Abstract
Nuclear gene(s) have been shown to modulate the phenotypic expression of mitochondrial DNA mutations. We report here the identification and characterization of the yeast nuclear gene MTO2 encoding an evolutionarily conserved protein involved in mitochondrial tRNA modification. Interestingly, mto2 null mutants expressed a respiratory-deficient phenotype when coexisting with the C1409G mutation of mitochondrial 15 S rRNA at the very conservative site for human deafness-associated 12 S rRNA A1491G and C1409T mutations. Furthermore, the overall rate of mitochondrial translation was markedly reduced in a yeast mto2 strain in the wild type mitochondrial background, whereas mitochondrial protein synthesis was almost abolished in a yeast mto2 strain carrying the C1409G allele. The other interesting feature of mto2 mutants is the defective expression of mitochondrial genes, especially CYTB and COX1, but only when coexisting with the C1409G allele. These data strongly indicate that a product of MTO2 functionally interacts with the decoding region of 15 S rRNA, particularly at the site of the C1409G or A1491G mutation. In addition, we showed that yeast and human Mto2p localize in mitochondria. The isolated human MTO2 cDNA can partially restore the respiratory-deficient pheno-type of yeast mto2 cells carrying the C1409G mutation. These functional conservations imply that human MTO2 may act as a modifier gene, modulating the phenotypic expression of the deafness-associated A1491G or C1409T mutation in mitochondrial 12 S rRNA.
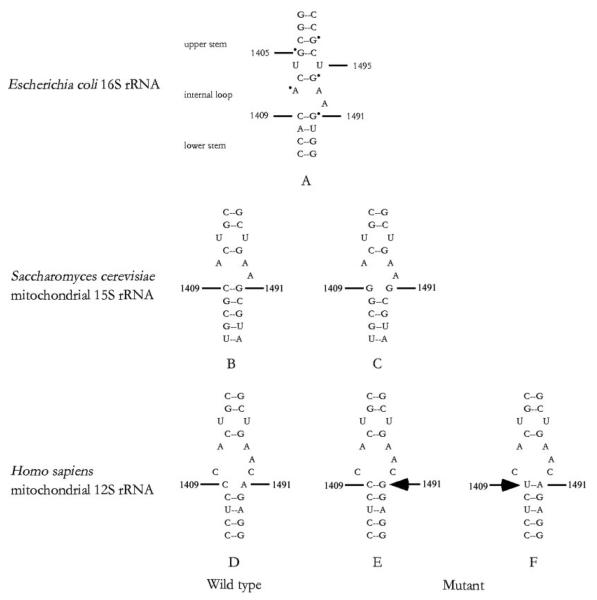
Interaction between the nuclear and mitochondrial genomes is necessary for controlling the phenotypic expression of mtDNA mutation(s). In humans, nuclear modifier gene(s) have been shown to modulate the phenotypic expression of the mitochondrial 12 S rRNA A1491G or C1409T mutation associated with aminoglycoside-induced and nonsyndromic deafness (1-5). As shown in Fig. 1, these mtDNA mutations are located at the A-site of the small ribosomal subunit, which is highly conserved from bacteria to mammals (6). The homologous region of 16 S rRNA in Escherichia coli is an essential part of the decoding site of the ribosome (7, 8) and is crucial for subunit association either by an RNA-protein or RNA-RNA interaction (9). This region is also an important locus of action for aminoglycosides (10, 11). Mutations, which disrupted the 1409–1491 base pair of E. coli 16 S rRNA or Saccharomyces cerevisiae mitochondrial 15 S rRNA, confer aminoglycoside resistance (8, 12-14). In human, the G-C or U-A pair in the mitochondrial 12 S rRNA created by the A1491G or C1409T mutation facilitates the binding of aminoglycoside (4-5, 15, 16). Our previous investigations revealed that the A1491G or C1409T mutation in human mtDNA is the primary contributor underlying the development of deafness but is not sufficient to produce a clinical phenotype (2-5). However, the product of nuclear modifier gene(s), which may functionally interact with the mutated 12 S rRNA, influences the phenotypic manifestation of the A1491G or C1409T mutation by enhancing or suppressing the effect of these mutations (2-5).
Fig. 1. Secondary structure of the decoding site of small ribosomal RNAs.
A, the A-site of the E. coli 16 S rRNA oligonucleotide showing the dimethyl sulfate footprints, observed in the presence of the aminoglycosides neomycin or paromomycin, is marked with a dot (10, 11). The corresponding region of S. cerevisiae mitochondrial 15 S rRNA and human mitochondrial 12 S rRNA is shown as the wild type version (B and D) and in the version containing the C1409G (nt 1477 in yeast 15 S rRNA) (C), A1491G (nt 1555 in human 12 S rRNA) (E), and C1409T mutations (nt 1494 in human 12 S rRNA) (F), respectively.
An interesting model for nuclear-mtDNA interaction for the phenotypic expression of the A1491G or C1409T mutation has been proposed in S. cerevisiae. The mutant allele of MTO1 or MSS1, encoding mitochondrial proteins, manifests a respiratory-deficient phenotype only when coupled with the paromomycin-resistance mitochondrial 15 S rRNA C1409G mutation (PR454 or PR) (17, 18). These observations strongly indicate that Mss1p or Mto1p affects the phenotypic expression of the C1409G mutation by functionally interacting with the region of the C1409G mutation in mitochondrial 15 S rRNA. In E. coli, the products of mnmE (homolog of MSS1) (19), gidA (homolog of MTO1) (20), and trmU (21) have been shown to be involved in the biosynthesis of the hypermodified nucleoside 5-methylaminomethyl-2-thio-uridine (mnm5s2U34) (22). This modified nucleotide, found in the wobble position of several bacterial tRNAs specific for glutamate, lysine, arginine, and glutamine, has a pivotal role in the structure and function of tRNAs, including structural stabilization, aminoacylation, and codon recognition at the decoding site of small ribosomal RNAs (20, 22). Recently, we demonstrated that isolated human MTO1 or GTPBP3 (homolog of MSS1) cDNA can complement the respiratory-deficient phenotype of yeast mto1 or mss1 cells carrying the 15 S rRNA C1409G mutation, suggesting that the functions of those proteins are evolutionarily conserved (23, 24).
