, Yang L, Yang A, Zhu Y, Zhao J, Sun D, et al. Maternally inherited aminoglycoside-induced and nonsyndromic hearing loss is associated with the 12S rRNA C1494T mutation in three Han Chinese pedigrees. Gene. 2007;401:4–11. doi: 10.1016/j.gene.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rodriguez-Ballesteros M, Olarte M, Aguirre LA, Galan F, Galan R, Vallejo LA, et al. Molecular and clinical characterisation of three Spanish families with maternally inherited non-syndromic hearing loss caused by the 1494C- > T mutation in the mitochondrial 12S rRNA gene. J Med Genet. 2006;43:e54. doi: 10.1136/jmg.2006.042440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prezant TR, Agapian JV, Bohlman MC, Bu X, Oztas S, Qiu WQ, et al. Mitochondrial ribosomal RNA mutation associated with both antibiotic-induced and non-syndromic deafness. Nat Genet. 1993;4:289–294. doi: 10.1038/ng0793-289. [DOI] [PubMed] [Google Scholar]
- 7.Matthijs G, Claes S, Longo-Bbenza B, Cassiman J-J. Non-syndromic deafness associated with a mutation and a polymorphism in the mitochondrial 12S ribosomal RNA gene in a large Zairean pedigree. Eur J Hum Genet. 1996;4:46–51. doi: 10.1159/000472169. [DOI] [PubMed] [Google Scholar]
- 8.Usami SI, Abe S, Kasai M, Shinkawa H, Moeller B, Kenyon JB, Kimberling WJ. Genetic and clinical features of sensorineural hearing loss associated with the 1555 mitochondrial mutation. Laryngoscope. 1997;107:483–490. doi: 10.1097/00005537-199704000-00011. [DOI] [PubMed] [Google Scholar]
- 9.Estivill X, Govea N, Barcelo E, Badenas C, Romero E, Moral L, et al. Familial progressive sensorineural deafness is mainly due to the mtDNA A1555G mutation and is enhanced by treatment with aminoglycosides. Am J Hum Genet. 1998;62:27–35. doi: 10.1086/301676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Del Castillo FJ, Rodriguez-Ballesteros M, Martin Y, Arellano B, Gallo-Teran J, Morales-Angulo C, et al. Heteroplasmy for the 1555A > G mutation in the mitochondrial 12S rRNA gene in six Spanish families with non-syndromic hearing loss. J Med Genet. 2003;40:632–636. doi: 10.1136/jmg.40.8.632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li R, Greinwald JH, Yang L, Choo DI, Wenstrup RJ, Guan MX. Molecular analysis of mitochondrial 12S rRNA and tRNASer(UCN) genes in paediatric subjects with nonsyndromic hearing loss. J Med Genet. 2004;41:615–620. doi: 10.1136/jmg.2004.020230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li R, Xing G, Yan M, Cao X, Liu XZ, Bu X, Guan MX. Cosegregation of C-insertion at position 961 with A1555G mutation of mitochondrial 12S rRNA gene in a large Chinese family with maternally inherited hearing loss. Am J Med Genet. 2004;124A:113–117. doi: 10.1002/ajmg.a.20305. [DOI] [PubMed] [Google Scholar]
- 13.Li Z, Li R, Chen J, Liao Z, Zhu Y, Qian Y, et al. Mutational analysis of the mitochondrial 12S rRNA gene in Chinese pediatric subjects with aminoglycoside induced and non-syndromic hearing loss. Hum Genet. 2005;117:9–15. doi: 10.1007/s00439-005-1276-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Young WY, Zhao L, Qian Y, Wang Q, Li N, Greinwald JH, Jr, Guan MX. Extremely low penetrance of hearing loss in four Chinese families with the mitochondrial 12S rRNA A1555G mutation. Biochem Biophys Res Commun. 2005;328:1244–1251. doi: 10.1016/j.bbrc.2005.01.085. [DOI] [PubMed] [Google Scholar]
- 15.Zhao L, Wang Q, Qian Y, Li R, Cao J, Hart LC, et al. Clinical evaluation and mitochondrial genome sequence analysis of two Chinese families with aminoglycoside-induced and nonsyndromic hearing loss. Biochem Biophys Res Commun. 2005;336:967–973. doi: 10.1016/j.bbrc.2005.08.199. [DOI] [PubMed] [Google Scholar]
