Abstract
Inhaled formaldehyde is classified as a known human and animal carcinogen, causing nasopharyngeal cancer. Additionally, limited epidemiological evidence for leukemia in humans is available; however, this is inconsistent across studies. Both genotoxicity and cytotoxicity are key events in formaldehyde nasal carcinogenicity in rats, but mechanistic data for leukemia are not well established. Formation of DNA adducts is a key event in initiating carcinogenesis. Formaldehyde can induce DNA monoadducts, DNA-DNA cross-links, and DNA protein cross-links. In this study, highly sensitive liquid chromatography-tandem mass spectrometry-selected reaction monitoringmethods were developed and [13CD2]-formaldehyde exposures utilized, allowing differentiation of DNA adducts and DNA-DNA cross-links originating from endogenous and inhalation-derived formaldehyde exposure. The results show that exogenous formaldehyde induced N2-hydroxymethyl-dG monoadducts and dG-dG cross-links in DNA from rat respiratory nasal mucosa but did not form [13CD2]-adducts in sites remote to the portal of entry, even when five times more DNA was analyzed. Furthermore, no N6-HO13CD2-dA adducts were detected in nasal DNA. In contrast, high amounts of endogenous formaldehyde dG and dA monoadducts were present in all tissues examined. The number of exogenous N2-HO13CD2-dG in 1- and 5-day nasal DNA samples from rats exposed to 10-ppm [13CD2]-formaldehyde was 1.28 ± 0.49 and 2.43 ± 0.78 adducts/107 dG, respectively, while 2.63 ± 0.73 and 2.84 ± 1.13 N2-HOCH2-dG adducts/107 dG and 3.95 ± 0.26 and 3.61 ± 0.95 N6-HOCH2-dA endogenous adducts/107 dA were present. This study provides strong evidence supporting a genotoxic and cytotoxic mode of action for the carcinogenesis of inhaled formaldehyde in respiratory nasal epithelium but does not support the biological plausibility that inhaled formaldehyde also causes leukemia.
Keywords: formaldehyde, DNA adduct, DNA-DNA cross-link, cancer, leukemia
Formaldehyde is a known human and animal carcinogen according to the International Agency for Research on Cancer (IARC, 2006), causing nasopharyngeal cancer in humans and squamous cell carcinomas in the nasal respiratory epithelium of rats and mice (Kerns et al., 1983; Monticello et al., 1996; Swenberg et al., 1980). Associations between formaldehyde exposure and the induction of leukemia were identified in epidemiology studies of professional and industrial workers. However, this finding was only positive when peak exposure but not cumulative exposure was used as the exposure metric and was not consistent across studies (Coggon et al., 2003; Hauptmann et al., 2003, 2004). A recent follow-up of the epidemiology study of U.S. industrial workers also found an association between peak exposure to formaldehyde with the induction of leukemia, especially myeloid leukemia (Beane Freeman et al., 2009). Whether or not formaldehyde causes leukemia remains debatable, as no consistent experimental data provide mechanistic insight on the induction of leukemia, although several possible mechanisms have been proposed (Zhang et al., 2009a; Zhang et al., 2009b) and some possible support for the induction of leukemia in humans has been published (Zhang et al., 2010).
The association between formaldehyde exposure and the induction of leukemia has raised the question whether inhaled formaldehyde can reach remote tissues such as bone marrow to cause genotoxicity. Formation of DNA adducts and increased cell proliferation secondary to cytotoxicity are well-established key events that drive the induction of nasal cancer in rats by formaldehyde (IARC, 2006). Formaldehyde readily induces DNA protein cross-links (DPCs) in nasal epithelial DNA of rats (Casanova-Schmitz and Heck, 1983), the site of contact following inhalation exposure; however, DPC have not been demonstrated in tissues remote from the portal of entry (Casanova-Schmitz et al., 1984). DPC measurements have utilized non–chemical-specific methods, primarily based on physical chemistry. Chemical-specific DNA biomarkers have not been evaluated following inhalation exposure to formaldehyde, a primary route of exposure. Thus, the present study is the first study to examine inhalation-specific DNA adducts of formaldehyde.
Formaldehyde's well-known toxicity and carcinogenicity and wide spread human exposure has raised public concern over its safety. Formaldehyde can directly enter into the body through environmental exposure such as vehicle emissions, off-gassing of building materials, food, tobacco smoke and indoor air, such as living in FEMA trailers (http://www.cdc.gov/nceh/ehhe/trailerstudy). Formaldehyde can also be indirectly generated through metabolism of a variety of compounds including nutrients, drugs, and proteins by demethylation. Of equal importance, formaldehyde is an essential metabolic intermediate in all living cells and is endogenously produced from serine, glycine, methionine, and choline. The endogenous concentration of formaldehyde in the blood of human subjects is about 0.1mM (IARC, 2006). Therefore, the presence of both endogenous and exogenous formaldehyde makes developing chemical-specific biomarkers to monitor the molecular dose of inhaled formaldehyde exposure and understand its toxicity especially challenging.
Several previous in vitro studies demonstrated that N6-hydroxymethyl-dA (N6-HOCH2-dA) was a primary formaldehyde-DNA monoadduct (McGhee and von Hippel, 1975; Zhong and Hee, 2004; Zhong and Que Hee, 2004). Recently, the Hecht laboratory reported increased amounts of N6-HOCH2-dA in multiple tissues of rats treated with N-nitrosodimethylamine (NDMA) or 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) (Cheng et al., 2008; Wang et al., 2007), which were attributed to the production of formaldehyde during the metabolism of NDMA and NNK. More recently, they have demonstrated clear differences in the amounts of formaldehyde-induced N6-HOCH2-dA in leukocytes of smokers and nonsmokers (Wang et al., 2009). Thus, N6-HOCH2-dA may serve as a good biomarker for formaldehyde exposure after inhalation. In preliminary cell culture experiments, we found that N2-hydroxymethyl-dG was also a suitable DNA biomarker for formaldehyde exposure. Therefore, in the present study, we measured both N2-hydroxymethyl-dG and N6-hydroxymethyl-dA in tissues of rats exposed by inhalation to 10-ppm [13CD2]-formaldehyde for 6 h or for 5 days (6 h/day). Exposures for the present study were modeled after the 10-ppm exposure data from a previous cancer bioassay where it was clearly carcinogenic (Monticello et al., 1996), induced marked increases in sustained cell proliferation, yet was less cytotoxic than 15-ppm formaldehyde. The use of [13CD2]-formaldehyde permitted the simultaneous measurement of both endogenous and exogenous formaldehyde-DNA adducts in any tissue using sensitive liquid chromatography-electrospray ionization-tandem mass spectrometry-selected reaction monitoring (LC-ESI-MS/MS-SRM) methods to detect and quantify formaldehyde-induced DNA adducts and DNA-DNA cross-links (Fig. 1). The data generated in this study provide critical information to understand formaldehyde's toxicity, carcinogenicity, and the biological plausibility of leukemia induction following inhalation exposure to formaldehyde.
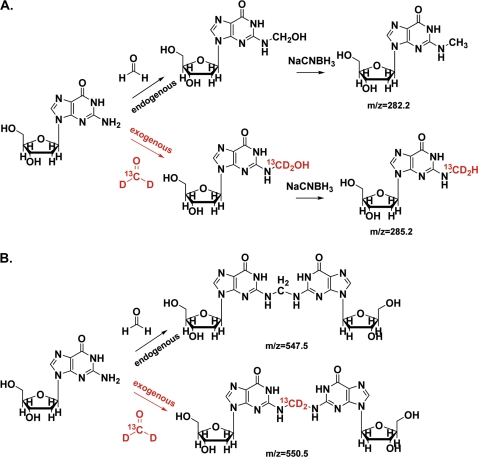
FIG. 1.
