SUMMARY
Understanding the production and function of specialized cells during development requires the isolation of individual cell types for analysis, but this is currently a major technical challenge. Here we describe a method for cell type-specific RNA and chromatin profiling that circumvents many of the limitations of current methods for cell isolation. We used in vivo biotin labeling of a nuclear envelope protein in individual cell types followed by affinity isolation of labeled nuclei to measure gene expression and chromatin features of the hair and non-hair cell types of the Arabidopsis root epidermis. We identified hundreds of genes that are preferentially expressed in each cell type and show that genes with the largest expression differences between hair and non-hair cells also show differences between cell types in the trimethylation of histone H3 at lysines 4 and 27. This method should be applicable to any organism that is amenable to transformation.
INTRODUCTION
Growth and development of multicellular organisms requires the production of many specialized cell types that make up the tissues and organs of the adult body. The generation of a differentiated cell from an undifferentiated progenitor involves epigenetic reprogramming of the stem cell genome to establish the appropriate lineage-specific transcription program. Initial establishment and subsequent maintenance of this transcriptional program is effected through chromatin-based gene silencing and activation mechanisms involving the dynamic interplay of transcription factors, post-translational modification of histones, the deposition of histone variants, DNA methylation, and nucleosome remodeling (Brien and Bracken, 2009; Muller and Leutz, 2001; Ng and Gurdon, 2008). Defining precisely how cellular differentiation is imposed and maintained is a central goal of developmental biology, and is also critical to understanding how the process can go awry, leading to disease states such as cancer. Despite the importance of this problem, our knowledge of the mechanics of differentiation processes in vivo is still quite limited, in large part due to the technical difficulty associated with isolating pure cell types from a tissue for transcriptional and epigenomic profiling.
Current methods for the study of pure individual cell types include the use of cultured cell lines (Mito et al., 2005; Rao and Stice, 2004; Rivolta and Holley, 2002), ex vivo differentiation from progenitor cells (Bhattacharya et al., 2009; Irion et al., 2008), laser capture microdissection (LCM) of sectioned tissues (Brunskill et al., 2008; Jiao et al., 2009; Nakazono et al., 2003), and fluorescence-activated cell sorting (FACS) of fluorescently labeled cell lines or protoplasts (Birnbaum et al., 2003; de la Cruz and Edgar, 2008; Gifford et al., 2008; Zhang et al., 2002). Of these techniques, LCM and FACS are the only ones applicable to in vivo studies, but both are limited in that they involve extensive tissue manipulation, require complex and highly expensive equipment, and offer relatively low throughput. Several new methods, such as cell type-specific chemical modification of RNA (Miller et al., 2009) and affinity tagging of ribosomal proteins or poly(A)-binding proteins (Heiman et al., 2008; Mustroph et al., 2009; Roy et al., 2002) have also been successfully employed to measure the gene expression profiles of individual cell types, but these approaches cannot be used to study chromatin features.
In order to circumvent the limitations of current methods and to make the study of cell differentiation and function more accessible, we sought to develop a simple and generally applicable method for studying gene expression and chromatin in individual cell types. To avoid the need for dissociating or mechanically separating cells, we developed a strategy to transgenically tag nuclei in specific cell types and then use affinity isolation to purify them from the total pool of nuclei derived from a tissue. A similar strategy has been used to isolate chloroplasts from specific cell types (Truernit and Hibberd, 2007), and fluorescently labeled phloem cell nuclei have been purified by FACS and used for gene expression analysis (Zhang et al., 2008). Furthermore, it has been shown that the nuclear and total cellular mRNA pools are generally comparable, making nuclei a reasonable source of mRNA for gene expression measurements (Barthelson et al., 2007; Jacob et al., 2007) Thus, affinity purified nuclei should be easy to obtain and could be used for the measurement of the gene expression and chromatin profiles of individual cell types. Our strategy to achieve this was to express a fusion protein consisting of a nuclear envelope targeting sequence, green fluorescent protein (GFP), and the biotin ligase recognition peptide (BLRP), in the presence of E. coli biotin ligase (BirA) in individual cell types in order to generate biotinylated nuclei specifically in those cells. These nuclei could then be purified from the total nuclear pool by virtue of the interaction between biotin and streptavidin. We refer to this strategy as INTACT, for isolation of nuclei tagged in specific cell types.
As a proof-of-concept we employed the INTACT system to study the two cell types of the Arabidopsis root epidermis: hair cells and non-hair cells. These two cell types originate from a common progenitor and make up the entire epidermal layer of the root, arising in alternating vertical cell files along the axis of this organ. The hair cells form long tubular outgrowths that are involved in water and nutrient uptake, anchorage, and interaction with soil microbes, while the non-hair cells do not produce such outgrowths (Grierson and Schiefelbein, 2003). The formation of these cell types has been extensively studied at the genetic and cell biological levels (Ishida et al., 2008) and many genes that are expressed preferentially in each cell type have been identified (Birnbaum et al., 2003; Brady et al., 2007; Won et al., 2009), providing us with a point of comparison for our gene expression studies using the INTACT method.
Our results demonstrate that the INTACT method results in high yield and purity of nuclei from each of the tested cell types and is suitable for both gene expression and chromatin profiling studies. We identified hundreds of genes that are preferentially expressed in each cell type, including nearly all of the previously confirmed hair cell-specific genes. We further show that the preferential expression of a gene in one cell type often correlates with major differences between the cell types in the trimethylation of histone H3 at lysines 4 and 27, demonstrating that chromatin differences exist between hair and non-hair cells and these can be readily monitored in nuclei purified using this method. The INTACT method is simple, fast, and should be widely applicable.
RESULTS
Establishment of the INTACT method
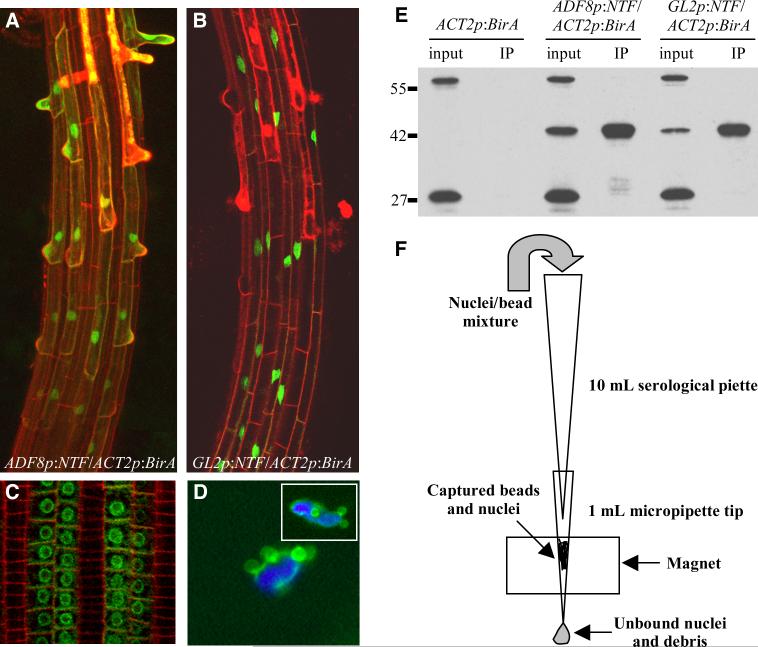
We first developed a system for biotin tagging of nuclei using an outer nuclear envelope-targeted fusion protein that also served as a substrate for biotinylation. This nuclear targeting fusion (NTF) protein consisted of three parts: the WPP domain of Arabidopsis RAN GTPASE ACTIVATING PROTEIN 1 (RanGAP1), which is necessary and sufficient for envelope association (Rose and Meier, 2001), green fluorescent protein (GFP) for visualization, and the biotin ligase recognition peptide (BLRP), which acts as a substrate for the E. coli biotin ligase BirA (Beckett et al., 1999). Thus, expression of the NTF and BirA in the same cell type should produce biotinylated nuclei exclusively in that cell type. We drove cell type-specific expression of the NTF protein in hair cells using the ACTIN DEPOLYMERIZING FACTOR 8 (ADF8) promoter (Ruzicka et al., 2007) in one transgenic line, and in non-hair cells using the GLABRA2 (GL2) promoter (Masucci et al., 1996) in another line. Both of these transgenic lines also expressed BirA from the constitutive ACTIN2 (ACT2) promoter (An et al., 1996) to provide biotinylation of the NTF in the hair or non-hair cell types. Fluorescence microscopic examination of the ADF8p:NTF/ACT2p:BirA and GL2p:NTF/ACT2p:BirA lines showed that both promoters were expressed exclusively in the expected cell type and that the NTF did indeed accumulate on the nuclear envelope (Figure 1A-C). Furthermore, we observed that nuclei isolated from these lines retained the NTF on their surface and could be specifically bound by streptavidin-coated magnetic beads (Figure 1D and Figure S1). As a further confirmation that the NTF was biotinylated we performed streptavidin western blotting on whole cell extracts, on anti-GFP immunoprecipitates from the roots of each transgenic line, and on extracts from a line expressing only ACT2p:BirA. As shown in Figure 1E, a biotinylated protein of the expected 42 kD size is detected only in plants that express the NTF, and this protein can be immunoprecipitated with an anti-GFP antibody. Thus, the NTF is expressed properly in each line and is biotinylated.