- 16.Jacobs HT, Hutchin TP, Käppi T, Gillies G, Minkkinen K, Walker J, et al. Mitochondrial DNA mutations in patients with postlingual, nonsyndromic hearing impairment. Eur J Hum Genet. 2005;13:26–33. doi: 10.1038/sj.ejhg.5201250. [DOI] [PubMed] [Google Scholar]
- 17.Guan MX, Fischel-Ghodsian N, Attardi G. Biochemical evidence for nuclear gene involvement in phenotype of non-syndromic deafness associated with mitochondrial 12S rRNA mutation. Hum Mol Genet. 1996;5:963–997. doi: 10.1093/hmg/5.7.963. [DOI] [PubMed] [Google Scholar]
- 18.Guan MX, Fischel-Ghodsian N, Attardi G. A biochemical basis for the inherited susceptibility to aminoglycoside ototoxicity. Hum Mol Genet. 2000;9:1787–1793. doi: 10.1093/hmg/9.12.1787. [DOI] [PubMed] [Google Scholar]
- 19.Guan MX, Fischel-Ghodsian N, Attardi G. Nuclear background determines biochemical phenotype in the deafness-associated mitochondrial 12S rRNA mutation. Hum Mol Genet. 2001;10:573–580. doi: 10.1093/hmg/10.6.573. [DOI] [PubMed] [Google Scholar]
- 20.Zhao H, Young WY, Yan Q, Li R, Cao J, Wang Q, et al. Functional characterization of the mitochondrial 12S rRNA C1494T mutation associated with aminoglycoside-induced and nonsyndromic hearing loss. Nucl Acids Res. 2005;33:1132–1139. doi: 10.1093/nar/gki262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guan MX, Yan Q, Li X, Bykhovskaya Y, Gallo-Teran J, Hajek P, et al. Mutation in TRMU related to transfer RNA modification modulates the phenotypic expression of the deafness-associated mitochondrial 12S ribosomal RNA mutations. Am J Hum Genet. 2006;79:291–302. doi: 10.1086/506389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Young WY, Zhao L, Qian Y, Li R, Chen J, Yuan H, et al. Variants in mitochondrial tRNAGlu, tRNAArg and tRNAThr may influence the phenotypic manifestation of deafness-associated 12S rRNA A1555G mutation in three Chinese families with hearing loss. Am J Med Genet. 2006;140:2188–2197. doi: 10.1002/ajmg.a.31434. [DOI] [PubMed] [Google Scholar]
- 23.Dai P, Liu X, Han D, Qian Y, Huang D, Yuan H, et al. Extremely low penetrance of deafness associated with the mitochondrial 12S rRNA mutation in 16 Chinese families: implication for early detection and prevention of deafness. Biochem Biophys Res Commun. 2006;340:194–199. doi: 10.1016/j.bbrc.2005.11.156. [DOI] [PubMed] [Google Scholar]
- 24.Yuan H, Qian Y, Xu Y, Cao J, Bai L, Shen W, et al. Cosegregation of the G7444A mutation in the mitochondrial COI/tRNASer(UCN) genes with the 12S rRNA A1555G mutation in a Chinese family with aminoglycoside-induced and non-syndromic hearing loss. Am J Med Genet. 2005;138A:133–140. doi: 10.1002/ajmg.a.30952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang X, Yang L, Zhu Y, Wang J, Qian Y, Wang X, et al. Very low penetrance of hearing loss in seven Han Chinese pedigrees carrying the deafness-associated 12S rRNA A1555G mutation. Gene. 2007;393:11–19. doi: 10.1016/j.gene.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 26.Abe S, Kelley PM, Kimberling WJ, Usami SI. Connexin 26 gene (GJB2) mutation modulates the severity of hearing loss associated with the 1555A– > G mitochondrial mutation. Am J Med Genet. 2001;103:334–338. [PubMed] [Google Scholar]
- 27.Rieder MJ, Taylor SL, Tobe VO, Nickerson DA. Automating the identification of DNA variations using quality-based fluorescence re-sequencing: analysis of the human mitochondrial genome. Nucl Acids Res. 1998;26:967–973. doi: 10.1093/nar/26.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 1999;23:147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- 29.Woischnik M, Moraes CT. Pattern of organization of human mitochondrial pseudogenes in the nuclear genome. Genome Res. 2002;12:885–893. doi: 10.1101/gr.227202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller G, Lipman M. Release of infectious Epstein–Barr virus by transformed marmoset leukocytes. Proc Natl Acad Sci U S A. 1973;70:190–194. doi: 10.1073/pnas.70.1.