The formation of N2-hydroxymethyl-dG (A) and dG-dG cross-links (B) originating from both endogenous and exogenous formaldehyde.
MATERIALS AND METHODS
Chemicals and materials.
Deoxyguanosine, deoxyadenine, potassium phosphate, Tris-HCl, MgCl2, formic acid, NaCNBH3, methanol, acetonitrile, high-performance liquid chromatography (HPLC)–grade water, and 10× PBS were all purchased from Sigma (St Louis, MO); 20% formaldehyde in water was procured from Tousimis (Rockville, MD). DNaseI, alkaline phosphatase, and phosphodiesterases were purchased from Sigma. [15N5]-dA and [13C1015N5]-dG were ordered from Cambridge Isotope Laboratories, Inc. (Cambridge, MA). N6-CH3-dA and N2-CH3-dG were obtained from Sigma and Berry & Associates (Dexter, MI), respectively. All chemicals were used as received unless otherwise stated.
High-performance liquid chromatography.
The purification of formaldehyde-DNA adducts was carried out on an Agilent 1200 series HPLC system equipped with a diode-array detector (Santa Clara, CA). Analytes in 30- to 200-μg DNA hydrolysate were separated by reverse-phase chromatography using a 150- × 2.5-mm T3 analytical column from Waters (Milford, MA). The mobile phases were 0.1% formic acid (A) and methanol (B). A linear gradient was run from 2% methanol to 30% methanol over 30 min, at a flow rate of 200 μl/min and monitored at 254 nm. N6-Me-dA and N2-Me-dG eluted at 22.2 and 25.2 min on the column in this system, respectively, and dG-dG cross-links eluted at 34.3 min with a linear gradient from 2 to 60% methanol over 60 min, at a flow rate of 200 μl/min.
The LC-MS.
LC-MS analyses were performed on a triple-quadrupole mass spectrometer Ultra TSQ-Quantum (Thermo Electron, Waltham, MA) operating in SRM mode to detect and quantify formaldehyde-DNA adducts. The mass spectrometer was interfaced with an ultra-performance liquid chromatography (UPLC) system from Waters. A 150- × 1.0-mm T3 column (3-μm particle size) from Waters was used. A linear gradient was run from 2% methanol in 0.1% aqueous formic acid to 60% methanol over 10 min, at 40 μl/min. For the analysis of monoadducts, the ESI source was set as follows: spray voltage, 4.0 kV; capillary temperature, 300°C; sheath gas pressure, 40 au; aux gas pressure, 10 au. For the analysis of dG-CH2-dG, the mass spectrometer was coupled with a Waters nanoAcquity LC system. A 100-mm × 100-μm nanoAcquity UPLC HSS T3 column was used. A linear gradient was run from 2% acetonitrile in 0.1% acetic acid to 50% in 10 min, at a flow rate at 0.6 or 1 μl/min. The ESI source was set as follows: spray voltage, 2.2–2.5 kV; capillary temperature, 280°C.
Preparation of internal standards.
10 mM [13C1015N5]-dG and [15N5]-dA solution was treated by 100mM formaldehyde in phosphate buffer (pH = 7.2) overnight at 37°C. The reaction mixture was separated by HPLC using a 150- × 2.5-mm C18 T3 analytical column. N2-hydroxymethyl-dG and N6-hydroxymethyl-dA eluted at 20.5 and 24.3 min, respectively. N2-hydroxymethyl-dG and N6-hydroxymethyl-dA were collected and incubated with 50mM NaCNBH3 (pH = 7.1) overnight at 37°C, followed by further separation using HPLC. [13C1015N5]-N2-CH3-dG and [15N5]-N6-CH3-dA eluted at 27.2 and 33.5 min on a 150- × 2.5-mm T3 column using 10mM ammonium acetate in 0.1% acetic acid and methanol as mobile phases, separately. The concentration of [13C1015N5]-N2-CH3-dG and [15N5]-N6-CH3-dA was determined by HPLC with unlabeled N6-CH3-dA and N2-CH3-dG as references. The usual conversion rate from hydroxymethyl group to methyl group was 65–85%. The internal standard of dG-dG cross-links was prepared by incubating 10mM [13C1015N5]-dG with 100mM formaldehyde in phosphate buffer (pH = 7.2) for 96 h at 37°C. The dG-dG cross-links eluted at 34.3 min using a linear gradient from 2% methanol in 0.1% formic acid to 60% methanol over 60 min, at a flow rate of 200 μl/min.
Animal exposures.
Test atmospheres of formaldehyde were generated by the thermal depolymerization of [13CD2]-paraformaldehyde (CAS No. 30525-89-4) obtained from Cambridge Isotopes Laboratories, Inc. Confirmation of the [13CD2]-paraformaldehyde purity and identity was accomplished using gas chromatography-mass spectrometry. The test atmospheres of [13CD2]-formaldehyde were generated from an 80-l Teflon bag using a peristaltic pump to deliver vaporized [13CD2]-formaldehyde into the nose-only chamber supply airflow. [13CD2]-Formaldehyde exposure atmospheres were measured by a calibrated infrared spectrophotometer (IR) (MIRAN 1A; The Foxboro Co., Foxboro, MA) with the IR sampling the concentration prior to entering the nose-only chamber. The IR was calibrated using the chromotropic acid assay; a wet chemistry method for the measurement of formaldehyde concentrations. The estimated limit of detection for the IR was 0.02 ppm/0.005 V.
Animal use in this study was approved by the Institutional Animal Use and Care Committee of The Hamner Institutes for Health Sciences and was conducted in accordance with the National Institutes of Health guidelines for the care and use of laboratory animals. Animals were housed in fully accredited American Association for Accreditation of Laboratory Animal Care facilities. Male Fischer 344 rats were exposed to 10-ppm [13CD2]-formaldehyde atmospheres for 1 or 5 days (6 h/day) using a single nose-only unit (Lab Products, Seaford, DE). One nose-only unit was used for all exposures. The air supply was controlled with a mass flow controller to maintain a total air flow that assured at least 2.5 times the estimated minute ventilation (0.20 ml/min) for a 250-g rat or ∼0.5 l/min at each open port. The air supply for 30 rats was maintained at ∼15 l/min. Temperature was measured near the center top of the chamber by a Pre-Con All Purpose Thermister Sensor, Model ST-A (Kele & Associates, Memphis, TN), connected to the Andover Building Automation System. Calibration was performed by comparing the thermister probe to a certified mercury thermometer. The mean of the hourly readings and SD for the 1-day exposure was 74.7 ± 0.6°F, while the mean of the daily mean and SD for the 5-day exposure was 69.2 ± 1.1°F for the target of 72°F. Relative humidity was measured near the center top of the chamber by a Rotronic Humidity Sensor (Series 1200; Rotronic Instrument Corp., Huntington, NY) connected to the Andover Building Automation System. The relative humidity sensor was calibrated by immersing the probe in an atmosphere of known humidity generated from saturated salt solutions. The mean of the hourly readings and SD for the 1-day exposure was 40.3 ± 1.4%, while the mean of the daily mean and SD for the 5-day exposure was 50.6 ± 3% for the target of 50%.