In E. coli, trmU has been shown to be responsible for the 2-thiolation of mnm5s2U34 in tRNALys, tRNAGlu, and tRNAGln (20-22). In S. cerevisiae, mto2/mtu1 (homolog of trmU) null mutants conferred defects in 2-thiouridylation in mitochondrial tRNALys, tRNAGlu, and tRNAGln but not cytoplasmic tRNALys (25). To examine whether yeast MTO2 (mitochondrial translational optimization) mediates the phenotypic expression of the 15 S rRNA C1409G mutation, this nuclear gene has been identified, and a null mto2 mutation in the wild type mitochondrial background (paromomycin-sensitive PS) or C1409G (PR) allele has been generated. These mto2 null mutants have been characterized by examining mitochondrial gene expression, mitochondrial protein synthesis, and functional complementation of human MTO2 cDNA in a yeast mto2 mutant carrying the mitochondrial C1409G (PR) allele. Furthermore, the yeast and human Mto2p were examined for subcellular localization by immunofluorescence analysis.
EXPERIMENTAL PROCEDURES
Yeast Strains, Media, and Genetic Techniques
The genotypes and sources of strains of S. cerevisiae used in this investigation are listed in Table I. The media used to grow yeast have been described elsewhere (27-28). Standard procedures were used for crossing and selecting diploids, including sporulation and dissecting tetrads (29).
Table I.
Genotype and sources of yeast strains
| Strains | Genotype | Source |
|---|---|---|
| W303–1B | α ade2-1, his3-1,15, leu2-3, trp1-1, ura3-1 | Ref. 17 |
| W1021–7C | a ade2-1, his3-1,15, leu2-3, ura3-1 | Ref. 26 |
| M12 | a ilv5, trp2 [ρ+, PS] | Ref. 18 |
| M12–54 | a ilv5, trp2 [ρ+, PR454] | Ref. 18 |
| A18 | α, lys2, met6, mss1-18 [PR454] | Ref. 18 |
| W303ΔMTO1(PR) | a ade2-1,his3-1, trp1–1, ura3-1,mto1::URA3[PR454] | Ref. 17 |
| W303ΔMTO2(PS) | α ade2-1, his3-1,15, leu2-3,trp1-1,ura3-1,mto2::HIS3 | This study |
| W303ΔMTO2(ρo) | α ade2-1, his3-1,15, leu2-3,trp1-1,ura3-1,mto2::HIS3 | This study |
| W1021ΔMTO2(PS) | a ade2-1, his3-1, leu2-3, ura3-1, mto2::HIS3 | This study |
| W1021ΔMTO2(ρo) | a ade2-1, his3-1, leu2-3, ura3-1, mto2::HIS3 | This study |
| QY1 | a/α ade2-1/+, his3-1/+, leu2-3/+, ura3-1/+, lys2/+, met6/+, mss1-18/MSS1,mto2::HIS3/MTO2, [PR454] | This study |
| W1021ΔMTO2(PR) | a ade2-1,his3-1,lys2, ura3-1,mto2::HIS3[PR454] | This study |
| W1021ΔMTO2/mss1(PR) | a his3-1,ura3-1, mss1-18, mto2::HIS3[PR454] | This study |
Cloning of Yeast MTO2
The peptide sequence of the E. coli trmU sequence (30) was subjected to a BLAST search of the publicly available S. cerevisiae sequence databases (NCBI/Genbank™/EMBL). This search led to the identification of one open reading frame encoding a protein (YDL033C, GenBank™ accession number Z71781) with a high degree of homology to E. coli TrmU. To obtain the full-length coding region of MTO2 DNA, PCR was performed using the high fidelity Pfu DNA polymerase (Promega) and total yeast genomic DNA isolated from W303–1B cells as a template, with primers 5′-AATTTTAAGAGCGCCGGG-3′ (nucleotides (nt)1 143–163) and 5′-ACATGATTCAAGGGAAAAGACC-3′ (nt 1713–1734) (GenBank™ accession number AY624369). The predominant PCR product was purified by agarose gel electrophoresis and subsequently cloned into a PCR 2.1-TOPO vector (Invitrogen). Sequencing was done using a Dye Terminator cycle sequencing kit (PerkinElmer Life Sciences) and an ABI PRISM™ 3100 genetic analyzer. The resultant plasmid carrying the full-length coding region of yeast MTO2 was designed as pYMTO2.
Construction of mto2 Null Mutants
mto2 null strains were generated by the one-step gene disruption technique of Rothstein (31). A 1.6-kb HindIII/XbaI fragment containing the full-length coding region of the MTO2 gene was isolated by digesting pYMTO2 with HindIII and XbaI and was ligated to the vector pGEM-7Zf(+) (Promega) to produce the resultant plasmid pGEM-MTO2. A 452-base-pair EcoRV fragment (positions 614–1066) spanning the coding region of MTO2 was deleted by digesting with EcoRV. The 1.2-kb fragment containing the full-length coding region of HIS3 was obtained from plasmid pRS303 by digesting with EcoRI and filled in with Klenow PolI. The resultant fragment was ligated into the EcoRV site of pGEM-MTO2 to replace the EcoRV fragment containing the MTO2 coding region. The resultant plasmid containing the MTO2::HIS3 allele was digested with HindIII and XbaI, and the fragment was introduced into wild type yeast strains W1021–7C and W303–1B by the method of Gietz and Schestl (32). The integrated deletion construct was selected for the cells by growing on glucose-minimal medium in the absence of histidine. Disruption was verified by PCR amplification using primers 5′-ATGCTGGCAAGATATTTAAA-3′ (nt 305–324) and 5′-AAGGACTCATCGTCGAT-3′ (nt 1493–1510).
Mitochondrial Gene Expression Analysis
Total cellular RNA was obtained using a Totally RNA™ kit (Ambion) from midlog phase yeast cultures (2.0 × 107 cells) according to the manufacturer's instructions. Equal amounts (20 μg) of total RNA were fractionated by electrophoresis through a 1.8% agarose-formaldehyde gel, transferred onto a positively charged membrane (Roche Applied Science), and initially hybridized with the CYTB-specific RNA probe. The probe was synthesized on the corresponding restriction enzyme-linearized plasmid using a DIG RNA labeling kit (Roche Applied Science). RNA blots were then stripped and rehybridized with DIG-labeled COX1, 15 S rRNA and 21 S rRNA probes, respectively. As an internal control, RNA blots were stripped and rehybridized with a DIG-labeled nuclear 25 S rRNA probe. The plasmids used for mtDNA probes were constructed by PCR-amplifying fragments of CYTB (positions 36595–36964), COX1 (positions 26211–26689), 15 S rRNA (positions 7532–8280), and 21 S rRNA (positions 58484–59580) (Gen-Bank™ accession number AJ011856) (33), as well as a nuclear 25 S rRNA probe (position 631–1665) (GenBank™ accession number U53879) and cloning the fragments into the pCRII-TOPO vector carrying SP6 and T7 promoters (Invitrogen).