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li X, Fischel-Ghodsian N, Schwartz F, Yan Q, Friedman RA, Guan MX. Biochemical characterization of the mitochondrial tRNASer(UCN) T7511C mutation associated with nonsyndromic deafness. Nucl Acids Res. 2004;32:867–877. doi: 10.1093/nar/gkh226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guan MX, Enriquez JA, Fischel-Ghodsian N, Puranam R, Lin CP, Marion MA, Attardi G. The Deafness-associated mtDNA 7445 mutation, which affects tRNASer(UCN) precursor processing, has long-range effects on NADH dehydrogenase ND6 subunit gene expression. Mol Cell Biol. 1998;18:5868–5879. doi: 10.1128/mcb.18.10.5868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Enríquez JA, Attardi G. Analysis of aminoacylation of human mitochondrial tRNAs. Methods Enzymol. 1996;264:183–196. doi: 10.1016/s0076-6879(96)64019-1. [DOI] [PubMed] [Google Scholar]
- 34.Wang X, Yan Q, Guan MX. Deletion of the MTO2 gene related to tRNA modification causes a failure in mitochondrial RNA metabolism in the yeast Saccharomyces cerevisiae. FEBS Lett. 2007;581:4228–4234. doi: 10.1016/j.febslet.2007.07.067. [DOI] [PubMed] [Google Scholar]
- 35.Brandon MC, Lott MT, Nguyen KC, Spolim S, Navathe SB, Baldi P, Wallace DC. MITOMAP: a human mitochondrial genome database – 2004 update. Nucl Acids Res. 2005;33:D611–D613. doi: 10.1093/nar/gki079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bibb MJ, Van Etten RA, Wright CT, Walberg MW, Clayton DA. Sequence and gene organization of mouse mitochondrial DNA. Cell. 1981;26:167–180. doi: 10.1016/0092-8674(81)90300-7. [DOI] [PubMed] [Google Scholar]
- 37.Gadaleta G, Pepe G, De Candia G, Quagliariello C, Sbisa E, Saccone C. The complete nucleotide sequence of the Rattus norvegicus mitochondrial genome: cryptic signals revealed by comparative analysis between vertebrates. J Mol Evol. 1989;28:497–516. doi: 10.1007/BF02602930. [DOI] [PubMed] [Google Scholar]
- 38.Roe A, Ma DP, Wilson RK, Wong JF. The complete nucleotide sequence of the Xenopus laevis mitochondrial genome. J Biol Chem. 1985;260:9759–9774. [PubMed] [Google Scholar]
- 39.Florentz C, Sohm B, Tryoen-Toth P, Putz J, Sissler M. Human mitochondrial tRNAs in health and disease. Cell Mol Life Sci. 2003;60:1356–1375. doi: 10.1007/s00018-003-2343-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sprinzl M, Horn C, Brown M, Ioudovitch Steinberg S. Compilation of tRNA sequences and sequences of tRNA genes. Nucl Acids Res. 1998;26:148–153. doi: 10.1093/nar/26.1.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Torroni A, Petrozzi M, D'Urbano L, Sellitto D, Zeviani M, Carrara F, et al. Haplotype and phylogenetic analyses suggest that one European-specific mtDNA background plays a role in the expression of Leber hereditary optic neuropathy by increasing the penetrance of the primary mutations 11778 and 14484. Am J Hum Genet. 1997;60:1107–1121. [PMC free article] [PubMed] [Google Scholar]
- 42.Li R, Ishikawa K, Deng JH, Yang L, Tamagawa Y, Bai Y, et al. Maternally inherited nonsyndromic hearing loss is associated with the mitochondrial tRNASer(UCN) 7511C mutation in a Japanese family. Biochem Biophys Res Commun. 2005;328:32–37. doi: 10.1016/j.bbrc.2004.12.140. [DOI] [PubMed] [Google Scholar]
- 43.Tanaka M, Cabrera VM, González AM, Larruga JM, Takeyasu T, Fuku N, et al. Mitochondrial genome variation in eastern Asia and the peopling of Japan. Genome Res. 2004;14:1832–1850. doi: 10.1101/gr.2286304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Taylor RW, Giordano C, Davidson MM, d'Amati G, Bain H, Hayes CM, et al. A homoplasmic mitochondrial transfer ribonucleic acid mutation as a cause of maternally inherited hypertrophic cardiomyopathy. J Am Col Cardiol. 2003;41:1786–1796. doi: 10.1016/s0735-1097(03)00300-0. [DOI] [PubMed] [Google Scholar]