Abstract
Some aminoacyl-tRNA synthetases (AARSs) employ an editing mechanism to ensure the fidelity of protein synthesis. Leucyl- (LeuRS), isoleucyl- (IleRS) and valyl-tRNA synthetase (ValRS), share a common insertion, called the CP1 domain, which is responsible for clearing misformed products. This discrete domain is connected to the main body of the enzyme via two β-strand tethers. The CP1 hydrolytic editing active site is located ~30 Å from the aminoacylation active site in the canonical core of the enzyme requiring translocation of mischarged amino acids for editing. An ensemble of crystal and co-crystal structures for LeuRS, IleRS, and ValRS suggest that the CP1 domain rotates via its flexible β-strand linkers relative to the main body along various steps in the enzyme’s reaction pathway. Computational analysis suggested that the end of the N-terminal β-strand acted as a hinge. We hypothesized that a molecular hinge could specifically direct movement of the CP1 domain relative to the main body. We introduced a series of mutations in both β-strands in attempts to hinder movement and alter fidelity of LeuRS. Our results have identified specific residues within the β-strand tethers that selectively impact enzyme activity, supporting that β-strand orientation is crucial for LeuRS canonical core and CP1 domain functions.
INTRODUCTION
Aminoacyl-tRNA synthetases (AARSs) are ancient enzymes that are responsible for covalently linking specific amino acids to their cognate tRNAs (1). The family of AARSs “charge” tRNA by covalently linking it to an amino acid via a two-step aminoacylation reaction. The enzyme binds adenosine triphosphate (ATP) and amino acid (AA) to form an aminoacyl-adenylate intermediate with release of pyrophosphate (PPi). The activated amino acid is then transferred to the 2′ or 3′-ribose hydroxyl of the tRNA’s terminal adenosine to form an aminoacyl-tRNA Charged tRNA is delivered to the ribosome where the attached amino acid is incorporated into protein.
The AARSs have evolved different strategies to achieve high fidelity and prevent the misincorporation of an incorrect amino acid during translation (2, 3). Certain amino acids, such as cysteine and tyrosine, have unique side chains that are readily recognized at the molecular level (4). However, isosteric sets of amino acids that differ by a single methyl group (i.e., isoleucine and valine or alanine and glycine) are not easily distinguished (5). Thus, ten of the AARSs have evolved amino acid editing mechanisms to clear mistakes (2, 3). A noncognate amino acid mischarged to the wrong tRNA isoacceptor can be cleared via a post-transfer editing mechanism which cleaves the amino acid from the tRNA. Alternatively, pre-transfer editing hydrolyzes misactivated adenylate intermediates (6, 7).
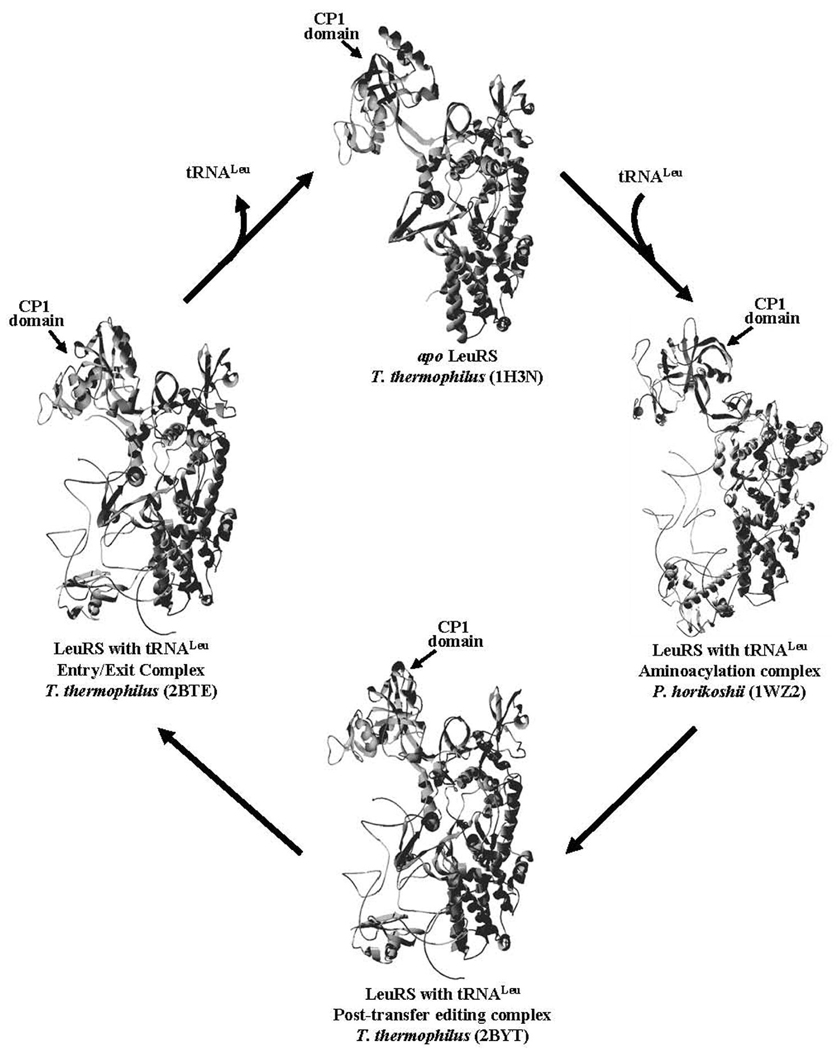
The class I LeuRS, IleRS, and ValRS have highly homologous aminoacylation and editing domains (8). Within their canonical Rossmann fold cores where the aminoacylation active site resides, these enzymes have a ~190 residue insert that folds into a large independent domain (9–11), termed CP1 (connective polypeptide 1)(12). The CP1 domain is tethered to the main body of the enzyme via two flexible β-strand linkers (Figure 1). This insert houses the enzyme’s editing active site, which is responsible for clearing mischarged amino acids.
Figure 1.
Tertiary Structures of LeuRS. The enzyme cycle of LeuRS shows the apo X-ray crystal structure (top, PDB code: 1H3N), aminoacylation co-crystal structure complex with tRNA (right, PDB code: 1WZ2), and the post-transfer editing co-crystal complex (bottom, PDB code: 2BYT). The exit complex, which has also been proposed to be an entry complex for tRNA binding (29), is shown at the left (PDB code: 2BTE). The LeuRS protein structure is displayed as a ribbon, while the bound tRNA is indicated as a distinct line. The CP1 domain is oriented at the top left of each structure as indicated by marked arrow.
Over 25 crystal structures of the homologous LeuRS, IleRS, and ValRS display different orientations that the CP1 domain adopts to accommodate aminoacylation and post-transfer editing complexes as well as intermediate states. These multiple orientations of the CP1 domain during various stages of the enzyme’s cycle would require flexibility of the β-strands linking the CP1 domain to the canonical core. The co-crystal structure of the aminoacylation complex of P. horikoshii LeuRS with tRNALeu showed that the CP1 domain swings ~20° relative to the canonical core of the enzyme to prevent a clash with the bound 5′ terminus of tRNA (13). Subsequent to aminoacylation, the 3′ end of the tRNA is translocated ~30 Å from the synthetic active site to the amino acid editing site for proofreading, requiring the editing domain to rotate by ~35° compared to the apo state of T. thermophilus LeuRS (14). The exit complex shows the 3′ end of the tRNA displaced from the editing active site primed for release from the enzyme.