Analysis of Mitochondrial Protein Synthesis
Yeast strains were pulse-labeled for 2.5 min with [35S]methionine in methionine-free medium in the presence of cycloheximide to inhibit cytoplasmic protein synthesis, as described elsewhere (34). The radiolabeled proteins were separated on SDS-exponential polyacrylamide gradient gels (2, 35). The gels were treated with Me2SO/2,5-diphenyloxazole, dried, and exposed for fluorography.
Subcellular Localization of Yeast Mto2p
The coding sequence for MTO2 lacking its natural stop codon was obtained by PCR using pYMTO2 as a template. The primers 5′-CCGGAATTCCGGATGCTGGCAAGATATTTA-3′ (nt 305–322) and 5′-CCCAAGCTTGGGTGCATGGGTGTCATTATT-3′ (nt 1538–1555) were used for PCR amplification. PCR products were first cloned into the pCR 2.1-TOPO vector (Invitrogen). After sequence determination, the insert was subsequently subcloned into the expression vector pGFP-C-FUS under the control of the MET25 promoter (36). The MTO2-GFP fusion construct or the vector pGFP-C-FUS alone was transformed into the wild type strain W303–1B. Resultant transformants were grown at 30 °C to midlog phases in 10 ml of glucose-minimal medium with 2% galactose and the appropriate auxotrophic requirements. To stain mitochondria, cells were incubated with 0.05 μm of MitoTracker™ Red CMXRos (Molecular Probes, Portland, Oregon) for 1 h at 30 °C. The cells were then examined under a Carl Zeiss confocal fluorescence microscope.
Cloning and Expression of Human MTO2 cDNA
The coding region of human MTO2 cDNAs lacking its natural stop codon were obtained by reverse transcription-PCR using the high fidelity Pfu polymerase (Promega). Total RNA was extracted from 143B cells to use as a template. The primers 5′-CCGCTCGAGCGGATGCAGGCCTTGCGGCAC-3′ (nt 52–69) and 5′-CGGAATTCCGAGCAAGGGACTCAGGCC-3′ (nt 1297–1314) (GenBank™ accession number AY062123) were used for the PCR amplification. The PCR products were digested with XhoI and EcoRI and cloned into pBluescript II KS+ (Promega). After sequence determination, the inserts were subcloned into pEGFP-N1 (Clontech) to generate pEGFP-N1-MTO2. The resultant construct or vector alone was transfected into the human 143B osteosarcoma cell line using the SuperFect™ transfection reagent (Qiagen) according to the manufacturer's protocol. Immunofluorescence analysis was performed as detailed elsewhere (23, 24).
S. cerevisiae wild type and mto2 strains used for this study were W301–1B and W303ΔMTO2(PR), respectively. A yeast expression shuttle vector pDB20 was used for the expression of human MTO2 in S. cerevisiae (37). A human MTO2 cDNA was obtained by PCR amplification using pEGFP-N1-MTO2 as the template. Primers used for PCR amplification were 5′-AAGCTTAGCTGCAGCTGGCGAAGT-3′ (nt 20–38) and 5′-AAGCTTAGCAGCAAGCTGGCCCTT-3′ (nt 1338–1355). PCR products were cloned into PCR2.1-TOPO vector (Invitrogen). After sequence determination, the insert was subsequently subcloned into pDB20 to generate pDB20-MTO2. These constructs were transformed into the yeast W303ΔMTO2(PR) strain. Ura3+ transformants were selected at 30 °C on glucose-minimal medium. Transformants were then spotted on glucose and glycerol plates and incubated at 30 °C for 3–5 days. Colonies growing on glycerol medium were subjected to further analysis.
RESULTS
Identification of Yeast MTO2
The product of the E. coli trmU sequence (30) was subjected to a BLAST search of the publicly available S. cerevisiae sequence databases (NCBI/Gen-bank™/EMBL). This search led to the identification of one open reading frame encoding a protein with a high degree of homology to E. coli TrmU. The S. cerevisiae homolog MTO2 on chromosome IV (YDL033C, GenBank™ accession number Z71781) is predicted to encode a 417-amino-acid protein with a molecular mass of 47,049 Da. The predicated yeast Mto2 polypeptide revealed an extensive conservation of amino acid sequences and similarity in size by an alignment with homologs, from Homo sapiens, Mus musculus (38), Schizosaccharomyces pombe, Drosophila melanogaster, E. coli (30) to Bacillus subtilis. In particular, the overall identity of the predicted amino acid sequence of S. cerevisiae Mto2p with homologs of H. sapiens, M. musculus, D. melanogaster, S. pombe, B. subtilis, and E. coli is 35, 35, 36, 39, 41, and 39%, respectively, whereas the similarity is 47, 47, 47, 48, 51, and 49%, respectively. Mto2p is likely a mitochondrial protein, due to the presence of a typical mitochondrial target presequence (39).
The mto2 Null Mutant Expresses a Respiratory-deficient Phenotype when Combined with the 15 S rRNA C1409G Allele
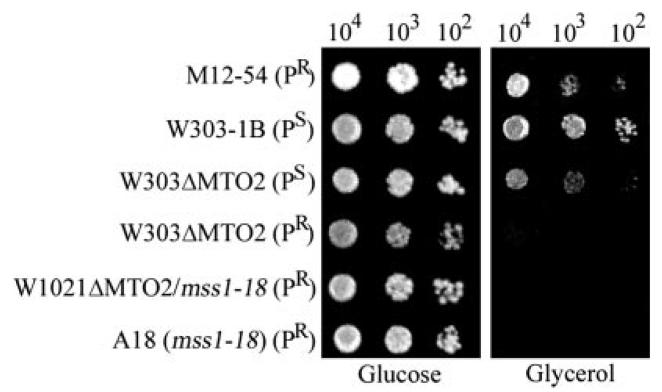
To determine whether MTO2 is essential for mitochondrial function, a single copy of the gene was disrupted in a haploid strain. The resulting mto2 null strains were tested for growth on glycerol-complete medium. As can been seen in Fig. 2, the mto2 null cells in the wild type mitochondrial background (PS) were able to grow on glycerol-complete medium, indicating that these cells were respiratory-competent. To test whether the mitochondrial genetic background affects the phenotypic expression of mto2 null mutants, the W303ΔMTO2(PS) strain was treated with ethidium bromide to cause the loss of mtDNA (ρo) (28). ρo derivatives were then crossed with the M12–54 strain carrying the 15 S rRNA C1409G (PR) mutation or with the M12 strain carrying the identical nuclear genetic background and the PS allele (18). The resulting diploids were sporulated and products of meiosis were dissected onto glucose medium. Meiotic progeny derived from the cross between the mto2 null ρo strain and M12–54 showed 2:2 segregation of the respiratory-competent phenotype. In all cases, the respiratory-deficient phenotype coincided with histidine independence (data not shown). By contrast, all four meiotic progeny from the cross between the mto2 null ρo strain and the wild type strain M12 (PS) were respiratory-competent, even though the His3+ phenotype showed 2:2 segregation patterns. Furthermore, the W1021ΔMTO2 ρo strain was also crossed with the mss1 strain A18 carrying the 15 S rRNA C1409G allele and sporulated. The resulting mss1/mto2 double mutants were unable to grow on glycerol medium. The double mutants were then examined for the presence of mtDNA by the PCR amplification of mitochondrial 15 S rRNA. All meiotic progeny from the cross retained the 15 S rRNA (data not shown), suggesting that the strains still retained mtDNA, despite some of them showing the respiratory-deficient phenotype. These data strongly suggested that the expression of respiratory deficiency in the mto2 null mutant was fully dependent on the presence of the 15 S rRNA C1409G mutation.