Figure 1. Components and performance of the INTACT system.
(A) Confocal projection of the differentiation zone of an ADF8p:NTF/ACT2p:BirA transgenic root showing expression of the NTF in hair cells. GFP signal is shown in green and propidium iodide staining of cell walls is shown in red. (B) Confocal projection of the differentiation zone of an GL2p:NTF/ACT2p:BirA transgenic root showing expression of the NTF in non-hair cells. (C) Confocal section of the post-meristematic region of a GL2p:NTF/ACT2p:BirA transgenic root. (D) Fluorescence micrograph of nuclei (one is shown in inset) isolated from ADF8p:NTF/ACT2p:BirA transgenic roots and incubated with streptavidin Dynabeads. GFP and beads are shown in green and DAPI staining of DNA is shown in blue. (E) Streptavidin western blot of whole cell extracts (input) and anti-GFP immunoprecipitates (IP) from roots of ACT2p:BirA, ADF8p:NTF/ACT2p:BirA, and GL2p:NTF/ACT2p:BirA plants. Top and bottom bands in each lane are endogenous biotinylated proteins and the middle band is the 42 kD NTF. (F) Diagram of the apparatus used for purification of biotinylated nuclei. See also supplementary figures 1, 2, and 3.
To purify labeled nuclei from hair and non-hair cells we extracted total nuclei from the fully differentiated root hair zone of young seedlings in each transgenic line and incubated the nuclei with streptavidin-coated magnetic beads. We then employed a simple liquid flow-based system to capture the bead-bound nuclei on a magnet as the solution of bound and unbound nuclei flowed past. This apparatus was constructed from common laboratory supplies and a Dynal Mini-MACS magnet, as diagrammed in Figure 1F. Using two successive rounds of flow purification we were able to isolate an average (+/- SD) of 150,000 +/- 45,000 hair cell nuclei from ADF8p:NTF/ACT2p:BirA and 250,000 +/- 65,000 non-hair cell nuclei from GL2p:NTF/ACT2p:BirA, starting with 3 g of root segments from each line. The consistently higher yield of non-hair cell nuclei from the GL2p:NTF/ACT2p:BirA line was expected given that there are generally 10-14 non-hair cell files in the epidermis and only 8 hair cell files (Dolan et al., 1993; Grierson and Schiefelbein, 2003). The average purity (+/- SD) of the nuclei obtained was found to be 92.8 +/- 1.6% for hair cell nuclei and 95 +/- 2.2% for non-hair cell nuclei.
Gene expression profiling using INTACT-purified nuclei
Having successfully purified nuclei from fully differentiated hair and non-hair cells, we then sought to measure gene expression profiles of each cell type using nuclear RNA. We prepared and amplified cDNA from the total nuclear RNA of each cell type, and this material was Cy dye-labeled and hybridized to Roche NimbleGen whole-genome tiling microarrays along with fragmented genomic DNA labeled with the complementary Cy dye. Expression scores for the 26,992 annotated genes represented on the array were calculated using data from each of two biological replicates per cell type, and these datasets were then compared. A gene was defined as preferentially expressed in a given cell type if it showed a fold difference between cell types of >1.3 with a Bayes p value of <0.02 (Baldi and Long, 2001). Using these criteria we found 946 genes that were enriched in hair cells and 118 genes enriched in non-hair cells (Table S1).
To determine whether the hair and non-hair cell-enriched genes identified by INTACT correspond to genes identified using other methods, we compared our cell type-enriched gene lists to those obtained in previous expression studies. When we compared our hair cell-enriched gene list to a list of 24 reporter-confirmed hair cell-specific genes (Won et al., 2009), we found that 19 of the 24 known hair cell-specific genes were present in the INTACT hair cell gene set, and none were found in the non-hair gene set. Therefore, most of the previously confirmed hair cell-enriched genes were found using INTACT, and these genes were found throughout the range of expression levels in our dataset, indicating that INTACT can identify cell type-specific genes regardless of expression level (Table S2).
We compared our cell type-enriched gene lists to those from earlier studies that performed expression profiling using FACS-purified protoplasts of hair and non-hair cells (Birnbaum et al., 2003; Brady et al., 2007). We found that only about 20% of the genes previously defined as specific to each cell type were present in our corresponding gene lists. In addition, only 11 of the 24 confirmed hair cell-specific genes were found in the FACS-based hair cell-enriched gene list. The discrepancies between INTACT and FACS-based expression profiles of each cell type could be attributable to technical differences between the studies, such as cDNA amplification methods, microarray platforms used, and methods for defining cell-type specific expression. However, a major source of variation may also arise from differences in the purity of target cells or nuclei achieved with each of the methods. While the INTACT method is shown here to give nearly 100% purity of the desired nuclei, using a published FACS protocol (Birnbaum et al., 2005) we were unable to achieve a purity of greater than 50% for hair or non-hair cell protoplasts from our transgenic lines (Figure S2). Thus, differences in the expression profiles could also result from a higher level of contamination from other cell types that seems to be inherent to FACS purification of plant protoplasts.
Another possible explanation for the discrepancies between INTACT and FACS-based expression profiles is that differences in the total and nuclear RNA pools could be prevalent in the tissue used for these experiments. In order to address this issue we performed whole-genome expression profiling of nuclear and total RNA from the same tissue used for INTACT purification of hair and non-hair cell nuclei. We found a very high degree of similarity (R = 0.94) in the composition of these two RNA pools (Figure S3). Therefore, expression profiles derived from pure nuclei or protoplasts of a given cell type should be comparable.