We hypothesize that these rotations and orientations of the CP1 domain at various stages of the reaction pathway are dependant on the dynamic connecting β-strand linkers. Flexibility of the β-strands would also be required for translocation of the tRNA from the aminoacylation to editing active site. In order to identify specific sites within the β-strand linkers that are important to the different steps of the enzyme’s reaction cycle, we computationally investigated each of the linker peptides for specific hinge sites. We also created a series of two-amino acid deletions and tested each for alterations in aminoacylation, mischarging and amino acid editing. We found that different sites within the C- and N-terminal β-strands preferentially impact aminoacylation over amino acid editing and vice versa.
EXPERIMENTAL PROCEDURES
Mutagenesis
The polymerase chain reaction (PCR) was carried out for deletion mutations and point mutations. Plasmid p15ec3-1 (15), which encodes the gene for wild type E. coli LeuRS, was used as template. The 50 µL PCR reaction contained 200 ng plasmid template, 125 ng of each forward and reverse primer (Integrated DNA Technologies, Coralville, IA), 0.2 mM dNTPs and 0.05 U Pfu DNA polymerase (Stratagene, La Jolla, CA) in commercially prepared buffer. The PCR reaction mixture was restriction digested with 0.4 U Dpn I (Promega, Madison, WI) for 6 h at 37 °C and then used to transform E. coli DH5α (Stratagene). The mutated gene sequence was confirmed by DNA sequencing (UIUC core sequencing facility, Urbana, IL).
Purification of E. coli LeuRS
Wild-type and mutant protein expression was induced with 1 mM IPTG (isopropyl-β-D-thiogalactopyranoside) in E. coli strain BL21 (DE3) plysS (Novagen, Madison, WI). The cells were harvested at 6000 rpm for 15 min in an Avanti J-E centrifuge (Beckman Coulter, Fullerton, CA). The cell pellet was resuspended in 5 ml of HA-I buffer [20 mM sodium phosphate (NaPi), 10 mM tris(hydroxymethyl) aminomethane (Tris), pH 8.0, 300 mM NaCl, and 5% glycerol]. Following sonication on ice for 2 min at 50% power using a Vibra Cell sonicator (Sonics, Newtown, CT), the lysate was centrifuged at 6000 rpm for 15 min at 4 °C. The supernatant was combined with 4 mLs of resuspended HIS-Select HF Nickel Affinity gel (Sigma, St. Louis. MO), pre-equilibrated with HA-I buffer, and incubated for 1 h at 4 °C on a rocker. The resin was washed multiple times with a total of 100 ml of HA-I buffer that contained 5 mM imidazole. The protein was eluted with 10 ml of HA-I buffer containing 200 mM imidazole and concentrated in a centricon-50 (Amicon, Bedford, MA). The final concentration was determined using a Bio-Rad protein assay according to the commercial protocol (Bio-Rad laboratories, Hercules, CA).
RNA Preparation
The plasmid ptDNALeu14 (16, 17) which harbors the gene for E. coli tRNALeuUAA (tRNALeu), was linearized by digesting 450 µg of the plasmid with Bst NI (New England Biolabs Inc., Beverly, MA) at 60 °C overnight and then used as a template for T7 RNA polymerase run-off transcription (18). The in vitro transcription reaction contained 40 mM Tris (pH 8.0), 30 mM MgCl2, 5 mM dithiothreitol (DTT), 0.01 % Triton X-100, 50 µg/ml bovine serum albumin (BSA), 4 mM of each NTP, 80 mg/ml PEG8000, 0.02 U/µl RNasin (Eppendorf, Hamburg, Germany), 2 mM spermidine, 0.4 mg/ml plasmid template, 0.01 mg/ml pyrophosphatase (Sigma), and 0.6 µM T7 RNA polymerase (18, 19). Reactions were incubated at 42 °C for 3 h, followed by addition of another 0.6 µM of T7 RNA polymerase and further incubation for 3 h. The reactions were quenched with 100% ethanol and precipitated at −80 °C for at least one-half hour.
The pelleted RNA was purified on a 10% denaturing polyacrylamide (19:1), 8 M urea gel by electrophoresis. The tRNALeu band was detected by UV shadowing, excised, crushed, and the RNA extracted three times in a solution of 0.5 M NH4OAc and 1 mM ethylenediaminetetraacetic acid (EDTA) (pH 8.0). After butanol-extraction to concentrate the RNA, it was ethanol precipitated and then the tRNA pellet was resuspended in 100 µl of nuclease free water (Ambion, Austin, TX). The concentration was calculated using an extinction coefficient of 840,700 (L/mol•cm) for tRNALeu (20).
Aminoacylation Assays
Reactions containing 60 mM Tris (pH 7.5), 10 mM MgCl2, 150 mM KCl, 1 mM DTT, 22 µM [3H]-leucine (167 µCi/ml; Amersham Pharmacia Biotech, Piscataway, NJ), 4 µM in vitro transcribed E. coli tRNALeu and catalytic amounts of enzyme were initiated with 4 mM ATP. Aliquots of 10 µl were quenched on Whatman filter pads, pre-wet with 5% trichloroacetic acid (TCA), at varying time intervals. The pads were washed three times for 10 min each with cold 5% TCA and then once with 70% ethanol. The washed pads were dried in anhydrous ether and then under a heat lamp. Radioactivity was quantified in a Beckman LS 6500 scintillation counter (Beckman Coulter, Inc., Fullerton, CA). Apparent kinetic rate constants were measured by incorporating six different in vitro transcribed tRNALeu concentrations ranging from 0.1 to 10 µM. The kinetic parameters were calculated and error analysis carried out using SigmaPlot (Systat Software, Inc., San Jose, CA).
Hydrolysis Assays
In vitro transcribed E. coli tRNALeu was misaminoacylated with [3H]-isoleucine (93 µCi/µl) (Amersham Pharmacia Biotech, Piscataway, NJ) by 1 µM of an E. coli LeuRS editing-deficient mutant as described above for 3 h at 37 °C. The reactions were quenched with 0.18% acetic acid (21). Protein was removed by extraction with a phenol/chloroform/isoamyl alcohol (125:24:1) at pH 4.3. The RNA was ethanol precipitated and resuspended in 50 mM potassium phosphate buffer (pH 5.0).
Post-transfer editing activity was tested in reactions containing 60 mM Tris (pH 7.5), 10 mM MgCl2, and approximately 4 µM [3H]-Ile-tRNALeu. Reactions were initiated with 100 nM enzyme. Aliquots of 5 µl were quenched on filter pads pre-soaked with 5% TCA, washed, and analyzed as described above.
Inorganic Pyrophosphate (PPi) Exchange Assays
Reactions containing 50 mM N-(2-hydroxyethyl)-piperazine-N′-2-ethanesulfonic acid (HEPES), pH 8.0, 10 mM MgCl2, 1 mM DTT, 1 mM [32P]-PPi (78 mCi/ml; Amersham Pharmacia Biotech, Piscataway, NJ), 1 mM leucine and 100 nM enzyme were initiated with 1 mM ATP (22). Aliquots of 2 µl were quenched on cellulose polyethyleneimine (PEI) thin-layer chromatography (TLC) plates (Scientific Adsorbents Inc., Atlanta, GA) that were pre-run in water. The reaction components were separated by developing the TLC plates in 750 mM KH2PO4, pH 3.5 and 4 mM urea. Separated radiolabelled bands were detected using a phosphorimager and FUJIX BAS 1000 film (FUJIFILM Medical Systems U.S.A., Stanford, CT). Bands were quantitated with a Storm 840 Molecular Dynamics Imager (GE Healthcare Piscataway, NJ).