Animals were anesthetized with Nembutal (50–60 mg/kg) i.p. within 2 h of the end of exposure; 3–5 ml of blood was collected by cardiac puncture for lymphocyte isolation. Nasal respiratory epithelium from the right and left sides of the nose and from the septum was collected, as were entire tissues of spleen, thymus, lung, and liver. Bone marrow was collected from both femurs by saline extrusion with a large bore needle. Tissue samples were collected and immediately frozen on dry ice followed by storage at −80°C.
DNA isolation and digestion.
DNA was isolated from the tissues of rats using a NucleoBond DNA isolation kit (Bethlehem, PA), as instructed by the manufacturer with small modifications. The resultant DNA was quantified and stored at −80°C for further analysis. DNA isolated from each nose sample (∼30–50 μg) was incubated with 50mM NaCNBH3 at 37°C for 6 h in phosphate buffer (pH = 7.1). Then, DNA was treated by DNaseI (200 U) for 10 min in the digestion buffer (80mM Tris-HCl, 20mM MgCl2, pH = 7.2), followed by the addition of 25 μl of alkaline phosphatase and 25 μl of phosphodiesterases for an additional hour. Enzymes and undigested DNA were removed by a Millipore Microcon YM-10 spin column for 45 min at 23°C, and the resultant solution was separated by HPLC to collect the fractions containing the corresponding DNA adducts. For monoadduct analysis of liver, lung, thymus, bone marrow, and spleen, 200 μg of DNA was used for digestion using the same methods. For monoadduct analysis of blood samples, DNA isolated from two to three animals (60–100 μg) was pooled for further analysis. Typically, 10 μl of hydrolysate was used to quantify dG and dA generated from DNA enzymatic digestion.
Sample workup for DNA-DNA cross-links.
For the analysis of dG-dG cross-links in nose samples, 30–50 μg isolated nasal DNA was digested enzymatically as described above in Tris buffer for 70 min at 37°C, followed by a cleaning procedure with a Millipore Microcon YM-10 spin column for 45 min at 23°C. NaCNBH3 was not used in DNA isolation for dG-dG cross-link analysis as it labilized the cross-links. The hydrolysate, after removing 10 μl for dG quantitation, was frozen at −20°C and separated by HPLC after 12 h storage. A parallel control experiment was carried out, to determine the extent of artifact. We spiked an equal amount of isotope-labeled [13C1015N5]-dG, as was produced from DNA digestion (determined by HPLC with UV detector using a small aliquot of DNA solution), into the sample before hydrolysis. Thus, the signal corresponding to cross-links dG-CH2-[13C1015N5]-dG accounted for approximately half of the artifacts formed during sample workup and storage, which demonstrated the amount of artifactual dG-CH2-dG formed during sample workup and storage. The extent of artifact was determined using the integrated peak area of dG-CH2-[13C1015N5]-dG divided by the peak area of dG-CH2-dG. For the analysis of dG-dG cross-links in other tissues, 200 μg DNA was digested for 70 min at 37°C, followed by a cleaning procedure with a Millipore Microcon YM-10 spin column for 60 min at 4°C. The resultant solution was frozen on dry ice immediately, followed by the storage in −80°C.
Quantitation of formaldehyde-DNA adducts.
The adducts were quantified by a triple-quadrupole mass spectrometer Ultra TSQ-Quantum (Thermo Electron) using SRM mode. N2-HOCH2-dG was quantified as N2-CH3-dG after reduction with NaCNBH3 using the transition of m/z 282.2 → m/z 166.1 and N2-HO13CD2-dG was quantified as N2-13CD2H-dG with the transition of m/z 285.2 → m/z 169.1. In addition, transitions were applied simultaneously to monitor possible precursors including m/z 284.2 and 283.2 which could arise from hydrogen-deuterium exchange of [13CD2]-formaldehyde. N6-HOCH2-dA was detected as N6-CH3-dA after treatment by NaCNBH3 with the transition of m/z 266.2→ m/z 150.1, while N6-HO13CD2-dA was monitored with the transition of m/z 269.2 → m/z 153.1 after reduction. Similarly, two additional transitions for m/z 268.2 and 267.2 were used to monitor possible ions arising as a consequence of hydrogen-deuterium exchange. The collision energy was set at 17 V after optimization. dG-CH2-dG and dG-13CD2-dG was detected and quantified using the transition of m/z 547.5 to m/z 152.1 and m/z 550.5 to m/z 152.1, with the collision energy set at 23V. The calibration curves for quantitation were obtained using the integrated peak area and amount of injected analytical standard and internal standard.
Statistical analysis of data.
Data represent mean ± SD. Unpaired Student's t-tests were performed using the SAS statistical package (SAS Institute, Cary, NC) with the sample size ranging from 5 to 8 for monoadduct analysis. Differences were considered statistically significant if p < 0.05.
RESULTS
Determination of Atmospheric Concentration of Formaldehyde for Animal Exposures
The mean of the hourly readings and SD for the 1-day exposure was 10.2 ± 0.5 ppm, while the mean of the daily mean and SD for the 5-day exposure was 10.6 ± 0.2 ppm for the target concentration of 10.0-ppm [13CD2]-formaldehyde in air.
Method Development for Analysis of Formaldehyde-Induced Monoadducts
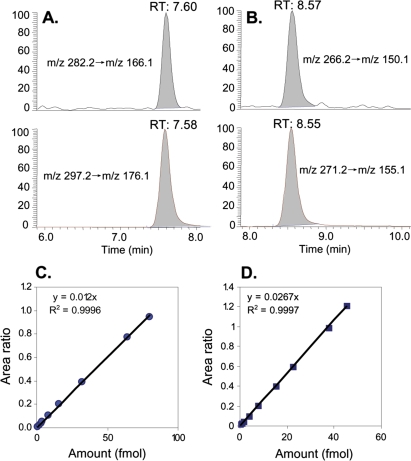
Highly sensitive methods for the analysis of N2-hydroxymethyl-dG and N6-hydroxymethyl-dA were developed that exhibited excellent precision and accuracy (Fig. 2 and Table 1). Hydroxymethyl adducts were detected and quantified as the corresponding stable methyl adducts after reduction with NaCNBH3. The limit of detection for N2-CH3-dG was ∼240 amol on the column (S/N = 3), while the limit of detection for N6-CH3-dA was ∼75 amol on the column (S/N = 3). Satisfactory accuracy and precision of this assay were shown by adding known amounts of N2-CH3-dG and N6-CH3-dA analytical standards to 50 μg of DNA, as listed in Table 1.
FIG. 2.
Typical LC-ESI-MS/MS-SRM chromatograms of 0.8 fmol of N2-CH3-dG and 80 fmol of internal standard [13C1015N5]-N2-CH3-dG loaded on the column (A). 0.15 fmol of N6-CH3-dA and 37.5 fmol of internal standard [15N5]-N6-CH3-dA (B). Typical calibration curve used for the quantitation of N2-Me-dG adducts (C). Typical calibration curves used for the quantitation of N6-Me-dA adducts (D).
TABLE 1.
Analysis of Rat Hepatic DNA with Added N2-CH3-dG and N6-CH3-dA
| N2-CH3-dG (fmol) |
N6-CH3-dA (fmol) |
||||||
| Added | Detecteda | Accuracy (%) | CV (%) | Added | Detected | Accuracy (%) | CV (%) |
| 10 | 10.8 ± 0.7b | 108 | 6 | 10 | 10.2 ± 0.5 | 102 | 5 |
| 20 | 22.5 ± 0.9 | 112 | 4 | 20 | 21.6 ± 1.2 | 108 | 5 |
| 40 | 42.3 ± 0.4 | 105 | 0.1 | 40 | 40.9 ± 1.4 | 102 | 3 |
Sample size n = 3.