Fig. 2. Growth properties of mto2 mutants.
Series dilutions of the MTO2 strains W303–1B(PS) and M12–54(PR), two null mto2 strains W303ΔMTO2(PR) and W303ΔMTO2(PS), mss1 strain (A18) (18) and W1021ΔMTO2(PR)/mss1 double mutant strain were spotted on glucose and glycerol plates. The plate was then incubated at 30 °C in glucose medium for 3 days and in glycerol medium for 5 days.
mto2 Mutation Affects the Expression of Mitochondrial Genome
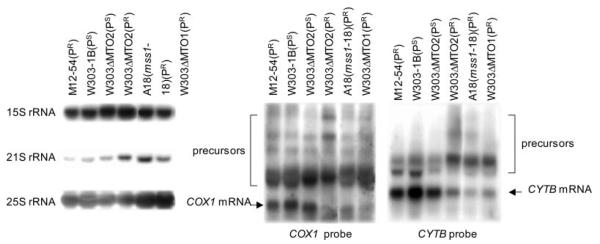
Previous studies revealed that the mss1 or mto1 mutation had effects on the expression of mitochondrial genes when carrying the C1409G allele (17, 18). We therefore examined whether the mto2 null mutation also affected the expression of mitochondrial genes by Northern blot analysis. RNA blots were hybridized with DIG-labeled probes for exon regions of CYTB, COX1, 15 S rRNA, and 21 S rRNA, respectively. In these mitochondrial genes, the 15 S rRNA gene lacks any intron, whereas the 21 S rRNA gene harbors two exons and one intron (33). Furthermore, CYTB consists of six exons and five introns, whereas COX1 contains 8 exons and 7 introns (33). As an internal control, RNA blots were stripped and rehybridized with the DIG-labeled nuclear-encoded 25 S rRNA probe.
As shown in Fig. 3, the 15 S rRNA and 21 S rRNA did not show obvious size changes and precursor accumulation in mto2 strains, as well as in mss1 and mto1 strains. However, there were increasing expressions of 15 S rRNA and 21 S rRNA genes in the mto2 cells in the PR background when compared with the wild type strain W303–1B. Deletion of MTO2 in the wild type mtDNA background had no obvious effect on CYTB processing, while it led to a slight accumulation of COX1 precursors. By contrast, in the presence of the 15 S rRNA PR allele, the mto2 mutant, similar to mss1 and mto1 mutants, had lower levels of mature CYTB mRNA and accumulated unprocessed or partially processed precursors. Furthermore, the COX1 probe detected an extremely low concentration of the mature mRNA and a high accumulation of unprocessed or partially processed precursors in mto2 mutants carrying the 15 S rRNA PR allele. It appeared that there were slightly different precursor accumulations of COX1 between mto2 and mss1 or mto1 mutants.
Fig. 3. Expression analysis of mitochondrial genes.
Equal amounts (20 μg) of total cellular RNA from various mutant and control strains were electrophoresed through a 1.8% agarose-formaldehyde gel and transferred onto a positively charged membrane. The blot was hybridized with a DIG-labeled CYTB RNA probe. After stripping the blot, it was hybridized with DIG-labeled COX1, 15 S RNA, and 21 S rRNA probes, respectively. Subsequently, after restripping of the blot, it was hybridized with a DIG-labeled nuclear 25 S RNA probe as an internal control.
Mitochondrial Protein Synthesis Defect in the mto2 Strains
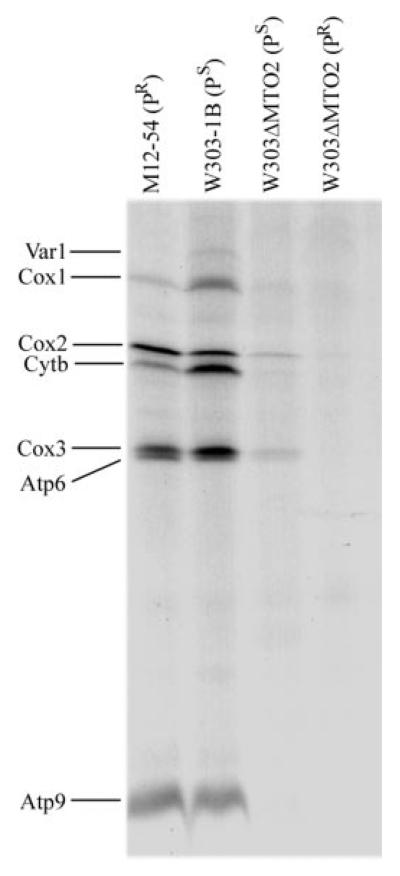
To examine whether the mto2 null mutation impairs mitochondrial protein synthesis, mutant and control strains were labeled for 2.5 min with [35S]methionine in a methionine-free medium in the presence of cycloheximide to inhibit cytoplasmic protein synthesis. Fig. 4 shows SDS-PAGE electro-phoretic patterns of the organelle-specific products of mutant and wild type strains. Interestingly, the patterns of the mitochondrial translational products from mto2 mutants carrying the PS or PR allele differ from those of the wild type MTO2 strains. Very strikingly, the mitochondrial translation was almost abolished in the mto2 null mutant carrying the mitochondrial C1409G (PR) allele, whereas there was a marked reduction in the overall labeling of the mitochondrial translation products in the mto2 mutant carrying the wild type mitochondrial genome (PS) when compared with wild type strain W303–1B. Furthermore, it is worthwhile to note that there were some differences in the mitochondrial protein labeling between the strain M12–54 carrying the PR allele and a wild type strain, W303–1B. In particular, several polypeptides, especially Var1, Cox1, and Cytb, were less labeled in the strain M12–54 carrying the PR allele than in the wild type strain W303–1B carrying the PS allele.