“Hinge” Prediction
We utilized the Morph Server (23) in the database of molecular motions (MolMovDB: www.molmovdb.org/)(24). This server characterizes macromolecular motions, by comparing two similar crystal structures as input. The tRNA coordinates in the crystal structures for T. thermophilus LeuRS in the post-transfer editing (PDB code 2BYT) and exit (PDB code 2BTE) complexes (14) were removed for comparative studies and the remaining protein coordinates were uploaded into the single-chain Morph Server (25). A hinge predictor graph was developed using the output of FlexOracle (26), TLSMD (Translation/Liberation/Screw Motion Determination)(27) as well as a combined HingeMaster prediction.
Circular Dichroism
CD measurement was carried out using a Jasco J-720 spectropolarimeter. A sample containing 0.8 µM protein in 5 mM KPi, pH 7.5, was measured in the far-ultraviolet region using a 0.1 cm path cell. Background signals from the cell and the buffer were subtracted from each spectrum.
RESULTS
Deletions Affect Aminoacylation Activity of E. coli LeuRS
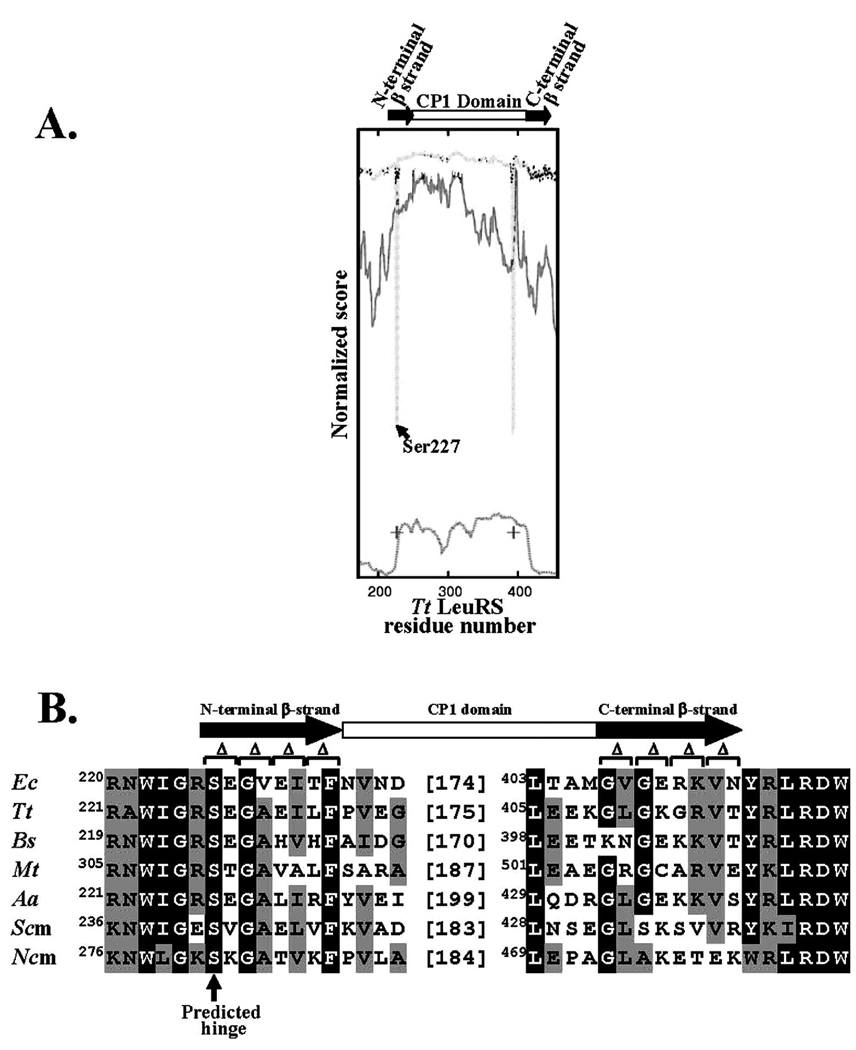
Multiple X-ray crystal structures and models suggest that the LeuRS CP1 domain adopts different conformations relative to the enzyme’s canonical core as it moves between the aminoacylation and editing complexes with tRNA during its reaction cycle (Figure 1). We hypothesized that this rigid body movement of the CP1 domain would require coordinated movement of the flexible β-strand linkers. We compared the crystal structures of T. thermophilus LeuRS (PDB codes 2BTE and 2BYT) using the database of molecular motions (24) to identify potential residues that are flexible enough to act as possible hinges. Three algorithms that predict highly flexible protein sites were employed to investigate LeuRS for hinge sites which might help drive the CP1 domain and canonical core into alternate conformations that were competent for aminoacylation and editing. Within the β-strands, a single hinge was predicted that corresponded to Ser227 at the N-terminus of the N-terminal β-strand (Figure 2A). This N-terminal β-strand serine site is highly conserved (Figure 2B). A second hinge was also predicted that was near the C-terminal β-strand, but actually located within the CP1 domain.
Figure 2.
Hinge predictor graph and sequence alignment for LeuRS. A. Hinge predictor graph for T. thermophilus LeuRS. The post-transfer editing (PDB code 2BYT) and exit (PDB code 2BTE) crystal structures for T. thermophilus LeuRS in the absence of tRNALeu were compared using the Morph Server (23). The lines on the resulting graph are represented as follows: combined HingeMaster predictor by the upper trace where minima correspond to suggested “hinge” regions; FlexOracle prediction (26) in the middle with minima corresponding to hinges; NSHP hinge predictor shown at the bottom. The TLSMD predicted hinge residues (27) are indicated by (+). The approximate locations of the CP1 and the connective linkers are depicted in the cartoon above the graph as an open rectangle and shaded flanking arrows, respectively. B. Sequence alignment of the CP1 domain and β-strand linkers. The N- and C-terminal β-strands are highlighted with shaded arrows. Two amino acid deletions for E. coli LeuRS are marked by the “Δ” signs. Black and gray shading represents highly conserved and homologous residues, respectively. Abbreviations are as follows: Ec, Escherichia coli; Tt, Thermus thermophilus; Ncm, Neurospora crassa (mitochondria); Scm, Saccharomyces cerevisiae (mitochondria); Bs, Bacillus subtilis; Hsm, Homo sapiens (mitochondria); Mt, Mycobacterium tuberculosis; Aa, Aquifex aeolicus (α subunit). Sequence alignments were generated using the Baylor College of Medicine Search Launcher ClustalW 1.8 multiple sequence alignment program (33).