Mean ± SD.
Exogenous and Endogenous N2-Hydroxymethyl-dG Is Formed in Nasal Respiratory DNA, but Exogenous N2-HO13CD2-dG Is Not Formed in DNA at Sites Distant to the Portal of Entry
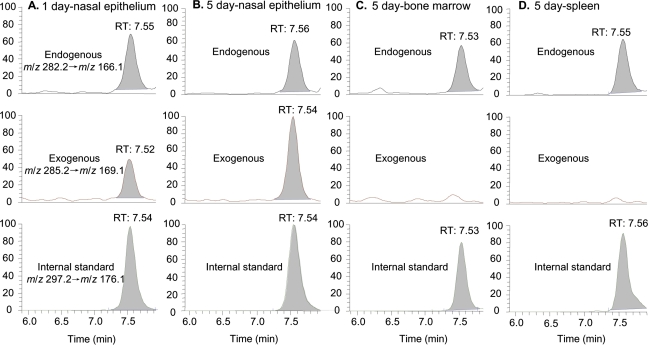
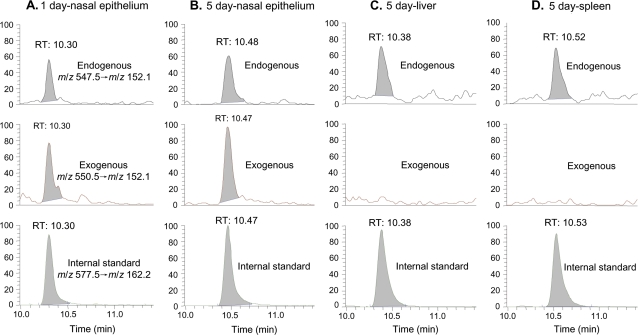
Figure 3A shows the LC-ESI-MS/MS chromatogram of N2-Me-dG in nasal DNA from a 1-day exposed rat. The peak corresponding to the specific transition of m/z 282.2 → m/z 166.1 and the same retention time with the [13C1015N5]-N2-CH3-dG internal standard unambiguously identified the formation of N2-HOCH2-dG from endogenous formaldehyde, as shown by the peak at 7.55 min in the top panel of Figure 3A. In addition to the peak of endogenous adducts, a new peak corresponding to the transition of m/z 285.2 → m/z 169.1 coeluted with the internal standard, which is consistent with N2-HO13CD2-dG formed from exogenous [13CD2]-formaldehyde. Similarly, both endogenous and exogenous dG adducts could be clearly identified and quantified in 30–50 μg of nasal epithelial DNA after 5 days of exposure, as shown in Figure 3B. Compared with Figure 3A, the amount of exogenous N2-HO13CD2-dG in nasal DNA increased as a consequence of extended exposure. In all other tissues including liver, lung, thymus, bone marrow, and spleen of 1- or 5-day exposed rats, only endogenous formaldehyde-induced N2-HOCH2-dG could be observed in 200 μg DNA. As shown by the middle panels of Figures 3C and 3D, no signal corresponding to the transition of m/z 285.2 → m/z 169.1 of exogenous adducts was present at the same retention time with the internal standard. In addition, two additional transitions to monitor m/z 284.2 and 283.2 did not detect any exogenous dG adducts arising from hydrogen-deuterium exchange. Therefore, there were no detectable amounts of N2-HO13CD2-dG in sites remote to the portal of entry after 1 or 5 days of exposure, despite the utilization of fivefold more DNA for these tissues in an assay with a limit of detection of ∼240 amol on column for N2-HO13CD2-dG.
FIG. 3.
LC-ESI-MS/MS-SRM chromatograms of N2-Me-dG in typical tissues: 1-day exposed nasal respiratory epithelium (A), 5-day exposed nasal respiratory epithelium (B), bone marrow (C), and spleen (D).
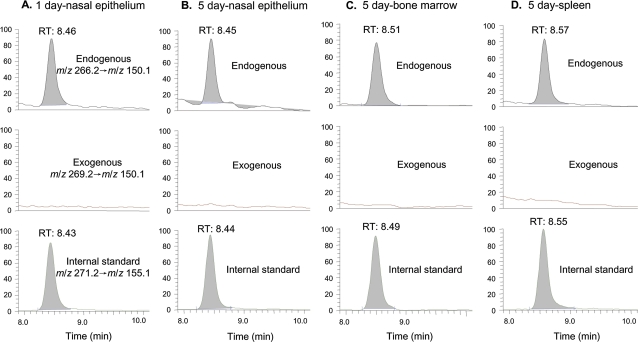
Endogenous N6-Hydroxymethyl-dA Is Present in All Tissues, but Exogenous N6-HO13CD2-dA Is Not Present in Any Tissue
We then analyzed N6-hydroxymethyl-dA adducts in rats exposed to 10-ppm [13CD2]-formaldehyde, as shown in Figure 4. Figures 4A and 4B illustrate the LC-ESI-MS/MS-SRM chromatograms of N6-Me-dA in noses of rats exposed to [13CD2]-formaldehyde for 1 or 5 days. The peak corresponding to the specific transition of m/z 266.2 → m/z 150.1 and the same retention time as the internal standard identified the formation of N2-HOCH2-dA from endogenous formaldehyde. However, there was no signal for N6-HO13CD2-dA (m/z 269.2 → m/z 153.1). Figures 4C and 4D show the chromatograms of N6-Me-dA in bone marrow and spleen of 5-day exposed rats. Moreover, additional transitions to monitor m/z 268.2 and 267.2 did not identify any exogenous dA adducts generated from hydrogen-deuterium exchange. Therefore, we did not detect a peak for N6-HO13CD2-dA after [13CD2]-formaldehyde exposure in any tissue using an assay with a limit of detection of ∼75 amol on column, while we clearly observed the peak of N6-HOCH2-dA from endogenous formaldehyde in all tissues. These data show that N6-HOCH2-dA is not a good biomarker for inhaled formaldehyde.
FIG. 4.
LC-ESI-MS/MS-SRM chromatograms of N6-Me-dA of typical tissues of rats: nasal respiratory epithelium of a 1-day exposed rat (A), nasal respiratory epithelium of a 5-day exposed rat (B), bone marrow of a 5-day exposed rat (C), and spleen of a 5-day exposed rat (D).