Fig. 4. Mitochondrial protein labeling analysis.
Electrophoretic patterns of the mitochondrial translation products of wild type and mutant cells labeled for 2.5 min with [35S]methionine in the presence of 150 μg of cycloheximide/ml. Samples containing equal amounts of total cellular protein (50 μg) were run in SDS/polyacrylamide gradient gels. Var1 (a ribosomal protein), Cox1, -2, and -3 (subunits I, II, and III of cytochrome c oxidase), Atp6 and Atp9 (subunits 6 and 9 of the H+-ATPase), and Cytb (apocytochrome b) are shown.
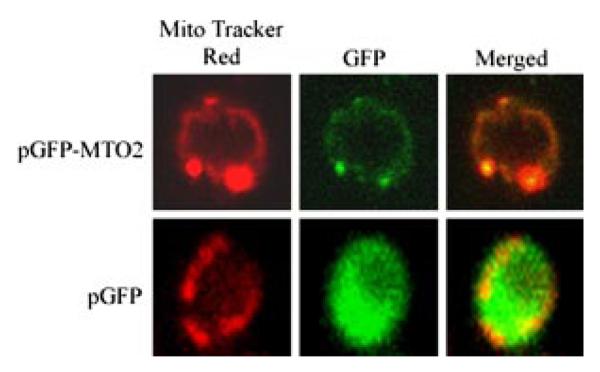
Subcellular Location of the Yeast Mto2p
To determine the subcellular localization of Mto2p, a DNA fragment spanning the entire coding region (but lacking its natural stop codon) was obtained by PCR amplification using pYMTO2 as the template. After sequence determination, the insert was subsequently subcloned into the pGFP-C-Fus to fuse yeast MTO2 with the GFP cDNA under the control of the MET25 promoter. This vector allows the expression of the GFP-MTO2 fusion protein in methionine-free medium. The resultant construct pGFP-CFUS-MTO2 was then introduced into the yeast W303–1B strain, and the resultant Ura+ transformants were subsequently stained with MitoTracker Red™. As can been seen in Fig. 5, MTO2-GFP-expressing cells showed a green fluorescence typical of a mitochondrial pattern. With the mitochondrial-specific dye MitoTracker Red™, the same cells showed a pattern of red fluorescence that coincides with MTO2-GFP expression. The superimposition of the two panels showed almost a complete overlap of the two patterns, demonstrating that the yeast Mto2p localizes to mitochondria.
Fig. 5. Yeast Mto2p is a mitochondrial protein.
Wild type W303–1B cells were transformed with the plasmid pGFP-C-FUS-MTO2 or pGFP-C-FUS, and the resultant transformant cells were stained with MitoTracker Red™ and then examined under a Carl Zeiss confocal fluorescence microscope, as detailed under “Experimental Procedures.” Mto2p-GFP, MitoTracker Red™, and merged images with both Mto2p and MitoTracker Red™ are shown.
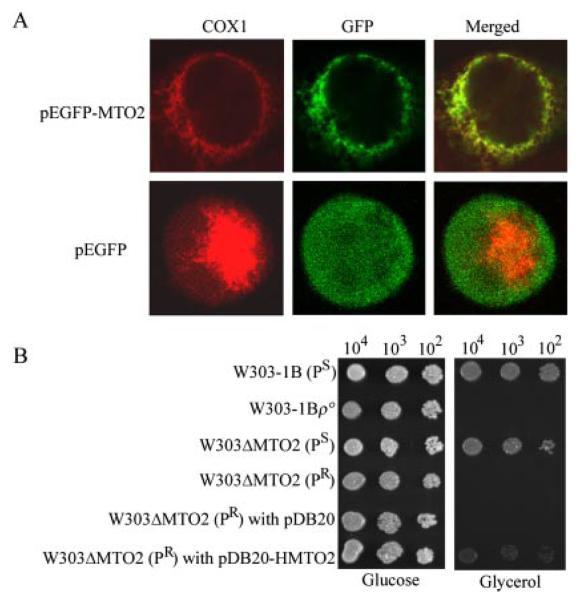
Functional Complementation of the Yeast mto2 Mutant by Human MTO2 cDNA
We have cloned the putative human homolog MTO2. The subcellular location of human Mto2p was examined by transient expression of the protein fused with GFP in human 143B cells. Fig. 6A shows the immunofluorescence pattern of transfected 143B cells double-labeled with rabbit monoclonal antibody specific for GFP and mouse monoclonal antibody to COXI, a subunit of the cytochrome c oxidase (COX) complex in the mitochondrial inner membrane. Typical mitochondrial staining patterns were observed with both antibodies, and superimposition of the two panels showed complete overlap of the two patterns. These data demonstrated that the product of human MTO2 localizes exclusively to mitochondria.
Fig. 6. Properties and expression of human gene homolog of yeast MTO2.
A, subcellular localization of hMto2p in human 143B cells. Cells were transiently transfected with a MTO2 cDNA fused with GFP or pEGFP. The fusion protein was visualized by indirect immunofluorescence using antibodies to human COX1 and to GFP. A merged image is shown on the right. B, series dilutions of the wild type and mutant strains (the null mto2 strain carrying the C1409G allele transformed with the pDB20-HMTO2 and pDB20 vector) were spotted on glucose and glycerol media, respectively. The plates were then incubated at 30 °C in glucose medium for 3 days and in glycerol medium for 5 days.
To examine the functional conservation of yeast MTO2, we tested whether human MTO2 cDNA was able to complement the respiratory defect of the yeast mto2 mutant carrying the mitochondrial C1409G allele. The entire coding region of human MTO2 cDNA was cloned into an expression vector, pDB20, designed as pDB20-HMTO2. These constructs were then transformed into a yeast mto2 strain W303ΔMTO2 (PR) carrying the C1409G allele, and the resultant transformants were tested for their growth on rich glycerol medium. As illustrated in Fig. 6B, the growth defect of the mto2 null mutant on glycerol medium was partially restored by the entire coding region of human MTO2 cDNA (pDB20-HMTO2).