We hypothesized that the predicted hinge site at Ser227 within the N-terminal β-strand might facilitate conformational changes of the LeuRS that promoted aminoacylation or editing or both. We introduced a series of two-amino acid deletions on the N-terminal β-strand of E. coli LeuRS beginning at the hinge site, Ser227 at its N-terminus (Figure 2B). These included a two amino acid deletion, ΔS227E228 at the predicted serine hinge site, as well as ΔG229V230, ΔE231I232, and ΔT233F234, which spanned Ser227 to Phe234 on the N-terminal β-strand. We also generated two amino acid deletions on the C-terminal β-strand that encompassed ΔG407V408, ΔG409E410, ΔR411K412, and ΔV413N414 spanning Gly407 to Asn414 (Figure 2B).
Each of the β-strand deletion mutants of LeuRS were stably expressed in E. coli and purified by affinity chromatography using an N-terminal six-histidine tag. The purified deletion mutants were then tested for aminoacylation activity with in vitro transcribed tRNALeu. Only one mutant, the ΔS227E228 LeuRS deletion that included the predicted hinge site on the N-terminal β-strand, failed to aminoacylate tRNALeu (Table 1), even at high concentrations of 2 µM enzyme (data not shown). This mutant enzyme also did not activate amino acid in pyrophosphate exchange assays suggesting that the conformation of the N-terminal β-strand influenced the aminoacylation active site directly (Figure 3A).
Table 1.
Apparent Kinetic Parameters for Aminoacylationa
| Deletion mutants |
KM(µM) | kcat (s−1) |
kcat/KM (µM1 s−1) |
Relative kcat/KM |
|---|---|---|---|---|
| Wild type | 1.2 ± 0.2 | 14.3 ± 0.3 | 11.9 | 1 |
| N-terminal β-strand | ||||
| ΔS227E228 | NDb | NDb | NDb | - |
| ΔS227 | NDb | NDb | NDb | - |
| ΔE228 | NDb | NDb | NDb | - |
| ΔG229V230 | 0.94 ± 0.4 | 9.2 ± 2.7 | 9.7 | 0.8 |
| ΔE231I232 | 1 ± 0.1 | 10.2 ± 4 | 10.2 | 0.9 |
| ΔT233F234 | 2.3 ± 0.5 | 39.1 ± 3.3 | 17 | 1.4 |
| C-terminal β-strand | ||||
| ΔG407V408 | 0.9 ± 0.2 | 12.5 ± 1 | 13.8 | 1.2 |
| ΔG409E410 | 0.6 ± 0.1 | 6.9 ± 1.2 | 11.5 | 1 |
| ΔR411K412 | 2.1 ± 0.5 | 35.2 ± 3.5 | 16.7 | 1.4 |
| ΔV413N414 | 1.7 ± 0.6 | 4.5 ± 0.3 | 2.7 | 0.2 |
| ΔV413 | 0.7 ± 0.2 | 0.2 ± 0.05 | 0.3 | 0.02 |
| ΔN414 | 2.8 ± 0.5 | 0.6 ± 0.3 | 0.2 | 0.02 |
The kinetic parameters that are reported are apparent values.
ND – Not determined because of low activity.
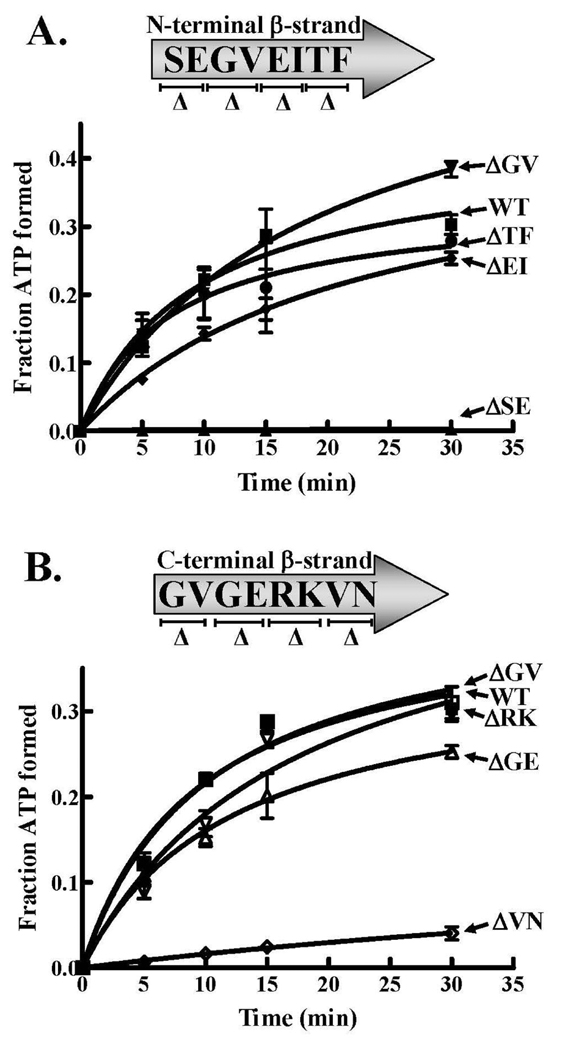
Figure 3.
Leucine-dependant pyrophosphate exchange activities of LeuRS wild type and β-strand two-amino acid deletion mutants. The reactions contained 100 nM wild type or mutant LeuRS. A. N-terminal β-strand deletion mutant activities. B. C-terminal β-strand deletion mutant activities. Symbols are represented as follows: wild type (■), ΔS227E228 (▲), ΔG229V230 (▼), ΔE231I232 (◆), ΔT233F234 (●), ΔG407V408 (□), ΔG409E410(△), ΔR411K412 (▽) and ΔV413N414 (◊). Error bars are the result of reactions repeated in triplicate and are indicated for each point.
The ΔG229V230 and ΔE231I232 mutant LeuRSs retained significant aminoacylation activity compared to wild-type LeuRS, with a kcat/KM of 9.7 and 10.2 µM−1s−1, respectively (Table 1). The ΔT233F234 LeuRS mutant activity was enhanced compared to that of wild-type LeuRS, largely due to an increase in kcat. As would be expected and in contrast to the ΔS227E228 mutation, each of these other deletion mutants maintained robust amino acid-dependent pyrophosphate exchange activity (Figure 3A).
Amongst the C-terminal β-strand deletion mutants, the kcat/KM for the ΔV413N414 deletion mutant at the C-terminus of the C-terminal β-strand was the most significantly decreased compared to the wild type enzyme. The fourfold decrease was largely due to a modest decrease in the kcat (Table 1). The ΔV413N414 LeuRS deletion mutant displayed lowered amino acid activation that correlated to its decrease in aminoacylation activity (Figure 3B). The ΔR411K412 deletion mutant exhibited an elevated kcat/KM relative to the wild type enzyme because of an increase in the kcat. The ΔG407V408 also had a slightly increased kcat/KM, while the ΔG409E410 deletion mutant activity was similar to wild type LeuRS.
Deletions Affect Post-transfer Editing Activity of E. coli LeuRS
Each of the two-amino acid deletion mutants were tested for amino acid editing activity of mischarged Ile-tRNALeu. Although it was inactive in aminoacylation, the ΔS227E228 deletion mutant at the predicted hinge site on the N-terminal β-strand exhibited some amino acid editing activity, albeit significantly reduced compared to the wild type LeuRS activities (Figure 4A). We hypothesized that deletion of the Ser227 and Glu228 residues “kinks” the β-strands in such a way that allows tRNA binding in the editing complex, but not the aminoacylation complex. The other N-terminal β-strand deletion mutants exhibited slightly decreased hydrolysis rates that reached wild type enzyme plateau levels. As previously determined for other LeuRS mutations that had only small decreases in editing activity (28), the N-terminal β-strand deletion mutants did not confer mischarging (data not shown). Although the ΔS227E228 LeuRS had significantly decreased editing activity, its lack of mischarging activity was likely due to its failure to aminoacylate in general.