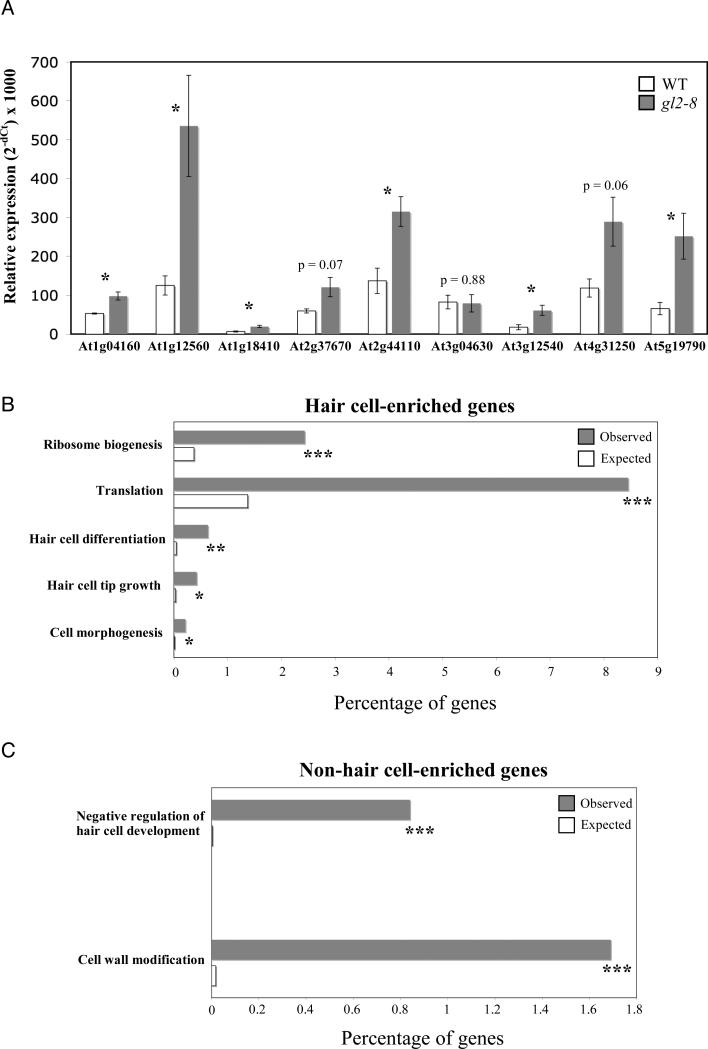
As an independent measure of the accuracy of our expression profiles we selected 27 genes from our hair cell-enriched set and analyzed their expression levels in wild-type and gl2-8 mutant roots. Given that all epidermal cells are converted to hair cells in a gl2 mutant (Di Cristina et al., 1996; Masucci et al., 1996), we reasoned that true hair cell-specific genes should show higher relative expression levels in gl2-8 roots as compared to wild-type roots. In total, 21 of the 27 genes (78%) tested were found to have a higher relative expression level in gl2-8 roots, and 10 of these 21 were found only in the INTACT hair cell dataset and not in the FACS-based dataset (Brady et al., 2007) (Table S3). Expression levels for a representative subset of the tested genes is shown in Figure 2A.
Figure 2. Validation of expression profiles generated from INTACT-purified nuclei.
(A) RT-PCR analysis of selected INTACT hair (H) cell-enriched genes in wild type and gl2-8 roots, where all epidermal cells are H cells. Expression of true H cell-specific genes is expected to be higher in gl2-8 given that the H cell type has a higher relative abundance in this genotype. Data represent the average of two biological replicates +/- SD. Asterisks indicate p values < 0.05. P values higher than 0.05 are indicated on the graph. See also Tables S1 and S3. (B) Observed versus expected percentage of genes in each Gene Ontology (GO) annotation category for hair cell-enriched genes. Chi squared tests were performed for each comparison. P values are indicated as *** < 0.001, ** < 0.01, and * < 0.03. (C) Same as in (B) but for non-hair cell-enriched genes. See also supplementary table 4.
Given the high purity of our cell-type specific population of nuclei, we suspect that our inability to detect increases in expression for 6/27 hair-cell enriched genes has a biological basis. It is unknown how closely the hair-like cells induced in the mutant resemble normal hair cells in terms of their global gene expression profile. It is possible that these hair-like cells express only a part of the hair cell transcriptome, certainly enough to cause polarized growth and secondary cell wall thickening, but perhaps not all of it. Therefore genes that are at significantly higher levels in gl2-8 are very likely to be hair cell-specific, but those that do not increase are not necessarily false positives. Furthermore, because our expression profile comparisons were only between hair and non-hair cells, genes are categorized as hair-cell specific only relative to non-hair cells, but some of these genes might also be expressed in other root cell types. In the case of such genes, an expression increase in the mutant could be obscured by signals from other root cell types.
To test for biological functions known to be associated with the hair and non-hair cell types, we analyzed each cell type-enriched gene set for overrepresentation of Gene Ontology (GO) terms (Ashburner et al., 2000). In the hair cell gene set we found significant enrichment of multiple GO terms at all levels, including those associated with protein translation, actin and tubulin cytoskeletal systems, cell wall modification, and hair cell differentiation and growth (Figure 2B and Table S4). Within the non-hair cell gene set we observed significant overrepresentation of GO terms for cell wall modification and negative regulation of hair cell specification (Figure 2C and Table S3). Thus, in each case we were able to detect overrepresentation of terms that correspond to biological functions known to be relevant to each cell type (Grierson and Schiefelbein, 2003; Masucci et al., 1996).
Chromatin profiling using INTACT-purified nuclei
In order to gain insight into the chromatin changes that accompany the differentiation of hair and non-hair cells from a common progenitor, we profiled two different histone modifications in each cell type: the transcription-associated mark trimethylation of H3 lysine 4 (H3K4me3) (Santos-Rosa et al., 2002) and the Polycomb silencing-associated mark trimethylation of H3 lysine 27 (H3K27me3) (Nekrasov et al., 2007).
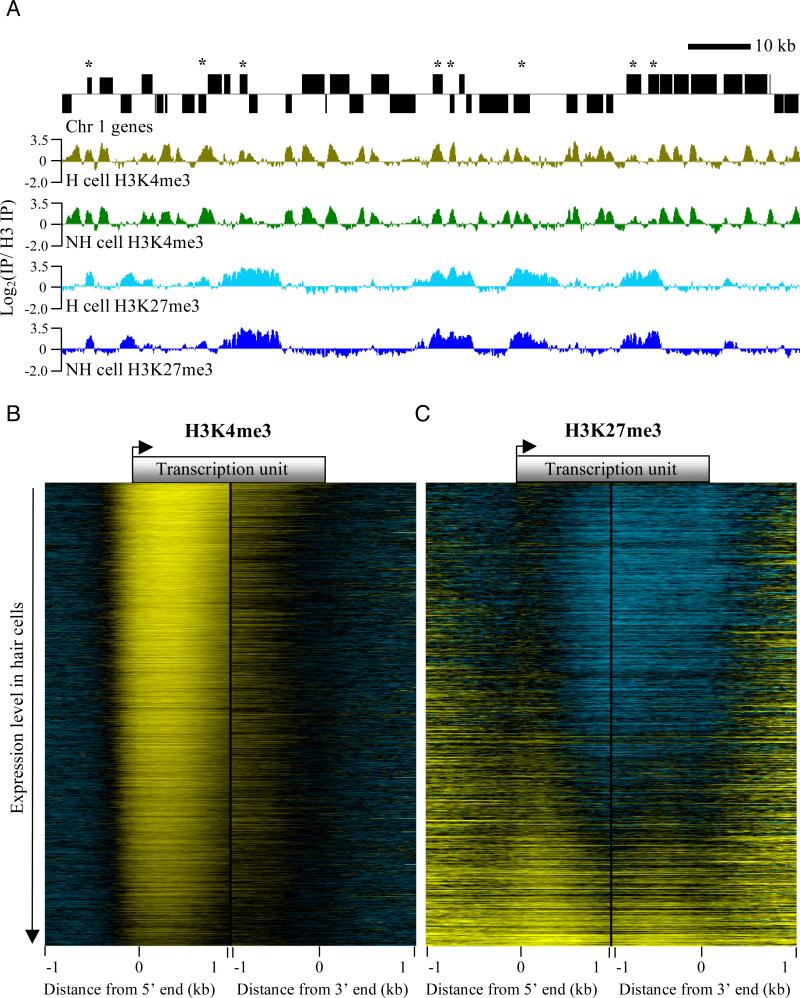
Chromatin immunoprecpitation (ChIP) was performed by shearing crosslinked chromatin from purified hair and non-hair cell nuclei to an average size of 500 bp, followed by immunoprecipitation with an antibody against either H3K4me3 or H3K27me3. To equalize for nucleosome occupancy, a sample of each input chromatin was also immunoprecipitated with an antibody against the C-terminus of H3, which should precipitate all nucleosomes irrespective of their post-translational modifications. Each amplified and labeled H3K4me3 or H3K27me3 ChIP DNA was co-hybridized to tiling arrays along with amplified and labeled H3 ChIP DNA from the same input chromatin. Two biological replicates of each ChIP were performed for each of the two cell types. As shown in Figure 3A, examination of a gene-rich region of chromosome 1 indicated that the ChIP experiments were highly reproducible and showed a high level of similarity between cell types for both modifications.