DISCUSSION
In this study, we have identified and characterized yeast MTO2, a nuclear gene encoding a mitochondrial protein involved in tRNA modification. The first 23 amino acids of the N-terminal sequence of yeast Mto2p make up a characteristic mitochondrial targeting sequence (39). The MitoProt program predicts that the probability for yeast Mto2p to import into mitochondria is 0.87, suggesting that Mto2p has a strong mitochondrial targeting signal. Fluorescence analysis of yeast cells expressing the MTO2-GFP fusion protein demonstrated the co-localization of Mto2p with mitochondrial specific dye (MitoTracker Red™), indicating that Mto2p functions in mitochondria. The alignments of yeast Mto2p with its homologs from other species, including human, mouse, S. pombe, E. coli, and D. melanogaster, showed a high amino acid sequence conservation as well as similarity in size (38). In E. coli, TrmU (the homolog of Mto2p) has been shown to be responsible for the biosynthesis of hypermodified 2-thiouridine (s2U) in the wobble position of tRNALys, tRNAGlu, and tRNAGln (21, 40-42). 2-Thiouridine modification was also found in the wobble position of the yeast mitochondrial tRNAs (43). In human mitochondria, 5-taurinomethyl-2-thiouridine (τm5s2U) has been shown to exist at position 34 of tRNALys, tRNAGlu, and tRNAGln (25, 44). τm5s2U is further modified to mnm5s2U34 in the same position of those tRNAs in E. coli and human (20, 22). Indeed, this modified nucleotide plays a pivotal role in the structure and function of tRNAs, including the stabilization of anticodon structure, the ribosome binding ability to tRNA, and the improvement of reading frame maintenance (22). Thus, the localization to mitochondria and the degree of amino acid conservation between yeast Mto2p and its homologs imply that yeast Mto2p is responsible for the 2-thiouridylation of the nucleotide at position 34 of mitochondrial tRNAs. In fact, the disruption of yeast MTO2/MTU1 or siRNA down-regulation of human MTO2/TMU1/TRMT1 leads to a defect in 2-thiouridylation in yeast and human mitochondrial tRNALys, tRNAGlu, and tRNAGln (but not cytoplasmic tRNALys), respectively (25). These observations demonstrated the role of yeast Mto2p in the 2-thiouridylation of the nucleotide at position 34 of these mitochondrial tRNAs.
Unmodified tRNAs caused by the deletion of MTO2, similar to those in cell lines carrying the A10S mutation in human MTO2/TRMT1,2 likely leave the tRNA more exposed to degradation, thus reducing the levels of mitochondrial tRNAs. Alternatively, these unmodified tRNAs may not function accurately with the mitochondrial ribosomes, especially affecting the efficiency and accuracy of codon-anticodon interaction. Indeed, a considerable number of unmodified tRNAs bind their cognate or wobble codon very poorly in the A-site of the small ribosomal subunit in E. coli (41, 46, 47). It has also been shown that a lack of tRNA modification can enhance translational frameshift (20, 48), and a defect in the synthesis of this mnm5s2U34 of tRNAGlu gives rise to an impairment of aminoacylation (49). Furthermore, it has been suggested that the wobble modification defect is the primary factor for mutant tRNALys losing its cognate codon binding affinity, which results in the mutant tRNALys becoming translationally inactive, which subsequently results in mitochondrial dysfunction (50). Thus, a failure in mitochondrial tRNA metabolism most probably accounts for a marked decrease in the overall rate of mitochondrial translation in the mto2 strain in the wild type mtDNA background. However, it appears that the defect caused by the deletion of MTO2 itself is not sufficient to produce a respiratory-deficient phenotype, as mto2 mutants in the wild type mtDNA background were able to grow on glycerol medium. On the other hand, in the presence of the 15 S rRNA C1409G mutation, mto2 mutants exhibited a complete loss of a total mitochondrial protein labeling, in contrast to the fact that there was only the complete absence of subunit 1 of cytochrome oxidase (the product of COX1 in the mto1 or mss1 mutant (17, 18). As a result, the mto2 null mutation confers the respiratory-deficient phenotype when its mtDNA carries the C1409G allele. These observations strongly indicate that the products of MTO2, similar to Mss1p and Mto1p (17, 18), functionally interact with the decoding region of 15 S rRNA, particularly at the site of the C1409G mutation.
The C1409G mutation, as shown in Fig. 1, disrupted a highly conserved C1409-G1491 base pairing at the decoding site of mitochondrial ribosome where the codon-anticodon recognition occurs (7, 10, 11, 12, 18). This base pair is adjacent to the A-site tRNA binding bases, including A1408, A1492, and A1493 (10, 11). Thus, the C1409G mutation may result in a local conformational change in the A-site of mitochondrial 15 S rRNA, thereby affecting the efficiency and accuracy of codon-anticodon interaction. In particular, the hypermodified tRNAs (synthesized by the participation of Mss1p, Mto1p, and Mto2p) are less efficient for the decoding of the codon ending in G than C of the 15 S rRNA (14, 24). In fact, those ribosomes carrying the mutated 15 S rRNA had a decreased level of natural frame-shifting, suggesting a more stringent proofreading (14). In addition, a mild defect in the mitochondrial protein labeling was observed in the yeast strain M12–54 carrying the C1409G allele. However, this strain was able to respire, implying that the C1409G mutation itself is insufficient to lead to a respiratory-deficient phenotype. These observations are consistent with our previous data indicating that there was a mild reduction in the rate of mitochondrial protein synthesis in human cell lines carrying the deafness-associated 12 S rRNA A1491G (2, 3) or C1409T mutation (5). On the other hand, unmodified tRNAs caused by the deletion of MTO2 may be much less efficient in binding to the A-site nucleotides, including A1492 and A1493. In the case of E. coli, tRNALys(UUU)-mnm5U34 protected 16 S rRNA bases A1492 and A1493 from chemical modification, whereas the protection of A-site bases was not evident when unmodified tRNALys(UUU) was present (51). Therefore, the C1409G mutation in 15 S rRNA, in combination with the mto2 mutation, led to the nearly complete loss of mitochondrial protein synthesis. Such translational defects apparently alter the synthesis of the maturases encoded by the mitochondrial genome (45, 52, 53). Maturases are required for the removal of introns from mitochondrial genes, including CYTB and COX1 (17, 18). Thus, introns in mitochondrial genes (especially for CYTB and COX1) were not spliced completely, thereby causing the accumulation of unprocessed or partially processed precursors of CYTB and COX1 in the mto2 mutants in the context of the 15 S rRNA C1409G allele, as in the case of mss1 and mto1 mutants (17, 18). These data strongly support the idea that the products of MTO2, MSS1, and MTO1 are members of the same pathway for the biosynthesis of hypermodified nucleoside 5-methyl-aminomethyl-2-thio-uridine (mnm5s2U34) of mitochondrial tRNAs (17, 20, 25).