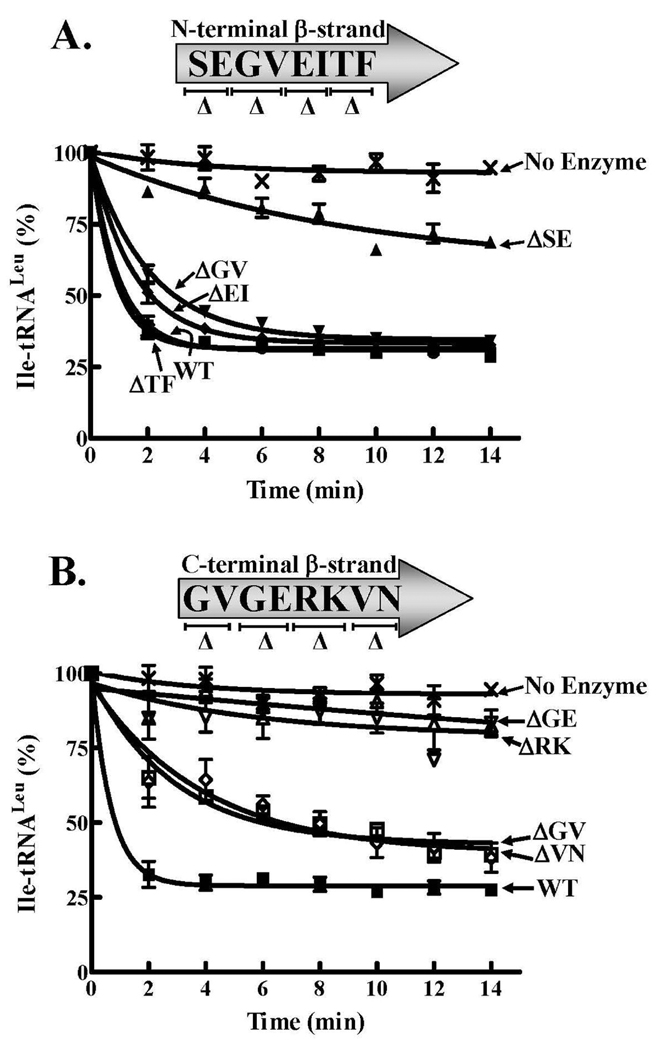
Figure 4.
Amino acid editing activities of LeuRS wild type and β-strand linker two-amino acid deletion mutants. The reactions containing approximately 4 µM [3H]-Ile-tRNALeu were initiated with 100 nM wild type or mutant LeuRS. A. N-terminal β-strand deletion mutant activities. B. C-terminal β-strand deletion mutant activities. Symbols are represented as follows: wild type (■), ΔS227E228 (▲), ΔG229V230 (▼), ΔE231I232 (◆), ΔT233F234 (●), ΔG407V408 (□), ΔG409E410(△), ΔR411K412 (▽), ΔV413N414 (◊) and no enzyme (X). Error bars are based on reactions that were repeated at least in triplicate.
In contrast to the N-terminal mutants, each of the C-terminal deletion mutants exhibited reduced amino acid editing activity relative to the wild type enzyme. Although ΔR411K412 on the C-terminal β-strand had wild type-like aminoacylation activity, amino acid editing activity was significantly reduced (Figure 4B). The editing activity of ΔG409E410 was also decreased similar to the ΔR411K412 deletion mutant. While the ΔG407V408 and ΔV413N414 deletion mutants retained editing activity, neither attained plateau levels of hydrolysis of Ile-tRNALeu that correlated to the wild type LeuRS.
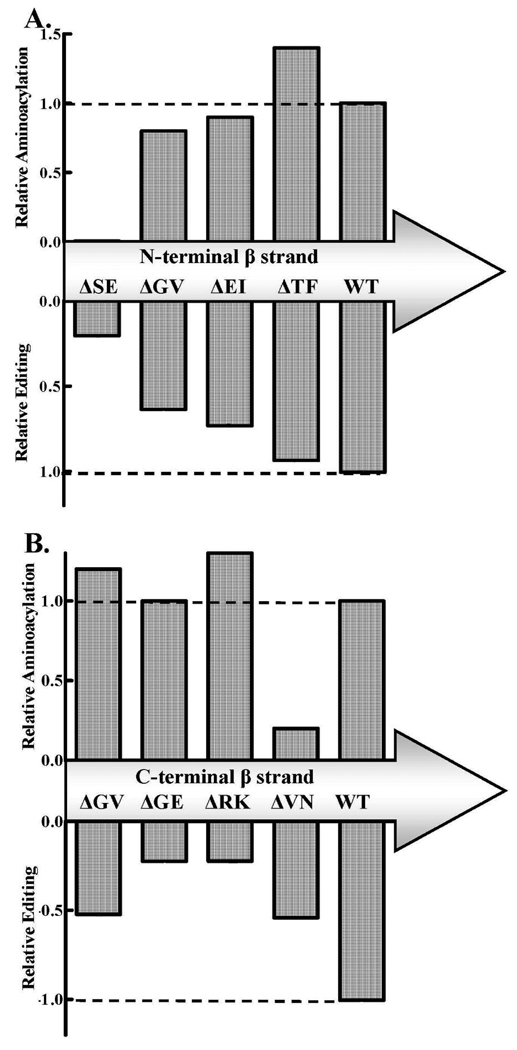
Spatially Diverse Effects of β-strand Linkers on Enzyme Activity
We compared the relative activities for aminoacylation and/or editing for each of the LeuRS deletion mutants and wild type. We found spatial “hot spots” that influenced aminoacylation and editing on the N- and C-terminal β-strands. These sensitive areas on each β-strand linker were strikingly distinct. Deletions across the N-terminal β-strand determined that sites that were closer to the predicted hinge site near the canonical core of E. coli LeuRS more dramatically influenced both aminoacylation and editing activities (Figure 5A). We hypothesize that LeuRS requires more sequence specific flexibility at the β-strand end that is near the canonical core.
Figure 5.
Relative aminoacylation and editing activities of deletion mutants on the β-strands of E. coli LeuRS. A. N-terminal β-strand. B. C-terminal β-strand. The activity of aminoacylation (top) and amino acid editing (bottom) were normalized to wild type aminoacylation activity based on the kcat/KM (Table 1) and are shown in bar graph form. Estimates of the post-transfer editing activities were normalized by comparing the hydrolyzed Ile-tRNALeu at the two minute time point. The horizontal dashed line represents the activity of wild type E. coli LeuRS.
In contrast, deletions of the C-terminal β-strand showed diverse effects on aminoacylation and amino acid editing (Figure 5B). Each of the deletion mutants significantly impacted amino acid editing. Only the ΔV413N414 LeuRS mutant at the end of the C-terminal β-strand near the canonical core influenced aminoacylation. The ΔG409E410 and ΔR411K412 LeuRS deletion mutants significantly reduced the editing activity of the enzyme, but retained wild type-like aminoacylation activity. It is possible that the C-terminal β-strand linker requires a flexible molecular hinge at the Val413-Asn414 site to form the aminoacylation complex.