Figure 3. Overview of ChIP data obtained from INTACT-purified nuclei.
(A) Representative euchromatic chromatin landscapes of H3K4me3 and H3K27me3 in hair (H) and non-hair (NH) cells. Chromosome 1 genes are shown above the chromatin tracks. Genes on the top strand are shown above the black line and those below the line are on the bottom strand. Asterisks indicate genes where H3K4me3 and H3K27me3 overlap in both cell types. Each chromatin landscape is the average of two biological replicates, displayed on the same log-ratio scale. (B) Heat map of the H3K4me3 chromatin landscape relative to gene ends in H cells (-1 kb to +1 kb relative to transcription start and end sites). Genes are ranked from highest to lowest expression level in H cells. Yellow indicates positive, black indicates zero, and blue represents negative log2 ratios. (C) Same as in (B) but for the H3K27me3 chromatin landscape. Heatmaps made the same way for each modification in non-hair cells showed a nearly identical pattern to those in hair cells, as shown in Figure S4. See also supplementary table 5.
To visualize the relationship between gene expression and each of the modifications, we made heat maps by aligning the profiles for each modification at the 5′ and 3′ ends of each annotated gene on the array, and then ranking genes by decreasing expression level in the corresponding cell type. H3K4me3 is maximal just downstream of the transcription start site, and decreases with decreasing gene expression level in both hair and non-hair cells (Figure 3B and Figure S4A, respectively), as described previously in other organisms (Bernstein et al., 2005; Krogan et al., 2003; Roh et al., 2006). Conversely, H3K27me3 is generally excluded from the most highly expressed genes, is found in promoters of genes with mid-level expression, and covers the entire body of genes with the lowest expression levels in both cell types (Figure 3C and Figure S4B). We also found that many genes showed an overlap of H3K4me3 and H3K27me3 (Figure 4A and Table S5), as has been described in mammalian stem cell lines and isolated primary cells (Bernstein et al., 2006; Roh et al., 2006).
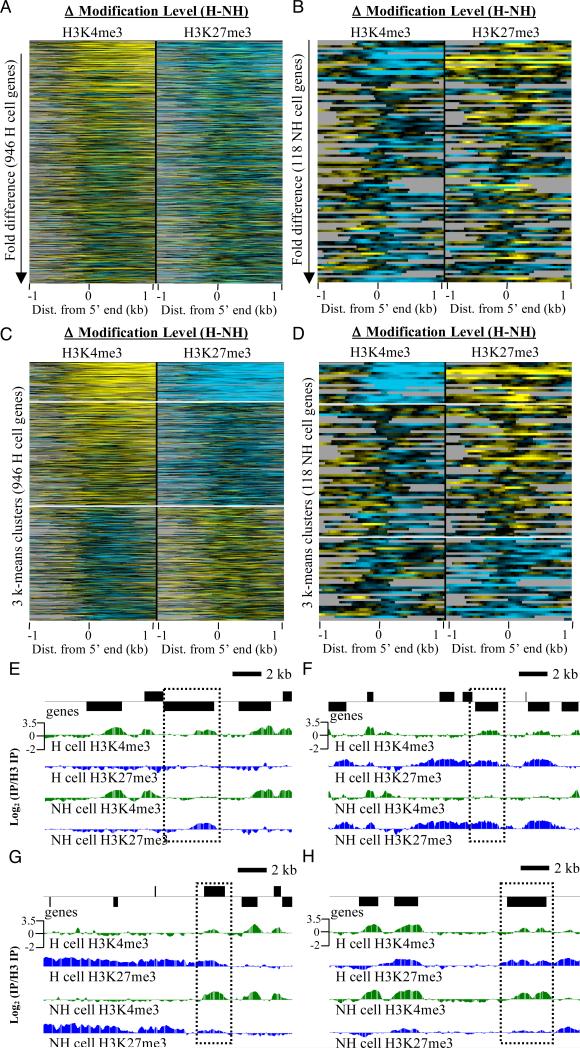
Figure 4. Cell type-specific differences in histone methylations and their relationship to gene expression.
(A) Heat map showing H3K4me3 and H3K27me3 differences between hair (H) and non-hair (NH) cell types (H cell profile minus NH cell profile) around the 5′ end of genes (-1 kb to +1 kb from transcription start site). The 946 H cell-enriched genes are ranked from highest to lowest fold difference in expression level between H and NH cells. Yellow represents higher modification levels in H cells while blue indicates lower levels in H cells, black represents no difference, and gray indicates no data where analysis was stopped when another genomic feature was encountered. (B) Same as in (A) except that the 118 NH cell-enriched genes were used, and these were ranked from highest to lowest fold difference between H and NH cells. Since the subtraction of profiles was done in the same direction as in (A), blue represents higher modification levels in NH cells while yellow indicates lower levels in NH cells. (C) Same heat map rows in (A) clustered into 3 groups (kmeans = 3) over -1 kb to +1 kb. White bars delineate the three clusters. (D) Same heat map rows as in (B) clustered into 3 groups. All heat maps are shown at the same contrast level. (E and F) Comparison of H3K4me3 and H3K27me3 profiles in H and NH cells on two H cell-enriched genes, At5g70450 and At3g49960, respectively. Genes above the x-axis are on the top strand while those below the line are on the bottom strand. Dotted boxes surround the gene of interest in each case. (G and H) Comparison of H3K4me3 and H3K27me3 profiles in H and NH cells on two NH cell-enriched genes, At1g66800 and At5g42591, respectively. See also supplementary figure 5.
In order to determine whether differences in the H3K4me3 and H3K27me3 profiles between cell types might correspond to genes that were preferentially expressed in each cell type, we subtracted each non-hair cell profile from the corresponding hair cell profile. Heat maps were generated from the subtracted profiles for each modification by aligning them at the 5′ ends of genes and ranking each list of cell type-enriched genes based on the fold difference in expression level between the cell types, from largest to smallest. We found that cell type-enriched genes with the largest fold differences between cell types often showed both higher H3K4me3 and lower H3K27me3 levels in the cell type where they were preferentially expressed (Figure 4A-B). K-means clustering of the same heat maps into 3 clusters showed that many genes enriched in a given cell type show this pattern, and this was particularly evident in hair cells. However, many of the cell-type enriched genes show no distinct chromatin differences between cell types, while others show subtle chromatin differences in the opposite direction (Figure 4C-D), indicating that a change in the balance of H3K4me3 and H3K27me3 identifies some, but not all, genes with preferential expression in a given cell type. Using larger numbers of clusters showed that the class of genes with higher H3K4me3 and lower H3K27 remained a coherent group (Figure S5). This higher-level clustering also revealed that there were genes on which only H3K4me3 was higher or only H3K27me3 was lower in the cell type where the gene was preferentially expressed (Figure S5). Examination of the H3K4me3 and H3K27me3 chromatin landscapes over individual hair or non-hair cell-enriched genes indeed showed that these genes often display differences in both modifications, as exemplified in Figure 4E-H.