The observation that human MTO2 cDNA can functionally complement the respiratory-deficient phenotype of the yeast mto2 allele carrying the C1409G mutation implicates that the products of yeast and human MTO2 have a similar function in mitochondrial tRNA modification. Modified mitochondrial tRNAs (synthesized by the participation of the product of MTO2) apparently interact with the decoding region of small ribosomal RNA, particularly in the vicinity of C1409T and A1491G. Thus, the product of human MTO2 may regulate the translational efficiency and accuracy of codon-anticodon base pairings in the mitochondrial ribosomes. Therefore, the defect in the expression or mutation(s) of human MTO2 could act as a nuclear modifier factor and then contribute to the phenotypic variability of deafness-associated A1491G or C1409T mutation by enhancing or suppressing the phenotypic manifestation of the A1491G or C1409T mutation.
Acknowledgments
We thank Dr. Alex Tzagoloff (Columbia University) for yeast strains, Dr. Mark Johnston (Washington University, St. Louis, MO) for the pEGFP-Fus-C vector, and Dr. Leonard Guarente (Massachusetts Institute of Technology, Cambridge, MA) for pDB20 vector. We are grateful to Li Yang, Chuck Loftice, and Terri Wallace for technical and clerical support.
Footnotes
This work was supported by Public Health Service Grant RO1DC05230 from the NIDCD, National Institutes of Health, Grant RO1NS44015 from the NINDS, National Institutes of Health, and a grant from the National Basic Research Priorities Program of China (2004CCA02200) (to M.-X. G.).
The nucleotide sequence(s) reported in this paper has been submitted to the GenBank™/EBI Data Bank with accession number(s) AY062123 and AY624369.
The abbreviations used are: nt, nucleotide(s); GFP, green fluorescent protein; DIG, digoxigenin.
Q. Yan, X. Li, Y. Bykhovskaya, P. Hajek, N. Umeda, E. Mengesha, R. Li, J. L. Peters, T. Suzuki, M. Shohat, Y. Qian, X. Estivill, K. Watanabe, N. Fischel-Ghodsian, and M. X. Guan, submitted for publication.
REFERENCES
- 1.Guan MX. Ann. N. Y. Acad. Sci. 2004;1011:259–271. doi: 10.1007/978-3-662-41088-2_25. [DOI] [PubMed] [Google Scholar]
- 2.Guan MX, Fischel-Ghodsian N, Attardi G. Hum. Mol. Genet. 2001;10:573–580. doi: 10.1093/hmg/10.6.573. [DOI] [PubMed] [Google Scholar]
- 3.Guan MX, Fischel-Ghodsian N, Attardi G. Hum. Mol. Genet. 1996;5:963–971. doi: 10.1093/hmg/5.7.963. [DOI] [PubMed] [Google Scholar]
- 4.Zhao H, Li R, Wang Q, Yan Q, Deng JH, Han D, Bai Y, Young WY, Guan MX. Am. J. Hum. Genet. 2004;74:139–152. doi: 10.1086/381133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao H, Young WY, Yan Q, Li R, Cao J, Wang Q, Li X, Peters JL, Han D, Guan MX. Nucleic Acids Res. 2005;33:1132–1139. doi: 10.1093/nar/gki262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neefs JM, Van de Peer Y, De Rijik P, Goris A, De Wachter R. Nucleic Acids Res. 1991;19(suppl.):1987–2018. doi: 10.1093/nar/19.suppl.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zimmermann RA, Thomas CL, Wower J. In: The Ribosome: Structure, Function, and Evolution. Hill WE, Moore PB, Dahlberg A, Schlessinger D, Garrett RA, Warner JR, editors. American Society for Microbiology; Washington, D. C.: 1990. pp. 331–347. [Google Scholar]
- 8.De Stasio EA, Dahlberg AE. J. Mol. Biol. 1990;212:127–133. doi: 10.1016/0022-2836(90)90309-A. [DOI] [PubMed] [Google Scholar]
- 9.Zwieb C, Jemiolo DK, Jacob WF, Wagner R, Dahlberg AE. Mol. Gen. Genet. 1986;203:256–264. doi: 10.1007/BF00333963. [DOI] [PubMed] [Google Scholar]
- 10.Moazed D, Noller HF. Nature. 1987;327:389–394. doi: 10.1038/327389a0. [DOI] [PubMed] [Google Scholar]
- 11.Purohit P, Stern S. Nature. 1994;370:659–662. doi: 10.1038/370659a0. [DOI] [PubMed] [Google Scholar]
- 12.Gregory ST, Dahlberg AE. Nucleic Acids Res. 1995;23:4234–4238. doi: 10.1093/nar/23.21.4234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li M, Tzagoloff A, Underbrink-Lyon K, Martin NC. J. Biol. Chem. 1982;257:5921–5928. [PubMed] [Google Scholar]
- 14.Weiss-Brummer B, Huttenhofer A. Mol. Gen. Genet. 1989;217:362–369. doi: 10.1007/BF02464905. [DOI] [PubMed] [Google Scholar]
- 15.Hamasaki K, Rando RR. Biochemistry. 1997;36:12323–12328. doi: 10.1021/bi970962r. [DOI] [PubMed] [Google Scholar]
- 16.Guan MX, Fischel-Ghodsian N, Attardi G. Hum. Mol. Genet. 2000;9:1787–1793. doi: 10.1093/hmg/9.12.1787. [DOI] [PubMed] [Google Scholar]
- 17.Colby G, Wu M, Tzagoloff A. J. Biol. Chem. 1998;273:27945–27952. doi: 10.1074/jbc.273.43.27945. [DOI] [PubMed] [Google Scholar]
- 18.Decoster E, Vassal A, Faye G. J. Mol. Biol. 1993;232:79–88. doi: 10.1006/jmbi.1993.1371. [DOI] [PubMed] [Google Scholar]
- 19.Cabedo H, Macian F, Villarroya M, Escudero JC, Martinez-Vicente M, Knecht E, Armengod ME. EMBO J. 1999;18:7063–7076. doi: 10.1093/emboj/18.24.7063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brégeon D, Colot V, Miroslav M, Radman M, Taddei F. Genes Dev. 2001;15:2295–2306. doi: 10.1101/gad.207701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kambampati R, Lauhon CT. Biochemistry. 2003;42:1109–1117. doi: 10.1021/bi026536+. [DOI] [PubMed] [Google Scholar]