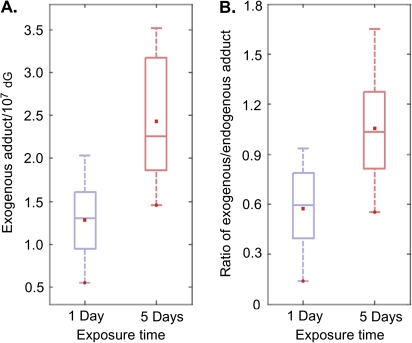
Table 2 summarizes the number of adducts in all the tissues measured in this study. For 1-day exposed samples, endogenous N2-HOCH2-dG ranged from 1.05 to 2.66 adducts/107 dG depending on tissues, while endogenous N6-HOCH2-dA was from 1.85 to 3.95 adducts/107 dA. For 5-day exposed rats, the numbers of endogenous N2-HOCH2-dG and N6-HOCH2-dA were 1.10–3.24 adducts/107 dG and 2.23–3.66 adducts/107 dA, respectively. It is of interest to note that endogenous N6-HOCH2-dA was present in control rat liver at 1.96 ± 1.86 adducts/107 dA (Wang et al., 2007), which is similar to our 1-day (2.62 ± 0.46 adducts/107 dA) and 5-day (2.87 ± 0.65 adducts/107 dA) endogenous N6-HOCH2-dA in rat liver DNA. Exogenous [13CD2]-formaldehyde exposure only induced DNA adducts with dG, and this adduct was only detected in nasal epithelium from 1- or 5-day exposed rats. The mean amounts of exogenous N2-HO13CD2-dG in 1- or 5-day exposed nasal epithelium samples were 1.28 ± 0.49 and 2.43 ± 0.78 adducts/107 dG, respectively (Fig. 5A, p < 0.05). The ratio of exogenous versus endogenous N2-hydroxymethyl-dG is shown in Figure 5B. There was a statistically significant difference between 1-day and 5-day exposed samples (p < 0.05). The former had a ratio of 0.57 ± 0.28, while the ratio of 5-day exposed sample was 1.06 ± 0.40.
TABLE 2.
Formaldehyde-Induced Monoadducts and dG-dG Cross-links in Rats Exposed to 10-ppm Formaldehyde for 1 day or 5 daysa
| N2-HOCH2-dG (adducts/107 dG) |
N6-HOCH2-dA (adducts/107 dA) |
dG-CH2-dG (adducts/107 dG) |
|||||
| Exposure period | Tissues | Exogenous | Endogenous | Exogenous | Endogenous | Exogenous | Endogenous |
| 1 day | Nose | 1.28 ± 0.49b | 2.63 ± 0.73 | nd | 3.95 ± 0.26 | 0.14 ± 0.06c | 0.17 ± 0.05 |
| Lung | ndd | 2.39 ± 0.16e | nd | 2.62 ± 0.24 | nd | 0.20 ± 0.04f | |
| Liver | nd | 2.66 ± 0.53 | nd | 2.62 ± 0.46 | nd | 0.18 ± 0.05 | |
| Spleen | nd | 2.35 ± 0.31 | nd | 1.85 ± 0.19 | nd | 0.15 ± 0.06 | |
| Bone Marrow | nd | 1.05 ± 0.14 | nd | 2.95 ± 1.32 | nd | 0.09 ± 0.01 | |
| Thymus | nd | 2.19 ± 0.36 | nd | 2.98 ± 1.11 | nd | 0.10 ± 0.03 | |
| Bloodg | nd | 1.28 ± 0.38 | nd | 3.80 ± 0.29 | nd | 0.12 ± 0.09 | |
| 5 day | Nose | 2.43 ± 0.78 | 2.84 ± 1.13 | nd | 3.61 ± 0.95 | 0.26 ± 0.07 | 0.18 ± 0.06 |
| Lung | nd | 2.61 ± 0.35 | nd | 2.47 ± 0.55 | nd | 0.20 ± 0.03 | |
| Liver | nd | 3.24 ± 0.42 | nd | 2.87 ± 0.65 | nd | 0.21 ± 0.08 | |
| Spleen | nd | 2.35 ± 0.59 | nd | 2.23 ± 0.89 | nd | 0.16 ± 0.08 | |
| Bone Marrow | nd | 1.17 ± 0.35 | nd | 2.99 ± 0.08 | nd | 0.11 ± 0.03 | |
| Thymus | nd | 1.99 ± 0.30 | nd | 2.48 ± 0.11 | nd | 0.19 ± 0.03 | |
| Bloodg | nd | 1.10 ± 0.28 | nd | 3.66 ± 0.78 | nd | 0.10 ± 0.07 | |
The limit of detection for dG monoadducts, dA monoadducts, and dG-dG cross-links was ∼240, ∼75, and ∼60 amol, respectively.
n = 5–8 nose samples for the analysis of monoadducts in 30–50 μg of DNA; data represent mean ± SD.
n = 4–5 nose samples for the analysis of cross-links, artifacts have been subtracted from the data.
Not detectable (nd) in 200 μg of DNA.
n = 4–5 for distant tissues.
n = 3 for distant tissues.
60–100 μg of DNA was typically used for analysis of white blood cells isolated from blood.
FIG. 5.
The amount of exogenous N2-HO13CD2-dG in nasal respiratory epithelial DNA of rats exposed to 10-ppm formaldehyde for 1 day or 5 days (A). The ratio of exogenous versus endogenous N2-hydroxymethyl-dG for 1-day and 5-day exposed nasal respiratory epithelial DNA (B).
Endogenous dG-CH2-dG Cross-links Are Present in All Tissues, but Exogenous dG-13CD2-dG Cross-links Are Only Formed in Nasal DNA
A sensitive nano-LC-MS/MS-SRM method was also developed, with a limit of detection of ∼60 amol on column, to detect dG-CH2-dG cross-links in the tissue samples. Some typical chromatograms are shown in Figure 6. Similar to the monoadducts, the exogenous dG-13CD2-dG only could be detected in nasal DNA (Figs. 6A and 6B) and not in DNA from any distant tissue such as liver and spleen (Figs. 6C and 6D). These data demonstrate that no detectable amounts of exogenous dG-dG cross-links were induced at distant sites in rats by inhalation exposure to formaldehyde.
FIG. 6.
LC-ESI-MS/MS-SRM chromatograms of dG-dG cross-links in typical tissues of rats: nasal respiratory epithelium of a 1-day exposed rat (A), nasal respiratory epithelium of a 5-day exposed rat (B), liver of a 5-day exposed rat (C), and spleen of a 5-day exposed rat (D). The exogenous dG-13CD2-dG only could be detected in nasal respiratory epithelial DNA and not in DNA from any tissue remote from the portal of entry.
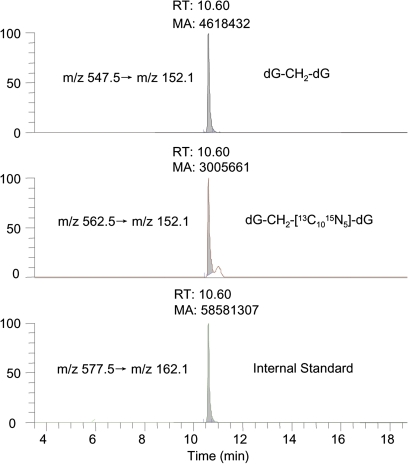
The numbers of dG-dG cross-links in individual tissues are also summarized in Table 2. The amount of endogenous cross-links ranged from 0.09 to 0.21 adducts/107 dG across all tissues. In contrast, the amounts of exogenous dG-13CD2-dG were 0.14 ± 0.06 and 0.26 ± 0.07 adducts/107 dG in DNA from nasal epithelium of 1-day and 5-day exposed rats, respectively, but were not detectable in other tissues. These numbers were roughly 10% of the amount of corresponding monoadducts. Since N2-hydroxymethyl-dG can further react with dG, potential artifacts could form during sample workup and storage. Therefore, we carried out a parallel control experiment adding amounts of [13C1015N5]-dG equal to the amount of dG in the sample and determined that ∼65% of the cross-links were artifact by calculating the peak ratio under the conditions used to analyze nose samples (Fig. 7). The data in Table 2 were corrected to remove such artifact. Therefore, the data on dG-dG cross-links were considered less rigorous than monoadduct data due to the issue of potential artifacts. Special caution is needed when generating or using quantitative data on formaldehyde-induced dG-dG cross-links as the molecular dosimeter. Nevertheless, no exogenous dG-dG cross-links were detected in tissues remote to the portal of entry, even though we utilized fivefold more DNA which increased our ability to quantify true cross-link formation and formation from artifact.