Molecular Analysis of Aminoacylation “Hot Spots” on the β-strand Linkers
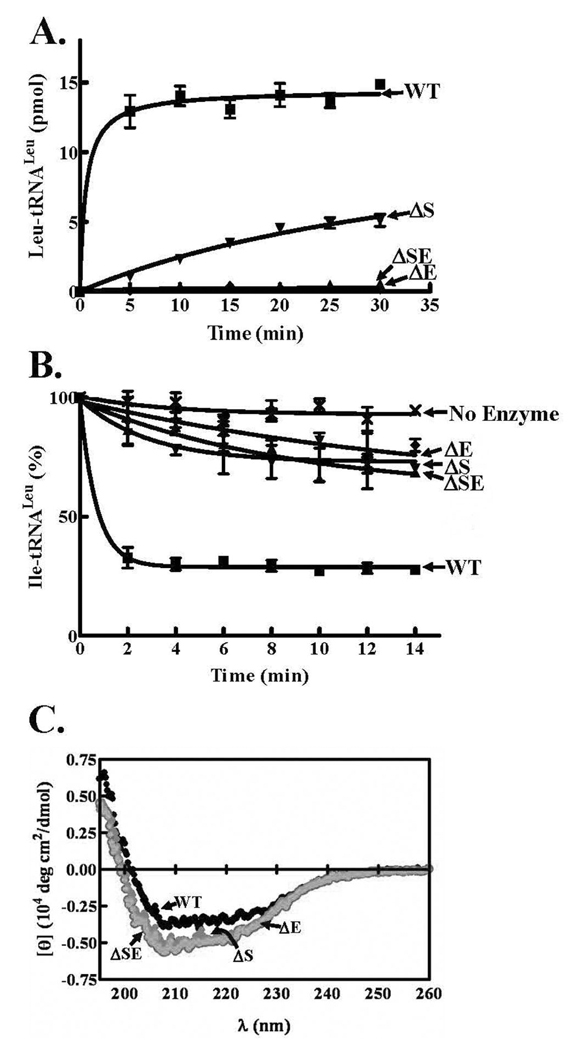
We mutationally analyzed the two amino acid deletion sites that exhibited the greatest impact on LeuRS activity. As found for the ΔS227E228 deletion, single deletions of either amino acid reduced both amino acid activation and aminoacylation. The ΔE228 LeuRS deletion mutant abolished aminoacylation while the ΔS227 mutant retained some activity, albeit significantly reduced compared to wild type LeuRS (Figure 6A). Hydrolysis of Ile-tRNALeu by each of the single deletion mutants was also substantially decreased similar to the double ΔS227E228 LeuRS deletion mutant (Figure 6B). We tested whether mutations at this critical hinge site cause structural deformities. However, the CD spectrum of each of the deletion mutants at Ser227 and Glu228 showed that their secondary structures compared well to the wild type enzyme (Figure 6C).
Figure 6.
Enzymatic activities of LeuRS wild type and N-terminal β-strand mutants. A. Leucylation activities were measured using enzyme and tRNALeu concentrations of 50 nM and 4 µM, respectively. B. Amino acid editing activities were determined using approximately 4 µM [3H]-Ile-tRNALeu and were initiated with 100 nM enzyme. Symbols are represented as follows: wild type (■), ΔS227E228 (▲), ΔS227 (▼), ΔE228 (◆) and no enzyme (X). Error bars are the result of reactions repeated in triplicate and are indicated for each point. C. CD spectra. The dark symbols represent the wild type LeuRS spectrum, while the light symbols overlap for the spectra of the three deletion mutants.
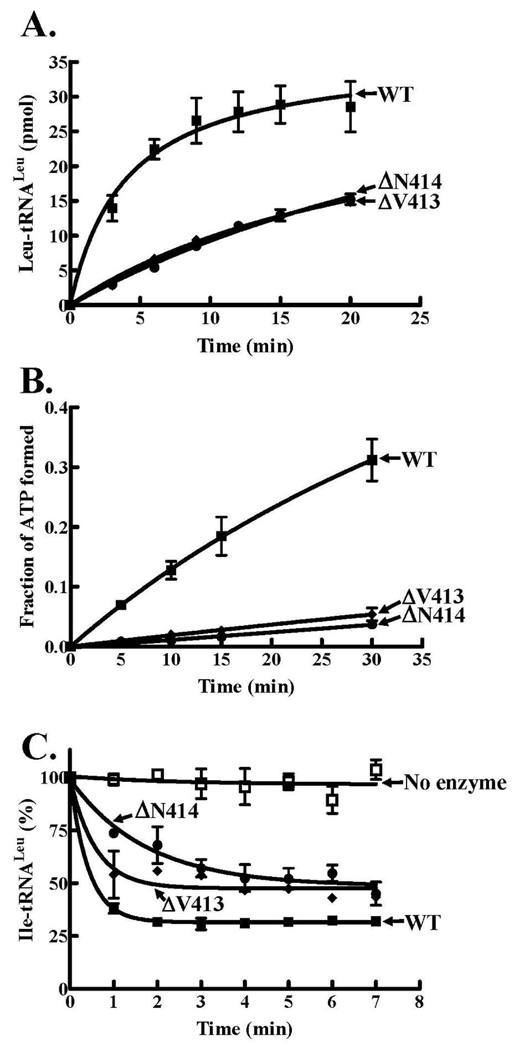
We also constructed single deletion mutations at the Val413 and Asn414 sites within the C-terminal β-strand. While these mutants retained a robust amino acid editing activity of Ile-tRNALeu, individual deletions of Val413 and Asn414 resulted in significantly lowered aminoacylation activities to greater than 80% (Figure 7). The amino acid activation activity was also greatly decreased similar to the ΔV413N414 double LeuRS mutant (Figure 7B). Although the KM values were comparable to wild type, the kcat values for both the ΔV413 and ΔN414 LeuRS single deletion mutants were considerably reduced by six-fold (Table 1).
Figure 7.
Enzymatic activities of LeuRS wild type and C-terminal single deletion β-strand mutants. A. Leucylation activities were measured using enzyme concentrations of 50 nM. B. Leucine-dependent pyrophosphate exchange activities were determined using 100 nM enzyme. C. Amino acid editing activities were determined using approximately 4 µM [3H]-Ile-tRNALeu and were initiated with 100 nM enzyme. Symbols are represented as follows: wild type (■), ΔV413 (◆), ΔN414 (●), and no enzyme (□). Error bars are the result of reactions repeated in triplicate and are indicated for each point.
DISCUSSION
Translocation of the 3′ charged end between the aminoacylation active site in the canonical core and the CP1 domain-based hydrolytic site occurs via a mechanism that remains virtually undefined. The accumulation of numerous LeuRS crystal structures (2) shows clearly that the enzyme undergoes significant conformational changes to accommodate the aminoacylation and the editing complexes. This is also accompanied by changes in the tRNA, particularly at the 3′ end that is charged, and its interactions with the AARS.