DISCUSSION
We have developed the INTACT method to allow analysis of the specific gene expression programs and underlying epigenetic factors that define a given cell type. This approach is based on the premise that affinity-tagged nuclei can be produced in vivo in a specific cell type, and these can then be isolated by standard affinity purification techniques. In this study we have shown that this method is easy to perform, does not require sophisticated instrumentation or specialized skills, and can produce large quantities of the desired nuclei at very high purity, in contrast to FACS and LCM-based methods for cell isolation. For example, INTACT provided recovery of >105 nuclei at nearly 100% purity, whereas we recovered <10% of hair cell-specific protoplasts with only 50% purity using FACS based on GFP fluorescence (Figure S2). INTACT is also clearly suitable for isolating nuclei from relatively rare cell types, given that hair and non-hair cells each represent only about 10% of cells in the primary root (Dolan et al., 1993). Given the high specificity and avidity of the biotin-streptavidin interaction, nuclei from cells with even lower abundance should also be possible to purify in sufficient quantities simply by starting with a larger amount of whole tissue. In addition, this approach should be applicable to any organism that can be transformed, and is limited only by the need for a suitable nuclear envelope-targeting domain and a promoter that is expressed in the cell type of interest and not in nearby cells. The RanGAP1 WPP domain is likely to be useful for many other, if not all, plant cell types, although it is not likely to work for non-plant cells given differences in the targeting of RanGAP to the nuclear envelope between plants and other organisms (Meier et al., 2008). For adaptation of the method to non-plant systems, the C-terminus of RanGAP, or perhaps certain nuclear pore complex proteins could be used in place of the WPP domain for nuclear targeting. Thus, INTACT represents a universal strategy for cell type-specific profiling.
Gene expression profiling using INTACT-purified hair and non-hair cell nuclei revealed a large number of genes that are preferentially expressed in each of these cell types. Among the genes we classified as hair cell-enriched, we identified most of the reporter-confirmed hair cell-specific genes and observed increased expression of many of our putative hair cell genes in the gl2-8 mutant roots as compared to wild-type roots. Analysis of overrepresentation of GO terms within our gene sets revealed genes that were previously characterized as being involved in the specification of each of these cell types. In the case of hair cells we also observed an overabundance of genes involved in structural and physiological processes known to be important for the function of this cell type, such as translation, energy generation, cell expansion, vacuole function, and cytoskeletal dynamics. Furthermore, because nuclear and total RNA pools have a very similar composition, and INTACT provides nuclei at nearly 100% purity, the expression profiles generated from INTACT-purified nuclei should accurately represent the transcriptome of the cell type from which they were purified.
Profiling of two histone modifications, H3K4me3 and H3K27me3, in hair and non-hair cell nuclei showed that it is possible to produce robust and highly reproducible ChIP data from the number of nuclei obtained using INTACT. We found that both of these histone modifications showed distributions similar to those recently described in Arabidopsis (Oh et al., 2008; Zhang et al., 2009; Zhang et al., 2007). In addition, we show that in each cell type the level of H3K4me3 within a gene decreases with decreasing expression level and the H3K27me3 modification increases decreasing expression (Figures 3 and S4), as expected. These correlations between expression levels and well-studied chromatin modifications serve as an independent confirmation of the accuracy of our gene expression profiles for each cell type.
Previous profiling of H3K4me3 and H3K27me3 in Arabidopsis suggested that many plant genes have overlapping regions of H3K4me3 and H3K27me3, as observed in mammalian cells, but because whole plant tissues were used in these experiments it was not clear whether these overlaps were in individual cells or were an artifact of amalgamation of signals from multiple cell types (Oh et al., 2008; Zhang et al., 2009; Zhang et al., 2007). By profiling chromatin landscapes at cell type-resolution we are able to show that these modifications do indeed coexist in the same cell type, as has been observed in mammalian cells (Bernstein et al., 2006; Roh et al., 2006).
A comparison of each histone modification profile by subtraction of the non-hair cell profile from that of the hair cell showed that the largest expression differences between cell types often corresponded to an increase in H3K4me3 and a decrease in H3K27me3 in the cell type showing preferential expression of a given gene. This suggests that a balance between the activities of Trithorax group protein-mediated H3K4 trimethylation and Polycomb group protein-mediated trimethylation of H3K27 is involved in establishing cell type-specific expression. However, many differentially expressed genes showed little difference in histone modification levels between cell types over cell type-enriched genes, indicating that there are mechanisms for generating cell type-specific expression that are unrelated to the H3K4me3/H3K27me3 balance.
In conclusion, we have demonstrated that the INTACT method is a robust and simple technique for gene expression and chromatin profiling of individual cell types within a complex tissue, and this approach has many advantages over currently available methods. The INTACT method is far gentler than FACS or LCM in that extensive tissue manipulation is not required and the procedure is much faster, therefore minimizing the possibility of artifacts arising from tissue manipulation and the time required to obtain sufficient material for epigenomic profiling. An additional advantage is that because nuclei can be isolated simply by freezing and grinding of tissue, the INTACT method will be easier to apply to cells embedded within deep internal tissues as well as cells that are recalcitrant to dissociation from a tissue for other reasons. INTACT, being an affinity-based method, also has the distinct advantage of being insensitive to autofluorescence and other optical disturbances that will interfere with FACS-based purification. In addition, the INTACT method requires no specialized skills beyond those common to any molecular biologist, obviates the need for sophisticated and expensive equipment, and should be widely applicable.
EXPERIMENTAL PROCEDURES
Constructs and transgenic plants for INTACT
The nuclear envelope targeting protein used for INTACT consisted of a fusion of the WPP domain of Arabidopsis RanGAP1 (At3g63130; amino acids 1-111, inclusive) (Rose and Meier, 2001) at the N-terminus, followed by the enhanced green fluorescent protein (eGFP) (Zhang et al., 1996) and the biotin ligase recognition peptide (BLRP) (Beckett et al., 1999) at the C-terminus. The WPP domain of RanGAP1 was separated from GFP by 3 alanine residues, and GFP was separated from BLRP by 5 alanine residues. The fusion gene encoding this protein (called NTF, for nuclear tagging fusion) was cloned under control of the ADF8 (At4g00680) promoter (Ruzicka et al., 2007) for hair cell expression and the GL2 (At1g79840) promoter (Masucci et al., 1996) for non-hair cell expression. Each of these constructs was co-transformed into Arabidopsis ecotype Col-0 along with the E. coli biotin ligase (BirA) gene driven from the constitutive ACT2 (At3g18780) promoter (An et al., 1996; Zilberman et al., 2008). First-generation double transgenic plants were selfed to produce plants that were homozygous for both the NTF and BirA transgenes. Multiple individual NTF/BirA double transgenic lines showing the expected expression patterns were combined and used in all subsequent experiments.
Plant growth and harvesting of root tissue
Plants were grown under fluorescent light for 16 hours per day at 22° C on agar-solidified 1/2 strength MS media (Murashige and Skoog, 1962). Plates were kept in a nearly vertical orientation, such that the roots grew along the surface of the media. When plants reached 7 days of age a 1.25 cm section of the roots, from within the fully differentiated root hair zone but below the position of the first lateral roots, was harvested with a razor blade. This region of root tissue was used in all experiments.
Purification of biotinylated nuclei
For each purification 3 g of root tissue was frozen in liquid nitrogen, ground to a fine powder and resuspended in 10 mL of nuclei purification buffer (NPB: 20 mM MOPS, 40 mM NaCl, 90 mM KCl, 2 mM EDTA, 0.5 mM EGTA, 0.5 mM spermidine, 0.2 mM spermine, pH = 7) containing Roche Complete protease inhibitors. Nuclear suspensions were then filtered through 70 μM nylon mesh and pelleted at 1000 × g for 5 min at 4° C. Nuclei were washed with 1 mL of NPB, pelleted again, and finally resuspended in 1 mL of NPB. Twenty-five microliters of Invitrogen M-280 streptavidin-coated dynabeads (~1.5 × 107 beads) were added to the nuclear suspensions and this mixture was rotated at 4° C for 30 min to allow binding of beads to the biotinylated nuclei.