FIG. 7.
Formation of artifacts under the conditions used to analyze dG-dG cross-links in nasal DNA samples. The extent of artifacts (∼65%) was determined by the area ratio of peak562.5→152.1 (middle panel) versus peak547.5→152.1 (top panel) in parallel control experiments by adding amounts of [13C1015N5]-dG equal to the amount of dG in the sample during sample workup and storage.
DISCUSSION
This study addressed several critical issues related to formaldehyde toxicity and carcinogenicity. Exogenous N2-HO13CD2-dG was only detected in DNA from nasal respiratory epithelium of rats after inhalation. The same was true for exogenous dG-dG cross-links. The absence of exogenous formaldehyde-induced DNA adducts and cross-links in other tissues supports the conclusion that genotoxic effects of inhaled formaldehyde are implausible at sites remote to the portal of entry. Endogenous formaldehyde-induced N6-HOCH2-dA was detected in all tissues, but N6-HO13CD2-dA was not detectable in DNA from any tissue examined in rats exposed by inhalation to [13CD2]-formaldehyde. It is important to note that previous methods used to measure DPC (Casanova et al., 1994; Casanova-Schmitz and Heck, 1983; Casanova-Schmitz et al., 1984; Kato et al. 2001) and specific DNA adducts (Cheng et al., 2008) could not differentiate between endogenous and exogenous DNA damage. It was only through the use of the [13CD2]-formaldehyde and mass spectrometry that endogenous and exogenous adducts could specifically be measured simultaneously in the same tissue. Furthermore, by monitoring the transitions that would occur if there were any hydrogen-deuterium exchange, we have demonstrated that neither inhaled formaldehyde nor methanediol derived from inhaled formaldehyde reaches sites distant to the portal of entry.
In this study, we found that exogenous formaldehyde-induced N2-hydroxymethyl-dG adducts and corresponding dG-dG cross-links only formed in nasal DNA of rats exposed to 10-ppm [13CD2]-formaldehyde by inhalation (Figs. 3 and 6). These data support causation of nasal carcinoma following exposure to formaldehyde by inhalation. Formaldehyde was first identified as a carcinogen when rats exposed to 0, 2, 6, or 15 ppm for up to 24 months developed squamous cell carcinomas of the nasal passages (Kerns et al., 1983; Swenberg et al., 1980). Nasal carcinomas developed in 51 males and 52 females exposed to 15-ppm rats but only in 1 male and 1 female rat exposed to 6-ppm formaldehyde. No malignant tumors of the nose occurred in rats exposed to 2 ppm or in controls (Kerns et al., 1983). A second carcinogenicity study was conducted that expanded the exposure response to include 0.7-, 2-, 6-, 10-, and 15-ppm formaldehyde. Formaldehyde induced nasal squamous cell carcinomas in a highly nonlinear fashion, with no neoplasms at the lowest two concentrations (0/90 animals for each group), and 1/90, 20/90, and 69/147 carcinomas in rats exposed to 6-, 10-, and 15-ppm formaldehyde (Monticello et al., 1996). In addition to the squamous cell carcinomas, polypoid adenomas were present in 0/90 of the control, 0.7-, 2-, and 6-ppm exposed rats and 5/90 and 14/147 animals exposed to 10- and 15-ppm formaldehyde. Cell proliferation was also greatly increased at 10- and 15-ppm exposures.
IARC recently classified formaldehyde as a known human carcinogen based on sufficient evidence for nasopharyngeal carcinoma (IARC, 2006). Extensive research has demonstrated that both genotoxicity and cytotoxicity contribute to the carcinogenesis of formaldehyde in nasal tissues (IARC, 2006). As mentioned above, marked and sustained increases in cell proliferation in the noses of rats have been observed after exposure to ≥ 10-ppm formaldehyde (Conolly et al., 2003a; Monticello et al., 1996). In this study, we found that exogenous formaldehyde-induced N2-hydroxymethyl-dG adducts and corresponding dG-dG cross-links formed in nasal DNA of rats after 10-ppm [13CD2]-formaldehyde exposure. These findings provide new insight regarding how exposure to formaldehyde may cause nasal cancer. Several guanine point mutations have been identified by DNA sequence analysis of p53 cDNA from formaldehyde-induced squamous cell carcinomas in nasal passages of rats, including 398G → T, 638G → T, 812G → A, and 842G → C (Recio et al., 1992). Earlier studies of Heck and Casanova demonstrated a linear response for DPC between formaldehyde exposures of 6 and 15 ppm but reduced numbers of DPC per ppm formaldehyde at 2 and 0.7 ppm (Casanova-Schmitz et al., 1984). The present study quantitated endogenous dG adducts and dG-dG cross-links in numerous tissues, including the nasal respiratory epithelium. It also quantitated the number of endogenous dA adducts in these tissues. Surprisingly, the number of total endogenous formaldehyde adducts was greater than the number of exogenous adducts in nasal DNA following exposures to 10-ppm [13CD2]-formaldehyde. The number of endogenous formaldehyde adducts was similar across tissues. This suggests that the marked increases in cell proliferation induced by exposure to 10 and 15 ppm may play a critical role in converting both endogenous and exogenous labile but promutagenic adducts into mutations. At low exposures (< 0.1 ppm), minimal numbers of exogenous formaldehyde adducts will be formed relative to the high number of endogenous formaldehyde adducts. Likewise, no increase in cell proliferation has been associated with such low exposures, so fewer adducts would result in mutations. Nasal squamous cell carcinomas are rare in nonexposed rats but were very prominent in rats exposed to 10- or 15-ppm formaldehyde. This improved understanding of the mechanisms involved strengthens the role of cytotoxicity-induced cell proliferation in the mutagenesis and carcinogenesis of inhaled formaldehyde.
We have demonstrated that exogenous [13CD2]-formaldehyde only induced DNA lesions at the site of contact but not in DNA of distant organs (Figs. 3 and 6). This result has strong implications regarding the plausibility of formaldehyde causing leukemia. Evidence for an association between leukemia and formaldehyde exposure have been identified in several epidemiology studies but were not present in others (Coggon et al., 2003). Leukemia was increased in the U.S. formaldehyde worker cohort only when the highest versus lowest peak exposure were compared but not when cumulative exposure was the metric (Beane Freeman et al., 2009). Likewise, a meta-analysis found an increase in lymphohematopoietic malignancies, with the highest relative risk for myeloid leukemia (Zhang et al., 2009b). In contrast, Coggon found no association with leukemia in the British cohort, even though 28% of the workers had exposures > 2.0 ppm (Coggon et al., 2003). Previous studies suggested that it was not possible to identify a mechanism for the induction of myeloid leukemia in humans. For example, no increase has been found in the formaldehyde concentration in the blood of exposed humans and animals, there were no detectable protein adducts or DPC in the bone marrow of rats exposed to as high as 15-ppm formaldehyde, no DPC were detectable in the bone marrow of Rhesus monkeys exposed to formaldehyde as high as 6 ppm, and no chromosomal aberrations were found in the bone marrow of rats exposed to as high as 15-ppm formaldehyde (Heck and Casanova 2004). Recently, Zhang et al. hypothesized three possible mechanisms for the induction of leukemia by formaldehyde (Zhang et al., 2009b). First, formaldehyde may act directly on the bone marrow. The present study demonstrates that this does not occur. They then suggested that hematopoietic stem cells/early progenitor cells in the circulation, or residing in the nasal passages, could be exposed in the nose, travel to the bone marrow, and be transformed into leukemia cells. However, in this study, we did not detect any exogenous formaldehyde-induced DNA adducts in white blood cell samples although two times more DNA was used to increase our ability to detect exogenous DNA damage. The numbers of endogenous N2-HOCH2-dG and N6-HOCH2-dA in white blood cells were 1.10 to 1.28 adducts/107 dG and 3.66 to 3.80 adducts/107 dA, which is similar to other tissues we examined. Our methods would have been able to detect ∼3 N2-HO13CD2-dG adducts/109 dG, which is ∼30 times less than the number of endogenous N2-HOCH2-dG adducts present in white blood cells. The fact that lung, spleen, liver, thymus, and bone marrow did not have detectable HO13CD2-dG adducts in 200 μg of DNA suggests that the likelihood that this adduct would be formed is even lower.