It is likely that LeuRS translocation is pseudo-reversible such that the tRNA’s 3′ end can move from the editing active site to the aminoacylation active site, and naturally occurs as an obligatory part of the enzyme’s reaction cycle. Recently, a boron containing inhibitor that binds in the LeuRS editing active site, AN2690, was shown to covalently trap the 3′ end of the tRNA in the editing complex (29). Surprisingly, AN2690 cross-linking occurs quite effectively with uncharged tRNA in the absence of leucine and/or ATP. This suggests that the uncharged 3′ end of the tRNA sweeps through the editing site enroute to binding to the aminoacylation active site. This result also raises the possibility that the so-called exit complex structure (14) of LeuRS bound to tRNALeu also serves as an entrance complex for tRNA binding (29). In the exit complex, the charged 3′ end of the tRNA resides near the editing active site (14). If the entrance complex resembles the exit complex, then we might expect the tRNA’s 3′ end to transiently occupy the editing active site as it transitions to the aminoacylation active site. Once charged, the tRNA would be translocated back to the editing complex for proofreading, and if necessary editing. Subsequent to screening by the hydrolytic editing active site, the correctly charged tRNA would transition to the exit complex, prior to release.
These dynamic transitions would be expected to require coordinated movement by the enzyme and tRNA. We hypothesized that LeuRS, ValRS and IleRS require the flexible β-strand linkers to act as hinges to direct these conformational changes. Computational analysis indicated that the N-terminus of the N-terminal β-strand linker to the mobile CP1 domain may act as such a hinge. Deletion mutations at positions Ser227 and Glu228 drastically affected aminoacylation, but retained small amounts of amino acid editing activity. In addition, neighboring two-amino acid deletions appear to have an inter-related effect on aminoacylation and post-transfer editing efficiency that declined as the disruptions sequentially moved away from the main body of the enzyme on the N-terminal β-strand and toward the CP1 domain (Figure 5).
In the co-crystal structure of T. thermophilus bound to tRNALeu, the ribose of Cyt74 interacts with Ser227 in the exit complex (14). Along with reorientation of the 3′ acceptor stem of tRNA, this is accompanied by a 5° rotation of the CP1 domain relative to the core of the enzyme (14). The deletion of Ser227 would disrupt this interaction near the editing active site. If the tRNA entry complex resembles the tRNA exit complex, then we would also expect tRNA binding to be disrupted precluding formation of an active aminoacylation complex.
The N-terminal β-strand is flanked by the mobile zinc-containing ZN-1 domain, which appears to sterically impede a conformational transition of the tRNA to the aminoacylation complex (14). This domain adopts open and closed conformations, where the closed position blocks entry of the 3′ end of tRNA into the catalytic site for aminoacylation, but does not impede post-transfer editing (14). Deletions at Ser227 and Glu228 could result in a structural conformation that hampers movement of the adjacent ZN-1 domain. Thus, an active aminoacylation complex would be blocked, albeit an editing complex could be formed that stimulates at least some deacylation activity as found for ΔS227 and ΔS227E228 mutant enzymes.
The C-terminal β-strand is flanked by the CP1 editing domain and the conserved “RDW” region on the canonical core of the enzyme (30). This RDW peptide interacts with tRNA in various conformations and may position tRNA for different enzyme activities (30). A model based on the T. thermophilus and P. horikoshii enzyme-tRNA complexes suggests that the RDW peptide positions the 3′ end of tRNA for transition to an aminoacylation complex (14). Disruption of this interaction could account for the aminoacylation deficient ΔV413N414 LeuRS deletion mutant that retains amino acid editing and is located the closest to the RDW peptide. In addition, in the exit/entry complex of T. thermophilus LeuRS, Arg418 (Arg416 in E. coli) of this peptide interacts with Ade73 of the tRNA (14). The robust aminoacylation activity of deletion mutants spanning Gly407 to Lys412 could come at a cost to the enzyme’s editing activity by “trapping” the CP1 editing domain in an aminoacylation conformation.
Hinge sites to facilitate the movement of editing substrates between the aminoacylation and editing active site have also been characterized for IleRS (31). Substitutions at proposed hinge sites within the canonical core of the homologous IleRS (Lys183 and Trp421) affect translocation and thereby, the overall editing of the enzyme (31). It was proposed that these mutations impeded the CP1 domain movement that would be required for editing to occur. This “hinge” lysine is also conserved in LeuRS and plays a clear role in the fidelity mechanisms of E. coli LeuRS (32). It was hypothesized that this surface lysine on the canonical core of LeuRS interacts with negatively charged amino acids on the CP1 domain surface to stabilize one or more of the enzyme’s conformations during the reaction cycle.
Communication between the CP1 domain and the core of the enzyme, at various phases of enzyme activity, is dependant on the β-strands that link them. Structural evidence suggests that the concerted movements of the ZN-1 and CP1 domains relative to the canonical core are likely orchestrated by the 3′ end of tRNA (14). The different orientations adopted by the enzyme would require that movement of the ZN-1 and CP1 domains coincides with changes at flexible “hinge” sites within the β-strands.
ACKNOWLEDGEMENT
We are grateful to Dr. Stephen Cusack for valuable discussions and technical advice. We acknowledge the National Institutes of Health (GM063789) for support.
REFERENCES
- 1.Ibba M, Francklyn C, Cusack S, editors. Aminoacyl-tRNA Synthetases. Georgetown, TX: Landes Bioscience/Eurekah.com; 2005. [Google Scholar]
- 2.Mascarenhas AP, An S, Rosen AE, Martinis SA, Musier-Forsyth K. Fidelity mechanisms of the aminoacyl-tRNA synthetases. In: RajBhandary UL, Kohrer C, editors. Protein Engineering. Springer Verlag; 2007. pp in press. [Google Scholar]
- 3.Hendrickson TL, Schimmel P. Transfer RNA-dependent amino acid discrimination by aminoacyl-tRNA synthetases. In: Lapointe J, Brakier-Gingras L, editors. Translation Mechanisms. Kluwer Academic/Plenum Publishers; 2003. pp. 35–64. [Google Scholar]
- 4.Zhang CM, Perona JJ, Hou YM. Amino acid discrimination by a highly differentiated metal center of an aminoacyl-tRNA synthetase. Biochemistry. 2003;42:10931–10937. doi: 10.1021/bi034812u. [DOI] [PubMed] [Google Scholar]
- 5.Pauling L. The probability of errors in protein synthesis, in Festschrift Arthur Stoll Siebzigsten Geburtstag. 1958:597–602. [Google Scholar]