The 1 mL suspension of beads and nuclei was diluted to 10 mL volume with NPB containing 0.1% Triton X-100 (NPBt) and drawn into a plastic 10 mL serological pipette. We then employed a MiniMACS separator magnet (Miltenyi Biotec, catalog # 130-042-102) to capture the dynabead-bound nuclei using a flow-based setup (Figure 1F). This was accomplished by inserting a 1 mL micropipette tip into the groove running the length of the magnet and then inserting the narrow end of the serological pipette, containing the nuclei and bead suspension, into the wide end of the 1 mL pipette tip and allowing the suspension to flow past the magnet at a rate of 0.75 mL/min. As the suspension flowed past the magnet, beads and nuclei were captured on the wall of the 1 mL pipette tip, and all of the solution was allowed to drain out. Beads and nuclei were then eluted from the wall of the tip by placing it on a pipette and repeatedly drawing 1 mL of NPBt into and out of the tip. This suspension was again brought up to a final volume of 10 mL with NPBt, and the magnetic purification was repeated just as before. Beads and nuclei were again released into 1 mL of NPBt, then collected by centrifugation, decanted and used immediately or resuspended in 20 μL NPB and frozen at -20° prior to use. The 1 mL pipette tips used in the purification were pre-treated with NPB + 1% BSA for 10 minutes to prevent the beads from sticking too firmly to the wall of the tip. Typical yields from 3 g of tissue were 1-3 × 105 nuclei, and this amount was used for each RNA isolation or chromatin immunoprecipitation experiment as described below. Purity and yield of nuclei after purification were determined by staining of total nuclei with DAPI prior to purification and subsequent counting of the number of bead-bound nuclei and unbound nuclei in the purified preparation, considering bead-bound nuclei to be the target nuclei and non bead-bound nuclei as contaminating nuclei from other cell types.
Immunoprecipitation and western blotting
Whole cell extracts were prepared from transgenic roots by grinding in liquid N2 and resuspension in 2 volumes of RIPA buffer (50 mM Tris, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate, pH = 7.5) containing Roche Complete protease inhibitors. This extract was cleared by centrifugation to give the input fraction. An aliquot of input was treated with an anti-GFP polyclonal antibody (Santa Cruz Biotechnology, catalog # GFP-FL), followed by incubation with protein A agarose (Millipore, catalog # 16-157) to immunoprecipitate the NTF protein. Bead-bound proteins were washed twice for 5 minutes with RIPA buffer and eluted with 2X SDS loading buffer (100 mM Tris, 10% sodium dodecyl sulfate, 30% glycerol, 1% β-mercaptoethanol, 0.2% bromophenol blue, pH = 7.5). Input and immunoprecipitated fractions were electrophoresed on a 12% SDS polyacrylamide gel and transferred to a nitrocellulose membrane. The membrane was blocked in PBSt (11.9 mM sodium phosphate, 137 mM NaCl, 2.7 mM KCl, 0.1% Triton X-100, pH = 7.4) with 10% milk for 30 minutes, washed twice for 5 minutes with PBSt, and incubated with a 1:2000 dilution of streptavidin-HRP (GE, catalog # RPN1231) in PBSt with 1% BSA for 30 minutes. The membrane was then washed three times for 5 minutes with PBSt and biotinylated proteins were detected using ECL detection reagents (Pierce, catalog # 34075).
Gene expression profiling using nuclear RNA
Total RNA was isolated from purified nuclei using the Qiagen RNeasy Micro kit. RNA was first treated with RNase-free Dnase I and then cDNA was prepared and amplified using the Sigma Whole Transcriptome Amplification Kit (Sigma, catalog # WTA2). This synthesis/amplification method begins with a cDNA synthesis using primers with a random 3′ end and defined 5′ end, followed by PCR using primers that match the 5′ end of the primers used for cDNA synthesis. The amplified cDNA was labeled in a random priming reaction using Cy dye-containing random 9mers as directed in the Roche NimbleGen protocol supplied with the arrays. Sheared genomic DNA was labeled with the complementary Cy dye and was then cohybridized along with labeled cDNA to a custom-designed Arabidopsis 1.9 million feature tiling array from Roche NimbleGen, which was described previously (Bernatavichute et al., 2008). This array covers the entire sequenced portion of the Arabidopsis genome with an isothermal probe design. All array hybridizations and scanning were performed by the Genomics Shared Resource lab at the Fred Hutchinson Cancer Research Center.
Two biological replicates of the experiment were performed for each cell type and the raw log2 ratio data from each of these were processed by conversion to standard deviates on a probe-by-probe basis. An expression score was then calculated for each gene by averaging the log2 ratios of the first 100 exonic probes, starting at the 3′ end of the gene and moving toward the 5′ end. In order to define the set of genes enriched in each cell type we compared the data sets from each cell type using the program CyberT (Baldi and Long, 2001). Within CyberT we performed a Bayesian analysis using with a window size of 101 and a confidence level of 10. Genes were classified as enriched in a given cell type if they showed a fold difference between cell types of >1.3 and a Bayes p value of <0.02.
Gene Ontology (GO) analysis was performed on each set of cell type-enriched genes using the GeneCodis 2.0 program (Carmona-Saez et al., 2007; Nogales-Cadenas et al., 2009) with a hypergeometric test and false discovery rate calculation to correct the p values for multiple testing. The full set of genes present on the array was used as the background set in these analyses. Chi squared tests were also performed on the observed versus expected percentage of genes in selected GO categories.
qRT-PCR analysis
WT Col-0 and gl2-8 mutant seedlings (T-DNA insertion line SALK_130213) (Alonso et al., 2003) were grown on plates of agar-solidified 1/2 strength MS as described above, and RNA was prepared from the root hair zone of 7-day-old seedlings using the Qiagen RNeasy Plant Mini kit. Each RNA sample was treated with RNase-free DNAse I and cDNA was prepared using the Superscript III kit (Invitrogen, catalog # 18080-051) with oligo dT primers according to the manufacturer's instructions. Real-time PCR was performed on an Applied Biosystems 7900HT instrument using SYBR green detection chemistry. Relative quantities of each transcript were calculated using the 2-ddCt method (Livak and Schmittgen, 2001) with At1g13320 serving as the endogenous control transcript in each case (Czechowski et al., 2005). Primer sequences are given in Table S3.
Chromatin profiling by chromatin immunoprecipitation
For chromatin immunoprecipitation (ChIP) experiments we first treated the excised root tissue with 1% formaldehyde in NPB for 15 minutes prior to extraction and purification of biotinylated nuclei as described above. Our ChIP protocol is based on that of Gendrel et al (Gendrel et al., 2005), but was modified for smaller amounts of starting material. Purified nuclei were lysed in 120 μL of nuclei lysis buffer (50 mM Tris, 10 mM EDTA, 1% sodium dodecyl sulfate, pH = 8) and sonicated using a Diagenode Bioruptor to yield chromatin fragments with an average size of ~500 bp. Sonicated chromatin was cleared by centrifugation and diluted to 1.3 mL final volume with ChIP dilution buffer (16.7 mM Tris, 1.2 mM EDTA, 1.1% Triton X-100, 167 mM NaCl, pH = 8). Diluted chromatin was pre-treated with 20 μL (bed volume) of protein A agarose beads (Millipore, catalog # 16-157) for 30 minutes at 4° C and then cleared by centrifugation. This chromatin was then divided into 2-3 aliquots of equal volume and 1-3 μg of antibody was added to each aliquot. The following antibodies were used in the experiments: H3, Abcam ab1791; H3K4me3, Abcam ab8580; H3K27me3, Millipore 07-449. Antibodies were incubated with chromatin at 4° overnight on a rocking platform, then 20 μL (bed volume) of protein A agarose beads were added with rocking at 4° for an additional 2 hours. Beads were washed once for 5 minutes at 4° in 0.5 mL of each of the following buffers: low salt wash buffer (20 mM Tris, 150 mM NaCl, 0.1% sodium dodecyl sulfate, 1% Triton X-100, 2 mM EDTA, pH = 8), high salt wash buffer (20 mM Tris, 500 mM NaCl, 1% sodium deoxycholate, 1% NP-40, 1 mM EDTA, pH = 8), LiCl wash buffer (10 mM Tris, 250 mM LiCl, 0.1% sodium dodecyl sulfate, 1% Triton X-100, 2 mM EDTA, pH = 8), and TE (10 mM Tris, 1 mM EDTA, pH = 7.5). Chromatin was eluted from the beads in 200 μL of elution buffer (100 mM NaHCO3, 1% sodium dodecyl sulfate) with vortexing for 5 minutes, then NaCl was added to 0.5 M and eluted chromatin was heated to 100° for 15 minutes to reverse crosslinks. DNA was isolated by treating the chromatin with RNase A, Proteinase K, and purification using the Qiagen MinElute kit. Amplification of ChIP DNA was performed with the Sigma Single Cell Whole Genome Amplification kit (Sigma, catalog # WGA4) as directed, and the amplified material was labeled with Cy3 or Cy5 dye as described above. For each experiment the H3K4me3 or H3K27me3 ChIP DNA was cohybridized to the tiling array (same array as used for expression analysis) along with H3 ChIP DNA from the same starting chromatin to equalize for nucleosome occupancy.