There are no data that support these later two mechanisms for the induction of leukemia by any chemical. Furthermore, circulating myeloid cells do not typically return to bone marrow and proliferate (McKinney-Freeman and Goodell, 2004). Finally, the exogenous formaldehyde-DNA adducts are labile and would not be expected to persist for more than a day or so, based on our data for 1 versus 5 days of exposure and in vitro experiments (Lu et al., 2010; Zhong and Hee, 2004). The cells containing labile adducts would need to travel to the bone marrow and undergo cell division shortly after being adducted. A recent paper by Zhang et al. (2010) suggests that formaldehyde exposure disrupts hematopoietic function and induces aneuploidy in cultured myeloid progenitor cells from a small group of exposed workers. However, the methods used in this study do not differentiate whether the chromosomal changes were induced in vivo or in vitro. The possibility that formaldehyde induces leukemia in humans cannot be totally ruled out by this study due to well-known species differences in airway architecture, ventilation rate, and breathing mode. Humans could be differentially exposed to formaldehyde through oronasal breathing, while rats are exposed through obligatory nasal breathing. Although DPC were detected in some regions of the upper respiratory tract in addition to the turbinates and anterior lateral wall/septum of monkeys after exposure to 6-ppm formaldehyde, no DPC were found in the bone marrow, the sinuses, or in proximal lung of monkeys (Casanova et al., 1991). Moreover, when using DPC as an end point, the yield of DPC in the noses of monkeys was about an order of magnitude lower than in the noses of rats. This was attributed to species differences in minute volume and quantity of exposed tissue (Casanova et al., 1991). In addition, comprehensive analysis of nasal airflow and formaldehyde transport in rat, monkey, and human by computational fluid dynamic models coupled with dose-response predications derived from a combined rodent and human data set indicated that human cancer risks associated with inhaled formaldehyde would be significantly lower at relevant human exposures (Conolly et al., 2003b, 2004; Kimbell et al., 2001). Therefore, our finding that exogenous N2-HO13CD2-dG adducts were only formed in nasal DNA, that the DNA adducts are labile, and that high numbers of endogenous formaldehyde adducts are always present indicate that inhaled formaldehyde is unlikely to cause leukemia. The data from this study may also be useful in constructing a quantitative analysis of the likely flux in humans at higher ventilation rates (Conolly et al., 2004).
In summary, the results of this study clearly demonstrated that endogenous and exogenous formaldehyde-induced DNA adducts and DNA-DNA cross-links could be unambiguously differentiated utilizing [13CD2]-formaldehyde exposures and mass spectrometry. This approach allowed us to quantitatively examine the molecular dose of endogenous and exogenous formaldehyde-DNA adducts in a multitude of tissues following inhalation exposure. Moreover, we demonstrated that exogenous formaldehyde-induced N2-HO13CD2-dG is the primary DNA monoadduct in rats exposed by inhalation to 10-ppm [13CD2]-formaldehyde for 1 day or 5 days. This provides clear evidence that inhaled formaldehyde induced exposure-specific DNA adducts at the target site for carcinogenesis. More importantly, we have shown that exogenous formaldehyde-induced DNA monoadducts and dG-dG cross-links only occur in rat nasal mucosa, providing clear evidence that genotoxic effects at sites remote to the portal of entry of inhaled formaldehyde are implausible. The data generated in this study add greatly to our understanding of the toxicity and carcinogenicity of formaldehyde, a high-volume chemical of intense public health concern.
FUNDING
National Institutes of Health (P30-ES10126 and P42-ES05948 to J.A.S.); the Formaldehyde Council, Inc. (to J.A.S.).
Acknowledgments
The authors thank Drs Louise Ball, Avram Gold, David Kaufman, and Jun Nakamura for critical reading and valuable suggestions on the manuscript. The authors thank Ms Lina Gao for her valuable assistance with the nano-UPLC-MS/MS.
References
- Beane Freeman LE, Blair A, Lubin JH, Stewart PA, Hayes RB, Hoover RN, Hauptmann M. Mortality from lymphohematopoietic malignancies among workers in formaldehyde industries: the National Cancer Institute cohort. J. Natl. Cancer Inst. 2009;101:751–761. doi: 10.1093/jnci/djp096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova M, Morgan KT, Gross EA, Moss OR, Heck HA. DNA-protein cross-links and cell replication at specific sites in the nose of F344 rats exposed subchronically to formaldehyde. Fundam. Appl. Toxicol. 1994;23:525–536. doi: 10.1006/faat.1994.1137. [DOI] [PubMed] [Google Scholar]
- Casanova M, Morgan KT, Steinhagen WH, Everitt JI, Popp JA, Heck HD. Covalent binding of inhaled formaldehyde to DNA in the respiratory tract of rhesus monkeys: pharmacokinetics, rat-to-monkey interspecies scaling, and extrapolation to man. Fundam. Appl. Toxicol. 1991;17:409–428. doi: 10.1016/0272-0590(91)90230-2. [DOI] [PubMed] [Google Scholar]
- Casanova-Schmitz M, Heck HD. Effects of formaldehyde exposure on the extractability of DNA from proteins in the rat nasal mucosa. Toxicol. Appl. Pharmacol. 1983;70:121–132. doi: 10.1016/0041-008x(83)90185-0. [DOI] [PubMed] [Google Scholar]
- Casanova-Schmitz M, Starr TB, Heck HD. Differentiation between metabolic incorporation and covalent binding in the labeling of macromolecules in the rat nasal mucosa and bone marrow by inhaled [14C]- and [3H]formaldehyde. Toxicol. Appl. Pharmacol. 1984;76:26–44. doi: 10.1016/0041-008x(84)90026-7. [DOI] [PubMed] [Google Scholar]
- Cheng G, Wang M, Upadhyaya P, Villalta PW, Hecht SS. Formation of formaldehyde adducts in the reactions of DNA and deoxyribonucleosides with alpha-acetates of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK), 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (NNAL), and N-nitrosodimethylamine (NDMA) Chem. Res. Toxicol. 2008;21:746–751. doi: 10.1021/tx7003823. [DOI] [PubMed] [Google Scholar]