- 6.Fersht AR. Editing mechanisms in protein synthesis. Rejection of valine by the isoleucyl-tRNA synthetase. Biochemistry. 1977;16:1025–1030. doi: 10.1021/bi00624a034. [DOI] [PubMed] [Google Scholar]
- 7.Hati S, Ziervogel B, Sternjohn J, Wong FC, Nagan MC, Rosen AE, Siliciano PG, Chihade JW, Musier-Forsyth K. Pre-transfer editing by class II prolyl-tRNA synthetase: Role of aminoacylation active site in "selective release" of noncognate amino acids. J Biol Chem. 2006;281:27862–27872. doi: 10.1074/jbc.M605856200. [DOI] [PubMed] [Google Scholar]
- 8.O'Donoghue P, Luthey-Schulten Z. On the evolution of structure in aminoacyl-tRNA synthetases. Microbiol Mol Biol Rev. 2003;67:550–573. doi: 10.1128/MMBR.67.4.550-573.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cusack S, Yaremchuk A, Tukalo M. The 2 Å crystal structure of leucyl-tRNA synthetase and its complex with a leucyl-adenylate analogue. EMBO J. 2000;19:2351–2361. doi: 10.1093/emboj/19.10.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nureki O, Vassylyev DG, Tateno M, Shimada A, Nakama T, Fukai S, Konno M, Hendrickson TL, Schimmel P, Yokoyama S. Enzyme structure with two catalytic sites for double-sieve selection of substrate. Science. 1998;280:578–582. doi: 10.1126/science.280.5363.578. [DOI] [PubMed] [Google Scholar]
- 11.Fukai S, Nureki O, Sekine S, Shimada A, Tao J, Vassylyev DG, Yokoyama S. Structural basis for double-sieve discrimination of L-valine from L-isoleucine and L-threonine by the complex of tRNA(Val) and valyl-tRNA synthetase. Cell. 2000;103:793–803. doi: 10.1016/s0092-8674(00)00182-3. [DOI] [PubMed] [Google Scholar]
- 12.Starzyk RM, Burbaum JJ, Schimmel P. Insertion of new sequences into the catalytic domain of an enzyme. Biochemistry. 1989;28:8479–8484. doi: 10.1021/bi00447a031. [DOI] [PubMed] [Google Scholar]
- 13.Fukunaga R, Yokoyama S. Aminoacylation complex structures of leucyl-tRNA synthetase and tRNALeu reveal two modes of discriminator-base recognition. Nat Struct Mol Biol. 2005;12:915–922. doi: 10.1038/nsmb985. [DOI] [PubMed] [Google Scholar]
- 14.Tukalo M, Yaremchuk A, Fukunaga R, Yokoyama S, Cusack S. The crystal structure of leucyl-tRNA synthetase complexed with tRNALeu in the post-transfer-editing conformation. Nat Struct Mol Biol. 2005;12:923–930. doi: 10.1038/nsmb986. [DOI] [PubMed] [Google Scholar]
- 15.Martinis SA, Fox GE. Non-standard amino acid recognition by Escherichia coli leucyl-tRNA synthetase. Nucleic Acids Symp Ser. 1997;36:125–128. [PubMed] [Google Scholar]
- 16.Tocchini-Valentini G, Saks ME, Abelson J. tRNA leucine identity and recognition sets. J Mol Biol. 2000;298:779–793. doi: 10.1006/jmbi.2000.3694. [DOI] [PubMed] [Google Scholar]
- 17.Normanly J, Ogden RC, Horvath SJ, Abelson J. Changing the identity of a transfer RNA. Nature. 1986;321:213–219. doi: 10.1038/321213a0. [DOI] [PubMed] [Google Scholar]
- 18.Sampson JR, Uhlenbeck OC. Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. Proc Natl Acad Sci U S A. 1988;85:1033–1037. doi: 10.1073/pnas.85.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grodberg J, Dunn JJ. ompT encodes the Escherichia coli outer membrane protease that cleaves T7 RNA polymerase during purification. J Bacteriol. 1988;170:1245–1253. doi: 10.1128/jb.170.3.1245-1253.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhai Y, Martinis SA. Two conserved threonines collaborate in the Escherichia coli leucyl-tRNA synthetase amino acid editing mechanism. Biochemistry. 2005;44:15437–15443. doi: 10.1021/bi0514461. [DOI] [PubMed] [Google Scholar]
- 21.Schreier AA, Schimmel PR. Transfer ribonucleic acid synthetase catalyzed deacylation of aminoacyl transfer ribonucleic acid in the absence of adenosine monophosphate and pyrophosphate. Biochemistry. 1972;11:1582–1589. doi: 10.1021/bi00759a006. [DOI] [PubMed] [Google Scholar]
- 22.Lincecum TL, Martinis SA. The tRNA synthetase proofreading and editing active sites: a novel antibiotic target. In: Ballal SK, editor. SAAS Bull. Biochem. Biotechnol. Vol. 13. 2000. pp. 25–33. [Google Scholar]
- 23.Flores S, Echols N, Milburn D, Hespenheide B, Keating K, Lu J, Wells S, Yu EZ, Thorpe M, Gerstein M. The Database of Macromolecular Motions: new features added at the decade mark. Nucleic Acids Res. 2006;34:D296–D301. doi: 10.1093/nar/gkj046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gerstein M, Krebs W. A database of macromolecular motions. Nucleic Acids Res. 1998;26:4280–4290. doi: 10.1093/nar/26.18.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krebs WG, Gerstein M. The morph server: a standardized system for analyzing and visualizing macromolecular motions in a database framework. Nucleic Acids Res. 2000;28:1665–1675. doi: 10.1093/nar/28.8.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Flores SC, Gerstein MB. FlexOracle: predicting flexible hinges by identification of stable domains. BMC Bioinformatics. 2007;8:215. doi: 10.1186/1471-2105-8-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Painter J, Merritt EA. A molecular viewer for the analysis of TLS rigid-body motion in macromolecules. Acta Crystallogr D Biol Crystallogr. 2005;61:465–471. doi: 10.1107/S0907444905001897. [DOI] [PubMed] [Google Scholar]
- 28.Mursinna RS, Martinis SA. Rational design to block amino acid editing of a tRNA synthetase. J Am Chem Soc. 2002;124:7286–7287. doi: 10.1021/ja025879s. [DOI] [PubMed] [Google Scholar]
- 29.Rock FL, Mao W, Yaremchuk A, Tukalo M, Crepin T, Zhou H, Zhang YK, Hernandez V, Akama T, Baker SJ, Plattner JJ, Shapiro L, Martinis SA, Benkovic SJ, Cusack S, Alley MR. An antifungal agent inhibits an aminoacyl-tRNA synthetase by trapping tRNA in the editing site. Science. 2007;316:1759–1761. doi: 10.1126/science.1142189. [DOI] [PubMed] [Google Scholar]
- 30.Nawaz MH, Pang YL, Martinis SA. Molecular and functional dissection of a putative RNA-binding region in yeast mitochondrial leucyl-tRNA synthetase. J Mol Biol. 2007;367:384–394. doi: 10.1016/j.jmb.2006.12.051. [DOI] [PubMed] [Google Scholar]
- 31.Bishop AC, Beebe K, Schimmel PR. Interstice mutations that block site-to-site translocation of a misactivated amino acid bound to a class I tRNA synthetase. Proc Natl Acad Sci U S A. 2003;100:490–494. doi: 10.1073/pnas.0237335100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williams AM, Martinis SA. Mutational unmasking of a tRNA-dependent pathway for preventing genetic code ambiguity. Proc Natl Acad Sci USA. 2006;103:3586–3591. doi: 10.1073/pnas.0507362103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith RF, Wiese BA, Wojzynski MK, Davison DB, Worley KC. BCM Search Launcher--an integrated interface to molecular biology data base search and analysis services available on the World Wide Web. Genome Res. 1996;6:454–462. doi: 10.1101/gr.6.5.454. [DOI] [PubMed] [Google Scholar]
- 34.Silvian LF, Wang J, Steitz TA. Insights into editing from an Ile-tRNA synthetase structure with tRNAIle and mupirocin. Science. 1999;285:1074–1077. [PubMed] [Google Scholar]