Two biological replicates of each ChIP were performed and the log2 ratios from each replicate array were converted to standard deviates, averaged, and smoothed using triangular smoothing as described previously (Ooi et al., 2009). These data were used for all analyses. Cluster analysis was performed with Cluster 3 (Eisen et al., 1998) and results were viewed using Java Treeview 1.1.0 (Saldanha, 2004). Ends analysis was performed as previously described (Henikoff et al., 2009), and the analysis of each gene was stopped at the point where another genomic feature (gene or transposable element) was encountered. All microarray data are available from GEO (Accession Number GSE19654).
Supplementary Material
ACKNOWLEDGEMENTS
We are especially grateful to Jorja Henikoff for assistance with data processing and analysis. We also thank Takehito Furuyama, Mary Gehring, and Florian Steiner for reading and discussions of the manuscript, Daniel Zilberman for the ACT2p:BirA construct, Andy Marty and Jimiane Ashe at the Hutchinson Center Genomics Resource lab for microarray processing, and the Arabidopsis Biological Resource Center at Ohio State University for providing the gl2-8 mutant seed. This research was supported by funding from the Howard Hughes Medical Institute to S.H. and a Ruth L. Kirschstein Postdoctoral Fellowship from the National Institutes of Health to R.B.D.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- An YQ, McDowell JM, Huang S, McKinney EC, Chambliss S, Meagher RB. Strong, constitutive expression of the Arabidopsis ACT2/ACT8 actin subclass in vegetative tissues. Plant J. 1996;10:107–121. doi: 10.1046/j.1365-313x.1996.10010107.x. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldi P, Long AD. A Bayesian framework for the analysis of microarray expression data: regularized t -test and statistical inferences of gene changes. Bioinformatics. 2001;17:509–519. doi: 10.1093/bioinformatics/17.6.509. [DOI] [PubMed] [Google Scholar]
- Barthelson RA, Lambert GM, Vanier C, Lynch RM, Galbraith DW. Comparison of the contributions of the nuclear and cytoplasmic compartments to global gene expression in human cells. BMC Genomics. 2007;8:340. doi: 10.1186/1471-2164-8-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckett D, Kovaleva E, Schatz PJ. A minimal peptide substrate in biotin holoenzyme synthetase-catalyzed biotinylation. Protein Sci. 1999;8:921–929. doi: 10.1110/ps.8.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernatavichute YV, Zhang X, Cokus S, Pellegrini M, Jacobsen SE. Genome-wide association of histone H3 lysine nine methylation with CHG DNA methylation in Arabidopsis thaliana. PLoS ONE. 2008;3:e3156. doi: 10.1371/journal.pone.0003156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ, 3rd, Gingeras TR, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Bhattacharya B, Puri S, Puri RK. A review of gene expression profiling of human embryonic stem cell lines and their differentiated progeny. Curr Stem Cell Res Ther. 2009;4:98–106. doi: 10.2174/157488809788167409. [DOI] [PubMed] [Google Scholar]
- Birnbaum K, Jung JW, Wang JY, Lambert GM, Hirst JA, Galbraith DW, Benfey PN. Cell type-specific expression profiling in plants via cell sorting of protoplasts from fluorescent reporter lines. Nat Methods. 2005;2:615–619. doi: 10.1038/nmeth0805-615. [DOI] [PubMed] [Google Scholar]
- Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN. A gene expression map of the Arabidopsis root. Science. 2003;302:1956–1960. doi: 10.1126/science.1090022. [DOI] [PubMed] [Google Scholar]
- Brady SM, Orlando DA, Lee J-Y, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science. 2007;318:801–806. doi: 10.1126/science.1146265. [DOI] [PubMed] [Google Scholar]
- Brien GL, Bracken AP. Transcriptomics: unravelling the biology of transcription factors and chromatin remodelers during development and differentiation. Semin Cell Dev Biol. 2009;20:835–841. doi: 10.1016/j.semcdb.2009.07.010. [DOI] [PubMed] [Google Scholar]
- Brunskill EW, Aronow BJ, Georgas K, Rumballe B, Valerius MT, Aronow J, Kaimal V, Jegga AG, Yu J, Grimmond S, et al. Atlas of gene expression in the developing kidney at microanatomic resolution. Dev Cell. 2008;15:781–791. doi: 10.1016/j.devcel.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmona-Saez P, Chagoyen M, Tirado F, Carazo JM, Pascual-Montano A. GENECODIS: a web-based tool for finding significant concurrent annotations in gene lists. Genome Biol. 2007;8:R3. doi: 10.1186/gb-2007-8-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005;139:5–17. doi: 10.1104/pp.105.063743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz AF, Edgar BA. Flow cytometric analysis of Drosophila cells. Methods Mol Biol. 2008;420:373–389. doi: 10.1007/978-1-59745-583-1_24. [DOI] [PubMed] [Google Scholar]
- Di Cristina M, Sessa G, Dolan L, Linstead P, Baima S, Ruberti I, Morelli G. The Arabidopsis Athb-10 (GLABRA2) is an HD-Zip protein required for regulation of root hair development. Plant J. 1996;10:393–402. doi: 10.1046/j.1365-313x.1996.10030393.x. [DOI] [PubMed] [Google Scholar]
- Dolan L, Janmaat K, Willemsen V, Linstead P, Poethig S, Roberts K, Scheres B. Cellular organisation of the Arabidopsis thaliana root. Development. 1993;119:71–84. doi: 10.1242/dev.119.1.71. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel AV, Lippman Z, Martienssen R, Colot V. Profiling histone modification patterns in plants using genomic tiling microarrays. Nat Methods. 2005;2:213–218. doi: 10.1038/nmeth0305-213. [DOI] [PubMed] [Google Scholar]
- Gifford ML, Dean A, Gutierrez RA, Coruzzi GM, Birnbaum KD. Cell-specific nitrogen responses mediate developmental plasticity. Proc Natl Acad Sci U S A. 2008;105:803–808. doi: 10.1073/pnas.0709559105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grierson, Schiefelbein Root Hairs. Arabidopsis Book. 2003:1–22. doi: 10.1199/tab.0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiman M, Schaefer A, Gong S, Peterson JD, Day M, Ramsey KE, Suarez-Farinas M, Schwarz C, Stephan DA, Surmeier DJ, et al. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 2008;135:738–748. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Henikoff JG, Sakai A, Loeb GB, Ahmad K. Genome-wide profiling of salt fractions maps physical properties of chromatin. Genome Research. 2009;19:460–469. doi: 10.1101/gr.087619.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irion S, Nostro MC, Kattman SJ, Keller GM. Directed differentiation of pluripotent stem cells: from developmental biology to therapeutic applications. Cold Spring Harb Symp Quant Biol. 2008;73:101–110. doi: 10.1101/sqb.2008.73.065. [DOI] [PubMed] [Google Scholar]
- Ishida T, Kurata T, Okada K, Wada T. A genetic regulatory network in the development of trichomes and root hairs. Annu Rev Plant Biol. 2008;59:365–386. doi: 10.1146/annurev.arplant.59.032607.092949. [DOI] [PubMed] [Google Scholar]