- 22.Björk GR. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt FC, Curtiss R III, Ingraham JL, Lin ECC, Low BK, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE, editors. American Society for Microbiology; Washington, D. C.: 1996. pp. 861–886. [Google Scholar]
- 23.Li X, Guan MX. Mol. Cell. Biol. 2002;22:7701–7711. doi: 10.1128/MCB.22.21.7701-7711.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li X, Li R, Lin X, Guan MX. J. Biol. Chem. 2002;277:27256–27264. doi: 10.1074/jbc.M203267200. [DOI] [PubMed] [Google Scholar]
- 25.Umeda N, Suzuki T, Yukawa M, Ohya Y, Shindo H, Watanabe K, Suzuki T. J. Biol. Chem. 2005;280:1613–1624. doi: 10.1074/jbc.M409306200. [DOI] [PubMed] [Google Scholar]
- 26.Qiu J, Guan MX, Bailis AM, Shen BH. Nucleic Acids Res. 1998;26:3077–3083. doi: 10.1093/nar/26.13.3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen XJ, Guan MX, Clark-Walker GD. Nucleic Acids Res. 1993;21:3473–3477. doi: 10.1093/nar/21.15.3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guan MX. Mol. Gen. Genet. 1997;255:525–532. doi: 10.1007/s004380050525. [DOI] [PubMed] [Google Scholar]
- 29.Sherman F, Fink GR, Hicks JB. Methods in Yeast Genetics. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1983. [Google Scholar]
- 30.Green SM, Malik T, Giles IG, Drabble WT. Microbiology (N. Y.) 1996;142:3219–3230. doi: 10.1099/13500872-142-11-3219. [DOI] [PubMed] [Google Scholar]
- 31.Rothestein RJ. Methods Enzymol. 1993;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- 32.Gietz RD, Schiestl RH. Yeast. 1991;7:253–263. doi: 10.1002/yea.320070307. [DOI] [PubMed] [Google Scholar]
- 33.Foury F, Roganti T, Lecrenier N, Purnelle B. FEBS Lett. 1998;440:325–331. doi: 10.1016/s0014-5793(98)01467-7. [DOI] [PubMed] [Google Scholar]
- 34.Barrientos A, Korr D, Tzagoloff A. EMBO J. 2002;21:43–52. doi: 10.1093/emboj/21.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chomyn A. Methods Enzymol. 1996;264:197–211. doi: 10.1016/s0076-6879(96)64020-8. [DOI] [PubMed] [Google Scholar]
- 36.Niedenthal RK, Riles L, Johnston M, Hegemann JH. Yeast. 1996;12:773–786. doi: 10.1002/(SICI)1097-0061(19960630)12:8%3C773::AID-YEA972%3E3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 37.Becker DM, Fikes JD, Guarente L. Proc. Natl. Acad. Sci. U. S. A. 1991;88:1968–1972. doi: 10.1073/pnas.88.5.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yan Q, Guan MX. Biochim. Biophys. Acta. 2004;1676:119–126. doi: 10.1016/j.bbaexp.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 39.Hartl FU, Neupert W. Science. 1990;247:930–938. doi: 10.1126/science.2406905. [DOI] [PubMed] [Google Scholar]
- 40.Sullivan MA, Cannon JF, Webb FH, Bock RM. J. Bacteriol. 1985;161:368–376. doi: 10.1128/jb.161.1.368-376.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Elseviers D, Petrullo LA, Gallagher PJ. Nucleic Acids Res. 1984;12:3521–3534. doi: 10.1093/nar/12.8.3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hagervall TG, Pomerantz SC, McCloskey JA. J. Mol. Biol. 1998;284:33–42. doi: 10.1006/jmbi.1998.2162. [DOI] [PubMed] [Google Scholar]
- 43.Nakai Y, Umeda N, Suzuki T, Nakai M, Hayashi H, Watanabe K, Kagamiyama H. J. Biol. Chem. 2004;279:12363–12368. doi: 10.1074/jbc.M312448200. [DOI] [PubMed] [Google Scholar]
- 44.Suzuki T, Suzuki T, Wada T, Saigo K, Watanabe K. EMBO J. 2002;21:6581–6589. doi: 10.1093/emboj/cdf656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rodeheffer MS, Shadel GS. J. Biol. Chem. 2003;278:18695–18701. doi: 10.1074/jbc.M301399200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ashraf SS, Sochacka E, Cain R, Guenther R, Malkiewicz A, Agris PF. RNA (N. Y.) 1999;5:188–194. doi: 10.1017/s1355838299981529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yarian C, Marszalek M, Sochacka E, Malkiewicz A, Guenther R, Miskiewicz A, Agris PF. Biochemistry. 2000;39:13390–13395. doi: 10.1021/bi001302g. [DOI] [PubMed] [Google Scholar]
- 48.Urbonavicius J, Qian Q, Durand JM, Hagervall TG, Björk GR. EMBO J. 2001;20:4863–4873. doi: 10.1093/emboj/20.17.4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krüger MK, Sørensen MA. J. Mol. Biol. 1998;284:609–620. doi: 10.1006/jmbi.1998.2197. [DOI] [PubMed] [Google Scholar]
- 50.Yasukawa T, Suzuki T, Ishii N, Ohta S, Watanabe K. EMBO J. 2001;20:4794–4802. doi: 10.1093/emboj/20.17.4794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yarian C, Townsend H, Czestkowski W, Sochacka E, Malkiewicz AJ, Guenther R, Miskiewicz A, Agris PF. J. Biol. Chem. 2002;277:16391–16395. doi: 10.1074/jbc.M200253200. [DOI] [PubMed] [Google Scholar]
- 52.Costanzo MC, Fox TD. Annu. Rev. Genet. 1990;24:91–108. doi: 10.1146/annurev.ge.24.120190.000515. [DOI] [PubMed] [Google Scholar]
- 53.Islas-Osuna MA, Ellis TP, Marnell LL, Mittelmeier TM, Dieckmann CL. J. Biol. Chem. 2002;277:37987–37990. doi: 10.1074/jbc.M206132200. [DOI] [PubMed] [Google Scholar]