- Coggon D, Harris EC, Poole J, Palmer KT. Extended follow-up of a cohort of British chemical workers exposed to formaldehyde. J. Natl. Cancer Inst. 2003;95:1608–1615. doi: 10.1093/jnci/djg046. [DOI] [PubMed] [Google Scholar]
- Conolly RB, Kimbell JS, Janszen D, Schlosser PM, Kalisak D, Preston J, Miller FJ. Biologically motivated computational modeling of formaldehyde carcinogenicity in the F344 rat. Toxicol. Sci. 2003a;75:432–447. doi: 10.1093/toxsci/kfg182. [DOI] [PubMed] [Google Scholar]
- Conolly RB, Kimbell JS, Janszen D, Schlosser PM, Kalisak D, Preston J, Miller FJ. Biologically motivated computational modeling of formaldehyde carcinogenicity in the F344 rat. Toxicol. Sci. 2003b;75:432–447. doi: 10.1093/toxsci/kfg182. [DOI] [PubMed] [Google Scholar]
- Conolly RB, Kimbell JS, Janszen D, Schlosser PM, Kalisak D, Preston J, Miller FJ. Human respiratory tract cancer risks of inhaled formaldehyde: dose-response predictions derived from biologically-motivated computational modeling of a combined rodent and human dataset. Toxicol. Sci. 2004;82:279–296. doi: 10.1093/toxsci/kfh223. [DOI] [PubMed] [Google Scholar]
- Hauptmann M, Lubin JH, Stewart PA, Hayes RB, Blair A. Mortality from lymphohematopoietic malignancies among workers in formaldehyde industries. J. Natl. Cancer Inst. 2003;95:1615–1623. doi: 10.1093/jnci/djg083. [DOI] [PubMed] [Google Scholar]
- Hauptmann M, Lubin JH, Stewart PA, Hayes RB, Blair A. Mortality from solid cancers among workers in formaldehyde industries. Am. J. Epidemiol. 2004;159:1117–1130. doi: 10.1093/aje/kwh174. [DOI] [PubMed] [Google Scholar]
- Heck H, Casanova M. The implausibility of leukemia induction by formaldehyde: a critical review of the biological evidence on distant-site toxicity. Regul. Toxicol. Pharmacol. 2004;40:92–106. doi: 10.1016/j.yrtph.2004.05.001. [DOI] [PubMed] [Google Scholar]
- International Agency for Research on Cancer (IARC) Formaldehyde, 2-butoxyethanol and 1-tert-butoxypropan-2-ol. IARC Monogr. Eval. Carcinog. Risks Hum. 2006;88:1–287. [PMC free article] [PubMed] [Google Scholar]
- Kato S, Burke PJ, Koch TH, Bierbaum VM. Formaldehyde in human cancer cells: detection by preconcentration-chemical ionization mass spectrometry. Anal. Chem. 2001;73:2992–2997. doi: 10.1021/ac001498q. [DOI] [PubMed] [Google Scholar]
- Kerns WD, Pavkov KL, Donofrio DJ, Gralla EJ, Swenberg JA. Carcinogenicity of formaldehyde in rats and mice after long-term inhalation exposure. Cancer Res. 1983;43:4382–4392. [PubMed] [Google Scholar]
- Kimbell JS, Overton JH, Subramaniam RP, Schlosser PM, Morgan KT, Conolly RB, Miller FJ. Dosimetry modeling of inhaled formaldehyde: binning nasal flux predictions for quantitative risk assessment. Toxicol. Sci. 2001;64:111–121. doi: 10.1093/toxsci/64.1.111. [DOI] [PubMed] [Google Scholar]
- Lu K, Ye W, Zhou L, Collins BL, Chen X, Gold A, Ball LM, Swenberg JA. Structural characterization of formaldehyde-induced cross-links between amino acids and deoxynucleosides and their oligomers. J. Am. Chem. Soc. 2010 doi: 10.1021/ja908282f. Advance Access published on Februrary 23, 2010; doi: 10.1021/ja908282f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee JD, von Hippel PH. Formaldehyde as a probe of DNA structure. I. Reaction with exocyclic amino groups of DNA bases. Biochemistry (Mosc). 1975;14:1281–1296. doi: 10.1021/bi00677a029. [DOI] [PubMed] [Google Scholar]
- McKinney-Freeman S, Goodell MA. Circulating hematopoietic stem cells do not efficiently home to bone marrow during homeostasis. Exp. Hematol. 2004;32:868–876. doi: 10.1016/j.exphem.2004.06.010. [DOI] [PubMed] [Google Scholar]
- Monticello TM, Swenberg JA, Gross EA, Leininger JR, Kimbell JS, Seilkop S, Starr TB, Gibson JE, Morgan KT. Correlation of regional and nonlinear formaldehyde-induced nasal cancer with proliferating populations of cells. Cancer Res. 1996;56:1012–1022. [PubMed] [Google Scholar]
- Recio L, Sisk S, Pluta L, Bermudez E, Gross EA, Chen ZC, Morgan K, Walker C. p53 Mutations in formaldehyde-induced nasal squamous cell carcinomas in rats. Cancer Res. 1992;52:6113–6116. [PubMed] [Google Scholar]
- Swenberg JA, Kerns WD, Mitchell RI, Gralla EJ, Pavkov KL. Induction of squamous cell carcinomas of the rat nasal cavity by inhalation exposure to formaldehyde vapor. Cancer Res. 1980;40:3398–3402. [PubMed] [Google Scholar]
- Wang M, Cheng G, Balbo S, Carmella SG, Villalta PW, Hecht SS. Clear differences in levels of a formaldehyde-DNA adduct in leukocytes of smokers and nonsmokers. Cancer Res. 2009;69:7170–7174. doi: 10.1158/0008-5472.CAN-09-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Cheng G, Villalta PW, Hecht SS. Development of liquid chromatography electrospray ionization tandem mass spectrometry methods for analysis of DNA adducts of formaldehyde and their application to rats treated with N-nitrosodimethylamine or 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone. Chem. Res. Toxicol. 2007;20:1141–1148. doi: 10.1021/tx700189c. [DOI] [PubMed] [Google Scholar]
- Zhang L, Freeman LE, Nakamura J, Hecht SS, Vandenberg JJ, Smith MT, Sonawane BR. Formaldehyde and leukemia: epidemiology, potential mechanisms, and implications for risk assessment. Environ. Mol. Mutagen. 2009a doi: 10.1002/em.20534. Advance Access published on September 29, 2009; doi: 10.1002/em.20534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Steinmaus C, Eastmond DA, Xin XK, Smith MT. Formaldehyde exposure and leukemia: a new meta-analysis and potential mechanisms. Mutat. Res. 2009b;681:150–168. doi: 10.1016/j.mrrev.2008.07.002. [DOI] [PubMed] [Google Scholar]
- Zhang L, Tang X, Rothman N, Vermeulen R, Ji Z, Shen M, Qiu C, Guo W, Liu S, Reiss B, et al. Occupational exposure to formaldehyde, hematotoxicity, and leukemia-specific chromosome changes in cultured myeloid progenitor cells. Cancer Epidemiol. Biomarkers Prev. 2010;19:80–88. doi: 10.1158/1055-9965.EPI-09-0762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong W, Hee SQ. Quantitation of normal and formaldehyde-modified deoxynucleosides by high-performance liquid chromatography/UV detection. Biomed. Chromatogr. 2004;18:462–469. doi: 10.1002/bmc.337. [DOI] [PubMed] [Google Scholar]
- Zhong W, Que Hee SS. Formaldehyde-induced DNA adducts as biomarkers of in vitro human nasal epithelial cell exposure to formaldehyde. Mutat. Res. 2004;563:13–24. doi: 10.1016/j.mrgentox.2004.05.012. [DOI] [PubMed] [Google Scholar]