- Jacob Y, Mongkolsiriwatana C, Veley KM, Kim SY, Michaels SD. The nuclear pore protein AtTPR is required for RNA homeostasis, flowering time, and auxin signaling. Plant Physiol. 2007;144:1383–1390. doi: 10.1104/pp.107.100735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Tausta SL, Gandotra N, Sun N, Liu T, Clay NK, Ceserani T, Chen M, Ma L, Holford M, et al. A transcriptome atlas of rice cell types uncovers cellular, functional and developmental hierarchies. Nat Genet. 2009;41:258–263. doi: 10.1038/ng.282. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Dover J, Wood A, Schneider J, Heidt J, Boateng MA, Dean K, Ryan OW, Golshani A, Johnston M, et al. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol Cell. 2003;11:721–729. doi: 10.1016/s1097-2765(03)00091-1. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Masucci JD, Rerie WG, Foreman DR, Zhang M, Galway ME, Marks MD, Schiefelbein JW. The homeobox gene GLABRA2 is required for position-dependent cell differentiation in the root epidermis of Arabidopsis thaliana. Development. 1996;122:1253–1260. doi: 10.1242/dev.122.4.1253. [DOI] [PubMed] [Google Scholar]
- Meier I, Xu XM, Brkljacic J, Zhao Q, Wang H-J. Going green: plants’ alternative way to position the Ran gradient. Journal of microscopy. 2008;231:225–233. doi: 10.1111/j.1365-2818.2008.02038.x. [DOI] [PubMed] [Google Scholar]
- Miller MR, Robinson KJ, Cleary MD, Doe CQ. TU-tagging: cell type-specific RNA isolation from intact complex tissues. Nat Methods. 2009;6:439–441. doi: 10.1038/nmeth.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mito Y, Henikoff JG, Henikoff S. Genome-scale profiling of histone H3.3 replacement patterns. Nat Genet. 2005;37:1090–1097. doi: 10.1038/ng1637. [DOI] [PubMed] [Google Scholar]
- Muller C, Leutz A. Chromatin remodeling in development and differentiation. Curr Opin Genet Dev. 2001;11:167–174. doi: 10.1016/s0959-437x(00)00175-1. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue culture. Plant Physiol. 1962;15:473–497. [Google Scholar]
- Mustroph A, Zanetti M, Jang C, Holtan H, Repetti P, Galbraith D, Girke T, Bailey-Serres J. Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proc Natl Acad Sci USA. 2009 doi: 10.1073/pnas.0906131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazono M, Qiu F, Borsuk LA, Schnable PS. Laser-capture microdissection, a tool for the global analysis of gene expression in specific plant cell types: identification of genes expressed differentially in epidermal cells or vascular tissues of maize. Plant Cell. 2003;15:583–596. doi: 10.1105/tpc.008102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasov M, Klymenko T, Fraterman S, Papp B, Oktaba K, Kocher T, Cohen A, Stunnenberg HG, Wilm M, Muller J. Pcl-PRC2 is needed to generate high levels of H3-K27 trimethylation at Polycomb target genes. EMBO J. 2007;26:4078–4088. doi: 10.1038/sj.emboj.7601837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng RK, Gurdon JB. Epigenetic inheritance of cell differentiation status. Cell Cycle. 2008;7:1173–1177. doi: 10.4161/cc.7.9.5791. [DOI] [PubMed] [Google Scholar]
- Nogales-Cadenas R, Carmona-Saez P, Vazquez M, Vicente C, Yang X, Tirado F, Carazo JM, Pascual-Montano A. GeneCodis: interpreting gene lists through enrichment analysis and integration of diverse biological information. Nucleic Acids Res. 2009;37:W317–322. doi: 10.1093/nar/gkp416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S, Park S, van Nocker S. Genic and global functions for Paf1C in chromatin modification and gene expression in Arabidopsis. PLoS Genet. 2008;4:e1000077. doi: 10.1371/journal.pgen.1000077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi SL, Henikoff JG, Henikoff S. A native chromatin purification system for epigenomic profiling in Caenorhabditis elegans. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao RR, Stice SL. Gene expression profiling of embryonic stem cells leads to greater understanding of pluripotency and early developmental events. Biol Reprod. 2004;71:1772–1778. doi: 10.1095/biolreprod.104.030395. [DOI] [PubMed] [Google Scholar]
- Rivolta MN, Holley MC. Cell lines in inner ear research. J Neurobiol. 2002;53:306–318. doi: 10.1002/neu.10111. [DOI] [PubMed] [Google Scholar]
- Roh T-Y, Cuddapah S, Cui K, Zhao K. The genomic landscape of histone modifications in human T cells. Proc Natl Acad Sci USA. 2006;103:15782–15787. doi: 10.1073/pnas.0607617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose A, Meier I. A domain unique to plant RanGAP is responsible for its targeting to the plant nuclear rim. Proc Natl Acad Sci U S A. 2001;98:15377–15382. doi: 10.1073/pnas.261459698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy PJ, Stuart JM, Lund J, Kim SK. Chromosomal clustering of muscle-expressed genes in Caenorhabditis elegans. Nature. 2002;418:975–979. doi: 10.1038/nature01012. [DOI] [PubMed] [Google Scholar]
- Ruzicka DR, Kandasamy MK, McKinney EC, Burgos-Rivera B, Meagher RB. The ancient subclasses of Arabidopsis Actin Depolymerizing Factor genes exhibit novel and differential expression. Plant J. 2007;52:460–472. doi: 10.1111/j.1365-313X.2007.03257.x. [DOI] [PubMed] [Google Scholar]
- Saldanha AJ. Java Treeview--extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- Truernit E, Hibberd JM. Immunogenic tagging of chloroplasts allows their isolation from defined cell types. Plant J. 2007;50:926–932. doi: 10.1111/j.1365-313X.2007.03113.x. [DOI] [PubMed] [Google Scholar]
- Won S-K, Lee Y-J, Lee H-Y, Heo Y-K, Cho M, Cho H-T. cis-Element- and Transcriptome-Based Screening of Root Hair-Specific Genes and Their Functional Characterization in Arabidopsis. PLANT PHYSIOLOGY. 2009;150:1459–1473. doi: 10.1104/pp.109.140905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Barthelson RA, Lambert GM, Galbraith DW. Global characterization of cell-specific gene expression through fluorescence-activated sorting of nuclei. PLANT PHYSIOLOGY. 2008;147:30–40. doi: 10.1104/pp.107.115246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Gurtu V, Kain SR. An enhanced green fluorescent protein allows sensitive detection of gene transfer in mammalian cells. Biochem Biophys Res Commun. 1996;227:707–711. doi: 10.1006/bbrc.1996.1573. [DOI] [PubMed] [Google Scholar]
- Zhang X, Bernatavichute YV, Cokus S, Pellegrini M, Jacobsen SE. Genome-wide analysis of mono-, di- and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana. Genome Biol. 2009;10:R62. doi: 10.1186/gb-2009-10-6-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Clarenz O, Cokus S, Bernatavichute YV, Pellegrini M, Goodrich J, Jacobsen SE. Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol. 2007;5:e129. doi: 10.1371/journal.pbio.0050129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Ma C, Delohery T, Nasipak B, Foat BC, Bounoutas A, Bussemaker HJ, Kim SK, Chalfie M. Identification of genes expressed in C. elegans touch receptor neurons. Nature. 2002;418:331–335. doi: 10.1038/nature00891. [DOI] [PubMed] [Google Scholar]
- Zilberman D, Coleman-Derr D, Ballinger T, Henikoff S. Histone H2A.Z and DNA methylation are mutually antagonistic chromatin marks. Nature. 2008;456:125–129. doi: 10.1038/nature07324. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.